Abstract
Microbes use signaling factors for intraspecies and interspecies communications. While many intraspecies signaling factors have been found and characterized, discovery of factors for interspecies communication is lagging behind. To facilitate the discovery of such factors, we explored the potential of a mixed microbial culture (MMC) derived from wheatgrass, in which heterogeneity of this microbial community might elicit signaling factors for interspecies communication. The stability of Wheatgrass MMC in terms of community structure and metabolic output was first characterized by 16S ribosomal RNA amplicon sequencing and liquid chromatography/mass spectrometry (LC/MS), respectively. In addition, detailed MS analyses led to the identification of 12-hydroxystearic acid (12-HSA) as one of the major metabolites produced by Wheatgrass MMC. Stereochemical analysis revealed that Wheatgrass MMC produces mostly the (R)-isomer, although a small amount of the (S)-isomer was also observed. Furthermore, 12-HSA was found to modulate planktonic growth and biofilm formation of various marine bacterial strains. The current study suggests that naturally derived MMCs could serve as a simple and reproducible platform to discover potential signaling factors for interspecies communication. In addition, the study indicates that hydroxylated long-chain fatty acids, such as 12-HSA, may constitute a new class of interspecies signaling factors.
Keywords: Mixed culture, Microbial communication, Hydroxy fatty acid, Planktonic growth, Biofilm
Graphical Abstract

1. Introduction
Microbes use signaling factors (SFs) for intraspecies and interspecies communications.1 Over the course of the past three decades, a series of SFs have been discovered, which include autoinducers, such as N-acyl homoserine lactones (AHL),2 furanosyl borate esters called AI-2,3 diffusible signal factors (DSF),4 3-hydroxypalmitic methyl esters,5 indole and its derivatives,6 and cyclic peptides, such as autoinducing peptides and diketopiperazines.7–9 While many SFs have been characterized, the chemical lexicon of microbes as we know today is limited because only a small number of microbes have been studied thus far.
One limiting factor in the discovery of microbial SFs is the cultivation method. Most known SFs were discovered from axenic cultures.10–14 While axenic culture is a tractable and reproducible system, its use could limit our ability to discover interspecies SFs because microbes in axenic cultures do not need to communicate with other microbial species. In fact, most known SFs are for intraspecies communication. Another approach that could be used for SF discovery is co-cultivation. Many bioactive small molecules have been found from artificial mixtures of known microbes.15–18 Yet, co-cultivation might not be an ideal system to discover interspecies SFs, because it does not reflect natural microbial interactions.
One untested, but potentially useful, system is mixed microbial culture (MMC). MMCs can be generated by cultivating microbial communities obtained from the environment, such as plants and soil samples. Since microbes in each MMC are derived from the same environment, they are expected to “speak” the common chemical language. Therefore, MMCs could serve as a useful system to uncover potential interspecies SFs. At present, however, little is known about the stability of MMCs in terms of community structure and metabolic output. This is an important problem because, if we are to use MMCs as a new platform to find interspecies SFs, they must be stable and reproducible.
As a first step to explore the utility of MMCs, the stability of an MMC derived from wheatgrass (Wheatgrass MMC) was examined. Wheatgrass MMC was used as a model system for several reasons. Plants, including wheatgrass, are known to harbor diverse microbial communities.19 Yet, plant-derived MMCs have not been extensively studied in terms of metabolite production. While some SFs of plant pathogens, such as indole derivatives and DSF, have been characterized,4,20,21 exploration of non-pathogenic plant symbionts remains largely understudied. Thus, the purpose of this study was to characterize the stability of Wheatgrass MMC in terms of community structure and metabolic output. Furthermore, our study examined the metabolites produced by Wheatgrass MMC, which uncovered a potentially new class of interspecies SFs.
2. Results
2.1. Preparation of Wheatgrass MMC
Figure 1A summarizes the procedure to prepare Wheatgrass MMC. Wheatgrass was first rinsed in running water to remove loosely attached microbes on the surface. Then, the microbial community associated with wheatgrass was obtained by sonication in water. The resulting aqueous suspension was used to inoculate Reasoner’s 2A (R2A) liquid medium, which is a low-nutrient medium suitable for the cultivation of slow-growing species in environmental samples. The resulting culture was used to prepare frozen stock. To test the stability of Wheatgrass MMC over time, cultures at three different time points were prepared as follows. R2A was inoculated with the frozen stock and incubated for one week to obtain the first time point (Week 1). The Week 1 culture was subcultured and grown for another week to obtain the second time point (Week 2), which, in turn, was subcultured and grown for another week to obtain the third time point (Week 3). Figure 1B shows the image of Wheatgrass MMC under the transmission electron microscope (TEM).
Figure 1.

Preparation of Wheatgrass MMC. (A) Schematic overview. To examine the stability of Wheatgrass MMC over time, cultures at three different time points (Weeks 1, 2, 3) were prepared. (B) The image of Wheatgrass MMC under the transmission electron microscope (TEM).
As a background control, the whole procedure was repeated without wheatgrass to obtain Control MMC. This control was necessary because the initial steps of our procedure, namely, rinsing of wheatgrass in running water and placing wheatgrass in a tube prior to sonication, were done outside of a biosafety cabinet. Thus, Control MMC, which accounted for the microbes that could be introduced from indoor dusts, was prepared to determine whether microbes in Wheatgrass MMC were unique to wheatgrass.
