Abstract
It has been well-established that cancer cells often display altered metabolic profiles, and recent work has concentrated on how cancer cells adapt to serine removal. Serine can be either taken exogenously or synthesized from glucose, and its regulation forms an important mechanism for nutrient integration. One of several important metabolic roles for serine is in the generation of bioactive sphingolipids since it is the main substrate for serine palmitoyltransferase, the initial and rate limiting enzyme in the synthesis of sphingolipids. Previously, serine deprivation has been connected to the action of the tumor suppressor p53, and we have previously published on a role for p53 regulating sphingosine kinase 1 (SK1), an enzyme that phosphorylates sphingosine to form sphingosine-1-phosphate (S1P). SK1 is a key enzyme in sphingolipid synthesis that functions in pro-survival and tumor promoting pathways and whose expression is also often elevated in cancers. Here we show that SK1 was degraded during serine starvation in a time and dose dependent manner, which led to sphingosine accumulation. This was independent of effects on p53 but required the action of the proteasome. Furthermore, we show that overexpression of SK1, to compensate for SK1 loss, was detrimental to cell growth under conditions of serine starvation, demonstrating that the suppression of SK1 under these conditions is adaptive. Mitochondrial oxygen consumption decreased in response to SK1 degradation, and this was accompanied by an increase in intracellular reactive oxygen species (ROS). Suppression of ROS with N-acteyl-cysteine (NAC) resulted in suppression of the metabolic adaptations and in decreased cell growth under serine deprivation. The effects of SK1 suppression on ROS were mimicked by D-erythro sphingosine, whereas S1P was ineffective, suggesting that the effects of loss of SK1 were due to accumulation of its substrate sphingosine. This study reveals a new mechanism for regulating SK1 levels, and a link of SK1 to serine starvation as well as mitochondrial function.
Introduction
Altered metabolic profiles are commonly observed in cancer cells (1). While the mechanisms behind such changes can vary, the changes are generally a result of the metabolic needs of rapid cell division, nutrient limiting conditions, and/or to counteract the metabolic stress of oncogene activation or tumor suppressor loss (2). An emerging important effector of metabolic reprogramming is serine loss (3). Serine is a non-essential amino acid biosynthesized from glucose and plays an important role in the biosynthesis of nucleotides, NADPH generation, methylation reactions and sphingolipid synthesis, and a precursor to several amino acids such as cysteine and glycine (3, 4). It was previously shown that serine deprivation in the HCT-116 cell line results in growth arrest in a p53 dependent manner, allowing for metabolic remodeling (5). This work has been extended by studying the implications of the ‘relative’ dependence of cancer cells on external serine.
Serine is an essential component of sphingolipids, being a starting molecule in the synthesis of sphingolipids through the action of serine palmitoyltransferase (SPT). Bioactive sphingolipids include ceramides which are often (but by no means exclusively) associated with apoptosis, S1P which is often associated with cellular survival mechanisms (6), and sphingosine, which is also involved in cell growth regulation. Sphingosine is generated from the hydrolysis of ceramides through the action of ceramidases, and it is phosphorylated to S1P by that action of sphingosine kinase (SK) 1 and 2 (7). There are two major isoforms (SK1 and SK2) that are encoded by separate genes and which share 80% similarity of their amino acid sequences, both of which have been associated with cellular survival pathways (8).
Studies of serine deprivation have implicated a role for mitochondria in their downstream effects. Serine donates a carbon to the one carbon cycle which is important in nucleotide synthesis, glycine synthesis, and the integration of nutrient status, many of the enzymes needed are found in the mitochondria (9). Interestingly, it has been reported that serine synthesis is necessary for cellular respiration, vital for the assembly of Complex I (10). In addition, mitochondrial DNA depletion has been shown to increase serine synthesis, while disruption of the mitochondrial respiratory could also inhibit the one carbon cycle (11).
Sphingolipids, including ceramides, have also been previously shown to be intimately involved in the regulation of mitochondrial functions (12). Increased ceramide generation in the mitochondrial membranes has been observed in isolated rat liver mitochondria (13). During mitochondrial-mediated apoptosis, increased ceramides allow for pore formation and the insertion of BAX (14, 15). In yeast, it was observed that the increase of mitochondrial sphingosine species compromise mitochondrial activity and shorten yeast life span (16). Other studies have shown that ceramide species can affect the electron transport chain (17). On the other hand, inhibition of S1P receptor 2 promoted mitochondrial dysfunction in human renal glomerular endothelial cells (18). Additionally, oxidative stress has been reported to inhibit SK1 activity, while conversely inhibition of SK1 activity has been reported to increase ROS (19, 20).
In addition to their role in generating ATP, mitochondria play an important role in the generation of metabolites and their byproducts (such as reactive oxygen species, ROS) and in the coordination of cellular responses to metabolic stress (21). Mitochondria generate ATP through electron transport, polarizing mitochondrial membranes in order to pump protons into the inner mitochondrial membrane (22). Therefore, regulation and maintenance of mitochondrial activity are vital for normal cellular function, and utilize many biochemical and biophysical strategies (23). Changes in mitochondrial function are commonly observed in cancer cells such as membrane depolarization, that can lead to reduced electron transport, reduced oxygen consumption and mitophagy (24). Many of these mitochondrial changes are often selected for by suboptimal tumor microenvironmental conditions (25).
We have previously shown that induction of p53 results in degradation of SK1 and modulation of levels of bioactive sphingolipids (increase ceramides and sphingosine and decreased S1P) (26). Therefore, we set out to determine whether serine starvation had any effect on SK1 activity. The results show that serine deprivation does induce loss of SK1 but in a p53-independent mechanism. In contrast to previous studies, we show that in the context of serine starvation, SK1 expression is detrimental to cell survival, through a mechanism involving mitochondria.
Materials and Methods
Cell lines and experiments
The human colon carcinoma cell line HCT 116 was obtained from the ATCC. Homozygous p53 knockout HCT 116 was a gift from Dr. Ute Moll and first described by Bunz et al (27). The human embryonic lung cell line HeL 299, human embryonic kidney cell line HEK 293T and HCT-116 cells were grown in DMEM, while the human myelogenous leukemia cell line K562 was grown in RPMI 1640. Cell lines were verified by ATCC STR profiling and checked monthly for Mycoplasma. For experiments, cells were initially seeded in high glucose DMEM (11965) with 10% FBS and 2 mM L-glutamine and kept at 37 °C in humidified 5% CO2 in air. For experiments on the effects of serine starvation, complete medium was formulated to closely match the nutrient composition of DMEM (which contains 0.4 mM serine and 0.4 mM glycine); complete medium consisted of MEM (21090) supplemented with additional 1× MEM vitamins (11120), 1xMEM amino acids (11130), 10% dialyzed-FBS (HyClone, Thermo Scientific, Bridgewater, NJ, USA), L-glutamine 2 mM, additional D-glucose (to 25 mM), serine 0.4 mM (42mg/l) and glycine 0.4 mM. For starvation experiments, cells were fed the same medium formulation without serine and glycine. Bortezumib was purchased from Selleckchem. Caspase 2 (Z-VDVAD-FMK) and caspase-7 inhibitors (Z-DEVD-FMK) were obtained through R&D Systems (Minneapolis, MN, USA). The PHGDH inhibitor CBR 5884 (Sigma) was used at 50μM. Sphingolipids were purchased from Avanti Polar Lipids Inc (Alabaster, AL, USA). Gene silencing of SK2 was carried out using siRNA directed against human SK2 (Hs_SPHK2_5 FlexiTube siRNA SI00288561; Qiagen, Germantown, MD, USA); wit, with Qiagen AllStar (AStar) siRNA as a negative control. Gene silencing of Kelch-like protein 5 was carried out using ON-TARGETplus SMARTpool siRNAs (Horizon Discovery, Lafayette, CO, USA)) Transfections were carried out using Lipofectamine RNAiMax according to the manufacturer’s protocol.
SK1 RT-PCR
RNA extraction and cDNA synthesis were carried out using PureLink® RNA Mini Kit (Life Technologies, Carlsbad, CA, USA) and Quanta qScript cDNA SuperMix (Quanat Biosciences, Beverly, MA, USA), respectively, and according to the manufacturer’s instructions. PCR analysis of cDNA was performed using the Applied Biosystems 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) using Taqman probe (Life Technologies) human SK1 (Hs00184211_m1). Cycle threshold (C t) values were obtained for each gene of interest and β -actin. ΔC t values were calculated and the relative gene expression normalized to control samples was calculated from ΔΔ C t values.
SK1 stable overexpression
To generate HCT-116 cell lines overexpressing empty vector or SK1, lentiviral particles were produced by transfecting HEK-293T cells (ATCC) with 2μg each of VSV-G, dVPR, and target constructs using X-tremeGENE 9 (Roche, Basel, Switzerland). Viral containing media was harvested 72h post-transfection, filtered (0.45μm PVDF filter), and stored at −80C. HCT-116 cells were transduced at 70% confluency using 8μg/ml polybrene (Millipore, Burlington, MA, USA) and selected with antibiotic (50μg/ml blasticidin, InVivoGen, San Diego, CA, USA) for 8 days.
