Abstract
Despite vast diversity in non-human hosts and conspicuous recent spillover events, only a small number of coronaviruses have been observed to persist in human populations. This puzzling mismatch suggests substantial barriers to establishment. We detail hypotheses that might contribute to explain the low numbers of endemic coronaviruses, despite their considerable evolutionary and emergence potential. We assess possible explanations ranging from issues of ascertainment, historically lower opportunities for spillover, aspects of human demographic changes, and features of pathogen biology and pre-existing adaptive immunity to related viruses. We describe how successful emergent viral species must triangulate transmission, virulence, and host immunity to maintain circulation. Characterizing the factors that might shape the limits of viral persistence can delineate promising research directions to better understand the combinations of pathogens and contexts that are most likely to lead to spillover.
Keywords: viral ecology, cross-reactivity, landscape of immunity, antigenic space
Emergent and endemic coronavirus diversity
Transmissible and pathogenic novel human coronavirus species have emerged three times since 2002 [severe acute respiratory syndrome coronavirus-1 (SARS-CoV-1) 2002, Middle East respiratory syndrome (MERS) 2012, and SARS-CoV-2 2019]. Within species, rapid evolution has been observed, both in terms of the appearance of new variants after emergence (e.g., for SARS-CoV-2 [1]) and immune escape (see Glossary) for established endemic coronavirus species (e.g., human coronaviruses HCoV-229E and HCoV-OC43 [2,3]). Given this evidence of frequent spillover, rapid evolution, and the millennia of human history over which coronavirus diversity could have accumulated, one would expect a diverse abundance of coronaviruses in the human pathogen pool. However, only four endemic species are known to persist in human populations (HCoV-229E, -NL63, -OC43, and -HKU1), which harbor limited genetic diversity within species [4,5].
What could explain this apparent mismatch between the pace of emergence and the low diversity of endemic coronaviruses? The coronaviruses provide a timely example, but this pattern is not without precedent: frequent emergence is seen but without evidence of historical accumulation of a diverse endemic community for an array of pathogen groups (Box 1 ). We evaluate here the multiple factors that may govern the diversity of the endemic pathogen pool of a host, using coronaviruses as an example; we note in particular how population variation in immunity resulting from multi-pathogen dynamics might influence the prospects for pathogen emergence and persistence.
Box 1. The comparative context of coronavirus diversity.
Approximately 20–30 viral families and 200–300 viral species are known to infect humans [27]. Comparing viral diversity across lower (e.g., serotype, genogroup, or strain) or higher (e.g., species or family) taxonomic levels is complicated by differences in taxonomy conventions. However, species level and intra-species diversity is low for some human viral groups. For example, most viral families have fewer than 10 known human viruses, of which not all have demonstrated stable transmission. This is compared with estimates that 1–2 million viral species may be circulating in mammals and birds alone [102], and >10 000 are thought to have the potential to infect humans [103]. Likewise, some non-viral pathogen groups are similar to coronaviruses in that evidence of recent emergence events does not correspond to high rates of endemism. In malaria, for example, two new species (Plasmodium knowlesi [104], and to a lesser extent Plasmodium cynomolgi [105., 106., 107.]), have been observed to infect humans since the 1990s, but there are only four other long-known human malaria-causing parasite species [108,109]. Conversely, some pathogen groups exhibit high circulating diversity. Examples include enteroviruses, for which more than 100 serotypes are thought to co-circulate [77], and the Bunyaviridae, for which >40 human-infecting viruses have been described [27]. Bunyaviruses that are known to infect humans include a high number of species, but are often limited by the distribution of their arthropod vectors [110]. The high diversity of circulating enterovirus serotypes might be explained by the lack of cross-reactivity among enteroviruses lineages [77]; because immunity is highly specific for this group, many related viruses may be able to spread independently without being affected by immunity previously generated against other lineages. Characterizing the viral diversity of influenza virus, and others that evolve antigenically in response to immune pressure must account for the facts that, on the one hand, strains accumulate antigenic diversity rapidly via antigenic drift, but on the other hand, only a small fraction of that diversity of strains circulates at a given time [72].
Alt-text: Box 1
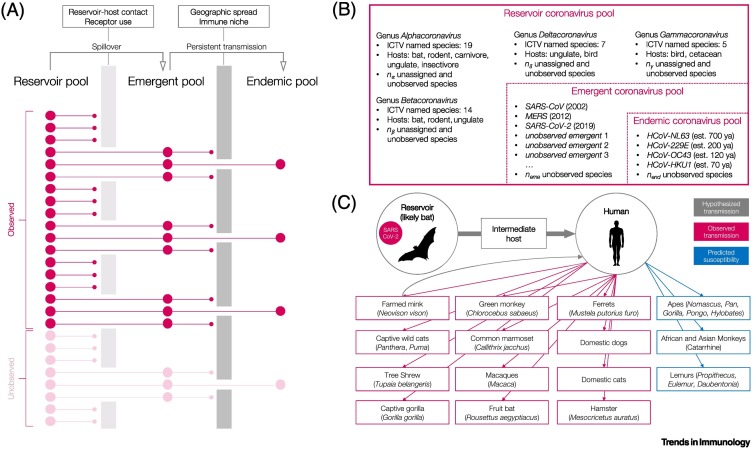
For a pathogen with emergence potential, barriers to becoming endemic may act at both the reservoir-to-emergent and emergent-to-endemic transitions (Figure 1A). Wildlife-infecting coronaviruses are rich in diversity, with ~50 classified species, and likely many more unassigned or unobserved lineages that infect a broad range of mammalian hosts [6]. A subset of this diversity – the four endemic human coronaviruses – are thought to have entered human populations between 50 and 700 years ago (Figure 1B) and are observed as common respiratory infections globally [7., 8., 9., 10., 11.]. The endemic species are distantly related and may have emerged from different reservoirs, or in some cases via different intermediate hosts, suggesting that, for coronaviruses, there might be multiple pathways to becoming endemic, although this remains conjectural.
Figure 1.
Coronaviruses and the pathway to endemism.
(A) In this schematic, a subset of pathogens in the diverse reservoir pool enters the emergent pool via spillover, and a subset of emergent pathogens attain sufficiently stable transmission to enter the pool of persistently circulating endemic pathogens. Owing to incomplete sampling, a fraction of each pool remains unobserved (lighter colors). (B) Depicted are the reservoir, emergent, and endemic pools for coronaviruses, and again a fraction of each remains unobserved. Classification follows the International Committee on Taxonomy of Viruses (ICTV) [6]. (C) Spillover and cross-species transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from a likely bat reservoir, to a possible intermediate host, to humans, and then to secondary human–animal transmission. Bats have been hypothesized to be unusually prolific hosts for pathogens [111], but many other hosts are also noted (including non-human primates, wild and domestic carnivores, and ungulates). Note that human–mink–human transmission is suspected from fur farms, and susceptibility based on angiotensin-converting enzyme 2 (ACE2) receptor sequence variation was predicted, but unconfirmed, for some non-human primates [15., 16., 17., 18.]. Abbreviation: est. n ya, established n years ago; MERS, Middle East respiratory syndrome.
