Abstract
In this paper, a progressive global perception and local polishing (PCPLP) network is proposed to automatically segment the COVID-19-caused pneumonia infections in computed tomography (CT) images. The proposed PCPLP follows an encoder-decoder architecture. Particularly, the encoder is implemented as a computationally efficient fully convolutional network (FCN). In this study, a multi-scale multi-level feature recursive aggregation (mmFRA) network is used to integrate multi-scale features (viz. global guidance features and local refinement features) with multi-level features (viz. high-level semantic features, middle-level comprehensive features, and low-level detailed features). Because of this innovative aggregation of features, an edge-preserving segmentation map can be produced in a boundary-aware multiple supervision (BMS) way. Furthermore, both global perception and local perception are devised. On the one hand, a global perception module (GPM) providing a holistic estimation of potential lung infection regions is employed to capture more complementary coarse-structure information from different pyramid levels by enlarging the receptive fields without substantially increasing the computational burden. On the other hand, a local polishing module (LPM), which provides a fine prediction of the segmentation regions, is applied to explicitly heighten the fine-detail information and reduce the dilution effect of boundary knowledge. Comprehensive experimental evaluations demonstrate the effectiveness of the proposed PCPLP in boosting the learning ability to identify the lung infected regions with clear contours accurately. Our model is superior remarkably to the state-of-the-art segmentation models both quantitatively and qualitatively on a real CT dataset of COVID-19.
Keywords: Coronavirus disease 2019 (COVID-19), Global perception, Local polishing, Feature recursive aggregation, Multiple supervision
1. Introduction
The spread of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) becomes a global public health crisis known as the coronavirus disease 2019 (COVID-19) pandemic. SARS-CoV-2 continues threatening the world due to its high infectivity and extreme lethality. To stop the spread of SARS-CoV-2, a robust, accurate, and rapid testing protocol plays a critical role. The gold standard for detecting COVID-19 infections in clinics is reverse transcription-polymerase chain reaction (RT-PCR) [1]. Unfortunately, the shortage of equipment and long operation time (i.e., a typical 24-hour sample-to-result turnaround time) dramatically limits our ability to screen suspected patients rapidly. In addition, the reduced sensitivity of RT-PCR tests caused by inappropriate sample collection, storage, transfer, purification, and processing yield high false-negative errors, becoming a hazardous factor for stopping the COVID-19 pandemic.
Radiological techniques, including chest X-ray [2], [3], [4] and computed tomography (CT) [5], [6], [7], offer critical imaging tools for the detection of COVID-19-related pneumonia infections and in evaluating the respiratory complications related to coronavirus. Due to its high spatial resolution, chest CT is more effective than X-ray radiography for detecting COVID-19. This is because CT can be used to detect small lesions showing signs of infections on the lung with a higher degree of sensitivity than X-ray radiography.
When CT imaging is used to diagnose COVID-19-caused pneumonia infections, one important task is to delineate the suspected infections in the lung. The identification (also known as image segmentation) of COVID-19-caused infections through CT images is crucial for further quantitative analysis of the disease. Also, the segmentation of the COVID-19-caused lesions can be used to monitor and determine the severity of the disease over time, which allows doctors to predict risks and prognostics in a "patient-specific" fashion. Currently, the segmentation of the COVID-19-caused pneumonia infections has been done manually by experienced radiologists. However, such a task is labor-intensive and prone to inter- and intra-observer variability. Automated segmentation of COVID-19-caused infections from CT images, will relieve clinicians' workloads and make radiologic diagnoses more reliable and reproducible. However, there are several challenges: 1) compared with classification tasks, the segmentation of infection regions in the lung needs to process more discriminative information; 2) texture of the infected regions on CT images, especially the tiny infection regions, is devilishly complicated and detailed CT image characteristics vary dramatically, leading to false-negative; 3) the segmentation is sensitive to several intrinsic factors in the images, e.g., the difference of the locations of the lesions, intensity inhomogeneity of the infected regions, high variation of the infection characteristics, soft tissue appearance within the proximity of a suspected infection, etc.
Our image segmentation method is inspired by practices in the clinical workflow. More specifically, during the diagnosis of pulmonary infections, clinicians first identify the overall infected regions and then zoom in to the details of those detected pulmonary lesions, e.g., local appearances on CT images. In other words, clinicians use both global (a rough lesion contour) and local (detailed imaging characteristics) information to distinguish the infected lesions from normal tissues. Our algorithm first predicts the rough infection areas and then refines the suspected lesion boundaries by taking imaging details into considerations. However, the combination of imaging artifacts (e.g., motion artifacts) and insufficient image contrast between the infections and their surroundings (soft tissue) makes the automated image segmentation of COVID-19 infection regions challenging.
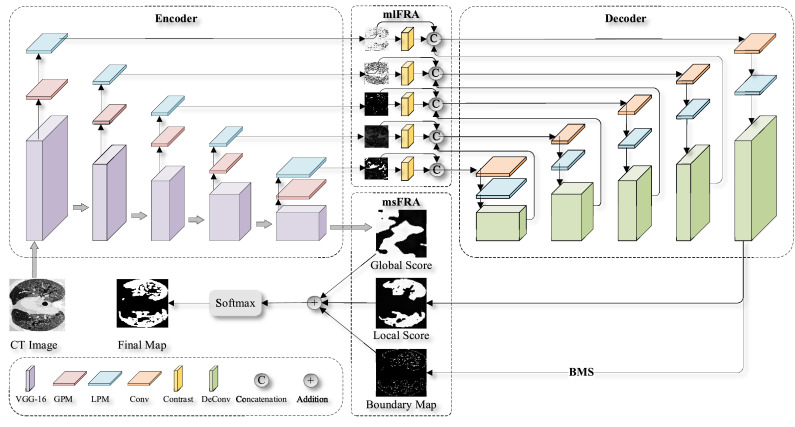
This paper proposes a progressive global perception and local polishing (PCPLP) deep network to overcome the technological challenges mentioned above. As illustrated in Fig. 1 , the proposed PCPLP network follows an encoder-decoder architecture. The encoder structure uses a VGG-16 framework, which starts by extracting multi-level Pyramid features, i.e., high-level semantic features, middle-level comprehensive features, and low-level detailed features. Two additional modules: a global perception module (GPM) and a local polishing module (LPM), are added to extend the classical VGG model. GPM is used to extract the global knowledge to find the infected regions' rough locations. In contrast, the LPM is used to extract the local information to obtain the infected regions' internal details and external contours. After the image encoding, local features obtained by the LPM models at various scales, the discriminant features of the contrast layers, and the upsampling features of the previous decoder layer are hierarchically integrated to obtain the enriched feature representations (i.e., local score map in Fig. 1). After that, the local refinement features, which are generated through multi-level feature recursive aggregation (mlFRA) by multistage parallel fusion in a recurrent manner, are combined with the global guidance features (i.e., a global score map in Fig. 1) via multi-scale feature recursive aggregation (msFRA) to produce the initial segmentation map. Finally, we propose to train the multi-scale multi-level feature recursive aggregation (mmFRA) network by exploiting the boundary-aware multiple supervision (BMS), i.e., segmentation cross-entropy loss (SCEL), boundary cross-entropy loss (BCEL), and boundary-refinement loss (BRL).
Fig. 1.
An illustrative flowchart showing major components of the proposed PCPLP network.
In summary, our main contributions of this work are fourfold:
-
1)
We propose a novel deep fully encoder-decoder convolutional network, PGPLP, for COVID-19 lung infected region segmentation from chest CT images.
-
2)
The entire network comprises a pair of msFRA and mlFRA networks, which progressively aggregate the global guidance information and local refinement information in a coarse-to-fine fashion to get the initial segmentation map.
-
3)
We build GPM and LPM modules to guide the message through the network to learn accurate positioning information and extensive detailed knowledge of the lung infected regions.
