Summary
Training and experiences are usually required to successfully culture and differentiate human embryonic stem cells (hESCs). Here, we describe a simple but highly efficient protocol to induce endoderm differentiation of hESCs with crotonate, a precursor of crotonyl-CoA for histone crotonylation deposition on endodermal genes. In this protocol, adding crotonate in different endoderm differentiation media significantly increases the differentiation efficiency and substantially reduces the amount of required reagents.
For complete details on the use and execution of this protocol, please refer to Fang et al. (2021).
Subject areas: Cell Biology, Cell culture, Stem Cells, Cell Differentiation, Tissue Engineering
Graphical abstract

Highlights
-
•
Crotonate increases the efficiency of endoderm differentiation from different hESCs
-
•
Crotonate promotes homogeneous endoderm differentiation of hESCs
-
•
Crotonate reduces the amount of Activin A required for endoderm differentiation
Training and experiences are usually required to successfully culture and differentiate human embryonic stem cells (hESCs). Here, we describe a simple but highly efficient protocol to induce endoderm differentiation of hESCs with crotonate, a precursor of crotonyl-CoA for histone crotonylation deposition on endodermal genes. In this protocol, adding crotonate in different endoderm differentiation media significantly increases the differentiation efficiency and substantially reduces the amount of required reagents.
Before you begin
Timing: 1h–16h
The following protocol focuses on a commercial STEMDIFF endoderm differentiation medium (Stemcell Technologies) with 5 mM crotonate in mel1 hESCs, and on a RPMI 1640 based endoderm differentiation medium containing 25 ng/mL Activin A, 2 μM CHIR99021, 1 × MEM NEAA, 0.2% defined FBS (dFBS), and 5 mM crotonate (we refer to it as RACC medium) in H9 hESCs.
Prepare media and reagents
-
1.
Prepare and obtain the required medium and reagents as indicated in the key resources table, Table 1, and Table 2.
Note: Prepare 1M stock solution of crotonate in dH2O and adjust pH to 7.4. Please also prepare 10 mM Y-27632 stock solution and 0.5 mM EDTA in DPBS. It is important to keep all the reagents sterile for cell culture, and prewarm the medium to 37°C before adding to cells.
Table 1.
Stock and working concentrations of reagents
| Reagent | Solvent | Stock concentration | Working concentration | Storage conditions | Aliquot |
|---|---|---|---|---|---|
| Crotonate, pH 7.4 | dH2O | 1 M | 5 mM | 18°C–25°C for 1 year | |
| Y-27632 | dH2O | 10 mM | 10 μM | −20°C for 1 month | 0.1 mL/tube |
| 0.5 mM EDTA | DPBS | 0.5 mM | 18°C–25°C for 6 months | 100 mL/bottle | |
| CHIR99021 | DMSO | 5 mM | 2 μM | −80°C for 2 years | 0.1 mL/tube |
| Activin A | 0.1% dFBS in DPBS | 100 ng/mL | 25 ng/mL | −80°C for 3 months | 20 μL/tube |
Table 2.
RACC endoderm differentiation medium
| Reagent | Stock concentration | Working concentration | Volume |
|---|---|---|---|
| RPMI 1640 | N/A | N/A | 49.1 mL |
| 0.2% dFBS | 100% | 0.2% | 100 μL |
| Activin A | 100 ng/mL | 25 ng/mL | 12.5 μL |
| CHIR99021 | 5 mM | 2 μM | 20 μL |
| MEM NEAA | 100 × | 1 × | 0.5 mL |
| Crotonate, PH 7.4 | 1 M | 5 mM | 0.25 mL |
| Total | n/a | n/a | 50 mL |
The following protocol is for a 6-well plate. For detailed information on other plate sizes, please see Table 3.
Table 3.
Cell number and medium volume in different plates
| Plate | Area (cm2) | Matrigel (mL) | Mel1 cells (1.05 × 104/cm2 | H9 cells (2.1 × 104/cm2) | STEMDIFF endoderm differentiation medium plus 5 mM crotonate (0.13–0.26 mL/cm2) | RACC endoderm differentiation medium (0.19 mL/cm2) |
|---|---|---|---|---|---|---|
| 24 well | 1.9 | 0.35–0.5 | 2.0 × 104 | 4.0 × 104 | 0.3–0.5 mL | 0.36 mL |
| 12 well | 3.8 | 0.5–1 | 4.0 × 104 | 8.0 × 104 | 0.5–1 mL | 0.73 mL |
| 6 well | 9.5 | 1.5–2 | 10.0 × 104 | 20.0 × 104 | 1.3–2 mL | 1.8 mL |
Coat plates with Matrigel
hESCs will be cultured and differentiated in feeder-free conditions with Matrigel. This step describes the preparation of Matrigel-coated plates.
-
2.
Thaw a new vial of Matrigel (5 mL) at 4°C for 18–24 h.
-
3.
On the next day, precool 1 mL filtered tips, 25 mL pipettes, and a 50 mL sterile centrifuge tube at −20°C for at least 20 min and place some sterile 1.5 mL Eppendorf tubes on ice.
-
4.
Place the Matrigel vial on ice inside a tissue culture hood, aliquot Matrigel to precooled sterile Eppendorf tubes with the precooled tips, and immediately put the tubes containing Matrigel on ice. Keep one Eppendorf tube of Matrigel on ice and store the rest of aliquoted Matrigel in −80°C freezer.
