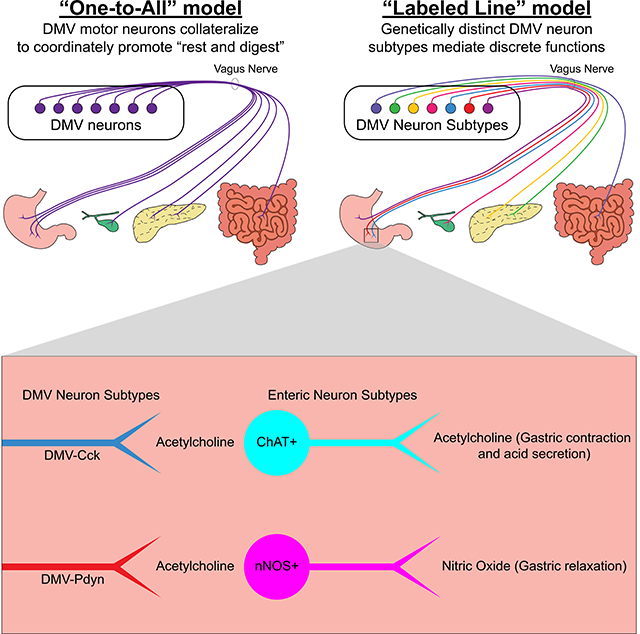
Summary
The vagus nerve innervates many organs, and most if not all of its motor fibers are cholinergic. However, no one knows its organizing principles; whether or not there are dedicated neurons with restricted targets that act as “labeled lines” to perform certain functions – including two opposing ones (gastric contraction versus relaxation). By performing unbiased transcriptional profiling of DMV cholinergic neurons, we discovered seven molecularly-distinct subtypes of motor neurons. Then, by using subtype-specific Cre driver mice, we go on to show that two of these subtypes exclusively innervate the glandular domain of the stomach where, remarkably, they contact different enteric neurons releasing functionally opposing neurotransmitters (acetylcholine versus nitric oxide). Thus, the vagus motor nerve communicates via genetically-defined labeled lines to control functionally unique enteric neurons within discrete subregions of the gastrointestinal tract. This discovery reveals that the parasympathetic nervous system utilizes a striking division of labor to control autonomic function.
Graphical Abstract
etoc blurb
The dorsal motor nucleus of the vagus is the primary source of parasympathetic input to the digestive system. Tao et al. demonstrate a labeled line model of the DMV by showing that Cck- and Pdyn-expressing DMV neurons exclusively innervate the glandular stomach where they target functionally-opposing enteric neurons.
Introduction
Langley first coined the term “autonomic nervous system” (ANS) in 1897 to describe the nerve fibers that innervate tissues other than skeletal muscle (Langley, 1897; Langley, 1921). The parasympathetic nervous system (PNS) is a major component of the ANS and is often characterized as mediating the “rest and digest” response (Gibbons, 2019). In accordance with the “rest and digest” function of the PNS, vagal motor neurons originating from the dorsal motor nucleus of the vagus (DMV) provide parasympathetic motor input to the stomach, small intestine, large intestine, gallbladder and pancreas. These DMV neurons engage enteric neurons to regulate multiple digestive processes, including gastric contraction, relaxation, and acid secretion, pancreatic endocrine and exocrine secretions, gallbladder contraction, and intestinal motility (Rogers and Hermann, 2012; Liddle, 2018; Mawe et al., 2018; Schubert and Peura, 2008).
However, the organizational principles of the DMV are unknown and it is unclear how vagal motor neurons accomplish brain control of visceral function. Are there discrete vagal neurons that target narrow domains within the GI tract, and then functionally distinct enteric neurons within these narrow domains (a highly selective labeled line view), or alternatively, consistent with the widely talked about general “rest and digest” function of the PNS, do vagal motor neurons broadcast their projections broadly? Some have speculated that “labeled lines” of communication should exist to account for the varied functions carried out by the vagus (Huang, Tork, and Paxinos, 1993; Rogers, Hermann, and Travagli, 1999; Chang, Mashimo, and Goyal, 2003). To date there have been no demonstrations that this “labeled line” view is true, and if true, the field has lacked any means of experimentally “accessing” such labeled lines. This gap in knowledge and the lack of tools have prevented progress in the field.
Recombinase-expressing mouse lines, in which a DNA recombinase (e.g., Cre) is expressed from a targeted gene locus, can be used to genetically access specific neuron subtypes (Luo, Callaway, and Svoboda, 2018) for mapping axon projections, manipulating and measuring neuronal activity, and determining postganglionic targets. However, this approach requires a priori knowledge of genetic markers for each neuron subtype, which does not exist for DMV neurons. In this study, we investigate the hypothesis that functionally distinct subtypes of DMV neurons express different genes, and that such knowledge provides the means for selectively accessing each subtype. Using single-cell transcriptomics, we identified seven subtypes of DMV neurons and genetic markers for each subtype. Remarkably, we found two DMV subtypes that exclusively innervate the glandular stomach and target neurochemically-distinct enteric neurons. In total, our findings strongly support the “labeled line” model of communication between the DMV and the gut, and provide an experimental approach for interrogating these discrete brain-to-gut circuits.
Results
Classification of DMV Neuron Subtypes by Genome-Wide Expression Profiles
We used single-nucleus RNA-seq (sNuc-Seq) (Habib et al., 2016; Lake et al., 2016; Todd et al., 2020) to transcriptionally profile cholinergic (Chat+) DMV neurons and systemically identify their subtypes and markers (Figure 1A). A Cre-dependent nuclear reporter adeno-associated virus (AAV), AAV-DIO-H2b-mCherry, was injected into the brainstem of four adult male Chat-IRES-Cre mice. Tissue samples containing the DMV were dissected from the brains of these mice and used to collect H2b-mCherry+ single nuclei by fluorescence-activated cell sorting. The samples were processed into cDNA libraries with Smart-Seq2 (Todd et al., 2020; Picelli et al., 2014) and sequenced. Samples expressing <5,000 genes were considered low quality and excluded from further analysis. This left 306 samples with 11,231 +/− 1,471 genes detected per nucleus. Clustering the samples by their gene expression profiles identified eight neuron subtypes that differed in their gene expression profiles, including marker genes (Figures 1B and 1C; Table S1). Gene expression profiles at the single cell and cluster levels can be readily visualized and further analyzed via the publicly available Broad Institute Single Cell Portal (https://singlecell.broadinstitute.org/single_cell/study/SCP1345/dmvchat-neuron2021#study-summary). Primary sequencing data FASTQ files are available both through the Single Cell Portal and at GEO (GEO accession number GSE172411). As expected, all clusters expressed the housekeeping gene, Actb, the neuronal marker, Tubb3, and Chat. Glial specific genes (Gfap, Olig1, Aif1, and Cspg4) on the other hand were lowly expressed or not detected (Figure 1C). These results confirmed that the isolated samples were cholinergic neurons. We cross-referenced the top marker genes for each cluster with in situ hybridization data from the Allen Mouse Brain Atlas (Lein et al., 2007). We identified one neuron cluster (cluster c1) that did not originate from the DMV. This cluster came from cholinergic neurons in the neighboring hypoglossal nucleus as it expressed Calca but not Phox2b (Pattyn et al., 1997; Brunet and Pattyn, 2002) or Adcyap1 (Figure 1D). Note that Calca is expressed in the hypoglossal nucleus while Phox2b and Adcyap1 are expressed in the DMV (Allen Brain Atlas) (Lein et al., 2007). The remaining seven clusters represent DMV neuron subtypes, as all were positive for Phox2b and Adcyap1 but negative for Calca. We hypothesized that the molecular subtypes of DMV neurons form distinct circuits with visceral organs.
Figure 1. Classification of DMV Neuron Subtypes by Genome-Wide Expression Profiles.
