Summary
Volatolomics is gaining consideration as a viable approach to diagnose several diseases, and it also shows promising results to discriminate COVID-19 patients via breath analysis. This paper extends the study of the relationship between volatile compounds (VOCs) and COVID-19 to blood serum. Blood samples were collected from subjects recruited at the emergency department of a large public hospital. The VOCs were analyzed with a gas chromatography mass spectrometer (GC/MS). GC/MS data show that in more than 100 different VOCs, the pattern of abundances of 17 compounds identifies COVID-19 from non-COVID with an accuracy of 89% (sensitivity 94% and specificity 83%). GC/MS analysis was complemented by an array of gas sensors whose data achieved an accuracy of 89% (sensitivity 94% and specificity 80%).
Subject areas: Health sciences, Diagnostics, Biotechnology, Medical biotechnology
Graphical abstract

Highlights
-
•
Volatile compounds released from human blood serum were analyzed
-
•
Blood serum of COVID-19 patients is identified by a pool of 17 volatile compounds
-
•
An array of gas sensors detects COVID-19 from serum volatile compounds analysis
Health sciences; Diagnostics; Biotechnology; Medical biotechnology
Introduction
Since late 2019, COVID-19 spread rapidly worldwide until reaching the status of a pandemic disease (Wu et al., 2020; Xu et al., 2020). The diagnosis of CO2VID-19 is based on the direct detection in patients' samples of its etiological agent known as SARS-COV-2. Reverse transcription real-time PCR assays performed on nasal-pharyngeal swabs are, to date, the gold standard for diagnosis (Udugama et al., 2020). These tests are carried out on a massive scale daily worldwide. In the European Union, from March 2020 to February 2021 about 360 million tests were performed (www.ecdc.europa.eu/en/publications-data/covid-19-testing). Molecular tests require specialized staff, and they are costly and time consuming. This last feature is in conflict with the needs in emergency rooms to promptly isolate COVID-19 patients.
Alternative rapid tests based on the detection of viral proteins (antigens) have been quickly developed by several companies in order to offer a faster and cheaper detection of the SARS-COV-2 infection. These tests are currently in use in different countries (https://ec.europa.eu/health/sites/health/files/preparedness_response/docs/covid-19_rat_common-list_en.pdf). Nevertheless, antigens rapid tests are usually less sensitive than molecular tests being reliable only at high viral loads (Ciotti et al., 2021). Moreover the performance variability of different tests produced by different companies is very high.
A strategy to further reduce the complexity of tests and to improve their massive diffusion is the incorporation of SARS-COV-2 molecular receptors in engineered biosensors (Mattioli et al., 2020). Alternative approaches could be oriented rather than to the direct detection of SARS-COV-2 to the detection of the evidence produced by COVID-19 (Lin et al., 2020). This is the approach of metabolomics which considers the systematic study of the chemical fingerprints, the metabolome, that specific processes leave in the organism (Wishart, 2016).
Among the metabolites the small molecules which are volatile or semi volatile, and can be found in gas phase, are attracting a large attention. This fraction of the metabolome is sometimes called volatolome. Human volatolome is very rich, thousands of different molecules, representing a wide variety of chemical diversity, are present in different human samples (Amann et al., 2014). The volatolome has been shown to be related to a vast range of phenomena observable in vitro, even at single cell level, and in vivo (Boots et al., 2012; Broza et al., 2015; Serasanambati et al., 2019).
Gas chromatography and mass spectrometers are the main analytical equipment for volatolomics, and they provide a thorough investigation into the volatolome composition. On the other hand, portable and easy to use instruments based on sensors arrays have been demonstrated to be sufficiently sensitive and selective to identify diseases analyzing various human samples such as breath, urine, and sweat (Haick et al., 2020; Di Natale et al., 2014).
Since COVID-19 affects mainly the lungs, breath has been considered the most straightforward sample to analyze (Lamote et al., 2020). Attempts to study COVID-19 induced alteration of breath composition appeared in literature. Proton transfer reaction mass spectrometry has been used to compare the breath of COVID-19 and non-COVID mechanically ventilated patients in a French hospital (Grassin-Delyle et al., 2021). Another study used a gas chromatography ion mass spectrometer to investigate the breath of COVID and non COVID patients recruited at the emergency department of two hospitals in UK and Germany (Ruszkiewicz et al., 2020). Both studies indicate that COVID-19 can be identified by the analysis of breath. As usually happens in volatolomics, these studies evidenced that it is not a single compound that can be univocally related to COVID infection but rather a pattern of volatile compounds (VOCs).
The feasibility of sensors approach was investigated with an array of nanostructured sensors that was used to analyze the breath of subjects in two chinese hospitals (Shan et al., 2020). The results of this study show that sensors can discriminate COVID-19 respect to healthy subjects and to subjects affected by other lung pathologies.
Studies on breath analysis carried out with different technologies indicate that volatolomics offer an alternative route for COVID-19 detection. On the other hand, breath is also the major vehicle of propagation of SARS-COV-2, so breath analysis raises safety issues for medical operators and for the analytical equipment. This concern creates interest in the diagnostic properties of other human samples (e.g. urine and serum) that are accessible for volatolomic studies.
Serum is particularly interesting. Proteomic, metabolomics, and lipidomic profiles of serum have been found to be strongly affected by COVID-19 (Bruzzone et al., 2020; Shen et al., 2020; Shi et al., 2021). Additionally, serum samples are always available since blood analysis is part of routine admission to hospitals and emergency departments.
Blood serum volatolome is not frequently studied. In most metabolomics studies, the target metabolites are not volatile and they need to be derivatized in order to become volatile and to be detected with gas analyzers (Dunn et al., 2011). The analysis of VOCs released by serum without the addition of derivatizating agents has been rarely considered. For instance, it has been investigated to search for biomarkers of acute myocardial infarction (Ali et al., 2016).
The above mentioned reasons prompted us to investigate the relationship between the COVID-19 and the volatolome of blood serum. The study was carried out with a gas chromatography mass spectrometer (GC/MS) and a gas sensor array. In this paper, the VOCs directly released by the serum of 97 patients in an emergency department have been analyzed. The classification of GC/MS and sensor array data show that COVID-19 can be identified with accuracies of about 90%.
Results and discussion
Ninety seven samples of serum, one per patient, were analyzed by GC/MS and gas sensor array as described in the STAR Methods section. Table 1 shows the demographic data and major comorbidities.
