Abstract
Background
Ethnic differences in testicular cancer rates (TCRs) are recognized internationally. Cannabis is a known risk factor for testicular cancer (TC) in multiple studies with dose-response effects demonstrated, however the interaction between ancestral and environmental mutagenic effects has not been characterized. We examined the effects of this presumed gene-environment interaction across US states.
Methods
State based TCR was downloaded from the Surveillance Epidemiology and End Results (SEER) website via SEERStat. Drug use data for cigarettes, alcohol use disorder, analgesics, cannabis and cocaine was taken from the National Survey of Drug Use and Health a nationally representative study conducted annually by the Substance Abuse and Mental Health Services Administration (SAMHSA) with a 74.1% response rate. Cannabinoid concentrations derived from Drug Enforcement Agency publications. Median household income and ethnicity data (Caucasian-American, African-American, Hispanic-American, Asian-American, American-Indian-Alaska-Native-American, Native-Hawaiian-Pacific-Islander-American) was from the US Census Bureau. Data were processed in R using instrumental regression, causal inference and multiple imputation.
Results
1975–2017 TCR rose 41% in African-Americans and 78.1% in Caucasian-Americans; 2003–2017 TCR rose 36.1% in Hispanic-Americans and 102.9% in Asian-Pacific-Islander-Americans. Ethnicity-based scatterplot-time and boxplots for cannabis use and TCR closely mirrored each other. At inverse probability-weighted interactive robust regression including drugs, income and ethnicity, ethnic THC exposure was the most significant factor and was independently significant (β-estimate = 4.72 (2.04, 7.41), P = 0.0018). In a similar model THC, and cannabigerol were also significant (both β-estimate = 13.87 (6.33, 21.41), P = 0.0017). In additive instrumental models the interaction of ethnic THC exposure with Asian-American, Hispanic-American, and Native-Hawaiian-Pacific-Islander-American ethnicities was significant (β-estimate = − 0.63 (− 0.74, − 0.52), P = 3.6 × 10− 29, β-estimate = − 0.25 (− 0.32, − 0.18), P = 4.2 × 10− 13, β-estimate = − 0.19 (− 0.25, − 0.13), P = 3.4 × 10− 9). After multiple imputation, ethnic THC exposure became more significant (β-estimate = 0.68 (0.62, 0.74), P = 1.80 × 10− 92). 25/33 e-Values > 1.25 ranging up to 1.07 × 105. Liberalization of cannabis laws was linked with higher TCR’s in Caucasian-Americans (β-estimate = 0.09 (0.06, 0.12), P = 6.5 × 10− 10) and African-Americans (β-estimate = 0.22 (0.12, 0.32), P = 4.4 × 10− 5) and when dichotomized to illegal v. others (t = 6.195, P = 1.18 × 10− 9 and t = 4.50, P = 3.33 × 10− 5).
Conclusion
Cannabis is shown to be a TC risk factor for all ethnicities including Caucasian-American and African-American ancestries, albeit at different rates. For both ancestries cannabis legalization elevated TCR. Dose-response and causal relationships are demonstrated.
Keywords: Testicular cancer, Cannabis, Other drugs, Ethnicity, Pathways, Mechanisms, Casual inference
Background
Testicular Cancer (TC) is the commonest cancer in men aged 15–44 years, and rates have increased two to three times in many nations in recent decades [1]. Indeed TC is the leading cause of individual ‘years of life lost’ of any adult cancer [1].
Both genetic and environmental factors are believed to be significant, with 25% of the risk ascribed to genetic factors [1, 2]. This includes eight to ten- fold elevation in risk of brothers of cases, and four- to six- fold elevation in their sons [1]. 2–5% of cases are bilateral [1, 2]. Well-described differing rates by ethnicity have also been reported from many continents including Europe, New Zealand and USA [3–5], with a twenty-fold variation in TC rates (TCRs) in nationals of northern Europe (Denmark and Norway) compared to various African nations [1, 5, 6].
Data from the US SEER*Explorer website reveals that the all-age all-stages age-adjusted TCR in American males rose 83.45% from 3.4415 to 6.3136 / 100,000 in the years 1976–2017 [3]. The commonest age for TC is in males aged 30–34 years which is represented in the official statistics by the 15–39 year age group whose age-adjusted rate rose 92.16% from 6.29 to 12.09 /100,000 from 1975 to 2017 across all ethnicities combined [3].
However, these US TCR trends for “all ethnicities combined” conceal vastly different rates of TCR amongst different ethnic groups. For example amongst the US 15–39 year age-group TCR for Caucasian-Americans vs. African-Americans for 1975 was 7.01, and 3.09/100,000 respectively. However, this TCR for each ethnic group has not remained static over time.
Amongst the 15–39 year age-group the TCR amongst Caucasian-Americans rose 111.61% from 7.01 to 14.81 / 100,000 across the years 1975–2016. In contrast, amongst African-Americans in this age group the TCR fell 14.24% from 3.09 to 2.65/100,000. Accordingly across this time-period the mean TCR in Caucasian-Americans increased to a mean of 11.7 (+ 0.3374 S.E.M.), whereas in African Americans decreased to 2.82 (+ 0. 0.0737 S.E.M.) / 100,000. That is the rate of TCR amongst African Americans was decreased to 24.05% of the rate in Caucasian-Americans suggesting a strong gene-environment interaction. SEER*Explorer data show that the rate amongst Caucasian-Americans 15–39 years rose 3.7% annually 1975–1987 and 1.0% from 1987 to 2017 [3]. No annual percent change was listed on this site for African-American ancestry.
Interestingly the nature of the different genetic risk between African-Americans and Caucasian-Americans has been precisely defined as being due to a differing allelic frequency at an anomalous P53 response element on Chromosome 9 which drives cellular proliferation rather than the usual suppression of growth-related activities in cells experiencing genotoxic stress which is more generally associated with P53 activation [7, 8]. This effect is described further in the Discussion.
Curiously, international reports include documentation of very different case rates within the same ethnic group further indicating a strong role for environmental components in TC, with current data suggesting the prospect of one or more gene-environment interactions [7, 9]. For example some ethicities in neighbouring nations across Europe show significantly different rates of TC [1, 5, 6]. Similarly, geographic TC clusters amongst some ethnic populations have also been reported in northern Netherlands [10], while TCR in the Hispanic-American community has been reported as rising most rapidly in recent years [1]. Moreover, a man of Caucasian descent who moves from a low to a high incidence area assumes the risk of the high incidence area in the following generation [1]. This is consistent with data suggesting TC risk is 25% genetic [1, 2], and 75% environmental [7, 9].
It is generally agreed that TC likely results from in utero germ cell anomalies which then become activated by the hormonal surge of adolescence. Important classification and treatment insights have flowed from the increasing recent elucidation of the biology of testicular cancer and its close relationship to disordered primordial germ cell and spermatogonial development often from antenatal germ cell neoplasia in situ (GCNIS) [1, 11–14]. For example, on occasion immune checkpoint inhibition with PD-L1 inhibitors is recommended in selected patients [14].
That higher potency varieties of cannabis have been increasingly used in many countries over the past decades, couple with published data that cannabis exposure is both genotoxic [15], and results in genomic and chromosomal damage [13, 16] raises the possibility that cannabinoid exposure in utero may be a risk factor for TC.
However, notwithstanding the role of genetics and likely in-utero cannabis exposure, all four studies to examine the association of TC and cannabis have noted that personal cannabis use is linked with rising testicular cancer rates [17–20]. Importantly, dose-response effects have been described [17–19] which was also confirmed in meta-analysis [6]. Two meta-analyses have been published on this data which found pooled odds ratios for exposure of 2.59 (95%C.I. 1.60, 4.19) for chronic, current and frequent cannabis use [6] and 1.71 (1.12, 2.60) [21].
Thus while TC may have an genetic component and is believed to arise a result of in utero germ cell anomalies in which cannabis exposure may be a factor, all four studies linking testicular cancer with cannabis use describe personal use in the preceding twenty years. This suggests both that exposure through personal use may be an important environmental component and that the usual pathophysiological processes of testicular cancerogenesis are greatly accelerated by personal cannabinoid exposure. Multiple pathways and mechanisms for cannabis damage to the primordial germ cell are reviewed in the Discussion section.
This evident temporal compression of the natural history of testicular carcinogenesis by cannabis exposure may offer further pathogenetic insights into the accelerated development of this tumour type.
As cannabis use has been linked with TCR in all four studies to examine the association [17–20], and as several reviews of TC epidemiology have noted its likely significance [1, 9, 13, 14, 22], it seemed important to examine the association between cannabis and testicular cancer across both space and time, along with ethnicity and other drug and cannabis exposures. It was also important to determine if any potential relationship satisfied the quantitative criteria for causality in all ethnicities. As data on known risk factors such as cryptorchidism, inguinal herniae, industrial pollution and sedentary lifestyles across space and time was not available to the present investigators it was not possible to include these covariates in the present analysis. The USA formed a suitable setting for this epidemiological investigation as the required data on TCR and other drug and cannabis use by State and different ethnicities over time is readily publicly available.
Methods
Data
National data on age-adjusted TCR, age-specific TCR and ethnic TCR were downloaded from the SEER*Explorer website [3]. State based data both as overall age-adjusted TCR and by ethnicity was downloaded using the SEERStat software from the National Program of Cancer Registries (NPCR) and Surveillance Epidemiology and End Results (SEER) Incidence File from the US Cancer Statistics Public Use Database, Submission 2001–2017 [23]. Drug use data for the years 2003–2017 was taken from the Restricted Use Data Analysis System (RDAS) of the Substance Abuse and Mental Health Data Archive (SAMHDA) of the National Survey of Drug Use and Health (NSDUH) from the Substance Abuse and Mental Health Services Administration (SAMHSA) [24]. The major drugs of interest were last year cocaine use (Cocaine), last year non-medical use of pain relievers (Analgesics), last month cigarette use (Cigarettes) and last year abuse or dependence on Alcohol (Alcohol Use Disorder, AUD). Data was supplemented by median household income and state-based ethnicity data from the US Census bureau focussing on Caucasian-American, African-American, Hispanic-American, Asian-American, American Indian / Alaskan Native (AIAN) -American and Native Hawaiian / Pacific Islander (NHPI) -American ethnicities. This ethnicity data was paired as closely as possible with the ethnicity data from the SEER database which sometimes used a slightly different categorization system. Cannabinoid concentration data was obtained from publications of Drug Enforcement Agency listing the various concentrations found in Federal seizures [25–27]. Data relating to the legal status of cannabis in US states was derived from an internet search [28].
Derived data
Data on intensity of use of cannabis was taken from a variable called “mrjmdays” denoting the number of days in the past month for which cannabis had been used. The responses to this variable are categorized as 0, 1–2, 3–5, 6–19 and 20–30 days per month. For each ethnicity and for each year of the NSDUH the percentage responding in each category was multiplied by the mean number of days in that category and summed to provide an ethnic cannabis use score for that year. This ethnic cannabis use intensity score was then multiplied by the percent of cannabis used last month in that state and the concentration for that year of THC in Federal seizures to provide an estimate of ethnic-specific THC exposure. State-based cannabinoid concentrations were estimated as the product of the concentration of various cannabinoids in Federal seizures multiplied by the last month cannabis use in each State.
Statistics
Data was processed using R version 4.0.2 and R-Studio 1.3.1093 in October 2020. Data was read in and reconfigured using dplyr from the tidyverse suite [29]. Maps were drawn in ggplot2, sf and RColorBrewer [30–32]. Graphs drawn in Microsoft Excel, ggplot2 and lattice [30, 33]. Data was log transformed as indicated by the results of the Shapiro-Wilks test. All regression models were manually serially reduced by elimination of the least significant term as is performed in the classical technique of model reduction. Mixed effects was conducted using the nlme package [34]. Two-step instrumental variable regression was conducted using the ivreg function from the AER package [35].
Inverse probability weighting was calculated from the ipw package [36] and was used to control for cannabis exposure across all the groups by other substance exposure. This was applied in mixed effects repeated measures, two-step instrumental variable regression and robust regression. Each of these forms of multivariable regression was used for different purposes. Mixed effects and instrumental variable models all provided model standard deviations from which e-Values could be calculated. Robust generalized linear regression from the survey package was used to perform robustified regression [37]. Mixed effects models with state as the random effect were used to account for the repeated measures nature of the data. Instrumental variable regression was used to test directly for cannabinoid effects underlying the effects of the primary covariates as described in the Results section.
SEER data is suppressed for cell counts less than 15 or very low rates. Hence a high rate of missing data was noted particularly for TCR by ethnicity. The problem was most marked for ethnic minorities. This issue was addressed by multiple imputation by chained equations using the mice package in R [38]. In view of the size of the missing data problem 256 imputations were used each with 20 iterations. The initial seed used was 59. The imputation method was by the classification tree (“cart”) method which provided the best fit for the ethnically grouped data. Linear models were used to investigate this data. The results from models were pooled appropriately using Rubin’s rules. Using these techniques the fraction of missing information obtained in simple linear models regressing TCR against ethnic THC exposure was reduced to 2.6%.
e-Values were calculated using the e-Value algorithms of the EValue package [39].
All t-tests were two sided. P < 0.05 was considered significant.
Data sharing and availability
Data has been deposited in the Mendeley data repository along with the software programming code in R and may be found at URL 10.17632/yxy3dg2wt6.1.
Ethics
All methods were carried out in accordance with relevant guidelines and regulations. This study was approved by the Human Research Ethics Committee of the University of Western Australia on 7th January 2020 RA/4/20/7724.
Results
The outline plan for the results section is as follows. We first present the univariate ethnic and testicular cancer rate data upon which the analysis rests. We then examine various bivariate relationships including different ethnic time trends. Various forms of multivariable regression are used to determine the impact of multivariate adjustment on the bivariate relationships described. All multivariable models are inverse probability weighted and E-Values are freely used to allow formal causal inferences to be drawn both qualitative and quantitatively. Multiple imputation is employed to complete ethnic TCR data where missing data is severely problematical. Finally the effects of legal status on the TCR in various ethnicities is considered and investigated by the tools of causal inference. Each of these steps in this procedure has several component steps which are included along the way as is mandatory in a formal presentation of such analyses.
