Abstract
We report the largest and most diverse genetic study of type 1 diabetes (T1D) to date (61,427 participants), yielding 78 genome-wide significant (P < 5 × 10−8) regions, including 36 novel. We define credible sets of T1D-associated variants and show they are enriched in immune cell-accessible chromatin, particularly CD4+ effector T cells. Using chromatin accessibility profiling of CD4+ T cells from 115 individuals, we map chromatin accessibility quantitative trait loci (caQTLs) and identify five regions where T1D risk variants colocalize with caQTLs. We highlight rs72928038 in BACH2 as a candidate causal T1D variant leading to decreased enhancer accessibility and BACH2 expression in T cells. Finally, we prioritize potential drug targets by integrating genetic evidence, functional genomic maps, and immune protein-protein interactions, identifying 12 genes implicated in T1D that have been targeted in clinical trials for autoimmune diseases. These findings provide an expanded genomic landscape for T1D.
Type 1 diabetes (T1D) is characterized by an autoimmune attack on insulin-producing β cells in the pancreatic islets, driven by diverse genetic1–6 and environmental7 factors. Genetic screening and autoantibody surveillance can detect islet autoimmunity before overt progression to T1D8–10, providing an opportunity for prevention. Multiple immune therapies have been explored in clinical trials11. Recently, a 14-day course of teplizumab, an anti-CD3 monoclonal antibody, delayed T1D in high genetic-risk individuals by a median of two years12. This success shows that appropriately timed immune-modulating therapy can alter the autoimmune process preceding disease onset. Defining the genetic variants contributing to T1D risk and how they disrupt immune pathways may lead to more precise therapeutic targets, better characterization of their role in disease initiation and progression, and improved opportunities for safe and effective intervention and, ultimately, prevention of T1D13,14.
Approximately 60 genomic regions have been associated with T1D risk in individuals of European ancestry1–3,15–21. However, less is known in non-European ancestry groups, despite recent increases in T1D diagnoses in these understudied populations22. Additionally, the mechanisms underlying most T1D associations are unknown. We showed previously that T1D credible variants are most strongly enriched in lymphocyte and thymic enhancers3. Yet, resolving causal variants, mapping them to genes, and determining causal mechanisms remains a challenge.
Here, we double the sample size from the previous largest T1D study, genotype ancestrally diverse T1D cases, controls, and affected families, and impute additional variants23. Using this expanded data set, we perform discovery and fine-mapping analyses. In T1D-associated regions, we use chromatin accessibility quantitative trait loci (caQTL) to prioritize credible variants for interrogation of molecular mechanisms underlying T1D association. We present a compelling hypothesis of genetic regulatory mechanism in the T1D locus encoding the transcription factor BACH2. Finally, by integrating implicated genes with immune protein networks, we identify drugs that target T1D candidate genes and networks.
Results
Thirty-six new genome-wide significant regions.
After quality filtering, 61,427 participants (Supplementary Table 1 and Supplementary Fig. 1) and 140,333 genotyped ImmunoChip variants (Online Methods) were included in analyses, providing dense coverage in 188 autosomal regions (“ImmunoChip regions”)24 and sparse genotyping in other regions (Supplementary Tables 2 and 3). Each participant was assigned to one of five ancestry groups using principal component analysis (Online Methods, Supplementary Fig. 2): European (EUR, n = 47,319), African Admixed (AFR, n = 4,290), Finnish (FIN, n = 6,991), East Asian (EAS, n = 588) and Other Admixed (AMR, n = 2,239). Association analyses included 16,159 T1D cases, 25,386 controls and 6,143 trio families (i.e., an affected child and both parents) (Supplementary Tables 4 and 5 and Supplementary Fig. 3). Genotypes at additional variants were imputed using the Trans-Omics for Precision Medicine (TOPMed)23 multi-ethnic reference panel to improve discovery and fine-mapping resolution (Online Methods). After imputation, the number of variants in ImmunoChip regions with imputation R-squared > 0.8 and minor allele frequency (MAF) > 0.005 in each ancestry group was 166,274 (EUR), 322,084 (AFR), 163,612 (FIN), 137,730 (EAS), and 188,550 (AMR). We compared imputed genotypes to whole genome sequencing data from a subset of individuals and observed high concordance (Online Methods, Supplementary Note, Supplementary Figs. 4 and 5).
Initially, we analyzed unrelated cases and controls (n = 41,545), assuming an additive inheritance model. With minimal evidence of artificial inflation of association statistics due to population structure (Supplementary Note, Supplementary Fig. 6, and Supplementary Table 6), we identified 64 T1D-associated regions outside the major histocompatibility complex (MHC, including the HLA loci), including 24 regions associated with T1D at genome-wide significance (P < 5 × 10−8) for the first time. Following conditional analysis, 78 independent associations were identified (P < 5 × 10−8; Supplementary Table 7). On the X chromosome, the most T1D-associated variant was rs4326559 (A>C, C allele OR = 1.09, P = 4.5 × 10−7).
We extended the discovery analysis to incorporate T1D trio families (n = 6,143 trios, some trio families were multiplex and analyzed as multiple trios; Online Methods). Meta-analysis of case-control and trio results identified 78 chromosome regions associated with T1D (P < 5 × 10−8), including 42/43 chromosome regions previously identified in an ImmunoChip-based study3 (rs4849135 (G>T) was P = 2.93 × 10−7). When comparing these 78 regions to previous T1D studies1–3,15–21, 36 novel regions associated with T1D at genome-wide significance for the first time (Table 1). In the remaining 42 regions, the lead variant was within 250 kb of the lead variant in a previous T1D study. The 1q21.3 region, which contains the gene encoding the interleukin-6 receptor (IL-6R), was among the regions associated with T1D at genome-wide significance for the first time. Interestingly, the lead variant in this region was rs2229238 (NC_000001.11:g.154465420T>C, P = 3.02 × 10−9), not the nonsynonymous variant rs2228145 (NC_000001.11:g.154454494A>C; NP_000556.1:p.Asp358Ala; P = 2.20 × 10−4), which was previously suggested to be causal for T1D in targeted analysis25 and remains a candidate causal variant for rheumatoid arthritis26.
Table 1 |. Regions of association with T1D, identified to genome-wide significance (P < 5 × 10−8) for the first time.
Of these 36 regions, 13 had a lead variant that was in strong linkage disequilibrium (r2 > 0.95 in 1000 Genomes Project European population) with variants that are associated with at least one related trait.
| Chr | Position (bp)† | Lead variant rsID | A1 | A2 | Putative candidate gene* | AFEUR (A2) | OR**META | PMETA | Traits with shared association*** |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 63643100 | rs2269241 | T | C | PGM1 | 0.196 | 1.111 | 4.67 × 10−12 | |
| 1 | 92358141 | rs34090353 | G | C | RPAP2 | 0.361 | 1.078 | 1.10 × 10−8 | |
| 1 | 119895261 | rs2641348 | A | G | NOTCH2 | 0.107 | 1.113 | 1.61 × 10−8 | Crohn’s disease, T2D |
| 1 | 154465420 | rs2229238 | T | C | IL6R | 0.813 | 0.896 | 1.38 × 10−12 | |
| 1 | 172746562 | rs78037977 | A | G | FASLG | 0.124 | 0.884 | 2.41 × 10−9 | Asthma, vitiligo, allergic sensitization |
| 1 | 192570207 | rs2816313 | G | A | RGS1 | 0.719 | 1.090 | 4.57 × 10−9 | |
| 1 | 212796238 | rs11120029 | G | T | TATDN3 | 0.147 | 1.102 | 1.82 × 10−8 | |
| 2 | 12512805 | rs10169963 | C | T | AC096559.1 | 0.580 | 1.074 | 2.78 × 10−8 | |
| 2 | 100147438 | rs12712067 | G | T | AFF3 | 0.358 | 0.925 | 4.12 × 10−9 | |
| 2 | 191105394 | rs7582694 | C | G | STAT4 | 0.773 | 0.916 | 2.83 × 10−9 | SLE, hypothyroidism, celiac disease, RA |
| 2 | 241468331 | rs10933559 | A | G | FARP2 | 0.208 | 1.109 | 2.39 × 10−11 | |
| 4 | 973543 | rs113881148 | C | A | TMEM175 | 0.626 | 1.082 | 5.72 × 10−9 | Body fat percentage |
| 4 | 38602849 | rs337637 | G | A | KLF3 | 0.364 | 0.919 | 2.57 × 10−10 | White blood cell count |
| 5 | 40521603 | rs1876142 | G | T | PTGER4 | 0.658 | 0.905 | 2.18 × 10−14 | |
| 5 | 56146422 | rs10213692 | T | C | ANKRD55/IL6ST | 0.241 | 0.912 | 2.85 × 10−9 | RA, Crohn’s disease, MS |
| 6 | 424915 | rs9405661 | C | A | IRF4 | 0.514 | 1.080 | 2.26 × 10−9 | |
| 6 | 137682468 | rs12665429 | T | C | TNFAIP3 | 0.370 | 0.907 | 1.36 × 10−13 | |
| 6 | 159049210 | rs212408 | G | T | TAGAP | 0.638 | 1.112 | 1.42 × 10−15 | MS, Crohn’s disease, eczema |
| 7 | 20557306 | rs17143056 | A | G | ABCB5 | 0.183 | 0.909 | 2.44 × 10−8 | |
| 7 | 28102567 | rs10245867 | G | T | JAZF1 | 0.331 | 0.928 | 3.15 × 10−8 | Eczema, hay fever, MS, SLE, monocyte percentage |
| 8 | 11877675 | rs2250903 | G | T | CTSB | 0.283 | 0.905 | 1.35 × 10−10 | |
| 9 | 99823263 | rs1405209 | T | C | NR4A3 | 0.375 | 1.075 | 3.45 × 10−8 | |
| 10 | 33137219 | rs722988 | T | C | NRP1 | 0.367 | 1.108 | 3.21 × 10−15 | |
| 11 | 35267496 | rs11033048 | C | T | SLC1A2 | 0.366 | 1.091 | 1.53 × 10−10 | Vitiligo |
| 11 | 60961822 | rs79538630 | G | T | CD5/CD6 | 0.035 | 1.213 | 1.14 × 10−9 | |
| 11 | 61828092 | rs968567 | C | T | FADS2 | 0.177 | 0.903 | 8.42 × 10−9 | RA, neutrophil percentage |
| 11 | 64367826 | rs645078 | A | C | CCDC88B | 0.385 | 0.925 | 3.34 × 10−9 | |
| 11 | 128734337 | rs605093 | G | T | FLI1 | 0.470 | 1.077 | 4.25 × 10−9 | |
| 12 | 8942630 | rs1805731 | T | C | M6PR | 0.389 | 1.073 | 4.16 × 10−8 | Eosinophil count |
| 12 | 53077434 | rs7313065 | C | A | ITGB7 | 0.162 | 1.101 | 3.28 × 10−9 | |
| 13 | 42343795 | rs74537115 | C | T | AKAP11 | 0.141 | 1.109 | 5.41 × 10−9 | |
| 14 | 68286876 | rs911263 | C | T | RAD51B | 0.710 | 1.083 | 1.69 × 10−8 | PBC, SLE, RA |
| 16 | 20331769 | rs4238595 | T | C | UMOD | 0.687 | 0.912 | 2.43 × 10−11 | |
| 17 | 45996523 | rs1052553 | A | G | MAPT | 0.232 | 0.879 | 1.65 × 10−15 | Parkinson’s disease |
| 17 | 47956725 | rs2597169 | A | G | PRR15L | 0.348 | 1.081 | 3.35 × 10−9 | |
| 21 | 44204668 | rs56178904 | C | T | ICOSLG | 0.187 | 0.898 | 6.48 × 10−11 |
Genome build 38
Closest gene or gene with mechanistic support from the literature.
Additive odds ratio for the addition of an A2 allele.
Related traits (https://genetics.opentargets.org) where the lead variant is in strong LD (r2 > 0.95 in 1000 Genomes Project European population) with T1D lead variant.
RA, rheumatoid arthritis; T2D, type 2 diabetes; SLE, systemic lupus erythematosus; MS, multiple sclerosis; IBD, inflammatory bowel disease; PBC, primary biliary cholangitis; AF, allele frequency; OR, odds ratio.
Additional regions identified using alternative inheritance models and metric of statistical significance.
Applying the Benjamini-Yekutieli false discovery rate (FDR) < 0.0127 to assess statistical significance, 143 regions were associated with T1D (Supplementary Table 8). Their lead variants overlapped substantially with lead variants for 14 immune-mediated diseases from published studies, but the direction of effects frequently differed between traits (Supplementary Fig. 7). Associated variants with FDR < 0.01 but not meeting genome-wide significance (P < 5 × 10−8) had smaller absolute effect sizes but similar MAFs to those satisfying genome-wide significance (median (IQR) OR = 1.07 (1.06, 1.09) vs. 1.11 (1.09, 1.13); median (IQR) MAF = 0.301 (0.152, 0.397) vs. 0.306 (0.184, 0.374)). These results indicate that remaining regions associated with T1D may have increasingly smaller effect sizes (Supplementary Fig. 8), requiring genome-wide coverage and larger sample sizes for detection.