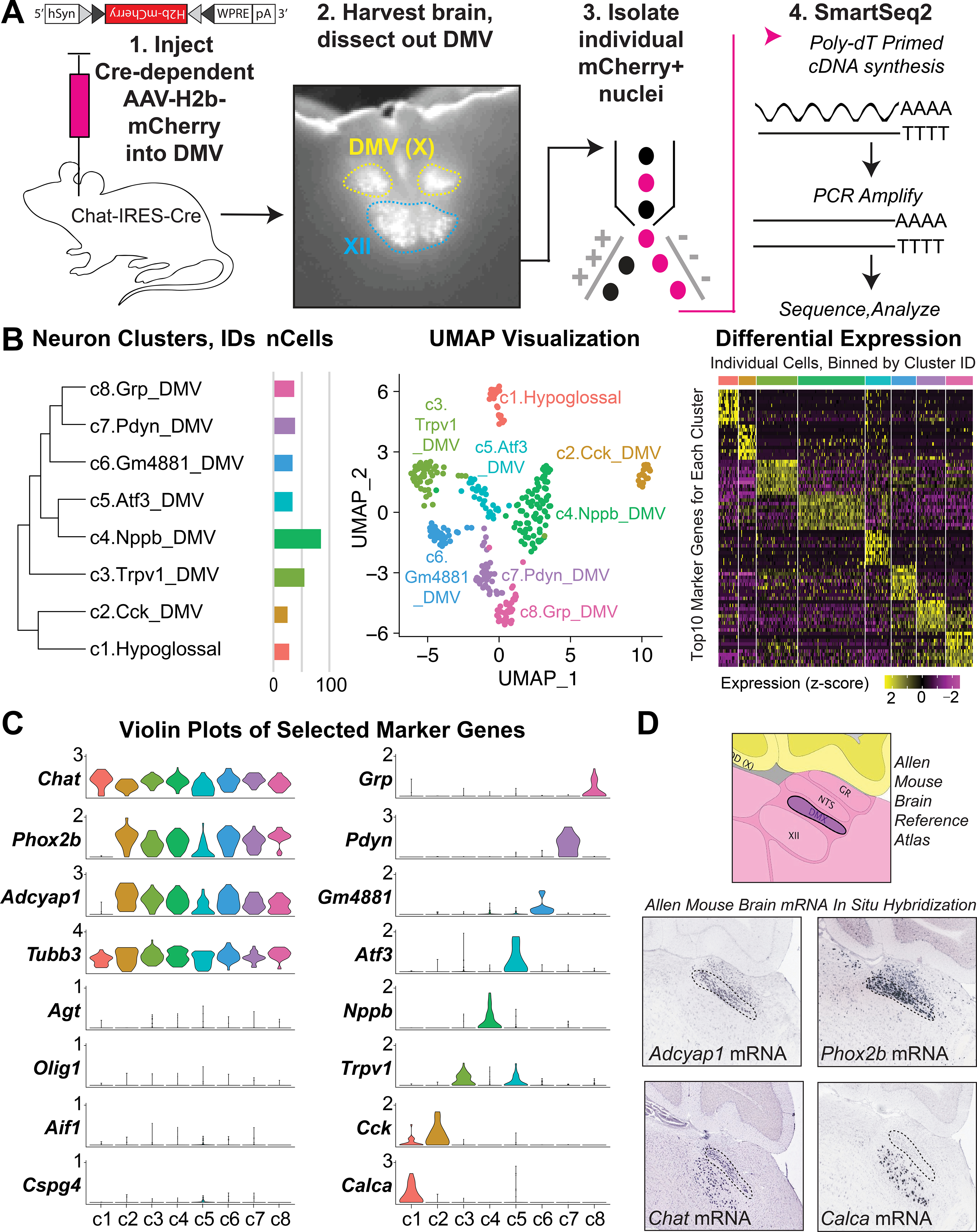
(A) Strategy for isolating and sequencing DMV neurons from Chat-IRES-Cre mice (n = 4).
(B) Neuron clusters, UMAP plots, and differential expression of top marker genes in each cluster.
(C) Relative expression of relevant genes in each cluster.
(D) in situ hybridization of marker genes in sagittal brain sections from the Allen Mouse Brain Atlas.
Cck and Pdyn Mark Separate DMV Neurons With Different Anatomical Distributions
To test our hypothesis, we selected marker genes for two DMV neuron subtypes (Cck for cluster c2, Pdyn for cluster c7, Figure 1C) for further analysis. We used in situ hybridization to assess the expression of these genes in the DMV. We labeled DMV neurons with systemic injections of the retrograde tracer, Fluorogold. Fluorogold is taken up by axon terminals and, when injected systemically, it efficiently labels CNS neurons that project beyond the blood brain barrier, including vagal motor neurons (Leong and Ling, 1990; Chang et al., 2015; Powley, Fox, and Berthoud, 1987). Cck and Pdyn were each expressed in a small proportion of fluorogold labeled DMV neurons (240 Cck+ neurons and 232 Pdyn+ neurons of 2,904 fluorogold labeled DMV neurons, Figure 2B). Co-localization of the two markers was minimal (2 Cck+ Pdyn+ neurons of 2,904 fluorogold labeled DMV neurons), confirming that these two genes mark separate DMV neuron subtypes. Interestingly, we observed that Cck+ and Pdyn+ DMV neurons differ in their distributions along the rostral-caudal axis of the DMV (Figure 2A). Most Cck+ DMV neurons were found in the rostral and intermediate DMV while most of the Pdyn+ DMV neurons were found in the intermediate and caudal DMV (Figures 2B and 2C). These results show that Cck and Pdyn expressions mark distinct neurons located in different regions of the DMV.
Figure 2. Cck and Pdyn Mark Separate DMV Neurons With Different Anatomical Distributions.
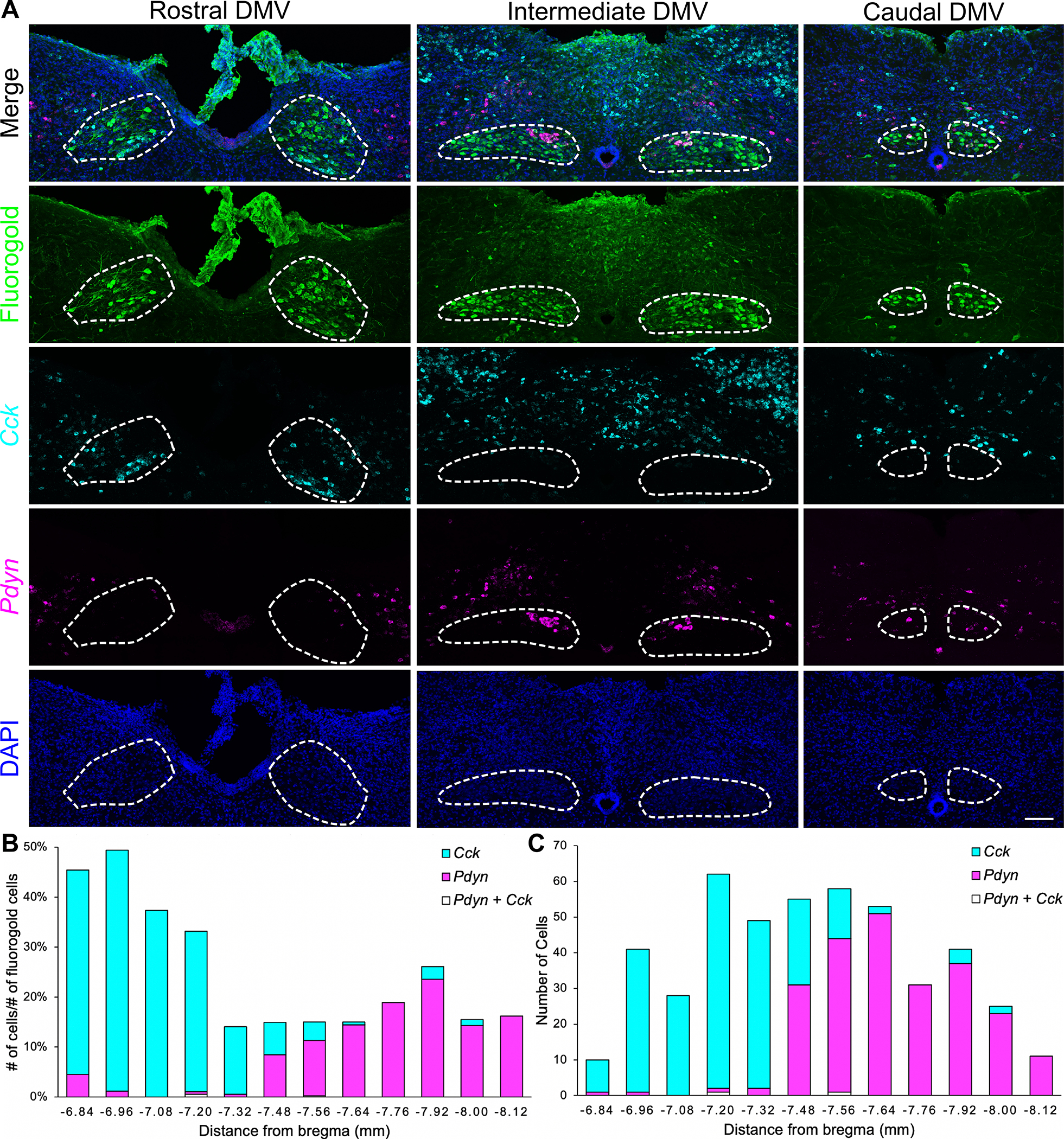
(A) Representative images of in situ hybridization for Cck and Pdyn and fluorogold immunohistochemistry in the rostral DMV (rostral to area postrema, bregma −6.84 to −7.20 mm), intermediate DMV (level of area postrema, bregma −7.32 to −7.76 mm), and caudal DMV (caudal to area postrema, bregma −7.92 to −8.12 mm) from adult C57BL/6J mice (n = 3); scale bar: 100 μm.