CRISPR/Cas9-mediated gene knockouts
To knockout SK1 from HCT-116 cells (SK1 KO), the LentiCRISPR v2.0 system was used as we have previously described (28). The LentiCRISPR v2 plasmid was a gift from Feng Zhang (Addgene plasmid #52961). Briefly, guide RNAs targeting the SK1 gene (SK1-A gRNA target sequence: GGTTTTGTTTTCTCAGCGGG, SK1-B gRNA target sequence: CGTGCAGCCCCTTTTGGCTG) were designed using the ChopChop algorithm (http://chopchop.cbu.uib.no/) and cloned into the plasmid according to the manufacturer’s conditions. To generate lentivirus, plasmids were co-transfected with VSV-G and dVPR into 293T cells and virus-containing medium was harvested and filtered (0.22μm PVDF membrane) after 72 h. HCT-116 cells (100,000) were infected with 1 ml virus in the presence of polybrene (8 μg/ml). After 48 h, cells were selected in puromycin (2 μg/ml) for 7 days. Subsequently, cells were maintained in normal growth medium. Validation of genetic modification was performed with the GeneArt genomic cleavage detection kit (Thermo Scientific) according to manufacturer’s instructions. Loss of cellular SK1 was verified using both RT-PCR and Western blot. The SK1 expression status was validated as described by Pulkoski-Gross et al. (28).
Proliferation assays
HCT 116 cells were seeded in 6-well plates at 105 per well and allowed to grow for 24 h. The medium was aspirated, and the cells were washed with PBS, before replacement with complete or serine/glycine-free media. Cells were counted daily using a Countess automated cell counter (Invitrogen, Carlsbad, CA, USA). Each condition was plated in triplicate and read twice. Data are presented as a mean of each reading ± s.d.m.
Determination of ROS release
Two methods for ROS detection were employed. MitoSOX (ThermoFisher Scientific) was used to detect ROS (being more reactive to superoxide, and less reactive to singlet oxygen and peroxide). Cells were plated onto sterilized glass slides before the medium was changed 24 h before microscopy. MitoSOX reagent (Absorption/emission maxima: ~510/580 nm) was used according to the manufacturer’s directions and visualized using a Zeiss Axio Imager M2. Images were quantified using the FiJi extension to ImageJ. For quantification of ROS-induced fluorescence, cells were plated in a 96 well plate in triplicate. After loading with DCFH-DA (Cell Biolabs Inc, San Diego, CA, USA) and washing, the cells were lysed and specific fluorescence (Ex/Em = 495/529 nm) measured using a Spectramax M5 (Molecular Devices, San Jose, CA, USA). N-acetyl cysteine (NAC) (Sigma, Saint Louis, MO, USA) was made up fresh before each usage, filter sterilized and used at a final concentration of 0.5 mM.
Western blotting
Whole cell lysates were prepared in RIPA buffer containing protease inhibitors (Santa Cruz) and quantified using the BCA Protein Assay (Thermo Scientific). Equal amounts of protein were loaded and separated on pre-cast Novex 4–20% SDS–PAGE midi gels (Life Technologies, Carlsbad, CA, USA), transferred to nitrocellulose membranes, and probed with antibodies against SK1 (12071), p53 (9282), p21waf1/cip1 (2947), and pERM (3726) from Cell Signaling Technology (Danvers, MA, USA); SK2 (ab37977) from Abcam; and Actin (A5441) from Sigma. Secondary HRP-conjugate Abs were purchased from Jackson Laboratories (Bar Harbor, ME, USA). Protein bands were revealed using Enhanced Chemiluminescence reagent (Thermo Scientific).
Live cell oxygen consumption
An XFe96 Extracellular Flux Analyzer (Agilent Technologies, Santa Clara, CA, USA) was used to measure oxygen consumption rates (OCR) as described (29). In brief, HCT-116 cells were seeded at a density of 12,000 cells per well of an XFe96 cell culture microplate and incubated for 24 h to ensure attachment. Before the assay, cells were equilibrated for 1 h at 37oC in unbuffered XF assay medium supplemented with 25 mM glucose, and 2 mM glutamine, in a non-CO2 incubator. Mitochondrial processes were examined through sequential injections of oligomycin (0.5 μg ml−1), carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP, 1 μM), and rotenone (0.5 μM)/antimycin A (1 μM). Each value was normalized to total nucleic acid quantified using CyQuant assay (Invitrogen). Results were presented as mean ± s.d.m.
Sphingolipid analysis
Cells were scraped and pelleted, and lipid extraction and analysis by LC/MS mass spectrometry were performed as described previously (30). Lipids were normalized to total lipid phosphate levels of the selected sample.
13C isotope tracing experiments.
Cells were treated for 24 h before the medium was changed to one that was pre-warmed to 37°C and containing [13C6]-glucose (Cambridge Isotope Laboratories, Tewksbury, MA, USA). Cells were incubated for a further 12 h before the medium was removed and cells were scraped into 50% ice-cold methanol. Before scraping, 1μl of a 5 mM solution of Adonitol was added as an internal standard. Cells were lysed in methanol by three freeze/thaw cycles (transfer of samples from liquid nitrogen to 37oC water bath). The samples were dried under nitrogen and resuspended in 50 μl 2% MOX reagent (Pierce #45950). After 1 h incubation at 42°C, samples were dried and TMS-derivatized in 100 μl N, O-Bistrifluoroacetamide (Sigma Aldritch) for 1 h at 75°C. Derivatized samples were analyzed by gas chromatography electron impact mass spectrometry. Sample peaks were identified by matching retention times and EI fragmentation patterns to neat standards using Agilent MassHunter Software.
Statistical analysis
All experiments have been independently repeated at least three times. One-way ANOVA was used for three or more group comparisons and Student’s t-test was used for two group comparisons. A P value less than 0.05 was considered significant.
Results
Serine deprivation causes downregulation of SK1 protein.
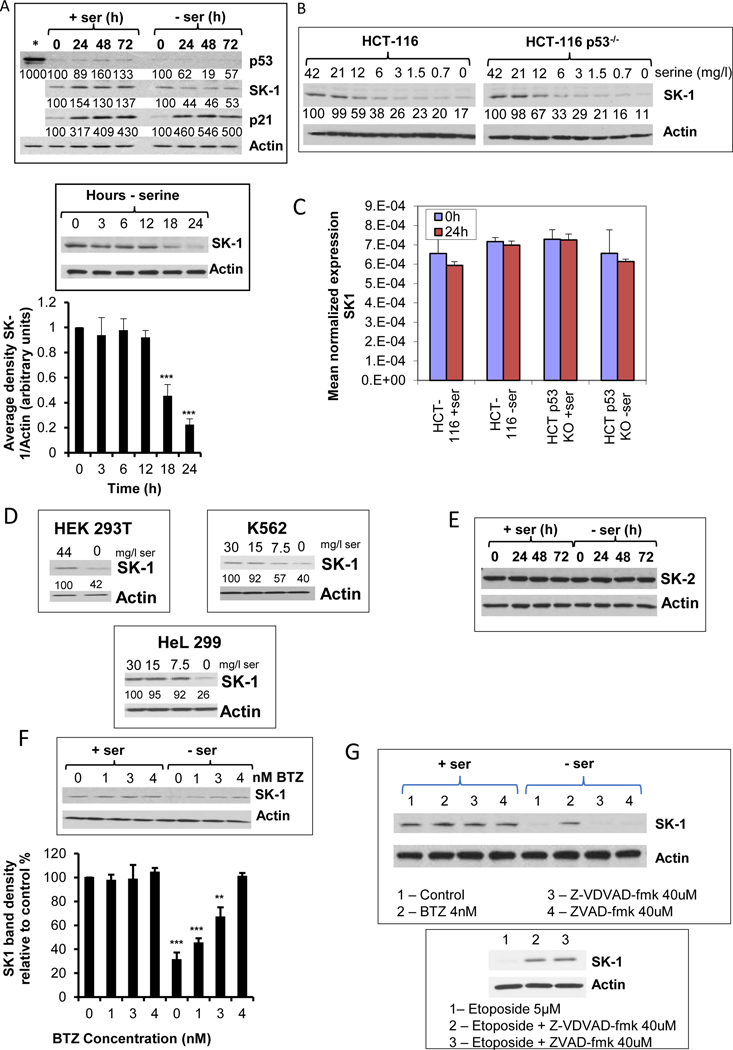
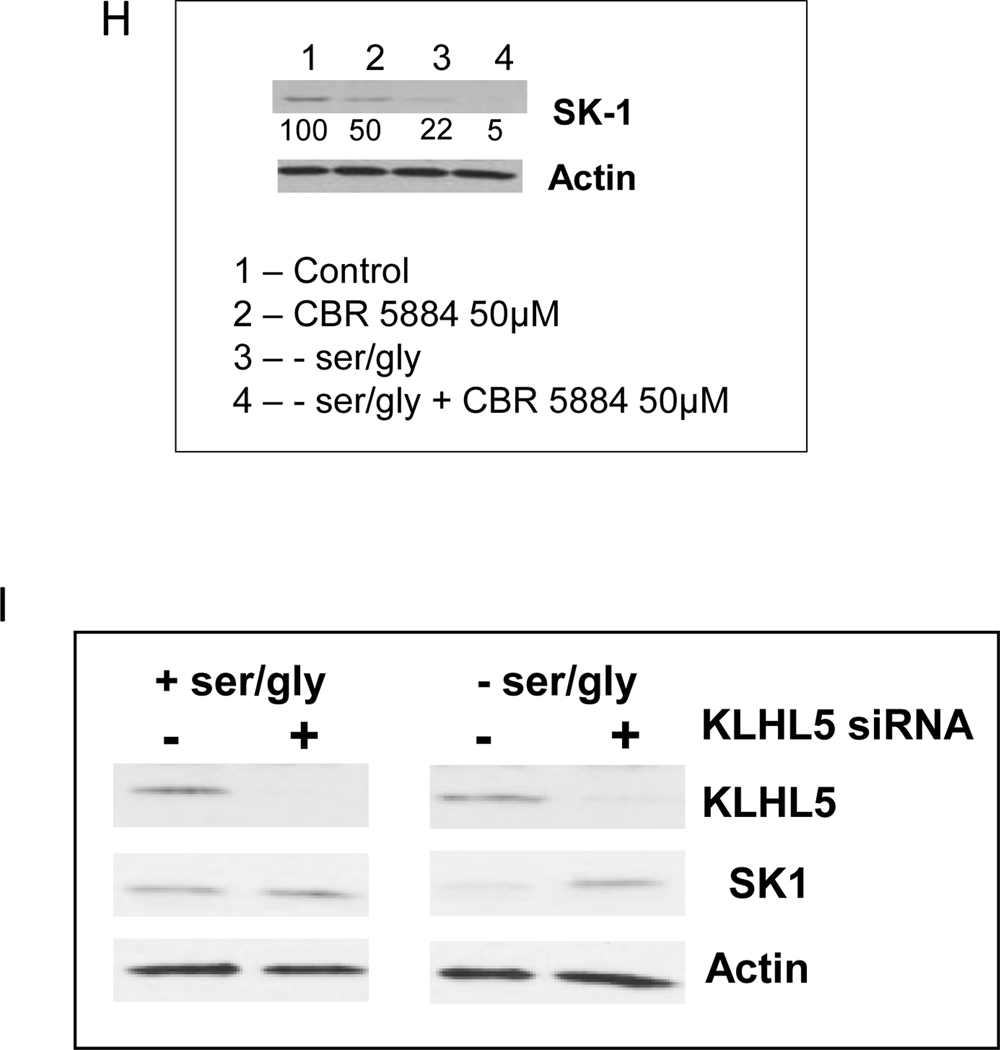
