Abstract
The individual physiological response to high-altitude hypoxia involves both genetic and non-genetic factors, including epigenetic modifications. Epigenetic changes in hypoxia factor pathway (HIF) genes are associated with high-altitude acclimatization. However, genome-wide epigenetic changes that are associated with short-term hypoxia exposure remain largely unknown. We collected a series of DNA samples from 15 participants of European ancestry trekking to Everest Base Camp to identify DNA methylation changes associated with incremental altitude ascent. We determined genome-wide DNA methylation levels using the Illumina MethylationEPIC chip comparing two altitudes: baseline 1,400 m (day 0) and elevation 4,240 m (day 7). The results of our epigenome-wide association study revealed 2,873 significant differentially methylated positions (DMPs) and 361 significant differentially methylated regions (DMRs), including significant positions and regions in hypoxia inducible factor (HIF) and the renin–angiotensin system (RAS) pathways. Our pathway enrichment analysis identified 95 significant pathways including regulation of glycolytic process (GO:0006110), regulation of hematopoietic stem cell differentiation (GO:1902036), and regulation of angiogenesis (GO:0045765). Lastly, we identified an association between the ACE gene insertion/deletion (I/D) polymorphism and oxygen saturation, as well as average ACE methylation. These findings shed light on the genes and pathways experiencing the most epigenetic change associated with short-term exposure to hypoxia.
Keywords: epigenetics (DNA methylation), genome-wide DNA methylation analysis, high altitude acclimatization, HIF pathway, hypoxia
Introduction
Altitude acclimatization in humans is characterized by complex physiological responses, which include the cardiovascular, hemopoietic, respiratory, and metabolic systems [for review, see Palmer (2010)]. Each system responds uniquely to low oxygen environments. For example, cardiovascular output increases (i.e., increased heart rate and stroke volume) upon initial altitude exposure and returns to pre-altitude baseline after several days of acclimatization [for review, see Naeije (2010)]. The respiratory system’s response is to initiate hyperventilation. The hypoxic ventilatory response (HVR) is elicited shortly upon exposure to high altitude, with ventilatory acclimatization emerging following 5–7 days of sustained exposure to hypoxia (Powell et al., 1998). Lastly, the hemopoietic response in the form of increased erythrocyte production is evident after several days to weeks of exposure (Rodriguez et al., 2000). Each of these responses facilitates acute acclimatization to the low ambient oxygen tension present at high altitudes, allowing humans to acclimatize to hypoxic conditions.
Epigenetic change is one mechanism through which physiological acclimatization may occur. Epigenetic modifications can affect gene expression and include DNA methylation, histone tail modifications, and short RNA regulation. The most well-studied epigenetic mark is DNA methylation, the addition of a methyl group primarily to cytosine bases. DNA methylation patterns can change upon exposure to various environmental conditions, including exposure to different diets, stress, and toxicants (Dolinoy et al., 2007; Baccarelli et al., 2009; Colacino et al., 2012; Childebayeva et al., 2019b). Previous studies have demonstrated that changes in DNA methylation are associated with exposure to the low oxygen environment of high altitude (Alkorta-Aranburu et al., 2012; Childebayeva et al., 2019a, b, 2020). These studies show that genes in the hypoxia inducible factor (HIF) pathway exhibit changes in DNA methylation associated with high-altitude exposure.
The HIF pathway is the main oxygen sensing pathway that regulates cellular homeostasis in metazoans (Bigham and Lee, 2014). The pathway takes its name after the master transcriptional regulator HIF, a heterodimeric transcription factor that is formed by one of three α-subunits (HIF-1α, HIF-2α, or HIF-3α) and a β-subunit (also known as ARNT). In normoxia, HIF1A is hydroxylated and subsequently degraded by the ubiquitin–proteosome pathway. Under hypoxia, this hydroxylation is inhibited by the lack of oxygen availability, leading to the dimerization of HIF and activation of target genes. HIF is responsible for transducing changes in oxygen tension to changes in gene expression through hypoxia response elements (HREs) (Wang and Semenza, 1995; Kaelin and Ratcliffe, 2008; Semenza, 2012). The renin–angiotensin system or RAS is a second pathway that is involved in the response to hypoxia. It is one of the body’s most important regulators of blood pressure and inflammation (Muller et al., 1997; Rupert et al., 2003). The RAS protein, angiotensin converting enzyme (ACE), is a central peptide in blood-pressure regulation responsible for converting angiotensin-I to the vasoconstrictor, angiotensin-II. An insertion/deletion (I/D) polymorphism in ACE is associated with physical performance at high altitude (Woods et al., 2002; Tsianos et al., 2005). The I-allele has been associated with higher levels of submaximal oxygen saturation (SaO2) among Andean Quechua (Bigham et al., 2008), and in trekkers of European ancestry (Woods et al., 2002).
Previous research by our group has shown that acclimatization to hypoxia is associated with DNA methylation changes in HIF pathway genes including EPAS1, EPO, PPARa, and RXRA (Childebayeva et al., 2019a). However, it is not well understood what other genes and pathways display DNA methylation changes upon exposure to hypoxia. To understand how acclimatization to hypoxia affects genome-wide DNA methylation patterns, we performed an epigenome-wide association study in individuals trekking to Everest Base Camp. Our analysis compared baseline methylation measured in Kathmandu, Nepal at 1,400 m (day 0) with methylation measured at a high-altitude location, Pheriche, Nepal at 4,240 m (day 7 of the trek).
Materials and Methods
Ethics Statement
Ethical approval was received from the Syracuse University Institutional Review Board (Protocol 18-006) and the University of Michigan Institutional Review Board (HUM00141118). The study abided by the Canadian Government Tri-Council policy on research ethics with human participants (T2) and the Declaration of Helsinki, except for registration in a database. Ethical approval was received also from the Mount Royal University Human Research Ethics Board (Protocol 100012 and 101361) and harmonized with the Nepal Health Research Council (Protocol 109-2017).
