Abstract
Loss of the fragile X protein FMRP is a leading cause of intellectual disability and autism1,2, but the underlying mechanism remains poorly understood. We report that FMRP deficiency results in hyperactivated nonsense-mediated mRNA decay (NMD)3,4 in human SH-SY5Y neuroblastoma cells and fragile X syndrome (FXS) fibroblast-derived induced pluripotent stem cells (iPSCs). We examined the underlying mechanism and found that the key NMD factor UPF1 binds directly to FMRP, promoting FMRP binding to NMD targets. Our data indicate that FMRP acts as an NMD repressor. In the absence of FMRP, NMD targets are relieved from FMRP-mediated translational repression so that their half-lives are decreased and, for those NMD targets encoding NMD factors, increased translation produces abnormally high factor levels despite their hyperactivated NMD. Transcriptome-wide alterations caused by NMD hyperactivation have a role in the FXS phenotype. Consistent with this, small-molecule-mediated inhibition of hyperactivated NMD, which typifies iPSCs derived from patients with FXS, restores a number of neurodifferentiation markers, including those not deriving from NMD targets. Our mechanistic studies reveal that many molecular abnormalities in FMRP-deficient cells are attributable—either directly or indirectly—to misregulated NMD.
We established transcriptome-wide RNA sequencing (RNA-seq) methodologies5,6 to detect and quantify nonsense-mediated messenger RNA decay (NMD) targets in neuronal cells. Using SH-SY5Y neuroblastoma cells as an abundant source of human neural cells, we identified high-confidence NMD targets based on the satisfaction of two verified criteria: upregulation upon UPF1 knockdown (KD) using RNA-seq (Fig. 1a and Extended Data Fig. 1a,b); and binding by hyperphosphorylated UPF1 (hereafter called p-UPF1) using RNA immunoprecipitation sequencing (RIP-seq) footprinting (Fig. 1a and Extended Data Fig. 1a,c). Based on these criteria, we identified 1,277 high-confidence neuronal NMD targets (hereafter called NMD targets; Fig. 1a and Supplementary Table 1). This was consistent with findings for other mammalian cells that ~5–10% of protein-encoding cellular genes produce NMD targets3,4. As expected, we found that the majority of these NMD targets are also upregulated upon KD of another NMD factor, UPF3X (Extended Data Fig. 1d-f and Supplementary Table 1), consistent with UPF3X mediating the NMD of most but not all NMD targets in HeLa cells7,8.
Fig. 1 ∣. NMD targets encode neuronal cell proteins that function in diverse neurogenic programs.
a, Venn diagram defining SH-SY5Y NMD targets based on upregulation by UPF1 siRNA (UPF1 KD) relative to control siRNA (log2[fold change] > 0 and control siRNA reads per kilobase of transcript, per million mapped reads >1) and increased abundance in p-UPF1 immunoprecipitations (IPs) relative to both the input sample and rabbit IgG immunoprecipitations (log2[fold change] > 1 and reads in p-UPF1 immunoprecipitation > 10). n represents the number of mRNAs. See also Supplementary Table 1. b, Gene Ontology term enrichment analysis using defined NMD targets. Only the top ten ranked functional terms are shown. P values were calculated using Fisher’s exact test. c, Western blots of lysates of SH-SY5Y cells transiently transfected with control siRNA or two different UPF1 siRNAs. Here and elsewhere, the left-most lanes under the wedge analyse serial threefold dilutions of the lysate. The results represent three independent biological replicates. Ctl, control. d, Histogram of the RT-qPCR quantitations of the specified mRNAs, each normalized to the level of its pre-mRNA, using lysates from siRNA-transfected SH-SY5Y cells. The results are presented as means with s.d. (n = 3 independent biological replicates). P values pertain to comparisons with control siRNA-transfected samples (*P < 0.05; **P < 0.01; ***P < 0.001) and were determined by two-sided t-test. e, RT-qPCR quantitations of the specified NMD targets, normalized to quantitations of GAPDH mRNA, in SY-SY5Y cells transiently transfected with the specified siRNA and cultured 3 d later in the presence (5 μg ml−1) of the transcriptional inhibitor actinomycin D. Note the biphasic nature of the NMD target decay curves, where NMD is largely confined to newly synthesized mRNA. All of the results are shown as means and s.d. values (n = 3 independent biological replicates at each time point). Statistical source data and unprocessed blots are provided online.
Tissue expression analysis showed that neuronal NMD targets are expressed throughout the brain (Extended Data Fig. 1g). In particular, >90% (1,184/1,277) of NMD targets identified are expressed in the cerebral cortex, which governs motor control and cognitive abilities such as attention, language and responses to fear and pleasure. Gene Ontology term enrichment analysis for biological processes revealed that NMD targets are significantly enriched to encode proteins forming highly interconnected networks associated with neuronal function, such as axon guidance, neuron development and neuron projection morphogenesis (Fig. 1b). Biological pathway analysis showed that proteins encoded by neuronal NMD targets are enriched in signalling pathways that mediate neuronal function, including axon growth, attraction and repulsion, and many types of synaptic signalling pathways (Extended Data Fig. 1h,i and Supplementary Fig. 1). Many of the NMD targets whose expression was increased by UPF1 KD (Fig. 1c,d) or whose half-life was increased by UPF1 KD upon actinomycin D treatment (Fig. 1e and Extended Data Fig. 2a) encode proteins mutated in neuronal diseases. These diseases include spastic paraparesis9 (for example, adhesion G protein-coupled receptor B2 (BAI2)), amyotrophic lateral sclerosis10 (for example, dipeptidyl peptidase-like 6 (DPP6)), frontotemporal dementia with parkinsonism11 (for example, microtubule-associated protein tau (MAPT)) and autism spectrum disorder12,13 (for example, HECT and RLD domain-containing E3 ubiquitin protein ligase 2 (HERC2) and the RNA-splicing factor serine/arginine repetitive matrix protein 4 (SRRM4/nSR100) that promotes neural-specific microexon inclusion in ~2,500 transcripts (Fig. 1d,e)). Moreover, NMD targets are often misregulated in neurodevelopmental, psychiatric or degenerative diseases (Extended Data Fig. 2b).
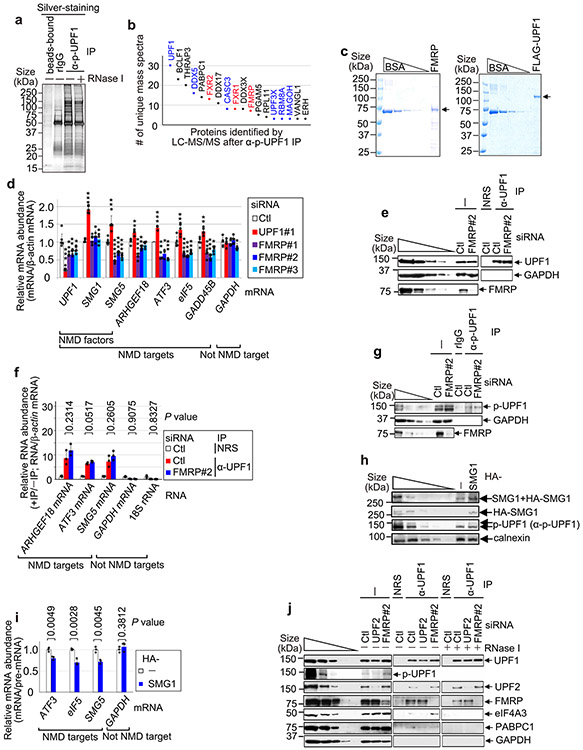
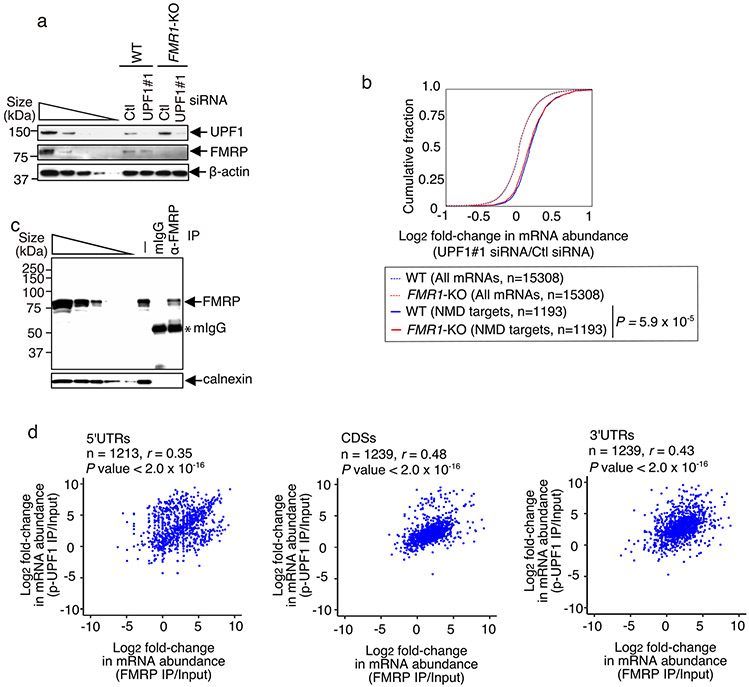
Given the extensive influence of NMD on human neuronal cell functions, we next sought to identify proteins that may play a direct role in NMD of neuronal transcripts. To this end, we performed mass spectrometry of proteins that co-immunoprecipitate with NMD-activated p-UPF1 in the presence of RNase I. Using human embryonic kidney 293T (HEK293T) cells, which are technically more facile to use than SH-SY5Y cells and share ~90% of the RNAs found in the human brain14, top hits unexpectedly included the fragile X mental retardation protein (FMRP; the protein deficient in fragile X syndrome (FXS)) and its two paralogues FXR1 and FXR2 (Extended Data Fig. 3a,b and Supplementary Table 2). Next, we corroborated that FMRP co-immunoprecipitates with both UPF1 and p-UPF1 in a largely RNase I-insensitive manner in lysates of HEK293T cells. This occurred irrespective of whether the cells were treated with okadaic acid, which upregulates the cellular abundance of p-UPF1 by inhibiting protein phosphatase activity5,15 (Fig. 2a,b). Therefore, FMRP behaved like the known NMD factor SMG7, which co-immunoprecipitated with UPF1 and p-UPF1 in a largely RNase I-insensitive manner, unlike the co-immunoprecipitation of the exon junction complex constituent eukaryotic initiation factor 4A3 (eIF4A3) or the cytoplasmic poly(A)-binding protein PABPC1. β-actin was absent from all immunoprecipitations (Fig. 2a,b). In addition, we found that human FMRP purified from Escherichia coli was pulled down with FLAG-tagged human UPF1 purified from baculovirus (Fig. 2c and Extended Data Fig. 3c). These findings provide evidence that FMRP interacts directly with the NMD factor UPF1.
Fig. 2 ∣. Evidence that FMRP binding to NMD targets is promoted by direct binding to UPF1.
