Abstract
With or without sex chromosomes, sex determination is a synthesis of many molecular events that drives a community of cells towards a coordinated tissue fate. In this review, we will consider how a sex determination pathway can be engaged and stabilized without an inherited genetic determinant. In many reptilian species, no sex chromosomes have been identified, yet a conserved network of gene expression is initiated. Recent studies propose that epigenetic regulation mediates the effects of temperature on these genes through dynamic post-transcriptional, post-translational and metabolic pathways. It is likely that there is no singular regulator of sex determination, but rather an accumulation of molecular events that shift the scales towards one fate over another until a threshold is reached sufficient to maintain and stabilize one pathway and repress the alternative pathway. Investigations into the mechanism underlying sex determination without sex chromosomes should focus on cellular processes that are frequently activated by multiple stimuli or can synthesize multiple inputs and drive a coordinated response.
This article is part of the theme issue ‘Challenging the paradigm in sex chromosome evolution: empirical and theoretical insights with a focus on vertebrates (Part I)’.
Keywords: environmental sex determination, temperature-dependent sex determination, epigenetics
1. Introduction
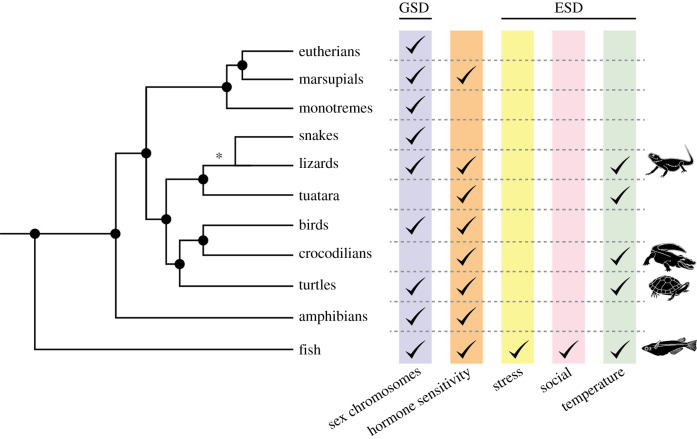
Sex determination is classically considered to be the process by which an organism initiates the development of ovaries or testes from the embryonic gonad. Because differentiation of the gonad into an ovary or testis typically drives the development of other sex-specific traits such as genitalia through hormone production, the outcome of this developmental decision is central to the phenotypic sex of the organism. For this reason, this review will use the terms ‘female’ and ‘male’ to describe the pathways that drive the gonad towards an ovary or testis fate. Gonadal sex determination is classified as being either genetically activated (genetic sex determination, GSD) or environmentally driven (environmental sex determination, ESD). GSD mechanisms include the presence or dosage of sex chromosomes or the accumulation of specific gene products that initiate development of one sex or the other. In ESD, exogenous forces such as temperature, stress or social dynamics initiate molecular cascades that bias the system towards a female or male fate. Modes of ESD have arisen independently in multiple lineages (figure 1).
Figure 1.
Environmental sex determination (ESD) has evolved repeatedly in multiple vertebrate lineages. Multiple sex determining systems can operate simultaneously. Specific examples discussed in this review are indicated by an animal icon. We have broken the Squamates node (indicated by an asterisk) into lizards and snakes to highlight the observation that while ESD has evolved multiple times in many lizard species, this phenomena has not yet been described in snakes. Adapted from [1] and [2].
Evidence is accumulating that sex chromosomes directly influence the fate of other, non-gonadal tissues in GSD species [3,4]. However, in species where sex chromosomes are absent, ESD forces control the differentiation of all sex traits through the effects of steroid hormones and other factors produced by the gonad. Thus, ESD is a fascinating example of how the environment directly influences molecular pathways that drive the development and subsequent life history of an individual.
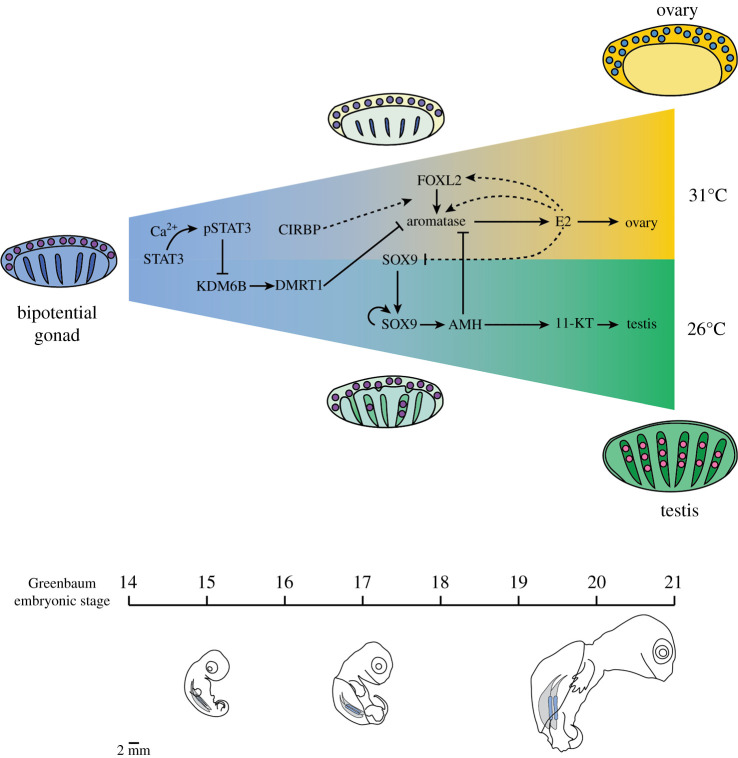
The bipotential early gonad arises from the mesonephros, usually during the middle third of embryonic development. In most vertebrates, this early gonad has a coelomic epithelium encompassing a medullary compartment. However, the organization of these regions, and the future gonad structures they give rise to, are variable [5–7]. Despite the fact that the process of morphogenesis varies, the adult structure of the ovary and testis is strikingly similar across vertebrates. While no single factor is responsible for activation of ovary or testis development in all vertebrate species, sex determination pathways seem to converge on the same cohort of genetic regulators that maintain and stabilize gonad fate. For example, in all vertebrates investigated so far: SF1 and WT1 are involved in establishment of gonadal cells; SOX9 is the defining marker of the supporting cells of testis cords; AMH inhibits the development of the female reproductive tract; Aromatase (CYP19A1) is required for production of oestrogen from androgens; and FOXL2 is required for female-supporting cell differentiation (figure 2) (reviewed by [1]). In mammals, the Y-chromosome-linked gene Sry, is the dominant male determinant [9]. DMRT1 plays a dominant role at the top of the hierarchy in birds, turtles, frogs and fishes, whereas, its role in mammals seems to be to maintain male fate (reviewed by [10]). Curiously, other genes that are part of the female and male pathways are not always activated in the same order [11,12]. While the reason for this is not yet understood, it suggests that the overall network rather than a fixed sequence of expression is important. In many species, manipulation of steroid hormones during embryonic development can partially or completely redirect sexual trajectory of the embryo [13–16]. Therefore, the regulatory control of aromatase (CYP19A1) expression is of critical importance to understanding gonad differentiation.