(B) Numbers of Cck+ and Pdyn+ DMV neurons divided by the number of fluorogold labeled DMV neurons along the rostral-caudal axis.
(C) Numbers of Cck+ and Pdyn+ DMV neurons along the rostral-caudal axis.
Cck+ and Pdyn+ DMV Neurons Exclusively Innervate the Glandular Stomach
Given that Cck and Pdyn mark distinct populations of DMV neurons, we wondered if Cck+ and Pdyn+ DMV neurons have different axon projection profiles. We used AAVs expressing either a Cre-dependent tdTomato (AAV-FLEX-tdTomato) or a placental alkaline phosphatase (AAV-FLEX-PLAP) to map the axon projections of DMV neurons. For genetic access to Chat+ (i.e., all DMV cholinergic neurons), and the Cck+ and Pdyn+ DMV subtypes, we used Chat-IRES-Cre, Cck-IRES-Cre, and Pdyn-IRES-Cre knockin mice, respectively. Two-color in situ hybridization validated that Cre recombinase was expressed in the correct neurons of Cck-IRES-Cre and Pdyn-IRES-Cre mice (Figure S1). Following AAV injections into the brainstem, we observed tdTomato expression in virtually all DMV neurons in Chat-IRES-Cre mice and only a subset of DMV neurons in Cck-IRES-Cre and Pdyn-IRES-Cre mice (Figure S2). In accordance with previous studies (Rogers and Hermann, 2012; Liddle, 2018; Mawe et al., 2018; Travagli et al., 2006), we observed that Chat+ DMV neurons innervate the entire GI tract, gallbladder, and pancreas (Figures 3A–3F). The rodent stomach is divided into two domains: (1) the nonglandular stomach, or forestomach, which has a non-secretory squamous epithelium similar to that of the esophagus and serves as a reservoir for food contents and (2) the glandular stomach, or corpus and antrum, which has a thick muscular wall for mixing food contents and a columnar glandular epithelium for secreting acid, proteins, and hormones. Remarkably, while Chat+ DMV neurons innervate both domains of the stomach, the Cck+ and Pdyn+ DMV neurons both exclusively innervate the glandular stomach and not the nonglandular stomach (Figures 3B and 3C). Furthermore, Cck+ and Pdyn+ DMV neurons do not innervate the pancreas, gallbladder, or lower GI tract (Figures 3D, 3E, and 3F). Because our studies focused on the above-mentioned visceral targets of the DMV (i.e., GI tract, gallbladder, and pancreas), we cannot exclude the possibility that Cck+ or Pdyn+ DMV neurons might also send projections to a site that was not studied. In total, to our knowledge this is the first demonstration that genetically-distinct DMV neuron subtypes can exclusively and selectively innervate a very specific region of a GI organ, in this case the glandular portion of the stomach.
Figure 3. Cck+ and Pdyn+ DMV Neurons Exclusively Innervate the Glandular Stomach.
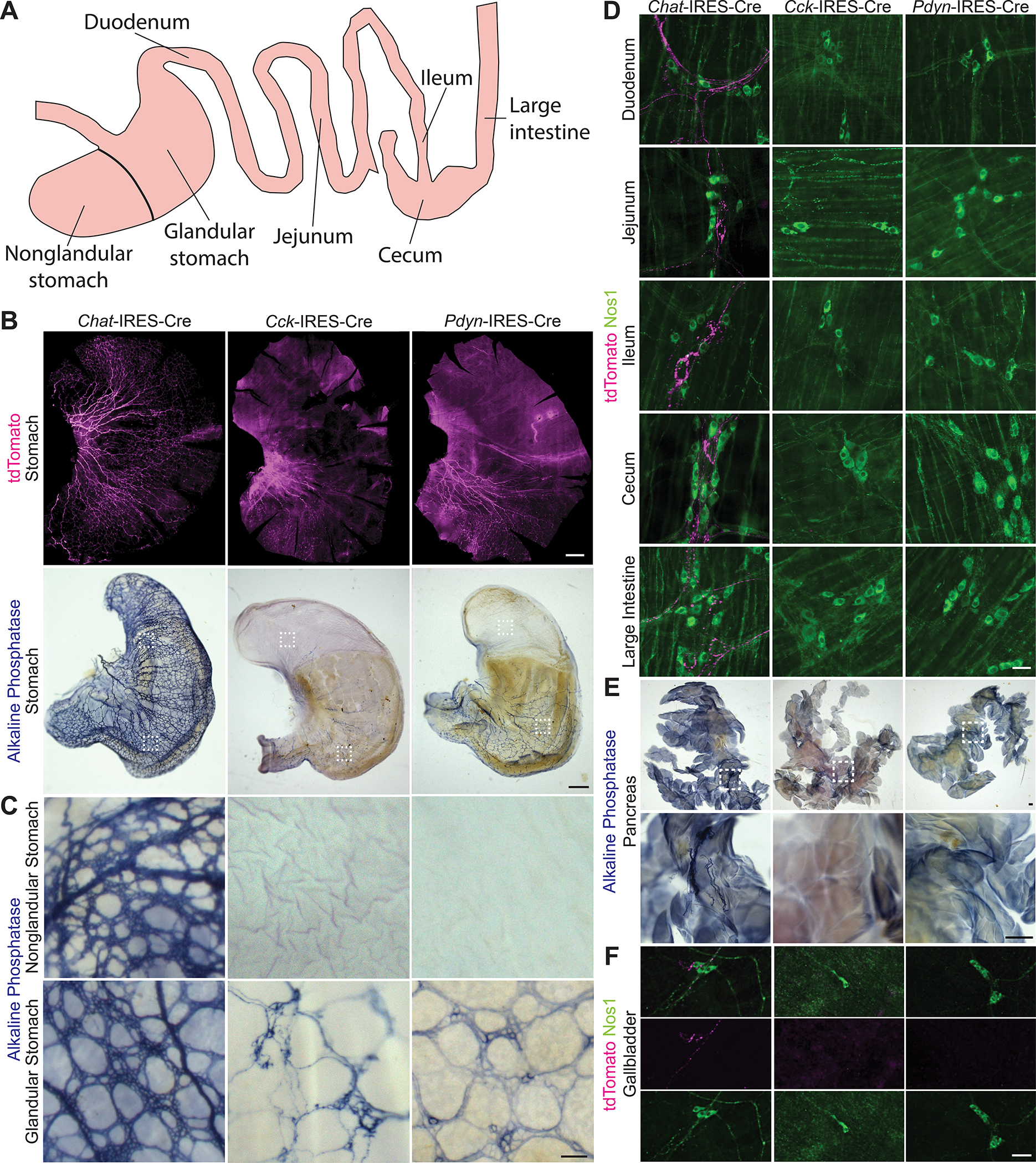
(A) Schematic of the gastrointestinal tract
(B) DMV axon projections in the stomach using Chat-IRES-Cre (n = 10), Cck-IRES-Cre (n = 8), and Pdyn-IRES-Cre (n = 8) mice; scale bar: 1 mm. White boxes indicate regions of the glandular and nonglandular stomach regions highlighted below.
(C) Axon terminals from Chat+, Cck+, and Pdyn+ DMV neurons in the glandular and nonglanular stomach; scale bar: 100 μm.
(D) DMV axon projections in the lower GI tract; scale bar: 50 μm.
(E) DMV axon projections in the pancreas; scale bars: 500 μm.
(F) DMV axon projections in the gallbladder; scale bar: 200 μm.
