Abstract
This article reviews deep learning applications in biomedical optics with a particular emphasis on image formation. The review is organized by imaging domains within biomedical optics and includes microscopy, fluorescence lifetime imaging, in vivo microscopy, widefield endoscopy, optical coherence tomography, photoacoustic imaging, diffuse tomography, and functional optical brain imaging. For each of these domains, we summarize how deep learning has been applied and highlight methods by which deep learning can enable new capabilities for optics in medicine. Challenges and opportunities to improve translation and adoption of deep learning in biomedical optics are also summarized.
Keywords: biomedical optics, biophotonics, deep learning, machine learning, computer aided detection, microscopy, fluorescence lifetime, in vivo microscopy, optical coherence tomography, photoacoustic imaging, diffuse tomography, functional optical brain imaging, widefield endoscopy
INTRODUCTION
Biomedical optics is the study of biological light-matter interactions with the overarching goal of developing sensing platforms that can aid in diagnostic, therapeutic, and surgical applications [1]. Within this large and active field of research, novel systems are continually being developed to exploit unique light-matter interactions that provide clinically useful signatures. These systems face inherent trade-offs in signal-to-noise ratio (SNR), acquisition speed, spatial resolution, field of view (FOV), and depth of field (DOF). These trade-offs affect the cost, performance, feasibility, and overall impact of clinical systems. The role of biomedical optics developers is to design systems which optimize or ideally overcome these trade-offs in order to appropriately meet a clinical need.
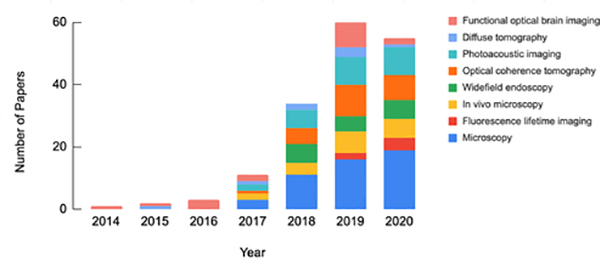
In the past few decades, biomedical optics system design, image formation, and image analysis have primarily been guided by classical physical modeling and signal processing methodologies. Recently, however, deep learning (DL) has become a major paradigm in computational modeling and demonstrated utility in numerous scientific domains and various forms of data analysis [2, 3]. As a result, DL is increasingly being utilized within biomedical optics as a data-driven approach to perform image processing tasks, solve inverse problems for image reconstruction, and provide automated interpretation of downstream images. This trend is highlighted in Fig. 1, which summarizes the articles reviewed in this paper stratified by publication year and image domain.
Fig 1:
Number of reviewed research papers which utilize DL in biomedical optics stratified by year and imaging domain.
This review focuses on the use of DL in the design and translation of novel biomedical optics systems. While image formation is the main focus of this review, DL has also been widely applied to the interpretation of downstream images, as summarized in other review articles [4, 5]. This review is organized as follows. First, a brief introduction to DL is provided by answering a set of questions related to the topic and defining key terms and concepts pertaining to the articles discussed throughout this review. Next, recent original research in the following eight optics-related imaging domains is summarized: (1) microscopy, (2) fluorescence lifetime imaging, (3) in vivo microscopy, (4) widefield endoscopy, (5) optical coherence tomography, (6) photoacoustic imaging, (7) diffuse tomography, and (8) functional optical brain imaging. Within each domain, state-of-the-art approaches which can enable new functionality for optical systems are highlighted. We then offer our perspectives on the challenges and opportunities across these eight imaging domains. Finally, we provide a summary and outlook of areas in which DL can contribute to future development and clinical impact biomedical optics moving forward.
DEEP LEARNING OVERVIEW
What is deep learning?
To define DL, it is helpful to start by defining machine learning (ML), as the two are closely related in their historical development and share many commonalities in their practical application. ML is the study of algorithms and statistical models which computer systems use to progressively improve their performance on a specified task [6]. To ensure the development of generalizable models, ML is commonly broken in two phases: training and testing. The purpose of the training phase is to actively update model parameters to make increasingly accurate predictions on the data, whereas the purpose of testing is to simulate a prospective evaluation of the model on future data.
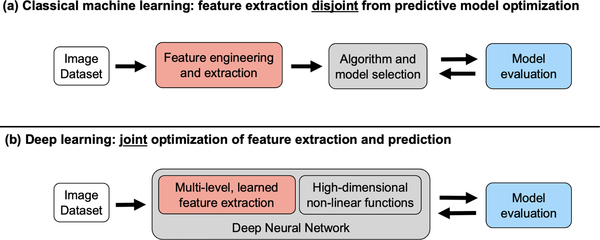
In this context, DL can be considered a subset of ML, as it is one of many heuristics for development and optimization of predictive, task-specific models [7]. In practice, DL is primarily distinguished from ML by the details of the underlying computational models and optimization techniques utilized. Classical ML techniques rely on careful development of task-specific image analysis features, an approach commonly referred to as “feature engineering” (Fig. 2(a)). Such approaches typically require extensive manual tuning and therefore have limited generalizability. In contrast, DL applies an “end-to-end” data-driven optimization (or “learning”) of both feature representations and model predictions [2] ((Fig. 2(b)). This is achieved through training a type of general and versatile computational model, termed deep neural network (DNN).
Fig 2:
(a) Classical machine learning uses engineered features and a model. (b) Deep learning uses learned features and predictors in an “end-to-end” deep neural network.
DNNs are composed of multiple layers which are connected through computational operations between layers, including linear weights and nonlinear “activation” functions. Thereby, each layer contains a unique feature representation of the input data. By using several layers, the model can account for both low-level and high-level representations. In the case of images, low-level representations could be textures and edges of the objects, whereas higher level representations would be object-like compositions of those features. The joint optimization of both feature representation at multiple levels of abstraction and predictive model parameters is what makes DNNs so powerful.
How is deep learning implemented?
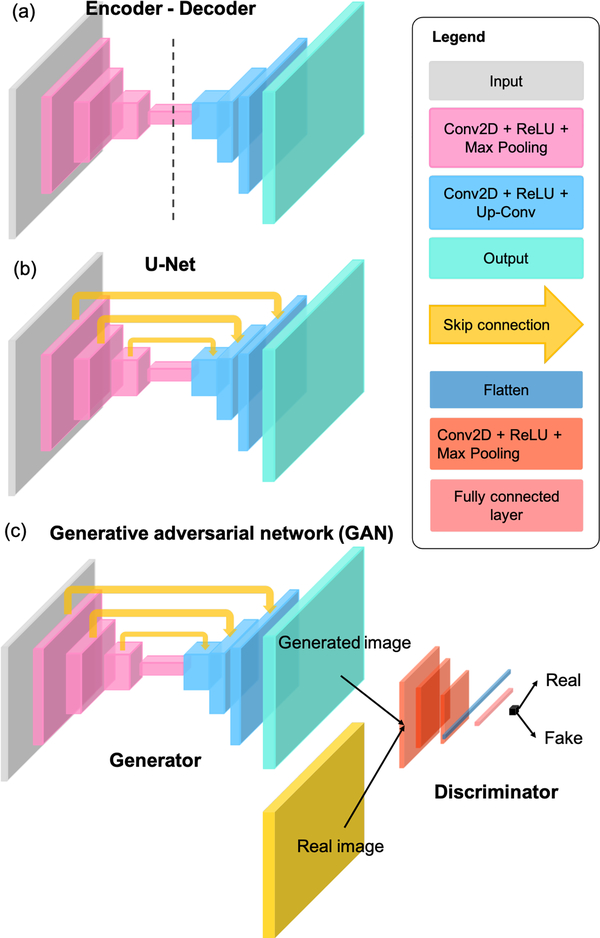
The majority of the existing DL models in biomedical optics are implemented using the supervised learning strategy. At a high-level, there are three primary components to implement a supervised DL model: 1) labeled data, 2) model architecture, and 3) optimization strategy. Labeled data consist of the raw data inputs as well as the desired model output. Large amounts of labeled data are often needed for effective model optimization. This requirement is currently one of the main challenges for utilizing DL on small-scale biomedical data sets, although strategies to overcome this are an active topic in the literature, such as unsupervised [8], self-supervised [9], and semi-supervised learning [10]. For a typical end-to-end DL model, model architecture defines the hypothesis class and how hierarchical information flows between each layer of the DNN. The selection of a DNN architecture depends on the desired task and is often determined empirically through comparison of various state-of-the-art architectures. Three of the most widely used DNN architectures in current biomedical optics literature are illustrated in Fig. 3.
Fig 3:
Three of the most commonly-used DNN architectures in biomedical optics: (a) Encoder-decoder, (b) U-Net, and (c) GAN.
The encoder-decoder network [11] shown in Fig. 3(a) aims to establish a mapping between the input and output images using a nearly symmetrically structure with a contracting “encoder” path and an expanding “decoder” path. The encoder consists of several convolutional blocks, each followed by a down-sampling layer for reducing the spatial dimension. Each convolutional block consists of several convolutional layers (Conv2D) that stacks the processed features along the last dimension, among which each layer is followed by a nonlinear activation function, e.g. the Rectified Linear Unit (ReLU). The intermediate output from the encoder has a small spatial dimension but encodes rich information along the last dimension. These low-resolution “activation maps” go through the decoder, which consists of several additional convolutional blocks, each connected by a upsampling convolutional (Up-Conv) layer for increasing the spatial dimension. The output of the network typically has the same dimension as the input image.
The U-Net [12] architecture shown in Fig. 3(b) can be thought of as an extension to the encoder-decoder network. It introduces additional “skip connections” between the encoder and decoder paths so that information across different spatial scales can be efficiently tunneled through the network, which has shown to be particularly effective to preserve high-resolution spatial information [12].
The generative adversarial network (GAN) [13] shown in Fig. 3(c) is a general framework that involves adversarially training a pair of networks, including the “generator” and “discriminator”. The basic idea is to train the generator to make high-quality image predictions that are indistinguishable from the real images of the same class (e.g. H&E stained lung tissue slices). To do so, the discriminator is trained to classify that the generator’s output is fake, while the generator is trained to fool the discriminator. Such alternating training steps iterate until a convergence is met when the discriminator can hardly distinguish if the images produced from the generator are fake or real. When applying to biomedical optics techniques, the generator is often implemented by the U-Net. The discriminator is often implemented using an image classification network. The input image is first processed by several convolutional blocks and downsampling layers to extract high-level 2D features. These 2D features are then “flattened” to a 1D vector, which is then processed by several fully connected layers to perform additional feature synthesis and make the final classification.
Once labeled data and model architecture have been determined, optimization of model parameters can be undertaken. Optimization strategy includes two aspects: 1) cost function, and 2) training algorithm. Definition of a cost function (a.k.a. objective function, error, or loss function) is needed to assess the accuracy of model predictions relative to the desired output and provide guidance to adjust model parameters. The training algorithm iteratively updates the model parameters to improve model accuracy. This training process is generally achieved by solving an optimization problem, using variants of the gradient descent algorithm, e.g. stochastic gradient descent and Adam [14]. The optimizer utilizes the gradient of the cost function to update each layer of the DNN through the principle of “error backpropagation” [15]. Given labeled data, a model architecture, and the optimization guides the model parameters towards a local minimum of the cost function, thereby optimizing model performance.
With the recent success of DL, several software frameworks have been developed to enable easier creation and optimization of DNNs. Many of the major technology companies have been active in this area. Two of the front-runners are TensorFlow and PyTorch, which are open-source frameworks published and maintained by Google and Facebook, respectively [16, 17]. Both frameworks enable easy construction of custom DNN models, with efficient parallelization of DNN optimization over high-performance graphics computing units (GPUs). These frameworks have enabled non-experts to train and deploy DNNs and have played a large role in the spread of DL research into many new applications, including the field of biomedical optics.
What is deep learning used for in biomedical optics?
There are two predominant tasks for which DL has been utilized in biomedical optics: 1) image formation and 2) image interpretation. Both are important applications; however, image formation is a more central focus of biomedical optics researchers and consequently is the focus of this review.
With regards to image formation, DL has proven very useful for effectively approximating the inverse function of an imaging model in order to improve the quality of image reconstructions. Classical reconstruction techniques are built on physical models with explicit analytical formulations. To efficiently compute the inverse of these analytical models, approximations are often needed to simplify the problem, e.g. linearization. Instead, DL methods have shown to be very effective to directly “learn” an inverse model, in the form of a DNN, based on the training input and output pairs. This in practice has opened up novel opportunities to perform image formation that would otherwise be difficult to formulate an analytical model. In addition, the directly learned DL inverse model can often better approximate the inverse function, which in turn leads to improved image quality as shown in several imaging domains in this review.
Secondly, DL has been widely applied for modeling image priors for solving the inverse problems across multiple imaging domains. Most image reconstruction problems are inherently ill-posed in that the reconstructed useful image signal can be overwhelmed by noise if a direct inversion is implemented. Classical image reconstruction techniques rely on regularization using parametric priors for incorporating features of the expected image. Although being widely used, such models severely limit the type of features that can be effectively modeled, which in turn limit the reconstruction quality. DL-based reconstruction bypasses this limitation and does not rely on explicit parameterization of image features, but instead represents priors in the form of a DNN which is optimized (or “learned”) from a large data set that is of the same type of the object of interest (e.g. endoscopic images of esophagus). By doing so, DL enables better quality reconstructions.
Beyond achieving higher quality reconstructions, there are other practical benefits of DNNs in image formation. Classical inversion algorithms typically require an iterative process that can take minutes to hours to compute. Furthermore, they have stringent sampling requirements, which if lessened, make the inversion severely ill-posed. Due to more robust “learned” priors, DL-based techniques can accommodate highly incomplete or undersampled inputs while still providing high-quality reconstructions. Additionally, although DNNs typically require large datasets for training, the resulting models are capable of producing results in real time with a GPU. These combined capabilities allow DL-based techniques to bypass physical trade-offs (e.g., acquisition speed and imaging quality) and enable novel capabilities beyond existing solutions.
By leveraging these unique capabilities of DL methods, innovative techniques have been broadly reported across many imaging domains in biomedical optics. Examples include improving imaging performance, enabling new imaging functionalities, extracting quantitative microscopic information, and discovering new biomarkers. These and other technical developments have the potential to significantly reduce system complexity and cost, and may ultimately improve the quality, affordability, and accessibility of biomedical imaging in health care.
DEEP LEARNING APPLICATIONS IN BIOMEDICAL OPTICS
Microscopy
Overview.
Microscopy is broadly used in biomedical and clinical applications to capture cellular and tissue structures based on intrinsic (e.g. scattering, phase, and autofluorescence) or exogenous contrast (e.g. stains and fluorescent labels). Fundamental challenges exist in all forms of microscopy because of the limited information that can be extracted from the instrument. Broadly, the limitations can be categorized based on two main sources of origin. The first class is due to the physical tradeoffs between multiple competing performance parameters, such as SNR, acquisition speed, spatial resolution, FOV, and DOF. The second class is from the intrinsic sensitivity and specificity of different contrast mechanisms. DL-augmented microscopy is a fast-growing area that aims to overcome various aspects of conventional limitations by synergistically combining novel instrumentation and DL-based computational enhancement. This section focuses on DL strategies for bypassing the physical tradeoffs and augmenting the contrast in different microscopy modalities.
Overcoming physical tradeoffs.
An ideal microscopy technique often needs to satisfy several requirements, such as high resolution in order to resolve the small features in the sample, low light exposure to minimize photo-damage, and a wide FOV in order to capture information from a large portion of the sample. Traditional microscopy is fundamentally limited by the intrinsic tradeoffs between various competing imaging attributes. For example, a short light exposure reduces the SNR; a high spatial resolution requires a high-magnification objective lens that provides a small FOV and shallow DOF. This section summarizes recent achievements in leveraging DL strategies to overcome various physical tradeoffs and expand the imaging capabilities.
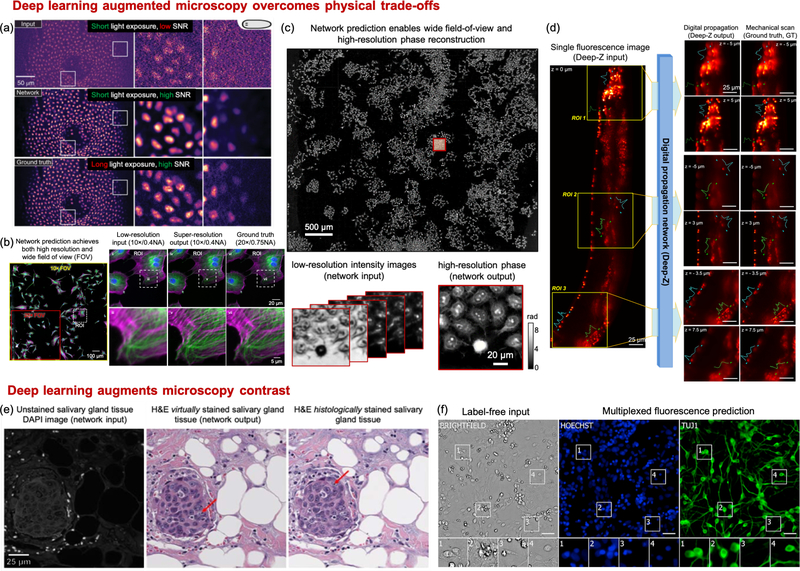
Denoising. Enhancing microscopy images by DL-based denoising has been exploited to overcome the tradeoffs between light exposure, SNR, and imaging speed, which in turn alleviates photo-bleaching and photo-toxicity. The general strategy is to train a supervised network that takes a noisy image as the input and produces the SNR-enhanced image output. Weigert et al. [18] demonstrated a practical training strategy of a U-Net on experimental microscopy data that involves taking paired images with low and high light exposures as the noisy input and high-SNR output of the network (Fig. 4(a)). This work showed that the DNN can restore the same high-SNR images with 60-fold fewer photons used during the acquisition. Similar strategies have been applied to several microscopy modalities, including widefield, confocal, light-sheet [18], structured illumination [24], and multi-photon microscopy [25].
Image reconstruction. Beyond denoising, the imaging capabilities of several microscopy techniques can be much expanded by performing image reconstruction. To perform reconstruction by DL, the common framework is to train a DNN, such as the U-Net and GAN, that takes the raw measurements as the input and the reconstructed image as the output. With this DL framework, three major benefits have been demonstrated. First, Wang et al. showed that GAN-based super-resolution reconstruction allows recovering high-resolution information from low-resolution measurements, which in turn provides an enlarged FOV and an extended DOF [19] (Fig. 4(b)). For widefield imaging, [19] demonstrated super-resolution reconstruction using input images from a 10×/0.4-NA objective lens and producing images matching a 20×/0.75-NA objective lens. In a cross-modality confocal-to-STED microscopy transformation case, [19] showed resolution improvement from 290 nm to 110 nm. Similar results have also been reported in label-free microscopy modalities, including brightfield [26], holography [27], and quantitative phase imaging [20] (Fig. 4(c)).
Fig 4:
DL overcomes physical tradeoffs and augments microscopy contrast. (a) CARE network achieves higher SNR with reduced light exposure (with permission from the authors [18]). (b) Cross-modality super-resolution network reconstructs high-resolution images across a wide FOV [19] (with permission from the authors). (c) DL enables wide-FOV high-resolution phase reconstruction with reduced measurements (adapted from [20]). (d) Deep-Z network enables digital 3D refocusing from a single measurement [21] (with permission from the authors). (e) Virtual staining GAN transforms autofluorescence images of unstained tissue sections to virtual H&E staining [22] (with permission from the authors) (f) DL enables predicting fluorescent labels from label-free images [23] (Reprinted from Cell, 2018 Apr 19;173(3):792–803.e19, Christiansen et al., In Silico Labeling: Predicting Fluorescent Labels in Unlabeled Images, Copyright (2020), with permission from Elsevier).
Second, DL-based 3D reconstruction technique allows drastically extending the imaging depth from a single-shot and thus bypasses the need for physical focusing. In [21], Wu et al. demonstrated 20× DOF extension in widefield fluorescence microscopy using a conditional GAN (Fig. 4(d)). Recent work on DL-based extended DOF has also shown promising results on enabling rapid slide-free histology [28].