In recent years there have been several emergent coronaviruses. SARS-CoV emerged in 2002–2004 and infected >8000 people across 30 countries [12]. To date, MERS has infected >2000 people across 27 countries and has repeatedly emerged and become extinct in human populations [13]. SARS-CoV-2 has infected >150 million humans globally since emerging in 2019 [14]. Supporting a broad host competency, SARS-CoV-2 has also infected a broad range of hosts, including various carnivores, bats, non-human primates, and rodents [15., 16., 17.], and is likely capable of infecting many more [18] (Figure 1C). Other historical emergences may have gone undocumented [19., 20., 21.].
A range of possible hypotheses might contribute to explaining why the evolution and emergence of coronaviruses – via different pathways – are commonly observed, but with limited species and intraspecies diversity. We first summarize evidence suggesting that difficulties in detecting viral species are alone unlikely to explain this mismatch. Next, multiple lines of evidence suggest that barriers to spillover, or limits to host competency, are unlikely to explain the observed pattern of coronavirus diversity. This motivates the exploration of the downstream processes by which emergent viruses (post-spillover) can become, or fail to become, persistent endemic pathogens. We conclude by noting the urgency for achieving a better understanding of the role that cross-pathogen immunity plays in enabling pathogen emergence, and spotlight future empirical work that might yield insight in this area.
Hypothesis 1: limits of coronavirus detection
A first potential explanation for the apparent mismatch between coronavirus emergence and endemism rates might simply be underestimated endemic viral diversity. For instance, many previous studies on endemic coronaviruses have occurred in wealthy, high-latitude nations [22., 23., 24., 25., 26.], suggesting a general bias in viral discovery [27]. In addition, although existing surveillance efforts identified SARS-CoV-1, MERS, and SARS-CoV-2 soon after their emergence, it is less likely that the timely observation of an endemic virus that is not associated with severe clinical manifestations will occur. However, there is evidence that the rate of viral discovery has slowed, suggesting that current estimates likely capture the broad scale of endemic viral diversity [27,28]. Further, gaps in surveillance likely comprise missing emergent as well as endemic pathogens. For example, emergent pathogen diversity may be underestimated due to the paucity of viral and immunological surveillance in populations where the incursion of humans into previously forested areas is occurring (see hypothesis 2 below). It seems unlikely that global sampling efforts systematically over-detect transient emergence events and also under-detect common pathogens; thus, the rarity of endemic events relative to the frequency of emergence is not likely to be solely an issue of ascertainment.
Hypothesis 2: nonlinear rates of coronavirus spillover in human history
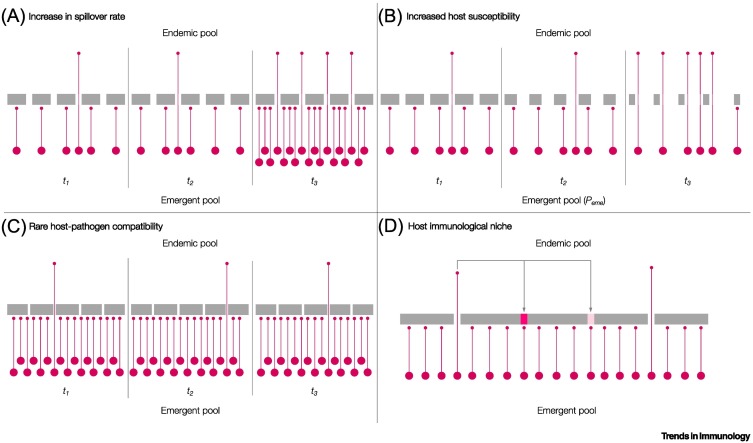
Another explanation may be that viral spillover could have recently accelerated. If the context of human-zoonotic pathogen transmission in the past few decades is substantially different from previous periods in human history, the currently limited endemic pathogen pool might simply reflect historical lower rates of pathogen emergence compared with those seen currently (Figure 2A, Key figure). Notable emergence events, including Zika, Ebola, and influenza H1N1 viruses [29], have occurred often in the past few decades and have been tied to a nonlinear increase in the rate of anthropogenic disruptions of natural systems [30., 31., 32.]. Alternatively, historical spillovers may have occurred frequently, but perhaps remained local, because of a much lower scale of travel relative to current rates [33].
Figure 2.
Key figure. Possible hypotheses to explain the limits of endemic viral diversity.
(A) An increase in spillover over time leads to the movement of more pathogens (shown as pink circles) from the emergent pool to the endemic pool. The filters or barriers to establishment as an endemic pathogen (shown as a gray bar with gaps) are constant; in other words the probability of an emergent pathogen being established remains fixed over time. (B) Host susceptibility increases over time such that a greater proportion of emergent pathogens can exploit widening gaps in immunity. (C) Emergent pathogens are numerous and diverse but the probability of establishment is small owing to narrow requirements for host and pathogen factors that are compatible with persistent transmission. (D) Opportunities for viral emergence depend on previously established pathogens that block or partially block immune niches or gaps via cross-reactive immune responses (e.g., for coronaviruses [84]).
However, epidemics associated with zoonoses going back hundreds of years provide ample evidence of historical spillovers that pre-date the scale of extractive behavior and human contact seen today. The 1918 influenza virus pandemic, as well as many large-scale epidemics in the pre-industrial world (e.g., smallpox [34], measles [35], and others; e.g., the plague [36,37]) indicate that sufficient human connectivity for pathogen spread at large spatial scales has long been present. Indeed, although air-traffic enabled SARS-CoV and SARS-CoV-2 global spread, data indicating the establishment of some endemic coronaviruses hundreds of years ago [9] show that consistent coronavirus spread was possible prior to the rates of (air) travel seen in the recent past.
In the past two decades, coronaviruses have emerged more than once per decade. Even if earlier times were less favorable for viral emergence, a lower rate maintained over the many thousands of years of pre-modern history would still have resulted in many emerging viruses [19,21]. If these emerging viruses persisted, even at low rates, there would be many endemic coronaviruses around today. As a result, lower spillover rates and lower travel rates in the past do not seem to fully explain why only a small number of coronaviruses are endemic in the present day.