2.2. Stability of microbial community in Wheatgrass MMC
The microbial communities in Control and Wheatgrass MMCs were characterized by 16S ribosomal RNA (rRNA) amplicon sequencing. Figure 2 shows taxonomic distributions of Control and Wheatgrass MMCs at the order level over three weeks (see Fig S1 for other taxonomic levels). The comparison between the two MMCs revealed more taxa in Wheatgrass MMC than in Control MMC. Furthermore, observed taxa in the two MMCs did not overlap except for Burkholderiales, suggesting that most taxa in Wheatgrass MMC were derived uniquely from wheatgrass. It is also noted that Rhizobiales, which was the predominant order (>90%) in Control MMC, was not observed in Wheatgrass MMC.
Figure 2.

Taxonomic distributions of Control and Wheatgrass MMCs at the order level over three weeks. (A) Control MMC. (B) Wheatgrass MMC. The x-axis represents time after the initial inoculation, and the y-axis represents relative abundance of order taxon. Legend shows the orders observed in each culture; the “Others” category represents the orders whose relative abundance never exceeded 1% during the three-week period.
In both Wheatgrass and Control MMCs, community structures shifted slightly during the three-week period. Wheatgrass MMC contained five orders that exceeded 1% in relative abundance during the three-week period. The most abundant order was Lactobacillales, which shifted from 78.4% in Week 1 to 69.4% (Week 3), followed by Enterobacteriales (from 4.4% in Week 1 to 15.4% in Week 3), Pseudomonadales (from 11.2% in Week 1 to 5.2% in Week 3), Xanthomonadales (from 2.5% in Week 1 to 2.7% in Week 3), and Burkholderiales (from 0.6% in Week 1 to 6.4% in Week 3). The five orders persisted over the three-week period, even though Wheatgrass MMC was subcultured twice.
2.3. Stability of metabolic output of Wheatgrass MMC
To examine metabolic output, culture broth of Wheatgrass MMC was extracted with ethyl acetate and subjected to liquid chromatography/mass spectrometry (LC/MS). As a control, culture broth of Control MMC was also analyzed. Total ion chromatograms from the blank injection (for background measurement), Control MMC, and Wheatgrass MMC were compared to identify peaks unique to Wheatgrass MMC (Fig 3). The analysis revealed four peaks that were consistently observed only in Wheatgrass MMC (peaks i-iv, highlighted in the yellow background, Fig 3C).
Figure 3.

LC/MS profiles of MMC samples. MS was measured in the positive ion mode. (A) Blank. (B) Control MMC. (C) Wheatgrass MMC. The four peaks (i-iv) unique to Wheatgrass MMC are highlighted in the yellow background. Shown are representative data of triplicate experiments.
Next, the stability of LC/MS profiles over time was examined. Wheatgrass MMC samples at Weeks 1–3 were subjected to LC/MS, which gave nearly identical chromatograms (Fig 4). Thus, the metabolic output of Wheatgrass MMC appeared to be stable over the three-week period.
Figure 4.

LC/MS profiles of Wheatgrass MMC extracts over time. MS was measured in the positive ion mode. (A) Week 1. (B) Week 2. (C) Week 3. The four unique peaks (i-iv) of Wheatgrass MMC are highlighted in the yellow background. Shown are representative data of triplicate experiments.
2.4. Identification of 12-hydroxystearic acid (12-HSA) in Wheatgrass MMC
To gain structural information of the four unique peaks (i-iv), their MS data were further analyzed. High resolution MS revealed that molecular formulas of peaks i, ii, iii, and iv were C18H33N, C18H35N, C20H37N, and C18H36O3, respectively. Tandem MS (MS/MS) showed that smaller fragment ions from peaks i-iii were identical (data not shown), suggesting that they are a series of structurally related amines. However, their exact chemical structures remain to be clarified. On the other hand, the molecular formula of peak iv (C18H36O3) seemed to be consistent with a hydroxylated stearic acid. Further analysis of peak iv by tandem MS (MS/MS) in the negative ion mode revealed fragment ions at m/z 253.2533 ([C17H33O]−), 169.1595 ([C11H21O]−), and 113.0973 ([C7H13O]−) (Fig S2). These fragment ions matched with previously reported characteristic fragments of 12-hydroxystearic acid (12-HSA) (Fig 5 and Fig S3).22 The identity of peak iv was subsequently confirmed as 12-HSA based on the comparisons of peak iv and a commercial 12-HSA sample using LC/MS retention time (Fig S4) and MS/MS fingerprints (Fig S5).
Figure 5.