-
4)
To make the network focus on the infected regions and the boundary pixels in the training phase, we employ BMS to predict the lung infection regions and the corresponding contours simultaneously.
The remainder of this paper is organized as follows: The related work is presented in Section 2. The architecture of the proposed PGPLP network is described in Section 3. Extensive experimental evaluations are provided in Section 4 followed by some closing remarks in Section 5.
2. Related Works
In this section, image analysis techniques with applications in the COVID-19 pandemic, which are most relevant to our work, are discussed.
2.1. Artificial Intelligence for Combating COVID-19
Many researchers proposed various strategies based on artificial intelligence (AI) technology to contribute to the combat against the COVID-19 epidemic [8]. For instance, Jiang et al. [9] presented an AI architecture with the prophetic ability for COVID-19 detection to support rapid clinical decision-making. Li et al. [10] developed a fully automatic COVID-19 detection neural network (COVNet) to distinguish SARS-CoV-2 infections from community-acquired pneumonia on chest CT images. Harmon et al. [11] employed an AI algorithm to detect COVID-19 pneumonia on chest CT datasets from multi-institutions. Their method reached an accuracy of 90.8%. Besides utilizing chest CT images, Salman et al. [12] exploited a deep learning model for COVID-19 pneumonia detection in X-ray images, which achieved comparable performance to expert radiologists. Wang et al. [13] developed a tailored deep convolutional neural network for detecting COVID-19 utilizing chest X-ray images. Adhikari et al. [14] introduced a two-staged DenseNet to diagnose COVID-19 from CT and X-ray images to alleviate the workloads of radiologists. All those radiography-based algorithms can decide whether a patient is infected by SARS-CoV-2 and contribute to the control of the COVID-19 epidemic, especially when RT-PCR tests are not available.
2.2. Segmentation of COVID-19 Infections from Chest CT Images
In addition to AI-based detection of COVID-19-caused pneumonia [15], [16], [17], research efforts have also been devoted to the segmentation of COVID-19 caused infections [18], [19], [20], [21]. Image segmentation enables subsequent quantitative analysis for patients infected by SARS-CoV-2. A critical application of infection segmentation is to provide a comprehensive prediction of the disease severity; this can be done by visualizing and stratifying the lesion distribution utilizing the percentage of infection (POI). As mentioned before, image segmentation algorithms must overcome several challenges, including intensity inhomogeneity, interference of imaging artifacts, lack of image contrast among different tissue types, etc. Some innovative strategies have been proposed to overcome the above-said challenges. The most prominent neural network architectures of this line of research are U-Net [18] and its variants (e.g., residual attention U-Net [19], spatial and channel attention U-Net [20], 3D U-Net [21], etc.). Particularly, Zhou et al. [22] designed a rapid, accurate, and machine-agnostic model to segment the infection regions in COVID-19 CT scans. Wang et al. [23] presented a noise-robust deep segmentation network to learn the pneumonia lesions of COVID-19. Fan et al. [24] proposed a lung infection segmentation network to segment COVID-19 infected regions on chest CT images automatically. More recently, Laradji et al. [25] put forward a weakly supervised learning framework for COVID-19 segmentation in CT images, which utilized the consistency-based loss to enhance the performance of the segmentation.
Our proposed automatic deep segmentation model was inspired by observed clinical practices, making it different from other published methods. Our method first learns multi-level discriminant information of lesions via a global perception strategy. It then progressively refines the infection regions with additional local detailed image characteristics through the local polishing method. As described in the proceeding sections, the proposed method possesses enormous potentiality to assist experts in analyzing and interpreting COVID-19 CT images.
3. The Proposed Lung Infection Segmentation Network
Anecdotally, experienced radiologists usually adopt a two-step procedure to segment an infected region caused by SARS-CoV-2: 1) roughly locate an overall infection region, and 2) complete an accurate delineation of the infection region characterizing local tissue structures and considering detailed imaging characteristics. Inspired by this diagnostic process, a global perception and local polishing guided deep network is proposed to automatically identify the infected regions so that both rough global structure information and fine local boundary information can be progressively integrated. The proposed neural network model can extract the lung infected regions in CT images, even with the presence of low image contrast, image blurring, drastic local changes, and complex local patterns.
In this section, we first present an overview of the architecture of the proposed automatic lung infected region segmentation network. Then, we describe both the global perception module (GPM) and the local polishing module (LPM) in detail. After that, we provide the design of multi-scale multi-level feature recursive aggregation (mmFRA). To the end, a boundary-aware multiple supervision (BMS) strategy and the training loss of the network are elucidated.
3.1. Overview of Network Architecture
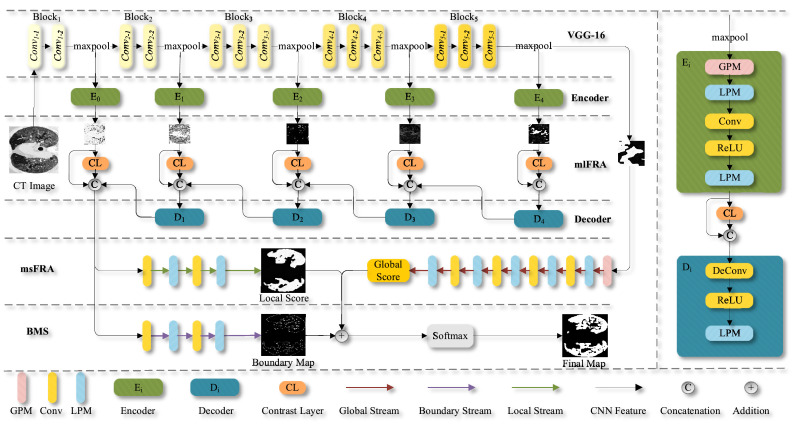
The proposed network follows the encoder-decoder architecture, as displayed in Fig. 2 . The encoder part is based on VGG-16 and is used to extract multi-level coarse features. In contrast, the decoder part recursively integrates the multi-scale fine features to generate the infected region segmentation map through supervised learning. The proposed network consists of four key components: GPM, LPM, mmFRA (viz. mlFRA and msFRA), and BMS.
Fig. 2.
An overview of the proposed PCPLP network architecture.
Our encoder configuration is derived from the classic VGG-16 architecture due to its simplicity and elegance. It is also worth noting that VGG-16 still has state-of-the-art performance and good generalization properties for image segmentation. The comparisons among four deep-learning neural networks frequently used as the backbones of various segmentation architectures are shown in Table 1 . We modified the VGG-16 [26] architecture by removing all the fully connected layers. Hence, the encoder becomes a fully convolutional network (FCN), and the removal of fully connected layers allows us to improve computational efficiency. However, our encoder still allows pixel-wise characterization of CT imaging data.
Table 1.
A summary of four commonly used classic neural network models for image segmentation.
| AlexNet [27] | VGG [26] | GoogLeNet [28] | ResNet [29] | |
|---|---|---|---|---|
| Input Size | 227 × 227 | 224 × 224 | 224 × 224 | 224 × 224 |
| Number of Layers | 8 | 19 | 22 | 152 |
| Number of Conv. Layers | 5 | 16 | 21 | 151 |
| Filter Sizes | 3, 5, 11 | 3 | 1, 3, 5, 7 | 1, 3, 5, 7 |
| Strides | 1, 4 | 1 | 1, 2 | 1, 2 |
| Fully Connected Layers | 3 | 3 | 1 | 1 |
| TOP-5 Test Accuracy | 84.6% | 92.7% | 93.3% | 96.4% |
| Contributions | ReLU, Dropout | Small filter kernel | 1 × 1 Conv. | Residual learning |
| Advantages | Increase training speed and prevent overfitting | Suitable for parallel acceleration, nonlinear | Reduce the amount of computation | Overcome gradient vanishing |
| Disadvantages | Low accuracy | Small receptive field | Overfitting, vanishing gradient | Many parameters, long training time |
We denote the remaining layers (13 convolutional layers and 5 pooling layers) of VGG-16 as ~ to extract the features (i=0, 1, 2, 3, 4) at five different levels. The details of each block are provided in Table 2 .