Third, DL significantly improves both the imaging acquisition and reconstruction speeds and reduces the number of measurements for microscopy modalities that intrinsically require multiple measurements for the image formation, as shown in quantitative phase microscopy [20, 29, 30] (Fig. 4(c)), single molecule localization microscopy [31–33], and structured illumination microscopy [24]. For example, in [20], a 97% data reduction as compared to the conventional sequential acquisition scheme was achieved for gigapixel-scale phase reconstruction based on a multiplexed acquisition scheme using a GAN.
Augmenting contrasts.
The image contrast used in different microscopy modalities can be broadly categorized into endogenous and exogenous. For example, label-free microscopy captures endogenous scattering and phase contrast, and is ideal for imaging biological samples in their natural states, but suffers from lack of molecular specificity. Specificity is often achieved by staining with absorbing or fluorescent labels. However, applications of exogenous labeling are limited by the physical staining/labeling process and potential perturbation to the natural biological environment. Recent advances in DL-augmented microscopy have the potential to achieve the best of both label-free and labeled microscopy. This section summarizes two most widely used frameworks for augmenting microscopy contrast with DL.
-
Virtual staining/labeling. The main idea of virtual staining/labeling is to digitally transform the captured label-free contrast to the target stains/labels. DL has been shown to be particularly effective to perform this “cross-modality image transformation” task. By adapting this idea to different microscopy contrasts, two emerging applications have been demonstrated. First, virtual histological staining has been demonstrated for transforming a label-free image to the brightfield image of the histologically-stained sample (Fig. 4(e)). The label-free input utilized for this task include autofluorescence [22, 34], phase [35, 36], multi-photon and fluorescence lifetime [37]. The histological stains include H&E, Masson’s Trichrome and Jones’ stain. Notably, the quality of virtual staining on tissue sections from multiple human organs of different stain types was assessed by board-certified pathologists to show superior performance [22]. A recent cross-sectional study has been carried out for clinical evaluation of unlabeled prostate core biopsy images that have been virtually stained [38]. The main benefits of the virtual staining approach include saving time and cost [22], as well as facilitating multiplexed staining [34]. Interested readers can refer to a recent review on histopathology using virtual staining [39].
Second, digital fluorescence labeling has been demonstrated for transforming label-free contrast to fluorescence labels [23, 40–43] (Fig. 4(f)). In the first demonstration [23], Christiansen et al. performed 2D digital labeling using transmission brightfield or phase contrast images to identify cell nuclei (accuracy quantified by Pearson correlation coefficient PCC = 0.87–0.93), cell death (PCC = 0.85), and to distinguish neuron from astrocytes and immature dividing cells (PCC = 0.84). A main benefit of digital fluorescence labeling is digital multiplexing of multiple subcellular fluorescence labels, which is particularly appealing to kinetic live cell imaging. This is highlighted in [40], 3D multiplexed digital labeling using transmission brightfield or phase contrast images on multiple subcellular components are demonstrated, including nucleoli (PCC~0.9), nuclear envelope, microtubules, actin filaments (PCC~0.8), mitochondria, cell membrane, Endoplasmic reticulum, DNA+ (PCC~0.7), DNA (PCC~0.6), Actomyosin bundles, tight junctions (PCC~0.5), Golgi apparatus (PCC~0.2), and Desmosomes (PCC~0.1). Recent advances further exploit other label-free contrasts, including polarization [41], quantitative phase map [43], and reflectance phase-contrast microscopy [42]. Beyond predicting fluorescence labels, recent advances further demonstrate multiplexed single-cell profiling using the digitally predicted labels [42].
In both virtual histopathological staining and digital fluorescence labeling, the U-Net forms the basic architecture to perform the image transformation. GAN has also been incorporated to improve the performance [22, 38].
Classification Instead of performing pixel-wise virtual stain/label predictions, DL is also very effective in holistically capturing complex ‘hidden’ image features for classification. This has found broad applications in augmenting the label-free measurements and provide improved specificity and classify disease progression [44, 45] and cancer screening [46–48], as well as detect cell types [49, 50], cell states [44, 51], stem cell lineage [52–54], and drug response [55]. For example, in [44], Eulenberg et al. demonstrated a classification accuracy of 98.73% for the G1/S/G2 phase, which provided 6× improvement in error rate as compared to the previous state-of-the-art method based on classical ML techniques.
Opportunities and challenges.
By overcoming the physical tradeoffs in traditional systems, DL-augmented microscopy achieves unique combinations of imaging attributes that are previously not possible. This may create new opportunities for diagnosis and screening. By augmenting the contrast using virtual histological staining techniques, DL can open up unprecedented capabilities in label-free and slide-free digital pathology. This can significantly simplify the physical process and speed up the diagnosis. By further advancing the digital fluorescence labeling techniques, it can enable high-throughput and highly multiplexed single-cell profiling and cytometry. Beyond clinical diagnoses, this may find applications in drug screening and phenotyping.
In addition, several emerging DL techniques can further enhance the capabilities of microscopy systems. First, DL can be applied to optimize the hardware parameters used in microscopy experiments. In quantitative phase microscopy, DL was applied to optimized the illumination patterns to reduce the data requirement [30, 56]. In single molecule localization microscopy, DL was used to optimize the point spread functions to enhance the localization accuracy [33, 57]. DL has also been used to optimize the illumination power [58] and focus positions [59–61].
Second, new DL frameworks are emerging to significantly reduce the labeled data requirements in training, which is particularly useful in biomedical microscopy since acquiring a large-scale labeled training data set is often impractical. For example, a novel denoising approach, known as Noise2Noise [62], has been developed that can be trained using only independent pairs of noisy images, and bypasses the need for ground-truth clean images. Following this work, self-supervised denoising DL approaches have been advanced to further alleviate the training data requirement. Techniques, such as Noise2Void, Noise2Self and their variants, can be directly trained on noisy data set without the need for paired noisy images [63–65]. In addition, semi-supervised and unsupervised DL approaches have also been developed to reduce or completely remove the need for labeled training data during training, which have been demonstrated for vessel segmentation [66, 67]. Lastly, physics-embedded DL opens up a new avenue for reducing training requirements by incorporating the physical model of the microscopy technique [68, 69].
Finally, uncertainty quantification techniques address the need for assessing the reliability of the DL model by quantifying the confidence of the predictions, and has recently been applied in quantitative phase reconstruction [20].
Fluorescence Lifetime Imaging
Overview.
Fluorescence imaging has become a central tool in biomedical studies with high sensitivity to observe endogenous molecules [70, 71] and monitor important biomarkers [72]. Increasingly, fluorescence imaging is not limited to intensity-based techniques but can extract additional information by measuring fluorophore lifetimes [73–75]. Fluorescence lifetime imaging (FLI) has become an established technique for monitoring cellular micro-environment via analysis of various intracellular parameters [76], such as metabolic state [77, 78], reactive oxygen species [79] and/or intracellular pH [80]. FLI is also a powerful technique for studying molecular interactions inside living samples, via Förster Resonance Energy Transfer (FRET) [81], enabling applications such as quantifying protein-protein interactions [82], monitoring biosensor activity [83] and ligand-receptor engagement in vivo [84]. However, FLI is not a direct imaging technique. To quantify lifetime or lifetime-derived parameters, an inverse solver is required for quantification and/or interpretation.
To date, image formation is the main utility of DL in FLI. Contributions include reconstructing quantitative lifetime image from raw FLI measurements, enabling enhanced multiplexed studies by leveraging both spectral and lifetime contrast simultaneously, and facilitating improved instrumentation with compressive measurement strategies.
Lifetime quantification, representation, and retrieval.
Conventionally, lifetime quantification is obtained at each pixel via model-based inverse-solvers, such as least-square fitting and maximum-likelihood estimation [85], or the fit-free phasor method [86, 87]. The former is time-consuming, inherently biased by user-dependent a priori settings, and requires operator expertise. The phasor method is the most widely-accepted alternative for lifetime representation [87]. However, accurate quantification using the phasor method requires careful calibration, and when considering tissues/turbid-media in FLI microscopy (FLIM) applications, additional corrections are needed [87, 88]. Therefore, it has largely remained qualitative in use.
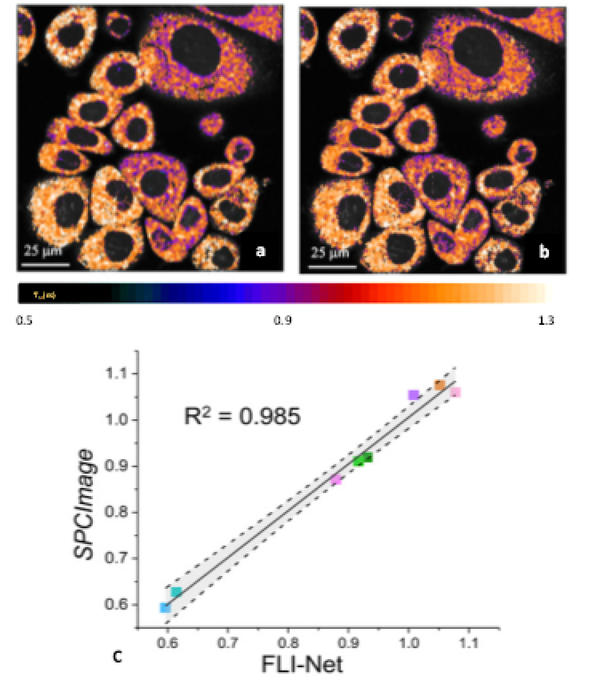
Wu et al. [89] demonstrated a multilayer perceptron (MLP) for lifetime retrieval for ultrafast bi-exponential FLIM. The technique exhibited an 180-fold faster speed then conventional techniques, yet it was unable to recover the true lifetime-based values in many cases due to ambiguities caused by noise. Smith et al. [90] developed an improved 3D-CNN, FLI-Net, that can retrieve spatially independent bi-exponential lifetime parameter maps directly from the 3D (x, y, t) FLI data. By training with a model-based approach including representative noise and instrument response functions, FLI-Net was validated across a variety of biological applications. These include quantification of metabolic and FRET FLIM, as well as preclinical lifetime-based studies across the visible and near-infrared (NIR) spectra. Further, the approach was generalized across two data acquisition technologies – Time-correlated Single Photon Counting (TCSPC) and Intensified Gated CCDs (ICCD). FLI-Net has two advantages. First, it outperformed classical approaches i’n the presence of low photon counts, which is a common limitation in biological applications. Second, FLI-Net can output lifetime-based whole-body maps at 80 ms in wide-field pre-clinical studies, which highlights the potential of DL methods for fast and accurate lifetime-based studies. In combination with DL in silico training routines that can be crafted for many applications and technologies, DL is expected to contribute to the dissemination and translation of FLI methods as well as to impact the design and implementation of future-generation FLI instruments. An example FLI-Net output for metabolic FLI is shown in Fig. 5.
Fig 5:
Example of quantitative FLI metabolic imaging as reported by NADH tm for a breast cancer cell line (AU565) as obtained (a) with SPCImage and (b) FLI-Net. (c) Linear regression with corresponding 95% confidence band (gray shading) of averaged NADH Tm values from 4 cell line data (adapted from [90]).
Emerging FLI applications using DL.
The technologies used in FLI have not fundamentally shifted over the last two decades. One bottleneck for translation is a lack of sensitive, widefield NIR detectors. Advances in computational optics have sparked development of new approaches using structured light [91], such as single-pixel methods [92]. These methods are useful when widefield detectors are lacking, such as in applications with low photon budget and when higher dimensional data are sought [93] (e.g., hyperspectral imaging [94]). However, these computational methods are based on more complex inverse models that require user expertise and input.
Yao et al. [95] developed a CNN, NetFLICS, capable of retrieving both intensity and lifetime images from single-pixel compressed sensing-based time-resolved input. NetFLICS generated superior quantitative results at low photon count levels, while being four orders of magnitude faster than existing approaches. Ochoa-Mendoza et al. [96] further developed the approach to increase its compression ratio to 99% and the reconstruction resolution to 128×128 pixels. This dramatic improvement in compression ratio enables significantly faster imaging protocols and demonstrates how DL can impact instrumentation design to improve clinical utility and workflow [97].
Recent developments have made hyperspectral FLI imaging possible across microscopic [98] and macroscopic settings [99]. Traditionally, combining spectral and lifetime contrast analytically is performed independently or sequentially using spectral decomposition and/or iterative fitting [100]. Smith et al. [101] proposed a DNN, UNMIX-ME, to unmix multiple fluorophore species simultaneously for both spectral and temporal information. UNMIX-ME takes a 4D voxel (x, y, t, λ) as the input and outputs spatial (x, y) maps of the relative contributions of distinct fluorophore species. UNMIX-ME demonstrated higher performance during tri- and quadri-abundance coefficient retrieval. This method is expected to find utility in applications such as autofluorescence imaging in which unmixing of metabolic and structural biomarkers is challenging.
Although FLI has shown promise for deep tissue imaging in clinical scenarios, FLI information is affected by tissue optical properties. Nonetheless, there are several applications that would benefit from optical property-corrected FLI without solving the full 3D inverse problem. For optical guided surgery, Smith et al. [102] proposed a DNN that outputs 2D maps of the optical properties, lifetime quantification, and the depth of fluorescence inclusion (topography). The DNN was trained using a model-based approach in which a data simulation workflow incorporated “Monte Carlo eXtreme” [103] to account for light propagation through turbid media. The method was demonstrated experimentally, with real-time applicability over large FOVs. Both widefield time-resolved fluorescence imaging and Spatial Frequency Domain Imaging (SFDI) in its single snapshot implementation were performed with fast acquisition [91] and processing speeds [104]. Hence, their combination with DL-based image processing provides a possible future foundation for real-time intraoperative use.
While recent advances in FLI-based classification and segmentation are limited to using classical ML techniques [105–107], Sagar et al. [108] used MLPs paired with bi-exponential fitting for label-free detection of microglia. However, DL approaches often outperform such “shallow learning” classifiers. Although reports using DL for classification based on FLI data are currently absent from the literature, it is expected that DL will play a critical role in enhancing FLI classification and semantic segmentation tasks in the near future.
In vivo microscopy
Overview.
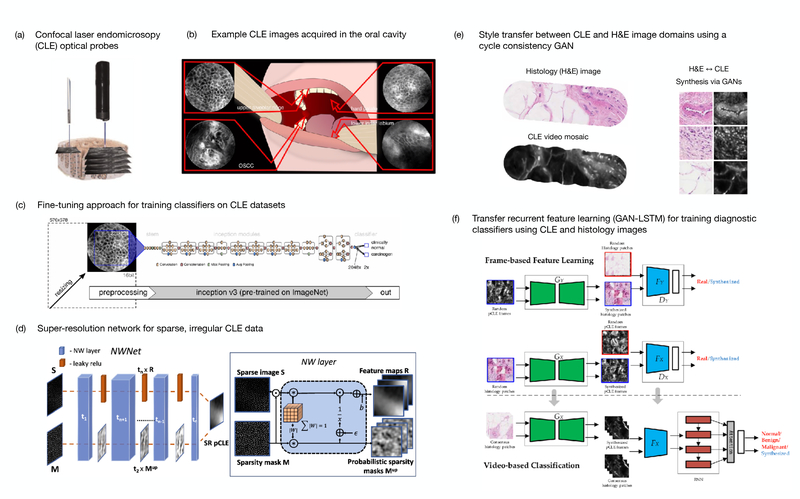
In vivo microscopy (IVM) techniques enable real-time assessment of intact tissue at magnifications similar to that of conventional histopathology [113]. As high-resolution assessment of intact tissue is desirable for many biomedical imaging applications, a number of optical techniques and systems have been developed which have trade-offs in FOV, spatial resolution, achievable sampling rates, and practical feasibility for clinical deployment [113]. However, a commonality of IVM systems used for clinical imaging is the need for image analysis strategies to support intraoperative visualization and automated diagnostic assessment of the high-resolution image data. Currently, three of the major IVM techniques for which DL is being utilized are optical coherence tomography (OCT) [114], confocal laser endomicroscopy (CLE, Fig. 6) [115], and reflectance confocal microscopy (RCM) [116]. This section focuses on DL approaches for CLE and RCM. More specifically, endoscopic imaging using probe-based CLE (pCLE) and dermal imaging for RCM. OCT is discussed in a subsequent section.
Fig 6:
DL approaches to support real-time, automated diagnostic assessment of tissues with confocal laser endomicroscopy. (a) Graphical rendering of two confocal laser endomicroscopy probes (left: Cellvizio, right: Pentax) (adapted from [109]). (b) Example CLE images obtained from four different regions of the oral cavity (adapted from [110]) (c) Fine-tuning of CNNs pre-trained using ImageNet is utilized in the majority of CLE papers reported since 2017 (adapted from [110]). (d) Super-resolution networks for probe-based CLE images incorporate novel layers to better account for the sparse, irregular structure of the images (adapted from [111]). (e) Example H&E stained histology images with corresponding CLE images. Adversarial training of GANs to transfer between these two modalities has been successful (adapted from [112]). (f) Transfer recurrent feature learning utilizes adversarially trained discriminators in conjunction with an LSTM for state-of-the-art video classification performance (adapted from [112]).
Automated diagnosis.
Automated diagnostic classification has been the earliest and most frequent application of DL within IVM. Most commonly, histopathology analysis of imaged specimens provides a ground truth categorization for assessing diagnostic accuracy. The limited size of pCLE and RCM datasets and logistical challenges in precisely correlating them with histopathology remain two ongoing challenges for training robust classifiers. To address these challenges, a variety of strategies have been applied which range from simple classification schemes (benign vs malignant) using pre-trained CNNs [110] to more complicated tasks, such as cross-domain feature learning and multi-scale encoder-decoder networks [112, 117]. The following section contrasts recent reports and methods utilizing DL for diagnostic image analysis of pCLE and RCM image datasets.
-
CNNs and transfer learning approaches. Early reports on DL-based image classification for CLE and RCM have demonstrated that transfer learning using pre-trained CNNs can outperform conventional image analysis approaches, especially when data is limited as is often the case for CLE and RCM [110, 118–120].
Aubreville et al. published an early and impactful study comparing the performance of two CNN-based approaches to a textural feature-based classifier (random forest) on pCLE video sequences acquired during surgical resection of oral squamous carcinoma (Fig. 6b) [110]. Of their two CNN-based approaches, one was a LeNet-5 architecture and was trained to classify sub-image patches whereas the other utilized transfer learning of a pre-trained CNN (Fig. 6c) for whole image classification. Using leave-one-out cross validation on 7,894 frames from 12 patients, the two CNN-based approaches both outperformed the textural classifier.
Transfer learning is one strategy to overcome limited dataset sizes, which remains a common challenge for CLE and RCM. As larger CLE and RCM datasets are obtainable in the future, transfer learning is unlikely to be an optimal strategy for image classification; however, it can remain a useful benchmark for the difficulty of image classification tasks on novel, small-scale datasets moving forward. The subsequent sections introduce alternatives to transfer learning which utilize video data as well as cross-domain learning.
Recurrent convolutional approaches. CLE and RCM are typically used in video recording while the optical probe is physically or optically scanned to obtain images over a larger tissue area or at varying depths. Some reports have utilized recurrent convolutional networks to account for spatial and/or temporal context of image sequences [121–123]. The additional spatial/temporal modeling provided by recurrent networks is one promising approach to leverage video data. [121–123].
Cross-domain learning. A novel approach, termed “transfer recurrent feature learning”, was developed by Gu et al. which leveraged cross-domain feature learning for classification of pCLE videos obtained from 45 breast tissue specimens [112]. Although this method relied on data acquired ex vivo, the data itself is not qualitatively different from other pCLE datasets and still provides a proof-of-principle. Their model utilized a cycle-consistent GAN (CycleGAN) to first learn feature representations between H&E microscopy and pCLE images and to identify visually similar images (Fig. 6e). The optimized discriminator from the CycleGAN is then utilized in conjunction with a recurrent neural network to classify video sequences (Fig. 6f). The method outperformed other DL methods and achieved 84% accuracy in classifying normal, benign, and malignant tissues.