Hypothesis 3: host susceptibility to coronavirus infection and changing human populations
Another dramatic change in recent decades has been the rapid increase in the average age of human populations owing to increased lifespan and declining birth rates [38]. All else equal, novel spillover pathogens are more likely to encounter older human hosts. Populations with a higher number of older individuals might be more susceptible to pathogen colonization for two reasons. First, because of original antigenic sin, the immune response to a novel pathogen could be modulated by previous exposure to other, often related, pathogens [39]. During the first challenge with a pathogen-specific antigen, memory B cell responses are generated. Upon subsequent host infection with other pathogens bearing cross-reactive antigens, naïve B cells may be less able to develop new, more specific memory responses to these cross-reactive antigens compared to the first antigen from the first encounter. From the host perspective, these responses might be less effective in preventing infection or onward transmission relative to de novo priming responses. From the pathogen perspective, novel pathogens are likely to encounter heterogeneous host immunity, with adaptive and innate immune responses varying depending on factors such as host age, geography, and historical exposure to other infections (e.g., influenza virus [40]). Second, for instance in humans and mouse model systems, as the host ages, immunosenescence has been associated with depletion of naïve B and T cells capable of recognizing new pathogens; this in turn can lower the defense barrier against pathogen colonization and/or onward transmission (Figure 2B) [41].
Individuals with chronic diseases associated with different immunological states, as well as immunosuppressed individuals, are increasingly prevalent (e.g., because of treatments associated with chemotherapy or owing to HIV-1 infection). The presence of these groups within populations might modify the context of viral emergence by altering immune system functioning and changing the evolutionary selection pressure for mutations associated with transmission or immune escape, as suggested for SARS-CoV-2 [42]. However, various confounding factors could limit the effects of ill, immunosenescent, or immunosuppressed individuals. For example, as in the case of hunters, there might be an association between a given occupation and exposure to a potential spillover – the latter remaining constant regardless of overall demographic trends [43]; this in turn might contribute to minimizing age-related differences in the observed immune responses to pathogens. Nevertheless, although the scale of influence remains uncertain, shifting demographics may have relevant implications for immune patterns at the population level, potentially altering environments for viral emergence.
Hypothesis 4: pathogen-driven limits to endemic infection
Host preference and receptor binding
If contact between two host species occurs, a first requirement for viral spillover is the ability of the virus to invade cells in the new host. A second requirement is the ability to overcome host innate immune defenses such as the secretion of type I interferon (IFN) from immune cells [44,45] to impair completion of a viral replication cycle within the new host. Could these requirements for host–pathogen compatibility explain the limited number of endemic coronavirus species in human populations? (Figure 2C). Many lines of evidence suggest that this is unlikely. Coronaviruses have repeatedly demonstrated the ability to cross the species barrier and replicate successfully; for example, betacoronavirus 1 can replicate in dogs, humans, and numerous ungulate species [46]. In addition, SARS-CoV-2 spillback to pets such as dogs [17], farmed minks [47], and tigers and lions in zoos [48] is unsurprising given the broad host potential for the virus, as evidenced from analyses on the structure of the SARS-CoV-2 receptor-binding domain and its potential to bind to mammalian host cell receptors [49]. In addition, broad and overlapping host ranges may allow genome recombination among viral lineages, which can further diversify the pathogen pool [50,51].
The coronaviruses known to infect humans are relatively phylogenetically distant [9], suggesting that there is no evolutionarily rare, 'special' feature that is necessary to invade human populations. They also use an array of different receptors: MERS-CoV uses dipeptidyl peptidase 4 (DPP4, a ubiquitous cell-surface protease [46]); HCoV-NL63, SARS-CoV-1, and SARS-CoV-2 use angiotensin-converting enzyme 2 (ACE2); HCoV-OC43 and -HKU1 bind to 9-O-acetylated sialic acids [52]; and HCoV-229E uses aminopeptidase N [53]. These receptors have in common that they are widely expressed in human cells, and tend to be evolutionarily conserved among hosts [18,54].
Together, the evidence for considerable host promiscuity among coronaviruses, and the wide range of origins for different coronaviruses infecting humans, suggest that host species boundaries pertaining to receptor binding, cellular tropism, evasion of innate immunity, and replication are not the only limiting factors for coronavirus spillover. If the virus can also successfully complete a replicative cycle in host cells (further discussed under hypothesis 5) the next set of barriers to viral emergence include how viral replication translates into host survival and host-to-host transmission.
Suitable transmission virulence
An ability to successfully invade and replicate in new host cells is still not sufficient to ensure that a virus can successfully persist. It must also achieve sustained onward transmission (Figure 2C). This implies that the virus possesses a set of adaptations that are necessary not only to invade and transmit (e.g., present in droplets exhaled by the host) but also to avoid killing the host too swiftly either from damage inflicted by the virus or from a robust immune reaction [55]. Of note, early morbidity and mortality with a potential to curtail transmission were characteristic of SARS-CoV-1 infections in humans; indeed, early symptomatic presentation was an important feature for the successful containment of this pathogen [56]. By contrast, SARS-CoV-2-associated morbidity and mortality in humans seem to occur after much of the transmission has occurred [57]. This suggests that a wide range of transmission and virulence features are possible among coronaviruses.
The number of new infections per infected individual in a completely susceptible population, R 0, provides one lens to understand the trade-offs between loss of hosts due to mortality and rate of transmission. R 0, also termed the net reproductive value, is defined by the rate of transmission (β) that reflects the rate of increase of infectious individuals, divided by the rates of loss via recovery (γ) or via mortality (μ 1): R 0 = β/(γ + μ 1) [55]. For an emergent pathogen to be established R 0 > 1 is required, and high values of μ 1 will tend to reduce R 0. Because human coronavirus infections are predominantly respiratory (despite being predominantly enteric in animal reservoirs [58]), and disruption of the lung can be rapidly fatal, high virulence may be a particular barrier to coronavirus emergence in human hosts. Indeed, endemic coronaviruses are minimally virulent (i.e., have little effect on mortality or on symptoms that are sufficient to curtail transmission), thus contributing to an R0 estimated to oscillate between 1.7 and 2.2 [59], which enables persistence of the viruses in the population. Of the emergent coronaviruses, R0 has been estimated to be in the range 0.8–1.3 for MERS [60] and between 2–3 for SARS-CoV-1 [61]; these magnitudes, together with their relatively short durations of infectiousness but high virulence, may be in part what has restricted the spread of these two emergent pathogens. However, with an R0 of 2–3, as well as considerable asymptomatic transmission, SARS-CoV-2 has established itself broadly [62]. Furthermore, this magnitude of transmission and duration of infection are such that susceptible host numbers are unlikely to rapidly decrease in frequency. The loss of susceptibles can constitute another potential barrier for emergent pathogens, especially for those that are highly transmissible, because the pathogen may become extinct by chance, when available susceptible hosts become rare after the first wave of infection [63].