One exception underscores the need for inclusion of understudied populations to enhance biological insight, even with limited sample sizes, and suggests the potential value of considering alternative metrics for defining statistical significance in genetic studies28. On chromosome 1p22.1 near the Metal Response Element Binding Transcription Factor 2 (MTF2) gene, rs190514104 (NC_000001.11:g.93145882G>A) had a large effect on T1D risk (OR (95% CI) = 2.9 (1.9–4.5); P = 6.6 × 10−7) in the AFR ancestry group. The minor allele (A) at rs190514104:G>A was common in the AFR ancestry group (> 1%) but rare in the others (< 0.1%). Considering the limited sample size, potential heterogeneity of the AFR cohort, and possible over-estimation of effect sizes due to “the winner’s curse”, this association requires replication in an independent cohort.
Use of recessive and dominant models of inheritance identified 35 regions (25 dominant, 10 recessive) with a better fit than the additive model (lower Akaike Information Criterion (AIC) in Europeans) at FDR < 0.01, including nine regions that did not reach FDR < 0.01 under the additive model (Supplementary Table 9). Thus, a total of 152 regions were associated with T1D at FDR < 0.01, 143 under an additive model and nine under recessive or dominant models.
Fine mapping reveals over a third of T1D loci contain more than one independent association.
To define the local architecture of T1D regions, we applied a Bayesian stochastic search method (GUESSFM29) to the European ancestry case-control data (Online Methods, Statistical fine mapping). Of 52 ImmunoChip regions (Supplementary Table 2) associated with T1D, GUESSFM predicted 21 (40%) to contain more than one causal variant (Fig. 1a), compared to nine regions using stepwise conditional regression. In four regions, the lead variant in the discovery analysis was not prioritized by fine mapping (posterior probability < 0.5): 2q33.2 (CTLA4), 4q27 (IL2), 14q32.2 (MEG3) and 21q22.3 (UBASH3A). In these regions, the lead variant likely tags two or more T1D-associated haplotypes that can be identified using GUESSFM but not stepwise logistic regression, a phenomenon observed previously29,30. For example, although stepwise regression analysis in the UBASH3A locus supported a single causal variant (Supplementary Table 7), GUESSFM fine mapping and haplotype analyses indicated that the lead variant in this region, rs11203203 (NC_000021.9:g.42416077G>A), is unlikely to be causal. GUESSFM fine mapping supported a three-variant model (rs9984852 (NC_000021.9:g.42408836T>C), rs13048049 (NC_000021.9:g.42418534G>A) and rs7276555 (NC_000021.9:g.42419803T>C)) (Fig. 1b), which had a better fit than the single variant model (AIC 45073 vs. 45138, Fig. 1c). Haplotype analysis (Online Methods) demonstrated that when rs11203203:G>A is present without the GUESSFM-prioritized variants, there is no effect of rs11203203:G>A on T1D risk (Fig. 1d). Resampling experiments consistently supported two or more causal variants in the region, with at least one of the three GUESSFM-prioritized variants more likely to be causal than rs11203203:G>A (Supplementary Table 10). Given the complexity of association in the UBASH3A region, and likely at many loci, statistical methods designed to use univariable summary statistics alone are not sufficient to explore the genetic architecture of T1D. We provide the comprehensive list of T1D credible variants and haplotype analyses for all 52 fine-mapped regions (Supplementary Table 11, https://github.com/ccrobertson/t1d-immunochip-2020).
Figure 1 |. Fine-mapping T1D regions using a Bayesian stochastic search algorithm.
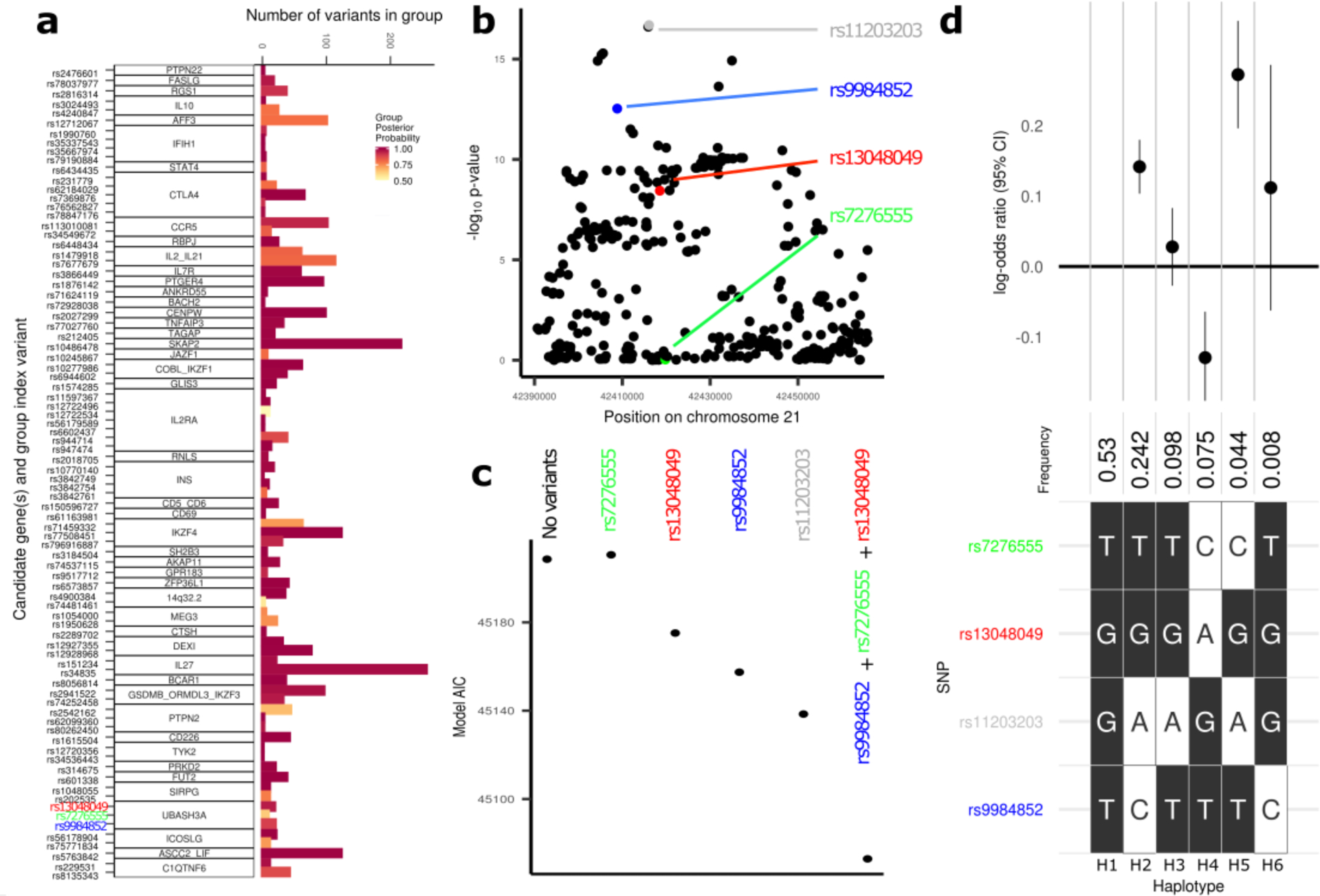
a, Number of variants in GUESSFM-prioritized groups with group posterior probability > 0.5. Candidate gene names and lead variants for each group are shown on the y-axis. b, Manhattan plot of the UBASH3A region from the EUR case-control analysis, highlighting the lead variant from the univariable analysis, rs11203203:G>A (grey), and the three variants prioritized using GUESSFM, rs9984852:T>C (blue), rs13048049:G>A (red) and rs7276555:T>C (green). c, Comparison of model AIC in the UBASH3A region for models fit using EUR cases and controls only, comparing combinations of alleles prioritized either in univariable (grey) or GUESSFM analyses (red, green and blue). d, Analysis of haplotypes associated with T1D in the UBASH3A region. The most common haplotype (H1: T-G-G-T for rs7276555-rs13048049-rs11203203-rs9984852) is presented on the far left; alternative haplotypes (H2-H6) are shown with white squares highlighting the differentiating alleles (C, A, A, or C, respectively). The frequency and effect estimates for association with T1D relative to the baseline haplotype (H1) are shown above the grid (the point and error bars represent the log odds ratio and 95% confidence interval of the log odds ratio, respectively); for example, the log odds ratio for T1D risk for haplotype H3 (T-G-A-T) relative to the baseline haplotype (H1) is close to zero and the 95% confidence interval crosses zero. Haplotype analyses were performed based on n = 33,601 unrelated EUR individuals (13,458 T1D cases and 20,143 controls).
Differences in linkage disequilibrium (LD) between ancestry groups can be advantageous in prioritizing causal variants31. In the 30 regions where analysis suggested a single causal variant, we performed multi-ethnic fine-mapping using PAINTOR32. Eight regions identified an associated variant (P < 5 × 10−4) in more than one ancestry group: five with associations in EUR and FIN, and three with associations in EUR and AFR. In three regions, the number of variants prioritized was markedly reduced by including multiple ancestry groups: 4p15.2 (RBPJ), 6q22.32 (CENPW) and 18q22.2 (CD226) (Fig. 2a, Extended Data Figs. 1 and 2, and Supplementary Table 12). In the chromosome 4p15.2 (RBPJ) region, the credible set from EUR ancestry contained 24 variants. In contrast, using PAINTOR with EUR and AFR summary statistics, only five variants were prioritized with a posterior probability > 0.1 (Fig. 2a). Among these prioritized variants, rs34185821 (NC_000004.12:g.26083858A>G) and rs35944082 (NC_000004.12:g.26093692A>G), both located in the non-coding transcript LINC02357, have the potential to disrupt multiple transcription factor binding motifs33. rs35944082:A>G also overlaps open chromatin in multiple adaptive immune cell types (Fig. 2b) and resides in a FANTOM enhancer site34. Further, rs34185821:A>G is one of three prioritized variants flanking an activation-dependent Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) peak in lymphocytes and a stable response element in human islets35, with potential to perturb an extended TATA box motif36.
Figure 2 |. Fine-mapping of the chromosome 4p15.2 region.
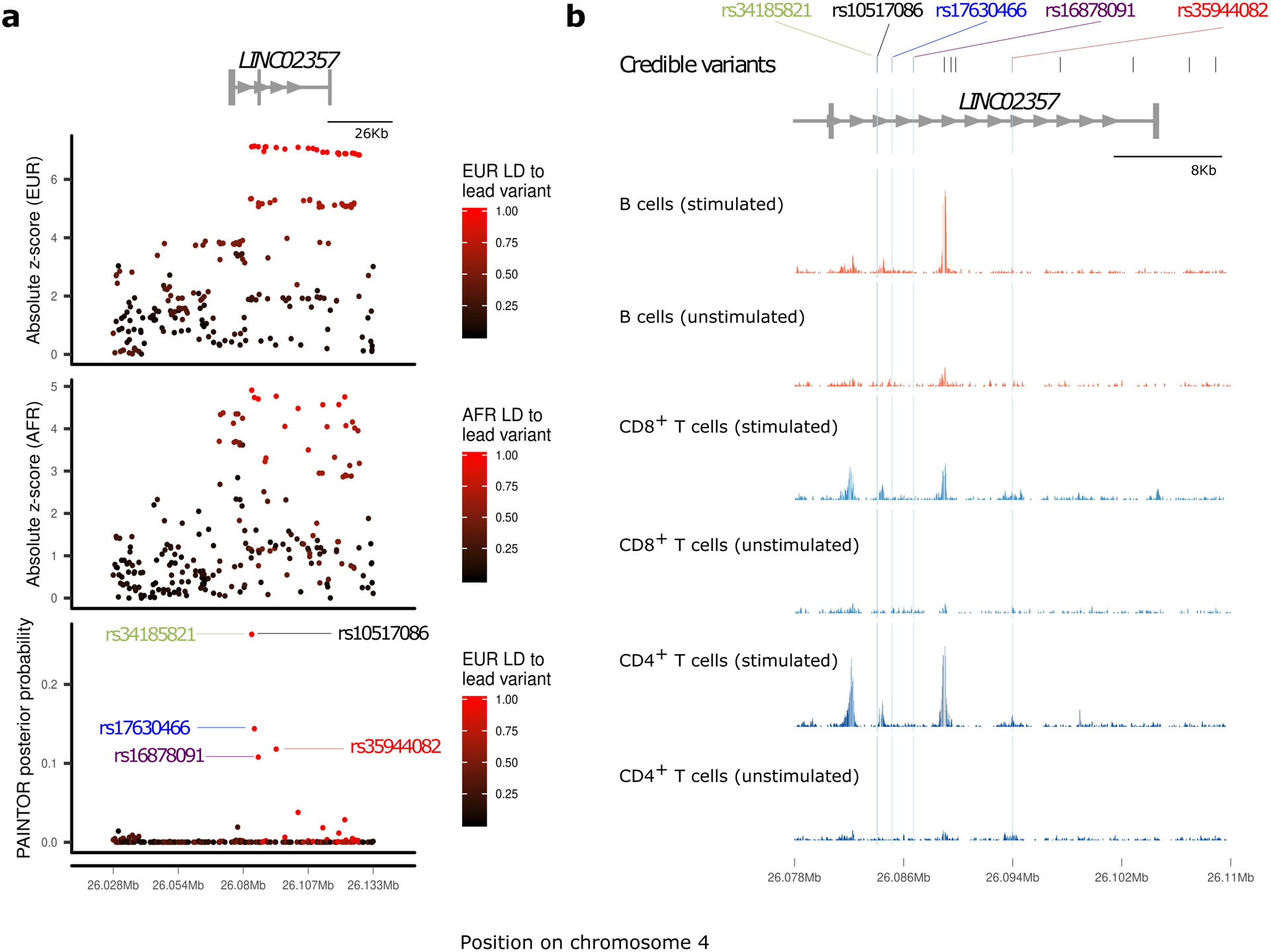
a, European (EUR, top panel) and African (AFR, middle panel) ancestry group association z-score statistics; posterior probabilities (bottom panel) from multi-ethnic fine-mapping of EUR and AFR using PAINTOR; z-scores are colored by linkage disequilibrium (LD) to the lead PAINTOR-prioritized variant. b, Overlay of T1D-credible variants with open chromatin ATAC-seq peaks in immune cells, with variants prioritized by PAINTOR (posterior probability > 0.1) highlighted with blue dashed lines. Normalized ATAC-seq read count shown for effector CD4+ T cells, B cells, and CD8+ T cells, under stimulated and non-stimulated conditions.
