Abstract
This study aims to perform population analysis of the rumen ciliated protozoa of the free-living European bison (wisent, Bison bonasus, Linnaeus). The samples of the rumen fluid from the 18 bison subjected to the controlled culls within the free-ranging population in the Bialowieza primeval forest in Poland were collected and examined. The examined ciliates population consisted of the species of the families Isotrichidae and Ophryoscolecidae. There were 12 genera (Isotricha, Dasytricha, Diplodinium, Elytroplastron, Entodinium, Eodinium, Epidinium, Eremoplastron, Eudiplodinium, Metadinium, Ophryoscolex, and Ostracodinium) and 32 morphospecies of the ciliates. We observed the prevalence of a type B protozoan population (56% animals) with the typical Epidinium and Eudiplodinium genera members. Other examined animals possessed the mixed A–B population with Ophryoscolex genus, distinct for type A ciliate population. The average total ciliates count was 2.77 ± 1.03 × 105/ml (mean ± SD). The most abundant genera were Entodinium, 83%, and Dasytricha, 14%. The abundance of other genera was <1% of the total count. Within the 16 Entodinium species determined, the most abundant species was Entodinium nanellum (16.3% of total ciliates count). The average Shannon–Wiener diversity index was 2.1 ± 0.39, evenness was 0.7 ± 0.11, and species richness was 24 ± 3.0 (mean ± SD). Our study is the first report on the population composition and diversity of rumen ciliates of European bison. The composition and counts of ciliate genera and species were similar to the composition and counts of the rumen ciliated protozoa of American bison and many other kinds of free-living and domestic ruminants. Our European bison ciliate population analysis has shown medium ciliate density and high diversity typical for large free-living ruminants with mixed feeding behavior.
Keywords: Bison bonasus, ciliates, population diversity, protozoa, rumen
Introduction
The European bison (wisent, Bison bonasus L.) is the largest terrestrial mammal in Europe. The European bison has been successfully restored after the extinction in the wild at the beginning of the twentieth century. The total area of European bison habitat covers about 130,000 ha in Poland. Due to herd management of growing free-ranging wisent population in the Bialowieza Forest (northeast Poland), the annual culls have been conducted since 1971 (Krasińska and Krasiński, 2013). They aim to reduce the population on average by 11%. The most frequent reasons for bison selections were various injuries, entering fields, aggression toward people, and, after 1980, changes in the genitourinary system caused by posthitis/balanoposthitis illness (Krasińska and Krasiński, 2013). The free-living ruminants are interesting from a scientific perspective because few studies describe their complex rumen microbial ecosystems (Ishaq et al., 2015). The complex rumen microbial population comprises prokaryotes (eubacteria and archaea) and eukaryotes (fungi and protozoa). The dominant component of rumen protozoa is ciliated protozoa (Williams and Coleman, 1992), which contribute to the breakdown of plant and microbial carbohydrates and proteins. Protozoa influence the rumen microbial population by predation on other protozoa, prokaryotes, and fungi, suggesting that protozoa are still attractive from a scientific point of view (Williams et al., 2020). In general, the number of rumen ciliates species in different host animals is about 45 or less, influenced mainly by host feeding behavior and seasonal variation in the diet (Dehority, 2004). The studies on the rumen ciliates population revealed two main types, types A and B. Type A population is characterized by the presence of genera of Polyplastron and Ophryoscolex (Eadie, 1962, 1967). Type B population is characterized by the presence of genera of Epidinium and Eudiplodinium. On the other hand, some other genera are commonly present in most ruminants (Entodinium spp. and Isotrichids). Rumen ciliates were described in many ruminants' species; however, their description of the genus Bison is limited only to American bison (A. bison, Bison bison). All protozoan species found in A. bison have also been reported in domestic livestock. The type B population predominated in free-living A. bison without contacts with cattle. The mixed A–B population occurred in bison in areas inhabited by domestic livestock (Towne et al., 1988b). The percentage distribution of rumen ciliate species among A. bison varies among the geographical regions, depending on the type and quantity of consumed feed and on the contact with other animals in the group. The present study aimed to examine and describe the ciliate population of the rumen fluid of European bison obtained from culled animals of the free-ranging population in Bialowieza. The description of rumen ciliate protozoa of the European bison contributes to our knowledge of the ecology and diversity of rumen ciliates of the free-living European ruminants.
Materials and Methods
The material was obtained from culled European bison originating from the free-ranging population in the Bialowieza Forest (northeast Poland; longitude between 23°31′ and 24°21′ E and latitude 52°29′ and 52°57′ N). Samples were collected immediately after death (1–3 h) from 18 animals of both sexes of various ages during February and March 2007 (Table 1). For the microscopic counts, about 10 g/animal of the rumen contents were preserved with an equal amount of 8% formaldehyde solution (w/w), strained through four layers of cheesecloths into 10 ml polypropylene tubes with screw cups and stored at 8°C in a refrigerator until analysis. Ciliates were counted microscopically in an aliquot of the suitably diluted sample (Williams and Coleman, 1992). At least four replicates were counted per sample and per ciliate species. The protozoan genera and species were identified according to the size and the shape of cells, skeletal plates (if present), macronucleus, and ciliature arrangement (Dogiel, 1927; Ogimoto and Imai, 1981; Williams and Coleman, 1992; Ito and Imai, 1998; Ito et al., 2001; Cedrola et al., 2017a,b, 2018). Different staining procedures were used to stain skeletal plates (iodine solution), nuclei (methyl green-formalin-saline and chrome-alum-carmine), and infraciliature (pyridinated silver carbonate method) (Ogimoto and Imai, 1981). The pictures of ciliates were taken under bright field illumination by Moticam Pro CCD Camera (Motic Incorporation Ltd., Hong Kong) mounted on a BA400 microscope (Motic Incorporation Ltd., Hong Kong). The images were processed and analyzed using ImageJ software according to ImageJ software documentation (Abramoff et al., 2004; Siritantikorn et al., 2012; Choudhry, 2016). Morphometric measurements were performed by image analysis of at least 20 cells. Only ciliates of the families Isotrichidae and Ophryoscolecidae were present in the samples. Differentiated count of species of genus Entodinium was performed in 13 samples (animals). Direct bacterial count estimated the total bacteria count through image analysis of pictures taken under bright field illumination of dried smears of formaldehyde-fixed samples (Siritantikorn et al., 2012; Choudhry, 2016). Two smears stained with methylene blue and known dimensions and known volumes per sample (animal) were prepared according to the Breed method (Horáková, 1988). Twenty randomly selected pictures per smear were taken at an objective magnification of ×100 by the Moticam Pro CCD Camera (Motic Incorporation Ltd., Hong Kong) mounted on a BA400 microscope (Motic Incorporation Ltd., Hong Kong). The images were processed and analyzed using ImageJ software (Selinummi et al., 2005). Ciliates and bacteria counts per milliliter were expressed as geometric means of log natural transformed values ± geometric standard deviation and arithmetic means ± standard deviations. We evaluated bison age and sex effects on the total count of bacteria and ciliates by nonparametric Kruskal–Wallis test (GraphPad Prism, GraphPad Software, Inc., San Diego, CA, USA). The biodiversity indices (Shannon–Wiener diversity index, evenness, and species richness) (Spellerberg and Fedor, 2003; Jost, 2006) were calculated with an Excel calculator (Microsoft Office Professional, 2007, Microsoft, Redmond, WA, USA). The Shannon–Wiener (SW) diversity index was computed to explain the entropy, taking into account the species richness and evenness of the community. The species richness was evaluated by counting the number of taxa per sample (animal). Correlation analysis on counts of ciliates genera was calculated with Prism 5 (GraphPad Software, Inc., San Diego, CA, USA). Probability value p < 0.05 was considered significant. Principal component analysis was performed on counts of ciliates genera with the aid of the STATISTICA (Data Analysis Software System), version 9.0. (StatSoft, Inc., Tulsa, OK, USA, 2009).