Cck+ and Pdyn+ DMV Neurons Target Different Enteric Neurons
Given that the vast majority of DMV neurons release the same neurotransmitter, acetylcholine, it has been proposed that DMV neurons exert different and sometimes opposing effects (e.g., contraction vs. relaxation) by engaging neurochemically-distinct types of postganglionic enteric neurons (Powley et al., 2019; Rogers and Hermann, 2012; Chang, Mashimo, and Goyal, 2003). Enteric neurons in the stomach release either nitric oxide for gastric relaxation or acetylcholine for gastric contraction and acid secretion (Schubert and Peura, 2008; Travagli et al., 2006; Furness, 2000). While previous studies have established that DMV neurons form pericellular arborizations around enteric neurons (Powley et al., 2019) that result in synaptic contacts (Hayakawa et al., 2006), and that an individual DMV axon appears to selectively target either cholinergic or nitrergic enteric neurons (Powley et al., 2019), it is unknown whether genetically-defined DMV neuron subtypes, as classes of DMV neurons, selectively engage either cholinergic or nitrergic (nitric oxide-releasing) enteric neurons. As the Cck+ and Pdyn+ DMV neuron subtypes both exclusively innervate the glandular stomach, we hypothesized that these two DMV neuron subtypes target different enteric neuron types. We visualized cholinergic enteric neurons using Chat-GFP mice and nitrergic enteric neurons using immunohistochemistry. Consistent with previous studies showing that enteric neurons in the stomach are either cholinergic or nitrergic (Schubert and Peura, 2008; Travagli et al., 2006; Furness, 2000), and validating our approach for visualizing the two types of enteric neurons, we found that all enteric neurons in the glandular stomach are marked by either Chat-GFP or Nos1 IHC, and that essentially none are marked by both (Figure S3A). Of interest, we found that Cck+ DMV neurons selectively form pericellular arborizations around cholinergic enteric neurons and avoid nitrergic enteric neurons. In striking contrast, Pdyn+ DMV neurons selectively form pericellular arborizations around nitrergic enteric neurons and avoid cholinergic enteric neurons (Figures 4A and 4B). We also observed that Cck+ DMV neurons formed pericellular arborizations around a subset of cholinergic enteric neurons and that Pdyn+ DMV neurons formed pericellular arborization with most but not all of the nitrergic enteric neurons (Figure S3B).
Figure 4. Cck+ and Pdyn+ DMV Neurons Target Different Enteric Neurons.
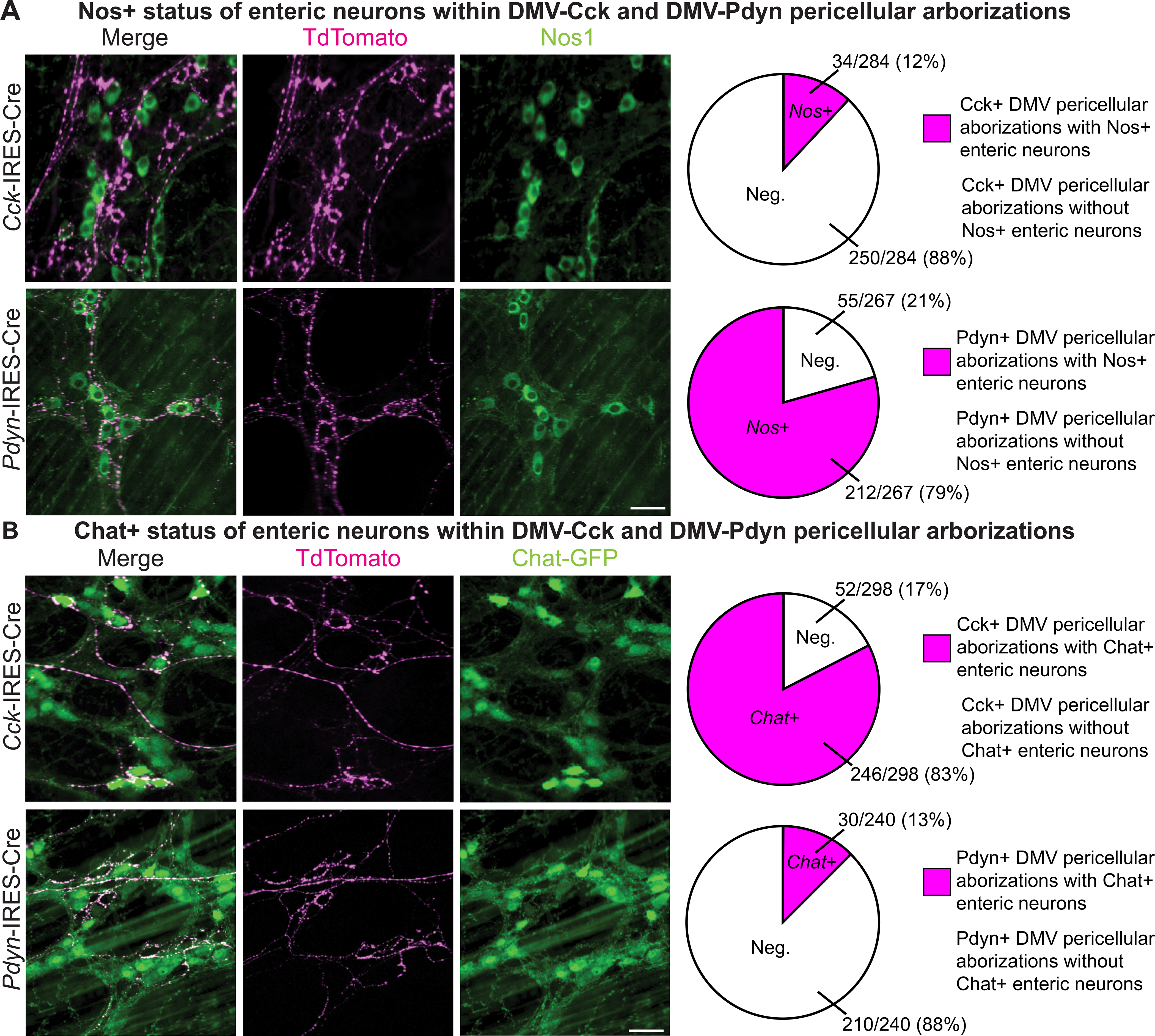
(A-B) Pericellular arborizations of Cck+, and Pdyn+ DMV neurons in the glandular stomach with immunohistochemistry for nitrergic (Nos1) or cholinergic (Chat-GFP) enteric neurons in Cck-IRES-Cre (n = 3), Cck-IRES-Cre; Chat-GFP (n = 3), Pdyn-IRES-Cre (n = 3), and Pdyn-IRES-Cre;Chat-GFP (n = 3) mice; scale bars: 50 μm. The numbers of pericellular arborizations with and without enteric neurons were counted (right).
Discussion
The DMV is the primary source of parasympathetic input to the gastrointestinal (GI) system and plays a major role in regulating multiple digestive processes, including gastric contraction, relaxation, and acid secretion, pancreatic endocrine and exocrine secretions, gallbladder contraction, and intestinal motility (Rogers and Hermann, 2012; Liddle, 2018; Mawe et al., 2018; Schubert and Peura, 2008). While activation of the parasympathetic nervous system is typically described as promoting the general “rest and digest” response, the variety of digestive functions controlled by the DMV suggests a division of labor mediated by distinct neuron subtypes. Here, we used single-cell transcriptomics to systematically classify seven Chat+ DMV neuron subtypes by their genome-wide expression profiles. We show that expression of two genes, Cck and Pdyn, specially marks two transcriptionally distinct DMV neuron subtypes whose somas lie in different anatomical domains of the DMV. Then using respective Cre driver mice, we show that both of these DMV neuron subtypes very selectively innervate the glandular stomach, and not the nonglandular stomach or any other GI organ. Finally, we show that Cck+ and Pdyn+ DMV neurons selectively form pericellular arborizations around different enteric neurons releasing functionally opposite neurotransmitters. Our studies confirm that single-cell transcriptomics is a powerful approach for elucidating highly specific lines of brain-to-gut parasympathetic communication. Of interest, it was recently shown that cholinergic sympathetic preganglionic neurons in the spinal cord, which are analogous to DMV parasympathetic preganglionic neurons, are also transcriptionally heterogenous (Blum et al., 2021). Perhaps like DMV neurons, these transcriptional differences similarly predict specificity in targeting of sympathetic postganglionic neurons and hence sympathetically regulated tissues/organs.
