Abstract
The tempo of sex chromosome evolution—how quickly, in what order, why and how their particular characteristics emerge during evolution—remains poorly understood. To understand this further, we studied three closely related species of African clawed frog (genus Xenopus), that each has independently evolved sex chromosomes. We identified population polymorphism in the extent of sex chromosome differentiation in wild-caught Xenopus borealis that corresponds to a large, previously identified region of recombination suppression. This large sex-linked region of X. borealis has an extreme concentration of genes that encode transcripts with sex-biased expression, and we recovered similar findings in the smaller sex-linked regions of Xenopus laevis and Xenopus tropicalis. In two of these species, strong skews in expression (mostly female-biased in X. borealis, mostly male-biased in X. tropicalis) are consistent with expectations associated with recombination suppression, and in X. borealis, we hypothesize that a degenerate ancestral Y-chromosome transitioned into its contemporary Z-chromosome. These findings indicate that Xenopus species are tolerant of differences between the sexes in dosage of the products of multiple genes, and offer insights into how evolutionary transformations of ancestral sex chromosomes carry forward to affect the function of new sex chromosomes.
This article is part of the theme issue ‘Challenging the paradigm in sex chromosome evolution: empirical and theoretical insights with a focus on vertebrates (Part I)’.
Keywords: comparative transcriptomics, recombination suppression, heterogamy, dosage tolerance
1. Introduction
Many vertebrates have separate male and female individuals rather than hermaphrodites, with sex-specific traits in each sex being regulated by transcripts with sex-biased expression. In species with genetic sex determination, DNA variation that triggers sexual differentiation is present on sex chromosomes. This is important because sex-specific genetic variation establishes a genomic compartment—which may expand—that has unique features as compared to autosomes (copy number, mode of inheritance, rates of mutation and recombination) that strongly influence evolution in important ways [1,2]. For example, compared to autosomes, non-recombining portions of sex chromosomes tend to have a higher abundance of repetitive elements and more extensive pseudogenization [3], and distinctive evolutionary dynamics of non-neutral mutations that depend on the degree of dominance [4,5]. Sex chromosomes evolve from autosomes and, in theory, genetic recombination near the genomic region that contains the master regulator of sex determination can decline or become absent owing to several non-exclusive factors. For example, it is conceivable that neutral processes may drive recombination suppression [4]. It is also possible that inheritance (or non-inheritance) of a non-recombined genomic region may be advantageous in order to avoid sterile progeny [6], genetic linkage to a sex-determining locus may resolve genomic conflict associated with mutations with opposite fitness effects in each sex [5,7], or recombination suppression could be favoured to maintain heterozygosity, especially in inbred populations [8]. Mechanistically, recombination suppression may be achieved by inversion or translocation [9,10], or potentially without genomic rearrangement [11] owing to genetic mutations that alter recombination (recombination modifiers [12]).
There are costs of recombination suppression in sex-linked regions that are associated with Hill-Robertson effects, which reduce the efficacy of natural selection [13]. Some of these costs may be curtailed by the origin of and gene conversion among duplicates [14]. Benefits of recombination suppression also exist, which are related to sex-specific or sex-biased modes of inheritance. This can help resolve genomic conflict, and these regions can also foster the elaboration of sex-specific genetic functions, such as the many genes that are crucial for spermatogenesis on the male-specific portion of the human Y-chromosome [14]. Gene loss on the sex-specific sex chromosome (the Y- or W-) creates hemizygous genotypes, allowing natural selection to respond more efficiently to recessive alleles in the heterogametic sex (XY males or WZ females); the associated rapid evolution is known as the faster-X (or faster-Z) effect [2,4]. One of the consequences of their unique evolution is that sex chromosomes play an oversized role in the establishment of reproductive incompatibilities among species [2,4,15].
(a) . Signs of rapid sex chromosome evolution
In species that have separate sexes, sexual differentiation is usually a necessary prelude to reproduction, and disruption of this developmental process can have dire fitness consequences. Consequently, one might expect purifying selection on sex-determination pathways—including the trigger for sex determination—to be strong. Contrary to this expectation, components of the developmental cascade that govern sexual differentiation actually vary considerably among species, including and especially the triggers for sex determination and the chromosomes on which they reside. This variation is sometimes evidenced by a known trigger for sex determination that is present in one lineage but not another. An example of this is the Sry locus, which triggers male differentiation in most placental and marsupial mammals, but not in monotremes [16]. Another line of evidence for rapid evolution comes from studies that document variation among species or populations in whether females or males are heterogametic. In Diptera, for instance, the heterogametic sex in some species is male but female in others, and sex-linked regions in several species are not homologous [17]. Another example of rapid evolution involves dramatic differences in the extent of sex chromosome heteromorphy—microscopic differences in the size of the sex chromosomes or genomic differences in the magnitude and genomic extent of differentiation between sex chromosomes—with some species having non-diverged (homomorphic) sex chromosomes and others with heteromorphic sex chromosomes. These differences are owing to variation in the genomic extent of recombination suppression surrounding a trigger for sex determination; an example of this variation is found in livebearing teleost fishes [18]. Some aspects of rapid evolution eventually impede further change. For example, sex chromosome hemizygosity associated with loss of alleles on the non-recombining portion of the sex-specific sex chromosome creates imbalances in the stoichiometries of interacting sex-linked and autosomal genes. This has the potential to arrest sex chromosome divergence [19], favour the origin of new sex chromosomes [20] or spur the evolution of systems for dosage compensation [21]. Over time, the origin of systems for dosage compensation and sex-specific viability alleles may act to dampen the evolutionary meanderings of sex chromosomes [22].
(b) . Young sex chromosomes of Xenopus frogs
How can we make sense of the diverse and sometimes correlated or counterposed drivers that choreograph the evolutionary fates of sex chromosomes? One approach is to examine young sex chromosomes with an eye on identifying early steps in evolution before they become blurred or confounded by subsequent change. To this end, the sex chromosomes of African clawed frogs (genus Xenopus)—and particularly the Marsabit clawed frog, Xenopus borealis—provide compelling subjects. Within Xenopus, all of the examples of rapid sex chromosome evolution discussed above are present (figure 1). Variation in heterogamy exists within Xenopus tropicalis, with a variation on chromosome 7 (hereafter chr7) defining W-, Y- and Z-sex chromosomes [23]. In Xenopus laevis and several other closely related species, a recently evolved genetic trigger for female sex determination called dmw established chromosome 2L (chr2L) as the sex chromosomes [24]. Xenopus tropicalis diverged before dmw arose [25], whereas X. borealis lost dmw and evolved new sex chromosomes on chromosome 8L (chr8L; [26]). Though all Xenopus species have homomorphic sex chromosomes [27], there exists considerable variation in recombination suppression around the sex-linked portions of sex chromosomes in different Xenopus. Xenopus laevis and X. tropicalis have a very small region of recombination suppression, whereas this region in X. borealis comprises almost half of the sex chromosomes; the other half of chr8L undergoes recombination in both sexes, and, therefore, is a pseudoautosomal region [28,29].
Figure 1.
Evolutionary relationships among focal species (a) and their orthologues and homeologues (b) in this study. Allopolyploidization involved whole-genome duplication (WGD) via the merger of two extinct ancestral species (daggers) that generated two subgenomes (L and S, red and blue) in the tetraploid species X. borealis and X. laevis. Sex chromosomes in each species are indicated on the right above branches in grey, pink or blue. The pink line corresponds with the origin of the female-determining gene dm-w on chr2L. A dashed blue line indicates the unknown timing of the origin of the X. tropicalis sex chromosomes; branches not to scale. (Online version in colour.)
Xenopus borealis is an allotetraploid species, meaning that it was formed via the fusion of two diploid ancestral species (figure 1; reviewed in [30]). For this reason, its genome contains autosomal chromosomes that are homeologous to the sex chromosomes (chromosome 8S; chr8S). The closely related species X. laevis is also allotetraploid, shares a most recent common allotetraploid ancestor with X. borealis approximately 18–34 million years ago (Ma) [31] or older [32] and has autosomal chromosomes that are orthologous (chr8L) and homeologous (chr8S) to the sex chromosomes of X. borealis. Likewise, the sex chromosomes of X. laevis are orthologous and homeologous to autosomes of X. borealis (chr2L and chr2S, respectively). Xenopus tropicalis, a diploid, has autosomal chromosomes (chr8 and chr2) that are each, respectively, orthologous to the sex chromosomes of X. borealis and of X. laevis, and the sex chromosomes of X. tropicalis (chr7) are orthologous to pairs of homeologous autosomes in X. borealis and also in X. laevis (chr7L and chr7S—both of these homeologous chromosomes are in both of these allotetraploid species). Xenopus tropicalis, X. laevis and X. borealis share a most recent common ancestor approximately 60 Ma [32]. Together, these three sets of alternatively sex-linked, orthologous and/or homeologous chromosomes in these frogs provide informative contrasts with which to consider the evolution of sex-linked regions (figure 1). We follow the X. laevis chromosome nomenclature of [33] by postfixing an ‘L’ to the name of chromosome pairs from the ‘long’ subgenome and an ‘S’ to the names of chromosome pairs from the ‘short’ subgenome; these postfixes thus refer to entire chromosome pairs and not to the long or short arms of individual chromosomes. For X. tropicalis, we follow the chromosomal nomenclature of [34] which uses the same numbers for orthologous chromosomes as [33] except for X. tropicalis chr9 and chr10 which are orthologous to fused chromosomes in X. laevis and X. borealis (chr9_10L, chr9_10S).
