Abstract
SPTBN1 encodes βII-spectrin, the ubiquitously expressed β-spectrin that forms micrometer-scale networks associated with plasma membranes. Mice deficient in neuronal βII-spectrin have defects in cortical organization, developmental delay, and behavioral deficiencies. These phenotypes, while less severe, are observed in haploinsufficient animals, suggesting that individuals carrying heterozygous SPTBN1 variants may also present with measurable compromise of neural development and function. Here we identify heterozygous SPTBN1 variants in 29 individuals who present with developmental, language and motor delays, mild to severe intellectual disability, autistic features, seizures, behavioral and movement abnormalities, hypotonia, and variable dysmorphic facial features. We show that these SPTBN1 variants lead to effects that affect βII-spectrin stability, disrupt binding to key molecular partners, and disturb cytoskeleton organization and dynamics. Our studies define SPTBN1 variants as the genetic basis of a neurodevelopmental syndrome, expand the set of spectrinopathies affecting the brain, and underscore the critical role of βII-spectrin in the central nervous system.
Spectrins are ubiquitously expressed, elongated polypeptides that bind membrane lipids and ankyrins to line the plasma membrane1,2. The spectrin meshwork is formed by heterodimeric units of α-spectrin and β-spectrin assembled side-to-side in antiparallel fashion, which then form head-to-head tetramers that crosslink F-actin to form spectrin-actin arrays1,2. Mammalian neurons express the most diverse repertoire of spectrins, which includes αII-spectrin and all five β-spectrins (βI–V spectrins)3. Together with ankyrins, spectrins self-assemble with both remarkable long-range regularity and micro- and nanoscale specificity to precisely position and stabilize cell adhesion molecules, membrane transporters, ion channels, and other scaffolding proteins3. Some spectrins also enable intracellular organelle transport3. Unsurprisingly, deficits in spectrins underlie several neurodevelopmental and neurodegenerative disorders4–6. For example, inherited autosomal dominant variants in βIII-spectrin (encoded by SPTBN2) cause late onset spinocerebellar ataxia type 5 (SCA5)5, while pathogenic de novo variants have been associated with early childhood ataxia, intellectual disability (ID), and developmental delay (DD)7–12. Similarly, autosomal recessive SPTBN2 variants13–15 are associated with childhood ataxia, which may also present with ID and DD13. De novo pathogenic variants in SPTAN1, which encodes αII-spectrin, cause childhood onset epileptic syndromes16–20 including West syndrome, an early-infantile epileptic encephalopathy (EIEE) characterized by frequent severe seizures and persistent abnormality of cortical function5. Some individuals co-present with spastic quadriplegia, DD, and various brain defects5. In addition, dominantly inherited SPTAN1 nonsense variants cause juvenile onset hereditary motor neuropathy21. Biallelic alterations in βIV-spectrin (encoded by SPTBN4) result in congenital hypotonia, neuropathy, and deafness, with and without ID6,22,23.
Neuronal βII-spectrin, encoded by SPTBN1, is the most abundant β-spectrin in the brain and forms tetramers with αII-spectrin, which intercalate F-actin rings to build a sub-membranous periodic skeleton (MPS)24. A cytosolic pool of βII-spectrin promotes bidirectional axonal organelle transport25,26. We previously reported that mice lacking βII-spectrin in all neural progenitors (Sptbn1flox/flox;Nestin-Cre; referred to as βIISp-KO) show early postnatal lethality, reduced long-range cortical and cerebellar connectivity, spontaneous seizures, and motor deficits26. However, the impact of human genetic variation in SPTBN1 on βII-spectrin function and its association with disease has not been studied. Here we describe a cohort of 29 individuals carrying rare, mostly de novo variants in SPTBN1 affected by an autosomal dominant neurologic syndrome presenting with global developmental, language and motor delays, mild to severe ID, autistic features, seizures, behavioral abnormalities, hypotonia, and variable dysmorphisms. This suggests conserved roles for βII-spectrin in neuronal development and function. Our functional studies indicate that SPTBN1 variants affect protein stability, disrupt binding to key protein partners, and affect cytoskeleton organization and dynamics. Consequently, histology and behavioral studies in brain βII-spectrin-deficient mice recapitulated developmental and behavioral phenotypes of individuals with SPTBN1 variants. Collectively, our data strongly support pathogenic mechanisms of SPTBN1 variants as the genetic cause of a neurodevelopmental syndrome and underscores the multifaceted role of βII-spectrin in the nervous system.
Results
SPTBN1 variants carriers present with a neurodevelopmental syndrome.
A cohort of 29 individuals with a neurodevelopmental disorder from 28 families (one pair of siblings) who harbor heterozygous variants in SPTBN1 was identified through whole genome (WGS) or exome (WES) sequencing (Fig. 1, Supplementary Table 1 and Supplementary Note). Twenty-eight unique variants are described (P10 has two de novo variants in cis), of which 22 are missense, three are nonsense, and three are canonical splice-site variants, with two predicted by SpliceAI27 to lead to in-frame deletions and one to a frameshift that introduces a premature stop codon (Fig. 1a and Supplementary Table 2). Missense variants in codons Gly205, Thr268, Arg411 and Arg1003 were identified in multiple individuals (Fig. 1a). Approximately half of the variants cluster in the second CH domain (CH2), with the rest distributed in various spectrin repeats (SR) (Fig. 1a). Twenty-four individuals carry de novo variants, with proband P10 having two de novo variants in cis (p.(Thr268Ala) and p.(Phe344Leu)). Mosaicism in P17 (13.3% of reads) suggests that the p.(Glu491Gln) variant occurred de novo. Two maternal half-siblings (P21 and P22) inherited the p.(Arg1003Trp) variant from their unaffected mother, who is mosaic for the variant at a low level (1.8% of next generation sequencing reads).
Fig. 1 |. SPTBN1 variants found in individuals with neurodevelopmental disorders.

a, Schematic representation of functional domains of βII-spectrin. CH1, calponin homology domain 1 (teal); CH2, calponin homology domain 2 (red); SR, spectrin repeat (green); PH, pleckstrin homology domain (purple). The locations of SPTBN1 variants are indicated. b, Alignment of protein sequences for βII-spectrin and orthologs show that missense variants identified in affected individuals in this study are located at highly conserved residues across species from humans to Drosophila. Accession numbers: human (Homo Sapiens, NP_003119.2), chimp (Pan troglodytes, XP_001154155.1), mouse (Mus musculus, NP_787030.2), frog (Xenopus tropicalis, NP_001362280.1), zebrafish (Danio rerio, XP_009304586.2), worm (C. elegans, NP_001024053.2), fly (Drosophila melanogaster, NP_001259660.1). The position of SPTBN1 variants analyzed in the sequenced of human βII-spectrin is shown for reference. c, Photos of individuals with SPTBN1 variants. Ages at the time of photograph are: P8, 7y8m; P9, 16y; P12, 11y; P13, 6y; P21 left, unknown; P21 right, 11y; P22, 15y; P28, 3y11m. d, Examples of brain MRI findings: diffuse cerebral parenchymal volume loss (L>R) and asymmetric appearance of hippocampi (P1, acquired at <1y), white matter disease in the supratentorial and infratentorial regions (P18, acquired at 7y), thinning of the posterior body of the corpus callosum without significant volume loss (P28, acquired at 10m).
SPTBN1 is intolerant to both missense (Z = 4.54) and loss-of-function (LOF) variants (pLI = 1, LOEUF = 0.09) (gnomAD v2.1.128, https://gnomad.broadinstitute.org/) and its haploinsufficiency score (%HI = 2.62) suggests haploinsufficiency29. Sequence alignment of human βII-spectrin and its orthologs shows a high evolutionary conservation of the residues impacted by these variants (Fig. 1b). Consistent with their implied functional relevance, all variants are absent or extremely rare in the population (gnomAD v2.1.1)28, with most predicted to be likely damaging to protein function. Complete variant details with inheritance and in silico prediction scores can be found in Supplementary Table 2 and the Supplementary Note.
Phenotypic findings are summarized in Supplementary Table 1, and detailed clinical and family histories are provided in the Supplementary Note. The cohort included 17 male and 12 female probands (P) spanning from six months to 26 years of age at last evaluation. All but one proband had early onset of symptoms with primarily DD presentations, with 21 reporting ID. Developmental regression was noted in P9, P10, and P12. P29 exhibited mild delays; however, the primary symptom, dystonia, was observed at age 1330. Similarly, while delays were noted for P23, the primary phenotype was liver-related. Only partial phenotype information was obtained for previously reported P4 and P2531, as well as for P3 from the Deciphering Developmental Disorders (DECIPHER) database32.
Nine individuals have a history of seizures, four of whom were diagnosed with frontal lobe or generalized epilepsy. Seven individuals had abnormal brain MRI findings, including P2, P10, and P28 with thinning of the corpus callosum (CC) (Fig. 1d), P1 and P16 with ventriculomegaly (Fig. 1d), P2 and P22 with delayed myelination, and P1 and P10 respectively showing diffuse cerebral parenchymal (Fig. 1d) and mild cerebellar and vermian atrophy. Other brain MRI findings were unique (Fig. 1d, Supplementary Table 1, and Supplementary Note). Behavioral concerns were common within the cohort. Six individuals displayed autistic features or had an autism spectrum disorder (ASD) diagnosis, including P19 and P24 previously reported in a WES study of ASD individuals33. P25 was identified in a WES study of Tourette syndrome cohorts31. Fifteen individuals presented with other behavioral concerns, including attention deficit and hyperactivity disorder (ADD/ADHD) (n = 12), anxiety (n = 3), obsessive behavior (n = 3), emotional liability (n = 8), and aggressive or self-injurious behaviors (n = 7). Seven individuals experienced sleep disturbances, in some cases co-occurring with seizure episodes. Additional findings include changes in muscle tone, movement abnormalities, hearing impairments, dysmorphic features (Fig. 1c), and head size and shape anomalies (Supplementary Note).
Individuals with the same or similar genetic variation display phenotypic variability. The c.3007C>T (p.(Arg1003Trp)) variant was identified in P20 and in maternal half-siblings P21 and P22 of an unrelated family. While all three have DD features, only P21 reported seizures, only P22 reported abnormal brain MRI, and P21 did not report behavioral or emotional concerns. Similarly, variants in unrelated duos P4 and P5, and P15 and P16 affect p.Gly205 and p.Arg411 residues, respectively, but result in different amino acid substitutions. All these individuals have DD, P4 and P5 had an abnormal EEG, and P15 and P16 had hypotonia, but each also has some distinct features consistent with variability in the cohort. Likewise, unrelated P10, P11, and P12 carry different amino acid substitutions in p.Thr268 and present with overlapping phenotypes. Notably, P10 has two βII-spectrin variants in cis (p.(Thr268Ala) and p.(Phe344Leu)), which may contribute to the more severe phenotype. Of note, P19 was diagnosed with neurofibromatosis and has a variant in NF1 (c.3449C>G; p.(Ser1150*)), which likely would not explain the behavioral challenges and autism in this individual. P2734 has a variant in GNB1 (NM_001282539.1:c.700–1G>T) inherited from her mother also affected with delays.
In sum, the above clinical presentations suggest that pathogenic SPTBN1 variants cause a neurodevelopmental syndrome with a wide range of neurological and behavioral manifestations. These observations are consistent with pleiotropic functions and critical roles of βII-spectrin in brain development and function26.
SPTBN1 variant classification.
Classification of the 28 unique SPTBN1 variants using the functional evidence described below and the 2015 ACMG Guidelines35 and interpretation recommendations36–38 resulted in 17 variants classified as pathogenic, nine as likely pathogenic, and two as variants of uncertain significance (VUS) (Supplementary Table 3). Importantly, P10 has two de novo SPTBN1 variants in cis (p.(Thr268Ala) and p.(Phe344Leu)). p.(Thr268Ala) has two allelic variants p.(Thr268Asn) and p.(Thr268Ser) and the functional studies support a pathogenic classification. p.(Phe344Leu) is classified as a VUS since it is in cis with a pathogenic variant and showed no functional deficits; thus, its contribution to the phenotype of this individual is unclear.
βII-spectrin variants alter cell morphology and protein distribution.
To assess the pathogenic mechanisms of SPTBN1 variants, we introduced a subset of these variants in GFP-tagged human βII-spectrin (GFP-βIISp), transfected the constructs into HEK 293T/17 cells, either alone or with pmCherry-C1, and monitored their effects on GFP-βIISp levels, localization, and stability. Of the 25 variants tested, protein levels of seven were changed relative to control (Fig. 2a and Extended Data Fig. 1a). Nonsense variants (p.C183*, p.E892* and p.W1787*) yielded GFP-βIISp fragments of the expected size, suggesting that the truncated products are structurally stable. The p.G205D and p.G205S variants reduced GFP-βIISp protein levels (Fig. 2a and Extended Data Fig. 1a) and solubility (Extended Data Fig. 1b,c). Transduction of βII-spectrin null cortical neurons with lentivirus expressing selected RFP-βII-spectrin variants resulted in protein expression trends similar to the ones observed in HEK 293T/17 cells (Fig. 2b and Extended Data Fig. 1d). Endogenous levels of full-length (250 kDa) βII-spectrin were not changed in human induced pluripotent stem cell (iPSC) lines reprogrammed from peripheral blood mononuclear cells (PBMCs) from P12 (p.(Thr268Ser)), P21 (p.(Arg1003Trp)), and P28 (p.(E1886Q)) relative to its expression in iPSC from a 13 year old healthy subject39 (Fig. 2c,d and Extended Data Fig. 1e). In contrast, iPSCs from P27 (p.(Trp1787*)) expressed more full-length βII-spectrin and slightly reduced levels of the 205 kDa truncation (Fig. 2d and Extended Data Fig. 1e). This result suggests that the transcript harboring p.(Trp1787*) does not undergo efficient non-sense mediated decay (NMD), which is supported by RNA-seq of blood RNA showing allelic expression bias with 28% of reads carrying the variant (Extended Data Fig. 1f).
Fig. 2 |. SPTBN1 variants alter protein expression and subcellular distribution.

a, Levels of mutant GFP-βIISp in HEK 293T/17 relative to WT GFP-βIISp. b, Levels of RFP-βIISp proteins in cortical βIISp-KO neurons transduced with indicated RFP-βIISp lentivirus. c, (left) Pluripotency assessment of iPSCs harvesting SPTBN1 variants reprogrammed from PBMCs. Representative bright field images and immunofluorescence staining for pluripotency markers of reprogrammed iPSCs (n = 1 line per variant) collected from one independent experiment. Scale bar, 125 μm. c, (right) TaqMan ScoreCard assessment of pluripotency and trilineage differentiation potential of undifferentiated (top) and differentiated (bottom) p.E1886Q iPSCs. The box plot displays the sample score (color dot) (n = 1) against the internal control reference set (gray box and whiskers) provided by the manufacturer. d, Endogenous βIISp expression in iPSCs of the indicated genotypes. α-tubulin is a loading control. Data in a were compiled from n = 3 biological replicates from three experiments. Data in b (n = 3 biological replicates) and d (n = 1 biological replicate) were collected from three and four independent experiments, respectively. All data represent mean ± SEM. One-way ANOVA with Dunnett’s post hoc test for multiple comparisons. a, *P = 0.0441, ****P < 0.0001. b, *P = 0.0136, **P = 0.0011, ****P < 0.0001. d, *P = 0.0103. e, Immunofluorescence images of HEK 293T/17 cells expressing GFP-βIISp plasmids and stained for actin (phalloidin) and DAPI. Scale bar, 10 μm. White arrowheads indicate GFP-positive aggregates. White asterisk mark cells with increased density of membrane protrusions. Data in e are representative of six independent experiments. See statistics summary in Source Data Figure 2.
Wild-type (WT) GFP-βIISp localized throughout the cytosol and the cell membrane of HEK 293T/17 cells, whereas p.I159_Q160del, p.C183*, p.Y190_R216del, p.G205D, p.G205S, p.L247H, and p.L250R GFP-βIISp formed cytosolic aggregates (Fig. 2e, white arrowheads, and Extended Data Fig. 1g). Interestingly, CH domain variants that caused GFP-βIISp aggregates also produced an additional 70-kDa band in HEK 293T/17 cells and mouse neurons (Extended Data Fig. 1a,d, red arrowheads), which could represent degradation or cleavage products, possibly promoted by structural instability of the CH1–CH2 domain. p.T268A/N/S, p.V271M, and p.H275R variants resulted in normal GFP-βIISp distribution but caused enlarged cells with increased membrane protrusions (Fig. 2e, asterisks and Extended Data Fig. 1h,i).