Note: All the supplies used need to be precooled and sterile. Matrigel should be placed on ice. Matrigel will solidify when temperature is over 10°C. Each batch of Matrigel has a specific protein concentration or a dilution factor included on the Certificate of Analysis. Generally, it is about 275 μl Matrigel for 25 mL DMEM/F-12 medium. We usually aliquot the amount of Matrigel required for 25 mL medium in each tube.
-
5.
Transfer 25 mL DMEM/F-12 medium at 4°C to the precooled 50 mL tubes, and immediately add an appropriate amount of Matrigel to DMED/F-12 medium as indicated by the dilution factor of each batch of Matrigel.
-
6.
Immediately mix the medium, and transfer 1.5–2 mL Matrigel-containing medium to each well of a 6-well plate, or 0.5–1 mL to each well of a 12-well plate. Gently swirl plates to allow medium to cover the whole surface and then incubate plates at a 37°C incubator for 30–60 min.
-
7.
Aspirate the Matrigel-containing medium, then add 2 mL fresh DMEM/F-12 medium to each well.
-
8.
The resulting Matrigel-coated plates, which are incubated with DMEM/F-12 medium, can be kept at a 37°C incubator for up to 2 weeks before use.
Note: It is important that supplies and DMEM/F-12 medium are precooled in this step, and the coating procedure is performed as quickly as possible. Do not leave the Matrigel-containing DMEM/F-12 medium in 50 mL centrifuge tube at room temperature (20°C–25°C) for too long before coating, as the temperature will quickly rise above 10°C. If more time is needed, place the indicated medium on ice. Additionally, do not leave the plates with Matrigel-containing medium at a 37°C incubator for too long, as Matrigel will form 3D structures which are not suitable for hESC culture. No feeder cells are needed when Matrigel is used.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-FOXA2∗ (1:200 dilution) | Cell Signaling Technology | Cat#: 8186T, RRID:AB_10891055 |
| Goat polyclonal anti-human SOX17∗ (1:200 dilution) | R&D Systems | Cat#: AF1924-SP, RRID:AB_355060 |
| Donkey Anti-Rabbit IgG H&L (Alexa Fluor 488) (1:400–500 dilution) | Abcam | Cat#: ab150073, RRID: AB_2636877 |
| Donkey Anti-Goat IgG H&L (Alexa Fluor 594) (1:400–500 dilution) | Abcam | Cat#: ab150132, RRID: AB_2810222 |
| Anti-Oct3/4 antibody (C-10)∗ (1:200 dilution) | Santa Cruz | Cat#: sc-5279, RRID:AB_628051 |
| Chemicals, peptides, and recombinant proteins | ||
| Matrigel hESC-qualified | Corning | Cat#: 354277 |
| Y-27632 dihydrochloride (ROCK inhibitor)∗ | Tocris | Cat#: 1254 |
| Crotonic acid∗ | Sigma | Cat#: 113018 |
| TesRTM-E8TM | STEMCELL Technologies, Inc. | Cat#: 05990 |
| DMEM/F-12 | Thermo Fisher Scientific | Cat#: 11320033 |
| Embryonic Stem Cell FBS | Thermo Fisher Scientific | Cat#: 10439024 |
| Accutase | Innovative Cell Technologies, Inc. | Cat#: AT104 |
| HyClone DPBS/Modified | Cytiva | Cat#: SH30028.02 |
| Cell culture plate | Corning | Cat#: 353046; Cat#: 3512 |
| Recombinant human Activin A∗ | Cell Guidance Systems | Cat#: GFH6 |
| iQ SYBR Green Supermix | Bio-Rad | Cat#: 170886 |
| HyClone Defined Fetal Bovine Serum (dFBS), US Origin∗ | Cytiva | Cat#: SH30070.02 |
| MEM non-essential amino acid solution (NEAA; 100×) | Invitrogen | Cat#: 11140-050 |
| RPMI 1640 Medium∗ | Thermo Fisher Scientific | Cat#: 11875-093 |
| VECTASHIELD HardSet Antifade Mounting Medium with DAPI | Vector Laboratories | Cat#: H-1500 |
| Qiagen RNeasy Mini Kit | QIAGEN | Cat#: 74104 |
| HyClone USDA-tested Fetal Bovine Serum (FBS) | Cytiva | Cat#: SH30910.03 |
| High-Capacity cDNA Reverse Transcription Kit | Applied Biosystems | Cat#: 4368813 |
| CHIR99021 (CT99021) HCL∗ | Selleck Chemicals | Cat#: S2924 |
| DEPC dH2O | KsD Scientific | Cat#: W1137-1000 |
| 0.5 mM EDTA solution | Sigma | Cat#: E8008 – 100 mL |
| Critical commercial assays | ||
| STEMdiff Trilineage Differentiation Kit∗ | STEMCELL Technologies, Inc. | Cat#: 05230 |
| Experimental models: cell lines | ||
| Mel1 hESCs | Dr. Andrew Elefanty and Edouard Stanley at the University of Queensland, Australia | NIH Registration number: 0139 |
| H9 hESCs | Dr. James A. Thomson at the University of Wisconsin-Madison (via WiCell Research Institute) | NIH Registration number: 0062 RRID:CVCL_9773 |
| Oligonucleotides | ||
|
POU5F1 5′-CTTGCTGCAGAAGTGGGTGGAG GAA-3’; 5′-CTGCAGTGTGGGTTTCGGGCA-3′ |
This paper | N/A |
|
NANOG 5′-CAAAGGCAAACAACCCACTT-3’; 5′-TCTGCTGGAGGCTGAGGTAT-3′ |
This paper | N/A |
|
FOXA2 5′-GACAAGTGAGAGAGCAAGTG-3’; 5′-ACAGTAGTGGAAACCGGAG-3′ |
This paper | N/A |
|
SOX17 5′-GTGGACCGCACGGAATTTG-3’; 5′-GGAGATTCACACCGGAGTCA-3′ |
PrimerBank MGH-PGA | https://pga.mgh.harvard.edu/cgi-bin/primerbank/new_search2.cgi |
|
GSC 5′-AACGCGGAGAAGTGGAACAAG-3’; 5′-CTGTCCGAGTCCAAATCGC-3′ |
PrimerBank MGH-PGA | https://pga.mgh.harvard.edu/cgi-bin/primerbank/new_search2.cgi |
Critical reagents
Materials and equipment
CRITICAL: To keep the activity of the Activin A solution, aliquot the solution in a small amount and store at −80°C. Avoid repeated freezing and thawing.