Table 1.
Average weight, age, rumen ciliates, and bacteria counts of European bison (wisent, Bison bonasus, L.).
| Bison counts | Weight (kg) | Age (year) | Ciliate count (Ln/ml) | Bacteria count (Ln/ml) | |||||
|---|---|---|---|---|---|---|---|---|---|
| n | Mean | SD | Mean | SD | Geom-mean | Geom-SD | Geom-mean | Geom-SD | |
| Males | 5 | 309 | 92.3 | 2.6 | 0.89 | 12.32 | 1.714 | 23.81 | 1.410 |
| Females | 6 | 395 | 73.1 | 12.4 | 8.32 | 12.25 | 1.679 | 23.81 | 1.254 |
| Young females | 7 | 108 | 16.1 | 0.6 | 0.18 | 12.34 | 2.498 | 23.92 | 1.168 |
Results
Age, weight, and sex of hosts and the total count of ciliates and bacteria are summarized in Table 1. The abundance of bacteria was estimated to be 235.108 ± 43 × 108/ml (arithmetic mean ± SD). The mean count of ciliates was 2.77 × 105 ± 1.03 × 105/ml (arithmetic mean ± SD). We observed no effects of sex and age of host on the abundance of bacteria (P = 0.809) and ciliates (P = 0.412). We determined 12 genera and 32 ciliates species by microscopic examination (Tables 2, 3). All examined bison ciliates populations can be classified as type B, although the Ophryoscolex genus was present in eight individuals. We observed the following genera and morphospecies of the family Isotrichidae: Isotricha (Isotricha prostoma, Isotricha intestinalis) and Dasytricha (Dasytricha ruminantium). We observed the following genera and morphospecies of the family Ophryoscolecidae: Diplodinium (Diplodinium dentatum), Entodinium (Entodinium brevispinum, Entodinium caudatum, Entodinium dubardi, Entodinium exiguum, Entodinium furca, Entodinium lobosospinosum, Entodinium longinucleatum, Entodinium nanellum, Entodinium nanum, Entodinium ovinum, Entodinium ovoideum, Entodinium parvum, Entodinium rostratum, Entodinium simplex, Entodinium triacum, and Entodinium yunnense), Elytroplastron (Elytroplastron bubali), Eodinium (Eodinium posterovesiculatum), Epidinium (Epidinium caudatum, Epidinium ecaudatum, Epidinium parvicaudatum, Epidinium quadricaudatum, and Epidinium tricaudatum), Eremoplastron (Eremoplastron rostratum), Eudiplodinium (Eudiplodinium maggii), Metadinium (Metadinium esalqum), Ostracodinium (Ostracodinium gracile), and Ophryoscolex (Ophryoscolex purkyniei), (Figures 1, 2). The most abundant genera were Entodinium (83%, 2.22 × 105/ml) and Dasytricha (13%, 0.33 × 105/ml). The remaining genera were <1% abundant. The genera of Entodinium, Epidinium, Diplodinium, Eudiplodinium, Elytroplastron, and Dasytricha were present in all rumen samples. The E. posterovesiculatum was infrequent and not countable. The Epidinium species of E. caudatum, E. ecaudatum, E. quadricaudatum, and E. tricaudatum were infrequent and not countable. The dominant Epidinium species was E. parvicaudatum. Within other large ciliates, the least abundant species were E. bubali and O. gracile. Within the Entodinium species, the most numerous were E. nanellum, with 16.3% of total ciliates count (Table 3). Other Entodinium species were abundant <5% of the total ciliates count. Table 4 shows the evaluation of the population variability of rumen ciliates of European bison with diversity (Shannon–Wiener index) of 2.1, evenness (Peliou index) of 0.7, and species richness of 24. Correlation analysis is summarized in Table 5 (the numbers within brackets are r and P, respectively). Analysis revealed the positive correlation of Isotricha counts with counts of Entodinium (0.73, 0.001), total counts (0.72, 0.001), Ostracodinium (0.68, 0.02), Eudiplodinium (0.63, 0.007), Eremoplastron (0.66, 0.01), and Elytroplastron (0.62, 0.008). Entodinium counts correlated positively with total counts (0.94, 0.001), counts of Eremoplastron (0.83, 0.001), Isotricha (0.73, 0.01), Eudiplodinium (0.72, 0.01), and Ostracodinium (0.62, 0.04). Epidinium counts correlated positively with counts of Eremoplastron (0.66, 0.01) and negatively with animal weight (−0.47, 0.05). Eremoplastron counts correlated with counts of Entodinium (0.83, 0.001), totals (0.78, 0.002), Eudiplodinium (0.75, 0.003), Ostracodinium (0.75, 0.03), Epidinium (0.66, 0.01), and Isotricha (0.66, 0.01). Eudiplodinium counts correlated with counts of Eremoplastron (0.75, 0.003), Entodinium (0.72, 0.001), Isotricha (0.63, 0.01), and totals (0.63, 0.005). Elytroplastron counts correlated with the counts of Ostracodinium (0.93, 0.001) and Isotricha (0.62, 0.008). Ophryoscolex counts correlated only with animal age (0.73, 0.04). Ostracodinium counts correlated with the counts of Elytroplastron (0.93, 0.001), Eremoplastron (0.75, 0.03), Isotricha (0.68, 0.02), and Entodinium (0.62, 0.04). No significant correlations were observed on counts of Dasytricha, Diplodinium, and Metadinium. No correlations were observed on animal gender. Animal weight was associated with animal age (0.70, 0.001). These relationships are illustrated with a PCoA plot of the variables, which shows similar relationships (Figure 3).
Table 2.
The analysis of rumen ciliated protozoa population of European bison (Bison bonasus. L.).
| Family, genus, species | Prevalence (%) | Percentage of total count(%) | Count (C/ml) | |||||
|---|---|---|---|---|---|---|---|---|
| Mean | SD | n | Median | Min | Max | |||
| Ophryoscolecidae | ||||||||
| Entodinium spp. | 100 | 82.86 | 229,537 | 94,228.29 | 18 | 217,333 | 90,666 | 404,000 |
| Epidinium | ||||||||
| Epidinium parvicaudatum | 100 | 0.29 | 812 | 1,116.28 | 18 | 402 | 13 | 4,870 |
| Eremoplastron | ||||||||
| Eremoplastron rostratum | 72 | 0.30 | 831 | 1,188.05 | 13 | 350 | 100 | 3,950 |
| Diplodinium | ||||||||
| Diplodinium dentatum | 100 | 0.86 | 2,390 | 1,578.42 | 18 | 2,120 | 13 | 5,180 |
| Metadinium | ||||||||
| Metadinium esalqum | 100 | 0.13 | 366 | 331.67 | 18 | 264 | 60 | 1,307 |
| Eudiplodinium | ||||||||
| Eudiplodinium maggii | 100 | 0.23 | 636 | 598.50 | 18 | 310 | 13 | 2,040 |
| Elytroplastron | ||||||||
| Elytroplastron bubali | 100 | 0.12 | 345 | 409.64 | 18 | 270 | 7 | 1,727 |
| Ostracodinium | ||||||||
| Ostracodinium gracile | 61 | 0.01 | 32 | 44.21 | 11 | 20 | 7 | 160 |
| Ophryoscolex | ||||||||
| Ophryoscolex purkynei | 44 | 0.03 | 92 | 135.33 | 8 | 30 | 5 | 400 |
| Isotrichidae | ||||||||
| Dasytricha | ||||||||
| Dasytricha ruminantium | 100 | 14.36 | 39,787 | 33,637.57 | 18 | 42,800 | 400 | 144,000 |
| Isotricha | ||||||||
| Isotricha prostoma | 78 | 0.48 | 1,317 | 1,399.83 | 14 | 707 | 253 | 4,013 |
| Isotricha intestinalis | 83 | 0.39 | 1,071 | 1,543.91 | 15 | 440 | 40 | 5,520 |
| Total count | 100 | 100 | 277,022 | 103,503.93 | 18 | 274,620 | 105,279 | 463,181 |
Table 3.