Although the similar scale of R0 across all these exemplars hints that there might be biological constraints on transmission (β), evidence of rapid transmission within other reservoirs (e.g., pigs [64,65]) suggests that this is not necessarily the case for all coronavirus infections. Overall, it is difficult to characterize the degree to which this combination of pathogen features presents an important barrier to coronavirus spillover. Among the coronaviruses that have emerged over the past two decades, one (SARS-CoV-2) has had the right combination of features for establishment. This suggests that obtaining the right combination of pathogen features is not an insuperable barrier.
Hypothesis 5: host immunological niche and limits to establishment
Successful emergent pathogens must persistently evade innate and adaptive immunity to establish endemically. Some aspects of coronavirus biology in reservoir hosts may have contributed to the preadaptation of SARS-CoV-2 to evading human innate immunity. For instance, sequence analysis indicates that SARS-CoV-2 and its closest relatives have the lowest CpG dinucleotide rates among all known bat and human coronaviruses, which has been hypothesized to result from selection within particular bat host species or tissues. This low CpG content has been suggested to be a factor that might facilitate human innate immune system evasion by SARS-CoV-2 because CpG-rich sequences, which are relatively rare in the human genome, are targeted by the innate immune system [66,67]. Moreover, similar historical selection of the virus in non-human hosts may have resulted in the ability of SARS-CoV2 to evade IFN pathways within replicative cell cycles, as evidenced from in vitro infection experiments using epithelial cell lines [68]. Presumably, not all spillover viruses have this capacity. Indeed, humans living near caves harboring bats – known reservoirs of non-human SARS-CoV-related viruses – were found to be seropositive but without evidence of continued circulation of those non-human coronaviruses; this in turn indicated that some spillovers do not achieve onward transmission [20]. Likewise, the recent detection of porcine deltacoronavirus (PDCoV) in three children in Haiti further suggests that there may be independent coronavirus spillovers [69].
Beyond innate immunity, a distinctive feature of adaptive immunity is the highly specific targeting of pathogens to which the host has been previously exposed – the principle of immunological memory. This feature might make adaptive immunity initially negligible to novel pathogens, while placing strong selective pressure on endemic pathogens, as evidenced from antigenic drift [70,71]. Antibodies that can neutralize circulating viruses based on their current configuration can drive the spread of variants that are able to escape, or partially escape, this immunity [72]. Well-characterized for influenza virus, similar processes have been reported for endemic coronaviruses [2]. Changes that enhance immune escape can also increase receptor binding in influenza viruses [70] and coronaviruses [3,73], thus increasing the challenge for achieving effective host immune defenses.
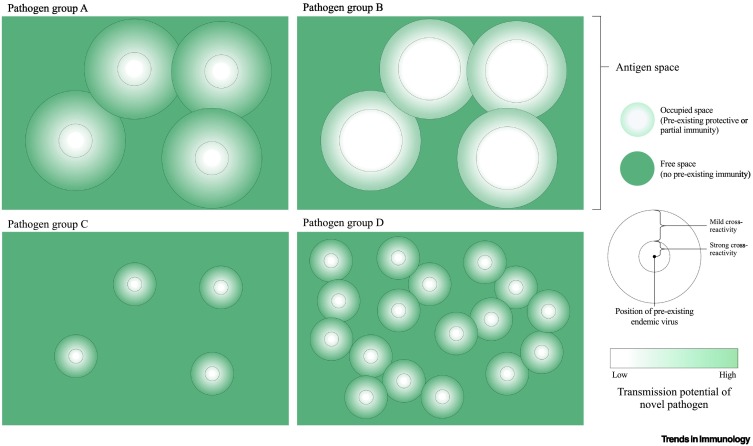
However, cross-reactivity is also a signature of adaptive immunity: an adaptive immune response raised against one pathogen may be protective against another (Figure 2D). For example, reduced endemic coronavirus infections in human populations have been associated with prior influenza virus infection [74], and rhinovirus infections in human bronchial epithelial cells can block SARS-CoV-2 replication [75]; this suggests that prior infection with another pathogen might reduce the colonization and/or establishment of an emergent coronavirus. Pathogens can thus compete with each other for host invasion, limiting the prospects for emergence: early growth of spillover pathogens will no longer be governed by R0 but by R0 × S, where S constitutes the fraction of the population that is left susceptible by the competing species (Figure 3 ) [55]. Indeed, the barrier to monkeypox spillover created by cross-protective immunity associated with smallpox vaccination is a remarkable example of this phenomenon: recent cohorts of human hosts not receiving smallpox vaccination lack this barrier, such that the frequency of monkeypox spillover into these human cohorts is currently increasing over time [76]. Conversely, the absence of cross-protective immunity removes this possible immune barrier to spillover: this may also potentially constitute one explanation for the rich strain diversity that is observed among enteroviruses in humans (Box 1) [77] (also assuming limited behavioral interactions) [78].
Figure 3.
The landscape of variable immune responses against coronaviruses and the concept of 'free antigen space'.
'Antigenic space' can be represented in 2D [112]. Above, a large proportion of possible antigenic phenotypes are within areas of antigenic space featuring some degree of cross-reactivity to existing endemic pathogens. The potential for transmission (e.g., in terms of R0) of a novel pathogen situated near an existing pathogen can vary from low (white) to high (green), and the proportion of 'occupied' space where transmission is strongly reduced can vary from small (group A) to large (group B). Below, for different pathogen groups, each existing pathogen generates an immune response that occupies a smaller proportion of antigenic space, either because of more specific immunity or a larger possible antigenic space. As a result, a greater proportion of antigenic space is 'free' (group C) or more pathogens can occupy the antigenic space without much overlap (group D).
The issue of cross-protective immunity may also provide another angle on why the number of endemic coronaviruses is currently four: competition between species can also influence the species that coexist. Specifically, if cross-protection is broadly symmetrical, then, among those coronaviruses eliciting overlapping crossprotection, the virus with the highest R0 may exclude all the other coronaviruses, including spillover species [79], resulting in a single endemic viral lineage. If differences in R0 are minor (as suggested for coronaviruses [59]), then persistent co-circulation among host populations may depend on how coexisting species avoid sharing a given immunological niche – for example, by evolving away from existing cross-protective immunity – or on how they tolerate sharing the immunological niche – for example, by shifting infection to a different time of year or to a different age group [22].