Univariate data
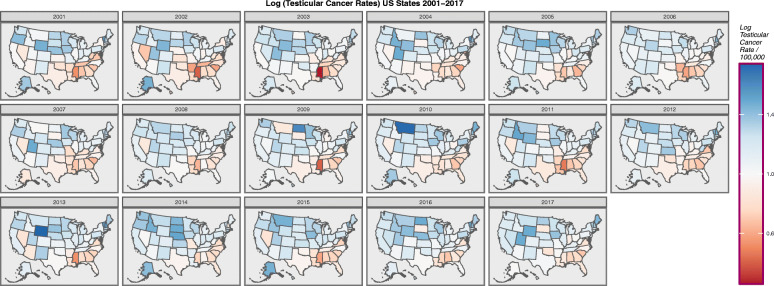
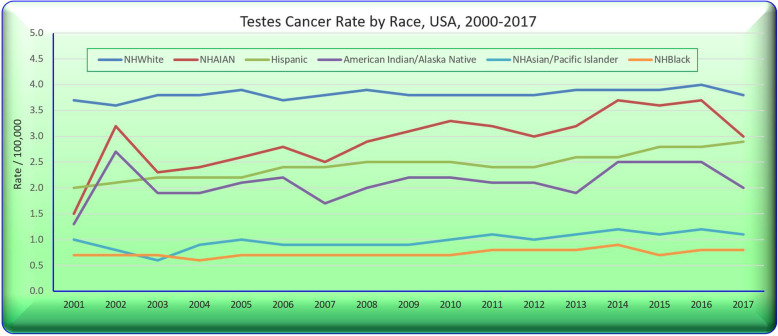
Figure 1 shows map-graphically the distribution of the age-adjusted TCRs across USA states over time 2001–2017 across all ages. The maps are notable because cases seem to be most concentrated in the upper midwest and northeast and to be negatively associated with the south-eastern corner.
Fig. 1.
Choropleth map of log(testicular cancer incidence rates) across USA by Year. Maps were drawn in ggplot2, sf and RColorBrewer
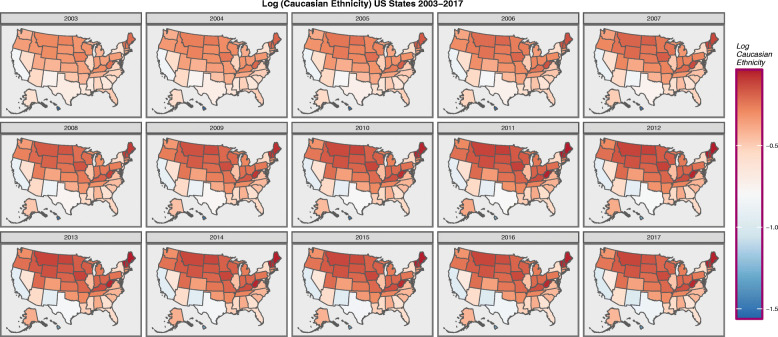
Figure 2 sets out in a similar format the ethnic composition of USA states as the log (Caucasian-American) population.
Fig. 2.
Choropleth map of log(Caucasian-American Population) across USA by Year. Maps were drawn in ggplot2, sf and RColorBrewer
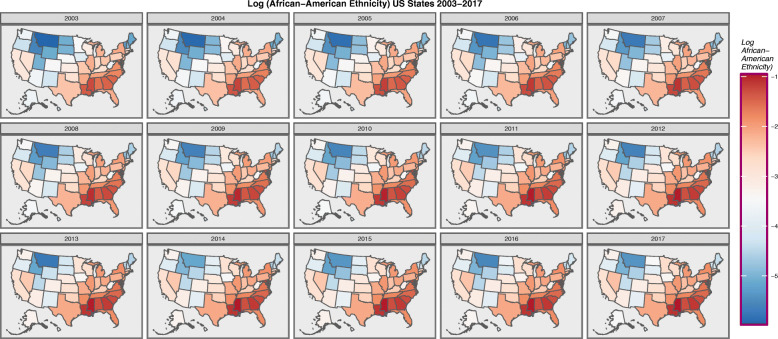
Figure 3 does the same for the African-American population.
Fig. 3.
Choropleth map of log(African-American Population) across USA by Year. Maps were drawn in ggplot2, sf and RColorBrewer
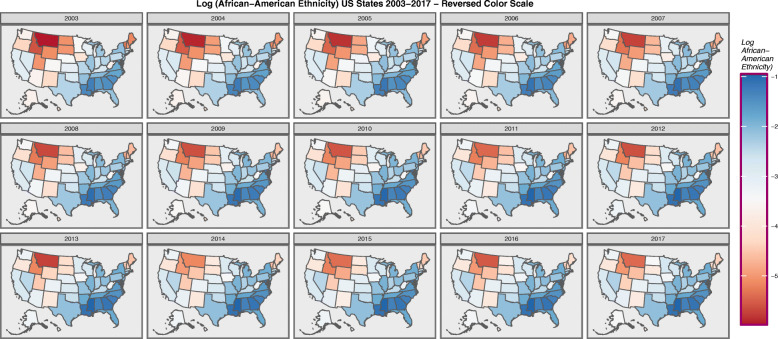
When the colour coding for Fig. 3 is reversed, as shown in Fig. 4, the map resembles Fig. 1 of the TCRs rather closely. This is consistent with the above described preponderance of cases in the Caucasian-American community. One presumes that in these maps ethnicity is acting as a surrogate marker for TC incidence.
Fig. 4.
Choropleth map of log(Caucasian-American Population) across USA by Year – Reversed Colouring. Maps were drawn in ggplot2, sf and RColorBrewer
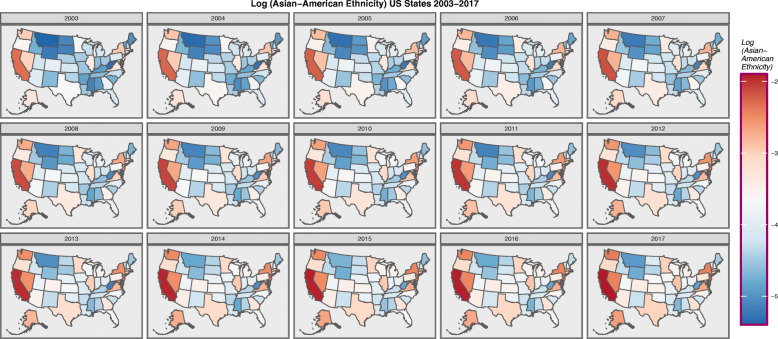
Figure 5 presents the log of the Asian-American ethnicity.
Fig. 5.
Choropleth map of log(Asian-American Population) across USA by Year. Maps were drawn in ggplot2, sf and RColorBrewer
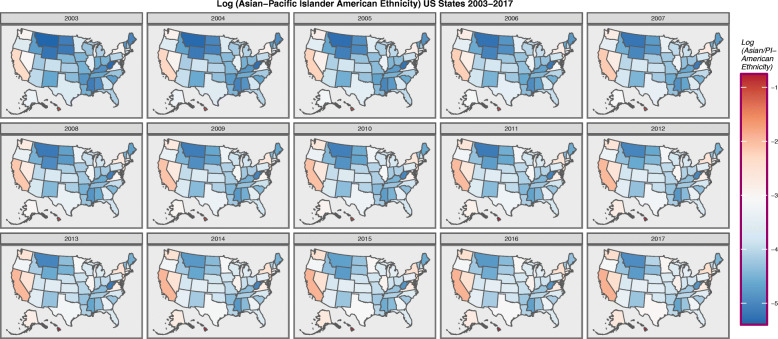
Figure 6 presents the log of the Asian-Pacific Islander -American ethnicity. Hawaii is noted to be a particular hot spot.
Fig. 6.
Choropleth map of log(Asian-Pacific Islander-American Population) across USA by Year. Maps were drawn in ggplot2, sf and RColorBrewer
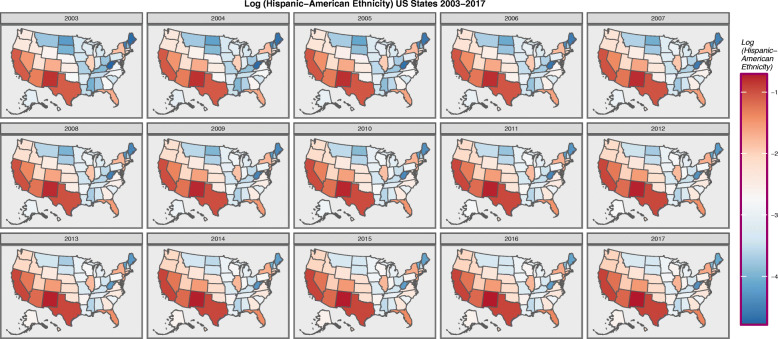
Figure 7 shows the log of the Hispanic-American population density. It seems to be concentrated along the southern border states.
Fig. 7.
Choropleth map of log(Hispanic-American Population) across USA by Year. Maps were drawn in ggplot2, sf and RColorBrewer
Bivariate Relationships and Ethnic Differentials.
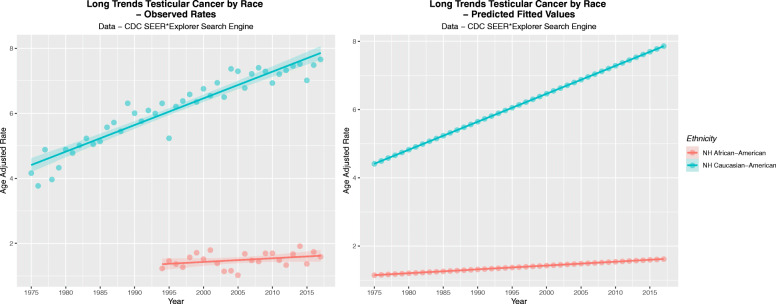
Figure 8A shows the observed age-adjusted TCRs 15–60 years for the Caucasian-American and African-American populations. Figure 8B shows the linear projections of this data.
Fig. 8.
(A) Observed and (B) Predicted Incidence rates of testicular cancer for two ethnicities
Table 1 sets out the observed and predicted values from which these graphs were drawn.
Table 1.
Long Term Testicular Cancer Rates by Ethnicity
| Year | Caucasian-American - Observed | African-American - Observed | NH Caucasian-American Predicted | NH African-American Predicted |
|---|---|---|---|---|
| 1975 | 4.1621 | 4.4103 | 1.1457 | |
| 1976 | 3.7714 | 4.4924 | 1.1570 | |
| 1977 | 4.879 | 4.5744 | 1.1682 | |
| 1978 | 3.9667 | 4.6565 | 1.1795 | |
| 1979 | 4.3295 | 4.7385 | 1.1908 | |
| 1980 | 4.8836 | 4.8205 | 1.2020 | |
| 1981 | 4.7748 | 4.9026 | 1.2133 | |
| 1982 | 5.0104 | 4.9846 | 1.2245 | |
| 1983 | 5.2268 | 5.0667 | 1.2358 | |
| 1984 | 5.0458 | 5.1487 | 1.2470 | |
| 1985 | 5.1316 | 5.2307 | 1.2583 | |
| 1986 | 5.567 | 5.3128 | 1.2696 | |
| 1987 | 5.7151 | 5.3948 | 1.2808 | |
| 1988 | 5.444 | 5.4769 | 1.2921 | |
| 1989 | 6.306 | 5.5589 | 1.3033 | |
| 1990 | 6.0005 | 5.6410 | 1.3146 | |
| 1991 | 5.7543 | 5.7230 | 1.3258 | |
| 1992 | 6.0864 | 5.8050 | 1.3371 | |
| 1993 | 5.9933 | 5.8871 | 1.3484 | |
| 1994 | 6.305 | 1.2295 | 5.9691 | 1.3596 |
| 1995 | 5.2299 | 1.4628 | 6.0512 | 1.3709 |
| 1996 | 6.2072 | 1.3649 | 6.1332 | 1.3821 |
| 1997 | 6.3764 | 1.267 | 6.2152 | 1.3934 |
| 1998 | 6.5804 | 1.573 | 6.2973 | 1.4046 |
| 1999 | 6.3472 | 1.7145 | 6.3793 | 1.4159 |
| 2000 | 6.7535 | 1.5164 | 6.4614 | 1.4272 |
| 2001 | 6.5388 | 1.7943 | 6.5434 | 1.4384 |
| 2002 | 6.9373 | 1.3865 | 6.6254 | 1.4497 |
| 2003 | 6.4961 | 1.1416 | 6.7075 | 1.4609 |
| 2004 | 7.3667 | 1.16 | 6.7895 | 1.4722 |
| 2005 | 7.2916 | 1.0217 | 6.8716 | 1.4834 |
| 2006 | 6.7774 | 1.6811 | 6.9536 | 1.4947 |
| 2007 | 7.2126 | 1.4774 | 7.0357 | 1.5060 |
| 2008 | 7.399 | 1.4451 | 7.1177 | 1.5172 |
| 2009 | 7.2871 | 1.6959 | 7.1997 | 1.5285 |
| 2010 | 6.9299 | 1.6984 | 7.2818 | 1.5397 |
| 2011 | 7.2031 | 1.4834 | 7.3638 | 1.5510 |
| 2012 | 7.3214 | 1.3336 | 7.4459 | 1.5622 |
| 2013 | 7.4551 | 1.6739 | 7.5279 | 1.5735 |
| 2014 | 7.5153 | 1.9154 | 7.6099 | 1.5848 |
| 2015 | 7.0112 | 1.3706 | 7.6920 | 1.5960 |
| 2016 | 7.4819 | 1.7396 | 7.7740 | 1.6073 |
| 2017 | 7.6552 | 1.5911 | 7.8561 | 1.6185 |
Table 2 shows the relative long term rises for all ages by ethnicity and the applicable periods for which they have been monitored. Only nine cancer registries contribute to long term cancer data in the SEER database. More recent data 2001–2017 is contributed by 21 registries.
Table 2.
Long Term Testicular Cancer Incidence Trends by Ethnicity
| Ethnicity | Start Year | End Year | Initial Rate | End Rate | Change | Rise |
|---|---|---|---|---|---|---|
| Observed Rates | ||||||
| NH_Caucasian-American | 2001 | 2017 | 5.0632 | 5.0834 | 0.0203 | 0.40% |
| Hispanic-American | 2003 | 2016 | 3.3823 | 4.6036 | 1.2213 | 36.11% |
| NH_African-American | 2002 | 2017 | 1.1420 | 1.1571 | 0.0151 | 1.32% |
| Asian-Pacific Islander-American | 2003 | 2017 | 0.9368 | 1.9098 | 0.9730 | 103.87% |
| Modelled Rates | ||||||
| NH_African-American | 1975 | 2017 | 1.1457 | 1.6185 | 0.4728 | 41.27% |
| NH_Caucasian-American | 1975 | 2017 | 4.4103 | 7.8561 | 3.4457 | 78.13% |
| Relative Elevation of Rates | 3.8495 | 4.8539 | 7.2879 | 1.8932 | ||
Table 2 is notable for marked ethnic disparities in the rate of rise of the TCR. For example across the period cited the Non-Hispanic Caucasian American population TCR grew only 0.4% compared to the growth in the Hispanic-American TCR of 36.11% and in the Asian-Pacific Islander -American population where it grew 103.87% albeit from a lower starting point. This may imply differing exposures to some environmental intoxicant.
The long term data appears at the bottom of this table. One notes that the growth of the TCR in the African-American community of 41.27% was only 52.82% of the growth of the Caucasian-American community which was 78.13%.
Figure 9 sets out these trends by ethnicity across all ages.
Fig. 9.
