Summary
Inflammaging is associated with an increased risk of chronic disease. Monocytes are the principal immune cells for the production of inflammatory cytokines and contribute to inflammaging in the elderly. However, the underlying mechanisms remain largely unknown. Here, we found that monocytes from aged individuals contained high levels of lipid droplets (LDs), and this increase was correlated with impaired fatty acid oxidation. Downregulated peroxisome proliferator-activated receptor (PPAR)-α may be responsible for the pro-inflammatory phenotype of monocytes in aged individuals, as it was positively correlated with LD accumulation and increasing TNF-α concentration. Interestingly, interventions that result in PPAR-α upregulation, such as fenofibrate treatment, TNF-α neutralization, or calorie restriction, reversed the effect of aging on monocytes. Thus the downregulation of PPAR-α and LD levels in monocytes represents a novel biomarker for inflammaging. Furthermore, PPAR-α activation in the elderly may also alleviate long-term inflammaging, preventing the development of life-limiting chronic diseases.
Subject areas: human metabolism, Immunology, Cell biology
Graphical abstract

Highlights
-
•
Monocytes from aged individuals contained high levels of lipid droplets (LDs)
-
•
Downregulated PPAR-α is responsible for the aged monocytes profiles
-
•
TNF-α might accelerate monocyte aging by downregulating PPAR-α expression
-
•
PPAR-α activation in the elderly may also alleviate long-term inflammaging
Human metabolism; Immunology; Cell biology
Introduction
As we age, the efficiency of basic cellular processes declines and the production of pro-inflammatory cytokines increases. This change is thought to be caused in part by the complex process of cellular senescence and is characterized by the development of a senescence-associated secretory phenotype. The result is sterile, systemic, chronic, low-level inflammation (SCI), which is known as inflammaging (Franceschi et al., 2018). Inflammaging can, in turn, accelerate aging and contribute to age-associated diseases (Ferrucci and Fabbri, 2018). Monocytes, the precursors of most tissue macrophages, are likely to play key roles in inducing inflammaging (Linehan and Fitzgerald, 2015; Oishi and Manabe, 2016): during normal aging, the percentage of non-classical monocytes markedly increases and the resulting elevation in pro-inflammatory cytokine levels promotes cellular senescence (Ong et al., 2018) (Fagiolo et al., 1993). Therefore, understanding monocyte profiles in aging and investigating strategies to reduce inflammaging in the elderly is of great interest (Bouchlaka et al., 2013) (Fink et al., 2009).
Recent progress in the study of immunometabolism has indicated that macrophage lipid metabolism and inflammatory activation are closely intertwined. For example, saturated fatty acids (SFAs) activate Toll-like receptor (TLR) signaling in both human and murine macrophages to elicit inflammatory responses (Milanski et al., 2009; Rocha et al., 2016). Mechanistically, SFAs induce endoplasmic reticulum (ER) stress by activating the c-Jun N-terminal kinase (JNK) and nuclear factor-κB signaling pathways, which can lead to inflammation (Hu et al., 2006; Urano et al., 2000). Conversely, fatty acid (FA) catabolism, including fatty acid oxidation (FAO), sustains anti-inflammatory macrophage differentiation and attenuates ER stress and inflammatory responses in mice and humans (Namgaladze and Brüne, 2016). Therefore, maintaining an optimal FA level is necessary to ensure efficient mitochondrial respiration and prevent macrophage-mediated inflammation through mitochondrial dysfunction (Jana et al., 2019). In addition, aging is accompanied by modifications to nutrient sensing and metabolic pathways (Saare et al., 2020). These findings have led many to consider whether lipid metabolism reprogramming might alleviate inflammaging by altering lipid metabolism in aging monocytes.
The steroid receptor superfamily of peroxisome proliferator-activated receptors (PPARs) has emerged as a link between lipid metabolism and innate immunity. For example, PPAR-α agonists inhibit interferon-γ production and increase interleukin (IL)-4 cytokine secretion (Gervois et al., 2000). Meanwhile, Ppara−/− mice exhibit enhanced monocyte chemoattractant protein and tumor necrosis factor (TNF)-α production and an increased number of neutrophils and macrophages in the bronchoalveolar lavage fluid (Delayre-Orthez et al., 2005). Further supporting a link between PPARs and monocytes, long-chain FAs that accumulate with healthy aging have been found to promote pro-inflammatory monocyte polarization via PPAR-γ (Pararasa et al., 2016). These results suggest that PPAR activation may mediate the anti-inflammatory function of monocytes (Masternak and Bartke, 2007).
Targeting the inflammaging process by mediating the altered metabolic profiles of monocytes is of great interest. However, whether changes in lipid metabolism mediate the pro-inflammatory function of monocytes during aging or the underlying mechanisms remains largely unknown. To address these questions, we characterized lipid metabolism and inflammatory cytokine production in monocytes isolated from healthy old and young individuals. We then evaluated the effects of PPAR-α manipulation on markers of inflammaging in mice and humans. Finally, we investigated the monocyte response to PPAR-α activation as a therapeutic strategy.
Results
Altered FAO and LD accumulation in elderly-derived monocytes
Normal aging is accompanied by high serum levels of TNF-α and IL-6. We first confirmed that serum TNF-α and IL-6 levels were positively correlated with increasing age in healthy human donors (Figure S1A). Consistent with these results, we also detected increased expression levels of the pro-inflammatory genes TNFA, IL6, and CXCL1 in human monocytes (Figure S1B). Similar to the human data, TNF-α and IL-6 serum and mRNA levels in old mice were significantly higher than those in young mice (Figures S1C and S1D). The human and murine monocytes were isolated and identified via their specific makers (Figure S2). We detected high levels of Ly6G+CD11b+ neutrophils in the blood of old mice, indicative of an inflammatory status (Figures S3A and S3B). Taken together, these results suggest that aging monocytes exhibit a pro-inflammatory phenotype even before infiltrating into tissues.
The central pathways of cell anabolism and catabolism, the AMPK and mTOR pathway, respectively, have critical roles in the regulation of monocyte function. We found that AMPK signaling was markedly suppressed, whereas the mTOR pathway was unaffected by aging (Figure 1A), suggesting that only catabolism was altered in monocytes during aging. We also confirmed that monocyte metabolism was altered during aging, as indicated by decreased ATP production and a suppressed oxygen consumption rate (OCR) (Figures 1B and 1C). Specifically, we observed that the expression of key FAO enzymes, ACADVL and HADHA, was inhibited (Figures 1D and 1E). It seems that monocytes exhibit a pro-inflammatory phenotype during aging, as well as an impaired FAO capacity.
Figure 1.
Impaired FAO and LD accumulation in elderly-derived monocytes
(A–I) Blood monocytes were isolated from young and old mice. (A) Western blot analysis of p-AMPK and p-mTOR expression in each group. Representative blots of three independent experiments are shown. (B) ATP concentrations were tested using an ATP colorimetric/fluorometric assay and normalized by the protein concentration (n = 4 per group). (C) The OCR was tested in XF-96 assay medium and normalized by protein concentration. Continuous OCR values (pmoles/min/μg protein) are shown. Each repetition involved three mice per group, and three replicates were performed. (D) Western blot analysis of ACADVL, HADHA, and ACADM expression in each group. Representative blots of three independent experiments are shown. (E) FAO enzyme abundance in monocytes was detected by flow cytometry. Representative mean fluorescent intensity (MFIs) and the mean MFIs are shown (n = 4 per group). (F) Morphological analysis of young and old murine monocytes by electron microscopy. (G) LDs in monocytes were stained with BODIPY 493/503 dye and then imaged under a confocal microscope before 3D rendering using Imaris software (version 9.0). (H) The mean LD volume of 50 analyzed cells. (I) LD density in monocytes from young (red line) and old (blue line) mice were analyzed by flow cytometry.
(J) Human blood monocytes were isolated by using two-step procedures, and LDs were identified in CD14+ cells by BODIPY 493/503 immunofluorescence staining.
(K) BODIPY 493/503 MFI values in CD14+/CD16+ blood monocytes isolated from young and elderly human subjects.
(L) Correlation analysis between age and monocytes, BODIPY MFI values. (n = 30).
The resulting Spearman's correlation coefficient (r) and corresponding p value are reported. All data represent mean ± SD. ∗∗p < 0.01, ∗∗∗p < 0.001; t test (B, E, and H).
Redundant FA and lipid droplet (LD) accumulation may activate pro-inflammatory signaling in macrophages. We next investigated whether reduced FAO in aged monocytes causes excessive FA and LD accumulation. Using electron microscopy, we observed that monocytes isolated from old mice contained a lower number of mitochondria and increased LD levels compared with monocytes from young mice (Figure 1F). We quantified LD levels using BODIPY staining, immunofluorescence microscopy, and 3D rendering. The average LD volume in isolated monocytes from young and old mice was 2.12 ± 0.22 versus 15 ± 1.1 μm3, respectively (Figures 1G–1I and S4).