T1D-associated protein-altering variants.
Only 34/2,732 (1.2%) credible variants (group posterior probability > 0.5) were protein-altering (nonsynonymous, frameshift, stop-gain, or splice-altering) with 12 having support for a role in T1D (Online Methods, Supplementary Table 13). We identified several previously unreported protein-altering variants as highly prioritized in T1D credible sets (posterior probability > 0.1): a protective missense variant in UBASH3A, rs13048049:G>A (NP_061834.1:p.Arg324Gln; OR = 0.84; AFEUR = 0.051); two low-frequency splice donor variants in IFIH1, rs35732034 (NC_000002.12:g.162268086C>T; OR = 0.63; AFEUR = 0.0089) and rs35337543 (NC_000002.12:g.162279995C>G; OR = 0.61; AFEUR = 0.0099); and a missense variant in CTLA4, rs231775 (NC_000002.12:g.203867991A>G; NP_001032720.1:p.Thr17Ala; OR = 1.20; AFEUR = 0.36).
T1D credible variants are over-represented in accessible chromatin in T and B cells.
ATAC-seq offers a high-resolution map of accessible chromatin with potential regulatory function37. Using publicly available38–40 and newly generated ATAC-seq data from healthy donors, we assessed enrichment (Online Methods) of 2,431 T1D credible variants (group posterior probability > 0.8) in accessible chromatin across diverse immune and non-immune cell types (including 25 primary immune cell types, pancreatic islets, and, as control cell types unlikely to be central to T1D etiology, fetal and adult cardiac fibroblasts). T1D credible variants were enriched in open chromatin in multiple primary immune cell types based on two complementary enrichment analysis approaches (Online Methods, Supplementary Fig. 9), with strong enrichment observed in stimulated CD4+ effector T cells (Supplementary Fig. 9b). There was no enrichment in pancreatic islets (P = 0.14), the primary target of autoimmunity in T1D, even after exposure to proinflammatory cytokines (P = 0.05) or in cardiac fibroblasts (P > 0.60) (Supplementary Fig. 9). We also examined enrichment for T1D credible variants in condition-specific accessible chromatin and observed the largest enrichment in stimulation-specific peaks from effector CD4+ T cells (Supplementary Note, Supplementary Table 14, and Supplementary Fig. 10).
Colocalization of T1D association with QTLs in immune cells.
Chromatin accessibility profiles were generated across 115 participants (nEUR = 48, nAFR = 67) in primary CD4+ T cells, the cell type in which accessible chromatin is most strongly enriched for T1D credible variants (Supplementary Figs. 9 and 10). We examined additive effects of genotype on local chromatin accessibility (cis window < 1 Mb), identifying 11 “peaks” of chromatin accessibility significantly (P < 5 × 10−5) associated with T1D credible variants. Colocalization analysis of T1D association and caQTLs (R package coloc41, Online Methods) identified five regions supporting a common causal variant underlying association with T1D and chromatin accessibility (PP.H4.abf > 0.8; Table 2). In all five regions, at least one T1D credible variant overlapped the caQTL-associated peak. Six of these “within-peak” credible variants were directly genotyped on the ImmunoChip, allowing us to examine allele-specific accessibility in heterozygous participants (Online Methods). At all six variants, the proportion of ATAC-seq reads from heterozygotes containing the alternative allele was consistent with the direction of the caQTL effect (Supplementary Table 15). When integrated with whole blood cis-eQTLs41,42, colocalization identified T1D candidate genes in four of five T1D-caQTL regions (PP.H4.abf > 0.8; Table 2).
Table 2 |. T1D-associations colocalizing with caQTLs in CD4+ T cells.
Five regions show colocalization between T1D and a caQTL with a colocalization posterior probability > 0.8. In all of these regions, at least one T1D credible variant overlaps the caQTL peak itself. In four regions, the T1D association also colocalizes with an eQTL for expression of one or more genes in whole blood.
| T1D lead variant* | BetaT1D ** | Peak | T1D-credible variants in peak | caQTL lead variant* | BetacaQTL ** | PcaQTL | PP | Whole blood cis-eQTLs*** |
|---|---|---|---|---|---|---|---|---|
| rs71624119 (chr5:56144903:G:A) |
−0.099 | chr5:56147972-56149111 | rs7731626 | rs7731626 (chr5:56148856:G:A) |
−0.5 | 2.4 × 10−9 | 0.97 |
ANKRD55 (z = −58; PP = 0.98) IL6ST (z = −10; PP = 0.98) |
| rs72928038 (chr6:90267049:G:A) |
0.172 | chr6:90266766-90267747 | rs72928038 | rs72928038 (chr6:90267049:G:A) |
−1.0 | 3.9 × 10−16 | 1.00 | BACH2 (z = −21; PP = 1) |
| rs2027299 (chr6:126364681:G:C) |
0.147 | chr6:126339725-126340580 | rs9388486 | rs1361262 (chr6:126380821:T:C) |
−0.4 | 2.0 × 10−16 | 0.87 | CENPW (z = −9.8; PP = 0.82) |
| rs61555617†
(chr12:56047884:TA:T) |
0.257 | chr12:56041256-56042638 | rs705704 rs705705 |
rs705704 (chr12:56041628:G:A) |
−0.2 | 1.1 × 10−15 | 0.97 | GDF11 (z = −7.5††; PP = 0.97) |
| rs4900384 (chr14:98032614:A:G) |
0.118 | chr14:98018322-98019163 | rs11628807 rs4383076 rs11628876 rs11160429 |
rs11628807 (chr14:98018774:T:G) |
0.7 | 1.8 × 10−21 | 0.95 | - |
T1D lead variant is the most associated variant in the credible set, as defined by fine mapping (Supplementary Table 11); caQTL lead variant is the most associated variant with chromatin accessibility at the peak of interest. Variants are provided as rsid (chromosome:hg38_position:reference:alternative).
BetaT1D refers to the effect size for the alternative allele of the T1D lead variant; BetacaQTL refers to the effect size for the alternative allele of the caQTL lead variant.
Whole blood cis-eQTL statistics from eQTLGen for the T1D lead variant and colocalization with the T1D association.
rs61555617 is referred to as rs796916887 in supplementary tables
cis-eQTL statistics for rs61555617 are missing in eQTLGen; the reported GDF11 cis-eQTL z-score is for the highly correlated variant rs705704.
PP, posterior probability of colocalization between the QTL (eQTL or caQTL) and the T1D association (referred to in coloc documentation as “PP.H4.abf”); caQTL, chromatin accessibility quantitative trait locus; eQTL, expression quantitative trait locus.
Functional annotation of T1D-associated variants in the BACH2 region.
Fine mapping of the BACH2 locus refined the T1D association to two intronic variants, rs72928038 (NC_000006.12:g.90267049G>A) and rs6908626 (NC_000006.12:g.90296024G>T) (Fig. 3a). The EUR minor alleles of rs72928038:G>A and rs6908626:G>T are associated with increased T1D risk (OR = 1.18; P < 1 × 10−20, MAFEUR = 0.18). Chromatin-state annotations across cell types from the BLUEPRINT Consortium and NIH Roadmap Epigenomics Project annotate rs72928038:G>A as overlapping a T cell-specific active enhancer and rs6908626:G>T as lying in the ubiquitous BACH2 promoter (Fig. 3b). Promoter-capture Hi-C data from diverse immune cell types43 indicates that the enhancer region containing rs72928038:G>A contacts the BACH2 promoter in T cells (Fig. 3c). Although weak interactions were observed in multiple T cell subtypes, only naïve CD4+ T cells had a significant interaction score.
Figure 3 |. Functional annotation of T1D-associated variants in the BACH2 region.
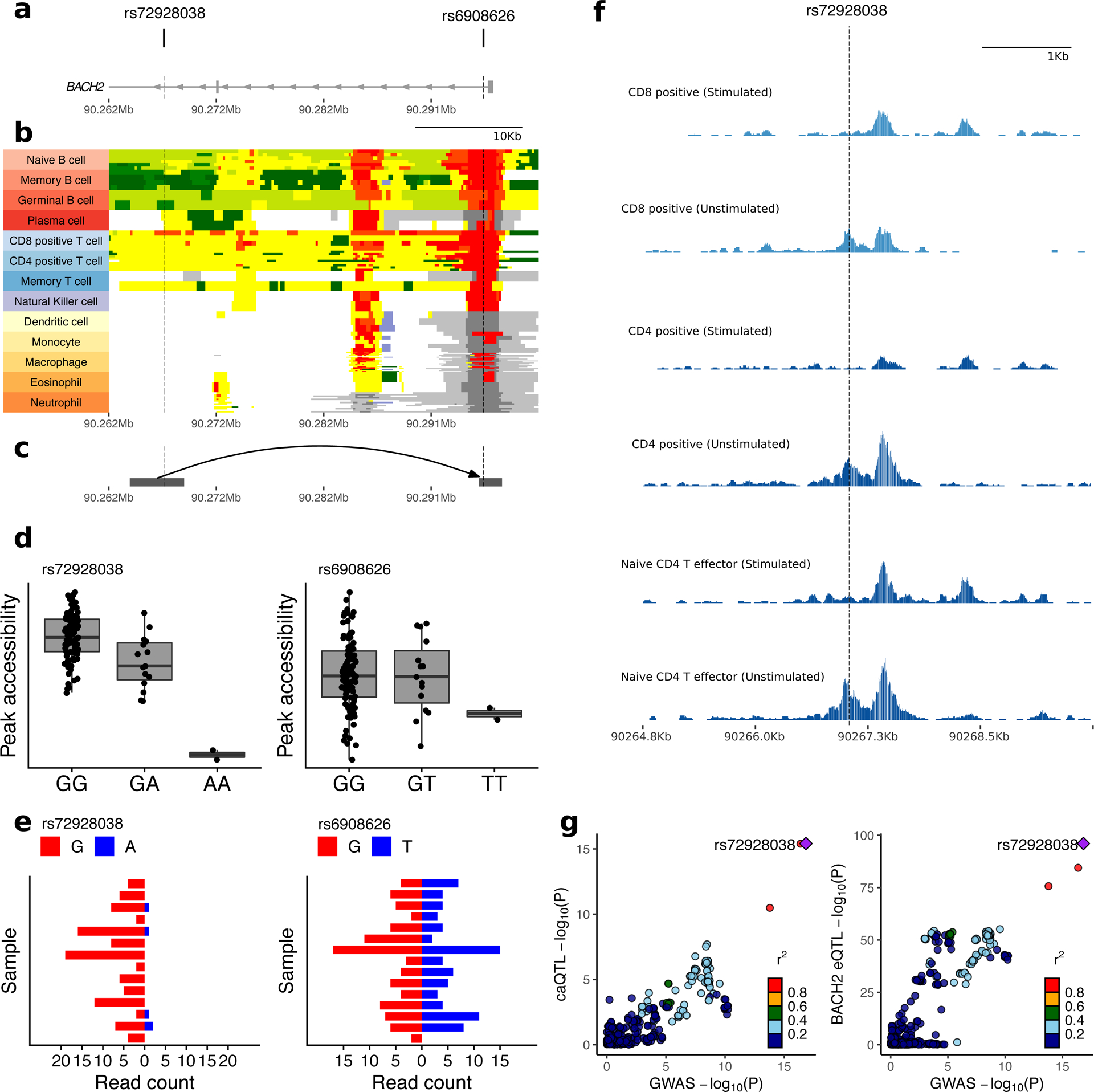
a–c, Position of T1D credible variants (rs72928038:G>A and rs6908626:G>T) relative to introns and exons of BACH2 (a), chromHMM tracks across diverse immune cell types from the BLUEPRINT consortium (red, active promoter; orange, distal active promoter; dark green, transcription; light green, genic enhancer; yellow, enhancer; white, quiescent; light grey, Polycomb repressed; dark grey, repressed; blue, heterochromatin) (b), and interactions with the BACH2 promoter in published PCHi-C data from naïve CD4+ T cells43 (grey squares indicate boundaries of target (left) and bait (right)) (c). Chromatin coordinates and scale are identical and aligned in figures a–c. d, Accessibility of regions overlapping rs72928038:G>A and rs6908626:G>T by genotype; peak accessibility is quantified as normalized transposase cut frequency (Online Methods); center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range (n = 115 individuals). e, Allele-specific accessibility of chromatin within heterozygous individuals at rs72928038:G>A (n = 14 heterozygous individuals) and rs6908626:G>T (n = 15 heterozygous individuals). f, Chromatin accessibility profiles in the region overlapping rs72928038:G>A across resting and activated CD4+ and CD8+ T cells (published data38). Height of tracks represent transposase cut frequency; all tracks are plotted using the same vertical scale. g, LocusCompare plots showing colocalization between T1D association, the caQTL for chr6:90266766–90267715 (left), and the eQTL for BACH2 (right).