Testicular cancer rates by selected ethnicities
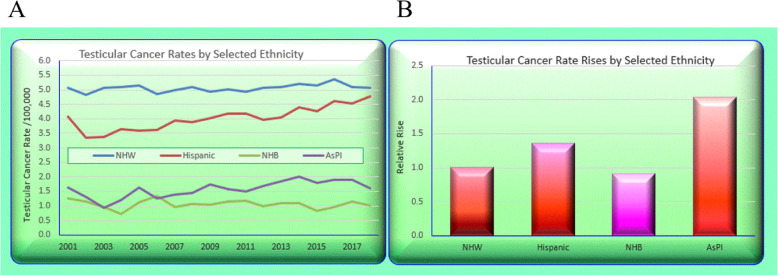
Figure 10A again sets out the ethnic trends and the bar chart in Panel B shows the relative rises in each of the four groups.
Fig. 10.
Testicular cancer rates derived from state data by selected ethnicity. (A) Over time. (B) Rises across periods
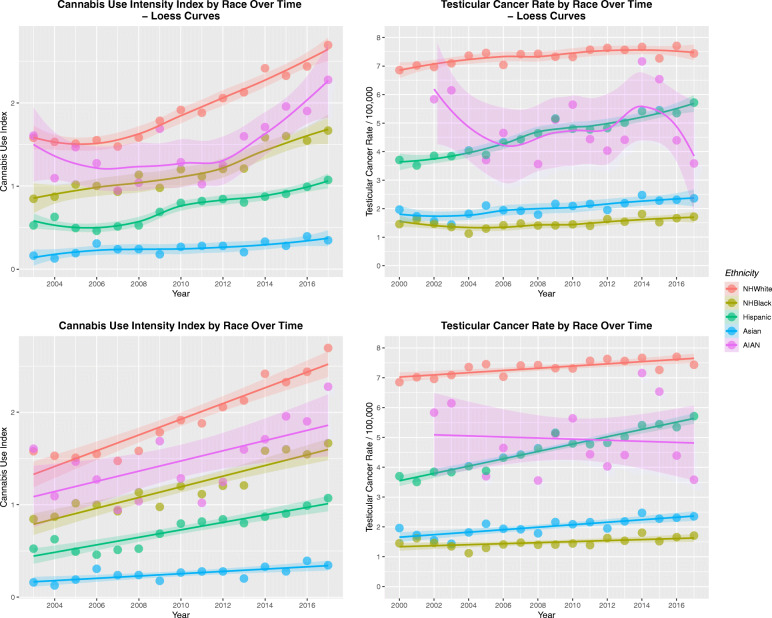
Figure 11 shows four scatterplots of the cannabis use intensity index by ethnicity (Panels A and C) and the TCRs by ethnicity (Panels B and D) as loess curves (A, B) and regression lines of best fit (C, D). One notes a very striking resemblance between the two sets of graphs.
Fig. 11.
Cannabis use and testicular cancer rates by time and by aggregate time. (A and C) cannabis use, (B and D) testicular cancer rates. (A and B) loess curves of best fit. (C and D) Least squares regression lines
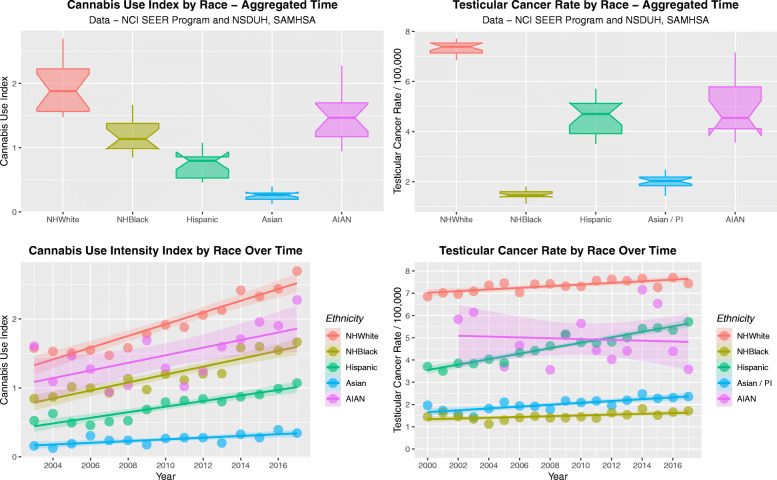
Figure 12 sets out these data as boxplots in panels A and B and time-dependent regression lines in panels C and D. One reads the boxplot graphs by noting that groups which do not have overlapping notches are statistically significantly different from each other. The broad parallels between the two sets of plots is again apparent.
Fig. 12.
Cannabis use and testicular cancer rates by time and by aggregate time. (A and C) cannabis use, (B and D) testicular cancer rates. (A and B) Boxplots; (C and D) Regression lines
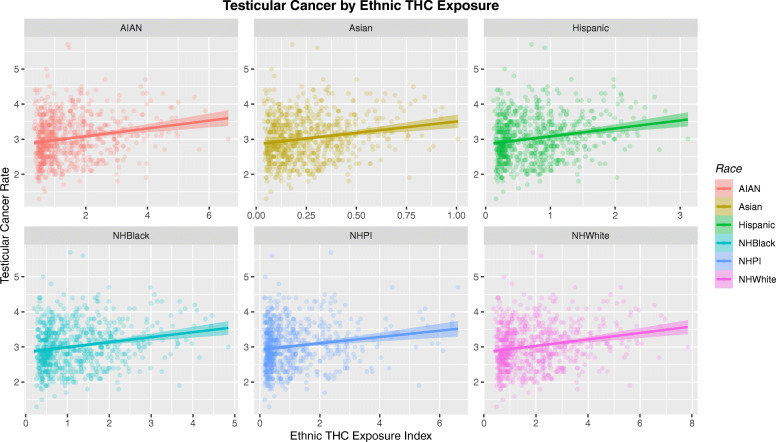
Figure 13 sets out the relationship of the TCR to the ethnic THC exposure index by ethnicity. Whilst each of these six plots look similar careful comparison shows that the scales on the horizontal axis are very different.
Fig. 13.
Testicular cancer rates by ethnic exposure to THC. Note variable scales on horizontal-axis
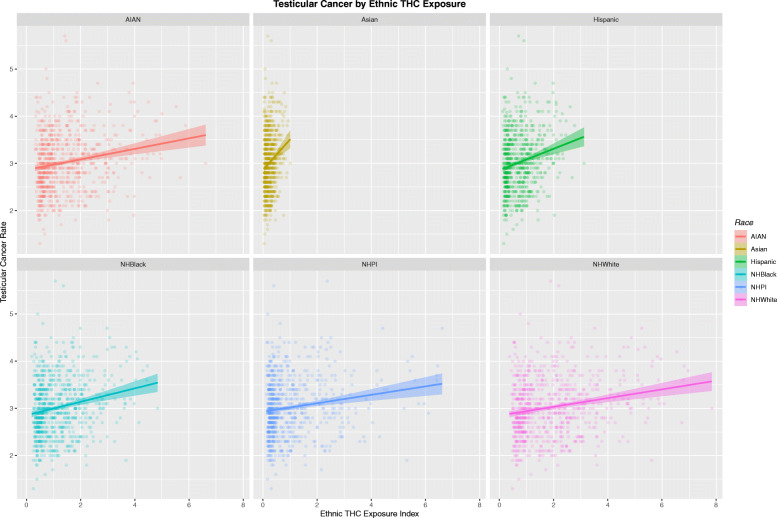
These data are therefore re-plotted with comparable axis scales in Fig. 14. Now two striking trends appear. Firstly the much higher THC exposure in the Caucasian-American and AIAN-American groups is very apparent. Also the slope of the regression lines in each case is very different. Hence the regression line for the Asian-American group is very short in horizontal scope, but very steep. This graph is very thought-provoking and has far reaching implications indeed. These differences clearly indicate major impacts of cannabis exposure by ethnic background and may potentially be related to one or several genomic or epigenomic processes acting in concert.
Fig. 14.
Testicular cancer rates by ethnic exposure to THC. Note fixed scales on horizontal-axis and variable slope of regression lines
Although this paper focusses on the ethnic disparities it should be pointed out that each ethnicity is not one homogenous block. The heatmap in Fig. 15 shows the log (TCR) in the Caucasian-American community and notes an obvious hotspot in Hawaii which has rates in each of the nominated years of 9.4, 11.2, 12.7, 8.8, 10.4 and 9.4 (10.32 + 0.59 mean + S.E.M.) which is much higher than the comparable rates for the Caucasian-American ethnicity in all other states which is 5.351 + 0.033 (t = 115.12, df = 738.37, P = 0.0000; for comparison t = 67, df = 738.37, P = 2.44 × 10− 316).
Fig. 15.
Heatmap of Caucasian-American testicular cancer incidence rates by state and year. Note very high rate for Hawaii near the top
We now turn to the formal analysis of the state-based data for ages 15–60 years derived from the most recent datafiles held in the National Program of Cancer Registries and the SEER Incidence – US Cancer Statistics Public Use Database 2019 submission 2001–2017 which includes data for the whole country.
Inverse probability weights may be calculated for this data which control for the effect of cannabis exposure as a function of the other substance exposures.
Table 3 presents the results of linear regressions of the TCR against ethnicity and the ethnic THC exposure in both wide and long datasets. In the latter all the ethnicities are collated into one column and the data-table becomes longer by a factor of the number of ethnicities.
Table 3.
Introductory Linear Regressions
| Parameter Estimates | Model Parameters | ||||||
|---|---|---|---|---|---|---|---|
| Parameter | Estimate (C.I.) | P-Value | S.D. | R-Squared | F | dF | P |
| Race | |||||||
| (lm(Rate ~ Race) | |||||||
| Caucasian-American | 4.6441 (4.2, 5.08) | 8.30E-31 | 0.8828 | 0.8657 | 426.3 | 1,65 | 2.98E-30 |
| (lm(Rate ~ Year * Race) | |||||||
| Caucasian-American | − 137 (− 175.61, − 98.39) | 2.47E-09 | 0.3069 | 0.9838 | 1334 | 3,63 | 6.27E-57 |
| Year: Caucasian-American | 0.08 (0.07, 0.09) | < 2.2E-16 | |||||
| lm(TC_Rate ~ EthnTHCExp * Race) | |||||||
| log(EthnTHCExp) | 0.06 (0.04, 0.08) | 1.17E-07 | 0.2017 | 0.0293 | 13.34 | 11,4488 | < 2E-16 |
| RaceAsian | 0.09 (0.05, 0.12) | 1.46E-05 | |||||
| RaceHispanic | 0.04 (0.01, 0.06) | 0.0021 | |||||
| RaceNHPI | 0.02 (0, 0.04) | 0.0456 | |||||
| In Ethnic Dataset, Long | |||||||
| (lm(Rate ~ Cannabis) | |||||||
| Cannabis | 0.47 (0.41, 0.52) | 1.70E-69 | 0.5922 | 0.0663 | 320.2 | 14,498 | 3.10E-69 |
| lm(Rate ~ Time * Cannabis) | |||||||
| Cannabis | 0.49 (0.44, 0.55) | 9.89E-64 | 0.5919 | 0.0671 | 162.6 | 24,497 | 1.31E-36 |
| Year | −0.0049 (− 0.0093, − 0.0004) | 0.03169 | |||||
In each case ethnicity and the ethnic THC exposure is noted to be a highly significant covariate of TCR.
Multivariable adjustment
Table 4 presents the results of inverse probability weighted mixed effects regression all from the long dataset. Many terms involving ethnicity and ethnic THC exposure are highly significant. In an additive model with the other four drugs ethnic THC exposure is significant (β-estimate = 0.05 (0.04, 0.06), P = 5.80 × 10− 31). In a four-way interactive model with other drugs ethnic cannabis exposure is significant (β-estimate = 0.64 (0.17, 1.10), P = 0.0072).
Table 4.
Mixed Effects Model Regressions
| Parameters | Model Parameters | |||||
|---|---|---|---|---|---|---|
| Parameter | Estimate (C.I.) | P-Value | SD | AIC | BIC | logLik |
| lme(TestCa ~ Race) | ||||||
| Asian-Am. | − 0.2 (− 0.39, − 0.02) | 0.0320 | 1.141 | 3566.977 | 3595.930 | − 1777.489 |
| Hispanic-Am. | 2.82 (2.65, 3) | 2.5E-150 | ||||
| NHCaucasian-Am. | 4.17 (4, 4.34) | 1.3E-246 | ||||
| lme(TestCa ~ Ethnic_THC_Exposure) | ||||||
| Ethnic_THC_Exp | 0.05 (0.04, 0.06) | 4.40E-31 | 0.202 | − 1595.682 | −1570.036 | 801.841 |
| lme(TestCa ~ Race + Ethnic_THC_Exposure) | ||||||
| NHCaucasian-Am. | 4.06 (3.88, 4.23) | 1.13E-234 | 1.126 | 3546.406 | 3580.177 | − 1766.203 |
| Hispanic-Am. | 2.93 (2.76, 3.11) | 4.23E-155 | ||||
| Ethnic_THC_Exp | 0.24 (0.15, 0.32) | 2.13E-07 | ||||
| lme(TestCa ~ Race * Ethnic_THC_Exposure) | ||||||
| NHCaucasian-Am. | 4.12 (3.95, 4.3) | 9.4E-236 | 1.115 | 3539.109 | 3587.320 | − 1759.554 |
| Hispanic-Am. | 3.17 (2.96, 3.37) | 1.5E-141 | ||||
| Hispanic-Am.: Ethnic_THC_Exp | 0.58 (0.4, 0.77) | 6.6E-10 | ||||
| NHCaucasian-Am.: Ethnic_THC_Exp | 0.16 (0.05, 0.27) | 0.0044 | ||||
| Asian-Am.: Ethnic_THC_Exp | 0.15 (−0.12, 0.43) | 0.2753 | ||||
| Asian-Am. | 0.06 (− 0.42, 0.54) | 0.8075 | ||||
| NHAfrican-Am.: Ethnic_THC_Exp | −0.05 (− 0.35, 0.25) | 0.7381 | ||||
| lme(TestCa ~ Year * Race) | ||||||
| Year: Hispanic-Am. | 0.2 (0.17, 0.22) | 2.9E-38 | 1.036 | 3427.797 | 3476.008 | − 1703.898 |
| Year: NHCaucasian-Am. | 0.02 (0.01, 0.04) | 0.0032 | ||||
| Hispanic-Am. | −396.69 (− 504.97, −288.41) | 1.5E-12 | ||||
| lme(TestCa ~ Year * Ethnic_THC_Exposure) | ||||||
| Ethnic_THC_Exp | 1.39 (1.25, 1.53) | 1.51E-72 | 2.110 | 4659.713 | 4683.846 | − 2324.857 |
| Year | −0.14 (−0.18, − 0.11) | 2.21E-17 | ||||
| Additive Model | ||||||
| lme(Rate ~ Cigarettes + AUD + Ethnic_THC_Exp + Analgesics + Cocaine) | ||||||
| AUD | 5.49 (5.11, 5.87) | 6.57E-162 | 0.180 | − 2610.768 | − 2559.484 | 1313.384 |
| Ethnic_THC_Exp | 0.05 (0.04, 0.06) | 5.80E-31 | ||||
| Cocaine | −2.46 (−3.3, −1.62) | 1.17E-08 | ||||
| Analgesics | −0.13 (− 0.16, − 0.1) | 3.09E-21 | ||||
| Cigarettes | − 1.12 (− 1.27, − 0.97) | 7.42E-49 | ||||
| Interactive Model - 3-Way | ||||||
| lme(Rate ~ Cigarettes * AUD * Ethnic_THC_Exp + Analgesics + Cocaine) | ||||||
| Cigarettes: AUD | 23.11 (21.54, 24.67) | 2.14E-169 | 0.179 | − 2675.013 | − 2617.321 | 1346.507 |
| Cigarettes: Ethnic_THC_Exp | 0.44 (0.31, 0.57) | 5.22E-11 | ||||
| Ethnic_THC_Exp | −0.05 (− 0.08, − 0.01) | 3.80E-03 | ||||
| Cocaine | −1.96 (−2.8, − 1.11) | 6.09E-06 | ||||
| Analgesics | − 0.13 (− 0.16, − 0.11) | 2.56E-22 | ||||
| Cigarettes | − 2.4 (− 2.59, − 2.2) | 2.39E-124 | ||||
| Interactive Model - 4-Way | ||||||
| lme(Rate ~ Cigarettes * AUD * Ethnic_THC_Exp * Analgesics + Cocaine) | ||||||
| Cigarettes: AUD | 23.48 (21.9, 25.05) | 6.73E-172 | 0.177 | − 2725.041 | − 2648.125 | 1374.520 |
| Analgesics | 0.31 (0.15, 0.48) | 0.0002 | ||||
| Ethnic_THC_Exp: Analgesics | 0.22 (0.07, 0.37) | 0.0036 | ||||
| Ethnic_THC_Exp | 0.64 (0.17, 1.1) | 0.0072 | ||||
| Cigarettes: Ethnic_THC_Exp | −3.8 (−5.8, −1.79) | 0.0002 | ||||
| Cigarettes: Ethnic_THC_Exp: Analgesics | −1.38 (−2.03, −0.73) | 3.19E-05 | ||||
| Cocaine | −1.87 (−2.71, − 1.02) | 1.62E-05 | ||||
| Cigarettes: Analgesics | −2.21 (− 2.96, − 1.47) | 6.05E-09 | ||||
| Cigarettes | −9.3 (− 11.61, −6.99) | 3.98E-15 | ||||
Table 5 presents the results of an inverse probability weighted two-step instrumental variable regression model on the long dataset. The table is very interesting. When race alone is considered in the first model with African-American race as the comparator, no significant changes are seen. However when ethnic THC exposure is considered it is very highly significant. When race and ethnic THC exposure are considered in an additive model all parameters are significant and ethnic THC exposure is the most significant.