Table 1.
Demographic data of involved patients and co-morbidities
| Characteristic | COVID-19 | Non-COVID |
|---|---|---|
| Male | 34 | 19 |
| Female | 20 | 24 |
| Age (mean ± standard deviation) | 65 ± 15 | 55 ± 14 |
| Hypertension | 25 (45%) | 4 (9%) |
| Diabetes | 12 (22%) | 3 (7%) |
| Obesity | 2 (4%) | 0 (0%) |
| Hearth diseases | 15 (27%) | 3 (7%) |
| Cancer | 8 (14%) | 2 (4%) |
| Lung diseases | 8 (14%) | 1 (2%) |
The measurement setup was adapted from previous literature studies about volatolome analysis of serum (Ali et al., 2016). Figure 1 shows the observational design and the data analysis flow followed for both GC/MS and gas sensor array data.
Figure 1.
Observational design, and data treatment and analysis plan
Gas chromatography/mass spectrometer
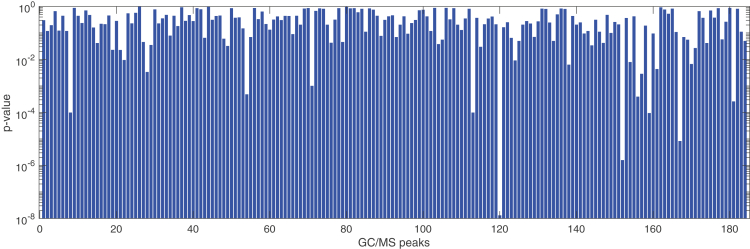
GC/MS analysis detected a total pool of 184 different VOCs differently distributed among the samples. The objective of this study was to investigate the correlation between VOCs and the COVID-19 infection. Thus, GC/MS data were first scrutinized for their capability to discriminate COVID-19 patients from the others. Chromatograms are shown in Figure S1 in the supplemental information. Peaks abundance extends for a range of about 3 orders of magnitude, then neither the presence of all peaks nor the differences between COVID-19 and controls are easily appreciable in Figure S1. To better evaluate the discrimination capability of GC/MS data, the Kruskal-Wallis rank test was applied to the abundance of each peak detected by GC/MS.
Figure 2 shows the probability of null-hypothesis (p value) of all detected VOCs. Seventeen VOCs, listed in Table 2, show a p value less than 0.05. These VOCs have been putatively identified by comparing the mass fragmentation spectra with those in libraries. They are aromatics, methylated alkanes, alkenes, and alcohols which are common elements of human volatolome (Amann et al., 2014) and whose concentration may be altered by cancer and other diseases (Einoch Amor et al., 2019). The presence of ketones among the selected variables is corroborated by a preliminary study that found an excess of ketones in urines of COVID-19 patients (Li et al., 2020). Ketones in general are supposed to be linked to protein metabolism and to an increased rate of fatty acid oxidation while a possible source of alcohols is the metabolism of hydrocarbons (Haick et al., 2014).
Figure 2.
Null hypothesis probability of GC/MS peaks in separating COVID from non-COVID samples
The difference between groups, expressed by the p-value, has been estimated with a Kruskal-Wallis rank sum test.
Table 2.
List of GC/MS peaks in serum with p value <0.05 respect to COVID-19
| Volatile compound | Covid-19 vs non-covid p value | Variation vs. non-covid | Sex p value | Spearman's correlation coefficient with age | Hypertension p value | |
|---|---|---|---|---|---|---|
| 1 | Toluene | 0.001 | ↓ | 0.242 | 0.404 | 0.212 |
| 2 | 4-Heptanone | 0.041 | ↑ | 0.805 | 0.129 | 0.216 |
| 3 | 3-Heptanol | 0.023 | ↑ | 0.051 | 0.222 | 0.071 |
| 4 | Ethanol, 2-butoxy- | 0.022 | ↓ | 0.281 | 0.299 | 0.332 |
| 5 | Heptane,3,3,5,-trimethyl | 0.003 | ↑ | 0.465 | 0.286 | 0.176 |
| 6 | 3-Hepten-2-one | 0.035 | ↑ | 0.519 | 0.179 | 0.489 |
| 7 | 1-Hexanol, 2-ethyl- | 0.032 | ↑ | 0.054 | 0.084 | 0.074 |
| 8 | Octane, 3,6-dimethyl- | 0.001 | ↓ | 0.404 | 0.441 | 0.074 |
| 9 | Hexane, 2,4,4-trimethyl- | 0.030 | ↑ | 0.985 | 0.065 | 0.109 |
| 10 | Benzene, 1,3,5-triethyl- | <0.001 | ↓ | 0.137 | 0.549 | 0.107 |
| 11 | Heptane, 3,3,5-trimethyl- | 0.009 | ↓ | 0.990 | 0.357 | 0.960 |
| 12 | Heptane, 2,2,4,6,6-pentamethyl- | <0.001 | ↓ | 0.105 | 0.472 | 0.075 |
| 13 | 3-Hexanone, 2,2-dimethyl- | 0.008 | ↑ | 0.142 | 0.229 | 0.537 |
| 14 | 1-Pentanol, 2-ethyl-4-methyl- | 0.029 | ↑ | 0.073 | 0.077 | 0.441 |
| 15 | 1-Hexene, 2,4,4-triethyl- | 0.001 | ↓ | 0.147 | 0.334 | 0.202 |
| 16 | 2,4,4-Trimethyl-1-pentanol | 0.004 | ↑ | 0.904 | 0.173 | 0.506 |
| 17 | 3-Hexanol, 3,5-dimethyl- | <0.001 | ↑ | 0.054 | 0.352 | 0.074 |
Some of these compounds have been found in human breath. In particular, toluene, heptane, 2,2,4,6,6-pentamethyl, benzene, 1,3,5-triethyl, octane, 3,6-dimethyl- (Phillips et al., 2013) (Phillips et al., 1999), and 4-heptanone (Mochalski et al., 2013). Ethanol 2-butoxy-, besides to be a component of breath, is also correlated with gastric cancer (Xu et al., 2013). Furthermore, 1-hexanol 2-ethyl- is a component of the headspace of human gastric cancer cell lines (Leiherer et al., 2021).
The correspondence of compounds in serum and breath supports the hypothesis of VOCs transfer from blood to breath at the air/blood interface in the lung (Haick et al., 2014). Furthermore, it suggests that these compounds could also act as COVID-19 biomarkers in breath.
