Abstract
The aim of this work was to study the value of the main allergen Asp n 3 of Aspergillus niger as a molecular marker of allergenicity and pathogenicity with the potential to be used in the identification of A. niger as a contaminant and cause of spoilage of Mangifera indica. Real-time polymerase chain reaction (RT-PCR) was used for the amplification of Asp n 3 gene. Two pairs of primers were designed: one for the amplification of the entire sequence and another one for the amplification of the most conserved region of this peroxisomal protein. The presence of A. niger was demonstrated by the early detection of the allergenic protein Asp n 3 coding gene, which could be considered a species-specific marker. The use of primers designed based on the conserved region of the Asp n 3 encoding gene allowed us to identify the presence of the closely related fungal species Aspergillus fumigatus by detecting Asp n 3 homologous protein, which can be cross-reactive. The use of conserved segments of the Asp n 3 gene or its entire sequence allows us to detect phylogenetically closely related species within the Aspergilaceae family or to identify species-specific contaminating fungi.
Keywords: food spoilage, Aspergillus, Asp n 3, DNA-marker, fungal food pathogen
Introduction
Several species belonging to the fungal genus Aspergillus are well known as plant pathogens that may cause damage in plants as well as spoil postharvest fruit, vegetables, and cereals, resulting in serious agriculture and economic losses (Perrone et al., 2007; Samson et al., 2010; Mahfooz et al., 2017). Although the main substrate of black aspergilli (Aspergillus section Nigri) is soil, these fungi are among the most common fungi causing food spoilage and biodeterioration of other materials. Several authors report that the Aspergillus niger species complex can be responsible for the postharvest decay of different fresh fruits and some vegetables (Gautam et al., 2011; Sharma, 2012). It has been demonstrated that Aspergillus spores are one of the most frequently identified fungal conidia in the atmosphere (Kasprzyk, 2008; Dijksterhuis, 2019). Its extensive ubiquity in nature probably justifies why this species is one of the most common contaminating fungal genera recorded in various agricultural commodities, such as fruit, nuts, beans, cereals, and vegetables (Samson et al., 2010). In particular, A. niger can produce rotting of numerous fruits and vegetables, causing a substantial economic loss. Onions, mangoes, grapes, or tomatoes are the most cited fruits to which A. niger can be the major cause of rot phenotype plant disease (Gautam et al., 2011). On the other hand, selected A. niger strains are also used in agriculture as promising beneficial microbes (Lubna et al., 2018; Pertile et al., 2021).
Aspergillus niger is described as a producer of a wide variety of toxic metabolites, including ochratoxins, a group of secondary metabolites that are classified as possible human carcinogens (Blumenthal, 2004; Tian et al., 2015). In addition to producing toxins, Aspergillus species are an important cause of allergy as a relevant source of allergen proteins that are involved in the development of allergic symptoms, particularly in atopic individuals (Simon-Nobbe et al., 2008; Gabriel et al., 2016b).
Among the several allergens described in this mold, the major A. niger allergen, Asp n 3, which is homologous to Asp f 3 of Aspergillus fumigatus (peroxisomal protein), has been demonstrated to be a clinically relevant component because 72% of the Aspergillus sensitized population shows reactive IgE specific to this allergen (Hemmann et al., 1997). This component is also a key marker to use in the differential diagnosis of some different types of aspergillosis (Crameri et al., 1998).
The fungal contamination of fruit with toxins or allergens can lead to a significant health risk to humans. Fruit colonization with these fungi might be used as a direct indicator of fruit or plant damage, food quality, and safety for human consumption (Battilani et al., 2008). Thus, to minimize the fungal diseases associated with both economic losses and health risk, it is essential to detect and identify pathogens at an early stage of the infection process (Amalaradjou and Venkitanarayanan, 2008). In fact, the early and accurate detection and identification of pathogens will allow the control of the spread of fungal infections as well as the implementation of disease management strategies.
The traditional identification of fungi by means of methodologies based on morphological characteristics has a low degree of sensitivity and requires considerable expertise. To address these shortcomings, in recent years, molecular methods such as polymerase chain reaction (PCR) have been considered suitable alternatives for the rapid and early diagnosis of fungal contamination (Gabriel et al., 2015, 2017).
With this in mind, the objective of this work was to study the value of the main allergen Asp n 3 of A. niger as a molecular marker of allergenicity and pathogenicity, with the potential to be used in the identification of A. niger as a contaminant and cause of spoilage of Mangifera indica. Its ability to identify closely related species of the genus Aspergillus was also analyzed. The specific detection of Asp n 3 was evaluated in an experimental infection of M. indica fruit with A. niger, according to Gabriel et al. (2017), by a recently developed PCR method based on the Alt a 1 coding gene for Alternaria alternata.
Materials and Methods
Fungal Strains and Culture Conditions
The A. niger strain CECT 20156 Spanish Type Culture Collection (CECT, University of Valencia, Spain) and the A. fumigatus AF54 strain (Laboratory of Parasitology and Allergy, Lascaray Research Centre, University of the Basque Country, Vitoria-Gasteiz, Spain) were used in this study.
Aspergillus niger was grown on malt extract agar plates for 4 days at 25°C. The spores were collected in 0.2% agar and 0.05% Tween 80 solution. Then, the spore suspension was diluted 1:100, counted in a Neubauer chamber, and adjusted to a final concentration of 1 × 106 spores/ml with sterile phosphate-buffered saline (PBS) and 0.05 Tween 80 (Gabriel et al., 2017).
Fruit Inoculation and Incubation
Twenty fresh mangoes were sterilized by immersion in a solution of 1% (v/v) sodium hypochlorite for 1 h at room temperature. Later, they were rinsed out with sterile distilled water and dried out. Sixteen fruits were inoculated with 100 μl of the spore suspension and four mangoes with 100 μl of sterile distilled water as the negative control under sterile conditions. Inoculation was performed by the injection of the spore suspension at 3 mm deep under the fruit skin. Then, the mangoes were placed in polypropylene bags which were closed and incubated at 25°C for 2, 4, 8, and 14 days.
The infection areas in the fruit were measured daily. Contours of the infected zones were drawn into transparent adhesive tape, and the areas were measured by AutoCAD. The negative controls were also checked daily to discard any spontaneous fungal contamination growth.