Table 5.
Mixed Effects Model Regressions
| Parameters | Model Parameters | |||||||
|---|---|---|---|---|---|---|---|---|
| Instrumental Variable | Parameter | Estimate (C.I.) | P-Value | S.D. | R-Squared | Wald ChiSqu. | dF | P |
| ivreg(TestCa ~ Race) | ||||||||
| Race_Asian | 0.26 (−25.24, 25.77) | 0.984 | 132.073 | 0.8888 | 2463 | 3921 | < 2.2E-16 | |
| Race_Hispanic | 2.7 (−13.73, 19.12) | 0.748 | ||||||
| Race_NHWhite | 3.5 (−12.93, 19.92) | 0.677 | ||||||
| ivreg(TestCa ~ Ethnic_THC_Exposure) | ||||||||
| Ethnic_THC_Exp | 0.89 (0.88, 0.91) | << 2.3E-320 | 102.3782 | 0.9332 | 1.29E+ 04 | 1923 | << 2.3E-320 | |
| ivreg(Rate ~ Ethn_THC_Exposure + Race) | ||||||||
| Ethn_THC_Exposure: NHCaucasian-Am._THC_Exposure | 0.08 (0.02, 0.13) | 0.0095 | 8.4472 | 0.0361 | 29.06 | 64,493 | << 1.0E-320 | |
| Ethn_THC_Exposure: NHAfrican-Am._THC_Exposure | −0.09 (−0.14, −0.03) | 0.0032 | ||||||
| Ethn_THC_Exposure: NHPI_THC_Exposure | −0.19 (− 0.25, − 0.13) | 3.4E-09 | ||||||
| Ethn_THC_Exposure: Hispanic-Am._THC_Exposure | −0.25 (− 0.32, − 0.18) | 4.2E-13 | ||||||
| Ethn_THC_Exposure: Asian-Am._THC_Exposure | − 0.63 (− 0.74, − 0.52) | 3.6E-29 | ||||||
| Ethnic_THC_Exp | − 0.35 (− 0.4, − 0.3) | 1.5E-38 | ||||||
| ivreg(TestCaRt ~ log(EthnTHCExp) * Race) | ||||||||
| NHWhite: Ethn_THC_Exp | 1.22 (1.16, 1.27) | 6.9E−128 | 95.9894 | 0.9430 | 3703 | 4920 | < 2e-16 | |
| Hispanic: Ethn_THC_Exp | 1.48 (1.38, 1.59) | 5.6E-214 | ||||||
| ivreg(TestCaRt ~ log(EthnTHCExp) * Race) | ||||||||
| THC | RaceAsian | 3.88E+ 05 (−8.93E+ 07, 9.01E+ 07) | 0.993 | 130,195 | -1.1E-05 | 9.013E+ 05 | 3921 | 1.0000 |
| Cannabigerol | RaceHispanic | 1.47E+ 05 (−3.38E+ 07, 3.40E+ 07) | 0.993 | |||||
| Cannabinol | RaceNHWhite | 1.47E+ 05 (− 3.38E+ 07, 3.41E+ 07) | 0.993 | |||||
| ivreg(TestCa ~ Cigarettes * AUD * Ethn_THC_Exposure + Race + Cocaine + MHY + Analgesics) | ||||||||
| AUD | 138.37 (126.96, 149.78) | < 2e-16 | 24.2846 | 0.9962 | 2.04E+ 04 | 12,912 | < 2.2E-16 | |
| AUD: Ethnic_THC_Exp | 312.53 (290.54, 334.53) | < 2e-16 | ||||||
| Cigarettes: Ethnic_THC_Exp | 150.41 (139.67, 161.15) | < 2e-16 | ||||||
| Analgesics | 1.07 (0.67, 1.47) | 1.8E-07 | ||||||
| NHCaucasian-Am. | 3.25 (0.22, 6.28) | 0.0361 | ||||||
| Hispanic-Am. | 2.91 (−0.12, 5.93) | 0.0597 | ||||||
| Cocaine | − 132.56 (−150.11, −115.01) | < 2e-16 | ||||||
| Cigarettes | −45.52 (−50.67, −40.37) | < 2e-16 | ||||||
| Ethnic_THC_Exp | −22.08 (−24, −20.17) | < 2e-16 | ||||||
| MHY | −12.03 (− 12.98, − 11.08) | < 2e-16 | ||||||
| Cigarettes: AUD: Ethnic_THC_Exp | − 2032.86 (− 2169.16, − 1896.55) | < 2e-16 | ||||||
| ivreg(TestCa ~ Cigarettes * AUD * Ethnic_THC_Exp * Race + Cocaine + MHY + Analgesics) | ||||||||
| THC | Cigarettes | 30.94 (21.36, 40.52) | 3.9E-10 | 16.2755 | 0.0138 | 20.04 | 3921 | 1.37E-12 |
| Cannabigerol | Ethnic_THC_Exp | 0.55 (0.05, 1.05) | 0.0298 | |||||
| Cannabinol | AUD | −49.84 (−64.89, −34.78) | 1.4E-10 | |||||
| ivreg(Rate ~ Cigarettes * AUD + Analgesics + Cocaine + MHY + Race * Ethnic_THC_Exposure) | ||||||||
| Cocaine | 61.73 (56.9, 66.56) | 6.1E-130 | 6.8060 | 0.3801 | 173.4 | 16,4483 | << 1.0E-320 | |
| Cigarettes: AUD | 142.1 (126.47, 157.73) | 5.2E-69 | ||||||
| NHPI | 0.52 (0.34, 0.7) | 1.8E-08 | ||||||
| Ethnic_THC_Exp | 0.32 (0.2, 0.43) | 4.7E-08 | ||||||
| Asian-Am. | 0.44 (0.25, 0.63) | 6.8E-06 | ||||||
| Hispanic-Am. | 0.24 (0.06, 0.42) | 0.00752 | ||||||
| Asian-Am.: Ethnic_THC_Exp | −0.41 (−0.56, −0.26) | 1.68E-07 | ||||||
| NHPI: Ethnic_THC_Exp | −0.37 (− 0.5, − 0.24) | 2.96E-08 | ||||||
| Analgesics | −1.13 (−1.29, − 0.97) | 7.65E-44 | ||||||
| Cigarettes | −38.49 (−42.35, −34.63) | 3.90E-82 | ||||||
| MHY | −9.58 (−10.06, −9.1) | 4.1E-289 | ||||||
When Race and ethnic THC exposure are considered in an interactive model Caucasian-American and Hispanic-American races both in interaction with ethnic THC exposure are highly significant (β-estimate = 1.48 (1.38, 1.59), P = 5.62 × 10− 214 and β-estimate = 1.22 (1.16, 1.27), P = 65.86 × 10− 128 respectively).
However, when the same regression is performed with THC, cannabigerol and cannabinol as instrumental variables significance amongst both the terms and the model is lost and adjusted R-squared falls from 0.9430 to − 0.0000108.
Similarly when all substances and income are considered along with ethnic THC exposure, two interactive terms including ethnic THC exposure are positive and highly significant.
However when THC, cannabigerol and cannabinol are included as instrumental variables again there is a marked collapse of significant findings, adjusted R-squared falls from 0.9962 (which is very high) to 0.0138 (quite low) and the model Wald coefficient falls from 20,000 to 20.04.
In the final model in this table using a different interaction structure the ethnic THC exposure is again independently highly statistically significant (β-estimate = 0.32 (0.20, 0.43), P = 4.7 × 10− 8).
Table 6 presents the results of inverse probability weighted robust regressions in the long dataset. Ethnic THC exposure is clearly highly significantly prominent. As model complexity increases so the significance of terms including the cannabinoids increases.
Table 6.
Robust Inverse Probability Weighted Regression
| Parameter | Estimate (C.I.) | P-Value |
|---|---|---|
| Additive Model with State Cannabis | ||
| svyglm(TestCaRt ~ Cigarettes + Cannabis + + AUD + Analgesics + Cocaine) | ||
| AUD | 64.55 (56.89, 72.22) | < 2.2E-16 |
| Analgesics | 11.01 (6.24, 15.77) | 5.7E-05 |
| NHWhite | 5.44 (2.36, 8.53) | 0.0013 |
| Hispanic | 4.63 (1.54, 7.71) | 0.0055 |
| Cannabis | −9.38 (−12.76, −5.99) | 3.4E-06 |
| Cocaine | −131.14 (− 138.96, − 123.33) | < 2.2E-16 |
| Additive Model with Ethnic THC Exposure | ||
| svyglm(TestCaRt ~ Cigarettes * Ethn_THC_Exposure * + AUD + Analgesics + Cocaine) | ||
| Cigarettes | 28.96 (27.87, 30.05) | < 2E-16 |
| Hispanic | 2.55 (2.05, 3.04) | 1.8E-12 |
| Asian | 4.89 (1.08, 8.69) | 0.0160 |
| Ethn_THC_Exposure | 2.9 (0.41, 5.38) | 0.0281 |
| Cocaine | −56.03 (−110.14, − 1.93) | 0.0492 |
| Interactive Model with Ethnic_THC_Exposure | ||
| svyglm(TestCaRt ~ Cigarettes * Ethn_THC_Exposure * + AUD + Analgesics + Cocaine + Race + Income) | ||
| Ethn_THC_Exposure | 4.72 (2.04, 7.41) | 0.0018 |
| Asian | 3.93 (1.64, 6.22) | 0.0022 |
| Cigarettes | 11.09 (3.77, 18.42) | 0.0060 |
| Cigarettes: NHWhite | 109.87 (24.24, 195.5) | 0.0177 |
| Cigarettes: Ethn_THC_Exposure: Asian | 17.32 (1.64, 33.01) | 0.0388 |
| NHWhite | −19.53 (−39.11, 0.05) | 0.0603 |
| Ethn_THC_Exposure: Asian | −4.45 (−8.41, −0.5) | 0.0352 |
| Cocaine | −88.11 (−161.51, − 14.7) | 0.0257 |
| Cigarettes: Ethn_THC_Exposure | −20.11 (− 32.44, −7.78) | 0.0034 |
| Cigarettes: Asian | −16.35 (−26.11, −6.6) | 0.0027 |
| Interactive Model with State Cannabis | ||
| svyglm(TestCaRt ~ Cigarettes * Cannabis * + AUD + Analgesics + Cocaine + Race + Income) | ||
| AUD | 13.58 (7.69, 19.47) | 0.0001 |
| Cigarettes: Cannabis: NHWhite | 398.68 (214.58, 582.79) | 0.0002 |
| Cigarettes: NHWhite | 933.24 (493.05, 1373.44) | 0.0003 |
| Cigarettes: Hispanic | 1372.45 (672.78, 2072.11) | 0.0007 |
| Cigarettes: Cannabis: Hispanic | 635.8 (305.36, 966.24) | 0.0008 |
| Analgesics | 19.88 (9.19, 30.57) | 0.0011 |
| Cannabis | 42.63 (18.65, 66.61) | 0.0017 |
| Cigarettes: Cannabis | −199.68 (− 315.91, −83.44) | 0.0023 |
| Cigarettes | − 578.67 (− 907.11, − 250.23) | 0.0018 |
| Cocaine | −75.51 (− 117.23, −33.78) | 0.0015 |
| Cannabis: Hispanic | − 144.06 (− 218.53, −69.59) | 0.0008 |
| Hispanic | − 308.36 (− 466.63, − 150.09) | 0.0007 |
| NHWhite | −214 (− 315.14, − 112.87) | 0.0003 |
| Cannabis: NHWhite | −93.06 (− 135.37, −50.75) | 0.0002 |
| Interactive Model with State Cannabinoids | ||
| svyglm(TestCaRt ~ Cigarettes * THC * Cannabigerol * + AUD + Analgesics + Cocaine + Race + Income) | ||
| Cigarettes: NHWhite | 127.24 (92.37, 162.1) | 4.7E-07 |
| Cigarettes: THC: Cannabigerol: NHBlack | 13.87 (6.33, 21.41) | 0.0017 |
| Cigarettes: NHBlack | 53.77 (18.14, 89.41) | 0.0075 |
| Cigarettes: THC: NHBlack | 49.95 (14.93, 84.96) | 0.0108 |
| Cigarettes: Cannabigerol: NHBlack | 14.62 (3.67, 25.57) | 0.0161 |
| Cigarettes: THC: Cannabigerol: NHWhite | 35.97 (7.97, 63.97) | 0.0200 |
| Cigarettes: THC: Hispanic | 214.68 (31.83, 397.53) | 0.0317 |
| Cigarettes: Hispanic | −310.38 (− 540.45, −80.31) | 0.0152 |
| Cigarettes: Cannabigerol: Hispanic | −89.37 (−155.46, −23.28) | 0.0150 |
| AUD | −43.07 (−68.76, −17.38) | 0.0035 |
| THC: Cannabigerol: NHWhite | −8.18 (−11.39, − 4.96) | 6.2E-05 |
| NHWhite | −16.98 (−23.16, − 10.81) | 2.4E-05 |
Multiple imputation of missing ethnicity data
It would be useful to study these ethnic effects in further detail. However if one simply considers the Caucasian-American and African-American datasets it is noted that 1782 of 2700 datapoints are absent, or 66.0%. This is a severe limitation on further detailed analysis.
