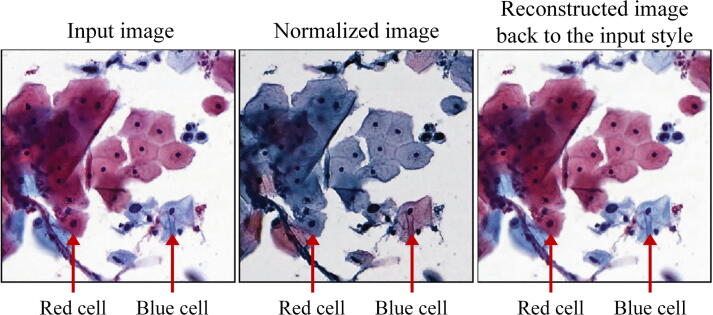
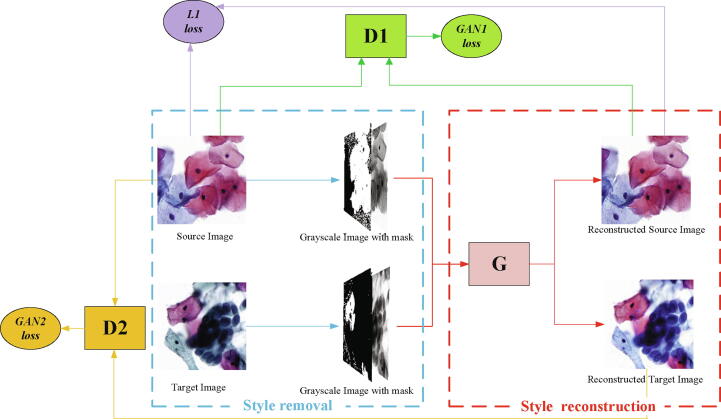
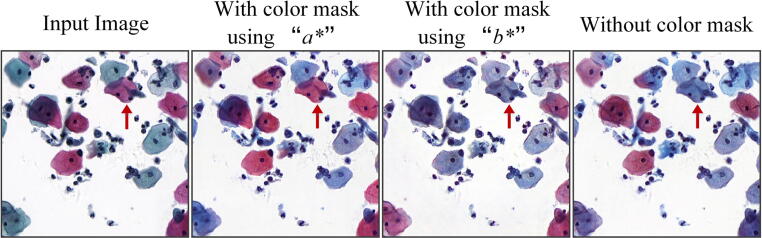
Graphical abstract
Keywords: Cytopathology images, Unsupervised image style normalization, Generative adversarial learning, Domain adversarial networks
Abstract
Diverse styles of cytopathology images have a negative effect on the generalization ability of automated image analysis algorithms. This article proposes an unsupervised method to normalize cytopathology image styles. We design a two-stage style normalization framework with a style removal module to convert the colorful cytopathology image into a gray-scale image with a color-encoding mask and a domain adversarial style reconstruction module to map them back to a colorful image with user-selected style. Our method enforces both hue and structure consistency before and after normalization by using the color-encoding mask and per-pixel regression. Intra-domain and inter-domain adversarial learning are applied to ensure the style of normalized images consistent with the user-selected for input images of different domains. Our method shows superior results against current unsupervised color normalization methods on six cervical cell datasets from different hospitals and scanners. We further demonstrate that our normalization method greatly improves the recognition accuracy of lesion cells on unseen cytopathology images, which is meaningful for model generalization.
1. Introduction
The discipline of medicine relies on inductive logic, empirical learning and evidence-based usage. In recent years, artificial intelligence has played an increasingly important role in medical applications. In the field of cytopathology, the accumulation of digital slide images provides a huge database for cytopathology image analysis where artificial intelligence is increasingly used [1], [2], [3], [4]. Assisted screening systems based on big data and artificial intelligence decrease workload and reduce subjective errors in manual interpretation. This is of great significance in popularizing cytopathology screening and reducing the incidence of cervical cancer.
However, the actual deployment of algorithm-based screening systems faces great challenges in model generalization on diverse cytopathology images [5], [6], [7]. Due to variations in scanning instruments and each hospital’s specific procedure of staining, there is inherent difference in cytopathology image styles [8], as shown in Fig. 1. Such image style inconsistency indicates there is a numerical distribution change between images of different styles including brightness, saturation and hue. For most of screening systems, their core automated analysis models based on machine learning are usually trained on a finite dataset [9], [10]. Nevertheless, machine learning methods generally work well under a common assumption: training and testing data are drawn from the same distribution [11]. This assumption is violated as the training data and testing data have different image styles and thus the performance of analysis models degrades in actual deployment process [12]. Mixing training and transfer learning are effective for improving the model performance on data with new image styles [11], but it requires the manual labeling of new style data, which is resource-intensive. An ideal way to solve this problem without extra manual annotations is to normalize the input images.
Fig. 1.
Cytopathology image style diversity. Image style and visual appearance can differ greatly depending on slide staining procedures and scanning devices. Different rows of patches stand for different hospitals while different columns represent different scanning devices.
A lot of traditional color normalization methods are proposed for eliminate pathology image style difference caused by staining procedure, mainly including color deconvolution [13], [14], [15], [16], [17], [18] or stain spectral matching [19], [20], color transfer [21], [22], histogram matching [23], and some other methods [24], [25]. Color deconvolution methods were widely studied in past years. The principle of color deconvolution is below: firstly deconvolve an image into stain components, then match the components between target images and template images, finally convolve the matched individual components to obtain normalized target images [25], [26]. Some methods further improve the color normalization performance by detecting cell nucleus, cell cytoplasm and background to better identify stain components [14], [27]. Color transfer methods generally match color distributions between target images and template images by equalizing the mean and standard deviation for each color channel in the perceptual color space “L*a*b*” [21]. Histogram matching is a classic image processing technique to transform the intensity distribution in all color channels of target images to match that of template images by histogram specification [23]. These traditional color normalization methods generally are manually defined models with empirical parameters and don’t depend on paired images of different staining protocols. Thus, they can be directly applied to various images of different color distributions. However, there are two shortcomings in these methods, which affect their practical application performance. The first one is that the predefined models and parameters limit color normalization performance for images with huge differences in staining and imaging characteristics. The second is that the identified several template images with optimal staining and visual appearance may not represent the color distribution of the entire reference dataset. In particular, the content of a cytopathology image with limited pixels is usually much sparser than that of a histopathology image. It’s quite difficult to find several template images representative of all cell categories to maintain the color normalization performance.