It was previously shown that serine deprivation in HCT-116 cells results in growth arrest in a p53 dependent manner, allowing for metabolic remodeling (5). Additionally, in previous studies we have shown that p53 activation results in caspase-mediated degradation of SK1 (26). In order to determine whether a p53-dependent mechanism regulates SK1 expression during ser/gly starvation, we first evaluated the ability of ser/gly starvation to regulate p53 in HCT-116 cells. Fig. 1A shows that there was no appreciable increase in p53 protein following ser/gly starvation. On the other hand, a steady increase in p21waf1/cip1 protein, over a period of up to three days, was observed as cells grew to confluence either in presence or absence of serine (Fig. 1A). Of note, however, was the observation that during ser/gly starvation, SK1 protein levels significantly decreased (Fig. 1A), and this was initiated between 12 h and 18 h after ser/gly starvation (Fig. 1A). This was in stark contrast to the increase in the levels of SK1 during growth in the presence of serine. SK1 levels were not changed by the addition of L-glycine (data not shown), nor did L-glycine starvation alone change SK1 protein levels (data not shown), demonstrating that this effect was specific to serine. Next, we evaluated the role of p53, we employed the p53-knockout HCT-116 cell line (HCT116 p53 KO), which is isogenic to HCT 116 that express wild-type p53. The results showed that SK1 levels were decreased in both HCT 116 and HCT 116 p53 KO cells during ser/gly starvation (Fig. 1B), ruling out p53-mediated loss of SK1. Levels of SK1 mRNA in ser/gly starved cells remained mostly unchanged when compared to normal media (Fig. 1C), suggesting the loss of SK1 protein was likely through proteolytic degradation. Interestingly, SK1 loss was also observed in a range of different cell lines subjected to growth conditions with suboptimal levels of serine (Fig. 1D). On the other hand, levels of SK2 protein and mRNA remained unchanged in ser/gly starved cells, suggesting a specific mechanism for SK1 removal (Fig. 1E).To confirm proteolysis of SK1 during ser/gly starvation, cells were incubated with the proteasomal inhibitor bortezomib (31). At doses as low as 4 nM, bortezomib was able to inhibit SK1 proteolysis 24 h after the initiation of ser/gly starvation (Fig. 1F). Caspase 2 and caspase 7 inhibitors, previously shown to inhibit p53-induced SK1 proteolysis in response to genotoxic drugs (26, 32, 33), had no effect on ser/gly-starvation-induced SK1 proteolysis (Fig. 1G), although both caspase inhibitors were able to protect SK1 from proteolysis induced by etoposide (Fig. 1G, bottom) suggesting a previously unappreciated mechanism of controlling SK1 protein levels. In an attempt to ascertain the mechanism behind the downregulation of SK1, the role of Kelch-like protein 5 (KLHL5) was investigated. It has been previously shown by Powell et al, that KLHL5 could mediate the ubiquitination and proteolysis of SK1 (34). Therefore, siRNA knock down of KLHL5 was employed (Fig. 1I).Treatment with siRNA to KLHL5 could inhibit SK1 proteolysis in response to ser/gly starvation. Thus, serine deprivation in media induces proteolysis of SK1 via a KLHL5-mediated mechanism.
Figure 1. Culture of cell with sub optimal doses of serine causes downregulation of SK1 protein.
A) Effects of ser/gly starvation on SK1, p53, and p21 protein levels. HCT-116 cells were grown with either ser/gly (0.4 mM each) in the media or without serine and glycine. The effects of ser/gly starvation on SK1, p21, and p53 were evaluated using western blotting at the indicated time points. The starred lane indicates doxorubicin (0.5 μM) treated sample as a positive inducer of p53. An earlier time course is shown for Western blot analysis of SK1 and its densitometry adjusted to levels of actin. B) Effect of decreasing serine levels on SK1 protein in both HCT-116 with WT p53 and p53 KO. Cells were incubated for 24 h with decreasing levels of serine before SK1 levels were evaluated by Western blot. C) Measurement of SK1 mRNA during serine starvation. Samples were prepared after 24h serine starvation, and SK1 levels were measured by RTPCR. Levels of mRNA were normalized to actin mRNA, and shows the mean average of 3 separate measurements +/− s.d, D) Expression levels of SK1 during serine starvation in other cell lines. Numbers below the SK-1 blot show densitometry of the SK-1 band divided by the Actin density, and shown as a percentage of the cells grown in serine-replete medium. E) Expression of SK2 during serine starvation. F) Effect of the proteasomal inhibitor bortezomib (BTZ) on SK1 levels. Increasing concentrations of BTZ were added concurrently to the change to serine-free or serine-replete media. Densitometry of the bands is also shown as % of intensity relative to 0 μM BTZ. Cells were medium changed to either serine-replete, or the complete medium was diluted with serine-free medium to obtain progressively lower levels of serine. After 24h incubation, the cells were prepared for Western blot and SK-1 levels were determined. G) Effect of caspase inhibitors on SK-1 degradation during serine starvation. Directly below is the positive control showing caspase inhibition of SK1 proteolysis after incubation for 24h with 5μM etoposide for 24h. H) Effect of PHGDH inhibition on SK1 expression. CBR-5884 (50μM) was added concurrently with a media change to either serine-replete or serine-deficient. After 24h, SK1 levels were examined by Western blot. I) KLHL5 knockdown and the effect on SK1 expression. Cells were transfected with siRNA to KLHL5 for 24h before the medium was changed to either one with ser/gly and one without. A further 24h later the cells were prepared for Western blot. + indicates siRNA to KLHL5, and – indicates treatment with the control All Star. Densitometry of the bands is also shown as % of intensity relative to the control. Representative scans were used for Western blots. Data in A, C, F and H indicate mean ± s.d., n = 3.
Serine is a “non essential’ amino acid as it can be generated endogenously. To evaluate the effects of decreasing synthesis of endogenous serine, we employed CBR-5884, an inhibitor of phosphoglycerate dehydrogenase (PHGDH), the first committed enzyme in de novo synthesis of endogenous serine. The results showed that CBR-5884 (in the presence of serine in media) also caused a loss of SK1 (Fig. 1H). Interestingly, addition of CBR-5884 together with ser/gly starvation caused a further decrease in SK1 protein levels, suggesting that overall cellular serine levels, whether from exogenous or endogenous sources, can affect SK1 protein levels. Taken together, these results demonstrate that serine starvation results in a p53-independent decrease in SK1 protein and enzymatic function.