With an aim of better understanding the evolutionary dynamics of sex chromosomes, we used genome-wide approaches to test for population structure and variation in sex chromosome differentiation in natural populations of X. borealis sampled in their native range in Kenya. We then evaluated whether and how the expression of genes in the sex-linked region of the sex chromosomes differs from the rest of the genome, and when these changes emerged during Xenopus diversification. We considered whole transcriptome data from X. borealis adult liver and gonad/mesonephros tissues from two tadpole stages during the onset of gonadal differentiation, and we also contextualize findings in X. borealis with similar comparisons with adult liver transcriptomes of X. laevis and X. tropicalis.
2. Material and methods
(a) . Samples, analyses of population structure in Xenopus borealis
Xenopus borealis naturally occurs in Kenya and northern Tanzania. We analysed a total of 93 wild-caught X. borealis specimens that were sampled through the central portion of its range in Kenya in 2018. Information on the sampling localities, voucher specimens and sex of these samples is provided in the electronic supplementary material, table S1; figure 2; specimens from this collection, including genetic samples, are accessioned at the Museum of Comparative Zoology at Harvard University, USA, and the National Museums of Kenya in Nairobi, Kenya. As described in the electronic supplementary material, we performed phylogenetic analysis on a portion of mitochondrial DNA from a subset of these samples (84 specimens) and also two laboratory specimens. We also collected and analysed reduced representation genome sequences (RRGS, [35]) from a subset of these samples (51 specimens, 26 females, 25 males) combined with whole-genome sequencing data from the two laboratory specimens. These analyses allowed us to discern population structure and sex chromosome differentiation within X. borealis.
Figure 2.
Sampling locations for X. borealis (a) and estimated relationships among four mitochondrial clades in X. borealis with respect to other species in the muelleri species group (b); bootstrap values are above branches and a scale bar indicates substitutions per site. The size of the circles corresponds to the number of mitochondrial sequences analysed from each locality; pie charts from two localities indicate the proportions of each clade sampled with colours matching the phylogeny. Stars indicate locations of major mountains and grey shading indicates location of the eastern arm of the Great Rift Valley. (Online version in colour.)
(b) . Comparative transcriptomics of Xenopus borealis, Xenopus laevis, and Xenopus tropicalis
We sequenced and analysed transcriptomes from gonads at two juvenile developmental stages and adult liver from our laboratory strain of X. borealis. In order to gain an evolutionary perspective on these transcriptomes, we also analysed transcriptomes from adult liver from X. laevis and X. tropicalis. Xenopus borealis and X. laevis specimens were obtained directly or were offspring of animals that were purchased from Xenopus Express (Brooksville, Florida, USA) and the X. tropicalis adult liver data were obtained from the NCBI-SRA (from two males: SRR5412273 and SRR5412274, and two females: SRR5412275 and SRR5412276). Details of RNA extraction and sequencing, and the transcriptome assembly and analysis of sex-biased expression are provided in the electronic supplementary material.
Genomic locations of assembled transcripts from each tetraploid species (X. borealis and X. laevis) were assessed in the X. laevis genome assembly version 9.2 [31], which were obtained from Xenbase [36], using the splice-aware aligner GMAP [37] with parameters: -A -B 5 -t 20 -f samse. The X. tropicalis transcripts were mapped to the v. 10 X. tropicalis genome assembly (NCBI BioProject AAMC00000000.4; GenBank Assembly submission GCA_000004195.4) using the same approach. The parameter cross-species was used for the X. borealis transcripts to accommodate divergence from the X. laevis reference genome.
In our laboratory strain of X. borealis, the sex-linked region of the sex chromosomes corresponds to positions 4 605 306 to 56 690 925 base pairs (bp) on chr8L of the X. laevis genome assembly v. 9.2 [29]. For this study, we assume that the sex-linked region starts from the beginning of chr8L because no RRGS data from [29] mapped to less than 4 605 306 bp on chr8L. In order to establish approximate margins of regions of orthology and homeology for sex-linked regions in each of our three focal species, we generated dot plots from chromosomes in the genome assemblies of X. laevis and X. tropicalis using Gopard version 1.40 [38] and re-DOTable v. 1.0 [39]. Based on these plots, we concluded that the sex-linked region of X. borealis is homeologous to 16–45 and 51–68 Mb of X. laevis chr8S, and orthologous to 0–12 and 20–73 Mb of the X. tropicalis chromosome 8 (e.g. electronic supplementary material, figure S1). The sex-linked region of the X. laevis sex chromosomes is above 178 Mb on chr2L [29], and based on dotplots, this region is homeologous to 154–158 Mb on X. laevis chr2S and orthologous to 172–177 Mb on chr2 of X. tropicalis. The sex-linked region of the X. tropicalis sex chromosome is less than 11 Mb on chr7 [28,40]. This region is orthologous to less than 9 Mb and less than 6 Mb on X. laevis chr7L and chr7S, respectively. This information allows us to compare homologous (i.e. either orthologous or homeologous) genomic regions that are and that are not sex-linked.
For each tetraploid transcriptome, we used a generalized linear model to test whether a binary response variable—whether or not a transcript was significantly differentially expressed (1) or not (0) according to false detection rate (FDR) corrected p-values from edgeR (significance)—was affected by the interaction between two predictor variables: whether a transcript was from the L (1) or S (0) subgenome (subgenome), and whether a transcript was encoded by a gene in the sex-linked region or the region that is homeologous to the sex-linked region (1), or by a gene from elsewhere in the genome (0) (SL_homeologous):
We had a one-sided expectation that there would be a positive interaction between subgenome and SL_homeologous, which would indicate that odds of sex-biased expression are higher for transcripts encoded by genes in the sex-linked portion of chr8L compared to the rest of the genome, after controlling for the subgenome effects. It is important to consider subgenome effects because in several Xenopus allotetraploids the S subgenome has more rearrangements, shorter coding regions, and more pseudogenization than the L subgenome, and transcripts from the S subgenome tend to be more lowly expressed than homeologous transcripts from the L subgenome [31,41]. For the diploid transcriptome of X. tropicalis, there was no subgenome parameter, and the model simplifies to a logistic regression with one predictor variable (sex_linked)—that indicates whether or not a transcript was from a sex-linked (1) or non-sex-linked gene (0).
For juvenile tissues from X. borealis, we were only able to make intraspecific comparisons, for example between the sex-linked portion of chr8L and the homeologous portion of chr8S (the latter of which is autosomal). For adult liver tissues, we had comparable data from all three focal species. In order to compare evolutionary trends across these species for adult liver, we performed three different logistic regressions, each with sex-linked and homeologous or orthologous to sex-linked regions across each species defined to match each one of our three focal species. For example, when the adult liver of X. borealis was the focal species, SL_homeologous and sex_linked were set to a value of one for transcripts encoded by genes in the sex-linked, homeologous and orthologous regions of chr8L, chr8S and chr8 for transcriptome data from each of the three focal species (even though this region is only sex-linked in X. borealis), and transcripts encoded by genes in the rest of each of these three genomes were set to zero. The same was done for models in each species for chr2L, chr2S and chr2 (when X. laevis was the focal species) or chr7L, chr7S and chr7 (when X. tropicalis was the focal species). These tests allowed us to test whether sex-linked changes in one species arose concurrently with sex linkage or whether they were present ancestrally. More specifically, if the sex-biased expression of transcripts encoded by sex-linked genes evolved independently in the different sex-linked regions of each species, we expected a significant coefficient when the SL_homeologous or sex_linked matched their sex-linked region of the focal species but not the non-focal species. Alternatively, if the sex-biased expression was present in an ancestor prior to a region becoming sex-linked, we expected significant coefficients for some non-focal species even when the SL_homeologous and sex_linked parameters did not match their sex-linked region.
Analysis of individual transcripts allowed us to evaluate the sex-biased expression of splice variants or possibly individual alleles. We also analysed differential expression at the gene level based on summed normalized expression levels of all transcripts from individual genes. For this gene-level analysis, expression levels were summed for transcripts that mapped within or overlapped with (by at least 200 bp) annotated genes in v. 9.2 of the X. laevis (for X. borealis and X. laevis in this study) and version 10 of the X. tropicalis genome (for X. tropicalis in this study). We also included in this analysis the summed expression levels of transcripts in unannotated portions of these genomes that overlapped by at least 200 bp.
3. Results
(a) . Population variation in sex chromosome differentiation in Xenopus borealis
Partial mitochondrial DNA sequences from wild-caught X. borealis cluster into four closely related, geographically structured clades. Some of these clades have only modest bootstrap support, and relationships among them are not well resolved (figure 2). At two localities in central Kenya (Njoro and Nairobi), two of these four clades were present; at all other localities, only one clade was present in our sample. In Kenya, a geographical boundary between the most diverged intraspecific mitochondrial lineages of X. borealis may roughly correspond to the eastern arm of the Great Rift Valley. The two sequences from our laboratory strain clustered in different clades: the male (BJE3896) clustered in the black clade in figure 2, whereas the female (BJE3897) clustered in the blue clade, with no divergent sites compared to four other sequences, all of which were from Nairobi specimens. To our knowledge, sequences from the western (red) clade and the Kitobo (purple) clade have not been previously reported. The black and blue clades (figure 2) have been previously reported and are estimated to have diverged from one another about 2 Ma [33,41].