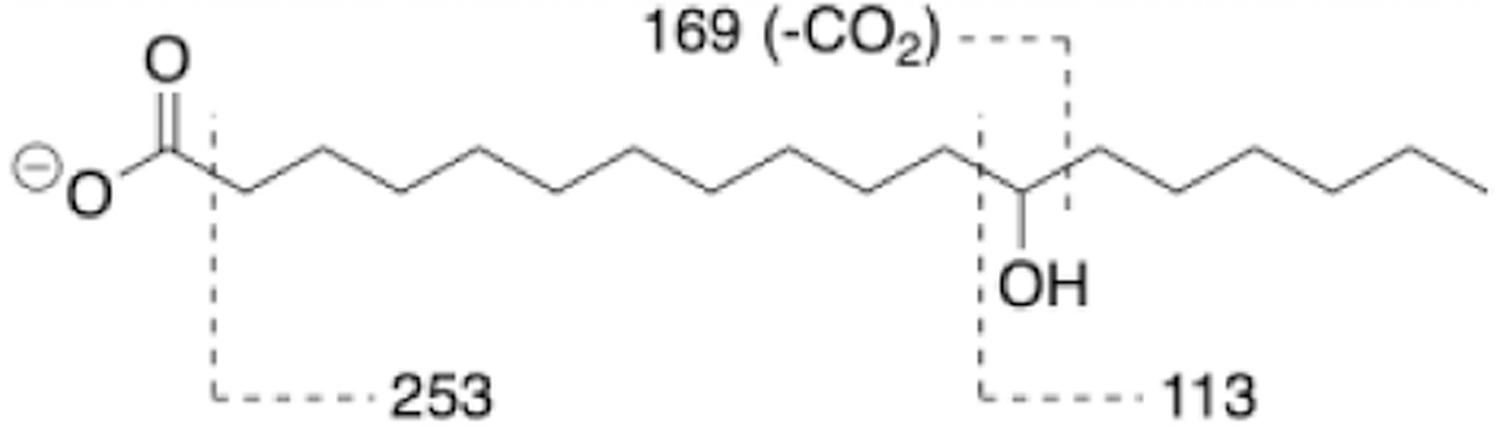
Characteristic MS/MS fragments of 12-hydroxystearic acid (12-HSA).22
2.5. Stereochemical characterization of 12-HSA in Wheatgrass MMC
Although commercial 12-HSA, which is purified from castor oil, is known to have the (R)-configuration,a the stereochemistry of 12-HSA from microbial sources has never been characterized. Since the enantiopurity of 12-HSA from Wheatgrass MMC was not known, we chose not to use standard methods for stereochemical analysis, such as Mosher’s method, that require pure derivatives. A possible alternative approach was the method that was originally developed for the stereochemical analysis of 10-hydroxystearic acid (10-HSA),25 in which the methyl ester of 10-HSA was derivatized with (S)-(+)-O-acetyl mandelic acid and subjected to 1H-NMR analysis. While this method can determine the enantiomeric purity of 10-HSA based on the 1H signals near 5.88 ppm (the methine proton of the mandelic acid moiety) and 3.67 ppm (the methyl ester),26 it was unclear whether the same approach could be applicable to 12-HSA.
To examine the utility of this mandelate-based method, the (S)-(+)-O-acetyl mandelic acid derivative (4) of racemic 12-HSA methyl ester was prepared from commercial (R)-12-HSA (1) (Fig 6A). Although 1H-NMR did not clearly differentiate the two stereoisomers in the racemic 12-HSA derivative (Fig S7), 13C-NMR showed signals that are clearly different between the two stereoisomers, such as C-16 and C-17 (Fig 6b, the middle spectrum). The 13C-NMR spectrum of (R)-12-HSA derivative (Fig 6b, the top spectrum) was used for the stereochemical assignment of C-16 and C-17 signals. The 13C-NMR spectrum of Wheatgrass MMC 12-HSA derivative (Fig 6b, the bottom spectrum), on the other hand, revealed (R)-12-HSA as the predominant (~90%) stereoisomer, although a small amount (~10%) of the (S)-isomer was also detected.
Figure 6.

Stereochemical characterization of 12-HSA in Wheatgrass MMC. (a) Preparation of (S)-(+)-O-acetylmadelic acid derivative of racemic 12-HSA methyl ester. (b) 13C-NMR spectra of (S)-(+)-O-acetylmandelic acid derivatives of (R)-12-HSA methyl ester (top), racemic 12-HSA methyl ester (middle), and Wheatgrass MMC 12-HSA methyl ester (bottom).
2.6. 12-HSA modulates planktonic growth and biofilm formation of marine bacteria
The discovery of 12-HSA in a heterogeneous community like Wheatgrass MMC opened a possibility that 12-HSA might be an interspecies SF sensed by a broad range of bacterial species. As a first step to characterize how widely 12-HSA might be recognized by different bacterial species, the effects of 12-HSA were examined with a series of marine bacterial strains, which were not observed in Wheatgrass MMC. Since the supply of 12-HSA from Wheatgrass MMC was limited, this study used commercial (R)-12-HSA, which was the major stereoisomer of 12-HSA in Wheatgrass MMC.
First, Brevundimonas mediterranea (SDH6), which is an Alphaproteobacterial strain previously shown to produce biofilm,27 was incubated with 12-HSA and examined for biofilm formation and planktonic growth. As a control, the strain was also treated with indole, which is a well-known interspecies SF capable of modulating biofilm formation positively or negatively depending on the bacterial strain.20 Figure 7 summarizes the effects of indole and 12-HSA on biofilm formation and planktonic growth of B. mediterranea (SDH6) at 27°C. Indole inhibited both biofilm formation and planktonic growth of this strain (Fig 7A). On the other hand, 12-HSA inhibited its biofilm formation but promoted planktonic growth slightly (Fig 7B). When the experiments were repeated at a lower temperature (19°C), which could influence the extent of biofilm formation,28 the inhibitory effect of indole on biofilm formation was less pronounced, whereas 12-HSA retained its effects on biofilm formation and planktonic growth regardless of the temperature (Fig S8a).
Figure 7.

Box-and-whisker plots of the effects of indole (control) (A) and 12-HSA (B) on biofilm formation and planktonic growth of B. mediterranea at 27°C. The effects are normalized by the DMSO control. Blue boxes show the effects of compounds on biofilm formation at different concentrations. Adjacent gray boxes indicate the effects on planktonic growth. x = Mean, __ = Median, o = Data points, n=12.