Immunity against coronaviruses is suggested to be relatively short (e.g., reinfection has been often observed at 12 months for endemic human coronaviruses from multi-decade longitudinal studies) [80., 81., 82.]. In addition, immunity against endemic coronaviruses can be cross-reactive, and (based on analyzing the time-series for cases of coexisting species [59], in conjunction with systems serology approaches identifying convalescent plasma viral neutralization capacity [83]) can be to some extent cross-protective [2,84]. If the immune response elicited by each virus reduces viral growth more than it does for other competing viruses, then coexistence may ensue – assuming similar R0 magnitudes. In addition, differences in R0 might be offset by differences in susceptibility to cross-reactive immunity; specifically, a lineage that is less transmissible but is able to avoid cross-reactive immunity, might harbor an advantage over other lineages subjected to cross-reactive immunity. Moreover, slight differences in seasonal patterns of transmission, associated with small heterogeneities in climate dependencies [85,86], might enable coexistence; for example, by enabling multi-annual cycles where pairs of viruses exhibiting the highest cross-reactivity might reach their maximum incidence in different years. For HCoV-229E and -NL63, this pattern was suggested in time-series studies of human endemic coronaviruses undertaken in Scotland [22], Sweden [23], and Michigan [25]. For larger differences in R 0, differences in the duration and specificity of immune reactions against different viruses might enable partitioning of the immunological niche across human age ranges [87]. However, to date, this remains highly conjectural for human coronaviruses and warrants further investigation [22., 23., 24., 25., 26.,88., 89., 90.].
The evidence of seasonal offsets [22], antigenic drift [2,91], and cross-neutralizing antibodies [24,84,92,93] among endemic coronaviruses is consistent with evolutionary pressure acting on endemic coronaviruses as a result of competition among themselves (Figure 3). That endemic coronaviruses are not entirely offset temporally or antigenically (indicating that opportunities for competition continue) suggests that there might be an upper limit to the degree to which antigenic variation and immune escape can occur, while other aspects of viral fitness are maintained (epistasis). This remains an important open question that is closely related to the issue of which combined circumstances might have created an opportunity for SARS-CoV2 spillover into humans in late 2019. Perhaps it was simply a case of bad luck – the random local extinction of an endemic coronavirus [22] could have created a local, temporary gap in immunity at the place where spillover occurred. It may have occurred by chance: events in the evolutionary history of the virus in the nonhuman reservoir could have led to the antigenic profile of SARS-CoV-2 being such that it evaded existing cross-neutralizing immunity in human populations. Indeed, although antibody cross-reactivity between endemic coronaviruses and SARS-CoV-2 is often detected in human populations [94], no effect on hospitalization has been identified [95], suggesting that existing antibodies might not be protective against severe disease. Similarly, memory T cells in humans without prior exposure to SARS-CoV-2 do cross-react with the virus [96], but there is little evidence that this has slowed the spread of the pandemic [97]; furthermore, cross-reactivity does not seem to be inevitable because T cell responses to endemic coronaviruses do not cross-react with MERS [98]. Overall, multiple lines of evidence suggest opportunities for immune-driven competition among coronaviruses. This competition might act to filter out potential emergent viruses that, unlike successful emergent species (e.g., SARS-CoV-2), occupy an immunological niche for which strong protective cross-reactivity is already common in the population.
Concluding remarks
The existence of four coronavirus species endemic to the human population seems to be a small number compared with the three spillovers that have been observed in the past two decades. Assuming that this is not simply an issue of ascertainment (hypothesis 1) or of greatly changed recent geodemographic conditions (hypotheses 2 and 3), the answer to the question of why there so few coronavirus species endemic to human populations must lie at the intersection of at least three factors: transmission, virulence, and any existing immune cross-reactivity elicited from other coronaviruses. Indeed, to successfully emerge, a new species must thread the needle in terms of virulence and transmission, while also successfully evading existing immunity in the human population.
Of note, techniques for measuring population-level heterogeneity in immunity are rapidly advancing [99,100]. Combined with expanding synthetic biology approaches (e.g., constructing a spectrum of potential coronavirus spike domains), such data might yield insight into the potential viral spillover events that the immune system might successfully keep at bay. Conversely, it may also be possible to assess vulnerabilities, such as gaps in human immunity around other non-human coronaviruses with the potential to emerge (see Outstanding questions). In turn, these data could assist in identifying locations and specific viral lineages for which increased surveillance efforts or pre-emptive vaccine development could be considered. Collectively, such information may be valuable in assessing the SARS-CoV-2 pandemic and any future putative coronavirus spillover/pandemic events; it may also allow better dissection of the relevance and applicability of the five hypotheses presented in this opinion article.
Outstanding questions.
What proportion of coronavirus lineages in the reservoir pool are at a position in antigenic space that is occupied by cross-reactivity to an existing human endemic coronavirus?
Before emergence, can we identify existing wildlife coronaviruses that are (i) able to avoid protective cross-reactivity from endemic coronaviruses, and (ii) have transmission and virulence parameters that permit persistent circulation in human populations?
Why are human coronavirus infections respiratory (at least predominantly) whereas most major animal coronavirus infections are enteric (e.g., swine, canine, and feline CoVs) or pneumoenteric (bovine CoV)? Given the greater fragility of the lung in comparison to the gut, how do tissue tropism and virulence interact to shape emergence potential? Do viral groups infecting different tissue systems have different patterns of species or intraspecies diversity?
Comparing across viral groups, do groups for which immunity against existing pathogens occupies a smaller proportion of antigenic space have more diverse endemic pathogen pools (i.e., do endemic viruses cast narrower cross-reactivity shadows for some groups)?
Alt-text: Outstanding questions
More broadly, this discussion is relevant to fundamental questions in viral biology, such as why only ~200–300 viral species infect humans, out of the large number that are circulating; also, a pressing question concerns why some subgroups of human viruses (e.g., the enteroviruses) are largely diverse, whereas others (e.g., respiratory syncytial virus, RSV), are more restricted (Box 1 and see Outstanding questions). Broadening the comparative context of these analyses might also inform questions in viral ecology. These include examining the role of environmental context in enhancing viral diversity (e.g., a shared wetland habitat can act as a reservoir for influenza virus transmission among waterbirds [101]) and the role of tissue tropism (e.g., the different limits to immunity and pathogen tolerance in the gut versus the lung). Lastly, the non-pharmaceutical interventions that were deployed globally in 2020–2021 may yield additional and fascinating information regarding the coexistence and persistence of endemic coronaviruses in humans.
Acknowledgments
Acknowledgments
This work was supported in part by the PREMISE initiative at the Vaccine Research Center, the intramural program of the NIAID, the NIH, and the Flu Lab.
Declaration of interests
None are declared.
Glossary
- Antigenic drift
evolution, often rapid, of the genetic sequence encoding a pathogen antigen driven by host antibodies against the original sequence.
- Cross-reactivity
an adaptive immune response raised against an antigen from one pathogen which recognizes and may be protective against another.
- Emergent-to-endemic transition
following emergence, the process of attaining sustained and widespread transmission such that the pathogen establishes itself as endemic in a host population.
- Endemic coronavirus species
also referred to as 'common-cold' coronaviruses, HCoV-229E, -NL63, -OC43, and -HKU1 are globally distributed, regularly observed, directly transmitted acute viral species hat infect humans.