Morphological changes induced by SR variants varied. Cells expressing p.R411W, located in SR1, which is required for dimerization with αII-spectrin40 and actin binding41, were enlarged and had more membrane protrusions (Fig. 2e, asterisk and Extended Data Fig. 1h,i). More membrane protrusions were also detected in cells expressing the p.E491Q variant in SR2, p.A850G, p.R1003W, and p.E1110D variants within SR6–8, and the p.E1886Q variant in SR15 (Fig. 2e and Extended Data Fig. 1h,i). Surprisingly, no cellular phenotypes were observed with expression of p.E892* and p.W1787* GFP-βIISp, lacking polypeptide portions from SR6 to C-terminus and SR14 to C-terminus, respectively, which contain the ankyrin binding42, the tetramerization43, and pleckstrin homology (PH) domains44. Together, these data indicate that βII-spectrin variants alter cellular morphology, likely through changes in cytoskeleton architecture and dynamics.
βII-spectrin variants affect interaction with the cytoskeleton.
A subcortical network of F-actin- and ankyrin-bound βII-/αII-spectrin tetramers promotes membrane stability and organizes specialized membrane microdomains1–3. SPTBN1 variants could impair neuronal function by altering βII-spectrin binding to or localization of cytoskeletal partners. Correspondingly, expression of CH domain variants in HEK 293T/17 cells resulted in depletion of membrane-bound GFP-βIISp and in cytosolic aggregates containing actin and mCherry-αIISp (Fig. 3a, arrowheads and Extended Data Fig. 2a). These CH domain variants also caused GFP-βIISp aggregation in βII-SpKO cortical neurons (Fig. 3b,c), independently of endogenous βII-spectrin levels (Extended Data Fig. 2b), and sequestered endogenous αII-spectrin and actin (Fig. 3c, arrowheads) within the aggregates.
Fig. 3 |. SPTBN1 variants alter interaction with critical cytoskeleton partners.
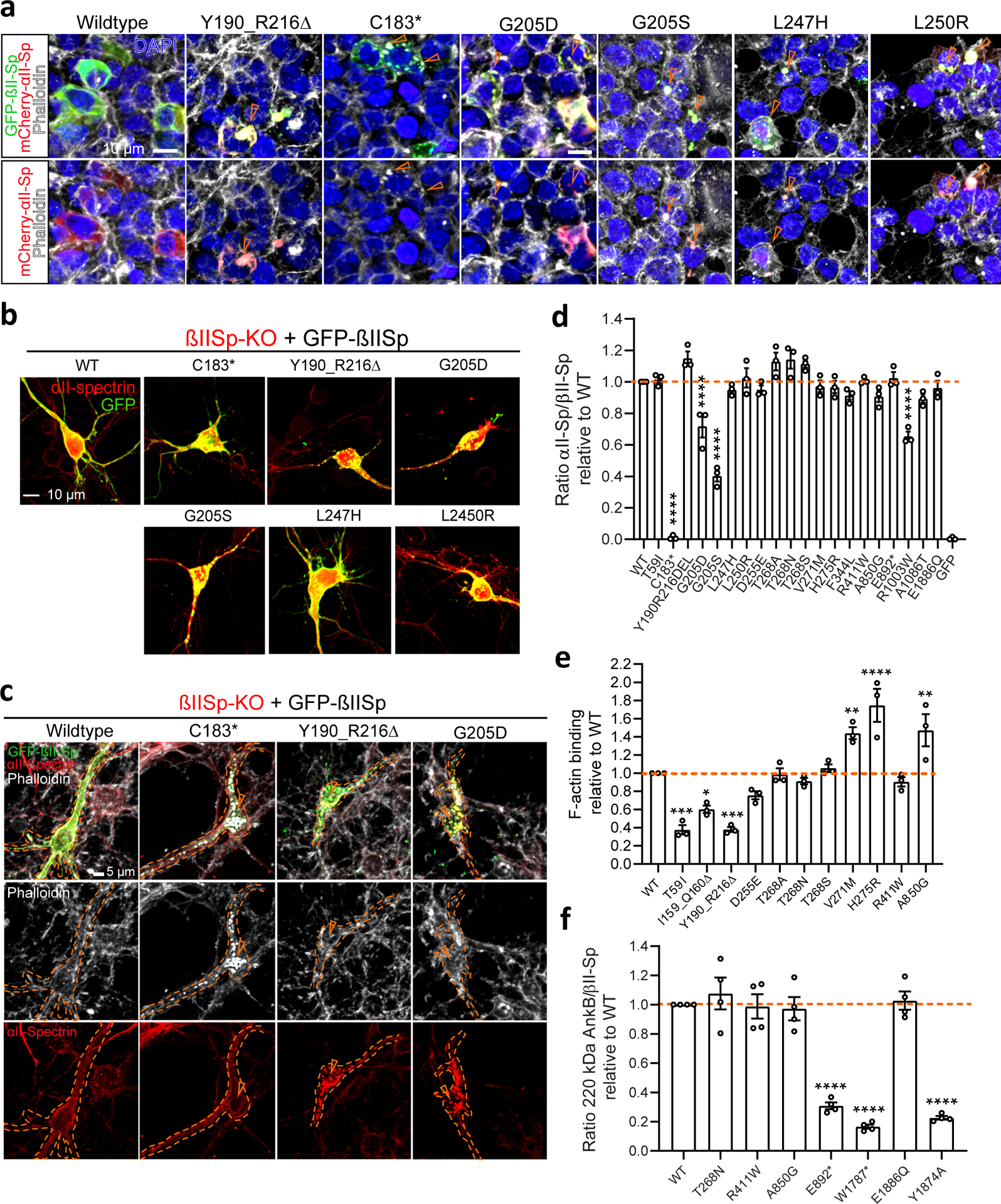
a, Immunofluorescence images of HEK 293T/17 cells transfected with mCherry-αIISp and with either WT or mutant GFP-βIISp plasmids. Cells were stained for actin (phalloidin) and DAPI. Scale bar, 10 μm. b, Immunofluorescence images of DIV8 mouse βIISp-KO cortical neurons transfected with indicated GFP-βIISp plasmids and stained for endogenous αII-spectrin. Scale bar, 10 μm. c, Immunofluorescence images of DIV8 mouse βIISp-KO cortical neurons transfected with indicated GFP-βIISp plasmids and stained for actin (phalloidin) and endogenous αII-spectrin. Scale bar, 5 μm. In a and c GFP-positive aggregates (orange arrowheads) also contain either actin or αII-spectrin proteins, or both. d, Quantification of binding of mCherry-αIISp to GFP-βIISp proteins relative to the abundance of mCherry-αIISp/WT GFP-βIISp complexes. e, Binding of purified βII-spectrin proteins to purified F-actin assessed through an actin co-sedimentation assay. f, Binding of GFP-βIISp proteins to 220-kDa AnkB-3xHA assessed via co-IP from HEK 293T/17 cells. The Y1874A βII-spectrin variant known to disrupt the formation of AnkB/βII-spectrin complexes was used as control. Graphs in d and e summarize results from three independent experiments. Data in f summarize four independent experiments. All data represent mean ± SEM. One-way ANOVA with Dunnett’s post hoc analysis test for multiple comparisons. d, ****P < 0.0001. e, *P = 0.0222, **P = 0.0098 (V271M), **P = 0.0051 (A850G), ***P = 0.0003, ****P < 0.0001. f, ****P < 0.0001. See statistics summary in Source Data Figure 3.
We evaluated whether βII-spectrin variants affect binding to cytoskeletal partners based on their position in domains that are critical for interaction with specific partners. We first assessed whether variants disrupted the formation of βII-spectrin/αII-spectrin complexes by incubating GFP beads coupled to WT or mutant GFP-βIISp with mCherry-αIISp lysates and measuring mCherry-αIISp levels in pulldown eluates. As expected, C183* GFP-βIISp, which lacks the SR1–SR2 heterodimerization domain40, neither associated with nor sequestered mCherry-αIISp or endogenous αII-spectrin into GFP-βIISp aggregates (Fig. 3a–d and Extended Data Fig. 2c). Pulldown of mCherry-αII-spectrin with p.G205D and p.G205S GFP-βIISp baits yielded less αII-/βII-spectrin complexes, partly due to the lower expression of these variants, but also indicating lower affinity for αII-spectrin (Fig. 3d and Extended Data Fig. 2c). Except for p.R1003W, none of the other variants tested affected αII-spectrin binding (Fig. 3d and Extended Data Fig. 2c). The weaker αII-spectrin binding of p.R1003W GFP-βIISp could result from local or long-range conformational changes that might weaken interactions along the dimer.
Next, we evaluated whether CH domain variants affect binding to F-actin using a co-sedimentation assay. GFP-βII-spectrin containing a PreScission protease (PP) cleavage site between GFP and βII-spectrin (GFP-PP-βIISp) was captured on GFP beads. Purified WT and mutant proteins were recovered from beads upon PP cleavage and mixed with purified F-actin. The partition of βII-spectrin between soluble (S) and actin-containing pellet (P) fractions was used to assess binding between both proteins. p.T59I, p.I159_Q160del, p.Y190_R216del, and p.D255E variants reduced F-actin binding (Fig. 3e and Extended Data Fig. 2d). In contrast, p.V271M and p.H275R variants increased F-actin binding, while p. T268A/N/S variants bound F-actin at levels similar to WT (Fig. 3e and Extended Data Fig. 2d). This affinity range is likely due to both local and CH domain-wide conformational changes caused by modified intramolecular interactions that impact intermolecular contacts at the βII-spectrin/F-actin interface. Surprisingly, the p.A850G variant, located several SR away from the CH domains, also increased F-actin binding (Fig. 3e and Extended Data Fig. 2d), which may explain its effects on cell morphology (Fig. 2a,b,e and Extended Data Fig. 2h,i).
Finally, we evaluated if βII-spectrin variants affected interaction with ankyrins by co-expressing 220 kDa HA-ankyrin-B with WT or mutant GFP-βIISp in HEK 293T/17 cells and measuring HA signal in GFP eluates. As reported42, expression of p.Y1874A GFP-βIISp in SR15 (ankyrin-binding domain) almost entirely abrogated binding to ankyrin-B (Fig. 3f and Extended Data Fig. 2e). Truncated βII-spectrin lacking SR15 caused by p.E892* and p.W1787* variants also disrupted binding (Fig. 3f and Extended Data Fig. 2e). Interestingly, the SR15 p.E1886Q variant did not affect ankyrin-B binding despite its proximity to the p.Y1874 binding site42.
Modeling of βII-spectrin variants predicts molecular defects.
We further assessed the impact of SPTNB1 variants through molecular modeling. We first modeled the 10 missense variants involving seven residues in the CH1–CH2 domain. The CH domain is a protein module of around 100 residues composed of four alpha helices45 found in cytoskeletal and signal transduction actin-binding proteins (ABP)46. Biochemical studies using ABP, including spectrin superfamily members α-actinin-4 (ACTN4) and utrophin (UTRN), suggest dynamic transitions between “closed” and “open” configurations of the tandem domains, whereas the open state exposes CH1 residues to enable its predominant role of binding actin, with CH2 regulating the conformational state through autoinhibition46. The electrostatic surface profile of βII-spectrin CH1 and CH2 domains modeled using an available crystal structure of utrophin47 indicates that they each have one electrically active side complementary to each other and one neutral side, consistent with an energetically balanced closed conformation (Fig. 4a–c). This model indicates that six mutated CH domain residues reside at the CH1–CH2 dimer interface, potentially impacting interdomain helix-helix interactions, thereby dysregulating the natural autoinhibition (Fig. 4b,c).
Fig. 4 |. βII-spectrin CH domain variants likely alter CH1–CH2 dimer stability.
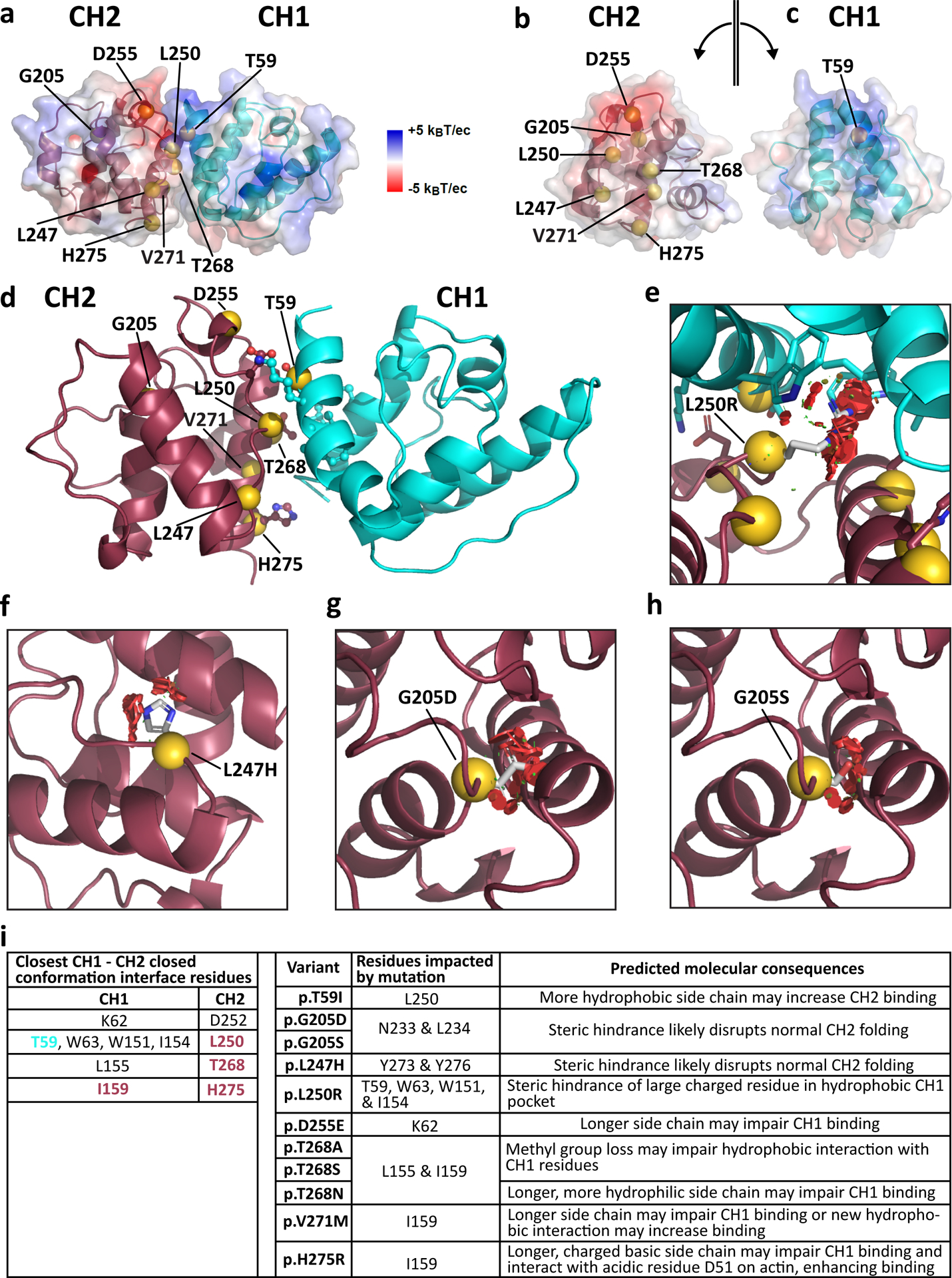
a, Closed conformation of the βII-spectrin CH1–CH2 dimer modeled after utrophin47 showing the sites of βII-spectrin variants and the electrostatic surface of each domain calculated independently. Electrostatic surface scale from negatively (red) to positively (blue) charged. b,c, Electrostatic complementarity shows that both CH domains have a polar side, where CH2 is negatively charged (red) (b) and CH1 is positively charged (blue) (c), and both have a neutral side. d, Closed conformation of the βII-spectrin CH1–CH2 dimer modeled by docking the CH2 domain of βII-spectrin48 onto the CH1 domain modeled after βIII-spectrin49. e, The p.L250R variant introduces a large, positively charged residue that clashes with a hydrophobic CH1 pocket through steric hindrance and electric instability. f, p.L247H introduces a large aromatic amino acid and likely disrupts normal CH2 folding. g,h, Steric hindrance and negative charge introduced by p.G205D (g) and p.G205S (h) in the interior of CH2 likely disrupts normal CH2 folding. i, Key interactions at the CH1–CH2 interface (site of variants in CH1 (teal) and CH2 (red)) and likely molecular perturbations caused by STPBN1 variants.