Figure 2.
Sex determination is a cumulative process. Female- and male-sex determination pathways are initiated in the bipotential gonad and are maintained over the course of multiple embryonic stages until ovary or testis fate is stabilized. Proposed mechanisms of sex determination in the red-eared slider turtle T. scripta are diagrammed in relation to onset of expression per Greenbaum embryonic stage [8].
Female and male pathways are mutually antagonistic, with ovary regulatory loops promoting differentiation of ovary fates while simultaneously repressing testis differentiation genes and vice versa, driving the gonad towards a coordinated fate [17–20]. Adding another level of complexity, the gonad is composed of multiple cell types, including germ cells (future oogonia or spermatogonia) and somatic cells, which include supporting cells (granulosa/Sertoli) and steroidogenic cells (theca/Leydig). All three of these cell types are reported to be drivers of sex determination in different vertebrate species (reviewed by [1]). While supporting cells initiate sex determination in mammals [21–26], steroidogenic pathways seem to be entry points in sex-reversing fishes (reviewed by [27]), and the number of germ cells can drive sex determination in zebrafish and medaka [28,29]. Because of the feedback regulatory loops that coordinate a single ovary or testis gonadal fate, anything that biases the regulatory loop in favour of one pathway over the other could be considered the sex determining switch, regardless of the cell type in which it occurs or whether that bias is inherited (like a sex chromosome) or is the result of environmental input (like temperature) [30].
The large number of genetic and cellular processes that can contribute to sex determination suggests that the process is cumulative. This is most evident in ESD species, where the developmental window during which the environment can influence sex is quite long, extending over a period of weeks. For example, in species with temperature-dependent sex determination (TSD) (first described by [31,32]), shifting eggs between the female and male temperatures can switch the sexual trajectory up until when morphogenesis of the ovary and testis diverge days to weeks later [33]. The later the shift occurs during this window, the fewer animals undergo a change in trajectory, consistent with the idea that (i) the process is cumulative and (ii) the outcome varies across the population, perhaps because individuals either experience different inputs or respond differently to those inputs. When no sex chromosomes or dominant genetic determinants are present to provide a genetic bias towards one pathway or another, what inputs drive the gonad to commit to becoming an ovary or testis? How do environmental cues intersect with sex determination pathways, and how do gonadal cells interpret potentially conflicting environmental cues, like fluctuating temperature?
In this review, we will consider how one of the two sex determination pathways can be engaged without an inherited genetic determinant. Although we will draw from multiple ESD systems, we will focus primarily on the red-eared slider turtle. In this TSD species, a histone demethylase regulated by environmental temperature controls the epigenetic state of a key sex determining gene that is critical to the activation of the testis pathway [34,35]. In this case, temperature affects a post-translational modification, but we will also review evidence that temperature regulates allelic expression, protein stability and RNA-splicing—all of which may contribute to an expression bias in female or male pathways that leads to the resolution of sexual fate over time.
2. Epigenetic control of sex determination
The epigenetic state of genes in the sex determination cascade appears to be critical in the activation of ovary or testis-specific pathways in both GSD and ESD (reviewed by [36]). In the mouse GSD system, a Jumonji histone demethylase KDM3A (JMJD1A) is required for activation of Sry, the Y-chromosome-linked sex determining gene at the top of the male pathway [37]. Another study showed that at the bipotential stage, supporting cell progenitors in the XX and XY mouse gonad carry active and repressive histone marks (H3K4me3 and H3K27me3) on many genes associated with both the female and male pathways. This suggests that genes of both pathways are bivalently poised for activation or repression in the early embryonic gonad. After sex determination has occurred, genes associated with the pathway that is activated lose their repressive marks while genes associated with the alternative pathway remain bivalent [38,39]. These results suggest that maintenance of—or changes to—the epigenetic state of a gene is a critical step in activation of ovary- or testis-specific development.
Several studies proposed that epigenetic regulation mediates the effects of environmental forces on gene expression to initiate female or male-sex determination pathways. Aromatase was an early focus of epigenetic studies, owing to its crucial role in driving and maintaining female sexual fate [40,41]. Indeed, exposure to high temperatures led to increased methylation at a gonad-specific aromatase promoter and correlated with male development in the European sea bass [40]. When eggs at the male producing temperature (MPT) were shifted to the female producing temperature (FPT), less CpG methylation was observed near FOXL2 and SF1 binding sites in the 5′ flanking region of the Cyp19a1 promoter. However, temperature-dependent methylation patterns disappeared after the end of the temperature-sensing period, suggesting that DNA methylation may serve as a temporary mark that helps maintain activation of the female pathway through expression of aromatase, after which other regulatory mechanisms take over [42]. There is also evidence that environmental toxins such as polychlorinated biphenyl (PCB) exposure can interfere with oestrogen signalling and affect DNA methylation at key sex determination genes. Exposure to PCBs biased sex ratios towards female fates but DNA methylation around the aromatase promoter was atypical for FPT embryos, which maintained an MPT-typical pattern. These data suggest that DNA methylation is not required for initiation of female or male pathways [43]. Whether changes in methylation are causative or simply reflective of a change in the activity of the gene has been difficult to determine. In support of a causative role, one study in the half-smooth tongue sole Cynoglossus semilaevis indicated that changes in methylation at Dmrt1 caused by temperature exposure were heritable and capable of over-riding chromosomal sex in the next generation [44]. How temperature might be linked to changes in methylation has not been elucidated.
More recently, attention has shifted to the role of histone modifications driven in part by the discovery that transcriptional changes in two Jumonji-family histone demethylases, JARID2 and KDM6B, show a high correlation with temperature of incubation in multiple reptile species, including the red-eared slider turtle Trachemys scripta elegans, the American alligator Alligator mississippiensis and the Australian central bearded dragon, Pogona vitticeps [12,35,45,46].