Study Design and Sample Collection
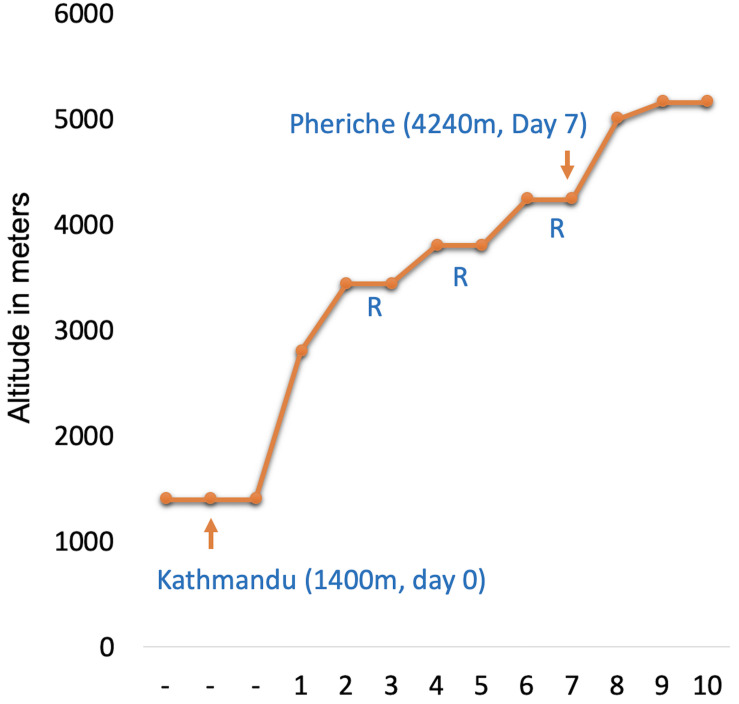
Thirty-two samples (16 samples at 1,400 m and 16 samples at 4,240 m) corresponding to 16 unique individuals were selected from a larger participant cohort from the research expedition to Everest Base Camp in the Nepal Himalaya (Childebayeva et al., 2019a). Briefly, study participants and researchers flew from Kathmandu (baseline) to Lukla from where the research group trekked for 10 days from 2,800 to 5,160 m (Figure 1). In the morning between 06:00 and 08:00 local time at 1,400 m (Kathmandu; day 0) and 4,240 m (Pheriche; day 7), saliva samples for DNA and physiological measures were taken following one night of sleep at each altitude. Physiological measurements included hemoglobin concentration [Hb], patient end-tidal carbon dioxide (PETCO2), a measure of CO2 partial pressure in expired air, which reflects the CO2 level in the arterial blood, and peripheral oxygen saturation (SaO2). Detailed information on phenotype collection and sampling is provided in Childebayeva et al. (2019a). All participants were healthy, non-pregnant, non-lactating, non-smokers between 19 and 41 years of age. All participants were of self-reported European ancestry and had at least 1 year since their last altitude experience. Participant characteristics can be found in Table 1.
FIGURE 1.
Ascent profile with sample collection altitudes indicated with arrows and labels. Study participants flew from baseline (day 0) Kathmandu to 2,800 m to begin the trek. Three non-trekking rest days are indicated by “R.” Epigenome wide association study was performed on matched samples collected at 1,400 m (day 0) and 4,240 m (day 7).
TABLE 1.
Participant characteristics.
| 1,400 m (day 0) | 4,240 m (day 7) | |
| Hemoglobin (mg/L)* | 13.1 (1.8) | 14.8 (1.4) |
| BMI (kg/m2)# | 22.6 (2.4) | 22.4 (2.2) |
| PETCO2 (Torr)** | 30.7 (3.2) | 22.1 (2.9) |
| SaO2 (%)** | 97.1 (1.1) | 89.8 (2.4) |
| % Female | 53% | |
| Age, year | 23.6 (6.0) | |
Data are means (SD). Significance symbols denote the difference between Kathmandu baseline and each altitude. #p-value < 0.10; *p-value < 0.01; and **p-value < 0.001.
Phenotype Testing
We performed linear mixed models using the R package lmerTest to test for significant differences in phenotypes between Kathmandu and Pheriche (Table 1). The following model was tested: Phenotype ∼ Altitude + Sex + Age + (1 | ID).
DNA Methylation
We generated DNA methylation data for ∼850,000 CpG sites using the Illumina Infinium® MethylationEPIC BeadChip assay for 32 samples in our study. We used the EZ-96 DNA MethylationTM Kit (Zymo Research, Irvine, CA, United States) to bisulfite convert each DNA sample following the standard protocol with alternative incubation conditions optimized for the Illumina Infinium® MethylationEPIC BeadChip assay. We used R for data processing and analysis implementing the packages minfi, ChAMP, and SmartSVA (Aryee et al., 2014; Morris et al., 2014; Chen et al., 2017). Based on QC metrics, two samples from the same participant failed and were excluded from all analyses; thus, the final sample size was n = 30 (15 at 1,400 m and 15 at 4,240 m).
Data normalization was performed using the funnorm normalization function in minfi (Aryee et al., 2014). We removed all probes that were above the 10e5 detection p-value threshold (N = 8,126) in more than 5% of the samples, all cross-reactive probes, probes associated with sex chromosomes, probes containing SNPs with MAF > 5% at target CpG sites, single base extension sites of type I probes, and in the body of the probe (Chen et al., 2013). All analyses were performed with N = 657,569 sites after normalization and probe removal. Samples were tested for batch effects using singular value decomposition (SVD) analysis in champ. SmartSVA (Chen et al., 2017) was used to perform a surrogate variable test, and the surrogate variable 1 was used for correcting for any saliva cell type differences associated with altitude. SmartSVA is a surrogate variable analysis method that can be used for reference-free adjustment for cell mixtures (Chen et al., 2017).
Differential Methylation Testing
Fully processed M-values were tested for differential methylation using the package limma (Ritchie et al., 2015). The following model was used to test for the differentially methylated positions (DMPs): DNA methylation ∼ Sample ID + Altitude + Surrogate Variable 1 (from smartSVA). P-values were adjusted for multiple testing using the false-discovery rate (FDR) following the Benjamini–Hochberg procedure (Hochberg and Benjamini, 1990) to produce FDR-corrected q-values. Differentially methylated regions (DMRs) were determined using DMRcate with default parameters (lambda = 1,000, C = 2, min.CpG sites = 2) (Peters et al., 2015). Pathway enrichment was performed using the package methylGSA (Ren and Kuan, 2019).
Angiotensin converting enzyme genotyping was performed using the same protocol as in Bigham et al. (2008). We extracted ACE CpG sites from the MethylationEPIC array to assess its methylation status independent from the epigenome-wide association analysis. We tested the relationship between ACE I/D status and SaO2 separately for Kathmandu and Pheriche using linear modeling and adjusting for age and sex. The relationship between ACE genotype and phenotypes, as well as ACE genotype and ACE DNA methylation, was tested using linear mixed modeling in R using the lmerTest package (Kuznetsova et al., 2017). The linear mixed model was adjusted for altitude, age, sex, and individual IDs. Plotting was performed using the ggplot2 package (Wickham, 2009).
Results
Participant Demographics
Our study group included n = 15 participants of self-reported European ancestry, with 53% females and the average BMI of 22.60 (SD 2.36) at baseline. Participant characteristics can be found in Table 1.
Physiological Changes With Altitude Exposure
We detected significant physiological changes between altitude 1,400 m (day 0) and 4,240 m (day 7) (henceforth physiological variables are referred to as phenotypes in this manuscript) in arterial oxygen saturation (SaO2), hemoglobin concentration [(Hb)], and end-tidal carbon dioxide partial pressure (PETCO2) (Table 1). Briefly, we observed a significant increase in [Hb] and a significant decrease in SaO2 and PETCO2 with increasing altitude. The physiological responses we have reported are expected at high altitude, i.e., lower arterial oxygen saturation due to decreased ambient PO2, a decrease in PETCO2 indicating an increase in alveolar ventilation, and higher [Hb], reflecting the body’s physiological response to low-oxygen conditions by increasing hemoglobin production.