The counts of rumen Entodinium species of European bison (Bison bonasus. L.).
| Species | Prevalence % | Percentage of total count % | Count C/ml | |||||
|---|---|---|---|---|---|---|---|---|
| Mean | SD | n | Median | Min | Max | |||
| E. brevispinum | 92 | 2.6 | 7,300 | 5,345 | 12 | 5,000 | 400 | 17,200 |
| E. caudatum | 77 | 0.7 | 1,880 | 1,544 | 10 | 1,600 | 200 | 4,800 |
| E. dubardi | 100 | 3.5 | 9,660 | 6,144 | 13 | 9,200 | 1,000 | 19,200 |
| E. exiguum | 100 | 4.6 | 12,740 | 8,060 | 13 | 9,600 | 3,800 | 32,000 |
| E. furca monolobum | 92 | 1.6 | 4,430 | 2,885 | 12 | 3,800 | 200 | 8,800 |
| E. lobosospinosum | 92 | 1.4 | 4,020 | 2,680 | 12 | 3,900 | 600 | 9,400 |
| E. longinucleatum | 100 | 1.3 | 3,520 | 2,385 | 13 | 3,600 | 1,000 | 8,200 |
| E. nanellum | 100 | 16.3 | 45,060 | 25,686 | 13 | 41,800 | 11,800 | 95,800 |
| E. nanum | 100 | 5.0 | 13,862 | 7,744 | 13 | 12,800 | 3,600 | 28,200 |
| E. ovinum | 8 | 0.1 | 200 | 1 | 200 | 200 | 200 | |
| E. ovoideum | 92 | 2.7 | 7,450 | 4,408 | 12 | 6,500 | 2,000 | 17,000 |
| E. parvum | 100 | 4.6 | 12,750 | 7,683 | 13 | 11,400 | 800 | 23,800 |
| E. rostratum | 77 | 1.4 | 3,980 | 6,275 | 10 | 1,900 | 400 | 21,600 |
| E. simplex | 100 | 1.7 | 4,830 | 3,513 | 13 | 4,200 | 200 | 11,600 |
| E. triacum | 85 | 1.3 | 3,530 | 3,501 | 11 | 1,400 | 200 | 11,200 |
| E. yunnense | 92 | 0.3 | 820 | 679 | 12 | 600 | 200 | 2,000 |
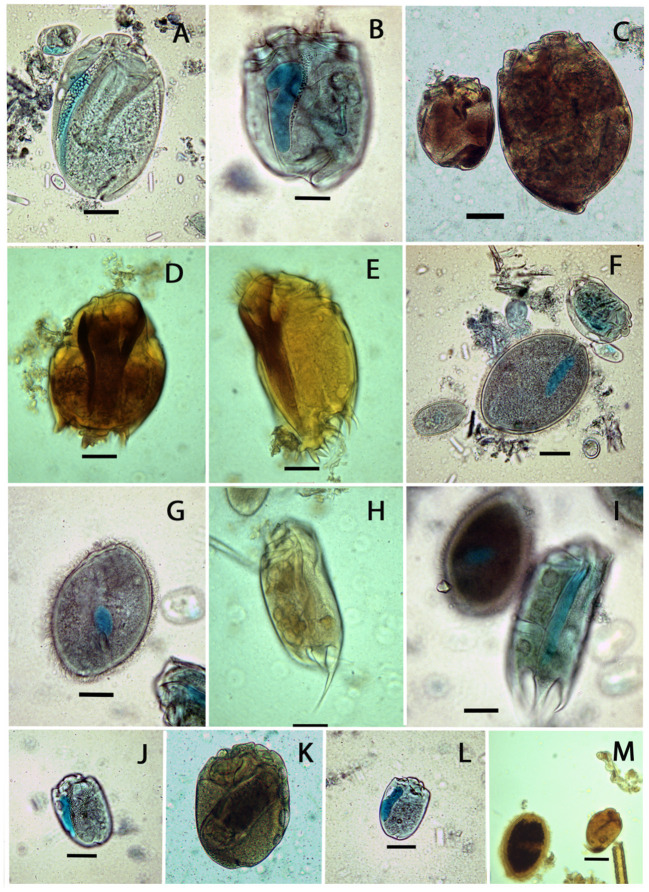
Figure 1.
Photomicrographs of some ciliates observed in the rumen of European bison. (A) Elytroplastron bubali; (B) Eudiplodinium maggii; (C) the illustration of two extreme sizes of E. maggii; (D,E) Ophryoscolex purkyniei; (F) (from left to right) Dasytricha ruminantium, Isotricha prostoma, Diplodinium dentatum, and Entodinium exiguum; (G,I) Isotricha intestinalis; (H,I) Epidinium parvicaudatum; (J) Metadinium esalqum; (K) Elytroplastron with engulfed Epidinium; (L) Eodinium posterovesiculatum; (M) Dasytricha ruminantium and Eremoplastron rostratum. Samples were colorized by (A,B,F,G,I–L) methyl-green formalin, (D,E,H,M) iodine solutions, and (C) pyridinated silver carbonate. Scale bars are 20 μm in all pictures.
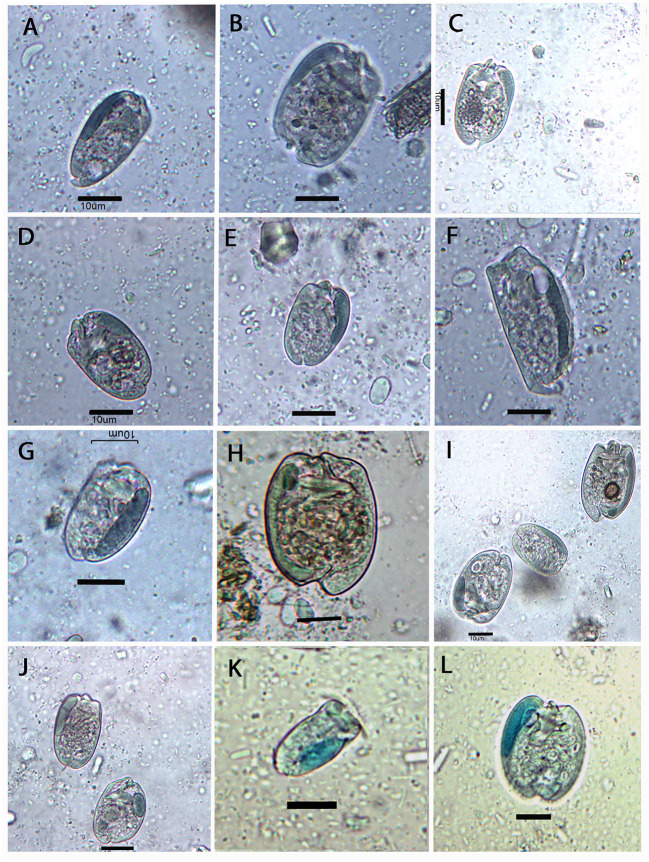
Figure 2.
Photomicrographs of some entodiniid ciliates observed in the rumen of European bison. (A) Entodinium brevispinum; (B) E. caudatum; (C) E. lobosospinosum; (D) E. simplex; (E) E. nanellum; (F) E. rostratum; (G) E. parvum; (H) E. yunense; (I) (from left to right) E. simplex, E. nanellum, and E. yunense; (J) E. orbicularis; (K) E. exiguum; and (L) E. dubardi. Samples were colorized by (A–J) chrome-alum-carmine and (K,L) methyl-green formalin solutions. Scale bars are 10 μm in all pictures.