To tackle the issues of traditional methods, color normalization methods based on deep learning [26], [28], [29], [30], [31], [32], [33], [34], [35] especially generative adversarial networks (GANs) [36] are proposed. In contrast to traditional color normalization methods, GAN-based methods consider the overall dataset of target style as the template and approach the problem of color normalization as image-to-image translation [28], [29], [30]. GAN-based color normalization methods can be divided into supervised and unsupervised. Supervised methods require paired images of different staining protocols, then use mean absolute error (L1 loss) and adversarial losses to optimize generative networks like pix2pix GAN [37]. Supervised methods usually have good performance, but obtaining paired images of different styles requires extra multiple staining and imaging, increasing the difficulty of data acquisition. Unsupervised methods do not require paired images, and most of them are based on CycleGAN [38]. Shaban et al. [28] proposed StainGAN based on standard CycleGAN for pathology images. Zhou et al. [34] proposed an enhanced CycleGAN color normalization method by computing a stain color matrix for stabilizing the cycle consistency loss. De Bel et al. [30] further developed residual CycleGAN to learn residual mapping for color domain transformation instead of regular image-to-image mapping. Generally, the cycle consistency loss is designed to keep the structure of generated images not be distorted, but this purpose cannot be strictly guaranteed since the cycle consistency loss is indirect. For cytopathology images, generated images by CycleGAN-based methods may face problems such as cell cross-color (i.e., hue inconsistency). As shown in Fig. 2, the red cell cytoplasm in the input image was transformed into blue in the normalized image, and meanwhile the blue cells also had cross-color phenomenon. The hue inconsistency will affect manual interpretation or automatic analysis, since the cell color represents types of superficial, middle and basal cells according cytopathology staining protocol [39]. Besides, Tellez et al. [26] and Cho et al. [32] proposed unsupervised color normalization methods by applying heavy color augmentation or gray-scale transformation to input images and using a convolutional neural network to reconstruct the original appearance of the input images. Li et al. [31] also utilized the color normalization strategy in pathology image super resolution. Compared with CycleGAN-based method, the color normalization strategy does not require training for each target domain. But it is difficult for color augmentation or gray-scale transformation to fully imitate or eliminate the style diversity of practical staining and imaging, limiting the effectiveness of the color normalization strategy. In fact, the style diversity of staining and imaging is reflected in both color distribution and intensity distribution, and simple gray-scale transformation cannot normalize images of diverse styles.
Fig. 2.
Cell cross-color phenomenon of CycleGAN-based normalization methods. Results of CycleGAN for style normalization: the target image to be normalized (left), the normalized target image with user-selected style (middle) and the image reconstructed back to the input style (right).
To address the above demand and challenges, we propose a novel unsupervised style normalization method for cytopathology images without using CycleGAN. As shown in Fig. 3 below, our method begins with a style removal module in which we convert each colorful image from both source and target domain into a grayscale image with a color-encoding mask. Then, we build a reconstruction module to map them back to a colorful image with the source domain style. Our method normalizes cytopathology images at both pixel-level and style-level through direct per-pixel regression and two types of domain adversarial learning. We enforce both hue and structure consistency before and after normalization by using the color-encoding mask and L1 loss of per-pixel regression. An intra-domain adversarial learning is applied to ensure the style of normalized images consistent with the source domain style. An inter-domain adversarial learning is proposed to make the reconstruction network robust for input images of different domains. To evaluate our method, we formed cervical cell datasets with different image styles from multiple hospitals and scanners. Experimental results demonstrate that our method outperforms current traditional and deep learning-based unsupervised color normalization methods. Besides, we further demonstrate that our normalization method greatly improves the recognition accuracy of lesion cells on unseen cytopathology images, which is critical for model generalization. The main contributions are summarized as follows:
-
•
We propose an unsupervised generative method to normalize cytopathology image styles without using regular the cycle consistency strategy.
-
•
Our method enforces both hue and structure consistency by the color-encoding mask and L1 loss, and ensures the style of normalized images from different domains consistent with the user-selected style by domain adversarial style reconstruction.
-
•
Our method yields superior color normalization results quantitatively and qualitatively against current traditional and deep learning-based color normalization methods on multiple cervical cell datasets, and greatly improves the recognition accuracy of lesion cells on unseen cytopathology images.
Fig. 3.
Image style normalization process with two-stage domain adversarial style normalization framework. The proposed framework consists of a style reconstruction network G(.), an intra-domain discriminator D1(.) and an inter-domain discriminator D2(.). It normalizes the target image with style removal and style reconstruction. In the process of style reconstruction, we take multiple losses (including GAN1 Loss ( ), GAN2 Loss (
), GAN2 Loss ( ) and L1 Loss (
) and L1 Loss ( )) to ensure the reconstructed style consistent with the source style and the reconstructed structure identical with its origin.al input images.
)) to ensure the reconstructed style consistent with the source style and the reconstructed structure identical with its origin.al input images.
2. Materials
2.1. Cervical image style normalization datasets
We used six cervical cytopathology slides datasets which were gathered from 2 hospitals (denoted as Hospital1 and Hospital22 separately) and scanned with 4 types of scanners (denoted as Device1, Device2, Device3 and Device43 separately). As these datasets were produced by different hospitals or scanned with different scanners, they had variant image styles and were suitable to evaluate our method for image style normalization across domains.The slide preparation protocols of Hospital1 and Hospital2 are both liquid-based cytology preparation, but Hospital1 is membrane-based and Hospital2 is sedimentation. The dyeing schemes include fixing, clearing and dehydrating. The imaging magnification and imaging resolution of Devices1-4 are 20 – 0.243 m/pixel, 20 – 0.243 m/pixel, 20 – 0.293m/pixel and 40 – 0.180 m/pixel separately. The dataset produced by Hospital1 and scanned with Device1 was considered as the source domain and denoted as S. The other datasets were considered as the target domain and denoted as T1T5 4. We randomly split the slides of each dataset into training and test sets and then extracted 512512 patches with pixel size of 0.586 m from slides. The details of datasets are shown in Table 1.
Table 1.
Cervical image style normalization datasets.
| Name | Hospital | Scanner | Training slides | Training patches | Test slides | Test patches |
|---|---|---|---|---|---|---|
| S | Hospital1 | Device1 | 170 | 100,000 | 40 | 6,000 |
| T1 | Hospital1 | Device2 | 343 | 50,000 | 119 | 5,000 |
| T2 | Hospital1 | Device3 | 169 | 50,000 | 26 | 5,000 |
| T3 | Hospital1 | Device4 | 60 | 50,000 | 20 | 5,000 |
| T4 | Hospital2 | Device2 | 48 | 50,000 | 16 | 5,000 |
| T5 | Hospital2 | Device4 | 39 | 50,000 | 10 | 5,000 |
2.2. Cervical lesion cell recognition datasets
To evaluate our method’s improvement to the generalization ability of analysis models, we organized cervical lesion cell recognition datasets. For both the source domain and target domain (T is one of T1, T2 and T3), we invited cytopathologists to label the lesion cells in positive slides. For S, T1, T2 and T3, they are the same as the datasets used in the previous datasets but with lesion cell annotations. All cells in negative slides were considered as normal cells. We used the annotations of the source domain to train a analysis model whose task is to classify the lesion and normal cells. To train for cell classification in source domain, we generated 200,000 positive patches by sampling from 29,360 annotations of lesion cells and 200,000 negative patches by randomly sampling from the negative slides. The test patches of different target domains were generated by the same way. The details of datasets are shown in Table 2.
Table 2.