For this reason formal data imputation by multiple imputation of chained equations was performed using the R-package mice [38].
Table 7 shows the impact of missing data by ethnicity. Mean and median data and missing data rates are indicated.
Table 7.
Missing Data Considerations
| Ethnicity | Mean Rate Overall | Median Rate Overall | Mean Rate from Population Totals | Mean Rate from Population Rates |
|---|---|---|---|---|
| NHWhite | 5.37 | 5.40 | 5.39 | 5.39 |
| NHBlack | 1.09 | 1.00 | 1.23 | 1.24 |
| Hispanic | 3.31 | 3.35 | 3.63 | 3.70 |
| Asian-PI | 1.26 | 1.20 | 1.92 | 2.03 |
| Ethnicity | Observations | Total | Missing Values | % Missing |
| NHWhite | 644 | 675 | 31 | 4.59% |
| NHBlack | 63 | 675 | 612 | 90.67% |
| Hispanic | 177 | 675 | 498 | 73.78% |
| Asian-PI | 34 | 675 | 641 | 94.96% |
Following [40] 256 imputations were performed with 20 iterations each. Figure 16 shows the density plot of the multiply imputed data. One notes the obvious peaks in the lower areas corresponding to the smaller values of the ethnic minorities.
Fig. 16.
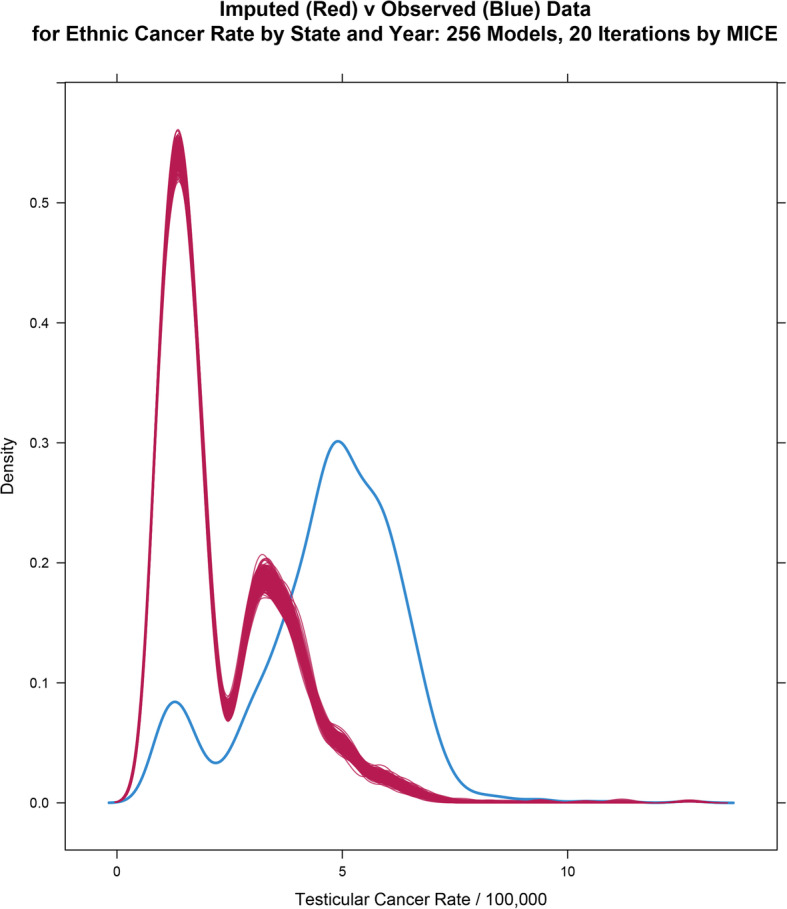
Density plot for imputed values for ethnicity data. Note lower peaks for ethnic minority rates
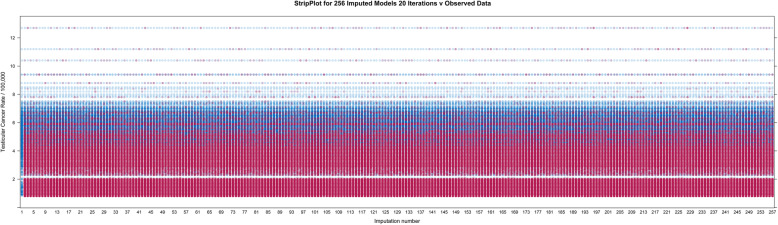
Figure 17 is a strip plot illustrating how the imputed values nicely follow the values of the known observed data including their distribution pattern.
Fig. 17.
Stripplot for multiply imputed data. Note that the imputed values are all taken from the values of the available data
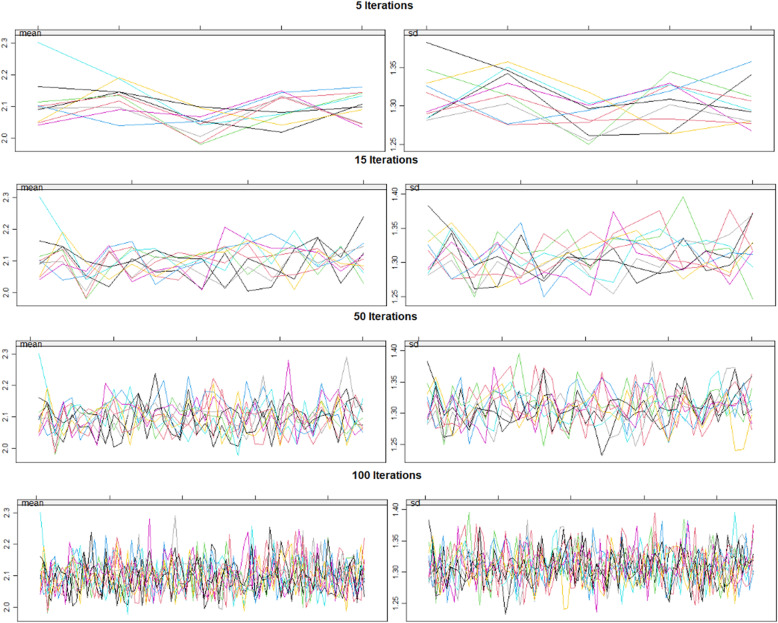
Figure 18 demonstrates the manner in which the first 100 imputations nicely converge.
Fig. 18.
Imputation convergence. Note increasing convergence of data with increasing iterations of imputation algorithm
Multiple imputation in this manner allows the calculation of various regression equations from the data with pooling of the final models into a meaningful outcome by applying Rubin’s rules for pooling of such chained models.
Table 8 shows the result of various linear model formulae performed in this way on the imputed long dataset. Ethnic cannabis exposure is noted to continue to be highly significant. Quintile effects are also demonstrated.
Table 8.
Linear Regressions on Multiply Imputed Data
| Parameters | Model | |||||
|---|---|---|---|---|---|---|
| Parameter | Estimate (C.I.) | P-Value | No. Imputa- tions | S.D. | Lambda | FMI |
| Ethnic_THC_Exposure Alone | ||||||
| lm(TestCaRt ~ Ethnic_THC_Exposure) | ||||||
| Ethnic_THC_Exposure | 0.68 (0.62, 0.74) | 1.80E-92 | 256 | 1.7000 | 0.0292 | 0.0299 |
| Ethnic_THC_Exposure * Race | ||||||
| lm(TestCaRt ~ Ethnic_THC_Exposure + Race) | ||||||
| NHWhite-American | 4.17 (4.08, 4.26) | << 9.8E-324 | 256 | 0.8183 | 0.0560 | 0.0568 |
| Hispanic-American | 2.63 (2.53, 2.73) | << 2.4E-315 | 0.1681 | 0.1690 | ||
| Asian-American | 1.1 (0.98, 1.21) | 2.45E-70 | 0.1018 | 0.1026 | ||
| Ethnic_THC_Exposure | 0.19 (0.14, 0.24) | 7.99E-15 | 0.1521 | 0.1530 | ||
| Additive - Drugs | ||||||
| lm(TestCaRt ~ Cigarettes + AUD + Ethnic_THC_Exposure + Analgesics + Cocaine) | ||||||
| Ethnic_THC_Exposure | 0.72 (0.66, 0.79) | 9.10E-99 | 256 | 1.6893 | 0.0313 | 0.0321 |
| AUD | 12.64 (8.37, 16.91) | 7.28E-09 | 0.0391 | 0.0399 | ||
| Interactive Full Model | ||||||
| lm(TestCaRt ~ Cigarettes + AUD + Ethnic_THC_Exposure + Analgesics + Cocaine + 5_Races + MHY + Cigarettes.AUD + Cigarettes.EthnTHCExpRt + EthnTHCExpRt.AUD) | ||||||
| NHWhite-American | 4.2 (4.11, 4.29) | << 3.1E-322 | 256 | 0.8017 | 0.0636 | 0.0644 |
| Hispanic-American | 2.59 (2.5, 2.69) | << 8.9E-323 | 0.1727 | 0.1737 | ||
| Asian-American | 0.99 (0.87, 1.11) | 1.99E-52 | 0.1129 | 0.1138 | ||
| AUD | 6.45 (4.21, 8.68) | 2.01E-08 | 0.1491 | 0.1500 | ||
| Ethnic_THC_Exposure | 0.12 (0.07, 0.18) | 1.89E-05 | 0.1658 | 0.1667 | ||
| Analgesics | −5.31 (−8.82, −1.8) | 3.04E-03 | 0.1167 | 0.1175 | ||
| Cigarettes | −3.72 (−4.64, −2.8) | 3.55E-15 | 0.1336 | 0.1345 | ||
| Quintiles | ||||||
| lm(TestCaRt ~ Quintile) | ||||||
| Quintile 2 | 0.02 (−0.2, 0.24) | 0.8801 | 256 | 1.8379 | 0.0261 | 0.0268 |
| Quintile 3 | 0.1 (−0.12, 0.32) | 0.3566 | 0.0295 | 0.0302 | ||
| Quintile 4 | 0.2 (−0.02, 0.42) | 0.0755 | 0.0247 | 0.0255 | ||
| Quintile 5 | 0.29 (0.06, 0.51) | 0.0118 | 0.0331 | 0.0338 | ||
| Dichotomized Quintiles | ||||||
| lm(TestCaRt ~ Dichotomized_Quintiles) | ||||||
| Upper_2_Quintiles | 0.2 (0.06, 0.35) | 0.0058 | 256 | 1.8375 | 0.0309 | 0.0316 |
Causal inference
Table 9 sets out the e-Values relating to the various calculations presented above. Minimum e-Values are set out in Table 10 listed in descending order. One notes that 29/37 e-Values are > 1.25 which is said to be the cut-off for causal attribution [41].
Table 9.
e-Values
| Parameter | Estimate (C.I.) | R.R. (C.I.) | E-Values |
|---|---|---|---|
| LINEAR MODELS | |||
| lm(TC_Rate ~ EthnTHCExp * Race) | |||
| Ethnic_THC_Exposure | 0.06 (0.04, 0.08) | 1.31 (1.18, 1.45) | 1.95, 1.66 |
| In Ethnic Dataset, Long | |||
| (lm(Rate ~ Race) | |||
| Cannabis | 0.47 (0.41, 0.52) | 2.04 (1.68, 2.47) | 3.05, 2.76 |
| lm(Rate ~ Time * mrjmon) | |||
| Cannabis | 0.49 (0.44, 0.55) | 2.19 (1.95, 2.32) | 3.68, 3.31 |
| MIXED EFFECTS MODELS | |||
| lme(TestCa ~ Ethnic_THC_Exposure) | |||
| Ethnic_THC_Exp | 0.05 (0.04, 0.06) | 1.23 (1.19, 1.28) | 1.78, 1.68 |
| lme(TestCa ~ Race + Ethnic_THC_Exposure) | |||
| Ethnic_THC_Exp | 0.24 (0.15, 0.32) | 1.21 (1.13, 1.30) | 1.72, 1.50 |
| lme(TestCa ~ Race * Ethnic_THC_Exposure) | |||
| Hispanic-Am.: Ethnic_THC_Exp | 0.58 (0.4, 0.77) | 1.67 (1.27, 2.26) | 2.74, 1.85 |
| lme(TestCa ~ Year * Ethnic_THC_Exposure) | |||
| Ethnic_THC_Exp | 1.39 (1.25, 1.53) | 1.82 (1.72, 1.93) | 3.05, 2.83 |
| Additive Model - Drugs | |||
| Ethnic_THC_Exp | 0.05 (0.04, 0.06) | 1.29 (1.23, 1.34) | 1.89, 1.77 |
| Interactive Model - 3-Way | |||
| Cigarettes: Ethnic_THC_Exp | 0.44 (0.31, 0.57) | 949 (4.86, 18.54) | 18.8, 9.20 |
| Interactive Model - 4-Way | |||
| Ethnic_THC_Exp: Analgesics | 0.22 (0.07, 0.37) | 3.14 (1.45, 6.79) | 5.74, 2.28 |
| Ethnic_THC_Exp | 0.64 (0.17, 1.1) | 26.50 (2.44, 287.17) | 52.52, 4.33 |
| IV-REGRESSION | |||
| ivreg(TestCa ~ Ethnic_THC_Exposure) | |||
| Ethnic_THC_Exposure | 1.03 (0.89, 1.17) | 1.07 (1.06, 1.07) | 1.33, 1.30 |
| ivreg(TestCa ~ Ethnic_THC_Exposure) | |||
| Ethnic_THC_Exposure | 0.89 (0.88, 0.91) | ||
| ivreg(TestCaRt ~ log(EthnTHCExp): Race) | |||
| Ethnic_THC_Exp: Asian-Am. | 2.72 (0.84, 4.61) | 1.21 (1.06, 1.38) | 1.72, 1.31 |
| Ethnic_THC_Exp: NHCaucasian-Am. | 0.44 (0.3, 0.59) | 1.03 (1.02, 1.04) | 1.21, 1.17 |
| ivreg(TestCa ~ Ethnic_THC_Exp * Race) | |||
| NHCaucasian-Am.: Ethnic_THC_Exp | 1.22 (1.16, 1.27) | 1.01 (1.01, 1.02) | 1.12, 1.12 |
| Hispanic-Am.: Ethnic_THC_Exp | 1.48 (1.38, 1.59) | 1.01 (1.01, 1.02) | 1.14, 1.13 |
| ivreg(Rate ~ Cigarettes * AUD + Analgesics + Cocaine + MHY + * Ethnic_THC_Exposure) | |||
| Ethnic_THC_Exp | 0.32 (0.2, 0.43) | 1.04 (1.03, 1.06) | 1.25, 1.19 |
| ivreg(Rate ~ Ethn_THC_Exposure + Race) | |||
| Ethn_THC_ExposureNHCaucasian-Am._THC_Exposure | 0.08 (0.02, 0.13) | 1.01 (1.00, 1.01) | 1.10, 1.05 |
| ivreg(TestCa ~ Cigarettes * AUD * Cannabis + + Cocaine + MHY + Analgesics) | |||
| AUD: Ethnic_THC_Exp | 312.53 (290.54, 334.53) | 1.22E+ 05 (5.36E+ 04, 2.77E+ 05) | 2.43E+ 05, 1.07E+ 05 |
| Cigarettes: Ethnic_THC_Exp | 150.41 (139.67, 161.15) | 280.39 (187.63, 419.03) | 560.29, 374.76 |
| ivreg(TestCa ~ Cigarettes * AUD * Ethnic_THC_Exp * + Cocaine + MHY + Analgesics) | |||
| Ethnic_THC_Exp | 0.55 (0.05, 1.05) | 1.03 (1.00, 1.06) | 1.21, 1.06 |
| MULTIPLE IMPUTATION | |||
| Ethnic_THC_Exposure Alone | |||
| Ethnic_THC_Exposure | 0.68 (0.62, 0.74) | 1.44 (1.39, 1.49) | 2.35, 2.13 |
| Ethnic_THC_Exposure * Race | |||
| Ethnic_THC_Exposure | 0.19 (0.14, 0.24) | 1.24 (1.17, 1.30) | 1.77, 1.62 |
| Additive - Drugs | |||
| Ethnic_THC_Exposure | 0.72 (0.66, 0.79) | 1.48 (1.43, 1.53) | 2.31, 2.20 |
| Interactive Full Model | |||
| Ethnic_THC_Exposure | 0.12 (0.07, 0.18) | 1.14 (1.08, 1.22) | 1.56, 1.37 |
| Quintiles | |||
| Quintile 5 | 0.29 (0.06, 0.51) | 1.15 (1.03, 1.28) | 1.57, 1.21 |
| Dichotomized Quintiles | |||
| Upper_2_Quintiles | 0.2 (0.06, 0.35) | 1.10 (1.03, 1.18) | 1.44, 1.21 |
| LEGAL STATUS | |||
| Whites | |||
| Liberal | 0.09 (0.06, 0.12) | 1.57 (1.36, 1.81) | 2.51, 2.07 |
| Blacks | |||
| Liberal | 0.22 (0.12, 0.32) | 2.83 (1.78, 4.84) | 5.10, 2.96 |
| Both_Races | |||
| Medical | 0.62 (0.43, 0.8) | 1.79 (1.51, 2.13) | 2.99, 2.39 |
| Decriminalized | 0.33 (0.15, 0.51) | 1.36 (1.15, 1.62) | 2.07, 1.56 |
| Legal | 0.61 (0.24, 0.98) | 1.78 (1.25, 2.55) | 2.97, 1.82 |
| Dichotomized_Status | |||
| Liberal | 0.48 (0.34, 0.62) | 1.58 (1.38, 1.81) | 2.54, 2.11 |
Table 10.