Multiscale segmentation. Kose et al. [117] developed a novel segmentation architecture, “multiscale encoder-decoder network” (MED-Net), which outperformed other state-of-the-art network architectures for RCM mosaic segmentation. In addition to improving accuracy, MED-Net produced more globally consistent, less fragmented pixel-level classifications. The architecture is composed of multiple, nested encoder-decoder networks and was inspired by how pathologists often examine images at multiple scales to holistically inform their image interpretation.
Image quality assessment. A remaining limitation of many studies was some level of manual or semi-automated pre-processing of pCLE and RCM images/videos to exclude low-quality and/or non-diagnostic image data. Building on the aforementioned reports for diagnostic classification, additional work utilized similar techniques for automated image quality assessment using transfer learning [124, 125] as well as MED-Net [126].
Super-resolution.
Several IVM techniques, including pCLE, utilize flexible fiber-bundles as contact probes to illuminate and collect light from localized tissue areas [127]. Such probes are needed for minimally invasive endoscopic procedures and can be guided manually or via robotics. The FOV of fiber-optic probes is typically <1 mm2 and lateral resolution is limited by the inter-core spacing of individual optical fibers, which introduce a periodic image artifact (“honeycomb patterns”) from the individual fibers.
Shao et al. [128] developed a novel super-resolution approach which outperformed maximum a posteriori (MAP) estimation using a two-stage CNN model which first estimates the motion of the probe and then reconstructs a super-resolved image using the aligned video sequence. The training data was acquired using a dual camera system, one with and one without a fiber-bundle in the optical setup, to obtain paired data.
Others have taken a more computational approach to pCLE super-resolution by using synthetic datasets. For example, Ravì et al. [129] demonstrated super-resolution of pCLE images using unpaired image data via a CycleGAN, and Szczotka et al. [111] introduced a novel Nadaraya-Watson layer to account for the irregular sparse artifacts introduced by the fiber-bundle (Fig. 6d).
Future directions.
Beyond automatic diagnosis and super-resolution approaches in IVM, recent advances also highlight ways in which DL can enable novel instrumentation development and image reconstructions to enable new functionalities for compact microscopy systems. Such examples include multispectral endomicroscopy [130], more robust mosaicking for FOV expansion [131], and end-to-end image reconstruction using disordered fiber-optic probes [132, 133]. We anticipate that similarly to ex vivo microscopy, in the coming years DL will be increasingly utilized to overcome physical constraints, augment contrast mechanisms, and enable new capabilities for IVM systems.
Widefield endoscopy
Overview.
The largest application of optics in medical imaging, by U.S. market size, is widefield endoscopy [134]. In this modality, tissue is typically imaged on the >1 cm scale, over a large working distance range, with epi-illumination and video imaging via a camera. Endoscopic and laparoscopic examinations are commonly used for screening, diagnostic, preventative, and emergency medicine. There has been extensive research in applying various DL tools for analyzing conventional endoscopy images for improving and automating image interpretation [135–138]. This section instead reviews recent DL research in image formation tasks in endoscopy, including denoising, resolution enhancement, 3D scene reconstruction, mapping of chromophore concentrations, and hyperspectral imaging.
Denoising.
A hallmark of endoscopic applications is challenging geometrical constraints. Imaging through small lumens such as the gastrointestinal tract or “keyholes” for minimally-invasive surgical applications requires optical systems with compact footprints–often on the order of 1-cm diameter. These miniaturized optical systems typically utilize small-aperture cameras with high pixel counts, wide FOVs and even smaller illumination channels. Consequently, managing the photon budget is a significant challenge in endoscopy, and there have been several recent efforts to apply DL to aid in high-quality imaging in these low-light conditions. A low-light net (LLNET) with contrast-enhancement and denoising autoencoders has been introduced to adaptively brighten images [139]. This study simulated low-light images by darkening and adding noise and found that training on this data resulted in a learned model that could enhance natural low-light images. Other work has applied a U-Net for denoising on high-speed endoscopic images of the vocal folds, also by training on high-quality images that were darkened with added noise [140]. Brightness can also be increased via laser-illumination, which allows greater coupling efficiency than incoherent sources, but results in laser speckle noise in the image from coherent interference. Conditional GANs have been applied to predict speckle-free images from laser-illumination endoscopy images by training on image pairs acquired of the same tissue with both coherent and incoherent illumination sources [141].
Improving image quality.
In widefield endoscopy, wet tissue is often imaged in a perpendicular orientation to the optical axis, and the close positioning of the camera and light sources leads to strong specular reflections that mask underlying tissue features. GANs have been applied to reduce these specular reflections [142]. In this case, unpaired training data with and without specular reflections were used in a CycleGAN architecture with self-regularization to enforce similarity between the input specular and predicted specular-free images. Other work has found that specular reflection removal can be achieved in a simultaneous localization and mapping elastic fusion architecture enhanced by DL depth estimation [143]. Lastly, Ali et al. [144] introduced a DL framework that identifies a range of endoscopy artifacts (multi-class artifact detection), including specular reflection, blurring, bubbles, saturation, poor contrast, and miscellaneous artifacts using YOLOv3-spp with classes that were hand-labeled on endoscopy images. These artifacts were then removed and the image restored using GANs.
Resolution enhancement.
For capsule endoscopy applications, where small detectors with low pixel counts are required, DL tools have been applied for super-resolution with the goal of obtaining conventional endoscopy-like images from a capsule endoscope [145]. In this study, a conditional GAN was implemented with spatial attention blocks, using a loss function that included contributions of pixel loss, content loss, and texture loss. The intuition behind the incorporation of spatial attention blocks is that this module guides the network to prioritize the estimation of the suspicious and diagnostically relevant regions. This study also performed ablation studies and found that the content and texture loss components are especially important for estimating high-spatial frequency patterns, which becomes more important for larger upsampling ratios. With this framework, the resolution of small bowel images was increased by up to 12× with favorable quantitative metrics as well as qualitative assessment by gastroenterologists. Though this study demonstrated that the resolution of gastrointestinal images could be enhanced, it remains to be seen if preprocessing or enhancing these images provides any benefit to automated image analysis.
3D imaging and mapping.
The three dimensional shape of the tissue being imaged via endoscopy is useful for improving navigation, lesion detection and diagnosis, as well as obtaining meaningful quality metrics for the effectiveness of the procedure [146]. However, stereo and time-of-flight solutions are challenging and expensive to implement in an endoscopic form factor. Accordingly, there has been significant work in estimating the 3D shape of an endoscopic scene from monocular images using conditional GANs trained with photo realistic synthetic data [147, 148]. Domain adaptation can be used to improve the generalizability of these models, either by making the synthetic data more realistic, or by making the real images look more like the synthetic data that the depth-estimator is trained on [149]. Researchers have also explored joint conditional random fields and CNNs in a hybrid graphical model to achieve state-of-the-art monocular depth estimation [150]. A U-Net style architecture has been implemented for simultaneously estimating depth, color, and oxygen saturation maps from a fiber-optic probe that sequentially acquired structured light and hyperspectral images [151]. Lastly, DL tools have been applied to improve simultaneous localization and mapping (SLAM) tasks in endoscopic applications, both by incorporating a monocular depth estimation prior into a SLAM algorithm for dense mapping of the gastrointestinal tract [143], and by developing a recurrent neural network to predict depth and pose in a SLAM pipeline [152].
Widefield spectroscopy.
In addition to efforts to reconstruct high-quality color and 3D maps through an endoscope, DL is also being applied to estimate bulk tissue optical properties from wide FOV images. Optical property mapping can be useful for meeting clinical needs in wound monitoring, surgical guidance, minimally-invasive procedures, and endoscopy. A major challenge to estimating optical properties in turbid media is decoupling the effects of absorption, scattering, and the scattering phase function, which all influence the widefield image measured with flood illumination. Spatial frequency domain imaging can provide additional inputs to facilitate solving this inverse problem by measuring the attenuation of different spatial frequencies [153]. Researchers have demonstrated that this inverse model can be solved orders of magnitude faster than conventional methods with a 6-layer Perceptron [154]. Others have shown that tissue optical properties can be directly estimated from structured light images or widefield illumination images using content-aware conditional GANs [155]. In this application, the adversarial learning framework reduced errors in the optical property predictions by more than half when compared to the same network trained with an analytical loss function. Intuitively, the discriminator learns a more sophisticated and appropriate loss function in adversarial learning, allowing for the generation of more-realistic optical property maps. Moreover, this study found that the conditional GANs approach resulted in an increased performance benefit when data is tested from tissue types that were not spanned in the training set. The authors hypothesize that this observation comes from the discriminator preventing the generator from learning from and overfitting to the context of the input image. Optical properties can also be estimated more quickly using a lighter-weight twin U-Net architecture with a GPU-optimized look-up table [104]. Further, chromophores can be computed in real-time with reduced error compared to an intermediate optical property inference by directly computing concentrations from structured illumination at multiple wavelengths using conditional GANs [156].
Going beyond conventional color imaging, researchers are also processing 1D hyperspectral measurement through an endoscope using shallow CNNs to classify pixels into the correct color profiles, illustrating the potential to classify tissue with complex absorbance spectra [157]. The spectral resolution can be increased in dual-modality color/hyperspectral systems from sparse spectral signals with CNNs [151]. To enable quantitative spectroscopy measurements in endoscopic imaging, it may be necessary to combine hyperspectral techniques with structured illumination and 3D mapping [104, 151, 155, 158].
Future directions.
Future research in endoscopy and DL will undoubtedly explore clinical applications. Imaging system for guiding surgery are already demonstrating clinical potential for ex-vivo tissue classification: a modified Inception-v4 CNNs was demonstrated to effectively classify squamous cell carcinoma versus normal tissue at the cancer margin from ex-vivo hyperspectral images with 91 spectral bands [159]. For in-vivo applications, where generalizability may be essential and training data may be limited, future research in domain transfer [149] and semi-supervised learning [160] may become increasingly important. Moreover, for clinical validation, these solutions must be real-time, easy-to-use, and robust, highlighting the need for efficient architectures [104] and thoughtful user interface design [161].
Optical coherence tomography
Overview.
Optical coherence tomography (OCT) is a successful example of biophotonic technological translation into medicine [162, 163]. Since its introduction in 1993, OCT has revolutionized the standard-of-care in ophthalmology around the world, and continued thriving in technical advances and other clinical applications, such as dermatology, neurology, cardiology, oncology, gastroenterology, gynecology, and urology [164–171].
Image segmentation.
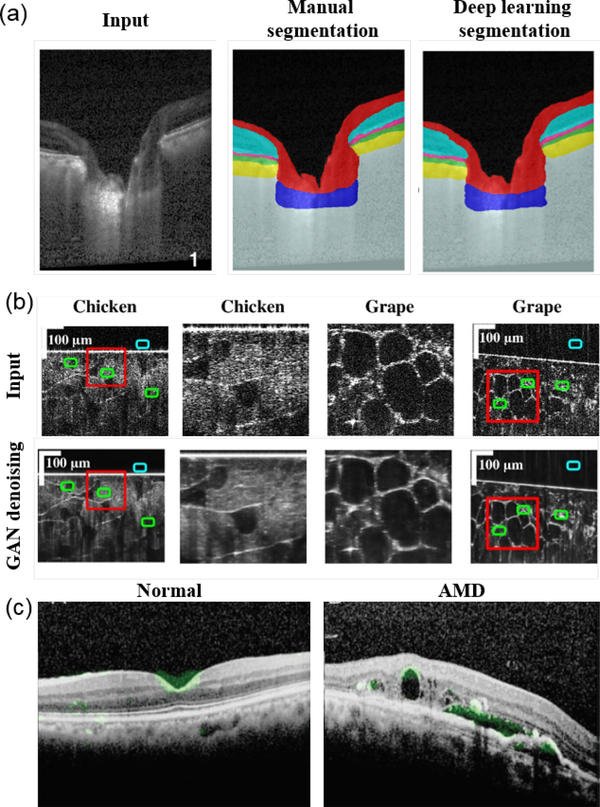
The most common use of OCT is to quantify structural metrics via image segmentation, such as retinal anatomical layer thickness, anatomical structures, and pathological features. Conventional image processing is challenging in the case of complex pathology where tissue structural alteration can be complex and may not be fully accounted for when designing a rigid algorithm. Image segmentation is the earliest application of DL explored in OCT applications. Several DNNs have been reported for OCT segmentation in conjunction with manual annotations (Fig. 7(a)), including U-Net [172–174], ResNet [175], and fully-convolutional network (FCN) [176, 177]. Successful implementation of DNNs have been broadly reported in different tissues beyond the eye [178–180]. In all areas of applications, the DNN showed superior segmentation accuracy over conventional techniques. For example, Devalla et al. [175] quantified the accuracy of the proposed DRUNET(Dilated-Residual U-Net) for segmenting the retinal nerve fiber layer (RNFL), retinal Layers, the retinal pigment epithelium (RPE), and choroid on both healthy and glaucoma subjects, and showed that the DRUNET consistently outperformed alternative approaches on all the tissues measured by dice coefficient, sensitivity, and specificity. The errors of all the metrics between DRUNET and the observers were within 10% and the patch-based neural network always provided greater than 10% error irrespective of the observer chosen for validation. In addition, the DRUNET segmentation further allowed automatic extraction of six clinically relevant neural and connective tissue structural parameters, including the disc diameter, peripapillary RNFL thickness (p-RNFLT), peripapillary choroidal thickness (p-CT), minimum rim width (MRW), prelaminar thickness (PLT), and the prelaminar depth (PLD).
Fig 7:
(a) Example automatic retinal layer segmentation using DL compared to manual segmentation (reprinted from [175]). (b) GAN for denoising OCT images (adapted from [181]). (c) Attention map overlaid with retinal images indicated features that CNN used for diagnosing normal versus age-related macular degeneration (AMD) [182] (Reproduced from Detection of features associated with neovascular age-related macular degeneration in ethnically distinct data sets by an optical coherence tomography: trained deep learning algorithm, Hyungtaek et al., Br. J. Ophthalmol. bjophthalmol-2020–316984, 2020 with permission from BMJ Publishing Group Ltd.).
Denoising and speckle removal.
OCT images suffer from speckle noise due to coherent light scattering, which leads to image quality degradation. There exist other sources of noise to further degrade the image quality when the signal level is low. Denoising and despeckling are important applications of DNNs, which are often trained with the averaged reduced-noise image as the ‘ground truth’ in a U-Net and ResNet [183, 184]. GAN has also been applied and provided improved visual perception than the DNNs trained with only the least-squares loss function [181] (Fig. 7(b)). For example, Dong et al. [181] showed that the GAN-based denoising network outperformed state-of-the-art image processing based (e.g. BM3D and MSBTD) and a few other DNNs (e.g. SRResNet and SRGAN) in terms of contrast-to-noise ratio (CNR) and peak signal-to-noise ratio (PSNR).
Clinical diagnosis and classification.
In clinical applications using DL, a large body of literature over the past 3 years emerges particularly in ophthalmology. Most of the studies use a CNN to extract image features for diagnosis and classification [185]. A clear shift of attention recently is to interpret the DNN, for example using the attention map [182, 186] (Fig. 7(c)). The purpose is to reveal the most important structural features that the DNN used for making the predictions. This addresses the major concern from the clinicians on the “black-box” nature of DL. Another emerging effort is to improve the generalization of a trained DNN to allow process images from different devices, with different image qualities and other possible variations. Transfer learning has been reported to refine pre-trained DNNs to other dataset, with much reduced training and data burdens [187, 188]. Domain adaptation is another method to generalize the DNN trained on images taken by one device to another [189, 190]. We expect more innovations for addressing the generalization in clinical diagnosis and prediction.
Emerging applications.
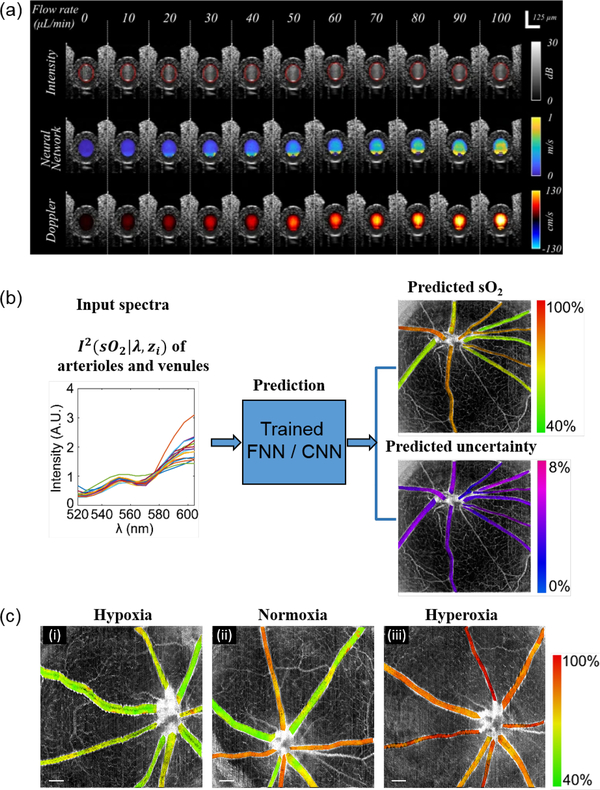
Beyond segmentation, denoising, and diagnosis/classification, there are several emerging DL applications for correlating the OCT measurements with vascular functions. OCT angiography (OCTA) and Doppler OCT (DOCT) are two advanced methods to measure label-free microangiography and blood flows. While normally requiring specific imaging protocols, the raw OCT measurements contain structural features that may be recognized by a CNN. Reports have shown that angiographic image and blood flows can be predicted by mere structural image input without specific OCTA or DOCT protocols [191, 193, 194]. For example, Braaf et al. [191] showed that DL enabled accurate quantification of blood flow from OCT intensity time-series measurements, and was robust to vessel angle, hematocrit levels, and measurement SNR. This is appealing for generating not only anatomical features, but also functional readouts using the simplest OCT imaging protocols by any regular OCT device (Fig. 8(a)). Recent work also reports the use of a fully connected network and a CNN to extract the spectroscopic information in OCT to quantify the blood oxygen saturation (sO2) within microvasculature, as an important measure of the perfusion function [192] (Fig. 8(b-c)). The DL models in [192] demonstrated more than 60% error reduction for predicting sO2 as compared to the standard nonlinear least-squares fitting method. These advances present emerging directions of DL applied to OCT to extract functional metrics beyond structures.
Fig 8:
(a) Examples of using DL to predict blood flow based on structural OCT image features (reprinted from [191]). (b) Example of deep spectral learning for label-free oximetry in visible light OCT (reprinted from [192]). (c) The predicted blood oxygen saturation and the tandem prediction uncertainty from rat retina in vivo in hypoxia, normoxia and hyperoxia (reprinted from [192]).
Photoacoustic imaging and sensing
Overview.
Photoacoustic imaging relies on optical transmission, followed by sensing of the resulting acoustic response [195, 196]. This response may then be used to guide surgeries and interventions [197, 198] (among other possible uses [199]). In order to guide these surgeries and interventions, image maps corresponding to structures of high optical absorption must be formed, which is a rapidly increasing area of interest for the application of DL to photoacoustic imaging and sensing. This section focuses on many of the first reports of DL for photoacoustic source localization, image formation, and artifact removal. Techniques applied after an image has been formed (e.g., segmentation, spectral unmixing, and quantitative imaging) are also discussed, followed by a summary of emerging applications based on these DL implementations.
Source localization.