To refine our prediction of the closed conformation of the βII-spectrin CH1–CH2 domain and to identify interactions at the interface, we docked the CH2 domain (residues 173–278) of βII-spectrin48 onto the modeled structure of the βIII-spectrin CH1 domain (residues 55–158) (95% homologous with βII-spectrin)49 and chose the top docking pose (Fig. 4d). This pose was the same compared to the closed conformations of actinin-4 (PDB ID 6oa6) (unpublished) and utrophin (PDB ID 1qag)47 (Extended Data Fig. 3a–c). Figure 4i summarizes the predicted key interacting residues at the CH1–CH2 interface and the structural consequences of variants in those domains. Residues affected by variants p.T59 in CH1 and p.L250, p.T268, and p.H275 in CH2 are predicted to participate in interdomain interactions (Fig. 4d,i). The missense variants in these and in the two other interface residues p.D255 and p.V271 likely introduce destabilizing effects (Fig. 4i). For example, substitutions of T268 by alanine (A) (smaller and more hydrophobic), serine (S) (loss of methyl group), and asparagine (N) (larger and more hydrophilic) likely alter the hydrophobic interaction of T268 with p.L155 in CH1 and p.I159 in the CH1–CH2 linker differently. However, it appears that any potential conformational changes in the CH1–CH2 domain that may result from these amino acid changes in p.T268 are not sufficient to cause appreciable changes in F-actin binding (Fig. 3e and Extended Data Fig. 2d). Similarly, the p.D255E variant causes a relatively small residue change that does not alter F-actin affinity. Conversely, the p.V271M (larger and hydrophobic) and the p.H275R (longer and significantly more hydrophilic) substitutions may impair CH1 binding to cause a shift towards the open CH1–CH2 conformation and higher F-actin affinity. This is also expected for the p.L250R variant, which likely causes significant steric hindrance by the clashing of the large, charged residue with a hydrophobic CH1 pocket (Fig. 4e). In line with this prediction, p.L250R GFP-βIISp aggregates in cells (Figs. 2e and 3a,b and Extended Data Fig. 2b). Conversely, p.T59I introduces a slightly longer but more hydrophobic group that might promote a stronger interaction with p.L250 in CH2, potentially shifting the equilibrium to a CH1–CH2 closed configuration consistent with less F-actin binding (Fig. 3e and Extended Data Fig. 2d).
p.G205D/S and p.L247H substitutions in the interior of CH2 are predicted to cause instability due to significant steric hindrance (Fig. 4f–h). p.G205D and p.G205S introduce destabilization by positioning an interior negative charge and steric hindrance against the neighbor N233 and L234 side chains (Fig. 4g,h), which likely underlies protein aggregation in cells (Figs. 2e and 3a–c and Extended Data Fig. 2b). The in-frame deletion p.Y190_R216del also results in βII-spectrin aggregation and diminished F-actin binding (Figs. 2e and 3a–c,e and Extended Data Fig. 2b,d). In these cases, the autoinhibitory interactions will also be lost if the structure of the CH2 domain is compromised. To explore whether some of the mutants are involved in binding F-actin, we independently docked the CH1 and CH2 domains onto an F-actin model built from chains A–F of 6anu48 using ClusPro50,51 (Extended Data Fig. 3d–f). The top eight CH1 docking poses predicted by the balanced and electrostatic scoring algorithms almost all corresponded to the location and orientation of CH1 molecules on F-actin as defined by the cryo-EM structure 6anu (Extended Data Fig. 3d, dark blue). For CH2 docking onto F-actin, the top eight docking poses predicted by the balanced and electrostatic scoring algorithms almost all correspond to symmetry-related locations and poses on F-actin (Extended Data Fig. 3e). In addition, the predicted orientation of CH2 molecules on F-actin is consistent with the known binding site of the CH1 domain, as judged by the length of the linker that would be required to join the C-terminus of the docked CH1 domain to the N-terminus of the docked CH2 domain (Extended Data Fig. 3f). Our model predicts that neither the T59 residue nor its mutated version are directly involved in F-actin binding (Extended Data Fig. 3d). The p.H275R variant may interact more strongly with negatively charged D51 in F-actin (Extended Data Fig. 3e) to contribute to its higher actin binding propensity (Fig. 3e and Extended Data Fig. 2d). We also modeled the missense variants in the SR domains (Extended Data Fig. 3g,h). Except for p.F344L, all SR variants face outwards, to the solvent, and could be involved in protein binding at the interface. Interestingly, all variants within the second and third helices of the spectrin fold result in neutral or more hydrophobic residues, and those in the first helix of the SR are more hydrophilic. Given the consistency of this trend, it possibly underlies a conserved functional role important for heterodimerization and larger order assemblies.
In sum, our modeling results provide a strong molecular rationale for several of the biochemical and cellular observations described above, which implicate protein stability, abnormal assembly and dynamics of the βII-spectrin-F-actin skeleton, and potential disruptions of βII-spectrin binding to other molecular partners, consistent with similar changes caused by variants in other members of the spectrin superfamily45,48,49.
βII-spectrin variants disrupt neuron architecture and function.
Individuals with SPTBN1 variants display a wide range of neurological presentations that are consistent with phenotypes of neural progenitor-specific βII-spectrin null mice26. Neurons from these mice show disruption of the spectrin-actin MPS24, impaired axon initial segment (AIS) organization and axonal growth, and reduced axonal organelle transport26,52. These reports and our initial cellular and molecular observations suggest that mutant βII-spectrin may disrupt the organization and dynamics of the neuronal cytoskeleton, and the morphology and function of neurons. Thus, we next investigated the effects of disease-linked variants using a structure-function rescue approach in βII-SpKO cortical neurons.
First, we expressed WT and mutant GFP-βIISp together with mCherry in day in vitro (DIV) 3 βII-SpKO cortical neurons26 and evaluated axonal growth and AIS morphology at DIV8. We also evaluated WT (Sptbn1flox/flox/+) and heterozygous (Sptbn1flox/+;Nestin-Cre; henceforth abbreviated as βII-SpHet) neurons. Axonal length was impaired in βII-SpKO neurons but restored upon expression of WT GFP-βIISp (Fig. 5a and Extended Data Fig. 4). βII-SpHet axons grew to only half the length of WT axons but were almost twice as long as βII-SpKO axons (Fig. 5a and Extended Data Fig. 4). Most mutant GFP-βIISp proteins failed to rescue axonal length to WT levels, while p.A1086T and p.E1110D restored length to heterozygous levels (Fig. 5a and Extended Data Fig. 4). AIS from βII-SpKO cortical neurons exhibited normal length but abnormal ankyrin-G (AnkG) clustering as fragmented puncta as previously observed52, which was restored by WT GFP-βIISp and by a subset of the variants (Fig. 5b,c). p.Y190_R216del, p.T268S, p.H275R, p.R411W, and p.G1398S GFP-βII failed to rescue AIS AnkG clustering (Fig. 5b,c). In contrast, p.T59I, p.C183*, p.G205D/S, p.L247H, p.E892*, p.R1003W, and p.W1787* GFP-βIISp did not rescue AnkG clustering and led to changes in AIS length (Fig. 5b,c).
Fig. 5 |. SPTBN1 variants affect neuronal axonal growth, AIS morphology and organelle transport.

a, Axonal length of DIV8 neurons (n = 12–34 neurons/genotype) from three experiments. Data represent mean ± SEM. One-way ANOVA with Tukey’s multiple comparisons test, ****P < 0.0001. Orange and blue lines indicate average length of βII-SpWT and βII-SpHet axons, respectively. b, Images representative of three independent experiments show AnkG clustering at the AIS. Scale bar, 10 μm. c, AIS length (n = 10–80 neurons/genotype) compiled from three experiments. Data represent mean ± SEM. d, Kymographs of RFP-LAMP1 motion in axons. Trajectories are shown in green for anterograde, red for retrograde, and blue for static vesicles. Scale bar, 10 μm and 60 s. e, Percent of motile axonal LAMP1-RFP cargo. f,g, Quantification of the anterograde and retrograde velocity (f) and distance traveled (g) of LAMP1-RFP cargo. For e-g the box plots show all data points from minimum to maximum. Boxes represent data from the lower (25th percentile) to the upper (75th percentile) quartiles. The box center corresponds to the 50th percentile. The median is indicated by a horizontal line. Whiskers extends from the largest dataset number smaller than 1.5 times the interquartile range (IQR) to the smallest dataset number larger than 1.5IQR. Data was collected in n=9–13 axons from three independent experiments. Data in c and e–g were analyzed by one-way ANOVA with Tukey’s (c) and Dunnett’s (e–g) post hoc analysis tests, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. See statistics summary in Source Data Figure 5.
Organelle transport is essential for the maintenance of neuronal processes and neuron viability, and defects in transport can contribute to the pathology of several neurological diseases53. We previously showed that βII-spectrin promotes axonal organelle transport independently of its role assembling the MPS26. Expression of WT βII-spectrin in βII-spectrin null cortical neurons rescues the processivity, motility, and flux of synaptic vesicles and lysosomes26. To evaluate the effects of selected βII-spectrin variants on axonal transport, we tracked the dynamics of the endosome/lysosome marker RFP-LAMP1. Loss of βII-spectrin impaired the bidirectional motility of LAMP1-RFP cargo and caused deficits in their run length and retrograde velocity (Fig. 5d–g). Remarkably, βII-spectrin haploinsufficiency caused similar deficits (Fig. 5d–g), indicating that 50% reduction of βII-spectrin levels is insufficient to maintain normal organelle transport. As expected26, deficient lysosome dynamics in βII-SpKO neurons was rescued by expression of WT GFP-βIISp. However, selected variants that do not rescue axonal length also fail to restore lysosome dynamics (Fig. 5d–g), including p.E892* and p.W1787* GFP-βIISp, which lack the PH domain required for βII-spectrin coupling to organelle membranes25. It is possible that the abnormal binding to molecular partners observed in other mutants unable to rescue organelle dynamics interfere with the formation of complexes between βII-spectrin and molecular motors, its coupling to organelle membranes, or its cytosol to MPS partitioning.
We also evaluated the effect of βII-spectrin deficiency or expression of GFP-βIISp variants on dendritic morphology of DIV18 cortical neurons. Both βII-spectrin reduction (βII-SpHet) and total loss (βII-SpKO) resulted in shorter dendritic processes relative to WT (Extended Data Fig. 5a,b), but the number of primary and secondary dendrites remained unchanged (Extended Data Fig. 5a,c). Dendritic morphology of βII-SpKO neurons was rescued by WT GFP-βII-Sp (Extended Data Fig. 5). In contrast, CH domain variants that caused GFP-βIISp aggregation (Figs. 2e and 3a–c) also led to a significant decrease in dendrite number and length, whereas variants p.E892* and p.W1787* reduced dendrite number but not length (Extended Data Fig. 5). The other variants evaluated led to a range of alterations in dendritic morphology (Extended Data Fig. 5). CH domain variants often produced extensive aberrant membrane features in the form of lamellipodia and filopodia around the cell body and along the neuronal processes (Extended Data Fig. 6a). Neuronal membrane expansion was accompanied by a shift in the boundaries of actin and αII-spectrin distribution (Extended Data Fig. 6a). Together, these results confirm that clinically relevant βII-spectrin variants can cause marked disruptions in neuronal architecture, likely driven by changes in submembrane cytoskeleton organization and dynamics, which may be a pathogenic factor in SPTBN1-associated syndrome.
βII-spectrin haploinsufficiency affects neuronal connectivity.
βII-spectrin is expressed in all brain cells54, and its loss in neurons and glial cells in βII-SpKO mice disrupts the development of long-range cerebellar axons, and tracts connecting cerebral hemispheres, including the corpus callosum (CC)26. Consistent with a diminished axonal growth in vitro (Fig. 5a and Extended Data Fig. 4), PND25 βII-SpHet mice exhibited callosal hypoplasia (Fig. 6a,c). CC thinning was also detected by MRI in probands P2, P10, and P28 (Fig. 1d, Supplementary Table 1 and Supplementary Note), which further implicates βII-spectrin in regulating brain cytoarchitecture. Deficient connectivity of long axonal tracts can also result from defects in neuronal migration and axonal pathfinding, which is affected by glial cells55. To determine the neuron-specific effects of βII-spectrin depletion on CC wiring, we generated mice selectively lacking βII-spectrin in projection neurons driven by Nex-Cre56 (Sptbn1flox/flox;Nex-Cre; henceforth βIISp-Nex KO). βII-spectrin loss or haploinsufficiency only in cortical projection neurons is sufficient to induce CC hypoplasia (Fig. 6b,d and Extended Data Fig. 6b). Finally, CC malformations could arise from deficits in the development of cortical layers given that callosal axons originate primarily from projection neurons of layer II/III and layer V of the neocortex, which are specified by Satb2 and Ctip2 expression, respectively57. Consistent with this prediction, PND0 βII-SpKO brains show reductions in the thickness of cortical layers II/III and V relative to overall cortical thickness (Fig. 6e,f). A trend towards a significant deficit in the formation of Sabt2 and Ctip2 layers was observed in βII-SpHet mice brains (Fig. 6e,f). Combined, these results suggest that partial βII-spectrin LOF can produce neuronal miswiring in the cortex and those defects are at least in part neuron-autonomous.
Fig. 6 |. βII-spectrin deficiency disrupts proper cortical development.
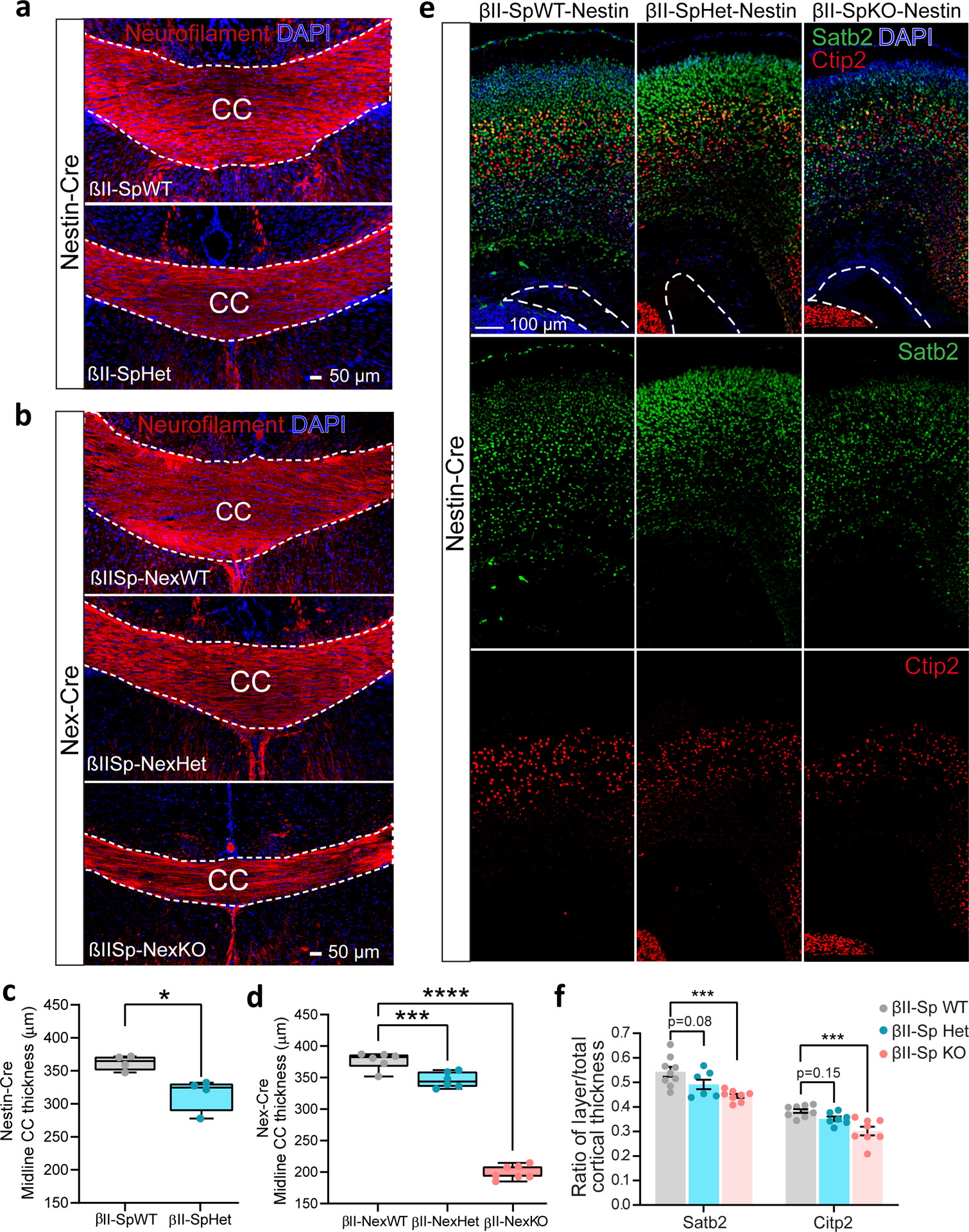
a,b, Images of coronal sections from PND25 mice expressing Nestin-Cre (a) or Nex-Cre (b) collected from n = 2 litters and stained for neurofilament and DAPI in one independent experiment. Scale bar, 50 μm. White lines indicate the corpus callosum (CC). c, Midline CC thickness of mice expressing Nestin-Cre (n = 4 mice/genotype). Data represent mean ± SEM. Two-tailed unpaired t-test, *P = 0.0134. d, Midline CC thickness assessed from βII-SpWT (n = 6), βII-SpHet-Nex (n = 6) and βII-SpKO-Nex (n = 7) brains. For c and d, the box plots show all data points from minimum to maximum. Boxes represent data from the lower (25th percentile) to the upper (75th percentile) quartiles. The box center corresponds to the 50th percentile. The median is indicated by a horizontal line inside the box. Whiskers extends from the largest dataset number smaller than 1.5 times the interquartile range (IQR) to the smallest dataset number larger than 1.5IQR. e, Images of PND0 βII-SpWT, βII-SpHet and βII-SpKO brains expressing Nestin-Cre stained for Satb2 and Ctip2 to label neocortical layers and DAPI. A white line indicates the position of the left ventricle. Scale bar, 100 μm. f, Quantification of Sabt2- and Ctip2-positive cortical layer thickness relative to total cortical thickness assessed from βII-SpWT (n = 9), βII-SpHet (n = 8) and βII-SpKO (n = 7) brains expressing Nestin-Cre. Data in d and f represent mean ± SEM and were analyzed by one-way ANOVA with Dunnett’s post hoc test for multiple comparisons. d, ***P = 0.0003, ****P < 0.0001. f, Satb2 (***P = 0.0008), Ctip2 (***P = 0.0002). See statistics summary in Source Data Figure 6.
