Abstract
Klebsiella pneumoniae is an important pathogen causing hospital‐acquired infections in human beings. Samples from suspected patients of K pneumoniae associated with respiratory and urinary tract infections were collected at Bolan Medical Complex, Quetta, Balochistan. Clinical samples (n = 107) of urine and sputum were collected and processed for K pneumoniae isolation using selective culture media. Initially, 30 of 107 isolates resembling Klebsiella spp. were processed for biochemical profiling and molecular detection using gyrase A (gyrA) gene for conformation. The K pneumoniae isolates were analysed for the presence of drug resistance and virulence genes in their genomes. The 21 of 107 (19.6%) isolates were finally confirmed as K pneumoniae pathogens. An antibiogram study conducted against 17 different antibiotics showed that a majority of the isolates are multidrug resistant. All the isolates (100%) were resistant to amoxicillin, cefixime, amoxicillin‐clavulanic acid, cefotaxime, and ceftriaxone followed by tetracycline (95.2%), ciprofloxacin and gentamicin (76.2%), sulphamethoxazol (66.7%), nalidixic acid (61.9%), norfloxacine (42.9%), piperacillin‐tazobactam (23.8%), cefoperazone‐sulbactam (19%), and cefotaxime‐clavulanic acid (33.3%), whereas all the isolates showed sensitivity to amikacin, chloramphenicol, and imipenem. The presence of tetracycline, sulphamethoxazol‐resistant genes, and extended‐spectrum beta‐lactamase was reconfirmed using different specific genes. The presence of virulence genes fimH1 and EntB responsible for adherence and enterobactin production was confirmed in the isolates. The high virulence and drug resistance potential of these Klebsiella isolates are of high public health concern. Multidrug resistance and virulence potential in K. pneumoniae are converting these nosocomial pathogens into superbugs and making its management harder.
Keywords: antibiotics, antimicrobial, nosocomial infections, opportunistic pathogens, pathogenicity
1. INTRODUCTION
The genus Klebsiella falls under Enterobacteriaceae family, a Gram‐negative bacteria with rod shape, lysine decarboxylase but not ornithine decarboxylase producers, and Voges‐Proskauer positive. 1 Klebsiella species are ubiquitous in nature and can be frequently found in different environments such as water, soil, 2 and dirt. This bacteria can colonise in nasopharynx and gastrointestinal tract, 3 which makes them able to act as an opportunistic pathogen in human being. 4 This bacteria has also been reported in insects 5 and mammals. 6 Gastrointestinal colonisation is likely a common and significant reservoir among body sites in terms of risk of transmission and infection. 7
Klebsiella pneumoniae is a clinically important member of the genus Klebsiella, reportedly being responsible for around 86% of human infections due to Klebsiella, 8 which makes it the most significant pathogen of this genus. However, species of Klebsiella oxytoca are reported as the second most widespread species, responsible for about 26% of infections. 1 , 9 Klebsiella spp. are considered as an important member of hospital‐acquired pathogens, which are not only responsible for numerous infections of respiratory and urinary tract, but can also cause the infection of soft tissues, wounds, sepsis, and septicaemia. 9 , 10 The presence of virulence factor and drug resistance enhances the colonisation of Klebsiella, particularly in the case of nosocomial infections. The most common pathogenicity factors in this species are iron acquisition capability, possession of fimbriae, lipopolysaccharide, and so on. 11 Types 1 and 3 fimbriae present in Klebsiella enhanced the urinary tract infections. 12 Virulence genes enterobactin synthase component B (entB), aerobactin siderophore receptor (iutA), putative salicylate synthetase (ybtS), iron regulatory protein 1 and 2 (irp‐1, irp‐2), and ferric yersiniabactin uptake receptor (fyuA) responsible for siderophores production, which are present in this species, can make them able to acquire iron from human host. 13 The lipopolysaccharide and capsule increase its pathogenicity and help them avoiding phagocytosis, resulting sepsis and septic shock. 14
Drug resistance to multiple antibiotics in pathogens is the increasing cause of morbidity and mortality. 15 K pneumoniae has been reported with resistance to fluoroquinolones, aminoglycosides, and beta‐lactam antibiotics. The production of beta‐lactamase enzyme by Klebsiella species is making them resistant to a wide group of antibiotics. 16 In a majority of K pneumoniae isolates, the β‐lactamase is encoded by chromosomal‐encoded SHV‐1 gene. 6 Pathogens with increased pathogenicity factors and multidrug resistance act as a superbug and are a growing public health threat to the developing and developed countries. A study was designed to record the prevalence of K pneumoniae in urinary tract infection (UTI) and respiratory tract infections (RTI), and to determine their multidrug resistance and virulence potential of Balochistan patients in Quetta City.