Table 2.
Details of the Block1~Block5 to extract image features in a multi-level Pyramid scheme.
| Block | Layer | Filter Size/Channels | Stride | Padding |
|---|---|---|---|---|
| Block1 | Conv1-1 | 3 × 3/64 | 1 | Yes |
| Conv1-2 | 3 × 3/64 | 1 | Yes | |
| Maxpool | 2 × 2/64 | 2 | No | |
| Block2 | Conv2-1 | 3 × 3/128 | 1 | Yes |
| Conv2-2 | 3 × 3/128 | 1 | Yes | |
| Maxpool | 2 × 2/128 | 2 | No | |
| Block3 | Conv3-1 | 3 × 3/256 | 1 | Yes |
| Conv3-2 | 3 × 3/256 | 1 | Yes | |
| Conv3-3 | 3 × 3/256 | 1 | Yes | |
| Maxpool | 2 × 2/256 | 2 | No | |
| Block4 | Conv4-1 | 3 × 3/512 | 1 | Yes |
| Conv4-2 | 3 × 3/512 | 1 | Yes | |
| Conv4-3 | 3 × 3/512 | 1 | Yes | |
| Maxpool | 2 × 2/512 | 2 | No | |
| Block5 | Conv5-1 | 3 × 3/512 | 1 | Yes |
| Conv5-2 | 3 × 3/512 | 1 | Yes | |
| Conv5-3 | 3 × 3/512 | 1 | Yes | |
| Maxpool | 2 × 2/512 | 2 | No |
The features obtained by are used as the input to the corresponding encoder block (i=0, 1, 2, 3, 4). Each contains one GPM and two LPMs, which are used to capture the overall shape information and the complement detail information of the potential lung infection regions, respectively. More details regarding GPM and LPM will be introduced in the proceeding sections below. A contrast layer is applied to every feature map that is an output of each to measure the dissimilarity between each patch and its local average. The contrast feature can be obtained by conducting an average pooling operation with a 3 × 3 kernel as follows
| (1) |
The contrast layer contributes to making the infected regions standing out from their normal surroundings. The feature maps combined from each decoder and contrast layer are fed into decoder block ~ for further refinement, as shown in Fig. 2.
Now referring to the decoder configuration, we develop a supervised learning framework that enables recursive integration of multi-scale high-level features for generating the predications of the lung infection regions. The four decoder blocks ~ share a similar structure, which contains a ReLU activation function and an LPM. Each decoder block is designed to merge the features from a contrast layer, an encoder block, and a previous decoder block, as shown in Fig. 2 (far right column). Of note, features from the previous decoder are upsampled via a deconvolution layer before the above-referenced feature merge. For instance, the coarse-level features from the encoder block , the discriminant features from the contrast layer, and the upsampled output from are concatenated to produce the fine-level features. The fine-level features are then passed through two convolution layers and two LPMs to obtain the final local refinement information (see the local score map in Fig. 2) that is used to precisely determine the boundaries of lung infection regions.
After , a GPM followed by a series of LPMs and convolution layers is placed to capture the global guidance information (i.e., Global Score; rough estimation of the suspected lung infection regions). Hence, the initial lung infected region segmentation map can be successfully generated by integrating the complementary global guidance information (Global Score) and local refinement information (Local Score). Furthermore, the fine-level features are further utilized to refine the determination of the boundaries of the suspected lung infection regions (see BMS component in Fig. 2). The details regarding each layer of the encoder-decoder framework are shown in Table 3 .
Table 3.
Details of the proposed encoder-decoder convolutional network.
| Module | Block | Layer | Filter Size/Channels | Stride | Padding | |
|---|---|---|---|---|---|---|
| Encoder | GPM | GPMleft | Conv | 7 × 1/128 | 1 | Yes |
| Conv | 1 × 7/256 | 1 | Yes | |||
| GPMright | Conv | 1 × 7/128 | 1 | Yes | ||
| Conv | 7 × 1/256 | 1 | Yes | |||
| LPM | Conv | 3 × 3/256 | 1 | Yes | ||
| ReLU | ||||||
| Conv | 3 × 3/256 | 1 | Yes | |||
| Conv | Conv | 3 × 3/128 | 1 | Yes | ||
| ReLU | ReLU | |||||
| LPM | Conv | 3 × 3/128 | 1 | Yes | ||
| ReLU | ||||||
| Conv | 3 × 3/128 | 1 | Yes | |||
| Contrast Layer | Contrast Layer | avg_pool | 3 × 3/128 | 1 | No | |
| Decoder | DeConv | DeConv | 3 × 3/384 | 2 | Yes | |
| ReLU | ReLU | |||||
| LPM | Conv | 3 × 3/384 | 1 | Yes | ||
| ReLU | ||||||
| SConv | 3 × 3/384 | 1 | Yes | |||
| ReLU | ReLU | |||||
3.2. Global Perception Module (GPM)
In the past, when a CNN is designed (e.g., VGGNet [26], AlexNet [27], GoogLeNet [28], ResNet [29], LeNet [30], DenseNet [31]), small convolutional kernels (e.g., or ) are commonly used to extract hierarchical features because they are less computationally expensive (low time and memory requirements). However, small kernel sizes ( or ) limit the network's overall representation ability. It is desirable to explore a more effective strategy for handling large receptive field problems, which aims at capturing the global shape and appearance information of the lung infected regions. Research efforts have been devoted to enlarging the receptive field; strategies include dilated convolutions [32], deepening the convolutional layers [33], and adding a series of downsampling [34]. However, these operations pose new challenges as explained below. Conducting dilated convolutions and network deepening will lead to an increase in storage space and a decrease in computing efficiency. Performing multiple downsampling results in the loss and distortion of high-level feature information, thereby negatively impacting the determination of the gross lung infection regions. We are motivated to introduce a global perception module (GPM) that can enlarge the range of the receptive field but has low memory requirements and computation demands.
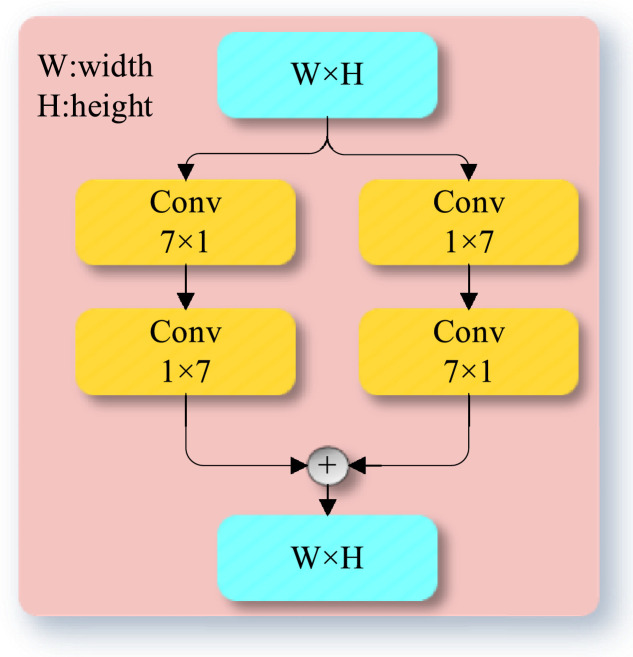
GPM can enlarge the receptive field while maintaining the resolution of the feature map so that the loss of spatial information in the feature maps can be avoided. In particular, using a convolution operation followed by a convolution operation is equivalent to a convolution with an receptive field [35]. Let denote a 2-d image, and denote two 1-d kernels along x-dimension and y-dimension, respectively. This mechanism can be represented as follows:
| (2) |
where is a 2-d kernel, and are the width and height of the 2-d image, respectively. As a result, the number of parameters is dramatically reduced and the performance degradation is minimal. Furthermore, let denote the number of channels in the input layer and output layer of GPM, the computational cost of adopting two 1-d convolution operations is , which is less than that of a 2-d convolution operation .