These findings, coupled with data showing that removal of H3K27me3 is associated with activation of the male pathway in mice [39] led to a functional experiment to knock-down the H3K27me3 demethylase, Kdm6b, in T. scripta. This experiment resulted in a failure to demethylate Dmrt1 and the development of greater than 80% females at the male producing temperature, suggesting that epigenetic regulation is an intersection point between temperature and molecular pathways [35].
3. Molecular signalling changes that capture temperature cues
In T. scripta, at the earliest stages of gonad development, expression of Kdm6b is specific to the male-producing temperature, suggesting a primary effect of temperature on activation or silencing of the gene. This finding led to an investigation of known regulators of Kdm6b in other contexts. One such regulator is signal transducer and activator of transcription 3 (STAT3) which can activate or repress Kdm6b depending on its phosphorylation status and binding partners [47,48]. pSTAT3 is activated through phosphorylation of the tyrosine 705 residue and forms a dimer that enters the nucleus, binds DNA and controls gene expression [49]. To test whether STAT3 regulates Kdm6b in T. scripta, several inhibitors were used to block its phosphorylation and dimerization. Results showed that inhibition of pSTAT3 did not affect the male pathway but caused upregulation of Kdm6b and Dmrt1 expression at the female-producing temperature, indicating that during sex determination in T. scripta, pSTAT3 inhibits expression of Kdm6b at female-producing temperatures [34].
Interestingly, phosphorylation of STAT3 can be regulated by calcium signalling pathways known to be activated in response to temperature and other environmental signals [49–55]. Experiments showed that higher female temperatures led to a robust elevation of calcium in T. scripta gonad cells and phosphorylation of STAT3, an effect that could be mimicked by stimulation of turtle gonad cells with an ionophore [34]. These experiments implicate ion channels and calcium signalling in the regulation of STAT3 activity, but it is not yet clear which ion channels are involved, or whether calcium acts directly through a calmodulin-dependent protein kinase (CAMK) or by initiating downstream changes in calcium stores in the cell, a cascade which could trigger STAT activation indirectly. It has been proposed that both calcium and redox status (CaRe) serve as cellular sensors of environmental cues in ESD species and work through common ancient cellular pathways such as the JAK/STAT pathway [56].
A class of well-characterized environmentally sensitive proteins, transient receptor potential channels (TRPs), may function as temperature-sensors in TSD. TRP channels are a family of ion channels that open or close in response to temperature and other external stimuli including pain, pressure and taste [57]. They are also permeable to a number of cations such as Ca2+ [57]. TRP-V and TRP-M classes are most frequently associated with temperature-sensing and are expressed all over the vertebrate body [57], and specifically in the T. scripta gonad [12]. TRPV4, a warm-sensing channel typically activated between 27̊C and 35̊C, similar to the nest temperature for many reptiles with TSD, was investigated in the American alligator A. mississippiensis through the use of specific agonists and antagonists. In these experiments, TRPV4 activity was correlated with activation of Sox9 and Amh at warm temperatures (which are male-producing in crocodilians) [58]. Another group of researchers investigated TRPV1 in thermosensation and sex ratios in Maremys reevesii, another freshwater turtle with TSD, and found that inhibition of TRPV1 during embryonic development resulted in fewer female hatchlings [59]. Further experiments are required to establish these links in the pathway.
4. Post-transcriptional modification and RNA processing
Alternative splicing is a key regulator of sex-specific gene expression in Drosophila melanogastor, many other insects, and Caenorhabditis elegans [60–63]. Splicing may play a significant role in the initiation or maintenance of vertebrate sex determination as well. Deveson et al. [45] reported sexually dimorphic patterns of alternative splicing in three different reptiles with TSD, notably in the Jumonji-family chromatin modifiers Kdm6b and Jarid2. In all three cases, alternative splicing led to intron retention (IR), introducing a premature stop codon and protein truncation expected to lead to a non-functional protein. This work raised a number of questions. In species with TSD, IR predominantly occurred at cooler temperatures, which in T. scripta (26°C) is male-producing, but in A. mississippiensis (30°C) is female-producing. However, IR was also observed in ZZ dragons sex-reversed to female at higher temperature. Together, these data showed that specific splicoforms of Jumonji-family chromatin modifiers are consistently present, but are not consistently associated with one sex or one temperature, at least in the three species studied [45]. It is possible that Jumonji-family chromatin regulators have been co-opted repeatedly and used in different contexts in different sex determination networks. Although loss of Kdm6b led to a failure to demethylate Dmrt1 and to initiate testis development in T. scripta [35], the functional role of splicing variants has not been investigated.
Adding further complexity, JARID2 and KDM6B have opposite functions. JARID2 is part of the polycomb repression complex (PRC), which catalyses the deposition of H3K27me3 at target loci [64], whereas KDM6B, an H3K27me3 demethylase, removes the H3K27me3 repressive mark and activates silent genes [65]. The effects of splicing on protein structure/function, may provide some answers that reconcile details of previous studies. For example, truncation of either of these proteins could affect specificity, binding, or stability. A recent study showed that JARID2 can exist as a lower molecular weight protein lacking the N-terminal PRC2 interacting domain (N-JARID2). In this form, it can serve as an activator [66]. Additionally, histone demethylases such as Kdm6b may have roles in the recruitment of activators independent of their demethylase activity [67]. Further experiments are required to develop a coherent idea of how splicing differences of these chromatin regulators affects the establishment or maintenance of female and male pathways.
In addition to a direct role in regulating histone modifiers, splicing probably contributes to broader changes in the transcriptional landscape. In ESD species, alternative splicing and mRNA processing could provide the initial bias that drives one pathway over another or reinforce chromatin and gene expression changes that must be maintained to stabilize gonad fate. Interestingly, the activity of CDC-like kinases (CLKs) responds to physiological temperature: lower body temperature results in elevated phosphorylation of splicing-activating proteins which results in alternative splicing in response to temperature. CLK4 has been proposed to regulate splicing in reptilian species with TSD. One alternatively spliced target of CLK4 is the cold-inducible RNA binding protein, CIRBP, which has been implicated in TSD (see below). Splicoform differences in this transcript may control RNA stability [68]. A global analysis of splicing events in reptiles with TSD will be highly informative. How alternative splicing events contribute to sex determination pathways and changes in the chromatin landscape remain open questions.