2. MATERIALS AND METHODS
2.1. Sampling
A total of 107 samples, of which 72 were urine and 35 sputum, were collected from different patients suffering UTI and RTI in Bolan Medical Complex, Quetta. All the samples were collected aseptically in sterile sampling bottles, following safety protocols and procedures, with the patient inform consent and according to the will and satisfaction of patients. The samples' inclusion criteria were strictly limited to UTI and RTI patients. Samples were carried to the laboratory in the University of Balochistan and processed within 1 to 2 hours of collection, not later than 6 hours.
2.2. Isolation of the target pathogen
The collected samples were aseptically inoculated to pre‐prepared sterile MacConkey agar (Oxoid, UK) media and incubated at 37°C for 24 hours. Mucoid, circular, and lactose‐fermenting colonies were subculture to Eosin Methylene Blue agar (EMB) (Oxoid, UK) for conformation and differentiation with Escherichia coli.
2.3. Biochemical characterisation
Presumptively selected isolates were Gram stained and biochemically characterised using biochemical tests, such as catalase, oxidase, urease, sulphur, indole, motility, methyl red, Vogues‐Proskauer, and citrate utilisation. 17
2.4. Molecular conformation of the isolates
The preliminary conformed Klebsiella isolates were confirmed with the help of genotyping using Klebsiella‐specific gene gyrA (F‐CGCGTACTATACGCCATGAACGTA and R‐ACCGTTGATCACTTCGGTCAGG) 18 following standard protocol for DNA extraction. 19 Polymerase chain reaction (PCR) (25 μL) was prepared by mixing 12.5 μL master mix with 1 μL of each forward and reverse primers, 7.5 μL nuclease‐free PCR water and DNA sample (3 μL). The mixture was processed at 94°C denaturing for 4 minutes. 94°C for 30 seconds, 55°C for 40 seconds, 72°C for 60 seconds, and 30 cycle. Final extension was performed at 72°C for 10 minutes and at a final temperature of 4°C.
2.5. Antibiogram studies
The antibiogram studies of the conformed K pneumoniae were conducted by using the Kirby‐Bauer technique of disc diffusion with Mueller‐Hinton agar (MHA) (Oxide, UK). 20 The target bacteria were spread over the surface of sterile MHA plates using sterile cotton swab. Antibiotics discs amoxicillin (AML 10 μg), cefixime (CFM 5 μg), sulphamethoxazol (SXT 25 μg), tetracycline (TE 30 μg), nalidixic acid (NA10 μg), ciprofloxacin (CIP 5 μg), norfloxacin (NOR 10 μg), imipenem (IMP 10 μg), amikacin (Ak 20 μg), gentamycin (CN 10 μg), chloramphenicol (C30 μg), cefotaxime (CTX 30 μg), amoxicillin‐clavulanic acid (AMC 30 μg), piperacillin‐tazobactam (TZP 110 μg), cefoperazone‐sulbactam (SCF 105 μg), ceftriaxone (CRO 30 μg), and cefotaxime‐clavulanic acid (CTX + CLA 30/10 μg) were placed over the surface of the bacterial lawn and incubated at 37°C for 16 to 24 hours. The zone of inhibitions of each antibiotic was recorded in millimetre (mm) and it corresponds to the CLSI standard values of respective antibiotics. 21
2.6. Extended‐spectrum beta‐lactamase production
The extended‐spectrum beta‐lactamase (ESBL) production test was performed by double discs ceftriaxone (CRO 30 μg) and cefotaxime‐clavulanic acid 30/10 μg (BD). 22
2.7. Molecular conformation of drug resistance genes in the isolates
The phenotypically evaluated resistance in Klebsiella isolates was confirmed against specific genes corresponding resistance to selected antibiotics through PCR. Tetracycline resistance was confirmed by gene tetB using primers F‐CCTTATCAT GCCAGTCT TGC, R‐ACTGCCGT TTTTTCGC C, 23 while Sul1 gene using primer F‐CGGCGTGGGCTACCT GAACG and R‐GCCGATCGCGTGAAGTTCCG responsible for sulphonamide‐resistant 24 and SHV gene for ESBL production using primer F‐ATTTGTCGCTTCTTTACTCGC and R‐TTTATGGCGTTACCTTTGACC. 25
2.8. Molecular detection of virulence factors
The virulence factors associated with K pneumoniae were determined by targeting specific gene fimH encoding for type 1 fimbriae, the main virulence factor of the bacteria, using primer F′‐ATGAACGCCTGGTCCTTTGC, R‐GCTGAACGCCTATCCCCTGC, 26 and EntB gene responsible for enterobactin production using primer F‐ATTTCCTCAACTTCTGGGGC and R‐AGCATCGGTGGCGGTGGTCA. 27
3. RESULTS
Thirty isolates were preliminary identified as K pneumoniae out of the total (107) samples based on initial screening. Of which 21 of 107 isolates were biochemically and molecularly confirmed as K pneumoniae with the help of Gyrase A (gyrA) gene (Figure 1), making 19.6% prevalence of the pathogens in the patients. All the isolates were collected from the urine (72) and sputum (35) samples of the patient suspected with K pneumoniae associated RTI and UTI. The K pneumoniae isolates of 20.8% (15/72) were from urine and of 17.1% (6/35) from sputum.