Sample Collection and Processing
Four samples from four different fruits corresponding to 2, 4, 8, and 14 days of incubation were analyzed. Samples of 2 cm2 × 1 cm deep were obtained from the fruit area where the inoculation was performed. Half of the sample was frozen at −80°C for further PCR analysis, and the other half was incubated in Czapek Dox agar medium at 25°C until visible growth. The negative controls were subjected to the same procedure (Gabriel et al., 2017). A. niger positive control and A. fumigatus specificity control for PCR were grown on Czapek Dox broth.
RNA Extraction
The samples were ground in liquid nitrogen with a mortar and pestle. Previously, the material was treated with 70% ethanol, and RNAses were removed by RNAse Zap solution (Thermo Fisher Scientific, Waltham, MA, United States). Then, 4.5 μl of 2-mercaptoethanol was added to 100 mg of the pulverized sample, and Qiagen RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) was used to extract the total RNA according to the instructions provided by the manufacturer (Gabriel et al., 2017).
cDNA Synthesis, Primer Design, and PCR Amplification
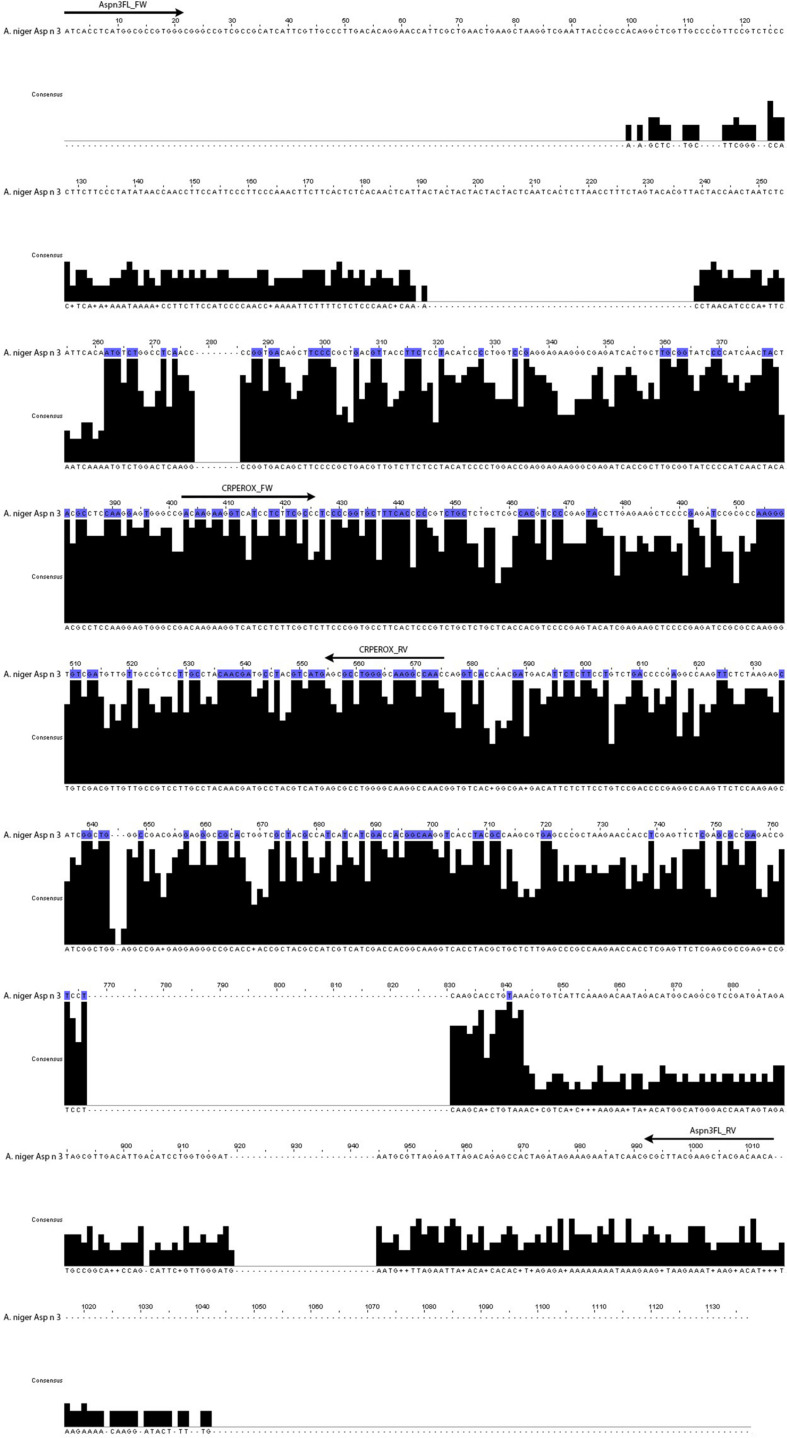
The reverse transcription reactions were carried out with RevertAid First Strand cDNA Synthesis Kit (Fermentas, Sankt Leon-Rot, Germany). Two pairs of primers were manually designed by comparison of the complete sequence that encodes Asp n 3 with the consensus sequences of the related fungal species considered in the design (Figure 1). The GenBank accession numbers of fungal species used for the multiple alignment analysis by the ClustalW program are shown in Table 1.
FIGURE 1.
Complete sequence of the gene that codifies for the allergen Asp n 3 compared to the consensus sequence of the fungal species considered in the design (A. niger, A. fumigatus, A. terreus, A. clavatus, A. nidulans, A. flavus, A. oryzae, Penicillium citrium, P. chrysogenum, Trichophyton rubrum, and Neurospora crassa). The arrows indicate the position of the two pairs of primers corresponding to complete cDNA gene and conserved region.
TABLE 1.
GenBank accession numbers of the sequences used in multiple alignment analysis.
| Species name | GenBank accession |
| Aspergillus niger CBS 513.88 | XM 001395871.2 |
| Aspergillus terreus NIH2624 | XM 001209908.1 |
| Aspergillus fumigatus ATCC 42202 | U58050.1 |
| Aspergillus clavatus NRRL 1 | XM 001270302.1 |
| Aspergillus nidulans FGSC A4 | XM 676869.1 |
| Aspergillus flavus NRRL3357 | XM 002380885.1 |
| Aspergillus oryzae RIB40 | AB226160.1 |
| Penicillium chrysogenum Wisconsin 54-1255 | XM 002566221.1 |
| Penicillium citrium 52-2 | AF144753.1 |
| Trichophyton rubrum CBS 118892 | XM 003238084.1 |
| Neurospora crassa OR74A | XM 959107.3 |
The primers named Aspn3FL_FW (ATCACCTCATG GCGCCGTGGG) and Aspn3FL_RV (TGTTGTCGTA GCTTCGTAAGCGC) were designed to amplify the complete Asp n 3 gene of A. niger, including the non-conserved regions of the gene.