5. Proteins whose activity is influenced by temperature or other environmental signals
Another class of responders to environmental cues involve proteins whose expression or activity (owing to structural changes or protein modifications) is influenced by temperature. Heat-shock proteins (HSPs) are classic sensors of heat stress, but are indicated in a number of other physiological responses including cold, UV light and inflammation [69,70]. Several HSPs demonstrate sexually dimorphic expression in T. scripta [12,71], and could act as cellular chaperones to facilitate differential protein folding and stability of sex-specific proteins under different environmental conditions [69,70].
Sensitivity to environmental forces also may result from allelic variants that alter the response of proteins to differences in temperature or other environmental conditions. For example, an inherited thermosensitive single nucleotide polymorphism (SNP) in CIRBP was identified in gonads from the common snapping turtle Chelydra serpentina [72]. In other species, CIRBP responds to a number of environmental stimuli including temperature, UV light and hypoxia and contributes to mRNA processing and stabilization [51,73]. In C. serpentina, the CIRBP A allele was induced at warmer, female-producing temperatures and AA (or to a lesser extent AC) genotypes were more likely to develop ovaries. Different CIRBP allele frequencies were observed in proximate turtle populations, which could influence TSD patterns and subsequent sex ratios in a population-specific way [74]. Additionally, there is evidence that CIRBP regulates STAT3 phosphorylation through NF-κβ and JAK signalling [75,76].
Several investigators have hypothesized that temperature influences sexual fate indirectly through activation of stress response pathways and changing the redox status of the cells [45,56,74]. The stress hormone cortisol is thought to be central to environmental sex determination and sex change in reef fishes [77]. In sequentially hermaphroditic reef fishes, social changes can trigger sexual transitions via intersection of the hypothalamic-pituitary-interrenal pathway and hypothalamic-pituitary-gonad pathways: perceived social changes induce changes in cortisol, which in turn affects steroid hormone synthesis via gonadotropins released from the pituitary. For example, in protogynous species, the death of the dominant male triggers a drop in cortisol in the alpha female and transition into a fertile male, whereas addition of cortisol can have the opposite effect [77]. Further support for the idea that these pathways intersect come from studies in birds showing that sex ratios can be manipulated through modulation of yolk corticosterone levels [78].
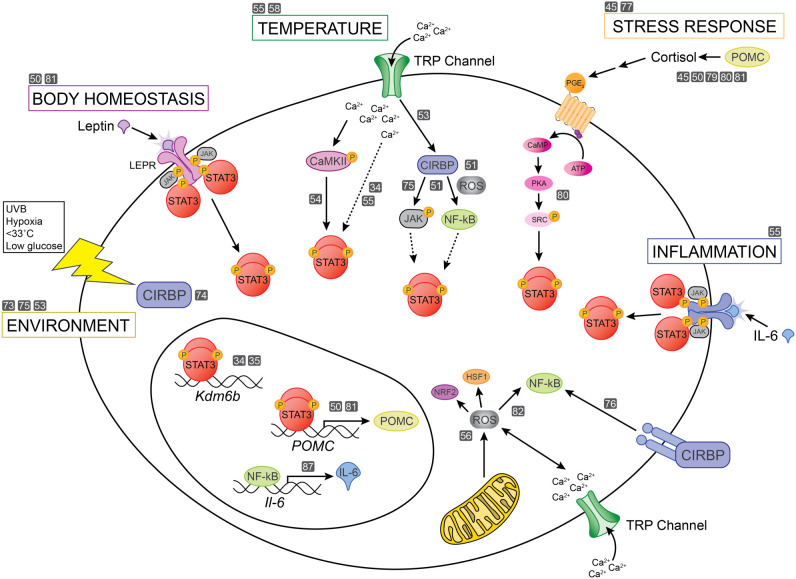
Deveson et al. [45] proposed that the pituitary stress response pathways could be working together with CIRBP to mediate environmental stress in species with TSD. Intriguingly, propiomelanocortin (POMC), the precursor of adrenocorticotropic hormone (ACTH) which regulates levels of cortisol, is upregulated in sex-reversed P. vitticeps. In P. vitticeps, this stress response correlates with changes in alternative splicing of Jumonji-family histone modifiers, suggesting a relationship between cellular stress and sex-specific gene expression changes [45]. Fascinatingly, CIRBP can also regulate POMC transcription and promote cortical stress response [79,80]. Redox-regulated signalling triggered by stress responses or other inputs can also work through the JAK-STAT signalling pathway [81,82] and could act through epigenetic regulators or other pathways to control ESD. Many of these potential interactions are summarized in figure 3.
Figure 3.
Sex determination is a complex synthesis of many molecular events. Without sex chromosomes, initiation and stabilization of sexual fate probably involves post-transcriptional, post-translational and metabolic pathways that affect gene regulatory events. An accumulation of molecular events will be sufficient to maintain and stabilize one pathway and repress the alternative pathway. Here, we show how many environmental pathways converge upon STAT3 activation, which critically regulates epigenetic activation of male gene expression. Grey boxes with numbers refer to the Reference list.
6. How is the temperature-dependent sex determination system robust to fluctuating temperature in the wild?
In the laboratory, reptilian eggs are typically incubated at a constant temperature, but this is obviously unlike the wild, where temperatures fluctuate day to night, in response to variable rainfall throughout the incubation period, and across the species environmental range. How is sex determination robust to constant environmental changes?
In general, this may have to do with the cumulative input over a long temperature sensitive period combined with the multiple feedback loops that stabilize one pathway and repress the alternative pathway. However, the key to understanding how this works in T. scripta (the system that we know the most about at present) may be learning more about how phosphorylation of STAT3 is regulated. pSTAT3 is reported to have a long half-life greater than 12 h [83]. A long half-life of the protein could mean that FPT temperatures need only reach high levels periodically to repress transcription of Kdm6b, and activation of the male pathway. Proteins active at MPT could also control pSTAT3 through de-phosphorylation, active repression of the STAT3 activator(s), or phosphorylation of alternative STAT3 sites. For example, protein tyrosine phosphatase (PTP) family members are known to negatively regulate STAT3 activation through dephosphorylation of the tyrosine 705 residue that is required for STAT3 activity [84]. Most of the PTPs known to dephosphorylate STAT3 are expressed in the T. scripta embryonic gonad during the TSP. Intriguingly, one of these candidates, PTPRD, is significantly upregulated at MPT [12] and can directly interact with STAT3 [85]. While there are no reports that PTPRD is itself temperature sensitive, PTP family members are known to be regulated by oxidation [86]. Therefore, an intriguing possibility is that PTPs mediate changes in cellular calcium and redox regulation (CaRe) [56] by controlling STAT3 activity. Male-specific upregulation of PTPRD past stage 16 could act to stabilize commitment to male fate.