Sphingolipid changes as a consequence of serine-starvation mediated SK1 degradation
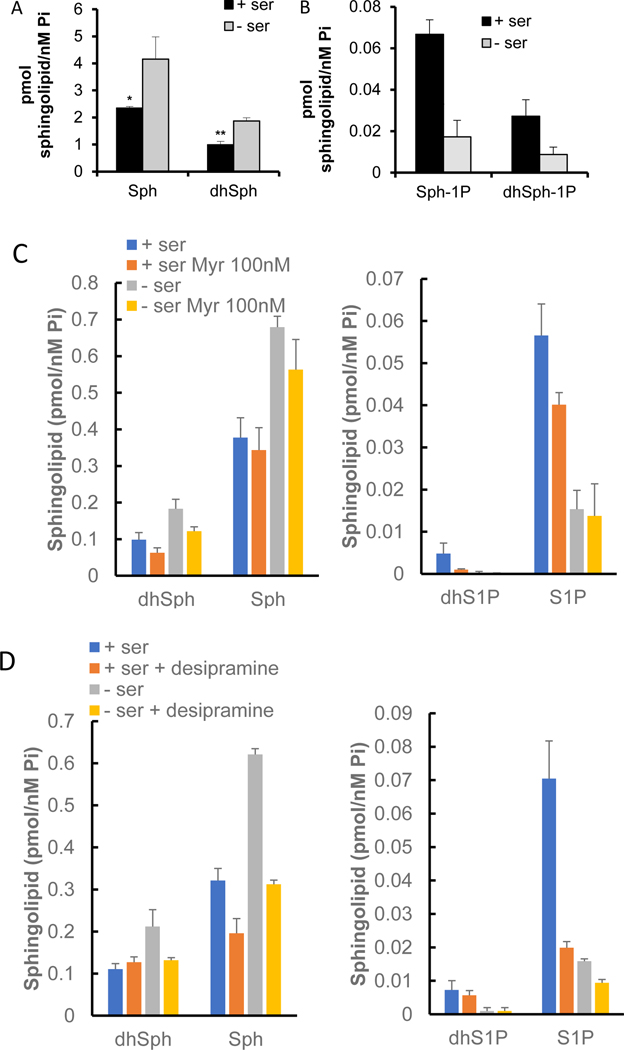
Since changes in SK1 protein were observed, the effect of ser/gly deprivation on sphingolipid levels were analyzed. Sphingolipids were analyzed with quantitative LC/MS/MS following 24h ser/gly starvation. Levels of sphingosine and sphinganine (substrates of SK1) increased in response to ser/gly removal (Fig. 2A). This is in contrast to one study (35) that showed a decrease in sphingosine following ser/gly deprivation (although that was a single measurement using qualitative rather than quantitative analysis). Also consistent with loss of SK1, levels of both respective products, S1P and dhS1P, decreased over the same time period (Fig. 2B), thus demonstrating significant effects of SK1 loss on intracellular levels of its bioactive substrates and products. Sphingosine is generated from the catabolism of ceramides, which in turn can derive (in substantial amounts) from catabolism of complex sphingolipids and not only from de novo synthesis. To verify the source of sphingosine, HCT-116 cells were preincubated with either 100 nM of the SPT-inhibitor myriocin (Fig. 2C) or 10 μM of the indirect inhibitor (functional inhibitor) of acid sphingomyelinase, desipramine (Fig. 2D). Myriocin had no significant effect on sphingosine levels (although sphingosine levels were consistently repressed by a small amount). On the other hand, desipramine significantly inhibited sphingosine levels following ser/gly starvation, indicating that inhibition of lysosomal recycling of sphingolipids by desipramine could reduce the amount of sphingosine. Therefore, hydrolysis of lysosomal ceramides to sphingosine is the main source of the sphingosine observed.
Figure 2. Sphingolipid changes as a consequence of serine-starvation mediated SK1 degradation.
A) Levels of sphingosine (Sph) and dihydrosphingosine (dhSph) after 24 h serine starvation, B) Levels of S1P and dihydro S1P after 24 h serine starvation, C) Effect of inhibiting de novo sphingolipid synthesis and D) ceramide hydrolysis on sphingosine, dihydrosphingosine, and their respective phosphorylated products following serine starvation. HCT-116 cells were cultured with and without serine for 24 h prior to sphingolipid analysis by HPLC MS/MS. Results were then normalized according to total lipid phosphate present.
Effects of SK1 on cell growth during serine starvation
In order to evaluate the functional consequences of loss of SK1, the growth of HCT-116 cells with and without serine was examined. As can be seen (Fig. 3A), ser/gly starvation significantly slowed growth of HCT-116. To determine the role of loss of SK1 in this process, HCT-116 cells were generated that overexpress SK1, and, interestingly, the levels of SK1 did not significantly change during serine starvation (inset Fig. 3A), suggesting that overexpressed SK1 overcomes loss of endogenous SK1. Unexpectedly, considering the usual ‘pro-growth’ functions of SK1, the results showed that in the context of ser/gly deprivation, over a period of 3 days, SK1 overexpression caused a significant growth disadvantage, when compared to control empty vector transduced cells (Fig. 3A).These results suggest that loss of SK1 functions as an adaptive/protective mechanism to serine deprivation such that the persistence of SK1 levels and activity would be detrimental to the cell’s response to serine starvation. To discount any possibility of a compensatory effect of SK2, cells were pretreated with siRNA to SK2 before serine starvation (Fig. 3B). No effect of SK2 downregulation was observed on cell growth. To further establish the role of SK1 in regulating the sphingosine rise during serine starvation, HCT-116 cells overexpressing SK1 were serine starved for 24h (Fig. 3C). Cells overexpressing SK1 showed suppression of the sphingosine increase following serine removal, further underlining the importance of SK1 loss to increased sphingosine levels. These results reveal an unexpected protective role for the loss of SK1 in the context of serine deprivation.
Figure 3. Effects of SK1 on cell growth during serine starvation.
A) Effect of SK1 expression on the growth (cell count) of ser/gly-starved cells. HCT-116 cells overexpressing SK1 and control cells were cultured with and without ser/gly for 3 days. Western blots show the differences in the effect of serine-starvation on levels of overexpressed SK1 and on endogenous SK1. Triplicate samples of cells were counted every 24 h. Results show mean ± s.d. n = 3. Statistics shown comparing the -ser/gly condition with and without SK1 overexpression. * P<0.05. B) Effect of SK2 downregulation on cell growth during ser/gly starvation. Cells were pretreated with siRNA to SK2 before being cultured with and without ser/gly for 3 days. Inset shows the effect of siRNA to SK2 on SK2 expression. C) Effect of over expressed SK1 on sphingosine levels following serine starvation. Stably over expressing SK1 HCT-116 cells incubated for 24h in either serine-replete or serine-deficient media before sphingosine levels were measured by HPLC MS/MS. Data indicates mean ± s.d., n = 3
SK1 regulates ROS during serine starvation.
The above results prompted us to investigate possible cellular functions that connect SK1 to growth regulation under serine starvation. Published data have suggested a link between SK1 expression and levels of reactive oxygen species (ROS) (36, 37), and it has been recently shown that phytosphingosine (yeast sphingosine) accumulation due to upregulation of alkaline ceramidases could induce mitochondrial dysfunction in yeast (16). Since mitochondria are the primary producers of intracellular ROS (38), and that increased ROS can both negatively influence and reflect mitochondrial function (39), levels of ROS were examined in Ser/Gly starved cells. To visualize changes in ROS, mitoSOX dye staining was employed, which is sensitive to superoxide as well as other types of ROS. The results showed that ser/gly starved cells had increased levels of mitochondrial ROS (Fig. 4A). Interestingly, SK1 overexpression prevented the increase in ROS during ser/gly starvation (Fig. 4A), suggesting that SK1 suppresses mitochondrial ROS levels. To corroborate these results, we generated CRISPR-Cas9-mediated deletion of SK1 in HCT-116 cells (28). The results showed that SK1 KO resulted in elevation of ROS levels when compared to the control cells (Fig. 4B). Thus, the current data suggest that the loss of SK1 allows for an increase in intracellular ROS levels.
Figure 4. SK1 regulates ROS during serine starvation.
A) Effects of serine starvation and of SK1 expression on mitochondrial ROS. Cells were starved of ser/gly for 24 h then pre-treated with mito-SOX for up to 2 h before fluorescence was visualized. The left panel images are representative of multiple fields from three independent results. The right panel shows quantification of each individual image. B) Levels of intracellular ROS in SK1 KO cells. Cells were incubated for 24 h before intracellular ROS levels were measured by DCFH-DA staining. Triplicate samples of cells were counted every 24 h. Results show mean ± s.d. n = 3. C) Growth effects of N-acetylcysteine (NAC)-treatment on ser/gly starved cells. NAC was added at the same time as media was changed to either ser/gly replete (left panel) or deficient (right panel). HCT-116 cells were incubated with 1 mM NAC and with or without serine for up to 72 h and cells were enumerated every 24 h. ROS was measured using DCFD-HA and quantified by fluorescent spectrometry. * P<0.05, ** P<0.005.
To determine the functional consequences of ROS generation, cells were treated with N-acetyl cysteine (NAC), which quenches ROS. The results showed that while the addition of NAC had no significant effect on the cell growth of non-serine starved cells (Fig. 4C, left panel), a slight but significant growth retardation was observed in NAC-treated serine starved cells (Fig. 4C, right panel). These results are consistent with a protective role for ROS in the context of serine starvation, consistent with other studies that demonstrate ‘pro-growth’ functions for moderate/limited increases in ROS (40, 41).