To characterize how 12-HSA affects different strains of a same bacterial species, the assays were repeated with three strains (SD3, SD8, SD9) of Alteromonas oceani, which is a Gammaproteobacterial species of marine origin (Fig S8b–g).27 The results are summarized as tile plots in Figure 8. Overall, the effects of 12-HSA on the planktonic growth of SD3, SD8, and SD9 were marginal. On the other hand, 12-HSA exhibited differential effects on their biofilm formation in a manner dependent on strain and temperature. Specifically, 12-HSA strongly promoted biofilm formation of SD3 and SD9 at 19°C and 27°C, respectively, whereas it hardly affected biofilm formation of SD8 at both temperatures. Collectively, our results indicated that 12-HSA could differentially regulate biofilm formation of diverse bacterial species.
Figure 8.

Tile plots summarizing the effects of indole (A and B) and 12-HSA (C and D) on biofilm formation and planktonic growth of marine bacterial species at 19°C and 27°C. Concentrations of the compound are listed on the horizontal axis in µg/mL. SD3, SD8, SD9: Alteromonas oceani strains (Gammaproteobacteria). SDH6: Brevundimonas mediterranea (Alphaproteobacteria).
3. Discussion
Our 16S amplicon sequencing analysis revealed core microbial communities of both Control and Wheatgrass MMCs, which persisted through repeated propagation. Given the fact that MMCs were subcultured twice during the three-week period, the observed stability is rather remarkable. The LC/MS analysis corroborated the 16S results. MS peaks, which reflect metabolites present in the crude culture extracts, were stable over the three-week period. The consistent LC/MS profile indicated that Wheatgrass MMC could serve as a reproducible system to look for potential SFs for microbial communication.
Our search for potential SFs in Wheatgrass MMC led to the identification of 12-HSA. Similar hydroxystearic acids (C18H36O3) have also been detected in MMCs derived from cabbage, bamboo, and ant mound (Fig S9), although their full structures remain to be characterized. Furthermore, there are many reports of structurally diverse hydroxy fatty acids of microbial origin.29,30 Fatty acids are certainly not strangers to microbial communication. They have been implicated in the gut microbiome gut-brain axis.31 Diffusible signal factors (DSF), a family of mainly cis-2-unsaturated long chain fatty acids, are known to modulate microbial mass behaviors, such as growth, biofilm formation, and virulence.1,32,33 However, little is currently known about the roles of hydroxylated long-chain fatty acids in microbial communication. While 12-HSA is known as an organogelator with applications in food and cosmetic industries,34 its role in microbial communication has not been characterized. Perhaps, the most well-characterized hydroxy fatty acids of bacterial origin are β-hydroxy acids, such as 3-hydroxypalmitic acid, which originate from the lipid A moiety of lipopolysaccharide.35–37 There is a report of the methyl ester of 3-hydroxypalmitic acid acting as a quorum sensing SF in Ralstonia solanacearum.38 Yet, it remains to be determined whether hydroxylated long chain fatty acids in general play a broader role in microbial communication.
The discovery of 12-HSA in Wheatgrass MMC raised a question of its stereochemistry. Although commercial 12-HSA is believed to have the (R)-configuration, which we have verified with the Mosher’s method (Fig. S6), the stereochemistry and optical purity of 12-HSA in Wheatgrass MMC had to be characterized. To that end, we established a simple protocol based on (S)-(+)-O-acetyl mandelate derivatization, followed by 13C-NMR. Our analysis revealed that 12-HSA in Wheatgrass MMC consists mostly of the (R)-isomer but a small amount (~10%) of the (S)-isomer also exists. While some insects are known to use mixtures of stereoisomeric pheromones to transmit specific messages,39 it is currently unknown whether microbial communities also use stereoisomeric SF mixtures for communications.
The current study also demonstrates that 12-HSA, which was found in a terrestrial, plant-based bacterial community, affected the biofilm formation of marine bacteria. As such, the finding opens a new possibility that hydroxylated long chain fatty acids, such as 12-HSA, may constitute an as-yet uncharacterized group of SFs for interspecies communication. Importantly, the differential effects on the three A. oceani strains suggest that the effects of 12-HSA are not mediated by a non-specific mechanism, such as modulation of membrane fluidity. Instead, the observed differential effects suggest the presence of a distinct signaling system. Comparisons of the three A. oceani strains by genomic, transcriptomic, and/or proteomic tools might provide mechanistic insights into the mode of action of 12-HSA, which remains to be characterized. As such, further investigation is warranted to clarify the roles of 12-HSA in interspecies communication.
4. Conclusion
The current study demonstrates that MMCs can serve as a simple and reproducible platform for the discovery of potential interspecies SFs. In fact, although this was our first study to explore the utility of MMCs, 12-HSA was rapidly identified and characterized as a potential SF for interspecies communication. As such, the use of MMCs could greatly facilitate the discovery of as-yet uncharacterized interspecies signals.
5. Experimental section
5.1. Preparation of cryogenic glycerol stocks of Wheatgrass MMC
Fresh, organic wheatgrass was purchased from a local Whole Foods Market and rinsed with tap water to dislodge any loosely held contaminating bacteria. Blades of rinsed grass weighing ~0.5 grams were combined with 10 mL of reverse osmosis (RO) water in a 15 mL Falcon tube and sonicated for 10 minutes to dislodge resident bacteria. A control tube was also prepared with 10 mL of RO water and sonicated for 10 minutes. 1 mL aliquots of this water from the control sample and from the wheatgrass were inoculated into separate 50 mL Falcon tubes containing 25 mL sterile Reasoner’s 2A (R2A) liquid media, capped loosely and left to grow at room temperature (25°C) without shaking for 7 days. 500 µL of this culture was added to 500 µL of sterile 50% glycerol (aq.). The resulting mixture was stored at −80°C.
