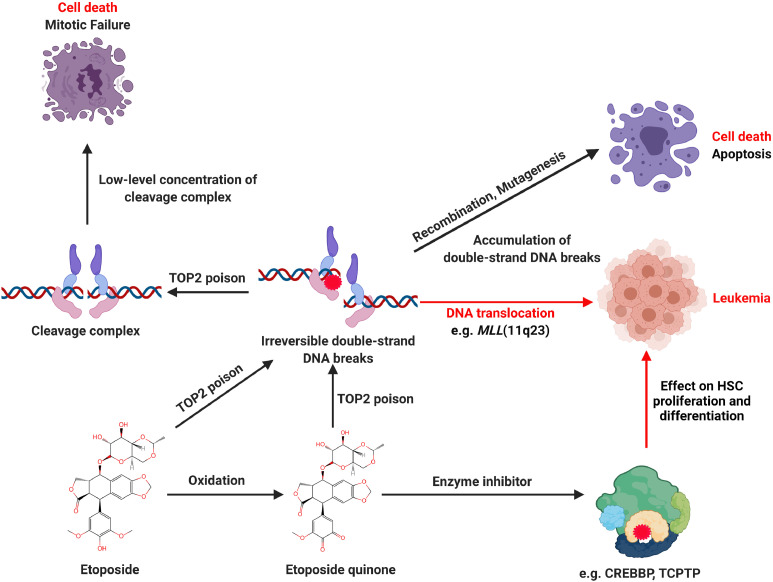
Abstract
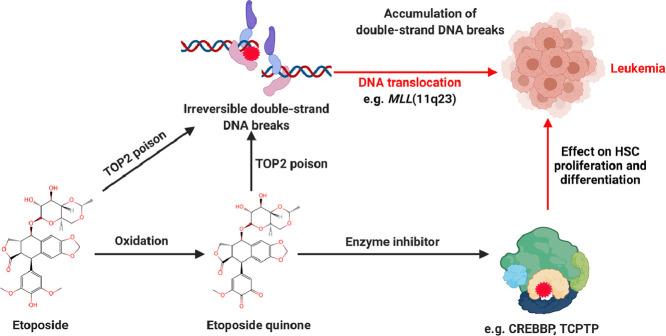
Etoposide is a semi-synthetic glycoside derivative of podophyllotoxin, also known as VP-16. It is a widely used anticancer medicine in clinics. Unfortunately, high doses or long-term etoposide treatment can induce therapy-related leukemia. The mechanism by which etoposide induces secondary hematopoietic malignancies is still unclear. In this article, we review the potential mechanisms of etoposide induced therapy-related leukemia. Etoposide related leukemogenesis is known to depend on reactive oxidative metabolites of etoposide, notably etoposide quinone, which interacts with cellular proteins such as topoisomerases II (TOP2), CREB-binding protein (CREBBP), and T-Cell Protein Tyrosine Phosphatase (TCPTP). CYP3A4 and CYP3A5 metabolize etoposide to etoposide catechol, which readily oxidizes to etoposide quinone. As a poison of TOP2 enzymes, etoposide and its metabolites induce DNA double-stranded breaks (DSB), and the accumulation of DSB triggers cell apoptosis. If the cell survives, the DSB gives rise to the likelihood of faulty DNA repair events. The gene translocation could occur in mixed-lineage leukemia (MLL) gene, which is well-known in leukemogenesis. Recently, studies have revealed that etoposide metabolites, especially etoposide quinone, can covalently bind to cysteines residues of CREBBP and TCPTP enzymes, . This leads to enzyme inhibition and further affects histone acetylation and phosphorylation of the JAK-STAT pathway, thus putatively altering the proliferation and differentiation of hematopoietic stem cells (HSC). In brief, current studies suggest that etoposide and its metabolites contribute to etoposide therapy-related leukemia through TOP2 mediated DSB and impairs specific enzyme activity, such as CREBBP and TCPTP.
Keywords: Topoisomerase II poison, Etoposide, Oxidative metabolite, Double-stranded breaks, Therapy-related leukemia
Graphical abstract
Etoposide is a commonly used anticancer drug
Etoposide is a fundamental and essential part of combination chemotherapies for treating numerous cancers, such as lung cancer, lymphoma, leukemia [1]. Like most anti-neoplastic drugs, etoposide treatment cancer has limited single-agent activity [2,3]. It is mainly associated with cisplatin, carboplatin, and cyclophosphamide in combination chemotherapy [4], [5], [6]. Currently, 4 to 6 cycles etoposide and cisplatin combination is the standard therapy commonly used for most small cell lung cancer in the clinic, with a 50%−80% objective response rate [7,8]. In combination with several drugs (lomustine, methotrexate, and prednisone), etoposide is also proposed to be first-line therapy in patients with non-Hodgkin's lymphoma with no major cardiotoxicity [9,10]. Similarly, in Hodgkin's disease, etoposide is a first-line chemotherapeutic agent in combination with other chemotherapeutic agents (vincristine, chloramphenicol, and prednisolone), was positive (77% response rate) [11]. In addition, it is reported that etoposide was active against gestational trophoblastic disease [12]. Etoposide was used for breast cancer treatment, the single-agent trial in untreated patients showed a response rate of approximately 15% in untreated patients [13]. Oral etoposide has been investigated in many clinical trials for the treatment of ovarian cancer response rates have varied in different studies (20.4% to about 30%), and it seems that the activity of this drug depends to a large extent on the degree of prior treatment [14].
Although etoposide has been widely and successfully used to treat many types of cancer, patients treated with etoposide may develop secondary leukemia. Due to the increases in the overall cure rate of patients, interest has arisen on the adverse effects, and special attention has been focused on the potential risk of therapy-related secondary leukemia [15], [16], [17] and the FDA (U.S. Food and Drug Administration) and EMA (European Medicines Agency) recommend different doses and treatment periods for different cancers as well [18,19].
Metabolism of etoposide in human body
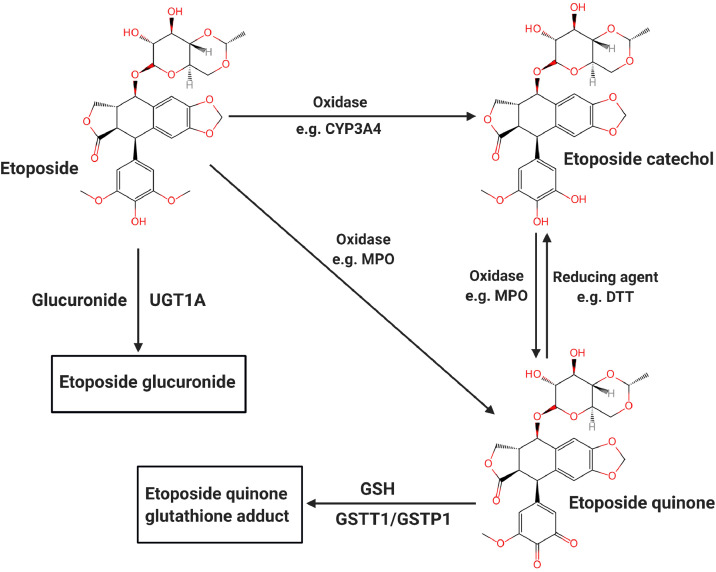
In cells, the oxidation of etoposide mainly involves cytochrome P450 3A family enzymes and/or peroxidases, such as myeloperoxidase (MPO). CYP3A4 and CYP3A5 are members of the cytochrome P450 superfamily of enzymes (encoded by the CYP3A4 and CYP3A5 genes). CYP3A4 and CYP3A5 metabolize etoposide to etoposide catechol, which is readily oxidized to etoposide quinone by cellular peroxidases [20].The induction of CYP3A4 and CYP3A5 can enhance the clearance rate of etoposide [21,22]. Etoposide catechol is oxidized by MPO in liver or bone marrow lysosomes [23,24]. Etoposide can be directly metabolized to etoposide quinone with the assistance of prostaglandin-endoperoxidases 1/2 (PGTS1/2) or by other peroxidases (such as MPO) [25]. Oxidative metabolites of etoposide have a more potent inhibitory activity on DNA TOP2 cleavage near the MLL translocation breakpoint and more remarkable oxidizing ability than etoposide [26]. Etoposide can be cleared by UGT1A1 (UDP-glucuronosyltransferase family 1 member A1) etoposide glucuronides. In addition, etoposide quinone may be transferred to glutathione via GSTT1/GSTP1 (glutathione S-transferase). These two conjugated metabolites seem to inactivate the biological properties of etoposide [27,28]. (Fig. 1)
Fig. 1.
The main metabolic pathway of etoposide in vivo.
Mechanism of action of etoposide
Early in the development of etoposide, its mechanism of action was considered similar to its parent compound, podophyllotoxin. These compounds inhibit microtubule assembly by preventing tubulin polymerization and then destroying spindle fibers. When the cells are exposed to podophyllotoxin, sister chromatids cannot be separated during mitosis because of a missing spindle [29]. As a result, cell division is arrested in mitosis with an increased cell number in metaphase. Nevertheless, cells exposed to etoposide show a decreased cell number in metaphase rather than an increase [30]. Simultaneously, studies reported that a low concentration of etoposide blocks the cell cycle in the late S or early G2 phase [31,32]. These are significant indications that etoposide has a different mechanism compared to podophyllotoxin.