Differential Methylation Analysis
We generated DNA methylation data for ∼850,000 CpG sites using the Illumina Infinium® MethylationEPIC BeadChip. After QC, we performed differential methylation analysis on 755,636 probes. We identified 2,873 DMPs at q-value < 0.10 (Supplementary Table 1) that differed between baseline 1,400 and 4,240 m genome-wide inflation factor λ = 1.2. Among these, we identified HIF pathway genes: ANGPT1, CREBBP, CUL2, HIF1A, HK1, HMOX1, PDK1 (two significant CpG sites), PIK3R3, PLCG1, PRKCG, RELA, and STAT3, and RAS pathway genes: ABL1, ANGPT1, EFNA3, FGFR1, GAB1, GNB1, GNB3, GNB4, GRB2, KITLG, KRAS, MAPK10, PAK1, PAK2, PDGFA, PIK3R3, PLCG1, PRKCG, PTPN11, RALA, RAP1A, RAP1B, RASA3 (five significant CpG sites), RASSF1, RELA, RGL2, and RIN1 (Table 2). We also identified genes associated with inflammation: IL12B, TRIM31, NLRP3, IL1RAP, among others, and genes associated with cognitive function: ASH1L and TNIK.
TABLE 2.
Significant CpG sites associated with HIF and RAS pathways.
| Pathway | Gene | CpG | p-value | q-value | Chr | Position (hg19) | Relation to island |
| HIF | ANGPT1 | cg09443479 | 1.96E-04 | 0.08 | 8 | 108,511,174 | OpenSea |
| CREBBP | cg16560077 | 7.48E-05 | 0.05 | 16 | 3,781,408 | Island | |
| CUL2 | cg09080721 | 1.76E-04 | 0.07 | 10 | 35,361,575 | OpenSea | |
| HIF1A | cg16788202 | 2.45E-04 | 0.08 | 14 | 62,162,340 | Island | |
| HK1 | cg06506461 | 3.14E-04 | 0.09 | 10 | 71,112,319 | OpenSea | |
| HMOX1 | cg15724965 | 1.87E-05 | 0.03 | 22 | 35,777,001 | Island | |
| PDK1 | cg13462525 | 7.98E-05 | 0.05 | 2 | 173,420,046 | N_Shore | |
| PDK1 | cg11703569 | 4.63E-05 | 0.04 | 2 | 173,421,320 | Island | |
| PIK3R3 | cg12800095 | 9.33E-05 | 0.06 | 1 | 46,594,087 | OpenSea | |
| PLCG1 | cg13312309 | 5.68E-05 | 0.05 | 20 | 39,799,964 | OpenSea | |
| PRKCG | cg14975881 | 3.28E-05 | 0.04 | 19 | 54,389,945 | N_Shelf | |
| RELA | cg04962756 | 2.35E-06 | 0.01 | 11 | 65,425,928 | OpenSea | |
| STAT3 | cg09804439 | 1.59E-04 | 0.07 | 17 | 40,540,457 | Island | |
| RAS | ABL1 | cg13609937 | 4.40E-05 | 0.04 | 9 | 133,588,314 | Island |
| ANGPT1 | cg09443479 | 1.96E-04 | 0.08 | 8 | 108,511,174 | OpenSea | |
| EFNA3 | cg06058618 | 6.65E-06 | 0.02 | 1 | 155,057,452 | Island | |
| FGFR1 | cg00676030 | 1.74E-05 | 0.03 | 8 | 38,307,962 | OpenSea | |
| GAB1 | cg24244452 | 3.54E-04 | 0.09 | 4 | 144,284,260 | OpenSea | |
| GNB1 | cg14953148 | 1.56E-04 | 0.07 | 1 | 1,792,846 | OpenSea | |
| GNB3 | cg06444189 | 2.38E-05 | 0.03 | 12 | 6,953,740 | OpenSea | |
| GNB4 | cg12872693 | 4.08E-04 | 0.10 | 3 | 179,168,798 | Island | |
| GRB2 | cg11495544 | 3.76E-04 | 0.10 | 17 | 73,402,155 | S_Shore | |
| KITLG | cg22688836 | 6.67E-05 | 0.05 | 12 | 88,967,594 | OpenSea | |
| KRAS | cg02850821 | 8.03E-06 | 0.02 | 12 | 25,403,680 | OpenSea | |
| MAPK10 | cg03886687 | 2.43E-04 | 0.08 | 4 | 87,281,409 | OpenSea | |
| PAK1 | cg26996201 | 2.86E-04 | 0.09 | 11 | 77,122,864 | Island | |
| PAK2 | cg02319016 | 1.34E-06 | 0.01 | 3 | 196,469,777 | S_Shelf | |
| PDGFA | cg22466784 | 2.41E-04 | 0.08 | 7 | 540,176 | OpenSea | |
| PIK3R3 | cg12800095 | 9.33E-05 | 0.06 | 1 | 46,594,087 | OpenSea | |
| PLCG1 | cg13312309 | 5.68E-05 | 0.05 | 20 | 39,799,964 | OpenSea | |
| PRKCG | cg14975881 | 3.28E-05 | 0.04 | 19 | 54,389,945 | N_Shelf | |
| PTPN11 | cg16207631 | 2.76E-04 | 0.09 | 12 | 112,856,603 | Island | |
| RALA | cg19104112 | 2.75E-04 | 0.09 | 7 | 39,663,043 | Island | |
| RAP1A | cg25355888 | 2.83E-04 | 0.09 | 1 | 112,162,642 | Island | |
| RAP1B | cg00758412 | 2.23E-04 | 0.08 | 12 | 69,033,023 | OpenSea | |
| RASA3 | cg21364828 | 1.18E-04 | 0.06 | 13 | 114,825,608 | OpenSea | |
| RASA3 | cg13818243 | 2.76E-04 | 0.09 | 13 | 114,789,734 | S_Shelf | |
| RASA3 | cg04421280 | 1.20E-04 | 0.06 | 13 | 114,898,225 | Island | |
| RASA3 | cg00427150 | 1.92E-04 | 0.07 | 13 | 114,770,568 | N_Shelf | |
| RASA3 | cg20028528 | 2.20E-04 | 0.08 | 13 | 114,812,184 | N_Shore | |
| RASSF1 | cg25486143 | 3.20E-04 | 0.09 | 3 | 50,378,527 | Island | |
| RELA | cg04962756 | 2.35E-06 | 0.01 | 11 | 65,425,928 | OpenSea | |
| RGL2 | cg08312215 | 4.75E-05 | 0.04 | 6 | 33,266,943 | Island | |
| RIN1 | cg15082918 | 2.81E-04 | 0.09 | 11 | 66,104,153 | S_Shore |
In order to detect biological pathways overrepresented among the significant CpG sites from the analysis of differential methylation, we performed a pathway enrichment analysis using the methylgometh function in the R package methylGSA (Ren and Kuan, 2019). Ninety-five significant pathways were identified by methylgometh including the GO pathways regulation of glycolytic process (GO:0006110), regulation of hematopoietic stem cell differentiation (GO:1902036), and regulation of angiogenesis (GO:0045765) (Supplementary Table 2). Other pathways of interest included brain development (GO:0007420), negative regulation of neuron differentiation (GO:0045665), and interleukin-1-mediated signaling pathway (GO:0070498).