In caQTL analysis, rs72928038:G>A is associated with decreased accessibility of the enhancer it overlaps (chr6:90266766–90267715) (Fig. 3d, left), while rs6908626:G>T does not affect accessibility at the BACH2 promoter (chr6:90294665–90297341) (Fig. 3d, right). Similarly, among 14 subjects heterozygous for rs72928038:G>A, only 4% (5/121) of ATAC-seq reads overlapping that site contain the T1D risk allele (A) (Fig. 3e, left, and Supplementary Table 15), suggesting it leads to restricted accessibility. In contrast, chromatin accessibility at rs6908626:G>T does not exhibit allelic bias in heterozygotes (Fig. 3e, right). These data help to prioritize rs72928038:G>A, rather than rs6908626:G>T, as functionally relevant in CD4+ T cells.
In eQTL studies, rs72928038:G>A is associated with decreased expression of BACH2 in whole blood42 and purified immune cell types44. In the DICE consortium44, rs72928038:G>A is associated with decreased expression of BACH2 in multiple cell types, with the strongest effects in naïve CD4+ and CD8+ T cells. This result is consistent with the observation that the enhancer region overlapping rs72928038:G>A is accessible specifically in unstimulated bulk CD4+, unstimulated bulk CD8+, and naïve CD4+ T effector cells (Fig. 3f). Both the enhancer caQTL and BACH2 eQTL colocalize with T1D association (Fig. 3g and Table 2).
The BACH2 rs72928038:G>A variant overlaps binding sites for STAT1 and the ETS family of transcription factors, based on canonical transcription factor binding motifs33. We performed super-shift electrophoretic mobility shift assay (EMSA) experiments of the DNA sequence flanking rs72928038:G>A that demonstrated allele-specific ETS1 binding, but no STAT1 binding (Supplementary Fig. 11). This result builds on experiments demonstrating allele-specific nuclear protein binding of rs72928038:G>A in Jurkat cells45. These data prioritize rs72928038:G>A as a likely functional variant in T cells and provide preliminary support for a candidate regulatory mechanism underlying the 6q15 region association with T1D. Specifically, we hypothesize that the rs72928038:G>A minor allele (A) disrupts ETS1 binding, which leads to decreased enhancer activity and BACH2 expression in naïve CD4+ T cells.
T1D drug target identification.
To identify potential T1D therapeutic targets with human genetic support, we used the Priority Index (Pi) algorithm46, which integrates genetic association results with genome annotations, regulatory maps, and protein-protein networks (Online Methods). Using improved T1D association statistics and additional eQTL resources from whole blood42, we identified 50 highly-ranked gene targets (Supplementary Table 16). These targets include 26 “seed genes” (implicated by T1D-associated loci through proximity, eQTL effects, or chromatin looping) and 24 non-seed genes (not in T1D regions but highly connected to T1D seed genes in immune protein networks). Although we excluded variants in the MHC region from algorithm input, the networks implicated by non-MHC seed genes led to prioritization of HLA-DRB1, an established T1D risk factor. Among the top 50 gene targets, 13 were not previously implicated by Pi analyses (STAT4, RGS1, CXCR6, IL23A, PTPN22, NFKB1, MAPK3, EPOR, DGKQ, GALT, IL12RB1, IL12RB2, IL6R)46, while 12 have been targeted in clinical trials for autoimmune diseases (IL2RA, IL6ST, IL6R, TYK2, IFNAR2, JAK2, IL12B, IL23A, IL2RG, JAK3, JAK1 and IL2RB). T1D susceptibility alleles may alter expression of gene targets in either direction, and gene regulatory effects may be seen across multiple major immune cell populations or be restricted to a single cell type (Supplementary Fig. 12). For example, T1D risk alleles are associated with increased expression of MAPK3 and DGKQ but decreased expression of TYK2 across multiple major immune cell populations. In contrast, risk alleles decrease expression of RGS1 across most immune cell types but increase expression specifically in CD8+ T cells. The directionality and cell type-specificity of gene regulatory effects associated with T1D risk alleles may inform therapeutic target considerations.
Discussion
In the largest genetic analysis of T1D to date, we identified 36 novel regions at genome-wide significance and implicated a total of 152 regions outside the MHC in T1D susceptibility at FDR < 0.01. We refined the set of putative causal variants and number of independent associations in many T1D regions through increased sample size, dense genotyping and imputation, inclusion of diverse ancestry groups, and optimized analytical approaches to fine mapping. We assessed the intersection of T1D-associated variants with regions of putative regulatory function with public and newly generated ATAC-seq data from diverse cell types and states, demonstrating that T1D credible variants were enriched in stimulation-responsive open chromatin peaks in CD4+ T cells. We assessed colocalization of T1D associations with CD4+ T cell caQTLs to generate mechanistic hypotheses centered on this highly relevant cell type. Finally, we identified potential T1D drug targets for use in prevention trials. Experimental follow-up studies are required to test these hypotheses and further dissect the mechanisms altering T1D risk in each region.
Despite enrichment of credible variants in CD4+ T cell open chromatin, only five of 52 fine-mapped T1D associations could be explained by a colocalized caQTL. This result is consistent with work exploring functional effects of variants associated with immune traits47. One explanation is limited power in QTL discovery due to small sample sizes or imprecise cell types47,48. Analysis of more refined cell types, for example using single cell approaches, for both enrichment analyses and QTL discovery may lead to additional discoveries49,50. Nevertheless, while this approach may lack sensitivity, the five regions showing colocalization between caQTL and T1D associations prioritize variants with regulatory effects that represent realistic targets for experimental follow-up. In particular, within-peak credible variants with consistent caQTL effects and allele-specific accessibility, while not definitively causal, provide high priority candidate variants for functional follow-up. As four of the five T1D associations that colocalize with caQTLs also colocalize with whole-blood eQTLs, these regions offer hypotheses for how causal variants influence disease risk through their effects on regulatory element activity and gene expression in T1D-relevent cell types.
In the 5q11.2 region, fine mapping and caQTL colocalization point to the within-peak variant, rs7731626 (NC_000005.10:g.56148856G>A), as a potential causal variant for T1D. This result complements a recent regulatory QTL fine-mapping study that highlighted the same variant as likely functional in T cells51. Additionally, the T1D association colocalizes with eQTLs for both ANKRD55 and IL6ST, mirroring results in multiple sclerosis, Crohn’s disease, and rheumatoid arthritis47. The region overlapping rs7731626:G>A loops to the IL6ST promoter in CD4+ T cells, according to promoter capture Hi-C data43. Although we did not find evidence that rs7731626:G>A loops to the canonical transcription start site for ANKRD55, nascent RNA-sequencing data suggest it overlaps the 5’ end of the transcriptionally active region of ANKRD55 in human T cells52, consistent with a potential regulatory role.
We highlight the BACH2 region on chromosome 6q15 as an example of unbiased QTL colocalization that leads to hypotheses for functional mechanisms driving variant-T1D association. We hypothesize that rs72928038:G>A, the T1D-associated allele, abolishes ETS1 binding at an enhancer that promotes BACH2 expression in naïve CD4+ T cells. BACH2 encodes the transcription factor from the BTB-basic leucine zipper family, BACH2, which has established roles in B and T cell biology, including maintaining the naïve T cell state53,54. BACH2 haploinsufficiency has been shown to cause congenital autoimmunity and immunodeficiency55, demonstrating that a functioning human immune system depends on BACH2 expression in a dose-dependent manner. In addition to cis-effects on BACH2 expression, rs72928038:G>A is associated with altered expression of 39 distal genes42 in whole blood, including seven genes in autoimmune disease-associated regions. These observations raise the hypothesis that the minor A allele at rs72928038:G>A increases T1D risk by reducing BACH2 expression in a precise cellular context (e.g., the naïve T cell state). This effect may lead to shifts in BACH2-regulated transcriptional programs, thereby altering T cell lineage differentiation in response to antigen exposure.
Previous studies demonstrated shared genetic risk across autoimmune diseases3,56 and suggest potential for repurposing drugs to treat or prevent T1D. Our Pi analysis identified 12 targets that have been the focus of clinical trials for treatment of autoimmune diseases. One example is IL23A, which has been successfully targeted in the treatment of inflammatory bowel disease (IBD)57 and psoriasis58. The IL-23 inhibitors are being explored for use in T1D (ClinicalTrials.gov identifiers NCT02204397 and NCT03941132). Our results provide genetic support for these trials. Similarly, JAK1, JAK2 and JAK3 were implicated in T1D etiology in our analysis. JAK inhibitors are safe and effective in the treatment of rheumatoid arthritis59 and ulcerative colitis60. Finally, this study presents the first well-powered, convincing genetic evidence linking interleukin-6 (IL-6), a cytokine with known roles in multiple autoimmune diseases, to T1D etiology. The IL-6 receptor complex consists of two essential subunits: the alpha subunit (encoded by IL6R) and the signal transducing subunit (encoded by IL6ST). Both the IL6ST and IL6R regions were identified here as T1D-associated at genome-wide significance for the first time (Table 1), and both IL6ST and IL6R were prioritized by the Pi analysis. IL6ST is implicated by QTL colocalization, and the lead T1D variant near IL6R (rs2229238:T>C) is an eQTL for IL6R expression in whole blood (formal colocalization was not assessed as the IL6R region is not densely covered by the ImmunoChip). We cannot say, based on current evidence, that IL6ST and IL6R are T1D causal genes. The associations in each region may be unrelated and due to different causal genes; for example, the association near IL6ST also colocalizes with an eQTL for ANKRD55. However, we note that the humanized IL-6 receptor antagonist monoclonal antibody, tocilizumab, is an approved treatment for rheumatoid arthritis and systemic juvenile idiopathic arthritis, both of which share substantial genetic effects with T1D3 (Supplementary Fig. 7), and a trial of this drug in recently diagnosed T1D cases is underway (ClinicalTrials.gov identifier NCT02293837). Surprisingly, we showed that the lead T1D variant near IL6R (rs2229238:T>C) tags a causal variant distinct from the nonsynonymous variant in IL6R, rs2228145:A>C (NP_000556.1:p.Asp358Ala), thought to drive the association in rheumatoid arthritis26, suggesting potentially different mechanisms altering disease risk in this region. The recent success of anti-CD3 therapy, after 40 years of study through experimental models and clinical trials targeting different patient subgroups and time points relative to disease diagnosis61, highlights both the challenges and hopes for translating target identification to efficacious clinical outcomes in T1D.
One limitation of this study is that genotyping was restricted to ImmunoChip content, which provides dense coverage in 188 immune-relevant genomic regions, as defined by previous largely European ancestry-based GWAS of immune-related traits. This design restricts the scope of discovery, fine mapping, and generalizability of subsequent functional enrichment analyses. This may explain the absence of T1D variant enrichment in open chromatin of non-immune cell types (e.g., pancreatic islets)62,63. Additionally, the effect sizes of novel loci are likely over-estimated due to winner’s curse, particularly those identified in non-European ancestry groups where sample sizes remain small, such as rs190514104:G>A near MTF1. We also acknowledge the possibility of results in non-European ancestry groups being confounded by admixture. While this analysis is the largest and most comprehensive study prioritizing novel gene targets in T1D according to genetic evidence, extension of future genetic studies to genome-wide analyses28 and continuing efforts to expand cohorts from diverse populations will further define the genetic landscape of T1D.
Online Methods
Genotyping and quality control.
DNA samples were genotyped on the Illumina ImmunoChip at University of Virginia (UVA) Genome Sciences Laboratory (n = 52,219), Sanger Institute (n = 4,347), University of Cambridge (n = 2,941), and Feinstein Institute (n = 1,811). Raw genotyping files were assembled at UVA. Genotype clusters were generated using the Illumina GeneTrain2 algorithm. Stringent SNP- and sample-level quality control filtering and data cleaning was performed to ensure high quality genotypes and accurate pedigrees (Supplementary Fig. 1). The following variant filters were applied: (1) re-annotated ImmunoChip variant positions by aligning probe sequences to GRCh37 and removed any variants with <100% match or multiple matches at different positions in the genome; (2) removed variants with call rates <98%; (3) removed variants with any discordance between duplicate or monozygotic twin samples, as confirmed by genotype-inferred relationships; (4) removed variants with Mendelian inconsistencies in >1% of informative trios or parent-offspring pairs, based on genotype-inferred relationships.
For sample filtering, we used X chromosome heterozygosity and Y chromosome missingness to identify and exclude participants with apparent sex chromosome anomalies or resolve inconsistencies with reported sex. Pedigree-defined and genotype-inferred sample relationships were compared using KING version 2.1.364. Samples were excluded when inconsistencies could not be resolved, including relationships between families, within and across cohorts. For each pair of related families observed, we randomly selected one to remove from association analysis. After resolving sex and relationship issues, samples with genotype call rate < 98% were removed. Variants with genotype frequencies deviating from Hardy-Weinberg Equilibrium (P < 5 × 10−5) in unrelated European ancestry controls were excluded before imputation.
Stratification of major ancestry groups and family trios.