Cervical lesion cell recognition datasets.
| Name | Positive Slides | Annotations | Positive Patches | Negative Slides | Negative Patches | |
|---|---|---|---|---|---|---|
| S-train | 84 | 29,360 | 200,000 | 86 | 200,000 | |
| S-test | 20 | 2,734 | 4,000 | 20 | 4,000 | |
| T1-test | 19 | 1,292 | 2,500 | 100 | 2,500 | |
| T2-test | 15 | 728 | 1,300 | 11 | 1,300 | |
| T3-test | 11 | 602 | 1,100 | 9 | 1,100 |
3. Methods
The proposed method performs the domain adversarial style normalization via the two-stage strategy including style removal and style reconstruction. To better explain our method, we assume that the source domain is the user-selected style dataset with annotations and the target domain is the dataset to be normalized without annotations. Variations in the process of staining and imaging result in that and present different image styles and a domain shift. Our goals are to normalize the target domain image style to the source domain image style both visually and numerically and thus mitigate the degradation of analysis models. Our method consists of three sub-networks, separately named style reconstruction network G(.), intra-domain discriminator D1(.) and inter-domain discriminator D2(.). The parameters of each sub-network are defined as , and . We refer to the style reconstructed image of a cytopathology image x as .
In the following, we give detailed descriptions of style removal (Section 3.1) and style reconstruction (Section 3.2), followed by the definition of loss functions (Section 3.3) and the description of network architectures (Section 3.4).
3.1. Style removal
The most intuitive discrepancy between the source and target domain is their different image styles. For the style reconstruction network G(.), our requirement is that: (i) G(.) has the same performance for and , i.e. it can reconstruct almost identical style for cytopathology images from different domains; (ii) G(.) ensures the pathological information of images is consistent before and after reconstruction. In essence, G(.) is similar to other convolutional neural networks and has a strong sensitivity to the different numerical distributions. If there is no additional operation or supervision to help G(.) overcome this distribution change, G(.) will struggle to exhibit ideal normalization performance. Therefore, we design a style removal module to bind the input images of G(.) from different domains to a roughly consistent distribution while preserving the cell morphology information and rough color information of cytopathology images. This module converts the natural RGB image into a two-channel image, which is formed by concatenating a grayscale image and a color-encoding mask (called gray-scale image with mask, GM), as the input of G(.).
The gray-scale image is obtained by the weighted addition of the R, G, B channels of the colorful image as follows [23]:
| (1) |
Eq. (1) is the standard formula to calculate the gray value of a pixel in RGB color space. This translation coarsely erases the style of RGB image to roughly normalize the input distribution of G(.) while preserving the morphology information (e.g. texture and structure). For cytopathology images, the color information representing some pathological information should be retained. In the process of producing a cytopathology slide, the staining agents dye the cell cytoplasm to red or blue according to the difference in acidity and alkalinity, which means most of cells in cytopathology images are red or blue [39]. It is necessary to use this color information for manual interpretation or automatic interpretation with the analysis models. However, if G(.) only obtains gray-scale image, the style reconstructed by G(.) will ignore such information and may result in hue inconsistency (shown in Fig. 2). To solve the above problem, we convert the original cytopathology image into a color-encoding mask as part of the input of G(.), thus remaining the rough color information of the cytoplasm. The specific color-encoding mask generation process is as follows: we turn the natural RGB image to “L*a*b*” color space which expresses color as three values: “L*” for the lightness from black (0) to white (100), “a*” from green (−127) to red (+128) and “b*” from blue (−127) to yellow (+128) [23]. Then we set a hyper-parameter and encode the pixels with values of “a*” channel more than as 1 (red) and encode the others as 0 (not red). With such color-encoding mask, our method would greatly overcome the problem of hue inconsistency. .
3.2. Style reconstruction
In order to reconstruct the source domain image style according to the input {GM} across domains and ensure the consistency before and after image translation, we impose an intra-domain adversarial loss, an L1 penalty and an inter-domain adversarial loss on the reconstruction objective.
3.2.1. Intra-domain adversarial and per-pixel regressive learning
The distribution of and is similar as the style removal module can roughly erase two domains’ different image styles while preserving the fine morphology information and coarse color information of the cytopathology image. It means that when we learn a mapping function G(.) from to , it can also map to to some extent.
In the intra-domain adversarial training, G(.) attempts to generate a as similar to as possible to fool the intra-domain discriminator D1(.), whereas D1(.) wants to distinguish the reconstructed image from the real image , thus forming a process of adversarial training between G(.) and D1(.). Naturally, this intra-domain adversarial loss should be defined as follow[36]:
| (2) |
In addition, as is obtained by removing the image style component of , G(.) should be encouraged to generate an image as the same as . Therefore, we impose a per-pixel penalty on reconstruction objective to achieve per-pixel regression, which is a much stronger supervision to enforce the content (including hue, texture and structure) consistency before and after reconstruction. Following [37], we adopt the L1 penalty, which offers strong supervision and encourages less blur rather than higher-order penalty. The specific loss function is defined as follows:
| (3) |
3.2.2. Inter-domain adversarial learning
In addition to color information such as hue, the images contain some other details including brightness, contrast and intensity. Although the style removal module can roughly normalize the input image of G(.) by erasing the detailed hue information, differences in other types of information still persist in the gray-scale image, causing certain discrepancy between the distributions of and . Due to such distribution change, G(.), which is only trained with two aforementioned intra-domain losses, may struggle to reconstruct a image style completely match the source image style .
Recent advances in adversarial domain adaptation [40], [41], [42], [43] proved that the domain adversarial learning could leverage deep networks to learn invariant representations across domains. Such benefit can be observed in the adversarial learning of GANs. The discriminator perceives the difference between the generated images and real images and encourage the generator to generate images more realistic and indistinguishable to the discriminator. Motivated by domain adversarial insight, we present a novel form of adversarial training between two domains, called inter-domain adversarial learning, to supervise G(.) to learn a domain-invariant reconstruction. In this loss, G(.) attempts to reconstruct a more realistic to fool the inter-domain discriminator D2(.), while D2(.) hopes to distinguish the reconstructed image from the real image , creating a new dynamic equilibrium and forming a new adversarial learning. The loss function of the inter-domain adversarial learning is defined as follows:
| (4) |
3.3. Loss functions
The goal of domain adversarial style normalization is to reconstruct a image style highly consistent with the source domain image style. Taking all above loss functions together, our complete objective of the method can be defined as follows:
| (5) |
where , , are hyper-parameters indicating the relative importance of the different loss functions in the objective. , , , were empirically set to 1, 1, 100, 21 in our work. This ultimately corresponds to solving for a style reconstruction network G(.) according to the optimization problem.
| (6) |
3.4. Network architecture
Similar to other image-to-image translation problems, there is a good deal of low-level information (such as structure, texture, etc) shared between the input and output of the style reconstruction network G(.), and it would be desirable to shuttle this information directly across the network. Therefore, we used the U-net with full convolution and skip connection [44] as our style reconstruction network. For considerations of calculation speed and GPU memory, G(.) does not downsample the input image to a vector before upsampling, like the common U-net architecture. Instead, we adjust the convolution stride size and downsample the feature maps to 1/64 of the original image size with six convolution layers, and then gradually upsample with six deconvolution layers [45] to the original image size. Note that the convolution (or deconvolution) layers refer to sequential operation modules: convolution (or deconvolution) – batch normalization – ReLU activation. For the intra-domain discriminator D1(.) and inter-domain discriminator D2(.), we use the PatchGAN proposed by [37], which only penalizes structure at the scale of patches and restricts attention to the content in local image patches.