Consistent with the mouse data, LD concentration in both CD14+ and/or CD16+ human monocytes was also positively correlated with age (Figures 1J–1L). Furthermore, human monocyte LD density was positively correlated with serum TNF-α concentration (Figure S5). These findings suggest that abnormal FA metabolism and LD density could reflect the pro-inflammatory features of monocytes that manifest with aging. Notably, LD density in bone marrow-derived macrophages, blood monocytes, splenic macrophages, and peritoneal macrophages (PMs) was also significantly higher in old mice compared with young mice (Figure S6). However, we found no significant difference in LD levels in B cells, T cells, and neutrophils isolated from young and old mice (Figure S7). We thus concluded that altered lipid metabolism occurs during the earliest stages of monocyte infiltration and might be maintained in the tissues during the aging process. Taken together, these results suggest that LD accumulation in aged monocytes indicates abnormal lipid metabolism in these cells and that high LD levels may serve as a biomarker for aging monocytes.
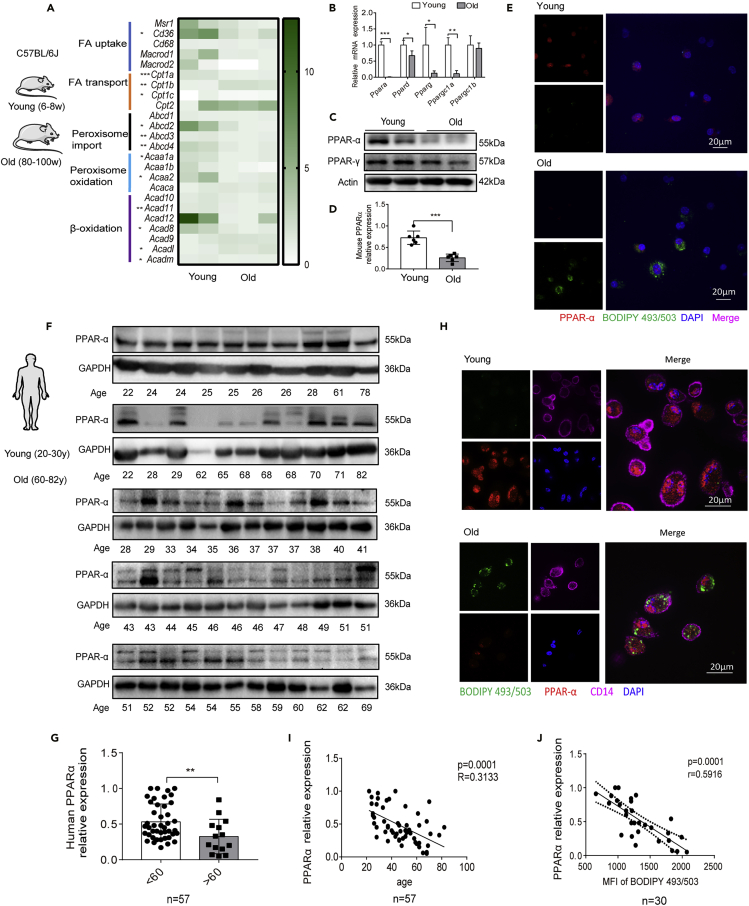
PPAR-α expression in monocytes decreases during aging
Having shown that FA catabolism and LD accumulation in monocytes is impaired with aging, we next investigated the underlying molecular mechanisms. Using RNA sequencing (RNA-seq) analysis, we identified 1,859 upregulated genes and 1,850 downregulated genes between monocytes isolated from young and old mice (Figures S8A–S8C). KEGG pathway enrichment analysis revealed several enriched pathways involved in inflammation and lipid metabolism (Table S1).
Among the cell metabolism pathways, 15.7% of the altered metabolic-related genes were involved in lipid metabolism (Figure S8D). Strikingly, the majority of the genes associated with FA metabolism were suppressed in the aged monocytes (Figure 2A).
Figure 2.
PPAR-α downregulation in elderly human and mouse monocytes
(A–D) Blood monocytes were isolated from young and old mice. (A) RNA-seq analysis was performed, and data for fatty acid metabolism-related genes are shown. (B) qRT-PCR analysis of Ppar mRNA expression (n = 5 per group). The data represent the means ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; unpaired t test. (C) Western blot analysis of PPAR-α and PPAR-γ protein expression in monocytes. Actin was used as a loading control. (D) The relative expression of PPAR-α based on western blot data was analyzed (n = 6 per group). ∗∗∗p < 0.001, t test.
(E) Immunofluorescence staining showing the LD (green) and PPAR-α (red) expression that were detected by BODIPY493/503 and PPAR-α antibody labeling. DAPI (blue) was used to stain the nuclei.
(F) Western blot analysis of PPAR-α expression in human monocytes isolated from healthy donors (see Table S2). GAPDH was used as a loading control.
(G) Relative PPAR-α expression levels (to GAPDH) were determined by gray scale analysis; the relative expression in young (<60 years, n = 43) and old (>60 years, n = 14) groups were analyzed.
(H and I) (H) Immunofluorescent labeling of LD (green) and PPAR-α (red) in young and old human CD14+ monocytes (violet). DAPI (blue) was used to stain the nuclei. (I) The correlation of PPAR-α expression with age was analyzed.
(J) LD levels in human monocytes were determined by BODIPY 493/503 staining and flow cytometric analysis. The correlation with PPAR-α relative expression is presented. (I and J)
The resulting Spearman's correlation coefficient (r) and corresponding p value are reported. The data represent the mean ± SD. ∗∗p < 0.01, t test.
In addition to the altered FA metabolism-associated genes, PPARs captured our attention. PPARs also have a central role in FA metabolism, so we hypothesized that the differences observed in FA metabolism between young and old monocytes might be driven by PPARs. Similar to our RNA-seq data, Ppara was the most significantly reduced gene among all the Ppars in mouse monocytes (Figure 2B), and we found consistent results at the protein level (Figures 2C–2E).
We next tested PPAR-α expression in isolated human monocytes (the details of human donor information can be found in Table S2, related to Figure 2). Interestingly, the PPAR-α relative expression was downregulated in the elderly (>60 years) by up to 1.6-fold (Figures 2F–2H). Pearson's correlation coefficient analysis showed that PPAR-α protein expression markedly decreased with age (Figure 2I). Furthermore, PPAR-α expression was negatively correlated with LD levels (Figure 2J). Taken together, these results suggest that PPAR-α expression decreases, whereas LD levels increase with age in both human and murine monocytes.
PPAR-α regulates FA aerobic oxidation and LD accumulation in monocytes
Having shown that with aging, monocytes show reduced FAO and LD accumulation, we next hypothesized that this effect might be mediated by PPAR-α downregulation. MK886, a specific inhibitor of PPAR-α, markedly decreased OCR and ATP production in young monocytes (Figures 3A–3C), whereas the LD levels were clearly increased (Figure 3D).
Figure 3.
PPAR-α mediates FAO and LD accumulation in monocytes
(A–D) Monocytes were isolated from young mice. (A) Western blot analysis of PPAR-α and HADHA protein levels in monocytes treated with the indicated concentrations of the PPAR-α inhibitor MK886. (B–D) Monocytes were treated with MK886 (0.5 nM). Continuous OCR values (B), ATP concentrations (C), and LD levels (D) were detected.
(E–H) MSC2 cells were transfected with Crispr-cas9 lentiviral-sg Ppara knockdown vectors. The transfection efficiency was assessed by western blot analysis (E). The continuous OCR (F), ATP levels, (G) and LD levels (H) in MSC2 and Ppara knockdown cells were examined.
(I–L) Western blot analysis of PPAR-α and HADHA expression (I), OCR values (J), ATP levels (K), and LD levels (L) in monocytes isolated from old mice treated with the indicated concentrations of fenofibrate.
(M–P) MSC2-C4 (Ppara−/−) cells were infected with lentiviral PPARA overexpression vectors. The transfection efficiency was examined by western blot analysis (M), and the OCR values (N), ATP levels, (O), and LD concentrations (P) were determined in MSC2-C4 and MSC2-C4-OE3 (PPAR-α overexpression) cells.
(Q–S) Blood monocytes were isolated from WT or Ppara−/- mice. PPAR-α protein levels were determined by western blot (Q), and LD levels (S) and Tnfa and Il6 mRNA expression was detected in monocytes isolated from 4- and 20-week-old mice. (n = 4 per group).
All experiments were replicated at least three times. The data represent the mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; t test.
To further confirm the involvement of PPAR-α, we established a Ppara knockout cell line MSC2 (myeloid suppressor cells-2, denoted as MSC2-C4). Compared with MSC2 control cells, MSC2-C4 cells showed significantly reduced OCR and ATP production but increased LD density (Figures 3E–3H). These findings suggest that PPAR-α helps to regulate FAO and to suppress LD accumulation.
We then conducted reciprocal experiments. Upon treating monocytes isolated from old mice treated with the PPAR-α agonist fenofibrate, we observed a dose-dependent upregulation of PPAR-α expression (Figure 3I). This upregulation led to increased OCR and ATP production and decreased LD levels in monocytes from old mice (Figures 3J–3L). We confirmed these findings by stably over-expressing Ppara in MSC2-C4 cells (denoted as MSC2-C4-OE3 cells and established from MSC2-C4 cells; Figures 3M–3P).