We then tested for DMRs, i.e., contiguous regions in the genome that show differential methylation between phenotypes or groups. We used DMRcate (Peters et al., 2015) to find DMRs between low- (1,400 m) and high-altitude (4,240 m) samples. Using this approach, we identified 361 significant DMRs out of 657,408 possible DMRs (Supplementary Table 3). These included DMRs near/in genes associated with the HIF pathway: HIF1A and ENO1 (glycolytic enzyme), and the RAS pathway: ABL1, FGFR3, KRAS, RASA3, and RGL2.
Phenotype Associations
To determine if changes in DNA methylation could be driving acclimatization, we performed association testing between significant genome-wide methylation positions and phenotypes associated with high-altitude acclimatization. To do so, we focused our analysis on significant CpG sites identified in the DMP analysis (N = 2,873) and phenotypes that were significantly different between the groups (Table 1) including SaO2, [Hb], and PETCO2. Two CpG sites, cg16546681 (chr1:155244518, q-value = 0.01, β regression coefficient = 6.46) in the gene CLK2 and cg14548038 (chr9:140178418, q-value = 0.03, β regression coefficient = 4.73) upstream of the gene TOR4A, were significantly positively associated with SaO2 (%). No significant associations were identified for [Hb] or PETCO2 after correcting for multiple comparisons.
ACE I/D, Oxygen Saturation, and DNA Methylation
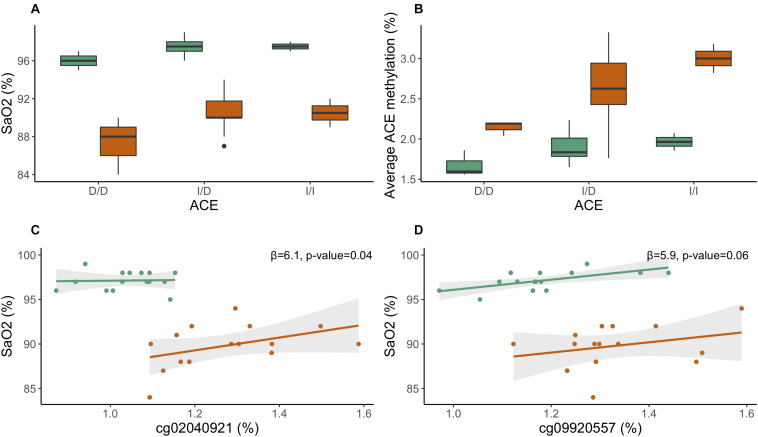
We tested the relationship between ACE, a gene associated with high-altitude performance, and high-altitude phenotypes [SaO2, PETCO2, (Hb)]. Individuals in this study were genotyped for the ACE I/D (rs4646994) polymorphism. We performed a genotypic test, wherein I/I and I/D genotypes were compared to D/D genotype, and identified a significant association between ACE genotype and SaO2. Individuals with genotypes I/D (β regression coefficient = 1.69, p-value < 0.01) and I/I (β regression coefficient = 1.85, p-value < 0.05) had significantly higher SaO2 than individuals with the D/D genotype at 1,400 m (Kathmandu); the relationship was not significant for 4,240 m (Pheriche) (Figure 2). In an additive model, the I-allele was associated with increased SaO2 (β regression coefficient = 1.03, p-value < 0.05) at 1,400 m; the relationship was also not significant for 4,240 m. In a dominant model, individuals who were either heterozygotes or homozygotes for the I-allele (grouped together) displayed higher SaO2 (β regression coefficient = 1.71, p-value < 0.01) at 1,400 m; the relationship was approaching significance (β regression coefficient = 2.79, p-value = 0.09) for 4,240 m. Our results suggest that the dominant model, wherein individuals carrying either the I/D or I/I alleles have higher oxygen saturation than individuals carrying the D/D allele, is best suited to explain the relationship between SaO2 ACE I/D in our study.
FIGURE 2.
Relationship between ACE I/D, ACE methylation, and SaO2. (A) Boxplot of arterial oxygen saturation by ACE I/D genotype. (B) Boxplot of average ACE DNA methylation by ACE I/D genotypes. (C) ACE CpG site cg02040921 plotted against SaO2. (D) ACE CpG site cg09920557 plotted against SaO2. Day 0: 1,400 m (K) is indicated in green and day 7: 4,240 m (P) is noted in orange.
Average ACE methylation was positively associated with the I-allele when we tested the relationship using an additive model (β regression coefficient = 0.31, p-value = 0.03) (Figure 2B). We also tested the relationship between individual ACE CpG sites and high-altitude phenotypes. ACE CpG sites, cg02040921 and cg09920557, were associated with SaO2 (cg02040921: p-value = 0.04; cg09920557: p-value = 0.06). Increased methylation of ACE CpGs was associated with increased SaO2 (Figures 2C,D). No significant associations were identified for [Hb] or PETCO2.
Discussion
The role of epigenetic change, including DNA methylation, in acclimatization to short-term hypoxia exposure is not well characterized. We aimed to fill this gap using genome-wide DNA methylation data from the same individuals measured at different altitudes during a trek to Everest Base Camp. We identified significant associations between genome-wide DNA methylation and short-term altitude exposure, among which were CpG sites and regions associated with HIF pathway, including HIF1A, and RAS pathway genes.
We identified both a significant CpG position (DMP) and a DMR associated with hypoxia inducible factor 1A or HIF1A, which is a central gene in the body’s hypoxic response (Slemc and Kunej, 2016). In normoxia, HIF1A is degraded via ubiquitination but is allowed to accumulate in hypoxic conditions. This allows its protein product to bind to a constitutively expressed HIF1B forming a heterodimer that activates downstream genes (Wenger, 2002). HIF1A activity is under epigenetic control in human cancer cells and hematopoietic cell lines (Walczak-Drzewiecka et al., 2010; Nguyen et al., 2013; Cimmino et al., 2019). Importantly, the HIF1A-associated DMR identified here overlaps with the promoter region of the gene, suggesting that methylation at this locus may be associated with changes in gene expression.