As illustrated in Fig. 3 , the proposed GPM comprises two sub-branches, each of which consists of two convolutional layers with kernel size and , respectively. After the convolution operations, the two sub-branches are integrated to enable the corresponding feature extraction layers to focus on a larger receptive field rather than on a small one. GPMs can capture deep high-level semantic information without significantly increasing the memory space and computation cost. A GPM has powerful global representation ability and can effectively avoid the loss of small-target information since its "effective" kernel size is . We consider that the adoption of GPM modules is essential for accurate pixel-wise segmentation of infected lung regions.
Fig. 3.
A schematic diagram showing the structure of GPM module.
3.3. Local Polishing Module (LPM)
Along with the increased feature levels, the holistic structure of the lung infection regions gradually appears with the help of the proposed GPMs. However, when deep convolutional networks are applied for image segmentation, they generally produce blurred boundaries and are not sensitive in terms of recognizing narrow non-infected regions that are (fully or partially) surrounded by their infected neighboring regions. This is not surprising because convolution layers (e.g., strides and pooling) may cause information loss. Recall that we import guidance information from the previous decoder layers (see Fig. 2), which contains less accurate information due to convolution and deconvolution layers. Particularly, around the contours of the infected regions, such information accuracy could deteriorate remarkably.
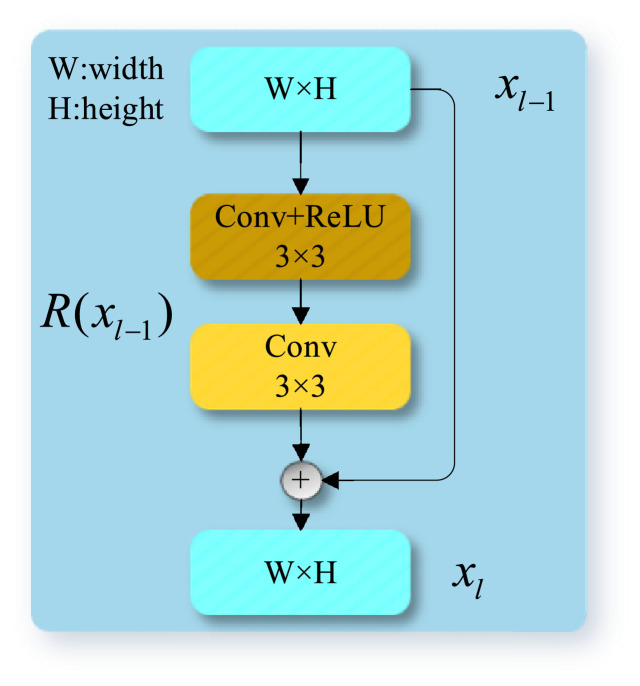
The local polishing module (LPM) possesses a simple but effective residual structure [29] and can be used to polish the feature representation, especially the boundary information. The idea of LPM is to promote the network to learn the residual representations of the input data via nonlinearity and it is implemented by introducing shortcut connections to skip the input layer to its output without extra parameters or computational complexity (see Fig. 4 ). LPM can be used together with any deep convolutional layers.
Fig. 4.
A schematic diagram showing the structure of LPM.
These skip connections help to retain spatial information at the network output so that the low-level knowledge contains rich edges, textures, and shapes can be effectively transmitted, making the proposed encoder-decoder framework more suitable for explicit segmentation. Specifically, LPMs keep the local details of the lung infection representations obtained by the CNNs, such as the backbone or preceding GPM, and learn to refine them with the residual connection. The output of LPM can be defined as:
| (3) |
where is the output of the previous layer, denotes the output after performing the convolution and activation operations.
As demonstrated in Fig. 4, an LPM is a localized residual net consisting of two convolution layers and one ReLU on . Since the residual connection directly propagates the input information to the output layer, the high-level and low-level information can be conveniently concatenated to preserve the integrity of the lung infection regions and detailed boundaries between the infected and non-infected regions.
3.4. Multi-Scale Multi-Level Feature Recursive Aggregation (mmFRA)
We propose two schemes to improve feature aggregation in this section. A multi-level feature recursive aggregation (mlFRA) scheme is to improve feature aggregation during the encoder step, while a multi-scale feature recursive aggregation (msFRA) scheme is to fine-tune information fusion prior to the generation of the initial segmentation map.
It is important to note that the execution of the proposed GPMs enables the global guidance information to be absorbed in the feature maps for different pyramid levels and the utilization of the LPMs allows the local refinement information to be maintained as much as possible during the knowledge transmission between convolutional layers. Now, our effort is steered towards the seamless aggregation of coarse-level features at different scales. Taking the proposed encoder (i.e., ~ in Fig. 2) as an example, the convolutional layers of different levels correspond to abstract features in different spatial scales: 1) for high-level features, there is abundant semantic information that is beneficial to positioning the lung infection regions and suppressing noise interference; 2) for middle-level features, both the semantic and detailed information can be adaptively prioritized, offering flexibility for feature utilization; 3) for low-level features, the fine-grained details can be preserved and are useful for predicting the boundaries of the lung infection regions. We strive to enhance multi-scale feature fusion by developing the innovative mlFRA scheme during the encoder step. Our goal is to fully integrate the high-level, middle-level, and low-level features by a strategy of recursive learning, which in turn generates a comprehensive and discriminative fine-level feature representation with local perception.
As shown in Fig. 2, the features at different levels, which are extracted by ~ of the VGG-16 backbone, are progressively refined in parallel to be more accurate and representative by passing through the encoder and contrast layers. In particular, the parallel encoding and decoding structure at different layers of VGG-16 can maintain the feature information at each level. Furthermore, the features at each level are updated with the integration of the encoder layer, contrast layer, and preceding decoder layer. Practically speaking, the proposed mlFRA can be achieved by concatenating the local feature , the contrast feature , and the higher level unpooled feature as follows,
| (4) |
where CAT is a concatenation operation.
As stated before, when VGG-16 is used to extract the features, two problems intrinsic to convolution neural networks arise, i.e., the inadequate expression of low-level features and the loss of feature information as the convolution layer deepens. The proposed mlFRA allows the semantic information of the high-level features, comprehensive information of the middle-level features, and detailed information of the low-level features to be gradually aggregated. Since three levels of features are complemental to each other, such aggregation can minimize information loss.
At a late step (i.e., prediction step), there are still fusion problems of global and local information from pyramid features extracted by the backbone network. We employ multi-scale feature recursive aggregation (msFRA) to finally integrate the global and local features for lung infected region segmentation. Instead of exploiting more complicated modules and sophisticated strategies to integrate the multi-scale features, our msFRA architecture is quite simple. It directly concatenates the global guidance feature map (which is obtained by feeding the backbone VGG-16 output into a series of GPM, LPM, and convolution with varying subsampling rates in the forward pass) and the local refinement feature map (which is obtained by feeding the output from mlFRA to a series of LPMs and convolutions with varying downsampling rates). In concrete terms, the proposed msFRA is implemented by integrating the global features with local features through a Softmax function:
| (5) |
where and denote the linear operators of the local feature map (denoted as ) and the global feature map (denoted as ), respectively. In effect, the probability of pixel belonging to the lung infection regions in the initial segmentation map can be predicted. In this way, the multi-scale features are aggregated to assist the localization of lung infection regions and the refinement of the local details and generate a rough segmentation map. The rough map will be used as the input to the BMS module (see Section 3.5 below).