Moreover, LD levels were significantly increased in Ppara whole-body knockout mouse monocytes isolated at 4 and 20 weeks of age, compared with littermate wild-type (WT) mice (Figures 3Q and 3R). Tnfa and Il-6 mRNA expression levels in Ppara−/- monocytes were also significantly increased (Figure 3S). These findings suggest that PPAR-α downregulation impairs FAO in aged monocytes. This leads to LD accumulation and elevated inflammatory cytokine production in monocytes with age.
TNF-α in the aging microenvironment promotes PPAR-α downregulation in monocytes
The local environment determines macrophage polarization (Sica and Mantovani, 2012) (Mahbub et al., 2012). Thus, we next explored whether the aged cell microenvironment could affect PPAR-α expression in monocytes. To do so, we established a bone marrow (BM) transplantation experiment (Figure 4A) and found that Ppara expression in young, donor-derived monocytes was significantly reduced in old recipient mice (Figures 4A and 4B). In contrast, Ppara expression in monocytes from old mice (which was already relatively low at the baseline) was prominently upregulated in the young blood microenvironment (Figures 4C and 4D). This result suggests that the local microenvironment does serve a role in reducing Ppara expression in aged monocytes.
Figure 4.
The aging microenvironment suppresses Ppara expression in murine monocytes
(A) EGFP+ BM cells (106) were isolated from young mice and transplanted into wild-type (WT) young or old mice. After 7 days, the CD45+ CD11b+ monocytes were sorted by fluorescence-activated cell sorting (FACS).
(B) qRT-PCR analysis of Ppara mRNA expression in EGFP+ (donor) or EGFP− (recipient) blood monocytes isolated from young and old mice (n = 5 per group).
(C and D) (C) EGFP+ BM cells (106) were isolated from old mice and transplanted into young and old WT mice. (D) qRT-PCR analysis of Ppara mRNA expression in monocytes (n = 5 per group).
(E and F) Relative Tnfa and Il6 mRNA expression in young and old recipient mice, determined by qRT-PCR (n = 5 per group). The data represent the means ± SD.
(G and H) Blood monocytes isolated from young mice were daily stimulated with either mouse IL-6 (50–500 ng/mL) or TNF-α (50–500 ng/mL) for 7 days; the relative expression of PPAR-α was detected by qRT-PCR (G) and western blot analysis (H); GAPDH was used as a loading control.
(I) The LD level in TNF-α-treated monocytes was detected. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; t test.
The local aged tissue environment also upregulated Tnfa and Il6 expression in young mice that were transplanted with monocytes isolated from old mice (Figures 4E and 4F). In contrast, when BM cells isolated from old mice were injected into young mice, the expression of Tnfa and Il6 in monocytes (which was initially high) was significantly reduced in the recipients (Figure 4F). These data further confirm that PPAR-α downregulation induces the polarization of a senescent, inflammatory monocyte type.
However, we still needed to understand how the aging microenvironment inhibited PPAR-α expression in monocytes. When we analyzed the correlation between PPAR-α and TNF-α/IL-6 expression in healthy human blood samples, we found that TNF-α, but not IL-6, showed a weak correlation with PPAR-α downregulation (p = 0.0148, r = 0.1943; Figure S9). To investigate the effect of TNF-α on the downregulation of PPAR-α, we stimulated blood monocytes from young mice with either TNF-α or IL-6. Interestingly, TNF-α stimulation significantly reduced PPAR-α expression in monocytes in a dose-dependent manner, whereas IL-6 had no such effect (Figure 4G, 4H, and S10A). Similarly, TNF-α downregulated PPARα expression of human monocytes (Figures S10B and S10C). Furthermore, TNF-α stimulation induced LD accumulation in monocytes (Figure 4I). These results suggest that TNF-α expression in the aging microenvironment might play a key role in the downregulation of PPAR-α.
Fenofibrate reprograms FAO in monocytes and alleviates aging-associated inflammation in mice and humans
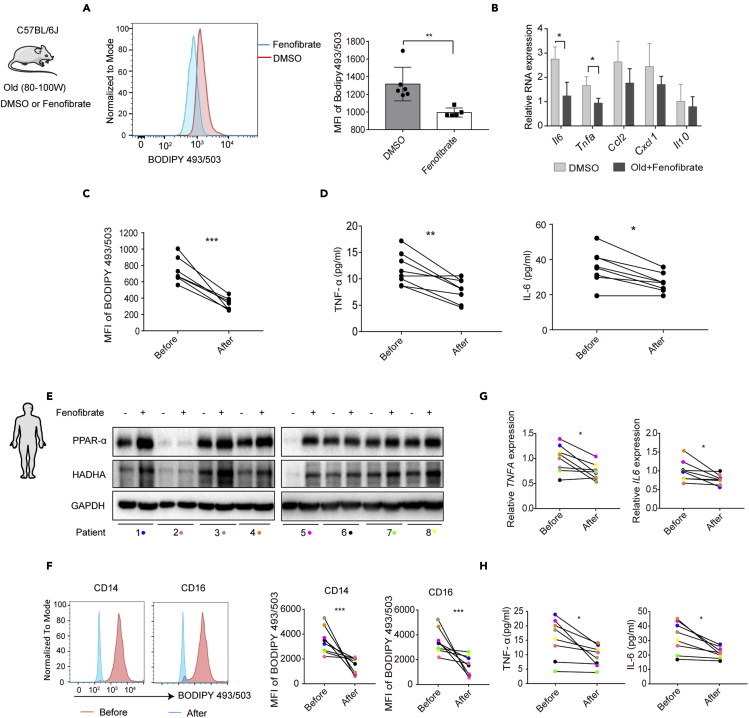
Thus far, our data suggest that monocytes develop pro-inflammatory properties with aging. To investigate whether PPAR-α activation could alleviate monocyte-derived inflammation, we treated old mice with fenofibrate, an activator of PPAR-α. We found that fenofibrate treatment significantly reduced LD accumulation and Il6 and Tnfa expression in monocytes (Figures 5A and 5B). Notably, after fenofibrate treatment, LD accumulation and TNF-α and IL-6 serum levels were significantly reduced in old mice (Figures 5C and 5D), supporting the anti-inflammatory effects of PPAR-α expression in vivo.
Figure 5.
Fenofibrate reverses the pro-inflammatory phenotype of aged monocytes
(A and B) Old mice were injected (intraperitoneally) with fenofibrate (200mg/kg/day for 7 days). (A) LD levels were detected by BODIPY 493/503 staining and flow cytometry (B) and Il6, Tnfa, Ccl2, Cxcl1, and Il10 expression in monocytes in aging was determined by qRT-PCR (n = 6 per group).
(C and D) Blood monocytes were isolated from aged mice before or after fenofibrate treatment for 7 days. The LD levels were assayed by FACS (C), and serum TNF-α and IL-6 levels were detected by ELISA (D) (n = 8).
(E–H) Patients with hypertriglyceridemia (n = 8) were treated with fenofibrate (200 mg/day) for 7 days, and blood samples were collected before or after treatment (see Table S3). (E) PPAR-α and HADHA expression was determined by western blot analysis. (F) LD levels were determined by BODIPY 493/503 staining and flow cytometry. The representative MFI of blood monocytes is shown. (G) TNFA and IL-6 mRNA levels were measured by qRT-PCR. (H) The serum TNF-α and IL-6 levels were detected by ELISA (n = 8).
The data represent the mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; t test.
In our next analysis, we aimed to determine whether fenofibrate could be used to effectively reduce inflammation in human patients. Here, we found that fenofibrate treatment (200 mg/day, 7 days, details of patients information are listed in Table S3, related to Figure 5) strongly upregulated PPAR-α expression in monocytes isolated from hypertriglyceridemia patients 1, 3, 4, and 5 (Figure 5E). This PPAR-α activation induced FAO renewal in aged monocytes, as indicated by the upregulation of HADHA expression (Figure 5E) and a reduction in LD accumulation (Figure 5F). Fenofibrate treatment also decreased TNF-α and IL-6 serum levels and mRNA expression in monocytes (Figures 5G and 5H). In summary, these findings suggest that fenofibrate treatment promotes FAO in monocytes and attenuates inflammatory cytokine production both in vitro and in vivo.
TNF-α neutralization and calorie restriction reverse the pro-inflammatory features of aged monocytes
Increased TNF-α expression in the aging cell microenvironment polarizes monocytes to an aged profile by downregulating PPAR-α expression. Interestingly, TNF-α neutralization significantly upregulated PPAR-α expression and reduced LD accumulation in aged monocytes (Figures 6A and 6B). Furthermore, the serum concentrations of TNF-α and IL-6 were also notably decreased after TNF-α neutralization (Figure 6C).
Figure 6.
TNF-α neutralization or CR reprograms the catabolism of aged monocytes
(A–C) Old mice were intravenously injected with 100 ng/day tmTNF-α or IgG for 7days. The blood monocytes were isolated and the PPAR-α expression was detected by western blot analysis, and actin was used as a loading control. (B) The LD level in monocytes was measured. (C) TNF-α and IL-6 serum concentration were measured by ELISA (n = 4 mice per group).