Given that most if not all DMV motor neurons are cholinergic, it has been proposed that DMV neurons exert different – and sometimes opposing – effects by engaging separate populations of enteric neurons (Powley et al., 2019; Rogers and Hermann, 2012; Chang, Mashimo, and Goyal, 2003). However, direct evidence for such a “labeled-line” model has been difficult to obtain due to the lack of methods for selectively and specifically visualizing discrete DMV neuron subtypes. Using the above-mentioned approaches, we show that two subtypes of DMV neurons, Cck+ and Pdyn+ neurons, innervate the same region of the digestive system but remarkably form pericellular arborizations around neurochemically-distinct enteric neurons. Specifically, Cck+ DMV neurons preferentially form pericellular arborizations around cholinergic enteric neurons while Pdyn+ DMV neurons do the opposite – form pericellular arborizations around nitrergic enteric neurons (Figure 4). As acetylcholine secreted from cholinergic enteric neurons results in gastric acid secretion and contraction, and nitric oxide released from nitrergic enteric neurons leads to gastric relaxation (Rogers and Hermann, 2012; Schubert and Peura, 2008; Travagli et al., 2006), our results are consistent with the view that separate and distinct DMV neuron subtypes, by engaging functionally unique enteric neurons, control different digestive functions.
Essentially all enteric neurons in the stomach receive DMV neuron pericellular arborizations (Chang, Mashimo, and Goyal, 2003; Berthoud, Carlson, and Powley, 1991). In this context, it is notable that Cck+ DMV neurons formed pericellular arborizations around only a subset of cholinergic enteric neurons and very few nitrergic enteric neurons, and that Pdyn+ DMV neurons formed arborizations around most nitrergic enteric neurons and very few cholinergic enteric neurons (Figure S3). There are two implications of this. First, there must be additional DMV subtypes that innervate the glandular stomach to engage cholinergic enteric neurons not innervated by Cck+ or Pdyn+ DMV neurons. These other DMV neurons are likely captured in our DMV single-cell transcriptomic analysis. Second, it suggests functional diversity in cholinergic enteric neurons in that different cholinergic enteric neurons are innervated by different DMV neuron subtypes. Consistent with this, some cholinergic neurons control gastric contraction while others control acid secretion (Furness, 2000). Of note, recent single-cell transcriptomic studies have shown that there are multiple subtypes of cholinergic and nitrergic enteric neurons (Zeisel et al., 2018; Morarach et al., 2021). In total, this suggests the possibility of a “labeled line” model where transcriptionally distinct DMV neuron subtypes control different digestive processes by targeting transcriptionally distinct enteric neuron subtypes.
The receptive relaxation reflex is a mechanism by which esophageal distension results in gastric relaxation to prevent buildup of intragastric pressure (Cannon and Leib, 1911). This reflex is an example of a vago-vagal reflex involving vagal afferent neurons that sense esophageal distension and transmit the signal to the brainstem where vagal efferent neurons in the DMV produce gastric relaxation (Chang, Mashimo, and Goyal, 2003). Using electrophysiology recordings, it was shown that esophageal distension activated caudal DMV neurons but inhibited rostral DMV neurons (Rogers, Hermann, and Travagli, 1999). These findings suggest that the DMV may have a functional organization, where rostral DMV neurons drive gastric contraction and caudal DMV neurons produce gastric relaxation (Chang, Mashimo, and Goyal, 2003). There are parallels between this study and our results in that we observed that Cck+ and Pdyn+ neurons are primarily located in the rostral and caudal halves of the DMV, respectively. Furthermore, Cck+ neurons target cholinergic enteric neurons which could contract the stomach while Pdyn+ neurons target nitrergic enteric neurons which could relax the stomach. Note that cholinergic and nitrergic enteric neurons, respectively, excite and inhibit smooth muscle activity (Powley et al., 2019; Chang, Mashimo, and Goyal, 2003). Thus, Cck+ and Pdyn+ DMV neurons could serve as vagal efferents of the receptive relaxation reflex. Alternatively, because these DMV neuron subtypes project to just the glandular domain of the stomach, and within this domain just a subset of enteric neurons, it is possible, or perhaps likely, that these subtypes play a more nuanced role in controlling certain aspects or phases of gastric contraction and relaxation.
Motor neurons in the primary motor cortex are organized in a motor homunculus where distinct cortical domains control different anatomical regions of the body (Penfield and Boldrey, 1937). Could vagal motor neurons be viscerotopically organized in the DMV? While DMV neuron somas are arranged into longitudinal columns based on their projections via the five subdiaphragmatic vagal branches (Fox and Powley, 1984), these columns don’t necessarily correspond to target sites in the gut because there is not a one-to-one correspondence between branches and sites of gut innervation (Berthoud, Carlson, and Powley, 1991). Given that Cck+ and Pdyn+ neurons project exclusively and very narrowly to just the glandular stomach, it is interesting and perhaps unexpected that they are distributed so very differently along the rostral-caudal axis of the DMV (Figure 2). Consistent with prior studies (Fox and Powley, 1984; Browning and Travagli, 2014), this strongly suggests that the DMV is not viscerotopically organized. The differences in the location of these similarly projecting Cck+ and Pdyn+ neurons in the DMV, on the other hand, may have more to do with their functionally opposite downstream enteric neurons, and thus presumed differences in their functions and therefore afferent control.
In summary, using single-cell transcriptomics, we identified seven molecular-distinct subtypes of DMV neurons and genetic markers for each subtype. We show that neurons from two subtypes are located in different regions of the DMV, send axon projections exclusively to the glandular stomach, and target neurochemically-distinct subtypes of enteric neurons. Our work provides three major steps forward - first, in the specific identification of two labeled line circuits to the stomach which provides insight into brain control of stomach function, second, in demonstrating the general “logic” by which the vagus is organized to control functions (labeled lines to discrete regions and functionally distinct enteric neurons), and third, in providing single-cell transcriptomics of the DMV vagus nerve which provides the means for elucidating all vagal motor “labeled lines” innervating all parts of the stomach, small intestine, large intestine, gallbladder, or pancreas.
STAR Methods
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed and will be fulfilled by the lead contact, Bradford Lowell (blowell@bidmc.harvard.edu).
Materials Availability
Plasmid generated for producing AAV-DIO-H2b-mCherry will be available from the lead contact.
Data and Code Availability
The raw and fully processed single-cell RNA-seq data and metadata are available at GEO accession number GSE172411.
Experimental Model and Subject Details
All animal care and experimental procedures were approved in advance by the National Institute of Health and Beth Israel Deaconess Medical Center Institutional Animal Care and Use Committee. The sNuc-Seq experiments used 12-week old male Chat-IRES-Cre mice (Rossi et al., 2011), on a C57BL/6J genetic background. The in situ hybridization experiments used C57BL/6J mice supplied by the Jackson Laboratory (JAX: 000664) as well as Cck-IRES-Cre (JAX: 012706) (Taniguchi et al., 2011) and Pdyn-IRES-Cre (Krashes et al., 2014) crossed to LSL-tdTomato mice (JAX: 007908) (Madisen et al., 2010). The axon tracing experiments used Chat-IRES-Cre, Cck-IRES-Cre, Pdyn-IRES-Cre, and Chat-GFP (JAX: 007902) (Tallini et al., 2006) mice that were maintained on a mixed background and have been described previously. Unless otherwise specified, all experiments used adult (8 – 12 week old) mice with approximately equal numbers of male and female mice. Mice were housed at 22–24 °C with a 12 h light: 12 h dark cycle and ad libitum access to standard mouse chow (Teklad F6 Rodent Diet 8664; 4.05 kcal per g, 3.3 kcal per g metabolizable energy, 12.5% kcal from fat; Harlan Teklad) and water.
Method Details
Viral injections
Stereotaxic injections were performed using previously described procedures (Resch et al., 2017). Mice were anesthetized with xylazine (5 mg/kg) and ketamine (75 mg/kg) diluted in saline (0.9%) and placed into a stereotaxic apparatus (Kopf). A pulled glass micropipette (20–40 μm diameter tip) was used for stereotaxic injections of adeno-associated virus (AAV), and coordinates for DMV injections were anterior ± 0.3 mm, lateral ± 0.2 mm, and ventral 0.25 and 0.5 mm from calamus scriptorius. Virus was injected (600 nl/side) by an air pressure system using picoliter air puffs through a solenoid valve (Clippard EV 24VDC) pulsed by a Grass S48 stimulator to control injection speed (40 nl min−1). The pipette was removed 3 minutes post-injection followed by wound closure using absorbable suture for muscle and silk suture for skin. Subcutaneous injection of sustained release Meloxicam (4 mg kg−1) was provided as postoperative care. To label the Chat+ neurons for the sNuc-seq studies (Figure 1A), an AAVDJ-hSyn-DIO-H2b-mCherry was prepared. The H2b-mCherry sequence was obtained from the targeting construct used to generate the TRAP mouse (Long et al., 2014) and cloned into the backbone of pAAV-hSyn-DIO-mCherry (Addgene #50459) between Asc I and Nhe I in reverse orientation. Axon tracing experiments (Figures 3, 4, and S2) utilized AAV9-CAG-FLEX-tdTomato (Oh et al., 2014) purchased from University of Pennsylvania School of Medicine Vector Core (donating investigator, Dr. Hongkui Zeng) and AAV9-CAG-FLEX-PLAP (Prescott et al., 2020) obtained from Harvard Medical School (donating investigator, Dr. Stephen Liberles). All subjects determined to be surgical “misses” based on little or absent reporter expression were excluded from analyses.