Role of SK1 in maintaining normal mitochondrial function.
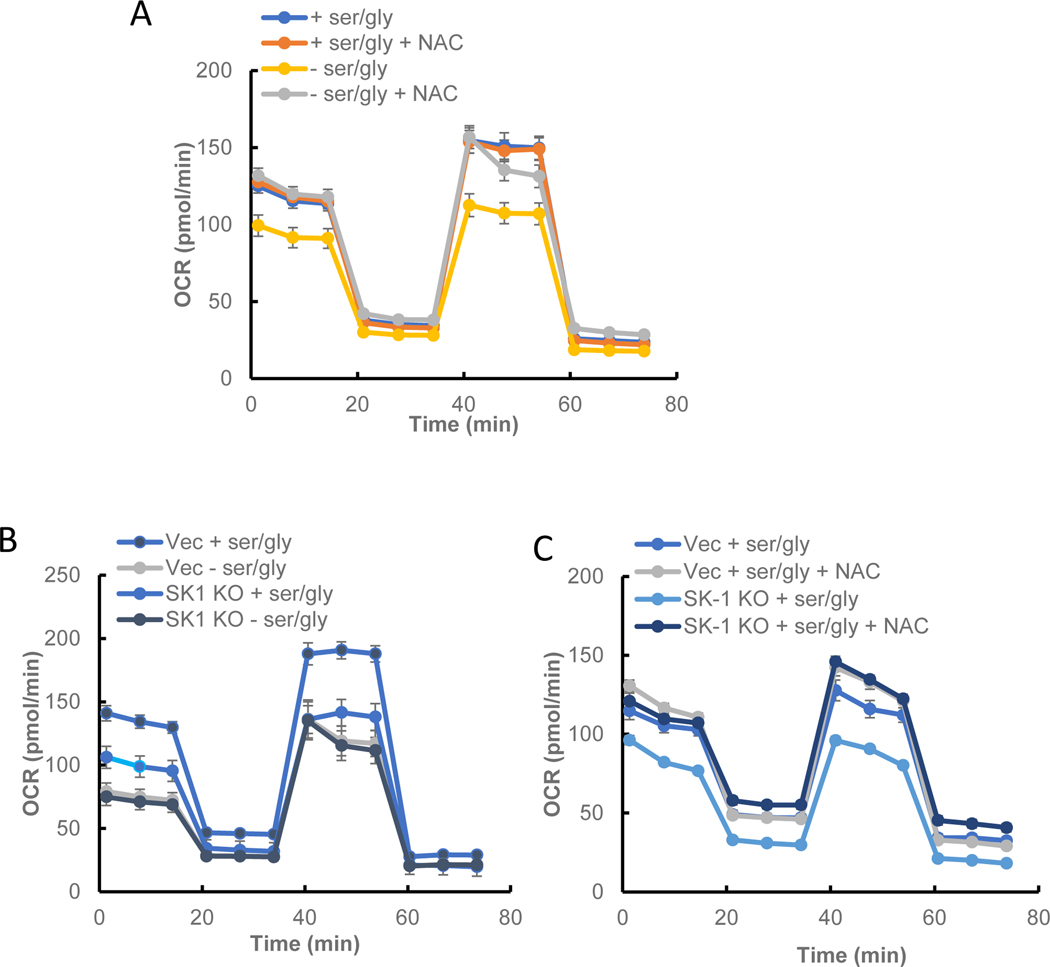
Given the previously established link between sphingosine, SK1, and mitochondrial function (12, 19), mitochondrial oxygen consumption rate (OCR) was measured in response to serine starvation. It was observed that after 24 h both basal and maximal peak stress OCR were decreased in serine starved cells (Fig. 5A). This drop in OCR could be mitigated by pretreatment with NAC, indicating the source of reduced mitochondrial activity as measured by OCR was due to increased ROS. Importantly, this was mimicked in SK1 KO cells (Fig. 5B). Moreover, loss of SK1 did not show an additive effect with serine deprivation, suggesting that they are on the ‘same pathway’. It is important to note that the OCR curves have similar profiles, but have different baselines, implying that the differences in mitochondrial OCR are due primarily to impaired basal mitochondrial function. These results support that loss of SK1 is necessary and sufficient to mediate suppression of mitochondrial function in response to serine deprivation.
Figure 5. Role of SK1 and ROS in maintaining normal mitochondrial function.
A) Effect of NAC on OCR in serine-starved HCT-116 cells. B) Effects of serine starvation and SK1 loss on maximum stress OCR. C) Effects of NAC-treatment on basal and maximal OCR in SK1 KO and WT HCT-116 cells. Cells were cultured with or without ser/gly. OCR was measured using Seahorse as recommended by the manufacturer. Quantification of OCR was performed at least 3 different times, and each value indicates mean average of 10 different measurements ± s.e.m.
Next, the relationship between induction of ROS and mitochondrial function was evaluated. Treatment with NAC counteracted the decrease in OCR following SK1 loss (Fig. 5C), underlining that increased ROS in ser/gly starvation can lead to altered mitochondrial function.
Together, the results suggest that loss of SK1 during serine deprivation promotes the generation of ROS, which in turn helps regulate mitochondrial OCR.
Effect of sphingosine accumulation on cell growth during serine starvation.
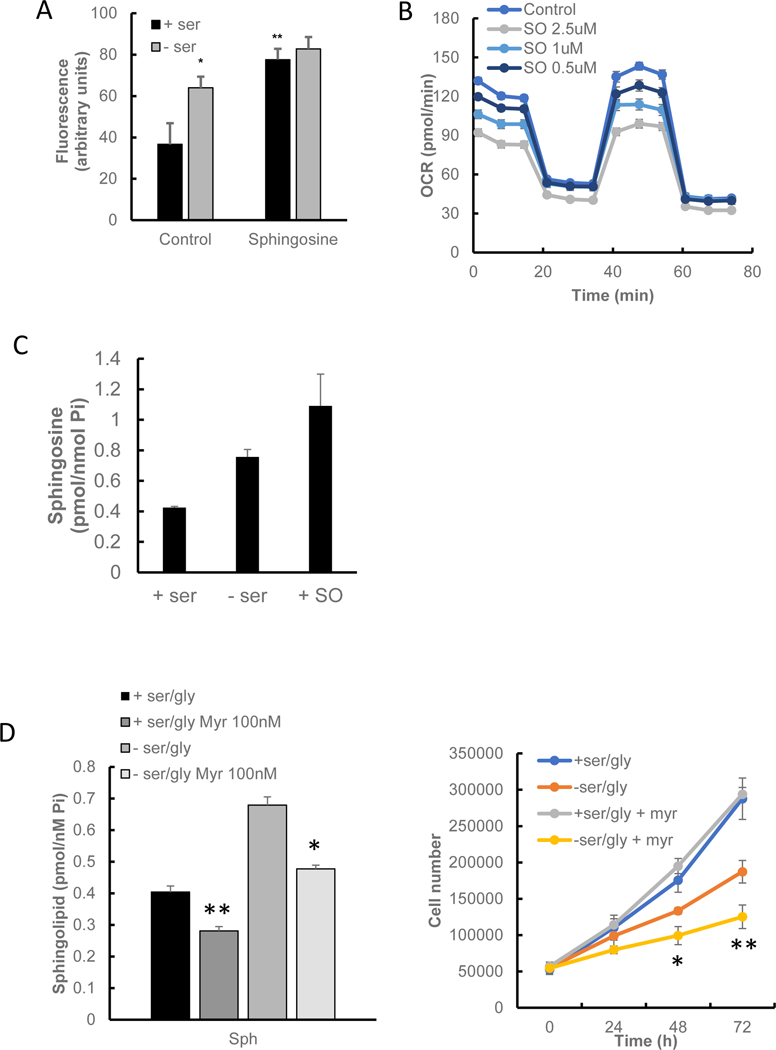
Given the above results and the established role of SK1 in regulating the levels of sphingoid bases and their phosphates, it became important to determine the lipid that mediates the effects of loss of SK1 on growth, ROS, and mitochondrial functions. Changes in mitochondrial sphingosine, either by reduction through knocking down neutral ceramidase (12) or by adding exogenous sphingosine to isolated mitochondria (42), can negatively affect mitochondrial function. These considerations prompted us to evaluate the effects of sphingosine. Addition of exogenous sphingosine increased intracellular ROS measured by H2DCFDA (Fig. 6A), while the addition of exogenous S1P did not have an effect on intracellular ROS levels (data not shown). Notably, the effects of sphingosine were not additive to those of serine deprivation. Importantly, sphingosine decreased OCR in a dose-dependent manner, with effects seen at concentrations as low as 0.5 μM (Fig. 6B). Measurement of intracellular sphingosine following exogenous addition showed that the levels taken were broadly in line with levels following serine starvation (Fig. 6C). Inhibition of sphingosine accumulation using myriocin (Fig 6D, left panel) significantly reduced HCT-116 growth during ser/gly starvation (Fig. 6D, right panel).
Figure 6. Effect of sphingosine accumulation on cell growth during serine starvation.
