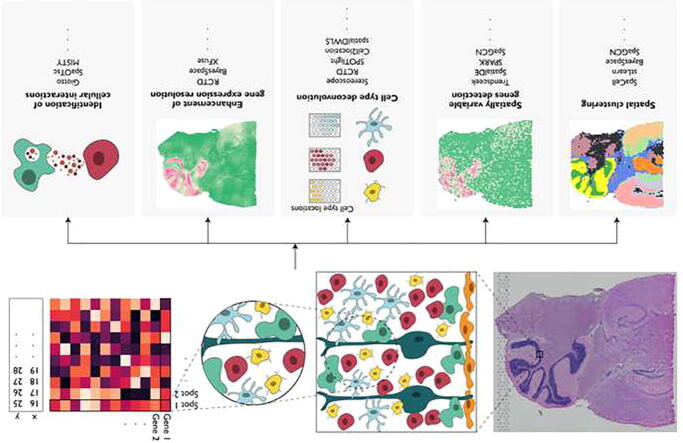
Graphical abstract
Keywords: Spatially resolved transcriptomics, Spatial clustering, Spatially variable genes, Celltype deconvolution, Cell-cell communications
Abstract
Recent developments in spatially resolved transcriptomics (SRT) technologies have enabled scientists to get an integrated understanding of cells in their morphological context. Applications of these technologies in diverse tissues and diseases have transformed our views of transcriptional complexity. Most published studies utilized tools developed for single-cell RNA sequencing (scRNA-seq) for data analysis. However, SRT data exhibit different properties from scRNA-seq. To take full advantage of the added dimension on spatial location information in such data, new methods that are tailored for SRT are needed. Additionally, SRT data often have companion high-resolution histology information available. Incorporating histological features in gene expression analysis is an underexplored area. In this review, we will focus on the statistical and machine learning aspects for SRT data analysis and discuss how spatial location and histology information can be integrated with gene expression to advance our understanding of the transcriptional complexity. We also point out open problems and future research directions in this field.
1. Introduction
The tissues in our body consist of diverse cell types with each cell type specialized to carry out a particular function. The behavior of a cell is influenced by its surrounding environment within a tissue. Knowledge of the relative locations of different cells in a tissue is critical for understanding the spatial organization of cell types and disease pathology. Although scRNA-seq has made it possible to characterize cell types and states and to study cellular mechanisms at an unprecedented resolution, the lack of physical relationship among cells has hindered the study of cell–cell interactions within tissue context. The maintenance of spatial context is critical for uncovering the complex transcriptional architecture of heterogenous tissues; for example, within a tumor, several subpopulations of cancer cells constituting a tumor can vastly differ from each other in their gene expression profiles and cellular properties due to residing in distinct tumor microenvironments.
Recent technology advances in spatially resolved transcriptomics (SRT) have enabled gene expression profiling with location information in tissues [1], [2], [3], [4], [5], [6]. Popular experimental methods to generate SRT data can be broadly classified into two categories. The first category is image-based in situ transcriptomics, termed single-molecule fluorescent in situ hybridization (smFISH) [7], [8], [9], [10], [11], which detects several mRNA transcripts simultaneously at subcellular resolution. Later efforts such as seqFISH [12], [13], seqFISH+ [14], and MERFISH [15], [16] have substantially increased the number of detectable mRNA species with multiplexed smFISH. With these new technologies, the expression level for hundreds to thousands of genes can be simultaneously measured with subcellular resolution in a single cell. The second category is based on spatial barcoding followed by next-generation sequencing-based techniques, such as Spatial Transcriptomics (ST) [17], SLIDE-seq [18], SLIDE-seq2 [19], and high-definition spatial transcriptomics (HDST) [20], which measure the expression level for thousands of genes in captured locations, referred to as spots. Recently, 10x Genomics commercialized the ST technology in their Visium Spatial Gene Expression platform, which reduced the diameter size from 100 µm per spot in ST to 55 µm. Although SLIDE-seq and HDST have higher resolution than ST and Visium, the number of unique molecules detected per spot by these technologies is lower than ST and Visium.
Sequencing-based methods for SRT are often complemented by high-resolution hematoxylin and eosin (H&E) stained histology images, which are invaluable for examining cellular morphology and how it changes over embryonic development or disease progression. Since sequencing-based technologies intend to capture mRNAs without the need to prespecify what genes to include, they can characterize both known and unknown molecular features in a tissue section. The combined gene expression and histological features within spatial context allow researchers to access additional dimensions of information helping to inform developmental trajectory and the origin and progression of complex disease. Using sequencing-based SRT technologies, Asp et al. profiled spatiotemporal gene expression patterns in developing human heart [21]; Maniatis et al. studied the progression of amyotrophic lateral sclerosis [22]; and Chen et al. identified transcriptional changes in tissue domains surrounding amyloid plaques in Alzheimer’s disease [23]. Sequencing-based SRT has also been employed to study various types of cancer, including prostate cancer [24], melanoma [25], breast cancer [26], pancreatic ductal adenocarcinomas [27], and squamous cell carcinoma [28].
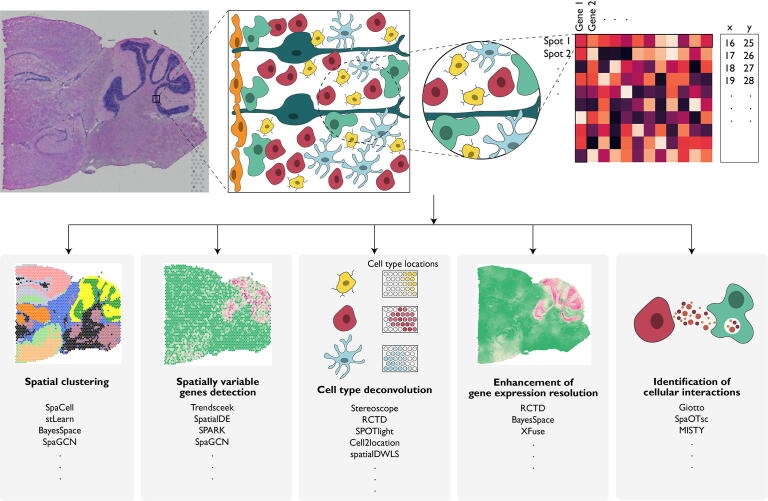
Due to the profound impact of SRT in advancing our views of transcriptional complexity, Nature Methods recently selected SRT as Method of the Year 2020 [29], [30], [31], [32], [33]. Many of the published studies on SRT used computational tools developed for scRNA-seq. However, SRT data have different properties from those from scRNA-seq e.g., sequencing-based SRT technologies often measure the transcriptomes of multiple cells per spot, and the gene expression levels of neighboring spots and cells are correlated. However, the spatial dependency of gene expression and the histological features are not modeled by tools that are developed for scRNA-seq. To fully harness the added spatial and histology information in SRT, new methods that can connect gene expression features with spatial location and histological features are needed. In this review, we will focus on statistical and machine learning methods for the analysis of SRT data with a particular emphasis on how histology image information can be jointly modeled with gene expression. Since numerous methods are available, we mainly focus on methods that can be applied to both imaging- and sequencing-based SRT data. We will discuss common tasks in data analysis and available methods for each of these tasks (Fig. 1). We will also point out open problems and future research directions.
Fig. 1.
Spatially resolved transcriptomics and its applications. Spatially resolved transcriptomics has enabled gene expression profiling with location information in tissues. In the experiment, the expression levels for thousands of genes in captured spots are measured, complemented by a high-resolution histology image for the same tissue section. In sequencing-based spatially resolved transcriptomics, the number of cells in each captured spot varies depending on the diameter of each spot. Some common tasks in spatially resolved transcriptomics data analysis include: 1) spatial clustering; 2) spatially variable gene detection; 3) cell-type deconvolution; 4) enhancement of gene expression resolution; 5) cell–cell communications.
2. Spatial clustering
In SRT studies, an important step is to cluster the spots and identify spatial domains, i.e., regions that are spatially coherent in both gene expression and histology. Identifying spatial domains requires methods that can jointly consider gene expression, spatial location, and histology. Traditional clustering methods used in scRNA-seq analysis, e.g., K-means [34] and Louvain’s method [35], only take gene expression data as input, but do not incorporate spatial location and histology information. As such, the resulting clusters may not be contiguous due to the lack of consideration of spatial and histology constraints during clustering [36]. To account for spatial dependency of gene expression, several new spatial clustering methods have been developed.