7. Conclusion
Sex determination is a complex synthesis of many molecular events over time that drives a community of cells towards a tissue fate. Temperature is the driving force behind sex determination in species like T. scripta, but we know that ovary and testis development in the turtle are also influenced by allelic differences [74] and hormone production (reviewed in [41]), and in other species such as sex-reversing fishes, by stress and interplay between the hypothalamic-pituitary-interrenal pathway and hypothalamic-pituitary-gonad pathways. For decades, the field has sought the molecular mechanism underlying TSD. However, as the data in this review demonstrate, this process is remarkably complex and dynamic and may involve post-transcriptional, post-translational and metabolic pathways (figure 3). It is likely that there is no singular regulator of sex determination, but rather an accumulation of molecular events that shift the scales towards one fate over another until a threshold is reached sufficient to maintain and stabilize one pathway and repress the alternative pathway.
In TSD species, calcium-sensitive channels are emerging as key transducers of environmental signals that intersect with epigenetic regulation of important genes in the pathways. Whether other environmental signals (such as stress and redox signalling) converge on STAT3 and epigenetic regulators remains to be seen. More details of the intracellular pathways that relay the external signal to the transcriptional network will probably be forthcoming in the next few years as genomes and transcriptomes become available and functional experiments come within reach in many different organisms. Investigations into the mechanism underlying TSD should focus on cellular processes that are frequently activated by multiple stimuli or can synthesize multiple inputs and drive a coordinated response.
Acknowledgements
We thank the Capel laboratory, our collaborators Chutian Ge and Guoying Qian, Zhejiang Wanli University, Ningbo, China, and our colleagues in the Department of Cell Biology for helpful discussions.
Data accessibility
This article has no additional data.
Authors' contributions
C.W. and B.C. both conceived of the ideas and contributed to the writing of this manuscript.
Competing interests
We declare we have no competing interests.
Funding
C.W. and B.C. were supported by National Science Foundation grant no. 1854642 and Duke University School of Medicine Bridge Funding.
References
- 1.Capel B. 2017. Vertebrate sex determination: evolutionary plasticity of a fundamental switch. Nat. Rev. Genet. 18, 675-689. ( 10.1038/nrg.2017.60) [DOI] [PubMed] [Google Scholar]
- 2.Greenbaum E. 2002. A standardized series of embryonic stages for the emydid turtle Trachemys scripta. Can. J. Zool. 80, 1350-1370. ( 10.1139/z02-111) [DOI] [Google Scholar]
- 3.Arnold AP, Xu J, Grisham W, Chen X, Kim Y-H, Itoh Y. 2004. Minireview: sex chromosomes and brain sexual differentiation. Endocrinology 145, 1057-1062. ( 10.1210/en.2003-1491) [DOI] [PubMed] [Google Scholar]
- 4.Bakker J. 2019. The sexual differentiation of the human brain: role of sex hormones versus sex chromosomes. Curr. Top. Behav. Neurosci. 43, 45-67. ( 10.1007/7854_2018_70) [DOI] [PubMed] [Google Scholar]
- 5.Estermann MA, Williams S, Hirst CE, Roly ZY, Serralbo O, Adhikari D, Powell D, Major AT, Smith CA. 2020. Insights into gonadal sex differentiation provided by single-cell transcriptomics in the chicken embryo. Cell Rep. 31, 107491. ( 10.1016/j.celrep.2020.03.055) [DOI] [PubMed] [Google Scholar]
- 6.Guioli S, Zhao D, Nandi S, Clinton M, Lovell-Badge R. 2020. In the chick embryo, estrogen can induce chromosomally male ZZ left gonad epithelial cells to form an ovarian cortex, which supports oogenesis. Development 147. ( 10.1242/dev.181693) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao HH, Capel B. 2005. Temperature, genes, and sex: a comparative view of sex determination in Trachemys scripta and Mus musculus. J. Biochem. 138, 5-12. ( 10.1093/jb/mvi097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ezaz T, Stiglec R, Veyrunes F, Marshall Graves JA. 2006. Relationships between vertebrate ZW and XY sex chromosome systems. Curr. Biol. 16, R736-R743. ( 10.1016/j.cub.2006.08.021) [DOI] [PubMed] [Google Scholar]
- 9.Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R. 1991. Male development of chromosomally female mice transgenic for Sry. Nature 351, 117-121. ( 10.1038/351117a0) [DOI] [PubMed] [Google Scholar]
- 10.Matson CK, Zarkower D. 2012. Sex and the singular DM domain: insights into sexual regulation, evolution and plasticity. Nat. Rev. Genet. 13, 163-174. ( 10.1038/nrg3161) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shoemaker CM, Queen J, Crews D. 2007. Response of candidate sex-determining genes to changes in temperature reveals their involvement in the molecular network underlying temperature-dependent sex determination. Mol. Endocrinol. 21, 2750-2763. ( 10.1210/me.2007-0263) [DOI] [PubMed] [Google Scholar]
- 12.Czerwinski M, Natarajan A, Barske L, Looger LL, Capel B. 2016. A timecourse analysis of systemic and gonadal effects of temperature on sexual development of the red-eared slider turtle Trachemys scripta elegans. Dev. Biol. 420, 166-177. ( 10.1016/j.ydbio.2016.09.018) [DOI] [PubMed] [Google Scholar]
- 13.Crews D, Bergeron JM. 1994. Role of reductase and aromatase in sex determination in the red-eared slider (Trachemys scripta), a turtle with temperature-dependent sex determination. J. Endocrinol. 143, 279-289. ( 10.1677/joe.0.1430279) [DOI] [PubMed] [Google Scholar]
- 14.Crews D, Bull JJ, Wibbels T. 1991. Estrogen and sex reversal in turtles: a dose-dependent phenomenon. Gen. Comp. Endocrinol. 81, 357-364. ( 10.1016/0016-6480(91)90162-Y) [DOI] [PubMed] [Google Scholar]