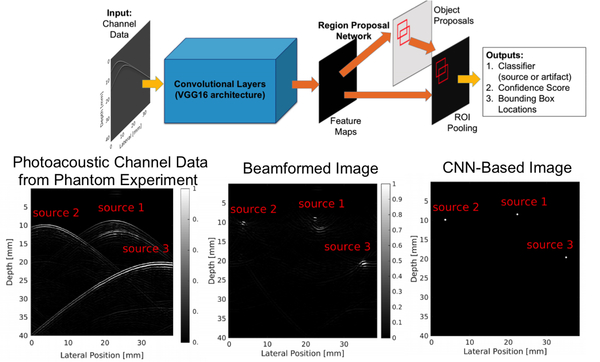
Localizing sources correctly and removing confusing artifacts from raw sensor data (also known as channel data) are two important precursors to accurate image formation. Three key papers discuss the possibility of using DL to improve source localization. Reiter and Bell [200] introduced the concept of source localization from photoacoustic channel data, relying on training data derived from simulations based on the physics of wave propagation. Allman et al. [201] built on this initial success to differentiate true photoacoustic sources from reflection artifacts based on wavefront shape appearances in raw channel data. Waves propagating spherically outward from a photoacoustic source are expected to have a unique shape based on distance from the detector, while artifacts are not expected to preserve this shape-to-depth relationship [201]. A CNN (VGG-16) was trained to demonstrate this concept, with initial results shown in Fig. 9. Johnstonbaugh et al. [202] expanded this concept by developing an encoder-decoder CNN with custom modules to accurately identify the origin of photoacoustic wavefronts inside an optically scattering deep-tissue medium. In the latter two papers [201, 202], images were created from the accurate localization of photoacoustic sources.
Fig 9:
Example of point source detection as a precursor to photoacoustic image formation after identifying true sources and removing reflection artifacts, modified from [201]. (©2018 IEEE. Adapted, with permission, from Allman et al. Photoacoustic source detection and reflection artifact removal enabled by deep learning, IEEE Transactions on Medical Imaging. 2018; 37:1464–1477.)
Image formation.
Beyond source localization, DL may also be used to form photoacoustic images directly from raw channel data with real-time speed [203, 204]. This section summarizes the application of DL to four technical challenges surrounding image formation: (1) challenges surrounding the limited view of transducer arrays [205–207] (in direct comparison to what is considered the “full view” provided by ring arrays), (2) sparse sampling of photoacoustic channel data [206, 208–210], (3) accurately estimating and compensating for the fluence differences surrounding a photoacoustic target of interest [211], and (4) addressing the traditional limited bandwidth issues associated with array detectors [212].
-
Limited view. Surgical applications often preclude the ability to completely surround a structure of interest. Historically, ring arrays were introduced for small animal imaging [213]. While these ring array geometries can also be used for in vivo breast cancer detection [214] or osteoarthritis detection in human finger joints [215], a full ring geometry is often not practical for many surgical applications [198]. The absence of full ring arrays often leads to what is known as “limited view” artifacts, which can appear as distortions of the true shape of circular targets or loss in the appearance of the lines in vessel targets.
DL has been implemented to address these artifacts and restore our ability to interpret the true structure of photoacoustic targets. For example, Hauptmann et al. [205] considered backprojection followed by a learned denoiser and a learned iterative reconstruction, concluding that the learned iterative reconstruction approach sufficiently balanced speed and image quality, as demonstrated in Fig. 10. To achieve this balance, a physical model of wave propagation was incorporated during the gradient of the data fit and an iterative algorithm consisting of several CNNs was learned. The network was demonstrated for a planar array geometry. Tong et al. [206] learned a feature projection, inspired by the AUTOMAP network [216], with the novelty of incorporating the photoacoustic forward model and universal backprojection model in the network design. The network was demonstrated for a partial ring array.
-
Sparse sampling. In tandem with limited view constraints, it is not always possible to sufficiently sample an entire region of interest when designing photoacoustic detectors, resulting in sparse sampling of photoacoustic responses. This challenge may also be seen as an extension of limited view challenges, considering that some of the desired viewing angles or spatial locations are missing (i.e., limited) due to sparse sampling, which often results in streak artifacts in photoacoustic images [217]. Therefore, networks that address limited view challenges can simultaneously address sparse sampling challenges [206, 218].
Antholzer et al. [208] performed image reconstruction to address sparse sampling with a CNN, modeling a filtered backprojection algorithm [219] as a linear preprocessing step (i.e., the first layer), followed by the U-Net architecture to remove undersampling artifacts (i.e., the remaining layers). Guan et al. [209] proposed pixel-wise DL (Pixel-DL) for limited-view and sparse PAT image reconstruction. The raw sensor data was first interpolated to window information of interest, then provided as an input to a CNN for image reconstruction. In contrast to previously discussed model-based approaches [205, 208], this approach does not learn prior constraints from training data and instead the CNN uses more information directly from the CNN and sensor data to reconstruct an image. This utilization of sensor data directly shares similarity with source localization methods [197, 201, 202].
The majority of methods discussed up until this point have used simulations in the training process for photoacoustic image formation. Davoudi et al. [210] take a different approach to address sparse sampling challenges by using whole-body in vivo mouse data acquired with a high-end, high-channel count system. This approach also differs from previously discussed approaches by operating solely in the image domain (i.e., rather than converting sensor or channel data to image data).
Fluence correction. The previous sections address challenges related to sensor spacing and sensor geometries. However, challenges introduced by the laser and light delivery system limitations may also be addressed with DL. For example, Hariri et al. [211] used a multi-level wavelet-CNN to denoise photoacoustic images acquired with low input energies, by mapping these low fluence illumination source images to a corresponding high fluence excitation map.
Limited transducer bandwidth. The bandwidth of a photoacoustic detector determines the spatial frequencies that can be resolved. Awasthi et al. [212] developed a network with the goal of resolving higher spatial frequencies than those present in the ultrasound transducer. Improvements were observable as better boundary distinctions in the presented photoacoustic data. Similarly, Gutta et al. [220] used a DNN to predict missing spatial frequencies.
Fig 10:
Example of blood vessel and tumor phantom results with multiple DL approaches. (Reprinted from [205].)
Segmentation.
After photoacoustic image formation is completed, an additional area of interest has been segmentation of various structures of interest, which can be performed with assistance from DL. Moustakidis et al. [221] investigated the feasibiliity of DL to segment and identify skin layers by using pretrained models (i.e., ResNet50 [222] and AlexNet [223]) to extract features from images and by training CNN models to classify skin layers directly using images, skipping the processing, transformation, feature extraction, and feature selection steps. These DL methods were compared to other ML techniques. Boink et al. [224] explored simultaneous photoacoustic image reconstruction and segmentation for blood vessel networks. Training was based on the learned primal-dual algorithm [225] for CNNs, including spatially varying fluence rates with a weighting between imaging reconstruction quality and segmentation quality.
Spectral unmixing and quantitative imaging.
Photoacoustic data and images may also be used to determine or characterize the content of identified regions of interest based on data obtained from a series of optical wavelength excitations. These tasks can be completed with assistance from DL. Cai et al. [226] proposed a DL framework for quantitative photoacoustic imaging, starting with the raw sensor data received after multiple wavelength transmisions, using a residual learning mechanism adopted to the U-Net to quantify chromophore concentration and oxygen saturation.
Emerging applications.
Demonstrated applications for image formation with DL has spanned multiple spatial scales, with applications that include cellular-level imaging (e.g., microscopy [227], label-free histology), molecular imaging (e.g., low concentrations of contrast agents in vivo [211]), small animal imaging [210], clinical and diagnostic imaging, and surgical guidance [197]. In addition to applications for image formation, other practical applications in photoacoustic imaging and sensing include neuroimaging [209, 228], dermatology (e.g., clinical evaluation, monitoring, and diagnosis of diseases linked to skin inflammation, diabetes, and skin cancer [221]), real-time monitoring of contrast agent concentrations, microvasculature, and oxygen saturation during surgery [203, 226], and localization of biopsy needle tips [229], cardiac catheter tips [229–231], or prostate brachytherapy seeds [201].
Diffuse Tomography
Overview.
Diffuse Optical Tomography (DOT), Fluorescence Diffuse Optical Tomography (fDOT, also known as Fluorescence Molecular Tomography - FMT) and Bioluminescence Tomography (BLT) are non-invasive and nonionizing 3D diffuse optical imaging techniques [232]. They are all based on acquiring optical data from spatially resolved surface measurements and performing similar mathematical computational tasks that involve the modeling of light propagation according to the tissue attenuation properties. In DOT, the main biomarkers are related to the functional status of tissues reflected by the total blood content (HbT) and relative oxygen saturation (StO2) that can be derived from the reconstructed absorption maps [233]. DOT has found applications in numerous clinical scenarios including optical mammography [234, 235], muscle physiology [236], brain functional imaging [237] and peripheral vascular diseases monitoring. In fDOT, the inverse problem aims to retrieve the effective quantum yield distribution (related to concentration) of an exogenous contrast agent [238, 239] or reporter gene in animal models [240] while illuminated by excitation light. In BLT, the goal is to retrieve the location and strength of an embedded bioluminescent source.
The highly scattering biological tissues lead to ill-posed nonlinear inverse problems that are highly sensitive to model mismatch and noise amplification. Therefore, tomographic reconstruction in DOT/fDOT/BLT is often performed via iterative approaches [241] coupled with regularization. Moreover, the model is commonly linearized using the Rytov (DOT) or Born (DOT/fDOT) methods [242]. Additional constraints, such as preconditioning [243] and a priori information are implemented [244–247]. Further experimental constraints in DOT/fDOT are also incorporated using spectral and temporal information [248]. Despite this progress, the implementation and optimization of a regularized inverse problem is complex and requires vast computational resources. Recently, DL methods have been developed for DOT/fDOT/BLT to either aid or fully replace the classical inverse solver. These developments have focused on two main approaches, including 1) learned denoisers and 2) end-to-end solvers.
Learned denoisers.
Denoisers can enhance the final reconstruction by correcting for errors from model mismatch and noise amplification. Long [249] proposed a 3D CNN for enhancing the spatial accuracy of mesoscopic FMT outputs. The spatial output of a Tikhonov regularized inverse solver was translated into a binary segmentation problem to reduce the regularization-based reconstruction error. The network was trained with 600 random ellipsoids and spheres as it only aimed to reconstruct simple geometries in silico. Final results displayed improved “intersection over union” values with respect to the ground truth. Since denoising approaches still involve inverting the forward model, it can still lead to large model mismatch. Hence, there has been great interest in end-to-end solutions that directly map the raw measurements to the 3D object without any user input.
End-to-end solvers.
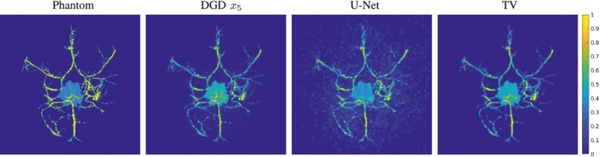
Several DNNs have been proposed to provide end-to-end inversion. Gao et al. [250] proposed a MLP for BLT inversion for tumor cells, in which the boundary measurements were inputted to the first layer that has a similar number of surface nodes as a standardized mesh built using MRI and CT images of the mouse brain, and output the photon distribution of the bioluminescent target. Similarly, Guo et al. [251] proposed “3D-En-Decoder”, a DNN for FMT with the encoder-decoder structure that inputs photon densities and outputs the spatial distribution of the fluorophores. It was trained with simulated FMT samples. Key features of the measurements were extracted in the encoder section and the transition of boundary photon densities to fluorophore densities was accomplished in the middle section with a fully connected layer. Finally, a 3D-Decoder outputted the reconstruction with better accuracy than L1-regularized inversion method in both simulated and phantom experiments.
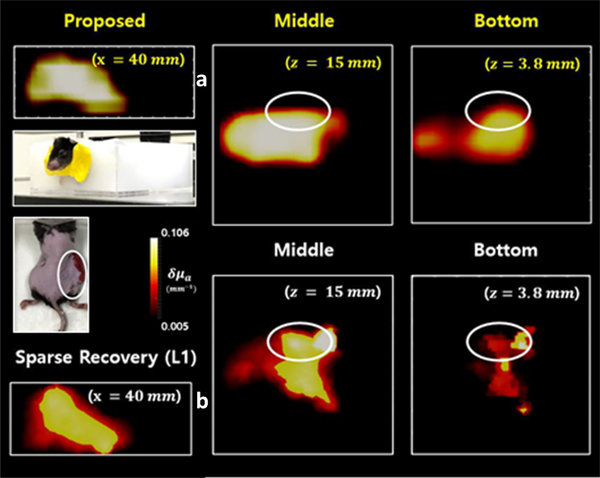
Huang et al. [253] proposed a similar CNN approach. After feature encoding, a “Gated Recurrent unit (GRU)” combines all the output features in a single vector, and the MLP (composed of two hidden layers with dropout and ReLu activations) outputs the fluorophores’ location. Simulated samples of a mouse model (with five organs and one fluorescent tumor target) were used. In silico results displayed comparable performance to an L1 inversion method. It was also validated with single-embeddings in silico by outputting only the positions since the network does not support 3D rendering. Yoo et al. [252] proposed an encoder-decoder DNN to invert the Lippmann–Schwinger integral photon equation for DOT using the deep convolutional framelet model [254] and learn the nonlinear scattering model through training with diffusion-equation based simulated data. Voxel domain features were learned through a fully connected layer, 3D convolutional layers and a filtering convolution. The method was tested in biomimetic phantoms and live animals with absorption-only contrast. Figure 11 shows an example reconstruction for an in vivo tumor in a mouse inside water/milk mixture media.
Fig 11:
Reconstruction for a mouse with tumor (right thigh) where higher absorption values are resolved (slices at z=15 and 3.8 mm) for the tumor area with the DNN in (a) compared to the L1-based inversion in (b). (adapted with permission from the authors from [252]).
Summary and future challenges.
DL has been demonstrated for improving (f)DOT image formation for solving complex ill-posed inverse problems. The DL models are often trained with simulated data, and in a few cases, validated in simple experimental settings. With efficient and accurate light propagation platforms such as MMC/MCX [255, 256], model-based training could become more efficient. Still, it is not obvious that such DL approaches will lead to universal solutions in DOT/FMT since many optical properties of tissues are still unknown and/or heterogeneous. Hence, further studies should aim to validate the universality of the architectures across different tissue conditions.
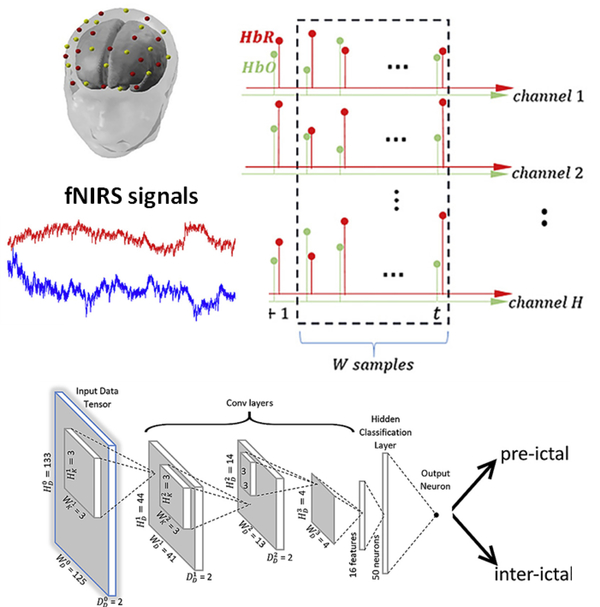
Functional optical brain imaging
Overview.
Functional optical brain imaging provides the opportunity to correlate neurobiological biomarkers with human behaviors, which impacts numerous fields, such as basic neuroscience, clinical diagnostics, brain computer interface (BCI) and social sciences. The two main established human functional optical brain imaging approaches are functional Near InfraRed Spectroscopy (fNIRS) and Diffuse Correlation Spectroscopy (DCS), both of which report brain activations via monitoring changes in optical signals as light reflected back to the detector while traveling through cortical areas. Classical neuroimaging studies are based on statistical analysis of biomarkers from a large group of subjects with different statuses (resting/active, stimuli/non-stimuli, disease/disease-free, etc.). However, the derivation of the biomarkers of interests are associated with data processing workflows that can be complex and computationally intensive. While numerous applications in neuroimaging inherently focus on classification of subjects based on spatiotemporal features, DL methods have two outstanding benefits. First, there is the potential to extract meaningful features from high-dimensional noisy data without expert knowledge required for the input/output mapping. Second, DL methods enable statistical inference at the single subject level which is critical for clinical practice. Hence, there has been a recent surge in DL solutions for functional optical brain imaging.
Classification based on cortical activations.
Most DL applications to functional optical brain imaging have focused on classification tasks based on fNIRS. Hiroyasu et al. [258] reported a DNN to perform gender classification on subjects performing a numerical memory task while subjecting to a white-noise sound environment to elicit gender-based differences in cortical activations. Using time series data of oxygenated hemoglobin of the inferior frontal gyrus on the left side of the head captured by 4 fNIRS channels, they reported a 81% accuracy in gender classification. The learned classifier identified the inferior frontal gyrus and premotor areas provide the highest discrimination accuracy. Mirbagheri et al. [259] developed a DNN to predict stress using fNIRS data collected on the prefrontal cortex regions, demonstrating 88% accuracy when following the Montreal Imaging Stress Task (MIST) protocols [260].
DL has also been used on fNIRS data for diagnostic and therapeutic applications [261]. Rosas-Romero et al. [257] developed a CNN to predict epilectic seizure (Fig. 12) and reported accuracy ranging between 97% and 100%, sensitivity between 95% and 100% and specificity between 98% and 100% using both oxy- and deoxy-hemoglobin time series as the input. Electroencephalography (EGG) data were acquired simultaneously, but fNIRS predictive features outperformed EGG predictive features. Another use of fNIRS is in psychological studies. Bandara et al. [262] reported a CNN with Long Short Term Memory (LSTM) to analyze spatiotemporal oxy- and deoxy- hemodynamics data from the prefrontal cortex for classifying human emotions and achieved 77% accuracy using both oxy- and deoxy- hemoglobin data and 1-s time steps. These results demonstrate that spatiotemporal features are desired for fNIRS based classification tasks, and the DL methods excel in feature exaction in such high dimensional data sets. However, all the reported studies followed well defined protocols that are prevalent in neuroimaging studies but are not always conducive for real-word applications.
Fig 12:
Hemodynamic time series for prediction of epileptic seizure using a CNN (with permission from the authors [257]) (Computers in Biology and Medicine, 11, 2019, 103355, Rosas-Romero et al., Prediction of epileptic seizures with convolutional neural networks and functional near-infrared spectroscopy signals, Copyright (2020), with permission from Elsevier).
Another thrust in fNIRS study is to evaluate mental workload from Human Computer Interaction (HCI) in scenarios, such as driving, air traffic control, and surgery. Benerradi et al. [263] reported a CNN for classifying mental workload using fNRIS data from HCI tasks and achieved an accuracy of 72.77% for 2 classes and 49.53% for 3 classes. The CNN was benchmarked against logistic regression and SVM, but no particular improvements were noted. Gao et al. [264] reported a BRAIN-Net to predict surgical skill levels within the Fundamental of Laparoscopic Surgery (FLS) program environment, demonstrating a ROC-AUC of 0.91 in predicting the FLS Score using fNIRS data collected on the prefrontal cortex of medical students performing the FLS pattern cutting task. BRAIN-Net outperformed classical ML techniques, including Kernel Partial Least Squares (KPLS), nonlinear SVM and Random Forest, when the data was larger than 600 samples. These results demonstrated the potential of DL for behavior prediction as reported by well-established metrics with freely mobile and unconstrained subjects performing challenging bimanual tasks. Hence, DL-enabled fNIRS methods have the potential for impacting real-world applications. In this regard, one of the most exciting applications of neuroimaging is BCI.
Brain computer interface.