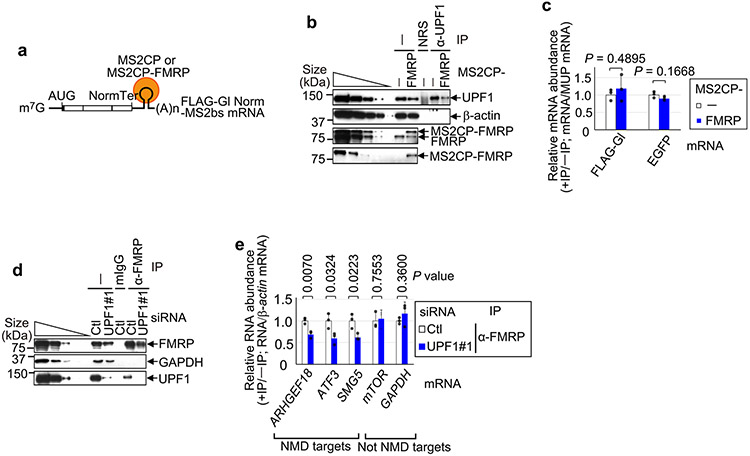
a, Western blots of lysates of HEK293T cells, with or without okadaic acid (OA), before (−) or after immunoprecipitation (IP) (±RNase I using anti-UPF1 or normal rabbit serum (NRS)). The results represent three biological replicates. b, As in a, but the immunoprecipitations used anti-p-UPF1 or rabbit IgG (rIgG). c, Pull-down of FLAG-UPF1 (Extended Data Fig. 3c) or FLAG-tagged bacterial alkaline phosphatase (FLAG-BAP) in the presence of FMRP (Extended Data Fig. 3c). The results represent three biological replicates. d, Western blots of lysates of siRNA-transfected HEK293T cells. The results represent three biological replicates. e, Quantitations of mRNAs, normalized to their pre-mRNA, using the lysates from d. The data represent means with s.d. (n = 3 biological replicates). P values pertain to comparisons with control siRNA samples and were determined by two-sided t-test. f, As in d, but transfections included plasmid expressing FLAG or FLAG-FMRP WTR, where R denotes siRNA resistance. g, As in e, but using the lysates from f. P values pertain to comparisons with control siRNA samples and were determined by two-sided t-test. h, Quantitations of RNA, normalized to β-actin mRNA, using the lysates from Extended Data Fig. 3g. The data represent means with s.d. (n = 3 biological replicates). P values are relative to control siRNA samples after anti-p-UPF1 immunoprecipitation (two-sided t-test). i, FLAG-Gl-MS2bs reporter mRNAs harbouring six MS2 coat protein (MS2CP; orange circle) binding sites (MS2bs), alone or fused to FMRP. j, Western blots of lysates of HEK293T cells transfected with plasmid expressing MS2CP or MS2CP-FMRP, pcDNA-EGFP and a pFLAG-Gl-MS2bs reporter. The results represent three biological replicates. k, Quantitations of FLAG-Gl-MS2bs Ter mRNA normalized to the level of FLAG-Gl-MS2bs Norm mRNA. The data represent means with s.d. (n = 3 biological replicates). The P values compare MS2CP-FMRP- versus MS2CP-transfected samples (two-sided t-test). l, FLAG-Gl-MS2bs Norm reporter as in i, but with MS2CP-UPF1. m, As in j, but before or after immunoprecipitation using anti-FMRP or mouse IgG (mIgG). n, As in k, but using the lysates from m. The data represent means with s.d. (n = 3 biological replicates). The P values compare MS2CP-UPF1- versus MS2CP-transfected samples (two-sided t-test). In all panels, *P < 0.05, **P < 0.01 and ***P < 0.001. Statistical source data and unprocessed blots are provided online.
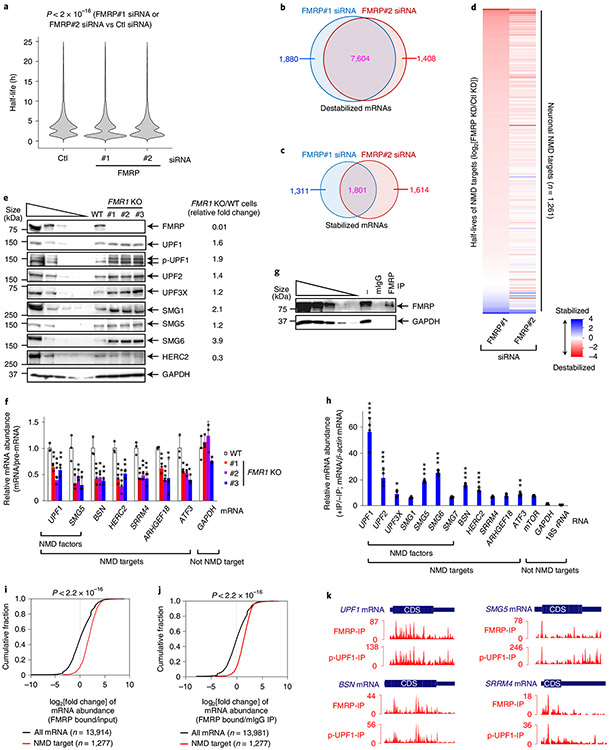
Based on evidence of a direct interaction between FMRP and UPF1, we hypothesized that loss of FMRP somehow influences NMD activity. Seven lines of evidence support the idea that FMRP functions as an NMD repressor. First, FMRP KD enhanced the efficiency of NMD, as evidenced by a ~2.6-fold increase in the level of p-UPF1 compared with in control small interfering RNA (siRNA)-treated cells (Fig. 2d). Second, FMRP KD downregulated the levels of previously characterized HEK293T cell NMD targets5 encoding, for example, Rho/Rac guanine exchange factor 18 (ARHGEF18, which functions in neuro-epithelial polarization and proliferation16), activating transcription factor 3 (ATF3, which functions in axon survival and regeneration17) and the NMD factor SMG5 (Fig. 2e and Extended Data Fig. 3d). Third, transiently expressing siRNA-resistant FMRP in FMRP KD cells negated the increased efficiency of NMD observed upon FMRP KD (Fig. 2f,g), indicating that it is FMRP itself that inhibits the NMD of these messenger RNAs (mRNAs). Fourth, compared with UPF1 binding (Extended Data Fig. 3e,f), FMRP KD increased the level of NMD-activated5,18 p-UPF1 binding to all NMD targets tested (Fig. 2h and Extended Data Fig. 3g). Taken together with the previous line of evidence, this indicates that the efficiency with which these mRNAs undergo NMD is augmented in the absence of FMRP. Augmentation is at least partially due to the observed ~4.3-fold upregulated expression of the UPF1 kinase SMG1 (Fig. 2d; see below): a roughly threefold upregulation of SMG1 expression alone is sufficient to increase the level of p-UPF1 roughly twofold (Fig. 2d and Extended Data Fig. 3h) and to increase the efficiency of NMD (Extended Data Fig. 3i). Fifth, tethering FMRP via six copies of the bacteriophage MS2 coat protein (MS2CP)-binding site (MS2bs) to the 3′ untranslated region (UTR) of a FLAG-β-globin (Gl) reporter transcript that either was (Ter) or was not (Norm) an NMD target (Fig. 2i) demonstrated that FMRP binding increased the level of FLAG-Gl-MS2bs Ter mRNA relative to FLAG-Gl-MS2bs Norm mRNA (Fig. 2j,k). Sixth, FMRP KD increased the co-immunoprecipitation of UPF2 with UPF1 (~1.3-fold) in an RNase I-insensitive manner, and of eIF4A3 with UPF1 (around twofold) in an RNase I-sensitive manner, both concomitantly with an increase in the level of cellular p-UPF1 (Extended Data Fig. 3j). Seventh, tethering UPF1 to the 3′ UTR of FLAG-Gl-MS2bs Norm mRNA (Fig. 2l), which binds FMRP at only background levels, increased the co-immunoprecipitation of FMRP with this mRNA (Fig. 2m,n), while tethering FMRP had no effect on the co-immunoprecipitation of UPF1 with this mRNA (Extended Data Fig. 4a-c). Consistent with UPF1/p-UPF1 stably associating with NMD targets but generally not non-NMD targets, downregulating UPF1 reduced the co-immunoprecipitation of NMD targets but not non-NMD targets with FMRP (Extended Data Fig. 4d,e). These data indicate not only that FMRP binding to an NMD target inhibits the NMD of that target via translational repression but also that inhibition generally typifies NMD targets since FMRP association is promoted by bound UPF1. Given the large number of NMD targets that influence neuronal cells, these findings have important implications for the loss of FMRP in FXS modelling.
The finding that loss of FMRP stimulates NMD raises the possibility that a significant proportion of transcriptome alterations in FXS are due to elevated NMD. This was supported by our finding that neuronal cells have hundreds of NMD targets (Fig. 1), all of which could potentially be downregulated in FXS due to more efficient NMD. To define neuronal mRNAs, including NMD targets, that are misregulated by FMRP deficiency, we developed a transcriptome-wide pulse-labelling method that we call 4-thio-uridine immunoprecipitation chase–deep sequencing (TRIC-seq) to measure mRNA stability in SH-SH5Y cells without changing the cell physiology by inhibiting gene transcription. The use of TRIC-seq (Extended Data Fig. 5a) was necessary since BRIC-seq6,19,20, which uses 5-bromo-uridine, inadequately labels RNAs in SH-SY5Y cells. In two independently performed TRIC-seq experiments, each involving six time points and using a different FMRP siRNA that downregulated FMRP cellular abundance to ~20–30% of normal (Extended Data Fig. 5b,c), 11,778 and 11,916 mRNAs, respectively, manifested a change in half-life relative to controls (Fig. 3a). The intersect of both experiments revealed 7,604 destabilized neuronal mRNAs (Fig. 3b), which was many more than the 1,801 that were stabilized (Fig. 3c). Among those destabilized in both experiments were 984/1,261 neuronal NMD targets (Fig. 3d, Extended Data Fig. 5d and Supplementary Table 3; not all 1,277 could be reliably analysed). As proof of principle, these included, 24/33 NMD targets (Extended Data Fig. 5e) that were defined using non-neuronal cells as enriched in p-UPF1 binding and manifesting a longer half-life upon UPF1 KD5,19,21-23. We conclude that FMRP deficiency prevalently causes destabilization of NMD targets.
Fig. 3 ∣. TRIC-seq and FMRP RIP-seq show that NMD targets are generally bound by FMRP in SH-SY5Y cells and destabilized upon FMRP KD.
a, Violin plot showing TRIC-seq-derived mRNA half-life changes binned in 1-h intervals. P values were determined by two-sided Wilcoxon signed-rank test (n = 11,778 and n = 11,916 in the presence of the control and FMRP siRNA, respectively). The results represent two biological replicates. b, Venn diagram of mRNAs destabilized in the presence of each FMRP siRNA. c, As in b, but for stabilized mRNAs. d, Heat maps of the half-life log2[fold change] for 1,261 neuronal cell NMD targets in the presence of FMRP#1 siRNA (sorted from the most destabilized to the most stabilized) and FMRP#2 siRNA (aligned to FMRP#1 siRNA values), each normalized to control siRNA values. e, Western blots of lysates of wild-type and FMR1 KO SH-SY5Y cell lines. The results represent three biological replicates. f, Quantitations of the specified mRNA, normalized to the level of its pre-mRNA, using the lysates analysed in e. The data represent means with s.d. (n = 3 biological replicates). The P values compare FMR1 KO cells with wild-type cells (two-sided t-test). g, Western blots of lysates of SH-SY5Y cells before (−) or after immunoprecipitation using anti-FMRP or mouse IgG. The results represent three biological replicates. h, As in f, but normalized to β-actin mRNA, using the lysates analysed in g. The data represent means with s.d. (n = 4 biological replicates). The P values are relative to GAPDH mRNA and were calculated using one-way analysis of variance (ANOVA) and Dunnett’s multiple comparisons test. i, Cumulative fraction of the log2[ratio] for mRNA enrichment of FMRP-bound NMD targets and all mRNAs in FMRP RIP-seq relative to input RNA. The P values were calculated by two-sided Wilcoxon rank-sum test. j, as in i, but using the mouse IgG immunoprecipitate as a normalization control. P values were calculated by two-sided Wilcoxon rank-sum test. k, Histograms denoting the positions and numbers of FMRP footprint reads (FMRP-IP) and p-UPF1-footprint reads (p-UPF1-IP) of exemplary NMD targets. The thick blue boxes represent CDSs, whereas the flanking thin boxes represent the 5′ and 3′ UTRs. In all panels, *P < 0.05, **P < 0.01 and ***P < 0.001. Statistical source data and unprocessed blots are provided online.