βII-spectrin deficiency affects development and behavior in mice.
SPTBN1 variant carriers exhibit a wide range of facial dysmorphisms, microcephaly, macrocephaly, and DD (Supplementary Table 1 and Supplementary Note). Embryonic day 19 (E19) βII-SpKO mice have enlarged head circumference and a trend towards increased distance between the eyes relative to head circumference (Fig. 7a–c), which is consistent with hypertelorism in some probands (Fig. 1c and Supplementary Note). In line with reported short stature of some probands, βII-SpKO mice show arrested growth (Fig. 7d,e)26 and βII-SpHet mice exhibit intermediate body size and weight (Fig. 7d–f). The global DD changes observed in βII-spectrin mice arise in part due to neuronal-autonomous effects, given that they are also observed in βIISp-Nex KO mice with selective deficits in projection neurons (Extended Data Fig. 7a).
Fig. 7 |. βII-spectrin deficiency causes developmental and behavioral deficits.

a, E19 male embryos. b, Head circumference (n = 5). c, Eye distance (βII-SpWT (n = 7), βII-SpHet (n = 5), βII-SpKO (n = 5) of E19 embryos. Box plots show data points from minimum to maximum. Boxes represent data from the lower (25th-percentile) to the upper (75th-percentile) quartiles. Center and horizontal line inside a box indicate the 50th-percentile and the median, respectively. Whiskers extends from the largest dataset number smaller than 1.5 times the interquartile range (IQR) to the smallest dataset number larger than 1.5IQR. One-way ANOVA with Tukey’s post hoc test. b, *P = 0.029, **P = 0.003. d, PND25 male mice. e, Body length at PND25. Data represent mean ± SEM (n = 12 mice/genotype). One-way ANOVA with Tukey’s post hoc test, ****P < 0.0001. f, Growth curve. Data represent mean ± SEM (n = 12 mice/genotype). g,h, Locomotor activity (g) and rearing (h) during an open-field test. Data in g and h represent mean ± SEM (n = 15 βII-SpWT, n = 5 βII-SpKO PND30 male mice). Data for f, g, i, and j were analyzed by Fisher’s PLSD tests following repeated measures ANOVA, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Statistical comparisons were not conducted for h due to zero scores in the βII-SpKO group. i, j, Locomotor activity (i) and rearing (j) during an open-field test. k, Social preference during a three-chamber choice task. Within-genotype repeated measures ANOVA, *P = 0.0452. l, Entries into a chamber with a stranger mouse. Fisher’s PLSD test following repeated measures ANOVA, *P = 0.0306. Data in i–l represent mean ± SEM (n = 12 mice/genotype). See statistics summary in Source Data Figure 7.
Since individuals carrying SPTBN1 variants present with ASD, ADHD, and learning and motor deficits (Supplementary Table 1 and Supplementary Note), we assessed behavioral effects of brain βII-spectrin deficiency in mice. First, we evaluated the effects of complete LOF using βII-SpKO mice, which do not survive longer than post-natal day (PND) 4026, and were only challenged with open field and acoustic startle tests at PND30. βII-SpKO mice had overt hyperactivity during the open field test (Fig. 7g) and profound deficits in rearing, a response requiring good hind limb function and balance (Fig. 7h). βII-SpKO mice also showed decreases in startle response amplitudes, but normal levels of prepulse inhibition (PPI) (Extended Data Fig. 7b,c), suggesting that reduced startle responses were due to motor deficits, rather than alterations in auditory function or sensorimotor gating. This is consistent with impaired motor abilities likely due to the severe loss of cerebellar connectivity26.
We next characterized behavioral phenotypes of βII-SpHet mice, whose normal lifespan allowed for an expanded battery of tests. In contrast to βII-SpKO mice, βII-SpHet animals had normal activity during an open field test (Fig. 7i,j), and normal performance in an acoustic startle test for PPI and in the rotarod test (Extended Data Fig. 7d–f), indicating that haploinsufficiency does not cause motor problems in young mice. βII-SpHet mice also exhibited normal spatial and reversal learning in the Morris water maze test (Extended Data Fig. 7g,h). Conversely, in the 3-chamber choice test, βII-SpHet demonstrated no preference for spending more time in proximity to a stranger mouse (stranger 1) versus an empty cage and made significantly fewer entries into the side containing the stranger (Fig. 7k,l). These genotype differences were not observed in the subsequent test for social novelty preference, in which βII-SpHet and βII-SpWT littermates demonstrated preference for the newly introduced mouse (stranger 2) (Extended Data Fig. 7k,l). Notably, βII-SpHet mice had a non-significant trend towards fewer entries in the social novelty test. The lack of sociability in the βII-SpHet mice was not associated with changes in anxiety-like behavior or olfactory function (Extended Data Fig. 7m). Overall, these results suggest that βII-spectrin LOF impairs global development and has a selective impact on social motivation and reward that may contribute to the autistic features and social behavior impairments manifested in some affected individuals.
Discussion
In this study, we report for the first time the identification of de novo SPTBN1 variants as a cause of a neurodevelopmental disorder most commonly characterized by DD, ID, and various neurologic and behavioral comorbidities. Twelve probands have been diagnosed with ADD/ADHD and six with ASD, with three having co-occurrence. This observation is consistent with a recent WES study of a Danish cohort of children with ASD and/or ADHD and controls that identified SPTBN1 as a top hit among genes with rare truncating variants co-occurring in these disorders58. SPTBN1 variants had previously been reported in individuals with ASD33, Tourette31, and DD32 (all included in our study). Notably, βII-spectrin’s canonical partner ankyrin-B is encoded by the high-confidence ASD gene ANK233, and some ASD individuals with ANK2 variants also exhibit ID59. Loss of ankyrin-B isoforms in mice result in axonal transport deficits60 and defects in brain connectivity59,60, two overlapping phenotypes observed in βII-spectrin mouse models. Although ankyrin-B and βII-spectrin independently modulate axonal transport26, they may converge through mechanisms that affect other neuronal functions. For example, ankyrin-B loss affects the polarized distribution of βII-spectrin in neurites, which causes its more even partitioning between axons and dendrites, and a higher prevalence of dendritic MPS61. The defects in dendrite development induced by several βII-spectrin variants suggest an additional pathogenic mechanism that can affect synaptic function. Conversely, disruption of the MPS due to loss of βII-spectrin24,26 may disrupt the periodic distribution of ankyrin-B and its membrane partners in axons60, which may be essential for signal transduction events62. Our results together with these observations support the association of SPTBN1 variants with ASD and ADHD.
Seizures and epilepsy were re-occurring phenotypes in our cohort. That SPTBN1 variants may have epileptogenic effects is not surprising given the strong association of de novo and inherited variants in SPTAN1 (αII-spectrin) with epileptic syndromes5,16–21. Although the precise pathogenic mechanism of SPTAN1 in epilepsy has not been fully elucidated, αII-spectrin aggregation has been reported for several disease variants16,20. As we show, αII-spectrin cellular distribution can be disrupted by mutant βII-spectrin to cause these partners to co-aggregate, or otherwise continue to associate in aberrant cellular patterns. On the other hand, variants such as p.(T59I) in P1, who presents with epilepsy, affect neither the levels nor the cellular distribution of αII-spectrin, which indicates the possibility of an epileptogenic mechanism independent of αII-spectrin. It is possible that AIS structural defects caused by βII-spectrin deficiencies alter the clustering of ion channels and action potential firing. Going forward, it will be critical to elucidate whether these tightly intertwined partners share pathways disrupted in channelopathies underlying seizures and epilepsy.
Besides the widely shared DD phenotype in our cohort, further evidence of the pathogenicity of SPTBN1 variants is the recurrence of de novo variants in the same amino acid position in unrelated individuals who share clinical manifestations, but also diverge in some presentations, likely due to differences in the amino acid substitution, sex, age, and genetic background. Another striking indicator of convergence in the pathogenic mechanism of the βII-spectrin variants is their clustering within the CH domains. The region of SPTBN1 encoding the CH domains has a higher degree of missense variant constraint than the rest of the protein in the population (ExAC v.10)63, indicating its importance for protein function and supporting the pathogenicity of the variants within. Our cellular and biochemical findings suggest that CH domain variants affect βII-spectrin’s interaction with F-actin and αII-spectrin and alter cytoskeleton dynamics and cellular morphology. The aberrant accumulation of mutant βII-spectrin within cytosolic aggregates suggests that a subset of the CH domain variants introduce destabilizing structural effects, which is supported by our modeling. These aggregates, which sequestered F-actin and αII-spectrin, likely contribute to deficits in neuronal development and morphology through dominant-negative effects. Similarly, altered βII-spectrin binding to F-actin, through gain-of-function or dominant-negative effects, may promote aberrant neuronal membrane morphology and changes in dendrite development. Interestingly, pathogenic CH domain variants have been reported in βI-spectrin64 and βIII-spectrin4,13, and shown to affect F-actin binding49. Together with our results, this evidence indicates that the abnormal modulation of F-actin binding by CH domain variants likely constitute a conserved pathogenic mechanism in spectrinopathies.
Like in other spectrinopathies4–23, missense variants affecting SR are likely to be disease-causing in the SPTBN1-associated syndrome, although the molecular mechanisms are not fully understood. For example, it is not clear how p.A850G phenocopies some of the cellular phenotypes caused by various CH domain variants. It is possible that p.A850G affects βII-spectrin/F-actin dynamics through allosteric mechanisms or dominant-negative effects due to overexpression. Alternatively, this and the other SR variants may disrupt βII-spectrin association with undefined binding partners or its coupling to organelles and motor proteins26.
Given the wide expression of βII-spectrin in non-neuronal brain cells, it will be important to assess if their function is affected by SPTBN1 variants. It is likely that the clinical variability observed in this cohort is at least partly rooted in the multifunctionality and ubiquitous expression of βII-spectrin, although some of the clinical manifestations may be caused by an alternate etiology. For example, the pathogenic NF1 variant in P19 may contribute to the learning disabilities, but likely not the behavioral challenges and autism in this individual. Finally, given the critical roles βII-spectrin plays in other organs65,66 and its association with other non-neurological disorders in probands from our cohort, the SPTBN1 syndrome warrants thorough clinical assessment and further studies in human iPSC-derived cellular systems, animal models and beyond the brain.
Methods
Identification of pathogenic SPTBN1 variants.
Pathogenic variants in SPTBN1 were identified by whole exome or genome sequencing performed on whole blood DNA from probands identified through diagnostic clinical practice or Institutional Review Board approved research studies. Affected individuals were identified through professional communication, connections through GeneMatcher67, and by searching the Undiagnosed Diseases Network (UDN) and the Deciphering Developmental Disorders (DDD) Research Study32 repositories. Variants were reported according to standardized nomenclature defined by the reference human genome GRCh37 (hg19) and SPTBN1 transcript GenBank: NM_003128.2. The minor-allele frequency of each variant was determined from genomic sequencing data derived from gnomAD.
Patient consent.
Human subject studies were approved by the local institutional review boards (IRB), including the Mayo Clinic (IRB 12-009346), the Institute for Genomic Medicine at Columbia University (protocol AAAO8410) and the Ethics Committee of the Medical Faculty of the University of Bonn (approvals 131/08 and 024/12). Patient consent for participation, phenotyping, and sample collection was obtained through the referring clinical teams and the appropriate institutional forms have been archived. Referring clinicians were requested to complete a comprehensive questionnaire that was based upon our current understanding of the phenotypic associations of SPTBN1. They included sections related to neurodevelopmental screening, behavior, dysmorphology, muscular, cardiac, and other systemic phenotypic features. The authors affirm that human research participants provided informed consent for publication of the images in Figure 1. Consent and collection of information conformed to the recognized standards of the Declaration of Helsinki.
Reprogramming of human iPSC lines from SPTBN1 variant carriers.
Peripheral blood mononuclear cells (PBMCs) were purified from fresh blood obtained from probands with SPTBN1 syndrome harboring the p.(Thr268Ser), p.(Arg1003Trp), p.(Trp1787*) and p.(Glu1886Gln), variants. Erythroblast expansion from PBMCs and reprogramming with Sendai viruses expressing the Yamanaka factors (CytoTune-iPS 2.0 Sendai Reprogramming, TermoFisher Scientific) was performed as previously described39. Established iPSCs were cultured in StemFlex medium (Thermo Fisher, A3349401) on Matrigel-coated dishes (Corning, 354277). iPSCs were passaged every 5–7 days for 7 passages using 5 mM EDTA buffer at a 1:6 ratio in the presence of 10 μM Y27632 (Peprotech, 1293823). Cells were maintained at 37 °C and 5% CO2 with daily medium changes.
Evaluation of pluripotency and differentiation capacity of iPSC lines from SPTBN1 variant carriers.
Pluripotency of reprogrammed iPSC lines were assessed after passage 7 by immunofluorescence staining for pluripotency markers. iPSCs were grown on Matrigel coated plates for 72 h, and cells were fixed with 4% formaldehyde (PFA) for 15 min followed by a permeabilization step with 0.3% Triton-X for 15 min, and incubation with a 5% BSA blocking solution at 23°C for 2 h. iPSCs were then subsequently incubated overnight with primary antibodies at 4 °C and with secondary antisera for 2 h at 23 °C, washed with PBS, and mounted with Prolong Gold Antifade reagent (Life Technologies). DAPI was used to contrastain the nucleus. The StemDiff Trilineage Differentiation kit (StemCell technologies, Cat # 05230) medium was used to test the capacity of the iPSCs to differentiate into ectoderm, mesoderm, and endoderm fates. iPSCs were collected with a 0.5 mM EDTA solution and plated as monolayer following the manufacture’s recommendations. Differentiated cells were collected from mesoderm and endoderm medium at day 5, and ectoderm medium at day 7, and RNA was extracted from pooled cells using the PureLink RNA extraction kit (Thermo Fisher Scientific, Cat# 12183018A). The differentiation potential was assessed using the qPCR-based assay TaqMan® hPSC Scorecard™ Panel and analyzed using the accompanied hPSC Scorecard analysis software. The algorithm assigns an individual score to each of the three germ layers based on an internal reference standard (gray box plots and whiskers), allowing to compare results against the same reference data.
RNA-seq from blood RNA.