The Kruskal-Wallis rank test and correlation tests were performed on GC/MS peaks to exclude the influence of sex, age, and hypertension. Variance analysis was performed on qualitative variables (sex and hypertension), and Spearman correlation coefficients were calculated respect to age (results are shown in Figures S2–S4 in the supplemental information). The p value of Kruskal-Wallis rank test was larger than 0.7 for both sex and hypertension hypotheses and the Spearman correlation coefficient with age was less than ±0.4. The results of rank-tests and Spearman correlation analysis for the 17 VOCs are listed in Table 2. The results of these analyses suggest that GC/MS data are not affected by sex, age, and hypertension condition.
Other conditions are numerically marginal and they do not allow for a reliable statistical analysis.
The abundance profiles of the set of compounds in Table 2 have been further investigated with multivariate analysis. It is important to note that the reduction of VOCs from 184 to 17 improves the reliability of multivariate data analysis (Hendriks et al., 2011).
Principal component analysis (PCA) offers a simple method to visualize the relationship between samples and to infer how each single variable, the peaks in case of GC/MS data, contributes to the multivariate data (Jolliffee, 2002).
The abundance of the seventeen GC/MS peaks were standardized (zero mean and unitary variance) before calculating the analysis. PCA results are shown in Figure 3. The scores plot of the first two principal components (Figure 3A) explains less than 48% of the total variance of the data. In Figure 3A the group of non-COVID subjects has been labeled according to the pathology: non-inflammatory pathologies, bacterial pneumonia, and other inflammatory pathologies. No evident separation between these three groups is observed. In particular, data of bacterial pneumonia do not overlap with those of COVID-19. Owing to the exiguity of the explained total variance, Figure 3A is only indicative of the whole distribution of data in the multivariate space. Nonetheless, the first two principal components show that COVID-19 samples tend to be segregated respect to the non-COVID. The Kruskal-Wallis rank test applied to the first principal component returns p < 10−6. Except the separation between COVID-19 and non-COVID samples, the scores plot do not show separations between sex, age and hypertension condition (see Figure S5 in the supplemental information).
Figure 3.
Plot of the first two principal components of selected GC/MS peaks
(A) scores plot;
(B) loadings plot. In scores plot, non-COVID data are separated in three subgroups.
The contribution of peaks to the principal components is described by the loadings. In Figure 3B scores and loadings are simultaneously plotted in order to compare the relationship between peaks and samples. Peaks are labeled according the numbers in Table 2. Peaks 2, 3, 6, 7, 9 13, 14, 16, and 17 contribute more to COVID-19, whereas the others are oriented toward the controls. The compounds oriented with COVID-19 include all ketones and alcohols while aromatics and alkanes point toward the non-COVID samples.
Sensor array
A similar analysis was carried out on sensor array data. Two samples were wrongly prepared, and then 95 samples were measured with the sensor array. Before each sample, to reduce the influence of background air, sensors were exposed to the air from an empty vial (see Figure S6 in supplemental information). The response of sensors was calculated as the difference between the frequency shift to serum sample and empty vial (see Figure S7 in supplemental information).
The capability of each sensor to capture the differences between the group of COVID-19 respect to non-COVID samples was tested with a Kruskal-Wallis rank test. Figure 4 shows the box-plots of sensors distributions. All sensors show a p value smaller than 0.01.
Figure 4.
Distribution of sensor response in the two groups of COVID and non-COVID
The p values, calculated with a Kruskal-Wallis rank test, of sensors are indicated in the header of each plot. Sensors are numbered according to the list given in Table S1. The distributions of responses are represented by a box-plots. In each box, the central mark is the median, the edges are the 25th and 75th percentiles, the whiskers extend to the most extreme data points not considered outliers. Outliers are labeled with a cross.
Most of the sensors show a larger response to non-COVID samples. In principle, the response of sensors should be predicted from the GC/MS data. However, this is practically impossible because the sensitivity of sensors to most of the compounds is unknown, and the abundances measured by GC/MS depend on the affinity of each compound with the Solid Phase Micro-Extraction (SPME) sampler. However, from a qualitative point the response of sensor can be compared with the total amount of VOCs in the sample. This quantity can be estimated by the total abundances of all VOCs detected in each sample. In agreement to sensors data, the distribution of the total abundance is slightly larger in non-COVID samples (see Figure S8 in supplemental information).
Variance analysis and correlation tests were performed on sensor's data to verify the lack of sensitivity of data respect to sex, age, and hypertension (results are shown in Figures S9–S11 in supplemental information). The p value of the Kruskal-Wallis rank test was larger than 0.3 for both sex and hypertension hypotheses, whereas the Spearman correlation coefficient with age was less than ±0.4. These results suggest that sex, age and hypertension do not affect sensors data.
To explore the multivariate distribution of sensors data the PCA was calculated. Figure 5 shows the scores plot of the first two principal components of sensor array data. The PCA was calculated on standardized data. The first two principal components account for more than 94% of the total variance of data. With respect to GC/MS data, sensor data are more correlated. This is a typical situation with sensor arrays and it is a consequence of the combinatorial selectivity principle (Hierlemann and Gutierrez-Osuna, 2008).
Figure 5.
Plot of the first two principal components of sensor array data
Non COVID data are separated in three subgroups.
As expected from the box-plots in Figure 4, the two groups are distinct but also rather overlapped.
The three sub-groups of non-COVID samples have been highlighted in the plot, no dependence between sensors data and the condition of non-COVID samples appears in Figure 4. Sensors' data confirm that non-COVID pneumonia is different from those of COVID-19. Eventually, the major discriminant among the data is the difference between COVID-19 and non-COVID.
Diagnostic performance evaluation
PCA is a powerful and simple algorithm to visualize multivariate data. Nevertheless, it is calculated by an unsupervised algorithm aimed at maximizing the variance of data. A more efficient indication of the diagnostic performance of multivariate data to capture the differences between groups is obtained by a supervised classification.
For this aim GC/MS sensor array data were analyzed with Partial Least Squares Discriminant Analysis (PLS-DA) (Barker and Rayens, 2003).
PCA shows that both GC/MS and sensors data are non-sensitive respect to the differences between the three sub groups of non-COVID subjects; thus, all non-COVID subjects were joined in a unique group of controls and PLS-DA was calculated in order to separate COVID-19 from non-COVID samples.