- Epistasis
interaction of different genes within the genome such that the overall effect of a mutation in one gene (e.g., a mutation that may allow immune escape) is dependent on other genes (e.g., that are associated with viral replication).
- Immune escape
the ability of new pathogen genotypes to avoid existing immunity.
- Original antigenic sin
a characteristic of the immune system where the response to the original virus infection affects, and often reduces the effectiveness of the antibody response to a second, related virus infection.
- Pathogen pool
the collection of pathogens regularly circulating in a host population.
- Reservoir
a non-human host for a pathogen.
- Reservoir-to-emergent transition
following spillover, the process of initial emergence and spread of a new pathogen in a host population.
- Spillover
exposure to the pathogen pool of an animal reservoir resulting in a new pathogen entering the human population.
References
- 1.Tegally H., et al. Sixteen novel lineages of SARS-CoV-2 in South Africa. Nat. Med. 2021;27:440–446. doi: 10.1038/s41591-021-01255-3. [DOI] [PubMed] [Google Scholar]
- 2.Eguia R.T., et al. A human coronavirus evolves antigenically to escape antibody immunity. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kistler K.E., Bedford T. Evidence for adaptive evolution in the receptor-binding domain of seasonal coronaviruses OC43 and 229e. Elife. 2021;10 doi: 10.7554/eLife.64509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corman V.M., et al. Hosts and sources of endemic human coronaviruses. Adv. Virus Res. 2018;100:163–188. doi: 10.1016/bs.aivir.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chinese SARS Molecular Epidemiology Consortium Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- 6.Walker P.J., et al. Changes to virus taxonomy and the statutes ratified by the International Committee on Taxonomy of Viruses (2020) Arch. Virol. 2020;165:2737–2748. doi: 10.1007/s00705-020-04752-x. [DOI] [PubMed] [Google Scholar]
- 7.Cui J., et al. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su S., et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forni D., et al. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huynh J., et al. Evidence supporting a zoonotic origin of human coronavirus strain NL63. J. Virol. 2012;86:12816–12825. doi: 10.1128/JVI.00906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vijgen L., et al. Evolutionary history of the closely related group 2 coronaviruses: porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43. J. Virol. 2006;80:7270–7274. doi: 10.1128/JVI.02675-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan Y., et al. Molecular epidemiology of the novel coronavirus that causes severe acute respiratory syndrome. Lancet. 2004;363:99–104. doi: 10.1016/S0140-6736(03)15259-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dudas G., et al. MERS-CoV spillover at the camel-human interface. Elife. 2018;7 doi: 10.7554/eLife.31257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong E., et al. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdel-Moneim A.S., Abdelwhab E.M. Evidence for SARS-CoV-2 Infection of animal hosts. Pathogens. 2020;9:529. doi: 10.3390/pathogens9070529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johansen M.D., et al. Animal and translational models of SARS-CoV-2 infection and COVID-19. Mucosal Immunol. 2020;13:877–891. doi: 10.1038/s41385-020-00340-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sit T.H.C., et al. Infection of dogs with SARS-CoV-2. Nature. 2020;586:776–778. doi: 10.1038/s41586-020-2334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melin A.D., et al. Comparative ACE2 variation and primate COVID-19 risk. Commun. Biol. 2020;3:641. doi: 10.1038/s42003-020-01370-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Enard D., Petrov D.A. Ancient RNA virus epidemics through the lens of recent adaptation in human genomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020;375 doi: 10.1098/rstb.2019.0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang N., et al. Serological evidence of bat SARS-related coronavirus infection in humans, China. Virol. Sin. 2018;33:104–107. doi: 10.1007/s12250-018-0012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Souilmi Y., et al. An ancient viral epidemic involving host coronavirus interacting genes more than 20,000 years ago in East Asia. Curr. Biol. 2021 doi: 10.1016/j.cub.2021.05.067. Published online June 17, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nickbakhsh S., et al. Epidemiology of seasonal coronaviruses: establishing the context for the emergence of coronavirus disease 2019. J. Infect. Dis. 2020;222:17–25. doi: 10.1093/infdis/jiaa185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dyrdak R., et al. Interactions between seasonal human coronaviruses and implications for the SARS-CoV-2 pandemic: a retrospective study in Stockholm, Sweden, 2009–2020. J. Clin. Virol. 2020;136 doi: 10.1016/j.jcv.2021.104754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aldridge R.W., et al. Seasonality and immunity to laboratory-confirmed seasonal coronaviruses (HCoV-NL63, HCoV-OC43, and HCoV-229E): results from the Flu Watch cohort study. Wellcome Open Res. 2020;5:52. doi: 10.12688/wellcomeopenres.15812.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monto A.S., et al. Coronavirus occurrence and transmission over 8 years in the HIVE cohort of households in Michigan. J. Infect. Dis. 2020;222:9–16. doi: 10.1093/infdis/jiaa161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heimdal I., et al. Human coronavirus in hospitalized children with respiratory tract infections: a 9-year population-based study from Norway. J. Infect. Dis. 2019;219:1198–1206. doi: 10.1093/infdis/jiy646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woolhouse M., et al. Human viruses: discovery and emergence. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2012;367:2864–2871. doi: 10.1098/rstb.2011.0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woolhouse M.E.J., et al. Temporal trends in the discovery of human viruses. Proc. Biol. Sci. 2008;275:2111–2115. doi: 10.1098/rspb.2008.0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Metcalf C.J.E., Lessler J. Opportunities and challenges in modeling emerging infectious diseases. Science. 2017;357:149–152. doi: 10.1126/science.aam8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plowright R.K., et al. Pathways to zoonotic spillover. Nat. Rev. Microbiol. 2017;15:502–510. doi: 10.1038/nrmicro.2017.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gibb R., et al. Zoonotic host diversity increases in human-dominated ecosystems. Nature. 2020;584:398–402. doi: 10.1038/s41586-020-2562-8. [DOI] [PubMed] [Google Scholar]
- 32.Myers S.S. Planetary health: protecting human health on a rapidly changing planet. Lancet. 2017;390:2860–2868. doi: 10.1016/S0140-6736(17)32846-5. [DOI] [PubMed] [Google Scholar]
- 33.Tatem A.J. Mapping population and pathogen movements. Int. Health. 2014;6:5–11. doi: 10.1093/inthealth/ihu006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duggan A.T., et al. 17th century variola virus reveals the recent history of smallpox. Curr. Biol. 2016;26:3407–3412. doi: 10.1016/j.cub.2016.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Düx A., et al. Measles virus and rinderpest virus divergence dated to the sixth century BCE. Science. 2020;368:1367–1370. doi: 10.1126/science.aba9411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alfani G., Murphy T.E. Plague and lethal epidemics in the pre-industrial world. J. Econ. Hist. 2017;77:314–343. [Google Scholar]