CRITICAL: RACC Medium can be stored at 4°C for up to 2 weeks. Avoid repeated warming at 37°C.
Note: We found that 0.13 mL/cm2 STEMDIFF endoderm differentiation medium plus 5 mM crotonate is more efficient than regular 0.26 mL/cm2 STEMDIFF endoderm differentiation medium in promoting endoderm differentiation.
Alternatives: In our protocols, hESCs are passaged into single-cell suspension for seeding before induction. In addition to EDTA, other enzyme-free passaging reagents, including ReLeSRTM (Stemcell Technologies) and Gentle Cell Dissociation Reagent (Stemcell Technologies) can be used for passaging cells growing in TeSRTM-E8TM medium. Alternatively, hESCs can also be maintained in mTeSRTM-1 medium and passaged by dispase (Stemcell Technologies). Alternative Matrigel for feeder-free hESC culture includes Vitronectin XFTM (Stemcell Technologies).
Step-by-step method details
Revive mel1 and H9 cells
Timing: 30 min
This step describes the procedure to revive human embryonic stem cell lines.
-
1.
Quickly thaw a vial of frozen mel1 or H9 cells (4×105 cells) by gently swirling the vial in a 37°C water bath.
-
2.
Once cells are thawed, immediately transfer cells to a 15 mL conical tube and add 5 mL DMEM/F-12 medium to the cells, drop by drop.
-
3.
Gently mix the cells.
-
4.
Centrifuge at 300 × g for 3 min at 20°C–25°C.
-
5.
Aspirate the supernatant and resuspend cells in 5 mL DMEM/F-12 medium.
-
6.
Centrifuge at 20°C–25°C at 300 × g for 3 min again.
-
7.
Remove the supernatant and resuspend cells with 2 mL TeSRTM-E8TM medium containing 10 μM Y-27632 which is prewarmed to 37°C.
CRITICAL: It is important to add Y-27632 in the medium for the first 24 h after reviving or splitting cells. Otherwise, the survival will be greatly impaired (Watanabe et al., 2007).
CRITICAL: If the freezing medium contains embryonic stem-cell FBS, cells must be washed at least twice with DMEM/F-12 medium. Otherwise, cells will differentiate in the presence of FBS.
-
8.
Aspirate the medium from Matrigel-coated 6-well plates, and seed 4×105 mel1 or H9 cells into them.
-
9.
Quickly shake the plate in short side-to-side and back-and-forth motions to evenly distribute the cells in the well.
Note: Because cells will quickly attach to Matrigel, it is important to shake the plate right away after adding cells.
-
10.
Place the plate in a 37°C incubator with 5% CO2.
-
11.
24 h later, replace Y-27632 containing TeSRTM-E8TM medium with 2 mL fresh warm Y-27632 free TeSRTM-E8TM medium.
Note: Y-27632 is not needed after cells grow on plates for 24 h.
-
12.
Change medium daily, and cells will reach confluency on day 3.
Note: Increase TeSRTM-E8TM medium to 3–4 mL per well when cells reach above 50% confluency.
Passaging of mel1 or H9 cells
Timing: 30 min
This step describes how to split and passage cells, which is essential for hESC maintenance and differentiation.
It is time to split cells when cells reach 70–80% confluency and colonies start to contact and fuse with each other.
-
13.
Aspirate TeSRTM-E8TM medium from cells.
-
14.
Add 2 mL warm 0.5 mM EDTA in DPBS to the cells and incubate at 37°C incubator for 10 min.
-
15.
Remove EDTA when cells are dissociated from each other but are still attached to plates.
-
16.Flush the surface of the wells with 1 mL prewarmed TeSRTM-E8TM containing 10 μM Y-27632.
 CRITICAL: It is important to include Y-27632 in the medium at this stage. Otherwise, the survival will be impaired.Note: Cells can be frozen for storage at this step.
CRITICAL: It is important to include Y-27632 in the medium at this stage. Otherwise, the survival will be impaired.Note: Cells can be frozen for storage at this step.-
a.Make 2 × freezing medium: 20% DMSO, 60% embryonic stem-cell FBS, 20% TeSRTM-E8TM medium with Y-27632 (Kent, 2009).
-
b.Mix cell suspension with equal volume of 2 × freezing medium.
-
c.Aliquot the cell suspension from 1 well of a 6-well plate to 5 cryogenic vials.