2.1. Hidden-Markov random field-based approach
Zhu et al. [37] developed a Hidden-Markov random field (HMRF) approach to model spatial dependency of gene expression. HMRF represents a stochastic process generated by a Markov random field whose state sequence cannot be observed directly but can be estimated through observations that are assumed to be a stochastic function of the state sequence. HMRF is commonly used to model the spatial distribution of signals, for example, segmentation of brain magnetic resonance images [38]. To utilize HMRF for clustering analysis of SRT data, Zhu et al. represented the spatial structure of cells as a set of nodes on a grid with neighboring nodes connected to each other. The cluster membership of each cell is hidden but can be estimated from the observed gene expression data. A critical assumption in HMRF is the Markov property, which assumes that the spatial dependency can be modeled by only considering the correlation between immediate neighboring nodes. With this assumption, the joint distribution of gene expression across all nodes can be decomposed as the product of much smaller components with each component defined on a fully connected subgraph. The HMRF framework constructs an undirected graph, which represents the spatial relationship among cells and enables clustering by systematically comparing the gene expression profile of each cell with its neighboring cells. Applying this approach to a seqFISH dataset generated from mouse visual cortex, they identified nine spatial domains, where some domains displayed a layered organization that resembled the anatomical structure of the visual cortex. Although these results are promising, the HMRF approach did not incorporate histology information. Thus, its performance for sequencing-based SRT data might be sub-optimal.
2.2. SpaCell
Pixel intensities in histology images contain informative features that can be used for diagnosing diseases such as cancer staging [39]. Effectively incorporating histology image information to gene expression data analysis is still an underexplored analysis area [40]. SpaCell [41] is the first paper that integrates histology and gene expression data in spatial clustering. SpaCell starts by dividing the whole tissue slide histology image into small tiles with each image tile resized to 299 × 299 pixels, containing one spot. Of note, as the diameter size for each spot varies in different sequencing-based SRT technologies, the pixel size also varies. To extract feature vectors for each image tile, the weights in a convolutional neural network (CNN) are initialized from the ResNet50 model that utilizes images in the ImageNet database [42] and then further fine-tuned. This CNN captures generic features of images and is used to find a latent embedding vector representing informative features in the image tile for each spot. After pretraining, SpaCell then trains two autoencoders, one for the tile images and one for the gene expression data. The obtained embedding layers for these two autoencoders are concatenated into one combined embedding layer, which is then used to perform clustering analysis using conventional clustering algorithms, e.g., K-means or Louvain. The authors showed that this integration approach outperformed methods that use gene expression data alone or histology imaging data alone in identifying cancer and non-cancer regions. Although SpaCell has shown promising performance, the spatial coordinates of the spots are not utilized in their autoencoders.
2.3. StLearn
Due to the technical limitations of the sequencing-based SRT technologies in detecting lowly expressed genes, some genes may not be measured, leading to excessive zeros, which may lead to difficulty in assigning cluster membership. However, the spatial dependency of gene expression across spots offers an opportunity to improve the gene expression quality. Recognizing this, stLearn uses the expression of neighboring spots as well as features extracted from a histology image to spatially normalize gene expression data before clustering [43]. The intuition behind this normalization is that spots that are physically close and share similar morphological features are expected to have similar gene expression. As such, stLearn normalizes the expression value for each gene in the center spot as the mean of morphological similarity-weighted expression values of its neighboring spots. A limitation of stLearn is that it uses an arbitrarily chosen radius to define the neighborhood of a given spot. Given the complexity of real data, it is unlikely that a fixed radius approach would work well. This histology guided normalization can help aggregate gene expression across closely related spots, not only by physical location but also by histological features. Consideration of histological features is an important step to ensure the appropriate aggregation of gene expression as physically close spots do not necessarily have similar expression patterns. StLearn uses a similar deep learning-based approach as described in SpaCell to extract histological features. Although histology information is useful, caution is needed to ensure histology-specific artifacts do not propagate to downstream analysis. More specifically, histological stains may demonstrate non-biological and spatially-determined variability due to differences in the ability of the stain to permeate different regions of the tissue [44]. Both SpaCell and stLearn use ImageNet to train their neural network. Since the images in ImageNet are not histology images, we caution that features extracted from ImageNet are not necessarily informative for histology images.
2.4. BayesSpace
BayesSpace employs a fully Bayesian approach for clustering analysis of SRT data [45]. To account for spatial dependency of gene expression, they model a low-dimensional representation of the gene expression matrix using a spatial prior. Specifically, they assume that given the unobserved cluster membership, the low-dimensional representation of the gene expression follows a multivariate normal distribution, where the mean vector and precision matrix follow a spatial prior that encourages neighboring spots to belong to the same cluster. BayesSpace estimates parameters using a Markov chain Monte Carlo method and infers cluster memberships via Metropolis-Hastings. While the spatial prior encourages spots that are physically close to be assigned to the same cluster, the spatial prior in their model does not explicitly use the spatial coordinates. Also, it does not consider information offered by histology images. As shown in both SpaCell [41] and stLearn [43], spots that are physically close to each other do not necessarily belong to the same spatial domain. For example, in the cortex, the tissue is organized in distinct tissue layers, ordered from L1 to L6, that are functionally distinct. Spots that are located at the boundaries of adjacent cortical layers are physically close but may belong to different layers and possess distinct gene expression profiles. Thus, failure to consider histology information may lead to misclustering of spots that belong to different tissue layers into the same cluster.
2.5. SpaGCN
More recently, Hu et al. developed SpaGCN, a graph convolutional network-based approach that considers both spatial location and histology information in clustering [36]. SpaGCN starts by building a weighted undirected graph in which each vertex represents a spot, and every two vertices are connected via an edge with prespecified weight that measures the degree of similarity between two spots. The distance between any two vertices is determined by both the physical locations of the corresponding spots and their histological features. The edge weight is negatively correlated with this distance; thus, two spots are considered similar if and only if they are physically close and have similar pixel intensities in the histology image. To define a distance metric considering both gene expression and histological features, SpaGCN extended the 2D space in the tissue slice into a 3D space that incorporates histology information as the third dimension. Next, SpaGCN utilizes a graph convolutional layer to aggregate gene expression from neighboring spots, where the expression in each spot is a weighted average across its neighboring spots with weights determined by spatial location and histology. Unlike stLearn which uses an arbitrarily chosen radius to define the neighborhood of a given spot, SpaGCN automatically weighs each spot in gene expression aggregation, which allows it to consider all spots simultaneously without arbitrarily defining a radius threshold. The graph convolutional layer in SpaGCN is connected to a clustering layer to iteratively cluster the spots into different spatial domains, where the filter parameters in the graph convolutional layer are also updated during this iterative clustering process. Each cluster identified from this analysis contains spots that are coherent in gene expression and histology. Rather than considering the corresponding histological features within each spot as an image as in SpaCell [41] and stLearn [43], SpaGCN extracts RGB values of each pixel and weighs the three-color channels according to their explained variation. This approach is less sensitive to artifacts in H&E stained histology images.
3. Identification of spatially variable genes
Another important task in SRT data analysis is to identify spatially variable genes (SVGs), i.e., genes that show spatial expression variation across a tissue section. Methods for SVG detection fall into two categories, where the first category aims to detect SVGs without the consideration of spatial domains, and the second category detects SVGs with the guidance of spatial domains identified from a spatial clustering algorithm. Recently developed methods, such as Trendsceek [46], SpatialDE [47], SPARK [48], belong to the first category, whereas SpaGCN [36] belongs to the second category. Consideration of spatial domains in SVG detection will help ensure the detected genes show enriched expression pattern in a spatial domain. These genes can serve as landmarks in helping to reconstruct the spatial locations of cells in scRNA-seq [49], [50].
3.1. Trendsceek
Trendsceek utilizes a marked point process to assess the significance of the spatial expression trend for each gene [46]. For all cell pairs within a particular radius, Trendsceek tests for a significant dependency between their gene expression levels and their 2D distance defined by the spatial coordinates of the cell pairs. To perform the test, four summary statistics of the pair distribution including conditional mean, conditional variance, Stoyan’s mark correlation, and the mark-variogram, are calculated and compared to the null distribution of the summary statistics derived from permuted gene expression labels. This test is non-parametric and does not need to pre-specify a spatial pattern or a spatial region of interest. As a result, the detected SVGs do not have a guaranteed spatial expression pattern. Since Trendsceek considers cells in a pairwise fashion, it may have limited power to detect the global expression pattern of a gene that spans beyond a cell pair.