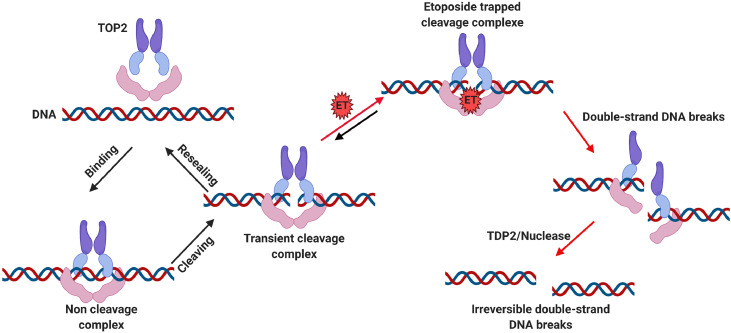
Currently, it is well known that etoposide is a TOP2 inhibitor, this inhibition being considered as a major anti-cancer mechanism of etoposide. TOP2 (alpha and beta isoforms) are ubiquitous enzymes that plays a vital role in many basic DNA processes and regulates the under- and over-winding of DNA during DNA replication. TOP2 inhibitors play an integral role in cancer treatment due to the collapse/collision of the resultant bifurcation and unsolved DSB, leading to cancer cell apoptosis. Generally, TOP2 enzymes create a transient TOP2-DNA cleavage complex (TOP2cc) and reseals TOP2cc rapidly. Once the inhibitor interacts with TOP2, the TOP2cc is trapped, leading to an accumulation of DSB. If the DSB cannot be repaired timely, the cell will initiate apoptosis and eventually die [33,34]. In contrast, if the DSB cell survives, it gives rise to the prospect of faulty DNA repair events, and the gene translocation could occur in the MLL gene, which is known for leukemogenesis. Chromosomal translocation causes abnormal growth and leads to the development of malignant tumors and neoplasms. More than 10,000 gene fusions have been identified in cancer [35,36]. Etoposide and other chemotherapeutic do not directly cause translocation. However, the DSB caused by these molecules must be repaired, and it is the aberrant repair that can result in a translocation. (Fig. 2)
Fig. 2.
Etoposide mechanism of action and poisons on TOP2. The normal catalytic activity of TOP2 creates and reseals the DSB, which involves three steps: TOP2 and DNA binding, TOP2 cleavages DNA double strands, and TOP2 reseal the transient cleaved DNA. Etoposide inhibits TOP2 by stabilizing the TOP2cc. If the trapped TOP2cc can not be adequately repaired, the TDP2 or nuclease will remove the TOP2 from the complex, leading to a persistent and irreversible double-strand DNA break.
Topoisomerase II and topoisomerase II poisons
Topoisomerases are classified according to whether they cut one (type I) or two strands (type II) of DNA. Human cells express two TOP2 subtypes, TOP2α and TOP2β. The two subtypes share 70% amino acid sequence identity and encoded by two different genes. TOP2α is encoded by TOP2A, which is located at chromosome 17. TOP2β is encoded by TOP2B, which is located on chromosome 13 [37], [38], [39]. TOP2α is highly expressed in proliferating cells and can be used as a biomarker of cell proliferation. While, TOP2α is poorly expressed in quiescent and differentiated cell populations [37]. In contrast, TOP2β expression was uniformly distributed in all cells [37,40]. Therefore, TOP2α is considered to be an effective target of TOP2 inhibitors, its impairment being considered as a contributor to the anticancer effects of etoposide.
TOP2 enzymes cleave DNA double-strands, which may cause DNA double-strand breaks [41,42]. Chemicals that inhibits TOP2 activity and decreases the levels of TOP2cc are termed TOP2 catalytic inhibitors. Chemicals that increase the levels of TOP2cc can convert the enzyme into a potential "toxin" that further produces DNA double-strand breaks are called TOP2 poisons [43].
TOP2 poisons can be grouped into two classes based on their mechanism of action. Interfacial poisons, such as etoposide, doxorubicin, or mitoxantrone bind non-covalently to the TOP2cc at the interface of protein-DNA complex and prevent TOP2 from rejoining the DNA ends [44,45]. Covalent poisons such as certain quinones (BQ or etoposide quinone) react with TOP2 at cysteine residues (albeit reaction with other amino acids cannot be ruled out) . Covalent poisons produce irreversible TOP2cc and increase the levels of DSB by altering the conformation of TOP2 [46].
All clinically used TOP2 targeted anticancer drugs impair activities of both TOP2α and TOP2β [37]. However, the extent to which any TOP2 poison targets TOP2α and TOP2β and the relative contribution of either isoform to the drug's therapeutic effect is not well understood [46,47]. It is worth noting that cellular and in vivo studies indicate that TOP2β is the primary enzyme responsible for gene breaks in MLL (see below).
Etoposide induced therapy-related leukemia
Leukemia is a carcinoma of the human hematopoietic system, comprising the bone marrow and lymphatic system. The precise causes of leukemia are not understood; cellular disorders and environmental factors are certainly involved [48]. Therapy-related leukemia has been under investigation for decades and is known to be induced by radiation and/or chemotherapy [49]. Drugs that induce secondary leukemia include alkylating agents, TOP2 poisons (for instance, etoposide and teniposide), cyclophosphamide, anthracyclines, and doxorubicin [15]. Etoposide is a well-known TOP2 poison and a commonly used anticancer drug. It is also recognized that etoposide can be leukemogenic, notably through MLL gene translocation [26,50].
Etoposide was approved for clinical use by the FDA in 1983, and a first report published in 1987 showed that etoposide treatment has a risk of induction of secondary leukemia in 1987 [51]. Subsequently, studies were published on etoposide and secondary leukemia, particularly acute myeloid leukemia (AML). Le Deley's study showed that a high concentration and continuous etoposide treatment of solid tumors give a higher risk of treatment-related acute myeloid leukemia (t-AML) [52]. The study performed by N. J. Winnick et al. revealed that within 23 to 68 months after etoposide treatment, 10 out of 205 children developed secondary AML, and there was a 5.9% ± 3.2% risk of developing leukemia in the next four years [53]. Sugita et al. reported a high incidence of etoposide-associated secondary leukemia in children with non-Hodgkin's lymphoma treated by etoposide. In order to reduce the associated secondary AML, the authors recommend giving etoposide treatment twice a week [54]. Kollmannsberger et al. demonstrated that patients receiving etoposide doses in excess of 2 g/m2 had a 1.3% likelihood of developing s-AML [55]. Meanwhile, Ratain et al. proposed that high-dose etoposide is leukemogenesis in non-small cell lung cancer treatment, and the median dose of etoposide is 6795 mg/m2 [51]. A study by M.A. Smith et al. found that the cumulative risk of secondary leukemia in six years was observed 3.3% in the low etoposide group, 0.7% in the medium etoposide group, and 2.2% in the high etoposide group, respectively (low: < 1.5 g/m2; Medium: 1.5 to 2.99 g/m2; high ≥3.0 g/m2), indicating that factors other than cumulative dose seem to be the main determinants of secondary leukemia risk [56]. Interestingly, it was revealed that receiving etoposide at doses greater than 4000 mg/m2 tended to increase secondary acute non-lymphoblastic leukemia (s-ANLL). Meanwhile, all leukemias described in the Italian Langerhans cell histiocytosis (LCH) group are acute promyelocytic leukemia, which are not identified in Austria, Germany, Netherlands, and Switzerland (AGDS) groups [57]. It is reported that secondary leukemia does occur after conventional doses of etoposide treatment, but the low incidence does not change the risk-benefit ratio of etoposide-based chemotherapy in germ cell carcinoma [58]. Based on these researches, there is evidence that high-dose and long-term doses are critical factors in secondary leukemia caused by etoposide treatment, while other factors cannot be ruled out.
Topoisomerase II-associated DNA double-strand breakage and repair mechanisms
A critical intermediate in topoisomerase activity is the cleavage complex. Once topoisomerases have cleaved the DNA, each subunit of the TOP2 becomes covalently attached to the broken end of the 5′-phosphate group [40]. The cleavage complex is usually transient and naturally results from circumstances that are not clear or induced by the presence of drugs that act as antitumor agents. Topoisomerase inhibitors poison the cleavage intermediates and lead to the formation of an aborted or irreversible TOP2cc [39,47].
In order to repair the TOP2cc captured by the TOP2 poisons, the non-homologous end joining (NHEJ) or homologous recombination (HR) repair pathway is activated. To proceed with NHEJ repair, the trapped TOP2cc is first degraded by the proteasome, and then the remaining tyrosine-linked end can be released by tyrosyl-DNA phosphodiesterase 2 (TDP2) [59,60]. Once the TOP2cc(which is trapped by etoposide), is captured by TDP2, the latter hydrolyzes the phosphodiester bond between TOP2 and DNA through proteasome degradation [61]. The collaboration between TDP2 and proteasome is not well understood. The study by Schellenberg et al. provided some novel findings [62]. First, zinc finger protein 451 (ZNF451) binds and reshapes TOP2cc, thereby opening the complex and allowing TDP2 to access and interact with the cleavage complex. Second, ZNF451 assists small ubiquitin like modifier 2 (SUMO2) protein in SUMOylation of trapped TOP2cc. The interaction of TDP2 with SUMO2 bound to TOP2cc further facilitates the interaction of TDP2 with the cleavage complex. This activity is instrumental in preventing the formation of the cell-deadly DSB that is typical of TOP2 poisons. To proceed with HR repair, the MRE11-RAD50-NBS1 (MRN) complex cooperates with other repair proteins (e.g., BRCA1 and CtIP) and can directly sever a small segment of DNA ends containing covalently bound TOP2. In case of DNA damage is recognized, the MRN complex recruits and activates ataxia-telangiectasia mutant kinase (ATM) dimers [63]. The activated ATM dimer interacts with the checkpoint kinase, causing the cell cycle to stop at G2/M [64]. Subsequently, the nuclease activity of the MRN complex cleaves the DNA ends, prompting HR repair. Meanwhile, it was demonstrated that MRN, CtIP, and BRCA1 are required to remove TOP2-DNA adducts induced by etoposide treatment, and subsequent excision of TOP2-induced DSB ends. It is revealed that the interaction between CtIP and BRCA1 is necessary for the resistance of cells to etoposide during genomic DNA replication [65]. In addition, the MRN complex is also involved in the repair of NHEJ, and is species-specific in the repair of the hairpin structure, but the exact mechanism is still unclear [66]. The degradation of TOP2 through these two processes will leave free DNA ends, which undergo continuous excision and final repair [67,68].