We found significant CpG sites associated with the RAS pathway, including ones in the genes ANGPT1 and RASA3 (RAS P21 protein activator 3). Angiopoietins 1 and 2 are regulated by HIF1, and ANGPT1 expression is associated with increased number of vessels without excessive permeability (Kelly et al., 2003). ANGPT1 can be activated and repressed by HIF1 in a cell-specific manner (Kelly et al., 2003). RASA3 (RAS P21 protein activator 3) is a Ras-GTPase activating protein that causes anemia and thrombocytopenia in mice when mutated (Blanc et al., 2012). RAS pathway is another canonical hypoxia-induced pathway. RAS has been linked to blood pressure (Fontes et al., 1994), cardiovascular disease (Lee et al., 1993), and primary hypertension (Frossard et al., 1998). The role of RAS in hypoxia has been explored in association with high-altitude pulmonary edema via the regulation of the pulmonary vascular tone (Stobdan et al., 2011).
We also found significant DNA methylation changes in genes outside of canonical pathways implicated in high-altitude acclimatization (i.e., HIF and RAS), including significant DNA methylation changes in genes associated with cognitive impairment [ASH1L (de Ligt et al., 2012; Crawley et al., 2016; Xi et al., 2020) and TNIK (Coba et al., 2012; Anazi et al., 2016)]. Cognitive decline is a common side effect of high-altitude hypoxia (Regard et al., 1989; Yan, 2014; Gao et al., 2015) that becomes apparent 1–2 weeks after initial exposure (Bolmont et al., 2000) and may improve to some degree upon acclimatization (Heinrich et al., 2019). This timing of the cognitive decline is consistent with our study design wherein we identified methylation changes in ASH1L and TNIK after 1 week of high-altitude exposure. In addition to methylation differences in genes associated with cognitive function, we also identified changes in several genes associated with inflammation. These included CpG sites in the genes IL12B (Glas et al., 2012; Liu et al., 2012) and TRIM31 (Song et al., 2016; Wang et al., 2018).
We specifically focused on the RAS pathway gene ACE as it is centrally involved in circulatory homeostasis, and the ACE I/D polymorphism has been linked to endurance performance (Myerson et al., 1999), adaptation of highland resident/native populations (Qadar Pasha et al., 2001; Bigham et al., 2008), and performance at altitude (all those other citations). The ACE I allele is associated with lower ACE activity (Costerousse et al., 1993) and higher SaO2 (Woods et al., 2002), potentially as a result of an increased HVR (Patel et al., 2003). We identified an association between ACE genotypes I/D and I/I with higher SaO2, which is consistent with previous research showing a significant relationship between ACE and SaO2 (Woods et al., 2002; Bigham et al., 2008).
We found that the ACE I-allele was associated with higher average ACE methylation, which has been shown before in a study of birth weight and ACE (Rangel et al., 2014). Notably, the ACE I-allele is associated with lower serum and tissue ACE activity (Rigat et al., 1990; Costerousse et al., 1993; Woods et al., 2000). Since methylation is commonly associated with gene silencing, the association between ACE I-allele and higher DNA methylation suggests that ACE methylation may be involved in mediating decreased ACE expression in individuals with the I-allele.
Individuals at high altitude displayed increased [Hb] and decreased SaO2 and PETCO2 compared to low altitude. We found two CpG sites, in the gene CLK2 and near the gene TOR4, that were associated with SaO2. CDC like kinase 2 or CLK2 suppresses PPARGC1A transcriptional activity on gluconeogenic genes (Sahu et al., 2019) and thus downregulates hepatic gluconeogenesis and glucose output. We found CLK2 methylation to be positively associated with SaO2, suggesting that CLK2 expression is potentially decreased in hypoxic conditions, given methylation is linked to gene repression. Interestingly, the CpG site in CLK2 is upstream of the gene PKLR that is significantly differentially methylated in high- compared to low-altitude Quechua (Childebayeva et al., 2020). We also found a CpG site upstream of TOR4A (Torsin family 4 member A), which is associated with dystonia (Cascalho et al., 2017). Dystonia is linked to hypoxic exposure, more specifically cerebral anoxia/hypoxia (Kuoppamaki et al., 2002; Kern et al., 2016), and our finding might indicate a potential epigenetic mechanism playing role in the development of this condition.
Tissue types can show different methylation profiles across the body, and the degree to which they correlate varies by study design, type of sample, or age (Langie et al., 2017). For example, there is evidence of a low correlation between salivary and blood global DNA methylation (Godderis et al., 2015). Here, we analyzed saliva. Saliva is an attractive tissue for the analysis of DNA methylation in field studies given its relative ease of collection compared to blood or other tissues (Langie et al., 2017). By focusing on a singular tissue type, our results may be restricted to salivary tissue alone. However, salivary DNA methylation patterns have been shown to correlate with DNA methylation from blood (Thompson et al., 2013; Langie et al., 2016), intestinal mucosa (Hearn et al., 2019), and the brain (Smith et al., 2015). Furthermore, saliva panels have shown proteomic changes upon hypoxic exposure in cell cultures (Jain et al., 2020), suggesting the relevance of this tissue for analyzing the overall hypoxic response. Therefore, we suggest that our analysis of saliva is an important first step in identifying DNA methylation changes to acute hypoxia that may be relevant to other bodily tissues.
Overall, our data demonstrate that various pathways and systems are affected by exposure to high altitude, including the HIF pathway, RAS pathway, cognitive performance, and inflammatory systems. Moreover, we identified a significant association between SaO2 and ACE I/D, and associations between ACE I/D and ACE methylation, further highlighting the connection between ACE and SaO2 as well as the role of ACE in altitude acclimatization.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author AC, upon request.
Ethics Statement
The studies involving human participants were reviewed and approved by Syracuse University Institutional Review Board (Protocol 18-006), University of Michigan Institutional Review Board (HUM00141118), Mount Royal University Human Research Ethics Board (Protocol 100012 and 101361), and Nepal Health Research Council (Protocol 109-2017). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
AC, TB, and AB: conceptualization. AC, TD, and JW: data curation. AC: formal analysis. TD and AC: funding acquisition. AC and TH: investigation. AC and AB: writing—original draft preparation. AB, AC, TB, TD, and TH: writing—review and editing. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to acknowledge our study participants from Syracuse University in Syracuse, NY, Mount Royal University in Calgary, Canada, and the MidSweden University in Sweden. This work would have not been possible without our Sherpa guides in Nepal: Nima Sherpa, Tsering Sherpa, Tashi Jangbu, and Fura Tashi Chuserwa, and their porter team.