Brain tissue preparation
Mice were terminally anesthetized with 7% choral hydrate (500 mg kg−1; Sigma Aldrich) diluted in saline and transcardially perfused first with 0.1 M phosphate-buffered saline (PBS) then 10% neutral-buffered formalin solution (NBF; Thermo Fisher Scientific). Brains were extracted and post-fixed overnight at 4° C in NBF. The next day brains were switched to PBS containing 30% sucrose for cryoprotection. Finally, brains were sectioned coronally at 30 μm on a freezing microtome (Leica Biosystems) into two equal series. A single series of sections per animal was used in the histological studies.
Gastrointestinal Organ Tissue Preparation
Mice were terminally anesthetized with 7% choral hydrate (500 mg kg−1; Sigma Aldrich) diluted in saline and transcardially perfused first with 0.1 M phosphate-buffered saline (PBS) then 10% neutral-buffered formalin solution (NBF; Thermo Fisher Scientific). Gastrointestinal organs were extracted and post-fixed overnight at 4° C in NBF. The next day, organs were switched to PBS and stored at 4° C before staining.
Immunofluorescence
Brain tissue sections were washed 4X in PBS prior to a blocking step containing 5% normal donkey serum and 0.5% Triton X-100 in PBS for one hour at room temperature. Primary antibody was prepared in the same blocking solution and incubated overnight at the following concentrations: Rat anti-mCherry (Invitrogen) 1:1,000, Rabbit anti-GFP (Invitrogen) 1:1,000, Goat anti-Chat (EMD Millipore) 1:500. The next day sections were washed 4X in PBS, then incubated for 2 hours at room temperature in Alexa Fluor fluorescent secondary antibody (Life Technologies; 1:1,000) prepared in blocking solution. Finally, sections were washed 4X in PBS, mounted on gelatin-coated slides, and coverslipped with Vectashield mounting media containing DAPI (Vector Labs). Fluorescent images were captured using an Olympus VS120 slide-scanning microscope.
Wholemount Immunofluorescence
Gastrointestinal organs were washed 4X in PBS contained 0.5% Triton-X 100 prior to a blocking step containing 5% normal donkey serum and 0.5% Triton X-100 in PBS for two hours at room temperature. Primary antibody was prepared in the same blocking solution and at the following concentrations: Rat anti-mCherry (Invitrogen) 1:1,000, Chicken anti-GFP (Invitrogen) 1:1,000, Rabbit anti-GFP (Invitrogen) 1:1,000, Rabbit anti-nNOS (Invitrogen) 1:500. After 48–72 hours of incubating at 4° C, organs were washed 4X in PBS containing 0.5% Triton X-100, then incubated for 24–48 hours at 4° C in Alexa Fluor fluorescent secondary antibody (Life Technologies; 1:1,000) prepared in blocking solution. Finally, the organs were washed 4X in PBS, mounted on gelatin-coated slides, and coverslipped with Vectashield mounting media containing DAPI (Vector Labs). Fluorescent images of all gastrointestinal organs except the gallbladder were captured using an Olympus VS120 slide-scanning microscope. Fluorescent images of the gallbladder were taken using a two-photon microscope from Dr. Mark Andermann’s lab.
Placental Alkaline Phosphatase Staining
Mice were terminally anesthetized with 7% choral hydrate (500 mg kg−1; Sigma Aldrich) diluted in saline and transcardially perfused first with 0.1 M phosphate-buffered saline (PBS). The stomach and pancreas were collected and fixed in 4% paraformaldehyde (Electron Microscopy Sciences) for 1 hour at room temperature. The tissues were then washed in PBS and incubated in alkaline phosphatase (AP) buffer (0.1 M Tris HCl pH 9.5, 0.1 M NaCl, 50 mM MgCl2, 0.1% Tween20, 5 mM levamisole) for two hours at 70 °C. Afterward, the tissues were washed twice in AP buffer. AP activity was visualized with NCT/BCIP solution (ThermoFisher Scientific 34042) and stained samples were post-fixed (4% paraformaldehyde, overnight, 4°C), dehydrated through a series of ethanol washes, and cleared using a 1:2 mixture of benzyl alcohol (Sigma-Aldrich 402834–500ML): benzyl benzoate (Sigma-Aldrich B6630–1L). Wholemount images were taken by light microscopy (Zeiss AxioZoom).
In situ Hybridization
Mice received three intraperitoneal injections of Fluorogold (30 mg/kg, Fluorochrome) on days 1, 3, and 5. On day 7, animals were terminally anesthetized with 7% choral hydrate (500 mg kg−1; Sigma Aldrich) diluted in saline and transcardially perfused first with 0.1 M phosphate-buffered saline (PBS) then 4% paraformaldehyde. Brains were extracted and post-fixed overnight at 4° C in 4% paraformaldehyde. The brains were then cryopreserved with 10%, 20%, and 30% sucrose for one day each at 4° C. On the next day, brains were sectioned coronally at 25 μm on a freezing microtome (Leica Biosystems). The sections were washed in PBS containing 0.5% Triton X-100 and mounted on Fisherbrand Superfrost Plus microscope slides. The slides were dried in an oven at 60° C for one hour. Sections were then fixed for 15 minutes in 4 % paraformaldehyde at 4° C, washed in PBS, dehydrated in an ethanol series, and then air dried. An ImmEdge Hydrophobic Barrier Pen was used to draw a barrier around the sections. The sections were then incubated in Protease Plus in a HybEZ II Oven for 30 minutes at 40° C, with target probes (Pdyn – 318771, Cck – 402271, and tdTomato – 317041) for 2 hours at 40° C, and then treated with Amp 1–3. Opal 570 and 650 dyes (Akoya Biosciences) were used for probe visualization. Immunofluorescence was performed as described above with Rabbit anti-fluorescent gold (EMD Millipore) 1:1,000 prepared in blocking solution. Fluorescent images were taken using a confocal microscope (Zeiss LSM 880).
sNuc-Seq
After allowing three weeks for DMV expression of the injected AAV-hSyn-DIO-H2b-mCherry, mice were sacrificed for brain extraction. To avoid stress-related changes in nuclear mRNA, the mice were rapidly decapitated immediately upon removal from their home cage in the vivarium. Tissue collection was performed in the morning, approximately three hours after the onset of the light cycle. Brain tissue was extracted, chilled in a DMEM/F12 media slush for two minutes, then manually blocked at 1 mm intervals through the hindbrain on an ice-cold, stainless steel mouse brain matrix. Coronal sections containing and adjacent to the DMV were incubated in ice-cold RNAlater (Qiagen catalog #76106) for at least thirty minutes prior to being imaged on a fluorescence stereoscope (Zeiss Discovery V8) in order to visualize H2b-mCherry-labeled cells in the DMV for dissection. Aided by fluorescence stereoscope imaging, H2b-mCherry+ DMV tissue was dissected using a microscalpel and pooled into a single sample. Individual H2b-mCherry+ nuclei were then isolated using fluorescence-activated cell sorting (FACS), processed into cDNA libraries, and sequenced by Next-Seq 500 (SR75, high-output kit) as previously described (Todd et al., 2020). Reads were demultiplexed by bcl2fastq2 v2.20.0 (Illumina) and aligned to the mouse genome by STAR v2.6.1 (Dobin et al., 2013). Duplicates were removed with Picard Tools v2.18.21. Aligned reads were processed into a digital gene expression (DGE) file with Drop-Seq Tools v2.3.0 and tagged using “GENCODE_M16_PRI” annotation. The DGE file was input to the single-cell analysis R package, Seurat (Stuart et al., 2019), version 3.2, for further analysis, using default settings except where noted. After filtering out genes expressed in fewer than three cells and cells expressing fewer than 5,000 genes, the dataset was normalized, scaled, and then analyzed for variable genes using Seurat’s NormalizeData, ScaleData, and FindVariableFeatures functions, respectively. The top 1,500 variable genes were dimensionally reduced with principal component (PC) analysis (RunPCA function), and the top 19 PCs were used for non-linear dimensionality reduction and clustering with Seurat’s FindNeighbors and FindClusters (resolution set to 1.2) functions. Seurat’s BuildClusterTree function was used to order clusters by their similarity in gene expression. Differential gene expression was analyzed with the FindAllMarkers function and the following settings: min.pct = 0.25, logfc.threshold = 0.25. Figures were generated with Seurat’s data visualization functions and Microsoft Excel.