(D–G) Young and old mice were fed a normal diet or underwent CR for 7 days before monocytes were isolated. (D) Tnfa and Il6 expression were analyzed by qRT-PCR (n = 6 mice per group). (E) p-AMPK and p-mTOR levels were detected by western blot analysis. Representative blots of three independent experiments are shown. (F) LD density was detected by BODIPY 493/503 staining and flow cytometry. The representative MFI of blood monocytes is shown. (G) PPAR-α protein expression was detected by western blot.
(H) Serum TNF-α and IL-6 levels in CR-induced old mice were measured by ELISA (n = 6 mice per group).
Data represent the mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; one-way ANOVA (D), t test (C, F, and H).
Calorie restriction (CR) has also been proposed to delay inflammaging, so we also investigated whether the pro-inflammatory characteristics of aged monocytes might be affected by CR. Compared with mice fed a normal diet, monocytes from CR-fed (60% of normal diet) old mice showed significantly reduced Tnfa and Il6 expression (Figure 6D). Moreover, CR reactivated the suppressed AMPK signaling in monocytes derived from old mice (Figure 6E). As expected, the mTOR pathway was not significantly altered in monocytes isolated from either CR or normal mice. We confirmed this reprogramming of CR-induced monocytes by detecting reduced LD density in monocytes taken from aged mice (Figure 6F). Together, these results suggest that CR might reverse the pro-inflammatory phenotype and reprogram the catabolism of monocytes in aging.
Having shown that CR attenuated Tnfa and Il6 mRNA expression, we finally investigated how CR affects PPAR-α expression. Interestingly, CR dramatically increased PPAR-α expression (Figure 6G) and reduced TNF-α and IL-6 secretion in old mice (Figure 6H). In addition, CR failed to rescue LD accumulation and Tnfa/Il6 high expression in Ppara−/- mice monocytes (Figure S11). These data suggest that CR might represent a viable strategy to target PPAR-α in a drug-free manner.
Discussion
Monocytes derived from aged individuals secreted more inflammatory cytokines than those taken from young individuals, which may contribute to inflammaging during aging. However, previous studies have failed to determine the underlying mechanisms. Here, we found that circulating monocytes exhibit a pro-inflammatory phenotype during aging that is characterized by LD accumulation. Further analyses suggested that this accumulation was induced by suppression of PPAR-α-driven FAO. Interestingly, TNF-α in the aged, inflammatory cell microenvironment inhibited PPAR-α expression in monocytes and accelerated the switch to a pro-inflammatory profile. Using PPAR-α activation, we were able to increase FA consumption by monocytes, and this effect might ultimately lead to a reduction in inflammaging during aging.
A dysregulated immune response that confers SCI is a major change known to occur during aging. This is the long-term result of chronic physiological stimulation of the innate immune system, which occurs in later life (van Beek et al., 2019). Monocytes are potent producers of pro-inflammatory cytokines (Puchta et al., 2016) and thus are speculated to play a key role in SCI and inflammaging. In our study, we confirmed that circulating IL-6 and TNF-α levels increased with age, and with this in mind, we speculated that targeting monocytes during aging may represent a useful strategy to alleviate inflammaging.
Macrophage polarization and functions are under metabolic control (Namgaladze and Brüne, 2016). FAO has a central role in regulating innate and adaptive immunity: it provides energy for anti-inflammatory macrophage polarization and modulates inflammatory signatures (Divakaruni et al., 2018). FAO can also reduce palmitate-mediated inflammation and ER stress by accelerating FA catabolism in macrophages (Namgaladze et al., 2014). Monocytes are precursors of tissue macrophages, and their polarization is mediated by metabolic processes. We revealed that circulating monocytes in old mice have a different metabolic gene expression profile than those in young mice. This altered profile is characterized by suppressed FA catabolism. Further experiments confirmed that ATP production is downregulated in aged monocytes, OCR is suppressed, and FAO capacity is reduced. We propose that this impairment is a key driver of monocyte dysfunction in aging.
Although FAO is characteristic of anti-inflammatory macrophages, FAO dysfunction and lipid overload can activate the pro-inflammatory profile of macrophages. For example, an increase in lipid uptake mediates mitochondrial dysfunction and FAO impairment by increasing reactive oxygen species production, which promotes macrophage inflammatory polarization and atherosclerosis (Namgaladze and Brüne, 2016). Lipid overload also promotes the polarization of inflammatory adipose tissue macrophages (Thomas and Apovian, 2017). Our data suggest that FAO dysfunction and lipid overload might induce the functional properties of both aged murine and human monocytes, although the underlying mechanisms still need to be fully investigated.
LD was initially described as an organelle found in adipocytes for fat storage, but increasing evidence suggests that LD has a vital role in lipid metabolism and immune regulation in other cell types (den Brok et al., 2018; Fujimoto and Parton, 2011; Olzmann and Carvalho, 2019). Indeed, a high level of cytoplasmic LDs in immune cells is considered an initiator of inflammation (Melo and Weller, 2016). For example, LD accumulation in tissue leukocytes has been observed in patients with inflammatory arthritis, acute respiratory distress syndrome, bacterial sepsis, and mycobacterial infection (Mattos et al., 2010). Similarly, our human data suggest that there is a high level of LDs in monocytes isolated from elderly subjects and high TNF-α/IL-6 levels in the serum. Thus, we conclude that LD density in monocytes might serve as an indicator of inflammaging in the elderly. Going forward, it might be worth screening for people who would benefit from intervention against inflammaging by detecting LD volume in monocytes isolated from their blood. Then, we could select the most appropriate cohort to investigate the therapeutic effects of anti-inflammaging approaches.
Based on our findings, we propose that reducing LD accumulation in aged monocytes might be a potential strategy to delay inflammaging. D'Avila et al. demonstrated that phagocytosis is neither essential nor sufficient for LD formation in macrophages (Bozza et al., 2007; D'Avila et al., 2006). Other groups have found that TLR-mediated pathogen recognition can induce LD formation in macrophages (Perlemuter et al., 2002). In this study, we found that the expression of FA catabolism-associated genes was decreased in aged monocytes. We thus speculate that FAO downregulation results in FA accumulation and ultimately, LD formation. However, whether LD directly induces inflammatory gene expression or whether this is mediated by other inflammatory pathways requires further investigation.
PPARs are the predominant regulators of lipid metabolism (Gervois et al., 2000). Both mitochondrial and peroxisomal FAO-related genes can be activated by PPAR-α, whereas PPAR-γ mainly targets FA uptake and storage (Han et al., 2017). Aging has also been reported to impair mitochondrial respiratory capacity in monocytes, which may cause increased oxidative stress and DNA damage (Pence and Yarbro, 2018). We also found that the number of mitochondria in elderly monocytes were reduced, which suggests that aging resulted in the impaired aerobic oxidation in monocytes. Furthermore, it was PPAR-α, rather than other isoforms, that was significantly reduced in aged monocytes, and the expression of PPAR-α in human monocytes was negatively correlated with both age and LD level. When PPAR-α was activated in mice using fenofibrate, LD density and pro-inflammatory cytokine production in old, isolated monocytes were significantly reduced. Although fenofibrate is not a specific agonist of PPAR-α, it still reversed the profile of aged monocytes and upregulated PPAR-α. More specific molecules that could activate PPAR-α without side effects need to be identified in future studies.
The extrinsic aged cell microenvironment contains high levels of pro-inflammatory factors that can affect macrophage function (Linehan and Fitzgerald, 2015). Our BM transfer experiments indicated that Ppara gene expression in monocytes is significantly downregulated in the aged cell microenvironment and, furthermore, that these expression levels recovered when the monocytes were injected into young mice. Previous studies have suggested that PPAR-α expression can be inhibited by TNF-α in the rat liver (Beier et al., 1997). It is possible, therefore, that TNF-α is involved in the pathway that ultimately triggers macrophage polarization with aging. Our results suggested the presence of a weak correlation between serum TNF-α concentration and monocytic PPAR-α level, and TNF-α stimulation reduced PPAR-α expression in monocytes. Furthermore, TNF-α neutralization in vivo significantly reduced lipid overload in monocytes taken from aging mice, as well as the concentration of circulating cytokines. We therefore speculate that increased TNF-α levels in aged mice might accelerate monocyte aging by downregulating PPAR-α expression; the resulting FAO impairment and pro-inflammatory cytokine elevation would subsequently aggravate inflammaging. However, the underlying mechanisms still need further exploration.
Our clinical data also confirmed that fenofibrate has great potential for attenuating monocyte-induced inflammation in aging. However, we only tested the therapeutic benefits of fenofibrate on cells isolated from patients with hyperlipidemia; its curative and adverse effects on healthy elderly people remain unknown. Thus, a method that increases PPAR-α activity would be highly beneficial.