RNA sequencing was performed from blood RNA from P27 bearing the p.(Trp1787*) variant by first isolating RNA using the miRNeasy Mini Kit (Qiagen) following the standard protocol from blood drawn in a PAXgene Blood RNA Tube (Qiagen). RNA libraries were prepared, and coding regions of the transcriptome were captured by pooling four of the cDNA libraries at 200 ng each according to the manufacturer’s instructions for the TruSeq® RNA Access Library Prep Kit (Illumina)68. Libraries were sequenced at ~65 million fragment reads per sample (4 samples/lane) following Illumina’s standard protocol using the Illumina cBot and HiSeq 3000/4000 PE Cluster Kit. The flow cells were sequenced as 100 × 2 paired-end reads on an Illumina HiSeq 4000 using HiSeq 3000/4000 sequencing kit and HCS v3.3.20 collection software. Base-calling was performed using Illumina’s RTA version 2.5.2. RNA-sequencing analysis was performed using MAP-RSeq69. Reads were aligned to the human genome (hg19) and transcriptome using Tophat270 running Bowtie (v1)71. Gene and exon level read counts were generated using HiSeq72 and BedTools73, respectively. Alignments were visualized using Integrative Genomics Viewer (IGV) (http://software.broadinstitute.org/software/igv/).
Variant interpretation and classification.
SPTBN1 variants were interpreted using the NM_003128.2 transcript and splice variants were evaluated using SpliceAI27 to predict the most likely mRNA splicing outcome. The SPTBN1 variants identified in this study were classified according to the ACMG 2015 Guidelines35. Based on the recommendations of PVS1 LOF criterion under the ACMG/AMP specifications36, PVS1_strong was used as a maximum weight of evidence. This is appropriate for this criterion as we have shown moderate clinical validity37, unrelated probands with a consistent phenotype, and robust functional evidence showing that these nonsense variants remove downstream portions of the protein known to be essential for protein function, and that both null and haploinsufficient mouse models recapitulate disease phenotypes. The maximum weight of functional evidence (PS3) used was moderate under the ACMG/ACMP specifications38.
Mouse lines and animal care.
Experiments were performed in accordance with the guidelines for animal care of the Institutional Animal Care and Use Committee of the University of North Carolina at Chapel Hill under animal protocol 19–209. All mouse (Mus musculus) lines were maintained in the C57BL/6J background by regular backcrossing to the C57BL/6J line (stock number 000664; The Jackson Laboratory). Male and female mice from E15-PND120 were used in experiments, unless otherwise indicated. To generate neural progenitor-specific βII-spectrin null (Sptbn1flox/flox/Nestin-Cre, βIISp-KO) and haploinsufficient (Sptbn1flox/+/Nestin-Cre, βIISp-Het) mice, Sptbn1flox/flox animals (a gift from Mathew Rasband52) were crossed with the Nestin-Cre mouse line (B6.Cg-Tg(Nes-cre)1Kln/J, stock number 003771; The Jackson Laboratory). Sptbn1flox/flox animals negative for the Cre transgene were used as littermate controls in all experiments. Mice lacking βII-spectrin in cortical projection neurons (Sptbn1flox/flox/Nex-Cre, βIISp-NexKO) were generated by crossing Sptbn1flox/flox and Nex-Cre (a gift from Klaus-Armin Nave55) animals for multiple generations. All mice were housed at 22 °C ± 2 °C on a 12-hlight/12-h-dark cycle and fed ad libitum regular chow and water.
Generation of human βII-spectrin variants.
The human βII-spectrin cDNA was subcloned into peGFP-C3 vector (Clontech) using HindIII and SacI sites to generate the peGFP-βIISp plasmid. For purification of full-length βII-spectrin proteins, both a PreScission protease site (LEVLFQGP) and a 6x histidine tag were respectively introduced between the GFP and start codon and before the C-terminal stop codon of peGFP-βII-spectrin using site-directed mutagenesis to generate the peGFP-PP-βII-Sp-6xHis construct. peGFP-βIISp and peGFP-PP-βII-Sp-6xHis plasmids bearing the human variants included in the study were generated using the In-Fusion HD Cloning Plus system (Takara) and primers specific for each variant site (Supplementary Table 4). Lentiviral plasmids carrying wild-type human βII-spectrin or a subset of SPTBN2 variants (pLV-hSyn-RFP-PP-βII-Sp) used for transduction of mouse cortical neurons were generated by introducing the human βII-spectrin cDNA in-frame with the RFP coding sequence in the lentiviral vector pLV-hSyn-RFP (Addgene plasmid #22909, gift from Edward Callaway). In details, full-length human βII-spectrin cDNA was amplified from peGFP-PP-βII-Sp-6xHis plasmids using primers PP-hSPTBN1-F and PP-hSPTBN1-R (Supplementary Table 4) and cloned into the pLV-hSyn-RFP vector (linearized with pLLV-Syn-F and pLLV-Syn-R primers, Supplementary Table 4) using In-fusion cloning. All plasmids were verified by full-length sequencing.
Plasmids.
Plasmid used in transfection experiments included: pLAMP1-RFP (Addgene plasmid #1817, gift from Walther Mothes), pmCherry-C1 (Clontech) and peGFP-C3 vector (Clontech). To generate mCherry-tagged αII-spectrin (pmCherry-αIISp), the cDNA sequence of human αII-spectrin (NM_001130438.3) was amplified by PCR as a BsrGI/XhoI fragment and cloned into the corresponding sites of pmCherry-C1 (Clontech). peGFP-C3-Y1874A-βII-spectrin and HA-tagged 220 kDa ankyrin-B (pAnkB-3xHA) plasmids were previously reported26. All plasmids were verified by full-length sequencing prior to transfection.
Antibodies.
Affinity-purified rabbit antibodies against GFP, ankyrin-G, and βII-spectrin, used at a 1:500 dilution for immunohistochemistry and 1:5,000 for western blot, were generated by Vann Bennett’s laboratory and have been previously described26,60,74. In addition, an affinity-purified anti-βII-spectrin antibody produced by immunization of rabbits with full-length brain βII-spectrin (a generous gift from Keith Burridge75) was used for detection of the 205 kDa truncated fragment produced by expression of the p.(W1787*) variant in human iPSCs. Other antibodies used for western blot analysis and immunoprecipitation included mouse anti-GFP (1:1,000, #66002-1-Ig), rabbit anti-GFP (1:1,000, #50430-2-AP), rabbit anti-HA tag (1:1,000, #51064-2-AP), mouse anti-alpha-tubulin (1:1,000, #66031-1-Ig) and mouse anti-6*His tag (1:1,000, #66005-1-Ig) all from Proteintech, rabbit anti-mCherry (1:2,000, #ab167453) and rabbit anti-RFP (1:500, #ab62341) from Abcam, and mouse anti-βIII-tubulin (1:1,000, clone TU-20, #MAB1637) from Millipore-Sigma. Commercial antibodies used for immunofluorescence included mouse anti-neurofilament (1:200, clone SMI-312, #837904) and mouse anti-αII-spectrin (1:200, clone D8B7, #803201) from BioLegend, chicken anti-GFP (1:1,000, #GFP-1020) from Aves, and rabbit anti-OCT4 (1:500, #ab19857), rabbit anti-SOX2 (1:500, #ab97959), rabbit anti-NANOG (1:500, #ab80892), mouse anti-Satb2 (1:200, clone SATBA4B10, # ab51502), and rat anti-Ctip2 (1:500, clone 25B6, # ab18465) all from Abcam. In addition, we used mouse anti-SSEA4 (1:200, #MA1–021) and mouse ant-TRA-1–60 (1:1,000, #41–1000) from Thermo Fisher Scientific, and rat anti-RFP (1:1,000, clone 5F8, #5F8–100) from Chromotek. Secondary antibodies purchased from Life Technologies were used at 1:400 dilution for fluorescence-based detection by confocal microscopy and at 1:1,000 for staining of iPSCs. Secondary antibodies included donkey anti-rabbit IgG conjugated to Alexa Fluor 568 (#A10042), donkey anti-mouse IgG conjugated to Alexa Fluor 488 (#A21202), goat anti-chicken IgG conjugated to Alexa Fluor 488 (#A11039), donkey anti-rat IgG conjugated to Alexa Fluor 647 (#A21247), goat anti-rat IgG conjugated to Alexa Fluor 568 (#A11077), donkey anti-mouse IgG conjugated to Alexa Fluor 568 (#A10037), donkey anti-rabbit IgG conjugated to Alexa Fluor 647 (#A31573), goat anti-rabbit IgG conjugated to Alexa Fluor 594 (#R37117), and goat anti-mouse IgG conjugated to Alexa Fluor 488 (#A11001). Fluorescent signals in western blot analysis were detected using goat anti-rabbit 800CW (1:15,000, #926–32211) and goat anti-mouse 680RD (1:15,000, #926–68070) from LiCOR.
Neuronal culture.
Primary cortical neuronal cultures were established from E17 mice. Cortices were dissected in Hibernate E (Life Technologies) and digested with 0.25% trypsin in HBSS (Life Technologies) for 20 min at 37 °C. Tissue was washed three times with HBSS and dissociated in DMEM (Life Technologies) supplemented with 5% fetal bovine serum (FBS, Genesee), and gently triturated through a glass pipette with a fire-polished tip. Dissociated cells were filtered through a 70-μm cell strainer to remove any residual non-dissociated tissue and plated onto poly-D-lysine-coated 1.5-mm coverglasses or dishes (MatTek) for transfection and time-lapse microscopy imaging. For all cultures, media was replaced 3 h after plating with serum-free Neurobasal-A medium containing B27 supplement (Life Technologies), 2 mM Glutamax (Life Technologies), and penicillin/streptomycin (Life Technologies) (neuronal growth media). 5 μM cytosine-D-arabinofuranoside (Sigma) was added to the culture medium to inhibit the growth of glial cells three days after plating. Neurons were fed twice a week with freshly made culture medium until use.
Lentiviral production and infection of primary cortical neurons.
pLV-hSyn-RFP-PP-βII-Sp constructs were packaged into lentiviral particles using pRSVRev, pMDLg/pRRE, and pCMV-VSVG plasmids in HEK 293T/17 cells (ATCC® CRL-11268™) transfected using the calcium phosphate transfection kit (Takara). This cell line was authenticated by ATCC based on its STR profile. Viral particles were harvested from culture media 48 h and 72 h post-transfection and concentrated by ultracentrifugation on a SW 28 Ti Swinging-Bucket rotor at 25,000 rpm for 90 min. βII-SpKO primary cortical neurons grown on 6-well plates were transduced at plating with virus and Polybrene (5 μg/mL). Sixteen hours later, neurons were washed to remove viral particles and then grown in neuronal growth media for an additional 96 h.
Plasmid transfection for time-lapse live imaging and immunofluorescence analysis.
For time-lapse imaging experiments, DIV5 cortical neurons were co-transfected with 1 μg of each pLAMP1-RFP and peGFP-βIISp plasmids using lipofectamine 2000 (Life Technologies) and imaged 48–96 h after transfection. For experiments that evaluate axonal length, DIV3 control and βIISp-Het neurons were transfected with 500 ng of pmCherry-C1 and 1 μg of peGFP-C3. βII-SpKO neurons were transfected with 500 ng of pmCherry-C1 and 1 μg of peGFP-βIISp recue plasmids bearing full-length wild-type or mutant β2-spectrin. Neurons were processed for immunofluorescence 5 days after transfection. Immunofluorescence evaluations of βII-spectrin distribution in HEK 293T/17 cells was conducted in cells transfected with 100 ng of peGFP-βIISp plasmids, or co-transfected with 100 ng of each peGFP-βIISp and pmCherry-αIISp plasmids 48 h post-transfection.
Plasmid transfection for biochemistry analysis.
All transfections were conducted in HEK 293T/17 cells grown in 10-cm culture plates using the calcium phosphate transfection kit (Takara). To purify full-length βII-spectrin proteins, cells were transfected with 8 μg of peGFP-PP-βII-Sp-6xHis plasmids. To determine levels and stability of βII-spectrin proteins, HEK 293T/17 cells were co-transfected with 8 μg of peGFP-PP-βII-Sp-6xHis and 4 μg of pmCherry-C1 plasmids. To determine interaction between ankyrin-B and βII-spectrin, cells were co-transfected with 8 μg of each peGFP-PP-βII-Sp-6xHis and pAnkB-3xHA plasmids. For assessment of binding between βII-spectrin and αII-spectrin, cells were separately transfected with 8 μg of peGFP-PP-βII-Sp-6xHis or 4 μg peGFP-C3 and 8 μg of pmCherry-αIISp.
Histology and immunohistochemistry.
Brains from mice two-weeks and older were fixed by transcardial perfusion with phosphate-buffered saline (PBS) and 4% PFA followed by overnight immersion in the same fixative. Brains from PND0-PND14 mice were fixed by direct immersion in 4% PFA for 36 h. After fixation, brains were rinsed with PBS, transferred to 70% ethanol for at least 24 h and paraffin-embedded. 7-μm coronal and sagittal brain sections were cut using a Leica RM2155 microtome and mounted on glass slides. Sections were analyzed by hematoxylin and eosin (H&E) staining or immunostaining. For antibody staining, sections were deparaffinized and rehydrated using a standard protocol of washes: 3 × 3-min Xylene washes, 3 × 2-min 100% ethanol washes, and 1 × 2-min 95%, 80%, and 70% ethanol washes followed by at least 5 min in PBS. Sections were then processed for antigen retrieval using 10 mM sodium citrate, pH 6 in the microwave for 20 min. Sections were allowed to cool, washed in PBS, and blocked using antibody buffer (2% bovine serum albumin (BSA), 1% fish oil gelatin, 5% donkey serum, and 0.02% Tween 20 in PBS) for 1 h at room temperature. Tissue sections were then subsequently incubated overnight with primary antibodies at 4 °C and with secondary antisera for 1.5 h at 4 °C, washed with PBS, and mounted with Prolong Gold Antifade reagent (Life Technologies). Neuronal cultures and HEK 293T/17 cells were washed with cold PBS, fixed with 4% PFA for 15 min, and permeabilized with 0.2% Triton-X100 in PBS for 10 min at room temperature. Neurons and HEK 293T/17 cells were blocked in antibody buffer for 1 h at room temperature and processed for fluorescent staining as tissue sections. For actin labeling, Alexa Fluor 568- or Alexa Fluor 633-conjugated phalloidin (1:100) was added to the secondary antibody mix. DAPI was added at a 1:1,000 dilution to the last PBS rinse for nuclei staining.
Immunoblots.
Protein homogenates from human iPSCs, mouse brains, and transduced or transfected cells were prepared in 1:9 (wt/vol) ratio of homogenization buffer (8 M urea, 5% SDS (wt/vol), 50 mM Tris pH 7.4, 5 mM EDTA, 5 mM N-ethylmeimide, protease and phosphatase inhibitors) and heated at 65 °C for 15 min to produce a clear homogenate. Total protein lysates were mixed at a 1:1 ratio with 5x PAGE buffer (5% SDS (wt/vol), 25% sucrose (wt/vol), 50 mM Tris pH 8, 5 mM EDTA, bromophenol blue) and heated for 15 min at 65 °C. Samples were resolved by SDS-PAGE on house-made 3.5–17.5% acrylamide gradient gels or 4–20% Mini-PROTEAN® TGX™ Precast Protein Gels (BioRad) in Fairbanks Running Buffer (40 mM Tris pH 7.4, 20 mM sodium acetate, 2 mM EDTA, 0.2%SDS (wt/vol)). Proteins were transferred overnight onto 0.45-μm nitrocellulose membranes (#1620115, BioRad) at 4 °C. Transfer efficiency was determined by Ponceau-S stain. Membranes were blocked in TBS containing 5% non-fat milk for 1 h at room temperature and incubated overnight with primary antibodies diluted in antibody buffer (TBS, 5% BSA, 0.1% Tween-20). After 3 washes in TBST (TBS, 0.1% Tween-20), membranes were incubated with secondary antibodies diluted in antibody buffer for 2 h at room temperature. Membranes were washed 3x for 10 min with TBST and 2x for 5 min in TBS. Protein-antibody complexes were detected by the Odyssey® CLx Imaging system (LI-COR) running Image Studio V5.2.
Immunoprecipitation.
For immunoprecipitation experiments, total protein homogenates from transfected HEK 293T/17 cells were prepared in TBS containing 150 mM NaCl, 0.32 M sucrose, 2 mM EDTA, 1% Triton X-100, 0.5% NP40, 0.1% SDS, and compete protease inhibitor cocktail (Sigma). Cell lysates were incubated with rotation for 1 h at 4 °C and centrifuged at 100,000 × g for 30 min. Soluble fractions were collected and precleared by incubation with Protein A/G magnetic beads (#88802, Life Technologies) for 1 h in the cold. Samples were subjected to immunoprecipitation in the presence of protein-G magnetic beads/antibody or protein-G magnetic beads/isotype control complexes overnight at 4 °C. Immunoprecipitation samples were resolved by SDS-PAGE and western blot and signal detected using the Odyssey® CLx imaging system.