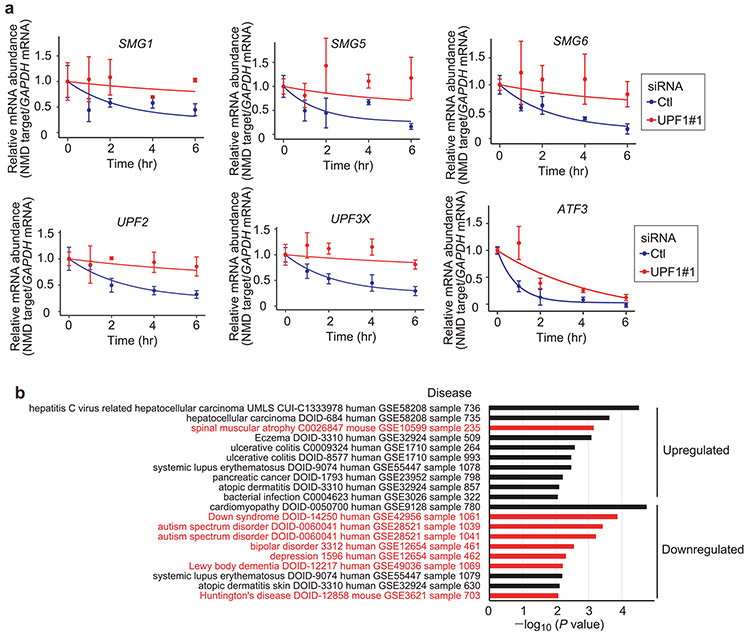
The efficiency of NMD was also upregulated in three FMR1 knockout (KO) SH-SY5Y cell lines that were generated to harbour different frameshift mutations in FMR1 (the gene encoding FMRP) using double-nicking CRISPR–Cas9n24 (Extended Data Fig. 5f). Supporting our findings using HEK239T cells (Fig. 2), FMR1 KO in SH-SY5Y cells, which eliminated FMRP production (Fig. 3e), increased by around twofold the abundance of p-UPF1 production (Fig. 3e), the level of which serves as a proxy for the efficiency of NMD5,6,18,25-29. FMR1 KO in SH-SY5Y cells also resulted in upregulation by ~1.2- to 3.9-fold of the steady-state levels of UPF1, UPF2, UPF3X, SMG1, SMG5 and SMG6 proteins, depending on the protein (Fig. 3e), all of which derive from NMD targets8 (Extended Data Fig. 2a), whose abundance was also reduced to ~30–60% of normal relative to their pre-mRNAs (Fig. 3f; see also Extended Data Fig. 5g). Transiently increasing the level of UPF1 in wild-type SH-SY5Y cells around twofold was alone sufficient to reproduce the hyperactivated NMD that was observed in FMR1 KO SH-SY5Y cells (Extended Data Fig. 5h,i), providing a second plausible mechanism by which NMD is globally hyperactivated in FMRP deficiency. Additional evidence that FMRP functions along with UPF1 in NMD was derived from transcriptome-wide RNA-seq data showing that UPF1 downregulation inhibited NMD comparably in wild-type SH-SY5Y cells relative to FMR1 KO SH-SY5Y cells (Extended Data Fig. 6a,b and Supplementary Table 4).
How, then, are the levels of the six NMD factors increased when they derive from mRNAs downregulated by hyperactivated NMD? In the case of these mRNAs, we hypothesize that FMR1 KO generates a relief from FMRP-mediated translational repression that is greater than their reduction in abundance by hyperactivated NMD. This phenomenon has been described for neural stem cells from Fmr1 KO mice as ‘translational buffering up’, which results in increased ribosome binding to mRNAs that are reduced in abundance30. Notably, not all NMD targets are translationally buffered up. For example, another NMD target, HERC2 mRNA (Fig. 1), which also co-immunoprecipitates with FMRP (Fig. 3g,h), produces abnormally low levels of HERC2 in the absence of FMRP (Fig. 3e,f).
Since FMRP binds to the coding sequences (CDSs) and 3′ UTRs of only some (~6,000) mRNAs in HEK293 cells14, it is reasonable to expect that only some mRNAs in SH-SY5Y cells would bind FMRP. However, we found that all NMD targets assayed were bound above background by FMRP in wild-type SH-SY5Y cells (Fig. 3g,h). Moreover, transcriptome-wide analyses of SH-SY5Y cell mRNAs bound by FMRP using RIP-seq footprinting demonstrated that NMD targets were significantly enriched in FMRP binding relative to the bulk of cellular mRNAs (Fig. 3i,j and Extended Data Fig. 6c). Thus, while FMRP can bind to mRNAs that are not NMD targets, as exemplified by mTOR mRNA (Fig. 3h), FMRP enrichment on NMD targets (Fig. 3I,j) is augmented by RNA-bound UPF1 and p-UPF1 (Fig. 2). In fact, RIP-seq footprinting demonstrated that FMRP enrichment positively correlated with p-UPF1 enrichment in both the protein-coding-regions (CDSs) and noncoding-regions (5′ UTRs and 3′ UTRs) of NMD targets (Fig. 3k and Extended Data Fig. 6d), consistent with p-UPF1 binding to both regions of NMD targets5,6.
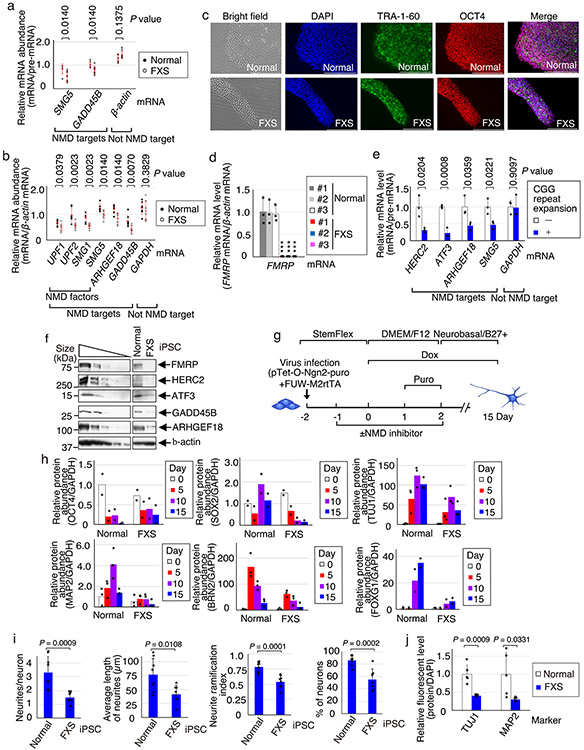
Our observation that the depletion or elimination of FMRP hyperactivates NMD in HEK293T and SH-SY5Y cells (Figs. 2 and 3) suggests that NMD may be hyperactivated in cells derived from patients with FXS, possibly contributing to the FXS disease mechanism. Indeed, relative to the corresponding cell type deriving from control individuals, using validated NMD targets (Fig. 1 and Extended Data Fig. 2a)5,19,21-23, NMD was hyperactivated in a number of cells derived from patients with FXS. These included fibroblasts derived from patients with FXS (Fig. 4a), lymphoblasts derived from patients with FXS (Extended Data Fig. 7a,b), induced pluripotent stem cells (iPSCs) that we developed from fibroblasts from patients with FXS (Fig. 4b and Extended Data Fig. 7c,d) and FXS iPSCs relative to their isogenic controls31 (Extended Data Fig. 7e). Notably, NMD targets encoding HERC2, ATF3, GADD45B and ARHGEF18 (Fig. 4b) provide additional examples of mRNAs that are not translationally buffered up (Extended Data Fig. 7f).
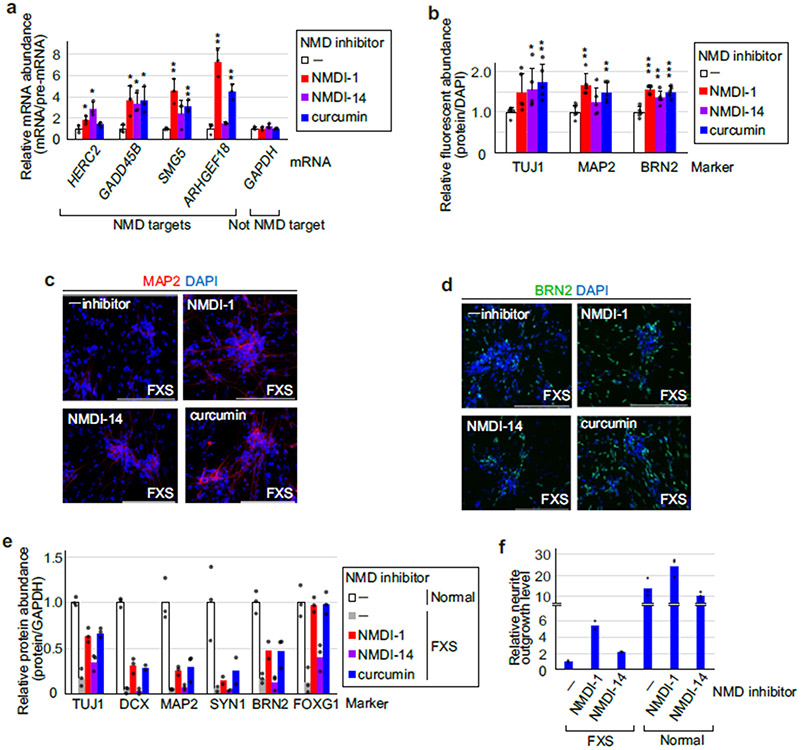
Fig. 4 ∣. Small molecules that inhibit hyperactivated NMD in FXS-derived iPSCs normalize neural differentiation.
a, Dot plot of RT-qPCR quantitations of the specified mRNAs, normalized to the levels of their pre-mRNAs, using lysates from healthy or FXS fibroblasts. The red bars represent means ± s.d. (n = 7 independent biological fibroblast cell lines). The P values represent comparisons of healthy versus FXS samples (two-sided Wilcoxon rank-sum test). b, Histogram of RT-qPCR quantitations of the specified mRNAs, normalized to the levels of their pre-mRNAs, using lysates from healthy human embryonic stem cells, healthy iPSCs or FXS iPSCs. The data represent means with s.d. (n = 3 biological replicates). *P < 0.05, **P < 0.01 and ***P < 0.001 compare FXS cells relative to healthy #1 cells (one-way ANOVA and Dunnett’s multiple comparison test). c, Western blots of representative healthy and FXS cell lysates on days 0, 5 10 and 15. The results represent three biological replicates. d, Immunofluorescence microscopy images of representative healthy and FXS neurons on day 15 after differentiation. DAPI (blue) stains the nuclei. Scale bars, 200 μm. The results represent three biological replicates. e, Immunofluorescence microscopy images of representative FXS neurons derived from iPSCs that were or were not exposed to the specified NMD inhibitor, essentially as in d. Scale bars, 200 μm. The results represent three biological replicates. f, Western blots of representative healthy and FXS neurons, with the latter from iPSCs that were or were not exposed to the specified NMD inhibitor. The results represent three biological replicates. g, Heat map of gene expression data for the specified neurons at day 15 of differentiation, as identified using RNA-seq, with three biological replicates. Shown are transcripts whose expression was significantly altered by NMDI-1 treatment. The colour key represents z score values of gene expression. h, Using genes in clusters #1 and #2 of g, Gene Ontology analysis identified top-ranked differentially expressed genes (P < 0.05; two-sided Fisher’s exact test) in neurons deriving from FXS iPSCs that were treated with NMDI-1 relative to those that were not treated with NMDI-1. i, Model for misregulated NMD in FXS. Statistical source data and unprocessed blots are provided online.