-
d.Freeze the vials at −80°C for 18–24 h in a 2-propanol-filled freezing container.
-
e.Transfer cells to a N2 tank for long-term storage.Note: Cells can also be frozen with 10% DMSO and 90% TeSRTM-E8TM medium with Y-27632 only. But adding embryonic stem-cell FBS in the freezing medium improves cell survival, especially after long-term storage. To remove FBS after revival, cells need to be washed with 5 mL DMEM/F-12 medium at least twice. hESCs can also be frozen with mFreSR cryopreservation medium (Stemcell Technologies).
-
a.
Seeding mel1 cells for endoderm differentiation using STEMdiff differentiation medium plus crotonate
Timing: 30 min
This step describes how to seed mel1 cells on Matrigel-coated plates for differentiation using STEMDIFF differentiation medium plus crotonate.
-
17.
Count cells, and seed 3.5–4 × 104 cells per well (0.92–1.05 × 104 cells/cm2) into a new Matrigel-coated well of a 12-well plate. – Day -1
CRITICAL: Cell density is important for efficient endoderm differentiation. We found that cells seeded at density ranging from 0.52 – 2.1 × 104 cells/cm2 can all efficiently differentiate into endoderm cells after a 3-day induction with crotonate (Figure 1). However, cells seeded at a density higher than 1.05 × 104 cells/cm2 experience increased cell death, and cells seeded at more than 4.21 × 104 cells/cm2 density dramatically reduce their differentiation efficiency (Figure 1).
Figure 1.
Endoderm differentiation of hESCs under different cell densities
(A) Phase contrast images of mel1 hESCs seeded at 2 × 104, 4 × 104, 8 × 104, or 16 × 104/well of a 12-well plate before induction (Day 0), and at 3 days after induction (Day 3). Mel1 cells were seeded at the indicated density. On day 0, cells were washed with DPBS once, and incubated with 1 mL (0.26 mL/cm2) STEMDIFF endoderm differentiation medium together with 5 mM crotonate for 3 days. Scale bar: 400 μm.
(B) High seeding cell density reduces endoderm differentiation. Mel1 cells were seeded at indicated densities and induced for endoderm differentiation for 3 days as described in (A). The total RNA was extracted, and the mRNA levels of SOX17 and FOXA2 were analyzed by qPCR and normalized with the mRNA levels of GAPDH (n = 3 biological replicates/group, ∗p<0.05, values are expressed as mean ± SEM).
-
18.
Grow cells at a 37°C incubator with 5% CO2.
Inducing mel1 cells for endoderm differentiation using STEMdiff differentiation medium plus crotonate
Timing: 10 min a day
This step describes the methods for endoderm differentiation using STEMDIFF differentiation medium plus crotonate.
-
19.
Remove TesRTM-E8TM medium on the next day. — Day 0
-
20.
Wash cells with DPBS once.
CRITICAL: It is important to wash cells once with DPBS to remove any remainder of TeSRTM-E8TM medium, as it maintains pluripotency and inhibits differentiation.
-
21.
Add prewarmed 1 mL (0.26 mL/cm2) endoderm differentiation medium from STEMDIFF trilineage differentiation kit, followed by adding crotonate to a final concentration of 5 mM.
CRITICAL: Please thaw and store the endoderm differentiation medium at 4°C to maintain the active differentiation factors in the medium. The medium can be stored at 4°C for up to 2 weeks. Only aliquot and warm enough medium for each use.
Note: 5 mM crotonate is sufficient to promote endoderm differentiation of mel1 cells.
-
22.
Gently move the plates to mix the medium and leave the plates at 37°C incubator with 5% CO2.
-
23.
Change endoderm differentiation medium containing crotonate daily for 2 days. — Day 1 and 2
Note: Cells do not grow within 24 h after induction. However, they start to proliferate and migrate after day 1, and reach confluency with 90.2% SOX17+ and FOXA2+ endoderm cells on day 3.
Pause point: At this step, cells can be frozen at −80°C for RNA isolation or kept at 4°C after fixation for several days before examining endoderm differentiation.
Seeding H9 cells for endoderm differentiation with 1 × MEM NEAA, 25 ng/mL Activin A, 2 μM CHIR99021, 0.2% defined FBS, and 5 mM crotonate (RACC medium)
Timing: 10–30 min per day
This step describes how to seed H9 cells into Matrigel-coated plates for endoderm differentiation with RACC medium.
-
24.
Count cells, and seed 20.0 × 104 cells per well (2.1 × 104 cells/cm2) into a new Matrigel-coated well of a 6-well plate with prewarmed TeSRTM-E8TM medium containing 10 μM Y-27632. – Day -2
CRITICAL: please seed the suggested cell number as described in Table 3.
-
25.
Quickly shake the plate to evenly distribute the cells in the well followed by incubating the plate in a 37°C incubator with 5% CO2.
-
26.
24 h later, gently aspirate old medium and replace it with 2 mL of new and warm TesRTM-E8TM medium. -- Day -1
-
27.
Place the dish in an incubator and grow cells at 37°C with 5% CO2.
Inducing H9 cells for endoderm differentiation with RACC medium
Timing: 10 min a day
This step describes the procedures to induce endoderm differentiation of H9 cells with RACC medium.
-
28.
18–24 h later, aspirate TesRTM-E8TM medium and wash with 2 mL of warm RPMI 1640 medium. — Day 0
CRITICAL: It is important to wash cells with warm RPMI 1640 medium before induction. Initiation of induction two days after H9 cells are seeded improves cell survival during differentiation.