Data obtained from various model systems suggest that CR can help extend lifespan. The underlying mechanisms seem to involve the downregulation of insulin and mTOR signaling, and concomitant activation of the NAD-dependent protein deacetylase sirtuin 1 (Lee and Longo, 2016). Alterations to these pathways activate autophagy, stress defenses, and survival pathways while attenuating pro-inflammatory responses (Barzilai et al., 2012). In macrophages, CR can decrease endotoxin-elicited pro-inflammatory mediator production in rat alveolar macrophages (Dong et al., 1998) and increase prostaglandin E2 secretion in mice PMs (Stapleton et al., 2001). Although CR is known to increase hepatic PPAR-α-mediated FAO and whole-body fat oxidation rates in rats (Takemori et al., 2011), its effect on monocytes in the context of aging was unknown before this study. Our results demonstrated that CR activates PPAR-α-induced catabolism in aged monocytes; thus, it seems that CR might also serve to suppress LD accumulation and pro-inflammatory production in aging. We speculate that CR could reduce FA and lipid synthesis and accelerate FA catabolism to provide energy and maintain monocyte function. Reducing free FA serum levels would likely suppress the activation of inflammatory pathways in monocytes, and the mechanistic details now require further investigation.
Taken together, our results show that with aging, monocytes accumulate LDs and start to produce high levels of pro-inflammatory cytokines. It seems that it is the downregulation of PPAR-α expression that drives this polarization of monocytes with age, and this ultimately impairs FAO. TNF-α neutralization reversed PPAR-α downregulation induced by the aged microenvironment, whereas fenofibrate and CR activated PPAR-α-induced FA catabolism. All these PPAR-α activation strategies ultimately attenuate monocyte-driven inflammation, and this suggests that reprogramming monocytic FA metabolism might be a useful therapeutic intervention to reduce inflammaging. The present study clears that the long-term effects of PPAR-α activation on anti-inflammaging in healthy, elderly subjects warrants a future longitudinal and large-scale analysis.
Limitations of the study
This study indicated that downregulated PPAR-α may be responsible for the pro-inflammatory phenotype of monocytes in aged individuals. In addition, increased TNF-α levels in aged mice might accelerate monocyte aging by downregulating PPAR-α expression. However, the underlying mechanisms of how TNF-α regulates PPAR-α need to be investigated. Furthermore, in the present study, fenofibrate did inhibit the pro-inflammatory features of monocytes by increasing their FAO capacity, although not all patients showed clear downregulation of inflammatory cytokines in response to fenofibrate treatment. Thus, more work is needed to understand how PPAR-α-induced FAO regulates inflammatory cytokine secretion in aged monocytes, and which patients might benefit from PPAR-α activation.
STAR★Methods
Key resources table
| REAGENTS or RESOURCES | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-phospho-AMPKa used at 1:1000 | Cell Signaling Technology | Cat # 2535; RRID: AB_331250 |
| Anti- AMPKa used at 1:1000 | Cell Signaling Technology | Cat #2603; RRID: AB_490795 |
| Anti-phospho-mTOR(Ser2481) antibody used at 1:1000 | Cell Signaling Technology | Cat # 2974; RRID: AB_2262884 |
| Anti-phospho-mTOR(Ser2448)Antibody used at 1:1000 | Cell Signaling Technology | Cat # 2971; RRID: AB_330970 |
| Anti-mTOR antibody used at 1:1000 | Cell Signaling Technology | Cat # 2972; RRID: AB_330978 |
| Anti-ACADM antibody used at 1:500 | Abcam | Cat # ab118183 |
| Anti-ACADVL antibody used at 1:500 | Abcam | Cat # ab118183 |
| Anti-HADHA antibody used at 1:500 | Abcam | Cat # ab118183 |
| Anti-PPAR-a antibody used at 1:1000 | Abcam | Cat # ab191226 |
| Anti-PPAR-a antibody used at 1:1000 | Abcam | Cat # ab24509; RRID: AB_448110 |
| Anti-PPAR-γ antibody used at 1:1000 | Cell Signaling Technology | Cat # 2435, RRID: AB_2166051 |
| Anti-GAPDH antibody used at 1:2000 | Cell Signaling Technology | Cat # 5174; RRID: AB_10622025 |
| Anti-ACTIN antibody used at 1:2000 | Cell Signaling Technology | Cat # 4970; RRID: AB_2223172 |
| Rabbit anti-mouse IgG mAb at 1:3000 | Cell Signaling Technology | Cat # 58,802; RRID: AB_2799549 |
| Mouse anti-rabbit IgG mAb at 1:3000 | Cell Signaling Technology | Cat # 45,262; RRID: AB_2799281 |
| Purified rat anti mouse CD16/CD32 | BD Bioscience | Cat # 560051 |
| Anti-mouse CD45 alexa fluor 700 use at 1:100 | Biolegend | Cat # 103128; RRID: AB_493715 |
| Anti-mouse F4/80 APC/Cyanine7 use at 1:100 | Biolegend | Cat # 123117; RRID: AB_893489 |
| Anti-mouse/human CD11b FITC use at 1:100 | BD Bioscience | Cat # 553310; RRID: AB_394774 |
| Anti-mouse CD4 APC/Cyanine7 use at 1:100 | Biolegend | Cat # 1004141; RRID: AB_312699 |
| Anti-mouse CD8b APC/Cyanine7 use at 1:100 | Biolegend | Cat # 126619; RRID: AB_2563950 |
| Anti-mouse Ly-6G APC use at 1:100 | Biolegend | Cat # 127614; RRID: AB_2227348 |
| Anti-human CD45 PE use at 1:100 | Biolegend | Cat # 304008; RRID: AB_314396 |
| Anti-human CD14 FITC use at 1:100 | Biolegend | Cat # 301804; RRID: AB_31,418 |
| Anti-human CD14 APC/Cyanine7 use at 1:100 | Biolegend | Cat # 325620; RRID: AB_830693 |
| Anti-human CD16 PE/Cy5 use at 1:100 | Biolegend | Cat # 302009; RRID: AB_314209 |
| DAPI | Boster | Cat # AR1176 |
| BODIPY™ 493/503 | Thermo Fisher | Cat #D3922 |
| Donkey anti-mouse IgG (H + L) secondary antibody alexa Fluor® 488 conjugate | Thermo Fisher | Cat # A-21202; RRID: AB_141607 |
| Donkey anti-mouse IgG (H + L) secondary antibody alexa Fluor® 555 conjugate | Thermo Fisher | Cat # A-31570; RRID: AB_2536180 |
| Biological samples | ||
| Healthy blood (n = 102) | The First affiliated Hospital of ZhengZhou University | http://fcc.zzu.edu.cn/ |
| Hypertriglyceridemia patient derived blood (n = 8) | The First affiliated Hospital of ZhengZhou University | http://fcc.zzu.edu.cn/ |
| Mouse blood samples (C57/6J background) | Animal center of ZhengZhou University | http://www5.zzu.edu.cn/dwsyzx/info/1012/1027.htm |
| Critical commercial assays | ||
| Fatty acid oxidation assay kit | Abcam | Cat # ab118183 |
| Human IL-6 ELISA MAX™ deluxe | Biolegend | Cat # 430504 |
| Human TNF-α ELISA MAX™ deluxe | Biolegend | Cat # 430204 |
| Mouse IL-6 ELISA MAX™ deluxe | Biolegend | Cat # 431304 |
| Mouse TNF-α ELISA MAX™ deluxe | Biolegend | Cat # 431904 |
| Oligonucleotides | ||
| sgRNAs target sequence Ppara1, CACCGCCCGTGATCCCCGCCAGCA, AAACTGCTGGCGGGGATCACGGGC | This paper | https://www.sangon.com/ |
| sgRNAs target sequence Ppara2, CACCGCCGTGCTGGCGGGGATCAC; AAACGTGATCCCCGCCAGCACGGC |
This paper | https://www.sangon.com/ |
| sgRNAs target sequence Ppara3, CACCGTAGACACCCTCTCTCCAGCTTC; AAACGAAGCTGGAGAGAGGGTGTCTAC |
This paper | https://www.sangon.com/ |
| sgRNAs target sequence Ppara4, CACCGCCTCTTCCATCTGTAGACACCC; AAACGGGTGTCTACAGATGGAAGAGGC |
This paper | https://www.sangon.com/ |
| TNFA forward: CTCTTCTGCCTGCTGCA CTTTG |
This paper | N/A |
| TNFA reverse: ATGGGCTACAGGCTTG TCACTC |
This paper | N/A |
| CXCL1 forward: GCCAGTGCTTGCAGACCCT | This paper | N/A |
| CXCL1 reverse: GGCTATGACTTCGGT TTGGG |
This paper | N/A |
| CCL2 forward: TCATAGCAGCCACCTT CATTC |
This paper | N/A |
| CCL2 reverse: TAGCGCAGATTCTTGGGTTG | This paper | N/A |
| IL6 forward: ACTCACCTCTTCAGAAC GAATTG |
This paper | N/A |
| IL6 reverse: CCATCTTTGGAAGGTTC AGGTTG |
This paper | N/A |
| IL8 forward: GAGAGTGATTGAGAGTGG ACCAC |
This paper | N/A |
| IL8 reverse: CACAACCCTCTGCACCCAGTTT | This paper | N/A |
| IL10 forward: GGTTGCCAAGCCTTGTCTGA | This paper | N/A |
| IL10 reverse: AGGGAGTTCACATGCGCCT | This paper | N/A |
| GAPDH forward: GCTCCCTCTTTCTTTG CAGC |
This paper | N/A |
| GAPDH reverse: ACCATGAGTCCTTCC ACGAT |
This paper | N/A |
|
Tnfa forward: AAAGACCAGGTGGAGTGG AAGAAC |
This paper | N/A |
|
Tnfa reverse: CTCAGTGCCGATGGAGTC CGAGTA |
This paper | N/A |
| Cxcl1 forward: CAAGGCTGGTCCATGCTCC | This paper | N/A |
|
Cxcl1 reverse: TGCTATCACTTCCTTTCT GTTGC |
This paper | N/A |
|
Ccl2 forward: TTTTGTCACCAAGCTCA AGAGA |
This paper | N/A |
|
Ccl2 reverse: ATTAAGGCATCACAGTCC GAGT |
This paper | N/A |
| Il6 forward: TGGGGCTCTTCAAAAGCTCC | This paper | N/A |
|
Il6 reverse: AGGAACTATCACCGGATC TTCAA |
This paper | N/A |
|
Il8 forward: ATGACTTCCAAGCTGGCC GTGGCT |
This paper | N/A |
|
Il8 reverse: TCTCAGCCCTCTTCAAAA ACTTCTC |
This paper | N/A |
| Il10 forward: CCCATTCCTCGTCACGATCTC | This paper | N/A |
|
Il10 reverse: TCAGACTGGTTTGGGAT AGGTTT |
This paper | N/A |
| Gapdh forward: TCTCTGCTCCTCCCTGTTCC | This paper | N/A |
|
Gapdh reverse: TACGGCCAAATCCGT TCACA |
This paper | N/A |
| Ppara forward: TGGTGTTCGCAGCTGTTTTG | This paper | N/A |
| Ppara reverse: AGATACGCCCAAATGCACCA | This paper | N/A |
| pEX5 (pGCMV/EGFP/MCS/Neo) plasmid | GENECAM | N/A |
| PPARa over-expression plasmid | GENECAM | N/A |
| Recombinant DNA | ||
| Software and algorithms | ||
| Seahorse Explorer analaysis program | - | https://www.agilent.com/zh-cn/products/cell-analysis/seahorse-analyzers |
| FlowJo v.10 | FlowJo | https://www.flowjo.com/solutions/flowjo |
| Imaris v.9 | OXFORD | https://imaris.oxinst.com/versions/9 |
| GraphPad prism version 7 for Windows | GraphPad software | https://www.graphpad.com/ |
| ImageJ | - | https://imagej.nih.gov/ij/ |
Resource availablity
Lead contact
Further information and requests for resources and regents should be directed to and will be fulfilled by the lead contact, Dr. Wang Ming (wangmingheda@163.com).