3.2. SpatialDE
SpatialDE is another recently developed method for SVG detection [47]. It is based on Gaussian process regression, a class of models used in geostatistics. Briefly, for each gene, SpatialDE decomposes its expression variability into spatial and nonspatial components using two random effect terms: a spatial variance term that parametrizes gene expression covariance by pairwise distances of samples and a noise term that models nonspatial variability. The ratio of the variance explained by these components quantifies the fraction of spatial variance. One can detect significant SVGs by comparing this full model to a model without the spatial variance component with a likelihood ratio statistic and P-value estimated by the chi-squared distribution with one degree of freedom. SpatialDE also provides a spatial clustering method to group genes that mark distinct expression patterns, which further allow them to uncover the hidden histological pattern of gene expression. Results from such analysis can elucidate the relationship between tissue structure and cell-type composition based on the expression patterns of the detected SVGs.
3.3. Spark
Both Trendsceek [46] and SpatialDE [47] transform gene expression count data into normalized expression before analysis. However, gene expression is count-based. Analyzing the normalized data may lead to loss of power because it fails to account for the mean–variance relationship that exists in raw counts. To directly model count data, Sun et al. developed SPARK [48], which is built upon a generalized linear spatial model with a variety of spatial kernels to accommodate count data generated from both smFISH- and sequencing-based SRT studies. Since the generalized linear spatial model likelihood consists of high-dimensional integral that has no closed-form solution, SPARK uses an approximate inference algorithm that is based on a penalized quasi-likelihood to make the computation more tractable. As a parametric based method, SPARK requires the pre-specification of spatial kernels. Since the spatial pattern varies from gene to gene, and it is often unknown without the consideration of spatial domains, SPARK considers multiple spatial kernels with each kernel representing a specific spatial gene expression pattern. To combine results across multiple spatial kernels together, SPARK uses a recently developed Cauchy combination approach to calculate a calibrated P-value [51]. Although SPARK has shown improved performance compared to Trendsceek [46] and SpatialDE [47], the reliance on pre-specified spatial kernels may limit its detection of genes whose expression patterns are not captured by their pre-specified kernels.
3.4. SpaGCN
Trendsceek [46], SpatialDE [47], and SPARK [48] examine each gene independently and return a P-value to represent the spatial variability of a gene. However, methods that independently test genes for spatial variability do not consider the highly correlated nature of gene expression. As a result, they may identify spurious, though statistically significant, spatial patterns of individual gene expression. To approach this limitation, Hu et al. proposed SpaGCN [36], which detects SVGs that are enriched in a spatial domain by domain-guided differential expression analysis. When a single gene cannot mark the expression pattern of a spatial domain, SpaGCN will construct a metagene, whose expression is a log-linear combination of the expression values of multiple SVGs, to represent the expression pattern of the domain. Briefly, SpaGCN first identifies a base gene that is weakly enriched in the target domain. Next, it detects some positive genes that are highly expressed in the target domain as well as other domains, and negative genes that are highly expressed in other domains but not the target domain. By adding positive genes to and subtracting negative genes from the base gene, SpaGCN can return a metagene that is uniquely expressed in the target domain. As the spatial domains are identified through joint consideration of gene expression, spatial location, and histology, SVGs detected by SpaGCN are guaranteed to have spatial patterns that match the spatial domains. Based on analysis across different species and tissues generated from both sequencing- and smFISH-based SRT technologies, Hu et al. showed that SpaGCN is more likely to detect genuinely biological SVGs and metagenes than SpatialDE and SPARK. They also showed that the SVGs and metagenes detected by SpaGCN are transferrable and can be utilized to study spatial variation of gene expression in other datasets.
4. Cell-type deconvolution in spatial transcriptomics spots
Although sequencing-based SRT technologies such as ST and Visium allow an unbiased survey of the transcriptome, the primary technological limitation of ST and Visium is the lack of single-cell resolution. The first-generation ST microarrays consist of ~1000 spots, each with a diameter of 100 μm and covering tens of cells. In the recent Visium platform, the throughput is increased to ~5000 spots and the diameter of each spot is reduced to 55 μm. Depending on tissue type, the number of cells per spot in Visium is about 1–10. Thus, the observed gene expression at each spot in ST and Visium may stem from a heterogeneous set of cells, not all necessarily of the same type. On the other hand, scRNA-seq profiles gene expression with single-cell resolution, although with the loss of spatial location information. Given the complementary information provided by sequencing-based SRT and scRNA-seq, one can use statistical approaches to integrate these two data types to infer the spatial locations of different cell types in a tissue. Cell-type deconvolution is not a new problem. Indeed, this approach has been employed in bulk RNA-seq to infer cell-type composition with cell-type-specific gene expression provided by scRNA-seq [52], [53], [54], [55]. However, traditional deconvolution methods do not work well for sequencing-based SRT data due to the lack of consideration of spatial dependency of gene expression. More recently, methods designed specifically for sequencing-based SRT data have emerged.
4.1. Stereoscope
Andersson et al. proposed a model-based method to infer cell-type proportions for sequencing-based SRT data [56]. For each gene, it first models the scRNA-seq gene expression count data using a Negative Binomial distribution, whose first parameter (the rate) is a product of a scaling factor that accounts for the library size of a cell and a cell-type-specific rate parameter, and whose second parameter (the success probability) that depends on the gene and is assumed to be shared across all cell types. The total number of transcripts for a given gene in each spot, which is the sum of transcripts across all cells within the spot, also follows a Negative Binomial distribution, with rate parameter equal to the sum of all contributing cells’ rates and the success probability remains unchanged. Next, the cell-type-specific rate parameter and the success probability are inferred from single-cell data and the library size scaling factor is adjusted for spot library size. For each spot, the proportion of cell types that best explains the spatial data is inferred using maximum a posteriori estimation. Applying Stereoscope to data generated from ST and Visium, cell types from mouse brain and developmental heart were mapped with expected cell-type arrangement.
4.2. RCTd
RCTD is a recently developed supervised learning approach for cell-type deconvolution in sequencing-based SRT [57]. RCTD models the observed gene expression counts using a Poisson-lognormal mixture hierarchical model. The gene counts are assumed to be Poisson-distributed with an expected rate determined by the spot’s total transcript count multiplied by a mixture of cell-type-specific expression profiles. The expected rate is modeled using a linear mixed-effects model comprised of the mean expression profile, the proportion of contribution for each cell type, a spot-specific fixed effect, a gene-specific platform random effect, and a random error term to account for additional variation, such as spatial effects. The cell-type proportions are inferred using maximum likelihood estimation. Cable et al. [57] demonstrated that platform effects between scRNA-seq reference and SRT target data can lead to challenges when transferring cell-type information. To correct for platform differences between scRNA-seq and SRT, RCTD performs a normalization procedure that relies on merging all spots into one pseudo-bulk measurement. After normalization, the proportion of each cell type in each spot is estimated using maximum likelihood estimation. Although RCTD accounts for cross-platform learning, this deconvolution method is limited in the use of spatial information and does not take into account the physical distance between spots, spatial dependency of gene expression, or histology imaging from the SRT data. If users have prior knowledge that more than two cell types per spot is rare, RCTD can run doublet mode to constrain the number of cell types per spot. This step can reduce overfitting, but may become too restrictive when more than two cell types are present.
4.3. SPOTlight
SPOTlight is a method for cell-type-deconvolution in SRT that is based on a seeded non-negative matrix factorization (NMF) regression framework [58]. SPOTlight first selects cell-type-specific marker genes and highly variables genes from scRNA-seq data and only uses their intersections with genes in the SRT data as input. Next, SPOTlight factorizes the normalized scRNA-seq gene expression matrix into two lower dimensionality matrices using NMF. The first output matrix is regarded as a gene-level topic distribution matrix while the second is regarded as a cell-level topic distribution matrix. Before running factorization, the two topic matrices are initialized with prior knowledge, thus guiding it towards a biologically relevant result. The gene-level topic distribution is used to map the normalized SRT gene expression matrix to a spot-level topic distribution matrix through a Non-Negative Least Squares regression (NNLS). Meanwhile, the cell-level topic distribution matrix is used to learn the cell-type specific topic profiles. SPOTlight then uses NNLS to find the weights of each cell-type-specific topic profile that can best reconstruct the spot-level topic distribution matrix, and the weights represent the cell-type proportions across all spots in the SRT data.