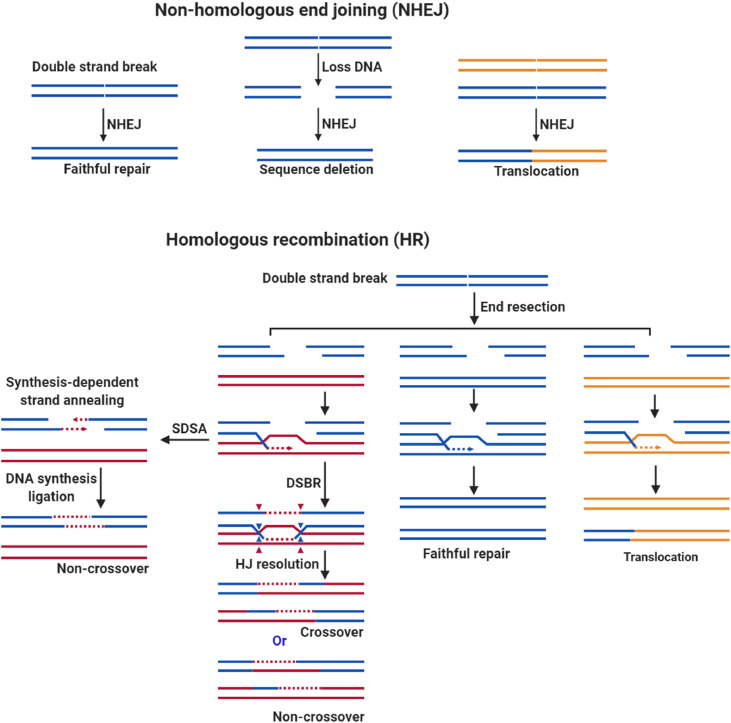
Once TOP2 is released from the DNA-TOP2 cross-linking adduct, NHEJ or HR repair is initiated. NHEJ repairs DSB directly rejoining DNA terminals, which may produce a perfect repair. If the sequence around the lesion is missing or presenting with other DSB, deletion or translocation may occur [69]. Compared to NHEJ, HR is a more precise mechanism where a homologous sequence is used as a template to guide the repair process. The DSB can be repaired by several homologous mediated pathways, including double-strand break repair (DSBR) and synthesis-dependent strand annealing (SDSA), both of the pathways are initiated by 3′ single-strand DNA [70]. The DSBR pathway produces a crossover or non-crossover product, while the SDSA pathway produces a non-crossover product. Crossover is essential for proper chromosome segregation during meiosis and interprets genetic variation. In fact, it does not rule out that a perfect repair can be produced if the template sequence is the same as the sequence with the break sequence. Rarely, if the template is a non-homologous sequence, it may produce a translocation as well [71]. NHEJ is usually involved in the development of lymphocytes, while HR is thought to be more involved in the catalytic recombination of meiosis [71,72], which suggests that the NHEJ pathway is mainly involved in DSB repair induced by TOP2 poisons. In this regard, several studies have emphasized that NHEJ pathway plays a vital role in the TOP2 poison induced DSB (etoposide was specifically listed) [73,74] (Fig. 3).
Fig. 3.
TOP2 poison induces translocation mediated by the DSB repair pathway. TOP2 poison-mediated DSB is likely to be repaired by two pathways, NHEJ or HR repair. NHEJ can repair precisely when the DSB at a low concentration level. If DSB accumulates to a high concentration level, it may cause sequence deletion and translocation. NHEJ mainly contains three types of repair products, faithful repair, sequence loss, and translocation. HR is mediated by a homologous sequence. According to the model sequence, it may produce a different product. In most cases, HR repair uses homologous sequences as templates. This situation generates mainly a crossover or a non-crossover product through two pathways, including SDSA and DSBR, which play an instrumental role in meiosis. In a few cases, precision repair occurs when HR repair uses the same sequence as a template, and translocations may happen when HR uses a non-homologous sequence as a template. The blue and the red chromosomes are homologous, while the orange indicates a non-homologous chromosome. The blue arrowheads are located at one of the Holliday junctions (HJ), and the red arrowheads are located at the other junctions.
Etoposide and chromosomal translocations
A distinct subset of t-AML is associated closely with TOP2 poisons. t-AML shows unique cytogenetic changes, the most common of which is the destruction of the MLL gene on chromosome 11q23 in the 8.3 kb breakpoint cluster region (BCR). The 7 to 13 exon of the MLL gene are located in the BCR, where most of the chromosomal translocation breakpoints are located [15,75]. Several lines of evidence suggested that etoposide caused TOP2cc cleavage activities induced site-specific DNA cleavage in the MLL BRC [76], [77], [78]. Likewise, Martin et al. reported that DNA cleavage within the MLL BCR is a specific event in the initial phase of apoptosis that is part of higher-order chromatin fragmentation leading to chromosomal translocations in the MLL and AML1 genes. In addition, in vitro incubation of etoposide with TOP2 enhanced the DNA-TOP2 cleavage complex near the translocation site within the DNA sequence containing MLL and partner genes identified in susceptible leukemia [77,79,80]. The partner genes found in MLL-related translocations include AF9, AF4, ENL, AF10, AF6, AF17, EPS15, GAS7, LOC100128568, CREBBP, and PTD [81].
Etoposide metabolites contribute to therapy-related leukemia: TOP2 poison
As described above, etoposide metabolites display activity against TOP2 enzymes, which is similar or greater than etoposide [82], [83], [84], [85]. However, studies have shown that etoposide quinone or etoposide catechol has a similar inhibitory activity towards TOP2, when performed with a buffer containing a large amount of reducing agents [26,86,87]. Studies with minimal reducing agents levels have shown that etoposide quinone is several times more effective than the parent compound in inducing TOP2-mediated DSB [83,84]. It was simultaneously demonstrated that the inhibition of TOP2 by etoposide quinone is not limited in the same manner as etoposide by blocking ligation as an interfacial poison [37,[82], [83], [84]]. Numerous studies imply that etoposide quinone is a redox-sensitive compound [84,[88], [89], [90]]. More significantly, etoposide quinone was reported to act as a covalent poison, a mechanism involving covalent adduction , which may lead to multiple effects: inactivation of the enzyme may occur due to blocking of DNA before binding to the enzyme or stabilizing N-terminal clap after DNA binding to the enzyme [82]. In addition, etoposide quinone is a covalent poison of TOP2α and TOP2β, and its reactivity to TOP2β is reported to be slightly higher than that of TOP2α [85]. Meanwhile, studies have shown that etoposide catechol is 2–3 folds more potent than etoposide and can induce high levels of double-stranded DNA cleavage under oxidative conditions [83]. Compared with etoposide, its metabolites are several times more potent on TOP2, and they induce higher levels of DSB under oxidative reaction conditions.
Benzene is a carcinogen that can induce AML in humans and various tumors in animals. According to several reports, benzene metabolites, including benzoquinone (BQ), are responsible for the genotoxic and leukemogenic effects of benzene . In particular, benzoquinone can directly modify DNA, causing genotoxicity and mutagenesis [91,92]. BQ is a TOP2 poison as well, similar to etoposide quinone, which blocks DNA linkage and produces TOP2-DNA adducts [47,93]. In addition, exposure to benzene metabolites in mouse fetal liver is known to increase reactive oxygen species (ROS) [94,95]. ROS has been shown to contribute to the modification of topoisomerases [96,97]. The c-Myb transcription factor has also been shown to be abnormally activated in hematopoietic cells exposed to BQ [98]. In addition, NAD (P)H quinone dehydrogenase 1 (NQO1) is an enzyme that metabolizes BQ into hydroquinone, which is a less reactive compound. C609T mutation encoding an inactive form of NQO1 has been suggested to increase the risk of leukemia with chromosome 11q23 translocations [99,100](particularly prominent in acute non-lymphoblastic leukemia, where the MLL gene is fused to the AF-4 gene on chromosome 4) [101]
Etoposide metabolites contribute to therapy-related leukemia: non-TOP2 factors
Exposure to benzene is correlated with the deregulation of the expression of specific genes associated with leukemia and DNA methylation changes [102,103]. Chromatin components, in particular histones may also react with etoposide (or its metabolite) as shown for histone H1 [104]. The benzene metabolite 1,2,4-benzenetriol can influence DNA methylation and histone acetylation in K562 cells [105]. Recently, etoposide quinone has been shown to inhibit certain enzymes, such as the histone acetyltransferase CREBBPwhich is known to be involved in the leukemogenesis [106]. CREBBP(also known as CBP or KAT6A) regulates both normal and malignant hematopoiesis [107], and the majority of Crebbp+/− or Crebbp−/− mice develop malignant hematopoietic pathologies indicating that CREBBP acts as a tumor suppressor and plays a crucial role in hematopoiesis [108], [109], [110]. We recently showed that etoposide quinone can inhibit CREBBP acetyltransferase activity by reacting with cysteine within zinc fingers domains that are key for CREBBP activity[106].