The primers named CRPEROX_FW [CAAGAAGGT(C/T) (A/G)T(C/T)CTC(T/G)TCGCC] and CRPEROX_RV (TGTT GTCGTAGCTTCGTAAGCGC) were designed on the basis of the highly conserved internal nucleotide region of the Asp n 3 gene sequence of A. niger and its homologs in the other Aspergillus species (Gabriel et al., 2017).
Polymerase chain reactions were carried out with a FastStart Taq DNA polymerase PCR kit (Roche, Mannheim, Germany), and a PCR-grade deoxynucleoside triphosphate set (Roche, Mannheim, Germany) was used, following the manufacturer’s instructions. Separate PCRs were performed for each pair of primers. The RT-PCR mixture contained 38.6 μl of water, 5 μl of buffer 10× with 20 mM MgCl2, 2 μl of each primer, 1 μl of dNTP mix (10 mM), 0.4 μl of FastStart Taq DNA polymerase, and 1 μl of cDNA. The program set for the Aspn3_FL primers started at 95°C for 4 min and then proceeded with 35 cycles consisting of the first 30 s at 95°C, 30 s at 62°C, and 75 s at 72°C. The program set for the CRPEROX primers started with natural denaturation at 95°C for 4 min followed by 35 amplification cycles consisting of 30 s at 95°C, 30 s at 53°C, and 45 s at 72°C. After that, there was a final extension phase at 72°C for 7 min in both cases. The RT-PCR detection limit was 1 × 104 spores of Aspergillus.
Sequencing reactions were performed using the BigDye Terminator, v.3, sequencing kit (Applied Biosystems) according to the manufacturer’s specifications, and the reactions were run in an ABI 3130XL sequencer (Servei de Genòmica, Universitat Autònoma de Barcelona). The amplicon sequence identity was confirmed with NCBI BLAST.
Alignment of Homologs to Asp n 3 Gene Sequences and Phylogenetic Tree
A BLAST sequence alignment of the Asp n 3 gene with the genes encoding peroxisomal homologous proteins from all expressing species included in the database was carried out. A phylogenetic tree of the nearest neighborhood of representatives constructed based on the nucleotide sequences of Asp n 3 homologs (peroxisomal protein) was constructed using Molecular Evolutionary Genetics Analysis (Tamura et al., 2007).
Results
Inoculation with A. niger resulted in a depressed, circular, brown, soft area surrounding the point of insertion of the pathogen. That area grew, softening the tissues of the mango and cracking the skin. The fungus was able to form spores in the areas where the flesh of the fruit was exposed. No new point of infection was observed during the experiment.
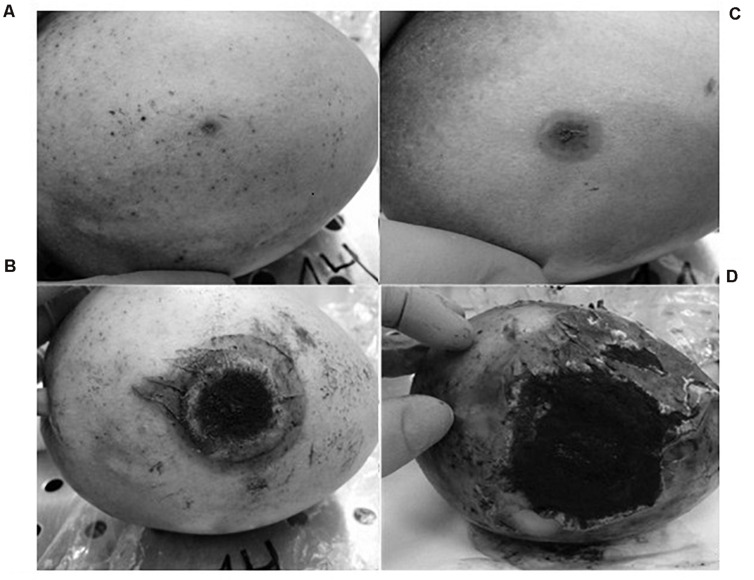
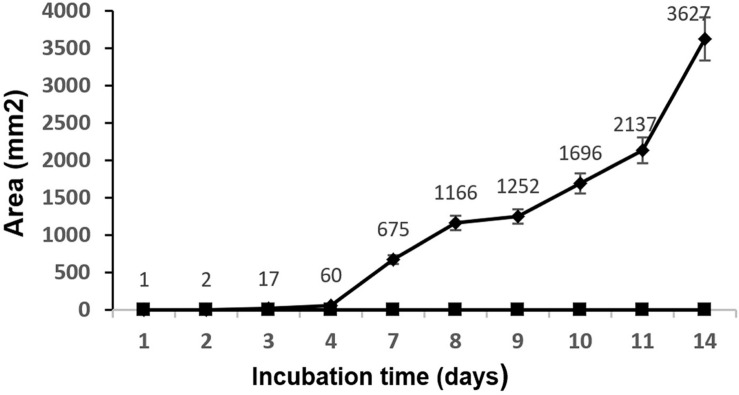
At 2 days post-inoculation, the lesions provoked by A. niger were evident in 100% of the inoculated mangoes. These lesions grew until they covered large areas of the fruit surface (Figure 2). The injuries caused by A. niger inoculation grew exponentially, showing lesions 0.8, 3.4, and 6 cm in diameter after 4, 8, and 14 days of incubation, respectively (Figure 3).
FIGURE 2.
Evolution of the injuries developed by A. niger in mango fruit: (A) 2 days post-infection (p.i.), (B) 4 days (p.i.), (C) 8 days (p.i.), and (D) 14 days (p.i.).
FIGURE 3.
Mean values (mm2) and standard deviation of the injured area by A. niger in mango fruit. ◆, samples infected with A. niger spores; ■, samples inoculated with sterile phosphate-buffered saline.