4.4. Cell2location
Kleshchevnikov et al. developed cell2location, a SRT deconvolution algorithm built on a hierarchical Bayesian framework [59]. The model assumes that the count for a gene in a given SRT spot follows a Negative Binomial distribution, with an unobserved rate parameter and a shared gene-specific over-dispersion parameter to represent the expression variance of that particular gene. The unobserved rate parameter for a given spot and a gene is defined as a linear function of the cell-type-specific gene expression signatures for that gene in the scRNA-seq data. This function incorporates a scaling parameter to account for the inherently different expression levels captured by different technologies and two additive shift parameters to account for differences in expression levels across spots and genes, respectively. Each cell-type-specific gene expression signature is weighted by a parameter specific to the spot of interest, which represents the abundance of cells in the spot expressing that signature. The method uses hierarchical Gamma priors for the scaling, weight, and additive shift parameters. In order to establish informative priors for these parameters, cell2location requires the user to define hyperparameters for the average number of cells, cell types, and tissue zones per spot and the average difference in technical sensitivity between the scRNA-seq and SRT data. While the authors specify that the choice of these hyperparameters greatly affects the performance of the model, some of the hyperparameters will often not be readily known by the user and accurate estimates may be unattainable or require thorough investigation of the SRT histology image.
4.5. spatialDWLS
spatialDWLS [60] is another recently developed method for cell-type-deconvolution in SRT. It is an extension of the dampened weighted least squares (DWLS) approach [55], but can account for the special properties of SRT data by restricting the analysis only to cell types that are likely to be present at each spot. DWLS is originally developed for bulk RNA-seq deconvolution. It estimates cell-type proportions by weighted least squares regression in which the weight is determined by minimizing the overall error rate. Since the number of cells in each spot in a typical sequencing-based SRT dataset is small, e.g., 5–10 cells in 10X Visium, traditional bulk RNA-seq deconvolution methods such as DWLS, MuSiC [54], and CIBERSORT [52] may not work well due to noise from unrelated cell types. As such, spatialDWLS only considers cell types that are likely to be present in deconvolution in which the cell types are identified by cell-type enrichment analysis implemented in Giotto [61]. Through benchmark evaluation, they demonstrate that spatialDWLS outperforms MuSiC, RCTD, SPOTlight, and Stereoscope. Interestingly, MuSiC outperforms RCTD, SPOTlight and Stereoscope, although the latter three methods are specifically designed for SRT deconvolution. Also, in terms of computational speed, spatialDWLS and MuSiC are both much faster than RCTD, SPOTlight and Stereoscope.
5. Enhancement of gene expression resolution
As described earlier, sequencing-based SRT data lack single-cell resolution. While cell-type deconvolution algorithms can infer the locations of cell types, the gene expression measured at each spot is still a mixture from different cells, possibly from different cell types. There is a need for spatial gene expression methods that address the relatively low resolution of the technology. Since gene expression in neighboring spots is correlated, it is possible to borrow information from neighboring spots to increase gene expression resolution. Furthermore, bright field histology images from H&E-stained tissue sections offer high-resolution information on cell morphology, which can be utilized to enhance gene expression resolution.
5.1. RCTd
In addition to cell-type deconvolution, RCTD is also able to compute the expected cell-type-specific gene expression for each spot [57]. This method computes the probability that a given unique molecular identifier comes from each cell type given the cell-type proportion estimates. Within a spot, the cell-type-specific gene expression is estimated using the expected gene expression measurement for a given cell type conditioned on the estimated cell-type proportions and the observed gene expression counts in that spot. However, for individual spots, this conditional expectation may have large variance due to sampling noise. Furthermore, this method of estimating cell-type-specific gene expression is limited to only using proportion estimates from RCTD, which may not be the most accurate or reliable method for deconvolution of SRT data. Additionally, the estimated cell-type proportions are treated as known parameters which introduce additional and unaccounted for variability due to the true proportions being unknown. This method also does not utilize shared spatial or histological information across spots when estimating the cell-type-specific gene expression. Lastly, these estimates are based on a strong modeling assumption that the random effects of gene expression are shared across all cell types. However, in the model, this random effect accounts for additional sources of variation including spatial effects, which are unlikely to be shared across all cell types.
5.2. BayesSpace
BayesSpace resolves expression at a subspot level by leveraging the spatial neighborhood structure [45]. It segments an observed spot into multiple equal-sized subspots, then infers the gene expression of each subspot while keeping the total expression of the original spot fixed. The performance of BayesSpace depends on the neighboring spots as without external information, subspot level gene expression can only be inferred from the original spots’ neighbors. For the current Visium data, the center-to-center distance between two adjacent spots is 100 μm. This means that a subspot and its neighbors are not immediately next to each other. Failure of considering this gap may lead to biased estimation. This also indicates that splitting the observed gene expression in a spot into subspots may run into the problem of identifiability as multiple solutions may exist. Without further constraint, it is not clear which splitting gives the optimal solution. Although their initial results are promising, further validation is needed to confirm the validity of the inferred gene expression at the subspot level.
5.3. XFuse
Bergenstråhle et al. developed XFuse [62], a deep generative model to infer high-resolution spatial gene expression from histology image data. XFuse assumes the histology image and gene expression share the same latent tissue state. Conditional on that state, the gene expression follows a Negative Binomial distribution, and the image pixel intensities follow a Gaussian distribution. The parameters of these two conditional distributions are mapped from the latent tissue state through a trainable convolutional generator network. Next, XFuse learns a posterior distribution of the latent tissue state from observed gene expression and histology data, and approximates the posterior using a tractable target distribution. The variational parameters along with the parameters of the generator network are learned and updated by minimizing the Kullback-Leibler divergence between the target and posterior distributions. Finally, XFuse infers the unseen high-resolution gene expression by estimating the posterior with Monte Carlo samples drawn from the variational distribution. Although XFuse has revealed fine-grained expression heterogeneity for a few genes in mouse olfactory bulb, further evaluation is needed to assess its generalizability to other genes. A limitation of XFuse is that it assumes the gene expression and histology image share the same latent state, which implies that only genes whose expression patterns are similar to the histology image will benefit from this approach, whereas genes whose expression patterns are not similar to the histology may not be inferred.
6. Cell-cell communications from gene expression
Previous gene expression studies have shown that intercellular communication contributes to organ function and other critical biological processes [63], [64], [65], [66]. After the locations of cell types are inferred by cell-type deconvolution, it is natural to ask how different cell types interact and how their interactions are influenced by their spatial proximity [67]. For example, when two cell types colocalize, cells in one cell type may secrete a signaling ligand molecule, whereas the other cell type may express a receptor molecule that recognizes the ligand. Binding of the ligand to the receptor allows the ligand to transmit a signal and change the molecular behavior of the receiver cell. Indeed, ligand-receptor pairs have been used to explore communications between cell types in scRNA-seq [65], [68]. Since the distance that the ligand signal travels is the main factor that determines the different types of cell–cell signaling, SRT offers richer information to study cell–cell communications in tissues with spatial structure. Recently, several methods have been developed to explore cell–cell communications from gene expression.
6.1. Giotto
Giotto is a comprehensive open-source toolbox for SRT data analysis and visualization [61]. It includes multiple modules that cover a wide range of algorithms for SRT data analysis. One of these modules can characterize how cells communicate within their microenvironment. Specifically, for any two cell types A and B, Giotto constructs an enrichment score, which is calculated as the weighted average expression of a ligand and the corresponding receptor in a subset of A and B cells that are proximal to each other. Then, by shuffling cell locations within each cell type, an empirical null distribution is constructed, which can be used to calculate the associated P-value. Finally, Giotto ranks all pairs of ligand-receptor genes in all pairs of neighboring cell types based on the score. By only considering cells proximal to each other between two cell types, however, Giotto cannot detect gene-gene interactions that are associated with complex interaction patterns.
6.2. SpaOTsc
SpaOTsc is a recently developed method that infers cell–cell communications by examining the interaction relationships between ligand-receptor pairs and their downstream genes [69]. It can be applied to both scRNA-seq and SRT data. Since scRNA-seq data do not have spatial location information, SpaOTsc uses external spatial information e.g., spatial marker genes, to calculate a spatial metric using the optimal transport algorithm, which returns a mapping that contains the probability distribution of each scRNA-seq cell over a spatial region. Then, it uses this metric to form an optimal transport plan from a probability distribution of signal “sender cells” to a target distribution of “receiver cells”, where the sender cell distribution is characterized by the expression levels of ligand genes, and the receiver cell distribution is distinguished by the paired receptor genes and the ligand-receptor downstream genes. Results from this analysis yield intercellular gene-gene regulatory information flows. SpaOTsc also adopts a random forest approach to estimate the spatial range of ligand-receptor signaling to further confine the inferred cell–cell communications. By doing so, long-distance connections between cells are eliminated. These cell–cell communication inference procedures can be directly applied to SRT data because spatial coordinates are available.