A) Effect of exogenous sphingosine on intracellular ROS. Cells were incubated for 24 h before the medium was changed to either serine-starved or serine-replete. Sphingosine (1μM) was added concurrently, and cells were incubated for a further 24 h. ROS was measured using DCFD-HA and quantified by fluorescent spectrometry. B) Dose response of sphingosine on OCR. Increasing doses of sphingosine (SO) were added 24 h before OCR was measured. C) Intracellular sphingosine levels following addition of exogenous Sph. D) Effect of sphingosine accumulation using myriocin. Cells had 100nM myriocin added at the same time as medium was changed to either serine-starved or serine-replete. After 24h sphingosine was measured using GC MS/MS. For growth cells were counted every 24h. Statistics shown for -ser/gly vs -ser/gly + myriocin. E) D-erythro-Sphingosine specificity compared to its enantiomer L-erythro-sphingosine on OCR. F) Effect of NAC on OCR in sphingosine-treated cells. OCR was measured using Seahorse as recommended. G) Effect of S1P on growth during ser/gly starvation. S1P (100nM) was added concurrently with serine-replete or serine-deficient media. After every 24h interval, cells were trypsinized and counted with trypan blue. Inset shows phosphorylated ERM following S1P addition for 10min, indicating that the S1P used was bioactive, as has been previously reported (66). Results show the average of 2 plates per time point, each counted twice. Results show mean ± s.d. H) Effect of S1P on ser/gly starvation induced OCR reduction. S1P was added concurrently with serine-replete or serine-deficient media, and incubated for 24h before OCR was measured as described in the Materials and Methods. I) Intracellular serine levels following addition of sphingosine. Data indicates mean ± s.e.m., n ≥ 8. * P<0.05, ** P<0.005.
Next, the specificity of sphingosine on OCR was evaluated, and the results showed that addition of the (unnatural) enantiomer L-erythro-sphingosine failed to show any impact on OCR (Fig. 6E). These significant effects, showing high potency (nM) and enantiomeric specificity, distinguish these effects of sphingosine from most other cellular effects attributed to sphingosine which usually require μM concentrations and do not demonstrate this level of stereospecificity. Thus, the accumulation of sphingosine following loss of SK1 is sufficient to mimic the effects of serine deprivation and the effects of loss of SK1 on ROS and on metabolic changes.
To evaluate the role of ROS and to determine if sphingosine was acting upstream of the changes in ROS, addition of NAC was found to rescue OCR changes following sphingosine (Fig. 6F). In contrast, exogenous S1P had no effect on cell growth during 4 days of serine starvation (Fig. 6G), despite it being causing the phosphorylation of ezrin radixin and moesin (ERM, Inset to Fig. 6G). In addition, S1P addition had no effect on mitochondrial activity during serine starvation (Fig. 6H). These results demonstrate that sphingosine accumulation during ser/gly starvation is an effector for reduced OCR and a non-lethal increase of ROS. Since sphingolipid generation played a role in HCT-116 growth during ser/gly starvation, levels of glucose-derived serine were measured after [13C6]-glucose labeling. Levels of de novo serine synthesized from labeled glucose (m+3) were increased in sphingosine-treated cells (Fig. 6I), demonstrating a key role for sph in driving serine synthesis.
Discussion
Although serine is a ‘founding’ metabolite for the class of sphingolipids, its functional effects on sphingolipids and possible roles of bioactive sphingolipids in regulating serine responses have not been studied. We demonstrate here that serine starvation causes the proteolysis of SK1, and this is a requirement for sustaining cellular growth. We also show that a modest increase in intracellular ROS is also necessary for growth during serine starvation, and for a small inhibition in the oxygen consumption rate of mitochondria. We also show that the expression of SK1 is important in ROS homeostasis, and fundamental in maintaining mitochondrial function.
A major conclusion from this study is that serine deprivation leads to loss of SK1 which in turns results in accumulation of sphingosine. This novel connection then launches an adaptive response of the cells, such that undoing the loss of SK1 (by overexpressing it) diminishes the levels of sphingosine and renders the cells more sensitive to growth suppressing effects of serine deprivation. This role of SK1 is ‘counter intuitive’ at multiple levels. First, SK1 has been generally associated with growth promotion; however, those studies have been performed under conditions of serine sufficiency. At the same time, sphingosine has been mostly associated with growth suppression, but our results show that sphingosine also drives the adaptation to serine deficiency. It should be noted that in this study, the concentrations of sphingosine are lower (0.5– 2 μM), than what is usually employed in cell biology studies, and its effects are highly stereospecific such that the L-erythro isomer did not mimic the actions of D-erythro sphingosine. Therefore, one possibility is that a modest increase in sphingosine can be adaptive whereas its high levels can lead to growth suppression. This is similar to the currently appreciated role of ROS as being positively adaptive at low increments but detrimental at higher levels. A recent study published by Muthusamy et al showed that inhibition of sphingolipid synthesis during serine starvation could rescue tumor growth (43). The authors concluded that sustained synthesis of certain sphingolipid species in response to serine starvation could lead to cell death. This would be consistent with studies that have suggested that chronic accumulation in deoxysphingolipids can be lead to deleterious effects such as cell death or inflammation (44) (45).
As to mechanisms of SK1 loss, our previous studies have shown that DNA damage through actinomycin D, etoposide, doxorubicin, or γ-radiation could cause SK1 proteolysis primarily through the induction of p53 (26). Inhibition of cathepsin B activity can both prevent SK1 degradation and rescue cells from apoptosis (26). For at least some chemotherapeutic drugs such as doxorubicin the proteolysis of SK1 occurs partly through a caspase 2-mediated mechanism (32). In another mechanism, the SK1 inhibitor SKI-II has been shown to cause SK1 degradation in a lysosomal-dependent mechanism, although the exact mechanism is still unknown (46). On the other hand ubiquitination of SK1 and degradation through the proteasome has been described in response to inhibitors of SK1 that are based on Sph (47, 48). Regulation of potential ubiquitination sites through acetylation has been shown to increase SK1 stability, having significant effects on cell size and cell growth (49). In our study, we ruled out a role for p53 and caspases, but the results support a role for the proteasomal degradation of SK1.
Our results show that increased sphingosine generation in response to serine deprivation decreases the OCR. A role for SK1 recruitment to mitochondria in the initiation of the unfolded protein response has been described in C elegans (50). A complex pathway of reciprocal regulation between both ROS and SK1 is therefore suggested, and has been at least partly described previously in a context that was deleterious to cell growth (36). This was supportive of the pro-survival role of SK1. The current study showed increased sphingosine accumulation as a result of SK1 degradation (Fig. 2A) causes decreased mitochondrial OCR, which is linked to maintaining cell growth during serine starvation. This strongly suggests that in conditions of normal serine bioavailability, SK1 plays a role in maintaining normal mitochondrial function.
Our studies connect SK1 loss to ROS generation, the latter having been connected to serine deprivation. There are many different metabolic pathways that can be affected by ROS (51). While cancer cells will upregulate antioxidant synthesis in order to counteract heightened ROS levels, higher levels of ROS are also associated with increased survival, through either limiting entry into the tricarboxylic acid cycle or affecting calcium signaling (52, 53). In fact, it has been observed that stimulation of receptor tyrosine kinases results in modest increases in ROS that have been implicated in the mitogenic response to these receptors (54). Additionally, higher intracellular ROS can affect the electron transport chain, but is probably not the primary mechanism leading to enhanced growth during serine starvation. However, measurement of OCR it is an accurate method to assess overall mitochondrial function (51).
The product of SK1 activity, S1P, has anti-apoptotic effects. By mediating oncogenic H-ras transformation, SK1 has been shown to act in some cases as an oncogene, despite rarely showing mutations in cancer cells (55, 56). The overexpression of SK1 has been linked to advanced disease progression, resistance to chemotherapeutic drugs like doxorubicin and also to a poor prognosis (57–62). Additionally, loss of SK1 in carcinoma cells could increase ROS (36). Another study has shown that oxidative-stress induced apoptosis of photoreceptor cells required sphingosine accumulation as a result of SK1 inhibition, again showing a link between SK1 activity and protection from high levels of ROS (63).
Increased serine biosynthesis has been described in many different cancers (9). Because of this, there has been significant recent interest in the use and development of serine synthesis inhibitors such as CBR 5448 (Fig. 1H) as an another approach for cancer treatment (64, 65). It is tempting to speculate that treatments that interfere with serine biosynthesis may be more effective in SK1-overexpressing tumors.
Together, these results suggest that sphingosine accumulation following serine deprivation initiates mitochondrial functional changes. This may be mediated either by changing the biophysical properties of the mitochondrial membranes and/or through altering of intracellular signaling through due to the increased intracellular ROS.
In summary, the results from this study reveal a novel link between serine deprivation and SK1, a key enzyme in regulating bioactive sphingolipids. The results also reveal a somewhat paradoxical role for SK1 in regulating mitochondrial function. In conditions of high serine bioavailability SK1 expression is linked to increased survival, while during serine starvation SK1 loss through a proteasome-associated mechanism is beneficial to cell survival. This reduced oxygen consumption of mitochondria due to SK 1 loss is likely responsible for such a beneficial effect during serine starvation. The role of novel sphingolipids induced by serine starvation is being extensively studied in our laboratory.