The cultures of samples of infected fruit in mycological media were positive for A. niger after 3–4 days of incubation. The samples from fruit inoculated with sterile PBS did not show fungal growth.
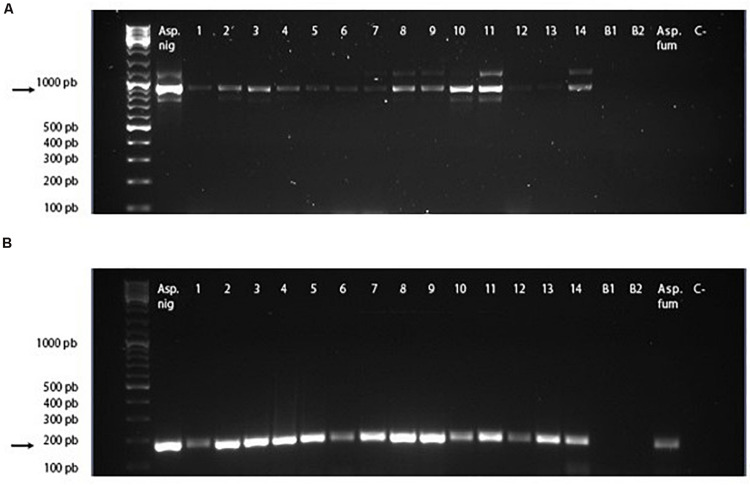
The PCR amplicons of the 913-bp complete gene and the 171-bp amplified fragment corresponding to the conserved region of the gene are shown in Figure 4. In 100% of the samples of mangoes infected with A. niger, the complete gene and the conserved gene region were amplified. In the sample infected by A. fumigatus, only the conserved region of the Asp n 3 gene was amplified, but not the complete region. There was no amplification in the mangoes used as the negative control.
FIGURE 4.
cDNA PCR-amplified regions of Asp n gene. (A) Full cDNA gene amplicon using primers Aspn3_FW and Aspn3_RV. (B) Conserved cDNA gene region amplified using primers CRPEROX_FW and CRPEROX_FW. Asp. Nig, Aspergillus niger (positive control). Lines 1–14: samples of mangoes inoculated with spores of A. niger. B1 and B2: samples of mangoes inoculated with phosphate-buffered saline. Asp. fum, Aspergillus fumigatus. C- :PCR negative control.
These results confirmed the specificity of the primers designed for the amplification of the Asp n 3 gene for detecting A. niger as well as the capability of detecting Asp n 3 cross-reactive proteins using the CRPEROX_FW/CRPEROX_RV primers. This technique allows detection of the presence of the allergen Asp n 3 and its homologs in infected mangoes from the early stages of infection (2 days after inoculation). It is known that the presence of inhibitory substances in food samples may often compromise DNA amplification and lead to the failure of PCR. To prevent false-negative results, positive controls using cDNA obtained from the mycelia of an A. niger reference strain (CECT 20156 Spanish Type Culture Collection University of Valencia, Spain) were also included. A sequence analysis of the amplicons allowed the confirmation of the identity of all DNA fragments as Asp n 3. No amplification products were detected in the PCRs using cDNA obtained from PBS-inoculated fruits (Figure 4, B1 and B2 lines).
The presence of A. niger was demonstrated by the early detection of the allergenic protein Asp n 3 coding gene, which could be considered a species-specific marker. The use of primers designed based on the conserved region of the Asp n 3 encoding gene allowed us to identify the presence of the closely related fungal species A. fumigatus by detecting Asp n 3 homologous protein, which can be cross-reactive. The detection of well-characterized fungal allergens could be considered a tool for identifying fruit rot by Aspergillus and to identify fruit contaminated by Aspergillus as an allergenic source.
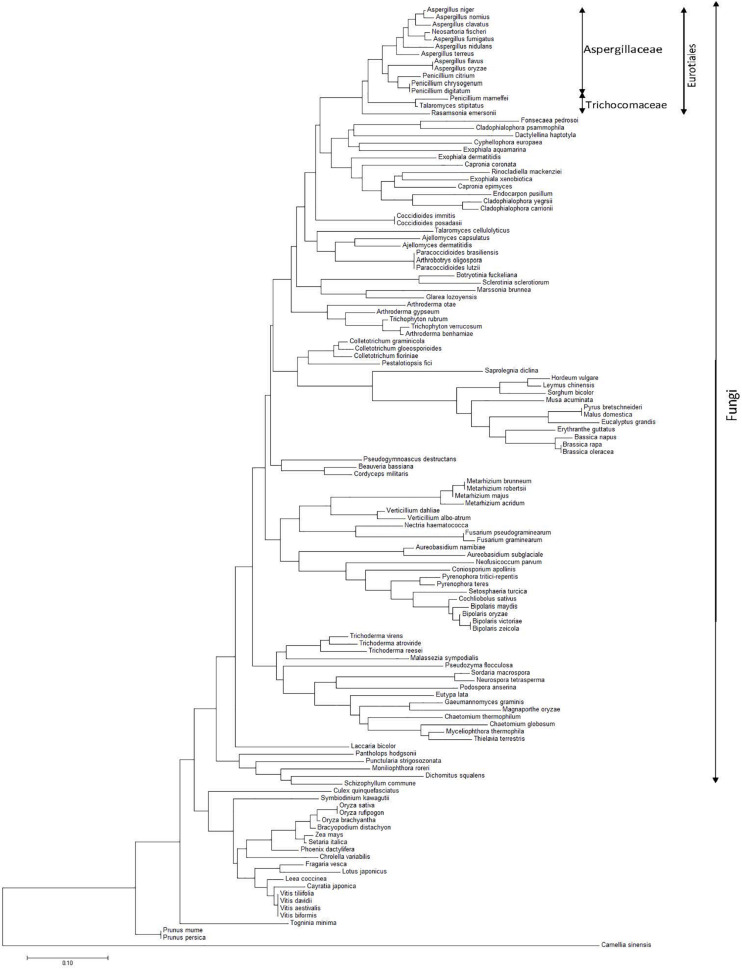
A phylogenetic tree (based on the nearest-neighbor method) of representatives constructed based on the nucleotide sequences of Asp n 3 homologs (peroxisomal protein) is shown in Figure 5. The sequence alignment of the Asp n 3 homologs by BLAST, belonging to the different species considered to construct the phylogenetic tree, A. niger, Aspergillus terreus, Neosartorya fischeri, A. fumigatus, Aspergillus clavatus, and Aspergillus nidulans, showed more than 90% identity; Aspergillus flavus and Aspergillus orizae revealed 86% identity; Penicillium species showed 81–84% identity; and Talaromyces and Ramsonia species showed 75–80% identity.