In summary, the progressive mlFRA significantly increases the representations of the multi-level features stage by stage, and the msFRA greatly improves the segmentation performance. As a result, with multi-scale multi-level feature recursive aggregation (mmFRA), the coarse and multi-level features can be rectified and combined to produce the fine-level features, the cluttered information can be removed to better sharpen the details of the lung infection regions.
3.5. Boundary-Aware Multiple Supervision (BMS) Module
To train the networks, the design of the loss functions is very important. For segmentation networks, cross-entropy loss (CEL) is often utilized to measure the dissimilarity between the segmentation map (SM, denoted by ) and ground truth (GT, denoted by ). Let represent the index of each pixel and indicate the number of the pixels, the proposed segmentation cross-entropy loss (SCEL, denoted by ) function is defined as:
| (6) |
| (7) |
where represents the binary cross-entropy loss, which is extensively used and can be robust in segmentation and binary classification tasks.
Although the proposed mmFRA encourages the overall network to predict the initial segmentation map with abundant structure and rich contour information, the pixels near the boundaries of the lung infection regions are difficult to be correctly classified. Given this, to incentivize the deep network to pay more attention to the boundary pixels during the training phase, a boundary cross-entropy loss (BCEL, denoted as ) and a boundary-refinement loss (BRL, denoted as ), serving to generate exquisite boundaries, are exploited to work together with the SCEL for segmenting the lung infected regions. Let and denote the estimated boundary map and the true boundary map, respectively, and can be defined as follow:
| (8) |
| (9) |
Mathematically, the proposed boundary-aware multiple supervision (BMS) can be given as the combination of the three losses:
| (10) |
| (11) |
With multiple supervision using , and in an intermingled manner, the final lung infected region segmentation map of the uniform highlighted foreground and finely sharpened contours can be effectively achieved.
4. Experimental Results
In this section, we demonstrate the effectiveness of the proposed PCPLP network by providing detailed qualitative and quantitative experimental results using publically available COVID-19 CT datasets [36]. The evaluation was performed by comparing it with other eight state-of-the-art models in terms of 12 metrics.
4.1. Experimental Setup
Evaluation dataset. The COVID-19 CT dataset [36], which consists of 100 axial CT images from more than 40 patients, was collected by the Italian Society of Medical and Interventional Radiology (SIRM). Each image was annotated by a radiologist using three different labels: ground-glass (the value is 1), consolidation (the value is 2), and pleural effusion (the value is 3). In our experiments, 50% of CT images of the dataset were randomly selected for training and the remaining 50% were used for tests. In addition, a semi-supervised COVID-19 dataset [24], which contained 1600 unlabeled CT images extracted from the COVID-19 image data collection in [37], was utilized for augmenting the training set.
Implementation Details. We implemented the proposed PCPLP network using TensorFlow [38] deep learning framework on an NVIDIA Ge-Force RTX 2070 GPU and Inter Core i3-9100F CPU (3.6GHz) processor with 32 GB RAM. Our model was trained by Adam optimizer with a batch size of 1 for 10 epochs, which took about nine hours. The initial learning rate is 10−6, the weights of all the convolution and deconvolution layers were randomly initialized using a truncated normal distribution (standard deviation is 0.01), and the biases were initialized to 0. During the training and testing, CT slides were resized to a fixed dimension of 416 × 416 pixels. We employed horizontal flips on the training images for data augmentation.
Evaluation models. The proposed PCPLP framework was compared with other eight state-of-the-art segmentation models including FCNet [39], UNet [18], AttUnet [40], BASNet [41], EGNet [42], PoolNet [43], UNet++ [44], and Inf-Net [24]. For the sake of fair comparisons, all these models were trained using the same COVID-19 CT dataset and the same settings.
Evaluation metrics. To comprehensively evaluate the performance of the proposed PCPLP architecture against the other eight algorithms, we exploited 12 performance metrics as shown in Table 4 . In Table 4, true-positive (TP) and true-negative (TN) represent the correct classification ratio of positives (lung infection pixels) and negatives (non-lung infection pixels), respectively, whereas false-positive (FP), and false-negative (FN) denote the incorrect prediction ratio of lung infection regions and non-lung infection regions, respectively.
Table 4.
The 12 metrics for evaluating the performance of various segmentation models.
| Metric | Formula | Description |
|---|---|---|
| Receiver operating characteristic (ROC) curve | TPR and FPR measure the proportion of correctly identified actual positives and actual negatives, respectively | |
| Precision-recall (PR) curve | PR curve mainly evaluates the comprehensiveness of the detected lung infection pixels | |
| F-measure curve |
is set to 0.3 to emphasize the effect of |
F-measure is computed by the weighted harmonic mean of precision and recall, which can reflect the quality of detection |
| DICE score | DICE score measures the similarity between the predicted map and the ground truth | |
| Sensitivity score | Sensitivity score measures the rate of missed detection | |
| Specificity score | Specificity score measures the rate of false detection | |
| Mean absolute error (MAE) score |
and denote the width and height of the image, respectively |
MAE score indicates the similarity between the segmentation map and the ground truth |
| Area under curve (AUC) score |
and denote the set of negative and positive examples, respectively |
AUC score gives an intuitive indication of how well the segmentation map predicts the true lung infection regions |
| Weighted F-measure (WF) score [45] |
and denote the weighted precision and weighted recall, respectively |
WP and WP measure the exactness and completeness, respectively |
| Overlapping ratio (OR) score |
denotes the binary segmentation map |
OR score measures the completeness of lung infection pixels and the correctness of non-lung infection pixels |
| Structure-measure (S-M) score [46] |
and denote the object-aware similarity and region-aware similarity, respectively |
S-M score measures the structural similarity between the segmentation map and the ground truth |
| Execution time | Average execution time per image (in second) | All experiments were performed with the same equipment and settings |
4.2. Experimental Results
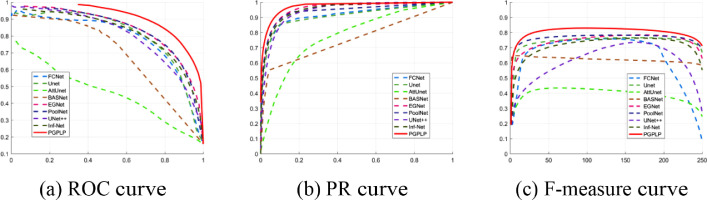
Quantitative results. The 12 metrics in Table 4 were used to measure the performance of the proposed method and the other eight methods. The obtained results are reported in Fig. 5 and Table 5 .
Fig. 5.
Performance comparisons of the proposed PCPLP framework with other models using the COVID-19 CT dataset [36].
Table 5.
Quantitative performance comparison of nine models in terms of different metrics. The best two results are highlighted in red and blue, respectively. The up-arrow ↑ indicates the higher value obtained, the better segmentation quality is, whereas the down-arrow ↓ implies the opposite.
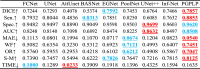 |
As depicted in Fig. 5, the curves of the proposed model, illustrated by the red solid lines, consistently lie above most of the other models, indicating that the predictions produced by our model had the minimum error in comparison with the ground truths. As can be seen in Table 5, it is evident that the scores of the DICE, Sensitivity, MAE, WF, OR, and S-M metrics, provided by the proposed PGPLP, outperformed all the compared state-of-the-art models. However, in terms of Specificity and AUC scores, our model only achieved the second-best results, which were slightly lower (0.0079 and 0.0124) than those obtained by UNet++ model. This indicates that our model is weaker than UNet++ in distinguishing non-infected regions but more outstanding in identifying segmented regions of interest.
We attributed the competitive performance to our progressive global perception and local polishing architecture, which yielded a robust feature representation with more complete structural information and finer detail information. In addition, by introducing the boundary-aware multi-supervised learning strategy into our framework, the segmentation accuracy can be further improved.