Purification of full-length βII-spectrin proteins.
Ten 10-cm plates of HEK 293T/17 cells expressing each peGFP-PP-βII-Sp-6xHis construct were used per purification. Total protein homogenates from transfected HEK 293T/17 cells were prepared in TBS containing 150 mM NaCl, 0.32 M sucrose, 2 mM EDTA, 1% Triton X-100, 0.5% NP40, 0.1% SDS, and complete protease inhibitor cocktail (Sigma) (IP buffer). Cell lysates were incubated with rotation for 1 h at 4 °C and centrifuged at 100,000 × g for 30 min. Soluble fractions were incubated overnight with Protein A/G magnetic beads (#88802, Life Technologies) coupled to GFP antibodies with rotation at 4 °C. Beads were extensively washed with IP buffer, followed by washes in TBS containing 300 mM NaCl, and TBS. Full-length βII-spectrin proteins were eluted from GFP-protein A/G magnetic beads by incubation with HRV-3C protease (#SAE0110, Sigma), which cleaves between GFP and the start codon of βII-spectrin in prescission protease buffer (25 mM HEPES, 150 mM NaCl, 1 mM EDTA, 1 mM DTT) for 36 h at 4 °C. The efficiency of cleavage and purity of the eluates was analyzed by western blot using validated antibodies specific for βII-spectrin, the GFP and 6*His tags, and by Coomassie blue stain. Eluates were concentrated using Pierce™ Protein Concentrators PES.
Pulldown assays.
For detection of βII-spectrin/αII-spectrin complexes, control and mutant GFP-βIISp proteins were coupled to GFP-bound Protein-A/G magnetic beads and incubated with lysates from HEK 293T/17 cells expressing mCherry-αIISp in IP buffer overnight at 4 °C. Beads complexes were washed sequentially with IP buffer, followed by washes in TBS containing 400 mM NaCl, and TBS. Proteins were eluted in 5x PAGE loading buffer and analyzed by SDS-PAGE and western blot.
Actin co-sedimentation assay.
Interaction between purified full-length βII-spectrin proteins and F-actin was evaluated using the Actin Binding Protein Spin-Down Biochem Kit (#BK001, Cytoskeleton) following the manufacturer’s recommendations. In brief, full-length βII-spectrin (1 mg/ml) and α-actinin (20 mg/ml, positive control) were prepared in general actin buffer (5 mM Tris-HCl pH 8.0 and 0.2 mM CaCl2) and centrifuged at 150,000 × g for 1 h at 4 °C. F-actin (1 mg/ml) was prepared by incubation of purified actin in general actin buffer for 30 min on ice followed by the actin polymerization step in actin polymerization buffer (50 mM KCl, 2 mM MgCl2, 1 mM ATP) for 1 h at 24 °C. F-actin (21 μM) was incubated with βII-spectrin (10 μM), α-actinin (2 μM), or BSA (2 μM, negative control) for 30 min at 24 °C. F-actin-protein complexes were pelleted by ultracentrifugation at 150,000 × g for 1.5 h at 24 °C. The presence of F-actin together with interacting proteins was assessed in the supernatant and pellet fractions by SDS-PAGE and Coomassie blue stain.
Fluorescence image acquisition and image analysis.
iPSCs were imaged with an Evos Auto FL microscope. The rest of the images were acquired using a Zeiss LSM780 confocal scope and 405-, 488-, 561-, and 633-nm lasers using the Zeiss ZEN 2.3 SP1 FP1 (black) V.14.0.9.201 acquisition software. Single images and Z-stacks with optical sections of 1-μm intervals and tile scans were collected using the 10× (0.4 NA) and 40× oil (1.3 NA) objective lens. Images were processed, and measurements taken and analyzed using NIH ImageJ software. Three-dimensional rendering of confocal Z-stacks was performed using Imaris (Bitplane).
Time-lapse video microscopy and movie analyses.
Live microscopy of neuronal cultures was carried out using a Zeiss 780 laser scanning confocal microscope (Zeiss) equipped with a GaAsP detector and a temperature- and CO2-controlled incubation chamber as previously reported76. Movies were taken in the mid-axon and captured at a rate of 1 frame/second for time intervals ranging from 60–300 seconds with a 40× oil objective (1.4NA) using the zoom and definite focus functions. Movies were processed and analyzed using ImageJ (http://rsb.info.nih.gov/ij). Kymographs were obtained using the KymoToolBox plugin for ImageJ (https://github.com/fabricecordelieres/IJ_KymoToolBox). In details, space (x axis in μm) and time (y axis in sec) calibrated kymographs were generated from video files. In addition, the KymoToolBox plugin was used to manually follow a subset of particles from each kymograph and report the tracked particles on the original kymograph and video files using a color code for movement directionality (red for anterograde, green for retrograde and blue for stationary particles). Quantitative analyses were performed manually by following the trajectories of individual particles to calculate dynamic parameters including, net and directional velocities and net and directional run length, as well as time of pause or movement in a direction of transport. Anterograde and retrograde motile vesicles were defined as particles showing a net displacement >3 μm in one direction. Stationary vesicles were defined as particles with a net displacement <2 μm.
Statistical analysis.
GraphPad Prism (GraphPad Software) was used for statistical analysis. Two groups of measurements were compared by unpaired Student’s t-test. Multiple groups were compared by one-way ANOVA followed by Tukey’s or Dunnett’s multiple comparisons test.
Molecular modeling of SPTBN1 variants.
We used the closed conformation of utrophin CH1–CH2 closed dimer (PDB 1qag)47 as a template for the analogous βII-spectrin conformation to estimate its electrostatic surface profile. Molecular structures from the 6.9 Å cryo-EM structure of the CH1 actin-binding domain of βIII-spectrin bound to F-actin (PDB 6anu)49 and the structure of the CH2 domain of βII-spectrin (PDB 1bkr)48 were used for protein-protein docking predictions. The ClusPro protein-protein docking webserver50,51 was used to (i) dock the CH1 domain of spectrin onto F-actin, (ii) dock the CH2 domain of spectrin onto F-actin, and (iii) dock the CH2 domain of spectrin onto the CH1 domain of spectrin. The CH1 structure used for the dockings reported here was the model of the CH1 domain of βIII-spectrin from 6anu (chain a)49. This CH1 model was built based on the crystal structure of plectin (PDB 1mb8)77 by I-TASSER78. The CH1 domain of βIII-spectrin shares 95% sequence identity with the CH1 domain of βII-spectrin. The actin model corresponded to chains A–F of 6anu, which in turn was generated from the cryo-EM structure of actin (PDB 5jlh)79. The molecular structure of the CH2 domain of βII-spectrin from 1bkr was of a 1.1 Å crystal structure48.
To identify the inactive closed conformation of the tandem domain (CH1–CH2) of βII-spectrin, the CH2 domain of βII-spectrin was docked onto the CH1 domain of βII-spectrin using the ClusPro webserver V2.0. The top 15 docking poses for each of the four scoring algorithms were evaluated for the placement of βII-spectrin residue L250 from the CH2 domain at the interface of the CH2/CH1 closed conformation. The top docking pose in the electrostatic scoring algorithm corresponded to a pose with a deeply buried L250 at the interface of the CH1/CH2 complex. The variant of the equivalent residue in βIII-spectrin (L253P) might disrupt the closed structure and drive the spectrin ensemble to a more open state suitable for binding to actin48. This same docking pose was also a top docking pose (pose 4) within the set of poses calculated by the balanced scoring algorithm. This pose was used for evaluation of the βII-spectrin mutants. It was also the same pose compared to the actinin-4 (PDB 6oa6) (unpublished) and utrophin (PDB 1qag)47 closed conformations.
For each of the three ClusPro protein docking analyses, the webserver provided up to 30 docking poses for each of four scoring algorithms (balanced; electrostatic-favored; hydrophobic-favored; VdW+Elec). The top 15 poses from each of the four scoring algorithms were included in the final analysis. For the dockings of the CH1 and CH2 domains from βII-spectrin onto F-actin, several of the top docking poses were to the ends of the actin segment defined as the receptor. These docking poses were immediately rejected as other actin molecules would be binding at those locations in F-actin and these sites would not be available for binding to spectrin. For CH1 docking onto F-actin, the remaining poses within the top eight docking poses predicted by the balanced and electrostatic scoring algorithms almost all corresponded to the location and orientation of CH1 molecules on actin as defined by the cryo-EM structure 6anu. For CH2 docking onto F-actin, the remaining poses within the top 8 docking poses predicted by the balanced and electrostatic scoring algorithms almost all corresponded to symmetry-related locations and poses on the F-actin. In addition, the predicted orientation of the CH2 molecules on F-actin was consistent with the known binding site of the CH1 domains, as judged by the length of the linker that would be required to join the C-terminus of the docked CH1 domain to the N-terminus of the docked CH2 domain.
βII-spectrin is a large multi-domain protein that requires a different approach for each type of domain. The SR have relatively low sequence identity to each other, and only a few have been experimentally solved, requiring independent models to be generated for each. We used RaptorX80 homology modeling to generate each model and assembled them into a linear conformation using Discovery Studio (Dassault Systèmes BIOVIA, Discovery Studio Modeling Environment, Release 2019, San Diego: Dassault Systèmes. 2019). We calculated protein electrostatics using APBS81 and visualized structures using PyMOL (The PyMOL Molecular Graphics System, Version 2.0.7 Schrödinger, LLC.). Individual spectrin repeats were also superimposed onto each other using a geometric algorithm82 as implemented in PyMOL, to investigate patterns across the fold.
Behavioral assessment.
Animals.
Because the Sptbn1flox/flox/Nestin-Cre (βII-SpKO) mice have early mortality (typically between PND30 and PND40), testing in these mice was conducted late in the juvenile period. We assessed 15 wild-type (Sptbn1flox/flox/+, βII-SpWT) and 5 βII-SpKO mice, taken from 5 litters. βII-SpKO mice were evaluated in two tests: open field (at PND 28–31) and acoustic startle (at PND 29–32). Sptbn1flox/+/Nestin-Cre (βII-SpHet) mice, which have normal survival rates, were subjected to a more expansive battery of tests. βII-SpHet mice (n = 12 per genotype, all males) underwent the tests below, with order planned so that more stressful procedures occurred closer to the end of the study (Supplementary Table 5).
Elevated plus maze.
A 5-min test for anxiety-like behavior was carried out on the plus maze (elevation, 50 cm H; open arms, 30 cm L; closed arms, 30 cm L, walls, 20 cm H). Mice were placed in the center (8 cm × 8 cm) at the beginning of the test. Measures were taken of percent open arm time and open arm entries, and total number of arm entries.
Open field.
Exploratory activity was evaluated by a 1-h test (30-min for βII-SpKO mice) in a novel open field chamber (41 cm × 41 cm × 30 cm) crossed by a grid of photobeams (VersaMax system, AccuScan Instruments). Counts were taken of photobeam breaks in 5-min intervals, with separate measures for locomotor activity (total distance traveled) and vertical rearing movements. Anxiety-like behavior was assessed by measures of time spent in the center region.
Accelerating rotarod.
Mice were first given three trials on the rotarod (Ugo Basile, Stoelting Co.), with 45 seconds between each trial. Two additional trials were conducted 48 h later, to evaluate consolidation of motor learning. RPM (revolutions per minute) progressively increased from 3 to a maximum of 30 rpm across 5 min (the maximum trial length), and latency to fall from the top of the rotating barrel was recorded.
Social approach in a three-chamber choice test.
Mice were evaluated for the effects of Sptbn1 deficiency on social preference. The procedure had three 10-min phases: habituation, sociability, and social novelty preference. In the sociability phase, mice were presented with a choice between proximity to an unfamiliar C57BL/6J adult male (“stranger 1”), versus an empty cage. In the social novelty phase, mice were presented with the already-investigated stranger 1 and a new unfamiliar mouse (“stranger 2”). The test was carried out in a rectangular, three-chambered Plexiglas box (60 cm L, 41. 5 cm W, 20 cm H). An automated image tracking system (Noldus Ethovision) provided measures of time spent within 5 cm proximity to each cage and entries into each side of the social test box.
Marble-burying.
Mice were tested for exploratory digging in a Plexiglas cage, placed inside a sound-attenuating chamber with ceiling light and fan. The cage floor had 5 cm of corncob bedding, with 20 black glass marbles (14 mm diameter) set up in a 5 × 4 grid on top of the bedding. Measures were taken of the number of marbles buried by the end of the 30-min test.
Buried food test.
Mice were presented with an unfamiliar food (Froot Loops, Kellogg Co.) in the home cage several days before the test. All home cage food was removed 16–24 h before the test. The assay was conducted in a tub cage (46 cm L, 23.5 cm W, 20 cm H), containing paper chip bedding (3 cm deep). One Froot Loop was buried in the cage bedding, and mice were given 15 min to locate the buried food. Latency to find the food was recorded.
Acoustic startle.
This procedure was used to assess auditory function, reactivity to environmental stimuli, and sensorimotor gating. The test was based on the reflexive whole-body flinch, or startle response, that follows exposure to a sudden noise. Mice were evaluated for startle magnitude and prepulse inhibition, which occurs when a weak prestimulus leads to a reduced startle in response to a subsequent louder noise. Startle amplitudes were measured by force displacement of a piezoelectric transducer (SR-Lab, San Diego Instruments). The test had 42 trials (7 of each type): no-stimulus trials, trials with the acoustic startle stimulus (40 msec; 120 dB) alone, and trials in which a prepulse stimulus (20 msec; either 74, 78, 82, 86, or 90 dB) occurred 100 msec before the onset of the startle stimulus. Levels of prepulse inhibition at each prepulse sound level were calculated as 100 − ((response amplitude for prepulse stimulus and startle stimulus together/response amplitude for startle stimulus alone) × 100).
Morris water maze.
The water maze (diameter = 122 cm) was used to assess spatial and reversal learning, swimming ability, and vision. The procedure had three phases: visible platform, acquisition in the hidden platform task, and reversal learning (with the platform moved to a new location). For each phase, mice were given four 60-sec trials per day. Measures were taken of time to find the escape platform (diameter = 12 cm) and swimming velocity by an automated tracking system (Noldus Ethovision). Criterion for learning was an average group latency of 15 sec or less to locate the platform. At the end of the acquisition and reversal phases, mice were given a 1-min probe trial in the maze without the platform. Selective quadrant search was evaluated by measuring number of crosses over the location where the platform (the target) had been placed during training, versus the corresponding areas in the other three quadrants.
Statistical analyses for behavioral tests.
All testing was conducted by experimenters blinded to mouse genotype. StatView 5.0.1 (SAS, Cary, NC) was used for data analyses. One-way or repeated measures analysis of variance (ANOVA) were used to determine effects of genotype. Post-hoc analyses were conducted using Fisher’s Protected Least Significant Difference (PLSD) tests only when a significant F value was found in the ANOVA. Within-genotype repeated measures analyses were used to determine side preference in the 3-chamber test and quadrant preference in the Morris water maze assay. For all comparisons, significance was set at P < 0.05.
Data Availability
The whole-genome and exome sequencing or transcriptomic data will not be made publicly available as they contain information that could compromise research participant privacy/consent. Information on the DNA and RNA sequencing raw data and other analyses supporting the findings of this study is available from the corresponding authors upon request. Source data are provided with this paper.
Extended Data
Extended Data Fig. 1. Expression of SPTBN1 variants alters protein expression, cellular distribution and morphology.