Although samples from Kitobo, which has a unique mitochondrial lineage, were not included in the RRGS data, available information from other samples is consistent with the findings from mitochondrial DNA in terms of evidencing geographical structure and also admixture between Nairobi and Njoro (figure 3). With two ancestry components, the geographical structure is evident, with specimens from the central Kenya localities of Nairobi and Njoro having a mixture of two ancestry components, specimens from Wundanyi (in the east) assigned to one ancestry component, and other specimens (all from west Kenya) assigned to another. With three ancestry components, the Wundanyi population is further distinguished from the others, and a gradient of ancestry components is still evident in the Nairobi and Njoro populations.
Figure 3.
Genetic cluster analysis of RRGS data from wild-caught specimens and homologous WGS data from two laboratory individuals. (a) Ancestry components of wild X. borealis from Kenya with K clusters from two (top) to five (bottom) exhibit geographical structure, and genetic affinities of a male (M) and female (F) laboratory animal (Lab, right side) suggest origins from central Kenya (Njoro, Nairobi). (b) log-likelihood of values of K from 1–8. (Online version in colour.)
Using RRGS data, we calculated FST between females and males for the wild samples of X. borealis. For the autosomes and pseudoautosomal regions, this statistic is expected to be influenced by polymorphism and differences in the number of males and females in different populations (because an unequal sample of each sex in different subdivided populations should increase FST between males and females across the sample). For the sex chromosomes, FST should be additionally influenced by the divergence between the W- and Z-sex chromosomes. We detected substantial differentiation in the region of recombination suppression on the sex chromosomes (chr8L, [27]) of the X. borealis specimens from central (Nairobi) and eastern (Wundanyi) Kenya, but not elsewhere on this chromosome (figure 4). In populations from western Kenya, there was no strong evidence of differentiation between the sexes in any substantially sized genomic region, including the sex-linked region of chr8L (figure 4). In both of these populations or when all samples were considered, there were no other substantially sized regions where FST was substantially elevated beyond the 95% confidence interval for the non-sex-linked portion of the genome (data not shown). In the sex-linked region of X. borealis specimens from central and eastern Kenya, there was a dip in FST around 20 Mb (figure 4); we suspect this could be because RRGS data from X. borealis chr8S inappropriately mapped to this part of chr8L in the X. laevis reference genome.
Figure 4.
FST between males and females is substantial on the sex-linked portion (red box) of the sex chromosomes (Chr8L) for the samples from central and east Kenya (bottom; nine females, six males) but not west Kenya (middle; 17 females, 19 males) and is slightly elevated when all samples are considered (top). Grey boxes indicate the 95% confidence interval for FST values in the non-sex-linked portion of the genome. (Online version in colour.)
Using Sanger sequencing, we checked whether two previously studied genes with sex-linked variation in our laboratory X. borealis [29] had sex-linked variation in the wild samples of X. borealis. One was SOX3-L, which is position approximately 34 Mb on X. laevis chr8L; the other is NR5A1, which is approximately 10 Mb on chr8L. We found that both had female-specific heterozygous sites in individuals from Wundanyi (SOX3: three females, two males; NR5A1: three females, four males), but not central or west Kenya for SOX3 (29 females, 19 males). For NR5A1, there also were heterozygous sites that were female-specific in central Kenya (Njoro, Nairobi and Kiambu; 13 females, six males) and almost female-specific in Kambu (present in four females and one male, not present in five other males), but no sex-specific variation was present in west Kenya (13 females, 10 males).
(b) . The genomic landscape of genes with sex-biased expression in Xenopus borealis
The transcriptome assembly for X. borealis tadpoles (a combined assembly for gonad/mesonephros from stages 46 and 48) had 523 313 transcripts; the assembly for X. borealis adult liver had 562 550, transcripts; independent assemblies for X. laevis adult liver and X. tropicalis adult liver had 391 563 and 207 184 transcripts, respectively. These transcripts include biological complexity stemming from splice variants and allelic variation and technical variation owing to incompletely assembled transcripts and assembly errors. We performed analyses of individual transcripts and also of groups of transcripts that were binned by genomic location into putative genes.
In the analysis of X. borealis transcripts from tadpole stages 46 and 48 gonad/mesonephros and adult liver, a total of 453, 763 and 614 transcripts, respectively, were significantly sex-biased (table 1). In all three of these X. borealis transcriptomes, there were significant and substantially higher densities of genes in the sex-linked region of the X. borealis sex chromosome that encoded sex-biased transcripts compared to (i) the homeologous region of chromosome 8S in X. borealis and (ii) the rest of the genome of X. borealis (p ≤ 0.001 for all comparisons, binomial tests, table 1 and figure 5). More specifically, between 31 and 50% of all of the sex-biased transcripts that mapped to chromosomes in these three X. borealis transcriptomes were encoded by genes in the sex-linked region of chr8L, which comprises only approximately 2% of the genome. With X. borealis as the focal species, the interaction terms from the logistic regression were significant for each X. borealis transcriptome ( for all three tests). The interaction coefficient of these models provides an estimate of the ratio of odds ratios of the probability a transcript is significantly differently expressed in the sex-linked region of chr8L compared to the rest of the L subgenome, divided by the probability that a transcript is significantly differently expressed in the region of chr8S that is homologous to the sex-linked region of chr8L compared to the rest of the S subgenome. Exponentiating the 95% confidence intervals for these interaction coefficients from each model indicates that the odds of significant sex-biased expression in the sex-linked region of chr8L is 2.9–8.6, 3.9–8.7 or 2.4–5.6 times higher than the rest of the genome after controlling for subgenome effects (for the gonad/mesonephros tadpole stages 46 and 48 and adult liver, respectively). In the X. borealis transcriptomes, the effect of subgenome was not significant in any of these models (p = 0.90, 0.12 and 0.85, respectively), and the results were essentially the same when the subgenome predictor and interaction term were not included in each model (data not shown).
Table 1.
The number of significantly sex-biased transcripts by chromosome in X. borealis tadpole stages 46 and 48 gonad/mesonephros (46, 48) and adult liver (L), including female-biased (FB), female-specific (FS), male-biased (MB) and male-specific (MS) transcripts; with biased transcripts being expressed in both sexes but significantly higher in one, and specific transcripts being expressed exclusively in one sex. (For the sex chromosome (chr8L), numbers in parentheses refer to the number of differentially expressed transcripts in X. borealis that are encoded by genes within the sex-linked region. For the chromosome that is homeologous to the sex chromosome (chr8S), numbers in parentheses indicate differentially expressed transcripts that are encoded by genes within the portion of this chromosome that is homeologous to the sex-linked region of the sex chromosome. For four other chromosomes, numbers in parentheses indicate the number of differentially expressed transcripts in X. borealis that are encoded by genes in genomic regions that are homologous to sex-linked regions of X. laevis (chr2L, chr2S) and X. tropicalis (chr7L, chr7S), respectively. Some sex-biased transcripts mapped to scaffolds whose chromosomal locations are unknown (scaffolds) or did not map to the X. laevis reference genome (unmapped).)
| chr | FB46 | FS46 | MB46 | MS46 | FB48 | FS48 | MB48 | MS48 | FBL | FSL | MBL | MSL |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| chr1L | 4 | 6 | 8 | 16 | 3 | 6 | 9 | 3 | 7 | 9 | 5 | 4 |
| chr1S | 2 | 3 | 6 | 1 | 0 | 4 | 0 | 1 | 2 | 1 | 6 | 5 |
| chr2L | 1(0) | 3(0) | 1(0) | 1(0) | 0(0) | 5(0) | 6(0) | 3(0) | 7(0) | 10(0) | 3(0) | 1(0) |
| chr2S | 0(0) | 4(0) | 0(0) | 3(0) | 2(0) | 6(0) | 5(0) | 0(0) | 2(0) | 12(0) | 3(0) | 3(0) |
| chr3L | 4 | 8 | 4 | 7 | 6 | 6 | 9 | 2 | 4 | 9 | 1 | 1 |
| chr3S | 1 | 2 | 0 | 2 | 8 | 13 | 7 | 3 | 5 | 1 | 3 | 0 |
| chr4L | 2 | 5 | 2 | 2 | 3 | 3 | 9 | 3 | 11 | 6 | 2 | 3 |
| chr4S | 2 | 2 | 1 | 1 | 4 | 4 | 2 | 2 | 6 | 2 | 3 | 1 |
| chr5L | 3 | 1 | 2 | 4 | 4 | 7 | 9 | 2 | 12 | 22 | 10 | 8 |
| chr5S | 1 | 3 | 3 | 1 | 5 | 3 | 1 | 2 | 3 | 3 | 4 | 6 |
| chr6L | 3 | 3 | 2 | 1 | 3 | 3 | 1 | 4 | 5 | 4 | 3 | 1 |
| chr6S | 0 | 3 | 1 | 0 | 4 | 3 | 7 | 0 | 2 | 4 | 4 | 7 |
| chr7L | 1(0) | 1(0) | 2(0) | 0(0) | 1(0) | 5(0) | 5(1) | 1(0) | 7(0) | 2(0) | 1(0) | 5(0) |
| chr7S | 0(0) | 2(0) | 1(0) | 0(0) | 2(0) | 0(0) | 3(1) | 1(0) | 2(0) | 3(0) | 2(1) | 0(0) |
| chr8L | 53(52) | 99(95) | 10(9) | 12(9) | 74(69) | 206(195) | 31(28) | 14(11) | 41(34) | 123(102) | 12(12) | 16(14) |
| chr8S | 4(3) | 18(13) | 2(2) | 1(1) | 12(7) | 29(25) | 6(3) | 0(0) | 8(6) | 28(19) | 2(1) | 3(2) |
| chr9_10L | 1 | 0 | 1 | 1 | 2 | 3 | 5 | 7 | 0 | 5 | 11 | 4 |
| chr9_10S | 2 | 4 | 5 | 4 | 3 | 3 | 2 | 2 | 4 | 8 | 3 | 4 |
| scaffolds | 12 | 34 | 4 | 6 | 0 | 48 | 14 | 8 | 12 | 21 | 6 | 15 |
| unmapped | 11 | 19 | 2 | 4 | 15 | 40 | 13 | 13 | 6 | 22 | 1 | 1 |
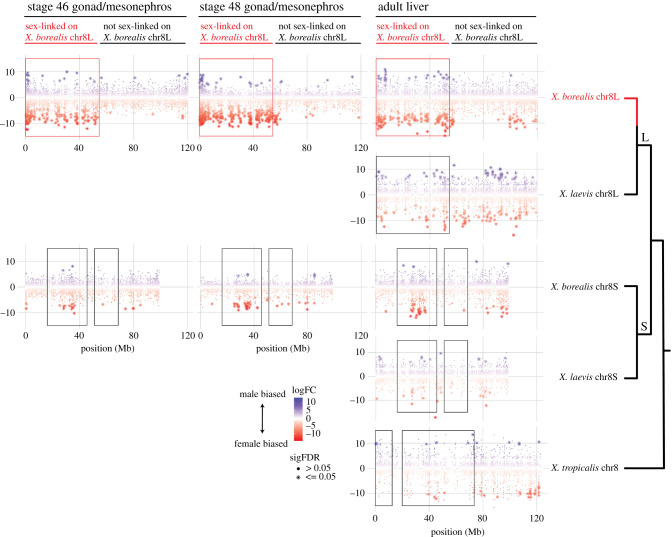
Figure 5.