TCPTP (also known as PTPN2) is a member of the protein tyrosine phosphatase (PTP) family, which negatively regulates JAK/STAT (Janus kinase/signaling transducer and activator of transcription) signaling, through dephosphorylation of different tyrosine phosphorylated JAK/STAT proteins, for instance, STAT1 or JAK1. TCPTP plays an essential role in normal and malignant hematopoiesis by regulating the JAK/STAT pathway [111,112]. Deletions or inactivating mutations of TCPTP were identified in non-Hodgkin's lymphoma and T-cell leukemia and associated with elevated STAT signaling and changes in gene expression [112], [113], [114]. The loss of TCPTP phosphatase activity may lead to excessive activation of the JAK/STAT pathway, which further changes the development of HSC. Etoposide and benzene metabolites, including etoposide quinone and benzoquinone, can irreversibly inhibit TCPTP activity, suggesting another new mechanisms for etoposide and benzene to induction of leukemia [115,116]. (Fig. 4)
Fig. 4.
The underlying mechanism of etoposide induced therapy-related leukemia. TOP2 is an essential as well as a genotoxic enzyme. Typically, the TOP2, DNA, and TOP2-DNA complex are maintained precisely to perform the relevant cellular functions. If the TOP2 enzyme activity is insufficient, the TOP2-DNA complex is lacking, resulting in the inability to release the torsional stress, leading to cell division failure and eventual death due to the failure to unwind the daughter chromosomes. In contrast, TOP2 poisons inactivate the enzymes via interfacial or covalent interaction, resulting in irreversible damage to the TOP2-DNA complex and ultimately creating DSB. Accumulation of DSB inhibits fundamental DNA processes and initiates recombination/repair pathways that produce chromosomal translocations and mutations. If the DSB overwhelms the cells, they will trigger apoptosis, which is the primary anticancer mechanism of etoposide. If the concentration of TOP2 mediated DSB is limited to induce apoptosis, mutations or chromosomal aberrations could occur, and it is possible to develop cancers. Etoposide caused therapy-related leukemia is a chromosomal rearrangement involving the MLL gene on chromosome 11q23. Meantime, etoposide quinone inhibits CREBBP or TCPTP, which has been reported to be associated with leukemogenesis.
Conclusion and further challenge
In summary, etoposide and its metabolite etoposide quinone can inhibit TOP2 enzymes, and the accumulation of DSB is the main cause of secondary leukemia. There is still a need to obtain more precise evidence to prove that CREBBP and TCPTP are involved treatment-related leukemia. However, it has been established that etoposide quinone could contribute more importantly to the development of leukemia than etoposide through TOP2 inhibition. It is likely that other mechanisms such as CREBBP and TCPTP inhibition, may also contribute to etoposide treatment-related leukemia.
Similarly, a large amount of evidence has been established that metabolites of benzene are involved in leukemogenesis through other mechanisms than TOP2 inhibition. For instance, chronic occupational exposure to benzene leads to an increase in the expression of genes involved in apoptosis in blood cells [117,118]. Benzene metabolite triggers apoptosis of hematopoietic progenitor cells in a dose-dependent manner in bone marrow [119]. Meanwhile, benzene affects cell apoptosis by inhibiting caspase-3 [120,121], which may promote the survival of cancer cells. It is also well known that inappropriate activation or expression of c-Myb is involved in the process of leukemia. The c-Myb transcription factor has also been shown to be abnormally activated in K562 and mouse models of hematopoietic cells exposed to 1,4-benzoquinone [122,123]. Moreover, the duration of exposure to benzene is associated with the dysregulation of specific gene expression associated with leukemia and changes in DNA methylation [102,103].
Based on the existence of homologous histone acetyltransferases or protein tyrosine phosphatases , it is likely that etoposide quinone may affect homologs of CREBBP and TCPTP. For example, p300, the homolog of CREBBP, is structurally and functionally similar to CREBBP, notably, on the acetyltransferase catalytic core , including RING andPHD domains. Etoposide quinone covalently binds to CREBBP in the RING and PHD domains [106]. Indeed, p300 has similar RING and PHD domains , It is thus likely that etoposide quinone may also inhibit the acetyltransferase activity of p300. Other histone acetyltransferases , such as TIP60, MOZ, and MOF, contain structural zinc fingers and may also be targets of etoposide quinone. Acetyltransferases, particularly CREBBP/p300, are involved in HR repair by acetylating Recombinant DNA repair protein 52 (RAD52) [124,125]. TIP60 plays a direct role in activating ATM (an essential DSB repair protein kinase) by acetylating it in response to DNA damage [126,127]. Therefore, the loss of acetyltransferase activity may directly affect DSB repair.
The catalytic cysteine 216 is an essential residue of TCPTP and participates to the enzymatic reaction.Cysteine 216 is a evolutionary conserved residue in the PTP family [128]. Therefore, etoposide quinone may inhibit other PTP family members, possibly leading to a broad loss of PTP activity in cells. This may affect JAK/STAT signal transductionwhich plays a key role inHSC differentiation and proliferation. In parallel, a study reported that deficiency in protein-tyrosine phosphatase 1B (PTP1B) is involved in the development of acute leukemia [129]. In addition, SHP2 (Src homology region 2 domain containing phosphatase 2, also known as PTPN11) is related to hematopoiesis and leukemia [130].
As suggested for benzene-induced leukemia, etoposide-induced secondary leukemia are likely to rely on different mechanisms of actions affecting several and distinct biological processes. More studies are needed to understand the pathogenesis of etoposide therapy-related leukemia. This mayfacilitates the determination of the optimal therapeutic administration methods, including the timing and dosage and thereby advances the prevention and treatment of etoposide therapy-related leukemia.
Declaration of Competing Interest
All authors declare no conflicts of interest.
Acknowledgments
This work was supported by running grants from Université de Paris, CNRS, ANSES (Agence Nationale de Sécurité Sanitaire de l’Alimentation, de l’Environnement et du Travail) and ITMO Cancer (plan Cancer-Environnement). WZ and PG were supported by the China Scholarship Council PhD fellowships. We thank Dr. Rose Ann Padua for English language manuscript reading and comments.
Author contributions
WZ, and PG designed the review. JMD, CC and FRL provided expertise. All authors contributed intellectually and to the writing of the manuscript.
Contributor Information
Wenchao Zhang, Email: zhangwch611@gmail.com.
Fernando Rodrigues-Lima, Email: fernando.rodrigues-lima@u-paris.fr.
References
- 1.Meresse P., Dechaux E., Monneret C., Bertounesque E. Etoposide: discovery and medicinal chemistry. Curr. Med. Chem. 2012;11:2443–2466. doi: 10.2174/0929867043364531. [DOI] [PubMed] [Google Scholar]
- 2.Carney D.N., Grogan L., Smit E.F., Harford P., Berendsen H.H., Postmus P.E. Single-agent oral etoposide for elderly small cell lung cancer patients. Semin. Oncol. 1990 doi: 10.1016/0169-5002(90)90229-f. [DOI] [PubMed] [Google Scholar]
- 3.Liang J., Bi N., Wu S., Chen M., Lv C., Zhao L., Shi A., Jiang W., Xu Y., Zhou Z., Wang W., Chen D., Hui Z., Lv J., Zhang H., Feng Q., Xiao Z., Wang X., Liu L., Zhang T., Du L., Chen W., Shyr Y., Yin W., Li J., He J., Wang L. Etoposide and cisplatin versus paclitaxel and carboplatin with concurrent thoracic radiotherapy in unresectable stage III non-small cell lung cancer: a multicenter randomized phase III trial. Ann. Oncol. 2017 doi: 10.1093/annonc/mdx009. [DOI] [PubMed] [Google Scholar]
- 4.Dasari S., Tchounwou P.Bernard. Cisplatin in cancer therapy: molecular mechanisms of action. Eur. J. Pharmacol. 2014 doi: 10.1016/j.ejphar.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y., Zou S., Zhao Z., Liu P., Ke C., Xu S. New insights into small-cell lung cancer development and therapy. Cell Biol. Int. 2020 doi: 10.1002/cbin.11359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saito T., Hatta Y., Hayakawa F., Takahashi T., Hagihara M., Iida H., Minauchi K., Yamazaki E., Sugiura I., Murayama T., Sakura T., Mori N., Imai K., Yahagi Y., Atsuta Y., Saito A.M., Hirakawa A., Kiyoi H., Matsumura I., Miyazaki Y. Combination of clofarabine, etoposide, and cyclophosphamide in adult relapsed/refractory acute lymphoblastic leukemia: a phase 1/2 dose-escalation study by the Japan adult leukemia study group. Int. J. Hematol. 2021 doi: 10.1007/s12185-020-03032-3. [DOI] [PubMed] [Google Scholar]
- 7.Qiu Z., Lin A., Li K., Lin W., Wang Q., Wei T., Zhu W., Luo P., Zhang J. A novel mutation panel for predicting etoposide resistance in small-cell lung cancer. Drug Des. Dev. Ther. 2019 doi: 10.2147/DDDT.S205633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rossi A., Di Maio M., Chiodini P., Rudd R.M., Okamoto H., Skarlos D.V., Früh M., Qian W., Tamura T., Samantas E., Shibata T., Perrone F., Gallo C., Gridelli C., Martelli O., Lee S.M. Carboplatin- or cisplatin-based chemotherapy in first-line treatment of small-cell lung cancer: the COCIS meta-analysis of individual patient data. J. Clin. Oncol. 2012 doi: 10.1200/JCO.2011.40.4905. [DOI] [PubMed] [Google Scholar]
- 9.Nemade H., Chaudhari U., Acharya A., Hescheler J., Hengstler J.G., Papadopoulos S., Sachinidis A. Cell death mechanisms of the anti-cancer drug etoposide on human cardiomyocytes isolated from pluripotent stem cells. Arch. Toxicol. 2018 doi: 10.1007/s00204-018-2170-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorigo A., Mansberg R., Kwan Y.L. Lomustine, etoposide, methotrexate and prednisone (LEMP) therapy for relapsed and refractory non-Hodgkin's lymphoma. Eur. J. Haematol. 1993 doi: 10.1111/j.1600-0609.1993.tb00072.x. [DOI] [PubMed] [Google Scholar]
- 11.Henwood R.B.JM. Etoposide. A review of its pharmacodynamic and pharmacokinetic properties, and therapeutic potential in combination chemotherapy of cancer. Drugs. 1990;39(3):438. doi: 10.2165/00003495-199039030-00008. [DOI] [PubMed] [Google Scholar]
- 12.Alifrangis C., Agarwal R., Short D., Fisher R.A., Sebire N.J., Harvey R., Savage P.M., Seckl M.J. EMA/CO for high-risk gestational trophoblastic neoplasia: good outcomes with induction low-dose etoposide-cisplatin and genetic analysis. J. Clin. Oncol. 2013;31:280–286. doi: 10.1200/JCO.2012.43.1817. [DOI] [PubMed] [Google Scholar]
- 13.Jr S.G. Etoposide in the management of metastatic breast cancer. Cancer. 1991;1(1 Sup) doi: 10.1002/1097-0142(19910101)67. 67https://doi.org/doi::1+<266::aid-cncr2820671310>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 14.RF O. Oral etoposide for the treatment of recurrent ovarian cancer. Drugs. 1999;58(Suppl 3) doi: 10.2165/00003495-199958003-00007. https://doi.org/doi. [DOI] [PubMed] [Google Scholar]
- 15.Ezoe S. Secondary leukemia associated with the anti-cancer agent, etoposide, a topoisomerase II inhibitor. Int. J. Environ. Res. Public Health. 2012;9:2444–2453. doi: 10.3390/ijerph9072444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pedersen-Bjergaard J., Daugaard G., Hansen S.W., R o rth M., Philip P., Larsen S.O. Increased risk of myelodysplasia and leukaemia after etoposide, cisplatin, and bleomycin for germ-cell tumours. Lancet. 1991 doi: 10.1016/0140-6736(91)90490-G. [DOI] [PubMed] [Google Scholar]
- 17.Shimada N., Ohno N., Tanosaki R., Fuji S., Suzuki Y., Yuji K., Uchimaru K., Tojo A. Therapy-related acute myeloid leukemia after the long-term administration of low-dose etoposide for chronic-type adult T-cell leukemia-lymphoma: a case report and literature review. Intern. Med. 2017 doi: 10.2169/internalmedicine.56.7763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The US Food and drug administration, Acute myeloid leukemia: developing drugs and biological products for treatment, (n.d.) 1–36. 2020. https://www.fda.gov/media/140821/download.