Attenuation of NMD is critical for the maturation of mouse neural progenitor cells32. Thus, we next characterized the consequence of hyperactivated NMD on the maturation of iPSCs from patients with FXS to neurons. We used rapid single-step neurogenin-2 (NGN2)-mediated induction33 to differentiate iPSCs from three fibroblast cell lines from patients with FXS and, as controls, iPSCs from two unaffected individuals as well as a commercially available human embryonic stem cell (hESC) line (Extended Data Fig. 7g). Relative to differentiated control neurons 5 d after doxycycline-induced NGN2 expression, cells deriving from FXS iPSCs manifested inefficient neuronal differentiation (Fig. 4c and Extended Data Fig. 7h). By day 7, FXS neurons exhibited ~60% fewer neurites per neuron and shorter and less branched projections, and were ~40% fewer in number relative to normal neurons (Extended Data Fig. 7i). By day 10, FXS neurons manifested abnormally low levels of the SRY-Box transcription factor (SOX2) marker for neural progenitors, the class III β-tubulin (TUJ1), which is expressed in early neuronal processes34, microtubule-associated protein 2 (MAP2), which is expressed in the soma and is a marker for more mature neurons34, and the POU domain class 3 transcription factor 2 (BRN2) and forkhead box G1 (FOXG1), both of which are markers for excitatory cortical neurons33 (Fig. 4c and Extended Data Fig. 7h). By day 15, FXS neurons showed ~60–70% less staining for both TUJ1 and MAP2 (Fig. 4d and Extended Data Fig. 7j). Our data are in keeping with the inefficient neurite outgrowth that has previously been described for FXS iPSC-derived neurons in vitro35-40. How this in vitro phenotype relates to dendritic spine abnormalities observed in patients remains unclear41. Notably, hyperactivated NMD no longer typifies FXS-derived iPSCs by day 5 of differentiation (Fig. 4c), possibly due to changes in the efficiency of NMD during neurodifferentiation. We suggest that the hyperactivation of NMD early in neurogenesis predisposes subsequent stages of neuronal differentiation to misregulation of gene expression. For example, hyperactivation of NMD that typifies undifferentiated FXS iPSCs (Fig. 4b,c) results in abnormal levels of differentiation markers, including SOX2, TUJ1, MAP2, BRN2 and FOXG1, none of which derives from an NMD target, by day 15 of differentiation, when NMD hyperactivation is less apparent.
Since we had found that a significant number of NMD targets encode proteins involved in neurological processes (Fig. 1 and Extended Data Fig. 1), we next examined whether inhibition of hyperactivated NMD in FXS iPSCs can restore neural properties. To control for possible off-target effects, FXS iPSCs were incubated for 72 h with one of three NMD inhibitors, each of which had a different mode of inhibition: NMD inhibitor-1 (NMDI-1) and NMDI-14 preclude the binding of SMG5 to UPF1 (ref. 42) and of SMG7 to UPF1 (ref. 43), respectively, while curcumin reduces the abundance of NMD factors44. Each small molecule effectively inhibited NMD after 24 h (Extended Data Fig. 8a) and was removed after 72 h since longer exposures resulted in cell toxicity. Immunofluorescence (Fig. 4e and Extended Data Fig. 8b-d) and western blotting (Fig. 4f and Extended Data Fig. 8e) showed that, by day 15 of differentiation, all three small molecules restored the expression of markers for early and mature neurons (for example, synaptic vesicle and excitatory cortical neuronal markers), where NMDI-14 was the least efficacious. None of these markers derive from an NMD target (Fig. 1). The inhibitors also promoted FXS neurite outgrowth two- to five-fold, restoring levels to ~20–40% of normal depending on the inhibitor, whereas neurite outgrowth of control cells was promoted either not at all or less than twofold (Extended Data Fig. 8f). RNA-seq analysis revealed that NMDI-1 treatment indeed normalized the expression of many genes by day 15 (Fig. 4g). Among the 847 transcripts whose expression was significantly altered by NMDI-1 treatment (Fig. 4g and Supplementary Table 5), Gene Ontology analysis revealed that upregulated transcripts included mRNAs encoding proteins that function in synaptic transmission and synaptic signalling, whereas downregulated transcripts included mRNAs encoding proteins that function in embryo development and cytoplasmic translation (Fig. 4h). Of the upregulated transcripts, 11% (24/226) were NMD targets, whereas 3% of NMD targets (17/621) were downregulated (Fig. 1 and Supplementary Table 5). Together, these results indicate that partial NMD suppression of hyperactivated NMD via small molecules may be an effective route to obviating neuronal differentiation defects seen in FXS cells. Future experiments that define optimal small-molecule exposure times and doses will probably improve the degree to which neuronal markers for each stage of differentiation are normalized in FXS cells.
In summary, using a battery of different cell lines to demonstrate that NMD is hyperactivated by the loss of FMRP function in most human cells tested (but apparently not in neurons differentiated from iPSCs derived from patients with FXS), we propose two non-mutually exclusive models, each of which results in an increase in p-UPF1 abundance. In one, FMRP deficiency results in the loss of FMRP recruitment to NMD targets via UPF1 so that the consequential upregulation of NMD target translation results in hyperactivated NMD (Fig. 4i). In the other, included among NMD targets are those encoding NMD factors, whose increased abundance also hyperactivates the cellular efficiency of NMD (Fig. 4i). The increased NMD factor abundance, despite these factors deriving from NMD targets, can be explained by buffering up30, whereby, in the absence of FMRP, the relief from translational repression overcompensates for the decrease in mRNA abundance by hyperactivated NMD.
There is evidence that NMD is hyperactivated in the fibroblasts of patients with amyotrophic lateral sclerosis who harbour the p.Arg521Gly or p.Pro525Arg substitution in the RNA-binding protein FUS45. While the underlying mechanism is unknown, the levels of UPF1, p-UPF1 and UPF3X were reported to be elevated. Thus, our finding that small-molecule NMD inhibitors ameliorate some of the neuronal cell dysfunctions that typify the process by which FXS iPSCs undergo neural differentiation may be useful when considering the mechanistic biochemistry of not only FXS but also some forms of amyotrophic lateral sclerosis46-48 and other neurological disorders (Fig. 1d and Extended Data Fig. 2b). It remains to be seen how misregulated NMD, which impinges on many cell signalling pathways (Supplementary Fig. 1), influences the effects of drugs currently used in FXS studies to dampen the mTOR and ERK pathways49.
Methods
Cell lines.
For cell lines, see Supplementary Table 6.
Cell culture, transfections and generation of iPSCs.
Human SH-SY5Y neuroblastoma cells were propagated in Dulbecco’s modified Eagle’s medium (DMEM)/Nutrient Mixture F-12 (Gibco) supplemented with 10% foetal bovine serum (FBS) (VWR). When specified, cells (1–5 × 107) were transiently transfected with 180 pmol siRNA (Dharmacon) using the Neon Transfection System (Invitrogen) or 50 pmol siRNA using the TransIT-X2 Dynamic Delivery System (Mirus Bio). FMR1 KO SH-SY5Y cells were generated using double-nicking RNA-guided CRISPR–Cas9n24. Two days after electroporating cells with guide RNA-encoding pSpCas9n(bb)-2A-Puro plasmids, transfected SH-SY5Y cells were selected using puromycin (1.5 μg ml−1) for 5 d. The resulting clonal cell colonies were expanded and characterized using western blotting and genome sequencing.
HEK293T cells were propagated in DMEM supplemented with 10% FBS. When specified, cells were transiently transfected with 20–30 nM siRNA (Dharmacon/GE Healthcare) using Lipofectamine RNAiMAX (Invitrogen) and/or plasmid DNA using Lipofectamine 2000 (Invitrogen).
Lymphoblasts derived from healthy individuals (GM14926, GM14643, GM14648, GM14583, GM14687, GM14811 or GM14907; Coriell Institute for Medical Research) or patients with FXS (GM03200, GM04025, GM06897, GM07294, GM07862, GM09316 or GM09237; Coriell Institute for Medical Research) were propagated in RPMI 1640 Medium (Gibco) supplemented with 10% FBS.
Fibroblasts derived from healthy individuals (BJ ATCC CRL-2522; American Type Culture Collection; GM00498, GM03468, GM05659, GM05756, GM08333 or GM08680; Coriell Institute for Medical Research) or patients with FXS (GM04026, GM04926, GM05131, GM05185, GM05848, GM07730 or GM09497; Coriell Institute for Medical Research) were propagated in DMEM supplemented with 10% FBS. Three lines of FXS fibroblasts (GM05131, GM05185 and GM05848) were reprogrammed using RNAs following established protocols50-52. Briefly, after plating in NutriStem XF/FF Medium (ReproCELL) on iMatrix-511 (Funakoshi)-coated 6-well plates, fibroblasts derived from patients with FXS were transfected on four consecutive days with non-modified mRNAs encoding OCT4, KLF4, SOX2, LIN28 and c-MYC, according to the manufacturer’s instructions using the StemRNA-NM Reprogramming Kit (Stemgent) and Lipofectamine RNAiMAX (Thermo Fisher Scientific).
The control lines iPSC-5433 and iPSC-5417 were generated by Sendai-based reprograming of peripheral blood mononuclear cells obtained from healthy donors without a history of known neurological disorders. Peripheral blood cells were plated using feeder-free conditions and transduced with CytoTune-iPS 2.0 reprogramming vectors according to the manufacturer’s instructions (Thermo Fisher Scientific). Emergent iPSC clones were expanded on hESC-qualified, LDEV-free Matrigel (Corning) in feeder-free StemFlex medium (Thermo Fisher Scientific). iPSC colonies were identified by live staining for the pluripotent stem cell surface marker TRA-1-60, and expanded under feeder-free PSC culture conditions. Labelling with the appropriate pluripotency markers (Oct4, TRA-1-60, SSEA4, TRA-1-81, alkaline phosphatase and Nanog) was used to select iPSC lines for further characterization53.