FIGURE 1.
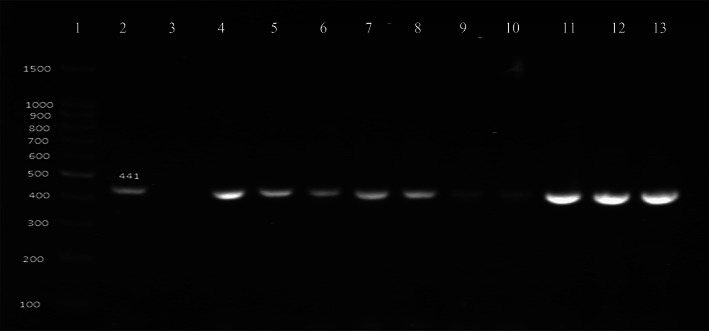
Polymerase chain reaction assay result for Klebsiella identification; line 1 = DNA size ladder 100 bp (GeneDireX); line 2 = reference strain for gyrA; line 3 = negative control; line 4–13 = positive samples
All the gyrase A gene‐positive K pneumonia isolates (21) were processed for antibiogram studies against 17 common antibiotics. The antibiogram studies revealed that all the isolates were multidrug resistant to the tested antibiotics (Figure 2). The antibiogram of the isolates showed 100% (21/21) resistance to CFM, AML, CTX, CRO, and AMC, whereas TET 95.2% (20/21), CIP and CN 76.2% (16/21), and SXT 66.7% (14/21). Resistance against NA were found to be 61.9% (13/21), while 42.9% (9/21), 33.33% (7/21), and 23.8% (5/21) against NOR, CTX+C, and TZP, respectively.
FIGURE 2.
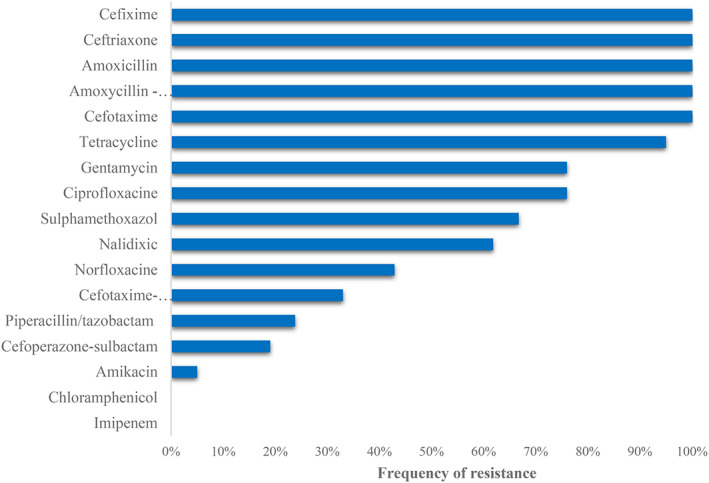
Drug resistance pattern of K pneumoniae clinical isolates against antibiotics used
The lowest percentage of resistance was found against SCF 19% (4/21), AK 4.8% (1/21), C, and IMP 0% (0/21). It was found that out of 21 isolates only 7 (33.3%) were positive for ESBL production. All of the isolates showed resistant to third‐generation cephalosporin.
In this study, imipenem and chloramphenicol were found to be highly active (100%) antibiotics against the K pneumoniae isolates, whereas only one (4.8%) isolate showed resistance to the drug amikacin. It was noted that out of 21 isolates, two isolates showed multidrug resistance to 14 and 13 antibiotics used in the study, while six isolates showed resistance to 11 antibiotics and four to 10 antibiotics used. Two isolates showed resistance to 12 and five isolates to 9 antibiotics, respectively. One isolate was found resistant to 8 antibiotics and the other to 6 antibiotics used. This study confirms the extensive multidrug resistance potential of the K pneumoniae clinical isolates.
The SXT, TET, and ESBL resistances were genotypically (Sul1, tetB, and SHV) reconfirmed using specific primers (Figure 3A‐C). The tetB gene was confirmed in all the isolates showing 100% resistance of the isolates against tetracycline, whereas Sul1 gene corresponding to sulphamethoxazol resistance was present in 14/21 (66.7%) isolates and SHV in 7/21 showing that 33.3% were positive for ESBL production. Pathogenicity of pathogens is dependent on the presence of virulence factors, which are directly proportional to the higher pathogenicity, resulting in more complicated infection.
FIGURE 3.