PLS-DA decomposes the multivariate data in a number of latent variables that, similarly to PCA scores, can be displayed to show the separation between groups. To be reliable, PLS-DA models need to be properly validated. Thus, the data sets have been split in two portions one used to train the model and the other to test. Random splitting of data in two groups can result in favorable conditions that could lead to over-optimistic conclusions. To avoid this drawback, PLS-DA was calculated 100 times, and each time a different partition of data was used to train and to test the model. At each step, a PLS-DA model was trained by a cross-validation procedure of training data and applied to the test data. The area under the curve (AUROC) was calculated as an indicator of the goodness of the model (Szymańska et al., 2012). Eventually, the model corresponding to the average AUROC was considered as representative for the classification. The confidence intervals of the areas under the ROC, specificity, and selectivity are calculated from the distribution of results obtained in the 100 random partitions in training and test sets.
In the case of GC/MS data, the large number of variables renders the application of classifiers unreliable, in particular PLS-DA. To this aim, for each training and test partition, a Kruskal-Wallis rank test was applied to the training data set and the variables returning the null-hypothesis probability (p value) less than 0.05 were selected for the PLS-DA calculation.
The dataset of VOCs corresponding to the average AUROC contains the same compounds listed in Table 2. In practice, with these data Kruskal-Wallis rank test gives similar results when calculated with 70% of samples.
The classification error of the PLS-DA model corresponding to the average AUROC is minimized by three latent variables. The three latent variables explain about 30% of the total variance of the abundance of the selected 17 peaks. Thus, 70% of the data variance does not contribute to COVID-19 identification; rather, it most likely describes the individual peculiarity of each subject. This is an indication of the complexity of the metabolic profile which only in a limited portion is affected by COVID-19. Figure S12 in the supplemental information shows the statistical distribution of data in each latent variable and the projection in the planes defined by the three latent variables.
The results of the classifier are summarized in the confusion matrices in Table 3. The total accuracy is 91% in training and 89% in test. The sensitivity of the classifier is 94% ± 1% and 94% ± 2% in training and test, respectively, while the specificity is 87% ± 2% and 83% ± 3%. Figure 6 shows the ROC curve. The areas under the ROC are 0.94 ± 0.02 and 0.94 ± 0.03 in training and test.
Table 3.
Confusion matrices of PLS-DA classifier in training and test
| True | |||||
|---|---|---|---|---|---|
| Training | Test | ||||
| COVID | Non-COVID | COVID | Non-COVID | ||
| Predicted | COVID | 35 | 4 | 16 | 2 |
| Non-COVID | 2 | 27 | 1 | 10 |
Figure 6.
Training and test ROCs of the PLS-DA model calculated with the selected 17 peaks of GC/MS
In the header the AUROCs in training and test are displayed.
The same procedure of calculation was applied to sensor's data.
Also in this case, the optimal PLS-DA model was made by three latent variables. The total variance explained by the three latent variables is larger than 93%. This difference with respect to GC/MS is due to the larger correlation between the sensors. As a consequence, the dominant difference between samples (COVID-19 vs. non-COVID) tends to incorporate the slight differences due to the peculiar behavior of each subject. Figure S13 in the supplemental information shows the box plots of the latent variables and the projection of data in the planes defined by the three latent variables.
In Table 4 the confusion matrices of the sensors data classifier are shown. The accuracy of the PLS-DA model of sensor array data is about 91% and 89% in training and test, respectively. Figure 7 shows the ROCs in training and test. The areas under the ROCs are 0.92 ± 0.02 in training and 0.91 ± 0.04 in test. The sensitivity of the classifier is 94% ± 2% in training and 94% ± 3% in test, and the specificity is 87% ± 2% in training and 80% ± 4% in test.
Table 4.
Confusion matrices of PLS-DA classifier of sensors data in training and test
| True | |||||
|---|---|---|---|---|---|
| Training | Test | ||||
| COVID | Non-COVID | COVID | Non-COVID | ||
| Predicted | COVID | 33 | 4 | 17 | 2 |
| Non-COVID | 2 | 28 | 1 | 8 |
Figure 7.
Training and test ROCs of the PLS-DA model calculated with the sensor array data
In the header the AUROCs in training and test are displayed.
The class of controls was made of patients with different conditions; in particular 14 of the non-COVID patients were affected by inflammatory pathologies. Figures 3 and 5 show that subjects with inflammatory pathologies in both GC/MS and sensors data tend to be distributed toward the group of non-COVID. To verify the assumption that inflammatory pathologies are not a confounding factor in COVID-9 identification, PLS-DA has been calculated to evaluate the discrimination of COVID-19 respect to inflammations. Data are numerically limited to perform a full training and test, hence the PLS-DA models have only been cross-validated with a leave-one-out procedure to estimate the classification performance (model details in Figures S14 and S15 in supplemental information). Results show that the accuracy of identification is 91% and 93% with GC/MS and sensor array, respectively. These values are similar to those obtained with the full set of controls. Thus, although limited by the lack of a proper test, this result suggests that inflammatory conditions should play a limited confounding factor in the identification of COVID-19.
Before this study, the serum VOCs involved in the discrimination of COVID-19 were unknown; thus, a general purpose sensor array made of porphyrinoids (porphyrins and corroles) coated quartz microbalances has been used (see STAR Methods section for a description of sensors and sensitive molecules). The discrimination of COVID-19 samples might be explained considering that the interactions between the compounds in Table 1 and sensors are expected to be driven by van der Waals forces and hydrogen bonds, and porphyrinoids coated quartz microbalances are known to be sensitive to these interactions (Paolesse et al., 2017). Nonetheless, the identification of target VOCs will drive the further design of porphyrinoids with improved selectivity toward COVID-19 related compounds.
The achieved classification performances are comparable with those obtained analyzing the VOCs in breath with different instruments such as proton transfer reaction (Grassin-Delyle et al., 2021), gas chromatography coupled with an ion mass detector (Ruszkiewicz et al., 2020) and sensor array (Shan et al., 2020) (see Table S2 in supplemental information). This result points out the diagnostic properties of serum suggesting that it is a valid alternative to breath in several practical situation where breath analysis may be not reliable for instance because of a lack of preparation of patients or for safety reasons as in the case of COVID-19.