Principal components (PC) were generated in 1000 Genomes phase 3 individuals using 8,297 autosomal ImmunoChip variants selected by excluding regions of long-range linkage disequilibrium (LD)65, pruning for short-range LD (r2 < 0.2 in 50-kb windows), and filtering for minor allele frequency (MAF) > 0.05). Participant genotypes were projected onto the 1000 Genomes PC space using PLINK v1.966. The first ten PCs were used in k-means clustering to define five clusters of ancestrally similar participants, European (EUR), African-American (AFR), East Asian (EAS), Finnish (FIN), and Admixed (AMR), labeled according to their closest 1000 Genomes super-population. For case-control analyses to be performed within each ancestry cluster, affected trios were excluded and a set of unrelated individuals was selected from the remaining subjects using KING version 2.1.3 software (“--unrelated” option)64. Cluster-specific PCs were calculated by performing PC analysis on unrelated controls and projecting the remaining subjects onto the resulting axes. Remaining population stratification within each ancestry cluster was assessed visually (Supplementary Fig. 3).
Defining targeted regions for discovery and fine-mapping analysis.
The ImmunoChip densely covered genetic variation in immune-associated genomic regions. Discovery analyses included all genotyped variants, as well as imputed variants from any 500-kb region that contained more than 50 genotyped variants (Supplementary Table 3). To define boundaries for fine-mapping regions, we mapped previously defined “ImmunoChip regions” (provided by the R package humarray) from GRCh36 to GRCh38 coordinates (Supplementary Table 2): for each region, we mapped all variants originally included in the region to GRCh38 to define boundaries as the lowest and highest observed GRCh38 positions among these variants (+/− 50 kb either side). Fine-mapping analyses were then restricted to densely genotyped regions overlapping these “ImmunoChip regions”.
Association analysis – Phase I (case-control analyses).
Genotypes were imputed with the NHLBI Trans-Omics for Precision Medicine (TOPMed) Freeze 5 (Supplementary Note) reference panel. We analyzed association with type 1 diabetes (T1D) for all genotyped variants and high-confidence imputed variants separately in the five ancestry groups (Supplementary Tables 2 and 3 and Supplementary Note). Assuming an additive mode of inheritance, we used logistic regression for unrelated case-control analyses, adjusting for five ancestry-specific PCs and using genotype posterior probabilities to account for uncertainty in imputed genotypes using the SNPTEST version 2.5.4 software67. Due to small sample size (38 cases and 106 controls), EAS subjects were excluded. We combined results using an inverse-variance weighted fixed-effects meta-analysis (METAL software version released on 2011-03-25)68. Forward stepwise logistic regression was performed to identify loci with more than one independent association with T1D. All conditionally independent associations (P < 5 × 10−8) were reported. Case-control analyses were performed under recessive and dominant models of inheritance. To evaluate the relative fit of the three models, we compared the Akaike Information Criterion (AIC) in EUR ancestry and identified the model providing the lowest AIC (best fit). On the X chromosome, only genotyped variants were examined for their association with T1D. The Y chromosome was not examined.
Association analysis – Phase II (trio families and combined analyses).
Trio families (two parents and an affected offspring) were analyzed within ancestry group using the transmission disequilibrium test (TDT)69. As TDT statistics are susceptible to substantial bias when applied to imputed genotypes70, a stringent variant filter was applied to imputed genotypes, removing all variants with Mendelian inconsistencies in >1% of trios with heterozygous offspring or parent-offspring pairs with homozygous offspring. From TDT summary statistics, we derived effect sizes and standard error estimates71 and meta-analyzed with Phase I results.
Statistical fine mapping.
Two complementary approaches were used to define credible variant sets within each T1D-associated ImmunoChip region. Fine mapping included high-confidence variants within 750 kb of the lead variant (1.5-Mb region total), usually consisting of imputed variants across the entire ImmunoChip region and genotyped variants adjacent to the ImmunoChip region.
Fine mapping using European case-control data only (GUESSFM).
Since forward stepwise model selection can fail to identify complex genetic architectures72, we applied a Bayesian method (GUESSFM, see Supplementary Note) in the EUR case-control data to identify the most likely combinations of variants explaining T1D risk29,73. In the results, we refer to groups of variants prioritized by GUESSFM as “credible sets” and variants within these groups as “credible variants”. Variants that failed quality control metrics (or were not genotyped or imputed in our data for other reasons) but are in LD (r2 > 0.9 in 1000 Genomes Phase 3) with a prioritized variant were included in the comprehensive list of credible variants (Supplementary Table 11).
Trans-ethnic fine mapping.
In regions where association signals were marginally associated (P < 5 × 10−4) in multiple ancestry groups and evidence from EUR ancestry only fine mapping suggested a single causal variant (marginal posterior probability for one causal variant in the region > 0.5), we applied the multi-ethnic fine-mapping method, PAINTOR32, to refine the association. PAINTOR uses association z-scores and population-level LD to identify the combination of alleles that best explain the phenotype, multiplying the posterior probability of the causal vector across ancestry groups, assuming the same variant(s) are causal in each ancestry group. Since loci examined were those with evidence of one causal variant in the region, we restricted the maximum model size to two variants in the region and enumerated the posterior of every model, rather than performing an MCMC search. The association z-scores used for each ancestry group were from a meta-analysis of case-controls and family trios in that ancestry cluster. PAINTOR input LD reference panels were generated separately for each ancestry group with LDstore version 1.174 using imputed genotype data from unrelated cases and controls.
Haplotype analyses.
Haplotype analyses were performed in the EUR ancestry cases and controls by taking “best-guess” genotype values for the variants included in the analysis and obtaining haplotype phase distribution estimates for each individual, using an expectation-maximization algorithm75. Each individual’s haplotype was sampled ten times and a logistic regression was fitted estimating the effect size of the haplotype relative to the most common haplotype in the population, with T1D status as the outcome and adjusting for five PCs. The estimates and standard errors for each haplotype relative to the most common were averaged over the ten logistic regression models to obtain overall haplotype effect sizes on T1D risk.
Annotating T1D-associated protein-altering variants.
The functional impacts of T1D credible variants (Supplementary Table 11) were annotated using ANNOVAR (version released on 16 April 2018)76 and the Ensembl and refGene annotation databases.
Generating representative cell type- and condition-specific chromatin accessibility profiles.
We downloaded publicly available ATAC-seq data from diverse immune cell types38, pancreatic islets35, and cardiac fibroblasts40 (see Data Availability statement).
We generated additional ATAC-seq data on CD4+ T cells (n = 6 donors) and CD19+ B cells (n = 4 donors), using different culture and stimulation conditions from Calderon et al.38. CD4+ T cells were enriched and stimulated as previously described77. B cells were positively selected from PBMCs using anti-CD19 beads (Miltenyi Biotec, GmbH) and cultured for 24 hours in X-VIVO 15 (Lonza, Switzerland) supplemented with 1% Human Ab Serum (Sigma) and penicillin/streptomycin (Thermo Fisher) and plated in 96-well CELLSTAR U–bottomed plates (Greiner Bio-One, Austria) at concentration of 2.5 × 105 cells/well. Cells were left untreated or stimulated with 10 μg/ml goat anti-human IgM/IgG/IgA antibody (109‐006‐064, Jackson Immunoresearch), 0.15 μg/ml rhCD40L (ALX-522-110-C010, ENZO Lifesciences), 20 ng/ml rhIL-21 and rhIL-4 (200-21 and 200-04 respectively, Peprotech) for 24 hours. ATAC-seq data was generated from 50,000 cells from each cell type and culture condition following the Omni-ATAC protocol78. ATAC-seq datasets were mapped to GRCh38.p1279 with minimap2 (version 2.17)80, except for GSE123404 (pancreatic islets dataset) where bowtie2 (version 2.3.5) was used. After mapping, the technical replicates (where available) were merged and PCR duplicated reads were detected with Picard tools (version 2.20.2). The percentage of detected duplicated reads was very low (mean value < 1%) in all datasets. bigWig files were generated with bamCoverage from the deeptools package (version 3.3.0), using reads per genome coverage (RPGC) normalization and ignoring allosomes and the mitochondrial chromosome. Peaks were called using macs281 (version 2.1.2) with the params “--nomodel --shift 37 --extsize 73 --keep-dup all”.
The immune cell ATAC-seq dataset GSE11818938 was used to create a consensus list of peaks. For each cell type, the donor contributing the fewest number of reads to that cell type was selected and the number of reads was divided by two. Reads were then randomly pooled by that number for each sample, creating a representative alignment file for that cell type. This procedure was performed twice in order to obtain two pseudo-replicates. Peaks were called with macs2 with the same parameters. Irreducible discovery rate (IDR) was calculated between the two pseudo replicates82, any peak with an IDR ≤ 0.05 were included in the consensus list of peaks. This list was then used as a feature reference and reads were counted per feature with featureCounts from the package subread83 (version 1.6.4). A similar approach was used for the other datasets in the analysis. IDR was used to obtain a reliable list of peaks. In these datasets, no feature reference was derived from the IDR, and counting was performed directly from the list obtained from GSE118189. Workflows were implemented using conda and snakemake.
ATAC-seq enrichment analyses.
To examine enrichment of T1D credible variants (group marginal posterior probability > 0.8 from GUESSFM) in open chromatin, for each cell type, two complementary approaches—SNP-matching and GoShifter84 (http://software.broadinstitute.org/mpg/goshifter/)—were employed. In the SNP-matching approach, variants were randomly sampled across the genome, matched on LD structure and gene density, to generate a null distribution of SNPs overlapping accessible chromatin (see below). GoShifter, in contrast, generates a null distribution within each locus (see Trynka et al.84).
SNP-matching enrichment analysis.
The number of T1D credible variants falling within open chromatin was compared to variants in regions of the genome with similar LD structure and gene density:
Using European individuals from 1000 Genomes Project data, identified all variants with a Pearson correlation > 0.8 with each other.
Binned the T1D credible variants with group marginal posterior probability > 0.8 with regards to their LD block size: 1–9, 10–19, 20–49, 50–74,75–99,100–149 or 150–249.
Binned the 1000 Genomes Project data variants with regards to LD block size, taking an LD block as the variants with Pearson correlation > 0.8 with an index variant.
For each T1D credible group, randomly selected an LD block from the 1000 Genomes Project data of the same bin size and with the same (or similar for large haplotypes) number of genes overlapping the credible group, therefore selecting a similar number of variants to the T1D credible group, with an approximately equivalent LD structure and gene density.
Repeated step (4) 100 times, yielding 100 randomly-sampled genome segments with approximately equivalent size and LD structure to the T1D credible variants.
For cell type “X”, counted the number of T1D credible SNPs overlapping ATAC-seq peaks. Compared this to the number overlapping ATAC-seq peaks from the first randomly sampled set of variants. Calculated a z-score (Fisher’s exact test) for the comparison of ATAC-seq peak overlap with T1D credible variants versus randomly sampled variants with equivalent size, gene density, and LD structure.
Repeated step (6) 100 times, one for each randomly sampled set of haplotypes across the genome, obtaining 100 z-scores.
Took the mean z-score from the 100 tests and compared it to a normal distribution to obtain an enrichment p-value for cell type “X”.
Steps (6) to (8) were performed for each cell type and condition.
Generating caQTL maps using T1DGC frozen samples.
We profiled chromatin accessibility in 115 individuals (57 controls and 58 T1D cases; 67 AFR and 48 EUR) from the Type 1 Diabetes Genetics Consortium (T1DGC). CD4+ T cells were purified from viably frozen PMBC samples using magnetic cell separation according to the manufacturer protocol, using either negative selection (n = 42; STEMCELL Technologies EasySep Human CD4+ T Cell Isolation Kit) or positive selection (n = 73; MACS Miltenyl Biotec). The selection approach was incorporated in data processing and analysis. After CD4+ T cell purification, the “Omni-ATAC-seq” protocol78 was followed for nuclei isolation, transposase incubation, and library preparation. Libraries were sequenced using 75 bp paired-end reads on an Illumina NextSeq and data were processed using the PEPATAC pipeline85. Briefly, reads were trimmed using Skewer (version0.2.2)86 and, after removing reads mapping to mitochondrial and human repeat regions, were mapped to GRCh38 using bowtie287. PCR duplicates were removed, enzymatic cut sites were inferred based on read alignment, and peaks called using macs281. Libraries with transcription start site (TSS) enrichment scores less than 6 million or fewer than 10 million aligned reads were excluded from analyses. A set of consensus peaks was determined by merging peaks across all samples using bedops (version 2.4.35)88. A matrix of peak counts was calculated by counting the number of cut sites within each consensus peak in each sample using the R package bigWig (https://github.com/andrelmartins/bigWig).
Peaks with low counts were excluded (required ≥ 10 reads in ≥ 50% of samples). Further peak quality filtering and normalization was performed using the R package edgeR89. These steps included:
filtering for peaks with ≥ 10 counts-per-million (CPM) across samples within each batch;
peak count normalization using the trimmed mean of M-values (TMM) method90;
mean-variance modeling-based transformation using the ‘voom’ function to enable linear modeling of peak counts assuming a normal distribution;
removing outlier peaks by clustering samples based on counts for each peak (one at a time using k-means with k = 2) and excluding any peak that results in one sample clustering separately from all other samples.
We confirmed matching sample identity between ATAC-seq libraries and genotyped subjects using the “Match BAM to VCF” (MBV) command in the software tool set QTLtools91. Association between imputed genotype dosage and chromatin accessibility (caQTL analysis) was tested with a linear model, adjusting for the first two genotype principal components, age at sample collection, TSS enrichment score, and CD4+ T cell purification approach using the R package MatrixEQTL92. The caQTL discovery analyses were performed separately by ancestry group (EUR and AFR) and combined in an inverse-variance weighted fixed effect meta-analysis (R package meta). All variant-peak combinations were tested where the accessibility peak was within 1 Mb of a T1D credible variant.
Colocalization analysis.