Comparative transcriptomics illustrates that female-biased expression (negative log(male/female fold change) on the y-axis, red asterisks) evolved contemporaneously with sex linkage in the new sex chromosomes (chr8L) of X. borealis. Results of analyses of differential expression for gonad/mesonephros tissue are presented for tadpole stages 46 (left) and 48 (centre) and adult liver (right). A high number of sex-biased transcripts are encoded by genes in the sex-linked portion of the X. borealis sex chromosomes (chr8L) (red boxes) but not in homeologous regions of X. borealis and X. laevis chr8S or orthologous regions of X. laevis chr8L or of X. tropicalis chr8 (black boxes). These differences indicate that these changes evolved recently in X. borealis, after allotetraploidization and speciation from X. laevis. On the right, the phylogeny depicts relationships among homeologues and orthologues as detailed in figure 1; branches corresponding to the L and S subgenomes are labelled. Plots of all chromosomes for each transcriptome and species in these analyses are provided in the electronic supplementary material, figures S2–S6. (Online version in colour.)
In the non-sex-linked portion of the X. borealis genome, the proportions of the sex-biased transcripts that mapped to chromosomes that were female-biased were fairly similar in all three transcriptomes (53%, 59% and 66%, respectively, for tadpole stages 46 and 48 gonad/mesonephros and adult liver), as was the density of sex-biased genes (0.07, 0.10 and 0.12 sex-biased transcripts Mb−1, respectively, assuming that the non-sex- linked portion of the genome is approximately 3.0 Gb). However, in the sex-linked region, the proportions of sex-biased transcripts with female-biased expression were uniformly higher than the rest of the genome (89%, 87% and 84% in tadpole stages 46 and 48 and adult liver, respectively) and the densities of sex-biased transcripts in the sex-linked region were 23–52 fold higher than the rest of the genome (2.89, 5.32 and 2.84 sex-biased transcripts Mb−1, respectively, assuming the sex-linked region of the X. borealis sex chromosomes is approximately 57 Mb in size [29]). This large disparity is qualitatively obvious when expression divergence of transcripts is visualized relative to the genomic position (red asterisks denote significantly negative log(male/female) fold change—indicating female-biased expression—in figure 5). It is also apparent when densities are calculated relative to the number of annotated genes in each region instead of Mb (electronic supplementary material, Results). Although the proportions of male-biased transcripts were lower in the sex-linked region of X. borealis compared to other genomic regions (owing to so many female-biased transcripts that are encoded by genes in this region), there was still a 10–16 times higher density of male-biased transcripts relative to the non-sex-linked part of the genome. Thus, the sex-linked region is enriched in female-biased as well as male-biased transcripts, but more so for the female-biased transcripts.
For adult liver, additional comparisons to orthologous transcriptomes of X. laevis and X. tropicalis were possible. These comparisons illustrate that the proportion of genes with differentially expressed transcripts in the sex-linked portion of the X. borealis sex chromosomes is higher than (i) the orthologous region of chr8L in X. laevis, (ii) the homeologous regions chr8S in X. laevis, and (iii) the orthologous region of chr8 in X. tropicalis. In contrast with the findings from X. borealis, the interaction term from the logistic regression was not significant for the X. laevis adult liver transcriptome when the sex-linked and homeologous regions were defined to match X. borealis (i.e. when X. laevis is a non-focal species and X. borealis is the focal species; p = 0.82). Likewise, in X. tropicalis, the logistic regression also found no significant increase in the odds of differential expression for transcripts in the genomic region that is orthologous to the sex-linked region of X. borealis as compared to other genomic regions (i.e. when X. tropicalis is a non-focal species and X. borealis is the focal species; p = 0.80). Previous work has demonstrated that the sex chromosomes of X. borealis evolved after divergence from the most recent common ancestor with X. laevis (figure 1, [27]). Thus, these results from X. laevis and X. tropicalis together with intraspecific comparisons within X. borealis demonstrate that rapid regulatory evolution of the sex-linked region of X. borealis corresponded in time with this region becoming sex-linked (i.e. after divergence from the most recent common ancestor with X. laevis), as opposed to this region of chr8L encoding a high number of differentially expressed transcripts in an ancestor prior to the origin of the new sex chromosomes of X. borealis. These transcriptome results demonstrate that the X. borealis transcriptome evolved to have sex-specific expression owing to evolution in cis- of the regulatory regions of sex-linked genes on the sex chromosomes of X. borealis. This effect was pronounced (figure 5) and significant, but affects only the small proportion of the transcriptome that is encoded by sex-linked genes.
On the homeologous chr8S of X. borealis, we observed a lower density of sex-biased genes than the sex chromosome (chr8L) for all three transcriptomes, although this was still higher than the rest of the genome (electronic supplementary material, figures S2–S4). A probable explanation is that some of these sex-biased transcripts erroneously mapped to chr8S of the X. laevis reference genome, even though they actually are encoded by genes on chr8L in X. borealis. Thus, we suspect the female-biased expression of sex-linked transcripts is even higher than our data suggests. In some of the X. borealis transcriptomes, various non-sex-linked genomic regions had a high density of female-biased transcripts (e.g. <15 Mb on chr3S in the gonad/mesonephros tissue of tadpole stage 48 transcriptome and 130–155 Mb on chr5L in the adult liver transcriptome, electronic supplementary material, figures S3 and S4). However, these patches were much smaller and had far lower densities of sex-biased transcripts than the sex-linked region of the new sex chromosomes of X. borealis.
(c) . The genomic landscapes of genes with sex-biased expression in Xenopus laevis and Xenopus tropicalis
Another question we were able to investigate is whether the sex-linked regions of X. laevis and X. tropicalis sex chromosomes also are enriched with transcripts with sex-biased expression (electronic supplementary material, figure S7). Similar to the findings in X. borealis, each of the independently evolved sex-linked regions of X. laevis and X. tropicalis also was significantly enriched with sex-biased transcripts compared to other homeologous and/or orthologous genomic regions. In X. laevis, the density of sex-biased transcripts in the sex-linked region was 27 sex-biased transcripts Mb−1, which is over 40 times the density elsewhere (0.57 sex-biased transcripts Mb−1; table 2; electronic supplementary material, figure S7). When sex-linked and homeologous regions were defined to match the sex chromosomes of X. laevis (i.e. portions of chr2L and chr2S respectively and X. laevis is the focal species), the interaction term of the linear model for X. laevis was significant and the 95% confidence interval of the odds that a transcript encoded by genes in the sex-linked region of X. laevis would have sex-biased expression was 1.9–5.7 times higher than the odds for transcripts encoded by genes in the rest of the genome after controlling for subgenome effects. When logistic regressions were performed on X. borealis and X. tropicalis transcriptomes but defining orthologous and homeologous to the sex-linked region to match X. laevis (i.e. these two species are the non-focal species and X. laevis is the focal species), there was no significant coefficient (for X. borealis for the interaction term (p = 0.953) or for X. tropicalis for the sex_linked predictor variable (p = 0.951)). These contrasts are consistent with the proposal that sex-biased expression of the portion of the X. laevis that is encoded by the sex-linked portion of chr2L arose after divergence from the most recent common ancestor with X. tropicalis and coincided with this region becoming sex-linked in X. laevis and then were lost in X. borealis after that species acquired a new sex chromosome (on chr8L).
Table 2.