Ordered Minimum e-Value List
| No. | Minimum E-Value |
|---|---|
| 1 | 1.07E+ 05 |
| 2 | 374.76 |
| 3 | 9.20 |
| 4 | 4.33 |
| 5 | 3.31 |
| 6 | 2.96 |
| 7 | 2.83 |
| 8 | 2.76 |
| 9 | 2.39 |
| 10 | 2.28 |
| 11 | 2.20 |
| 12 | 2.13 |
| 13 | 2.11 |
| 14 | 2.07 |
| 15 | 1.85 |
| 16 | 1.82 |
| 17 | 1.77 |
| 18 | 1.68 |
| 19 | 1.66 |
| 20 | 1.62 |
| 21 | 1.56 |
| 22 | 1.50 |
| 23 | 1.37 |
| 24 | 1.31 |
| 25 | 1.30 |
| 26 | 1.21 |
| 27 | 1.21 |
| 28 | 1.19 |
| 29 | 1.17 |
| 30 | 1.13 |
| 31 | 1.12 |
| 32 | 1.06 |
| 33 | 1.05 |
Cannabis legal status
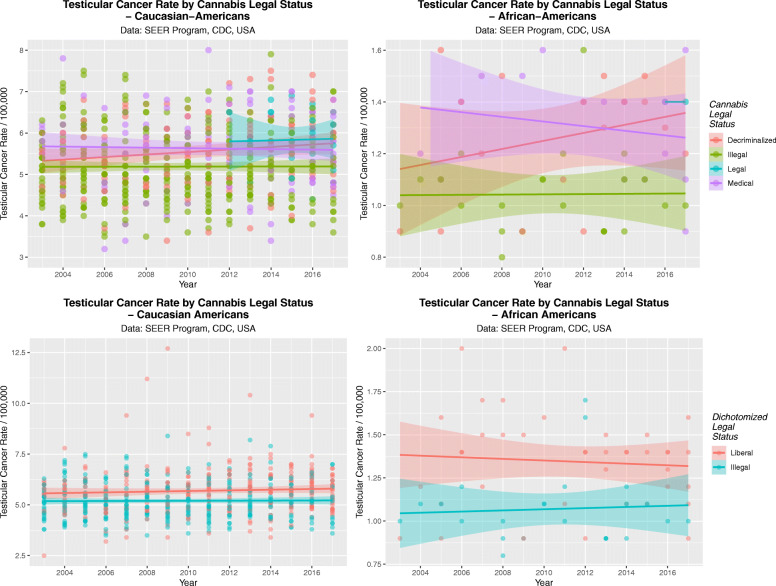
It was of interest to determine if cannabis legalization had an effect on the TCR by ethnic background. This data is presented in Fig. 19 as scatterplots for Caucasian-Americans (A, C) and African-Americans (B, D). Panels A and B present the TCR by legal status and in panels B and D legal status is dichotomized into liberal regimes v. illegal paradigms.
Fig. 19.
Scatterplots by time for the effects of cannabis legal status on testicular cancer rates. (A and C) Caucasian-Americans, (B and D) African-Americans. (A and B) Legal Status; (C and D) Dichotomized legal status, dichotomized as illegal v. not illegal status
These data are assessed quantitatively in Table 11A and B where the effect of cannabis legalization is noted to be highly significant and deleterious in both ethnic backgrounds by linear regression (β-estimate = 0.09 (0.06, 0.12), P = 6.5 × 10− 10 and (β-estimate = 0.22 (0.12, 0.32), P = 4.4 × 10− 5 in Caucasian-Americans and African-Americans respectively) and by mixed effects regression (Medical cannabis: β-estimate = 0.62 (0.43, 0.80), P = 2.5 × 10− 11 and Liberal cannabis (not illegal) paradigm: β-estimate = 0.48 (0.34, 0.62), P = 5.8 × 10− 11).
Table 11.
Linear Regressions on Cannabis Legal Status
| Linear Regression | |||||||
|---|---|---|---|---|---|---|---|
| Parameter Estimates | Model Parameters | ||||||
| Parameter | Estimate (C.I.) | P-Value | S.D. | R-Squared | F | dF | P |
| lm(Testicular_Cancer_Rate ~ Legal_Status) | |||||||
| Caucasian-Americans | |||||||
| Liberal | 0.09 (0.06, 0.12) | 6.5E-10 | 0.1770 | 0.0563 | 39.35 | 1642 | 6.50E-10 |
| African-Americans | |||||||
| Liberal | 0.22 (0.12, 0.32) | 4.40E-05 | 0.1961 | 0.2286 | 19.37 | 1,61 | 4.40E-05 |
| Mixed Effects Regression | |||||||
| Parameter Estimates | Model Parameters | ||||||
| Parameter | Estimate (C.I.) | P-Value | S.D. | AIC | BIC | logLik | |
| Status | |||||||
| lme(Testicular_Cancer_Rate ~ Legal_Status, Random_Effects = Race) | |||||||
| Medical | 0.62 (0.43, 0.8) | 2.5E-11 | 0.9544 | 1967.652 | 1994.985 | −977.8262 | |
| Decriminalized | 0.33 (0.15, 0.51) | 0.0004 | |||||
| Legal | 0.61 (0.24, 0.98) | 0.0014 | |||||
| Dichotomized_Status | |||||||
| lme(Testicular_Cancer_Rate ~ Dichotomized_Status, Random_Effects = Race) | |||||||
| Liberal | 0.48 (0.34, 0.62) | 5.8E-11 | 0.9580 | 1966.987 | 1985.22 | −979.4937 | |
The applicable e-Values for this data is presented at the foot of Table 9.
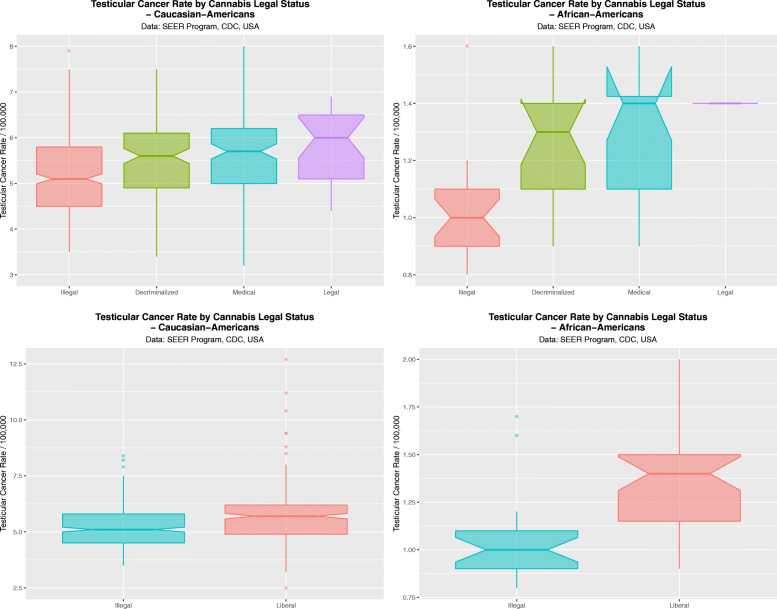
These data are aggregated by time and shown in Fig. 20. These data are significant for legal status in both races (Caucasian-Americans: ChiSqu.Trend = 179.2, df = 162, P = 0.0310; African-Americans: ChiSqu.Trend = 44.45, df = 30, P = 0.0434). When the data are dichotomized contrasting states where cannabis is illegal against those where it is not illegal these results become more significant (Caucasian-Americans: t = 6.1915, df = 538.95, P = 1.1820 × 10− 9; and African-Americans t = 4.50, df = 57.931, P = 3.327 × 10− 5).
Fig. 20.
Boxplots by aggregated time for the effects of cannabis legal status on testicular cancer rates. (A and C) Caucasian-Americans, (B and D) African-Americans. (A and B) Legal Status; (C and D) Dichotomized legal status, dichotomized as illegal v. not illegal status
Discussion
Main results
The present study assessed cannabis and other drug use as risk factors for testicular carcinogenesis and their potential to explain the well described ethnic differentials, and changes in TCR’s amongst ethnic populations across time. Data showed that exposure to THC and cannabigerol are risk factors for TC for all ethnicities investigated, and fulfil the criteria of causal relationships in all ethnicities studied. Data also confirmed the previously described four-fold elevation of TCR amongst Caucasian-Americans compared to African-Americans.
We confirm that time-based scatterplots and boxplots of intensity of cannabis use tend to follow TCR and the two are shown to be closely associated at multivariable regression by several different techniques. Different ethnicities demonstrate different sensitivities to the testicular oncogenic action of cannabinoids, and the pattern of TCR within each ethnicity is not necessarily constant e.g. Hawaii where it is much higher than elsewhere.
Since the relationships persist after inverse probability weighting and are accompanied by high e-Values, findings fulfil the quantitative criteria for causal relationships. These relationships were greatly strengthened when missing data are multiply imputed by chained equations. Legalization to make cannabis more available was also associated with higher TCRs. In that pro-cannabis legalization is associated with higher cannabis use and exposure [42] cannabis legalization may be said to exacerbate and contribute to higher TCRs.
Biological and mechanistic considerations
Description of biology of NSGCT
TC is believed to arise due to genotoxic and epigenotoxic insults incurred during utero life on the germ stem cells which then become activated postnatally by the hormonal surge of puberty [1, 11–14]. Rising rates therefore may imply a rising incidence of some in utero genotoxic or epigenotoxic insult which becomes apparent only later in life.
The four cohort studies of the cannabis - TGCT relationship all described adult / adolescent cannabis exposure [6, 17–21]. This implies a very significant truncation of the usual time course of TGCT by excluding the period of in utero exposure. It is not explicit in our data whether the major aetiological exposure occurs in utero or in later life – or indeed if both may be implicated. In the case of NSGCT which is oncogenically de-differentiated backwards this implies significant and relatively rapid genomewide demethylation [14].
Ethnic differential
P53 is known as the “guardian of the genome” since it is widely connected across the genomic machinery to strongly shut down aberrant DNA replication in the presence of any form of genotoxic stress [43]. In this context it is worth describing the genomic elucidation of the above-mentioned ethnogenomic variability. Investigators intersected 62,567 genomewide association study (GWAS) cancer-associated single nucleotide variations (SNP’s) with 17,118 unique positive signals for P53 activation response elements (P53-RE’s) in four different cell lines using seven P53 activators [7]. The base sequences surrounding the 86 positive hits were compared to assess their fit with the two canonical decameric DNA recognition sequences in P53. At position rs4590952 in the kit P53-RE on chromosome 9 the position weight matrix value dropped from 15.6 to 11.1 (median = 13.8) with guanine to adenosine substitution. Three nearby sites have been identified in three previous GWAS screens as being associated with TCR and to have a three-fold elevation per allele in TCR risk amongst Caucasians [44–46]. The Kit - Kit-ligand dimer is the key nearby receptor ligand pair which acts as the master transcription factor for primordial germ cell, controls their specification and prevents differentiation, and is highly and uniquely expressed in seminomas rather than other TGCT’s. Kit also plays a key controlling role in haemopoietic stem cells and melanoblasts [7].
This mutation has been identified as a risk factor for both seminomas and non-seminomatous germ cell tumours [7]. This site is unique as it is activated by P53 activation rather than suppressed as is more usual [43]. In a subsequent assay in testicular tumour cell lines the per allele activation of the P53-RE by P53 activation was 188-fold (range 93 to 373- fold) [7]. This allele was positively selected for in seminomas (21.7-fold) and also amongst Caucasian-Americans. It was thought that the allele was positively selected for in light skinned races as its effect to stimulate melanoblast activation and proliferation in the tanning response to UV radiation was protective of the skin from UV induced carcinogenesis [7]. This was thought to explain is relative prevalence amongst light skinned races.