We evaluated colocalization of T1D and caQTL for all peaks where at least one T1D credible variant (as defined by GUESSFM) was associated with peak accessibility (meta-analysis P < 5 × 10−5) using the R package coloc41, and visualized colocalized signals using the R package locuscomparer93. Conditional summary statistics were used in regions predicted to have more than one causal variant underlying the T1D association or regions with multiple, conditionally independent variants associated with accessibility of the same peak. When running coloc for T1D-caQTL colocalization, we used a prior probability of colocalization of 5 × 10−6 and provided association betas and standard errors as input data. When running coloc for T1D-eQTL colocalization, we used the same priors and supplied association z-scores. We considered GWAS and QTL signals were considered to be significantly colocalized when the posterior probability of colocalization was greater than 0.8 (‘PP.H4.abf’ > 0.8).
Allele-specific accessibility analysis.
For significant caQTLs that colocalized with T1D-associated variants, we tested for allele-specific accessibility of the caQTL peak. First, we identified individuals heterozygous for T1D credible variants overlapping the caQTL peak. Within each heterozygous individual, we then counted the number of reads overlapping the variant position containing the reference or alternative allele. We only performed this analysis if the T1D credible variant overlapping the caQTL peak was directly genotyped on the ImmunoChip, since uncertainty in the heterozygous status of an individual could lead to biased results. For peaks with at least 5 participants who had at least 5 reads overlapping the peak, we formally tested whether the proportion of reads containing an alternative allele significantly deviated from the expected null hypothesis proportion of 0.5. We calculated P-values for deviation from “allelic balance” (proportion = 0.5 for each read) by fitting a generalized linear mixed model where the dependent variable is the number of reads and follows a Poisson distribution and the independent variables include a fixed effect for the allele and a random effect for the participant.
EMSA supershift assay.
Jurkat cell line (E6–1) was purchased from ATCC and grown in Roswell Park Memorial Institute, RPMI (RPMI-1640; Gibco) supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin, 1% sodium pyruvate), at 37 °C and 5% CO2.
Labeled (5’ IRDye 700) and unlabeled 31-bp, single-stranded oligonucleotides containing rs72928038 were obtained from Integrated DNA Technologies (Reference Allele strand: 5’AGGGACGGATTTCCTGTAAGCTGATCTTGAA 3’ and Alternative Allele strand: 5’ AGGGACGGATTTCCTATAAGCTGATCTTGAA 3’) along with complementary oligonucleotides. Double-stranded oligonucleotides were generated by annealing equal amount of labeled or unlabeled complementary oligonucleotides at 95 °C for 5 min, followed by gradual cooling with a ramp rate of −1.2 °C/min for 1 h (Bio-Rad C1000 Touch Thermal Cycler). Nuclear extract from Jurkat cells was obtained by following the manufacturer’s protocol for NE-PER™ Nuclear and Cytoplasmic Extraction Reagents kit (Thermo Scientific) and the extracted nuclear protein was dialyzed with Slide-A-Lyzer MINI Dialysis Units, 10,000 MWCO (Thermo Scientific) against a 1 L buffer (10 mM Tris, pH 7.5, 50 mM KCl, 200 mM NaCl, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and 10% glycerol) for 16 h at 4 °C with slow stirring.
Binding reaction for the EMSA was carried out using 2 μL 10X binding buffer (100 mM Tris, 500 mM KCl, 10 mM DTT; pH 7.5), 2 μL 25 mM DTT (2.5% Tween 20), 1 μL Poly (dI-dC) (1 μg/μL in 10 mM Tris, 1 mM EDTA; pH 7.5), 1 μL 1% NP-40, 100 mM MgCl2, 20 fmol IRDye double-stranded oligonucleotide probe, and 16 μg Jurkat nuclear extract in a final volume of 20 μL. For supershift lanes, tested transcription-factor-binding antibodies (ETS-1 Rabbit mAb and Stat1 Rabbit mAb) were diluted 1:50 with ddH2O. Negative control Rabbit IgG was diluted to the same concentration as tested antibody. 1 μL of diluted antibody was added to the binding reaction mixture while maintaining a total volume of 20 ul. Binding reaction was incubated for 20 min at room temperature, after which 2 μL of 10X Orange Loading Dye was added. Electrophoresis was performed with binding reaction mixture on a pre-run 6% DNA retardation gel for 70 min at 70 V. To capture the image, the gel was placed directly on the Odyssey-CLx (Licor) scan bed. The gel was scanned with a thickness of 0.5 mm at 700 nm channel. The EMSA binding condition for rs72928038 was repeated three times to ensure reproducibility of the experiment.
Priority Index (Pi).
To prioritize drug targets implicated by T1D genetic associations, we ran the Prioritiy Index (Pi) algorithm, as implemented in the R package Pi46. Data used to identify eQTL colocalization (eGenes) included those from the initial publication (unstimulated monocytes94, n = 414; LPS-stimulated monocytes after 2 hours94, n = 261; LPS-stimulated monocytes after 24 hours94, n = 322; interferon-gamma-stimulated monocytes after 24 hours94, n = 367; unstimulated B cells95, n = 286; unstimulated NK cells (unpublished), n = 245; unstimulated neutrophils96, n = 114; unstimulated CD4+ T cells97, n = 293; unstimulated CD8+ T cells97, n = 283; and whole blood98, n = 5,311), as well as a larger whole blood study (n = 31,684)42. Hi-C data from monocytes, fetal thymus, naïve CD4+ T cells, total CD4+ T cells, activated total CD4+ T cells, non-activated total CD4+ T cells, naïve CD8+ T cells, total CD8+ T cells, naïve B cells, and total B cells43 were used to identify genes interacting with index variants (cGenes). Data used to define functional genes (fGenes, pGenes and dGenes) were those used in the initial publication. The STRING database99 was used to define protein-protein interaction networks, where confidence scores ≥ 700 were considered.
Statistical analyses.
Unless otherwise noted, all statistical analysis and data visualization was performed using R version 3.6100. All statistical tests based on symmetrically distributed test statistics were two-sided. No repeated measures data were analyzed in this study. All genotyped and ATAC-seq samples analyzed in association tests represent distinct individuals. The R packages ggplot2, cowplot, ggbio, GenomicRanges, gridExtra, RColorBrewer, and rtracklayer were used for data visualization.
Code availability
Code used to generate the results presented in this paper is available at https://github.com/ccrobertson/t1d-immunochip-2020. Pipelines for processing ATAC-seq data are available at https://github.com/dfloresDIL/MEGA and http://pepatac.databio.org.
Data Availability
All univariable summary statistics for genotype association with T1D (including imputed variants) are available through the NHGRI-EBI GWAS Catalog (GCST90013445 and GCST90013446). Chromatin accessibility QTL summary statistics are available through the Type 1 Diabetes Knowledge Portal (https://t1d.hugeamp.org).
Publicly available ATAC-seq.
Raw FASTQ files were obtained from Gene Expression Omnibus (GEO) accession number GSE118189. These data included four individuals and 25 immune cell types under resting conditions and after stimulation with anti-human CD3/CD28 dynabeads and human IL-2 (for 24 hours, T lymphocytes), F(ab)’2 anti-human IgG/IgM38 and human IL-4 (for 24 hours, B lymphocytes), human IL-2 (for 48 hours, NK cells), or LPS (for 6 hours, monocytes)37.
ATAC-seq data from pancreatic islets of five donors without glucose intolerance and five EndoCβH1 cell line replicates, under resting conditions and after stimulation with IFN-γ and IL-1β for 48 hours were downloaded from GEO, accession number GSE12340435.
ATAC-seq data from cardiac fibroblasts (two fetal and three adult) were downloaded from the European Nucleotide Archive (https://www.ebi.ac.uk/ena/data/view/SRX2843570 and https://www.ebi.ac.uk/ena/data/view/SRX2843571), as a control cell type that we did not expect to be involved in the etiology of T1D40.
Epigenome annotation tracks
Epigenome annotation tracks, chromHMM101 tracks from diverse primary human cells were obtained from the NIH Epigenome Roadmap, http://dcc.blueprint-epigenome.eu/#/md/secondary_analysis/Segmentation_of_ChIP-Seq_data_20140811 and additional immune-specific human primary and cell lines from the Blueprint consortium, https://egg2.wustl.edu/roadmap/data/byFileType/chromhmmSegmentations/ChmmModels/coreMarks/jointModel/final.
Whole blood eQTL summary statistics.
Summary statistics from whole blood cis eQTL analysis from 31,683 individuals42 were downloaded from https://eqtlgen.org.
Additional databases used in the Priority Index (Pi) drug target prioritization analysis were obtained through the relational database provided in the R package Pi (http://pi.well.ox.ac.uk:3010/download).
Extended Data
Extended Data Fig. 1. Fine mapping of the chromosome 6q22.32 region.
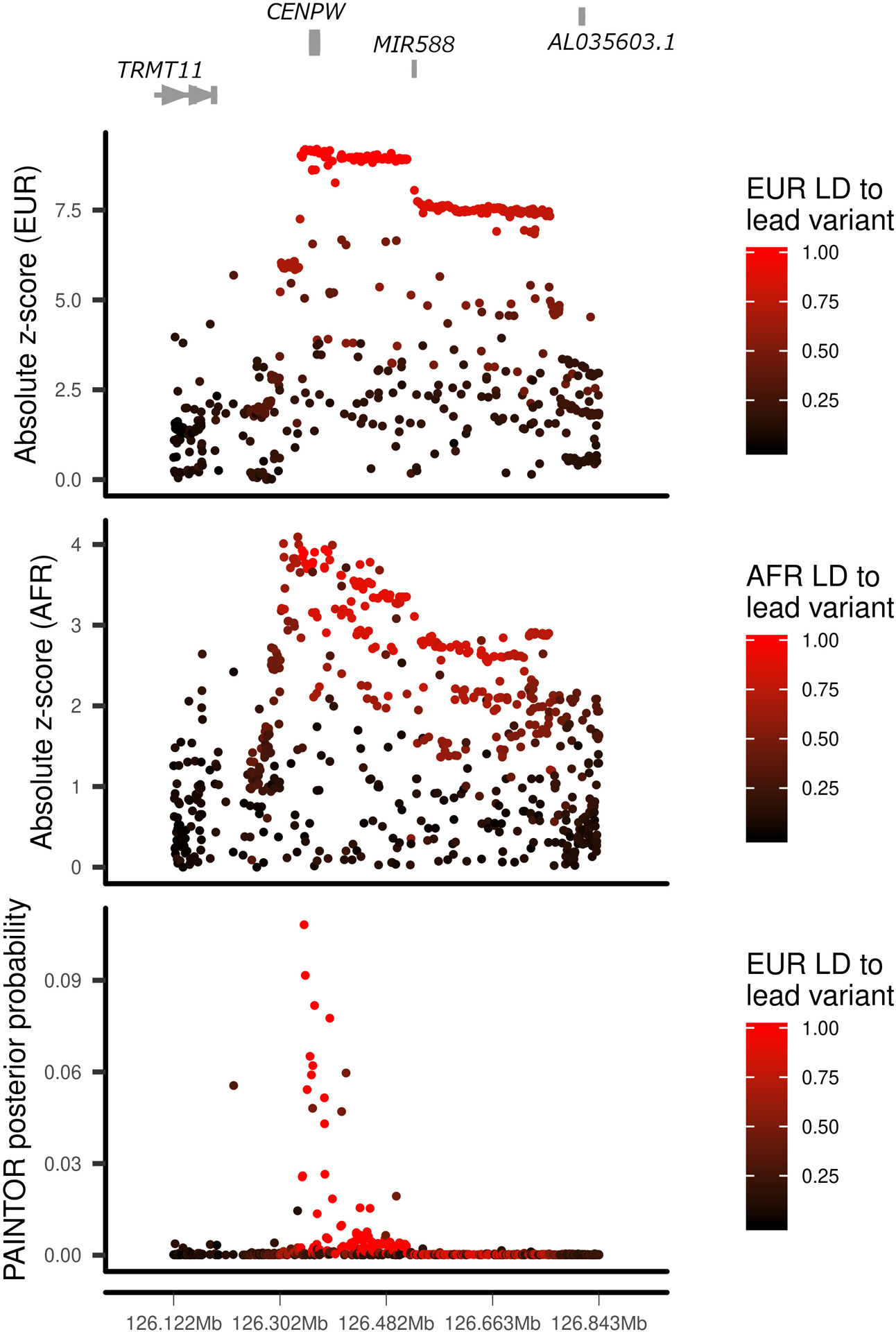
European (EUR, top panel) and African (AFR, middle panel) ancestry group association z-score statistics and posterior probabilities (bottom panel) from multi-ethnic fine mapping of EUR and AFR using PAINTOR. z-scores are colored by linkage disequilibrium (LD) to the lead PAINTOR-prioritized variant.
Extended Data Fig. 2. Fine mapping of the chromosome 18q22.2 region.