The number of differentially expressed transcripts by chromosome in X. laevis adult liver, with labelling following table 1. (For the sex chromosome (chr2L) and its homeologue (chr2S), numbers in parentheses refer to the number of differentially expressed transcripts that are encoded by genes within the genomic region that is sex-linked or homeologous to the sex-linked region, respectively, in X. laevis. For the orthologue to the X. borealis sex chromosome (chr8L), the homeologue to the X. borealis sex chromosome (chr8S) and both orthologues to the sex chromosome of X. tropicalis (chr7L, chr7S), numbers in parentheses indicate the number of differentially expressed transcripts in X. laevis that are encoded by genomic region that are homologous to the sex-linked regions of X. borealis or X. tropicalis, respectively. Numbers in parentheses comprise a large fraction of the total for the chromosome only for the sex chromosome of X. laevis (chr2L).)
| chr | FB | FS | MB | MS |
|---|---|---|---|---|
| chr1L | 45 | 97 | 28 | 33 |
| chr1S | 18 | 23 | 16 | 24 |
| chr2L | 21(18) | 28(22) | 8(7) | 42(33) |
| chr2S | 11(0) | 55(2) | 4(0) | 30(1) |
| chr3L | 34 | 63 | 29 | 87 |
| chr3S | 11 | 17 | 5 | 12 |
| chr4L | 63 | 76 | 13 | 47 |
| chr4S | 20 | 27 | 15 | 58 |
| chr5L | 13 | 12 | 4 | 15 |
| chr5S | 2 | 23 | 5 | 5 |
| chr6L | 23 | 27 | 7 | 13 |
| chr6S | 3 | 8 | 6 | 11 |
| chr7L | 44(1) | 121(1) | 18(1) | 41(2) |
| chr7S | 7(0) | 18(0) | 6(1) | 21(2) |
| chr8L | 44(19) | 55(12) | 15(6) | 45(16) |
| chr8S | 8(4) | 10(5) | 3(2) | 10(3) |
| chr9_10L | 20 | 13 | 11 | 6 |
| chr9_10S | 13 | 20 | 4 | 8 |
| scaffolds | 25 | 35 | 23 | 35 |
| unmapped | 1 | 3 | 0 | 4 |
There was also strong evidence for male-biased expression of transcripts encoded by the independently evolved sex-linked portion of chr7 in the genome of X. tropicalis. In this region, the density of sex-biased transcripts in the sex-linked portion of chr7 was 1.9 sex-biased transcripts Mb−1, which is 10 times the density of transcripts that map to other portions of chromosomes (0.18 sex-biased transcripts Mb−1; table 3; electronic supplementary material, figure S7). This enrichment is reflected by a significant coefficient in a logistic regression which indicates that the odds of sex-biased expression are 3.5–8.5 higher in the sex-linked region of chr7 than in other genomic regions (i.e. with X. tropicalis as the focal species). Although the comparison to X. laevis and X. borealis is complicated by genome duplication, we ran the linear models anyway, but defining the SL_homeologous predictor variable to match the sex-linked portions of X. tropicalis chr7 (i.e. with these two species as non-focal species and X. tropicalis as the focal species). In these analyses, there was no significant interaction between the SL_homeologous and subgenome predictor variables either for X. laevis or for X. borealis (p > 0.05 for both species). These contrasts are consistent with the proposal that the sex-biased expression of the sex-linked portion of the X. tropicalis transcriptome evolved in association with sex linkage in this species as opposed to being an ancestral condition, and that this enrichment was either lost when new sex chromosomes emerged in X. laevis and X. borealis or that it was never present if the most recent common ancestor of these three species had a sex chromosome elsewhere in the genome.
Table 3.
The number of differentially expressed transcripts by chromosome in X. tropicalis adult liver, with labelling following table 1. (For the sex chromosome (chr7), numbers in parentheses refer to the number of differentially expressed transcripts that are from genes within the genomic region that is sex-linked in X. tropicalis. For chr2 and chr8, numbers in parentheses refer to the number of differentially expressed transcripts in X. tropicalis that are encoded by genes within genomic regions that are orthologous to the sex-linked regions of X. laevis and X. borealis, respectively. There is a high number of male-biased genes in the sex-linked portion of the sex chromosome of X. tropicalis (chr7) even though this region is a small portion of this chromosome.)
| chr | FB | FS | MB | MS |
|---|---|---|---|---|
| chr1 | 13 | 30 | 5 | 7 |
| chr2 | 3(0) | 4(0) | 7(0) | 1(0) |
| chr3 | 6 | 10 | 1 | 12 |
| chr4 | 53 | 35 | 13 | 2 |
| chr5 | 6 | 2 | 6 | 4 |
| chr6 | 5 | 8 | 3 | 12 |
| chr7 | 2(1) | 1(0) | 7(3) | 24(17) |
| chr8 | 16(14) | 12(10) | 8(6) | 7(5) |
| chr9 | 4 | 1 | 3 | 2 |
| chr10 | 4 | 4 | 4 | 1 |
| scaffolds | 0 | 0 | 0 | 0 |
| unmapped | 1 | 0 | 0 | 0 |
In contrast with X. borealis, the sex-linked region of X. tropicalis has a higher proportion of male-biased or male-specific transcripts (n = 20 out of 21 sex-biased transcripts; 95%) than the rest of the genome, where 38% of sex-biased transcripts are male-biased (table 3). In X. laevis, there is a more similar proportion of female- and male-biased transcripts in the sex-linked region (50% female-biased) compared to the rest of the genome (60% female-biased; table 2), than in X. borealis.
One unexpected finding was that non-sex-linked transcripts in the X. laevis adult liver transcriptome were more strongly sex-biased compared to X. borealis and X. tropicalis (electronic supplementary material, figure S5 compared to electronic supplementary material, figures S4 and S6). In the X. laevis adult liver transcriptome, we detected 4.7 times as many sex-biased expressed transcripts in the non-sex-linked portion of the genome (n = 1718) compared to X. borealis (n = 368), although this disparity is magnified slightly by the larger size of the non-sex-linked region of the X. laevis genome. Even after excluding the sex-linked region, there were significantly more sex-biased expressed transcripts encoded by genes from the L (n = 1181) than the S subgenome in X. laevis (n = 537, , binomial test). After controlling for sex linkage, the partial effect of the subgenome predictor was significant in logistic regression for X. laevis when this species was the focal species (i.e. with SL_homeologous set to 1 for the ends of chr2L and chr2S, see Methods; ). Exponentiating the interaction coefficient indicates that the 95% confidence interval for the odds of sex-biased expression of transcripts encoded by genes in the L subgenome is 1.63–2.03 higher than for those encoded by genes in the S subgenome. Thus, not only are genes more highly expressed in the L subgenome [32], but it appears that they are more likely to encode transcripts with sex-biased expression, even after controlling for the effects of sex linkage on chr2L. One important caveat is that subgenome and significance are conflated because differential expression is statistically easier to detect for highly expressed transcripts (on the L), which violates assumptions of the linear model. Other caveats of this and other transcriptome comparisons are discussed in the electronic supplementary material.
Another unexpected observation relates to the gonad/mesonephros transcriptomes from tadpole stage 48 and adult liver, where the high density of sex-biased transcripts extends slightly beyond the sex-linked region identified by [29]. This was not the case in the corresponding transcriptome from tadpole stage 46 (figure 5). At this time, we do not have an explanation for this observation.
(d) . Sex-biased expression is less pronounced in gene-level than transcript-level analyses
When expression levels of transcripts that mapped to the same genomic region were summed into putative genes, analysis of the resulting genic expression recovered a less substantial or non-significant enrichment of sex-biased expression in the sex-linked region of these three frog species. With X. borealis as the focal species (i.e. when the SL_homeologous parameter defined to match the sex-linked and homeologous regions of X. borealis), the interaction term of the linear models was significant for X. borealis gonad/mesonephros tadpole stage 48 (p = 0.01) and adult liver (p = 0.003), but not gonad/mesonephros tadpole stage 46 (p = 0.16). Thus, after controlling for subgenome effects, the odds of sex-biased expression at the gene level were 1.6–20.4 and 1.5–8.1 times higher in the sex-linked region as compared to elsewhere in X. borealis gonad/mesonephros tadpole stage 48 and adult liver, respectively. When summed transcripts from X. laevis and X. tropicalis were analysed with each of these species as the focal species, the interaction coefficient for the X. laevis data was not significant, and the coefficient for sex linkage in X. tropicalis also was not significant (p > 0.05 for all coefficients). These results are consistent with plots of gene-level expression which illustrate that the summed (gene-level) sex-biased expression in the sex-linked regions is less distinguished from the rest of the respective genomes of these three focal species (electronic supplementary material, figures S8–S12) as compared to the analysis of individual transcripts (electronic supplementary material, figures S2–S4). Thus, it appears that much of the regulatory evolution that occurred on the sex-linked region influenced expression at the level of splice variants and alleles, as opposed to concurrently governing expression of all transcripts encoded by sex-linked genes.