- 15.MerchantLarios H, Ruiz-Ramirez S, Moreno-Mendoza N, Marmolejo-Valencia A. 1997. Correlation among thermosensitive period, estradiol response, and gonad differentiation in the sea turtle Lepidochelys olivacea. Gen. Comp. Endocrinol. 107, 373-385. ( 10.1006/gcen.1997.6946) [DOI] [PubMed] [Google Scholar]
- 16.Bull JJ, Gutzke WH, Crews D. 1988. Sex reversal by estradiol in three reptilian orders. Gen. Comp. Endocrinol. 70, 425-428. ( 10.1016/0016-6480(88)90117-7) [DOI] [PubMed] [Google Scholar]
- 17.Moreno-Mendoza N, Harley VR, Merchant-Larios H. 2001. Temperature regulates SOX9 expression in cultured gonads of Lepidochelys olivacea, a species with temperature sex determination. Dev. Biol. 229, 319-326. ( 10.1006/dbio.2000.9952) [DOI] [PubMed] [Google Scholar]
- 18.Kim Y, et al. 2006. Fgf9 and Wnt4 act as antagonistic signals to regulate mammalian sex determination. PLoS Biol. 4, e187. ( 10.1371/journal.pbio.0040187) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munger SC, Natarajan A, Looger LL, Ohler U, Capel B. 2013. Fine time course expression analysis identifies cascades of activation and repression and maps a putative regulator of mammalian sex determination. PLoS Genet. 9, e1003630. ( 10.1371/journal.pgen.1003630) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jameson SA, Lin YT, Capel B. 2012. Testis development requires the repression of Wnt4 by Fgf signaling. Dev. Biol. 370, 24-32. ( 10.1016/j.ydbio.2012.06.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jameson SA, Natarajan A, Cool J, Defalco T, Maatouk DM, Mork L, Munger SC, Capel B. 2012. Temporal transcriptional profiling of somatic and germ cells reveals biased lineage priming of sexual fate in the fetal mouse gonad. PLoS Genet. 8, e1002575. ( 10.1371/journal.pgen.1002575) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmer SJ, Burgoyne PS. 1991. In situ analysis of fetal, prepuberal and adult XX—XY chimaeric mouse testes: sertoli cells are predominantly, but not exclusively, XY. Development 112, 265-268. ( 10.1242/dev.112.1.265) [DOI] [PubMed] [Google Scholar]
- 23.McLaren A. 1981. The fate of germ cells in the testis of fetal sex-reversed mice. J. Reprod. Fertil. 61, 461-467. ( 10.1530/jrf.0.0610461) [DOI] [PubMed] [Google Scholar]
- 24.McLaren A. 1975. Sex chimaerism and germ cell distribution in a series of chimaeric mice. J. Embryol. Exp. Morphol. 33, 205-216. ( 10.1242/dev.33.1.205) [DOI] [PubMed] [Google Scholar]
- 25.Adams IR, McLaren A. 2002. Sexually dimorphic development of mouse primordial germ cells: switching from oogenesis to spermatogenesis. Development 129, 1155-1164. ( 10.1242/dev.129.5.1155) [DOI] [PubMed] [Google Scholar]
- 26.Albrecht KH, Eicher EM. 2001. Evidence that Sry is expressed in pre-Sertoli cells and Sertoli and granulosa cells have a common precursor. Dev. Biol. 240, 92-107. ( 10.1006/dbio.2001.0438) [DOI] [PubMed] [Google Scholar]
- 27.Todd EV, Liu H, Muncaster S, Gemmell NJ. 2016. Bending genders: the biology of natural sex change in fish. Sex Dev. 10, 223-241. ( 10.1159/000449297) [DOI] [PubMed] [Google Scholar]
- 28.Dranow DB, Tucker RP, Draper BW. 2013. Germ cells are required to maintain a stable sexual phenotype in adult zebrafish. Dev. Biol. 376, 43-50. ( 10.1016/j.ydbio.2013.01.016) [DOI] [PubMed] [Google Scholar]
- 29.Kurokawa H, Saito D, Nakamura S, Katoh-Fukui Y, Ohta K, Baba T, Morohashi K, Tanaka M. 2007. Germ cells are essential for sexual dimorphism in the medaka gonad. Proc. Natl Acad. Sci. USA 104, 16 958-16 963. ( 10.1073/pnas.0609932104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quinn AE, Sarre SD, Ezaz T, Marshall Graves JA, Georges A. 2011. Evolutionary transitions between mechanisms of sex determination in vertebrates. Biol. Lett. 7, 443-448. ( 10.1098/rsbl.2010.1126) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Charnier M. 1966. Action of temperature on the sex ratio in the Agama agama (Agamidae, Lacertilia) embryo. C R Seances Soc. Biol. Fil. 160, 620-622. [PubMed] [Google Scholar]
- 32.Pieau C. 1971. Sex ratio in the embryos of 2 chelonians (Testudo graeca L. and Emys orbicularis L.) born of artificially incubated ova. C R Acad. Hebd Seances Acad. Sci. D 272, 3071-3074. [PubMed] [Google Scholar]
- 33.Wibbels T, Bull JJ, Crews D. 1991. Chronology and morphology of temperature-dependent sex determination. J. Exp. Zool. 260, 371-381. ( 10.1002/jez.1402600311) [DOI] [PubMed] [Google Scholar]
- 34.Weber C, Zhou Y, Lee JG, Looger LL, Qian G, Ge C, Capel B. 2020. Temperature-dependent sex determination is mediated by pSTAT3 repression of Kdm6b. Science 368, 303-306. ( 10.1126/science.aaz4165) [DOI] [PubMed] [Google Scholar]
- 35.Ge C, Ye J, Weber C, Sun W, Zhang H, Zhou Y, Cai C, Qian G, Capel B. 2018. The histone demethylase KDM6B regulates temperature-dependent sex determination in a turtle species. Science 360, 645-648. ( 10.1126/science.aap8328) [DOI] [PubMed] [Google Scholar]
- 36.Garcia-Moreno SA, Plebanek MP, Capel B. 2018. Epigenetic regulation of male fate commitment from an initially bipotential system. Mol. Cell. Endocrinol. 468, 19-30. ( 10.1016/j.mce.2018.01.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuroki S, et al. 2013. Epigenetic regulation of mouse sex determination by the histone demethylase Jmjd1a. Science 341, 1106-1109. ( 10.1126/science.1239864) [DOI] [PubMed] [Google Scholar]