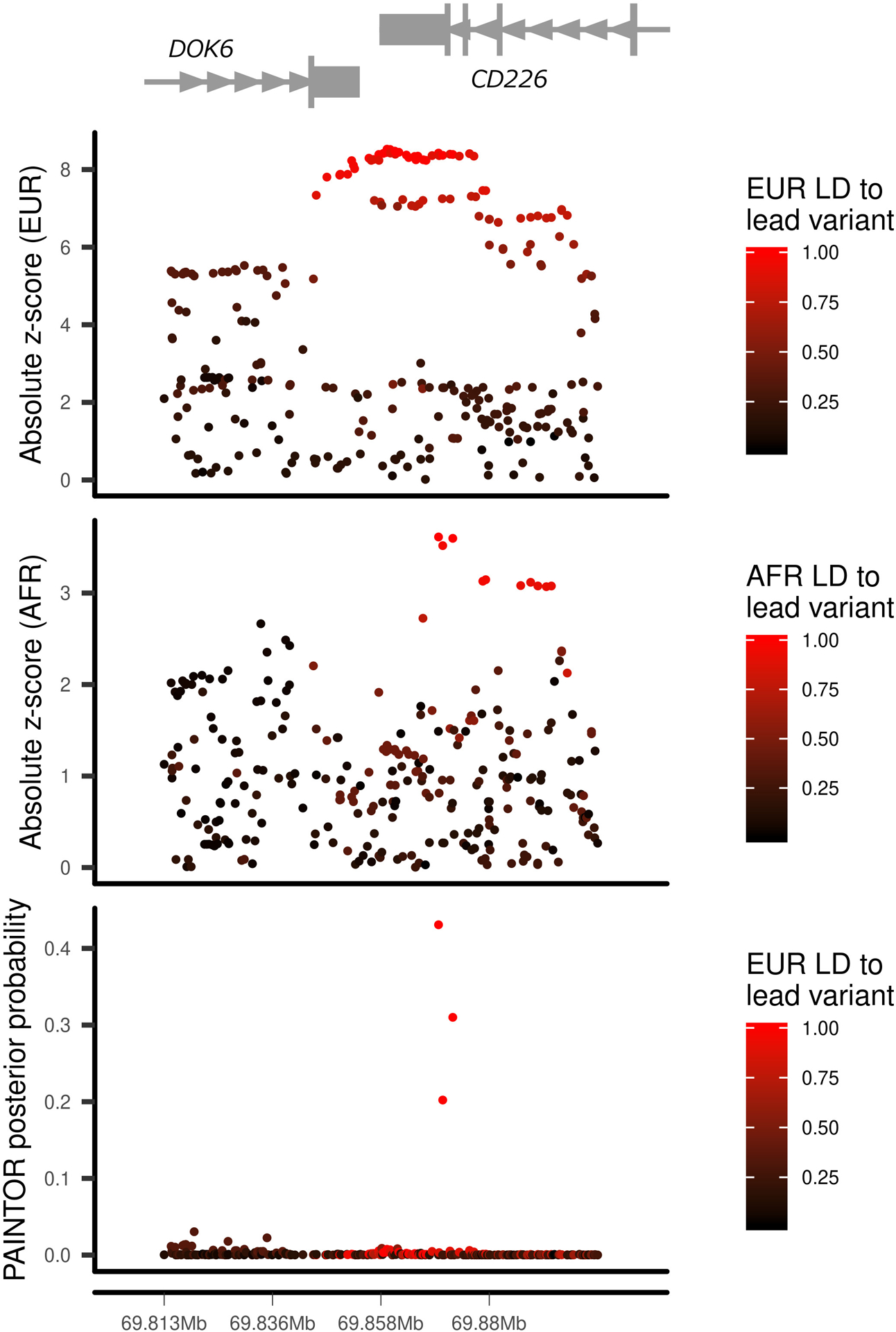
European (EUR, top panel) and African (AFR, middle panel) ancestry group association z-score statistics and posterior probabilities (bottom panel) from multi-ethnic fine mapping of EUR and AFR using PAINTOR. z-scores are colored by linkage disequilibrium (LD) to the lead PAINTOR-prioritized variant.
Supplementary Material
Acknowledgements
We thank the investigators and their studies for contributing samples and/or data to the current work, and the participants in those studies who made this research possible. These studies include the Type 1 Diabetes Genetics Consortium (T1DGC), British 1958 Birth Cohort, Genetic Resource Investigating Diabetes (GRID), Consortium for the Longitudinal Evaluation of African-Americans with Early Rheumatoid Arthritis (CLEAR), Epidemiology of Diabetes Interventions and Complications (EDIC), Genetics of Kidneys and Diabetes Study (GoKinD), New York Cancer Project (NYCP), SEARCH for Diabetes in Youth study (SEARCH), Type 1 Diabetes TrialNet study (TrialNet), Tyypin 1 Diabetekseen Sairastuneita Perheenjäsenineen (IDDMGEN), Tyypin 1 Diabeteksen Genetiikka (T1DGEN), Northern Ireland GRID Collection, Northern Ireland Young Hearts Project, Hvidoere Study Group on Childhood Diabetes (HSG), and International HapMap Project. Additional institutions contributing samples are: British Diabetes Association (BDA), NIHR Cambridge BioResource, UK Blood Service (UKBS), Benaroya Research Institute (BRI), National Institute of Mental Health (NIMH), University of Alabama at Birmingham (UAB), University of Colorado (UC), University of California San Francisco (UCSF), Medical College of Wisconsin (MCW), and Steno Diabetes Center. Samples and data can be obtained on T1DGC, EDIC, and GoKinD from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Central Repository.
This research utilizes resources provided by the T1DGC, a collaborative clinical study sponsored by the NIDDK, National Institute of Allergy and Infectious Diseases (NIAID), National Human Genome Research Institute (NHGRI), National Institute of Child Health and Human Development (NICHD), and Juvenile Diabetes Research Foundation (JDRF) and supported by U01 DK-062418. The generation of chromatin accessibility data on T1DGC samples was supported by grants from the NIDDK (DP3-111906 to S.S.R. and P.C., and DK-115694 to P.C.). Further support was provided by the National Institute of Allergy and Infectious Diseases (P01AI042288 to M.A.A.).
The JDRF/Wellcome Diabetes and Inflammation Laboratory was supported by grants from JDRF (4-SRA-2017-473-A-A) and the Wellcome (107212/A/15/Z). Computation used the Oxford Biomedical Research Computing (BMRC) facility, a joint development between the Wellcome Centre for Human Genetics and the Big Data Institute supported by Health Data Research UK and the NIHR Oxford Biomedical Research Centre. Financial support was provided by the Wellcome Core Award Grant Number 203141/Z/16/Z. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health.
While working on this project, C.C.R. was supported by a training grant from the U.S. National Library of Medicine (5T32LM012416) and the Wagner Fellowship from the University of Virginia.
This work made use of data and samples generated by the 1958 Birth Cohort (NCDS), which is managed by the Centre for Longitudinal Studies at the UCL Institute of Education, funded by the Economic and Social Research Council (grant number ES/M001660/1). Access to these resources was enabled via the Wellcome and MRC: 58FORWARDS grant [108439/Z/15/Z] (The 1958 Birth Cohort: Fostering new Opportunities for Research via Wider Access to Reliable Data and Samples). Before 2015 biomedical resources were maintained under the Wellcome and Medical Research Council 58READIE Project (grant numbers WT095219MA and G1001799).
We acknowledge use of DNA samples from the NIHR Cambridge BioResource. We thank volunteers for their support and participation in the Cambridge BioResource and members of the Cambridge BioResource Scientific Advisory Board (SAB) and Management Committee for their support of our study. We acknowledge the NIHR Cambridge Biomedical Research Centre for funding. Access to Cambridge BioResource volunteers and to their data and samples are governed by the Cambridge BioResource SAB. Documents describing access arrangements and contact details are available at http://www.cambridgebioresource.org.uk/.
The ethics for GRID were processed by the NRES Committee East of England Cambridge South MREC 00/5/44.
The authors thank the following CLEAR investigators who performed recruiting: Doyt Conn (Grady Hospital and Emory University, Atlanta, GA, USA), Beth Jonas and Leigh Callahan (University of North Carolina at Chapel Hill, Chapel Hill, NC, USA), Edwin Smith (Medical University of South Carolina, Charleston, SC, USA), Richard Brasington (Washington University, St. Louis, MO, USA), and Larry W. Moreland (University of Pittsburgh, Pittsburgh, PA, USA). The CLEAR Registry and Repository was funded by National Institutes of Health (NIH) Office of the Director grants N01-AR-0-2247 (9/30/2000-9/29/2006) and N01 AR-6-2278 (9/30/2006-3/31/2012) (S.L.B., PI).
Bio-samples and/or data for this publication were obtained from NIMH Repository & Genomics Resource, a centralized national biorepository for genetic studies of psychiatric disorders.
The SEARCH for Diabetes in Youth Study (www.searchfordiabetes.org) is indebted to the many youth and their families, as well as their health care providers, whose participation made this study possible. SEARCH for Diabetes in Youth is funded by the Centers for Disease Control and Prevention (PA numbers 00097, DP-05-069, and DP-10-001) and supported by the NIDDK. SEARCH Site Contract Numbers: Kaiser Permanente Southern California (U48/CCU919219, U01 DP000246, and U18DP002714), University of Colorado Denver (U48/CCU819241-3, U01 DP000247, and U18DP000247-06A1), Children’s Hospital Medical Center (Cincinnati) (U48/CCU519239, U01 DP000248, and 1U18DP002709), University of North Carolina at Chapel Hill (U48/CCU419249, U01 DP000254, and U18DP002708), University of Washington School of Medicine (U58/CCU019235-4, U01 DP000244, and U18DP002710-01), Wake Forest University School of Medicine (U48/CCU919219, U01 DP000250, and 200-2010-35171).
We acknowledge the support of the Type 1 Diabetes TrialNet Study Group (https://www.trialnet.org), which identified study participants and provided samples and follow-up data for this study. The Type 1 Diabetes TrialNet Study Group is a clinical trials network funded by the National Institutes of Health (NIH) through the National Institute of Diabetes and Digestive and Kidney Diseases, the National Institute of Allergy and Infectious Diseases, and The Eunice Kennedy Shriver National Institute of Child Health and Human Development, through the cooperative agreements U01 DK061010, U01 DK061016, U01 DK061034, U01 DK061036, U01 DK061040, U01 DK061041, U01 DK061042, U01 DK061055, U01 DK061058, U01 DK084565, U01 DK085453, U01 DK085461, U01 DK085463, U01 DK085466, U01 DK085499, U01 DK085505, U01 DK085509, and JDRF. The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the NIH or JDRF. Further support was provided by grants from the NIDDK (U01 DK103282 and U01 DK127404 to C.J.G.).
DNA samples from UAB were recruited, in part, with the support of P01-AR49084 (R.P.K.), UL1-TR001417 (R.P.K.), and UL1-TR003096 (R.P.K.)
We acknowledge the involvement of the Barbara Davis Center for Diabetes at the University of Colorado supported by grants from the NIH NIDDK to M.J.R.: DRC P30 DK116073 and R01 DK032493.
The collection of DNA samples at UCSF was supported by grant funding from the National Multiple Sclerosis Society (SI-2001-35701 to J.R.O.).
Whole genome sequencing (WGS) data production and variant calling was funded by an NHGRI Center for Common Disease Genomics award to Washington University in St. Louis (UM1 HG008853).
This study used the Trans-Omics in Precision Medicine (TOPMed) program imputation panel (version TOPMed-r2) supported by the National Heart, Lung and Blood Institute (NHLBI) (www.nhlbiwgs.org). TOPMed study investigators contributed data to the reference panel, which can be accessed through the Michigan Imputation
Server (https://imputationserver.sph.umich.edu). The panel was constructed and implemented by the TOPMed Informatics Research Center at the University of Michigan (3R01HL-117626-02S1; contract HHSN268201800002I). The TOPMed Data Coordinating Center (3R01HL-120393-02S1; contract HHSN268201800001I) provided additional data management, sample identity checks, and overall program coordination and support. We gratefully acknowledge the studies and participants who provided biological samples and data for TOPMed.
The individual members of the Type 1 Diabetes Genetics Consortium and the SEARCH for Diabetes in Youth Study are listed in the Supplementary Note.
Footnotes
Competing Interests statement
No authors have any competing interests to declare.