All iPSCs and hESC cultures were maintained at 5% O2/7% CO2 and in feeder-free StemFlex medium (Thermo Fisher Scientific) on LDEV-free Matrigel (Corning), with daily medium changes and manual removal of iPSC colonies that exhibited any spontaneously differentiating cells. Every 4–6 d, iPSCs were dissociated from dishes using ReLeSR (STEMCELL Technologies) and re-plated as small clumps at a density of 20,000–50,000 cells per well of a 6-well dish.
All newly generated iPSC lines were characterized by karyotype analysis (WiCell Characterization Laboratory, Madison, Wisconsin; Supplementary Table 7). A minimum of 20 mitoses were analysed with a band resolution of 400–575. All iPSC lines exhibited a normal karyotype without clonal abnormalities. The pluripotency of iPSC lines was tested using trilineage differentiation and adherent cell culture differentiation conditions (STEMdiff Trilineage Differentiation Kit, STEMCELL Technologies). RNA was extracted from differentiated cells and mixed in equal quantities before quantitative reverse transcription PCR (RT-qPCR) using Taqman Scorecard Panels54 (Thermo Fisher Scientific) on an ABI QuantStudio 12K Flex Real-Time PCR System. Ct values were analysed using hPSC Scorecard Analysis Software. Expression of ectodermal, endodermal and mesodermal lineage genes demonstrated pluripotency of all newly generated iPSC lines.
Neuronal differentiation and neurite outgrowth assays.
Reprogrammed iPSCs (including those derived from healthy individuals) and H7 human embryonic stem cells were cultured in StemFlex Medium and differentiated into neurons using NGN2-mediated neuro-induction33, which involves inducing pTet-O-Ngn2-puro and FUW-M2rtTA lentiviral plasmid vectors using doxycycline followed by puromycin selection. NGN2-induced iPSCs were further cultured in Neurobasal Plus Medium supplemented with 1 μg ml−1 brain-derived neurotrophic factor (R&D Systems) and 1 μg ml−1 neurotrophin-3 (R&D Systems). Neurite outgrowth was evaluated using a Neurite Outgrowth Staining Kit (Thermo Fisher Scientific) and a SpectraMax M4 Microplate Reader (Molecular Devices), averaging five fluorescent point measurements per well. Neuron morphology was also characterized by βIII tubulin or MAP2 immunofluorescent labelling of differentiated cells. 4′,6-diamidino-2-phenylindole (DAPI) staining was used to identify the nucleus/cell body. Replicate wells of differentiated cells were imaged using a CellInsight CX5 HCA plate scanner (Thermo Fisher Scientific). A minimum of ten random fields (900 μm × 900 μm) per sample were acquired at a resolution of 2,208 pixels × 2,208 pixels and analysed using HCS Studio version 6.6.1, NeuralProfiling version 4.2 (two-channel, fixed exposure with ISODATA background removal, triangular cell body recognition with nuclear segmentation, and median neurite detection). An average of over 200 neurons were analysed per condition. Pairwise comparisons of averages were performed using t-tests for two-tailed distribution with equal variance. Where specified, iPSCs were cultured in the presence of 0.5 μM NMDI-1 (provided by S. Velu) or NMDI-14 (MilliporeSigma) or 1.5 μM curcumin (MedChemExpress).
Plasmid constructions.
To generate pGEX-FMRP for human FMRP expression in E. coli, an FMRP-encoding fragment was first PCR amplified using the primer pair FMRP sense (S) and FMRP antisense (AS) (see Supplementary Table 6) and pFRT/TO/FLAG/HA-FMRP isoform 7 (ref. 14) as template DNA. The PCR product was digested using EcoRI and SalI, and the resulting ~1.8-kilobase pair (kbp) fragment was purified from an agarose gel and inserted into the EcoRI and SalI sites of pGEX6p-3 (GE Healthcare Biosciences).
To generate pFLAG-CMV2-FMRP WTR, pGEX-FMRP was digested using EcoRI and SalI, and the resulting ~1.8-kbp fragment was purified from an agarose gel and inserted into the EcoRI and SalI sites of pFLAG-CMV2. siRNA-resistant DNA fragments were PCR amplified using primer pairs (CMV forward and FMRP siRNAR reverse, and FMRP siRNAR forward and FMRP exon reverse) and pFRT/TO/FLAG/HA-FMRP isoform 7 as template DNA. The resulting ~1-kbp and ~0.7-kbp fragments were mixed and further PCR amplified using the primer pair CMV forward and FMRP exon reverse. The ~1.5-kbp PCR product was digested using EcoNI and KpnI to generate a ~1-kbp fragment, which was purified from an agarose gel and inserted into the EcoNI and KpnI sites of pFLAG-CMV2-FMRP.
To generate pcMS2-FMRP, pFRT/TO/FLAG/HA-FMRP isoform 7 (ref. 14) was digested using XhoI and NotI, and the resulting ~1.9-kbp fragment was purified from an agarose gel and inserted into the XhoI and NotI sites of pcMS2-HA55.
To construct pFLAG-Gl Norm-MS2bs or pFLAG-Gl Ter-MS2bs, pcDNA3-Gl Norm-MS2bs or pcDNA3-Gl Ter-MS2bs56 was digested using HindIII and XbaI, and the resulting ~1.8-kbp fragment was purified from an agarose gel and inserted into the HindIII and XbaI sites of pFLAG-CMV2 (Sigma–Aldrich).
To construct pcDNA-EGFP, pEGFP-N1 was digested using HindIII and XbaI, and the resulting ~0.8-kbp fragment was purified from an agarose gel and inserted into the HindIII and XbaI sites of pcDNA3.1 (Thermo Fisher Scientific).
To generate FMR1 KO SH-SY5Y cells using double nicking by RNA-guided CRISPR–Cas9 for enhanced genome editing specificity24, DNA oligo pairs (see Supplementary Table 6) FMR1 KO ex4 S1 and FMR1 KO ex4 AS1, FMR1 KO ex4 S2 and FMR1 KO ex4 AS2, FMR1 KO ex8 0S1 and FMR1 KO ex8 AS1 or FMR1 KO ex8 S2 and FMR1 KO AS2 were introduced into the BbsI restriction site of the pSpCas9n(bb)-2A-puro plasmid vector (Addgene).
The integrity of all constructs was validated using DNA sequencing.
Cell lysis and protein and RNA preparations.
Cell lysates were prepared using Hypotonic Gentle Lysis Buffer (10 mM Tris (pH 7.4), 10 mM NaCl, 10 mM EDTA and 0.5% wt/wt Triton X-100) supplemented with Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific). Proteins were quantified after the addition of NaCl to 150 mM using a protein assay dye reagent (Bio-Rad), and RNA was extracted and purified using TRIzol Reagent (Invitrogen).
Mass spectrometry.
HEK293T cells (8 × 107 per 150 mm dish) were cultured for 3 h in okadaic acid (200 nM; LC Laboratories) and subsequently collected and lysed using Hypotonic Gentle Lysis Buffer with Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific). Cellular p-UPF1 was immunoprecipitated using the rabbit polyclonal anti-p-UFP1 S1116 antibody (Millipore) in the presence of 1 U ml−1 RNase I (Ambion). Immunoprecipitated material was electrophoresed in 12% sodium dodecyl sulfate (SDS)–polyacrylamide, and bands stained silver using a SilverQuest Staining Kit (Invitrogen) were excised and subjected to in-gel trypsin digestion. Chromatographed peptides were identified at the Massachusetts Institute of Technology Whitehead Institute Proteomics Core Facility (http://massspec.wi.mit.edu/) using an Orbitrap Elite Hybrid Ion Trap-Orbitrap Mass Spectrometer with Dionex UltiMate 3000 Rapid Separation Liquid Chromatography (Thermo Fisher Scientific) and Scaffold Software (version Scaffold_4.2.1; Proteome Software).
siRNAs.
siRNAs consisted of Silencer Negative Control #1 siRNA (Ambion), UPF1#1 siRNA, UPF1#2 siRNA, FMRP#1 siRNA, FMRP#2 siRNA, FMRP#3 siRNA, UPF2 siRNA and UPF3X siRNA (see Supplementary Table 6).
Western blotting.
Proteins were electrophoresed in 6–14% polyacrylamide, transferred to a nitrocellulose (Bio-Rad) or polyvinylidene difluoride (Millipore) membrane and probed as described5 using the following antibodies (see Supplementary Table 6): anti-p-UPF1 S1116 (1:1,000), anti-UPF1 (1:2,000), anti-FMRP (1:2,000), anti-SMG7 (1:2,000), anti-eIF4A3 (1:1,000), anti-PABPC1 (1:2,000), anti-β-actin (1:5,000), anti-CBP80 (1:2,000), anti-eIF4E (1:2,00), anti-SMG5 (1:2,000), anti-SMG6 (1:1,000), anti-UPF2 (1:2,000), anti-UPF3X/UPF3B (1:2,000), anti-SMG1 (1:2,000), anti-mTOR (1:2,000), anti-HERC2 (1:2,000), anti-GADD45B (1:2,000), anti-ATF3 (1:2,000), anti-ARHGEF18/p114RhoGEF (1:2,000), anti-GAPDH (1:5,000), anti-OCT4 (1:2,000), anti-SOX2 (1:2,000), anti-β3-tubulin/TUJ1 (1:2,000), anti-MAP2 (1:2,000), anti-BRN2/POU3F2 (1:2,000), anti-FOXG1 (1:2,000), anti-doublecortin/DCX (1:2,000), anti-synapsin1/SYN1 (1:2,000), anti-calnexin (1:5,000), anti-MS2CP (1:2,000), anti-GFP (1:500), anti-HA HRP (1:1,000) or anti-FLAG HRP (1:1,000; Sigma–Aldrich). Western blots were quantitated using Image Studio Lite Version 4.0 (LI-COR Biosciences). Dilution standards assured that quantitations were in the linear range of analysis.
FLAG-tagged protein pull-downs.
FLAG-tagged human UPF1 was purified from baculovirus as described57. Human FMRP was purified from BL21-CodonPlus (DE3)-RIPL E. coli using a GSTrap HP Column (GE Healthcare Life Sciences) on which the GST tag was removed and FMRP was released using PreScission Protease (GE Healthcare Life Sciences). Protein purity and concentration were determined after electrophoresis in SDS–polyacrylamide and Coomassie blue staining, where serial dilutions of bovine serum albumin (Rockland) provided concentration standards. Pull-downs were performed essentially as described57 but using FLAG-tagged bacterial alkaline phosphatase (Sigma–Aldrich) as a negative control and anti-FLAG agarose beads (Sigma–Aldrich).
Immunoprecipitations.
Samples were generated before and after immunoprecipitation in the presence or absence of RNase I as described5 using 5–10 μg anti-FMRP, anti-p-UPF1 S1116, control rabbit immunoglobulin (IgG) (Sigma–Aldrich) or control mouse IgG (Sigma–Aldrich) per 0.5 ml lysate at 1–2 mg protein per ml. When determining immunoprecipitation efficiencies, 5% of immunoprecipitated samples was used in western blotting. When assaying for co-immunoprecipitated proteins, 20% of immunoprecipitated samples was used in western blotting.
RNA purification and RT-qPCR.