5.2. TEM
For each MMC, a bacterial suspension was prepared. 5 µl of bacterial suspension was placed on a carbon-coated nickel grid. After 1 minute, the sample was blotted off by touching the edge of the grid to filter paper. Once the grid was dry, images were taken with a Tecnai Spirit BioTwin electron microscope (FEI, Hillsboro, Oregon) operating at 120 kV, equipped with AMT BioSprint29 digital camera.
5.3. Culturing and propagation
200 µL of thawed glycerol stock of MMC was inoculated into a sterile tube containing 10 mL of R2A media and allowed to grow for seven days at room temperature (25°C) without shaking. After the seven-day growth period, 200 µL of culture was inoculated into 10 mL sterile R2A media to propagate the culture.
5.4. 16S rRNA amplicon sequencing
DNA extracts of MMCs at Week 1, 2, and 3 were prepared with Qiagen DNeasy PowerLyzer PowerSoil Kit. Purified DNA samples were quantified with Nanodrop and submitted to Integrated Microbiome Resource (IMR) at Dalhousie University in Halifax, Nova Scotia for 16S rRNA amplicon sequencing using a primer pair targeting bacterial V4-V5 (515F: GTGYCAGCMGCCGCGGTAA, 926R: CCGYCAATTYMTTTRAGTTT).40,41 The sequencing was conducted with Illumina MiSeq at IMR. The resulting paired-end Illumina reads were analyzed with Quantitative Insights into Microbial Ecology 2 (Qiime2) to characterize the microbial communities in MMCs.42 Briefly, sequence quality control and feature table construction were carried out with DADA2. Taxonomic analysis was conducted using a classifier that was trained on the Greengenes 13_8 99% OTUs, where sequences had been trimmed to the 16S rRNA region amplified by the 515F/926R primer pair.
5.5. LC/MS
The culture broths of Control and Wheatgrass MMCs were extracted with ethyl acetate. Solvent was removed in vacuo. Crude extracts were weighed and dissolved in 100 µL methanol: 500 µL acetonitrile. Average sample concentrations were 0.5 mg/mL. 10 µL of sample was diluted into 40 µL acetonitrile/water (1:1). Blank controls were prepared from the methanol/acetonitrile (1:5) mixture and diluted with acetonitrile/water (1:1) as described above. 5 µL sample was injected into Agilent iFunnel 6550 Q-ToF coupled to Agilent 1290 Infinity LC system equipped with Agilent Eclipse C18 column, 1.8 µm, 2.1 x 50 mm. Method: Solvent A, water with 0.1% formic acid, solvent B, acetonitrile with 0.1% formic acid; gradient of 2–98% acetonitrile in 18 min; flow rate of 0.4 mL/min; column temperature at 30°C. MS/MS settings: MS scan range 100–1200 m/z, where two most abundant precursors selected for MS/MS (range 50–800); collision energy of 50 eV (positive mode) and 35 eV (negative mode). Source settings: gas temperature at 250°C, drying gas at 17 L/min, nebulizer at 30 psi, sheath gas temperature at 250°C, and sheath gas flow at 12 L/min.
5.6. 12-HSA methyl ester
To a solution of commercial (R)-12-HSA (1) (50 mg, 0.17 mmol) in toluene/methanol (4:1) (1 mL) was added 200 µL of 2 M TMS diazomethane in hexane (Sigma-Aldrich). The mixture was concentrated in vacuo and the residue was purified by silica gel chromatography to obtain the purified product (40 mg, 0.13 mmol, 75%). 1H NMR (500 MHz, CDCl3) δ 3.64 (3H, s), 3.55 (1H, m), 2.27 (2H, t, J = 7.6 Hz), 1.59 (2H, m), 1.50–1.20 ppm (27H, m), 0.86 (3H, t, J = 7.0 Hz). 13C NMR (125 MHz, CDCl3) δ 174.61, 72.19, 51.69, 37.69, 37.67, 34.33, 32.07, 29.89, 29.79, 29.71, 29.62, 29.59, 29.45, 29.35, 25.86, 25.84, 25.16, 22.85, 14.33. HRMS (ESI; m/z) Calcd for C19H38O3Na [M+Na]+ 337.2719; found 337.2715.
5.7. Racemic 12-HSA methyl ester
To a solution of commercial (R)-12-HSA methyl ester (31 mg, 0.1 mmol) in dry dichloromethane (1 mL) was added Dess-Martin periodinane (42 mg, 0.1 mmol). The reaction mixture was stirred at room temperature for 3 h. The mixture was concentrated in vacuo and the residue was purified by silica gel chromatography to obtain the keto product 2 (25 mg, 0.08 mmol, 80 %). 1H NMR (500 MHz, CDCl3) δ 3.59 (3H, s), 2.31 (4H, m), 2.23 (2H, t, J = 7.5 Hz), 1.55 (2H, m), 1.48 (4H, m), 1.15–1.28 ppm (18H, m), 0.80 (3H, t, J = 7.0 Hz). 13C NMR (125 MHz, CDCl3) δ 211.82, 174.37, 51.48, 42.85, 42.81, 34.11, 31.63, 29.40, 29.25, 29.23, 29.14, 28.95, 24.95, 23.86, 23.85, 22.52, 14.06. HRMS (ESI; m/z) Calcd for C19H37O3 [M+H]+ 313.2737; found 313.2737. 2 (18 mg, 0.06 mmol) was then dissolved in dry methanol (1 mL). NaBH4 (3 mg, 0.081 mmol) was added to the solution, and the mixture was stirred at RT for 1 h. The mixture was concentrated in vacuo and the residue was purified by silica gel chromatography to obtain 3 (16 mg, 0.05 mmol, 85%).