Materials availability
All unique/stable reagents and mouse lines used in this study are available from the lead contact with a completed Materials Transfer Agreement.
Data and code availability
The datasets generated and analyzed during the study are included with the published manuscript (and supplemental information). All other data are available from the corresponding author upon request. The RNA sequencing data was uploaded in the Sequence Read Archive (SRA) database: SRR12130588-SRR12130592.
Experimental model and subject details
Human samples
All human samples were obtained from healthy donors (n = 102; male, 54; female, 48; age, 20–82 years; average age, 49.8 ± 19 years) or hypertriglyceridemia patients (n = 8, male, 4; female 4; average age, 55.8 ± 6.9 years) after physical examination in the First Affiliated Hospital of ZhengZhou University (ZhengZhou, China). Blood samples were collected following the standards of the Ethics Committee of ZhengZhou University (Ethics Number: KY249). Blood from patients with hypertriglyceridemia was collected before and after fenofibrate (200mg/day; Abbott Laboratories limited; Shanghai, China) administration for 7 days. All blood samples were stored in 7 mL anticoagulation tubes (BD Biosciences, Franklin Lakes, NJ, USA), and the serum and cells were separated for further experiments. Human donor information is listed in Table S2. Hypertriglyceridemia patient characteristics are summarized in Table S3.
Animals
Female C57BL/6J mice or EGFP mice were 8–80 weeks of age and purchased from the Vital River Laboratory Animal Technology Company (Beijing, China); Ppara knockout mice were generated at the Cyagen Biosciences (SuZhou, China). All the mice were housed in IVC cages on racks in a room with controlled temperature (22–25°C) and humidity (40–60%), and were subjected to 12 hr light/dark cycles in a specific pathogen-free (SPF) facility. All the mice had ad libitum (AL) access to sterilized water and maintenance diet, except the CR mice. CR mice received 60% of the food consumed by the AL animals. All animal experiments were in accordance with protocols approved by the Institutional Animal care and Use Committee of ZhengZhou University. Mice were euthanized with isoflurane, and tissues were collected for further experiments.
Methods details
Primary blood monocytes isolation and cell culture
Human or murine blood monocytes were collected and purified using gradient Ficoll-Hypaque (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and the Negative Selection Monocyte Isolation kit (STEMCELL Technologies, Vancouver, Canada). The fresh isolated monocytes were cultured in RPMI1640 medium (HyClone, Logan, UT, USA) supplemented with 10% FBS (PAN Biotech, Aidenbach germany), 100 IU/mL penicillin and 100 μg/mL streptomycin (Gibco, Waltham, MA, USA) at 37°C in a 5% CO2 incubator. To further test the effect of TNF-α and IL-6 on PPARa expression, monocytes were stimulated with 50ng/ml GM-CSF (BioLegend, San Diego, CA, USA), and different dose of TNF-α and IL-6 (Biolegend, San Diego, CA, USA) for 7 days. The macrophages cell lines RAW264.7 and MSC2 were also cultured in RPMI1640 complete medium.
Human and mouse serum ELISA
To determine serum TNF-α and IL-6 protein concentration, human or mouse blood samples were isolated rapidly after high-speed centrifugation (300g, 10 min) at 4°C. Human or mouse plasma concentrations of TNF-α and IL-6 were measured using their specific ELISA Ready -SET-GO-Assay according to the manufacturer's protocol (BioLegend, San Diego, CA, USA). The details of the ELISA Kits are provided in the Key Resources Table.
Flow cytometry
Anti-human CD45-PE; anti-human-CD14-APC/Cyanine7; anti-human-CD16-PE/Cy5; anti-mouse-CD45-Alexa Fluor 700; anti-mouse F4/80-APC/Cyanine7; anti-mouse/human CD11b-FITC; anti-mouse Ly-6G-APC; anti-mouse CD4-APC/Cyanine7; anti-mouse CD8b-APC/Cyanine7 were used for flow cytometric analysis in this study. Human monocytes were identified as CD45+CD14/CD16+. Mouse monocytes, neutrophils and T cells were identified as CD45+CD11b+, CD11b+ly6G+ and CD4+/CD8+ respectively. To analyze the LD concentration in blood monocytes or other immune cells, the isolated monocytes or immune cells were incubated with 2 μM Bodipy493/503 (D3922, Thermo Fisher Scientific, California, USA) staining solution in the dark for 15 min at 37°C. After washing 3 times with PBS, cells were stained with theirs surface markers for 30min at 4°C. Data were acquired on an FACSCanto II (BD Biosciences, Franklin Lakes, NJ, USA) and analyzed with FlowJo v10.
FAO enzyme measurements
FAO analysis was performed using a Fatty Acid Oxidation Flow Cytometry kit (ab118183, Abcam, Cambridge, UK). Levels of the FAO cycle enzymes ACADVL, ACADM and HADHA were determined according to the manufacturer's protocol. Briefly, the isolated mouse blood monocytes were washed with PBS, fixed in 4% Paraformaldehyde for 15 min, permeabilized with 0.1% Triton X- and unspecific binding was blocked (5% BSA). Primary antibodies (ACADVL, ACADM and HADHA) were applied 1 hr at 4°C, secondary antibodies (Alexa Fluor 488, Thermo Fisher Scientific) for 1 hr at 25°C. The fluorescence for each enzyme was measured in FACSCanto II (BD Biosciences, Franklin Lakes, NJ, USA) and analyzed with FlowJo v10.
Confocal microscopy and BODIPY staining
Monocytes were seeded on culture dishes (35mm, glass bottom, Thermo Fisher Scientific) 12 hr before staining. For LD staining, cells were stained by 2 μM Bodipy493/503(D3922, Thermo Fisher Scientific) for 30 min and then washed with PBS, fixed by 4% Paraformaldehyde for 10 min. Following stained with primary antibodies (CD14-APC/Cyanine7 and rabbit anti human PPAR-α) for 30 min at 4°C, monocytes were incubated with secondary antibodies (Alexa Fluor 555, Thermo Fisher Scientific) for 30 min at 4°C. Images were acquired using a Leica confocal microscope (Leica, Wetzlar, Germany). The LD density were measured by 3D rendering using Imaris 9 software (OXFORD).