- 19.Agency E.M. https://www.ema.europa.eu/en/documents/referral/etopophos-article-30-referral-chmp-assessment-report_en.pdf
- 20.Relling M.V., Nemec J., Schuetz E.G., Schuetz J.D., Gonzalez F.J., Korzekwa K.R. O-demethylation of epipodophyllotoxins is catalyzed by human cytochrome P450 3A4. Mol. Pharmacol. 1994 [PubMed] [Google Scholar]
- 21.Luo G., Guenthner T., Gan L.-.S., Humphreys W. CYP3A4 induction by xenobiotics: biochemistry, experimental methods and impact on drug discovery and development. Curr. Drug Metab. 2005 doi: 10.2174/1389200043335397. [DOI] [PubMed] [Google Scholar]
- 22.Kishi S., Yang W., Boureau B., Morand S., Das S., Chen P., Cook E.H., Rosner G.L., Schuetz E., Pui C.H., Relling M.V. Effects of prednisone and genetic polymorphisms on etoposide disposition in children with acute lymphoblastic leukemia. Blood. 2004 doi: 10.1182/blood-2003-06-2105. [DOI] [PubMed] [Google Scholar]
- 23.Fan Y., Schreiber E.M., Giorgianni A., Yalowich J.C., Day B.W. Myeloperoxidase-catalyzed metabolism of etoposide to its quinone and glutathione adduct forms in HL60 cells. Chem. Res. Toxicol. 2006 doi: 10.1021/tx0600595. [DOI] [PubMed] [Google Scholar]
- 24.Yang J., Bogni A., Schuetz E.G., Ratain M., Dolan M.Eileen, McLeod H., Gong L., Thorn C., Relling M.V., Klein T.E., Altman R.B. Etoposide pathway. Pharmacogenet. Genom. 2009 doi: 10.1097/FPC.0b013e32832e0e7f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haim N., Sinha B.K., Nemec J., Roman J. Peroxidase-catalyzed metabolism of etoposide (VP-16-213) and covalent binding of reactive intermediates to cellular macromolecules. Cancer Res. 1987 [PubMed] [Google Scholar]
- 26.Lovett B.D., Strumberg D., Blair I.A., Pang S., Burden D.A., Megonigal M.D., Rappaport E.F., Rebbeck T.R., Osheroff N., Pommier Y.G., Felix C.A. Etoposide metabolites enhance DNA topoisomerase II cleavage near leukemia-associated MLL translocation breakpoints. Biochemistry. 2001 doi: 10.1021/bi002361x. [DOI] [PubMed] [Google Scholar]
- 27.Wen Z., Tallman M.N., Ali S.Y., Smith P.C. UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human liver and intestinal microsomes: structural characterization of phenolic and alcoholic glucuronides of etoposide and estimation of enzyme kinetics. Drug Metab. Dispos. 2007 doi: 10.1124/dmd.106.012732. [DOI] [PubMed] [Google Scholar]
- 28.Watanabe Y., Nakajima M., Ohashi N., Kume T., Yokoi T. Glucuronidation of etoposide in human liver microsomes is specifically catalyzed by UDP-glucuronosyltransferase 1A1. Drug Metab. Dispos. 2003 doi: 10.1124/dmd.31.5.589. [DOI] [PubMed] [Google Scholar]
- 29.Hainsworth J.D., Greco F.A. Etoposide: twenty years later. Ann. Oncol. 1995;6:325–341. doi: 10.1093/oxfordjournals.annonc.a059180. [DOI] [PubMed] [Google Scholar]
- 30.Grieder A., Maurer R., Stähelin H. Effect of an Epipodophyllotoxin derivative (VP 16-213) on macromolecular synthesis and mitosis in mastocytoma cells in vitro. Cancer Res. 1974 [PubMed] [Google Scholar]
- 31.Colombo T., Broggini M., Vaghi M. Comparison between VP 16 and VM 26 in Lewis lung carcinoma of the mouse. Eur. J. Cancer Clin. Oncol. 1986 doi: 10.1016/0277-5379(86)90027-1. [DOI] [PubMed] [Google Scholar]
- 32.Roed H., Vindelov L.L., Christensen I.J., Spang-Thomsen M., Hansen H.H. The effect of the two epipodophyllotoxin derivatives etoposide (VP-16) and teniposide (VM-26) on cell lines established from patients with small cell carcinoma of the lung. Cancer Chemother. Pharmacol. 1987 doi: 10.1007/BF00296248. [DOI] [PubMed] [Google Scholar]
- 33.Kasahara K., Fujiwara Y., Sugimoto Y., Nishio K., Tamura T., Matsuda T., Saijo N. Determinants of response to the DNA topoisomerase II inhibitors doxorubicin and etoposide in human lung cancer cell lines. J. Natl. Cancer Inst. 1992;84:113–118. doi: 10.1093/jnci/84.2.113. [DOI] [PubMed] [Google Scholar]
- 34.Edwards C.M., Glisson B.S., King C.K., Smallwood-Kentro S., Ross W.E. Etoposide-induced DNA cleavage in human leukemia cells. Cancer Chemother. Pharmacol. 1987;20:162–168. doi: 10.1007/BF00253972. [DOI] [PubMed] [Google Scholar]
- 35.Mitelman F., Johansson B., Mertens F. The impact of translocations and gene fusions on cancer causation. Nat. Rev. Cancer. 2007 doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 36.Mertens F., Johansson B., Fioretos T., Mitelman F. The emerging complexity of gene fusions in cancer. Nat. Rev. Cancer. 2015 doi: 10.1038/nrc3947. [DOI] [PubMed] [Google Scholar]
- 37.M.P. Topoisomerase II and leukemia MaryJean. Ann N Y Acad Sci. 2015;1310:98–110. doi: 10.1111/nyas.12358.Topoisomerase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gómez-Herreros F. DNA Double Strand Breaks and Chromosomal Translocations Induced by DNA Topoisomerase II. Front. Mol. Biosci. 2019;6:1–7. doi: 10.3389/fmolb.2019.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deweese J.E., Osheroff N. The DNA cleavage reaction of topoisomerase II: wolf in sheep's clothing. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkn937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vos S.M., Tretter E.M., Schmidt B.H., Berger J.M. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 2011 doi: 10.1038/nrm3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pommier Y. Drugging topoisomerases: lessons and Challenges. ACS Chem. Biol. 2013 doi: 10.1021/cb300648v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McClendon A.K., Osheroff N. DNA topoisomerase II, genotoxicity, and cancer. Mutat. Res. - Fundam. Mol. Mech. Mutagen. 2007 doi: 10.1016/j.mrfmmm.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matias-Barrios V.M., Radaeva M., Song Y., Alperstein Z., Lee A.R., Schmitt V., Lee J., Ban F., Xie N., Qi J., Lallous N., Gleave M.E., Cherkasov A., Dong X. Discovery of new catalytic topoisomerase II inhibitors for anticancer therapeutics. Front. Oncol. 2021 doi: 10.3389/fonc.2020.633142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pommier Y., Marchand C. Interfacial inhibitors: targeting macromolecular complexes. Nat. Rev. Drug Discov. 2012 doi: 10.1038/nrd3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu C.C., Li T.K., Farh L., Lin L.Y., Lin T.S., Yu Y.J., Yen T.J., Chiang C.W., Chan N.L. Structural basis of type II topoisomerase inhibition by the anticancer drug etoposide. Science. 2011;(80) doi: 10.1126/science.1204117. [DOI] [PubMed] [Google Scholar]
- 46.Ketron A.C., Osheroff N. Phytochemicals as anticancer and chemopreventive topoisomerase II poisons. Phytochem. Rev. 2014 doi: 10.1007/s11101-013-9291-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nitiss J.L. Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer. 2009 doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar C.C. Genetic abnormalities and challenges in the treatment of acute myeloid Leukemia. Genes Cancer. 2011;2:95–107. doi: 10.1177/1947601911408076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ratain M.J., Rowley J.D. Review: therapy-related acute myeloid leukemia secondary to inhibitors of topoisomerase II: from the bedside to the target genes. Ann. Oncol. 1992;3:107–111. doi: 10.1093/oxfordjournals.annonc.a058121. [DOI] [PubMed] [Google Scholar]
- 50.Domer P.H., Head D.R., Renganathan N., Raimondi S.C., Yang E., Atlas M. Molecular analysis of 13 cases of MLL/11q23 secondary acute leukemia and identification of topoisomerase II consensus-binding sequences near the chromosomal breakpoint of a secondary leukemia with the t(4;11) Leukemia. 1995 [PubMed] [Google Scholar]
- 51.Ratain M.J., Kaminer L.S., Bitran J.D., Larson R.A., Le Beau M.M., Skosey C., Purl S., Hoffman P.C., Wade J., Vardiman J.W., Daly K., Rowley J.D., Golomb H.M. Acute nonlymphocytic leukemia following etoposide and cisplatin combination chemotherapy for advanced non-small-cell carcinoma of the lung. Blood. 1987 doi: 10.1182/blood.v70.5.1412.1412. [DOI] [PubMed] [Google Scholar]