4. Experimental results
We performed two types of experiments. First, we quantitatively and qualitatively evaluated the quality of normalized cytopathology images by our method and compared with other unsupervised normalization methods (Section 4.1). Then, we conducted a serious of ablation studies to verify the individual contributions of the color-encoding mask and the inter-adversarial learning (Section 4.2). Finally, we explored our method’s improvement to the generalization ability of analysis models (Section 4.3).
4.1. Style normalization
We evaluated our method together with other nine unsupervised normalization methods (SPCN [13], Macenko’s [15], Reinhard’s [21], Khan’s [14], Gupta’s [18], Zheng’s [17], CycleGAN [38], StainGAN [28] and Tellez’s [26]) on the cervical image style normalization datasets in Table 1.
4.1.1. Implementation details and evaluation metrics
The Adam optimizer [46] with a learning rate of 0.0001 and adaptive momentum with the parameters (=0.3, =0.999) was used to solve the objective. The networks including G(.), D1(.) and D2(.) were trained from scratch where their weights were initialized from a truncated Gaussian distribution of mean 0 and standard deviation 0.02. We separately trained these networks for each dataset. The hyper-parameters , , , were separately equal to 1, 1, 100, 21. Notably, the codes of Macenko’s, Khan’s and Reinhard’s were provided by Khan et al. (Stain Normalisation Toolbox for Matlab, BIALab, Department of Computer Science, University of Warwick) [14]. The codes of Tellez’s were rewritten according their principles. The codes of other methods were provided by their authors. We followed the recommended setting and hyper-parameters. Macenko’s method with default hyper-parameters had unreasonable results in the T2 target domain, thus we did our best to tune the hyper-parameters. The default stain classifier in Khan’s method [14] was used, which may be not suitable for cytology images.
We used the histogram distribution in “L*a*b*” color space to represent the image style and then used the similarity of distributions to evaluate and compare the normalization performance of different methods. The amount and kind of cells in cytopathology images are unbalanced and the similarity of two domains’ image styles could not be precisely measured with unbalanced proportions of compositions. We split each image into three compositions including background, red cells and blue cells 5. The threshold segmentation method was applied to separate the background and foreground while the color-encoding mask presented in 3.1 was applied to split the foreground region into red cells and blue cells. To balance the proportion, we randomly sampled 500 pixels from each composition on one domain’s each testing patch, and then counted this domain’s histogram distribution in “Lab” color space. We computed Bhattacharyya distance (lower scores indicate more similar distributions) and histogram intersection score [47] (higher scores indicate more similar distributions) between the distributions of source domain and each normalized target domain to measure the similarity of their image styles and evaluate the performance of different methods. Meanwhile, we compared different methods in the structure consistency before and after normalization of target images by computing structural similarity SSIM and peak signal noise ratio PSNR [48]. Because images before and after normalization had different color styles and influenced the metrics of SSIM and PSNR. We first conducted gray-scale transformation and foreground segmentation, then computed the metrics of SSIM and PSNR. The foreground segmentation is to avoid the interference of background pixels.
4.1.2. Results
Fig. 4 shows some typical normalized results of our method and other methods for image style normalization in the target domain T1. Generally, normalized images generated by methods based on deep learning match the image style of source domain better than traditional methods. Among traditional methods, SPCN, Macenko’s, Gupta’s and Zheng’s methods show better color similarity than Reinhard’s and Khan’s. For example, background pixels were incorrectly normalized in the normalized images by Khan’s. In terms of structure consistency before and after normalization, the traditional methods performed well except Reinhard’s. All deep learning based methods shows good structure consistency visually and color similarity. But we observe that CycleGAN probably generated images with cell hue inconsistency, in which red cells were translated into blue while blue cells were translated into red. And this can be attributed to that CycleGAN imposes no supervision to discover the hue mismatch of normalized and original images, which is one of the main drawbacks of unpaired image-to-image translation based on cycle consistency. With the color-encoding mask and strong hue and structure consistency supervision force (L1 penalty for per-pixel regression), our method avoids such inconsistency.
Fig. 4.
Examples of style normalized images of different normalization methods in the target domain T1. The first row refers to the typical examples of source domain. In general, normalized images generated by methods based on deep learning match the image style of source domain better than traditional methods. Phenomenon of hue inconsistency may be found in CycleGAN-based methods.
To better evaluate the color normalization performance of different methods, we provided the quantitative metrics of color distribution similarity and structure consistency in the five target domains T1-T5 in Table 3. The results indicate that deep learning based methods generally perform better than traditional methods, which confirms the superiority of learning based methods to capture the image style represented by whole domain rather than a single template. Our method outperforms all compared methods at all different source-target domain settings in color similarity metrics. The overall method () yields large boosts against the method without inter-domain adversarial loss (), which highlights the power of inter-adversarial learning to overcome the inherent inconsistency of gray-scale images across domains. All methods obtains high structure consistency metrics before and after normalization, except Reinhard’s and Khan’s. It can be seen that the inter-adversarial learning harms the structure consistency. This is because that adversarial learning helps style transfer and fine texture generation but introduces subtle artifacts. Thus, Tellez’s method with only L1 loss achieves almost best structure consistency metrics. Although the structure consistency metrics of our method are not the highest, they are still at a very high level, demonstrating the high structure consistency in color normalization. Besides, we visualized the distribution matching property in “L*a*b*” color space of different methods in Fig. 5. We can see that the color distributions of the source and target domains have large difference. After applying color normalization to the target domain, the result of our method shows the closest color distribution with the source domain.
Table 3.