Western blot analysis
Standard protocols were used for Western blot analyses. Briefly, monocytes were lysed using RIPA lysis buffer (20mM Tris-HCl pH7.8, 150mM NaCl, 1% (v/v) NP-40, 0.05% (w/v) Sodium Deoxycholate, 0.4% (w/v) SDS) and supplemented with 1% protease inhibitor cocktail (P8340, Sigma-Aldrich, St Louis, MO, USA) and 1 mM phenylmethylsulfonyl fluoride (PMSF) for 30 min. Protein concentrations were determined using the Pierce BCA Protein Assay Kit (233225, Thermo Scientific, Rockford, IL, USA). Total protein was separated by SDS-PAGE gel and transferred electrophoretically to a nitrocellulose filter membrane (HATF00010, Millipore, Bellerica, MA, USA). The details of the antibodies used are provided in the Key Resources Table.
RNA isolation and quantitative real-time RT-PCR
Total mRNA was isolated from human or mice monocytes with RNAiso Plus (9109, Takara, Tokyo, Japan), and cDNA was synthesized using the PrimeScript RT Master Mix (RR036A; Takara, Tokyo, Japan). cDNA was quantified by real-time PCR using TB Green Premix Ex Taq II (RR820A, Takara, Tokyo, Japan) on an ABI PRISM 7300HT Sequence Detection 542 System (Applied Biosystems, Foster City, CA, USA). The primer sequences are listed in Key Resources Table.
Bone marrow transplantation
For young and old mouse BM transplantation experiments, donor Enhanced Green Fluorescent Protein (EGFP) mice were sacrificed with isoflurane, and bone marrow cells (BMCs) were isolated from the femurs and tibias. Before transplantation into WT C576J mice, the BMCs were washed twice with PBS, filtered through a 70-micron filter, and then 2∗106 BMCs were intravenously injected into recipient mice via the tail vein. The donor/recipient combinations were: young EGFP to young WT; young EGFP to old WT; old EGFP to young WT; and old EGFP to old WT. After 1 week, blood monocytes were collected from the recipient mice and sorted for further experiments.
ATP assay
ATP levels in MSC2 cells or primary monocytes were determined using an ATP colorimetric/fluorometric assay kit according to the manufacturer's protocol (Novus Biologicals, Centennial, CO, USA).
OCR calculations using Seahorse
To measure mitochondrial function, 20,000 MSC2 cells or freshly isolated blood monocytes taken from young or old mice were seeded on Seahorse culture plates in assay media (DMEM, 1% BSA, 25 mM glucose, and 2 mM glutamine, 1 mM pyruvate) and analyzed using a Seahorse XFe96 system (Agilent Technologies, Inc., Santa Clara, CA, USA). Basal macrophage OCR was determined over 1 hr. To obtain the maximal respiratory and control values of cells, macrophages were stimulated with oligomycin (2 mM), carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP, 1 mM) and rotenone (0.5 mM)/antimycin A (0.5 mM), according to a standard protocol (Wu et al., 2019). The cell numbers were then normalized to the cell protein concentrations.
Electron microscopy
The isolated monocytes were fixed with 2.5% Glutaraldehyde in 0.1 M Sodium Cacodylate Buffer for 24 hr. Glutaraldehyde was removed, and then monocytes were washed 3 times with 0.1 M Sodium Cacodylate Buffer. The monocytes images were acquired using an JEOL-1200 EXII transmission electron microscope (JEOL GmbH, Freising, Germany).
RNA-seq
The RNA-sequencing experiments were performed by OE Biotech Co., Ltd. (Shanghai, China) using an Illumina HiseqTM 2500 system (Illumina, Inc., San Diego, CA, USA). RNA isolated from blood monocytes taken from young or old mice was used for RNA-seq. These RNA samples were converted into a library of cDNA fragments, Illumina sequencing adapters were added, and 50 bp single end read sequences were obtained using the Illumina HiSeq system. A quality check was performed on these sequence reads using FastQC. EdgeR software was used to analyze the data, and P < 0.05 was considered to indicate a statistically significant difference.
Cas 9 PPAR-α knockout
The CRISP-Cas 9 system was used to knock out PPAR-α in MSC2 cells. In brief, PPAR-α sgRNAs were designed using the following website: https://chopchop.cbu.uib.no/. The NDA target sequences of PPAR-α sgRNAs are shown in the Key Resources Table. DNA oligos of these sgRNAs were cloned into a pL-CRISPR.EFS.GFP plasmid, and lentiviral packaging and cell transduction were performed as previously described (Cribbs et al., 2013).
Quantification and statistical analysis
Statistical analysis
Statistical significance was calculated using Student's t-tests when comparing two groups, or one-way ANOVA when comparing more than two groups. P < 0.05 was considered to indicate a statistically significant difference. Pearson correlation coefficient was used to investigate the linear relationship between two factors.
Acknowledgments
We thank Dr. Jessica Tamanini from Insight Editing London for critical reading of the manuscript. We thank Dr. Chen Ni and Dr. Linyu Zhu for the project design. This work was supported by the National Natural Science Foundation of China (grant nos. 81630068, 31670881, and 81901466).
Author contributions
Conceptualization, M.W., Z.Q., Y.H., and H.W.; methodology, M.W., Y.Y., Z.Z., X.Y., X.D., and Z.J.; formal analysis, M.W., Y.Y., and X.Y.; resources, Z.Z. and P.Z.; supervision and fund acquisition, Z.Q.; writing – review & editing, M.W., R.G., J.A., Z.W., and Z.Q.
Declaration of interests
The authors declare no competing interests.
Published: July 23, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102766.
Supplemental information
References
- Barzilai N., Huffman D.M., Muzumdar R.H., Bartke A. The critical role of metabolic pathways in aging. Diabetes. 2012;61:1315–1322. doi: 10.2337/db11-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier K., Völkl A., Fahimi H.D. TNF-alpha downregulates the peroxisome proliferator activated receptor-alpha and the mRNAs encoding peroxisomal proteins in rat liver. FEBS Lett. 1997;412:385–387. doi: 10.1016/s0014-5793(97)00805-3. [DOI] [PubMed] [Google Scholar]
- Bouchlaka M.N., Sckisel G.D., Chen M., Mirsoian A., Zamora A.E., Maverakis E., Wilkins D.E., Alderson K.L., Hsiao H.H., Weiss J.M. Aging predisposes to acute inflammatory induced pathology after tumor immunotherapy. J. Exp. Med. 2013;210:2223–2237. doi: 10.1084/jem.20131219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozza P.T., Melo R.C., Bandeira-Melo C. Leukocyte lipid bodies regulation and function: contribution to allergy and host defense. Pharmacol. Ther. 2007;113:30–49. doi: 10.1016/j.pharmthera.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Cribbs A.P., Kennedy A., Gregory B., Brennan F.M. Simplified production and concentration of lentiviral vectors to achieve high transduction in primary human T cells. BMC Biotechnol. 2013;13:98. doi: 10.1186/1472-6750-13-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Avila H., Melo R.C., Parreira G.G., Werneck-Barroso E., Castro-Faria-Neto H.C., Bozza P.T. Mycobacterium bovis bacillus Calmette-Guerin induces TLR2-mediated formation of lipid bodies: intracellular domains for eicosanoid synthesis in vivo. J. Immunol. 2006;176:3087–3097. doi: 10.4049/jimmunol.176.5.3087. [DOI] [PubMed] [Google Scholar]
- Delayre-Orthez C., Becker J., Guenon I., Lagente V., Auwerx J., Frossard N., Pons F. PPARalpha downregulates airway inflammation induced by lipopolysaccharide in the mouse. Respir. Res. 2005;6:91. doi: 10.1186/1465-9921-6-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Brok M.H., Raaijmakers T.K., Collado-Camps E., Adema G.J. Lipid droplets as immune modulators in myeloid cells. Trends Immunol. 2018;39:380–392. doi: 10.1016/j.it.2018.01.012. [DOI] [PubMed] [Google Scholar]
- Divakaruni A.S., Hsieh W.Y., Minarrieta L., Duong T.N., Kim K.K.O., Desousa B.R., Andreyev A.Y., Bowman C.E., Caradonna K., Dranka B.P. Etomoxir inhibits macrophage polarization by disrupting CoA homeostasis. Cell Metab. 2018;28:490–503.e497. doi: 10.1016/j.cmet.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W., Selgrade M.K., Gilmour I.M., Lange R.W., Park P., Luster M.I., Kari F.W. Altered alveolar macrophage function in calorie-restricted rats. Am. J. Respir. Cell. Mol. Biol. 1998;19:462–469. doi: 10.1165/ajrcmb.19.3.3114. [DOI] [PubMed] [Google Scholar]
- Fagiolo U., Cossarizza A., Scala E., Fanales-Belasio E., Ortolani C., Cozzi E., Monti D., Franceschi C., Paganelli R. Increased cytokine production in mononuclear cells of healthy elderly people. Eur. J. Immunol. 1993;23:2375–2378. doi: 10.1002/eji.1830230950. [DOI] [PubMed] [Google Scholar]