5.8. (S)-(+)-O-acetylmandelic acid derivatives of 12-HSA methyl ester (4)
To a solution of 12-HSA methyl ester (4 mg, 12.7 µmol) dry dichloromethane (1 mL) was added (S)-(+)-O-acetylmandelic acid (14.6 mg, 75 µmol), EDC (14.4 mg, 78 µmol), DMAP (1.5 mg, 12.3 µmol) The reaction mixture was stirred at room temperature for 24 h. The mixture was concentrated in vacuo and the residue was purified by silica gel chromatography to obtain the (S)-(+)-O-acetylmandelic acid derivative (3 mg, 5.8 µmol, 46%). The racemic mixture (4): 13C NMR (125 MHz, CDCl3) δ 174.60, 174.59, 170.52, 168.95, 134.34, 129.31, 128.86, 127.82, 76.23, 74.96, 51.69, 34.35, 34.33, 34.08, 31.89, 31.77, 29.69, 29.66, 29.63, 29.61, 29.59, 29.56, 29.45, 29.37, 29.33, 29.23, 29.10, 25.37, 25.32, 25.17, 24.92, 24.85, 22.78, 22.64, 20.99, 14.31, 14.27. The (R)-isomer: 1H NMR (500 MHz, CDCl3) δ 7.45 (2H, m), 7.35 (3H, m), 5.85 (1H, s), 4.85 (1H, quint, J = 6.2 Hz), 3.65 (3H, s), 2.28 (2H, t, J = 7.6 Hz), 2.18 (3H, s), 1.61 (2H, m), 1.50 (2H, m), 1.35 (2H, m), 1.30–1.20 (16H, m), 1.12 (2H, m), 1.01 (4H, m), 0.86 (2H, m), 0.80 (3H, t, J = 7.4 Hz). 13C NMR (125 MHz, CDCl3) δ 174.61, 170.52, 168.95, 134.33, 129.31, 128.86, 127.82, 76.23, 74.97, 51.69, 34.36, 34.34, 34.08, 31.77, 29.70, 29.66, 29.63, 29.46, 29.36, 29.11, 25.37, 25.18, 24.85, 22.64, 20.99, 14.27. HRMS (ESI; m/z) Calcd for C29H47O6 [M+H]+ 491.3367; found 491.3371.
5.9. Assays for planktonic growth and biofilm formation
One isolate of B. mediterranea (SDH6) and three strains of A. oceani (SD3, SD8 and SD9) were recovered as described previously.27 Indole (Alfa Aesar A14427, Haverhill, MA) and 12-HSA (Alfa Aesar 44810, Haverhill, MA) were purchased to have sufficient quantities for our assays. Since compounds were dissolved in dimethyl sulfoxide (DMSO), DMSO was used as the vehicle control. Planktonic growth was quantified by OD600, whereas biofilm formation was characterized with crystal violet staining, as described previously.44 Assays were performed at two different temperatures (19°C and 27°C). In the assays at 19°C, bacteria were incubated with the compounds for 96 hours, whereas the assays at 27°C used a shorter incubation period (48 hours). In the planktonic growth assay of 12-HSA, the addition of the compound turned the culture broth turbid. As such, OD600 readings of cultures treated with 12-HSA were corrected by OD600 readings of 12-HSA in media without culture. For each treatment condition, 12 data points were obtained.
5.10. Data availability
Sequence files are available from the NCBI Sequence Read Archive (SRA) with accession number PRJNA672656. Mass spectrometric data sets are available from GNPS under accession number MSV000086394.
Supplementary Material
Acknowledgments
AK thanks PSC CUNY Grant TRADA-50-661 for the funding of this study. Purchase of the NEO-500 NMR spectrometer used to obtain results included in this publication was supported by the National Science Foundation under the award CHE MRI 1900509. VV and AV thank Hunter College MARC and RISE Programs, which are supposed by NIH/NIGMS awards T34GM007823 and R25GM060665, respectively. ACD thanks Early Research Initiative (ERI) Provost’s Pre-Dissertation Research Fellowship at The CUNY Graduate Center and the Japan Society for the Promotion of Science (JSPS) Short-Term Summer Fellowship. AK and ACD thank Ray Domzalski Jr. for his assistance in preparing graphic abstract. SDP thanks the Mustang Soil and Water Conservation District, Donald M. Allen for Scholarships. AO, DPH, and SDP thanks the University of Texas Permian Basin Laboratories Department for funding.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Competing Interest
The authors declare that they have no known competing financial interest or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at …
References
- 1.Greenberg EP, Whiteley M, Diggle SP. Progress in and promise of bacterial quorum sensing research. Nature. 2017;551(7680):313. doi: 10.1038/nature24624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eberhard A, Burlingame AL, Eberhard C, Kenyon GL, Nealson KH, Oppenheimer NJ. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry. 1981;20(9):2444–2449. doi: 10.1021/bi00512a013 [DOI] [PubMed] [Google Scholar]
- 3.Bassler BL, Wright M, Showalter RE, Silverman MR. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Molecular Microbiology. 1993;9(4):773–786. doi: 10.1111/j.1365-2958.1993.tb01737.x [DOI] [PubMed] [Google Scholar]
- 4.Barber CE, Tang JL, Feng JX, et al. A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Molecular Microbiology. 1997;24(3):555–566. doi: 10.1046/j.1365-2958.1997.3721736.x [DOI] [PubMed] [Google Scholar]