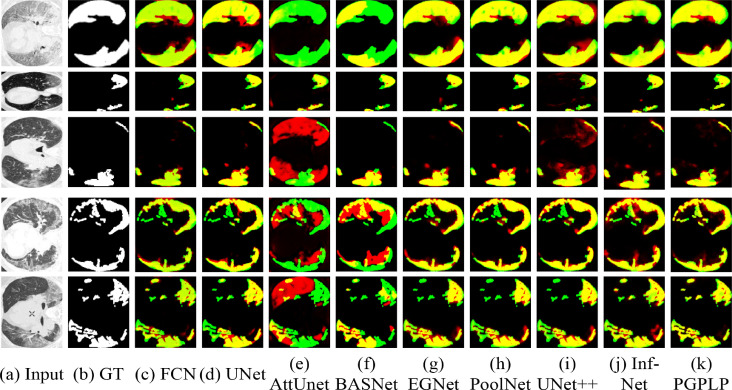
Qualitative results. As an assistant diagnostic means, the segmentation map is expected to offer more detailed information on the lung infection regions. The subjective performance comparisons between the manual annotations (ground truths) and nine AI-generated lung infection segmentation maps are shown in Fig. 6 . It can be seen clearly that the segmentation results of the proposed PGPLP model are highly consistent with the ground truth maps, which may supply a powerful guarantee for subsequent analysis. In contrast, the AttUnet and BASNet models generated unsatisfactory results, with many miss-segmented infection regions. FCN and UNet models were able to segment the large infection regions, but poor in identifying small regions. EGNet and UNet++ models over-segmented or improperly segmented some of the COVID-19 lesions. PoolNet and Inf-Net models produced relatively good results by aggregating the high-level features, but the complete structure and clear contour could not be predicted. The advantage of our PGPLP is mainly due to the coarse-to-fine deep FCN framework, in which the coarse-level Pyramid features are progressively integrated into fine-level features to roughly locate the lung infection regions and precisely segment the lesions respectively in an escalatory manner. This process mimics how a clinician visually segments the COVID-19 lung infected regions from CT slices and therefore can achieve good performance.
Fig. 6.
Visual comparisons of lung infection segmentation using different algorithms. The green and the yellow areas represent the undetected and detected true infection regions, respectively. The red areas indicate the false infection regions that are incorrectly detected. (a) The original CT images from the test set. (b) The corresponding ground truth for each image. (c-j) The corresponding segmentation results from the eight state-of-the-art models. (k) The segmentation maps of the proposed PGPLP.
4.3. Ablation Studies
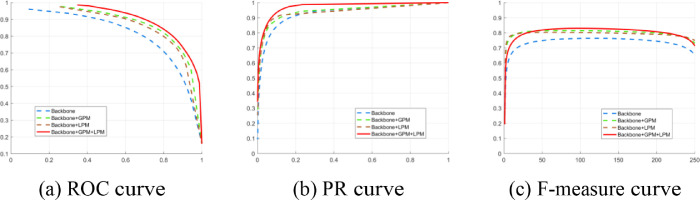
To assess the contribution of different modules of the proposed PGPLP architecture, an ablation study was performed under the same environment. The effectiveness of each component (GPM, LPM) in the proposed model is demonstrated in Fig. 7, Fig. 8 and Table 6 .
Fig. 7.
Performance comparisons using different variants of the proposed PCPLP model.
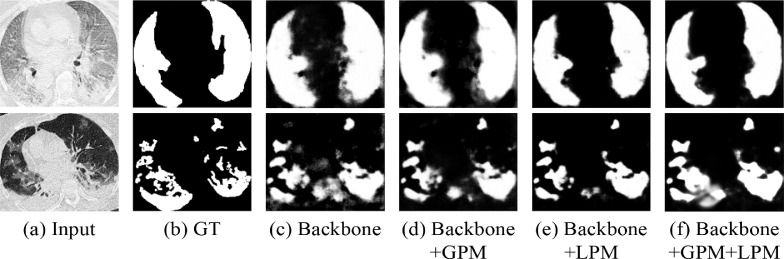
Fig. 8.
Qualitative performance comparisons using different variants of the proposed PCPLP model.
Table 6.
Quantitative performance comparisons using different variants of the proposed PCPLP model, the best two results are highlighted in red and blue colors, respectively.
In this part, we conducted an ablation study on three variants of the proposed PGPLP model: encoder-decoder model without GPM and LPM (Backbone), model without LPM (Backbone+GPM), and model without GPM (Backbone+LPM). The comparison results are shown in Fig. 7 and Table 6. As observed in Fig. 7, the embedded GPM and LPM are necessary for boosting performance. The advantages of GPM and LPM can also be found in Table 6. As can be seen in Table 6, the model with embedded GPM or LPM can perform competitively compared to the Backbone, and the complete model including both GPM and LPM can obtain more satisfactory performance than the model without LPM or GPM. To some extent, GPM helps the proposed model to obtain excellent results in localizing and counting the COVID-19 infection regions, and LPM also enables the model to achieve superior performance as it can accurately predict the contours of the infected regions. On the whole, the GPM and LPM improve the segmentation results in guiding the network to learn more informative features for the task of segmenting the COVID-19 infected regions.
Visual comparisons of the segmentation maps using different variants of the proposed PGPLP on the test set are presented in Fig. 8. It can be observed that the results of Backbone+GPM include complete infected regions, which demonstrates that the GPM component can enable the proposed model to exactly distinguish the true COVID-19 lesions. Meanwhile, the infection regions obtained by the Backbone+LPM model have sharpness boundaries, which verifies that the LPM component can significantly contribute to the fine-grained detail improvement. The optimum performance is gained with the joint GPM and LPM for higher emphasis on the structures and contours of the infected regions. Overall, both the GPM and LPM components play vital roles in the PGPLP model and bring lots of advantages to the segmentation results.
5. Conclusion
In this paper, a novel fully convolutional encoder-decoder network, named PGPLP, is proposed to help doctors quickly analyze the severity of pneumonia infections by segmenting the lung infected regions of COVID-19 from CT images. The proposed model employs a global perception module and a local polishing module to improve the localization and identification of the infection regions by more effectively retaining and assembling multi-level structure information and multi-scale detail information. Moreover, the multi-scale multi-level feature recursive aggregation strategy is exploited to integrate the multi-scale and multi-level features in a progressive manner, which not only substantially narrows the semantic gaps between the encoder and decoder blocks but also develops parallel inter-linking among multi-scale and multi-level features, thus greatly alleviates the vanishing gradient problem. Furthermore, our boundary-aware multiple supervision achieves considerable improvement over traditional single supervision methods for introducing more boundary information to produce finer details. Extensive experiments show that the proposed PGPLP architecture is capable of segmenting the infection regions of COVID-19 lesions under challenging conditions such as blurred infected interiors, diffusive infecting regions, and scattered boundaries.
Automated image segmentation enabled by our research offers opportunities to quantify the COVID-19 lesions, visualize the infection regions, and rapidly tracking the disease changes in the clinical workflow with minimal human intervention. Moreover, the proposed method has the potential to detect the abnormal areas between healthy tissues and lesions caused by other viruses.
Although promising results are achieved, one limitation of the proposed PGPLP model is the difficulty in detecting the small infection regions from the CT images with poor contrast. Those CT images require enhancement and more intelligent AI architectures so that additional features representing new knowledge can be learned and eventually applied to segmentation. In the future, the internal context information can be extracted using attention features and will be added to the training dataset to optimize the performance of the proposed model. We are optimistic that the limitation mentioned above can be overcome. Furthermore, our future plan also includes the integration of the segmentation data with the clinical presentation and laboratory results to help physicians better examine and diagnose COVID-19.