FIGURE 5.
Phylogenetic tree of the nearest neighborhood of representatives constructed on the basis of the nucleotide sequences of Asp n 3 homologs (peroxyredoxin proteins) encoding genes belonging to species from different kingdoms, with special accent on the fungi kingdom.
Discussion
Species from the genus Aspergillus are ubiquitous members of Ascomycotina, which are present in numerous and different habitats (Raper and Fennell, 1965; Christensen and Tuthill, 1986; Bennett, 2010).
Aspergillus niger is commonly isolated worldwide from soil, plant debris, air, and indoor environments, and it is considered a soil saprobe and a non-ligninolytic fungus able to produce numerous exoenzymes and organic acids involved in the decomposition cycle of organic matter. These features justify the capability of this fungus to cause the decay of fruits, vegetables, nuts, cereals, wood, or herbs. A. niger has also been described as a pathogen for humans and animals as an opportunistic parasite that causes allergic disorders or mycotoxin production (Gautam et al., 2011).
As a plant-pathogenic fungus, A. niger has been isolated from a variety of substrates, but its most prominent role is in the rotting or spoilage of numerous fruits, vegetables, and other food products, with relevant economic loss (Sharma, 2012).
The contamination of mango fruit by A. niger results in the qualitative deterioration of the product, showing a depressed, circular, brown, soft area surrounding the point where the pathogen starts the colonization (Palejwala et al., 1987; Al-Hindi et al., 2011).
The pathogenicity assay results obtained here showed that the mango fruit is very susceptible to contamination with A. niger, and the type of injuries caused by fungal colonization is compatible with that described by other authors (Kim et al., 2020; Shen et al., 2020). These results confirm the validity and applicability of this method for the study of the spoilage of mangoes by A. niger.
Because A. niger is a well-recognized producer of harmful toxins and significant allergenic proteins, exposure to A. niger-contaminated foodstuffs is likely to represent an inherent risk for human health (Simon-Nobbe et al., 2008; Gabriel et al., 2016a). This issue has been underestimated, and studies performed on it are very scarce (Simon-Nobbe et al., 2008; Gabriel et al., 2017). The precise and early detection of A. niger contamination would be a relevant added value to establish and prevent health risks and economic losses related to fungal colonization of foods such as fruits and vegetables.
The development of DNA technology has provided effective methods for microbial analysis. In recent decades, genetic studies have provided different nucleic acid markers for microbial identification. These markers are extensively used in PCR or chip-based detection. Based on 16S rDNA and internal transcribed spacer (ITS) sequencing, metagenomic approaches are now emerging technologies for analyzing the entire microbial community in a complex food and vegetable matrix (Shen et al., 2020). Recently, a multiplex RTPCR has been developed, which is able to identify common Aspergillus sections quantitatively and detect the presence of azole-resistant strains, allowing the successful detection of early stage fungal colonies within a day of incubation (Kim et al., 2020).
In recent years, several fungal allergens (Alt a 1, Alt a 6, Asp f 1, Asp f 3, Asp n 1, Asp n 3, and others) have been presented as valuable molecular markers of allergenicity and pathogenicity (Matricardi et al., 2016). Accordingly, it is expected that the use of approaches targeting the Asp n 3 gene and/or protein to unequivocally identify and detect the pathogen A. niger will yield privileged and powerful information regarding the quality and biosecurity control of foodstuffs (Gabriel et al., 2016b, 2017).
Previously, our research group developed a PCR-based method targeting the gene coding for A. alternata major allergen Alt a 1 using species-specific primer sets, and these tools were used to successfully detect the infection of citrus fruit by A. alternata early (Gabriel et al., 2017). With this in mind and applying the same concept to the A. niger major allergen Asp n 3, further work was conducted to take advantage of the potential of this early detection method to reduce and control postharvest losses during the storage and transportation of fresh mango fruits.
The PCR based on specific Asp n 3 DNA expression gene sequences was able to detect the contamination of mango fruit from the first week of fungal colonization. The amplicons of the complete gene and the amplified fragment corresponding to the conserved region of the gene revealed positive results in 100% of the samples of mangoes infected with A. niger. As expected, there was a positive result in the A. fumigatus sample for the primers that amplify the conserved region, but not for the primers that amplify the complete Asp n 3 gene.
These results confirmed the specificity of the primers designed for the amplification of the Asp n 3 gene for detecting A. niger as well as the capability of detecting proteins susceptible to present cross-reactivity with Asp n 3 using the primers designed to amplify the conserved gene region. This technique allows the detection of the presence of the allergen Asp n 3 and its homologs in infected mangoes from the early stages of the infection (2 days after inoculation). These results are similar to those found by us using the model of A. alternata infection in citrus fruit (Gabriel et al., 2017). These results shed light on ways to improve food and vegetable disease prevention, quality, and environmental control.
In the previous study, the gene encoding Alt a 1 defined a protein family associated only with the Pleosporaceae family of fungi (Chruszcz et al., 2012; Gabriel et al., 2017). In contrast, in this study, the peroxiredoxin Asp n 3 belongs to a large protein superfamily associated with numerous species from different kingdoms (Rhee, 2016; Harper et al., 2017). Thus, it could be expected that the specificity for the model studied in this work could be lower than that found previously for the Alt a 1 gene in the infection of citrus fruit with A. alternata (Sáenz-de-Santamaría et al., 2006; Gabriel et al., 2015, 2017).
Classically, the macro- and micromorphology of fungi have been the reference methodology for their identification (Visagie et al., 2014). Since the 1990s, DNA sequencing has become one of the most powerful tools for taxonomists because of creating new concepts for identifying the species and defining new tools to establish relationships among them, bringing the opportunity for sequence-based identification (Blaxter, 2003).