Table 4.
Evaluation of population variability of rumen ciliates of European bison.
| Mean | SD | Median | Min | Max | n | |
|---|---|---|---|---|---|---|
| Diversity (SW index) | 2.1 | 0.39 | 2.3 | 1.4 | 2.6 | 13 |
| Evenness (Peliou index) | 0.7 | 0.11 | 0.7 | 0.5 | 0.8 | 13 |
| Species richness | 24 | 3.0 | 24 | 15 | 27 | 13 |
Table 5.
Correlation matrix of animal gender, age, weight, and rumen ciliate counts of European bison.
| Gender | Age, years | Weight, kg | Entod | Epid | Eremopl. | Eudipl. | Elytropl. | Metad. | Ophryo. | Ostracod. | Dasytr. | Isotr. | Diplod. | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gender | ||||||||||||||
| Age, years | 0.32 (0.20) | |||||||||||||
| Weight, kg | −0.11 (0.67) | 0.70 (0.001) | ||||||||||||
| Entod | −0.03 (0.90) | −0.13 (0.62) | −0.25 (0.32) | |||||||||||
| Epid | 0.15 (0.56) | −0.33 (0.19) | −0.47 (0.05) | 0.02 (0.93) | ||||||||||
| Eremo | −0.12 (0.69) | 0.19 (0.54) | 0.33 (0.27) | 0.83 (0.001) | 0.66 (0.01) | |||||||||
| Eudipl | 0.07 (0.77) | 0.14 (0.59) | 0.14 (0.58) | 0.72 (0.001) | 0.74 (0.64) | 0.75 (0.003) | ||||||||
| Elytropl | 0.39 (0.11) | −0.14 (0.57) | −0.36 (0.15) | 0.40 (0.11) | 0.23 (0.35) | 0.25 (0.40) | 0.10 (0.68) | |||||||
| Metad | 0.10 (0.69) | −0.08 (0.76) | −0.07 (0.78) | 0.22 (0.38) | −0.01 (0.97) | 0.29 (0.34) | −0.04 (0.86) | 0.03 (0.92) | ||||||
| Ophryo | 0.30 (0.47) | 0.73 (0.04) | 0.43 (0.29) | 0.33 (0.43) | −0.38 (0.36) | 0.36 (0.43) | 0.69 (0.06) | 0.59 (0.13) | −0.35 (0.39) | |||||
| Ostrac | 0.20 (0.55) | −0.27 (0.42) | −0.40 (0.22) | 0.62 (0.04) | 0.03 (0.94) | 0.75 (0.03) | 0.17 (0.63) | 0.93 (0.001) | 0.02 (0.95) | −0.34 (0.51) | ||||
| Dasytr | −0.26 (0.30) | −0.07 (0.78) | 0.11 (0.67) | −0.24 (0.95) | −0.24 (0.33) | 0.02 (0.94) | −0.16 (0.52) | −0.12 (0.64) | 0.20 (0.44) | −0.10 (0.82) | −0.11 (0.75) | |||
| Isotr | 0.30 (0.24) | 0.08 (0.77) | −0.37 (0.15) | 0.73 (0.001) | 0.40 (0.11) | 0.66 (0.01) | 0.63 (0.007) | 0.62 (0.008) | −0.03 (0.92) | 0.34 (0.41) | 0.68 (0.02) | 0.01 (0.97) | ||
| Diplod | 0.03 (0.90) | 0.04 (0.32) | 0.04 (0.88) | 0.46 (0.05) | −0.13 (0.62) | 0.32 (0.29) | 0.25 (0.31) | −0.02 (0.94) | 0.14 (0.59) | 0.49 (0.22) | −0.14 (0.69) | 0.33 (0.18) | 0.41 (0.10) | |
| Totals | −0.10 (0.69) | −0.13 (0.61) | −0.20 (0.43) | 0.94 (0.001) | −0.05 (0.84) | 0.78 (0.002) | 0.63 (0.005) | 0.34 (0.17) | 0.28 (0.27) | 0.29 (0.48) | 0.59 (0.06) | 0.32 (0.20) | 0.72 (0.001) | 0.56 (0.02) |
The first value is correlation coefficient r, and a number in parentheses is probability value P. The significant values are highlighted.
Figure 3.
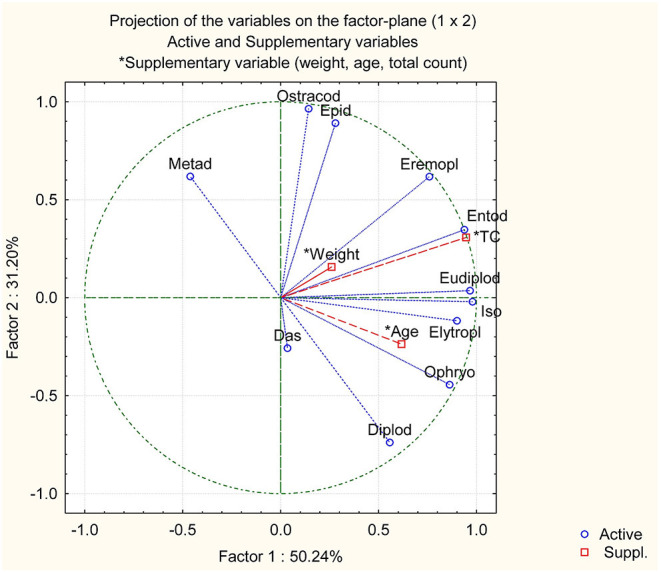
Principal component analysis of the European bison ciliates population. Metadinium (Metad), Ostracodinium (Ostracod), Epidinium (Epid), Eremoplastron (Eremopl), Entodinium (Entod), total count (TC), Eudiplodinium (Eudiplod), Isotricha (Iso), Elytroplastron (Elytropl), Ophryoscolex (Ophryo), Diplodinium (Diplod), and Dasytricha (Das).