Other similar loci have also been described including rs995030 and research in this area is on-going at the present time [8].
Cannabinoid pathophysiology
High dose marijuana smoking has been shown to grossly disturb human sperm morphology with shrunken and bent sperm heads, bent tails, multiple tails, bilobed heads, tangled tails, multiple heads, pyknotic heads and polymorphonuclear pus cells all described [47].
In mice exposure to Δ9THC was shown to induce ring and chain chromosome formation with chains up to four chromosomes long due to end-to-end fusion formation [48].
These authors also showed that when mouse sperm were exposed to the cannabinoids Δ9THC, cannabinol and cannabidiol there was a dramatic increase in chromosomal translocations from about 1% at control levels, to 4.95–6.48% comparable to the positive control which was the cytotoxic drug mitomycin C at 6.73% [48].
Cannabinoids have been shown to have marked effects on sperm function including reduction of sperm concentration in seminal fluid, induction of DNA fragmentation, defective sperm maturation, disorders of DNA packing within sperm and protamine-histone replacement, DNA nicking in sperm by Tnp2, defective DNA repair, defects of nuclear size and incomplete DNA packing by failure of the histone-protamine transition [49–51].
Cannabinoids induce collapse of the inner mitochondrial membrane potential by several routes [50, 51].
THC exposure has been shown to lead to marked demethylation of the genome of human sperm [52] a change which makes genes more susceptible to genomic damage and chromosomal rearrangements [13, 16].
Cannabis exposure of both lymphocytes and oocytes has been shown to induce 20% cell death with a single division and marked chromosomal bridging and nuclear bleb formation in surviving cells [53]. Cannabinoid exposure has also long been known to be associated with micronucleus formation and comet tail formation, which are two of the major genotoxicity assays implicating chromosomal mis-segregation and single- and double- stranded DNA breaks respectively [15].
Moreover low micromolar doses of cannabidiol and its propyl analogue cannabidivarin have been shown to cause micronucleus formation and prominent comet tails on formal testing, changes which are greatly exacerbated in an oxidizing environment [15].
Downs syndrome has been linked with cannabis exposure in Hawaii, Colorado, Canada and Australia [54–57] and early termination of pregnancy for anomaly-corrected rates of Downs syndrome, trisomies 18 and 13, Turners syndrome and Deletion 22q11.2 in a space time and odds ratio analysis in the USA [58].
Prenatal cannabis use has been linked with acute lymphoid leukaemia which is primarily a disease characterized genetically by chromosomal translocations (unpublished data).
Adult cannabis exposure is linked with TC [17–20] and present study, which is itself caused by whole genome duplication, isochromosome 12 formation, deletion and augmentation of many chromosomal arms, over 1200 micro-RNA’s [13, 14] and genomewide DNA demethylation.
Formation of ring and chain concatenation of chromosomes in rodents was mentioned above [47].
This list implicates cannabinoids in major chromosomal toxicity by many mechanisms including chromosomal deletion, reduplication, megabase scale reduplications, longitudinal and transverse duplications and gene amplification and oncogenic cellular de-differentiation.
All of this demonstrates that cannabinoid-exposed cells are clearly genomically stressed.
In the context of genomic stress P53 is activated – and the ethnogenomic differential mechanism described above becomes activated as a stimulus to tumour cell proliferation in light skinned races, and to an onco-protective block to mitosis and meiosis in darker skinned races.
Given the aforementioned, it is possible that cannabis exposure causes in utero germ cell damage which is associated with TC, suggesting that cannabis use during pregnancy should be cautioned. This is consistent with recommendations by both the American College of Obstetrics and Gynaecology (ACOG) and the American Academy of Pediatric (AAP) [59–63]. Notwithstanding this advice, a significant number of American women are reported as using cannabis whilst pregnant, perhaps explaining part of the rise in TC across many communities. In Colorado 69% of cannabis dispensaries contacted by a group of researchers recommended cannabis use to pregnant women [64], while in California 24% of pregnant teenagers either self-admitted to cannabis use whilst pregnant or tested positive for it during their gestation [65]. Nationwide it was estimated in 2017 that 161,000 pregnant women used cannabis whilst pregnant [24].
It is however important to stress that while TC is generally believed to arise as a result of in utero germ cell anomalies which may be impacted on by maternal cannabis use, all published studies on the association have identified an association between TC and personal cannabis use, suggesting that one or more likely gene / epigenome - environment interactions are at play whereby postnatal and adult cannabinoid exposure contribute to underlying genetic risk as environmental causal and exacerbating factors. Since the epigenetic state of the primordial germ cells / gonocytes is a key determinant of the differentiation block experienced by all NSGCT tumour cells, it follows that part of the effect of postnatal cannabinoid exposure must be to de-differentiate susceptible cells into a more immature foetal-like and pro-oncogenic state.
Generalizability
We feel that study findings are generalizable for many reasons. The SEER Cancer data is registry controlled and comes from most USA states and thus represents the great preponderance of the data from the population. The NSDUH survey has a good response rate at 74.1%. Moreover the effects we describe are often very strong. There is great internal consistency across results within this study with similar results being found for all ethnicities studied, and also good external consistency with all the published literature in this field. Moreover since data fulfil the criteria for causality we would expect that this causal relationship to hold widely across space and time.
Strengths and limitations
This study has a number of strengths and limitations. Its strengths include the use of a large population dataset and registry controlled data, and a variety of advanced statistical methods including inverse probability weighting, mixed effects models, robust regression, two-step instrumental variable regression, multiple imputation of missing data by chained equations and e-Values. Further, many levels of significance are very high as are their corresponding e-Values. Since the relationships described amply fulfill the quantitative criteria for causality we feel that the relationships described herein are transportable to other situations and other times. The main limitation of the present work is the absence from this dataset of individual exposure data which is a limitation commonly shared with most epidemiological studies. Also spatiotemporal data on known risk factors such as cryptorchidism, inguinal herniae, industrial pollution and sedentary lifestyles was not available to the present investigators. It would be a useful advance if future studies could be repeated with these factors included in the multivariable analysis.
Conclusion
Data analysis indicate that exposure to THC and cannabigerol is a risk factor for TC for all ethnicities. We have confirmed the four-fold elevation of TCR amongst Caucasian-Americans compared to African-Americans and data indicate that a likely gene-environment interaction is at play with cannabis the most likely environmental causal factor. In view of the high e-Values demonstrated we feel that the place of cannabis and cannabinoids is unlikely to be supplanted by other covariates with further research. All ethnicities are subject to an increase in testicular oncogenesis under a paradigm of increasing cannabinoid exposure with some ethnicities demonstrating marked differences in their apparent sensitivities. Time based plots and box pots of cannabis use and TCR generally move in parallel. Effects of cannabis, ethnic THC exposure and cannabinoid exposure are statistically highly significant, confirmed with a variety of multivariable techniques, and are independently significant. These relationships are strengthened by multiple imputation of missing ethnicity data. Findings fulfil the criteria of causal relationships in all ethnicities studied. Cannabis legalization significantly elevates the TCR for both African-American and Caucasian-American patients. In short, we feel that these findings are robust, fulfil the criteria for causal relatoinships and add an important transgenerational dimension to the present cannabis debate which applies to the major ethnic groups identified within the USA for which data is available.
Acknowledgements
Not applicable.
Authors’ contributions
ASR assembled the data, designed and conducted the analyses, and wrote the first manuscript draft. GKH provided technical and logistic support, co-wrote the paper, assisted with gaining ethical approval, provided advice on manuscript preparation and general guidance to study conduct. All authors have read and approved the manuscript.
Funding
No funding was provided for this study. No funding organization played any role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Availability of data and materials
No permissions are required to access the data which was used and collated in this study, e.g. NSDUH study. Data including shapefiles and R programming script is made publicly available on the Mendeley Data Archive at this URL: 10.17632/yxy3dg2wt6.1 .
Declarations
Ethics approval and consent to participate
The Human Research Ethics Committee of the University of Western Australia provided ethical approval for the study to be undertaken 7th June 2019 (No. RA/4/20/4724). Ethics approval was not required to access the data in the first instance. However Ethical approval provided permission to access, analyze and publish all the data obtained.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cheng L, Albers P, Berney DM, Feldman DR, Daugaard G, Gilligan T, Looijenga LHJ. Testicular cancer. Nat Rev Dis Primers. 2018;4(1):29. doi: 10.1038/s41572-018-0029-0. [DOI] [PubMed] [Google Scholar]
- 2.Czene K, Lichtenstein P, Hemminki K. Environmental and heritable causes of cancer among 9.6 million individuals in the Swedish family-Cancer database. Int J Cancer. 2002;99(2):260–266. doi: 10.1002/ijc.10332. [DOI] [PubMed] [Google Scholar]
- 3.SEER Explorer. https://seer.cancer.gov/explorer/application.html. Accessed 1 Oct 2020.
- 4.Gurney JK, Florio AA, Znaor A, Ferlay J, Laversanne M, Sarfati D, Bray F, McGlynn KA. International trends in the incidence of testicular Cancer: lessons from 35 years and 41 countries. Eur Urol. 2019;76(5):615–623. doi: 10.1016/j.eururo.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huyghe E, Matsuda T, Thonneau P. Increasing incidence of testicular cancer worldwide: a review. J Urol. 2003;170(1):5–11. doi: 10.1097/01.ju.0000053866.68623.da. [DOI] [PubMed] [Google Scholar]
- 6.Gurney J, Shaw C, Stanley J, Signal V, Sarfati D. Cannabis exposure and risk of testicular cancer: a systematic review and meta-analysis. BMC Cancer. 2015;15(1):897. doi: 10.1186/s12885-015-1905-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeron-Medina J, Wang X, Repapi E, Campbell MR, Su D, Castro-Giner F, Davies B, Peterse EF, Sacilotto N, Walker GJ, et al. A polymorphic p53 response element in KIT ligand influences cancer risk and has undergone natural selection. Cell. 2013;155(2):410–422. doi: 10.1016/j.cell.2013.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin JX, Cai ZL, Yang YF, Xing JC, Zhuang X. Association between KITLG gene polymorphisms and testicular germ cell tumors: a systematic review and meta-analysis. Curr Med Sci. 2019;39(5):702–706. doi: 10.1007/s11596-019-2095-2. [DOI] [PubMed] [Google Scholar]
- 9.McGlynn KA, Trabert B. Adolescent and adult risk factors for testicular cancer. Nat Rev Urol. 2012;9(6):339–349. doi: 10.1038/nrurol.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sonneveld DJ, Schaapveld M, Sleijfer DT, Meerman GJ, van der Graaf WT, Sijmons RH, Koops HS, Hoekstra HJ. Geographic clustering of testicular cancer incidence in the northern part of the Netherlands. Br J Cancer. 1999;81(7):1262–1267. doi: 10.1038/sj.bjc.6690839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilbert D, Rapley E, Shipley J. Testicular germ cell tumours: predisposition genes and the male germ cell niche. Nat Rev Cancer. 2011;11(4):278–288. doi: 10.1038/nrc3021. [DOI] [PubMed] [Google Scholar]
- 12.Hanna NH, Einhorn LH. Testicular cancer--discoveries and updates. N Engl J Med. 2014;371(21):2005–2016. doi: 10.1056/NEJMra1407550. [DOI] [PubMed] [Google Scholar]
- 13.Oosterhuis JW, Looijenga LHJ. Germ Cell Tumors from a Developmental Perspective: Cells of Origin, Pathogenesis, and Molecular Biology (Emerging Patterns) In: Nogales FF, Jimenez RE, editors. Pathology and Biology of Human Germ Cell Tumors. Berlin: Springer Berlin Heidelberg; 2017. pp. 23–129. [Google Scholar]
- 14.Shen H, Shih J, Hollern DP, Wang L, Bowlby R, Tickoo SK, Thorsson V, Mungall AJ, Newton Y, Hegde AM, Armenia J, Sánchez-Vega F, Pluta J, Pyle LC, Mehra R, Reuter VE, Godoy G, Jones J, Shelley CS, Feldman DR, Vidal DO, Lessel D, Kulis T, Cárcano FM, Leraas KM, Lichtenberg TM, Brooks D, Cherniack AD, Cho J, Heiman DI, Kasaian K, Liu M, Noble MS, Xi L, Zhang H, Zhou W, ZenKlusen JC, Hutter CM, Felau I, Zhang J, Schultz N, Getz G, Meyerson M, Stuart JM, Akbani R, Wheeler DA, Laird PW, Nathanson KL, Cortessis VK, Hoadley KA, Wang L, Xi L, Wheeler D, Hughes D, Covington K, Jayaseelan JC, Korchina V, Lewis L, Hu J, Doddapaneni HV, Muzny D, Gibbs R, Hoadley KA, Hollern D, Vincent BG, Chai S, Smith CC, Auman JT, Shi Y, Meng S, Skelly T, Tan D, Veluvolu U, Mieczkowski PA, Jones CD, Wilkerson MD, Balu S, Bodenheimer T, Hoyle AP, Jefferys SR, Mose LE, Simons JV, Soloway MG, Roach J, Parker JS, Hayes DN, Perou CM, Shih J, Cherniack AD, Meyerson M, Saksena G, Cibulskis C, Schumacher SE, Beroukhim R, Gabriel SB, Bowlby R, Mungall AJ, Brooks D, Kasaian K, Ally A, Balasundaram M, Carlsen R, Cheung D, Chuah E, Dhalla N, Holt RA, Jones SJM, Ma Y, Mayo M, Moore RA, Robertson AG, Schein JE, Sipahimalani P, Tam A, Thiessen N, Wong T, Marra MA, Shen H, Zhou W, Laird PW, Weisenberger DJ, van den Berg DJ, Lai PH, Berrios M, Holbrook A, Bootwalla MS, Maglinte DT, Armenia J, Sánchez-Vega F, Schultz N, Chakravarty D, Gao J, Heins Z, Kundra R, Ochoa A, Liu M, Sander C, Ladanyi M, Thorsson V, Radenbaugh AJ, Newton Y, Stuart JM, Cho J, Heiman DI, Noble MS, Zhang H, Getz G, Gehlenborg N, Saksena G, Voet D, Lin P, Frazer S, Kim J, Lawrence MS, Meier S, Defreitas T, Chin L, Hegde AM, Akbani R, Weinstein JN, Liu W, Mills GB, Lu Y, Pyle LC, Pluta J, Nathanson KL, Tickoo SK, Reuter VE, Mehra R, Looijenga L, Bryce AH, Cárcano FM, Carvalho AL, Cortessis VK, Feldman D, Godoy G, Ittmann M, Jones J, Kulis T, Lerner S, Lessel D, Nathanson KL, Shelley CS, Vidal DO, Leraas KM, Lichtenberg TM, Bowen J, Gastier-Foster JM, Gerken M, Helsel C, Ramirez NC, Wise L, Zmuda E, Cottingham S, Chesla D, Saller C, Tarvin K, Lopes LF, Scapulatempo-Neto C, Aredes NDA, Oosterhuis W, Gillis A, Stoop H, Eijkenboom W, Sandusky G, Martin SE, Aron M, Daneshmand S, Djaladat H, Quinn D, Dorff T, Lennerz JK, Thorne LB, Gamulin M, Kastelan Z, Hudolin T, Kubisch C, Boice L, Huang M, Perou AH, Rathmell WK, Pihl T, Wan Y, Sun Q, Naresh R, Chudamani S, Liu J, Lolla L, Wu Y, Ferguson ML, Zenklusen JC, Felau I, Zhang J(J), Sheth M, Demchok JA, Yang L, Wang Z, Tarnuzzer R, Hutter CM, Sofia HJ, Davidsen TM. Integrated molecular characterization of testicular germ cell tumors. Cell Rep. 2018;23(11):3392–3406. doi: 10.1016/j.celrep.2018.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russo C, Ferk F, Misik M, Ropek N, Nersesyan A, Mejri D, Holzmann K, Lavorgna M, Isidori M, Knasmuller S. Low doses of widely consumed cannabinoids (cannabidiol and cannabidivarin) cause DNA damage and chromosomal aberrations in human-derived cells. Arch Toxicol. 2019;93(1):179–188. doi: 10.1007/s00204-018-2322-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bröske AM, Vockentanz L, Kharazi S, Huska MR, Mancini E, Scheller M, Kuhl C, Enns A, Prinz M, Jaenisch R, Nerlov C, Leutz A, Andrade-Navarro MA, Jacobsen SEW, Rosenbauer F. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet. 2009;41(11):1207–1215. doi: 10.1038/ng.463. [DOI] [PubMed] [Google Scholar]
- 17.Daling JR, Doody DR, Sun X, Trabert BL, Weiss NS, Chen C, Biggs ML, Starr JR, Dey SK, Schwartz SM. Association of marijuana use and the incidence of testicular germ cell tumors. Cancer. 2009;115(6):1215–1223. doi: 10.1002/cncr.24159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Callaghan RC, Allebeck P, Akre O, McGlynn KA, Sidorchuk A. Cannabis use and incidence of testicular Cancer: a 42-year follow-up of Swedish men between 1970 and 2011. Cancer Epidemiol Biomark Prev. 2017;26(11):1644–1652. doi: 10.1158/1055-9965.EPI-17-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trabert B, Sigurdson AJ, Sweeney AM, Strom SS, McGlynn KA. Marijuana use and testicular germ cell tumors. Cancer. 2011;117(4):848–853. doi: 10.1002/cncr.25499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lacson JC, Carroll JD, Tuazon E, Castelao EJ, Bernstein L, Cortessis VK. Population-based case-control study of recreational drug use and testis cancer risk confirms an association between marijuana use and nonseminoma risk. Cancer. 2012;118(21):5374–5383. doi: 10.1002/cncr.27554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song A, Myung NK, Bogumil D, Ihenacho U, Burg ML, Cortessis VK. Incident testicular cancer in relation to using marijuana and smoking tobacco: A systematic review and meta-analysis of epidemiologic studies. Urol Oncol. 2020;38(7):642.e641–642.e649. doi: 10.1016/j.urolonc.2020.03.013. [DOI] [PubMed] [Google Scholar]
- 22.Rajpert-De Meyts E, McGlynn KA, Okamoto K, Jewett MA, Bokemeyer C. Testicular germ cell tumours. Lancet. 2016;387(10029):1762–1774. doi: 10.1016/S0140-6736(15)00991-5. [DOI] [PubMed] [Google Scholar]
- 23.All Cancer Sites Combined, Ages < 20 Years, Long Term Trends in SEER Incidence Rates 1975-2015, SEER Explorer [https://seer.cancer.gov/explorer/application.php?site=1&data_type=1&graph_type=1&compareBy=race&chk_sex_1=1&chk_sex_3=3&chk_sex_2=2&chk_race_1=1&chk_age_range_15=15&chk_data_type_1=1&advopt_precision=1&advopt_display=1&showDataFor=sex_1_and_age_range_15_and_data_type_1].