-
29.
Add 1.8 mL (0.19 mL/cm2) of RACC endoderm differentiation medium (RPMI 1640 medium containing 0.2% dFBS, 1 × MEM NEAA, 25 ng/mL Activin A, 2 μM of CHIR99021, and 5 mM of crotonate) to each well and grow the cells in a 37°C incubator with 5% CO2.
-
30.
24 h later, replace old medium with 1.8 mL of new endoderm differentiation medium. – Day 1
-
31.
24 h later, collect samples. – Day 2
Note: Upon differentiation, H9 cells quickly migrate and proliferate after differentiation induction (Figure 2).
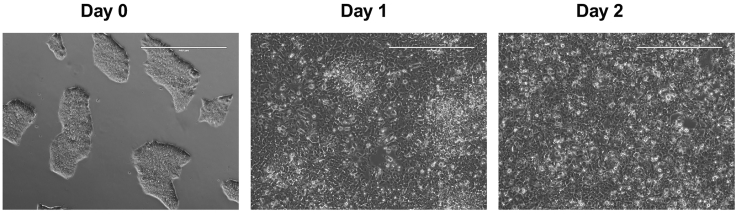
Figure 2.
Morphology of H9 cells undergoing endoderm differentiation in the RACC medium
20 × 104 H9 cells/well (2.1 × 104/cm2) of a 6-well plate were seeded, washed with warm RPMI 1640 medium once, and induced with 1.8 mL (0.19 mL/cm2) RACC endoderm differentiation medium for 48 h. Scale bar: 400 μm
Pause point: At this step, cells can be frozen at −80°C for RNA isolation or kept at 4°C after fixation for several days before examining endoderm differentiation.
Examining endoderm differentiations
Timing: 2 h
The expression of key endoderm markers like SOX17 and FOXA2 is examined and quantified by qPCR and flow cytometry to determine the efficiency of endoderm differentiation. CXCR4 is another widely used endoderm marker.
Immunofluorescence and/or flow cytometry analyses of endoderm cells using endodermal marker SOX17 and FOXA2. — Day 3 or 2
-
32.Immunofluorescence analysis:
-
a.On Day 3 or 2, take out endoderm cells differentiated on Matrigel-coated coverslips.
-
b.Aspirate medium, and wash with DPBS once.
-
c.Fix cells with 4% paraformaldehyde in DPBS at 20°C–25°C for 10 min.
-
d.Remove paraformaldehyde and wash coverslip with DPBS once.
-
e.Permeabilize cells with 0.5% Triton X-100 in DPBS at 20°C–25°C for 10 min, followed by wash with DPBS once.
-
f.Dilute anti-SOX17 antibody (1:200) and anti-FOXA2 antibody (1:200) in DPBS containing 10% FBS.
-
g.Spot 50 μL antibody solution for each coverslip on a clean parafilm and place a coverslip on the 50 μL antibody solution with cells facing down for 1 h at 20°C–25°C.
-
h.Float each coverslip with a drop of DPBS, and take all coverslips to a 12-well plate, wash with DPBS for 4–5 times.
-
i.Making secondary antibody solutions: 1:500 dilution for donkey anti-goat IgG (Alex Fluor 594) and 1:500 dilution for donkey anti-rabbit IgG (Alex Fluor 488) in DPBS containing 10% FBS.
-
j.Spot 50 μL secondary antibody solution for each coverslip on a clean parafilm, and place a coverslip on the 50 μL antibody solution with cells facing down for 30 min at 20°C–25°C.
-
k.Add a drop of DPBS under each coverslip to float it up, and take all of them to a 12-well plate, wash with DPBS for 4–5 times.
-
l.Mount coverslips on slides with antifade mounting medium with DAPI, and observe under fluorescence microscopy.
-
a.
-
33.Flow cytometry analysis of SOX17 and FOXA2 expression:
-
a.Remove differentiation medium from plates.
-
b.Add 1 mL Accutase to cells and incubate at 37°C for 1–2 min.
-
c.Remove Accutase.
-
d.Add 1 mL DPBS to flush the cells off the surface of the plate.
-
e.Centrifuge cells at 300 × g for 3 min at 4°C.
-
f.Remove the supernatant and resuspend cells with 1 mL DPBS.
-
g.Centrifuge cells at 300 × g for 3 min at 4°C, and remove the supernatant.
-
h.Resuspend cells in 500 μL 4% paraformaldehyde in DPBS, and incubate at 20°C–25°C for 10 min.
-
i.Centrifuge cells at 300 × g for 3 min at 4°C, and remove the supernatant.
-
j.Resuspend cells with 1 mL DPBS and centrifuge at 300 × g for 3 min at 4°C, and remove the supernatant.
-
k.Resuspend cells in 500 μL DPBS containing 0.5% Triton X-100, and incubate for 10 min at 20°C–25°C.
-
l.Centrifuge cells at 2350 × g for 5 min at 4°C, and remove the supernatant.
-
m.Wash cells once with DPBS containing 10% FBS, centrifuge at 2350 × g for 5 min at 4°C, and remove the supernatant.
-
n.Resuspend cells in 200 μL DPBS containing 10% FBS, anti-SOX17 (1:200 dilution) and anti-FOXA2 (1:200 dilution) antibodies, and incubate for 1 h at 20°C–25°C.