- 37.Bramanti B., et al. The third plague pandemic in Europe. Proc. Biol. Sci. 2019;286 doi: 10.1098/rspb.2018.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bongaarts J. Human population growth and the demographic transition. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2009;364:2985–2990. doi: 10.1098/rstb.2009.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Francis T. On the doctrine of original antigenic sin. Proc. Am. Philos. Soc. 1960;104:572–578. [Google Scholar]
- 40.Lessler J., et al. Evidence for antigenic seniority in influenza A (H3N2) antibody responses in southern China. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goronzy J.J., Weyand C.M. Understanding immunosenescence to improve responses to vaccines. Nat. Immunol. 2013;14:428–436. doi: 10.1038/ni.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kemp S.A., et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021;592:277–282. doi: 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wood J.L.N., et al. A framework for the study of zoonotic disease emergence and its drivers: spillover of bat pathogens as a case study. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2012;367:2881–2892. doi: 10.1098/rstb.2012.0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ströher U., et al. Severe acute respiratory syndrome-related coronavirus is inhibited by interferon- alpha. J. Infect. Dis. 2004;189:1164–1167. doi: 10.1086/382597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McNab F., et al. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peck K.M., et al. Coronavirus host range expansion and Middle East respiratory syndrome coronavirus emergence: biochemical mechanisms and evolutionary perspectives. Annu. Rev. Virol. 2015;2:95–117. doi: 10.1146/annurev-virology-100114-055029. [DOI] [PubMed] [Google Scholar]
- 47.Oude Munnink B.B., et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. 2021;371:172–177. doi: 10.1126/science.abe5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McAloose D., et al. From people to Panthera: natural SARS-CoV-2 infection in tigers and lions at the Bronx Zoo. MBio. 2020;11 doi: 10.1128/mBio.02220-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu L., et al. Broad host range of SARS-CoV-2 and the molecular basis for SARS-CoV-2 binding to cat ACE2. Cell Discov. 2020;6:68. doi: 10.1038/s41421-020-00210-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wells H.L., et al. The evolutionary history of ACE2 usage within the coronavirus subgenus Sarbecovirus. Virus Evol. 2021;7 doi: 10.1093/ve/veab007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li X., et al. Emergence of SARS-CoV-2 through recombination and strong purifying selection. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abb9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hulswit R.J.G., et al. Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A. Proc. Natl. Acad. Sci. U. S. A. 2019;116:2681–2690. doi: 10.1073/pnas.1809667116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yeager C.L., et al. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357:420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu K., et al. Cross-species recognition of SARS-CoV-2 to bat ACE2. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2020216118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anderson R.M., May R.M. Oxford University Press; 1992. Infectious Diseases of Humans: Dynamics and Control. [Google Scholar]
- 56.Fraser C., et al. Factors that make an infectious disease outbreak controllable. Proc. Natl. Acad. Sci. U. S. A. 2004;101:6146–6151. doi: 10.1073/pnas.0307506101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matheson N.J., Lehner P.J. How does SARS-CoV-2 cause COVID-19? Science. 2020;369:510–511. doi: 10.1126/science.abc6156. [DOI] [PubMed] [Google Scholar]
- 58.Saif L.J., Jung K. Comparative pathogenesis of bovine and porcine respiratory coronaviruses in the animal host species and SARS-CoV-2 in humans. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.01355-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kissler S.M., et al. Projecting the transmission dynamics of SARS-CoV-2 through the postpandemic period. Science. 2020;368:860–868. doi: 10.1126/science.abb5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cauchemez S., et al. Middle East respiratory syndrome coronavirus: quantification of the extent of the epidemic, surveillance biases, and transmissibility. Lancet Infect. Dis. 2014;14:50–56. doi: 10.1016/S1473-3099(13)70304-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.May R.M., et al. Epidemiology, transmission dynamics and control of SARS: the 2002–2003 epidemic. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2004;359:1091–1105. doi: 10.1098/rstb.2004.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bar-On Y.M., et al. SARS-CoV-2 (COVID-19) by the numbers. Elife. 2020;9 doi: 10.7554/eLife.57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Restif O., Grenfell B.T. Integrating life history and cross-immunity into the evolutionary dynamics of pathogens. Proc. Biol. Sci. 2006;273:409–416. doi: 10.1098/rspb.2005.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou P., et al. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556:255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edwards C.E., et al. Swine acute diarrhea syndrome coronavirus replication in primary human cells reveals potential susceptibility to infection. Proc. Natl. Acad. Sci. U. S. A. 2020;117:26915–26925. doi: 10.1073/pnas.2001046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nchioua R., et al. SARS-CoV-2 is restricted by zinc finger antiviral protein despite preadaptation to the low-CpG environment in humans. MBio. 2020;11 doi: 10.1128/mBio.01930-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takata M.A., et al. CG dinucleotide suppression enables antiviral defence targeting non-self RNA. Nature. 2017;550:124–127. doi: 10.1038/nature24039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lei X., et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11:3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lednicky J.A., et al. Emergence of porcine delta-coronavirus pathogenic infections among children in Haiti through independent zoonoses and convergent evolution. MedRxiv. 2021 doi: 10.1101/2021.03.19.21253391. Published online March 25, 2021. [DOI] [Google Scholar]
- 70.Hensley S.E., et al. Hemagglutinin receptor binding avidity drives influenza A virus antigenic drift. Science. 2009;326:734–736. doi: 10.1126/science.1178258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wiley D.C., et al. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981;289:373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- 72.Bedford T., et al. Integrating influenza antigenic dynamics with molecular evolution. Elife. 2014;3 doi: 10.7554/eLife.01914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Starr T.N., et al. Deep mutational ccanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nickbakhsh S., et al. Virus–virus interactions impact the population dynamics of influenza and the common cold. Proc. Natl. Acad. Sci. U. S. A. 2019;116:27142–27150. doi: 10.1073/pnas.1911083116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dee K., et al. Human rhinovirus infection blocks SARS-CoV-2 replication within the respiratory epithelium: implications for COVID-19 epidemiology. J. Infect. Dis. 2021;224:31–38. doi: 10.1093/infdis/jiab147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rimoin A.W., et al. Major increase in human monkeypox incidence 30 years after smallpox vaccination campaigns cease in the Democratic Republic of Congo. Proc. Natl. Acad. Sci. U. S. A. 2010;107:16262–16267. doi: 10.1073/pnas.1005769107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pons-Salort M., Grassly N.C. Serotype-specific immunity explains the incidence of diseases caused by human enteroviruses. Science. 2018;361:800–803. doi: 10.1126/science.aat6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rohani P., et al. Ecological interference between fatal diseases. Nature. 2003;422:885–888. doi: 10.1038/nature01542. [DOI] [PubMed] [Google Scholar]