- 52.Le Deley M.C., Vassal G., Taïbi A., Shamsaldin A., Leblanc T., Hartmann O. High cumulative rate of secondary leukemia after continuous etoposide treatment for solid tumors in children and young adults. Pediatr. Blood Cancer. 2005 doi: 10.1002/pbc.20380. [DOI] [PubMed] [Google Scholar]
- 53.Winick N.J., McKenna R.W., Shuster J.J., Schneider N.R., Borowitz M.J., Bowman W.P., Jacaruso D., Kamen B.A., Buchanan G.R. Secondary acute myeloid leukemia in children with acute lymphoblastic leukemia treated with etoposide. J. Clin. Oncol. 1993;11:209–217. doi: 10.1200/JCO.1993.11.2.209. [DOI] [PubMed] [Google Scholar]
- 54.Sugita K., Furukawa T., Tsuchida M., Okawa Y., Nakazawa S., Akatsuka J., Ohira M., Nishimura K. High frequency of etoposide (vp-16)-related secondary leukemia in children with non-hodgkin's lymphoma. J. Pediatr. Hematol. Oncol. 1993 doi: 10.1097/00043426-199302000-00013. [DOI] [PubMed] [Google Scholar]
- 55.Kollmannsberger C., Beyer J., Droz J.P., Harstrick A., Hartmann J.T., Biron P., Fléchon A., Schöffski P., Kuczyk M., Schmoll H.J., Kanz L., Bokemeyer C. Secondary leukemia following high cumulative doses of etoposide in patients treated for advanced germ cell tumors. J. Clin. Oncol. 1998;16:3386–3391. doi: 10.1200/JCO.1998.16.10.3386. [DOI] [PubMed] [Google Scholar]
- 56.Smith M.A., Rubinstein L., Anderson J.R., Arthur D., Catalano P.J., Freidlin B., Heyn R., Khayat A., Krailo M., Land V.J., Miser J., Shuster J., Vena D. Secondary leukemia or myelodysplastic syndrome after treatment with epipodophyllotoxins. J. Clin. Oncol. 1999 doi: 10.1200/jco.1999.17.2.569. [DOI] [PubMed] [Google Scholar]
- 57.Haupt R., Fears T.R., Heise A., Gadner H., Loiacono G., De Terlizzi M., Tucker M.A. Risk of secondary leukemia after treatment with etoposide (VP-16) for Langerhans’ cell histiocytosis in Italian and Australian-German populations. Int. J. Cancer. 1997 doi: 10.1002/(SICI)1097-0215(19970328)71. 1<9::AID-IJC3>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 58.Nichols C.R., Breeden E.S., Loehrer P.J., Williams S.D., Einhorn L.H. Secondary leukemia associated with a conventional dose of etoposide: review of serial germ cell tumor protocols. J. Natl. Cancer Inst. 1993 doi: 10.1093/jnci/85.1.36. [DOI] [PubMed] [Google Scholar]
- 59.Sciascia N., Wu W., Zong D., Sun Y., Wong N., John S., Wangsa D., Ried T., Bunting S.F., Pommier Y., Nussenzweig A. Suppressing proteasome mediated processing of topoisomerase II DNA-protein complexes preserves genome integrity. Elife. 2020 doi: 10.7554/eLife.53447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Szlachta K., Manukyan A., Raimer H.M., Singh S., Salamon A., Guo W., Lobachev K.S., Wang Y.H. Topoisomerase II contributes to DNA secondary structure-mediated double-stranded breaks. Nucleic Acids Res. 2020 doi: 10.1093/nar/gkaa483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsuda M., Kitamasu K., Hosokawa S., Nakano T., Ide H. Repair of trapped topoisomerase II covalent cleavage complexes: novel proteasome-independent mechanisms. Nucleosides, Nucleot. Nucleic Acids. 2020 doi: 10.1080/15257770.2019.1674332. [DOI] [PubMed] [Google Scholar]
- 62.Schellenberg M.J., Lieberman J.A., Herrero-Ruiz A., Butler L.R., Williams J.G., Muñoz-Cabello A.M., Mueller G.A., London R.E., Cortés-Ledesma F., Williams R.S. ZATT (ZNF451)–mediated resolution of topoisomerase 2 DNA-protein cross-links. Science. 2017;(80) doi: 10.1126/science.aam6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jackson S.P. Sensing and repairing DNA double-strand breaks. Carcinogenesis. 2002 doi: 10.1093/carcin/23.5.687. [DOI] [PubMed] [Google Scholar]
- 64.Anuranjani M.Bala. Concerted action of Nrf2-ARE pathway, MRN complex, HMGB1 and inflammatory cytokines - Implication in modification of radiationdamage. Redox Biol. 2014 doi: 10.1016/j.redox.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aparicio T., Baer R., Gottesman M., Gautier J. MRN, CtIP, and BRCA1 mediate repair of topoisomerase II-DNA adducts. J. Cell Biol. 2016 doi: 10.1083/jcb.201504005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qiu S., Huang J. MRN complex is an essential effector of DNA damage repair. J. Zhejiang Univ. Sci. B. 2021 doi: 10.1631/jzus.B2000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deshpande R.A., Lee J.H., Arora S., Paull T.T. Nbs1 converts the human Mre11/Rad50 nuclease complex into an endo/exonuclease machine specific for protein-DNA adducts. Mol. Cell. 2016 doi: 10.1016/j.molcel.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 68.Anand R., Jasrotia A., Bundschuh D., Howard S.M., Ranjha L., Stucki M., Cejka P. NBS1 promotes the endonuclease activity of the MRE11-RAD50 complex by sensing CtIP phosphorylation. EMBO J. 2019 doi: 10.15252/embj.2018101005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lieber M.R. The biochemistry and biological significance of nonhomologous DNA end joining: an essential repair process in multicellular eukaryotes. Genes to Cells. 1999;4:77–85. doi: 10.1046/j.1365-2443.1999.00245.x. [DOI] [PubMed] [Google Scholar]
- 70.A.T. Do, J.T. Brooks, M.K. Le Neveu, J.R. Larocque, Double-strand break repair assays determine pathway choice and structure of gene conversion events in drosophila melanogaster, 4 (2014) 425–432. https://doi.org/10.1534/g3.113.010074. [DOI] [PMC free article] [PubMed]
- 71.Ferguson D.O., Alt F.W. DNA double strand break repair and chromosomal translocation: lessons from animal models. Oncogene. 2001 doi: 10.1038/sj.onc.1204767. [DOI] [PubMed] [Google Scholar]
- 72.T. Hatkevich, D.E. Miller, J. Sekelsky, C.A. Turcotte, M.C. Miller, A pathway for error-free non-homologous end joining of resected meiotic double-strand breaks, 49 (2021) 879–890. https://doi.org/10.1093/nar/gkaa1205. [DOI] [PMC free article] [PubMed]
- 73.de Campos-Nebel M., Larripa I., González-Cid M. Topoisomerase ii-mediated DNA damage is differently repaired during the cell cycle by non-homologous end joining and homologous recombination. PLoS ONE. 2010;5:1–13. doi: 10.1371/journal.pone.0012541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Malik M., Nitiss K.C., Enriquez-Rios V., Nitiss J.L. Roles of nonhomologous end-joining pathways in surviving topoisomerase II-mediated DNA damage. Mol. Cancer Ther. 2006 doi: 10.1158/1535-7163.MCT-05-0263. [DOI] [PubMed] [Google Scholar]
- 75.Broeker P., Super H., Thirman M., Pomykala H., Yonebayashi Y., Tanabe S., Zeleznik-Le N., Rowley J. Distribution of 11q23 breakpoints within the MLL breakpoint cluster region in de novo acute leukemia and in treatment-related acute myeloid leukemia: correlation with scaffold attachment regions and topoisomerase II consensus binding sites. Blood. 1996 doi: 10.1182/blood.v87.5.1912.bloodjournal8751912. [DOI] [PubMed] [Google Scholar]
- 76.Winters A.C., Bernt K.M. MLL-rearranged leukemias- An update on science and clinical approaches. Front. Pediatr. 2017 doi: 10.3389/fped.2017.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stanulla M., Wang J., Chervinsky D.S., Thandla S., Aplan P.D. DNA cleavage within the MLL breakpoint cluster region is a specific event which occurs as part of higher-order chromatin fragmentation during the initial stages of apoptosis. Mol. Cell. Biol. 1997;17:4070–4079. doi: 10.1128/mcb.17.7.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Strissel P.L., Strick R., Rowley J.D., Zeleznik-Le N.J. An in vivo topoisomerase II cleavage site and a DNase I hypersensitive site colocalize near exon 9 in the MLL breakpoint cluster region. Blood. 1998 doi: 10.1182/blood.v92.10.3793.422a24_3793_3803. [DOI] [PubMed] [Google Scholar]