Discussion
The rumen protozoal population of free-living ruminants is influenced mainly by host feeding behavior and seasonal variation in the diet (Kamler, 1999; Dehority and Odenyo, 2003; Booyse and Dehority, 2011; Clauss et al., 2011; Obidziński et al., 2017). European bison can be considered mixed feeders with 68–97% of herbaceous plants in their natural diet (Gebczyńska et al., 1991; Kowalczyk et al., 2019). In some other references, the European bison are considered as grazers with 68% of grass in their natural diet (Pucek et al., 2002; Clauss et al., 2006; Przybyło et al., 2019). The population of rumen ciliated protozoa of grazers (e.g., cattle) is generally more diverse in comparison with typical browsers, selectors (e.g., roe deer and blue duiker), but this is not a rule (Kofoid and Christianson, 1934; Sládeček, 1946; Prins and Geelen, 1971; Giesecke and Gylswyk, 1975; Imai, 1988; Williams and Coleman, 1992; Dehority, 1994; Robbins et al., 1995; Dehority and Odenyo, 2003; Clauss et al., 2011). More generically diverse ciliate populations of grass and roughage eaters may result from a slower passage rate and higher rumen pH (Dehority and Odenyo, 2003). However, in the Bialowieza forest, other food (hay) is available to bison during winter (Gebczyńska et al., 1991; Pucek et al., 2002; Kowalczyk et al., 2011). Therefore, the differences in both summer and winter rumen ciliate populations are likely small. To our knowledge, this is the first study describing the population of protozoa in the rumen of European bison of the Białowieza region. Generally, the species composition of rumen ciliates of European bison was similar to the composition of rumen ciliates populations of many other species of free-living and domesticated ruminants (Kofoid and Christianson, 1934; Sládeček, 1946; Crha, 1972; Towne et al., 1988a,b; Ito et al., 1994; Moon-van der Staay et al., 2014). Recent research pointed to the existence of a core rumen microbiome across a wide geographical range, which is modified by the diet and the host (O'Kelly and Spiers, 1992; Guan et al., 2008; Shi et al., 2008; Moon-van der Staay et al., 2014; Henderson et al., 2015; Ishaq et al., 2015; Tapio et al., 2017; Reis et al., 2019; Furman et al., 2020; Xue et al., 2020). These studies indicate that dietary and animal feeding strategies dominate over host species. In the study of Henderson et al. (2015), the variability of protozoa between and within animal groups was much greater than that of bacteria and archaea. Analyses of different 18S ribosomal RNA (rRNA) genes showed extremely complex but related ciliate communities, which occur in the rumen of cattle, sheep, goats, and red deer (Moon-van der Staay et al., 2014). Although individual host genetic characteristics might influence the composition of the rumen prokaryotes (Shi et al., 2008; Xue et al., 2020), it seems that ciliates' feed preferences and relationship within ciliates contribute remarkably to their composition in the rumen (Eadie, 1967; Dehority, 1998; Martinele and D'Agosto, 2008). In vitro and in vivo experiments revealed the competitive relationship between certain starch preferring Entodinium species (Entodinium caudatum) and the fibrolytic ciliates Eudiplodinium maggii and Epidinium ecaudatum (Michalowski et al., 2003; Bełzecki et al., 2004; Zeitz et al., 2012). Both American bison and European bison have a similar ciliates genera composition with the prevalence of a type B protozoan population. Typical members of the type B population are Epidinium, Eudiplodinium, Metadinium, and Elytroplastron. On the other hand, typical antagonistic types A population members are Polyplastron, Ophryoscolex, and Diploplastron (Eadie, 1962, 1967). It is known that Polyplastron multivesiculatum grazes on Epidinium, resulting in the vanish of Epidinium from the rumen (Williams and Coleman, 1992). We noted 44% of animals with a mixed A–B population. The prevalence of type B protozoan population was observed in many species of free-living ruminants, e.g., deer, ibex, mouflon, chamois, moose, reindeer, muskox, gaur, antelopes, and giraffe (Kofoid and Christianson, 1934; Sládeček, 1946; Kleynhans and van Hoven, 1976; Kleynhans, 1982; Crha et al., 1985; Dehority, 1985, 1986; Dehority et al., 1999; Dehority and Odenyo, 2003; Karnati et al., 2003; Imai et al., 2004; Korchagina, 2006, 2012; de la Fuente et al., 2009). In contrast to A. bison, we observed only Ophryoscolex spp. (O. purkyniei) and no Polyplastron spp. in animals with mixed A–B protozoan population. Therefore, Epidinium species prevalence was not influenced in animals with mixed A–B protozoan populations (Table 2). Besides, the predatory behavior of rumen ciliates was observed not only in the type A population. In the type B population, the predatory behavior of E. bubali on Epidinium, Enoploplastron, and Entodinium was observed in sheep rumen (Martinele and D'Agosto, 2008). However, the predatory activity of E. bubali in our bison samples was low. Our correlation analysis revealed the prevalence of the positive correlations among the individual rumen protozoal genera counts. Most numerous positive correlations were observed on Isotricha spp. (with total counts, Entodinium, Eremoplastron, Eudiplodinium, Elytroplastron, and Ostracodinium counts). We can speculate that the growth of Isotricha can be promoted by soluble metabolic products of hydrolytic and proteolytic activities of Entodinium, Eremoplastron, Eudiplodinium, Elytroplastron, and Ostracodinium species. Isotricha is known to prefer soluble substrates (Williams, 1986). On the other hand, Entodiniomorphid ciliates prefer solid substrates (plant and bacterial particles) (Williams and Coleman, 1992). On the other hand, we have observed no correlations of Dasytricha and Metadinium with other ciliate genera. Differences in metabolic activities were observed between Isotricha and Dasytricha. They remarkably differ in carbohydrate fermentation (Howard, 1959). Dasytricha has greater metabolic versatility than Isotricha. Therefore, Dasytricha is probably less dependent on intermediate metabolic products of other members of the ciliate population. We can speculate on similar features of Metadinium species. For example, Metadinium medium could degrade starch, amylose, amylopectin, and hemicellulose (Naga and El-Shazly, 1968). The comparison with other studies is difficult because of different analysis methods, different animal diets, and different ciliate population structures observed. In Tan et al. (2020), the correlation of molecular data revealed the positive association of the Metadinium with Eudiplodinium, Isotricha with Dasytricha, and Polyplastron with Ostracodinium and Ophryoscolex. In our study, those associations were not observed. The influence of the season may also contribute to these differences. Our samples were taken in the winter when the animals eat a predominantly fibrous diet with a lack of green fodder rich in soluble nutrients. Our ciliate population analysis has shown medium ciliate density and high diversity typical for large free-living ruminants with mixed feeding behavior. We observed a similar total number of rumen ciliates of European bison (277.103/ml) and American bison (328.103/ml) (Towne et al., 1988a,b). A similar ciliate density was also observed in domestic ruminants (cattle and goats) (Imai, 1988; Ito et al., 1994, 1995; de la Fuente et al., 2009; Mishima et al., 2009). On the other hand, the domestic ruminants have the lower average number of ciliate species per host (8–18; Imai, 1988; Ito et al., 1994, 1995; de la Fuente et al., 2009; Mishima et al., 2009). It is considered that species evenness decreases in the ruminants on high concentrate feed (Ito et al., 1994). On the other hand, when ruminants are fed on high forage, the number of species per host increases to more than 30 (Mishima et al., 2009). There were considerable variations in the counts of all examined ciliates species (genera). The animal-to-animal variations in both the differential counts of ciliate species and total counts were also observed in A. bison and other ruminants (Towne et al., 1988a,b; Kittelmann and Janssen, 2011). Purser and Moir (Purser and Moir, 1966a,b) showed that rumen volume could be a factor involved in individual animal differences in rumen parameters of sheep fed the same diet. The significant differences in the total ciliates counts were removed after the counts' adjustment for rumen volume (Dehority, 1978). The same author observed decreased rumen volume of sheep fed concentrate diet than the forage (alfalfa) diet. The changes in the rumen volume regarding the feed changes were also observed in other ruminants (Kamler et al., 2003). Some studies also suggest the effects of animal age, sex, and weight on the ciliate population (Clauss et al., 2011; Duarte et al., 2018). Our correlation analysis revealed no effects of animal sex on ciliates counts. However, sex in our bison collection was not evenly represented among age groups. No effects of host age and sex on rumen protozoa were observed on Spanish ibex and domestic goats (de la Fuente et al., 2009). We have observed a positive correlation of Ophryoscolex counts with animal age and a negative correlation of Epidinium counts with host weight. Those relationships' physiological backgrounds are unclear, as analysis revealed the positive correlation of animal age and weight. It can point to the possible antagonistic relationship between O. purkyniei and E. parvicaudatum. This phenomenon needs more not only microscopic but also molecular and in vitro physiological studies.
Conclusion
Our study is the first report on the population composition and diversity of rumen ciliates of European bison. The population structure and counts of ciliate genera and species of European bison were similar to the composition and counts of the rumen ciliated protozoa of American bison and many other kinds of free-living and domestic ruminants. Our European bison ciliate population analysis has shown medium ciliate density and high diversity typical for large free-living ruminants with mixed feeding behavior.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Ethics Statement
The animal study was reviewed and approved by the Ethical Committee of the Institute of Animal Physiology of Centre of Biosciences of SAS approved the experimental protocol (resolution number Ro-3355/16-221).