Color distribution similarity and structure consistency metrics of style normalization methods at different source-target domain settings. “Our*” refers to , “Our” refers to .
| Method | T1S |
T2S |
T3S |
T4S |
T5S |
|---|---|---|---|---|---|
| Bhattacharyya distance/ Histogram intersection | |||||
| SPCN | 0.270/0.803 | 0.329/0.702 | 0.351/0.674 | 0.311/0.748 | 0.381/0.638 |
| Macenko’s | 0.127/0.867 | 0.312/0.657 | 0.234/0.764 | 0.191/0.833 | 0.368/0.615 |
| Reinhard’s | 0.320/0.674 | 0.336/0.654 | 0.392/0.572 | 0.358/0.643 | 0.381/0.603 |
| Khan’s | 0.374/0.654 | 0.447/0.576 | 0.403/0.616 | 0.384/0.639 | 0.404/0.612 |
| Gupta’s | 0.197/0.825 | 0.258/0.756 | 0.200/0.790 | 0.217/0.789 | 0.241/0.743 |
| Zheng’s | 0.155/0.864 | 0.298/0.675 | 0.311/0.706 | 0.206/0.799 | 0.356/0.660 |
| CycleGAN | 0.088/0.929 | 0.081/0.927 | 0.110/0.909 | 0.087/0.920 | 0.104/0.897 |
| StainGAN | 0.123/0.900 | 0.141/0.885 | 0.110/0.908 | 0.142/0.847 | 0.123/0.881 |
| Tellez’s | 0.121/0.896 | 0.367/0.625 | 0.234/0.748 | 0.173/0.847 | 0.225/0.766 |
| Our* | 0.092/0.905 | 0.107/0.891 | 0.161/0.844 | 0.112/0.882 | 0.120/0.881 |
| Our | 0.059/0.940 | 0.060/0.943 | 0.087/0.927 | 0.069/0.928 | 0.076/0.933 |
| SSIM/ PSNR | |||||
| SPCN | 0.890/21.06 | 0.874/16.18 | 0.895/21.91 | 0.878/20.96 | 0.870/18.11 |
| Macenko’s | 0.916/21.82 | 0.544/16.68 | 0.916/22.17 | 0.895/21.27 | 0.887/17.88 |
| Reinhard’s | 0.703/14.64 | 0.772/13.20 | 0.681/14.82 | 0.672/13.97 | 0.724/16.28 |
| Khan’s | 0.579/12.08 | 0.643/12.95 | 0.577/8.81 | 0.528/12.02 | 0.550/8.98 |
| Gupta’s | 0.947/23.92 | 0.966/22.47 | 0.970/23.90 | 0.931/22.11 | 0.954/19.84 |
| Zheng’s | 0.990/24.76 | 0.996/30.49 | 0.963/19.14 | 0.990/25.01 | 0.972/20.85 |
| CycleGAN | 0.831/21.14 | 0.943/22.97 | 0.884/17.09 | 0.858/19.92 | 0.734/14.45 |
| StainGAN | 0.956/27.60 | 0.958/24.44 | 0.891/16.95 | 0.854/20.05 | 0.777/15.23 |
| Tellez’s | 0.988/29.92 | 0.964/19.32 | 0.977/28.05 | 0.990/31.09 | 0.980/29.24 |
| Our* | 0.970/26.75 | 0.963/25.56 | 0.947/21.16 | 0.965/25.71 | 0.937/20.44 |
| Our | 0.962/26.87 | 0.929/22.93 | 0.880/17.03 | 0.892/22.42 | 0.842/16.29 |
Fig. 5.
Distribution matching property in “L*a*b*” color space of different normalization methods at T1S. Each curve in the plots represents the source domain S or the target domain T1 or its normalized target domain by different normalization methods. The closer two curves are, the better their color distribution similarity is.
4.2. Ablation studies
We conducted a serious of ablation studies to verify the individual contributions of the color-encoding mask, the inter-adversarial learning and the strategy of individual target domain learning.
4.2.1. The color-encoding mask
The color-encoding mask was proposed for eliminating the cell hue inconsistency problem in normalized images. To validate the role of the color-encoding mask, we did a comparison experiment: with the color mask generated by thresholding the channel “a*” in “L*a*b*” space, with the color mask generated by thresholding the channel “b*” and without the color mask. Table 4 shows the metrics of color distribution similarity and structure consistency. Fig. 6 shows the normalized images under the three conditions. The results indicate that a) the color mask is key for preventing the cell hue inconsistency and improving color distribution similarity of normalized images; b) generating the color mask by the channel “b*” is unsuitable and the incorrect mask harms the effect of color normalization. The goal of designing the color-encoding mask is to distinguish red cells and non-red cells. But the channel “b*” has values from blue (−127) to yellow (+128) and is not suitable to distinguish red cells.
Table 4.
Ablation experiment results of the color-encoding mask in the target domain T1.
| Metrics | With mask | With mask | Without mask |
|---|---|---|---|
| using “a*” | using “b*” | ||
| Bhattacharyya distance | 0.059 | 0.092 | 0.078 |
| Histogram intersection | 0.940 | 0.905 | 0.932 |
| SSIM | 0.962 | 0.920 | 0.948 |
| PSNR | 26.87 | 23.64 | 25.25 |
Fig. 6.
Ablation experiment results of the color-encoding mask in the target domain T1. The normalized images without using the color mask or with using unsuitable color mask have the cell hue inconsistency problem.
4.2.2. The strategy of individual target domain learning
The principle of our method supports simultaneous normalization of multiple target domains. We added a comparison experiment: training the individual target domains T1-T5 and training the union of all target domains T1-T5. Table 5 the results of color normalization and structure consistency metrics under the two conditions. The results indicate that the color normalization effect of training the individual target domains is obviously better than that of training the united target domains. The united domains of various color distributions may interfere the inter-domain adversarial learning and then harm the color reconstruction across multiple domains.
Table 5.
Ablation experiment results of training individual and united target domains T1-T5.
| Individual target domains |
|||||
|---|---|---|---|---|---|
| Metrics | T1S | T2S | T3S | T4S | T5S |
| Bhattacharyya distance | 0.059 | 0.060 | 0.087 | 0.076 | 0.070 |
| Histogram intersection | 0.940 | 0.943 | 0.927 | 0.928 | 0.933 |
| SSIM | 0.962 | 0.929 | 0.880 | 0.892 | 0.842 |
| PSNR | 26.87 | 22.93 | 17.03 | 22.42 | 16.29 |
| United target domains |
|||||
| Metrics | T1S | T2S | T3S | T4S | T5S |
| Bhattacharyya distance | 0.081 | 0.179 | 0.148 | 0.093 | 0.153 |
| Histogram intersection | 0.932 | 0.823 | 0.868 | 0.918 | 0.851 |
| SSIM | 0.915 | 0.774 | 0.895 | 0.912 | 0.889 |
| PSNR | 22.95 | 17.90 | 19.43 | 22.78 | 18.74 |
4.2.3. The inter-domain adversarial learning
The inter-domain adversarial learning is proposed to make the reconstruction network robust for input images of different domains. To validate its role in our method, we conducted a comparison experiment by setting different loss weight ratios of , and . Table 6 shows the results of color normalization and structure consistency metrics under different ratios. The results indicate that a) the inter-domain adversarial learning is important for improving color normalization effect of target domains; b) approximately equalizing the loss weights of the inter-domain and intra-domain adversarial learning is better for color normalization. Besides, the structure consistency metrics are all high under the four ratios, which illustrates the L1 loss is key for structure consistency.
Table 6.