- 79.Whitmarsh R.J., Saginario C., Zhuo Y., Hilgenfeld E., Rappaport E.F., Megonigal M.D., Carroll M., Liu M., Osheroff N., Cheung N.K.V., Slater D.J., Ried T., Knutsen T., Blair I.A., Felix C.A. Reciprocal DNA topoisomerase II cleavage events at 5′-TATTA-3′ sequences in MLL and AF-9 create homologous single-stranded overhangs that anneal to form der(11) and der(9) genomic breakpoint junctions in treatment-related AML without further processing. Oncogene. 2003 doi: 10.1038/sj.onc.1207052. [DOI] [PubMed] [Google Scholar]
- 80.Ng A., Taylor G.M., Eden O.B. Genotoxicity of etoposide: greater susceptibility of MLL than other target genes. Cancer Genet. Cytogenet. 2006;164:164–167. doi: 10.1016/j.cancergencyto.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 81.Meyer C., Burmeister T., Gröger D., Tsaur G., Fechina L., Renneville A., Sutton R., Venn N.C., Emerenciano M., Pombo-De-Oliveira M.S., Barbieri Blunck C., Almeida Lopes B., Zuna J., Trka J., Ballerini P., Lapillonne H., De Braekeleer M., Cazzaniga G., Corral Abascal L., Van Der Velden V.H.J., Delabesse E., Park T.S., Oh S.H., Silva M.L.M., Lund-Aho T., Juvonen V., Moore A.S., Heidenreich O., Vormoor J., Zerkalenkova E., Olshanskaya Y., Bueno C., Menendez P., Teigler-Schlegel A., Zur Stadt U., Lentes J., Göhring G., Kustanovich A., Aleinikova O., Schäfer B.W., Kubetzko S., Madsen H.O., Gruhn B., Duarte X., Gameiro P., Lippert E., Bidet A., Cayuela J.M., Clappier E., Alonso C.N., Zwaan C.M., Van Den Heuvel-Eibrink M.M., Izraeli S., Trakhtenbrot L., Archer P., Hancock J., Möricke A., Alten J., Schrappe M., Stanulla M., Strehl S., Attarbaschi A., Dworzak M., Haas O.A., Panzer-Grümayer R., Sedék L., Szczepa T., Caye A., Suarez L., Cavé H., Marschalek R. The MLL recombinome of acute leukemias in 2017. Leukemia. 2018 doi: 10.1038/leu.2017.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gibson E.G., King M.M., Mercer S.L., Deweese J.E. Two-mechanism model for the interaction of etoposide quinone with topoisomerase IIα. Chem. Res. Toxicol. 2016;29:1541–1548. doi: 10.1021/acs.chemrestox.6b00209. [DOI] [PubMed] [Google Scholar]
- 83.Jacob D.A., Gibson E.G., Mercer S.L., Deweese J.E. Etoposide catechol is an oxidizable topoisomerase II poison. Chem. Res. Toxicol. 2013;26:1156–1158. doi: 10.1021/tx400205n. [DOI] [PubMed] [Google Scholar]
- 84.Jacob D.A., Mercer S.L., Osheroff N., Deweese J.E. Etoposide quinone is a redox-dependent topoisomerase II poison. Biochemistry. 2011 doi: 10.1021/bi200438m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Smith N.A., Byl J.A.W., Mercer S.L., Deweese J.E., Osheroff N. Etoposide quinone is a covalent poison of human topoisomerase IIβ. Biochemistry. 2014;53:3229–3236. doi: 10.1021/bi500421q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gantchev T.G., Hunting D.J. Inhibition of the topoisomerase II-DNA cleavable complex by the ortho-quinone derivative of the antitumor drug etoposide (VP-16) Biochem. Biophys. Res. Commun. 1997 doi: 10.1006/bbrc.1997.7063. [DOI] [PubMed] [Google Scholar]
- 87.Gantchev T.G., Hunting D.J. The ortho-quinone metabolite of the anticancer drug etoposide (VP-16) is a potent inhibitor of the topoisomerase II/DNA cleavable complex. Mol. Pharmacol. 1998 doi: 10.1124/mol.53.3.422. [DOI] [PubMed] [Google Scholar]
- 88.Lindsey R.H., Bender R.P., Osheroff N. Stimulation of topoisomerase II-mediated DNA cleavage by benzene metabolites. Chem. Biol. Interact. 2005;153–154:197–205. doi: 10.1016/j.cbi.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 89.Bender R.P., Lehmler H.J., Robertson L.W., Ludewig G., Osheroff N. Polychlorinated biphenyl quinone metabolites poison human topoisomerase IIα: altering enzyme function by blocking the N-terminal protein gate. Biochemistry. 2006 doi: 10.1021/bi0524666. [DOI] [PubMed] [Google Scholar]
- 90.Mondrala S., Eastmond D.A. Topoisomerase II inhibition by the bioactivated benzene metabolite hydroquinone involves multiple mechanisms. Chem. Biol. Interact. 2010;184:259–268. doi: 10.1016/j.cbi.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 91.Bolton J.L., Trush M.A., Penning T.M., Dryhurst G., Monks T.J. Role of quinones in toxicology. Chem. Res. Toxicol. 2000 doi: 10.1021/tx9902082. [DOI] [PubMed] [Google Scholar]
- 92.Bolton J.L., Dunlap T. Formation and biological targets of quinones: cytotoxic versus cytoprotective effects. Chem. Res. Toxicol. 2017 doi: 10.1021/acs.chemrestox.6b00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lindsey R.H., Bromberg K.D., Felix C.A., Osheroff N. 1,4-Benzoquinone is a topoisomerase II poison. Biochemistry. 2004 doi: 10.1021/bi049756r. [DOI] [PubMed] [Google Scholar]
- 94.Philbrook N.A., Winn L.M. Benzoquinone toxicity is not prevented by sulforaphane in CD-1 mouse fetal liver cells. J. Appl. Toxicol. 2016 doi: 10.1002/jat.3251. [DOI] [PubMed] [Google Scholar]
- 95.Badham H.J., Renaud S.J., Wan J., Winn L.M. Benzene-initiated oxidative stress: effects on embryonic signaling pathways. Chem. Biol. Interact. 2010 doi: 10.1016/j.cbi.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 96.Lu H.R., Zhu H., Huang M., Chen Y., Cai Y.J., Miao Z.H., Zhang J.S., Ding J. Reactive oxygen species elicit apoptosis by concurrently disrupting topoisomerase II and DNA-dependent protein kinase. Mol. Pharmacol. 2005 doi: 10.1124/mol.105.011544. [DOI] [PubMed] [Google Scholar]
- 97.Meng L.H., Ding J. Salvicine, a novel topoisomerase II inhibitor, exerts its potent anticancer activity by ROS generation. Acta Pharmacol. Sin. 2007 doi: 10.1111/j.1745-7254.2007.00698.x. [DOI] [PubMed] [Google Scholar]
- 98.Singh R., Winn L.M. The effects of 1,4-benzoquinone on c-Myb and topoisomerase II in K-562 cells. Mutat. Res. - Fundam. Mol. Mech. Mutagen. 2008 doi: 10.1016/j.mrfmmm.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 99.Wiemels J.L., Pagnamenta A., Taylor G.M., Eden O.B., Alexander F.E., Greaves M.F. A lack of a functional NAD(P)H:quinone oxidoreductase allele is selectively associated with pediatric leukemias that have MLL fusions. Cancer Res. 1999 [PubMed] [Google Scholar]
- 100.Larson R.A., Wang Y., Banerjee M., Wiemels J., Hartford C., Le Beau M.M., Smith M.T. Prevalence of the inactivating 609C → T polymorphism in the NAD(P)H:quinone oxidoreductase (NQO1) gene in patients with primary and therapy-related myeloid leukemia. Blood. 1999 doi: 10.1182/blood.v94.2.803. [DOI] [PubMed] [Google Scholar]
- 101.Smith M.T., Wang Y., Skibola C.F., Slater D.J., Lo Nigro L., Nowell P.C., Lange B.J., Felix C.A. Low NAD(P)H: quinone oxidoreductase activity is associated with increased risk of leukemia with MLL translocations in infants and children. Blood. 2002 doi: 10.1182/blood-2001-12-0264. [DOI] [PubMed] [Google Scholar]
- 102.Li K., Jing Y., Yang C., Liu S., Zhao Y., He X., Li F., Han J., Li G. Increased leukemia-associated gene expression in benzene-exposed workers. Sci. Rep. 2014 doi: 10.1038/srep05369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bollati V., Baccarelli A., Hou L., Bonzini M., Fustinoni S., Cavallo D., Byun H.M., Jiang J., Marinelli B., Pesatori A.C., Bertazzi P.A., Yang A.S. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007 doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- 104.Chamani E., Rabbani-Chadegani A., Zahraei Z. Spectroscopic detection of etoposide binding to chromatin components: the role of histone proteins. Spectrochim. Acta - Part A Mol. Biomol. Spectrosc. 2014 doi: 10.1016/j.saa.2014.05.068. [DOI] [PubMed] [Google Scholar]
- 105.Yu C.H., Li Y., Zhao X., Yang S.Q., Li L., Cui N.X., Rong L., Yi Z.C. Benzene metabolite 1,2,4-benzenetriol changes DNA methylation and histone acetylation of erythroid-specific genes in K562 cells. Arch. Toxicol. 2019;93:137–147. doi: 10.1007/s00204-018-2333-6. [DOI] [PubMed] [Google Scholar]