Author Contributions
SK: project administration, investigation and methodology, validation, samples analysis and evaluation, data curation, manuscript writing, review, and editing. DD and KM: samples analysis and evaluation. ZV: investigation and methodology and manuscript review. JP: samples collection. MS-S: funding acquisition and supervision and manuscript review. AC: conceptualization, funding acquisition and supervision, and manuscript review. ZG: conceptualization, samples collection, and manuscript review. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This study was supported by the Grant Scientific Agency of the Ministry of Education, Science, Research and Sport of the Slovak Republic and the Slovak Academy of Sciences (VEGA 2/0009/08) and by the Faculty of Veterinary Medicine and Animal Science, Poznan University of Life Sciences, Poland, from the Department of Animal Nutrition (no. 506.533.04.00).
References
- Abramoff M. D., Magalhães P. J., Ram S. J. (2004). Image processing with ImageJ. Biophotonics Int. 11, 36–42. [Google Scholar]
- Bełzecki G., Miltko R., Michalowski T. (2004). Why does the establishment of the starch preferring Entodinium caudatum in the rumen decrease the numbers of the fibrolytic ciliate Eudiplodinium maggii?. Folia Microbiol. 49, 139–142. 10.1007/BF02931388 [DOI] [PubMed] [Google Scholar]
- Booyse D., Dehority B. A. (2011). Protozoa and digestive tract parameters of the impala. Onderstepoort J. Vet. Res. 78:327. 10.4102/ojvr.v78i1.327 [DOI] [PubMed] [Google Scholar]
- Cedrola F., Dias R. J. P., Martinele I., D'Agosto M. (2017a). Polymorphism and inconsistencies in the taxonomy of Diplodinium anisacanthum da Cunha, 1914 (Ciliophora, Entodiniomorphida, Ophryoscolecidae) and taxonomic notes on the genus Diplodinium Schuberg, 1888. Zootaxa 4306:249. 10.11646/zootaxa.4306.2.5 [DOI] [Google Scholar]
- Cedrola F., Rossi M. F., Martinele I., D'Agosto M., Dias R., Júnio P., et al. (2018). Morphology and description of infraciliary bands pattern in four Metadinium Awerinzew; Mutafowa, 1914 species (Ciliophora, Entodiniomorphida, Ophryoscolecidae) with taxonomic notes on the genus. Zootaxa 4500:574. 10.11646/zootaxa.4500.4.6 [DOI] [PubMed] [Google Scholar]
- Cedrola F., Senra M. V. X., D'Agosto M., Dias R. J. P. (2017b). Phylogenetic analyses support validity of genus Eodinium (Ciliophora, Entodiniomorphida, Ophryoscolecidae). J. Eukaryot. Microbiol. 64, 242–247. 10.1111/jeu.12355 [DOI] [PubMed] [Google Scholar]
- Choudhry P. (2016). High-throughput method for automated colony and cell counting by digital image analysis based on edge detection. PLoS ONE 11:e0148469. 10.1371/journal.pone.0148469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauss M., Hofmann R. R., Hummel J., Adamczewski J., Nygren K., Pitra C., et al. (2006). Macroscopic anatomy of the omasum of free-ranging moose (Alces alces) and muskoxen (Ovibos moschatus) and a comparison of the omasal laminal surface area in 34 ruminant species. J. Zool. 270, 346–358. 10.1111/j.1469-7998.2006.00148.x [DOI] [Google Scholar]
- Clauss M., Müller K., Fickel J., Streich W. J., Hatt J.-M., Südekum K.-H. (2011). Macroecology of the host determines microecology of endobionts: protozoal faunas vary with wild ruminant feeding type and body mass. J. Zool. 283, 169–185. 10.1111/j.1469-7998.2010.00759.x [DOI] [Google Scholar]
- Crha J. (1972). Rumen ciliates in Fallow Deer (Dama Dama L.) in Namest preserve. Acta Vet. Brno 41, 355–362. [Google Scholar]
- Crha J., Hrabě V., Koubek P. (1985). Rumen ciliate fauna in the chamois (Rupicapra rupicapra L.). Acta Vet. Brno 54, 141–147. 10.2754/avb198554030141 [DOI] [Google Scholar]
- de la Fuente G., Belanche A., Abecia L., Dehority B. A., Fondevila M. (2009). Rumen protozoal diversity in the Spanish ibex (Capra pyrenaica hispanica) as compared with domestic goats (Capra hircus). Eur. J. Protistol. 45, 112–120. 10.1016/j.ejop.2008.07.001 [DOI] [PubMed] [Google Scholar]
- Dehority B. A. (1978). Specificity of rumen ciliate protozoa in cattle and sheep*. J. Protozool. 25, 509–513. 10.1111/j.1550-7408.1978.tb04177.x [DOI] [Google Scholar]
- Dehority B. A. (1985). Rumen ciliates of musk-oxen (Ovibos moschatus Z.) from the canadian arctic. J. Protozool. 32, 246–250. 10.1111/j.1550-7408.1985.tb03045.x [DOI] [Google Scholar]
- Dehority B. A. (1986). “Microbes in the foregut of arctic ruminants,” in Control of Digestion and Metabolism in Ruminants, eds L. P. Milligan, W. L. Grovum, and A. Dobson (Prentice-Hall Englewood Cliffs), 307–325. [Google Scholar]
- Dehority B. A. (1994). Rumen ciliate protozoa of the blue duiker (Cephalophus monticola), with observations on morphological variation lines within the species Entodinium dubardi. J. Eukaryot. Microbiol. 41, 103–111. 10.1111/j.1550-7408.1994.tb01481.x [DOI] [PubMed] [Google Scholar]
- Dehority B. A. (1998). Microbial interactions in the rumen. Rev. Fac. Agron. 69–86. [Google Scholar]
- Dehority B. A. (2004). Rumen Microbiology. Thrumpton: Nottingham University Press. [Google Scholar]
- Dehority B. A., Demarais S., Osborn D. A. (1999). Rumen ciliates of white-tailed deer (Odocoileus virginianus), axis deer (Axis axis), sika deer (Cervus nippon) and fallow deer (Dama dama) from Texas. J. Eukaryot. Microbiol. 46, 125–131. 10.1111/j.1550-7408.1999.tb04595.x [DOI] [PubMed] [Google Scholar]
- Dehority B. A., Odenyo A. A. (2003). Influence of diet on the rumen protozoal fauna of indigenous African wild ruminants. J. Eukaryot. Microbiol. 50, 220–223. 10.1111/j.1550-7408.2003.tb00121.x [DOI] [PubMed] [Google Scholar]
- Dogiel V. A. (1927). Monographie der Familie Ophryoscolecidae, Teil I. Arch. fur Protistenkd. 59, 1–289. [Google Scholar]
- Duarte E. R., Abrão F. O., Oliveira Ribeiro I. C., Vieira E. A., Nigri A. C., Silva K. L., et al. (2018). Rumen protozoa of different ages of beef cattle raised in tropical pastures during the dry season. J. Appl. Anim. Res. 46, 1457–1461. 10.1080/09712119.2018.1530676 [DOI] [Google Scholar]
- Eadie J. M. (1962). Inter-relationships between certain rumen ciliate protozoa. J. Gen. Microbiol. 29, 579–588. 10.1099/00221287-29-4-579 [DOI] [Google Scholar]
- Eadie J. M. (1967). Studies on the ecology of certain rumen ciliate protozoa. J. Gen. Microbiol. 49, 175–194. 10.1099/00221287-49-2-175 [DOI] [PubMed] [Google Scholar]
- Furman O., Shenhav L., Sasson G., Kokou F., Honig H., Jacoby S., et al. (2020). Stochasticity constrained by deterministic effects of diet and age drive rumen microbiome assembly dynamics. Nat. Commun. 11:1904. 10.1038/s41467-020-15652-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebczyńska Z., Gebczyński M., Martynowicz E. (1991). Food eaten by the free-living European bison in białowieza forest. Acta Theriol. 36, 307–313. 10.4098/AT.arch.91-32 [DOI] [Google Scholar]