Total cell RNA was purified and treated with RQ01 DNase I (Promega), complementary DNA (cDNA) was synthesized using SuperScript III Reverse Transcriptase (Thermo Fisher Scientific), and RT-qPCR was undertaken using gene-specific primers (see Supplementary Table 6), using the 7500 Fast Real-Time PCR System (Applied Biosystems) and Fast SYBR Green Master Mix (Applied Biosystems) as described5. Alternatively, PCR was undertaken using the QuantStudio 5 Real-Time PCR System (Thermo Fisher Scientific) and either the Fast SYBR Green Master Mix or the SYBR Select Master Mix (Applied Biosystems).
Immunofluorescence microscopy.
iPSCs (2 × 104 per 24-well plate) were cultured on Matrigel-coated coverslips, fixed using 4% paraformaldehyde in phosphate-buffered saline (PBS) and subsequently permeabilized using five successive 2-min incubations at room temperature in PBS containing 0.2% Triton X-100. Coverslips were blocked with 3% bovine serum albumin (Rockland Immunochemicals) in Tris-buffered saline containing 0.1% Tween 20 (TBS-T) for 30 min at room temperature, washed once with TBS-T and incubated overnight at 4 °C in primary antibody that had been diluted in TBS-T using the following antibodies (see Supplementary Table 6): anti-p-UPF1 S1116 (1:250), anti-p-UPF1 S1089 (1:250), anti-UPF1 (1:500), anti-FMRP (1:250), anti-TRA-1–60 (1:500), anti-OCT4 (1:500), anti-MAP2 (1/250), anti-β3-Tubulin/TUJ1 (1:500) and anti-BRN2/POU3F2 (1:250). Coverslips were then washed extensively using TBS-T and subsequently incubated for 1 h at room temperature with DAPI (1 μg ml−1) and 1:1,000 (vol/vol) Alexa Fluor 488-labelled goat anti-rabbit IgG (Thermo Fisher Scientific) or Alexa Fluor 568-labelled goat anti-mouse IgG (Thermo Fisher Scientific). After further extensive washing using TBS-T, coverslips were mounted using ProLong Gold antifade reagent with DAPI (Thermo Fisher Scientific). Images were captured with an FluoView FV1000 confocal laser-scanning microscope (Olympus) or EVOS FL Imaging System (Thermo Fisher Scientific).
RNA processing for RNA-seq and RIP-seq footprinting.
To generate RNA-seq libraries, SH-SY5Y cells (3 × 107 cells per 150 mm dish) were lysed, and protein and RNA were purified as described above.
RIP-seq footprinting libraries were generated essentially as detailed5. Lysates of SH-SY5Y cells (1 × 108 cells per 4 mm × 150 mm dish) were immunoprecipitated using Dynabeads Protein A (Thermo Fisher Scientific) and either anti-p-UPF1 S1116 (Millipore) or, as a control, rabbit IgG (Sigma–Aldrich) or Dynabeads Protein G (Thermo Fisher Scientific) and either anti-FMRP (Millipore) or, as a control, mouse IgG (Sigma–Aldrich). For the anti-p-UPF1 and rabbit IgG immunoprecipitations, cells were cultured for 3 h in okadaic acid (200 nM) before harvesting. Cellular RNAs captured by bead-bound antibody complexes were digested to fewer than 100 nucleotides by incubating for 30 min at 4 °C with RNase I (1 U μl−1; Ambion). After extensive washing, bound complexes were eluted using immunoprecipitation elution buffer (125 mM Tris-HCl (pH 6.8), 4% vol/vol SDS, 20% vol/vol glycerol, 10% vol/vol 2-mercaptoethanol and bromophenol blue), and RNA fragments were purified using TRIzol Reagent and electrophoresed in 6 M urea–15% polyacrylamide in parallel with a DynaMarker Prestain Marker for Small RNA (BioDynamics Laboratory). Those in the ~25- to 50-nucleotide size range were excised, agitated overnight at 25 °C in RNA extraction buffer (20 mM Tris, 300 mM sodium acetate, 2 mM EDTA and 0.2% vol/vol SDS), purified using a Coaster Spin-X Centrifuge Tube Filter (Corning), extracted using TRIzol Reagent and concentrated by ethanol precipitation. Control immunoprecipitations using rabbit IgG were performed in parallel. Additionally, samples before immunoprecipitation were prepared using limited digestion with RNase I.
cDNA library constructions for RNA-seq and RIP-seq.
To generate SH-SY5Y cell libraries for RNA-seq, total cell RNA (500 ng) was subjected to two rounds of poly(A) selection using oligo-d(T)25 magnetic beads (New England Biolabs). cDNA libraries were constructed using the TrueSeq cDNA Library Prep Kit (Illumina). Briefly, poly(A)+ RNA was fragmented by heating at 94 °C for 15 min, followed by reverse transcription and second-strand cDNA synthesis. End-repaired cDNA fragments were ligated to double-stranded DNA adapters and purified using AMPure XP beads (Beckman Coulter). Adapter-ligated cDNA was PCR amplified using a universal forward primer and bar-coded reverse primer for 15 cycles. cDNA library sequencing was performed using the NextSeq 500 System (Illumina) in the single-read 1 × 75 cycles configuration.
iPSC-derived neuron libraries for RNA-seq were generated following essentially the same protocol. However, cDNA library sequencing was performed using a NovaSeq 6000 DNA sequencer (Illumina) and an S1 flow cell configuration with 100 cycles.
To generate libraries for RIP-seq, purified RNA fragments of ~25–50 nucleotides were treated with recombinant shrimp alkaline phosphatase (New England Biolabs) to remove 3′ phosphates, phosphorylated at their 5′ hydroxyl groups using T4 polynucleotide kinase (New England Biolabs) and purified using the miRNeasy Mini Kit (Qiagen). A 3′-adenylated DNA adapter (New England Biolabs) was then added to 3′ ends using truncated T4 RNA ligase 2 (New England Biolabs). After ligation of the 5′ RNA adapter (New England Biolabs) using T4 RNA ligase 1 (New England Biolabs), the reverse transcriptase primer was annealed to the 3′ adaptor to prevent adapter self-ligation. After reverse transcription of the adapter-ligated RNAs using SuperScript III reverse transcriptase (Thermo Fisher Scientific) and the primer (New England Biolabs), the resulting cDNAs were PCR amplified using Phusion High-Fidelity DNA Polymerase (New England Biolabs) for 15 cycles. PCR products were purified in 8% polyacrylamide, and cDNA quality and quantity was assessed using an Agilent Bioanalyzer and qPCR. cDNAs were then sequenced using the Illumina NextSeq 500 system. Adapter and primer sequences are provided in Supplementary Table 6.
Computational analyses of RNA-seq and RIP-seq data, tissue expression, Gene Ontology term enrichment, biological pathways and disease perturbation.
For RNA-seq and RIP-seq data processing, adapter sequences were trimmed from raw reads using Cutadapt (version 1.12)58. Bowtie 2 (ref. 59) was used to computationally remove repetitive elements and ribosomal RNAs. Processed reads were then aligned to the reference human genome (hg19) using STAR (version 2.5.2b)60. Genome-aligned reads were used for the RIP-enrichment analysis using featureCounts (version 1.5.0)61 and edgeR (version 3.14.0)62. Tissue expression analysis was performed using the Enrichr gene enrichment analysis tool based on the Jensen tissue expression database63,64. Gene Ontology term enrichment analysis for biological processes was performed using the Protein Analysis Through Evolutionary Relationships (PANTHER) classification system65. Biological pathway analysis was undertaken using the Kyoto Encyclopedia of Genes and Genomes database66. Disease perturbation analysis was performed using Enrichr based on the Gene Expression Omnibus database63.
To analyse RNA-seq data deriving from FMR1 KO SH-SY5Y cells transfected with control siRNA or UPF1 siRNA, or from neurons generated using iPSCs that were treated or not with NMDI-1, DESeq2-1.22.1 (ref. 67) within R-3.5.1 was used to perform data normalization and differential expression analysis, with an adjusted P value threshold of 0.05, on each set of raw expression measures. The lfcShrink method was applied, which moderates the log2[fold change] for lowly expressed genes. Heat maps were created using gplots (https://cran.r-project.org/web/packages/gplots/index.html) in R.
TRIC-seq data processing, including mRNA half-life quantitations.
While SH-SY5Y cell RNA was only inefficiently labelled using 5′-bromo-uridine, 4-thio-uridine, which allows shorter labelling times68, was efficiently incorporated into SH-SY5Y cell RNA. For TRIC-seq, siRNA-transfected cells were labelled with 150 mM 4-thio-uridine (Sigma–Aldrich) for 4 h, after which medium was replaced with fresh medium lacking 4-thio-uridine. Cells were harvested after an additional 0, 1, 2, 4, 7 or 10 h. Total cell RNA was isolated using RNAiso Plus reagent (TaKaRa). Subsequently, 4-thio-uridine-labelled RNA was isolated following a modification of Dölken69. Briefly, 4-thio-uridine-containing RNAs were converted to biotinyl RNAs using HPDP-Biotin (Thermo Fisher Scientific) in N,N-dimethylformamide. The resulting biotinyl RNAs were isolated using Dynabeads MyOne Streptavidin C1 (Thermo Fisher Scientific). Isolated RNAs were subjected to RNA-seq. RNA-seq data for individual 4-thio-uridine-labelled mRNAs was corroborated using RT-qPCR and mRNA-specific primers, and by normalizing the resulting mRNA levels to the level of GAPDH mRNA. For sequencing after cDNA library construction, mapped reads were quantified to estimate the RNA expression level for each gene and to perform differential expression analysis using Cufflinks (version 2.2.1)70. Reads were aligned to the reference human genome (hg19) using TopHat (version 2.1.1)71. mRNA half-lives were calculated using BridgeR2 (version 0.1.0)6,19,68,72.
Statistics and reproducibility.
All biochemical analyses, western blots, RT-qPCRs and immunofluorescence experiments were performed using at least three biologically independent samples unless otherwise stated. RT-qPCR quantitations are presented as means and s.d. values. Statistical analyses were performed using R (version 3.6.3).
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1 ∣. Identifying SH-SY5Y NMD targets and their possible contributions to neurological diseases.
a, Schematic of transcriptome-wide approaches used to define NMD targets in human SH-SY5Y neuroblastoma cells: RNA-seq to quantitate mRNAs upregulated by UPF1 siRNA relative to Ctl siRNA (left), and RIP-seq footprinting (right) to identify mRNAs enriched in anti(α)-p-UPF1 IP relative to rabbit (r)IgG IP after increasing cellular p-UPF1 abundance using okadaic acid. PAGE, polyacrylamide gel electrophoresis. b, Representative western blots showing downregulation of UPF1 (that is UPF1-KD) relative to Ctl siRNA in samples used for RNA-seq. Here and elsewhere, left-most lanes under the wedge analyze serial 3-fold dilutions of lysate, and results represent 3 independent biological replicates. c, Western blots of lysates of SH-SY5Y cells cultured with okadaic acid for 2 hr to increase the cellular abundance of p-UPF1 before (−) or after IP using anti-p-UPF1 S1116 or rIgG. Results are representative of 3 independent biological replicates. d, As in b, but using UPF3X siRNA. e, Scatter plot of the log2 fold-change in mRNA abundance in the presence of UPF1 siRNA normalized to the abundance in the presence of Ctl siRNA (x-axis) and UPF3X siRNA normalized to the abundance of Ctl siRNA (y-axis). r and P values were calculated using Pearson’s correlation coefficient test. f, Cumulative fraction of log2 fold-change in mRNA abundance upon UPF3X-KD relative to Ctl-KD of “All mRNAs” (black line) or neuronal NMD targets defined in Fig. 1a (red line). P values were calculated using the two-sided Wilcoxon rank-sum test. g, Number and localization in brain of human NMD targets identified in this study. h, List of 22 proteins encoded by human neuronal-cell NMD targets that function in axon guidance. i, Diagram of proteins encoded by human neuronal-cell NMD targets that function in synaptic signaling (pink). Statistical source data and unprocessed blots are provided in Source Data Extended Data Fig. 1.