Further studies are under development to extend the sample population and to include comorbidities and additional variables to the classification models. Additionally, the relationship between different stages of COVID-19 and VOCs should also be investigated to ascertain not only the diagnostics but also the prognostic properties of volatolomics.
As a final remark, it is important to consider that a full comprehension of the relationship between COVID-19 and VOCs, either in breath or in serum, is still not available. It will be particularly important to investigate the uniqueness of the relationship between VOC patterns and COVID-19 in order to improve the predictions of volatolomics based method toward the optimal goal of 100% sensitivity and specificity.
Limitations of the study
This paper provides a proof-of-concept demonstration that the analysis of the volatile compounds released by blood serum can be used for COVID-19 diagnosis. There were some limitations in the present study.
The first limitation is concerned with the fact that this pilot study was performed in one location, using one instrumentation, and for a given time interval. Environmental influences, repeatability, and long-time performance of the adopted instrumentation need to be verified in multicentric studies.
The second is concerned with the selection of patients. All subjects included in the study were recruited at the emergency department of a large hospital in Rome, Italy. In order to limit any influence of the stability of sensors it was decided to analyze samples from patients admitted in a range of two days. The group of COVID-19 patients show more co-morbidities with respect to non-COVID patients and the group of non-COVID patients included subjects with different pathologies; some of them with symptoms compatible with COVID-19. GC/MS and sensor array indicate that the largest separation among the data is due to the COVID-19 pathology; however, further studies with larger sample sizes are necessary to elucidate if and how co-morbidities influence the serum volatolome.
A third limitation is related to the sampling of VOCs for GC/MS. Sampling for GC/MS was made with a SPME sampler. SPMEs of different composition are available and each provides a different profile of VOCs. Here the same SPME fiber and measurement conditions adopted in a previous serum volatolome investigation was used (Ali et al., 2016). It was a wide range fiber aimed at compounds with a molecular weight in the range 40-275 suitable for untargeted analysis. Comparison between different SPMEs will be necessary for a thorough appraisal of the volatolome composition.
A fourth limitation concerns the sensor array. This was assembled with sensors used in previous in-vivo and in-vitro volatolomics studies. An optimal detection of COVID-19 will require the development of sensors that can maximize the sensitivity toward those compounds that can be associated with the disease.
Finally, a fifth limitation comes from the analysis of data. It is known that PLS-DA models tend to overfit when the number of variables exceeds the number of samples. For this reason the number of variables in GC/MS was reduced by selecting those with p < 0.05. However, with a larger sample the whole set of compounds could have been used for classification. This would lead to an optimal definition of the set of compounds relevant to the identification of COVID-19.
The overcoming of these limitations is expected to lead to the definition of a procedure for COVID-19 diagnosis based on the volatolomics analysis of blood serum. Furthermore, the detection of specific VOCs may shed additional light on the organisms response to COVID-19.
STAR★Methods
Key resource table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Software and algorithms | ||
| Matlab, Statistics and Machine Learning toolbox | Mathworks | matlab.mathworks.com |
| Others | ||
| Solid Phase Micro-Extraction : 50/30 μm Divinylbenzene/Carboxen/PDMS | Supelco | 57328-U |
| Gas Chromatography Mass Spectrometer -QP2010 | Shimadzu | GCMS-QP2010 SE |
| Quartz microbalances | KVG Quartz Crystal Technology GmbH | XA-3828 |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Corrado Di Natale, University of Rome Tor Vergata (dinatale@uniroma2.it)
Materials availability
This study did not generate new unique materials.
Data and code available
GC/MS, sensor array data and MATLAB code are available from the lead contact.
Methods details
Samples selection
97 serum samples were collected from subjects at the Emergency Department of the Policlinico Tor Vergata, a large public hospital in Rome; Italy. COVID-19 was diagnosed in 50 of them.
COVID-19 patients were hospitalized and tested several times by RTPCR to confirm the diagnosis together with the clinical and biochemical status, CT images and serological tests.
The group of non-COVID included four subjects with bacterial pneumonia, four with Central Nervous System (CNS) disorders, four with fever of unknown origin and one with cholecystis. The remnants of non-COVID subjects were affected by symptoms not related to inflammatory pathologies.
The study was approved by the Ethics Committee of Policlinico Tor Vergata (Rome, Italy)(#77.20).
Samples preparation
Blood samples were transferred in serum separator tubes, then centrifuged at 3000 rpm for 20 min and subsequently stored at −20°C. Prior the analysis, 2 mL of the supernatant serum was picked up and poured in two 20 mL crimp top HS glass vials (SUPELCO, Bellefonte, PA, USA) and tightly sealed with a crimp cap with PTFE/silica septum and used for GC/MS and sensor array analysis.3
GC/MS
Samples were left at room temperature for 5 min then immersed for 10 minutes in a water bath maintain ed at 50°C by the adjustable heater C-MAG HS 7 IKAMAG coupled to an ETS-D5 thermometer (IKA®-Werke GmbH & CO. KG, Staufen, Germany). The samples sequence was randomized to avoid memory effects and drift.
Volatile compounds were collected with a Solid Phase Micro-Extraction (SPME) sampler. It was a 50/30 μm Divinylbenzene/Carboxen/PDMS (SUPELCO, Bellefonte, PA, USA). The fiber was held in the vial for 60 minutes keeping the sample at 50°C.
The GC/MS was a GCMS-QP2010 (Shimadzu, Kyoto, Japan). The fiber, removed from the vial was manually inserted in the injection port and thermally desorbed for 3 minutes at 250°C in spitless mode. VOCs were separated in the capillary column EQUITY-5 (30 m × 0.25 mm × 0.25 μm) (SIGMA-Aldrich, St. Louis, MO, USA). The column oven temperature program was initially set at 40°C for 5 minutes, ramped up to 150°C at the rate of 5°C min−1, this temperature was maintained for 2 min, increased to 250°C for 7°C min−1, followed by a rise to 300°C and then kept for 2 min at the final temperature. Total analysis time was 48 min. The fiber was maintained in the injector for 15 min. The ion source temperature was 250°C. Ultra-high purity helium was used as carrier gas in a linear velocity flow mode, at a total flow rate of 5.9 ml/min.
Mass spectra were acquired at an ionization energy of 70 eV, through electron impact. The detector operated in the full scan monitoring mode, and the recorded mass range was between 30 and 400 m/z.