6.3. MISTy
Methods such as Giotto and SpaOTsc only focus on the local cellular niche, i.e., the expressions measured in the immediate neighborhood of each cell. Although methods that consider the broader tissue structure are available [70], the restriction on a fixed form of nonlinear relationship between markers and the high computational complexity make such methods less flexible. More recently, Tanevski et al. developed MISTy [71], a flexible machine learning framework that offers a range of cell–cell communication analysis in a scalable fashion. Using a late fusion multiview framework, MISTy constructs a domain-specific model for the expression of markers. Specifically, for each marker of interest, the cell–cell interactions are studied based on the spatial context — the “intrinsic view” models how other markers influence the given marker’s expression within the same location, the “jxtaview” models the local cellular niche and relates the expression from the immediate neighborhood of a cell to the observed expression within that cell, and the “paraview” captures the effect of tissue structure and relates the expression of markers measured in cells within a radius around a given cell. An appealing feature of MISTy is that it also allows the modeling of other views, e.g., interactions between different cell types, interactions within specific regions of interest or a higher-level functional organization. Since each MISTy view is considered as a potential source of variability in the measured marker expressions, results from MISTy allow the investigation of each view’s contribution to the total expression of each marker. The consideration of cell–cell communications from these different aspects allows an in-depth understanding of marker interactions.
7. Outlook and future research directions
SRT has shown enormous potential in biomedical research. Applications of SRT technologies in diverse tissue types and diseases have revealed fine-scale cellular heterogeneity within spatial context and transformed our understanding of the functional and structural foundations of tissue architecture. Using SRT, we can build a 3D transcriptome atlas of brain [72], delineate embryonic development [21], and elucidate disease progression [22], [23]. As new SRT analysis methods become available every month, it is impossible to include all available methods in this review. However, we have investigated common computational tasks for SRT data analysis, selected methods that are applicable to both imaging- and sequencing-based SRT data, and discussed how spatial location and histology information can be integrated with gene expression (Table 1). While we discussed the advantages and disadvantages of the reviewed methods, comprehensive benchmark evaluations are needed to fully understand the performance of each method. Evaluations should be conducted in terms of the reasonableness of the results and the uncertainty provided by each method, particularly those methods that involve unsupervised clustering. Since many methods reviewed in this paper may form individual components within a full analytical workflow, we suggest that a useful benchmark evaluation should also study how the uncertainty propagates and their potential effects on downstream analysis, e.g., identification of domain-specific marker genes. Although computational methods have evolved for SRT data analysis, better algorithms are still needed to leverage and integrate the rich information in SRT data, particularly scRNA-seq and high-resolution histology image data. Below we point out a few open problems and future research directions.
Table 1.
Summary of reviewed methods.
| Method | Category | Language | Software | Reference | Released Date | Advantages | Disadvantages | Use histology image |
|---|---|---|---|---|---|---|---|---|
| HMRF | Spatial clustering | R; Python; C | https://bitbucket.org/qzhud- fci/smfishhmrf-py | Identification of spatially associated subpopulations by combining scRNAseq and sequential fluorescence in situ hybridization data [37] | 2018-10-29 | Can simultaneously detect the combinatorial pattern of all profiled genes. | The classification of a small number of isolated cells as domains may be questionable. Cannot incorporate histology information in its model. |
No |
| SpaCell | Spatial clustering | Python | https://github.com/BiomedicalMachineLearning/SpaCell | SpaCell: integrating tissue morphology and spatial gene expression to predict disease cells [41] | 2020-04-01 | Can combine histology image data and spatial gene expression data for joint clustering. Can automatically and quantitatively identify cell types and disease stages. |
Spot location information is not utilized in the model. | Yes |
| stLearn | Spatial clustering | Python | https://github.com/BiomedicalMachineLearning/stLearn | stLearn: integrating spatial location, tissue morphology and gene expression to find cell types, cell–cell interactions and spatial trajectories within undissociated tissues [43] | 2020-05-31 | Can integrate gene expression and spatial distance information and histology image information. Applicable to any SRT data as long as tissue morphology, spatial location, and gene expression information are simultaneously captured. |
Cannot be applied to data without histology images. | Yes |
| BayesSpace | Spatial clustering; enhancement of gene expression resolution | R; C++ | https://github.com/edward130603/BayesSpace | BayesSpace enables the robust characterization of spatial gene expression architecture in tissue sections at increased resolution [45] | 2020-09-05 | Account for spatial dependency in clustering analysis. Can generate enhanced resolution gene expression data. |
Cannot incorporate histology information. | No |
| SpaGCN | Spatial clustering; identification of spatially variable genes | Python | https://github.com/jianhuupenn/SpaGCN | Integrating gene expression, spatial location and histology to identify spatial domains and spatially variable genes by graph convolutional network [36] | 2020–11-30 | Jointly consider spatial domain identification and SVG detection. Can integrate gene expression, spatial location and histology information (when available) in spatial domain identification.Computationally fast and memory efficient. |
Cannot account for cell type variations in spatially variable gene detection. | Yes |
| Trendsceek | Identification of spatially variable genes | R | https://github.com/edsgard/trendsceek | Identification of spatial expression trends in single-cell gene expression data [46] | 2018–03-19 | Perform a gene-level test that incorporates both spatial and expression-level information. | Cannot account for cell type variations in spatially variable gene detection. | No |
| SpatialDE | Identification of spatially variable genes | Python | https://github.com/Teichlab/SpatialDE | SpatialDE: identification of spatially variable genes [47] | 2018–03-19 | Use of a principled statistical approach to model spatial dependency of gene expression. | Rely on asymptotic normality and minimal P-value-combination rules for hypothesis testing, which may lead to false positives and loss of power. Cannot account for cell type variations in spatially variable gene detection. |
No |
| SPARK | Identification of spatially variable genes | R; C++ | https://github.com/xzhoulab/SPARK | Statistical analysis of spatial expression patterns for spatially resolved transcriptomic studies [48] | 2020–01-27 | Explicit modeling of gene expression as count data. Use of kernel approaches to model spatial dependency of gene expression. |
Computationally slow and memory consuming. Cannot account for cell type variations in spatially variable gene detection. |
No |
| Stereoscope | Cell-type deconvolution | Python | https://github.com/almaan/stereoscope | Spatial mapping of cell types by integration of transcriptomics data [56] | 2019–12-13 | First method for cell-type deconvolution in SRT. | Assume both SRT and scRNA-seq data follow a negative binomial distribution. Do not account for spatial dependency of gene expression. |
No |
| RCTD | Cell-type deconvolution; enhancement of gene expression resolution | R | https://github.com/dmcable/RCTD | Robust decomposition of cell type mixtures in spatial transcriptomics [57] | 2020–05-08 | Can correct for platform differences between SRT data and scRNA-seq reference. Can restrict deconvolution only to the most likely cell types. |
Do not explicitly model spatial dependency of gene expression. Assume platform effects are shared among all cell types. |
No |
| SPOTlight | Cell-type deconvolution | R | https://github.com/MarcElosua/SPOTlight_deconvolution_analysis | SPOTlight:Seeded NMF regression to Deconvolute Spatial Transcriptomics Spots with Single-Cell Transcriptomes [58] | 2020–06-04 | A small number of cells per cell-type is sufficient to train the model. | Need prior information of cell-type-specific marker genes. | No |
| Cell2location | Cell-type deconvolution | Python | https://github.com/BayraktarLab/cell2location | Comprehensive mapping of tissue cell architecture via integrated single cell and spatial transcriptomics [59] | 2020–11-15 | Can accurately infer the presence of rare cell types. Can provide estimates of relative cell type fractions along with additionally estimates of absolute cell type abundance. |
Do not explicitly model spatial dependency of gene expression. Require the user to define hyperparameters for the average number of cells, cell types, and tissue zones per spot and the average difference in technical sensitivity between scRNA-seq and SRT data. |
No |
| spatialDWLS | Cell-type deconvolution | R | https://github.com/RubD/Giotto | SpatialDWLS: accurate deconvolution of spatial transcriptomic data [60] | 2021–05-10 | Can restrict deconvolution only to the most likely cell types. | Do not account for spatial dependency of gene expression in deconvolution. | No |
| XFuse | Enhancement of gene expression resolution | Python | https://github.com/ludvb/xfuse | Super-resolved spatial transcriptomics by deep data fusion [62] | 2020–03-13 | Can infer spatial gene expression at the same resolution as the histology image data. | Assume the gene expression and histology image share the same latent state. | Yes |
| Giotto | Cell-cell communications | R | https://github.com/RubD/Giotto | Giotto, a pipeline for integrative analysis and visualization of single-cell spatial transcriptomic data [61] | 2020-05-30 | Can identify genes whose expression variation within a cell type is significantly associated with an interacting cell type. | Only focus on unsupervised correlation-based analysis, thus may fail to identify interactions that are limited to a specific area, specific cell types, or that are related to more complex patterns. | No |
| SpaOTsc | Cell-cell communications | Python | https://github.com/zcang/SpaOTsc | Inferring spatial and signaling relationships between cells from single cell transcriptomic data [69] | 2020–04-29 | Model both direct and indirect cell–cell communications. | The computation of the cell–cell distance inference can become intractable when the dataset is excessively large [69]. HYPERLINK "SPS:refid::bib69" | No |
| MISTy | Cell-cell communications | R | https://github.com/saezlab/mistyR | Explainable multi-view framework for dissecting inter-cellular signaling from highly multiplexed spatial data [71] | 2020–05-10 | Can build multiple views focusing on different spatial or functional contexts to dissect different effects. | Rely on a radius parameter to determine the number of cells to be included in each view. | No |
The methods reviewed in this paper are all developed for the analysis of a single tissue section. However, the field has now moved toward generating data with more complex structure. For example, when building spatially resolved molecular atlases of brains, e.g., Allen Brain Atlas, or whole organs, such as in the Human Cell Atlas [73] and the Human Biomolecular Atlas Program [74], multiple tissue samples from several subjects and multiple tissue sections per subject will be generated. Such complex data structure poses computational challenges. Effective modeling of such data requires new methods that can account for spatial dependency of gene expression in the 3D space, gene expression variability driven by spatial differences associated with biological variables such as sex, age, race, and body size. To account for these factors, the gene expression data from tissue sections across different individuals need to be registered to a common coordinate framework [75], [76]. Once the gene expression data are registered, methods that can account for variations across space and individuals can be utilized to identify marker genes that define the transcriptional landmarks. One of the earlier methods for such analysis is Splotch [77], which analyzes multiple tissue sections simultaneously and takes the experimental design into consideration to quantify biological and experimental variation at different levels. We anticipate that there will be greater needs for more powerful and efficient methods for the analysis of multiple tissue sections across different subjects in the next few years.