Acknowledgements:
The authors thank the Stony Brook Lipidomics Core for measurement and analysis of sphingolipids. This work was supported in part by NIH grants: GM130878, P01 CA097132, and 5R01GM130878–02.
This work is dedicated to the memory of Lina M Obeid.
Abbreviations
- LC/MS/MS
Liquid Chromatography with tandem mass spectrometry
- NAC
N-acetyl cysteine
- OCR
Oxygen consumption rate
- ROS
Reactive oxygen species
- S1P
Sphingosine 1 phosphate
- SK1
Sphingosine kinase 1
- Sph
Sphingosine
- SPT
Serine palmitoyl transferase
References
- 1.DeBerardinis RJ, and Chandel NS (2016) Fundamentals of cancer metabolism. Sci Adv 2, e1600200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boroughs LK, and DeBerardinis RJ (2015) Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol 17, 351–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amelio I, Cutruzzola F, Antonov A, Agostini M, and Melino G. (2014) Serine and glycine metabolism in cancer. Trends Biochem Sci 39, 191–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duan J, and Merrill AH Jr. (2015) 1-Deoxysphingolipids Encountered Exogenously and Made de Novo: Dangerous Mysteries inside an Enigma. J Biol Chem 290, 15380–15389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maddocks OD, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E, and Vousden KH (2013) Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature 493, 542–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hernandez-Corbacho MJ, Salama MF, Canals D, Senkal CE, and Obeid LM (2017) Sphingolipids in mitochondria. Biochim Biophys Acta Mol Cell Biol Lipids 1862, 56–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hannun YA, and Obeid LM (2018) Author Correction: Sphingolipids and their metabolism in physiology and disease. Nat Rev Mol Cell Biol 19, 673. [DOI] [PubMed] [Google Scholar]
- 8.Liu H, Sugiura M, Nava VE, Edsall LC, Kono K, Poulton S, Milstien S, Kohama T, and Spiegel S. (2000) Molecular cloning and functional characterization of a novel mammalian sphingosine kinase type 2 isoform. J Biol Chem 275, 19513–19520 [DOI] [PubMed] [Google Scholar]
- 9.Locasale JW (2013) Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer 13, 572–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lucas S, Chen G, Aras S, and Wang J. (2018) Serine catabolism is essential to maintain mitochondrial respiration in mammalian cells. Life Sci Alliance 1, e201800036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bao XR, Ong SE, Goldberger O, Peng J, Sharma R, Thompson DA, Vafai SB, Cox AG, Marutani E, Ichinose F, Goessling W, Regev A, Carr SA, Clish CB, and Mootha VK (2016) Mitochondrial dysfunction remodels one-carbon metabolism in human cells. Elife 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Novgorodov SA, Riley CL, Yu J, Borg KT, Hannun YA, Proia RL, Kindy MS, and Gudz TI (2014) Essential roles of neutral ceramidase and sphingosine in mitochondrial dysfunction due to traumatic brain injury. J Biol Chem 289, 13142–13154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bionda C, Portoukalian J, Schmitt D, Rodriguez-Lafrasse C, and Ardail D. (2004) Subcellular compartmentalization of ceramide metabolism: MAM (mitochondria-associated membrane) and/or mitochondria? Biochem J 382, 527–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Birbes H, Luberto C, Hsu YT, El Bawab S, Hannun YA, and Obeid LM (2005) A mitochondrial pool of sphingomyelin is involved in TNFalpha-induced Bax translocation to mitochondria. Biochem J 386, 445–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ganesan V, Perera MN, Colombini D, Datskovskiy D, Chadha K, and Colombini M. (2010) Ceramide and activated Bax act synergistically to permeabilize the mitochondrial outer membrane. Apoptosis 15, 553–562 [DOI] [PubMed] [Google Scholar]
- 16.Yi JK, Xu R, Jeong E, Mileva I, Truman JP, Lin CL, Wang K, Snider J, Wen S, Obeid LM, Hannun YA, and Mao C. (2016) Aging-related elevation of sphingoid bases shortens yeast chronological life span by compromising mitochondrial function. Oncotarget 7, 21124–21144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raichur S, Wang ST, Chan PW, Li Y, Ching J, Chaurasia B, Dogra S, Ohman MK, Takeda K, Sugii S, Pewzner-Jung Y, Futerman AH, and Summers SA (2014) CerS2 haploinsufficiency inhibits beta-oxidation and confers susceptibility to diet-induced steatohepatitis and insulin resistance. Cell Metab 20, 687–695 [DOI] [PubMed] [Google Scholar]
- 18.Chen W, Xiang H, Chen R, Yang J, Yang X, Zhou J, Liu H, Zhao S, Xiao J, Chen P, Chen AF, Chen S, and Lu H. (2019) S1PR2 antagonist ameliorate high glucose-induced fission and dysfunction of mitochondria in HRGECs via regulating ROCK1. BMC Nephrol 20, 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pchejetski D, Kunduzova O, Dayon A, Calise D, Seguelas MH, Leducq N, Seif I, Parini A, and Cuvillier O. (2007) Oxidative stress-dependent sphingosine kinase-1 inhibition mediates monoamine oxidase A-associated cardiac cell apoptosis. Circ Res 100, 41–49 [DOI] [PubMed] [Google Scholar]
- 20.Pyszko J, and Strosznajder JB (2014) Sphingosine kinase 1 and sphingosine-1-phosphate in oxidative stress evoked by 1-methyl-4-phenylpyridinium (MPP+) in human dopaminergic neuronal cells. Mol Neurobiol 50, 38–48 [DOI] [PubMed] [Google Scholar]
- 21.Spinelli JB, and Haigis MC (2018) The multifaceted contributions of mitochondria to cellular metabolism. Nat Cell Biol 20, 745–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DiMauro S, and Schon EA (2003) Mitochondrial respiratory-chain diseases. N Engl J Med 348, 2656–2668 [DOI] [PubMed] [Google Scholar]
- 23.Mishra P, and Chan DC (2016) Metabolic regulation of mitochondrial dynamics. J Cell Biol 212, 379–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Randow F, and Youle RJ (2014) Self and nonself: how autophagy targets mitochondria and bacteria. Cell Host Microbe 15, 403–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han Y, Cho U, Kim S, Park IS, Cho JH, Dhanasekaran DN, and Song YS (2018) Tumour microenvironment on mitochondrial dynamics and chemoresistance in cancer. Free Radic Res 52, 1271–1287 [DOI] [PubMed] [Google Scholar]
- 26.Taha TA, Osta W, Kozhaya L, Bielawski J, Johnson KR, Gillanders WE, Dbaibo GS, Hannun YA, and Obeid LM (2004) Down-regulation of sphingosine kinase-1 by DNA damage: dependence on proteases and p53. J Biol Chem 279, 20546–20554 [DOI] [PubMed] [Google Scholar]
- 27.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, and Vogelstein B. (1998) Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282, 1497–1501 [DOI] [PubMed] [Google Scholar]
- 28.Pulkoski-Gross MJ, Jenkins ML, Truman JP, Salama MF, Clarke CJ, Burke JE, Hannun YA, and Obeid LM (2018) An intrinsic lipid-binding interface controls sphingosine kinase 1 function. J Lipid Res 59, 462–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Folmes CD, Martinez-Fernandez A, Perales-Clemente E, Li X, McDonald A, Oglesbee D, Hrstka SC, Perez-Terzic C, Terzic A, and Nelson TJ (2013) Disease-causing mitochondrial heteroplasmy segregated within induced pluripotent stem cell clones derived from a patient with MELAS. Stem Cells 31, 1298–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bielawski J, Pierce JS, Snider J, Rembiesa B, Szulc ZM, and Bielawska A. (2010) Sphingolipid analysis by high performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS). Adv Exp Med Biol 688, 46–59 [DOI] [PubMed] [Google Scholar]
- 31.Momose I, and Kawada M. (2016) The therapeutic potential of microbial proteasome inhibitors. International immunopharmacology 37, 23–30 [DOI] [PubMed] [Google Scholar]
- 32.Carroll BL, Bonica J, Shamseddine AA, Hannun YA, and Obeid LM (2018) A role for caspase-2 in sphingosine kinase 1 proteolysis in response to doxorubicin in breast cancer cells - implications for the CHK1-suppressed pathway. FEBS Open Bio 8, 27–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taha TA, Kitatani K, El-Alwani M, Bielawski J, Hannun YA, and Obeid LM (2006) Loss of sphingosine kinase-1 activates the intrinsic pathway of programmed cell death: modulation of sphingolipid levels and the induction of apoptosis. FASEB J 20, 482–484 [DOI] [PubMed] [Google Scholar]
- 34.Powell JA, Pitman MR, Zebol JR, Moretti PAB, Neubauer HA, Davies LT, Lewis AC, Dagley LF, Webb AI, Costabile M, and Pitson SM (2019) Kelch-like protein 5-mediated ubiquitination of lysine 183 promotes proteasomal degradation of sphingosine kinase 1. Biochem J 476, 3211–3226 [DOI] [PubMed] [Google Scholar]