-
o.Centrifuge cells at 2350 × g for 5 min at 4°C, and remove the supernatant.
-
p.Resuspend cells in 0.5–1 mL DPBS containing 10% FBS, centrifuge at 2350 × g for 5 min at 4°C, and remove the supernatant.
-
q.Resuspend cells in 200 μL DPBS containing 10% FBS, donkey anti-goat IgG (Alex Fluor 594) (1:400 dilution) and donkey anti-rabbit IgG (Alex Fluor 488) (1:400 dilution), and incubate for 30 min at 20°C–25°C.
-
r.Centrifuge cells at 2350 × g for 5 min at 4°C, and remove the supernatant.
-
s.Wash with DPBS containing 10% FBS once, centrifuge and remove the supernatant.
-
t.Resuspend cells in 250 μL DPBS containing 5% FBS and analyze the cells with a flow cytometer.
-
a.
-
34.Quantitative Real-time PCR (qPCR) analysis of endoderm differentiation:
-
a.Aspirate medium and wash cells with DPBS once.
-
b.Extract RNA using Qiagen RNeasy mini kit according to the product manual.
-
c.Measure RNA concentrations using a nanodrop spectrophotometer.
-
d.Synthesize the first strand of cDNA using High capacity cDNA reverse transcription kit. For every 20 μL reaction, use 2 μg total RNA, 2 μL of 10 × RT buffer, 2 μL of 10 × RT random primers, 0.8 μL of 100 mM dNTP, 1 μL of MultiScribeTM reverse transcriptase, and 4.2 μL RNase-free DEPC dH2O.
-
e.Mix well, and centrifuge for 1 min to ensure that all the reagents are in the bottom of the tube.
-
f.Place the tube in a PCR machine programmed as: 25°C, 10 min, 50°C, 50 min, 85°C, 5 min.
-
g.Dilute 20 μL cDNA reaction 1:20 with DEPC water, mix it well, and pipet 9 μL as template to each well for every 20 μL RT-PCR reaction.
-
h.Mix well with 10 μL Bio-Rad iQ SYBR Green supermix and 0.6 μL of each primer (10 μM), and pipette 11 μL of the resulting mixture to the wells containing 9 μL cDNA.
-
i.Seal the PCR plate with film, centrifuge it for 1 min using a mini plate spinner.
-
j.Place the PCR plate in a RT-PCR machine using the following program: Start with a heating step at 95°C for 3 min, then 39 cycles of a step with 95°C for 10 s, 58°C for 30 s, and 72°C for 30 s. Finish.
-
a.
Expected outcomes
Crotonate promotes endoderm differentiation in the STEMDIFF endoderm differentiation medium. Firstly, crotonate dose-dependently promotes the expression of endodermal genes after endoderm differentiation of mel1 hESCs in the STEMDIFF endoderm differentiation medium. As shown in Figure 3A, after incubating 1.05 × 104/cm2 mel1 cells with 0.26 mL/cm2 endoderm differentiation medium (Stemcell technologies) and crotonate for 2 days, the expression of SOX17 and FOXA2 was markedly increased in a dose-dependent manner when compared to the gene expression in cells treated with endoderm differentiation medium only (Figure reprinted with permission from (Fang et al., 2021). Moreover, flow cytometry analysis showed that 90.2% cells were SOX17+FOXA2+ when induced together with 5 mM crotonate on day 3 (∼ 66 h) (Figure 3B, Endoderm + Crotonate). In contrast, although 86.2% cells were SOX17+FOXA2+ in samples induced without crotonate, the population was much more heterogeneous compared to cells induced with crotonate, with only 49.1% of cells expressing high levels of SOX17 and FOXA2 (Figure 3B, Endoderm). Further single-cell RNA-seq analysis revealed that when mel1 cells were induced with 0.13 mL/cm2 endoderm differentiation medium for 3 days (∼ 66 h), addition of 5 mM crotonate increased the fraction of differentiated endodermal cells from 63.8% to 93.5%, indicating that crotonate could reduce the amount of endoderm differentiation medium required for a high differentiation efficiency (Figure 3C) (Figure reprinted with permission from (Fang et al., 2021). Finally, crotonate also dose-dependently promotes the expression of endodermal genes in differentiated H9 hESCs using the STEMDIFF endoderm differentiation medium. When H9 cells (2.1 × 104/cm2) were induced with 0.26 mL/cm2 STEMDIFF endoderm differentiation medium and crotonate for 3 days, the expression of SOX17 and FOXA2 was increased in a dose-dependent manner compared to the gene expression levels in cells induced without crotonate (Figure 3D) (Figure reprinted with permission from (Fang et al., 2021). Together, these results demonstrate that crotonate can efficiently promote endoderm differentiation in a cell line-independent manner.
Figure 3.
Crotonate promotes endoderm differentiation in the STEMDIFF endoderm differentiation medium
(A) Crotonate dose-dependently promotes the expression of endodermal genes after endoderm differentiation of mel1 hESCs in the STEMDIFF endoderm differentiation medium. 4 × 104 mel1 cells/well (1.05 × 104/cm2) of a 12-well plate was induced with 1 mL (0.26 mL/cm2) STEMDIFF endoderm differentiation medium in the absence or presence of 5 mM crotonate for 48 h. RNA was collected, and the expression of the indicated genes was analyzed by qPCR with GAPDH as a loading control (n = 3 biological replicates/group, ∗p < 0.05, values are expressed as mean ± SEM). Figure reprinted with permission from Fang et al., 2021.