- Giesecke D., Gylswyk N. O. Van. (1975). A study of feeding types and certain rumen functions in six species of South African wild ruminants. J. Agric. Sci. 85, 75–83. 10.1017/S0021859600053430 [DOI] [Google Scholar]
- Guan L. L., Nkrumah J. D., Basarab J. A., Moore S. S. (2008). Linkage of microbial ecology to phenotype: correlation of rumen microbial ecology to cattle's feed efficiency. FEMS Microbiol. Lett. 288, 85–91. 10.1111/j.1574-6968.2008.01343.x [DOI] [PubMed] [Google Scholar]
- Henderson G., Cox F., Ganesh S., Jonker A., Young W., Global Rumen Census Collaborators G. R. C., et al. (2015). Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 5:14567. 10.1038/srep14567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horáková K. (1988). “Mikroskopické metódy (Microscopic methods),” in Mikrobiologické Laboratórne Metódy (Microbiological Laboratory Methods), eds V. Betina, H. Barátová, A. Fargašová, V. Frank, K. Horáková, and E. Šturdík (Bratislava: Alfa; ), 173–206. [Google Scholar]
- Howard B. H. (1959). The biochemistry of rumen protozoa. 1. Carbohydrate fermentation by Dasytricha and Isotricha. Biochem. J. 71, 671–675. 10.1042/bj0710671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S. (1988). Ciliate protozoa in the rumen of Kenyan zebu cattle, Bos taurus indicus, with the description of four new species. J. Protozool. 35, 130–136. 10.1111/j.1550-7408.1988.tb04092.x [DOI] [PubMed] [Google Scholar]
- Imai S., Oku Y., Morita T., Ike K., Guirong (2004). Rumen ciliate protozoal fauna of reindeer in Inner Mongolia, China. J. Vet. Med. Sci. 66, 209–212. 10.1292/jvms.66.209 [DOI] [PubMed] [Google Scholar]
- Ishaq S. L., Sundset M. A., Crouse J., Wright A.-D. G. (2015). High-throughput DNA sequencing of the moose rumen from different geographical locations reveals a core ruminal methanogenic archaeal diversity and a differential ciliate protozoal diversity. Microb. Genomics 1:e000034. 10.1099/mgen.0.000034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito A., Imai S. (1998). Infraciliary bands in the rumen Ophryoscolecid ciliate Ostracodinium gracile (Dogiel, 1925), observed by light microscopy. J. Eukaryot. Microbiol. 45, 628–636. 10.1111/j.1550-7408.1998.tb04559.x [DOI] [PubMed] [Google Scholar]
- Ito A., Imai S., Manda M., Ogimoto K. (1995). Rumen ciliates of Tokara native goat in Kagoshima, Japan. J. Vet. Med. Sci. 57, 355–357. 10.1292/jvms.57.355 [DOI] [PubMed] [Google Scholar]
- Ito A., Imai S., Ogimoto K. (1994). Rumen ciliate composition and diversity of Japanese beef black cattle in comparison with those of Holstein-Friesian cattle. J. Vet. Med. Sci. 56, 707–714. 10.1292/jvms.56.707 [DOI] [PubMed] [Google Scholar]
- Ito A., Miyazaki Y., ImaI S. (2001). Light microscopic observations of infraciliature and morphogenesis in six species of rumen Ostracodinium ciliates. J. Eukaryot. Microbiol. 48, 440–448. 10.1111/j.1550-7408.2001.tb00177.x [DOI] [PubMed] [Google Scholar]
- Jost L. (2006). Entropy and diversity. Oikos 113, 363–375. 10.1111/j.2006.0030-1299.14714.x [DOI] [Google Scholar]
- Kamler J. (1999). Infusorial Concentration in Rumen Fluid of Red Deer, Fallow Deer, Roe Deer and Mouflon. Acta Vet. Brno 68, 247–252. 10.2754/avb199968040247 [DOI] [Google Scholar]
- Kamler J., Dvorák J., Kamlerová K. (2003). Differences in relative volume and weight of forestomachs among four free living ruminants. Acta Vet. Brno 72, 33–39. 10.2754/avb200372010033 [DOI] [Google Scholar]
- Karnati S. K. R., Yu Z., Sylvester J. T., Dehority B. A., Morrison M., Firkins J. L. (2003). Technical note: Specific PCR amplification of protozoal 18S rDNA sequences from DNA extracted from ruminal samples of cows. J. Anim. Sci. 81, 812–815. 10.2527/2003.813812x [DOI] [PubMed] [Google Scholar]
- Kittelmann S., Janssen P. H. (2011). Characterization of rumen ciliate community composition in domestic sheep, deer, and cattle, feeding on varying diets, by means of PCR-DGGE and clone libraries. FEMS Microbiol. Ecol. 75, 468–481. 10.1111/j.1574-6941.2010.01022.x [DOI] [PubMed] [Google Scholar]
- Kleynhans C. J. (1982). The rumen ciliates of greater kudu Tragelaphus strepsiceros (Pallas) from South Africa and Zimbabwe with a description of one new species. South African J. Zool. 17, 11–14. 10.1080/02541858.1982.11447771 [DOI] [Google Scholar]
- Kleynhans C. J., van Hoven W. (1976). Rumen protozoa of the giraffe with a description of two new species. Afr. J. Ecol. 14, 203–214. 10.1111/j.1365-2028.1976.tb00164.x [DOI] [Google Scholar]
- Kofoid C. A., Christianson J. F. (1934). Ciliates from Bos gaurus H. Smith. Berkeley, CA: University of California Press. [Google Scholar]
- Korchagina T. A. (2006). Taxonomic diversity of endobiotic ciliates in different parts of the elk' stomach (Alces alces). (in Russian). Omsk. Nauchnyj Vestn. 46, 244–246. [Google Scholar]
- Korchagina T. A. (2012). Species diversity and population of ciliate endobionts in reindeer stomach (Rangifer tarandus L.). (in Russian). Perspekt. Nauk. 30, 5–10. [Google Scholar]
- Kowalczyk R., Taberlet P., Coissac E., Valentini A., Miquel C., Kamiński T., et al. (2011). Influence of management practices on large herbivore diet—Case of European bison in Białowieza Primeval Forest (Poland). For. Ecol. Manage. 261, 821–828. 10.1016/j.foreco.2010.11.026 [DOI] [Google Scholar]
- Kowalczyk R., Wójcik J. M., Taberlet P., Kamiński T., Miquel C., Valentini A., et al. (2019). Foraging plasticity allows a large herbivore to persist in a sheltering forest habitat: DNA metabarcoding diet analysis of the European bison. For. Ecol. Manage. 449:117474. 10.1016/j.foreco.2019.117474 [DOI] [Google Scholar]
- Krasińska M., Krasiński Z. A. (2013). European Bison. The Nature Monograph. Berlin; Heidelberg: Springer. 10.1007/978-3-642-36555-3 [DOI] [Google Scholar]
- Martinele I., D'Agosto M. (2008). Predação e canibalismo entre protozoários ciliados (Ciliophora: Entodiniomorphida: Ophryoscolecidae) no rúmen de ovinos (Ovis aries). Rev. Bras. Zool. 25, 451–455. 10.1590/S0101-81752008000300010 [DOI] [Google Scholar]
- Michalowski T., Belzecki G., Kwiatkowska E., Pajak J. J. (2003). The effect of selected rumen fauna on fibrolytic enzyme activities, bacterial mass, fibre disappearance and fermentation pattern in sheep. J. Anim. Feed Sci. 12, 45–64. 10.22358/jafs/67642/2003 [DOI] [Google Scholar]
- Mishima T., Katamoto H., Horii Y., Kakengi V. A. M. M., Ito A. (2009). Rumen ciliates from Tanzanian short horn zebu cattle, Bos taurus indicus, and the infraciliature of Entodinium palmare n.sp. and Enoploplastron stokyi (Buisson, 1924). Eur. J. Protistol. 45, 77–86. 10.1016/j.ejop.2008.07.002 [DOI] [PubMed] [Google Scholar]