As the resolution of histology images in SRT data is much higher than that of the companion gene expression data, an ideal approach should be able to effectively integrate histology information in analysis. Although progress has been made in integrating histology and gene expression, current methods mainly focus on the global pattern in histology images while the more granular information e.g., morphology of nucleus in each spot, is ignored. Nuclei segmentation in histopathology images is routinely done for pathology diagnosis [78], [79], [80]. However, such information has only been utilized to verify results after gene expression data are analyzed, but not directly used in analysis [27]. Information on the number of nuclei per spot and the associated morphology of each nucleus is invaluable in cell-type deconvolution because it can tell us how many cells and cell types are present in a spot. A deconvolution method that takes this detailed information into account will be able to infer cell-type proportions in each spot more precisely than existing methods, which in turn will also help estimate cell-type-specific gene expression. With these more accurate deconvolution results, we can get a better understanding of cell–cell communications and how they vary by spatial proximity.
Another open question is how to infer gene expression in blank regions between spots in SRT data. For example, in Visium, the diameter size of each spot is 55 µm and the center-to-center distance between spots is 100 µm. The size of these blank regions between two adjacent spots is 45 µm, which is larger than the diameter for cells of most cell types. The discontinuity of gene expression measurement may increase the uncertainty in gene-gene (e.g., ligand-receptor) and cell–cell interaction analysis. Although BayesSpace [45] can generate subspot level gene expression, these empty regions are still left unmeasured. XFuse [62] can use high-resolution histology images to fill in unmeasured gene expression, but XFuse is designed for genes whose expression patterns are highly correlated with histological features. Since the expression patterns for the majority of genes are not correlated with histological features, XFuse still leaves a large number of genes with unmeasured expression. New methods that can fill in the unmeasured gene expression in those blank regions are needed.
Batch effect is a common issue in the analyses of scRNA-seq data. Many methods have been developed for batch effect removal for scRNA-seq [81], [82], [83], [84], [85]. In SRT, the batch effect is even more complex, particularly for SRT data that have companion histology images in which batch effect can affect both gene expression and histology images across different tissue sections, subjects, and studies. This is still an unexplored area, but we envision that batch effect correction will become an important problem as the scale of SRT increases. Methods to evaluate and remove batch effects in both gene expression and histology images are needed.
The understanding of cellular behavior within spatial context is critical to our understanding of human disease. Although histopathology is the clinical gold-standard for the diagnosis of many diseases, interpretation of histology is still an art that makes pathologists essential for accurate disease diagnosis. In some cases e.g., rare diseases, histological assessment may be subject to diagnostic uncertainty due to the lack of knowledge of pathological changes. This uncertainty can be alleviated by expression information on genes with well-defined functions. The gene expression data together with high-resolution histology images in these new spatial technologies will help deepen our understanding of what is happening in tissue, which will be applicable to most areas of biomedical research. Since histology image and gene expression data provide complementary information, it will be desirable to have methods that can incorporate pathologist’s annotation as prior information in the analysis.
SRT has become the latest frontier for cutting-edge research in biomedicine, and new technologies are continued to be developed [86], [87]. In this review, we provide an overview of the current state and common computational tasks in SRT data analysis. A crucial aspect for the analysis of SRT data alongside histology images is the ability to visualize and work with such data, especially within a complex study design framework. To streamline the analysis, infrastructure such as SpatialExperiment [88] is needed. In addition, a few recently developed software packages, e.g., STUtility [75], Seurat [89], Giotto [61], Tangram [90], and SquidPy [91], have integrated many of the reviewed methods in their packages, which will facilitate the adoption of SRT analysis methods in real studies. As the scale and complexity of SRT data will continue to grow, software that can visualize and interrogate multimodel data and outputs will be needed. We hope this review will draw researchers with complementary expertise to collaborate and develop more effective computational methods to integrate information from gene expression and digital pathology to fully unleash the power of these spatial technologies.
8. Author statement
All authors have seen and approved the final version of the manuscript being submitted. The article is the authors’ original work, and hasn’t received prior publication and isn’t under consideration for publication elsewhere.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was funded in part by the following grants: R01GM125301, R01EY030192, R01EY031209 R01HL113147, and R01HL150359 (to M.L.).