Declaration of Competing Interest
We wish to confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
We also confirm that the manuscript has been read and approved by all named authors and that there are no other persons who satisfied the criteria for authorship but are not listed. We further confirm that the order of authors listed in the manuscript has been approved by all of us.
We have given due consideration to the protection of intellectual property associated with this work and that there are no impediments to publication, including the timing of publication, with respect to intellectual property. In so doing we confirm that we have followed the regulations of our institutions concerning intellectual property.
We understand that the Corresponding Author is the sole contact for the Editorial process (including Editorial Manager and direct communications with the office). He is responsible for communicating with the other authors about progress, submissions of revisions and final approval of proofs. We confirm that we have provided a current, correct email address that is accessible by the Corresponding Author and which has been configured to accept email from jinshant@mtu.edu.
Acknowledgments
Nan Mu was supported by the National Science Foundation of China (62006165).
Biographies
Nan Mu received the Ph.D. degree in the School of Computer Science and Technology from Wuhan University of Science and Technology, China, in 2019. Currently, he is a lecturer in the School of Computer Science, Sichuan Normal University, Chengdu, China. His research interests include image processing and deep learning.
Hongyu Wang received the B.E. degree in the School of Computer Science and Technology from Chengdu University of Technology, China, in 2019. She is currently working toward the M.E. degree at the School of Computer Science, Sichuan Normal University, Chengdu, China. Her research interests include computer vision and deep learning.
Yu Zhang received the B.E. degree in the school of business from Chengdu University, China, in 2020. She is currently working toward the M.E. degree at the School of Computer Science, Sichuan Normal University, Chengdu, China. Her research interests include deep learning and image processing.
Jingfeng Jiang received the B.S. and M.S. degrees in structural engineering from Zhejiang University, China, in 1995 and 1998, respectively, and the M.S. degree in computer science (image analysis) and the Ph.D. degree in civil engineering (computational mechanics) from the University of Kansas, in 2002 and 2003, respectively.,He was with the Department of Medical Physics, University of Wisconsin–Madison, from 2003 to 2012, first as a Postdoctoral Associate and then as a Research Scientist. He is currently an Associate Professor of biomedical engineering with Michigan Technological University, Houghton, MI, USA. He also holds affiliated/visiting appointments at the University of Wisconsin, Mayo Clinics, and SWPU. His overall research interests stride the borders among imaging, biology, and computational sciences. At the University of Wisconsin, he mainly worked on algorithm developments for ultrasound elastography. More recently, he has expanded his research into advanced open-source elastography simulations (https://github.com/jjiang-mtu/virtual-breast-project), cardiovascular flow analytics and medical image analysis, and machine learning and uncertainty quantification for computational biomechanics.
Dr. Jinshan Tang received his Ph.D degree in 1998 from Beijing University of Posts and Telecommunications, and got post-doctoral training in Harvard Medical School and National Institute of Health. Dr. Tang's research interests include biomedical image processing, biomedical imaging, and computer aided cancer detection. He has obtained more than three million dollars grants in the past years as a PI or Co-PI. He published more than 100 refereed journal and conference papers. He published two edited books on medical image analysis. One is Computer Aided Cancer Detection: Recent Advance and the other is Electronic Imaging Applications in Mobile Healthcare. Dr. Tang is a leading guest editor of several journals on medical image processing and computer aided cancer detection. He is a senior member of IEEE and Co-chair of the Technical Committee on Information Assurance and Intelligent Multimedia-Mobile Communications, IEEE SMC society. His research has been supported by USDA, DoD, NIH, Air force, DoT, and DHS.
References
- 1.Wong H.Y.F., Lam H.Y.S., Fong A.H.T., Leung S.T., Chin T.W.Y., Lo C.S.Y., Lui M.M.S., Lee J.C.Y., Chiu K.W.H., Chung T., Lee E.Y.P., Wan E.Y.F., Hung F.N.I., Lam T.P.W., Kuo M., Ng M.Y. Frequency and distribution of chest radiographic findings in COVID-19 positive patients. Radiology. 2020;296(2):E72–E78. doi: 10.1148/radiol.2020201160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of covid-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020;121:1–11. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Narin A., Kaya C., Pamuk Z. Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks. arXiv preprint arXiv:2003. 2020;10849:1–31. doi: 10.1007/s10044-021-00984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hemdan E.E.-D., Shouman M.A., Karar M.E. COVIDX-Net: A framework of deep learning classifiers to diagnose COVID-19 in X-Ray images. arXiv preprint arXiv:2003. 2020;11055:1–14. [Google Scholar]
- 5.Grillet F., Behr J., Calame P., Aubry S., Delabrousse E. Acute pulmonary embolism associated with COVID-19 pneumonia detected with pulmonary CT angiography. Radiology. 2020;296(3):E186–E188. doi: 10.1148/radiol.2020201544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shan F., Gao Y., Wang J., Shi W., Shi N., Han M., Xue Z., Shen D., Shi Y. Lung infection quantification of COVID-19 in CT images with deep learning. arXiv preprint arXiv:2003. 2020;04655:1–23. [Google Scholar]
- 7.Zhou L., Li Z., Zhou J., Li H., Chen Y., Huang Y., Xie D., Zhao L., Fan M., Hashmi S., Abdelkareem F., Eiada R., Xiao X., Li L., Qiu Z., Gao X. A rapid, accurate and machine-agnostic segmentation and quantification method for CT-based COVID-19 diagnosis. IEEE Trans. Med. Imaging. 2020;39(8):2638–2652. doi: 10.1109/TMI.2020.3001810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi F., Wang J., Shi J., Wu Z., Wang Q., Tang Z., He K., Shi Y., Shen D. Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for COVID-19. IEEE Rev. Biomed. Eng. 2020:1–13. doi: 10.1109/RBME.2020.2987975. [DOI] [PubMed] [Google Scholar]
- 9.Jiang X., Coffee M., Bari A., Wang J., Jiang X., Huang J., Shi J., Dai J., Cai J., Zhang T., Wu Z., He G., Huang Y. Towards an artificial intelligence framework for data-driven prediction of coronavirus clinical severity. Computers, Materials & Continua. 2002;62(3):537–551. [Google Scholar]
- 10.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., Bai J., Lu Y., Fang Z., Song Q., Cao K., Liu D., Wang G., Xu Q., Fang X., Zhang S., Xia J., Xia J. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 2020;296(2):E65–E71. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harmon S.A., Sanford T.H., Xu S., Turkbey E.B., Roth H., Xu Z., Yang D., Myronenko A., Anderson V., Amalou A., Blain M., Kassin M., Long D., Varble N., Walker S.M., Bagci U., Ierardi A.M., Stellato E., Plensich G.G., Franceschelli G., Girlando C., Irmici G., Labella D., Hammoud D., Malayeri A., Jones E., Summers R.M., Choyke P.L., Xu D., Flores M., Tamura K., Obinata H., Mori H., Patella F., Cariati M., Carrafiello G., An P., Wood B.J., Turkbey B. Artificial intelligence for the detection of COVID19 pneumonia on chest CT using multinational datasets. Nat. Commun. 2020;11(1):1–7. doi: 10.1038/s41467-020-17971-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salman F.M., Abu-Naser S.S., Alajrami E., Abu-Nasser B.S., Ashqar B.A. Covid-19 detection using artificial intelligence. International Journal of Academic Engineering Research. 2002;4(3):18–25. [Google Scholar]