Footnotes
Funding. AC was supported by the Marshall Weinberg Award (University of Michigan) and the National Geographic Early Career Award (EC-50834R-18). TD was supported by a Natural Sciences and Engineering Research Council of Canada Discovery grant (RGPIN-2016-04915).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2021.660906/full#supplementary-material
References
- Alkorta-Aranburu G., Beall C. M., Witonsky D. B., Gebremedhin A., Pritchard J. K., Di Rienzo A. (2012). The genetic architecture of adaptations to high altitude in ethiopia. PLoS Genet. 8:e1003110. 10.1371/journal.pgen.1003110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anazi S., Shamseldin H. E., AlNaqeb D., Abouelhoda M., Monies D., Salih M. A., et al. (2016). A null mutation in tnik defines a novel locus for intellectual disability. Hum. Genet. 135 773–778. 10.1007/s00439-016-1671-9 [DOI] [PubMed] [Google Scholar]
- Aryee M. J., Jaffe A. E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A. P., Hansen K. D., et al. (2014). Minfi: a flexible and comprehensive bioconductor package for the analysis of infinium DNA methylation microarrays. Bioinformatics 30 1363–1369. 10.1093/bioinformatics/btu049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A., Wright R. O., Bollati V., Tarantini L., Litonjua A. A., Suh H. H., et al. (2009). Rapid DNA methylation changes after exposure to traffic particles. Am. J. Respir. Crit. Care Med. 179 572–578. 10.1164/rccm.200807-1097oc [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigham A. W., Kiyamu M., Leon-Velarde F., Parra E. J., Rivera-Ch M., Shriver M. D., et al. (2008). Angiotensin-converting enzyme genotype and arterial oxygen saturation at high altitude in peruvian quechua. High Alt. Med. Biol. 9 167–178. 10.1089/ham.2007.1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigham A. W., Lee F. S. (2014). Human high-altitude adaptation: forward genetics meets the hif pathway. Genes Dev. 28 2189–2204. 10.1101/gad.250167.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc L., Ciciotte S. L., Gwynn B., Hildick-Smith G. J., Pierce E. L., Soltis K. A., et al. (2012). Critical function for the ras-gtpase activating protein rasa3 in vertebrate erythropoiesis and megakaryopoiesis. Proc. Natl. Acad. Sci. U. S. A. 109 12099–12104. 10.1073/pnas.1204948109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolmont B., Thullier F., Abraini J. H. (2000). Relationships between mood states and performances in reaction time, psychomotor ability, and mental efficiency during a 31-day gradual decompression in a hypobaric chamber from sea level to 8848 m equivalent altitude. Physiol. Behav. 71 469–476. 10.1016/s0031-9384(00)00362-0 [DOI] [PubMed] [Google Scholar]
- Cascalho A., Jacquemyn J., Goodchild R. E. (2017). Membrane defects and genetic redundancy: are we at a turning point for dyt1 dystonia? Mov. Disord. 32 371–381. 10.1002/mds.26880 [DOI] [PubMed] [Google Scholar]
- Chen J., Behnam E., Huang J., Moffatt M. F., Schaid D. J., Liang L., et al. (2017). Fast and robust adjustment of cell mixtures in epigenome-wide association studies with smartsva. BMC Genomics 18:413. 10.1186/s12864-017-3808-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. A., Lemire M., Choufani S., Butcher D. T., Grafodatskaya D., Zanke B. W., et al. (2013). Discovery of cross-reactive probes and polymorphic cpgs in the illumina infinium humanmethylation450 microarray. Epigenetics 8 203–209. 10.4161/epi.23470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childebayeva A., Goodrich J. M., Leon-Velarde F., Rivera-Chira M., Kiyamu M., Brutsaert T. D., et al. (2020). Genome-wide epigenetic signatures of adaptive developmental plasticity in the andes. Genome Biol. Evol. 13:evaa239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childebayeva A., Harman T., Weinstein J., Goodrich J. M., Dolinoy D. C., Day T. A., et al. (2019a). DNA methylation changes are associated with an incremental ascent to high altitude. Front. Genet. 10:1062. 10.3389/fgene.2019.01062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childebayeva A., Jones T. R., Goodrich J. M., Leon-Velarde F., Rivera-Chira M., Kiyamu M., et al. (2019b). Line-1 and epas1 DNA methylation associations with high-altitude exposure. Epigenetics 14 1–15. 10.1080/15592294.2018.1561117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino F., Avitabile M., Lasorsa V. A., Montella A., Pezone L., Cantalupo S., et al. (2019). Hif-1 transcription activity: Hif1a driven response in normoxia and in hypoxia. BMC Med. Genet. 20:37. 10.1186/s12881-019-0767-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coba M. P., Komiyama N. H., Nithianantharajah J., Kopanitsa M. V., Indersmitten T., Skene N. G., et al. (2012). Tnik is required for postsynaptic and nuclear signaling pathways and cognitive function. J. Neurosci. 32 13987–13999. 10.1523/jneurosci.2433-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colacino J. A., Arthur A. E., Dolinoy D. C., Sartor M. A., Duffy S. A., Chepeha D. B., et al. (2012). Pretreatment dietary intake is associated with tumor suppressor DNA methylation in head and neck squamous cell carcinomas. Epigenetics 7 883–891. 10.4161/epi.21038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerousse O., Allegrini J., Lopez M., Alhenc-Gelas F. (1993). Angiotensin i-converting enzyme in human circulating mononuclear cells: genetic polymorphism of expression in t-lymphocytes. Biochem. J. 290 33–40. 10.1042/bj2900033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley J. N., Heyer W. D., LaSalle J. M. (2016). Autism and cancer share risk genes, pathways, and drug targets. Trends Genet. 32 139–146. 10.1016/j.tig.2016.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ligt J., Willemsen M. H., van Bon B. W., Kleefstra T., Yntema H. G., Kroes T., et al. (2012). Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl. J. Med. 367 1921–1929. [DOI] [PubMed] [Google Scholar]
- Dolinoy D. C., Huang D., Jirtle R. L. (2007). Maternal nutrient supplementation counteracts bisphenol a-induced DNA hypomethylation in early development. Proc. Natl. Acad. Sci. U. S. A. 104 13056–13061. 10.1073/pnas.0703739104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontes M. A. P., Silva L. C. S., Campagnole-Santos M. J., Khosla M. C., Guertzenstein P. G., Santos R. A. S. (1994). Evidence that angiotensin-(1–7) plays a role in the central control of blood pressure at the ventro-lateral medulla acting through specific receptors. Brain Res. 665 175–180. 10.1016/0006-8993(94)91171-1 [DOI] [PubMed] [Google Scholar]