- 5.Clough SJ, Lee KE, Schell MA, Denny TP. A two-component system in Ralstonia (Pseudomonas) solanacearum modulates production of PhcA-regulated virulence factors in response to 3-hydroxypalmitic acid methyl ester. Journal of Bacteriology. 1997;179(11):3639–3648. doi: 10.1128/jb.179.11.3639-3648.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chant EL, Summers DK. Indole signalling contributes to the stable maintenance of Escherichia coli multicopy plasmids. Mol Microbiol. 2007;63(1):35–43. doi: 10.1111/j.1365-2958.2006.05481.x [DOI] [PubMed] [Google Scholar]
- 7.Abdelnour A, Arvidson S, Bremell T, Rydén C, Tarkowski A. The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infect Immun. 1993;61(9):3879–3885. Accessed September 11, 2020. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC281089/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ji G, Beavis R, Novick RP. Bacterial Interference Caused by Autoinducing Peptide Variants. Science. 1997;276(5321):2027–2030. Accessed September 11, 2020. https://www.jstor.org/stable/2892976 [DOI] [PubMed] [Google Scholar]
- 9.Holden MTG, Chhabra SR, Nys RD, et al. Quorum-sensing cross talk: isolation and chemical characterization of cyclic dipeptides from Pseudomonas aeruginosa and other Gram-negative bacteria. Molecular Microbiology. 1999;33(6):1254–1266. doi: 10.1046/j.1365-2958.1999.01577.x [DOI] [PubMed] [Google Scholar]
- 10.Fleming A On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. 1929. Bull World Health Organ. 2001;79(8):780–790. Accessed October 8, 2018. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2566493/ [PMC free article] [PubMed] [Google Scholar]
- 11.Tan SY, Tatsumura Y. Alexander Fleming (1881–1955): Discoverer of penicillin. Singapore Med J. 2015;56(7):366–367. doi: 10.11622/smedj.2015105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenberg EP. Acyl-Homoserine Lactone Quorum Sensing in Bacteria. Journal of microbiology (Seoul, Korea). 2000;38(3):117–121. Accessed June 21, 2018. https://iths.pure.elsevier.com/en/publications/acyl-homoserine-lactone-quorum-sensing-in-bacteria [Google Scholar]
- 13.Eberhard A, Burlingame AL, Eberhard C, Kenyon GL, Nealson KH, Oppenheimer NJ. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry. 1981;20(9):2444–2449. [DOI] [PubMed] [Google Scholar]
- 14.Shaw PD, Ping G, Daly SL, et al. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc Natl Acad Sci U S A. 1997;94(12):6036–6041. Accessed October 15, 2018. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC20996/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goers L, Freemont P, Polizzi KM. Co-culture systems and technologies: taking synthetic biology to the next level. J R Soc Interface. 2014;11(96). doi: 10.1098/rsif.2014.0065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Netzker T, Fischer J, Weber J, et al. Microbial communication leading to the activation of silent fungal secondary metabolite gene clusters. Front Microbiol. 2015;6. doi: 10.3389/fmicb.2015.00299 [DOI] [PMC free article] [PubMed]
- 17.Marmann A, Aly A, Lin W, Wang B, Proksch P. Co-Cultivation—A Powerful Emerging Tool for Enhancing the Chemical Diversity of Microorganisms. Marine Drugs. 2014;12(2):1043–1065. doi: 10.3390/md12021043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pettit RK. Mixed fermentation for natural product drug discovery. Appl Microbiol Biotechnol. 2009;83(1):19–25. doi: 10.1007/s00253-009-1916-9 [DOI] [PubMed] [Google Scholar]
- 19.Compant S, Samad A, Faist H, Sessitsch A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. Journal of Advanced Research. 2019;19:29–37. doi: 10.1016/j.jare.2019.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee J-H, Wood TK, Lee J. Roles of Indole as an Interspecies and Interkingdom Signaling Molecule. Trends in Microbiology. 2015;23(11):707–718. doi: 10.1016/j.tim.2015.08.001 [DOI] [PubMed] [Google Scholar]
- 21.Castro-Moretti FR, Gentzel IN, Mackey D, Alonso AP. Metabolomics as an Emerging Tool for the Study of Plant–Pathogen Interactions. Metabolites. 2020;10(2). doi: 10.3390/metabo10020052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moe MK, Strøm MB, Jensen E, Claeys M. Negative electrospray ionization low-energy tandem mass spectrometry of hydroxylated fatty acids: a mechanistic study. Rapid Communications in Mass Spectrometry. 2004;18(15):1731–1740. doi: 10.1002/rcm.1545 [DOI] [PubMed] [Google Scholar]
- 23.Kasai Y, Watanabe M, Harada N. Convenient method for determining the absolute configuration of chiral alcohols with racemic1H NMR anisotropy reagent, M?NP acid: Use of HPLC-CD detector. Chirality. 2003;15(4):295–299. doi: 10.1002/chir.10210 [DOI] [PubMed] [Google Scholar]
- 24.Kasai Y, Taji H, Fujita T, et al. MalphaNP acid, a powerful chiral molecular tool for preparation of enantiopure alcohols by resolution and determination of their absolute configurations by the (1)H NMR anisotropy method. Chirality. 2004;16(9):569–585. doi: 10.1002/chir.20077 [DOI] [PubMed] [Google Scholar]