Ablation experiment results of different loss weights (, , ) in the the target domain T1.
| Metrics | 100:0.5:1 | 100:1:1 | 100:1:0.5 | 100:1:0 |
|---|---|---|---|---|
| Bhattacharyya distance | 0.073 | 0.059 | 0.075 | 0.092 |
| Histogram intersection | 0.936 | 0.940 | 0.930 | 0.905 |
| SSIM | 0.936 | 0.962 | 0.946 | 0.970 |
| PSNR | 24.46 | 26.87 | 25.26 | 26.75 |
4.3. Improving recognition accuracy of unseen target domain images
Because of the image style inconsistency across domains, there would be performance degradation of automated analysis models trained in source domain when they are directly employed in target domain. We evaluated our method’s improvement to the generalization ability of analysis models on the cervical lesion cell recognition datasets in Table 2 while comparing with other normalization methods.
4.3.1. Implementation details and evaluation metrics
We used ResNet-50 [49] as the task network to classify the lesion and normal cells. At the top of , we used two fully connected layers (2048-256, 256-1) to produce the prediction output and adopted the sigmoid as the final activation function. On source domain training set, we fine-tuned from ImageNet pre-trained models [50] and used an Adam optimizer [46] with a learning rate of 0.001 and adaptive momentum with the parameters: = 0.9, = 0.999. We adopted data augmentation including image flipping and rotation. Finally, the average classification accuracy of in testing data of source domain arrived at 99.8%.
We used average classification accuracy to evaluate the performance of in different domains including source domain, each target domain and their normalized variant . To evaluate the performance of our method and other methods, we compared the performance of in target domain before and after applying these normalization methods and thus computed how much they mitigated the performance degradation of when applied across domains.
4.3.2. Results
Results in Table 7 confirm that the domain shift and distribution change caused by discrepant image styles can degrade the performance of task network in target domain (average precision drops from 99.8% on to 64.6% on wild ). However, the degradation could be effectively mitigated with our method (). For instance, average precision increases to 81.2% on T1S with our method. Besides, due to the superior color normalization performance, our method outperforms other normalization methods in improving the recognition task accuracy on unseen target domains. Another benefit of style normalization is that remained the original classification performance on source domain, since there is no refinement or retraining of .
Table 7.
Improved accuracies (%) of unseen target domain images.
| Method | T1S | T2S | T3S | Avg |
|---|---|---|---|---|
| Wild | 64.6 | 61.4 | 71.9 | 62.1 |
| SPCN | 66.7 | 49.8 | 71.6 | 62.7 |
| Macenko’s | 71.1 | 45.7 | 60.2 | 59.0 |
| Reinhard’s | 62.3 | 63.4 | 61.9 | 62.5 |
| Khan’s | 50.0 | 51.5 | 50.5 | 50.7 |
| Gupta’s | 77.5 | 62.1 | 57.9 | 65.8 |
| Zheng’s | 74.5 | 50.0 | 70.3 | 64.9 |
| Cycle-GAN | 75.3 | 67.6 | 60.4 | 67.8 |
| StainGAN | 66.4 | 66.2 | 72.9 | 68.5 |
| Tellez’s | 51.8 | 50.0 | 53.3 | 51.7 |
| Our | 81.2 | 81.5 | 83.2 | 82.0 |
We visualized the feature representations in the setting of T2S by t-SNE [51] in Fig.7. Before applying our method, the categories in source domain can be separated well, but the categories in target domain are not discriminated well. Besides, the feature representations from source and target domain are not aligned well, which means extracted different feature representations for the source and target domains. After using our method, the feature representations from the source and corresponding normalized target domains are aligned better and different categories in normalized target domain are discriminated better. It indicates that our color normalization method greatly mitigate the performance degradation.
Fig. 7.
T-SNE of the task network ’s feature representations from wild images (a) and corresponding normalized images by our method (b). Green and blue dots refer to positive and negative patches of S; purple and red dots refer to positive and negative patches of T2.
5. Conclusions
In this paper, we proposed a thorough domain adversarial style normalization method for cytopathology images. Our method is built from a style removal module followed by a style reconstruction module. The former binds the source and target images to a roughly consistent distribution while the latter reconstructs the distribution to source domain. We highlighted the superiority of such two-stage strategy to enforce the consistency of hue and structure. Extensive experiments shown that our method exhibited better style normalization performance than other unsupervised normalization methods and greatly improved lesion classification accuracy of unseen target domain images on multiple cervical cell datasets. These results illustrated the strong potential of our method by employing the two-stage strategy with thorough supervised learning including per-pixel regressive learning, inter-domain and intra-domain adversarial learning.
On the premise of maintaining the color normalization effect and structure consistency, generalization without retraining for different domains and real-time speed matching slide scanning are the key points for applying color normalization methods in practice. Traditional methods, Tellez’s method and our method without inter-domain adversarial learning () can be directly applied for different domains but have reduced color normalization effect. Meanwhile, CycleGAN-based methods and our method with inter-domain adversarial learning ( depend on retaining for different domains with better color normalization effect. Our method achieved superior results in both cases. In a computer with Intel Xeon Gold 6134 CPU (3.2 GHz), 256 GB memory and one Nvidia Titan V GPU (12 GB memory), the speed of our method is about 88 FPS (batch size = 10, without image I/O time), faster than other methods. Thus, processing a whole slide image of about 30 K30 K pixels needs about 90 s using common split-merge strategy with redundancy. Therefore, our method has the potential to be used in real time along with slide scanning.
In future work, we will apply the proposed method to more datasets, including histopathology images, and more specific tasks including cell segmentation and lesion detection to verify its general applicability and potential. About the binary color-encoding mask, adopting one hyper-parameter to approximately separate the red and blue compositions may be not the most ideal method. We will explore a more reasonable approach to replace the current style removal module such as a convolutional neural network.
CRediT authorship contribution statement
Xihao Chen: Methodology, Software, Validation, Writing - original draft. Jingya Yu: Methodology, Software, Validation, Writing - original draft. Shenghua Cheng: Conceptualization, Methodology, Writing - review & editing, Supervision. Xiebo Geng: Software, Validation. Sibo Liu: Software, Validation. Wei Han: Software. Junbo Hu: Data Acquisition, Annotation. Li Chen: Data Acquisition, Annotation. Xiuli Liu: Data Acquisition, Annotation. Shaoqun Zeng: Project administration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
The authors would like to thank Women and Children Hospital of Hubei Province and Tongji Hospital of Tongji Medical College, Huazhong University of Science and Technology for the support in data acquisition and lesion cells labeling. We gratefully acknowledge the support of Wuhan National Laboratory for Optoelectronics in slide scanning.
Footnotes
Hosipital1 and Hosipital2 were separately Women and Children Hospital of Hubei Province and Tongji Hospital of Tongji Medical College, Huazhong University of Science and Technology.
Device1 and Device2 were two different products from 3DHISTECH Ltd. Device3 and Device4 were separately developed by Wuhan National Laboratory for Optoelectronics and Wuhan Huaiguang Intelligent Technology Co., Ltd.
Each slide represented a unique patient’s cervical cancer thin prep testing.
In the process of producing a cytopathology slide, the staining agent dyes cell cytoplasm to red or blue according to the difference in acidity and alkalinity, which means most of cells in cytopathology images are red or blue.