DL is expected to advance BCI [265]. To date, DL methods for BCI have mainly focused on EGG and to a lesser extent to Magnetic resonance imaging (MRI) or Electromyography (EMG). About 70% of the current work use CNN as discriminative models, 20% use Recurrent neural network (RNN) [107], while generative models (e.g. GAN or VAE) are rarely employed. Impressive results have been reported for real time control of a robot arm using DL-based BCI [266]. Following these trends, a few studies have been reported on DL-enabled fNIRS BCI. Hennrich et al. [267] reported a DNN that offered similar accuracy as compared to conventional methods in mental task classification. Dargazany et al. [268] implemented a CNN to recognize activity response in fNIRS data for 5 different activities and reported a 77–80% accuracy in classifying these tasks. Trakoolwilaiwan et al. [269] developed a CNN to classify between rest, right- and left- hand motor execution tasks and achieved classification accuracy within 82–99% depending on the specific subject, which was 6.49% more accurate than SVM and 3.33% more accurate than ANN. As BCI is a challenging task due to noisy data, one current research direction is the implementation of multimodal systems, especially EGG-fNIRS systems, for improved performance. Saadati et al. [270] reported a DNN for processing multimodal input from the variation of oxy- and deoxy-hemoglobin from fNIRS and the event-related desynchronization (ERD) from EGG, achieving the highest accuracy when compared to methods using a single biomarker and 92% accuracy for the word generation task compared to 86% for SVM.
Denoising and fast data processing.
Data preprocessing in optical neuroimaging is critical and includes dynamic range correction, transforming light attenuation to chromophore concentration, regressing shallow hemodynamic response to increase the sensitivity to cortical tissues, identifying and removing noise, especially motion artefacts. These steps typically require user inputs and are computationally intensive. Gao et al. [271] demonstrated a DNN for suppressing motion artifacts in raw fNIRS signals and identified 100% of the motion artefacts almost in real time. Poon et al. [272] reported a DNN in DCS that was 23× faster in estimating the tissue blood flow index compared to the traditional nonlinear fitting method. Hence, DL methodologies may facilitate the adoption of DCS for neuroimaging studies by enabling real-time and accurate tissue blood flow quantification in deep tissues.
Future directions and associated challenges.
DL methods herald the potential for subject specific classification on the fly, leading to fast and direct feedback based on real-time monitoring of brain functions. It also has potential for neuro-feedback in numerous therapeutic scenarios or cognitive/skill learning programs. In addition, DL has been largely adopted in brain connectivity studies [273], which has become prevalent for deciphering the brain circuitry [274] and diagnostic purposes [275]. Similar to MRI [276], DL is expected to play a critical role in next generation functional brain connectivity studies [277]. Still, numerous challenges lie ahead to implement full end-to-end solutions in data processing and classification.
One main challenge is the size of the population needed for generating the data sets. As we are still far from being able to model the complexity of brain functions and dynamics, this challenge is complicated by the need to train and validate neuroimaging DL approaches with experimental data. In numerous fNIRS and DCS studies, subject recruitment is limited and no public database is readily available. Such limitations have been recognized in all existing work. For such emerging methodologies, great care should be directed to appropriate cross-validation of the DL methods. Hence, validation methods such as k-fold and/or leave-one-out (one refers to one subject out, one trial out, or one day out, etc.) are essential to convey confidence of the usefulness of the methodology [278].
In addition, numerous applications of optical neuroimaging involve environments and tasks that cannot be fully controlled and/or restricted. Thus, brain cortical activations and connectivity can reflect response to complex stimuli in which “ground truth” can be challenging to establish. For example, it would be ideal to use a standardized and accredited metric (i.e., the FLS score) in various neuro-based applications. However, such objective standards do not exist and labeling of the data can be problematic. These challenges also limit the potential of DL for discovery and mapping of the brain circuitry. If DL were to become preponderant in functional connectivity studies, it also faces the current challenge of being primarily employed in the brain at rest, which does not offer insight into active states of interest.
CHALLENGES AND OPPORTUNITIES ACROSS MULTIPLE IMAGING DOMAINS
Challenges
Data availability and bias.
Most DL models for biomedical optics rely on “supervised learning” that are trained on domain- and/or task-specific datasets, which need to be carefully curated to ensure high-quality predictions. As a result, there are several inherent challenges in the data generation process that need to be addressed, including data availability and data bias [279]. For many applications, it is often difficult and costly to acquire a large-scale dataset. Novel techniques that can better leverage small-scale dataset while still providing high-quality models are needed, such as unsupervised, semi-supervised, and self-supervised learning, transfer learning, and domain adaptation. In addition to the overall scale of the dataset, the data may also be skewed or biased [280] because it may be difficult to acquire data with a balanced distribution for each sub-group, such as gender, ethnicity, etc. DNNs trained on biased dataset can result in erroneous predictions in particular for under-represented populations and diseases. These obstacles may be mitigated to some extent with careful planning and data collection. However, there is a need to also identify and reduce data biases in the modeling step, such as data augmentation and balanced loss function design.
Interpretability.
A common challenge of DL models is that they are generally “black-boxes” and their predictions typically cannot be precisely explained. This is particularly problematic in health applications. To address this issue, “interpretable/explainable” DL techniques [281, 282] are needed. To this end, there are two general approaches that are actively being researched in the field [283]. The first is to develop an interpretable computational structure instead of DNNs [284, 285], so that the predictions are made based on the crafted logic in the DL model. The second approach is to provide post hoc model prediction interpretation, such as attention mechanism [42, 286] and uncertainty quantification [20, 287, 288], while keeping the same DNN structure.
Prospective and real-world validation.
In general, there is a need for prospective evaluations of DL-based systems in real clinical settings. The performance of DL models are commonly evaluated post hoc using metrics often not directly translatable to improving patient care. To critically evaluate the performance and move to clinical impact, these gaps must be bridged. First and foremost, large-scale prospective testing is needed, ideally with multiple sites, users, and instruments. Secondly, it is also important to develop quantitative metrics to relate those commonly used in DL model development to those most pivotal in improving the management of disease.
Opportunities
Exploiting multimodal data.
DNNs provide powerful frameworks for integrating multimodal and multidimensional data [15]. Biomedical optics systems often acquire measurements that augment traditional visualization or span a wide range of resolutions, imaging speeds, and sources of contrast. A fundamental barrier to the clinical translation of these technologies is that their benefit must outweigh the cost of additional training and time required to interpret and monitor these data. DL models can efficiently analyze these data together and transform them to actionable representations, reducing these training barriers while increasing the diagnostic power of multimodal imaging.
Lowering costs.
First, as shown in many examples herein, DL can enable new imaging capabilities that improve resolution, acquisition speed, FOV, and DOF often with minimal hardware modifications. This means that high-quality measurements can increasingly be made using relatively simple and lower cost systems. Second, DL technologies can enable more efficient workflows in healthcare and research, such as digital staining/labeling of tissues to reduce the cost and time associated with sample preparation.
Deskilling procedures.
Automated data processing and interpretation by DL may reduce the level of skill needed to obtain measurements and provide a diagnosis. A major benefit of DL based processing is that it is “end-to-end”. This means that once the DNN is trained, it enables automated reconstruction without any additional manual parameter tuning, potentially making it more generalizable and robust than classical approaches. This advantage must be balanced with great care and heightened responsibility to ensure ethical usage and unbiased outputs of these end-to-end DNN algorithms.
Increasing access to high-quality health care.
The ability of DL to lower cost and training requirements for diagnostic technologies holds tremendous potential for increasing access to high-quality health care in low-resource settings.
SUMMARY AND OUTLOOK
DL-based techniques have shown promise in addressing various technical challenges for developing novel biomedical optics systems, such as overcoming physical trade-offs, as well as enabling novel capabilities beyond existing solutions. Successful examples are available across multiple imaging domains, including microscopy, fluorescence lifetime imaging, in vivo microscopy, widefield endoscopy, optical coherence tomography, photoacoustic imaging, diffuse tomography, and functional optical brain imaging. Techniques are vast and varied, ranging from providing microscopic subcellular information to localizing image sources and offering macroscopic biomarkers. With the advances of DL techniques in many different biomedical optics domains, there are also some outstanding challenges that must be addressed in order to fully realize the impact of these techniques. As we are rapidly seeing across multiple biomedical optics modalities, DL techniques have promising potential to lower system costs, reduce required skill levels to carry out measurements, and ultimately increase the quality, affordability, and accessibility of health care.
Acknowledgments
This work was supported by the National Science Foundation 1813848, 1846784 (L.T.); the National Institutes of Health R21EB024700 (N.J.D.), R21EB025621 (M.A.L.B), R01NS108464 (J.Y.), R01EB019443, R01CA207725, R01CA237267, and R01CA250636 (X.I., J.T.S., M.O.); MTEC:20-05-IMPROVE-004 and W912CG2120001 (X.I.).
REFERENCES
- 1.Hyun Yun Seok, Kwok Sheldon JJ. Light in diagnosis, therapy and surgery Nature biomedical engineering. 2017;1:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yann LeCun, Yoshua Bengio, Geoffrey Hinton. Deep learning nature. 2015;521:436–444. [DOI] [PubMed] [Google Scholar]
- 3.Ian Goodfellow, Yoshua Bengio, Aaron Courville, Yoshua Bengio. Deep learning;1. MIT press Cambridge; 2016. [Google Scholar]
- 4.Geert Litjens, Thijs Kooi, Ehteshami Bejnordi Babak, et al. A Survey on Deep Learning in Medical Image Analysis Medical Image Analysis. 2017;42:60–88. arXiv: 1702.05747. [DOI] [PubMed] [Google Scholar]
- 5.Nichols James A, Herbert Chan Hsien W, Baker Matthew AB. Machine learning: applications of artificial intelligence to imaging and diagnosis Biophysical Reviews. 2019;11:111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jordan Michael I, Mitchell Tom M. Machine learning: Trends, perspectives, and prospects Science. 2015;349:255–260. [DOI] [PubMed] [Google Scholar]
- 7.The Difference Between AI, Machine Learning, and Deep Learning? | NVIDIA Blog [Google Scholar]
- 8.Juha Karhunen, Tapani Raiko, KyungHyun Cho. Chapter 7 - Unsupervised deep learning: A short review in Advances in Independent Component Analysis and Learning Machines (Ella Bingham, Samuel Kaski, Jorma Laaksonen, Jouko Lampinen., eds.):125–142Academic Press; 2015. [Google Scholar]
- 9.Longlong Jing, Yingli Tian. Self-supervised visual feature learning with deep neural networks: A survey IEEE Transactions on Pattern Analysis and Machine Intelligence. 2020. [DOI] [PubMed] [Google Scholar]
- 10.Xiaojin Zhu, Goldberg Andrew B. Introduction to Semi-Supervised Learning Synthesis Lectures on Artificial Intelligence and Machine Learning. 2009;3:1–130. Publisher: Morgan & Claypool Publishers. [Google Scholar]
- 11.Jonathan Masci, Ueli Meier, Dan Cireşan, Jürgen Schmidhuber. Stacked Convolutional Auto-Encoders for Hierarchical Feature Extraction in International conference on artificial neural networks Lecture Notes in Computer Science(Berlin, Heidelberg):52–59Springer; 2011. [Google Scholar]
- 12.Thorsten Falk, Dominic Mai, Robert Bensch, et al. U-Net: deep learning for cell counting, detection, and morphometry Nature Methods. 2019;16:67–70. Number: 1 Publisher: Nature Publishing Group. [DOI] [PubMed] [Google Scholar]
- 13.Goodfellow Ian J, Jean Pouget-Abadie, Mehdi Mirza, et al. Generative Adversarial Networks arXiv:1406.2661 [cs, stat]. 2014. arXiv: 1406.2661. [Google Scholar]
- 14.Kingma Diederik P, Jimmy Ba. Adam: A Method for Stochastic Optimization arXiv:1412.6980 [cs]. 2017. arXiv: 1412.6980. [Google Scholar]
- 15.Ramachandram D, Taylor GW. Deep Multimodal Learning: A Survey on Recent Advances and Trends IEEE Signal Processing Magazine. 2017;34:96–108. Conference Name: IEEE Signal Processing Magazine. [Google Scholar]
- 16.Martín Abadi, Ashish Agarwal, Paul Barham, et al. Tensorflow: Large-scale machine learning on heterogeneous distributed systems arXiv preprint arXiv:1603.04467. 2016. [Google Scholar]
- 17.Adam Paszke, Sam Gross, Francisco Massa, et al. Pytorch: An imperative style, high-performance deep learning library in Advances in neural information processing systems:8026–8037 2019. [Google Scholar]
- 18.Martin Weigert, Uwe Schmidt, Tobias Boothe, et al. Content-aware image restoration: pushing the limits of fluorescence microscopy Nature Methods. 2018;15:1090–1097. [DOI] [PubMed] [Google Scholar]
- 19.Hongda Wang, Yair Rivenson, Yiyin Jin, et al. Deep learning enables cross-modality super-resolution in fluorescence microscopy Nature Methods. 2019;16:103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yujia Xue, Shiyi Cheng, Yunzhe Li, Lei Tian. Reliable deep-learning-based phase imaging with uncertainty quantification Optica. 2019;6:618–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yichen Wu, Yair Rivenson, Hongda Wang, et al. Three-dimensional virtual refocusing of fluorescence microscopy images using deep learning Nature Methods. 2019:1–9. [DOI] [PubMed] [Google Scholar]
- 22.Yair Rivenson, Hongda Wang, Zhensong Wei, et al. Virtual histological staining of unlabelled tissue-autofluorescence images via deep learning Nature Biomedical Engineering. 2019;3:466–477. [DOI] [PubMed] [Google Scholar]
- 23.Christiansen Eric M, Yang Samuel J, Michael Ando D., et al. In Silico Labeling: Predicting Fluorescent Labels in Unlabeled Images Cell. 2018;173:792–803.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luhong Jin, Bei Liu, Fenqiang Zhao, et al. Deep learning enables structured illumination microscopy with low light levels and enhanced speed Nature Communications. 2020;11:1934. Number: 1 Publisher: Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sehyung Lee, Makiko Negishi, Hidetoshi Urakubo, Haruo Kasai, Shin Ishii. Mu-net: Multi-scale U-net for two-photon microscopy image denoising and restoration Neural Networks. 2020;125:92–103. [DOI] [PubMed] [Google Scholar]
- 26.Yair Rivenson, Zoltán Göröcs, Harun Günaydin, Yibo Zhang, Hongda Wang, Aydogan Ozcan. Deep learning microscopy Optica. 2017;4:1437–1443. [Google Scholar]
- 27.Tairan Liu, Kevin Haan, Yair Rivenson, et al. Deep learning-based super-resolution in coherent imaging systems Scientific Reports. 2019;9:3926. Number: 1 Publisher: Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lingbo Jin, Yubo Tang, Yicheng Wu, et al. Deep learning extended depth-of-field microscope for fast and slide-free histology Proceedings of the National Academy of Sciences. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thanh Nguyen, Yujia Xue, Yunzhe Li, Lei Tian, George Nehmetallah. Deep learning approach for Fourier ptychography microscopy Optics Express. 2018;26:26470–26484. Publisher: Optical Society of America. [DOI] [PubMed] [Google Scholar]
- 30.Michael Kellman, Emrah Bostan, Michael Chen, Laura Waller. Data-Driven Design for Fourier Ptychographic Microscopy arXiv:1904.04175 [cs, eess]. 2019. arXiv: 1904.04175. [Google Scholar]
- 31.Wei Ouyang, Andrey Aristov, Mickaël Lelek, Xian Hao, Christophe Zimmer. Deep learning massively accelerates super-resolution localization microscopy Nature Biotechnology. 2018;36:460–468. [DOI] [PubMed] [Google Scholar]
- 32.Elias Nehme, E. Weiss Lucien, Tomer Michaeli, Yoav Shechtman. Deep-STORM: super-resolution single-molecule microscopy by deep learning Optica. 2018;5:458. [Google Scholar]
- 33.Elias Nehme, Daniel Freedman, Racheli Gordon, et al. Deep-STORM3D: dense 3D localization microscopy and PSF design by deep learning Nature Methods. 2020:1–7. Publisher: Nature Publishing Group. [Google Scholar]
- 34.Yijie Zhang, Kevin Haan, Yair Rivenson, Jingxi Li, Apostolos Delis, Aydogan Ozcan. Digital synthesis of histological stains using micro-structured and multiplexed virtual staining of label-free tissue Light: Science & Applications. 2020;9:78. Number: 1 Publisher: Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yair Rivenson, Tairan Liu, Zhensong Wei, Yibo Zhang, Kevin Haan, Aydogan Ozcan. PhaseStain: the digital staining of label-free quantitative phase microscopy images using deep learning Light: Science & Applications. 2019;8:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nygate Yoav N, Mattan Levi, Mirsky Simcha K, et al. Holographic virtual staining of individual biological cells Proceedings of the National Academy of Sciences. 2020. Publisher: National Academy of Sciences Section: Physical Sciences. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Navid Borhani, Bower Andrew J, Boppart Stephen A, Demetri Psaltis. Digital staining through the application of deep neural networks to multi-modal multi-photon microscopy Biomedical Optics Express. 2019;10:1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aman Rana, Alarice Lowe, Marie Lithgow, et al. Use of Deep Learning to Develop and Analyze Computational Hematoxylin and Eosin Staining of Prostate Core Biopsy Images for Tumor Diagnosis JAMA Network Open. 2020;3:e205111–e205111. Publisher: American Medical Association. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yair Rivenson, Kevin Haan, Dean Wallace W., Aydogan Ozcan. Emerging Advances to Transform Histopathology Using Virtual Staining BME Frontiers. 2020. Publisher: Science Partner Journal Volume: 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chawin Ounkomol, Sharmishtaa Seshamani, Maleckar Mary M, Forrest Collman, Johnson Gregory R. Label-free prediction of three-dimensional fluorescence images from transmitted-light microscopy Nature Methods. 2018;15:917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Syuan-Ming Guo, Li-Hao Yeh, Jenny Folkesson, et al. Revealing architectural order with quantitative label-free imaging and deep learning eLife. 2020;9:e55502. Publisher: eLife Sciences Publications, Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shiyi Cheng, Sipei Fu, Mun Kim Yumi, et al. Single-cell cytometry via multiplexed fluorescence prediction by label-free reflectance microscopy Science Advances. 2021;7:eabe0431. Publisher: American Association for the Advancement of Science Section: Research Article. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kandel Mikhail E, He Yuchen R, Jae Lee Young, et al. Phase imaging with computational specificity (PICS) for measuring dry mass changes in sub-cellular compartments Nature Communications. 2020;11:6256. Number: 1 Publisher: Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Philipp Eulenberg, Niklas Köhler, Thomas Blasi, et al. Reconstructing cell cycle and disease progression using deep learning Nature Communications. 2017;8:1–6. Number: 1 Publisher: Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Minh Doan, Marian Case, Dino Masic, et al. Label-Free Leukemia Monitoring by Computer Vision Cytometry Part A. 2020;97:407–414. _eprint: https://onlinelibrary.wiley.com/doi/pdf/10.1002/cyto.a.23987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sixian You, Yi Sun, Lin Yang, et al. Real-time intraoperative diagnosis by deep neural network driven multiphoton virtual histology npj Precision Oncology. 2019;3:1–8. Number: 1 Publisher: Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tatsuya Matsumoto, Hirohiko Niioka, Yasuaki Kumamoto, et al. Deep-UV excitation fluorescence microscopy for detection of lymph node metastasis using deep neural network Scientific Reports. 2019;9:16912. Number: 1 Publisher: Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Jingfang K, He Yuchen R, Nahil Sobh, Gabriel Popescu. Label-free colorectal cancer screening using deep learning and spatial light interference microscopy (SLIM) APL Photonics. 2020;PHAI2020:040805. Publisher: American Institute of Physics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Christian Matek, Simone Schwarz, Karsten Spiekermann, Carsten Marr. Human-level recognition of blast cells in acute myeloid leukaemia with convolutional neural networks Nature Machine Intelligence. 2019;1:538–544. Number: 11 Publisher: Nature Publishing Group. [Google Scholar]