a, Western blot of total lysates from HEK 293T/17 cells co-transfected with GFP-βIISp and mCherry plasmids and blotted with anti-GFP and anti-mCherry antibodies. Results are representative of three independent experiments. b, Western blot of Triton-X100 soluble and insoluble fractions from HEK 293T/17 cell lysates transfected with GFP-βIISp plasmids and blotted with anti-GFP antibody. Images are representative of three independent experiments. c, Partition of indicated GFP-βIISp proteins expressed in HEK 293T/17 cells between Triton-X100 soluble and insoluble fractions relative to total GFP-βIISp levels. Data in c were collected from n = 3 biological replicates in three independent experiments. Data represent mean ± SEM. One-way ANOVA with Dunnett’s post hoc analysis test for multiple comparisons, ***P = 0.001, ****P < 0.0001. d, Western blot of total lysates from primary mouse cortical neurons from βIISp-KO mice transduced with lentivirus expressing RFP-PP-βIISp proteins driven by the neuronal-specific synapsin I promoter and blotted with anti-RFP and anti-βIII-tubulin antibodies. Red arrowheads and boxes mark the presence of an additional 70-kDa GFP-positive fragment in HEK 293T/17 (a) and mouse neuron (d) lysates expressing variants that result in GFP-positive aggregates. Blots are representative of three separate experiments. e, Western blot of total lysates from human iPSC lines reprogrammed from PBMCs carrying the indicated variants and blotted with anti-βII-spectrin and anti-α-tubulin antibodies. A red asterisk indicates the presence of a truncated 205-kDa βII-spectrin fragment in lysates from iPSCs reprogrammed from P27 (p.W1787*, c.5361G>A). Blots are representative of four independent experiments. Western blot images were cropped from Source Data Extended Data Figure 1. f, Analysis of sequencing reads from RNA-seq of blood RNA obtained from P27 (p.W1787*, c.5361G>A) indicate allelic expression bias, suggesting some level of nonsense mediated decay of the SPTBN1 allele transcript harboring the nonsense variant, and increased abundance of the major c.5361G SPTBN1 allele. g, Quantification of the percent of GFP-positive HEK 293T/17 cells with GFP aggregates for each of the indicated variants. Data were collected from n = 20 cells/genotype pooled from three independent experiments and the following number of transfection replicates: WT (n = 10), T59I (n = 3), I59Q_160Δ (n = 5), C183* (n = 6), Y190_R216Δ (n = 3), G205D (n = 4), G205S (n = 6), L247H (n = 5), L250R (n = 7), D255E (n = 5), T268A (n = 4), T268N (n = 4), T268S (n = 6), V271M (n = 4), H275R (n = 6), F344L (n = 4), R411W (n = 3), E491Q (n = 4), A850G (n = 3), E892* (n = 3), R1003W (n = 9), A1086T (n = 4), E1110D (n = 4), G1398S (n = 3), W1787* (n = 3), E1886Q (n = 4). h,i, Quantification of cell length (h) and filopodia density normalized to cell length (i) of GFP-positive HEK 293T/17 cells expressing the indicated variants. Data in h and i were collected from WT (n = 23), T59I (n = 13), I59Q_160Δ (n = 12), C183* (n = 12), Y190_R216Δ (n = 26), G205D (n = 11), G205S (n = 11), L247H (n = 22), L250R (n = 14), D255E (n = 18), T268A (n = 13), T268N (n = 15), T268S (n = 12), V271M (n = 10), H275R (n = 13), F344L (n = 12), R411W (n = 10), E491Q (n = 11), A850G (n = 11), E892* (n = 12), R1003W (n = 15), A1086T (n = 10), E1110D (n = 12), G1398S (n = 10), W1787* (n = 12), and E1886Q (n = 12) cells pooled from six independent experiments. All data represent mean ± SEM. One-way ANOVA with Dunnett’s post hoc analysis test for multiple comparisons. (g) ****P < 0.0001, nsP > 0.05. (h) *P = 0.0119 (T268S), *P = 0.0376 (H275R), *P = 0.0184 (R411W), *P = 0.0492 (A850G); **P = 0.0029 (T59I), **P = 0.0083 (V271M); ***P = 0.0009 (E1110D), ***P = 0.0005 (E1886Q); ****P < 0.0001. (i) *P = 0.0141 (G205D); **P = 0.0079 (C183*), **P = 0.0023 (Y190_R216Δ), **P = 0.0027 (G205S), **P = 0.0083 (R411W); ***P = 0.0006 (E491Q), ***P = 0.0002 (E1886Q); ****P < 0.0001. See statistics summary in Source Data Extended Data Figure 5.
Extended Data Fig. 2. SPTBN1 variants alter interaction with critical cytoskeleton partners.
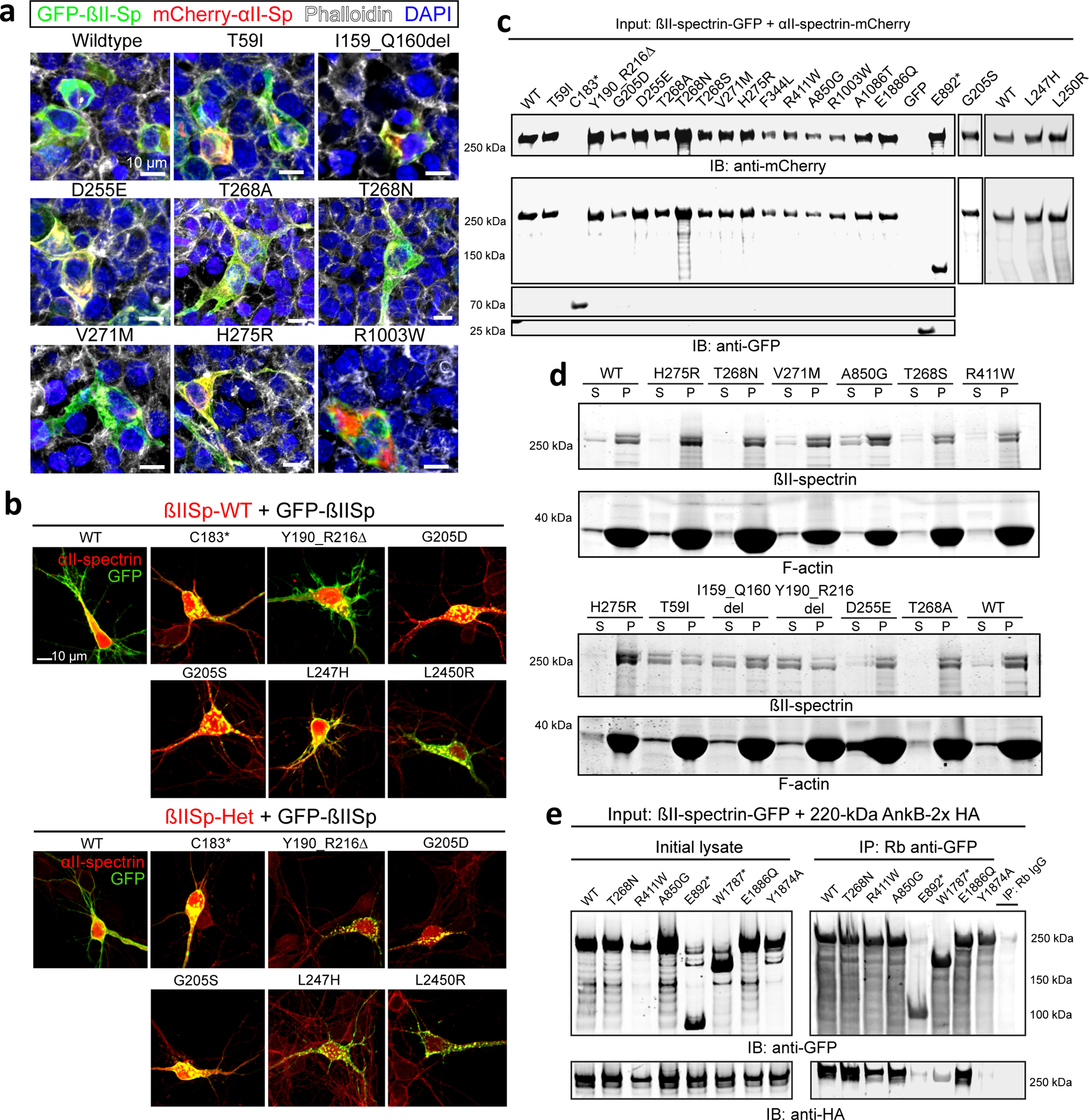
a, Immunofluorescence images, representative of three independent experiments, show HEK 293T/17 cells transfected with mCherry-αIISp and with either WT or mutant GFP-βIISp plasmids. Cells were stained for actin (phalloidin) and DAPI. Scale bar, 10 μm. b, Immunofluorescence images of DIV8 mouse βIISp-WT (top) and βIISp-Het (bottom) cortical neurons transfected with indicated GFP-βIISp plasmids. Scale bar, 10 μm. GFP-positive aggregates are detected in neurons expressing these subsets of CH domain variants regardless of the level of endogenous βII-spectrin. Images are representative of n = 15 neurons per transfection derived from three independent experiments. c, Western blot from a binding assay to assess interaction between mCherry-αIISp and GFP-βII-spectrin proteins representative of n = 3 biological replicates from three independent experiments. Lysates from HEK 293T/17 cells expressing mCherry-αII-spectrin were incubated with GFP-βII-spectrin proteins coupled to GFP beads. The presence of mCherry-αII-spectrin in eluates from GFP beads was evaluated by blotting with anti-GFP and anti-mCherry antibodies. d, Coomassie blue staining showing the presence of purified full-length βII-spectrin and F-actin in the supernatant (S) and pellet (P) fractions from an actin co-sedimentation assay. Blot is representative of three independent experiments each with n = 1 biological replicate. e, Co-IP assay in HEK 293T/17 cells to assess interaction between 220-kDa ankyrin-B (AnkB)-2HA and GFP-βII-spectrin proteins. The presence of 220-kDa AnkB-3xHA and GFP-βII-spectrin proteins in initial lysates and eluates from beads coupled to rabbit IgG isotype control of a rabbit anti-GFP antibody was detected by blotting with anti-GFP and anti-HA antibodies. Blot is representative of four independent experiments, each with n = 1 biological replicate. Western blot images were cropped from Source Data Extended Data Figure 2.
Extended Data Fig. 3. Modeling effects of SPTBN1 variants.

a–c, Potential coevolution of the closed conformation of the tandem calponin homology domain (CH1–CH2) of βII-spectrin (SPTBN1) (CH1 domain (teal), CH2 domain (red)), actinin-4 (ACTN4) (brown), and utrophin (UTRN) (purple)47. d,e, Top hits from docking simulations of βII-spectrin’s CH1 (d) and CH248 (e) onto F-actin (gray). Domains in dark blue correspond to cryo-EM structure of the CH1 domain of βIII-spectrin bound to F-actin49. f, Correct length of simulated interdomain linker (dark blue) in agreement with the orientation of the docked CH2 domain (red). g,h, Spatial distributions of the missense variants in βII-spectrin implicate disease mechanisms. g, Linear conformation of the entire 3D protein model is shown with the calponinhomology (CH) domains (CH1 and CH2) in the N-terminus (red), the spectrin repeats (SR) (green) and the pleckstrin homology (PH) domain in the C-terminus (purple). h, The 17 SR domains are superimposed with a minimal cartoon representation to emphasize the consistency of the 3D architecture despite high sequence diversity. The positions of the amino acid residues representing the missense variants are marked by gold-colored spheres.
Extended Data Fig. 4. Effects of SPTBN1 variants on axonal growth.

a, Images of DIV8 βII-SpWT, βII-SpHet, βII-SpKO, and GFP-βIISp rescued βII-SpKO neurons transfected at DIV3 with mCherry. Staining with an antibody specific for AnkG was used to label the AIS (yellow arrowhead) and to identify axonal processes. Scale bar, 30 μm. Images are representative of three independent experiments.
Extended Data Fig. 5. Effects of SPTBN1 variants on dendrites.

a, Images of DIV18 βII-SpWT, βII-SpHet, βII-SpKO, and GFP-βIISp rescued βII-SpKO neurons stained with an anti-GFP antibody. Scale bar, 30 μm. b,c, Quantification of length of primary dendrites (b) and of total number of primary and secondary dendrites (c) of βII-SpWT, βII-SpHet, βII-SpKO, and rescued βII-SpKO DIV18 neurons (n = 6–16 neurons/genotype) compiled from three independent experiments. Data represent mean ± SEM. One-way ANOVA with Dunnett’s post hoc analysis test for multiple comparisons, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. See statistics summary in Source Data Extended Data Figure 5.
Extended Data Fig. 6. Effects of βII-spectrin deficiency on neuronal morphology and brain development.

a, Image, representative from three independent experiments show DIV8 βII-SpKO cortical neurons rescued with WT GFP-βIISp or with GFP-βIISp bearing variants within the distal portion of the CH2 domain. Neurons were stained for actin (phalloidin) and endogenous αII-spectrin. Yellow dotted lines demark the cell edge. Scale bar, 5 μm. b, Images of PND25 βII-SpNexWT and βII-SpNexKO brains stained for neurofilament to label axons and DAPI. Staining for βII-spectrin show specific loss of the protein in axons from callosal projection neurons from βII-SpNexKO mice. Scale bar, 50 μm. White dotted lines denote the position and boundaries of the corpus callosum (CC). Brains were collected from two separate litters and processed for staining and imaging as part of one independent experiment.
Extended Data Fig. 7. Developmental and behavioral phenotypes of βII-spectrin deficient mice.
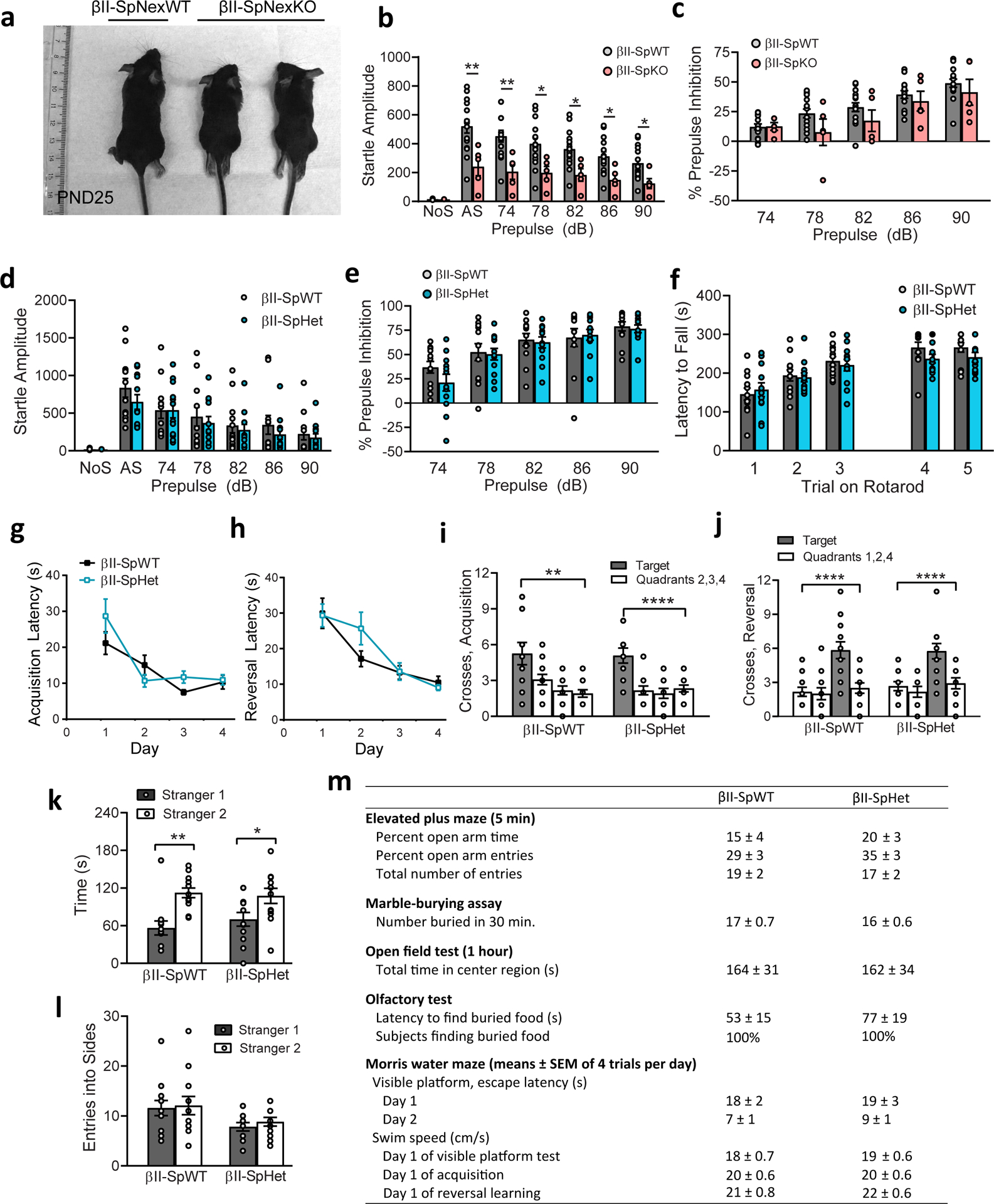
a, Images of male PND25 wildtype (βII-SpNexWT) mice and mice lacking βII-spectrin only in cortical and hippocampal projection neurons (βII-SpNexKO) driven by Nex-Cre. b–e, Magnitude of acoustic startle responses (b,d) and percent of prepulse inhibition (c,e) in βII-SpWT mice and mice with partial (βII-SpHet) and complete (βII-SpKO) loss of βII-spectrin in neural progenitors driven by Nestin-Cre. Trials included no stimulus (NoS) trials and acoustic startle stimulus (AS; 120 dB) alone trials. Data in b and c represent mean ± SEM (n = 15 βII-SpWT and n = 5 βII-SpKO male mice). Data in d and e represent mean ± SEM (n = 12 male mice/genotype). Fisher’s PLSD tests following repeated measures ANOVA. b,*P < 0.05, **P < 0.01. c–e, P > 0.05. f, Latency to fall from an accelerating rotarod. Trials 4 and 5 were given 48 h after the first three trials. g,h, Latencies to find the hidden escape platform during acquisition (g) and reversal (h) learning phases of the Morris water maze test for βII-SpWT and βII-SpHet mice. Data represent mean ± SEM of four trials per day. Fisher’s PLSD tests following repeated measures ANOVA. f–h, P > 0.05. i,j, Mice were given a one-minute probe trial without the platform following the acquisition and reversal phases of the Morris water maze test. Target indicates the site where the platform had been located in each phase. Measures were taken of swim path crossings over the target location or corresponding areas in the other quadrants. Within-genotype repeated measures ANOVA, effect of quadrant (the repeated measure), **P = 0.0012, ****P < 0.0001. k,l, Preference for social novelty during a three-chamber choice task. Within-genotype repeated measures ANOVA, *P = 0.0145, **P = 0.0052. Data in f–l represent mean ± SEM (n = 12 male mice/genotype). m, Lack of significant genotype effects on anxiety-like behavior in the elevated plus maze, marble-burying assay, and open field; sensory ability in the buried food test for olfactory function and hot plate test for thermal sensitivity; and vision and swimming ability in the Morris water maze. Data represent mean ± SEM (n = 12 male mice/genotype). Within-genotype repeated measures ANOVA, P > 0.05.