- 106.Zhang W., Berthelet J., Michail C., Bui L.C., Gou P., Liu R., Duval R., Renault J., Dupret J.M., Guidez F., Chomienne C., Lima F.R. Human CREBBP acetyltransferase is impaired by etoposide quinone, an oxidative and leukemogenic metabolite of the anticancer drug etoposide through modification of redox-sensitive zinc-finger cysteine residues, Free Radic. Biol. Med. 2021 doi: 10.1016/j.freeradbiomed.2020.11.027. [DOI] [PubMed] [Google Scholar]
- 107.Sun X.J., Man N., Tan Y., Nimer S.D., Wang L. The role of histone acetyltransferases in normal and malignant hematopoiesis. Front. Oncol. 2015 doi: 10.3389/fonc.2015.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rebel V.I., Kung A.L., Tanner E.A., Yang H., Bronson R.T., Livingston D.M. Distinct roles for CREB-binding protein and p300 in hematopoietic stem cell self-renewal. Proc. Natl. Acad. Sci. U. S. A. 2002 doi: 10.1073/pnas.232568499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zimmer S.N., Zhou Q., Zhou T., Cheng Z., Abboud-Werner S.L., Horn D., Lecocke M., White R., Krivtsov A.V., Armstrong S.A., Kung A.L., Livingston D.M., Rebel V.I. Crebbp haploinsufficiency in mice alters the bone marrow microenvironment, leading to loss of stem cells and excessive myelopoiesis. Blood. 2011 doi: 10.1182/blood-2010-09-307942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Qian M., Zhang H., Kham S.K.Y., Liu S., Jiang C., Zhao X., Lu Y., Goodings C., Lin T.N., Zhang R., Moriyama T., Yin Z., Li Z., Quah T.C., Ariffin H., Tan A.M., Shen S., Bhojwani D., Hu S., Chen S., Zheng H., Pui C.H., Yeoh A.E.J., Yang J.J. Whole-transcriptome sequencing identifies a distinct subtype of acute lymphoblastic leukemia with predominant genomic abnormalities of EP300 and CREBBP. Genome Res. 2017 doi: 10.1101/gr.209163.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dorritie K.A., McCubrey J.A., Johnson D.E. STAT transcription factors in hematopoiesis and leukemogenesis: opportunities for therapeutic intervention. Leukemia. 2014 doi: 10.1038/leu.2013.192. [DOI] [PubMed] [Google Scholar]
- 112.Pike K.A., Tremblay M.L. TC-PTP and PTP1B: regulating JAK-STAT signaling, controlling lymphoid malignancies. Cytokine. 2016 doi: 10.1016/j.cyto.2015.12.025. [DOI] [PubMed] [Google Scholar]
- 113.Kleppe M., Tousseyn T., Geissinger E., Atak Z.K., Aerts S., Rosenwald A., Wlodarska I., Cools J. Mutation analysis of the tyrosine phosphatase PTPN2 in Hodgkin's lymphoma and T-cell non-Hodgkin's lymphoma. Haematologica. 2011 doi: 10.3324/haematol.2011.041921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kleppe M., Lahortiga I., El Chaar T., De Keersmaecker K., Mentens N., Graux C., Van Roosbroeck K., Ferrando A.A., Langerak A.W., Meijerink J.P.P., Sigaux F., Haferlach T., Wlodarska I., Vandenberghe P., Soulier J., Cools J. Deletion of the protein tyrosine phosphatase gene PTPN2 in T-cell acute lymphoblastic leukemia. Nat. Genet. 2010 doi: 10.1038/ng.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nian Q., Berthelet J., Zhang W., Bui L.C., Liu R., Xu X., Duval R., Ganesan S., Leger T., Chomienne C., Busi F., Guidez F., Dupret J.M., Lima F.R. T-cell protein tyrosine phosphatase is irreversibly inhibited by etoposide-quinone, a reactive metabolite of the chemotherapy drug etoposide. Mol. Pharmacol. 2019 doi: 10.1124/mol.119.116319. [DOI] [PubMed] [Google Scholar]
- 116.Duval R., Bui L.C., Mathieu C., Nian Q., Berthelet J., Xu X., Haddad I., Vinh J., Dupret J.M., Busi F., Guidez F., Chomienne C., Rodrigues-Lima F. Benzoquinone, a leukemogenic metabolite of benzene, catalytically inhibits the protein tyrosine phosphatase PTPN2 and alters STAT1 signaling. J. Biol. Chem. 2019;294:12483–12494. doi: 10.1074/jbc.RA119.008666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McHale C.M., Zhang L., Lan Q., Li G., Hubbard A.E., Forrest M.S., Vermeulen R., Chen J., Shen M., Rappaport S.M., Yin S., Smith M.T., Rothman N. Changes in the peripheral blood transcriptome associated with occupational benzene exposure identified by cross-comparison on two microarray platforms. Genomics. 2009 doi: 10.1016/j.ygeno.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.McHale C.M., Zhang L., Lan Q., Vermeulen R., Li G., Hubbard A.E., Porter K.E., Thomas R., Portier C.J., Shen M., Rappaport S.M., Yin S., Smith M.T., Rothman N. Global gene expression profiling of a population exposed to a range of benzene levels. Environ. Health Perspect. 2011 doi: 10.1289/ehp.1002546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Moran J.L., Siegel D., Sun X.M., Ross D. Induction of apoptosis by benzene metabolites in HL60 and CD34+ human bone marrow progenitor cells. Mol. Pharmacol. 1996 [PubMed] [Google Scholar]
- 120.Ruiz-Ramos R., Cebrian M.E., Garrido E. Benzoquinone activates the ERK/MAPK signaling pathway via ROS production in HL-60 cells. Toxicology. 2005 doi: 10.1016/j.tox.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 121.Smith M.T. The mechanism of benzene-induced leukemia: a hypothesis and speculations on the causes of leukemia. Environ. Health Perspect. 1996;104:1219–1225. doi: 10.1289/ehp.961041219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ramsay R.G., Barton A.L., Gonda T.J. Targeting c-Myb expression in human disease. Expert Opin. Ther. Targets. 2003 doi: 10.1517/14728222.7.2.235. [DOI] [PubMed] [Google Scholar]
- 123.Wan J., Badham H.J., Winn L. The role of c-MYB in benzene-initiated toxicity. Chem. Biol. Interact. 2005 doi: 10.1016/j.cbi.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 124.Yasuda T., Kagawa W., Ogi T., Kato T.A., Suzuki T., Dohmae N., Takizawa K., Nakazawa Y., Genet M.D., Saotome M., Hama M., Konishi T., Nakajima N.I., Hazawa M., Tomita M., Koike M., Noshiro K., Tomiyama K., Obara C., Gotoh T., Ui A., Fujimori A., Nakayama F., Hanaoka F., Sugasawa K., Okayasu R., Jeggo P.A., Tajima K. Novel function of HATs and HDACs in homologous recombination through acetylation of human RAD52 at double-strand break sites. PLoS Genet. 2018 doi: 10.1371/journal.pgen.1007277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li S., Shi B., Liu X., An H.X. Acetylation and deacetylation of DNA repair proteins in cancers. Front. Oncol. 2020 doi: 10.3389/fonc.2020.573502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ikura T., Ogryzko V.V., Grigoriev M., Groisman R., Wang J., Horikoshi M., Scully R., Qin J., Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000 doi: 10.1016/S0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 127.K.K. Lee, Y. Zhang, R. Tirado- Magallanes, D. Rajagopalan, S.S. Bhatia, L. Ng, N. Desi, C.Y. Tham, W.S. Teo, M.M. Hoppe, A. Jeyasekharan, Y. Tay, W.J. Chng, D. Tenen, T. Benoukraf, S. Jha, TIP60 acetylates H2AZ and regulates doxorubicin-induced DNA damage sensitivity through RAD51 transcription, (2020). https://doi.org/10.1101/2020.06.10.145193.
- 128.Andersen J.N., Mortensen O.H., Peters G.H., Drake P.G., Iversen L.F., Olsen O.H., Jansen P.G., Andersen H.S., Tonks N.K., Møller N.P.H. Structural and evolutionary relationships among protein tyrosine phosphatase domains. Mol. Cell. Biol. 2001 doi: 10.1128/mcb.21.21.7117-7136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sommer S.Le, Morrice N., Pesaresi M., Thompson D., Vickers M.A., Murray G.I., Mody N., Neel B.G., Bence K.K., Wilson H.M., Delibegovic M. Deficiency in protein tyrosine phosphatase PTP1B shortens lifespan and leads to development of acute leukemia. Cancer Res. 2018 doi: 10.1158/0008-5472.CAN-17-0946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pandey R., Saxena M., Kapur R. Role of SHP2 in hematopoiesis and leukemogenesis. Curr. Opin. Hematol. 2017 doi: 10.1097/MOH.0000000000000345. [DOI] [PMC free article] [PubMed] [Google Scholar]