- 50.Maxim Lippeveld, Carly Knill, Emma Ladlow, et al. Classification of Human White Blood Cells Using Machine Learning for Stain-Free Imaging Flow Cytometry Cytometry Part A. 2020;97:308–319. _eprint: https://onlinelibrary.wiley.com/doi/pdf/10.1002/cyto.a.23920. [DOI] [PubMed] [Google Scholar]
- 51.Kandel Mikhail E, Marcello Rubessa, He Yuchen R, et al. Reproductive outcomes predicted by phase imaging with computational specificity of spermatozoon ultrastructure Proceedings of the National Academy of Sciences. 2020. Publisher: National Academy of Sciences Section: Physical Sciences. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Felix Buggenthin, Florian Buettner, Hoppe Philipp S, et al. Prospective identification of hematopoietic lineage choice by deep learning Nature Methods. 2017;14:403–406. Number: 4 Publisher: Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dai Kusumoto, Mark Lachmann, Takeshi Kunihiro, et al. Automated Deep Learning-Based System to Identify Endothelial Cells Derived from Induced Pluripotent Stem Cells Stem Cell Reports. 2018;10:1687–1695. Publisher: Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ariel Waisman, Alejandro La Greca, Möbbs Alan M, et al. Deep Learning Neural Networks Highly Predict Very Early Onset of Pluripotent Stem Cell Differentiation Stem Cell Reports. 2019;12:845–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hirofumi Kobayashi, Cheng Lei, Yi Wu, et al. Intelligent whole-blood imaging flow cytometry for simple, rapid, and cost-effective drug-susceptibility testing of leukemia Lab on a Chip. 2019;19:2688–2698. Publisher: The Royal Society of Chemistry. [DOI] [PubMed] [Google Scholar]
- 56.Kellman Michael R, Emrah Bostan, Repina Nicole A, Laura Waller. Physics-Based Learned Design: Optimized Coded-Illumination for Quantitative Phase Imaging IEEE Transactions on Computational Imaging. 2019;5:344–353. Conference Name: IEEE Transactions on Computational Imaging. [Google Scholar]
- 57.Eran Hershko, Weiss Lucien E, Tomer Michaeli, Yoav Shechtman. Multicolor localization microscopy and point-spread-function engineering by deep learning Optics Express. 2019;27:6158–6183. Publisher: Optical Society of America. [DOI] [PubMed] [Google Scholar]
- 58.Marcel Štefko, Baptiste Ottino, Douglass Kyle M, Suliana Manley. Autonomous illumination control for localization microscopy Optics express. 2018;26:30882–30900. [DOI] [PubMed] [Google Scholar]
- 59.Yang Samuel J, Marc Berndl, Michael Ando D, et al. Assessing microscope image focus quality with deep learning BMC Bioinformatics. 2018;19:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shaowei Jiang, Jun Liao, Zichao Bian, Kaikai Guo, Yongbing Zhang, Guoan Zheng. Transform- and multi-domain deep learning for single-frame rapid autofocusing in whole slide imaging Biomedical Optics Express. 2018;9:1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Henry Pinkard, Zachary Phillips, Arman Babakhani, Fletcher Daniel A, Laura Waller. Deep learning for single-shot autofocus microscopy Optica. 2019;6:794–797. [Google Scholar]
- 62.Jaakko Lehtinen, Jacob Munkberg, Jon Hasselgren, et al. Noise2Noise: Learning Image Restoration without Clean Data arXiv:1803.04189 [cs, stat]. 2018. arXiv: 1803.04189. [Google Scholar]
- 63.Alexander Krull, Tim-Oliver Buchholz, Florian Jug. Noise2Void - Learning Denoising from Single Noisy Images arXiv:1811.10980 [cs]. 2018. arXiv: 1811.10980. [Google Scholar]
- 64.Joshua Batson, Loic Royer. Noise2Self: Blind Denoising by Self-Supervision arXiv:1901.11365 [cs, stat]. 2019. arXiv: 1901.11365. [Google Scholar]
- 65.Broaddus C, Krull A, Weigert M, Schmidt U, Myers G. Removing Structured Noise with Self-Supervised Blind-Spot Networks in 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI):159–163 2020. ISSN: 1945–8452. [Google Scholar]
- 66.Waleed Tahir, Sreekanth Kura, Jiabei Zhu, et al. Anatomical modeling of brain vasculature in two-photon microscopy by generalizable deep learning BME Frontiers. 2021;2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shir Gur, Lior Wolf, Lior Golgher, Pablo Blinder. Unsupervised Microvascular Image Segmentation Using an Active Contours Mimicking Neural Network in 2019 IEEE/CVF International Conference on Computer Vision (ICCV)(Seoul, Korea (South)):10721–10730IEEE; 2019. [Google Scholar]
- 68.Emrah Bostan, Reinhard Heckel, Michael Chen, Michael Kellman, Laura Waller. Deep phase decoder: self-calibrating phase microscopy with an untrained deep neural network Optica. 2020;7:559–562. Publisher: Optical Society of America. [Google Scholar]
- 69.Zihui Wu, Yu Sun, Alex Matlock, Jiaming Liu, Lei Tian, Kamilov Ulugbek S. SIMBA: Scalable Inversion in Optical Tomography using Deep Denoising Priors IEEE Journal of Selected Topics in Signal Processing. 2020:1–1. Conference Name: IEEE Journal of Selected Topics in Signal Processing. [Google Scholar]
- 70.Lakowicz Joseph R, Henryk Szmacinski, Kazimierz Nowaczyk, Johnson Michael L. Fluorescence lifetime imaging of free and protein-bound NADH. Proceedings of the National Academy of Sciences. 1992;89:1271–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bidyut Das, Lingyan Shi, Yury Budansky, Adrian Rodriguez-Contreras, Robert Alfano. Alzheimer mouse brain tissue measured by time resolved fluorescence spectroscopy using single- and multi-photon excitation of label free native molecules Journal of Biophotonics. 2018;11:e201600318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alena Rudkouskaya, Nattawut Sinsuebphon, Marien Ochoa, et al. Multiplexed Non-Invasive Tumor Imaging of Glucose Metabolism and Receptor-Ligand Engagement using Dark Quencher FRET Acceptor Theranostics. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Laura Marcu. Fluorescence Lifetime Techniques in Medical Applications Annals of Biomedical Engineering. 2012;40:304–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.BECKER W. Fluorescence lifetime imaging - techniques and applications Journal of Microscopy. 2012;247:119–136. [DOI] [PubMed] [Google Scholar]
- 75.Rupsa Datta, Heaster Tiffany M, Sharick Joe T, Gillette Amani A, Skala Melissa C. Fluorescence lifetime imaging microscopy: fundamentals and advances in instrumentation, analysis, and applications Journal of Biomedical Optics. 2020;25:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Klaus Suhling, Hirvonen Liisa M, Levitt James A, et al. Fluorescence lifetime imaging (Flim): Basic concepts and recent applications in Advanced Time-Correlated Single Photon Counting Applications:119–188Springer; 2015. [Google Scholar]
- 77.Mengyan Wang, Feng Tang, Xiaobo Pan, et al. Rapid diagnosis and intraoperative margin assessment of human lung cancer with fluorescence lifetime imaging microscopy BBA Clinical. 2017;8:7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Irene Georgakoudi, Quinn Kyle P. Optical imaging using endogenous contrast to assess metabolic state. Annual review of biomedical engineering. 2012;14:351–67. [DOI] [PubMed] [Google Scholar]
- 79.Rupsa Datta, Alba Alfonso-García, Rachel Cinco, Enrico Gratton. Fluorescence lifetime imaging of endogenous biomarker of oxidative stress Scientific Reports. 2015;5:9848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hai-Jui Lin, Petr Herman, Lakowicz Joseph R. Fluorescence lifetime-resolved pH imaging of living cells Cytometry. 2003;52A:77–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shilpi Rajoria, Lingling Zhao, Xavier Intes, Margarida Barroso. FLIM-FRET for Cancer Applications Current Molecular Imaging. 2015;3:144–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yuansheng Sun, Day Richard N, Ammasi Periasamy. Investigating protein-protein interactions in living cells using fluorescence lifetime imaging microscopy Nature Protocols. 2011;6:1324–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sohila Zadran, Steve Standley, Kaylee Wong, Erick Otiniano, Arash Amighi, Michel Baudry. Fluorescence resonance energy transfer (FRET)-based biosensors: visualizing cellular dynamics and bioenergetics Applied Microbiology and Biotechnology. 2012;96:895–902. [DOI] [PubMed] [Google Scholar]
- 84.Alena Rudkouskaya, Nattawut Sinsuebphon, Jamie Ward, Kate Tubbesing, Xavier Intes, Margarida Barroso. Quantitative imaging of receptor-ligand engagement in intact live animals Journal of Controlled Release. 2018;286:451–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Christopher Thaler, Koushik Srinagesh V, Blank Paul S, Vogel Steven S. Quantitative Multiphoton Spectral Imaging and Its Use for Measuring Resonance Energy Transfer Biophysical Journal. 2005;89:2736–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Suman Ranjit, Leonel Malacrida, Jameson David M, Enrico Gratton. Fit-free analysis of fluorescence lifetime imaging data using the phasor approach Nature Protocols. 2018;13:1979–2004. [DOI] [PubMed] [Google Scholar]
- 87.Sez-Jade Chen, Nattawut Sinsuebphon, Alena Rudkouskaya, Margarida Barroso, Xavier Intes, Xavier Michalet. In vitro and in vivo phasor analysis of stoichiometry and pharmacokinetics using short-lifetime near-infrared dyes and time-gated imaging Journal of Biophotonics. 2019;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rinat Ankri, Arkaprabha Basu, Can Ulku Arin, et al. Single-Photon, Time-Gated, Phasor-Based Fluorescence Lifetime Imaging through Highly Scattering Medium ACS Photonics. 2020;7:68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gang Wu, Thomas Nowotny, Yongliang Zhang, Hong-Qi Yu, David Day-Uei Li. Artificial neural network approaches for fluorescence lifetime imaging techniques Optics Letters. 2016;41:2561. [DOI] [PubMed] [Google Scholar]
- 90.Smith Jason T, Ruoyang Yao, Nattawut Sinsuebphon, et al. Fast fit-free analysis of fluorescence lifetime imaging via deep learning Proceedings of the National Academy of Sciences. 2019;116:24019–24030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Angelo Joseph P, Chen Sez-Jade K, Marien Ochoa, Ulas Sunar, Sylvain Gioux, Intes Xavier. Review of structured light in diffuse optical imaging Journal of Biomedical Optics. 2018;24:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Edgar Matthew P, Gibson Graham M, Padgett Miles J. Principles and prospects for single-pixel imaging Nature Photonics. 2019;13:13–20. [Google Scholar]
- 93.Edgar Matthew P, Gibson Graham M, Padgett Miles J. Principles and prospects for single-pixel imaging Nature photonics. 2019;13:13–20. [Google Scholar]
- 94.Qi Pian, Ruoyang Yao, Nattawut Sinsuebphon, Xavier Intes. Compressive hyperspectral time-resolved wide-field fluorescence lifetime imaging Nature Photonics. 2017;11:411–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ruoyang Yao, Marien Ochoa, Pingkun Yan, Xavier Intes. Net-FLICS: fast quantitative wide-field fluorescence lifetime imaging with compressed sensing – a deep learning approach Light: Science & Applications. 2019;8:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Marien Ochoa Mendoza, Alena Rudkouskaya, Ruoyang Yao, Pingkun Yan, Margarida Barroso, Xavier Intes. High Compression Deep Learning based Single-Pixel Hyperspectral Macroscopic Fluorescence Lifetime Imaging In Vivo Biomedical Optics Express. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Arman Rahman, Chowdhury Jahangir, Lynch Seodhna M, et al. Advances in tissue-based imaging: impact on oncology research and clinical practice Expert Review of Molecular Diagnostics. 2020:1–11. [DOI] [PubMed] [Google Scholar]
- 98.Tokuko Haraguchi, Takeshi Shimi, Takako Koujin, Noriyo Hashiguchi, Yasushi Hiraoka. Spectral imaging fluorescence microscopy Genes to Cells. 2002;7:881–887. [DOI] [PubMed] [Google Scholar]
- 99.Qi Pian, Ruoyang Yao, Lingling Zhao, Xavier Intes. Hyperspectral time-resolved wide-field fluorescence molecular tomography based on structured light and single-pixel detection Optics Letters. 2015;40:431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wolfgang Becker, Axel Bergmann, Christoph Biskup. Multispectral fluorescence lifetime imaging by TCSPC Microscopy research and technique. 2007;70:403–409. [DOI] [PubMed] [Google Scholar]
- 101.Smith Jason T, Marien Ochoa, Xavier Intes. UNMIX-ME: spectral and lifetime fluorescence unmixing via deep learning Biomedical Optics Express. 2020;11:3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Smith Jason T, Enagnon Aguénounon, Sylvain Gioux, Xavier Intes. Macroscopic fluorescence lifetime topography enhanced via spatial frequency domain imaging Optics Letters. 2020;45:4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ruoyang Yao, Xavier Intes, Qianqian Fang. Direct approach to compute Jacobians for diffuse optical tomography using perturbation Monte Carlo-based photon “replay” Biomedical Optics Express. 2018;9:4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Enagnon Aguénounon, Smith Jason T, Mahdi Al-Taher, et al. Real-time, wide-field and high-quality single snapshot imaging of optical properties with profile correction using deep learning Biomedical Optics Express. 2020;11:5701–5716. Publisher: Optical Society of America. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Varun Mannam, Yide Zhang, Xiaotong Yuan, Cara Ravasio, Scott Howard. Machine learning for faster and smarter fluorescence lifetime imaging microscopy Journal of Physics: Photonics. 2020. [Google Scholar]
- 106.Walsh Alex J, Mueller Katherine P, Kelsey Tweed, et al. Classification of T-cell activation via autofluorescence lifetime imaging Nature Biomedical Engineering. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yide Zhang, Takashi Hato, Dagher Pierre C, et al. Automatic segmentation of intravital fluorescence microscopy images by K-means clustering of FLIM phasors Optics Letters. 2019;44:3928. [DOI] [PubMed] [Google Scholar]
- 108.Kader Sagar Md Abdul, Cheng Kevin P, Ouellette Jonathan N, Williams Justin C, Watters Jyoti J, Eliceiri Kevin W. Machine learning methods for fluorescence lifetime imaging (FLIM) based label-free detection of microglia Frontiers in neuroscience. 2020;14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Goetz M. Confocal laser endomicroscopy: Current indications and future perspectives in gastrointestinal diseases Endoscopia. 2012;24:67–74. [Google Scholar]
- 110.Marc Aubreville, Christian Knipfer, Nicolai Oetter, et al. Automatic classification of cancerous tissue in laserendomicroscopy images of the oral cavity using deep learning Scientific reports. 2017;7:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Szczotka Agnieszka Barbara, Ismail Shakir Dzhoshkun, Daniele Ravì, Clarkson Matthew J, Pereira Stephen P, Tom Vercauteren. Learning from irregularly sampled data for endomicroscopy super-resolution: a comparative study of sparse and dense approaches International Journal of Computer Assisted Radiology and Surgery. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yun Gu, Khushi Vyas, Jie Yang, Guang-Zhong Yang. Transfer recurrent feature learning for endomicroscopy image recognition IEEE transactions on medical imaging. 2018;38:791–801. [DOI] [PubMed] [Google Scholar]
- 113.Wells Wendy A, Michael Thrall, Anastasia Sorokina, et al. In vivo and ex vivo microscopy: moving toward the integration of optical imaging technologies into pathology practice Archives of pathology & laboratory medicine. 2019;143:288–298. [DOI] [PubMed] [Google Scholar]
- 114.Mehreen Adhi, Duker Jay S. Optical coherence tomography–current and future applications Current opinion in ophthalmology. 2013;24:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Martin Goetz. Confocal laser endomicroscopy: applications in clinical and translational science—a comprehensive review International Scholarly Research Notices. 2012;2012. [Google Scholar]
- 116.Milind Rajadhyaksha, Ashfaq Marghoob, Anthony Rossi, Halpern Allan C, Nehal Kishwer S. Reflectance confocal microscopy of skin in vivo: From bench to bedside Lasers in surgery and medicine. 2017;49:7–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kivanc Kose, Alican Bozkurt, Christi Alessi-Fox, et al. Segmentation of cellular patterns in confocal images of melanocytic lesions in vivo via a multiscale encoder-decoder network (med-net) Medical Image Analysis. 2021;67:101841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nils Gessert, Marcel Bengs, Lukas Wittig, et al. Deep transfer learning methods for colon cancer classification in confocal laser microscopy images International journal of computer assisted radiology and surgery. 2019;14:1837–1845. [DOI] [PubMed] [Google Scholar]
- 119.Cendre R, Mansouri A, Yannick Benezeth, Marzani F, Perrot JL, Cinotti E. Two Schemes for Automated Diagnosis of Lentigo on Confocal Microscopy Images in 2019 IEEE 4th International Conference on Signal and Image Processing (ICSIP):143–147IEEE; 2019. [Google Scholar]
- 120.Marek Wodzinski, Andrzej Skalski, Alexander Witkowski, Giovanni Pellacani, Joanna Ludzik. Convolutional Neural Network Approach to Classify Skin Lesions Using Reflectance Confocal Microscopy in 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC):4754–4757IEEE; 2019. [DOI] [PubMed] [Google Scholar]
- 121.Alican Bozkurt, Trevor Gale, Kivanc Kose, et al. Delineation of skin strata in reflectance confocal microscopy images with recurrent convolutional networks in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops:25–33 2017. [Google Scholar]
- 122.Yachun Li, Patra Charalampaki, Yong Liu, Guang-Zhong Yang, Stamatia Giannarou. Context aware decision support in neurosurgical oncology based on an efficient classification of endomicroscopic data International journal of computer assisted radiology and surgery. 2018;13:1187–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Marit Lucas, Liem Esmee IML, Dilara Savci-Heijink C, et al. Toward Automated In Vivo Bladder Tumor Stratification Using Confocal Laser Endomicroscopy Journal of Endourology. 2019;33:930–937. [DOI] [PubMed] [Google Scholar]
- 124.Marc Aubreville, Maike Stoeve, Nicolai Oetter, et al. Deep learning-based detection of motion artifacts in probe-based confocal laser endomicroscopy images International journal of computer assisted radiology and surgery. 2019;14:31–42. [DOI] [PubMed] [Google Scholar]
- 125.Marek Wodzinski, Miroslawa Pajak, Andrzej Skalski, Alexander Witkowski, Giovanni Pellacani, Joanna Ludzik. Automatic Quality Assessment of Reflectance Confocal Microscopy Mosaics using Attention-Based Deep Neural Network in 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC):1824–1827IEEE; 2020. [DOI] [PubMed] [Google Scholar]