References
- 1.Liao J., Lu X., Shao X., Zhu L., Fan X. Uncovering an organ's molecular architecture at single-cell resolution by spatially resolved transcriptomics. Trends Biotechnol. 2021;39(1):43–58. doi: 10.1016/j.tibtech.2020.05.006. [DOI] [PubMed] [Google Scholar]
- 2.Waylen L.N., Nim H.T., Martelotto L.G., Ramialison M. From whole-mount to single-cell spatial assessment of gene expression in 3D. Commun Biol. 2020;3:602. doi: 10.1038/s42003-020-01341-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burgess DJ. Spatial transcriptomics coming of age. Nat Rev Genet 20, 317, doi:10.1038/s41576-019-0129-z (2019). [DOI] [PubMed]
- 4.Asp M., Bergenstråhle J., Lundeberg J. Spatially resolved transcriptomes-next generation tools for tissue exploration. BioEssays. 2020;42(10):1900221. doi: 10.1002/bies.v42.1010.1002/bies.201900221. [DOI] [PubMed] [Google Scholar]
- 5.Crosetto N., Bienko M., van Oudenaarden A. Spatially resolved transcriptomics and beyond. Nat Rev Genet. 2015;16(1):57–66. doi: 10.1038/nrg3832. [DOI] [PubMed] [Google Scholar]
- 6.Moor A.E., Itzkovitz S. Spatial transcriptomics: paving the way for tissue-level systems biology. Curr Opin Biotechnol. 2017;46:126–133. doi: 10.1016/j.copbio.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Femino A.M., Fay F.S., Fogarty K., Singer R.H. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 8.Fan Y., Braut S.A., Lin Q., Singer R.H., Skoultchi A.I. Determination of transgenic loci by expression FISH. Genomics. 2001;71(1):66–69. doi: 10.1006/geno.2000.6403. [DOI] [PubMed] [Google Scholar]
- 9.Levsky J.M., Shenoy S.M., Pezo R.C., Singer R.H. Single-cell gene expression profiling. Science. 2002;297:836–840. doi: 10.1126/science.1072241. [DOI] [PubMed] [Google Scholar]
- 10.Raj A., Peskin C.S., Tranchina D., Vargas D.Y., Tyagi S., Schibler U. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4(10):e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raj A., van den Bogaard P., Rifkin S.A., van Oudenaarden A., Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5(10):877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lubeck E., Coskun A.F., Zhiyentayev T., Ahmad M., Cai L. Single-cell in situ RNA profiling by sequential hybridization. Nat Methods. 2014;11(4):360–361. doi: 10.1038/nmeth.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shah S., Lubeck E., Zhou W., Cai L. In situ transcription profiling of single cells reveals spatial organization of cells in the mouse hippocampus. Neuron. 2016;92(2):342–357. doi: 10.1016/j.neuron.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eng C.-H., Lawson M., Zhu Q., Dries R., Koulena N., Takei Y. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature. 2019;568(7751):235–239. doi: 10.1038/s41586-019-1049-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moffitt J.R., Bambah-Mukku D., Eichhorn S.W., Vaughn E., Shekhar K., Perez J.D. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science. 2018;362(6416):eaau5324. doi: 10.1126/science:aau5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090, doi:10.1126/science.aaa6090 (2015). [DOI] [PMC free article] [PubMed]
- 17.Ståhl P.L., Salmén F., Vickovic S., Lundmark A., Navarro J.F., Magnusson J. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353(6294):78–82. doi: 10.1126/science:aaf2403. [DOI] [PubMed] [Google Scholar]
- 18.Rodriques S.G., Stickels R.R., Goeva A., Martin C.A., Murray E., Vanderburg C.R. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science. 2019;363(6434):1463–1467. doi: 10.1126/science:aaw1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stickels R.R., Murray E., Kumar P., Li J., Marshall J.L., Di Bella D.J. Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2. Nat Biotechnol. 2021;39(3):313–319. doi: 10.1038/s41587-020-0739-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vickovic S., Eraslan G., Salmén F., Klughammer J., Stenbeck L., Schapiro D. High-definition spatial transcriptomics for in situ tissue profiling. Nat Methods. 2019;16(10):987–990. doi: 10.1038/s41592-019-0548-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asp M. et al. A Spatiotemporal Organ-Wide Gene Expression and Cell Atlas of the Developing Human Heart. Cell 179, 1647-1660 e1619, doi:10.1016/j.cell.2019.11.025 (2019). [DOI] [PubMed]
- 22.Maniatis S., Äijö T., Vickovic S., Braine C., Kang K., Mollbrink A. Spatiotemporal dynamics of molecular pathology in amyotrophic lateral sclerosis. Science. 2019;364(6435):89–93. doi: 10.1126/science:aav9776. [DOI] [PubMed] [Google Scholar]
- 23.Chen WT. et al. Spatial Transcriptomics and In Situ Sequencing to Study Alzheimer's Disease. Cell 182, 976-991 e919, doi:10.1016/j.cell.2020.06.038 (2020). [DOI] [PubMed]
- 24.Berglund E., Maaskola J., Schultz N., Friedrich S., Marklund M., Bergenstråhle J. Spatial maps of prostate cancer transcriptomes reveal an unexplored landscape of heterogeneity. Nat Commun. 2018;9(1) doi: 10.1038/s41467-018-04724-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thrane K., Eriksson H., Maaskola J., Hansson J., Lundeberg J. Spatially resolved transcriptomics enables dissection of genetic heterogeneity in stage III cutaneous malignant melanoma. Cancer Res. 2018;78:5970–5979. doi: 10.1158/0008-5472.CAN-18-0747. [DOI] [PubMed] [Google Scholar]
- 26.Yoosuf N., Navarro J.F., Salmen F., Stahl P.L., Daub C.O. Identification and transfer of spatial transcriptomics signatures for cancer diagnosis. Breast Cancer Res. 2020;22:6. doi: 10.1186/s13058-019-1242-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moncada R., Barkley D., Wagner F., Chiodin M., Devlin J.C., Baron M. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat Biotechnol. 2020;38(3):333–342. doi: 10.1038/s41587-019-0392-8. [DOI] [PubMed] [Google Scholar]
- 28.Ji AL. et al. Multimodal Analysis of Composition and Spatial Architecture in Human Squamous Cell Carcinoma. Cell 182, 497-514 e422, doi:10.1016/j.cell.2020.05.039 (2020). [DOI] [PMC free article] [PubMed]
- 29.Method of the Year 2020: spatially resolved transcriptomics. Nat Methods 18, 1, doi:10.1038/s41592-020-01042-x (2021). [DOI] [PubMed]
- 30.Larsson L., Frisén J., Lundeberg J. Spatially resolved transcriptomics adds a new dimension to genomics. Nat Methods. 2021;18(1):15–18. doi: 10.1038/s41592-020-01038-7. [DOI] [PubMed] [Google Scholar]
- 31.Zhuang X. Spatially resolved single-cell genomics and transcriptomics by imaging. Nat Methods. 2021;18(1):18–22. doi: 10.1038/s41592-020-01037-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Close J.L., Long B.R., Zeng H. Spatially resolved transcriptomics in neuroscience. Nat Methods. 2021;18(1):23–25. doi: 10.1038/s41592-020-01040-z. [DOI] [PubMed] [Google Scholar]
- 33.Marx V. Method of the Year: spatially resolved transcriptomics. Nat Methods. 2021;18(1):9–14. doi: 10.1038/s41592-020-01033-y. [DOI] [PubMed] [Google Scholar]
- 34.Lloyd S. Least squares quantization in PCM. IEEE Trans Inf Theory. 1982;28(2):129–137. [Google Scholar]
- 35.Blondel V.D., Guillaume J.-L., Lambiotte R., Lefebvre E. Fast unfolding of communities in large networks. J Stat Mech: Theory Exp. 2008;2008(10):P10008. doi: 10.1088/1742-5468/2008/10/P10008. [DOI] [Google Scholar]
- 36.Hu J. et al. Integrating gene expression, spatial location and histology to identify spatial domains and spatially variable genes by graph convolutional network. bioRxiv 405118; doi: 10.1101/2020.11.30.405118 (2020). [DOI] [PubMed]
- 37.Zhu Q., Shah S., Dries R., Cai L., Yuan G.-C. Identification of spatially associated subpopulations by combining scRNAseq and sequential fluorescence in situ hybridization data. Nat Biotechnol. 2018;36(12):1183–1190. doi: 10.1038/nbt.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y., Brady M., Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging. 2001;20(1):45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- 39.Coudray N., Ocampo P.S., Sakellaropoulos T., Narula N., Snuderl M., Fenyö D. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med. 2018;24(10):1559–1567. doi: 10.1038/s41591-018-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He B., Bergenstråhle L., Stenbeck L., Abid A., Andersson A., Borg Å. Integrating spatial gene expression and breast tumour morphology via deep learning. Nat Biomed Eng. 2020;4(8):827–834. doi: 10.1038/s41551-020-0578-x. [DOI] [PubMed] [Google Scholar]
- 41.Tan X, Su A, Tran M, Nguyen Q. SpaCell: integrating tissue morphology and spatial gene expression to predict disease cells. Bioinformatics 36, 2293-2294, doi:10.1093/bioinformatics/btz914 (2020). [DOI] [PubMed]
- 42.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. arXiv (2015).
- 43.Pham D. et al. stLearn: integrating spatial location, tissue morphology and gene expression to find cell types, cell-cell interactions and spatial trajectories within undissociated tissues. bioRxiv 125658; doi: 10.1101/2020.05.31.125658 (2020).
- 44.Chatterjee S. Artefacts in histopathology. J Oral Maxillofac Pathol. 2014;18:S111–116. doi: 10.4103/0973-029X.141346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao E. et al. BayesSpace enables the robust characterization of spatial gene expression architecture in tissue sections at increased resolution. bioRxiv 283812; doi: 10.1101/2020.09.04.283812 (2020).