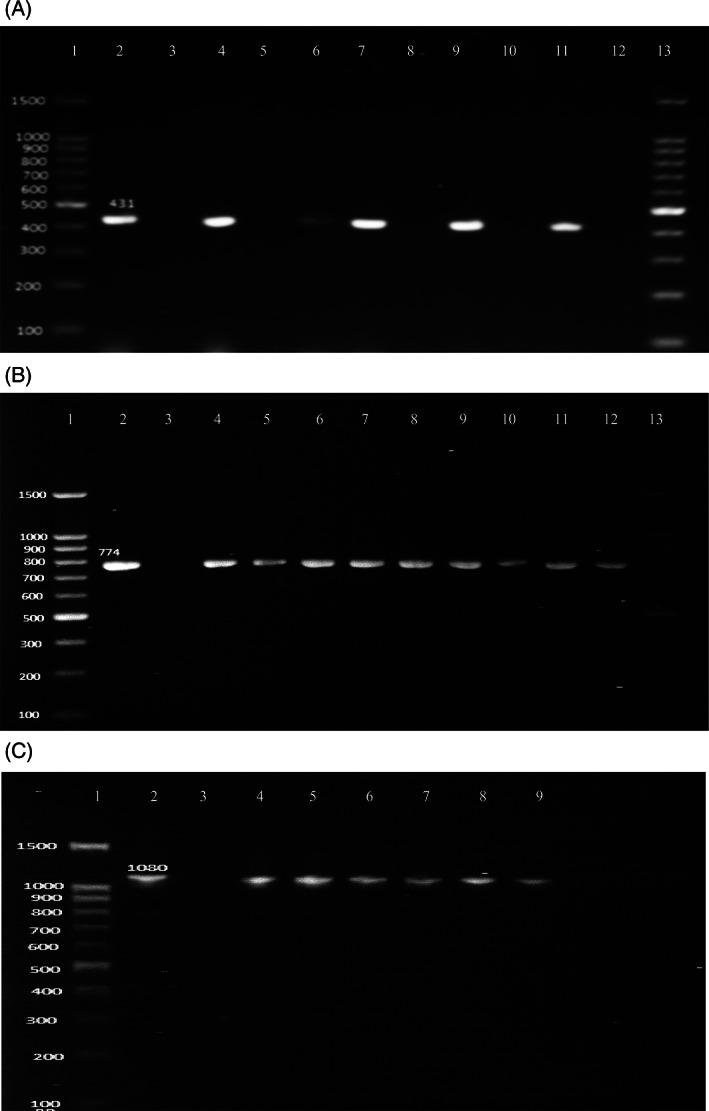
A, PCR results for Sul1 (431 bp) drug resistance gene; line 1 and 13 = DNA size ladder 100 bp (GeneDireX); line 2 = reference strain for Sul1 gene positive; lines 3, 5, 8, 10, and 12 = negative control; lines 4, 6, 7, 9, and 11 = positive samples. B, PCR results for tetB (774 bp) drug resistance gene; lines 1 and 13 = DNA size ladder 100 bp (GeneDireX); line 2 = reference strain for tetB gene positive; line 3 = negative control; lines 4–12 = positive samples. C, PCR results for SHV (1080 bp) ESBL resistance gene; line 1: DNA size ladder 100 bp (GeneDireX), line 2: reference strain for SHV gene positive; line 3: negative control; lines 4–9 = positive samples
In this study, we evaluated the presence of common and selected virulence factors associated with K pneumoniae clinical isolates. The virulence factors are important to show the clinical and nosocomial importance of the isolates. The 21 confirmed K pneumoniae isolates were analysed for the presence of fimH gene responsible for improved adherence properties and entB gene of enterobactin production. All the isolates found bearing the targeted genes, with 100% presence of type 1 fimbriae and enterobactin production (Figure 4A,B), showed the high pathogenicity potential of these isolates (Table 1).
FIGURE 4.
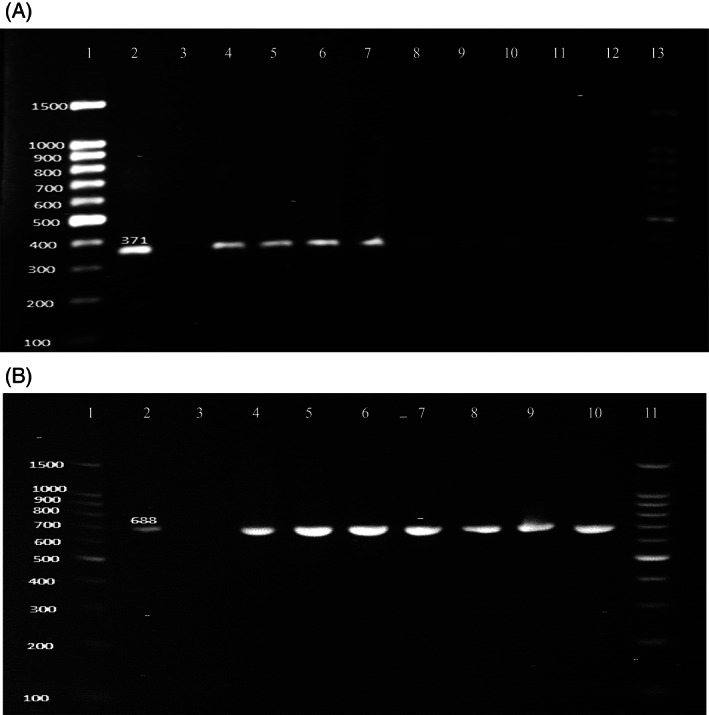
A, Polymerase chain reaction assay result for virulence gene EntB (371 bp); lines 1 and 13 = DNA size ladder 100 bp (GeneDireX); Line 2 = reference strain for EntB gene positive; line 3 = negative control; lines 4–12 = positive samples. B, PCR results for fimH‐1 (688 bp) virulence gene; lines 1 and 12 = DNA size ladder 100 bp (GeneDireX), line 2 = reference strain for fimH‐1 gene positive; line 3 = negative control; lines 4–11 = positive samples
TABLE 1.