- 38.Garcia-Moreno SA, Futtner CR, Salamone IM, Gonen N, Lovell-Badge R, Maatouk DM. 2019. Gonadal supporting cells acquire sex-specific chromatin landscapes during mammalian sex determination. Dev. Biol. 446, 168-179. ( 10.1016/j.ydbio.2018.12.023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcia-Moreno SA, Lin Y-T, Futtner CR, Salamone IM, Capel B, Maatouk DM. 2019. CBX2 is required to stabilize the testis pathway by repressing Wnt signaling. PLoS Genet. 15, e1007895. ( 10.1371/journal.pgen.1007895) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Navarro-Martin L, Viñas J, Ribas L, Díaz N, Gutiérrez A, Di Croce L, Piferrer F. 2011. DNA methylation of the gonadal aromatase (cyp19a) promoter is involved in temperature-dependent sex ratio shifts in the European sea bass. PLoS Genet. 7, e1002447. ( 10.1371/journal.pgen.1002447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramsey M, Crews D. 2009. Steroid signaling and temperature-dependent sex determination-reviewing the evidence for early action of estrogen during ovarian determination in turtles. Semin. Cell Dev. Biol. 20, 283-292. ( 10.1016/j.semcdb.2008.10.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsumoto Y, Buemio A, Chu R, Vafaee M, Crews D. 2013. Epigenetic control of gonadal aromatase (cyp19a1) in temperature-dependent sex determination of red-eared slider turtles. PLoS ONE 8, e63599. ( 10.1371/journal.pone.0063599) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsumoto Y, Hannigan B, Crews D. 2014. Embryonic PCB exposure alters phenotypic, genetic, and epigenetic profiles in turtle sex determination, a biomarker of environmental contamination. Endocrinology 155, 4168-4177. ( 10.1210/en.2014-1404) [DOI] [PubMed] [Google Scholar]
- 44.Shao C, et al. 2014. Epigenetic modification and inheritance in sexual reversal of fish. Genome Res. 24, 604-615. ( 10.1101/gr.162172.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deveson IW, Holleley CE, Blackburn J, Marshall Graves JA, Mattick JS, Waters PD, Georges A. 2017. Differential intron retention in Jumonji chromatin modifier genes is implicated in reptile temperature-dependent sex determination. Sci. Adv. 3, e1700731. ( 10.1126/sciadv.1700731) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holleley CE, O'meally D, Sarre SD, Marshall Graves JA, Ezaz T, Matsubara K, Azad B, Zhang X, Georges A. 2015. Sex reversal triggers the rapid transition from genetic to temperature-dependent sex. Nature 523, 79-82. ( 10.1038/nature14574) [DOI] [PubMed] [Google Scholar]
- 47.Przanowski P, et al. 2014. The signal transducers Stat1 and Stat3 and their novel target Jmjd3 drive the expression of inflammatory genes in microglia. J. Mol. Med. (Berl.) 92, 239-254. ( 10.1007/s00109-013-1090-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sherry-Lynes MM, Sengupta S, Kulkarni S, Cochran BH. 2017. Regulation of the JMJD3 (KDM6B) histone demethylase in glioblastoma stem cells by STAT3. PLoS ONE 12, e0174775. ( 10.1371/journal.pone.0174775) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhong Z, Wen Z, Darnell JE Jr. 1994. Stat3 and Stat4: members of the family of signal transducers and activators of transcription. Proc. Natl Acad. Sci. USA 91, 4806-4810. ( 10.1073/pnas.91.11.4806) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ernst MB, et al. 2009. Enhanced Stat3 activation in POMC neurons provokes negative feedback inhibition of leptin and insulin signaling in obesity. J. Neurosci. 29, 11 582-11 593. ( 10.1523/JNEUROSCI.5712-08.2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sakurai T, Yada N, Watanabe T, Arizumi T, Hagiwara S, Ueshima K, Nishida N, Fujita J, Kudo M. 2015. Cold-inducible RNA-binding protein promotes the development of liver cancer. Cancer Sci. 106, 352-358. ( 10.1111/cas.12611) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Si J, Collins SJ. 2008. Activated Ca2+/calmodulin-dependent protein kinase IIgamma is a critical regulator of myeloid leukemia cell proliferation. Cancer Res. 68, 3733-3742. ( 10.1158/0008-5472.CAN-07-2509) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fujita T, et al. 2017. TRPV4-dependent induction of a novel mammalian cold-inducible protein SRSF5 as well as CIRP and RBM3. Sci. Rep. 7, 2295. ( 10.1038/s41598-017-02473-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hulsurkar M, Quick AP, Wehrens XH. 2018. STAT3: a link between CaMKII-betaIV-spectrin and maladaptive remodeling? J. Clin. Invest. 128, 5219-5221. ( 10.1172/JCI124778) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoshida A, Furube E, Mannari T, Takayama Y, Kittaka H, Tominaga M, Miyata S. 2016. TRPV1 is crucial for proinflammatory STAT3 signaling and thermoregulation-associated pathways in the brain during inflammation. Sci. Rep. 6, 26088. ( 10.1038/srep26088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Castelli MA, Whiteley SL, Georges A, Holleley CE. 2020. Cellular calcium and redox regulation: the mediator of vertebrate environmental sex determination? Biol. Rev. Camb. Phil. Soc. 95, 680-695. ( 10.1111/brv.12582) [DOI] [PubMed] [Google Scholar]
- 57.Gees M, Colsoul B, Nilius B. 2010. The role of transient receptor potential cation channels in Ca2+ signaling. Cold Spring Harb. Perspect. Biol. 2, a003962. ( 10.1101/cshperspect.a003962) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yatsu R, et al. 2016. RNA-seq analysis of the gonadal transcriptome during Alligator mississippiensis temperature-dependent sex determination and differentiation. BMC Genomics 17, 1-3. ( 10.1186/s12864-016-2396-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ye YZ, Ma L, Sun B-J, Li T, Wang Y, Shine R, Du W-G. 2019. The embryos of turtles can influence their own sexual destinies. Curr. Biol. 29, 2597. ( 10.1016/j.cub.2019.06.038) [DOI] [PubMed] [Google Scholar]
- 60.Lalli E, Ohe K, Latorre E, Bianchi ME, Sassone-Corsi P. 2003. Sexy splicing: regulatory interplays governing sex determination from Drosophila to mammals. J. Cell Sci. 116, 441-445. ( 10.1242/jcs.00249) [DOI] [PubMed] [Google Scholar]