- 35.Gao X, Lee K, Reid MA, Sanderson SM, Qiu C, Li S, Liu J, and Locasale JW (2018) Serine Availability Influences Mitochondrial Dynamics and Function through Lipid Metabolism. Cell Rep 22, 3507–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huwiler A, Kotelevets N, Xin C, Pastukhov O, Pfeilschifter J, and Zangemeister-Wittke U. (2011) Loss of sphingosine kinase-1 in carcinoma cells increases formation of reactive oxygen species and sensitivity to doxorubicin-induced DNA damage. British journal of pharmacology 162, 532–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harijith A, Pendyala S, Reddy NM, Bai T, Usatyuk PV, Berdyshev E, Gorshkova I, Huang LS, Mohan V, Garzon S, Kanteti P, Reddy SP, Raj JU, and Natarajan V. (2013) Sphingosine kinase 1 deficiency confers protection against hyperoxia-induced bronchopulmonary dysplasia in a murine model: role of S1P signaling and Nox proteins. Am J Pathol 183, 1169–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphy MP (2009) How mitochondria produce reactive oxygen species. Biochem J 417, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diers AR, Broniowska KA, and Hogg N. (2013) Nitrosative stress and redox-cycling agents synergize to cause mitochondrial dysfunction and cell death in endothelial cells. Redox Biol 1, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheung EC, Lee P, Ceteci F, Nixon C, Blyth K, Sansom OJ, and Vousden KH (2016) Opposing effects of TIGAR- and RAC1-derived ROS on Wnt-driven proliferation in the mouse intestine. Genes Dev 30, 52–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahalingaiah PK, and Singh KP (2014) Chronic oxidative stress increases growth and tumorigenic potential of MCF-7 breast cancer cells. PLoS One 9, e87371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hassoun SM, Lancel S, Petillot P, Decoster B, Favory R, Marchetti P, and Neviere R. (2006) Sphingosine impairs mitochondrial function by opening permeability transition pore. Mitochondrion 6, 149–154 [DOI] [PubMed] [Google Scholar]
- 43.Muthusamy T, Cordes T, Handzlik MK, You L, Lim EW, Gengatharan J, Pinto AFM, Badur MG, Kolar MJ, Wallace M, Saghatelian A, and Metallo CM (2020) Serine restriction alters sphingolipid diversity to constrain tumour growth. Nature [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taha TA, Mullen TD, and Obeid LM (2006) A house divided: ceramide, sphingosine, and sphingosine-1-phosphate in programmed cell death. Biochim Biophys Acta 1758, 2027–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Norris GH, and Blesso CN (2017) Dietary and Endogenous Sphingolipid Metabolism in Chronic Inflammation. Nutrients 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ren S, Xin C, Pfeilschifter J, and Huwiler A. (2010) A novel mode of action of the putative sphingosine kinase inhibitor 2-(p-hydroxyanilino)-4-(p-chlorophenyl) thiazole (SKI II): induction of lysosomal sphingosine kinase 1 degradation. Cell Physiol Biochem 26, 97–104 [DOI] [PubMed] [Google Scholar]
- 47.Loveridge C, Tonelli F, Leclercq T, Lim KG, Long JS, Berdyshev E, Tate RJ, Natarajan V, Pitson SM, Pyne NJ, and Pyne S. (2010) The sphingosine kinase 1 inhibitor 2-(p-hydroxyanilino)-4-(p-chlorophenyl)thiazole induces proteasomal degradation of sphingosine kinase 1 in mammalian cells. J Biol Chem 285, 38841–38852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Byun HS, Pyne S, Macritchie N, Pyne NJ, and Bittman R. (2013) Novel sphingosine-containing analogues selectively inhibit sphingosine kinase (SK) isozymes, induce SK1 proteasomal degradation and reduce DNA synthesis in human pulmonary arterial smooth muscle cells. Medchemcomm 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu H, Shao Y, Gao L, Zhang L, Guo K, Wu C, Hu X, and Duan H. (2012) Acetylation of sphingosine kinase 1 regulates cell growth and cell-cycle progression. Biochem Biophys Res Commun 417, 1242–1247 [DOI] [PubMed] [Google Scholar]
- 50.Kim S, and Sieburth D. (2018) Sphingosine Kinase Activates the Mitochondrial Unfolded Protein Response and Is Targeted to Mitochondria by Stress. Cell Rep 24, 2932–2945 e2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Forrester SJ, Kikuchi DS, Hernandes MS, Xu Q, and Griendling KK (2018) Reactive Oxygen Species in Metabolic and Inflammatory Signaling. Circ Res 122, 877–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M, Bellinger G, Sasaki AT, Locasale JW, Auld DS, Thomas CJ, Vander Heiden MG, and Cantley LC (2011) Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science 334, 1278–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takahashi N, Chen HY, Harris IS, Stover DG, Selfors LM, Bronson RT, Deraedt T, Cichowski K, Welm AL, Mori Y, Mills GB, and Brugge JS (2018) Cancer Cells Co-opt the Neuronal Redox-Sensing Channel TRPA1 to Promote Oxidative-Stress Tolerance. Cancer cell 33, 985–1003 e1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bae YS, Oh H, Rhee SG, and Yoo YD (2011) Regulation of reactive oxygen species generation in cell signaling. Mol Cells 32, 491–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xia P, Gamble JR, Wang L, Pitson SM, Moretti PA, Wattenberg BW, D’Andrea RJ, and Vadas MA (2000) An oncogenic role of sphingosine kinase. Curr Biol 10, 1527–1530 [DOI] [PubMed] [Google Scholar]
- 56.Vadas M, Xia P, McCaughan G, and Gamble J. (2008) The role of sphingosine kinase 1 in cancer: oncogene or non-oncogene addiction? Biochim Biophys Acta 1781, 442–447 [DOI] [PubMed] [Google Scholar]
- 57.Van Brocklyn JR, Jackson CA, Pearl DK, Kotur MS, Snyder PJ, and Prior TW (2005) Sphingosine kinase-1 expression correlates with poor survival of patients with glioblastoma multiforme: roles of sphingosine kinase isoforms in growth of glioblastoma cell lines. Journal of neuropathology and experimental neurology 64, 695–705 [DOI] [PubMed] [Google Scholar]
- 58.Bonhoure E, Pchejetski D, Aouali N, Morjani H, Levade T, Kohama T, and Cuvillier O. (2006) Overcoming MDR-associated chemoresistance in HL-60 acute myeloid leukemia cells by targeting sphingosine kinase-1. Leukemia 20, 95–102 [DOI] [PubMed] [Google Scholar]
- 59.Sobue S, Iwasaki T, Sugisaki C, Nagata K, Kikuchi R, Murakami M, Takagi A, Kojima T, Banno Y, Akao Y, Nozawa Y, Kannagi R, Suzuki M, Abe A, Naoe T, and Murate T. (2006) Quantitative RT-PCR analysis of sphingolipid metabolic enzymes in acute leukemia and myelodysplastic syndromes. Leukemia 20, 2042–2046 [DOI] [PubMed] [Google Scholar]
- 60.Kawamori T, Osta W, Johnson KR, Pettus BJ, Bielawski J, Tanaka T, Wargovich MJ, Reddy BS, Hannun YA, Obeid LM, and Zhou D. (2006) Sphingosine kinase 1 is up-regulated in colon carcinogenesis. FASEB J 20, 386–388 [DOI] [PubMed] [Google Scholar]
- 61.Liu G, Zheng H, Zhang Z, Wu Z, Xiong H, Li J, and Song L. (2010) Overexpression of sphingosine kinase 1 is associated with salivary gland carcinoma progression and might be a novel predictive marker for adjuvant therapy. BMC cancer 10, 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Song L, Xiong H, Li J, Liao W, Wang L, Wu J, and Li M. (2011) Sphingosine kinase-1 enhances resistance to apoptosis through activation of PI3K/Akt/NF-kappaB pathway in human non-small cell lung cancer. Clin Cancer Res 17, 1839–1849 [DOI] [PubMed] [Google Scholar]
- 63.Abrahan CE, Miranda GE, Agnolazza DL, Politi LE, and Rotstein NP (2010) Synthesis of sphingosine is essential for oxidative stress-induced apoptosis of photoreceptors. Investigative ophthalmology & visual science 51, 1171–1180 [DOI] [PubMed] [Google Scholar]
- 64.Wang Q, Liberti MV, Liu P, Deng X, Liu Y, Locasale JW, and Lai L. (2017) Rational Design of Selective Allosteric Inhibitors of PHGDH and Serine Synthesis with Anti-tumor Activity. Cell Chem Biol 24, 55–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ravez S, Spillier Q, Marteau R, Feron O, and Frederick R. (2017) Challenges and Opportunities in the Development of Serine Synthetic Pathway Inhibitors for Cancer Therapy. J Med Chem 60, 1227–1237 [DOI] [PubMed] [Google Scholar]
- 66.Gandy KA, Canals D, Adada M, Wada M, Roddy P, Snider AJ, Hannun YA, and Obeid LM (2013) Sphingosine 1-phosphate induces filopodia formation through S1PR2 activation of ERM proteins. Biochem J 449, 661–672 [DOI] [PMC free article] [PubMed] [Google Scholar]