- Frossard P. M., Lestringant G. G., Elshahat Y. I., John A., Obineche E. N. (1998). An mboi two-allele polymorphism may implicate the human renin gene in primary hypertension. Hypertens. Res. 21 221–225. 10.1291/hypres.21.221 [DOI] [PubMed] [Google Scholar]
- Gao Y. X., Li P., Jiang C. H., Liu C., Chen Y., Chen L., et al. (2015). Psychological and cognitive impairment of long-term migrators to high altitudes and the relationship to physiological and biochemical changes. Eur. J. Neurol. 22 1363–1369. 10.1111/ene.12507 [DOI] [PubMed] [Google Scholar]
- Glas J., Seiderer J., Wagner J., Olszak T., Fries C., Tillack C., et al. (2012). Analysis of il12b gene variants in inflammatory bowel disease. PLoS One 7:e34349. 10.1371/journal.pone.0034349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godderis L., Schouteden C., Tabish A., Poels K., Hoet P., Baccarelli A. A., et al. (2015). Global methylation and hydroxymethylation in DNA from blood and saliva in healthy volunteers. Biomed Res. Int. 2015:845041. 10.1155/2015/845041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearn N. L., Coleman A. S., Ho V., Chiu C. L., Lind J. M. (2019). Comparing DNA methylation profiles in saliva and intestinal mucosa. BMC Genomics 20:163. 10.1186/s12864-019-5553-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich E. C., Djokic M. A., Gilbertson D., DeYoung P. N., Bosompra N.-O., Wu L., et al. (2019). Cognitive function and mood at high altitude following acclimatization and use of supplemental oxygen and adaptive servoventilation sleep treatments. PLoS One 14:e0217089. 10.1371/journal.pone.0217089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg Y., Benjamini Y. (1990). More powerful procedures for multiple significance testing. Stat. Med. 9 811–818. 10.1002/sim.4780090710 [DOI] [PubMed] [Google Scholar]
- Jain S., Paul S., Meena R. N., Gangwar A., Panjwani U., Ahmad Y., et al. (2020). Saliva panel of protein candidates: a comprehensive study for assessing high altitude acclimatization. Nitric Oxide 95 1–11. 10.1016/j.niox.2019.11.007 [DOI] [PubMed] [Google Scholar]
- Kaelin W. G., Jr., Ratcliffe P. J. (2008). Oxygen sensing by metazoans: the central role of the hif hydroxylase pathway. Mol. Cell 30 393–402. 10.1016/j.molcel.2008.04.009 [DOI] [PubMed] [Google Scholar]
- Kelly B. D., Hackett S. F., Hirota K., Oshima Y., Cai Z., Berg-Dixon S., et al. (2003). Cell type-specific regulation of angiogenic growth factor gene expression and induction of angiogenesis in nonischemic tissue by a constitutively active form of hypoxia-inducible factor 1. Circ. Res. 93 1074–1081. 10.1161/01.res.0000102937.50486.1b [DOI] [PubMed] [Google Scholar]
- Kern J., Bodek D., Niazi O. T., Maher J. (2016). Refractory case of paroxysmal autonomic instability with dystonia syndrome secondary to hypoxia. Chest 149 e39–e40. [DOI] [PubMed] [Google Scholar]
- Kuoppamaki M., Bhatia K. P., Quinn N. (2002). Progressive delayed-onset dystonia after cerebral anoxic insult in adults. Mov. Disord. 17 1345–1349. 10.1002/mds.10260 [DOI] [PubMed] [Google Scholar]
- Kuznetsova A., Brockhoff P. B., Christensen R. H. B. (2017). Lmertest package: tests in linear mixed effects models. J. Stat. Softw. 82 1–26. [Google Scholar]
- Langie S. A., Szarc Vel Szic K., Declerck K., Traen S., Koppen G., Van Camp G., et al. (2016). Whole-genome saliva and blood DNA methylation profiling in individuals with a respiratory allergy. PLoS One 11:e0151109. 10.1371/journal.pone.0151109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langie S. A. S., Moisse M., Declerck K., Koppen G., Godderis L., Vanden Berghe W., et al. (2017). Salivary DNA methylation profiling: aspects to consider for biomarker identification. Basic Clin. Pharmacol. Toxicol. 121 93–101. 10.1111/bcpt.12721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. A., Böhm M., Paul M., Ganten D. (1993). Tissue renin-angiotensin systems. Their role in cardiovascular disease. Circulation 87 IV7–13. [PubMed] [Google Scholar]
- Liu H., Irwanto A., Tian H., Fu X., Yu Y., Yu G., et al. (2012). Identification of il18rap/il18r1 and il12b as leprosy risk genes demonstrates shared pathogenesis between inflammation and infectious diseases. Am. J. Hum. Genet. 91 935–941. 10.1016/j.ajhg.2012.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris T. J., Butcher L. M., Feber A., Teschendorff A. E., Chakravarthy A. R., Wojdacz T. K., et al. (2014). Champ: 450k chip analysis methylation pipeline. Bioinformatics 30 428–430. 10.1093/bioinformatics/btt684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller D. N., Bohlender J., Hilgers K. F., Dragun D., Costerousse O., Menard J., et al. (1997). Vascular angiotensin-converting enzyme expression regulates local angiotensin ii. Hypertension 29 98–104. 10.1161/01.hyp.29.1.98 [DOI] [PubMed] [Google Scholar]
- Myerson S., Hemingway H., Budget R., Martin J., Humphries S., Montgomery H. (1999). Human angiotensin i-converting enzyme gene and endurance performance. J. Appl. Physiol. 87 1313– 1316. [DOI] [PubMed] [Google Scholar]
- Naeije R. (2010). Physiological adaptation of the cardiovascular system to high altitude. Prog. Cardiovasc. Dis. 52 456–466. 10.1016/j.pcad.2010.03.004 [DOI] [PubMed] [Google Scholar]
- Nguyen M. P., Lee S., Lee Y. M. (2013). Epigenetic regulation of hypoxia inducible factor in diseases and therapeutics. Arch. Pharm. Res. 36 252–263. 10.1007/s12272-013-0058-x [DOI] [PubMed] [Google Scholar]
- Palmer B. F. (2010). Physiology and pathophysiology with ascent to altitude. Am. J. Med. Sci. 340 69–77. 10.1097/maj.0b013e3181d3cdbe [DOI] [PubMed] [Google Scholar]
- Patel S., Woods D. R., Macleod N. J., Brown A., Patel K. R., Montgomery H. E., et al. (2003). Angiotensin-converting enzyme genotype and the ventilatory response to exertional hypoxia. Eur. Respir. J. 22 755–760. 10.1183/09031936.03.00086402 [DOI] [PubMed] [Google Scholar]
- Peters T. J., Buckley M. J., Statham A. L., Pidsley R., Samaras K., Vl R., et al. (2015). De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin 8:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell F. L., Milsom W. K., Mitchell G. S. (1998). Time domains of the hypoxic ventilatory response. Respir. Physiol. 112 123–134. 10.1016/s0034-5687(98)00026-7 [DOI] [PubMed] [Google Scholar]