(B) Crotonate promotes homogeneous endoderm differentiation of hESCs. 4 ×104 mel1 cells/well (1.05 × 104/cm2) of a 12-well plate were induced with 1 mL (0.26 mL/cm2) STEMDIFF endoderm differentiation medium with or without 5 mM crotonate for 3 days (∼66 h). Cells were stained with anti-SOX17 and anti-FOXA2 antibodies, then subjected to flow cytometry analysis. Cells stained with secondary antibodies only were used as negative controls.
(C) Crotonate promotes more hESCs to differentiate into endodermal cells in the STEMDIFF endoderm differentiation medium when analyzed at the single-cell transcriptome level. 82 × 104 mel1 cells/10-cm dish (1.05 ×104/cm2) were seeded and induced with 10 mL (0.13 mL/cm2) STEMDIFF endoderm differentiation medium with or without 5 mM crotonate for 3 days (∼66 h). Cells were collected and processed for single-cell RNAseq. Compared to the differentiation condition in (B), only half amount of endoderm differentiation medium was used in this experiment when considering volume per area). Figure reprinted with permission from Fang et al., 2021.
(D) Crotonate dose-dependently promotes the expression of endodermal genes after endoderm differentiation of H9 hESCs in the STEMDIFF endoderm differentiation medium. 8 × 104 H9 cells/well of a 12-well plate were induced with the STEMDIFF endoderm differentiation medium with or without crotonate for 3 days. RNA was collected, and the expression of the indicated genes was analyzed by qPCR with GAPDH as a loading control (n = 3 biological replicates/group, ∗p < 0.05, values are expressed as mean ± SEM). Figure reprinted with permission from Fang et al., 2021.
Crotonate promotes endoderm differentiation in the RACC endoderm differentiation medium. As shown in Figure 4, when 2.1 × 104/cm2 H9 cells were induced with 0.19 mL/cm2 RACC endoderm differentiation medium (containing 5 mM crotonate) for 48 h, 81.8% of cells were strong SOX17+FOXA2+. On the other hand, omitting crotonate from the RACC endoderm differentiation medium reduced the percentage of SOX17+FOXA2+ cells to 66.7% (Figure 4). Again, these results demonstrate that the crotonate containing RACC medium is a very robust endoderm differentiation medium that only needs a low concentration of Activin A.
Figure 4.
Crotonate promotes endoderm differentiation in the RACC endoderm differentiation medium
20 × 104 H9 cells/well (2.1 × 104/cm2) of a 6-well plate were induced with 1.8 mL (0.19 mL/cm2) RACC endoderm differentiation medium including or excluding 5 mM crotonate for 48 h. Cells were stained with anti-SOX17 and anti-FOXA2 antibodies for flow cytometry analysis, with cells stained with secondary antibodies only as negative controls. In the presence of crotonate, 81.8% cells were SOX17+FOXA2+. In the absence of crotonate, only 66.7% were SOX17+FOXA2+.
Quantification and statistical analysis
Flow cytometry and qPCR analyses were used for quantification of endoderm differentiation. In flow cytometry analysis, cells stained with secondary antibodies were used as negative controls. In real-time PCR analysis, the relative mRNA levels of endodermal genes were normalized with the mRNA levels of GAPDH.
For statistical analysis, values were expressed as mean ± standard error of mean (SEM) from at least three independent experiments, or three biological replicates/group. Two-tailed, unpaired, student’s t-test was used to determine whether the differences between the means is significant. The differences were considered significant at p < 0.05.
Limitations
This protocol uses STEMDIFF endoderm differentiation medium, and a differentiation protocol including 25 ng/mL Activin A, 2 μM CHIR99021, 1 × MEM NEAA, and 0.2% dFBS in RPMI 1640 medium in mel1 and/or H9 cells. We have not tested the impacts of crotonate on endoderm differentiation using other endoderm differentiation protocols (Borowiak et al., 2009; Broda et al., 2019; D'Amour et al., 2005; Diekmann et al., 2015; Diekmann et al., 2019; Jiang et al., 2013; Loh et al., 2014; McLean et al., 2007; Naujok et al., 2014; Teo et al., 2012). Further test and/or optimization on crotonate dose and treatment time are needed for these different endoderm differentiation media or protocols. It is also possible that different hES cell lines, which are maintained in different embryonic stem cell maintenance conditions, may respond differently to crotonate. Similar test or optimization will also be needed for different hESCs.
Troubleshooting
Problem 1
Low cell recovery after revival (step 1–7)
Potential solution
hESCs tend to have a reduced revival rate after long-term storage. As mentioned in the protocol, adding 30% Embryonic stem-cell FBS to freezing medium can improve survival after reviving. Increasing the cell numbers frozen in a vial will also help to obtain enough cells after reviving. Additionally, adding 10 μM Y-27632 into the culture medium for 2 days instead of 1 day after revival will also help to prevent massive cell death.
Problem 2
hESCs lose pluripotency (step 13–16)
Potential solution
To maintain hESC pluripotency, make sure to wash at least twice with 5 mL DMEM/F-12 medium to remove embryonic stem-cell FBS when reviving cells. Also only use Matrigel-coated plates within 2 weeks when stored at 37°C incubator, aliquot the included supplement medium and store at −20°C to keep the activity of growth factors in TeSRTM-E8TM medium, and store complete TeSRTM-E8TM medium at 4°C. Avoid repetitive warming of the medium.