References
- 1.Meijering E. A bird’s-eye view of deep learning in bioimage analysis. Comput Struct Biotechnol J. 2020;18:2312–2325. doi: 10.1016/j.csbj.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Komura D., Ishikawa S. Machine learning methods for histopathological image analysis. Comput Struct Biotechnol J. 2018;16:34–42. doi: 10.1016/j.csbj.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kleczek P., Jaworek-Korjakowska J., Gorgon M. A novel method for tissue segmentation in high-resolution HE-stained histopathological whole-slide images. Comput Med Imag Graphics. 2020;79 doi: 10.1016/j.compmedimag.2019.101686. [DOI] [PubMed] [Google Scholar]
- 4.Ma J., Yu J., Liu S., Chen L., Li X., Feng J., Chen Z., Zeng S., Liu X., Cheng S. PathSRGAN: multi-supervised super-resolution for cytopathological images using generative adversarial network. IEEE Trans Med Imag. 2020;39(9):2920–2930. doi: 10.1109/TMI.2020.2980839. [DOI] [PubMed] [Google Scholar]
- 5.Cai J., Zhang Z., Cui L., Zheng Y., Yang L. Towards cross-modal organ translation and segmentation: a cycle- and shape-consistent generative adversarial network. Med Image Anal. 2019;52:174–184. doi: 10.1016/j.media.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Conjeti S., Katouzian A., Roy A.G., Peter L., Sheet D., Carlier S., Laine A., Navab N. Supervised domain adaptation of decision forests: transfer of models trained in vitro for in vivo intravascular ultrasound tissue characterization. Med Image Anal. 2016;32:1–17. doi: 10.1016/j.media.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Cheng S, Liu S, Yu J, Rao G, Xiao Y, Han W, et al. Robust whole slide image analysis for cervical cancer screening using deep learning. [DOI] [PMC free article] [PubMed]
- 8.Lyon H.O., De Leenheer A.P., Horobin R.W., Lambert W.E., Schulte E.K., Van Liedekerke B., Wittekind D.H. Standardization of reagents and methods used in cytological and histological practice with emphasis on dyes, stains and chromogenic reagents. Histochem J. 1994;26(7):533–544. doi: 10.1007/BF00158587. [DOI] [PubMed] [Google Scholar]
- 9.Yap M.H., Pons G., Martí J., Ganau S., Sentís M., Zwiggelaar R., Davison A.K., Martí R. Automated breast ultrasound lesions detection using convolutional neural networks. IEEE J Biomed Health Inf. 2018;22(4):1218–1226. doi: 10.1109/JBHI.2017.2731873. [DOI] [PubMed] [Google Scholar]
- 10.Qiu J.X., Yoon H., Fearn P.A., Tourassi G.D. Deep learning for automated extraction of primary sites from cancer pathology reports. IEEE J Biomed Health Inf. 2018;22(1):244–251. doi: 10.1109/JBHI.2017.2700722. [DOI] [PubMed] [Google Scholar]
- 11.Pan S.J., Yang Q. A survey on transfer learning. IEEE Trans Knowl Data Eng. 2010;22(10):1345–1359. doi: 10.1109/TKDE.2009.191. [DOI] [Google Scholar]
- 12.Monaco J, Hipp J, Lucas D, Smith S, Balis U, Madabhushi A. Image segmentation with implicit color standardization using spatially constrained expectation maximization: detection of nuclei. In Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), vol. 7510 LNCS, Springer; 2012. pp. 365–372. doi:10.1007/978-3-642-33415-3_45. [DOI] [PubMed]
- 13.Vahadane A., Peng T., Sethi A., Albarqouni S., Wang L., Baust M., Steiger K., Schlitter A.M., Esposito I., Navab N. Structure-preserving color normalization and sparse stain separation for histological images. IEEE Trans Med Imag. 2016;35(8):1962–1971. doi: 10.1109/TMI.2016.2529665. [DOI] [PubMed] [Google Scholar]
- 14.Khan A.M., Rajpoot N., Treanor D., Magee D. A nonlinear mapping approach to stain normalization in digital histopathology images using image-specific color deconvolution. IEEE Trans Biomed Eng. 2014;61(6):1729–1738. doi: 10.1109/TBME.2014.2303294. [DOI] [PubMed] [Google Scholar]
- 15.Macenko M., Niethammer M., Marron J.S., Borland D., Woosley J.T., Guan X., Schmitt C., Thomas N.E. A method for normalizing histology slides for quantitative analysis. IEEE International Symposium on Biomedical Imaging: From Nano to Macro. 2009;2009:1107–1110. doi: 10.1109/ISBI.2009.5193250. [DOI] [Google Scholar]
- 16.Gavrilovic M., Azar J.C., Lindblad J., Wählby C., Bengtsson E., Busch C., Carlbom I.B. Blind color decomposition of histological images. IEEE Trans Med Imag. 2013;32(6):983–994. doi: 10.1109/TMI.2013.2239655. [DOI] [PubMed] [Google Scholar]
- 17.Zheng Y., Jiang Z., Zhang H., Xie F., Shi J., Xue C. Adaptive color deconvolution for histological wsi normalization. Comput Methods Programs Biomed. 2019;170:107–120. doi: 10.1016/j.cmpb.2019.01.008. URL: https://www.sciencedirect.com/science/article/pii/S0169260718312161. [DOI] [PubMed] [Google Scholar]
- 18.Gupta A., Duggal R., Gehlot S., Gupta R., Mangal A., Kumar L., Thakkar N., Satpathy D. Gcti-sn: geometry-inspired chemical and tissue invariant stain normalization of microscopic medical images. Med Image Anal. 2020;65 doi: 10.1016/j.media.2020.101788. URL: https://www.sciencedirect.com/science/article/pii/S1361841520301523. [DOI] [PubMed] [Google Scholar]
- 19.Tosta T.A.A., Rogrio P., Servato J.P.S., Neves L.A., Roberto G.F., Martins A.S., Zanchetta M. Unsupervised method for normalization of hematoxylin-eosin stain in histological images. Comput Med Imag Graphics. 2019;77 doi: 10.1016/j.compmedimag.2019.101646. URL: https://www.sciencedirect.com/science/article/pii/S0895611118305949. [DOI] [PubMed] [Google Scholar]
- 20.Tosta T.A.A., Rogrio P., Neves L.A., Zanchetta M. Color normalization of faded he-stained histological images using spectral matching. Comput Biol Med. 2019;111 doi: 10.1016/j.compbiomed.2019.103344. [DOI] [PubMed] [Google Scholar]
- 21.Reinhard E., Ashikhmin M., Gooch B., Shirley P. Color transfer between images. IEEE Comput Graphics Appl. 2001;21(5):34–41. doi: 10.1109/38.946629. [DOI] [Google Scholar]
- 22.Magee D, Treanor D, Crellin D, Shires M, Smith K, Mohee K, et al. Colour normalisation in digital histopathology images. In: Proc Optical Tissue Image analysis in Microscopy, Histopathology and Endoscopy (MICCAI Workshop), vol. 100, Citeseer; 2009. pp. 100–111.