- 24.McCantz-Katz E . Substance Abuse and Mental Health Services Administration, US Department of Health and Human Services, vol. 1. Silver Springs, Maryland: SAMHSA, US DHHS. 2020. The National Survey of Drug Use and Health: 2019; pp. 1–63. [Google Scholar]
- 25.ElSohly MA, Mehmedic Z, Foster S, Gon C, Chandra S, Church JC. Changes in Cannabis potency over the last 2 decades (1995-2014): analysis of current data in the United States. Biol Psychiatry. 2016;79(7):613–619. doi: 10.1016/j.biopsych.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chandra S, Radwan MM, Majumdar CG, Church JC, Freeman TP, ElSohly MA. New trends in cannabis potency in USA and Europe during the last decade (2008-2017) Eur Arch Psychiatry Clin Neurosci. 2019;269(1):5–15. doi: 10.1007/s00406-019-00983-5. [DOI] [PubMed] [Google Scholar]
- 27.ElSohly MA, Ross SA, Mehmedic Z, Arafat R, Yi B, Banahan BF., 3rd Potency trends of delta9-THC and other cannabinoids in confiscated marijuana from 1980-1997. J Forensic Sci. 2000;45(1):24–30. doi: 10.1520/JFS14636J. [DOI] [PubMed] [Google Scholar]
- 28.Legality of cannabis by U.S. jurisdiction. https://en.wikipedia.org/wiki/Legality_of_cannabis_by_U.S._jurisdiction. Accessed 1 Oct 2020.
- 29.Wickham H, Averick M, Bryan J, Chang W, McGowan LD, Francios R, Groelmund G, Hayes A, Henry L, Hester J, et al. Welcome to the Tidyverse. J Open Source Software. 2019;4(43):1686–1691. doi: 10.21105/joss.01686. [DOI] [Google Scholar]
- 30.Wickham H. ggplot2: elegant graphics for data analysis, vol. 1. New York: Springer-Verlag; 2016. [Google Scholar]
- 31.RColorBrewer: ColorBrewer Palettes. [https://CRAN.R-project.org/package=RColorBrewer]. Erich Neuwirth (2014).
- 32.Pebesma E. Simple features for R: standardized support for spatial vector data. R J. 2018;10(1):439–446. doi: 10.32614/RJ-2018-009. [DOI] [Google Scholar]
- 33.Sarkar D. Lattice: Multivariate Data Visualization with R, vol. 1: Springer. 2008. [Google Scholar]
- 34.Pinheiro J, Bates D, DebRoy S, Sarkar D, Team RC . nlme: Linear and Nonlinear Mixed Effects Models, vol. 1: R: Comprehensive R Archive Network. 2020. [Google Scholar]
- 35.Kliber C, Zeileis A. Applied econometrics with R. 2008. [Google Scholar]
- 36.Wal W. Geskus R: ipw: an R package for inverse probability weighting. J Stat Softw. 2011;43(13). 10.18637/jss.v043.i13.
- 37.Lumley T. Complex Surveys: a guide to analysis using R, vol. 1: Wiley. 2010. [Google Scholar]
- 38.Van Buuren S, Groothuis-Oudshoorn K. Mice: multivariate imputation by chained equations. J Stat Softw. 2011;45(3):1–67. doi: 10.18637/jss.v045.i03. [DOI] [Google Scholar]
- 39.VanderWeele TJ, Ding P. Sensitivity analysis in observational research: introducing the E-value. Ann Intern Med. 2017;167(4):268–74. [DOI] [PubMed]
- 40.Croissant Y, Millo G. Panel data econometrics with R, vol. 1. Oxford: Wiley; 2019. p. 1-299.
- 41.VanderWeele TJ, Ding P, Mathur M. Technical considerations in the use of the E-value. J Causal Inference. 2019;7(2):1–11. doi: 10.1515/jci-2018-0007. [DOI] [Google Scholar]
- 42.Hasin DS, Sarvet AL, Cerda M, Keyes KM, Stohl M, Galea S, Wall MM. US adult illicit Cannabis use, Cannabis use disorder, and medical marijuana Laws: 1991-1992 to 2012-2013. JAMA Psychiatry. 2017;74(6):579–588. doi: 10.1001/jamapsychiatry.2017.0724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358(6381):15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 44.Kanetsky PA, Mitra N, Vardhanabhuti S, Li M, Vaughn DJ, Letrero R, Ciosek SL, Doody DR, Smith LM, Weaver J, Albano A, Chen C, Starr JR, Rader DJ, Godwin AK, Reilly MP, Hakonarson H, Schwartz SM, Nathanson KL. Common variation in KITLG and at 5q31.3 predisposes to testicular germ cell cancer. Nat Genet. 2009;41(7):811–815. doi: 10.1038/ng.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rapley EA, Turnbull C, Al Olama AA, Dermitzakis ET, Linger R, Huddart RA, Renwick A, Hughes D, Hines S, Seal S, et al. A genome-wide association study of testicular germ cell tumor. Nat Genet. 2009;41(7):807–810. doi: 10.1038/ng.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turnbull C, Rapley EA, Seal S, Pernet D, Renwick A, Hughes D, Ricketts M, Linger R, Nsengimana J, Deloukas P, et al. Variants near DMRT1, TERT and ATF7IP are associated with testicular germ cell cancer. Nat Genet. 2010;42(7):604–607. doi: 10.1038/ng.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hembree WC, III, Nahas GG, Zeidenberg P, Huang HFS. Changes in Human Spermatozoa Associated with High Dose Marijuana Smoking. In: Nahas GG, Sutin KM, Harvey DJ, Agurell S, editors. vol. 1. Totowa: Humana Press; 1999. pp. 367–378. [Google Scholar]
- 48.Zimmerman AM, Zimmerman S, Raj AY. Effects of Cannabinoids on Spermatogensis in Mice. In: Nahas GG, Sutin KM, Harvey DJ, Agurell S, editors. Marijuana and Medicine. Volume 1, 1 edn. Totowa: Humana Press; 1999. pp. 347–358. [Google Scholar]
- 49.Chioccarelli T, Cacciola G, Altucci L, Lewis SE, Simon L, Ricci G, Ledent C, Meccariello R, Fasano S, Pierantoni R, et al. Cannabinoid receptor 1 influences chromatin remodeling in mouse spermatids by affecting content of transition protein 2 mRNA and histone displacement. Endocrinology. 2010;151(10):5017–5029. doi: 10.1210/en.2010-0133. [DOI] [PubMed] [Google Scholar]
- 50.Rossato M, Ion Popa F, Ferigo M, Clari G, Foresta C. Human sperm express cannabinoid receptor Cb1, the activation of which inhibits motility, acrosome reaction, and mitochondrial function. J Clin Endocrinol Metab. 2005;90(2):984–991. doi: 10.1210/jc.2004-1287. [DOI] [PubMed] [Google Scholar]
- 51.Rossato M, Pagano C, Vettor R. The cannabinoid system and male reproductive functions. J Neuroendocrinol. 2008;20(Suppl 1):90–93. doi: 10.1111/j.1365-2826.2008.01680.x. [DOI] [PubMed] [Google Scholar]
- 52.Murphy SK, Itchon-Ramos N, Visco Z, Huang Z, Grenier C, Schrott R, Acharya K, Boudreau MH, Price TM, Raburn DJ, Corcoran DL, Lucas JE, Mitchell JT, McClernon FJ, Cauley M, Hall BJ, Levin ED, Kollins SH. Cannabinoid exposure and altered DNA methylation in rat and human sperm. Epigenetics. 2018;13(12):1208–1221. doi: 10.1080/15592294.2018.1554521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morishima A. Effects of cannabis and natural cannabinoids on chromosomes and ova. NIDA Res Monogr. 1984;44:25–45. [PubMed] [Google Scholar]
- 54.Forrester MB, Merz RD. Risk of selected birth defects with prenatal illicit drug use, Hawaii, 1986-2002. J Toxicol Environ Health. 2007;70(1):7–18. doi: 10.1080/15287390600748799. [DOI] [PubMed] [Google Scholar]
- 55.Reece AS, Hulse GK. Canadian cannabis consumption and patterns of congenital anomalies: an ecological geospatial analysis. J Addiction Medicine. 2020;14(5):e195–e210. 10.1097/ADM.0000000000000638. [DOI] [PMC free article] [PubMed]
- 56.Reece AS, Hulse GK. Cannabis teratology explains current patterns of Coloradan congenital defects: the contribution of increased cannabinoid exposure to rising Teratological trends. Clin Pediatr. 2019;58(10):1085–1123. doi: 10.1177/0009922819861281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reece AS, Hulse GK. Broad Spectrum Epidemiological Contribution of Cannabis and Other Substances to the Teratological Profile of Northern New South Wales: Geospatial and Causal Inference Analysis. BMC Pharmacol Toxicol. 2020; In Press (Accepted: September 25th 2020):In Press. [DOI] [PMC free article] [PubMed]
- 58.Reece AS, Hulse GK. Cannabis in pregnancy – rejoinder, Exposition and Cautionary Tales. Psychiatric Times. 2020. [Google Scholar]
- 59.The impact of marijuana policies on youth: clinical, research, and legal update. Pediatrics. 2015;135(3):584–7. [DOI] [PubMed]
- 60.Ammerman S, Ryan S, Adelman WP. The impact of marijuana policies on youth: clinical, research, and legal update. Pediatrics. 2015;135(3):e769–e785. doi: 10.1542/peds.2014-4147. [DOI] [PubMed] [Google Scholar]
- 61.Dharmapuri S, Miller K, Klein JD. Marijuana and the pediatric population. Pediatrics. 2020;146(2):e20192629. 10.1542/peds.2019-2629. [DOI] [PubMed]
- 62.ACOG Committee Opinion: Marijuana Use During Pregnancy and Lactation. https://www.acog.org/en/Clinical/Clinical%20Guidance/Committee%20Opinion/Articles/2017/10/Marijuana%20Use%20During%20Pregnancy%20and%20Lactation. Accessed 1 Oct 2020.
- 63.Wong SS, Wilens TE. Medical cannabinoids in children and adolescents: a systematic review. Pediatrics. 2017;140(5):e20171818. 10.1542/peds.2017-1818. [DOI] [PubMed]
- 64.Dickson B, Mansfield C, Guiahi M, Allshouse AA, Borgelt LM, Sheeder J, Silver RM, Metz TD. Recommendations from Cannabis dispensaries about first-trimester Cannabis use. Obstet Gynecol. 2018;131(6):1031–1038. doi: 10.1097/AOG.0000000000002619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Young-Wolff KC, Tucker L, Alexeeff S, et al. Trends in self-reported and biochemically tested marijuana use among pregnant females in California from 2009-2016. JAMA. 2017;318(24):2490–2491. doi: 10.1001/jama.2017.17225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No permissions are required to access the data which was used and collated in this study, e.g. NSDUH study. Data including shapefiles and R programming script is made publicly available on the Mendeley Data Archive at this URL: 10.17632/yxy3dg2wt6.1 .