During the last decade, ITS rDNA was accepted as the official barcode for fungal identification. Since then, the ITS has been the most widely sequenced marker for fungi, and universal primers for this sequence are available (Schoch et al., 2012). After the introduction of ITS as the most relevant reference marker for taxonomy and phylogenetic relationship studies, in practice, several secondary markers have been analyzed for reliable species identification (Fitzpatrick et al., 2006; Houbraken et al., 2012; Samson et al., 2014; Visagie et al., 2014; Tsang et al., 2018). In A. niger, some genes, such as the collagen-like gene, beta-tubulin gene, and calmodulin gene, have been described as secondary markers (Bennett, 2010; Shen et al., 2020).
Peroxiredoxins make up a wide family of proteins associated with several biological functions, including fungal pathogenicity toward their host plants and animals (Chen et al., 2016; Rhee, 2016). The Aspergillus group 3 main allergens (Asp f 3, Asp fl 3, Asp n 3, Asp o 3, Asp t 3, Asp ni 3, and Neo fi 3) are peroxisomal proteins. Thus, it would be very interesting to use the peroxiredoxin Asp n 3 as a secondary marker in the identification of the species of this genus and to study the phylogenetic relationships among species (Gautam et al., 2011; Heller and Tudzynski, 2011; Sharma, 2012).
The results presented in this work demonstrate that the Asp n 3 peroxiredoxin gene is an effective tool to identify A. niger and establish phylogenetic differences with other species from different taxa. The Asp n 3 peroxiredoxin gene was also demonstrated to be an effective tool to identify pathogenic Aspergillus of plant-based foods as well as to identify sources of fungal allergenic sources growing in plant foods.
Fungi from Aspergillaceae play diverse and significant roles in biotechnology and human, animal, and plant pathology. Several genomes from Aspergillaceae have been studied, including the two iconic genera Aspergillus and Penicillium. However, their analysis does not fully reflect the diversity of the family. Understanding the evolution of technologically and medically significant fungi through their intrinsic features requires a robust phylogeny (Steenwyk et al., 2019). Other authors suggest that the conflicting results in the fungal classification are likely associated with various processes, such as incomplete lineage sorting, hybridization, or introgression as well as with analytical issues associated with poor taxon sampling (Yilmaz et al., 2014).
DNA markers such as ITS regions or conserved genes such as calmodulin, tubulin, or RNA polymerase are used to conduct molecular phylogenetic studies on Aspergillus species. Despite the fact that phylogenetic trees based on single genes are prone to errors, such analyses are robust and have contributed to improving taxonomic systems (Bennett, 2010).
The alignment of genes coding for peroxiredoxin proteins (homologous to Asp n 3) allowed the construction of an Asp n 3-based neighbor-joining phylogenetic tree of species expressing this family of proteins. A monophyletic clade containing species belonging to Eurotiales can be observed. In turn, this group is divided into two differentiate clades, each one containing species belonging to Aspergillaceae and Trichomonaceae.
In agreement with other authors, Aspergillus forms a monophyletic clade closely related to Penicillium (Aspergillaceae) (Samson et al., 2014). Within this clade, A. niger, A. nomius, A. clavatus, N. fisheri, and A. fumigatus form the closest Aspergillus species subgroup from the phylogenetic point of view. On the other branch, A. nidulans, A. terreus, A. flavus, Aspergillus oryzae, Penicillium citrinum, Penicillium chrysogenum, and Penicillium digitatum complete a second subgroup of species within the Aspergillaceae monophyletic clade closely related to Penicillium. Both subgroups matched Aspergillaceae.
Penicillium (Talaromyces) marneffei, Talaromyces stipitatus, and Ramsonia emersonii form a polyphyletic group matching Trichomonaceae, differentiating from the monophyletic clade containing Aspergillus and Penicillium species.
Some authors suggest the transference of Penicillium subgenus Biverticillum to Talaromyces, and others separate Ramsonia from the other genera of Trichocomaceae using a combination of phenotypic characters, extrolite patterns, ITS, and partial calmodulin and b-tubulin sequences (Houbraken et al., 2012; Yilmaz et al., 2014).
The other monophyletic clade, including the species from Fonsecaea pedrosoi to Cladophialophora carrionii, is the nearest phylogenetic group to the clade that contains Eurotiales species. This clade matches species belonging to Chaetothyriales. The results of this work suggest that Chaetothyriales is the closest related order to Eurotiales within the class Eurotiomycetes.
A phylogenetic analysis revealed a high level of divergence of Asp n 3 among filamentous fungi. The Asp n 3 peroxiredoxin DNA sequence is a marker able to identify A. niger species. The use of a highly conserved internal nucleotide region makes it possible to detect, but not to identify, other Aspergillus species.
The alignment of peroxiredoxin nucleotides showed more than 86% identity among species from the Aspergillus genus, 81–84% identity when Penicillium species were included, and 76–80% identity when the alignment was extended to Talaromyces and Ramsonia, taking into account that all these genera belong to Eurotiales.
These results justify the detection of Aspergillus species, but not their identification, when only highly conserved regions are used. Species identification requires tools based on full gene sequences.
Previously, some authors demonstrated that cytochrome B coding genes or small heat shock protein coding genes were useful for the most accurate classification of fungi (Yokoyama et al., 2001; Wu et al., 2016). In our case, we demonstrate that the peroxiredoxin Asp n 3 can be used in the analysis of phylogeny, classification, and identification of aspergilli.
Conclusion
The Asp n 3 DNA sequence is a reliable tool to detect and identify early fungal contamination by Aspergillus and postharvest damage in M. indica fruit. The use of conserved segments of the Asp n 3 gene or its entire sequence allows us to detect phylogenetically closely related species within the Aspergilaceae family or to identify species-specific contaminating fungi.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author Contributions
JM, IP, and ES designed the experiments. AN and MG performed the experiments. AN, AV-d-B, and PS conducted bioinformatics for the amplicon sequencing results and phylogenic studies. JM and IP wrote the manuscript. ES formatted the document. All the authors reviewed and revised the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the Government of the Basque Country, Department of Research and Education, Linguistic Policy and Culture in the Program: Grants for University Research Groups (project IT-1043-16).