To eliminate undesired compounds related to the ambient or the contamination of the SPME fiber, two blank analysis (not submitted to any extraction procedure) were executed with the same procedure before the exposure to the first serum sample.
Peaks were putatively identified using NIST 127, NIST 147, and NIST17 libraries.
Sensor array
The gas sensor array was an ensemble of twelve quartz microbalances (QMB). In these sensors, a mass change (Δm) on the quartz surface results in frequency changes (Δf) of the electrical output signal of an oscillator circuit to which each sensor is connected. In the low-perturbation regime, Δm and Δf are linearly proportional (Oprea and Weimar, 2019). QMBs had a fundamental frequency of 20 MHz, corresponding to a mass resolution of the order of a few nanograms. Each QMB was functionalized with a different layer of metalloporphyrins and corroles. These are versatile molecules that can host several interaction mechanisms from weak and non-selective dispersion forces to the more specific coordination on the central metal ion (Paolesse et al., 2017).
The sensor array used for these experiments was the last version of a series of electronic noses designed and manufactured, since 1996, at the University of Rome Tor Vergata.
These sensors have been used in several metabolomics studies. For instance, they have been used in breath analysis to detect lung cancer (Gasparri et al., 2016; Di Natale et al., 2003) and tuberculosis (Zetola et al., 2017), to study in vivo and in vitro the differentiation processes of stem cells (Capuano et al., 2017;(Capuano et al., 2018) ), to study the volatile compounds of a murine model of cancer related protein knockdown (Murdocca et al., 2019), and to analyze the volatile compounds released by malaria murine models (Capuano et al., 2017) and malaria infected human erythrocytes (Capuano et al., 2019).
The core of the instrument is an array of up to twelve 20 MHz Quartz Microbalances (QMBs). The chemical sensitivity of the sensors is provided by a molecular layer of porphyrinoids (see below). Sensors are placed in a measurement cell whose volume is approximately 8 cm3. The gas sensors are complemented by temperature and relative humidity sensors. Each QMB is connected to an oscillator circuit, the frequencies of the oscillators' outputs are measured taking advantage of a temperature compensated reference quartz that allows for a frequency resolution of 0.1 Hz. Electronics is implemented in a FPGA. Gaseous samples delivery is controlled by an embedded low noise, high-precision and durable miniature diaphragm pump (0-200 sccm) and an optimized PMMA CNC machined micro channel manifold.
The instrument is connected and powered via a single USB connection with a PC. The electronic nose functions and the data acquisition are controlled with an in-house written software running in Matlab.
Each QMB was functionalized with a molecular film made with porphyrinoids, such as porphyrins and corroles (Paolesse et al., 2017). Porphyrinoids have been synthesized and characterized according to literature methods (Bhyrappa and Krishnan, 1991; Brand and Arnold, 1995; Buchler, 1978; Paolesse et al., 2003)
Sensor array measurements
Quartz microbalance sensors must be operated at room temperature and with a constant flux. For these reasons the protocol used for GC/MS was not applicable to sensors measurements.
Serum samples were maintained at ambient temperature for 10 min before analysis, in order to reach a steady headspace. Sensor's measurements were performed with dynamic headspace sampling (Burlachenko et al., 2016). The headspace of the vial was transferred at the rate of 50 sccm and for 30 s to the sensors. To maintain pressure constant in the vial and to restore the volatile compounds, filtered laboratory air was reinjected in the vial. Laboratory air was filtered with a CaCl2 bed. The sensor's response was evaluated as the difference between the signals achieved with sample and filtered laboratory air. To remove any influence from changes in the laboratory air an empty vial was measured before each sample. The difference between the response to sample and the empty vial was considered as sensor response to the sample. For each serum sample the procedure was performed in triplicate and the average response was considered for successive analysis. The sequence of samples was randomized to avoid memory effects and sensor drift.
Quantification and statistical analysis
The statistical differences of GC/MS peaks abundances and sensors data in COVID-19 and non-COVID samples were evaluated with the Kruskal-Wallis rank sum test.
The absolute abundances of GC-MS identified peaks and the sensor responses were arranged in matrices and analyzed with multivariate data analysis (Trygg et al., 2007). Principal component analysis (PCA) (Jolliffee, 2002) and Partial Least Squares Discriminant Analysis (PLS-DA) (Barker and Rayens, 2003) were calculated on autoscaled data matrices. In autoscaling each variable of the matrices (GC/MS peaks and sensors signals) were normalized to null mean and unitary variance.
PLS-DA classifiers were optimized with a k-fold cross-validation procedure. Accuracy, sensitivity, specificity and area under the curve were evaluated to assess the performance of the classification. Since PLS-DA is prone to overfit, it is important to evaluate the performance of the classifier with a test data set not used in training. For this, the whole set of GC/MS and sensor array data were randomly split in training and test sets with 30% of samples used for tests. To avoid any bias in the split of data in the two groups, PLS-DA models were calculated 100 times on random training and test partitions, and the average results were considered as the performance of the classifiers.
All calculations were performed in Matlab R2020b, PCA and PLS-DA were calculated with the Statistics and Machine Learning toolbox of Matlab.
Role of funding source
This research did not receive funds.
Author contributions
C.D.N. and S.B. conceived the study; M.M. and J.M.L. collected epidemiological and clinical data; Y.K.M. and L.D.Z. performed the experiments; A.C. and R.P. prepared the protocol measurement for GC/MS and sensor array; R.C. and C.D.N. analyzed and interpreted the data; C.D.N. and S.B. drafted the manuscript; all authors read and approved the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: August 20, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102851.