- 46.Edsgärd D., Johnsson P., Sandberg R. Identification of spatial expression trends in single-cell gene expression data. Nat Methods. 2018;15(5):339–342. doi: 10.1038/nmeth.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Svensson V., Teichmann S.A., Stegle O. SpatialDE: identification of spatially variable genes. Nat Methods. 2018;15(5):343–346. doi: 10.1038/nmeth.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun S., Zhu J., Zhou X. Statistical analysis of spatial expression patterns for spatially resolved transcriptomic studies. Nat Methods. 2020;17(2):193–200. doi: 10.1038/s41592-019-0701-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33(5):495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Achim K., Pettit J.-B., Saraiva L.R., Gavriouchkina D., Larsson T., Arendt D. High-throughput spatial mapping of single-cell RNA-seq data to tissue of origin. Nat Biotechnol. 2015;33(5):503–509. doi: 10.1038/nbt.3209. [DOI] [PubMed] [Google Scholar]
- 51.Liu Y., Chen S., Li Z., Morrison A.C., Boerwinkle E., Lin X. ACAT: A fast and powerful p value combination method for rare-variant analysis in sequencing studies. Am J Hum Genet. 2019;104(3):410–421. doi: 10.1016/j.ajhg.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Newman A.M., Steen C.B., Liu C.L., Gentles A.J., Chaudhuri A.A., Scherer F. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37(7):773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang X., Park J., Susztak K., Zhang N.R., Li M. Bulk tissue cell type deconvolution with multi-subject single-cell expression reference. Nat Commun. 2019;10:380. doi: 10.1038/s41467-018-08023-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsoucas D., Dong R., Chen H., Zhu Q., Guo G., Yuan G.-C. Accurate estimation of cell-type composition from gene expression data. Nat Commun. 2019;10(1) doi: 10.1038/s41467-019-10802-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Andersson A., Bergenstråhle J., Asp M., Bergenstråhle L., Jurek A., Fernández Navarro J. Single-cell and spatial transcriptomics enables probabilistic inference of cell type topography. Commun Biol. 2020;3(1) doi: 10.1038/s42003-020-01247-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cable DM et al. Robust decomposition of cell type mixtures in spatial transcriptomics. bioRxiv 082750; doi: 10.1101/2020.05.07.082750 (2020). [DOI] [PMC free article] [PubMed]
- 58.Elosua-Bayes M, Nieto P, Mereu E, Gut I, Heyn H. SPOTlight: seeded NMF regression to deconvolute spatial transcriptomics spots with single-cell transcriptomes. Nucleic Acids Res, doi:10.1093/nar/gkab043 (2021). [DOI] [PMC free article] [PubMed]
- 59.Kleshchevnikov V. et al. Comprehensive mapping of tissue cell architecture via integrated single cell and spatial transcriptomics. bioRxiv 530378; doi: 10.1101/530378 (2020).
- 60.Dong R., Yuan G.C. SpatialDWLS: accurate deconvolution of spatial transcriptomic data. Genome Biol. 2021;22:145. doi: 10.1186/s13059-021-02362-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dries R et al. Giotto, a toolbox for integrative analysis and visualization of spatial expression data. bioRxiv 701680; doi: 10.1101/701680 (2020). [DOI] [PMC free article] [PubMed]
- 62.Bergenstrahle L et al. Super-resolved spatial transcriptomics by deep data fusion. bioRxiv 963413 doi: 10.1101/2020.02.28.963413 (2020). [DOI] [PubMed]
- 63.Arneson D., Zhang G., Ying Z., Zhuang Y., Byun H.R., Ahn I.S. Single cell molecular alterations reveal target cells and pathways of concussive brain injury. Nat Commun. 2018;9(1) doi: 10.1038/s41467-018-06222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ximerakis M., Lipnick S.L., Innes B.T., Simmons S.K., Adiconis X., Dionne D. Single-cell transcriptomic profiling of the aging mouse brain. Nat Neurosci. 2019;22(10):1696–1708. doi: 10.1038/s41593-019-0491-3. [DOI] [PubMed] [Google Scholar]
- 65.Skelly D.A., Squiers G.T., McLellan M.A., Bolisetty M.T., Robson P., Rosenthal N.A. Single-Cell transcriptional profiling reveals cellular diversity and intercommunication in the mouse heart. Cell Rep. 2018;22(3):600–610. doi: 10.1016/j.celrep.2017.12.072. [DOI] [PubMed] [Google Scholar]
- 66.Cohen M et al. Lung Single-Cell Signaling Interaction Map Reveals Basophil Role in Macrophage Imprinting. Cell 175, 1031-1044 e1018, doi:10.1016/j.cell.2018.09.009 (2018). [DOI] [PubMed]
- 67.Armingol E., Officer A., Harismendy O., Lewis N.E. Deciphering cell-cell interactions and communication from gene expression. Nat Rev Genet. 2021;22(2):71–88. doi: 10.1038/s41576-020-00292-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Efremova M., Vento-Tormo M., Teichmann S.A., Vento-Tormo R. Cell PhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc. 2020;15(4):1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 69.Cang Z., Nie Q. Inferring spatial and signaling relationships between cells from single cell transcriptomic data. Nat Commun. 2020;11:2084. doi: 10.1038/s41467-020-15968-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Arnol D, Schapiro D, Bodenmiller B, Saez-Rodriguez J, Stegle O. Modeling Cell-Cell Interactions from Spatial Molecular Data with Spatial Variance Component Analysis. Cell Rep 29, 202-211 e206, doi:10.1016/j.celrep.2019.08.077 (2019). [DOI] [PMC free article] [PubMed]
- 71.Tanevski J., Gabor A., Flores R.O.R., Shapiro D., Saez-Rodriguez J. Explainable multi-view framework for dissecting inter-cellular signaling from highly multiplexed spatial data. bioRxiv. 2020 doi: 10.1101/2020.05.08.084145. [DOI] [Google Scholar]
- 72.Ortiz C. et al. Molecular atlas of the adult mouse brain. Sci Adv 6, eabb3446, doi:10.1126/sciadv.abb3446 (2020). [DOI] [PMC free article] [PubMed]
- 73.Regev A. et al. The Human Cell Atlas. Elife 6, doi:10.7554/eLife.27041 (2017). [DOI] [PMC free article] [PubMed]
- 74.Hu B.C. The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature. 2019;574:187–192. doi: 10.1038/s41586-019-1629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bergenstrahle J., Larsson L., Lundeberg J. Seamless integration of image and molecular analysis for spatial transcriptomics workflows. BMC Genomics. 2020;21:482. doi: 10.1186/s12864-020-06832-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zeira R., Land M., Raphael B.J. Alignment and integration of spatial transcriptomics data. bioRxiv. 2021 doi: 10.1101/2021.03.16.435604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Aijo T. Splotch: robust estimation of aligned spatial temporal gene expression data. bioRxiv. 2020 doi: 10.1101/757096. [DOI] [Google Scholar]
- 78.Komura D., Ishikawa S. Machine learning methods for histopathological image analysis. Comput Struct Biotechnol J. 2018;16:34–42. doi: 10.1016/j.csbj.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bankhead P., Loughrey M.B., Fernández J.A., Dombrowski Y., McArt D.G., Dunne P.D. QuPath: open source software for digital pathology image analysis. Sci Rep. 2017;7(1) doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Humphries M.P., Maxwell P., Salto-Tellez M. QuPath: the global impact of an open source digital pathology system. Comput Struct Biotechnol J. 2021;19:852–859. doi: 10.1016/j.csbj.2021.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li X., Wang K., Lyu Y., Pan H., Zhang J., Stambolian D. Deep learning enables accurate clustering with batch effect removal in single-cell RNA-seq analysis. Nat Commun. 2020;11(1) doi: 10.1038/s41467-020-15851-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lakkis J. et al. A joint deep learning model for simultaneous batch effect correction, denoising and clustering in single-cell transcriptomics. bioRxiv 310003; doi: 10.1101/2020.09.23.310003 (2020). [DOI] [PMC free article] [PubMed]
- 83.Lopez R., Regier J., Cole M.B., Jordan M.I., Yosef N. Deep generative modeling for single-cell transcriptomics. Nat Methods. 2018;15(12):1053–1058. doi: 10.1038/s41592-018-0229-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stuart T. et al. Comprehensive Integration of Single-Cell Data. Cell 177, 1888-1902 e1821, doi:10.1016/j.cell.2019.05.031 (2019). [DOI] [PMC free article] [PubMed]
- 85.Korsunsky I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16(12):1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu Y. et al. High-Spatial-Resolution Multi-Omics Sequencing via Deterministic Barcoding in Tissue. Cell 183, 1665-1681 e1618, doi:10.1016/j.cell.2020.10.026 (2020). [DOI] [PMC free article] [PubMed]
- 87.Chen A. et al. Large field of view-spatially resolved transcriptomics at nanoscale resolution. bioRxiv 427004; doi: 10.1101/2021.01.17.427004 (2021).
- 88.Righelli D. SpatialExperiment: infrastructure for spatially resolved transcriptomics data in R using Bioconductor. bioRxiv. 2021 doi: 10.1101/2021.01.27.428431. [DOI] [Google Scholar]
- 89.https://satijalab.org/seurat/articles/spatial_vignette.html. Seurat. (2021).
- 90.Biancalani T. Deep learning and alignment of spatially-resolved whole transcriptomes of single cells in the mouse brain with Tangram. bioRxiv. 2021 doi: 10.1101/2020.08.29.272831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Palla G. Squidpy: a scalable framework for spatial single cell analysis. bioRxiv. 2021 doi: 10.1101/2021.02.19.431994. [DOI] [PMC free article] [PubMed] [Google Scholar]