- 13.Wang L., Lin Z.Q., Wong A. COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci. Rep. 2020;10(1):1–12. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adhikari N.C.D. Infection severity detection of CoVID19 from X-Rays and CT scans using artificial intelligence. International Journal of Computer. 2020;38(1):73–92. [Google Scholar]
- 15.Barstugan M., Ozkaya U., Ozturk S. Coronavirus (covid-19) classification using CT images by machine learning methods. arXiv preprint arXiv:2003. 2020;09424:1–10. [Google Scholar]
- 16.Jaiswal A., Gianchandani N., Singh D., Kumar V., Kaur M. Classification of the COVID-19 infected patients using DenseNet201 based deep transfer learning. J. Biomol. Struct. Dyn. 2020:1–8. doi: 10.1080/07391102.2020.1788642. [DOI] [PubMed] [Google Scholar]
- 17.Wang Z., Liu Q., Dou Q. Contrastive cross-site learning with redesigned net for COVID-19 CT classification. IEEE Journal of Biomedical and Health Informatics. 2020;24(10):2806–2813. doi: 10.1109/JBHI.2020.3023246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ronneberger O., Fischer P., Brox T. Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention. 2015. U-net: Convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- 19.Chen X., Yao L., Zhang Y. Residual attention U-Net for automated multi-class segmentation of COVID-19 chest CT images. arXiv preprint arXiv:2004. 2020;05645:1–7. [Google Scholar]
- 20.Zhou T., Canu S., Ruan S. Automatic COVID-19 CT segmentation using U-Net integrated spatial and channel attention mechanism. Int. J. Imaging Syst. Technol. 2020:1–12. doi: 10.1002/ima.22527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Müller D., Rey I.S., Kramer F. Automated Chest CT Image Segmentation of COVID-19 lung infection based on 3D U-Net. arXiv preprint arXiv:2007. 2020;04774:1–9. [Google Scholar]
- 22.Zhou L., Li Z., Zhou J., Li H., Chen Y., Huang Y., Xie D., Zhao L., Fan M., Hashmi S., Abdelkareem F., Eiada R., Xiao X., Li L., Qiu Z., Gao X. A rapid, accurate and machine-agnostic segmentation and quantification method for CT-based COVID-19 diagnosis. IEEE Trans. Med. Imaging. 2020;39(8):2638–2652. doi: 10.1109/TMI.2020.3001810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang G., Liu X., Li C., Xu Z., Ruan J., Zhu H., Meng T., Li K., Huang N., Zhang S. A noise-robust framework for automatic segmentation of COVID-19 pneumonia lesions from CT images. IEEE Trans. Med. Imaging. 2020;39(8):2653–2663. doi: 10.1109/TMI.2020.3000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan D.-P., Zhou T., Ji G.-P., Zhou Y., Chen G., Fu H., Shen J., Shao L. Inf-net: Automatic COVID-19 lung infection segmentation from CT images. IEEE Trans. Med. Imaging. 2020;39(8):2626–2637. doi: 10.1109/TMI.2020.2996645. [DOI] [PubMed] [Google Scholar]
- 25.Laradji I., Rodriguez P., Manas O., Lensink K., Law M., Kurzman L., Parker W., Vazquez D., Nowrouzezahrai D. Proceedings of the IEEE Winter Conference on Applications of Computer Vision. 2021. A weakly supervised consistency-based learning method for COVID-19 segmentation in CT images; pp. 2453–2462. [Google Scholar]
- 26.Simonyan K., Zisserman A. Proceedings of the International Conference on Learning Representations. 2015. Very deep convolutional networks for large-scale image recognition; pp. 1–14. [Google Scholar]
- 27.Krizhevsky A., Sutskever I., Hinton G.E. ImageNet classification with deep convolutional neural networks. Proceedings of Advances in Neural Information Processing Systems. 2012:1097–1105. [Google Scholar]
- 28.Szegedy C., Liu W., Jia Y., Sermanet P., Reed S., Anguelov D., Erhan D., Vanhoucke V., Rabinovich A. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2015. Going deeper with convolutions; pp. 1–9. [Google Scholar]
- 29.He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- 30.LeCun Y., Bottou L., Bengio Y., Haffner P. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 1998. Gradient-based learning applied to document recognition; pp. 2278–2324. [Google Scholar]
- 31.Huang G., Liu Z., van der Maaten L., Weinberger K.Q. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Densely connected convolutional networks; pp. 4700–4708. [Google Scholar]
- 32.Yu F., Koltun V. Multi-scale context aggregation by dilated convolutions. arXiv preprint arXiv:1511. 2015;07122:1–13. [Google Scholar]
- 33.Wang W., Shen J., Porikli F. Proceedings of the IEEE conference on computer vision and pattern recognition. 2015. Saliency-aware geodesic video object segmentation; pp. 3395–3402. [Google Scholar]
- 34.Wu Y.H., Liu Y., Zhang L., Cheng M.M. EDN: Salient object detection via extremely-downsampled network. arXiv preprint arXiv:2012. 2020;13093:1–10. doi: 10.1109/TIP.2022.3164550. [DOI] [PubMed] [Google Scholar]
- 35.Szegedy. V. Vanhoucke C., Ioffe S., Shlens J., Wojna Z. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Rethinking the inception architecture for computer vision; pp. 2818–2826. [Google Scholar]
- 36.H. B. Jenssen, COVID-19 CT segmentation dataset, https://medicalsegmentation.com/covid19/, 2020.
- 37.Cohen J.P., Morrison P., Dao L., Roth K., Duong T.Q, Ghassemi M. COVID-19 image data collection: Prospective predictions are the future. arXiv preprint arXiv:2006. 2020;11988:1–38. [Google Scholar]
- 38.Abadi M., Barham P., Chen J., Chen Z., Davis A., Dean J., Devin M., Ghemawat S., Irving G., Isard M., Kudlur M., Levenberg J., Monga R., Moore S., Murray D.G., Steiner B., Tucker P., Vasudevan V., Warden P., Wicke M., Yu Y., Zheng X. Proceedings of 12th USENIX Symposium on Operating Systems Design and Implementation. 2016. Tensorflow: A system for largescale machine learning; pp. 265–283. [Google Scholar]
- 39.Long J., Shelhamer E., Darrell T. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2015. Fully convolutional networks for semantic segmentation; pp. 3431–3440. [DOI] [PubMed] [Google Scholar]
- 40.Oktay O., Schlemper J., Folgoc L.L., Lee M., Heinrich M., Misawa K., Mori K., McDonagh S., Y Hammerla N., Kainz B., Glocker B., Rueckert D. Proceedings of the 1st Conference on Medical Imaging with Deep Learning. 2018. Attention u-net: Learning where to look for the pancreas; pp. 1–10. [Google Scholar]
- 41.Qin X., Zhang Z., Huang C., Gao C., Dehghan M., Jagersand M. Proceedings of the IEEE conference on computer vision and pattern recognition. 2019. BASNet: Boundary-aware salient object detection; pp. 7479–7489. [Google Scholar]
- 42.Zhao J.-X., Liu J.-J., Fan D.-P., Cao Y., Yang J., Cheng M.-M. Proceedings of the IEEE conference on computer vision and pattern recognition. 2019. EGNet: Edge guidance network for salient object detection; pp. 8779–8788. [Google Scholar]
- 43.Liu J.-J., Hou Q., Cheng M.-M., Feng J., Jiang J. Proceedings of the IEEE conference on computer vision and pattern recognition. 2019. A simple pooling-based design for real-time salient object detection; pp. 3917–3926. [Google Scholar]
- 44.Zhou Z., Siddiquee M.M.R., Tajbakhsh N., Liang J. Proceedings of Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. 2019. UNet++: A nested U-Net architecture for medical image segmentation; pp. 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Margolin R., Zelnik-Manor L., Tal A. Proceedings of the IEEE conference on computer vision and pattern recognition. 2014. How to evaluate foreground maps? pp. 248–255. [Google Scholar]
- 46.Fan D.-P., Cheng M.-M., Liu Y., Li T., Borji A. Proceedings of the IEEE International Conference on Computer Vision. 2017. Structure-measure: A new way to evaluate foreground maps; pp. 4548–4557. [Google Scholar]