Quantification and Statistical Analysis
No statistical analyses were performed except for sNuc-Seq data where detailed analysis is described above. No statistical methods were used to determine whether the data met the assumptions of the statistical approach. The number of mice (n) used in each experiment is indicated in the figure legends and here. Sample sizes from left to right: Figure 1 (4), Figure 2 (3), Figure 3B–C (10, 8 8), Figure 3D (5, 4, 4), Figure 3E (5, 4, 4), Figure 3F (5, 4, 4), Figure 4A (3, 3), Figure 4B (3, 3), Figure S1 (2, 2), Figure S3A (2), and Figure S3B (3, 3, 3, 3).
Supplementary Material
Table S1. Average Gene Expression in Each Neuron Cluster, Related to Figure 1
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken anti-GFP | Invitrogen | Cat #: A10262; RRID: AB_2534023 |
| Rabbit anti-GFP | Invitrogen | Cat #: A11122; RRID: AB_2307355 |
| Rat anti-mCherry | Invitrogen | Cat #: M11217; RRID: AB_2536611 |
| Rabbit anti-nNOS | Invitrogen | Cat #: 61–7000; RRID: AB_2313734 |
| Goat anti-Chat | MilliporeSigma | Cat #: AB144P; RRID: AB_2079751 |
| Rabbit anti-Fluorogold | MilliporeSigma | Cat #: AB153-I; RRID: AB_2632408 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Fluorogold | Fluorochrome | Cat #: Fluoro-gold; RRID: AB_2314408 |
| Opal 570 | Akoya Biosciences | Cat #: FP1488001KT |
| Opal 650 | Akoya Biosciences | Cat #: FP1496001KT |
| Critical Commercial Assays | ||
| RNAscope Multiplex Fluorescent Reagent Kit V2 | Advanced Cell Diagnostics | Cat #: 323100 |
| NexteraXT DNA Library Preparation Kit | Illumina | Cat #: FC-131–1096 |
| Nextera XT Index Kit v2 Set A | Illumina | Cat #: FC-131–2001 |
| Nextera XT Index Kit v2 Set B | Illumina | Cat #: FC-131–2002 |
| Nextera XT Index Kit v2 Set C | Illumina | Cat #: FC-131–2003 |
| Nextera XT Index Kit v2 Set D | Illumina | Cat #: FC-131–2004 |
| Deposited Data | ||
| Raw data from RNA sequencing | Gene Expression Omnibus | GEO: GSE172411 |
| Experimental Models: Organisms/Strains | ||
| Mouse/Chat-IRES-Cre | Jackson Laboratory; Rossi et al., 2011 | Cat #: 031661; RRID: IMSR_JAX:031661 |
| Mouse/Cck-IRES-Cre | Jackson Laboratory; Taniguchi et al., 2011 | Cat #: 012706; RRID: IMSR_JAX:012706 |
| Mouse/Pdyn-IRES-Cre | Jackson Laboratory; Krashes et al., 2014 | Cat #: 027958; RRID: IMSR_JAX:027958 |
| Mouse/Ai14(lox)-tdTomato | Jackson Laboratory; Madisen et al., 2010 | Cat #: 007914; RRID: IMSR_JAX:007914 |
| Mouse/Chat-GFP | Jackson Laboratory; Tallini et al., 2006 | Cat #: 007902; RRID: IMSR_JAX:007902 |
| Oligonucleotides | ||
| Mm-Pdyn | Advanced Cell Diagnostics | Cat #: 318771 |
| Mm-Cck-C2 | Advanced Cell Diagnostics | Cat #: 402271-C2 |
| tdTomato-C3 | Advanced Cell Diagnostics | Cat #: 317041-C3; |
| Recombinant DNA | ||
| AAVDJ-DIO-H2b-mCherry | This paper | N/A |
| AAV9-CAG-Flex-tdTomato | Penn Vector Core | Cat #: AV-9-ALL864; Addgene: 51503 |
| AAV9-CAG-Flex-PLAP | Prescott et al., 2020 | N/A |
| Software and Algorithms | ||
| FIJI | Schindelin et al., 2012 | https://fiji.sc/; RRID: RRID: SCR_002285 |
| R | R version 3.6 | https://www.r-project.org/; RRID: SCR_001905 |
| Illustrator | Adobe | https://www.adobe.com; RRID: SCR_010279 |
| Excel | Microsoft | https://www.microsoft.com/en-us/; RRID: SCR_016137 |
| bcl2fastq v2.20.0 | Illumina | https://support.illumina.com/sequencing/sequencing_software/bcl2fastq-conversion-software.html; RRID: SCR_015058 |
| STAR v2.6.1 | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| Picard Tools v2.18.21 | Broad Institute | http://broadinstitute.github.io/picard/; RRID: SCR_006525 |
| Drop-Seq Tools v2.3.0 | Broad Institute | https://github.com/broadinstitute/Drop-seq |
| Seurat v3.2 | Stuart et al., 2019 | https://github.com/satijalab/seurat/; RRID: SCR_007322 |
Highlights.
Single nucleus RNA-seq reveals seven molecularly-distinct vagal motor neuron subtypes
DMV-Pdyn and DMV-Cck neurons are located in different anatomical domains of the DMV
DMV-Pdyn and DMV-Cck neurons exclusively innervate the glandular stomach
DMV-Pdyn and DMV-Cck neurons target neurochemically-distinct enteric neurons
Acknowledgments
This work was supported by the following National Institutes of Health grants to B.BL.: P30DK046200, P30DK057521, R01DK122976, R01DK075632, R01DK089044, R01DK111401, R01DK096010; to J.T: F31DK122620. J.N.C. was supported with an American Heart Association Postdoctoral Fellowship (14POST20100011); an American Diabetes Association Pathway to Stop Diabetes award (1-18-INI-14); and pilot grant funding from the Boston Nutrition Obesity Research Center (under a grant from the National Institutes of Health, NIH; award number P30 DK046200) and from the Boston Area Diabetes Endocrinology Research Center (BADERC; NIH, under award number P30DK057521). S.D.L. is an investigator of the Howard Hughes Medical Institute. We thank Stephen Zhang from Mark Andermann’s lab for the two-photon imaging. We also thank the BNORC Bioinformatics Core (P30DK046200), the Bauer Core Facility at Harvard University, the Molecular Medicine Core of Beth Israel Deaconess Medical Center, and the ICCB-Longwood Screening Facility of Harvard Medical School for assistance with the single-nucleus RNA-Seq experiments, the Viral Core of Boston Children’s Hospital for AAV production, and the BNORC Transgenic Core (P30DK046200) for prior generation of Chat-IRES-Cre and Pdyn-IRES-Cre mice. Confocal imaging was performed at BIDMC’s Confocal Imaging Core. We thank the members of the B.B.L. and S.D.L. labs for helpful discussions.