Supplemental information
References
- Ali S.E., Farag M.A., Holvoet P., Hanafi R.S., Gad M.Z. A comparative metabolomics approach reveals early biomarkers for metabolic response to acute myocardial infarction. Sci. Rep. 2016;6:36359. doi: 10.1038/srep36359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann A., Costello B.D.L., Miekisch W., Schubert J., Buszewski B., Pleil J., Ratcliffe N., Risby T. The human volatilome: volatile organic compounds (VOCs) in exhaled breath, skin emanations, urine, feces and saliva. J. Breath Res. 2014;8:0340001. doi: 10.1088/1752-7155/8/3/034001. [DOI] [PubMed] [Google Scholar]
- Barker M., Rayens W. Partial least squares for discrimination. J. Chemom. 2003;17:166–173. [Google Scholar]
- Boots A.W., Van Berkel J.J.B.N., Dallinga J.W., Smolinska A., Wouters E.F., Van Schooten F.J. The versatile use of exhaled volatile organic compounds in human health and disease. J. Breath Res. 2012;6:027108. doi: 10.1088/1752-7155/6/2/027108. [DOI] [PubMed] [Google Scholar]
- Bhyrappa P., Krishnan V. Octabromotetraphenylporphyrin and its metal derivatives: electronic structure and electrochemical properties. Inorg. Chem. 1991;30:239–245. [Google Scholar]
- Brand H., Arnold J. Recent developments in the chemistry of early transition metal porphyrin compounds. Coord. Chem. Rev. 1995;140:137–168. [Google Scholar]
- Broza Y.Y., Mochalski P., Ruzsanyi V., Amann A., Haick H. Hybrid volatolomics and disease detection. Angew. Chem. Int. Ed. Engl. 2015;54:11036–11048. doi: 10.1002/anie.201500153. [DOI] [PubMed] [Google Scholar]
- Bruzzone C., Bizkarguenaga M., Gil-Redondo R., Diercks T., Arana E., García de Vicuña A., Seco M., Bosch A., Palazón A., San Juan I. SARS-CoV-2 infection dysregulates the metabolomic and lipidomic profiles of serum. IScience. 2020;23:101645. doi: 10.1016/j.isci.2020.101645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchler J.W. Synthesis and properties of metalloporphyrins. In: Dolphin D., editor. Volume 1. Academic Press; 1978. pp. 389–483. (The Porphyrins). [Google Scholar]
- Burlachenko J., Kruglenko I., Snopok B., Persaud K. Sample handling for electronic nose technology: state of the art and future trends. TrAC - Trends Anal. Chem. 2016;82:222–236. [Google Scholar]
- Capuano R., Domakoski A.C., Grasso F., Picci L., Catini A., Paolesse R., Sirugo G., Martinelli E., Ponzi M., Di Natale C. Sensor array detection of malaria volatile signature in a murine model. Sens. Actuators B Chem. 2017;245:341–351. [Google Scholar]
- Capuano R., Khomenko I., Grasso F., Messina V., Olivieri A., Cappellin L., Paolesse R., Catini A., Ponzi M., Biasioli F. Simultaneous Proton Transfer Reaction-Mass Spectrometry and electronic nose study of the volatile compounds released by Plasmodium falciparum infected red blood cells in vitro. Sci. Rep. 2019;9:12360. doi: 10.1038/s41598-019-48732-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capuano R., Spitalieri P., Talarico R., Catini A., Domakoski A., Martinelli E., Scioli M., Orlandi A., Cicconi R., Paolesse R. Volatile compounds emission from teratogenic human pluripotent stem cells observed during theirdifferentiation in vivo. Scientific Reports. 2018;8 doi: 10.1038/s41598-018-29212-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciotti M., Maurici M., Pieri M., Andreoni M., Bernardini S. Performance of a rapid antigen test in the diagnosis of SARS-CoV-2 infection. J. Med. Virol. 2021;93:2988–2991. doi: 10.1002/jmv.26830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Natale C., Paolesse R., Martinelli E., Capuano R. Solid-state gas sensors for breath analysis: a review. Anal. Chim. Acta. 2014;824:1–17. doi: 10.1016/j.aca.2014.03.014. [DOI] [PubMed] [Google Scholar]
- Di Natale C., Macagnano A., Martinelli E., Paolesse R., D’Arcangelo G., Roscioni C., Finazzi-Agrò A., D’Amico A. Lung cancer identification by the analysis of breath by means of an array of non-selective gas sensors. Biosens. Bioelectron. 2003;18:1209–1218. doi: 10.1016/s0956-5663(03)00086-1. [DOI] [PubMed] [Google Scholar]
- Dunn W.B., Broadhurst D., Begley P., Zelena E., Francis-Mcintyre S., Anderson N., Brown M., Knowles J.D., Halsall A., Haselden J.N. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011;6:1060–1083. doi: 10.1038/nprot.2011.335. [DOI] [PubMed] [Google Scholar]
- Einoch Amor R., Nakhleh M.K., Barash O., Haick H. Breath analysis of cancer in the present and the future. Eur. Respir. Rev. 2019;28:190002. doi: 10.1183/16000617.0002-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparri R., Santonico M., Valentini C., Sedda G., Borri A., Petrella F., Maisonneuve P., Pennazza G., D’Amico A., Di Natale C. Volatile signature for the early diagnosis of lung cancer. J. Breath Res. 2016;10:016007. doi: 10.1088/1752-7155/10/1/016007. [DOI] [PubMed] [Google Scholar]
- Grassin-Delyle S., Roquencourt C., Moine P., Saffroy G., Carn S., Heming N., Fleuriet J., Salvator H., Naline E., Couderc L.-J. Metabolomics of exhaled breath in critically ill COVID-19 patients: a pilot study. EBioMedicine. 2021;63:103154. doi: 10.1016/j.ebiom.2020.103154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haick H., Broza Y.Y., Mochalski P., Ruzsanyi V., Amann A. Assessment, origin, and implementation of breath volatile cancer markers. Chem. Soc. Rev. 2014;43:1423–1449. doi: 10.1039/c3cs60329f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haick H., Vishinkin R., Di Natale C., Marco S. Sensor systems. In: Beauchamp J., Davis C., Pleil J., editors. Breathborne Biomarkers and the Human Volatilome. Elsevier B.V.); 2020. pp. 202–220. [Google Scholar]
- Hendriks M.M.W.B., Eeuwijk F.A., Jellema R.H., Westerhuis J.A., Reijmers T.H., Hoefsloot H.C.J., Smilde A.K. Data-processing strategies for metabolomics studies. TrAC - Trends Anal. Chem. 2011;30:1685–1698. [Google Scholar]
- Hierlemann A., Gutierrez-Osuna R. Higher-order chemical sensing. Chem. Rev. 2008;108:563–613. doi: 10.1021/cr068116m. [DOI] [PubMed] [Google Scholar]