- 61.Planells B, Gómez-Redondo I, Pericuesta E, Lonergan P, Gutiérrez-Adán A. 2019. Differential isoform expression and alternative splicing in sex determination in mice. BMC Genomics 20, 202. ( 10.1186/s12864-019-5572-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Salz HK. 2011. Sex determination in insects: a binary decision based on alternative splicing. Curr. Opin Genet. Dev. 21, 395-400. ( 10.1016/j.gde.2011.03.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kerins JA, Hanazawa M, Dorsett M, Schedl T. 2010. PRP-17 and the pre-mRNA splicing pathway are preferentially required for the proliferation versus meiotic development decision and germline sex determination in Caenorhabditis elegans. Dev. Dyn. 239, 1555-1572. ( 10.1002/dvdy.22274) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Simon JA, Kingston RE. 2009. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell Biol. 10, 697-708. ( 10.1038/nrm2763) [DOI] [PubMed] [Google Scholar]
- 65.Hong S, Cho Y-W, Yu L-R, Yu H, Veenstra TD, Ge K. 2007. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc. Natl Acad Sci. USA 104, 18 439-18 444. ( 10.1073/pnas.0707292104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Al-Raawi D, Jones R, Wijesinghe S, Halsall J, Petric M, Roberts S, Hotchin NA, Kanhere A. 2019. A novel form of JARID2 is required for differentiation in lineage-committed cells. EMBO J. 38. ( 10.15252/embj.201798449) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miller SA, Mohn SE, Weinmann AS. 2010. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol. Cell 40, 594-605. ( 10.1016/j.molcel.2010.10.028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Haltenhof T, et al. 2020. A conserved kinase-based body-temperature sensor globally controls alternative splicing and gene expression. Mol. Cell 78, 57-69 e4. ( 10.1016/j.molcel.2020.01.028) [DOI] [PubMed] [Google Scholar]
- 69.Lindquist S. 1986. Heat-shock gene-expression. In Vitro Cell. Dev. Biol. 22, A43. [Google Scholar]
- 70.Schlesinger MJ. 1994. How the cell copes with stress and the function of heat-shock proteins. Pediatr. Res. 36, 1-6. ( 10.1203/00006450-199407001-00001) [DOI] [PubMed] [Google Scholar]
- 71.Kohno S, Katsu Y, Urushitani H, Ohta Y, Iguchi T, Guillette LJ Jr. et al. 2010. Potential contributions of heat shock proteins to temperature-dependent sex determination in the American alligator. Sex Dev. 4, 73-87. ( 10.1159/000260374) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rhen T, Schroeder A. 2010. Molecular mechanisms of sex determination in reptiles. Sex Dev. 4, 16-28. ( 10.1159/000282495) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nishiyama H, Higashitsuji H, Yokoi H, Itoh K, Danno S, Matsuda T, Fujita J. 1997. Cloning and characterization of human CIRP (cold-inducible RNA-binding protein) cDNA and chromosomal assignment of the gene. Gene 204, 115-120. ( 10.1016/S0378-1119(97)00530-1) [DOI] [PubMed] [Google Scholar]
- 74.Schroeder AL, Metzger KJ, Miller A, Rhen T. 2016. A novel candidate gene for temperature-dependent sex determination in the common snapping turtle. Genetics 203, 557-571. ( 10.1534/genetics.115.182840) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun W, Liao Y, Yi Q, Wu S, Tang L, Tong L. 2018. The mechanism of CIRP in regulation of STAT3 phosphorylation and bag-1/S expression upon UVB radiation. Photochem. Photobiol. 94, 1234-1239. ( 10.1111/php.12981) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brochu C, Cabrita MA, Melanson BD, Hamill JD, Lau R, Pratt MAC, Mckay BC. 2013. NF-κB-dependent role for cold-inducible RNA binding protein in regulating interleukin 1beta. PLoS ONE 8, e57426. ( 10.1371/journal.pone.0057426) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goikoetxea A, Todd EV, Gemmell NJ. 2017. Stress and sex: does cortisol mediate sex change in fish? Reproduction 154, R149-R160. ( 10.1530/REP-17-0408) [DOI] [PubMed] [Google Scholar]
- 78.Navara KJ. 2013. The role of steroid hormones in the adjustment of primary sex ratio in birds: compiling the pieces of the puzzle. Integr. Comp. Biol. 53, 923-937. ( 10.1093/icb/ict083) [DOI] [PubMed] [Google Scholar]
- 79.Jian F, et al. 2016. Cold inducible RNA binding protein upregulation in pituitary corticotroph adenoma induces corticotroph cell proliferation via Erk signaling pathway. Oncotarget 7, 9175-9187. ( 10.18632/oncotarget.7037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang W, et al. 2015. Phosphorylation of STAT3 mediates the induction of cyclooxygenase-2 by cortisol in the human amnion at parturition. Sci. Signal 8, ra106. ( 10.1126/scisignal.aac6151) [DOI] [PubMed] [Google Scholar]
- 81.Munzberg H, Huo L, Nillni EA, Hollenberg AN, Bjørbæk C. et al. 2003. Role of signal transducer and activator of transcription 3 in regulation of hypothalamic proopiomelanocortin gene expression by leptin. Endocrinology 144, 2121-2131. ( 10.1210/en.2002-221037) [DOI] [PubMed] [Google Scholar]
- 82.Morgan MJ, Liu ZG. 2011. Crosstalk of reactive oxygen species and NF-kappaB signaling. Cell Res. 21, 103-115. ( 10.1038/cr.2010.178) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Siewert E, Muller-Esterl W, Starr R, Heinrich PC, Schaper F. 1999. Different protein turnover of interleukin-6-type cytokine signalling components. Eur. J. Biochem. 265, 251-257. ( 10.1046/j.1432-1327.1999.00719.x) [DOI] [PubMed] [Google Scholar]
- 84.Kim M, Morales LD, Jang IS, Cho YY, Kim DJ. 2018. Protein tyrosine phosphatases as potential regulators of STAT3 signaling. Int. J. Mol. Sci. 19, 2708. ( 10.3390/ijms19092708) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Veeriah S, et al. 2009. The tyrosine phosphatase PTPRD is a tumor suppressor that is frequently inactivated and mutated in glioblastoma and other human cancers. Proc. Natl Acad. Sci. USA 106, 9435-9440. ( 10.1073/pnas.0900571106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Meng TC, Fukada T, Tonks NK. 2002. Reversible oxidation and inactivation of protein tyrosine phosphatases in vivo. Mol. Cell 9, 387-399. ( 10.1016/S1097-2765(02)00445-8) [DOI] [PubMed] [Google Scholar]
- 87.Libermann TA, Baltimore D. 1990. Activation of interleukin-6 gene expression through the NF-κB transcription factor. Mol. Cell. Biol. 10, 2327-2334. ( 10.1128/MCB.10.5.2327) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.