Percentage and number of genes detected among K pneumoniae clinical isolates
| Gene | Urine isolates | Sputum isolates | Total |
|---|---|---|---|
| fimH1 | 15/15 (100%) | 6/6 (100%) | 21/21(100%) |
| EntB | 15/15 (100%) | 6/6 (100%) | 21/21 (100%) |
| tetB | 15/15 (100%) | 6/6 (100%) | 21/21 (100%) |
| Sul1 | 9/15 (60%) | 5/6 (83.3%) | 14/21 (66.67%) |
| SHV | 3/15 (20%) | 4/6 (66.67%) | 7/21 (33.33%) |
| gyrA | 15/15 (100%) | 6/6 (100%) | 21/21 (100%) |
Abbreviations: EntB, Enterobactin synthase component B; fimH1, type 1 fimbrial adhesin; gyrA, gyrase A; SHV, sulphydryl variable β‐lactamas; Sul1, sulphamethoxazol; tetB, tetracycline.
4. DISCUSSION
K pneumoniae is an important member of ESKAPE (Enterococcus faecium, Staphylococcus aureus, K pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) and potential threat as a nosocomial infection. 28 In this study, we found that 19.6% samples from suspected UTI and RTI infections are positive for K pneumoniae pathogen, which can lead the patients to complicated infections in case of drug resistance. In a similar study, Younis 29 reported 73.3% K pneumoniae prevalence in clinically diseased chicken, which is higher than our study, whereas Martin et al 30 and Zhang et al 31 reported 23% and 73.9%, respectively, prevalence of K pneumoniae in clinical samples. Fatima et al 32 and Chakraborty et al 33 reported 17% and 24% prevalence of K pneumoniae in Pakistan and Bangladesh, respectively, from clinical samples. The antibiogram studies revealed that a number of isolates are multidrug resistance to commonly used antibiotics. Increasing drug resistance in pathogenic bacteria is a great human health concern. Our studies showed that 33.3% K pneumoniae is ESBL positive. Taneja et al 34 reported 70.7% ESBL prevalence in K pneumoniae isolates from Dehli. Hayat et al 35 detected ESBL gene in 56.5% of K pneumoniae isolates from clinical samples in Pakistan, while Chakraborty et al 33 reported 45% ESBL prevalence in K pneumoniae in a similar study in Bangladesh. In our study, we found a higher resistance of the clinical isolates towards antibiotics AMC, CFM, AML, CTX, and CRO. Significant resistance has been reported against cephalosporin by Nijssen et al. 36 Khamesipour and Tajbakhsh 37 claimed 87.8, 43.3, and 32.2% resistance to AMC, TET, and AK while 34.4% and 26.7% of CFM and CN, respectively. Compared to his study, resistance to these four antibiotics is higher in our study. Chakraborty et al 33 reported 56% multidrug resistance K pneumoniae in clinical isolates from Malaysia, where they showed 100% resistance to ampicillin; 90% to amoxicillin; 45% to ceftriaxone; 40% to ciprofloxacin; 45% to co‐trimoxazole; and 25%, 50%, and 35% to gentamicin, nalidixic acid, and tetracycline respectively.
In our study, we found that all the isolates were multidrug resistance to minimum 6 and maximum 14 antibiotics out of 17 used in the study. Lina et al 38 reported resistance in ceftazidime (36%), CN (27%), TET (27%), and CIP (45%) to K pneumoniae. It was found in a similar study that 61.2% of K pneumoniae are drug resistant, wherein 20.4% of the isolates showed 100% resistance to all cephalosporins (CFM, CRO, ceftazidime, and ceftizoxime), and 98% to carbenicillin, 55% to piperacillin, 32% to CTX, and 31% to ceftazidime with zero resistant to IMP. 39 These results are in agreement with our result where 100% resistance was found against cephalosporins and 0% against imipenem. In our study, the resistance rate against tetracycline, aminoglycosides, and fluoroquinolones was higher in comparison with ESBL‐producing isolates, similar results have been reported by Hashemi et al. 40 Similar to our study, Taitt et al 41 reported 100% resistance to tetracycline conformed with tetB and 60% Sul1 resistance genes in their study in Kenya. Gholipour et al 42 detected SHV genes responsible for ESBL production on 7.5% K pneumoniae isolates, which is less than that of our study (33.3%). The higher pattern of drug resistance in K pneumoniae isolates in our study can be linked to over the counter sale and extensive uses of antibiotics in animal farming. 20 Banning over the counter sale and abuse of antibiotics for animal farming is the possible solution to control the drug resistance in Pakistan.