- Ferrucci L., Fabbri E. Inflammageing: chronic inflammation in ageing, cardiovascular disease, and frailty. Nat. Rev. Cardiol. 2018;15:505–522. doi: 10.1038/s41569-018-0064-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink K., Ng C., Nkenfou C., Vasudevan S.G., van Rooijen N., Schul W. Depletion of macrophages in mice results in higher dengue virus titers and highlights the role of macrophages for virus control. Eur. J. Immunol. 2009;39:2809–2821. doi: 10.1002/eji.200939389. [DOI] [PubMed] [Google Scholar]
- Franceschi C., Garagnani P., Parini P., Giuliani C., Santoro A. Inflammaging: a new immune-metabolic viewpoint for age-related diseases. Nat. Rev. Endocrinol. 2018;14:576–590. doi: 10.1038/s41574-018-0059-4. [DOI] [PubMed] [Google Scholar]
- Fujimoto T., Parton R.G. Not just fat: the structure and function of the lipid droplet. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gervois P., Torra I.P., Fruchart J.C., Staels B. Regulation of lipid and lipoprotein metabolism by PPAR activators. Clin. Chem. Lab. Med. 2000;38:3–11. doi: 10.1515/cclm.2000.002. [DOI] [PubMed] [Google Scholar]
- Han L., Shen W.J., Bittner S., Kraemer F.B., Azhar S. PPARs: regulators of metabolism and as therapeutic targets in cardiovascular disease. Part II: PPAR-beta/delta and PPAR-gamma. Future Cardiol. 2017;13:279–296. doi: 10.2217/fca-2017-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P., Han Z., Couvillon A.D., Kaufman R.J., Exton J.H. Autocrine tumor necrosis factor alpha links endoplasmic reticulum stress to the membrane death receptor pathway through IRE1alpha-mediated NF-kappaB activation and down-regulation of TRAF2 expression. Mol. Cell. Biol. 2006;26:3071–3084. doi: 10.1128/mcb.26.8.3071-3084.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana B.A., Chintamaneni P.K., Krishnamurthy P.T., Wadhwani A., Mohankumar S.K. Cytosolic lipid excess-induced mitochondrial dysfunction is the cause or effect of high fat diet-induced skeletal muscle insulin resistance: a molecular insight. Mol. Biol. Rep. 2019;46:957–963. doi: 10.1007/s11033-018-4551-7. [DOI] [PubMed] [Google Scholar]
- Lee C., Longo V. Dietary restriction with and without caloric restriction for healthy aging. F1000Res. 2016;5 doi: 10.12688/f1000research.7136.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linehan E., Fitzgerald D.C. Ageing and the immune system: focus on macrophages. Eur. J. Microbiol. Immunol. 2015;5:14–24. doi: 10.1556/eujmi-d-14-00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahbub S., Deburghgraeve C.R., Kovacs E.J. Advanced age impairs macrophage polarization. J. Interferon Cytokine Res. 2012;32:18–26. doi: 10.1089/jir.2011.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masternak M.M., Bartke A. PPARs in calorie restricted and genetically long-lived mice. PPAR Res. 2007;2007:28436. doi: 10.1155/2007/28436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattos K.A., D'Avila H., Rodrigues L.S., Oliveira V.G., Sarno E.N., Atella G.C., Pereira G.M., Bozza P.T., Pessolani M.C. Lipid droplet formation in leprosy: toll-like receptor-regulated organelles involved in eicosanoid formation and Mycobacterium leprae pathogenesis. J. Leukoc. Biol. 2010;87:371–384. doi: 10.1189/jlb.0609433. [DOI] [PubMed] [Google Scholar]
- Melo R.C., Weller P.F. Lipid droplets in leukocytes: organelles linked to inflammatory responses. Exp. Cell Res. 2016;340:193–197. doi: 10.1016/j.yexcr.2015.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milanski M., Degasperi G., Coope A., Morari J., Denis R., Cintra D.E., Tsukumo D.M., Anhe G., Amaral M.E., Takahashi H.K. Saturated fatty acids produce an inflammatory response predominantly through the activation of TLR4 signaling in hypothalamus: implications for the pathogenesis of obesity. J. Neurosci. 2009;29:359–370. doi: 10.1523/jneurosci.2760-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namgaladze D., Brüne B. Macrophage fatty acid oxidation and its roles in macrophage polarization and fatty acid-induced inflammation. Biochim. Biophys. Acta. 2016;1861:1796–1807. doi: 10.1016/j.bbalip.2016.09.002. [DOI] [PubMed] [Google Scholar]
- Namgaladze D., Lips S., Leiker T.J., Murphy R.C., Ekroos K., Ferreiros N., Geisslinger G., Brüne B. Inhibition of macrophage fatty acid β-oxidation exacerbates palmitate-induced inflammatory and endoplasmic reticulum stress responses. Diabetologia. 2014;57:1067–1077. doi: 10.1007/s00125-014-3173-4. [DOI] [PubMed] [Google Scholar]
- Oishi Y., Manabe I. Macrophages in age-related chronic inflammatory diseases. NPJ Aging Mech. Dis. 2016;2:16018. doi: 10.1038/npjamd.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann J.A., Carvalho P. Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 2019;20:137–155. doi: 10.1038/s41580-018-0085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S.M., Hadadi E., Dang T.M., Yeap W.H., Tan C.T., Ng T.P., Larbi A., Wong S.C. The pro-inflammatory phenotype of the human non-classical monocyte subset is attributed to senescence. Cell Death Dis. 2018;9:266. doi: 10.1038/s41419-018-0327-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pararasa C., Ikwuobe J., Shigdar S., Boukouvalas A., Nabney I.T., Brown J.E., Devitt A., Bailey C.J., Bennett S.J., Griffiths H.R. Age-associated changes in long-chain fatty acid profile during healthy aging promote pro-inflammatory monocyte polarization via PPARγ. Aging Cell. 2016;15:128–139. doi: 10.1111/acel.12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pence B.D., Yarbro J.R. Aging impairs mitochondrial respiratory capacity in classical monocytes. Exp. Gerontol. 2018;108:112–117. doi: 10.1016/j.exger.2018.04.008. [DOI] [PubMed] [Google Scholar]
- Perlemuter G., Sabile A., Letteron P., Vona G., Topilco A., Chretien Y., Koike K., Pessayre D., Chapman J., Barba G., Brechot C. Hepatitis C virus core protein inhibits microsomal triglyceride transfer protein activity and very low density lipoprotein secretion: a model of viral-related steatosis. FASEB J. 2002;16:185–194. doi: 10.1096/fj.01-0396com. [DOI] [PubMed] [Google Scholar]
- Puchta A., Naidoo A., Verschoor C.P., Loukov D., Thevaranjan N., Mandur T.S., Nguyen P.S., Jordana M., Loeb M., Xing Z. TNF drives monocyte dysfunction with age and results in impaired anti-pneumococcal immunity. PLoS Pathog. 2016;12:e1005368. doi: 10.1371/journal.ppat.1005368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha D.M., Caldas A.P., Oliveira L.L., Bressan J., Hermsdorff H.H. Saturated fatty acids trigger TLR4-mediated inflammatory response. Atherosclerosis. 2016;244:211–215. doi: 10.1016/j.atherosclerosis.2015.11.015. [DOI] [PubMed] [Google Scholar]
- Saare M., Tserel L., Haljasmägi L., Taalberg E., Peet N., Eimre M., Vetik R., Kingo K., Saks K., Tamm R. Monocytes present age-related changes in phospholipid concentration and decreased energy metabolism. Aging Cell. 2020;19:e13127. doi: 10.1111/acel.13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sica A., Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J. Clin. Invest. 2012;122:787–795. doi: 10.1172/jci59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton P.P., Fujita J., Murphy E.M., Naama H.A., Daly J.M. The influence of restricted calorie intake on peritoneal macrophage function. Nutrition. 2001;17:41–45. doi: 10.1016/s0899-9007(00)00502-5. [DOI] [PubMed] [Google Scholar]
- Takemori K., Kimura T., Shirasaka N., Inoue T., Masuno K., Ito H. Food restriction improves glucose and lipid metabolism through Sirt1 expression: a study using a new rat model with obesity and severe hypertension. Life Sci. 2011;88:1088–1094. doi: 10.1016/j.lfs.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Thomas D., Apovian C. Macrophage functions in lean and obese adipose tissue. Metab. Clin. Exp. 2017;72:120–143. doi: 10.1016/j.metabol.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano F., Wang X., Bertolotti A., Zhang Y., Chung P., Harding H.P., Ron D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- van Beek A.A., Van den Bossche J., Mastroberardino P.G., de Winther M.P.J., Leenen P.J.M. Metabolic alterations in aging macrophages: ingredients for inflammaging? Trends Immunology. 2019;40:113–127. doi: 10.1016/j.it.2018.12.007. [DOI] [PubMed] [Google Scholar]
- Wu H., Han Y., Rodriguez Sillke Y., Deng H., Siddiqui S., Treese C., Schmidt F., Friedrich M., Keye J., Wan J. Lipid droplet-dependent fatty acid metabolism controls the immune suppressive phenotype of tumor-associated macrophages. EMBO Mol. Med. 2019;11:e10698. doi: 10.15252/emmm.201910698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the study are included with the published manuscript (and supplemental information). All other data are available from the corresponding author upon request. The RNA sequencing data was uploaded in the Sequence Read Archive (SRA) database: SRR12130588-SRR12130592.