4. Discussion
(a) . Among population variation of the sex chromosomes of Xenopus borealis
To help disentangle concurrent and confounding drivers of sex chromosome evolution, we scrutinized the independently evolved sex chromosomes of three closely related frog species: the Marsabit clawed frog X. borealis, the African clawed frog X. laevis and the western clawed frog X. tropicalis. Using field-collected animals and genomic and transcriptomic approaches, we demonstrate that: (i) X. borealis has a modest but significant population structure that is relatively recently evolved; (ii) there exists variation among X. borealis populations in the extent of sex chromosome differentiation that corresponds to a previously identified region of recombination suppression [27]; (iii) the sex-linked portion of the X. borealis sex chromosomes (chr8L) carry genes that encode a high number of transcripts with sex-biased expression, and especially female-biased expression; and (iv) sex-biased regulatory evolution evolved in X. borealis independently after divergence from X. laevis, and in association with the origin of sex linkage on the new sex chromosomes of this species. In the independently evolved sex-linked regions of X. laevis (chr2L) and X. tropicalis (chr7), we also identified a high density of sex-biased transcripts encoded by sex-linked transcripts, and we showed that sex-biased regulation evolved independently in association with sex linkage in each of these species as well. These findings are thus concordant across these three frog species, each with independently evolved sex chromosomes, and each with distinctive sex-linked genomic regions. These results point to independent evolution of cis- regulation in the sex-linked regions of each species that occurred contemporaneously with the onset of sex linkage, as opposed to having been present ancestrally in an autosomal precursor of each sex chromosome. When analysis was performed at the gene level on the transcriptome data from all three species, the signal of sex-biased expression by sex-linked genes was less substantial, which suggests that this effect is largely driven by allele-specific and splice variant-specific expression. Technical artefacts undoubtedly also affect our quantification of sex-biased expression, but we have no reason to suspect that this would result in an enrichment of sex-biased expression of transcripts encoded by genes in three different sex-linked regions of three species.
In the laboratory strain of X. borealis previously studied by Furman & Evans [26,29], sex chromosome differentiation was detected that coincided with recombination suppression [29]. Our study suggests that this strain originates from central Kenya (figures 2 and 3), shows that the extent of sex chromosome differentiation varies among populations of X. borealis (figure 4), and links recombination suppression and sex chromosome differentiation to the sex-biased expression of transcripts encoded by sex-linked genes in X. borealis from central and eastern Kenya. In a small sample of X. borealis from eastern Kenya, two previously identified loci on chr8L had female-specific heterozygous sites and a higher abundance of sex-linked markers in females than males (electronic supplementary material, figure S13), which is consistent with female heterogamy. By contrast, a lack of substantial differentiation between male and female X. borealis from western Kenya (figure 4) suggests that the sex chromosomes in these specimens are mostly pseudoautosomal regions that (by definition) recombine in both sexes. We were unable to identify sex-linked variation on chr8L in the western population of X. borealis (electronic supplementary material, figure S13), so it is conceivable that this population has a system for sex determination that is distinctive from X. borealis from central and eastern Kenya in terms of where in the genome the trigger for sex-determination resides, and/or whether males or females are the heterogametic sex.
At this time, we can only speculate about the mechanisms behind recombination suppression in the sex-linked portions of the sex chromosomes of X. borealis from central and eastern Kenya (figure 4). One possibility is that specimens from central and eastern Kenya (including our laboratory strain) have an inversion on the W-chromosome but not on the Z-chromosome and that suppresses recombination between this portion of these sex chromosomes. This inversion could be absent on both sex chromosomes of X. borealis from western Kenya—either because it was lost or because it was never present. If recombination suppression in the central and eastern populations is newly evolved compared to sex chromosomes in the western populations, this would suggest that sex-biased expression of sex-linked transcripts evolved quickly because divergence between these populations is relatively recent. Alternatively and more plausibly, X. borealis from west Kenya may have recently lost recombination suppression on chr8L, for example, if new sex-determining locus evolved on another chromosome in an ancestor of X. borealis from west Kenya. This scenario would be consistent with a more protracted period for sex-biased expression of sex-linked transcripts to arise in the central and eastern population, but it would still be more recently than the age of the most recent common ancestor of X. laevis and X. borealis (approximately 40 Ma [32]) because the sex chromosomes of X. borealis arose after this ancestor diversified [26]. Further study of sex linkage in X. borealis from west Kenya, and in other closely related species in the muelleri species group (figure 2) may help distinguish between these alternatives.
(b) . Effects of sex chromosome evolution on the transcriptome
In three frog species, we observed a higher density of sex-biased transcripts encoded by genes on the sex-linked portion of the sex chromosomes as compared to those encoded by genes located in the rest of the genome. In X. borealis, which has female heterogamy, this higher density of sex-biased transcripts was significantly skewed towards female-biased transcripts compared to the rest of the genome (table 1 and figure 5), but in X. tropicalis, a higher proportion of transcripts with male-biased expression are encoded in the sex-linked region. In X. laevis, there is also a high density of sex-biased transcripts in the sex-linked region, but no substantial skew towards these transcripts being more female- or male-biased as compared to sex-biased transcripts encoded by genes elsewhere. Several factors discussed below could account for strong sex-biased expression skews of transcripts encoded by sex-linked genes, including sex chromosome degeneration, and adaptive evolution of the sex-specific (W- or Y-) or sex-shared (X- or Z-) sex chromosome—including faster-X/Z effects.
(i) . Sex chromosome degeneration
In X. tropicalis, there is polymorphism in sex chromosomes, with W-, Z- and Y-chromosomes being present in different laboratory strains [23] and the Y-chromosome evolved from ancestral Z-chromosomes [28]. Male-biased expression of sex-linked genes is also observed in X. tropicalis tadpole gonad/mesonephros tissue, and patterns of nucleotide polymorphism in expressed transcripts X. tropicalis are consistent with degeneration of the W-chromosome as a primary mechanism driving male-biased expression [28]. The similarity in expression profiles reported here for adult liver tissue suggests that degeneration of the small sex-linked portion of the W-chromosome may account for the male-biased expression of sex-linked transcripts in many or all tissues throughout development.
In X. borealis, however, this explanation does not immediately explain the pattern of sex-biased expression that we observed because a strong skew towards female-biased expression is not predicted from W-chromosome degeneration. One hypothetical scenario that could explain these observations is that this biased expression is a consequence of degeneration of an ancestral Y-chromosome that eventually evolved into the contemporary Z-chromosome in association with an ancestral X-chromosome evolving to become the contemporary W-chromosome (table 1C in [42]). A transition from an XY to a ZW system is theoretically disfavoured by natural selection if it involves fixation of a degenerate Y-chromosome [20], but is still possible if the deleterious load of the ancestral Y-chromosome is modest. Amphibians are not known to have dosage compensation [43] and tolerance of dosage differences between the sexes is evidenced here by the sex-biased expression of transcripts encoded by genes in the sex-linked region (figure 5). However, molecular polymorphism of expressed sex-linked transcripts in male X. borealis is not substantially lower than that of expressed non-sex-linked transcripts (electronic supplementary material). This suggests that if a sex chromosome transition in X. borealis did occur, it did not occur recently because non-recombining portions of Y-chromosomes are expected to have much lower polymorphism than autosomes [1].
(ii) . Adaptive evolution on sex-specific sex chromosome
Another factor that potentially could contribute to higher expression in the heterogametic sex is adaptive evolution on the sex-specific sex chromosome, such as the W-chromosome of X. borealis or the Y-chromosome of X. tropicalis. While we do not have evidence for or against this possibility, adaptive evolution is generally expected to be faster on the X or Z-chromosome than on the Y- or W-chromosome [44,45].
(iii) . Adaptive evolution on sex-shared sex chromosome
In general, theory predicts adaptive evolution is generally faster on the autosomes than on the X-chromosome, but under certain conditions, this will be reversed with faster evolution on the X- (or Z-) chromosome [4]. Theoretical conditions for faster-X evolution include that (i) beneficial mutations are partially or completely recessive, (ii) fitness effects of mutations are similar in each sex, (iii) mutation rates and distributions of fitness effects of mutations are similar in each sex, (iv) the substitution rate is the product of the number of mutations per generation and their fixation probabilities (which are determined by selection coefficients in each sex), and (v) relative effective population size (Ne) on the X-chromosome is three-quarters that of the autosomes [4,46,47]. Departures from these assumptions, such as increased relative Ne of the X- as a consequence of sexual selection on males, can increase faster-X evolution, though this effect is diminished by high levels of random offspring mortality [48]. For faster Z-effects, expectations are similar, though faster-Z effects may be amplified by a higher male than female mutation rate, which is common in many species [49].
By way of comparison, in Drosophila, expression divergence of male-biased genes on the X-chromosome evolves faster than their counterparts in the autosome [50], and signatures of adaptive evolution are prominent in some male-biased genes on the X-chromosome [51]. It is also possible that faster-X (or faster-Z) effects benefit the homogametic sex by favouring the fixation of recessive alleles that are expressed in the heterogametic sex that benefit the homogametic sex. In fruit flies with male heterogamy, for example, there is evidence of faster-X effects in female-biased transcripts [52–54]. In fact, in species with male heterogamy, there is often a higher proportion of female- than male-biased expression in sex-linked transcripts [55–59], and in some species with female heterogamy, there can be an enrichment in sex-linked genes of transcripts with male-biased expression [60,61].
Could faster-Z effects account for male-biased expression in X. borealis? Relevant to this possibility is that adaptation which involves allele frequency changes at many sites throughout the genome (polygenic adaptation) or beneficial mutations that appear recurrently or at a high rate are not necessarily expected to have faster-X evolution [46]. This is because these conditions render the connection between the rate of substitution and the rate of beneficial mutation more complex (and idiosyncratic owing to effects of epistatic interactions for polygenic adaptation) as compared to the rate of adaptive evolution that occurs via a series of successive selective sweeps at individual sites [46]. Additionally, faster-X and faster-Z effects are expected to primarily exploit genetic variation that has been consistently beneficial since the mutations arose, as opposed to exploiting standing genetic variation where fitness effects of a mutation change as a consequence of changed environmental conditions [4,62]. Thus, if the sex-biased expression of transcriptomes encoded by sex-linked genes is owing to faster-X-like and faster-Z effects, this suggests that it should primarily be owing to new mutations, as opposed to standing variation that was previously at mutation-selection equilibrium. These factors collectively limit explanatory power of faster-Z effects as a major driver of widespread skew in the sex-biased expression of sex-linked transcripts in Xenopus or at least argue that it would take considerable time to manifest. Because we have no reason to suspect that mutation rates in Xenopus are particularly unusual, we prefer the hypothesis of degeneration of an ancestral Y-chromosome followed by a transition from a male (XY) to female (ZW) heterogamy.