The virulence factor such as enterobactin production helps the pathogens in biofilm formation and infection development. 43 Studies revealed that enterobactin biosynthesis is iron‐uptake proteins produced by Klebsiella 44 because of the presence of entB gene in its genome. 45 We found the presence of entB gene in 100% K pneumoniae isolates, which are in compliance with Aljanaby and Alhasani 44 study. El Fertas‐Aissani et al 27 also reported 100% detection of entB gene in K pneumoniae isolates in their study. Possessing fimbriae can increase the pathogenicity of pathogens. 46 fimH1 gene responsible for type 1 fimbriae can enhance the biofilm formation capability of Klebsiella to colonise in urinary and respiratory tract resulting in complicated infection. 44 In our study, we find 100% presence of fimH1 gene, and these results are supported by Xiao et al 47 study where they reported the high frequency of fimH1 gene identification in K pneumoniae. It was also reported that the presence of fimbriae can enhance the drug resistance of the pathogen. 48
5. CONCLUSIONS
It was concluded in this study that the increased and unnecessary uses of antibiotics produce superbugs within the common nosocomial pathogens such as K. pneumoniae, posing a serious threat to global public health. The emergence of multidrug resistance in common virulent species of pathogens will have a great impact over the health system and economy of developing countries. The K. pneumoniae isolates were found harbouring virulent drug‐resistant genes and were showing resistance to several common antibiotics, which are of great concern. Alternative management procedures and novel drug hunting along with wise uses of antibiotics are the possible solutions to fight the multidrug resistant pathogens.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Fatima S, Liaqat F, Akbar A, et al. Virulent and multidrug‐resistant Klebsiella pneumoniae from clinical samples in Balochistan. Int Wound J. 2021;18:510–518. 10.1111/iwj.13550
Sareeen Fatima and Faiza Liaqat contributed equally to the scientific work.
DATA AVAILABILITY STATEMENT
Data will be available on request.
REFERENCES
- 1. Martin R, Bachman M. Colonization, infection, and the accessory genome of Klebsiella pneumoniae . Front Cell Infect Microbiol. 2018;8(4):2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rock C, Thom KA, Masnick M, Johnson JK, Harris AD, Morgan DJ. Frequency of Klebsiella pneumoniae Carbapenemase (KPC)–producing and non‐KPC‐producing Klebsiella species contamination of healthcare workers and the environment. Infect Contr Hosp Epidemiol. 2014;35(4):426‐429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dao TT, Liebenthal D, Tran TK, et al. Klebsiella pneumoniae oropharyngeal carriage in rural and urban Vietnam and the effect of alcohol consumption. PLoS One. 2014;9(3):e91999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11(4):589‐603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dillon R, Vennard C, Charnley A. A note: gut bacteria produce components of a locust cohesion pheromone. J Appl Microbiol. 2002;92(4):759‐763. [DOI] [PubMed] [Google Scholar]
- 6. Liaqat F, Naeem W, Shafee M, et al. 2. Virulence factors and drug resistance in Klebsiella pneumoniae; an emerging superbug. Pure Appl Biol. 2019;8(2):1314‐1337. [Google Scholar]
- 7. Dorman MJ, Short FL. Genome watch: Klebsiella pneumoniae: when a colonizer turns bad. Nature. 2017;15(7):384. [DOI] [PubMed] [Google Scholar]
- 8. Sen P, Demirdal T. Evaluation of mortality risk factors in diabetic foot infections. Int Wound J. 2020;17:880‐889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Magill SS, Edwards JR, Fridkin SK. Survey of health care‐associated infections. New Engl J Med. 2014;370(26):2542‐2543. [DOI] [PubMed] [Google Scholar]