Extended Data Fig. 2 ∣. Confirmation of SH-SY5Y NMD targets and their change in abundance in neurological disease perturbation analyses.
a, As in Fig. 1e, but analyzing NMD targets that encode NMD factors or ATF3. Results derive from 3 independent biological replicates. b, Histogram representing disease perturbation analysis, illustrating that NMD targets are more often downregulated than upregulated in neurodevelopmental, psychiatric or neurodegenerative diseases (red). P values are calculated using the two-sided Fisher’s exact test. Statistical source data are provided in Source Data Extended Data Fig. 2.
Extended Data Fig. 3 ∣. Liquid chromatography-tandem mass spectrometry, p-UPF1 preferentially co-immunoprecipitates with NMD targets, and a ~3-fold increased SMG1 level typifying FMRP-KD cells recapitulates hyperactivated NMD.
a, Silver-stained polyacrylamide gel of lysates of HEK293T cells eluted from beads alone (beads-bound), or beads bound by rIgG- or anti-p-UPF1 (α-p-UPF1) used in LC-MS/MS. b, Proteins in anti-p-UFP1 IP identified using LC-MS/MS and ranked (x-axis) by the number of unique mass spectra (y-axis). FMRP and its FXR paralogs, red; proteins that function in NMD, blue. See also Supplementary Table 2. c, Coomassie Blue staining of human FMRP purified from E. coli (left) or FLAG-tagged human UPF1 purified from baculovirus (right). d, Quantitations of mRNAs, normalized to the level of ß-actin mRNA. Means with S.D., n = 3 independent biological replicates. (*)P < 0.05, (**)P < 0.01 or (***)P < 0.001 pertains to comparisons to Ctl-siRNA samples (two-sided t-test). e, Western blots of lysates of HEK293T cells transiently transfected with the denoted siRNA, eluted before (−) or after α-UPF1 or NRS IP. f, Quantitations of the specified RNA, normalized to the level of ß-actin mRNA, using lysates from e. Means with S.D., n = 3 independent biological replicates. P values compare Ctl siRNA- vs. specified siRNA-treated samples (two-sided t-test). g, Pertaining to Fig. 2h, western blots of lysates of HEK293T cells transfected with the specified siRNA, before or after α-p-UPF1 or rIgG IP. h, Western blot of lysates of HEK293T cells transfected with pcDNA-HA or pcDNA-HA-SMG1, the latter to approximate the increased level of SMG1 in FMRP siRNA-transfected cells. i, Quantitations of mRNAs, normalized to the level of their pre-mRNA, using lysates from h. Means with S.D., n = 3 independent biological replicates. P values compare HA-SMG1- vs. HA-transfected samples (two-sided t-test). j, Western blot of lysates of HEK293T cells transfected with the specified siRNA eluted before or after α-UPF1 or NRS IP. Results in a, e, g, h, j are representative of 3 independent biological replicates. Statistical source data and unprocessed blots are provided in Source Data Extended Data Fig. 3.
Extended Data Fig. 4 ∣. Tethering MS2CP-tagged FMRP fails to promote UPF1 recruitment, and UPF1 downregulation reduces FMRP binding to NMD targets.
a, As in Fig. 2l, but tethering MS2CP-FMRP in place of MS2CP-UPF1. b, Western blotting as in Fig. 2m but pertaining to a. c, Histogram representation as in Fig. 2n but using lysates analyzed in b. Results are represented as means with S.D., where n = 3 independent biological replicates. P values were calculated in comparisons of MS2CP-FMRP vs. MS2CP-transfected samples (two-sided t-test). d, Western blot of lysates of HEK293T cells, treated with the specified siRNA, before or after IP using anti-FMRP or, as a negative control, mIgG. Results are representative of 3 independent biological replicates. e, Histogram representation of RT-qPCR quantitations of the specified mRNA, as in Extended Data Fig. 3f, using lysates analyzed in d. Results are represented as means with S.D., where n = 3 independent biological replicates. P values were calculated in comparisons of Ctl siRNA vs. UPF1#1 siRNA-transfected samples (two-sided t-test). Statistical source data and unprocessed blots are provided in Source Data Extended Data Fig. 4.
Extended Data Fig. 5 ∣. Verification of FMRP-KD in TRIC-seq experiments, generation of FMR1-KO SH-SY5Y cells, and demonstration that ~2-fold overexpression of UPF1 in SH-SY5Y cells recapitulates the hyperactivated NMD that typifies FMR1-KO SH-SY5Y cells.
a, Scheme used to pulse-label SH-SY5Y-cell transcripts with 4-thiouridine (TU), chase for the indicated times in fresh medium lacking TU, biotinylate purified TU RNA, and capture biotinylated RNAs using streptavidin beads followed by RT-qPCR quantitations. b, Quantification of FMR1 mRNA relative to GAPDH mRNA in SH-SY5Y cells transfected with the specified siRNA, where n = 2 independent biological replicates. c, Western blots of lysates analyzed in b. Results are representative of 3 independent biological replicates. d, Cumulative fraction of log2 fold-change in mRNA half-life in FMRP#1-KD (upper) or FMRP#2-KD (lower) relative to Ctl-KD of “All mRNAs’’ (dotted line) or neuronal NMD targets (solid line). P values were calculated using the two-sided Wilcoxon rank-sum test. e, As in Fig. 3d, but for 33 NMD targets previously defined for non-neuronal cells. f, Diagram of relevant FMR1 gene exons, showing the sequences (blue) in exon 4 or 8 targeted by each pair of guide RNAs and CRISPR-Cas9n. Nicked sites, arrowheads; deleted or inserted sequences (red). g, As in Fig. 3f, but normalizations were to GAPDH mRNA. Means with S.D., n = 3 independent biological replicates. (*) P < 0.05 or (**) P < 0.01 is relative to WT samples (two-sided t-test). h, Western blots of lysates of WT SH-SY5Y cells expressing FLAG or FLAG-UPF1, the latter to bring the cellular abundance of UPF1 to approximate the increased level of UPF1 in FMR1-KO cells. Results are representative of 3 independent biological replicates. i, Quantitations of the specified mRNAs, normalized to the level of their pre-mRNA, using lysates from h. Means with S.D.,where n = 3 independent biological replicates. P values compare FLAG-UPF1 vs. FLAG-transfected samples (two-sided t-test). Statistical source data and unprocessed blots are provided in Source Data Extended Data Fig. 5.
Extended Data Fig. 6 ∣. Transcriptome-wide analyses of UPF1 siRNA downregulation in WT or FMR1-KO SHSY5Y cells show that NMD targets are comparably upregulated, and demonstration that the frequency of FMRP and p-UPF1 footprints on NMD targets is comparable.
a, Western blots of lysates of SH-SY5Y cells transiently transfected with Ctl or UPF1#1 siRNA using wild-type (WT) or FMR1-KO SH-SY5Y-cell lysates. b, Cumulative fraction of log2 fold-change in mRNA abundance upon UPF1#1-KD relative to Ctl-KD of “All mRNAs” (dotted line) or neuronal NMD targets (solid line) using WT (blue line) or FMR1-KO (red line) SH-SY5Y cell lysates, analyzed in a. P values were calculated using the two-sided Wilcoxon signed-rank test. c, Western blots of lysates of SH-SY5Y cells before (−) or after immunoprecipitation (IP) of using anti-FMRP or mouse (m)IgG. Results are representative of 3 independent biological replicates in a and c. d, Scatter plot of the abundance of FMRP footprints normalized to Input RNAs (x-axis) and p-UPF1 footprints normalized to Input RNAs (y-axis) on the specified regions (5′UTRs, CDSs and 3′UTRs) of NMD targets. r and P values were calculated using Pearson’s correlation coefficient test. Statistical source data and unprocessed blots are provided in Source Data Extended Data Fig. 6.
Extended Data Fig. 7 ∣. NMD is hyperactivated in FXS lymphoblasts, and characterization of iPSCs derived from healthy or FXS cells.
a, As in Fig. 4a, dot plots of RT-qPCR quantitations of the specified mRNA, normalized to the level of its pre-mRNA, using lysates from healthy (i.e. normal) or FXS lymphoblasts. Red bars represent means with S.D, where n = 6 independent biological cell lines. P values were calculated in comparisons of normal vs. FXS samples (two-sided Wilcoxon rank-sum test). b, as in a, but normalized to the level of ß-actin mRNA. Red bars, means with S.D., n = 6 independent biological cell lines. P values were calculated in comparisons of normal vs. FXS samples (two-sided Wilcoxon rank-sum test). c, Bright-field and IF images of iPSCs from a representative normal or a representative FXS patient-derived cell line for validation purposes. For IF, FXS iPSCs were stained for TRA-1-60 (green) or OCT4 (red), each of which is a pluripotent marker. Scale bar, 200 μm. Results are representative of 3 independent biological replicates. d, as in Fig. 4b, but normalized to the level of ß-actin mRNA. Means with S.D., where n = 3 biologically independent replicates. (***) P < 0.001 are relative to Normal #1 samples (one-way ANOVA and Dunnett’s multiple comparison test). e, As in Fig. 4b but for isogenic iPSCs + or − CGG repeat expansion. Means with S.D., where n = 3 independent biological replicates. P values were calculated in comparisons of + vs. − CGG repeat expansion (two-sided t-test). f, Western blots of lysates of normal and FXS iPSCs from Fig. 4b. Results are representative of 3 independent biological replicates. g, Protocol of neural differentiation − or + an NMD inhibitor. h, Quantitations of western blots shown in Fig. 4c. i, FXS iPSC-derived neurons exhibit deficient neurite formation on day 7 of differentiation. βIII-tubulin staining of normal and FXS neurons derived from iPSCs was used to assess neurite formation, average neurite length, neurite ramification, and neuronal differentiation efficacy. Means with S.D., where n = 8 independent biological replicates. P values were calculated in comparisons of normal vs. FXS samples (two-sided t-test). j, Quantitations of IF shown in Fig. 4d. Results are means with S.D., where n = 5 independent biological replicates. P values were calculated in comparisons of normal vs. FXS samples (two-sided t-test). Statistical source data and unprocessed blots are provided in Source Data Extended Data Fig. 7.