- Moon-van der Staay S. Y., Van der Staay G. W. M. M., Michalowski T., Jouany J.-P. P., Pristas P., Javorský P., et al. (2014). The symbiotic intestinal ciliates and the evolution of their hosts. Eur. J. Protistol. 50, 166–173. 10.1016/j.ejop.2014.01.004 [DOI] [PubMed] [Google Scholar]
- Naga M. A., El-Shazly K. (1968). The metabolic characterization of the ciliate protozoon Eudiplodinium medium from the rumen of buffalo. J. Gen. Microbiol. 53, 305–315. 10.1099/00221287-53-3-305 [DOI] [PubMed] [Google Scholar]
- Obidziński A., Miltko R., Bolibok L., Wajdzik M., Skubis J., Nasiadka P. (2017). Variation of natural diet of free ranging mouflon affects their ruminal protozoa composition. Small Rumin. Res. 157, 57–64. 10.1016/j.smallrumres.2017.09.019 [DOI] [Google Scholar]
- Ogimoto K., Imai S. (1981). Atlas of Rumen Microbiology. Tokyo: Scientific Societies Press. [Google Scholar]
- O'Kelly J., Spiers W. (1992). Possible contribution of protozoa to differences in rumen metabolism between cattle breeds. Aust. J. Agric. Res. 43:1795. 10.1071/AR9921795 [DOI] [Google Scholar]
- Prins R. A., Geelen M. J. H. (1971). Rumen Characteristics of Red Deer, Fallow Deer, and Roe Deer. J. Wildl. Manage. 35:673. 10.2307/3799772 [DOI] [Google Scholar]
- Przybyło M., Hummel J., Ortmann S., Codron D., Kohlschein G. M., Kilga D., et al. (2019). Digesta passage in nondomestic ruminants: separation mechanisms in “moose-type” and “cattle-type” species, and seemingly atypical browsers. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 235, 180–192. 10.1016/j.cbpa.2019.06.010 [DOI] [PubMed] [Google Scholar]
- Pucek Z., Belousova I. P., Krasiñska M., Krasiñski Z. A., Olech W. (2002). European bison, Bison bonasus: Current state of the Species and an Action Plan for its Conservation, ed Z. Pucek Bialowieza,: Mammal Research Institute, Polish Academy of Sciences. Available online at: https://ibs.bialowieza.pl/publications/1360.pdf (accessed May 30, 2021).
- Purser D. B., Moir R. J. (1966a). Rumen volume as a factor involved in individual sheep differences. J. Anim. Sci. 25, 509–515. 10.2527/jas1966.252509x [DOI] [Google Scholar]
- Purser D. B., Moir R. J. (1966b). Variations in rumen volume and associated effects as factors influencing metabolism and protozoa concentrations in the rumen of sheep. J. Anim. Sci. 25, 516–520. 10.2527/jas1966.252516x [DOI] [PubMed] [Google Scholar]
- Reis C. C., dos Maeda E. M., Cedrola F., Martins E. N., Paula F. M., De Martinele I. (2019). Diet and breed alter community structures of rumen protozoa in cattle subjected to different feeding systems. Semin. Ciências Agrárias 40:909. 10.5433/1679-0359.2019v40n2p909 [DOI] [Google Scholar]
- Robbins C. T., Spalinger D. E., van Hoven W. (1995). Adaptation of ruminants to browse and grass diets: are anatomical-based browser-grazer interpretations valid?. Oecologia 103, 208–213. 10.1007/BF00329082 [DOI] [PubMed] [Google Scholar]
- Selinummi J., Seppälä J., Yli-Harja O., Puhakka J. A. (2005). Software for quantification of labeled bacteria from digital microscope images by automated image analysis. Biotechniques 39, 859–863. 10.2144/000112018 [DOI] [PubMed] [Google Scholar]
- Shi P. J., Meng K., Zhou Z. G., Wang Y. R., Diao Q. Y., Yao B. (2008). The host species affects the microbial community in the goat rumen. Lett. Appl. Microbiol. 46, 132–135. 10.1111/j.1472-765X.2007.02274.x [DOI] [PubMed] [Google Scholar]
- Siritantikorn S., Jintaworn S., Noisakran S., Suputtamongkol Y., Paris D. H., Blacksell S. D. (2012). Application of ImageJ program to the enumeration of Orientia tsutsugamushi organisms cultured in vitro. Trans. R. Soc. Trop. Med. Hyg. 106, 632–635. 10.1016/j.trstmh.2012.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sládeček F. (1946). Ophryoscolecidae z bachoru jelena (Cervus elaphus L.), danka (Dama dama L.) a srnce (Capreolus L.). Ophryoscolecidae from the stomach of Cervus elaphus L., Dama dama L., and Capreolus capreolus L. Věstník Csl. Zool. společnosti 10, 201–231. [Google Scholar]
- Spellerberg I. F., Fedor P. J. (2003). A tribute to Claude Shannon (1916-2001) and a plea for more rigorous use of species richness, species diversity and the ‘shannon-wiener’ index. Glob. Ecol. Biogeogr. 12, 177–179. 10.1046/j.1466-822X.2003.00015.x [DOI] [Google Scholar]
- Tan C., Ramírez-Restrepo C. A., Shah A. M., Hu R., Bell M., Wang Z., et al. (2020). The community structure and microbial linkage of rumen protozoa and methanogens in response to the addition of tea seed saponins in the diet of beef cattle. J. Anim. Sci. Biotechnol. 11:80. 10.1186/s40104-020-00491-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapio I., Fischer D., Blasco L., Tapio M., Wallace R. J., Bayat A. R., et al. (2017). Taxon abundance, diversity, co-occurrence and network analysis of the ruminal microbiota in response to dietary changes in dairy cows. PLoS ONE 12:e0180260. 10.1371/journal.pone.0180260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towne G., Nagaraja T. G., Cochran R. C., Harmon D. L., Owensby C. E., Kaufman D. W. (1988a). Comparisons of ruminal fermentation characteristics and microbial populations in bison and cattle. Appl. Environ. Microbiol. 54, 2510–2514. 10.1128/AEM.54.10.2510-2514.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towne G., Nagaraja T. G., Kemp K. K. (1988b). Ruminal ciliated protozoa in bison. Appl. Environ. Microbiol. 54, 2733–2736. 10.1128/AEM.54.11.2733-2736.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A. G. (1986). Rumen holotrich ciliate protozoa. Microbiol. Rev. 50, 25–49. 10.1128/MMBR.50.1.25-49.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A. G., Coleman G. S. (1992). The Rumen Protozoa. New York, NY: Springer New York. 10.1007/978-1-4612-2776-2 [DOI] [Google Scholar]
- Williams C. L., Thomas B. J., McEwan N. R., Rees Stevens P., Creevey C. J., Huws S. A. (2020). Rumen protozoa play a significant role in fungal predation and plant carbohydrate breakdown. Front. Microbiol. 11:720. 10.3389/fmicb.2020.00720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue M. Y., Sun H. Z., Wu X. H., Liu J. X., Guan L. L. (2020). Multi-omics reveals that the rumen microbiome and its metabolome together with the host metabolome contribute to individualized dairy cow performance. Microbiome 8:64. 10.1186/s40168-020-00819-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitz J. O., Amelchanka S. L., Michałowski T., Wereszka K., Meile L., Hartnack S., et al. (2012). Effect of the rumen ciliates Entodinium caudatum, Epidinium ecaudatum and Eudiplodinium maggii, and combinations thereof, on ruminal fermentation and total tract digestion in sheep. Arch. Anim. Nutr. 66, 180–199. 10.1080/1745039X.2012.676817 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.