- 10. Bengoechea JA, Sa Pessoa J. Klebsiella pneumoniae infection biology: living to counteract host defences. FEMS Microbiol Rev. 2019;43(2):123‐144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fasciana T, Gentile B, Aquilina M, et al. Co‐existence of virulence factors and antibiotic resistance in new Klebsiella pneumoniae clones emerging in south of Italy. BMC Infect Dis. 2019;19(1):928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rosen DA, Hilliard JK, Tiemann KM, Todd EM, Morley SC, Hunstad DA. Klebsiella pneumoniae FimK promotes virulence in murine pneumonia. J Infect Dis. 2016;213(4):649‐658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Russo TA, Olson R, MacDonald U, et al. Aerobactin mediates virulence and accounts for increased siderophore production under iron‐limiting conditions by hypervirulent (hypermucoviscous) Klebsiella pneumoniae . Infect Immun. 2014;82(6):2356‐2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tsereteli M, Sidamonidze K, Tsereteli D, Malania L, Vashakidze E. Epidemiology of carbapenem‐resistant Klebsiella pneumoniae in intensive care units of multiprofile hospitals in Tbilisi, Georgia. Georgian Med News. 2018;2018(280–281):164‐168. [PubMed] [Google Scholar]
- 15. Naeem W, Liaqat F, Shafee M, Khan GI, Akbar A. 2. Multidrug resistance in pathogenic Escherichia coli; a public health concern. Pure Appl Biol. 2019;8(3):2104‐2118. [Google Scholar]
- 16. Navon‐Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017;41(3):252‐275. [DOI] [PubMed] [Google Scholar]
- 17. Akbar A, Anal AK. Occurrence of Staphylococcus aureus and evaluation of anti‐staphylococcal activity of Lactococcus lactis subsp. lactis in ready‐to‐eat poultry meat. Annals Microbiol. 2014;64(1):131‐138. [Google Scholar]
- 18. Brisse S, Verhoef J. Phylogenetic diversity of Klebsiella pneumoniae and Klebsiella oxytoca clinical isolates revealed by randomly amplified polymorphic DNA, gyrA and parC genes sequencing and automated ribotyping. Int J Syst Evolut Microbiol. 2001;51(3):915‐924. [DOI] [PubMed] [Google Scholar]
- 19. Shu Y, Wan‐Ting J, Ya‐Ning Y, Yu‐Han F. An optimized CTAB method for genomic DNA extraction from freshly‐picked pinnae of Fern Adiantum capillus‐veneris L. Bio Protocol. 2018;8(13):1‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Samad A, Abbas F, Ahmed Z, et al. Prevalence, antimicrobial susceptibility, and virulence of Campylobacter jejuni isolated from chicken meat. J Food Safety. 2019;39(2):e12600. [Google Scholar]
- 21. CLSI , ed. Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. CLSI approved standard M100‐S23 Wayne, PA: Clinical and Laboratory Standards Institute; 2013. [Google Scholar]
- 22. Sarojamma V, Ramakrishna V. Prevalence of ESBL‐producing Klebsiella pneumoniae isolates in tertiary care hospital. ISRN Microbiol. 2011;2011:1‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Maynard C, Bekal S, Sanschagrin F, et al. Heterogeneity among virulence and antimicrobial resistance gene profiles of extraintestinal Escherichia coli isolates of animal and human origin. J Clin Microbiol. 2004;42(12):5444‐5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gündoğdu A, Long YB, Vollmerhausen TL, Katouli M. Antimicrobial resistance and distribution of sul genes and integron‐associated intI genes among uropathogenic Escherichia coli in Queensland, Australia. J Med Microbiol. 2011;60(11):1633‐1642. [DOI] [PubMed] [Google Scholar]
- 25. Jemima S, Verghese S. Multiplex PCR for blaCTX‐M & blaSHV in the extended spectrum beta lactamase (ESBL) producing gram‐negative isolates. Indian J Med Res. 2008;128(3):313. [PubMed] [Google Scholar]
- 26. Carneiro V, Lessa D, Guttmann P, et al. Virulence, resistance, and genetic relatedness of Escherichia coli and Klebsiella sp. isolated from mule foals. Arquivo Brasileiro de Medicina Veterinária e Zootecnia. 2017;69(5):1073‐1082. [Google Scholar]
- 27. El Fertas‐Aissani R, Messai Y, Alouache S, Bakour R. Virulence profiles and antibiotic susceptibility patterns of Klebsiella pneumoniae strains isolated from different clinical specimens. Pathologie Biologie. 2013;61(5):209‐216. [DOI] [PubMed] [Google Scholar]
- 28. da Rosa TF, Coelho SS, Foletto VS, et al. Alternatives for the treatment of infections caused by ESKAPE pathogens. J Clin Pharm Therapeut. 2020;45:863‐873. [DOI] [PubMed] [Google Scholar]
- 29. Younis G, Awad A, El‐Gamal A, Hosni R. Virulence properties and antimicrobial susceptibility profiles of Klebsiella species recovered from clinically diseased broiler chicken. Adv Anim Vet Sci. 2016;4(10):536‐542. [Google Scholar]