It would be fascinating for future efforts to evaluate the functional significance of sex-biased expression patterns by comparing populations with distinctive sex chromosomes (e.g. the western versus eastern populations of X. borealis, including zones of intergradation if they exist). The observation of population variation in sex chromosomes of X. borealis (figure 4) opens the possibility that a sex chromosome turnover may have taken place in X. borealis from western Kenya. This raises the questions of what the sex-determination system of the western population of X. borealis is, where it is located in the genome, whether there is a strong skew of transcripts with female-biased expression encoded by sex-linked genes, and whether this population still has the Z-chromosome that is present in the central/eastern population and that potentially could be derived from a degenerate Y-chromosome.
5. Concluding remarks
In the Marsabit clawed frog, X. borealis, there exists population variation in the extent of differentiation between the W- and Z-chromosomes on a large sex-linked region of these sex chromosomes. This sex-linked region has a far higher density of transcripts with sex-biased expression—especially female-biased expression—as compared to orthologous and homeologous genomic regions, and to other recombining pseudoautosomal and autosomal genomic regions. This demonstrates that sex-biased expression of sex-linked genes occurred in concert with the origin of the new sex chromosomes of X. borealis. Similar findings were also recovered from transcriptomes of X. laevis and X. tropicalis, each of which has independently evolved sex chromosomes. Additionally, we found that transcripts encoded by sex-linked genes in X. borealis and X. tropicalis have a strong sex bias (female- or male-biased, respectively). A plausible explanation for the intense, sex-bias of transcripts that emerge from the sex-linked region is that it is owing to degeneration of a sex-specific sex chromosome (the W- in X. tropicalis and an ancestral Y- in X. borealis). For X. borealis, this scenario would involve a recent homologous sex chromosome turnover, wherein an ancestral sex-determination system on chromosome 8L with male (XY) heterogamy evolved into the current system on the same chromosome with female (ZW) heterogamy, with the ancestral X-chromosome becoming the contemporary W-chromosome and the ancestral Y-chromosome becoming the contemporary Z-chromosome. This hypothesis can be tested with comparative data from other populations and species. Still unknown are the functional consequences of extensive sex-biased expression of transcripts that are tethered to various sex-determining loci in different Xenopus species. This raises the question of whether and how this variation might clash or synergize in individuals in nature.
Acknowledgements
We thank Brian Golding for access to computational resources, Jonathan Dushoff and Ben Bolker for statistical advice, two anonymous reviewers for helpful comments, and our funding sources for supporting this research.
Ethics
This work was approved by the Animal Care Committee at McMaster University (AUP no. 17-12-43).
Data accessibility
The RRGS and RNAseq data from this study have been deposited in the Short Read Archive of NCBI (BioProject PRJNA616217). All new Sanger sequences were submitted to GenBank (accession nos MT998323–MT998434).
Authors' contributions
B.L.S.F., D.V.W. and B.J.E. performed the fieldwork. X.-Y.S., B.L.S.F., T.P., M.K., C.M.S.C., I.D. and B.J.E. collected and analysed data. X.-Y.S. and B.J.E. wrote the paper, and all authors edited the paper.
Competing interests
We declare have no competing interests.
Funding
This work was supported by the Natural Science and Engineering Research Council of Canada (RGPIN- 2017-05770, B.J.E.), Resource Allocation Competition awards from Compute Canada (B.J.E.), the Museum of Comparative Zoology at Harvard University (B.J.E.) and the National Research Foundation (NRF) of South Africa (NRF grant no. 87759, J.M.).
References
- 1.Charlesworth B. 2009. Effective population size and patterns of molecular evolution and variation. Nat. Rev. Gen. 10, 195-205. ( 10.1038/nrg2526) [DOI] [PubMed] [Google Scholar]
- 2.Vicoso B, Charlesworth B. 2009. Effective population size and the faster-X effect: an extended model. Evolution 63, 2413-2426. ( 10.1111/j.1558-5646.2009.00719.x) [DOI] [PubMed] [Google Scholar]
- 3.Charlesworth B, Charlesworth D. 2000. The degeneration of Y chromosomes. Phil. Trans. R. Soc. B 355, 1563-1572. ( 10.1098/rstb.2000.0717) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charlesworth B, Coyne JA, Barton NH. 1987. The relative rates of evolution of sex chromosomes and autosomes. Am. Nat. 130, 113-146. ( 10.1086/284701) [DOI] [Google Scholar]
- 5.Rice WR. 1987. The accumulation of sexually antagonistic genes as a selective agent promoting the evolution of reduced recombination between primitive sex chromosomes. Evolution 41, 991-914. ( 10.1111/j.1558-5646.1987.tb05872.x) [DOI] [PubMed] [Google Scholar]
- 6.Spigler RB, Lewers KS, Main DS, Ashman T-L. 2008. Genetic mapping of sex determination in a wild strawberry, Fragaria virginiana, reveals earliest form of sex chromosome. Heredity 101, 507-517. ( 10.1038/hdy.2008.100) [DOI] [PubMed] [Google Scholar]
- 7.Charlesworth B. 1991. The evolution of sex chromosomes. Science 251, 1030-1033. ( 10.1126/science.1998119) [DOI] [PubMed] [Google Scholar]
- 8.Charlesworth B, Wall JD. 1999. Inbreeding, heterozygote advantage and the evolution of neo–X and neo–Y sex chromosomes. Proc. R. Soc. Lond. B 266, 51-56. ( 10.1098/rspb.1999.0603) [DOI] [Google Scholar]
- 9.Charlesworth D, Charlesworth B, Marais G. 2005. Steps in the evolution of heteromorphic sex chromosomes. Heredity 95, 118-128. ( 10.1038/sj.hdy.6800697) [DOI] [PubMed] [Google Scholar]
- 10.Lahn BT, Page DC. 1999. Four evolutionary strata on the human X chromosome. Science 286, 964-967. ( 10.1126/science.286.5441.964) [DOI] [PubMed] [Google Scholar]
- 11.Sun Y, Svedberg J, Hiltunen M, Corcoran P, Johannesson H. 2017. Large-scale suppression of recombination predates genomic rearrangements in Neurospora tetrasperma. Nat. Comm. 8, 1140. ( 10.1038/s41467-017-01317-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brooks LD. 1988. The evolution of recombination rates. In The evolution of sex, pp. 87–105. Sunderland, MA: Sinauer Associates. [Google Scholar]
- 13.Hill WG, Robertson A. 1966. The effect of linkage on limits to artificial selection. Gen. Res. 8, 269-294. ( 10.1017/S0016672300010156) [DOI] [PubMed] [Google Scholar]
- 14.Skaletsky H, et al. 2003. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423, 825-837. ( 10.1038/nature01722) [DOI] [PubMed] [Google Scholar]
- 15.Masly JP, Presgraves DC. 2007. High-resolution genome-wide dissection of the two rules of speciation in Drosophila. PLoS Biol. 5, 1890-1898. ( 10.1371/journal.pbio.0050243) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wallis MC, Waters PD, Delbridge ML, Kirby PJ, Pask AJ, Grützner F, Rens W, Ferguson-Smith MA, Graves JAM. 2007. Sex determination in platypus and echidna: autosomal location of SOX3 confirms the absence of SRY from monotremes. Chr. Res. 15, 949. ( 10.1007/s10577-007-1185-3) [DOI] [PubMed] [Google Scholar]
- 17.Vicoso B, Bachtrog D. 2015. Numerous transitions of sex chromosomes in Diptera. PLoS Biol. 13, e1002078. ( 10.1371/journal.pbio.1002078) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darolti I, et al. 2019. Extreme heterogeneity in sex chromosome differentiation and dosage compensation in livebearers. Proc. Natl Acad. Sci. USA 116, 19 031-19 036. ( 10.1073/pnas.1905298116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adolfsson S, Ellegren H. 2013. Lack of dosage compensation accompanies the arrested stage of sex chromosome evolution in ostriches. Mol. Biol. Evol. 30, 806-810. ( 10.1093/molbev/mst009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saunders PA, Neuenschwander S, Perrin N. 2019. Impact of deleterious mutations, sexually antagonistic selection, and mode of recombination suppression on transitions between male and female heterogamety. Heredity 123, 419-428. ( 10.1038/s41437-019-0225-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu L, Walters JR. 2017. Evolution of sex chromosome dosage compensation in animals: a beautiful theory, undermined by facts and bedeviled by details. Genome Biol. Evol. 9, 2461-2476. ( 10.1093/gbe/evx154) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bachtrog D, et al. 2014. Sex determination: why so many ways of doing it? PLoS Biol. 12, e1001899. ( 10.1371/journal.pbio.1001899) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roco ÁS, Olmstead AW, Degitz SJ, Amano T, Zimmerman LB, Bullejos M. 2015. Coexistence of Y, W, and Z sex chromosomes in Xenopus tropicalis. Proc. Natl Acad. Sci. USA 112, E4752-E4761. ( 10.1073/pnas.1505291112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshimoto S, et al. 2008. A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proc. Natl Acad. Sci. USA 105, 2469-2474. ( 10.1073/pnas.0712244105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bewick AJ, Anderson DW, Evans BJ. 2011. Evolution of the closely related, sex-related genes DM-W and DMRT1 in African clawed frogs (Xenopus). Evolution 65, 698-712. ( 10.1111/j.1558-5646.2010.01163.x) [DOI] [PubMed] [Google Scholar]