- 126.Kivanc Kose, Alican Bozkurt, Christi Alessi-Fox, et al. Utilizing machine learning for image quality assessment for reflectance confocal microscopy Journal of Investigative Dermatology. 2020;140:1214–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wallace Michael B, Paul Fockens. Probe-based confocal laser endomicroscopy Gastroenterology. 2009;136:1509–1513. [DOI] [PubMed] [Google Scholar]
- 128.Jianbo Shao, Junchao Zhang, Rongguang Liang, Kobus Barnard. Fiber bundle imaging resolution enhancement using deep learning Optics express. 2019;27:15880–15890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Daniele Ravì, Barbara Szczotka Agnieszka, Pereira Stephen P, Tom Vercauteren. Adversarial training with cycle consistency for unsupervised super-resolution in endomicroscopy Medical image analysis. 2019;53:123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ziyi Meng, Mu Qiao, Jiawei Ma, Zhenming Yu, Kun Xu, Xin Yuan. Snapshot multispectral endomicroscopy Optics Letters. 2020;45:3897–3900. [DOI] [PubMed] [Google Scholar]
- 131.Sophia Bano, Francisco Vasconcelos, Marcel Tella-Amo, et al. Deep learning-based fetoscopic mosaicking for field-of-view expansion International Journal of Computer Assisted Radiology and Surgery. 2020:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Babak Rahmani, Damien Loterie, Georgia Konstantinou, Demetri Psaltis, Christophe Moser. Multimode optical fiber transmission with a deep learning network Light: Science & Applications. 2018;7:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jian Zhao, Yangyang Sun, Zheyuan Zhu, et al. Deep learning imaging through fully-flexible glass-air disordered fiber ACS Photonics. 2018;5:3930–3935. [Google Scholar]
- 134.Pogue Brian W. Optics of Medical Imaging SPIE Professional. 2018. [Google Scholar]
- 135.Pu Wang, Xiao Xiao, Glissen Brown Jeremy R, et al. Development and validation of a deep-learning algorithm for the detection of polyps during colonoscopy Nature Biomedical Engineering. 2018;2:741–748. Number: 10 Publisher: Nature Publishing Group. [DOI] [PubMed] [Google Scholar]
- 136.Byrne Michael F, Nicolas Chapados, Florian Soudan, et al. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model Gut. 2019;68:94–100. Publisher: BMJ Publishing Group Section: Endoscopy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Poon Carmen CY, Yuqi Jiang, Ruikai Zhang, et al. AI-doscopist: a real-time deep-learning-based algorithm for localising polyps in colonoscopy videos with edge computing devices npj Digital Medicine. 2020;3:1–8. Number: 1 Publisher: Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Catherine Le Berre, Sandborn William J, Sabeur Aridhi, et al. Application of Artificial Intelligence to Gastroenterology and Hepatology Gastroenterology. 2020;158:76–94.e2. [DOI] [PubMed] [Google Scholar]
- 139.Gwn Lore Kin, Adedotun Akintayo, Soumik Sarkar. LLNet: A deep autoencoder approach to natural low-light image enhancement Pattern Recognition. 2017;61:650–662. [Google Scholar]
- 140.Pablo Gómez, Marion Semmler, Anne Schützenberger, Christopher Bohr, Michael Döllinger. Low-light image enhancement of high-speed endoscopic videos using a convolutional neural network Medical & Biological Engineering & Computing. 2019;57:1451–1463. [DOI] [PubMed] [Google Scholar]
- 141.Bobrow Taylor L, Faisal Mahmood, Miguel Inserni, Durr Nicholas J. LSR Deep: a deep learning approach for laser speckle reduction Biomedical Optics Express. 2019;10:2869–2882. Publisher: Optical Society of America. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Isabel Funke, Sebastian Bodenstedt, Carina Riediger, Jürgen Weitz, Stefanie Speidel. Generative adversarial networks for specular highlight removal in endoscopic images in Medical Imaging 2018: Image-Guided Procedures, Robotic Interventions, and Modeling;10576:1057604.International Society for Optics and Photonics; 2018. [Google Scholar]
- 143.Chen Richard J, Bobrow Taylor L, Thomas Athey, Faisal Mahmood, Durr Nicholas J. SLAM Endoscopy enhanced by adversarial depth prediction arXiv:1907.00283 [cs, eess]. 2019. arXiv: 1907.00283. [Google Scholar]
- 144.Sharib Ali, Felix Zhou, Adam Bailey, et al. A deep learning framework for quality assessment and restoration in video endoscopy arXiv preprint arXiv:1904.07073. 2019. [Google Scholar]
- 145.Yasin Almalioglu, Bengisu Ozyoruk Kutsev, Abdulkadir Gokce, et al. EndoL2H: Deep Super-Resolution for Capsule Endoscopy arXiv:2002.05459 [cs, eess]. 2020. arXiv: 2002.05459. [Google Scholar]
- 146.Durr Nicholas J, Germán González, Parot Vicente. 3D imaging techniques for improved colonoscopy Expert Review of Medical Devices. 2014;11:105–107. [DOI] [PubMed] [Google Scholar]
- 147.Faisal Mahmood, Richard Chen, Sandra Sudarsky, Daphne Yu, Durr Nicholas J.. Deep learning with cinematic rendering: fine-tuning deep neural networks using photorealistic medical images Physics in Medicine and Biology. 2018;63:185012. [DOI] [PubMed] [Google Scholar]
- 148.Richard Chen, Faisal Mahmood, Alan Yuille, Durr Nicholas J. Rethinking Monocular Depth Estimation with Adversarial Training arXiv:1808.07528 [cs]. 2019. arXiv: 1808.07528. [Google Scholar]
- 149.Faisal Mahmood, Richard Chen, Durr Nicholas J. Unsupervised Reverse Domain Adaptation for Synthetic Medical Images via Adversarial Training IEEE transactions on medical imaging. 2018;37:2572–2581. [DOI] [PubMed] [Google Scholar]
- 150.Faisal Mahmood, Durr Nicholas J. Deep learning and conditional random fields-based depth estimation and topographical reconstruction from conventional endoscopy Medical Image Analysis. 2018;48:230–243. [DOI] [PubMed] [Google Scholar]
- 151.Jianyu Lin, Clancy Neil T, Ji Qi, et al. Dual-modality endoscopic probe for tissue surface shape reconstruction and hyperspectral imaging enabled by deep neural networks Medical Image Analysis. 2018;48:162–176. [DOI] [PubMed] [Google Scholar]
- 152.Ruibin Ma, Rui Wang, Stephen Pizer, Julian Rosenman, McGill Sarah K, Jan-Michael Frahm. Real-Time 3D Reconstruction of Colonoscopic Surfaces for Determining Missing Regions in Medical Image Computing and Computer Assisted Intervention – MICCAI 2019 (Dinggang Shen, Tianming Liu, Peters Terry M, et al., eds.)Lecture Notes in Computer Science(Cham):573–582Springer International Publishing; 2019. [Google Scholar]
- 153.Cuccia David J, Bevilacqua Frédéric P, Joseph Durkin Anthony, Ayers Frederick R, Jason Tromberg Bruce. Quantitation and mapping of tissue optical properties using modulated imaging Journal of biomedical optics. 2009;14:024012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yanyu Zhao, Yue Deng, Feng Bao, Hannah Peterson, Raeef Istfan, Darren Roblyer. Deep learning model for ultrafast multifrequency optical property extractions for spatial frequency domain imaging Optics Letters. 2018;43:5669–5672. Publisher: Optical Society of America. [DOI] [PubMed] [Google Scholar]
- 155.Chen Mason T, Faisal Mahmood, Sweer Jordan A, Durr Nicholas J. GANPOP: Generative Adversarial Network Prediction of Optical Properties From Single Snapshot Wide-Field Images IEEE transactions on medical imaging. 2020;39:1988–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chen Mason T, Durr Nicholas J. Rapid tissue oxygenation mapping from snapshot structured-light images with adversarial deep learning Journal of Biomedical Optics. 2020;25:112907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Alexandru Grigoroiu, Jonghee Yoon, Bohndiek Sarah E. Deep learning applied to hyperspectral endoscopy for online spectral classification Scientific Reports. 2020;10:3947. Number: 1 Publisher: Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chen Mason T, Melina Papadakis, Durr Nicholas J. Speckle illumination spatial frequency domain imaging for projector-free optical property mapping arXiv:2006.03661 [physics]. 2020. arXiv: 2006.03661. [Google Scholar]
- 159.Martin Halicek, Dormer James D, Little James V, et al. Hyperspectral Imaging of Head and Neck Squamous Cell Carcinoma for Cancer Margin Detection in Surgical Specimens from 102 Patients Using Deep Learning Cancers. 2019;11:1367. Number: 9 Publisher: Multidisciplinary Digital Publishing Institute. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Golhar M, Bobrow TL, Khoshknab MP, Jit S, Ngamruengphong S, Durr NJ. Improving Colonoscopy Lesion Classification Using Semi-Supervised Deep Learning IEEE Access. 2021;9:631–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Paul Thienphrapa, Torre Bydlon, Alvin Chen, et al. Interactive Endoscopy: A Next-Generation, Streamlined User Interface for Lung Surgery Navigation in Medical Image Computing and Computer Assisted Intervention – MICCAI 2019:83–91Springer, Cham: 2019. [Google Scholar]
- 162.Huang D, Swanson EA, Lin CP, et al. Optical coherence tomography Science. 1991;254:1178–1181. Publisher: American Association for the Advancement of Science Section: Reports. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zysk Adam M, Nguyen Freddy T, Oldenburg Amy L, Marks Daniel L, M.d Stephen A. Boppart. Optical coherence tomography: a review of clinical development from bench to bedside Journal of Biomedical Optics. 2007;12:051403. Publisher: International Society for Optics and Photonics. [DOI] [PubMed] [Google Scholar]
- 164.Jonas Olsen, Jon Holmes, Jemec Gregor BE. Advances in optical coherence tomography in dermatology—a review Journal of Biomedical Optics. 2018;23:040901. Publisher: International Society for Optics and Photonics. [DOI] [PubMed] [Google Scholar]
- 165.Jianfeng Wang, Yang Xu, Boppart Stephen A. Review of optical coherence tomography in oncology Journal of Biomedical Optics. 2017;22:121711. Publisher: International Society for Optics and Photonics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.James Fujimoto, David Huang. Foreword: 25 Years of Optical Coherence Tomography Investigative Ophthalmology & Visual Science. 2016;57:OCTi–OCTii. Publisher: The Association for Research in Vision and Ophthalmology. [DOI] [PubMed] [Google Scholar]
- 167.Tsung-Han Tsai, Leggett Cadman L, Trindade Arvind J, et al. Optical coherence tomography in gastroenterology: a review and future outlook Journal of Biomedical Optics. 2017;22:121716. Publisher: International Society for Optics and Photonics. [DOI] [PubMed] [Google Scholar]
- 168.Francesco Prati, Evelyn Regar, Mintz Gary S, et al. Expert review document on methodology, terminology, and clinical applications of optical coherence tomography: physical principles, methodology of image acquisition, and clinical application for assessment of coronary arteries and atherosclerosis European Heart Journal. 2010;31:401–415. Publisher: Oxford Academic. [DOI] [PubMed] [Google Scholar]
- 169.Axel Petzold, Boer Johannes F, Sven Schippling, et al. Optical coherence tomography in multiple sclerosis: a systematic review and meta-analysis The Lancet Neurology. 2010;9:921–932. [DOI] [PubMed] [Google Scholar]
- 170.McLean James P, Shuyang Fang, George Gallos, Myers Kristin M, Hendon Christine P Three-dimensional collagen fiber mapping and tractography of human uterine tissue using OCT Biomedical Optics Express. 2020;11:5518–5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lurie Kristen L, Smith Gennifer T, Khan Saara A, Liao Joseph C, Bowden Audrey K. Three-dimensional, distendable bladder phantom for optical coherence tomography and white light cystoscopy Journal of biomedical optics. 2014;19:036009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Venhuizen Freerk G, Bram Ginneken, Bart Liefers, et al. Robust total retina thickness segmentation in optical coherence tomography images using convolutional neural networks Biomedical Optics Express. 2017;8:3292–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Abhay Shah, Leixin Zhou, Abrámoff Michael D, Wu Xiaodong. Multiple surface segmentation using convolution neural nets: application to retinal layer segmentation in OCT images Biomedical Optics Express. 2018;9:4509–4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Donghuan Lu, Morgan Heisler, Sieun Lee, et al. Deep-learning based multiclass retinal fluid segmentation and detection in optical coherence tomography images using a fully convolutional neural network Medical Image Analysis. 2019;54:100–110. [DOI] [PubMed] [Google Scholar]
- 175.Krishna Devalla Sripad, Renukanand Prajwal K, Sreedhar Bharathwaj K, et al. DRUNET: a dilated-residual U-Net deep learning network to segment optic nerve head tissues in optical coherence tomography images Biomedical Optics Express. 2018;9:3244–3265. Publisher: Optical Society of America. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Jianxu Chen, Lin Yang, Yizhe Zhang, Mark Alber, Chen Danny Z. Combining Fully Convolutional and Recurrent Neural Networks for 3D Biomedical Image Segmentation Advances in Neural Information Processing Systems. 2016;29:3036–3044. [Google Scholar]
- 177.Milletari F, Navab N, Ahmadi S. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation in 2016 Fourth International Conference on 3D Vision (3DV):565–571 2016. [Google Scholar]
- 178.Dawei Li, Jimin Wu, Yufan He, et al. Parallel deep neural networks for endoscopic OCT image segmentation Biomedical Optics Express. 2019;10:1126–1135. Publisher: Optical Society of America. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lincan Li, Tong Jia. Optical Coherence Tomography Vulnerable Plaque Segmentation Based on Deep Residual U-Net Reviews in Cardiovascular Medicine. 2019;20:171–177. [DOI] [PubMed] [Google Scholar]
- 180.Sabina Stefan, Jonghwan Lee, Jonghwan Lee. Deep learning toolbox for automated enhancement, segmentation, and graphing of cortical optical coherence tomography microangiograms Biomedical Optics Express. 2020;11:7325–7342. Publisher: Optical Society of America. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zhao Dong, Guoyan Liu, Guangming Ni, Jason Jerwick, Lian Duan, Chao Zhou. Optical coherence tomography image denoising using a generative adversarial network with speckle modulation Journal of Biophotonics. 2020;13:e201960135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hyungtaek Rim Tyler, Lee Aaron Y, Ting Daniel S, et al. Detection of features associated with neovascular age-related macular degeneration in ethnically distinct data sets by an optical coherence tomography: trained deep learning algorithm British Journal of Ophthalmology. 2020. Publisher: BMJ Publishing Group Ltd Section: Clinical science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Bin Qiu, Zhiyu Huang, Xi Liu, et al. Noise reduction in optical coherence tomography images using a deep neural network with perceptually-sensitive loss function Biomedical Optics Express. 2020;11:817–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Zaixing Mao, Atsuya Miki, Song Mei, et al. Deep learning based noise reduction method for automatic 3D segmentation of the anterior of lamina cribrosa in optical coherence tomography volumetric scans Biomedical Optics Express. 2019;10:5832–5851. Publisher: Optical Society of America. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Fauw Jeffrey De, Ledsam Joseph R, Bernardino Romera-Paredes, et al. Clinically applicable deep learning for diagnosis and referral in retinal disease Nature Medicine. 2018;24:1342–1350. Number: 9 Publisher: Nature Publishing Group. [DOI] [PubMed] [Google Scholar]
- 186.Fang L, Wang C, Li S, Rabbani H, Chen X, Liu Z. Attention to Lesion: Lesion-Aware Convolutional Neural Network for Retinal Optical Coherence Tomography Image Classification IEEE Transactions on Medical Imaging. 2019;38:1959–1970. Conference Name: IEEE Transactions on Medical Imaging. [DOI] [PubMed] [Google Scholar]
- 187.Ryo Asaoka, Hiroshi Murata, Kazunori Hirasawa, et al. Using Deep Learning and Transfer Learning to Accurately Diagnose Early-Onset Glaucoma From Macular Optical Coherence Tomography Images American Journal of Ophthalmology. 2019;198:136–145. [DOI] [PubMed] [Google Scholar]
- 188.Jessica Loo, Leyuan Fang, David Cunefare, Jaffe Glenn J, Sina Farsiu. Deep longitudinal transfer learning-based automatic segmentation of photoreceptor ellipsoid zone defects on optical coherence tomography images of macular telangiectasia type 2 Biomedical Optics Express. 2018;9:2681–2698. Publisher: Optical Society of America. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Yufan He, Aaron Carass, Yihao Liu, Shiv Saidha, Calabresi Peter A, Prince Jerry L. Adversarial domain adaptation for multi-device retinal OCT segmentation in Medical Imaging 2020: Image Processing;11313:1131309.International Society for Optics and Photonics; 2020. [Google Scholar]
- 190.Yang S, Zhou X, Wang J, et al. Unsupervised Domain Adaptation for Cross-Device OCT Lesion Detection via Learning Adaptive Features in 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI):1570–1573 2020. ISSN: 1945–8452. [Google Scholar]
- 191.Boy Braaf, Sabine Donner, Néstor Uribe-Patarroyo, Bouma Brett E, Vakoc Benjamin J. A Neural Network Approach to Quantify Blood Flow from Retinal OCT Intensity Time-Series Measurements Scientific Reports. 2020;10:9611. Number: 1 Publisher: Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Rongrong Liu, Shiyi Cheng, Lei Tian, Ji Yi. Deep spectral learning for label-free optical imaging oximetry with uncertainty quantification Light: Science & Applications. 2019;8:102. Number: 1 Publisher: Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Lee Cecilia S, Tyring Ariel J, Yue Wu, et al. Generating retinal flow maps from structural optical coherence tomography with artificial intelligence Scientific Reports. 2019;9:5694. Number: 1 Publisher: Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Xi Liu, Zhiyu Huang, Zhenzhou Wang, et al. A deep learning based pipeline for optical coherence tomography angiography Journal of Biophotonics. 2019;12:e201900008. _eprint: https://onlinelibrary.wiley.com/doi/pdf/10.1002/jbio.201900008. Optics express. 2009;17:10489–10498. [DOI] [PubMed] [Google Scholar]
- 195.Minghua Xu, Wang Lihong V Photoacoustic imaging in biomedicine Review of Scientific Instruments. 2006;77:041101. [Google Scholar]
- 196.Paul Beard. Biomedical photoacoustic imaging Interface Focus. 2011;1:602–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Lediju Bell Muyinatu A. Deep learning the sound of light to guide surgeries in Advanced Biomedical and Clinical Diagnostic and Surgical Guidance Systems XVII;10868:108680.GInternational Society for Optics and Photonics; 2019. [Google Scholar]
- 198.Lediju Bell Muyinatu A Photoacoustic imaging for surgical guidance: Principles, applications, and outlook Journal of Applied Physics. 2020;128:060904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Andreas Hauptmann, Cox Ben T Deep Learning in Photoacoustic Tomography: Current approaches and future directions Journal of Biomedical Optics. 2020;25:112903. [Google Scholar]
- 200.Austin Reiter, Bell Muyinatu A Lediju. A machine learning approach to identifying point source locations in photoacoustic data in Photons Plus Ultrasound: Imaging and Sensing 2017;10064:100643.JInternational Society for Optics and Photonics; 2017. [Google Scholar]
- 201.Derek Allman, Austin Reiter, Bell Muyinatu A Lediju. Photoacoustic source detection and reflection artifact removal enabled by deep learning IEEE transactions on medical imaging. 2018;37:1464–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Kerrick Johnstonbaugh, Sumit Agrawal, Durairaj Deepit Abhishek, et al. A deep learning approach to photoacoustic wavefront localization in deep-tissue medium IEEE transactions on ultrasonics, ferroelectrics, and frequency control. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Woo Kim Min, Geng-Shi Jeng, Ivan Pelivanov, Matthew O Donnell. Deep-learning Image Reconstruction for Real-time Photoacoustic System IEEE Transactions on Medical Imaging. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Dufan Wu, Kyungsang Kim, Quanzheng Li. Computationally efficient deep neural network for computed tomography image reconstruction Medical physics. 2019;46:4763–4776. [DOI] [PubMed] [Google Scholar]
- 205.Andreas Hauptmann, Felix Lucka, Marta Betcke, et al. Model-based learning for accelerated, limited-view 3-d photoacoustic tomography IEEE transactions on medical imaging. 2018;37:1382–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Tong Tong, Wenhui Huang, Kun Wang, et al. Domain Transform Network for Photoacoustic Tomography from Limited-view and Sparsely Sampled Data Photoacoustics. 2020:100190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Dominik Waibel, Janek Gröhl, Fabian Isensee, Thomas Kirchner, Klaus Maier-Hein, Lena Maier-Hein. Reconstruction of initial pressure from limited view photoacoustic images using deep learning in Photons Plus Ultrasound: Imaging and Sensing 2018;10494:104942.SInternational Society for Optics and Photonics; 2018. [Google Scholar]