- 25.El-Sharkawy SH, Yang W, Dostal L, Rosazza JPN. Microbial Oxidation of Oleic Acid. APPL ENVIRON MICROBIOL. 1992;58:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang W, Dostal L, Rosazza JPN. Stereospecificity of Microbial Hydrations of Oleic Acid to 10-Hydroxystearic Acid. Appl Environ Microbiol. 1993;59(1):281–284. Accessed March 3, 2021. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC202091/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malalasekara L, Henderson DP, Oldham AL. A study of biofilm formation in marine bacteria isolated from ballast tank fluids. Trends in Research. 2020;3:1–7. doi: DOI: 10.15761/TR.1000169 [DOI] [Google Scholar]
- 28.Ryu J-H, Kim H, Beuchat LR. Attachment and Biofilm Formation by Escherichia coli O157:H7 on Stainless Steel as Influenced by Exopolysaccharide Production, Nutrient Availability, and Temperature. J Food Prot. 2004;67(10):2123–2131. doi: 10.4315/0362-028X-67.10.2123 [DOI] [PubMed] [Google Scholar]
- 29.Kim K-R, Oh D-K. Production of hydroxy fatty acids by microbial fatty acid-hydroxylation enzymes. Biotechnology Advances. 2013;31(8):1473–1485. doi: 10.1016/j.biotechadv.2013.07.004 [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Eser BE, Kristensen P, Guo Z. Fatty acid hydratase for value-added biotransformation: A review. Chinese Journal of Chemical Engineering. 2020;28(8):2051–2063. doi: 10.1016/j.cjche.2020.02.008 [DOI] [Google Scholar]
- 31.Nankova BB, Agarwal R, MacFabe DF, Gamma EFL. Enteric Bacterial Metabolites Propionic and Butyric Acid Modulate Gene Expression, Including CREB-Dependent Catecholaminergic Neurotransmission, in PC12 Cells - Possible Relevance to Autism Spectrum Disorders. PLOS ONE. 2014;9(8):e103740. doi: 10.1371/journal.pone.0103740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryan RP, Dow JM. Diffusible signals and interspecies communication in bacteria. Microbiology. 2008;154(7):1845–1858. doi: 10.1099/mic.0.2008/017871-0 [DOI] [PubMed] [Google Scholar]
- 33.Ryan RP, Dow JM. Communication with a growing family: diffusible signal factor (DSF) signaling in bacteria. Trends in Microbiology. 2011;19(3):145–152. doi: 10.1016/j.tim.2010.12.003 [DOI] [PubMed] [Google Scholar]
- 34.Mallia VA, George M, Blair DL, Weiss RG. Robust Organogels from Nitrogen-Containing Derivatives of (R)-12-Hydroxystearic Acid as Gelators: Comparisons with Gels from Stearic Acid Derivatives†. Langmuir. 2009;25(15):8615–8625. doi: 10.1021/la8042439 [DOI] [PubMed] [Google Scholar]
- 35.Sjögren J, Magnusson J, Broberg A, Schnürer J, Kenne L. Antifungal 3-Hydroxy Fatty Acids from Lactobacillus plantarum MiLAB 14. Appl Environ Microbiol. 2003;69(12):7554–7557. doi: 10.1128/AEM.69.12.7554-7557.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suda Y, Kim Y-M, Ogawa T, et al. Chemical structure and biological activity of a lipid A component from Helicobacter pylori strain 206. :10. [PubMed] [Google Scholar]
- 37.Zӓhringer U, Lindner B, Rietschel ETh. Molecular Structure of Lipid a, the Endotoxic Center of Bacterial Lipopolysaccharides11This article is dedicated to Professor Dr. Dr. med. h.c. Otto Westphal on the occasion of his 80th birthday (February 1st, 1993). In: Horton D, ed. Advances in Carbohydrate Chemistry and Biochemistry. Vol 50. Academic Press; 1994:211–276. doi: 10.1016/S0065-2318(08)60152-3 [DOI] [PubMed] [Google Scholar]
- 38.Shinohara M, Nakajima N, Uehara Y. Purification and characterization of a novel esterase (β-hydroxypalmitate methyl ester hydrolase) and prevention of the expression of virulence by Ralstonia solanacearum. Journal of Applied Microbiology. 2007;103(1):152–162. doi: 10.1111/j.1365-2672.2006.03222.x [DOI] [PubMed] [Google Scholar]
- 39.MORI K Stereochemical studies on pheromonal communications. Proc Jpn Acad Ser B Phys Biol Sci. 2014;90(10):373–388. doi: 10.2183/pjab.90.373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parada AE, Needham DM, Fuhrman JA. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environmental Microbiology. 2016;18(5):1403–1414. doi: 10.1111/1462-2920.13023 [DOI] [PubMed] [Google Scholar]
- 41.Walters W, Hyde ER, Berg-Lyons D, et al. Improved Bacterial 16S rRNA Gene (V4 and V4–5) and Fungal Internal Transcribed Spacer Marker Gene Primers for Microbial Community Surveys. mSystems. 2016;1(1):e00009–15. doi: 10.1128/mSystems.00009-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bolyen E, Rideout JR, Dillon MR, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology. 2019;37(8):852–857. doi: 10.1038/s41587-019-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: High resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581–583. doi: 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Toole GA. Microtiter dish biofilm formation assay. J Vis Exp. 2011;(47). doi: 10.3791/2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence files are available from the NCBI Sequence Read Archive (SRA) with accession number PRJNA672656. Mass spectrometric data sets are available from GNPS under accession number MSV000086394.