References
- Al-Hindi R. R., Al-Najada A. R., Mohamed S. A. (2011). Isolation and identification of some fruit spoilage fungi: screening of plant cell wall degrading enzymes. Afr. J. Microbiol. Res. 5 443–448. 10.5897/AJMR10.896 [DOI] [Google Scholar]
- Amalaradjou M. A. R., Venkitanarayanan K. (2008). “Chapter X: detection of Penicillium, Aspergillus and Alternaria species in fruits and vegetables,” in Mycotoxins in Fruits and Vegetables, eds Barkai-Golan R., Paster N. (Oxford: Academic Press, Elsevier; ), 225–247. 10.1016/B978-0-12-374126-4.00010-3 [DOI] [Google Scholar]
- Battilani P., Barbano C., Logrieco A. (2008). “Chapter I: risk assessment and safety evaluation of mycotoxins in fruits,” in Mycotoxins in Fruits and Vegetables, eds Barkai-Golan R., Paster N. (Oxford: Academic Press, Elsevier; ), 1–26. 10.1016/B978-0-12-374126-4.00001-2 [DOI] [Google Scholar]
- Bennett J. W. (2010). “An overview of the genus Aspergillus,” in Aspergillus: Molecular Biology and Genomics, ed. Gomi Machida K. (Portland, OR: Academic Press; ), 1–17. [Google Scholar]
- Blaxter M. (2003). Counting angels with DNA. Nature 421 1–3. 10.1016/j.simyco.2014.09.001 [DOI] [PubMed] [Google Scholar]
- Blumenthal C. Z. (2004). Production of toxic metabolites in A. niger and T. reesei: justification of mycotoxin testing in food grade enzyme preparations derived from the three fungi. Regul. Toxicol. Pharmacol. 39 214–228. 10.1016/j.yrtph.2003.09.002 [DOI] [PubMed] [Google Scholar]
- Chen X. L., Wang Z., Liu C. A. (2016). Roles of peroxisomes in the rice blast fungus. BioMed. Res. Int. 2016:9343417. 10.1155/2016/9343417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen M., Tuthill D. E. (1986). “Aspergillus: an overview,” in Advances in Penicillium and Aspergillus Systematics. NATO ASI Series (Series A: Life Sciences), eds Samson R. A., Pitt J. I. (Boston, MA: Springer; ), 102. [Google Scholar]
- Chruszcz M., Chapman M. D., Osinski T., Solberg R., Demas M., Porebski P. J., et al. (2012). Alternaria alternate allergen, Alt a 1-a unique, β-barrel protein dimer exclusively found in fungi. J. Allergy Clin. Immunol. 130 241–247. 10.1016/j.jaci.2012.03.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crameri C., Hemmann S., Ismail C., Menz G., Blaser K. (1998). Disease-specific recombinant allergens for the diagnosis of allergic bronchopulmonary. Int. Immunol. 10 1211–1216. 10.1093/intimm/10.8.1211 [DOI] [PubMed] [Google Scholar]
- Dijksterhuis J. (2019). Fungal spores: highly variable and stress-resistant vehicles for distribution and spoilage. Food Microbiol. 81 2–11. 10.1016/j.fm.2018.11.006 [DOI] [PubMed] [Google Scholar]
- Fitzpatrick D. A., Logue M. E., Stajich J. E., Butler G. (2006). A fungal phylogeny based on 42 complete genomes derived from super tree and combined gene analysis. BMC Evol. Biol. 6 99–113. 10.1186/1471-2148-6-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel M. F., Martínez J., Postigo I. (2016a). “Chapter XIII: fungal allergens: recent trends and future prospects,” in Medical Mycology: Current Trends and Future Prospects, eds Razzaghi-Abyaneh M., Shams-Ghahfarokhi M., Rai M. (Boca Raton, FL: CRC Press; ), 315–333. [Google Scholar]
- Gabriel M. F., Postigo I., Gutiérrez A., Suñén E., Tomaz C. T., Martínez J. (2015). Development of a PCR-based tool for detecting immunologically relevant Alt a 1 and Alt a 1 homologue coding sequences. Med. Mycol. 53 636–642. 10.1093/mmy/myv022 [DOI] [PubMed] [Google Scholar]
- Gabriel M. F., Postigo I., Tomaz C. T., Martínez J. (2016b). Alternaria alternata allergens: markers of exposure, phylogeny and risk of fungi-induced respiratory allergy. Environ. Int. 89-90 71–80. 10.1016/j.envint.2016.01.003 [DOI] [PubMed] [Google Scholar]
- Gabriel M. F., Uriel N., Teifoori F., Postigo I., Suñén E., Martínez J. (2017). The major Alternaria alternata allergen, Alt a 1: a reliable and specific marker of fungal contamination in citrus fruit. Int. J. Food Microbiol. 257 26–30. 10.1016/j.ijfoodmicro.2017.06.006 [DOI] [PubMed] [Google Scholar]
- Gautam A. K., Sharma S., Avasthi S., Bhadauria R. (2011). Diversity, pathogenicity and toxicology of A. niger: an important spoilage fungi. Res. J. Microbiol. 6 270–280. 10.3923/jm.2011.270.280 [DOI] [Google Scholar]
- Harper A. F., Leuthaeuser J. B., Babbitt P. C., Morris J. H., Ferrin T. E., Poole L. B., et al. (2017). An atlas of peroxiredoxins created using an active site profile-based approach to functionally relevant clustering of proteins. PLoS Comput. Biol. 13:e1005284. 10.1371/journal.pcbi.1005284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller J., Tudzynski P. (2011). Reactive oxygen species in phytopathogenic fungi: signaling, development, and disease. Annu. Rev. Phytopathol 49 369–390. 10.1146/annurev-phyto-072910-095355 [DOI] [PubMed] [Google Scholar]
- Hemmann S., Blaser K., Crameri R. (1997). Allergens of Aspergillus fumigatus and Candida boidinii share IgE-binding epitopes. Am. J. Respir. Crit. Care Med. 156 1956–1962. 10.1164/ajrccm.156.6.9702087 [DOI] [PubMed] [Google Scholar]
- Houbraken J., Spierenburg H., Frisvad J. C. (2012). Rasamsonia, a new genus comprising thermotolerant and thermophilic Talaromyces and Geosmithia species. AvL 101 403–421. 10.1007/s10482-011-9647-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasprzyk I. (2008). Aeromycology main research fields of interest during the last 25 years. Ann. Agric. Environ. Med. 15 1–7. [PubMed] [Google Scholar]