Footnotes
Declaration of Interests
S.D.L. is a consultant for Kallyope, Inc.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- • Berthoud HR, Carlson NR, and Powley TL (1991). Topography of efferent vagal innervation of rat gastrointestinal tract. Am J Physiol. 260, R200–R207. [DOI] [PubMed] [Google Scholar]
- • Blum JA, Klemm S, Shadrach JL, Guttenplan KA, Nakayama L, Kathiria A, Hoang PT, Gautier O, Kaltschmidt JA, Greenleaf WJ, et al. (2021). Single-cell transcriptomic analysis of the adult mouse spinal cord reveals molecular diversity of autonomic and skeletal motor neurons. Nat Neurosci. 24, 572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • Browning KN, and Travagli RA (2014). Central nervous system control of gastrointestinal motility and secretion and modulation of gastrointestinal functions. Compr Physiol. 4, 1339–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • Brunet JF, amd Pattyn A (2002). Phox2 genes - from patterning to connectivity. Curr Opin Genet Dev. 12, 435–40. [DOI] [PubMed] [Google Scholar]
- • Cannon WB, and Leib CW (1911). The receptive relaxation of the stomach. Am J Physiol. 29, 267–273. [Google Scholar]
- • Chang HY, Mashimo H, and Goyal RK (2003). Musings on the wanderer: what’s new in our understanding of vago-vagal reflex? IV. Current concepts of vagal efferent projections to the gut. Am J Physiol Gastrointest Liver Physiol. 284, G357–66. [DOI] [PubMed] [Google Scholar]
- • Chang RB, Strochlic DE, Williams EK, Umans BD, and Liberles SD (2015). Vagal sensory neuron subtypes that differentially control breathing. Cell. 161, 622–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • Dobin A, Davis CA, Schlesinger F, Denkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • Fox EA, and Powley TL (1984). Longitudinal columnar organization within the dorsal motor nucleus represents separate branches of the abdominal vagus. Brain Research. 341, 269–282. [DOI] [PubMed] [Google Scholar]
- • Furness JB (2000). Types of neurons in the enteric nervous system. J Auton Ner Syst. 81, 87–96. [DOI] [PubMed] [Google Scholar]
- • Gibbons CH (2019). Basics of autonomic nervous system function. Handb Clin Neurol. 160, 407–418. [DOI] [PubMed] [Google Scholar]
- • Habib N, Li Y, Heidenreich M, Swiech L, Avraham-Davidi I, Trombetta JJ, Hession C, Zhang F, Regev A (2016). Div-Seq: single-nucleus RNA-seq reveals dynamics of rare adult newborn neurons. Science. 353, 925–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • Hayakawa T, Kuwahara S, Maeda S, Tanaka K, and Seki M (2006). Direct synaptic contacts on the myenteric ganglia of the rat stomach from the dorsal motor nucleus of the vagus. J Comp Neurol. 498, 352–362. [DOI] [PubMed] [Google Scholar]
- • Huang XF, Tork I, and Paxinos G (1993). Dorsal motor nucleus of the vagus nerve: a cyto- and chemoarchitectonic study in the human. J Comp Neurol. 330, 158–182. [DOI] [PubMed] [Google Scholar]
- • Krashes MJ, Shah BP, Madara JC, Olson DP, Strochlic DE, Garfield AS, Vong L, Pei H, Watabe-Uchida M, Uchida N, et al. 2014. (2014). An excitatory paraventricular nucleus to AgRP neuron circuit that drives hunger. Nature. 507, 238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, Wildberg A, Gao D, Fung H-L, Chen S, et al. (2016). Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 352, 1586–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • Langley JN (1897). On the Regeneration of Pre-Ganglionic and of Post-Ganglionic Visceral Nerve Fibres. J Physiol. 22, 215–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • Langley JN (1921). The Autonomic Nervous System Part I (Cambridge: ). [Google Scholar]
- • Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. (2007) Genome-wide atlas of gene expression in the adult mouse brain. Nature. 445, 168–176. [DOI] [PubMed] [Google Scholar]
- • Leong SK, and Ling EA (1990). Labelling neurons with fluorescent dyes administered via intravenous, subcutaneous or intraperitoneal route. J Neurosci Methods. 32, 15–23. [DOI] [PubMed] [Google Scholar]
- • Liddle RA (2018). Regulation of Pancreatic Secretion. In Physiology of the Gastrointestinal Tract, Said HM, ed. (Elsevier/Academic Press; ), pp. 895–929. [Google Scholar]
- • Long JZ, Svensson KJ, Tsai L, Zeng X, Roh HC, Kong X, Rao RR, Lou J, Lokurkar I, Baur W, et al. (2014). A smooth muscle-like origin for beige adipocytes. Cell Metab 19, 810–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • Luo L, Callaway EM, and Svoboda K (2018). Genetic Dissection of Neural Circuits: A Decade of Progress. Neuron. 98, 256–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 13, 133–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • Mawe GM, Lavoie B, Nelson MT, and Pozo MJ (2018). Neuromuscular Function in the Biliary Tract. In Physiology of the Gastrointestinal Tract, Said HM, ed. (Elsevier/Academic Press; ), pp. 453–468. [Google Scholar]
- • Morarach K, Mikhailova A, Knoflach V, Memic F, Kumar R, Li W, Ernfors P, and Marklund U (2021). Diversification of molecularly defined myenteric neuron classes revealed by single-cell RNA sequencing. Nat Neurosci. 24, 34–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, Wang Q, Lau C, Kuan L, Henry AM, et al. (2014). A mesoscale connectome of the mouse brain. Nature. 508, 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • Pattyn A, Morin X, Cremer H, Goridis C and Brunet JF (1997) Expression and interactions of the two closely related homeobox genes Phox2a and Phox2b during neurogenesis. Development. 124, 4065–75. [DOI] [PubMed] [Google Scholar]
- • Penfield W, and Boldrey E (1937). Somatic motor and sensory representation in the cerebral cortex of man as studied by electrical stimulation. Brain. 60, 389–440. [Google Scholar]
- • Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, and Sandberg R (2014). Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181. [DOI] [PubMed] [Google Scholar]
- • Powley TL, Fox EA, amd Berthoud HR (1987). Retrograde tracer technique for assessment of selective and total subdiaphragmatic vagotomies. The American journal of physiology. 253, R361–370. [DOI] [PubMed] [Google Scholar]
- • Powley TL, Jaffey DM, McAdams J, Baronowsky EA, Black D, Chesney L, Evans C, and Philips RJ (2019). Vagal innervation of the stomach reassessed: brain-gut connectome uses smart terminals. Ann N Y Acad Sci. 1454, 14–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • Prescott SL, Umans BD, Williams EK, Brust RD, and Liberles SD (2020). An Airway Protection Program Revealed by Sweeping Genetic Control of Vagal Afferents. Cell. 181, 574–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • Resch JM, Fenselau H, Madara JC, Wu C, Campbell JN, Lyubetskaya A, Dawes BA, Tsai LT, Li MM, Livneh Y, et al. (2017). Aldosterone-sensing neurons in the NTS exhibit state-dependent pacemaker activity and drive sodium appetite via synergy with angiotensin II signaling. Neuron. 96, 90–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • Rogers RC, and Hermann GE (2012). Brainstem Control of the Gastric Function. In Physiology of the Gastrointestinal Tract, Johnson LR, Gishan FK, Kaunitz JD, Merchant JL, Said HM, and Wood JD, eds. (Academic Press; ), pp. 861–891. [Google Scholar]
- • Rogers RC, Hermann GE, and Travagli RA (1999). Brainstem pathways responsible for oesophageal control of gastric motility and tone in the rat. J Physiol. 514, 369–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • Rossi J, Balthasar N, Olson D, Scott M, Berglund E, Lee CE, Choi MJ, Lauzon D, Lowell BB, and Elmquist JK (2011). Melanocortin-4 receptors expressed by cholinergic neurons regulate energy balance and glucose homeostasis. Cell Metab. 13, 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • Schubert ML, and Peura DA (2008). Control of gastric acid secretion in health and disease. Gastroenterology. 134, 1842–60. [DOI] [PubMed] [Google Scholar]
- • Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R (2019). Comprehensive integration of single-cell data. Cell. 177, 1888–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • Tallini YN, Shui B, Greene KS, Deng KY, Doran R, Fisher PJ, Zipfel W, and Kotlikoff MI (2006). BAC transgenic mice express enhanced green fluorescent protein in central and peripheral cholinergic neurons. Physiol. Genomics. 27, 391–397. [DOI] [PubMed] [Google Scholar]
- • Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsiani D, Fu Y, Lu J, Lin Y, et al. (2011). A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 71, 995–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • Travagli RA, Hermann GE, Browning KN & Rogers RC (2006). Brainstem circuits regulating gastric function. Annu Rev Physiol. 68, 279–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • Todd WD, Venner A, Anaclet C, Broadhurst RY, De Luca R, Bandaru SS, Issokson L, Hablitz LM, Cravetchi O, Arrigoni E, et al. (2020). Suprachiasmatic VIP neurons are required for normal circadian rhythmicity and comprised of molecularly distinct subpopulations. Nat. Commun 11, 4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, Häring M, Braun E, Borm LE, La Manno G, et al. (2018). Molecular architecture of the mouse nervous system. Cell. 174, 999–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Average Gene Expression in Each Neuron Cluster, Related to Figure 1
Data Availability Statement
The raw and fully processed single-cell RNA-seq data and metadata are available at GEO accession number GSE172411.