- 208.Stephan Antholzer, Markus Haltmeier, Johannes Schwab. Deep learning for photoacoustic tomography from sparse data Inverse problems in science and engineering. 2019;27:987–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Steven Guan, Khan Amir A, Siddhartha Sikdar, Chitnis Parag V Limited-View and Sparse photoacoustic tomography for neuroimaging with Deep Learning Scientific Reports. 2020;10:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Neda Davoudi, Luís Deán-Ben Xosé, Daniel Razansky. Deep learning optoacoustic tomography with sparse data Nature Machine Intelligence. 2019;1:453–460. [Google Scholar]
- 211.Ali Hariri, Kamran Alipour, Yash Mantri, Schulze Jurgen P, Jokerst Jesse V. Deep learning improves contrast in low-fluence photoacoustic imaging Biomedical Optics Express. 2020;11:3360–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Navchetan Awasthi, Gaurav Jain, Kumar Kalva Sandeep, Manojit Pramanik, Yalavarthy Phaneendra K Deep Neural Network Based Sinogram Super-resolution and Bandwidth Enhancement for Limited-data Photoacoustic Tomography IEEE Transactions on Ultrasonics, Ferroelectrics, and Frequency Control. 2020. [DOI] [PubMed] [Google Scholar]
- 213.John Gamelin, Anastasios Maurudis, Andres Aguirre, et al. A real-time photoacoustic tomography system for small animals [Google Scholar]
- 214.Li Lin, Peng Hu, Junhui Shi, et al. Single-breath-hold photoacoustic computed tomography of the breast Nature communications. 2018;9:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Lei Xi, Huabei Jiang. High resolution three-dimensional photoacoustic imaging of human finger joints in vivo Applied Physics Letters. 2015;107:063701. [Google Scholar]
- 216.Bo Zhu, Liu Jeremiah Z, Cauley Stephen F, Rosen Bruce R, Rosen Matthew S. Image reconstruction by domain-transform manifold learning Nature. 2018;555:487–492. [DOI] [PubMed] [Google Scholar]
- 217.Yuan Xu, Wang Lihong V, Gaik Ambartsoumian, Peter Kuchment. Reconstructions in limited-view thermoacoustic tomography Medical physics. 2004;31:724–733. [DOI] [PubMed] [Google Scholar]
- 218.Tri Vu, Mucong Li, Hannah Humayun, Yuan Zhou, Junjie Yao. A generative adversarial network for artifact removal in photoacoustic computed tomography with a linear-array transducer Experimental Biology and Medicine. 2020;245:597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.David Finch, Haltmeier Markus Rakesh. Inversion of spherical means and the wave equation in even dimensions SIAM Journal on Applied Mathematics. 2007;68:392–412. [Google Scholar]
- 220.Sreedevi Gutta, Suryanarayana Kadimesetty Venkata, Kumar Kalva Sandeep, Manojit Pramanik, Sriram Ganapathy, Yalavarthy Phaneendra K Deep neural network-based bandwidth enhancement of photoacoustic data Journal of biomedical optics. 2017;22:116001. [DOI] [PubMed] [Google Scholar]
- 221.Serafeim Moustakidis, Murad Omar, Juan Aguirre, Pouyan Mohajerani, Vasilis Ntziachristos. Fully automated identification of skin morphology in raster-scan optoacoustic mesoscopy using artificial intelligence Medical physics. 2019;46:4046–4056. [DOI] [PubMed] [Google Scholar]
- 222.Kaiming He, Xiangyu Zhang, Shaoqing Ren, Jian Sun. Deep residual learning for image recognition in Proceedings of the IEEE conference on computer vision and pattern recognition:770–778 2016. [Google Scholar]
- 223.Alex Krizhevsky, Ilya Sutskever, Hinton Geoffrey E Imagenet classification with deep convolutional neural networks in Advances in neural information processing systems:1097–1105 2012. [Google Scholar]
- 224.Boink Yoeri E, Srirang Manohar, Christoph Brune. A Partially-Learned Algorithm for Joint Photo-acoustic Reconstruction and Segmentation IEEE transactions on medical imaging. 2019;39:129–139. [DOI] [PubMed] [Google Scholar]
- 225.Jonas Adler, Ozan Öktem. Learned primal-dual reconstruction IEEE transactions on medical imaging. 2018;37:1322–1332. [DOI] [PubMed] [Google Scholar]
- 226.Chuangjian Cai, Kexin Deng, Cheng Ma, Jianwen Luo. End-to-end deep neural network for optical inversion in quantitative photoacoustic imaging Optics letters. 2018;43:2752–2755. [DOI] [PubMed] [Google Scholar]
- 227.Xingxing Chen, Weizhi Qi, Lei Xi. Deep-learning-based motion-correction algorithm in optical resolution photoacoustic microscopy Visual Computing for Industry, Biomedicine, and Art. 2019;2:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Rayyan Manwar, Xin Li, Sadreddin Mahmoodkalayeh, Eishi Asano, Dongxiao Zhu, Kamran Avanaki. Deep Learning Protocol for Improved Photoacoustic Brain Imaging Journal of Biophotonics. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Derek Allman, Fabrizio Assis, Jonathan Chrispin, Lediju Bell Muyinatu A. Deep neural networks to remove photoacoustic reflection artifacts in ex vivo and in vivo tissue in 2018 IEEE International Ultrasonics Symposium (IUS):1–4IEEE; 2018. [Google Scholar]
- 230.Derek Allman, Fabrizio Assis, Jonathan Chrispin, Lediju Bell Muyinatu A. A deep learning-based approach to identify in vivo catheter tips during photoacoustic-guided cardiac interventions in Photons Plus Ultrasound: Imaging and Sensing 2019;10878:108785.EInternational Society for Optics and Photonics; 2019. [Google Scholar]
- 231.Derek Allman, Fabrizio Assis, Jonathan Chrispin, Lediju Bell Muyinatu A. Deep learning to detect catheter tips in vivo during photoacoustic-guided catheter interventions: Invited presentation in 2019 53rd Annual Conference on Information Sciences and Systems (CISS):1–3IEEE; 2019. [Google Scholar]
- 232.Yao Wang, Jeng-Hua Chang, Raphael Aronson, Barbour Randall L, Graber Harry L, Jack Lubowsky. Imaging of scattering media by diffusion tomography: an iterative perturbation approach in Proc. SPIE 1641, Physiological Monitoring and Early Detection Diagnostic Methods (Mang Thomas S., ed.):58–71 1992. [Google Scholar]
- 233.Boas DA, Brooks DH, Miller EL, et al. Imaging the body with diffuse optical tomography IEEE Signal Processing Magazine. 2001;18:57–75. [Google Scholar]
- 234.Gibson AP, Hebden JC, Arridge SR. Recent advances in diffuse optical imaging Physics in Medicine and Biology. 2005;50:R1–R43. [DOI] [PubMed] [Google Scholar]
- 235.Xavier Intes. Time-Domain Optical Mammography SoftScan Academic Radiology. 2005;12:934–947. [DOI] [PubMed] [Google Scholar]
- 236.Marco Ferrari. Progress of near-infrared spectroscopy and topography for brain and muscle clinical applications Journal of Biomedical Optics. 2007;12:062104. [DOI] [PubMed] [Google Scholar]
- 237.Eggebrecht Adam T, Ferradal Silvina L, Robichaux-Viehoever Amy, et al. Mapping distributed brain function and networks with diffuse optical tomography Nature Photonics. 2014;8:448–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Alper Corlu, Regine Choe, Turgut Durduran, et al. Three-dimensional in vivo fluorescence diffuse optical tomography of breast cancer in humans Optics Express. 2007;15:6696. [DOI] [PubMed] [Google Scholar]
- 239.Chinmay Darne, Yujie Lu, Sevick-Muraca Eva M. Small animal fluorescence and bioluminescence tomography: a review of approaches, algorithms and technology update Physics in Medicine and Biology. 2014;59:R1–R64. [DOI] [PubMed] [Google Scholar]
- 240.Rice William L, Shcherbakova Daria M, Verkhusha Vladislav V, Kumar Anand TN. In Vivo Tomographic Imaging of Deep-Seated Cancer Using Fluorescence Lifetime Contrast Cancer Research. 2015;75:1236–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Kak Avinash C, Malcolm Slaney, Ge Wang. Principles of Computerized Tomographic Imaging Medical Physics. 2002;29:107–107. [Google Scholar]
- 242.O’Leary MA, Boas DA, Chance B, Yodh AG. Experimental images of heterogeneous turbid media by frequency-domain diffusing-photon tomography Optics Letters. 1995;20:426. [DOI] [PubMed] [Google Scholar]
- 243.Ruoyang Yao, Qi Pian, Xavier Intes. Wide-field fluorescence molecular tomography with compressive sensing based preconditioning Biomedical Optics Express. 2015;6:4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Xiaofeng Zhang, Badea Cristian T, Allan Johnson G.. Three-dimensional reconstruction in free-space whole-body fluorescence tomography of mice using optically reconstructed surface and atlas anatomy Journal of Biomedical Optics. 2009;14:064010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Murat Guven, Birsen Yazici, Xavier Intes, Britton Chance. Diffuse optical tomography with a priori anatomical information Physics in Medicine and Biology. 2005;50:2837–2858. [DOI] [PubMed] [Google Scholar]
- 246.Ang Li, Miller Eric L, Kilmer Misha E, et al. Tomographic optical breast imaging guided by three-dimensional mammography Applied Optics. 2003;42:5181. [DOI] [PubMed] [Google Scholar]
- 247.Schulz RB, Angelique Ale, Athanasios Sarantopoulos, et al. Hybrid System for Simultaneous Fluorescence and X-Ray Computed Tomography IEEE Transactions on Medical Imaging. 2010;29:465–473. [DOI] [PubMed] [Google Scholar]
- 248.Vivek Venugopal, Jin Chen, Margarida Barroso, Xavier Intes. Quantitative tomographic imaging of intermolecular FRET in small animals Biomedical Optics Express. 2012;3:3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Feixiao Long. Deep learning-based mesoscopic fluorescence molecular tomography: an in silico study Journal of Medical Imaging. 2018;5:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Yuan Gao, Kun Wang, Yu An, Shixin Jiang, Hui Meng, Jie Tian. Nonmodel-based bioluminescence tomography using a machine-learning reconstruction strategy Optica. 2018;5:1451. [Google Scholar]
- 251.Lin Guo, Fei Liu, Chuangjian Cai, Jie Liu, Guanglei Zhang. 3D deep encoder–decoder network for fluorescence molecular tomography Optics Letters. 2019;44:1892. [DOI] [PubMed] [Google Scholar]
- 252.Jaejun Yoo, Sohail Sabir, Duchang Heo, et al. Deep Learning Diffuse Optical Tomography IEEE Transactions on Medical Imaging. 2020;39:877–887. [DOI] [PubMed] [Google Scholar]
- 253.Chao Huang, Hui Meng, Yuan Gao, Jiang ShiXin Wang Kung, Jie Tian. Fast and robust reconstruction method for fluorescence molecular tomography based on deep neural network in Imaging, Manipulation, and Analysis of Biomolecules, Cells, and Tissues XVII (Farkas Daniel L, Leary James F, Attila Tarnok., eds.):55SPIE; 2019. [Google Scholar]
- 254.Chul Ye Jong, Yoseob Han, Eunju Cha. Deep Convolutional Framelets: A General Deep Learning Framework for Inverse Problems SIAM Journal on Imaging Sciences. 2018;11:991–1048. [Google Scholar]
- 255.Qianqian Fang. Mesh-based Monte Carlo method using fast ray-tracing in Plücker coordinates Biomedical Optics Express. 2010;1:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Ruoyang Yao, Xavier Intes, Qianqian Fang. Generalized mesh-based Monte Carlo for wide-field illumination and detection via mesh retessellation Biomedical Optics Express. 2016;7:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Roberto Rosas-Romero, Edgar Guevara, Ke Peng, et al. Prediction of epileptic seizures with convolutional neural networks and functional near-infrared spectroscopy signals Computers in Biology and Medicine. 2019;111:103355. [DOI] [PubMed] [Google Scholar]
- 258.Tomoyuki Hiroyasu, Kenya Hanawa, Utako Yamamoto. Gender classification of subjects from cerebral blood flow changes using Deep Learning in 2014 IEEE Symposium on Computational Intelligence and Data Mining (CIDM):229–233IEEE; 2014. [Google Scholar]
- 259.Mahya Mirbagheri, Ata Jodeiri, Naser Hakimi, Vahid Zakeri, Kamaledin Setarehdan Seyed. Accurate Stress Assessment based on functional Near Infrared Spectroscopy using Deep Learning Approach in 2019 26th National and 4th International Iranian Conference on Biomedical Engineering (ICBME):4–10IEEE; 2019. [Google Scholar]
- 260.Katarina Dedovic, Catherine D Aguiar, Pruessner Jens C. What Stress Does to Your Brain: A Review of Neuroimaging Studies The Canadian Journal of Psychiatry. 2009;54:6–15. [DOI] [PubMed] [Google Scholar]
- 261.Sandra Vieira, Pinaya Walter HL, Andrea Mechelli. Using deep learning to investigate the neuroimaging correlates of psychiatric and neurological disorders: Methods and applications Neuroscience & Biobehavioral Reviews. 2017;74:58–75. [DOI] [PubMed] [Google Scholar]
- 262.Danushka Bandara, Leanne Hirshfield, Senem Velipasalar. Classification of affect using deep learning on brain blood flow data Journal of Near Infrared Spectroscopy. 2019;27:206–219. [Google Scholar]
- 263.Johann Benerradi, Horia A. Maior, Adrian Marinescu, Jeremie Clos, Max L. Wilson. Exploring Machine Learning Approaches for Classifying Mental Workload using fNIRS Data from HCI Tasks in Proceedings of the Halfway to the Future Symposium 2019(New York, NY, USA):1–11ACM; 2019. [Google Scholar]
- 264.Yuanyuan Gao, Uwe Kruger, Xavier Intes, Steven Schwaitzberg, Suvranu De. A machine learning approach to predict surgical learning curves Surgery. 2020;167:321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Hubert Cecotti, Axel Graeser. Convolutional Neural Network with embedded Fourier Transform for EEG classification in 2008 19th International Conference on Pattern Recognition:1–4IEEE; 2008. [Google Scholar]
- 266.Zied Tayeb, Juri Fedjaev, Nejla Ghaboosi, et al. Validating Deep Neural Networks for Online Decoding of Motor Imagery Movements from EEG Signals Sensors. 2019;19:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Johannes Hennrich, Christian Herff, Dominic Heger, Tanja Schultz. Investigating deep learning for fNIRS based BCI in 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC):2844–2847IEEE; 2015. [DOI] [PubMed] [Google Scholar]
- 268.Dargazany Aras R, Mohammadreza Abtahi, Kunal Mankodiya. An end-to-end (deep) neural network applied to raw EEG, fNIRs and body motion data for data fusion and BCI classification task without any pre-/post-processing arXiv preprint arXiv:1907.09523. 2019. [Google Scholar]
- 269.Thanawin Trakoolwilaiwan, Bahareh Behboodi, Jaeseok Lee, Kyungsoo Kim, Ji-Woong Choi. Convolutional neural network for high-accuracy functional near-infrared spectroscopy in a brain–computer interface: three-class classification of rest, right-, and left-hand motor execution Neurophotonics. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Marjan Saadati, Jill Nelson, Hasan Ayaz. Multimodal fNIRS-EEG Classification Using Deep Learning Algorithms for Brain-Computer Interfaces Purposes in Advances in Neuroergonomics and Cognitive Engineering:209–220Springer International Publishing; 2020. [Google Scholar]
- 271.Yuanyuan Gao, Lora Cavuoto, Pingkun Yan, et al. A deep learning approach to remove motion artifacts in fNIRS data analysis in Biophotonics Congress: Biomedical Optics 2020 (Translational, Microscopy, OCT, OTS, BRAIN)(Washington, D.C.):BM2C.7OSA 2020. [Google Scholar]
- 272.Chien-Sing Poon, Feixiao Long, Ulas Sunar. Deep learning model for ultrafast quantification of blood flow in diffuse correlation spectroscopy bioRxiv. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Heung-Il Suk, Chong-Yaw Wee, Seong-Whan Lee, Dinggang Shen. State-space model with deep learning for functional dynamics estimation in resting-state fMRI NeuroImage. 2016;129:292–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Fox Michael D, Snyder Abraham Z, Vincent Justin L, Maurizio Corbetta, Van Essen David C, Raichle Marcus E. From The Cover: The human brain is intrinsically organized into dynamic, anticorrelated functional networks Proceedings of the National Academy of Sciences. 2005;102:9673–9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Juan Zhou, Greicius Michael D, Gennatas Efstathios D, et al. Divergent network connectivity changes in behavioural variant frontotemporal dementia and Alzheimer’s disease Brain. 2010;133:1352–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Jeremy Kawahara, Brown Colin J, Miller Steven P, et al. BrainNetCNN: Convolutional neural networks for brain networks; towards predicting neurodevelopment NeuroImage. 2017;146:1038–1049. [DOI] [PubMed] [Google Scholar]
- 277.Bahareh Behboodi, Sung-Ho Lim, Miguel Luna, Hyeon-Ae Jeon, Ji-Woong Choi. Artificial and convolutional neural networks for assessing functional connectivity in resting-state functional near infrared spectroscopy Journal of Near Infrared Spectroscopy. 2019;27:191–205. [Google Scholar]
- 278.Sylvain Arlot, Alain Celisse. A survey of cross-validation procedures for model selection Statistics Surveys. 2010;4:40–79. [Google Scholar]
- 279.Kelly Christopher J, Alan Karthikesalingam, Mustafa Suleyman, Greg Corrado, Dominic King. Key challenges for delivering clinical impact with artificial intelligence BMC Medicine. 2019;17:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Ninareh Mehrabi, Fred Morstatter, Nripsuta Saxena, Kristina Lerman, Aram Galstyan. A Survey on Bias and Fairness in Machine Learning arXiv:1908.09635 [cs]. 2019. arXiv: 1908.09635. [Google Scholar]
- 281.Riccardo Guidotti, Anna Monreale, Salvatore Ruggieri, Franco Turini, Fosca Giannotti, Dino Pedreschi. A Survey of Methods for Explaining Black Box Models ACM Computing Surveys. 2018;51:1–42. [Google Scholar]
- 282.Wojciech Samek, Grégoire Montavon, Sebastian Lapuschkin, Anders Christopher J, Klaus-Robert Müller. Toward Interpretable Machine Learning: Transparent Deep Neural Networks and Beyond arXiv:2003.07631 [cs, stat]. 2020. arXiv: 2003.07631. [Google Scholar]
- 283.Amitojdeep Singh, Sourya Sengupta, Vasudevan Lakshminarayanan. Explainable deep learning models in medical image analysis arXiv:2005.13799 [cs, eess]. 2020. arXiv: 2005.13799. [Google Scholar]
- 284.Carlo Biffi, Cerrolaza Juan J, Giacomo Tarroni, et al. Explainable Anatomical Shape Analysis through Deep Hierarchical Generative Models arXiv:1907.00058 [cs, eess]. 2020. arXiv: 1907.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Cynthia Rudin. Stop Explaining Black Box Machine Learning Models for High Stakes Decisions and Use Interpretable Models Instead Nature Machine Intelligence. 2019;1:206–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Miriam Hägele, Philipp Seegerer, Sebastian Lapuschkin, et al. Resolving challenges in deep learning-based analyses of histopathological images using explanation methods Scientific Reports. 2020;10:6423. Number: 1 Publisher: Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Xiaoxiao Li, Yuan Zhou, Dvornek Nicha C, Yufeng Gu, Pamela Ventola, Duncan James S. Efficient Shapley Explanation for Features Importance Estimation Under Uncertainty in International Conference on Medical Image Computing and Computer-Assisted Intervention (Martel Anne L, Purang Abolmaesumi, Danail Stoyanov, et al., eds.)Lecture Notes in Computer Science(Cham):792–801Springer International Publishing; 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Reinhold Jacob C, Yufan He, Shizhong Han, et al. Finding novelty with uncertainty arXiv:2002.04626 [cs, eess]. 2020. arXiv: 2002.04626. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Most DL models for biomedical optics rely on “supervised learning” that are trained on domain- and/or task-specific datasets, which need to be carefully curated to ensure high-quality predictions. As a result, there are several inherent challenges in the data generation process that need to be addressed, including data availability and data bias [279]. For many applications, it is often difficult and costly to acquire a large-scale dataset. Novel techniques that can better leverage small-scale dataset while still providing high-quality models are needed, such as unsupervised, semi-supervised, and self-supervised learning, transfer learning, and domain adaptation. In addition to the overall scale of the dataset, the data may also be skewed or biased [280] because it may be difficult to acquire data with a balanced distribution for each sub-group, such as gender, ethnicity, etc. DNNs trained on biased dataset can result in erroneous predictions in particular for under-represented populations and diseases. These obstacles may be mitigated to some extent with careful planning and data collection. However, there is a need to also identify and reduce data biases in the modeling step, such as data augmentation and balanced loss function design.