- 23.Baxes G.A. John Wiley & Sons Inc; 1994. Digital image processing: principles and applications. [Google Scholar]
- 24.Janowczyk A., Basavanhally A., Madabhushi A. Stain normalization using sparse autoencoders (stanosa): application to digital pathology. Comput Med Imag Graphics. 2017;57:50–61. doi: 10.1016/j.compmedimag.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Onder D., Zengin S., Sarioglu S. A review on color normalization and color deconvolution methods in histopathology. Appl Immunohistochem Mol Morphol. 2014;22(10):713–719. doi: 10.1097/PAI.0000000000000003. [DOI] [PubMed] [Google Scholar]
- 26.Tellez D., Litjens G., Bandi P., Bulten W., Bokhorst J.-M., Ciompi F., van der Laak J. Quantifying the effects of data augmentation and stain color normalization in convolutional neural networks for computational pathology. Med Image Anal. 2019;58 doi: 10.1016/j.media.2019.101544. [DOI] [PubMed] [Google Scholar]
- 27.Ehteshami Bejnordi B., Litjens G., Timofeeva N., Otte-Höller I., Homeyer A., Karssemeijer N. Stain specific standardization of whole-slide histopathological images. IEEE Trans Med Imag. 2016;35(2):404–415. doi: 10.1109/TMI.2015.2476509. [DOI] [PubMed] [Google Scholar]
- 28.Shaban MT, Baur C, Navab N, Albarqouni S. Staingan: stain style transfer for digital histological images. In 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), IEEE; 2019. pp. 953–956.
- 29.BenTaieb A., Hamarneh G. Adversarial stain transfer for histopathology image analysis. IEEE Trans Med Imag. 2018;37(3):792–802. doi: 10.1109/TMI.2017.2781228. [DOI] [PubMed] [Google Scholar]
- 30.de Bel T., Bokhorst J.-M., van der Laak J., Litjens G. Residual cyclegan for robust domain transformation of histopathological tissue slides. Med Image Anal. 2021;70 doi: 10.1016/j.media.2021.102004. [DOI] [PubMed] [Google Scholar]
- 31.Li B., Keikhosravi A., Loeffler A.G., Eliceiri K.W. Single image super-resolution for whole slide image using convolutional neural networks and self-supervised color normalization. Med Image Anal. 2021;68 doi: 10.1016/j.media.2020.101938. URL: https://www.sciencedirect.com/science/article/pii/S1361841520303029. [DOI] [PubMed] [Google Scholar]
- 32.Cho H, Lim S, Choi G, Min H. Neural Stain-Style Transfer Learning using GAN for Histopathological Images arXiv:1710.08543.
- 33.Zheng Y., Jiang Z., Zhang H., Xie F., Hu D., Sun S., Shi J., Xue C. Stain standardization capsule for application-driven histopathological image normalization. IEEE J Biomed Health Inf. 2021;25(2):337–347. doi: 10.1109/JBHI.2020.2983206. [DOI] [PubMed] [Google Scholar]
- 34.Zhou N., Cai D., Han X., Yao J. Enhanced cycle-consistent generative adversarial network for color normalization of h&e stained images. In: Shen D., Liu T., Peters T.M., Staib L.H., Essert C., Zhou S., Yap P.-T., Khan A., editors. Medical Image Computing and Computer Assisted Intervention – MICCAI 2019. Springer International Publishing; Cham: 2019. pp. 694–702. [Google Scholar]
- 35.Lei G., Xia Y., Zhai D.-H., Zhang W., Chen D., Wang D. Staincnns: an efficient stain feature learning method. Neurocomputing. 2020;406:267–273. doi: 10.1016/j.neucom.2020.04.008. [DOI] [Google Scholar]
- 36.Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, et al. Generative adversarial nets. In Advances in Neural Information Processing Systems. vol. 27; 2014. pp. 2672–2680.
- 37.Isola P, Zhu JY, Zhou T, Efros AA. Image-to-image translation with conditional adversarial networks. In Proceedings – 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017, Vol. 2017-Janua; 2017. pp. 5967–5976. doi:10.1109/CVPR.2017.632.
- 38.Zhu J.Y., Park T., Isola P., Efros A.A. Proceedings of the IEEE International Conference on Computer Vision 2017-Octob. 2017. Unpaired image-to-image translation using cycle-consistent adversarial networks; pp. 2242–2251. [DOI] [Google Scholar]
- 39.Nayar R., Wilbur D. Springer International Publishing; 2015. The bethesda system for reporting cervical cytology: definitions, criteria, and explanatory notes. [Google Scholar]
- 40.Hoffman J, Tzeng E, Park T, Phillip J-yZ, Kate I, Alexei S, et al. CyCADA: Cycle-Consistent Adversarial Domain Adaptation arXiv:1711.03213.
- 41.Long M, Cao Z, Wang J, Jordan MI. Conditional adversarial domain adaptation. In Advances in Neural Information Processing Systems, vol. 2018-December; 2018. pp. 1640–1650. arXiv:1705.10667.
- 42.Tsai Y.H., Hung W.C., Schulter S., Sohn K., Yang M.H., Chandraker M. Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. 2018. Learning to adapt structured output space for semantic segmentation; pp. 7472–7481. [DOI] [Google Scholar]
- 43.Tzeng E, Hoffman J, Saenko K, Darrell T. Adversarial discriminative domain adaptation. In Proceedings – 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017, Vol. 2017-Janua; 2017. pp. 2962–2971. doi:10.1109/CVPR.2017.316.
- 44.Ronneberger O., Fischer P., Brox T. vol. 9351. Springer; 2015. U-net: convolutional networks for biomedical image segmentation; pp. 234–241. (Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics)). [DOI] [Google Scholar]
- 45.Zeiler MD, Krishnan D, Taylor GW, Fergus R. Deconvolutional networks. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, vol. 10; 2010. pp. 2528–2535. doi:10.1109/CVPR.2010.5539957.
- 46.Kingma DP, Ba J. Adam: A Method for Stochastic Optimization arXiv:1412.6980.
- 47.Barla A, Odone F, Verri A. Histogram intersection kernel for image classification. In IEEE International Conference on Image Processing. vol. 3. IEEE; 2003. pp. 513–516. doi:10.1109/icip.2003.1247294.
- 48.Horé A., Ziou D. Image quality metrics: Psnr vs. ssim. 2010:2366–2369. doi: 10.1109/ICPR.2010.579. [DOI] [Google Scholar]
- 49.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Vol. 2016-Decem; 2016. pp. 770–778. doi:10.1109/CVPR.2016.90.
- 50.Russakovsky O., Deng J., Su H., Krause J., Satheesh S., Ma S., Huang Z., Karpathy A., Khosla A., Bernstein M., Berg A.C., Fei-Fei L. ImageNet large scale visual recognition challenge. Int J Comput Vis. 2015;115(3):211–252. doi: 10.1007/s11263-015-0816-y. [DOI] [Google Scholar]
- 51.Van Der Maaten L., Hinton G. Visualizing data using t-SNE. J Mach Learn Res. 2008;9:2579–2625. [Google Scholar]