- 30. Martin RM, Cao J, Brisse S, et al. Molecular epidemiology of colonizing and infecting isolates of Klebsiella pneumoniae . MSphere. 2016;1(5):e00261‐e00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang Y, Wang Q, Yin Y, et al. Epidemiology of carbapenem‐resistant Enterobacteriaceae infections: report from the China CRE network. Antimicrobiol Agents Chemoth. 2018;62(2):e01882‐e01817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fatima S, Muhammad IN, Khan MN, Jamil S. Phenotypic expression and prevalence of multi drug resistant extended spectrum beta‐lactamase producing Escherichia coli and Klebsiella pneumoniae in Karachi. Pakistan Pak J Pharm Sci. 2018;31(4):1379‐1384. [PubMed] [Google Scholar]
- 33. Chakraborty S, Mohsina K, Sarker PK, Alam M, Karim M, Sayem S. Prevalence, antibiotic susceptibility profiles and ESBL production in Klebsiella pneumoniae and Klebsiella oxytoca among hospitalized patients. Periodicum Biologorum. 2016;118(1):53‐58. [Google Scholar]
- 34. Taneja J, Mishra B, Thakur A, Dogra V, Loomba P. Nosocomial blood‐stream infections from extended‐spectrum‐beta‐lactamase‐producing Escherichia coli and Klebsiella pneumonia from GB Pant Hospital, New Delhi. J Infect Dev Countr. 2010;4(08):517‐520. [DOI] [PubMed] [Google Scholar]
- 35. Hayat S, Siddique MH, Aslam B, et al. Extended‐spectrum‐β‐lactamase producing multidrug resistant Klebsiella pneumoniae isolates from pediatrics. Pak J Zool. 2019;51(4):1251‐1257. [Google Scholar]
- 36. Nijssen S, Florijn A, Bonten M, Schmitz F, Verhoef J, Fluit A. Beta‐lactam susceptibilities and prevalence of ESBL‐producing isolates among more than 5000 European Enterobacteriaceae isolates. Int J Antimicrobial Agents. 2004;24(6):585‐591. [DOI] [PubMed] [Google Scholar]
- 37. Khamesipour F, Tajbakhsh E. Analyzed the genotypic and phenotypic antibiotic resistance patterns of Klebsiella pneumoniae isolated from clinical samples in Iran. Biomedical Research. 2016;27(4):1017‐1026. [Google Scholar]
- 38. Lina TT, Rahman SR, Gomes DJ. Multiple‐antibiotic resistance mediated by plasmids and integrons in uropathogenic Escherichia coli and Klebsiella pneumoniae . Bangladesh J Microbiol. 2007;24(1):19‐23. [Google Scholar]
- 39. Leisy‐Azar S, Ebadi A. Examining the pattern of susceptibility and antibiotic resistance in Klebsiella pneumoniae strains isolated from urine samples of children with urinary tract infections from the children's hospital of Tabriz in 2015. Br Biomed Bull. 2017;5(307):10.21767. [Google Scholar]
- 40. Hashemi SH, Esna AF, Tavakoli S, Mamani M. The prevalence of antibiotic resistance of Enterobacteriaceae strains isolated in community‐and hospital‐acquired infections in teaching hospitals of Hamadan, west of Iran. J Res Health Sci. 2013;13(1):75‐80. [PubMed] [Google Scholar]
- 41. Taitt CR, Leski TA, Erwin DP, et al. Antimicrobial resistance of Klebsiella pneumoniae stool isolates circulating in Kenya. PLoS One. 2017;12(6):e0178880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gholipour A, Soleimani N, Shokri D, Mobasherizadeh S, Kardi M, Baradaran A. Phenotypic and molecular characterization of extended‐spectrum β‐lactamase produced by Escherichia coli, and Klebsiella pneumoniae isolates in an educational hospital. Jundishapur J Microbiol. 2014;7(10):1‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kang D, Kirienko NV. Interdependence between iron acquisition and biofilm formation in Pseudomonas aeruginosa . J Microbiol. 2018;56(7):449‐457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Aljanaby AAJ, Alhasani AHA. Virulence factors and antibiotic susceptibility patterns of multidrug resistance Klebsiella pneumoniae isolated from different clinical infections. Afr J Microbiol Res. 2016;10(22):829‐843. [Google Scholar]
- 45. May T, Okabe S. Enterobactin is required for biofilm development in reduced‐genome Escherichia coli . Environ Microbiol. 2011;13(12):3149‐3162. [DOI] [PubMed] [Google Scholar]
- 46. Huynh DTN, Kim A‐Y, Kim Y‐R. Identification of pathogenic factors in Klebsiella pneumoniae using impedimetric sensor equipped with biomimetic surfaces. Sensors. 2017;17(6):1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xiao X, Yao B, Zhou Q, Zhang J. Microbiological characteristics of hypermucoviscous Klebsiella pneumoniae isolates from different body fluids. J Infect Dev Countr. 2017;11(09):672‐678. [DOI] [PubMed] [Google Scholar]
- 48. Vizcarra IA, Hosseini V, Kollmannsberger P, et al. How type 1 fimbriae help Escherichia coli to evade extracellular antibiotics. Sci Rep. 2016;6(1):1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be available on request.