Problem 3
Cell death during differentiation (Protocol 1, step 23)
Potential solution
Reduce cell density when seeding since high cell density (more than 1.05 × 104/cm2) increases cell death during differentiation.
Problem 4
Cell death during differentiation (Protocol 2, step 29–31)
Potential solution
Avoid high cell density and only start differentiation induction on day 2 after seeding.
Problem 5
Low efficiency of endoderm cell differentiation
Potential solution
Low efficiency of endoderm cells differentiation can be caused by several issues. Firstly, hESCs should be fully pluripotent in order to have a high endoderm differentiation efficiency. Periodic checking hESCs by immunofluorescence staining with OCT4 antibody (1:200 dilution) is recommended to confirm their pluripotency. Secondly, inactivation of endoderm differentiation factors in endoderm differentiation medium could lead to failure to induce differentiation. Again, aliquoting the differentiation medium or Activin A, and storing at 4°C and/or −80°C are recommended. Avoid repetitively warming up medium. Thirdly, use an optimal cell density for differentiation induction. Please avoid overgrowth of hESCs before induction.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Xiaoling Li, lix3@niehs.nih.gov
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate/analyze datasets/code.
Acknowledgments
We thank Drs. Paul Wade and Guang Hu for critical reading of the manuscript. This research was supported by the Intramural Research Program of National Institute of Environmental Health Sciences of the NIH to X.L. (Z01 ES102205).
Author contributions
Y.F. designed the study, carried out experiments, analyzed data, and wrote the manuscript. X.L. guided and designed the study, analyzed data, and edited the manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Yi Fang, Email: yi.fang@nih.gov.
Xiaoling Li, Email: lix3@niehs.nih.gov.
References
- Borowiak M., Maehr R., Chen S., Chen A.E., Tang W., Fox J.L., Schreiber S.L., Melton D.A. Small molecules efficiently direct endodermal differentiation of mouse and human embryonic stem cells. Cell Stem Cell. 2009;4:348–358. doi: 10.1016/j.stem.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broda T.R., McCracken K.W., Wells J.M. Generation of human antral and fundic gastric organoids from pluripotent stem cells. Nat. Protoc. 2019;14:28–50. doi: 10.1038/s41596-018-0080-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amour K.A., Agulnick A.D., Eliazer S., Kelly O.G., Kroon E., Baetge E.E. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- Diekmann U., Lenzen S., Naujok O. A reliable and efficient protocol for human pluripotent stem cell differentiation into the definitive endoderm based on dispersed single cells. Stem Cells Dev. 2015;24:190–204. doi: 10.1089/scd.2014.0143. [DOI] [PubMed] [Google Scholar]
- Diekmann U., Wolling H., Dettmer R., Niwolik I., Naujok O., Buettner F.F.R. Chemically defined and xenogeneic-free differentiation of human pluripotent stem cells into definitive endoderm in 3D culture. Sci. Rep. 2019;9:996. doi: 10.1038/s41598-018-37650-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Xu X., Ding J., Yang L., Doan M.T., Karmaus P.W.F., Snyder N.W., Zhao Y., Li J.L., Li X. Histone crotonylation promotes mesoendodermal commitment of human embryonic stem cells. Cell Stem Cell. 2021;28:748–763.e7. doi: 10.1016/j.stem.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Zhang D., Bursac N., Zhang Y. WNT3 is a biomarker capable of predicting the definitive endoderm differentiation potential of hESCs. Stem Cell Rep. 2013;1:46–52. doi: 10.1016/j.stemcr.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent L. Freezing and thawing human embryonic stem cells. J. Vis. Exp. 2009;34:1555. doi: 10.3791/1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh K.M., Ang L.T., Zhang J., Kumar V., Ang J., Auyeong J.Q., Lee K.L., Choo S.H., Lim C.Y., Nichane M. Efficient endoderm induction from human pluripotent stem cells by logically directing signals controlling lineage bifurcations. Cell Stem Cell. 2014;14:237–252. doi: 10.1016/j.stem.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean A.B., D'Amour K.A., Jones K.L., Krishnamoorthy M., Kulik M.J., Reynolds D.M., Sheppard A.M., Liu H., Xu Y., Baetge E.E. Activin a efficiently specifies definitive endoderm from human embryonic stem cells only when phosphatidylinositol 3-kinase signaling is suppressed. Stem Cells. 2007;25:29–38. doi: 10.1634/stemcells.2006-0219. [DOI] [PubMed] [Google Scholar]
- Naujok O., Diekmann U., Lenzen S. The generation of definitive endoderm from human embryonic stem cells is initially independent from activin A but requires canonical Wnt-signaling. Stem Cell Rev. Rep. 2014;10:480–493. doi: 10.1007/s12015-014-9509-0. [DOI] [PubMed] [Google Scholar]
- Teo A.K., Ali Y., Wong K.Y., Chipperfield H., Sadasivam A., Poobalan Y., Tan E.K., Wang S.T., Abraham S., Tsuneyoshi N. Activin and BMP4 synergistically promote formation of definitive endoderm in human embryonic stem cells. Stem Cells. 2012;30:631–642. doi: 10.1002/stem.1022. [DOI] [PubMed] [Google Scholar]
- Watanabe K., Ueno M., Kamiya D., Nishiyama A., Matsumura M., Wataya T., Takahashi J.B., Nishikawa S., Nishikawa S., Muguruma K. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate/analyze datasets/code.