- 79.Shea K., Chesson P. Community ecology theory as a framework for biological invasions. Trends Ecol. Evol. 2002;17:170–176. [Google Scholar]
- 80.Edridge A.W.D., et al. Seasonal coronavirus protective immunity is short-lasting. Nat. Med. 2020;26:1691–1693. doi: 10.1038/s41591-020-1083-1. [DOI] [PubMed] [Google Scholar]
- 81.Galanti M., Shaman J. Direct observation of repeated infections with endemic coronaviruses. J. Infect. Dis. 2021;223:409–415. doi: 10.1093/infdis/jiaa392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Callow K.A., et al. The time course of the immune response to experimental coronavirus infection of man. Epidemiol. Infect. 1990;105:435–446. doi: 10.1017/s0950268800048019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Morgenlander W., et al. Antibody responses to endemic coronaviruses modulate COVID-19 convalescent plasma functionality. MedRxiv. 2020 doi: 10.1101/2020.12.16.20248294. Published online December 18, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Martinez D.R., et al. A broadly neutralizing antibody protects against SARS-CoV, pre-emergent bat CoVs, and SARS-CoV-2 variants in mice. BioRxiv. 2021 doi: 10.1101/2021.04.27.441655. Published online April 28, 2021. [DOI] [Google Scholar]
- 85.Baker R.E., et al. Susceptible supply limits the role of climate in the early SARS-CoV-2 pandemic. Science. 2020;369:315–319. doi: 10.1126/science.abc2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Baker R.E., et al. Assessing the influence of climate on wintertime SARS-CoV-2 outbreaks. Nat. Commun. 2021;12:846. doi: 10.1038/s41467-021-20991-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cobey S., Lipsitch M. Niche and neutral effects of acquired immunity permit coexistence of pneumococcal serotypes. Science. 2012;335:1376–1380. doi: 10.1126/science.1215947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kiyuka P.K., et al. Human coronavirus NL63 molecular epidemiology and evolutionary patterns in rural coastal Kenya. J. Infect. Dis. 2018;217:1728–1739. doi: 10.1093/infdis/jiy098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Petrie J.G., et al. Coronavirus occurrence in the HIVE cohort of Michigan households: reinfection frequency and serologic responses to seasonal and SARS coronaviruses. J. Infect. Dis. 2021;224:49–59. doi: 10.1093/infdis/jiab161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lau S.K.P., et al. Coronavirus HKU1 and other coronavirus infections in Hong Kong. J. Clin. Microbiol. 2006;44:2063–2071. doi: 10.1128/JCM.02614-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lau S.K.P., et al. Molecular epidemiology of human coronavirus OC43 reveals evolution of different genotypes over time and recent emergence of a novel genotype due to natural recombination. J. Virol. 2011;85:11325–11337. doi: 10.1128/JVI.05512-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ng K.W., et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020;370:1339–1343. doi: 10.1126/science.abe1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huang A.T., et al. A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat. Commun. 2020;11:4704. doi: 10.1038/s41467-020-18450-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wiens K., et al. Seroprevalence of severe acute respiratory syndrome coronavirus 2 IgG in Juba, South Sudan, 2020. Emerg. Infect. Dis. J. 2021;27:1598–1606. doi: 10.3201/eid2706.210568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Anderson E.M., et al. Seasonal human coronavirus antibodies are boosted upon SARS-CoV-2 infection but not associated with protection. Cell. 2021;184:1858–1864. doi: 10.1016/j.cell.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mateus J., et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lipsitch M., et al. Cross-reactive memory T cells and herd immunity to SARS-CoV-2. Nat. Rev. Immunol. 2020;20:709–713. doi: 10.1038/s41577-020-00460-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mok C.K.P., et al. T-cell responses to MERS coronavirus infection in people with occupational exposure to dromedary camels in Nigeria: an observational cohort study. Lancet Infect. Dis. 2021;21:385–395. doi: 10.1016/S1473-3099(20)30599-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mina M.J., et al. A Global lmmunological Observatory to meet a time of pandemics. Elife. 2020;9 doi: 10.7554/eLife.58989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xu G.J., et al. Viral immunology. Comprehensive serological profiling of human populations using a synthetic human virome. Science. 2015;348 doi: 10.1126/science.aaa0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rohani P., et al. Environmental transmission of low pathogenicity avian influenza viruses and its implications for pathogen invasion. Proc. Natl. Acad. Sci. U. S. A. 2009;106:10365–10369. doi: 10.1073/pnas.0809026106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Carroll D., et al. The Global Virome Project. Science. 2018;359:872–874. doi: 10.1126/science.aap7463. [DOI] [PubMed] [Google Scholar]
- 103.Carlson C.J., et al. Global estimates of mammalian viral diversity accounting for host sharing. Nat. Ecol. Evol. 2019;3:1070–1075. doi: 10.1038/s41559-019-0910-6. [DOI] [PubMed] [Google Scholar]
- 104.Singh B., Daneshvar C. Human infections and detection of Plasmodium knowlesi. Clin. Microbiol. Rev. 2013;26:165–184. doi: 10.1128/CMR.00079-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hartmeyer G.N., et al. Plasmodium cynomolgi as cause of malaria in tourist to Southeast Asia, 2018. Emerg. Infect. Dis. 2019;25:1936–1939. doi: 10.3201/eid2510.190448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ta T.H., et al. First case of a naturally acquired human infection with Plasmodium cynomolgi. Malar. J. 2014;13:68. doi: 10.1186/1475-2875-13-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Imwong M., et al. Asymptomatic natural human infections with the simian malaria parasites Plasmodium cynomolgi and Plasmodium knowlesi. J. Infect. Dis. 2019;219:695–702. doi: 10.1093/infdis/jiy519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Coatney, G.R. and National Institute of Allergy and Infectious Diseases (US) US National Institute of Allergy and Infectious Diseases; 1971. The Primate Malarias. [Google Scholar]
- 109.Escalante A.A., et al. Evolutionary origin of human and primate malarias: evidence from the circumsporozoite protein gene. Mol. Biol. Evol. 1995;12:616–626. doi: 10.1093/oxfordjournals.molbev.a040241. [DOI] [PubMed] [Google Scholar]
- 110.Gauci P.J., et al. Genomic characterisation of Trubanaman and Gan viruses, two bunyaviruses with potential significance to public health in Australia. Virol. Rep. 2016;6:1–10. [Google Scholar]
- 111.Brook C.E., et al. Accelerated viral dynamics in bat cell lines, with implications for zoonotic emergence. Elife. 2020;9 doi: 10.7554/eLife.48401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Smith D.J., et al. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305:371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]