References
- 1.Barrett JC et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet 41, 703–707 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Todd JA et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat. Genet 39, 857–864 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Onengut-Gumuscu S et al. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat. Genet 47, 381–386 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inshaw JRJ, Walker NM, Wallace C, Bottolo L & Todd JA The chromosome 6q22.33 region is associated with age at diagnosis of type 1 diabetes and disease risk in those diagnosed under 5 years of age. Diabetologia 61, 147–157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fortune MD et al. Statistical colocalization of genetic risk variants for related autoimmune diseases in the context of common controls. Nat. Genet 47, 839–846 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evangelou M et al. A Method for gene-based pathway analysis using genomewide association study summary statistics reveals nine new type 1 diabetes associations. Genet. Epidemiol 38, 661–670 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rewers M & Ludvigsson J Environmental risk factors for type 1 diabetes. Lancet 387, 2340–2348 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharp SA et al. Development and standardization of an improved type 1 diabetes genetic risk score for use in newborn screening and incident diagnosis. Diabetes Care 42, 200–207 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krischer JP et al. Predicting islet cell autoimmunity and type 1 diabetes: an 8-year TEDDY Study progress report. Diabetes Care 42, 1051–1060 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Onengut-Gumuscu S et al. Type 1 diabetes risk in African-ancestry participants and utility of an ancestry-specific genetic risk score. Diabetes Care 42, 406–415 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skyler JS Hope vs hype: where are we in type 1 diabetes? Diabetologia 61, 509–516 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Herold KC et al. An anti-CD3 antibody, teplizumab, in relatives at risk for type 1 diabetes. N. Engl. J. Med 381, 603–613 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.King EA, Davis JW & Degner JF Are drug targets with genetic support twice as likely to be approved? Revised estimates of the impact of genetic support for drug mechanisms on the probability of drug approval. PLoS Genet. 15, e1008489 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nelson MR et al. The support of human genetic evidence for approved drug indications. Nat. Genet 47, 856–860 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooper JD et al. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat. Genet 40, 1399–1401 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hakonarson H et al. A novel susceptibility locus for type 1 diabetes on Chr12q13 identified by a genome-wide association study. Diabetes 57, 1143–1146 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Grant SFA et al. Follow-up analysis of genome-wide association data identifies novel loci for type 1 diabetes. Diabetes 58, 290–295 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradfield JP et al. A genome-wide meta-analysis of six type 1 diabetes cohorts identifies multiple associated loci. PLoS Genet. 7, e1002293 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang J, Ellinghaus D, Franke A, Howie B & Li Y 1000 Genomes-based imputation identifies novel and refined associations for the Wellcome Trust Case Control Consortium phase 1 Data. Eur. J. Hum. Genet 20, 801–805 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu M et al. Identification of novel T1D risk loci and their association with age and islet function at diagnosis in autoantibody-positive T1D individuals: based on a two-stage genome-wide association study. Diabetes Care 42, 1414–1421 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Divers J et al. Trends in incidence of type 1 and type 2 diabetes among youths — selected counties and Indian reservations, United States, 2002–2015. Morb. Mortal. Wkly. Rep 69, 161–165 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taliun D et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature 590, 290–299 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cortes A et al. Promise and pitfalls of the Immunochip. Arthritis Res. Ther 13, 101 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferreira RC et al. Functional IL6R 358Ala allele impairs classical IL-6 receptor signaling and influences risk of diverse inflammatory diseases. PLoS Genet. 9, e1003444 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okada Y, Wu D, Trynka G & Towfique R Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506, 376–381 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benjamini Y & Yekutieli D The control of the false discovery rate in multiple testing under depencency. Ann. Stat 29, 1165–1188 (2001). [Google Scholar]
- 28.Crouch DJM et al. Enhanced genetic analysis of type 1 diabetes by selecting variants on both effect size and significance, and by integration with autoimmune thyroid disease. bioRxiv (2021). doi: 10.1101/2021.02.05.429962 [DOI] [Google Scholar]
- 29.Wallace C et al. Dissection of a complex disease susceptibility region using a Bayesian stochastic search approach to fine mapping. PLoS Genet. 11, e1005272 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asimit JL et al. Stochastic search and joint fine-mapping increases accuracy and identifies previously unreported associations in immune-mediated diseases. Nat. Commun 10, 3216 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wojcik GL et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature 570, 514–518 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kichaev G & Pasaniuc B Leveraging functional-annotation data in trans-ethnic fine-mapping studies. Am. J. Hum. Genet 97, 260–271 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boyle AP et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 22, 1790–1797 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lizio M et al. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol. 16, 22 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramos-rodríguez M et al. The impact of proinflammatory cytokines on the beta-cell regulatory landscape provides insights into the genetics of type 1 diabetes. Nat. Genet 51, 1588–1595 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ward LD & Kellis M HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 44, 877–881 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buenrostro JD, Wu B, Chang HY & Greenleaf WJ ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol 109, 21.29.1–21.29.9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Calderon D et al. Landscape of stimulation-responsive chromatin across diverse human immune cells. Nat. Genet 51, 1494–1505 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varshney A et al. Genetic regulatory signatures underlying islet gene expression and type 2 diabetes. Proc. Natl. Acad. Sci. USA 114, 2301–2306 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jonsson MKB et al. A Transcriptomic and epigenomic comparison of fetal and adult human cardiac fibroblasts reveals novel key transcription factors in adult cardiac fibroblasts. JACC Basic to Transl. Sci 1, 590–602 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giambartolomei C et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10, e1004383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Võsa U et al. Unraveling the polygenic architecture of complex traits using blood eQTL meta-analysis. bioRxiv (2018). 10.1101/447367/ [DOI] [Google Scholar]
- 43.Javierre BM et al. Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell 167, 1369–1384 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmiedel BJ et al. Impact of genetic polymorphisms on human immune cell gene expression. Cell 175, 1701–1715 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Westra HJ et al. Fine-mapping and functional studies highlight potential causal variants for rheumatoid arthritis and type 1 diabetes. Nat. Genet 50, 1366–1374 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fang H et al. A genetics-led approach defines the drug target landscape of 30 immune-related traits. Nat. Genet 51, 1082–1091 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chun S et al. Limited statistical evidence for shared genetic effects of eQTLs and autoimmune-disease-associated loci in three major immune-cell types. Nat. Genet 49, 600–605 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hukku A et al. Probabilistic colocalization of genetic variants from complex and molecular traits: promise and limitations. Am. J. Hum. Genet 108, 25–35 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chiou J et al. Large-scale genetic association and single cell accessible chromatin mapping defines cell type-specific mechanisms of type 1 diabetes risk. bioRxiv (2021). 10.1101/2021.01.13.426472 [DOI] [Google Scholar]
- 50.Benaglio P et al. Mapping genetic effects on cell type-specific chromatin accessibility and annotating complex trait variants using single nucleus ATAC-seq. bioRxiv (2020). 10.1101/2020.12.03.387894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kundu K et al. Genetic associations at regulatory phenotypes improve fine-mapping of causal variants for twelve immune-mediated diseases. bioRxiv (2020). 10.1101/2020.01.15.907436 [DOI] [PubMed] [Google Scholar]
- 52.Danko CG et al. Dynamic evolution of regulatory element ensembles in primate CD4+ T cells. Nat. Ecol. Evol 2, 537–548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsukumo S et al. Bach2 maintains T cells in a naive state by suppressing effector memory-related genes. Proc. Natl. Acad. Sci. USA 110, 10735–10740 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roychoudhuri R et al. BACH2 regulates CD8+ T cell differentiation by controlling access of AP-1 factors to enhancers. Nat. Immunol 17, 851–860 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Afzali B et al. BACH2 immunodeficiency illustrates an association between super-enhancers and haploinsufficiency. Nat. Immunol 18, 813–823 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cotsapas C et al. Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet. 7, e1002254 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Faegan BG et al. Risankizumab in patients with moderate to severe Crohn’s disease: an open-label extension study. Lancet Gastroenterol. Hepatol 3, 671–680 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Fotiadou C, Lazaridou E, Sotiriou E & Ioannides D Targeting IL-23 in psoriasis: current perspectives. Psoriasis Targets Ther. 8, 1–5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wollenhaupt J et al. Safety and efficacy of tofacitinib for up to 9.5 years in the treatment of rheumatoid arthritis: final results of a global, open-label, long-term extension study. Arthritis Res. Ther 21, 89 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sandborn WJ et al. Tofacitinib as induction and maintenance therapy for ulcerative colitis. N. Engl. J. Med 376, 1723–1736 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Gaglia J & Kissler S Anti-CD3 antibody for the prevention of type 1 diabetes: a story of perseverance. Biochemistry 58, 4107–4111 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aylward A, Chiou J, Okino M-L, Kadakia N & Gaulton KJ Shared genetic risk contributes to type 1 and type 2 diabetes etiology. Hum. Mol. Genet (2018). doi: 10.1093/hmg/ddy314 [DOI] [PubMed] [Google Scholar]
- 63.Dooley J et al. Genetic predisposition for beta cell fragility underlies type 1 and type 2 diabetes. Nat. Genet 48, 519–527 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 64.Manichaikul A et al. Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Price AL et al. Long-Range LD can confound genome scans in admixed populations. Am. J. Hum. Genet 83, 127–147 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chang CC et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marchini J & Howie B Genotype imputation for genome-wide association studies. Nat. Rev. Genet 11, 499–511 (2010). [DOI] [PubMed] [Google Scholar]
- 68.Willer CJ, Li Y & Abecasis GR METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Spielman RS, McGinnis RE & Ewens WJ Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am. J. Hum. Genet 52, 506–516 (1993). [PMC free article] [PubMed] [Google Scholar]
- 70.Taub MA, Schwender H, Beaty TH, Louis TA & Ruczinski I Incorporating genotype uncertainties into the genotypic TDT for main effects and gene-environment interactions. Genet. Epidemiol 36, 225–234 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kazeem GR & Farrall M Integrating case-control and TDT studies. Ann. Hum. Genet 69, 329–335 (2005). [DOI] [PubMed] [Google Scholar]
- 72.Asimit JL et al. Stochastic search and joint fine-mapping increases accuracy and identifies previously unreported associations in immune-mediated diseases. Nat. Commun 10, 3216 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bottolo L & Richardson S Evolutionary stochastic search for bayesian model exploration. Bayesian Anal. 5, 583–618 (2010). [Google Scholar]
- 74.Benner C et al. Prospects of fine-mapping trait-associated genomic regions by using summary statistics from genome-wide association studies. Am. J. Hum. Genet 101, 539–551 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Excoffier L & Slatkin M Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol. Biol. Evol 12, 921–927 (1995). [DOI] [PubMed] [Google Scholar]
- 76.Wang K, Li M & Hakonarson H ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38, e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Burren OS et al. Chromosome contacts in activated T cells identify autoimmune disease candidate genes. Genome Biol. 18, 165 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Corces MR et al. An improved ATAC- seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Harrow J et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li H Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gaspar JM Improved peak-calling with MACS2. bioRxiv (2018). 10.1101/496521 [DOI] [Google Scholar]
- 82.Li Q, Brown JB, Huang H & Bickel PJ Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat 5, 1752–1779 (2011). [Google Scholar]
- 83.Liao Y, Smyth GK & Shi W FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 84.Trynka G et al. Disentangling the effects of colocalizing genomic annotations to functionally prioritize non-coding variants within complex-trait loci. Am. J. Hum. Genet 97, 139–152 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Smith JP et al. PEPATAC: An optimized ATAC-seq pipeline with serial alignments. bioRxiv (2020). doi: 10.1101/2020.10.21.347054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jiang H, Lei R, Ding S & Zhu S Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics 15, 182 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Neph S et al. BEDOPS: high-performance genomic feature operations. Bioinformatics 28, 1919–1920 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Robinson MD, Mccarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Robinson MD & Oshlack A A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11, R25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fort A et al. MBV: a method to solve sample mislabeling and detect technical bias in large combined genotype and sequencing assay datasets. Bioinformatics 33, 1895–1897 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shabalin AA Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics 28, 1353–1358 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu B, Gloudemans MJ, Rao AS, Ingelsson E & Montgomery SB Abundant associations with gene expression complicate GWAS follow-up. Nat. Genet 51, 768–769 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fairfax BP et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science 343, 1246949 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fairfax BP et al. Genetics of gene expression in primary immune cells identifies cell type–specific master regulators and roles of HLA alleles. Nat. Genet 44, 502–510 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Andiappan AK et al. Genome-wide analysis of the genetic regulation of gene expression in human neutrophils. Nat. Commun 6, 7971 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kasela S et al. Pathogenic implications for autoimmune mechanisms derived by comparative eQTL analysis of CD4+ versus CD8+ T cells. PLoS Genet. 13, e1006643 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Westra H et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet 45, 1238–1243 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Szklarczyk D et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.R Core Team (2020). R: A language and environment for computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/. [Google Scholar]
- 101.Ernst J & Kellis M Chromatin-state discovery and genome annotation with ChromHMM. Nat. Protoc 12, 2478–2492 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All univariable summary statistics for genotype association with T1D (including imputed variants) are available through the NHGRI-EBI GWAS Catalog (GCST90013445 and GCST90013446). Chromatin accessibility QTL summary statistics are available through the Type 1 Diabetes Knowledge Portal (https://t1d.hugeamp.org).
Publicly available ATAC-seq.
Raw FASTQ files were obtained from Gene Expression Omnibus (GEO) accession number GSE118189. These data included four individuals and 25 immune cell types under resting conditions and after stimulation with anti-human CD3/CD28 dynabeads and human IL-2 (for 24 hours, T lymphocytes), F(ab)’2 anti-human IgG/IgM38 and human IL-4 (for 24 hours, B lymphocytes), human IL-2 (for 48 hours, NK cells), or LPS (for 6 hours, monocytes)37.
ATAC-seq data from pancreatic islets of five donors without glucose intolerance and five EndoCβH1 cell line replicates, under resting conditions and after stimulation with IFN-γ and IL-1β for 48 hours were downloaded from GEO, accession number GSE12340435.
ATAC-seq data from cardiac fibroblasts (two fetal and three adult) were downloaded from the European Nucleotide Archive (https://www.ebi.ac.uk/ena/data/view/SRX2843570 and https://www.ebi.ac.uk/ena/data/view/SRX2843571), as a control cell type that we did not expect to be involved in the etiology of T1D40.
Epigenome annotation tracks
Epigenome annotation tracks, chromHMM101 tracks from diverse primary human cells were obtained from the NIH Epigenome Roadmap, http://dcc.blueprint-epigenome.eu/#/md/secondary_analysis/Segmentation_of_ChIP-Seq_data_20140811 and additional immune-specific human primary and cell lines from the Blueprint consortium, https://egg2.wustl.edu/roadmap/data/byFileType/chromhmmSegmentations/ChmmModels/coreMarks/jointModel/final.
Whole blood eQTL summary statistics.
Summary statistics from whole blood cis eQTL analysis from 31,683 individuals42 were downloaded from https://eqtlgen.org.
Additional databases used in the Priority Index (Pi) drug target prioritization analysis were obtained through the relational database provided in the R package Pi (http://pi.well.ox.ac.uk:3010/download).