Supplementary Material
Acknowledgements
We thank all the families who participated in this study. We thank Matthew Rasband and Klaus-Armin Nave for the gift of the βII-spectrin conditional null and the Nex-Cre mice, respectively. We thank Natallia V. Riddick for her assistance with the behavioral studies. We thank Beverly Koller and Karen Mohlke for their insightful comments on this manuscript and James Bear for helpful discussions. M.A.C., L.E.S.-R., and E.W.K. were supported by the Center for Individualized Medicine at Mayo Clinic. D.N.L. was supported by the University of North Carolina at Chapel Hill (UNC-CH) School of Medicine as a Simmons Scholar, by the National Ataxia Foundation, and by the US National Institutes of Health (NIH) grant R01NS110810. E.E.E. was supported by the NIH grant MH101221. Microscopy was performed at the UNC-CH Neuroscience Microscopy Core Facility, supported, in part, by funding from the NIH-NINDS Neuroscience Center Grant P30 NS045892 and the NIH-NICHD Intellectual and Developmental Disabilities Research Center Support Grant U54 HD079124, which also supported the behavioral studies. Research reported in this manuscript was supported by the NIH Common Fund, through the Office of Strategic Coordination/Office of the NIH Director under Award Number U01HG007672 (Duke University to Vandana Shashi). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This research was made possible through access to the data and findings generated by the 100,000 Genomes Project. The 100,000 Genomes Project is managed by Genomics England Limited (a wholly owned company of the Department of Health and Social Care). The 100,000 Genomes Project is funded by the National Institute for Health Research and NHS England. The Wellcome Trust, Cancer Research UK and the Medical Research Council have also funded research infrastructure. The 100,000 Genomes Project uses data provided by patients and collected by the National Health Service as part of their care and support.
Footnotes
Competing interests
The authors declare the following competing interests: E.T., R.E.P., Y.S., E.A.N., and A.B. are employees of GeneDx, Inc.
References
- 1.Bennett V & Lorenzo DN Spectrin- and ankyrin-based membrane domains and the evolution of vertebrates. Curr. Top. Membr 72, 1–37 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Bennett V & Lorenzo DN An adaptable spectrin/ankyrin-based mechanism for long-range organization of plasma membranes in vertebrate tissues. Curr. Top. Membr 77, 143–184 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Lorenzo DN Cargo hold and delivery: Ankyrins, spectrins, and their functional patterning of neurons. Cytoskeleton 77, 129–148 (2020).32034889 [Google Scholar]
- 4.Ikeda Y et al. Spectrin mutations cause spinocerebellar ataxia type 5. Nat. Genet 38, 184–190 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Saitsu H et al. Dominant-negative mutations in alpha-II spectrin cause West syndrome with severe cerebral hypomyelination, spastic quadriplegia, and developmental delay. Am. J. Hum. Genet 86, 881–891 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang CC et al. βIV spectrinopathies cause profound intellectual disability, congenital hypotonia, and motor axonal neuropathy. Am. J. Hum. Genet 102, 1158–1168 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacob FD, Ho ES, Martinez-Ojeda M, Darras BT & Khwaja OS Case of infantile onset spinocerebellar ataxia type 5. J. Child Neurol 28, 1292–1295 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Parolin Schnekenberg R et al. De novo point mutations in patients diagnosed with ataxic cerebral palsy. Brain 138, 1817–1832 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nuovo S et al. Between SCA5 and SCAR14: delineation of the SPTBN2 p.R480W-associated phenotype. Eur. J. Hum. Genet 26, 928–929 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicita F et al. Heterozygous missense variants of SPTBN2 are a frequent cause of congenital cerebellar ataxia. Clin. Genet 96, 169–175 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Mizuno T et al. Infantile-onset spinocerebellar ataxia type 5 associated with a novel SPTBN2 mutation: A case report. Brain Development 41, 630–633 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Accogli A et al. Heterozygous missense pathogenic variants within the second spectrin repeat of SPTBN2 lead to infantile-onset cerebellar ataxia. J. Child Neurol 35, 106–110 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Lise S et al. Recessive mutations in SPTBN2 implicate β-III spectrin in both cognitive and motor development. PLoS Genet 8, e1003074 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yıldız Bölükbaşı E et al. Progressive SCAR14 with unclear speech, developmental delay, tremor, and behavioral problems caused by a homozygous deletion of the SPTBN2 pleckstrin homology domain. Am. J. Med. Genet. A 173, 2494–2499 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Al-Muhaizea M, et al. A novel homozygous mutation in SPTBN2 leads to spinocerebellar ataxia in a consanguineous family: Report of a new infantile-onset case and brief review of the literature. Cerebellum 17, 276–285 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Writzl K et al. Early onset West syndrome with severe hypomyelination and coloboma-like optic discs in a girl with SPTAN1 mutation. Epilepsia 53, e106–e110 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Hamdan FF et al. Identification of a novel in-frame de novo mutation in SPTAN1 in intellectual disability and pontocerebellar atrophy. Eur. J. Hum. Genet 20, 796–800 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nonoda Y et al. Progressive diffuse brain atrophy in West syndrome with marked hypomyalination due to SPTAN1 gene mutation. Brain Dev 35, 280–283 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Tohyama J et al. SPTAN1 encephalopathy: distinct phenotypes and genotypes. J. Hum. Genet 60, 167–173 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Syrbe S et al. Delineating SPTAN1 associated phenotypes: from isolated epilepsy to encephalopathy with progressive brain atrophy. Brain 140, 2322–2336 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beijer D et al. Nonsense mutations in alpha-II spectrin in three families with juvenile onset hereditary motor neuropathy. Brain 142, 2605–2616 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Knierim E et al. A recessive mutation in beta-IV-spectrin (SPTBN4) associates with congenital myopathy, neuropathy, and central deafness. Hum. Genet 136, 903–910 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Häusler MG et al. A novel homozygous splice-site mutation in the SPTBN4 gene causes axonal neuropathy without intellectual disability. Eur. J. Med. Genet 63, 103826 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Xu K, Zhong G & Zhuang X Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science 339, 452–456 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheney R, Hirokawa N, Levine J & Willard M Intracellular movement of fodrin. Cell Motility 3, 649–655 (1983). [DOI] [PubMed] [Google Scholar]
- 26.Lorenzo DN et al. βII-spectrin promotes mouse brain connectivity through stabilizing axonal plasma membranes and enabling axonal organelle transport. Proc. Natl. Acad. Sci. USA 116, 15686–15695 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaganathan K et al. Predicting splicing from primary sequence with deep learning. Cell 176, 535–548 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Karczewski KJ et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang N, Lee I, Marcotte EM & Hurles ME Characterising and predicting haploinsufficiency in the human genome. PLoS Genet 6, e1001154. (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zech M et al. Monogenic variants in dystonia: an exome-wide sequencing study. Lancet Neurol 19, 908–918 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willsey AJ et al. De novo coding variants are strongly associated with Tourette Disorder. Neuron 94, 486–499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Firth HV et al. DECIPHER: Database of chromosomal imbalance and phenotype in humans using ensembl resources. Am. J. Hum. Genet 84, 524–533 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iossifov I et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221. (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schultz-Rogers L et al. Haploinsufficiency as a disease mechanism in GNB1-associated neurodevelopmental disorder. Mol. Genet. Genomic Med 8, e1477 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richards S et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med 17, 405–424 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abou Tayoun AN et al. Recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion. Hum. Mutat 39, 1517–1524 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strande NT et al. Evaluating the clinical validity of gene-disease associations: An evidence-based framework developed by the Clinical Genome Resource. Am. J. Hum. Genet 100, 895–906 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brnich SE et al. Recommendations for application of the functional evidence PS3/BS3 criterion using the ACMG/AMP sequence variant interpretation framework. Genome Med 12, 3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Molina SG, Beltran AA & Beltran AS Generation of an integration-free induced pluripotent stem cell line (UNC001-A) from blood of a healthy individual. Stem Cell Res 49, 102015 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Speicher DW, Weglarz L & DeSilva TM Properties of human red cell spectrin heterodimer (side-to-side) assembly and identification of an essential nucleation site. J. Biol. Chem 267, 14775–14782 (1992). [PubMed] [Google Scholar]
- 41.Li X & Bennett V Identification of the spectrin subunit and domains required for formation of spectrin/adducin/actin complexes. J. Biol. Chem 271, 15695‐15702 (1996). [DOI] [PubMed] [Google Scholar]
- 42.Davis L et al. Localization and structure of the ankyrin-binding site on beta2-spectrin. J. Biol. Chem 284, 6982‐6987 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bignone PA & Baines AJ Spectrin alpha II and beta II isoforms interact with high affinity at the tetramerization site. Biochem. J 374, 613‐624 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hyvönen M et al. Structure of the binding site for inositol phosphates in a PH domain. EMBO J 14, 4676‐4685 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Korenbaum E & Rivero F Calponin homology domains at a glance. J. Cell Sci 115, 3543–3545 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Yin LM, Schnoor M & Jun CD Structural characteristics, binding partners and related diseases of the calponin homology (CH) domain. Front. Cell. Dev. Biol 8, 342 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keep NH et al. Crystal structure of the actin-binding region of utrophin reveals a head-to-tail dimer. Structure 7, 1539–1546 (1999). [DOI] [PubMed] [Google Scholar]
- 48.Bañuelos S, Saraste M & Djinović Carugo K Structural comparisons of calponin homology domains: implications for actin binding. Structure 6, 1419–1431 (1998). [DOI] [PubMed] [Google Scholar]
- 49.Avery AW et al. Structural basis for high-affinity actin binding revealed by a β-III-spectrin SCA5 missense mutation. Nat. Commun 8, 1350 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vajda S et al. New additions to the ClusPro server motivated by CAPRI. Proteins 85, 435–444 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kozakov D et al. The ClusPro web server for protein-protein docking. Nat. Protoc 12, 255–278 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Galiano MR et al. A distal axonal cytoskeleton forms an intra-axonal boundary that controls axon initial segment assembly. Cell 149, 1125‐1139 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sleigh JN, Rossor AM, Fellows AD, Tosolini AP & Schiavo G Axonal transport and neurological disease. Nat. Rev. Neurol 15, 691‐703 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Susuki K et al. Glial βII spectrin contributes to paranode formation and maintenance. J. Neurosci 38, 6063–6075 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gobius I et al. Astroglial-mediated remodeling of the interhemispheric midline is required for the formation of the corpus callosum. Cell Rep 17, 735–747 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goebbels S et al. Genetic targeting of principal neurons in neocortex and hippocampus of NEX-Cre mice. Genesis 44, 611–621 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Fame RM, MacDonald JL & Macklis JD Development, specification, and diversity of callosal projection neurons. Trends Neurosci 34, 41–50 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Satterstrom FK et al. Autism spectrum disorder and attention deficit hyperactivity disorder have a similar burden of rare protein-truncating variants. Nat. Neurosci 22, 1961–1965 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang R et al. ANK2 autism mutation targeting giant ankyrin-B promotes axon branching and ectopic connectivity. Proc. Natl. Acad. Sci. USA 116, 15262–15271 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lorenzo DN et al. A PIK3C3–ankyrin-B–dynactin pathway promotes axonal growth and multiorganelle transport. J. Cell Biol 207, 735–752 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhong G et al. Developmental mechanism of the periodic membrane skeleton in axons. Elife 3, e04581 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou R, Han B, Xia C & Zhuang X Membrane-associated periodic skeleton is a signaling platform for RTK transactivation in neurons. Science 365, 929–934 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lek M et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Park J et al. Mutational characteristics of ANK1 and SPTB genes in hereditary spherocytosis. Clin. Genet 90, 69–78 (2016). [DOI] [PubMed] [Google Scholar]
- 65.Baek HJ et al. Transforming growth factor-β adaptor, β2-spectrin, modulates cyclin dependent kinase 4 to reduce development of hepatocellular cancer. Hepatology 53, 1676–1684 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Derbala MH, Guo AS, Mohler PJ & Smith SA The role of βII spectrin in cardiac health and disease. Life Sci 192, 278–285 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sobreira N, Schiettecatte F, Valle D & Hamosh A GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat 36, 928–930 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zepeda-Mendoza CJ et al. An intragenic duplication of TRPS1 leading to abnormal transcripts and causing trichorhinophalangeal syndrome type I. Cold Spring Harb. Mol. Case Stud 5, a004655 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kalari KR et al. MAP-RSeq: Mayo analysis pipeline for RNA sequencing. BMC Bioinformatics 15, 224 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim D et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Langmead B Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinformatics 11, 7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Anders S, Pyl PT & Huber W HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics, 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Quinlan AR BEDTools: The Swiss-aramy tool for genome feature analysis. Curr. Protoc. Bioinformatics 47, 1–34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lorenzo DN & Bennett V Cell-autonomous adiposity through increased cell surface GLUT4 due to ankyrin-B deficiency. Proc. Natl. Acad. Sci. USA 114, 12743–12748 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Burridge K, Kelly T & Mangeat P Nonerythrocyte spectrins: actin-membrane attachment proteins occurring in many cell types. J. Cell Biol 95, 478–486 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Snouwaert JN et al. A mutation in the Borcs7 subunit of the lysosome regulatory BORC complex results in motor deficits and dystrophic axonopathy in mice. Cell Rep 24, 1254–1265 (2018). [DOI] [PubMed] [Google Scholar]
- 77.García-Alvarez B, Bobkov A, Sonnenberg A & de Pereda JM Structural and functional analysis of the actin binding domain of plectin suggests alternative mechanisms for binding to F-actin and integrin beta4. Structure 11, 615–625 (2003). [DOI] [PubMed] [Google Scholar]
- 78.Zhang Y I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.von der Ecken J, Heissler SM, Pathan-Chhatbar S, Manstein DJ & Raunser S Cryo-EM structure of a human cytoplasmic actomyosin complex at near-atomic resolution. Nature 534, 724–728 (2016). [DOI] [PubMed] [Google Scholar]
- 80.Källberg M, Margaryan G, Wang S, Ma J & Xu J RaptorX server: a resource for template-based protein structure modeling. Methods Mol. Biol 1137, 17–27 (2014). [DOI] [PubMed] [Google Scholar]
- 81.Jurrus E et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci 27, 112–128 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shindyalov IN & Bourne PE Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng 11, 739–747 (1998). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The whole-genome and exome sequencing or transcriptomic data will not be made publicly available as they contain information that could compromise research participant privacy/consent. Information on the DNA and RNA sequencing raw data and other analyses supporting the findings of this study is available from the corresponding authors upon request. Source data are provided with this paper.