- Qadar Pasha M. A., Khan A. P., Kumar R., Grover S. K., Ram R. B., Norboo T., et al. (2001). Angiotensin converting enzyme insertion allele in relation to high altitude adaptation. Ann. Hum. Genet. 65 531–536. 10.1046/j.1469-1809.2001.6560531.x [DOI] [PubMed] [Google Scholar]
- Rangel M., dos Santos J. C., Ortiz P. H., Hirata M., Jasiulionis M. G., Araujo R. C., et al. (2014). Modification of epigenetic patterns in low birth weight children: importance of hypomethylation of the ace gene promoter. PLoS One 9:e106138. 10.1371/journal.pone.0106138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regard M., Oelz O., Brugger P., Landis T. (1989). Persistent cognitive impairment in climbers after repeated exposure to extreme altitude. Neurology 39 210–213. 10.1212/wnl.39.2.210 [DOI] [PubMed] [Google Scholar]
- Ren X., Kuan P. F. (2019). Methylgsa: a bioconductor package and shiny app for DNA methylation data length bias adjustment in gene set testing. Bioinformatics 35 1958–1959. 10.1093/bioinformatics/bty892 [DOI] [PubMed] [Google Scholar]
- Rigat B., Hubert C., Alhenc-Gelas F., Cambien F., Corvol P., Soubrier F. (1990). An insertion/deletion polymorphism in the angiotensin i-converting enzyme gene accounting for half the variance of serum enzyme levels. J. Clin. Invest. 86 1343–1346. 10.1172/jci114844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., Shi W., et al. (2015). Limma powers differential expression analyses for rna-sequencing and microarray studies. Nucleic Acids Res. 43:e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez F. A., Ventura J. L., Casas M., Casas H., Pages T., Rama R., et al. (2000). Erythropoietin acute reaction and haematological adaptations to short, intermittent hypobaric hypoxia. Eur. J. Appl. Physiol. 82 170–177. 10.1007/s004210050669 [DOI] [PubMed] [Google Scholar]
- Rupert J. L., Kidd K. K., Norman L. E., Monsalve M. V., Hochachka P. W., Devine D. V. (2003). Genetic polymorphisms in the renin-angiotensin system in high-altitude and low-altitude native american populations. Ann. Hum. Genet. 67 17–25. 10.1046/j.1469-1809.2003.00004.x [DOI] [PubMed] [Google Scholar]
- Sahu B., Pani S., Swalsingh G., Bal N. C. (2019). Non and epigenetic mechanisms in regulation of adaptive thermogenesis in skeletal muscle. Front. Endocrinol. 10:517. 10.3389/fendo.2019.00517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza G. L. (2012). Hypoxia-inducible factors in physiology and medicine. Cell 148 399–408. 10.1016/j.cell.2012.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slemc L., Kunej T. (2016). Transcription factor hif1a: downstream targets, associated pathways, polymorphic hypoxia response element (hre) sites, and initiative for standardization of reporting in scientific literature. Tumour Biol. 37 14851–14861. 10.1007/s13277-016-5331-4 [DOI] [PubMed] [Google Scholar]
- Smith A. K., Kilaru V., Klengel T., Mercer K. B., Bradley B., Conneely K. N., et al. (2015). DNA extracted from saliva for methylation studies of psychiatric traits: evidence tissue specificity and relatedness to brain. Am. J. Med. Genet. B Neuropsychiatr. Genet. 168B 36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Liu B., Huai W., Yu Z., Wang W., Zhao J., et al. (2016). The e3 ubiquitin ligase trim31 attenuates nlrp3 inflammasome activation by promoting proteasomal degradation of nlrp3. Nat. Commun. 7:13727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stobdan T., Ali Z., Amjad Pervez K., Nejatizadeh A., Ram R., Thinlas T., et al. (2011). Polymorphisms of renin-angiotensin system genes as a risk factor for high-altitude pulmonary oedema. J. Renin Angiotensin Aldosterone Syst. 12 93–101. 10.1177/1470320310387177 [DOI] [PubMed] [Google Scholar]
- Thompson T. M., Sharfi D., Lee M., Yrigollen C. M., Naumova O. Y., Grigorenko E. L. (2013). Comparison of whole-genome DNA methylation patterns in whole blood, saliva, and lymphoblastoid cell lines. Behav. Genet. 43 168–176. 10.1007/s10519-012-9579-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsianos G., Eleftheriou K. I., Hawe E., Woolrich L., Watt M., Watt I., et al. (2005). Performance at altitude and angiotensin i-converting enzyme genotype. Eur. J. Appl. Physiol. 93 630–633. [DOI] [PubMed] [Google Scholar]
- Walczak-Drzewiecka A., Ratajewski M., Pulaski L., Dastych J. (2010). DNA methylation-dependent suppression of hif1a in an immature hematopoietic cell line hmc-1. Biochem. Biophys. Res. Commun. 391 1028–1032. 10.1016/j.bbrc.2009.12.011 [DOI] [PubMed] [Google Scholar]
- Wang G. L., Semenza G. L. (1995). Purification and characterization of hypoxia-inducible factor 1. J. Biol. Chem. 270 1230–1237. 10.1074/jbc.270.3.1230 [DOI] [PubMed] [Google Scholar]
- Wang H., Yao L., Gong Y., Zhang B. (2018). Trim31 regulates chronic inflammation via nf-kappab signal pathway to promote invasion and metastasis in colorectal cancer. Am. J. Transl. Res. 10 1247–1259. [PMC free article] [PubMed] [Google Scholar]
- Wenger R. H. (2002). Cellular adaptation to hypoxia: O2-sensing protein hydroxylases, hypoxia-inducible transcription factors, and o2-regulated gene expression. FASEB J. 16 1151–1162. 10.1096/fj.01-0944rev [DOI] [PubMed] [Google Scholar]
- Wickham H. (2009). Ggplot2 Elegant Graphics for Data Analysis Introduction. New York: Springer-Verlag. [Google Scholar]
- Woods D. R., Humphries S. E., Montgomery H. E. (2000). The ace i/d polymorphism and human physical performance. Trends Endocrinol. Metab. 11 416–420. 10.1016/s1043-2760(00)00310-6 [DOI] [PubMed] [Google Scholar]
- Woods D. R., Pollard A. J., Collier D. J., Jamshidi Y., Vassiliou V., Hawe E., et al. (2002). Insertion/deletion polymorphism of the angiotensin i-converting enzyme gene and arterial oxygen saturation at high altitude. Am. J. Respir. Crit. Care Med. 166 362–366. 10.1164/rccm.2103060 [DOI] [PubMed] [Google Scholar]
- Xi H., Peng Y., Xie W., Pang J., Ma N., Yang S., et al. (2020). A chromosome 1q22 microdeletion including ash1l is associated with intellectual disability in a chinese family. Mol. Cytogenet. 13:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X. (2014). Cognitive impairments at high altitudes and adaptation. High Alt. Med. Biol. 15 141–145. 10.1089/ham.2014.1009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author AC, upon request.