- Kim W. B., Park C., Cho S. Y., Chun H. S., Lee D. G. (2020). Development of multiplex real-time PCR for rapid identification and quantitative analysis of Aspergillus species. PLoS One 15:e0229561. 10.1371/journal.pone.0229561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubna, Asaf S., Hamayun M., Gul H., Lee I.-J., Hussain A. (2018). Aspergillus niger CSR3 regulates plant endogenous hormones and secondary metabolites by producing gibberellins and indoleacetic acid. J. Plant Interact. 1 100–111. 10.1080/17429145.2018.1436199 [DOI] [Google Scholar]
- Mahfooz S., Singh S. P., Mishra N., Mishra A. (2017). A comparison of microsatellites in phytopathogenic Aspergillus species in order to develop markers for the assessment of genetic diversity among its isolates. Front. Microbiol. 8:1774. 10.3389/fmicb.2017.01774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matricardi P. M., Kleine-Tebbe J., Hoffmann H. J., Valenta R., Ollert M. (2016). Molecular Allergology user’s Guide. European Academy of Allergy and Clinical Immunology. Hoboken, NJNJ: John Wiley & Sons. [Google Scholar]
- Palejwala V. A., Patki C. K., Bhatt S. V., Modi V. V. (1987). Post-harvest spoilage of mangoes by Aspergillus niger. Int. J. Food Microbiol. 5 111–116. 10.1016/0168-1605(87)90028-6 [DOI] [Google Scholar]
- Perrone G., Susca A., Cozzi G., Ehrlich K., Varga J., Frisvad J. C., et al. (2007). Biodiversity of Aspergillus species in some important agricultural products. Stud. Mycol. 59 53–66. 10.3114/sim.2007.59.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertile G., Lamorski K., Bieganowski A., Boguta P., Brzeziñska M., Polakowski C., et al. (2021). Immediate effects of the application of various fungal strains with urea fertiliser on microbiome structure and functions and their relationships with the physicochemical parameters of two different soil types. Appl. Soil Ecol. 163:103972. 10.1016/j.apsoil.2021.103972 [DOI] [Google Scholar]
- Raper K. B., Fennell D. I. (1965). The Genus Aspergillus. Baltimore, MD: Williams and Wilkins. 10.1126/science.150.3697.736-a [DOI] [Google Scholar]
- Rhee S. G. (2016). Overview on peroxiredoxins. Mol. Cells 39 1–5. 10.14348/molcells.2016.2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sáenz-de-Santamaría M., Postigo I., Gutiérrez-Rodríguez A., Cardona G., Guisantes J. A., Asturias A., et al. (2006). The major allergen of Alternaria alternata (Alt a 1) is expressed in other members of the Pleosporaceae family. Mycoses 49 91–95. 10.1111/j.1439-0507.2006.01195.x [DOI] [PubMed] [Google Scholar]
- Samson R. A., Houbraken J., Thrane U., Frisvald J. C., Andersen B. (2010). Food and Indoor Fungi. CBS Laboratory Manual Series. Utrecht: CBS-KNAW Fungal Diversity Centre. [Google Scholar]
- Samson R. A., Visagie C. M., Houbraken J., Hong S. B., Hubka V., Klaassen C. H. W., et al. (2014). Phylogeny, identification and nomenclature of the genus Aspergillus. Stud. Mycol. 78 141–173. 10.1016/j.simyco.2014.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch C. L., Seifert K. A., Huhndorf S., Robert V., Spouge J. L., Levesque C. A., et al. (2012). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc. Natl. Acad. Sci. U.S.A. 109 6241–6246. 10.1073/pnas.1117018109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R. (2012). Pathogenecity of Aspergillus niger in plants. CJM 1 47–45. [Google Scholar]
- Shen Y., Nie J., Kuang L., Zhang J., Li H. (2020). DNA sequencing, genomes and genetic markers of microbes on fruit and vegetables. Microb. Biotech. 14 323–362. 10.1111/1751-7915.13560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Nobbe B., Denk U., Pöll V., Rid R., Breitenbach M. (2008). The Spectrum of Fungal Allergy. Int. Arch. Allergy Immunol. 145 58–86. 10.1159/000107578 [DOI] [PubMed] [Google Scholar]
- Steenwyk J. L., Shen X. X., Lind A. L., Goldman G. H., Rokas A. (2019). A robust phylogenomic time tree for biotechnologically and medically important fungi in the genera Aspergillus and Penicillium. mBio 10:e00925-19. 10.1128/mBio.00925-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24 1596–1599. 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- Tian J., Wang Y., Zeng H., Li Z., Zhang P., Tessema A., et al. (2015). Efficacy and possible mechanisms of perillaldehyde in control of Aspergillus niger causing grape decay. Int. J. Food Microbiol. 202 27–34. 10.1016/j.ijfoodmicro.2015.02.022 [DOI] [PubMed] [Google Scholar]
- Tsang C. C., Tang J. Y. M., Lau S. K. P., Woo P. C. Y. (2018). Taxonomy and evolution of Aspergillus, Penicillium and Talaromyces in the omics era. Past, present and future. Comp. Struct. Biotech. J. 16 197–210. 10.1016/j.csbj.2018.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie C. M., Houbraken J., Frisvad J. C., Hong S. B., Klaassen C. H. W., Perrone G., et al. (2014). Identification and nomenclature of the genus Penicillium. Stud. Mycol. 78 343–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Wang M., Zhou L., Yu D. (2016). Small heat shock proteins, phylogeny in filamentous fungi and expression analyses in Aspergillus nidulans. Gene 575 675–679. 10.1016/j.gene.2015.09.044 [DOI] [PubMed] [Google Scholar]
- Yilmaz N., Visagie C. M., Houbraken J., Frisvad J. C., Samson R. A. (2014). Polyphasic taxonomy of the genus Talaromyces. Stud. Mycol. 78 175–341. 10.1016/j.simyco.2014.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama K., Wang L., Miyaji M., Nishimura K. (2001). Identification, classification and phylogeny of the Aspergillus section Nigri inferred from mitochondrial cytochrome b gene. FEMS Microbiol. Lett. 200 241–246. 10.1111/j.1574-6968.2001.tb10722.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.