- 26.Furman BLS, Evans BJ.. 2016. Sequential turnovers of sex chromosomes in African clawed frogs (Xenopus) suggest some genomic regions are good at sex determination. G3 6, 3625–3633. ( 10.1534/g3.116.033423) [DOI] [PMC free article] [PubMed]
- 27.Tymowska J. 1991. Polyploidy and cytogenetic variation in frogs of the genus Xenopus. In Amphibian cytogenetics and evolution (eds Green DM, Sessions SK), pp. 259-297. San Diego, CA: Academic Press. [Google Scholar]
- 28.Furman BLS, Cauret CM, Knytl M, Song, XY, Premachandra T, Ofori-Boateng C, Jordan DC, Horb ME, Evans BJ. 2020. A frog with three sex chromosomes that co-mingle together in nature: Xenopus tropicalis has a degenerate W- and a Y- that evolved from a Z-. PLoS. Gen. 16, e1009121. ( 10.1371/journal.pgen.1009121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Furman BLS, Evans BJ. 2018. Divergent evolutionary trajectories of two young, homomorphic, and closely related sex chromosome systems. Genome Biol. Evol. 10, 742-755. ( 10.1093/gbe/evy045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans BJ. 2008. Genome evolution and speciation genetics of clawed frogs (Xenopus and Silurana). Front Biosci. 13, 4687-4706. ( 10.2741/3033) [DOI] [PubMed] [Google Scholar]
- 31.Session AM, et al. 2016. Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538, 336-343. ( 10.1038/nature19840) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Evans BJ, et al. 2019. Xenopus fraseri: Mr. Fraser, where did your frog come from? PLoS ONE 14, e0220892. ( 10.1371/journal.pone.0220892) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuda Y, Uno Y, Kondo M, Gilchrist MJ, Zorn AM, Rokhsar DS, Schmid M, Taira M. 2015. A new nomenclature of Xenopus laevis chromosomes based on the phylogenetic relationship to Silurana/Xenopus tropicalis. Cytogenet Genome Res. 145, 187-191. ( 10.1159/000381292) [DOI] [PubMed] [Google Scholar]
- 34.Khokha MK, et al. 2009. Rapid gynogenetic mapping of Xenopus tropicalis mutations to chromosomes. Dev. Dyn. 238, 1398-1346. ( 10.1002/dvdy.21965) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA. 2008. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 3, e3376. ( 10.1371/journal.pone.0003376) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karimi K, et al. 2018. Xenbase: a genomic, epigenomic and transcriptomic model organism database. Nucleic Acids Res. 46(D1), D861-D868. ( 10.1093/nar/gkx936) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu TD, Watanabe CK. 2005. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21, 1859-1875. ( 10.1093/bioinformatics/bti310) [DOI] [PubMed] [Google Scholar]
- 38.Krumsiek J, Arnold R, Rattei T. 2007. Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics 23, 1026-1028. ( 10.1093/bioinformatics/btm039) [DOI] [PubMed] [Google Scholar]
- 39.Andrews S. 2019. re-DOT-able. Version: 1.1. See https://github.com/s-andrews/redotable.
- 40.Mitros T, et al. 2019. A chromosome-scale genome assembly and dense genetic map for Xenopus tropicalis. Dev. Biol. 452, 8-20. ( 10.1016/j.ydbio.2019.03.015) [DOI] [PubMed] [Google Scholar]
- 41.Furman BLS, Dang UJ, Evans BJ, Golding GB. 2018. Divergent subgenome evolution after allopolyploidization in African clawed frogs (Xenopus). J. Evol. Biol. 31, 1945-1958. ( 10.1111/jeb.13391) [DOI] [PubMed] [Google Scholar]
- 42.Bull JJ, Charnov EL. 1977. Changes in the heterogametic mechanism of sex determination. Heredity 39, 1. ( 10.1038/hdy.1977.38) [DOI] [PubMed] [Google Scholar]
- 43.Malcom JW, Kudra RS, Malone JH. 2014. The sex chromosomes of frogs: variability and tolerance offer clues to genome evolution and function. J. Genom. 2, 68. ( 10.7150/jgen.8044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orr HA, Kim Y. 1998. An adaptive hypothesis for the evolution of the Y chromosome. Genetics 150, 1693-1698. ( 10.1093/genetics/150.4.1693) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rice WR. 1996. Evolution of the Y sex chromosome in animals. Bioscience 46, 331-343. ( 10.2307/1312947) [DOI] [Google Scholar]
- 46.Connallon T, Singh ND, Clark AG. 2012. Impact of genetic architecture on the relative rates of X versus autosomal adaptive substitution. Mol. Biol. Evol. 29, 1933-1942. ( 10.1093/molbev/mss057) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meisel RP, Connallon T. 2013. The faster-X effect: integrating theory and data. Trends Gen. 29, 537-544. ( 10.1016/j.tig.2013.05.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pischedda A, Friberg U, Stewart AD, Miller PM, Rice WR. 2015. Sexual selection has minimal impact on effective population sizes in species with high rates of random offspring mortality: an empirical demonstration using fitness distributions. Evolution 69, 2638-2647. ( 10.1111/evo.12764) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson Sayres MA, Makova KD. 2011. Genome analyses substantiate male mutation bias in many species. Bioessays 33, 938-945. ( 10.1002/bies.201100091) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Llopart A. 2012. The rapid evolution of X-linked male-biased gene expression and the large-X effect in Drosophila yakuba, D. santomea, and their hybrids. Mol. Biol. Evol. 29, 3873-3886. ( 10.1093/molbev/mss190) [DOI] [PubMed] [Google Scholar]
- 51.Baines JF, Sawyer SA, Hartl DL, Parsch J. 2008. Effects of X-linkage and sex-biased gene expression on the rate of adaptive protein evolution in Drosophila. Mol. Biol. Evol. 25, 1639-1650. ( 10.1093/molbev/msn111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Connallon T, Clark AG. 2011. Association between sex-biased gene expression and mutations with sex-specific phenotypic consequences in Drosophila. Genome Biol. Evol. 3, 151-155. ( 10.1093/gbe/evr004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grath S, Parsch J. 2012. Rate of amino acid substitution is influenced by the degree and conservation of male-biased transcription over 50 myr of Drosophila evolution. Genome Biol. Evol. 4, 346-359. ( 10.1093/gbe/evs012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Müller L, Grath S, Von Heckel K, Parsch J. 2012. Inter-and intraspecific variation in Drosophila genes with sex-biased expression. Int. J. Evol. Biol. 2012, 963976. ( 10.1155/2012/963976) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Albritton SE, Kranz A-L, Rao P, Kramer M, Dieterich C, Ercan S. 2014. Sex-biased gene expression and evolution of the X chromosome in nematodes. Genetics 197, 865-883. ( 10.1534/genetics.114.163311) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khil PP, Smirnova NA, Romanienko PJ, Camerini-Otero RD. 2004. The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nat. Gen. 36, 642-646. ( 10.1038/ng1368) [DOI] [PubMed] [Google Scholar]
- 57.Leder EH, Cano JM, Leinonen T, O'hara RB, Nikinmaa M, Primmer CR, Merilä J. 2010. Female-biased expression on the X chromosome as a key step in sex chromosome evolution in threespine sticklebacks. Mol. Biol. Evol. 27, 1495-1503. ( 10.1093/molbev/msq031) [DOI] [PubMed] [Google Scholar]
- 58.Ranz JM, Castillo-Davis CI, Meiklejohn CD, Hartl DL. 2003. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science 300, 1742-1745. ( 10.1126/science.1085881) [DOI] [PubMed] [Google Scholar]
- 59.Reinius B, Johansson MM, Radomska KJ, Morrow EH, Pandey GK, Kanduri C, Sandberg R, Williams RW, Jazin E. 2012. Abundance of female-biased and paucity of male-biased somatically expressed genes on the mouse X-chromosome. BMC Genomics 13, 607. ( 10.1186/1471-2164-13-607) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaiser VB, Ellegren H. 2006. Nonrandom distribution of genes with sex-biased expression in the chicken genome. Evolution 60, 1945-1951. ( 10.1111/j.0014-3820.2006.tb00537.x) [DOI] [PubMed] [Google Scholar]
- 61.Pucholt P, Wright AE, Conze LL, Mank JE, Berlin S. 2017. Recent sex chromosome divergence despite ancient dioecy in the willow Salix viminalis. Mol. Biol. Evol. 34, 1991-2001. ( 10.1093/molbev/msx144) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Orr HA, Betancourt AJ. 2001. Haldane's sieve and adaptation from the standing genetic variation. Genetics 157, 875-884. ( 10.1093/genetics/157.2.875) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The RRGS and RNAseq data from this study have been deposited in the Short Read Archive of NCBI (BioProject PRJNA616217). All new Sanger sequences were submitted to GenBank (accession nos MT998323–MT998434).