- Jolliffee I. second edition. Springer US; 2002. Principal Component Analysis. [Google Scholar]
- Lamote K., Janssens E., Schillebeeckx E., Lapperre T.S., De Winter B.Y., Van Meerbeeck J.P. The scent of COVID-19: viral (semi-)volatiles as fast diagnostic biomarkers? J. Breath Res. 2020;14:042001. doi: 10.1088/1752-7163/aba105. [DOI] [PubMed] [Google Scholar]
- Leiherer A., Ślefarska D., Leja M., Heinzle C., Mündlein A., Kikuste I., Mezmale L., Drexel H., Mayhew C.A., Mochalski P. The volatilomic footprints of human HGC-27 and CLS-145 gastric cancer cell lines. Front. Mol. Biosci. 2021;7:607904. doi: 10.3389/fmolb.2020.607904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wang X., Chen J., Zuo X., Zhang H., Deng A. COVID-19 infection may cause ketosis and ketoacidosis. Diabetes Obes. Metab. 2020;22:1935–1941. doi: 10.1111/dom.14057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B., Liu J., Liu Y., Qin X. Progress in understanding COVID-19: insights from the omics approach. Crit. Rev. Clin. Lab. Sci. 2020;58:242–252. doi: 10.1080/10408363.2020.1851167. [DOI] [PubMed] [Google Scholar]
- Mattioli I.A., Hassan A., Oliveira O.N., Crespilho F.N. On the challenges for the diagnosis of SARS-CoV-2 based on a review of current methodologies. ACS Sensors. 2020;5:3655–3677. doi: 10.1021/acssensors.0c01382. [DOI] [PubMed] [Google Scholar]
- Mochalski P., Rudnicka J., Agapiou A., Statheropoulos M., Amann A., Buszewski B. Near real-time VOCs analysis using an aspiration ion mobility spectrometer. J. Breath Res. 2013;7:026002. doi: 10.1088/1752-7155/7/2/026002. [DOI] [PubMed] [Google Scholar]
- Murdocca M., Capuano R., Pucci S., Cicconi R., Polidoro C., Catini A., Martinelli E., Paolesse R., Orlandi A., Mango R. Targeting LOX-1 inhibits colorectal cancer metastasis in an animal model. Front. Oncol. 2019;9:927. doi: 10.3389/fonc.2019.00927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oprea A., Weimar U. Gas sensors based on mass-sensitive transducers part 1: transducers and receptors—basic understanding. Anal. Bioanal. Chem. 2019;411:1761–1787. doi: 10.1007/s00216-019-01630-7. [DOI] [PubMed] [Google Scholar]
- Paolesse R., Marini A., Nardis S., Froiio A., Mandoj F., Nurco D.J., Prodi L., Montalti M., Smith K.M. Novel routes to substituted 5,10,15-triarylcorroles. J. Porphyr. Phthalocyanines. 2003;7:25–36. [Google Scholar]
- Paolesse R., Nardis S., Monti D., Stefanelli M., Di Natale C. Porphyrinoids for chemical sensor applications. Chem. Rev. 2017;117:2517–2583. doi: 10.1021/acs.chemrev.6b00361. [DOI] [PubMed] [Google Scholar]
- Phillips M., Herrera J., Krishnan S., Zain M., Greenberg J., Cataneo R.N. Variation in volatile organic compounds in the breath of normal humans. J. Chromatogr. B Biomed. Sci. Appl. 1999;729:75–88. doi: 10.1016/s0378-4347(99)00127-9. [DOI] [PubMed] [Google Scholar]
- Phillips M., Cataneo R.N., Chaturvedi A., Kaplan P.D., Libardoni M., Mundada M., Patel U., Zhang X. Detection of an extended human volatome with comprehensive two-dimensional gas chromatography time-of-flight mass spectrometry. PLoS One. 2013;8:e75274. doi: 10.1371/journal.pone.0075274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruszkiewicz D.M., Sanders D., O’Brien R., Hempel F., Reed M.J., Riepe A.C., Bailie K., Brodrick E., Darnley K., Ellerkmann R. Diagnosis of COVID-19 by analysis of breath with gas chromatography-ion mobility spectrometry - a feasibility study. EClinicalMedicine. 2020;29–30:100609. doi: 10.1016/j.eclinm.2020.100609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serasanambati M., Broza Y.Y., Marmur A., Haick H. Profiling single cancer cells with volatolomics approach. iScience. 2019;11:178–188. doi: 10.1016/j.isci.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan B., Broza Y.Y., Li W., Wang Y., Wu S., Liu Z., Wang J., Gui S., Wang L., Zhang Z. Multiplexed nanomaterial-based sensor array for detection of COVID-19 in exhaled breath. ACS Nano. 2020;14:12125–12132. doi: 10.1021/acsnano.0c05657. [DOI] [PubMed] [Google Scholar]
- Shen B., Yi X., Sun Y., Bi X., Du J., Zhang C., Quan S., Zhang F., Sun R., Qian L. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182:59–72, e15. doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D., Yan R., Lv L., Jiang H., Lu Y., Sheng J., Xie J., Wu W., Xia J., Xu K. The serum metabolome of COVID-19 patients is distinctive and predictive. Metabolism. 2021;118:154739. doi: 10.1016/j.metabol.2021.154739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymańska E., Saccenti E., Smilde A.K., Westerhuis J.A. Double-check: validation of diagnostic statistics for PLS-DA models in metabolomics studies. Metabolomics. 2012;8:3–16. doi: 10.1007/s11306-011-0330-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trygg J., Holmes E., Lundstedt T. Chemometrics in metabonomics. J. Proteome Res. 2007;6:469–479. doi: 10.1021/pr060594q. [DOI] [PubMed] [Google Scholar]
- Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C., Chen H., Mubareka S., Gubbay J.B., Chan W.C.W. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- Wishart D.S. Emerging applications of metabolomics in drug discovery and precision medicine. Nat. Rev. Drug Discov. 2016;15:473–484. doi: 10.1038/nrd.2016.32. [DOI] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z.-Q., Broza Y.Y., Ionsecu R., Tisch U., Ding L., Liu H., Song Q., Pan Y.-Y., Xiong F.-X., Gu K.-S. A nanomaterial-based breath test for distinguishing gastric cancer from benign gastric conditions. Br. J. Cancer. 2013;108:941–950. doi: 10.1038/bjc.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetola N.M., Modongo C., Matsiri O., Tamuhla T., Mbongwe B., Matlhagela K., Sepako E., Catini A., Sirugo G., Martinelli E. Diagnosis of pulmonary tuberculosis and assessment of treatment response through analyses of volatile compound patterns in exhaled breath samples. J. Infect. 2017;74:367–376. doi: 10.1016/j.jinf.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.