Extended Data Fig. 8 ∣. RT-qPCR and IF demonstrating that NMD inhibitors partially normalize FXS-derived iPSC differentiation.
a, Quantitations demonstrating that NMD was inhibited by each of the 3 NMD inhibitors. Samples derived from day 0, that is 24-hr after culturing iPSCs in the presence of each inhibitor as shown in Fig. 4e, b, Results are means with S.D., where n = 3 independent biological replicates. (*) P < 0.05 or (**) P < 0.01 is relative to RNA samples without NMD inhibitor (two-sided t-test). b, As in Extended Data Fig. 7j, but with or without an NMD inhibitor. Means with S.D., where n = 5 independent biological replicates. (*) P < 0.05, (**) P < 0.01 or (***) P < 0.01 is relative to RNA samples without an NMD inhibitor (two-sided t-test). c, As in Fig. 4e, but staining for MAP2 (red). d, As in Fig. 4e, but staining for BRN2/POU3F2 (green). Results are representative of 3 independent biological replicates in (c) and (d). e, Histogram representations of quantitations of western blots shown in Fig. 4f. Results represent n = 3 independent biological replicates, except for SYN1 analyses in FXS neurons, BRN2 analysis in FXS neurons treated with NMDI-1, and DCX analysis in FXS neurons treated with curcumin, where n = 2 independent biological replicates. f, Histogram representation of neurite outgrowth manifested by representative normal or FXS neurons on day 15 after differentiation. Results represent 2 independent biological replicates for FXS neurons and 3 independent biological replicates for normal neurons. All cells were stained for viability, and fluorescent signals derive from minimally 4 fields per well, where the extent of fluorescence for FXS neurons in the absence (−) of inhibitor is defined as 1. Statistical source data are provided in Source Data Extended Data Fig. 8.
Supplementary Material
Acknowledgements
We thank M. Ascano for the pFRT/TO/Flag-HA-FMRP wild type, J. Lykke-Andersen for pcDNA3-Gl Norm-MS2bs and pcDNA3-Gl Ter-MS2bs, A. Paciorkowski for help generating iPSC lines 5417 and 5433, M. Lacagnina and N. Freitag for help maintaining the iPSCs, P. Jin for the FXS iPSC line and its isogenic control, F. Lejeune and D. Bedwell for helpful advice on the NMD inhibitors, S. Velu for synthesizing NMDI-1, M. Hoque (Genomics Center, Rutgers New Jersey Medical School) and the University of Rochester Genomic Research Center for generating libraries and performing RNA-seq and RIP-seq footprinting, K. Kawata for RNA-seq and RIP-seq data deposition, and M. Popp, X. Rambout, H. Sakano and J. Darnell for comments on the manuscript. Liquid chromatography/mass spectrometry analyses were performed by E. Spooner at the MIT Biopolymers and Proteomics Core Facility. This work was supported by R01 GM05696 (to L.E.M.) and MEXT KAKENHI (221S0002 to N.A.). C.P. was partially supported by 1R21NS104878 and Link Foundation. T.K. was partially supported by a post-doctoral fellowship from the FRAXA Research Foundation and funds from University of Rochester School of Medicine and Dentistry Pilot Funding in Stem Cell and Regenerative Medicine.
Footnotes
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41556-020-00618-1.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Code availability
All of the scripts used for data processing and statistical analysis were written in Python, Perl or R and are available upon request.
Competing interests
The authors declare no competing interests.
Extended data is available for this paper at https://doi.org/10.1038/s41556-020-00618-1.
Supplementary information is available for this paper at https://doi.org/10.1038/s41556-020-00618-1.
Peer review information Nature Cell Biology thanks Miles Wilkinson and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Data availability
Sequencing datasets (FASTQ files), including RNA-seq and RIP-seq, have been deposited in the DNA Data Bank of Japan under the accession number DRA005644. Proteomics datasets have been deposited in the ProteomeXchange Consortium under the accession number PXD014901. Source data are provided with this paper. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
References
- 1.Richter JD, Bassell GJ & Klann E Dysregulation and restoration of translational homeostasis in fragile X syndrome. Nat. Rev. Neurosci 16, 595–605 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hagerman RJ et al. Fragile X syndrome. Nat. Rev. Dis. Prim 3, 17065 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Jaffrey SR & Wilkinson MF Nonsense-mediated RNA decay in the brain: emerging modulator of neural development and disease. Nat. Rev. Neurosci 19, 715–728 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurosaki T, Popp MW & Maquat LE Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat. Rev. Mol. Cell Biol 20, 406–420 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kurosaki T et al. A post-translational regulatory switch on UPF1 controls targeted mRNA degradation. Genes Dev. 28, 1900–1916 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imamachi N, Salam KA, Suzuki Y & Akimitsu N A GC-rich sequence feature in the 3′ UTR directs UPF1-dependent mRNA decay in mammalian cells. Genome Res. 27, 407–418 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan W-K et al. An alternative branch of the nonsense-mediated decay pathway. EMBO J. 26, 1820–1830 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang L et al. RNA homeostasis governed by cell type-specific and branched feedback loops acting on NMD. Mol. Cell 43, 950–961 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Purcell RH, Purcell RH, Toro C, Gahl WA & Hall RA A disease-associated mutation in the adhesion GPCR BAI2 (ADGRB2) increases receptor signaling activity. Hum. Mutat 38, 1751–1760 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Es MA et al. Genetic variation in DPP6 is associated with susceptibility to amyotrophic lateral sclerosis. Nat. Genet 40, 29–31 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Spillantini MG & Goedert M Tau pathology and neurodegeneration. Lancet Neurol. 12, 609–622 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Puffenberger EG et al. A homozygous missense mutation in HERC2 associated with global developmental delay and autism spectrum disorder. Hum. Mutat 33, 1639–1646 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Irimia M et al. A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell 159, 1511–1523 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ascano M et al. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature 492, 382–386 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamashita A, Ohnishi T, Kashima I, Taya Y & Ohno S Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 15, 2215–2228 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herder C, Swiercz JM, Müller C, Peravali R & Quiring R ArhGEF18 regulates RhoA–Rock2 signaling to maintain neuro-epithelial apico-basal polarity and proliferation. Development 2797, 2787–2797 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Hunt D, Raivich G & Anderson PN Activating transcription factor 3 and the nervous system. Front. Mol. Neurosci 5, 7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durand S, Franks TM & Lykke-Andersen J Hyperphosphorylation amplifies UPF1 activity to resolve stalls in nonsense-mediated mRNA decay. Nat. Commun 7, 12434 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tani H et al. Identification of hundreds of novel UPF1 target transcripts by direct determination of whole transcriptome stability. RNA Biol. 9, 1370–1379 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamada T & Akimitsu N Contributions of regulated transcription and mRNA decay to the dynamics of gene expression. Wiley Interdiscip. Rev. RNA 10, e1508 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F & Dietz HC Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet 36, 1073–1078 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Lykke-Andersen S et al. Human nonsense-mediated RNA decay initiates widely by endonucleolysis and targets snoRNA host genes. Genes Dev. 28, 2498–2517 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lou C-H et al. Nonsense-mediated RNA decay influences human embryonic stem cell fate. Stem Cell Rep. 6, 844–857 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ran FA et al. Genome engineering using the CRISPR–Cas9 system. Nat. Protoc 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohnishi T et al. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol. Cell 12, 1187–1200 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Kashima I et al. Binding of a novel SMG-1–Upf1–eRF1–eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 20, 355–367 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Isken O et al. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell 133, 314–327 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee SR, Pratt GA, Martinez FJ, Yeo GW & Lykke-Andersen J Target discrimination in nonsense-mediated mRNA decay requires Upf1 ATPase activity. Mol. Cell 59, 413–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurosaki T, Miyoshi K, Myers JR & Maquat LE NMD-degradome sequencing reveals ribosome-bound intermediates with 3′-end non-templated nucleotides. Nat. Struct. Mol. Biol 25, 940–950 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu B et al. Regulatory discrimination of mRNAs by FMRP controls mouse adult neural stem cell differentiation. Proc. Natl Acad. Sci. USA 115, E11397–E11405 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie N et al. Reactivation of FMR1 by CRISPR/Cas9-mediated deletion of the expanded CGG-repeat of the fragile X chromosome. PLoS ONE 11, e0165499 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lou CH et al. Posttranscriptional control of the stem cell and neurogenic programs by the nonsense-mediated RNA decay pathway. Cell Rep. 6, 748–764 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 78, 785–798 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarnat HB Clinical neuropathology practice guide 5–2013: markers of neuronal maturation. Clin. Neuropathol 32, 340–369 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castren M et al. Altered differentiation of neural stem cells in fragile X syndrome. Proc. Natl Acad. Sci. USA 102, 17834–17839 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheridan SD et al. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS ONE 6, e26203 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Telias M, Segal M & Ben-Yosef D Neural differentiation of fragile X human embryonic stem cells reveals abnormal patterns of development despite successful neurogenesis. Dev. Biol 374, 32–45 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Doers ME et al. iPSC-derived forebrain neurons from FXS individuals show defects in initial neurite outgrowth. Stem Cells Dev. 23, 1777–1787 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Halevy T, Czech C & Benvenisty N Molecular mechanisms regulating the defects in fragile X syndrome neurons derived from human pluripotent stem cells. Stem Cell Rep. 4, 37–46 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Achuta VS et al. Functional changes of AMPA responses in human induced pluripotent stem cell-derived neural progenitors in fragile X syndrome. Sci. Signal 11, eaan8784 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Irwin SA et al. Abnormal dendritic spine characteristics in the temporal and visual cortices of patients with fragile-X syndrome: a quantitative examination. Am. J. Med. Genet 98, 161–167 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Durand S et al. Inhibition of nonsense-mediated mRNA decay (NMD) by a new chemical molecule reveals the dynamic of NMD factors in P-bodies. J. Cell Biol 178, 1145–1160 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin L et al. Identification and characterization of small molecules that inhibit nonsense-mediated RNA decay and suppress nonsense p53 mutations. Cancer Res. 74, 3104–3113 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng D et al. Increase of a group of PTC+ transcripts by curcumin through inhibition of the NMD pathway. Biochim. Biophys. Acta 1849, 1104–1115 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Kamelgarn M et al. ALS mutations of FUS suppress protein translation and disrupt the regulation of nonsense-mediated decay. Proc. Natl Acad. Sci. USA 115, E11904–E11913 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jackson KL et al. Preservation of forelimb function by UPF1 gene therapy in a rat model of TDP-43-induced motor paralysis. Gene Ther. 22, 20–28 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barmada S et al. Amelioration of toxicity in neuronal models of amyotrophic lateral sclerosis by hUPF1. Proc. Natl Acad. Sci. USA 112, 7821–7826 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun Y et al. C9orf72 arginine-rich dipeptide repeats inhibit UPF1-mediated RNA decay via translational repression. Nat. Commun 11, 3354 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gantois I, Popic J, Khourosky A & Sonenberg N Metformin for treatment of fragile X syndrome and other neurological disorders. Annu. Rev. Med 70, 167–181 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Warren L et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 7, 618–630 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miyoshi N et al. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell 8, 633–638 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Mandal PK & Rossi DJ Reprogramming human fibroblasts to pluripotency using modified mRNA. Nat. Protoc 8, 568–582 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Chan EM et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat. Biotechnol 27, 1033–1037 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Bock C et al. Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell 144, 439–452 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hosoda N, Kim YK, Lejeune F & Maquat LE CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat. Struct. Mol. Biol 12, 893–901 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Lykke-Andersen J, Shu MD & Steitz JA Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103, 1121–1131 (2000). [DOI] [PubMed] [Google Scholar]
- 57.Elbarbary RA, Miyoshi K, Hedaya O, Myers JR & Maquat LE UPF1 helicase promotes TSN-mediated miRNA decay. Gene. Dev 39, 1483–1493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martin M Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011). [Google Scholar]
- 59.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liao Y, Smyth GK & Shi W Sequence analysis featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuleshov MV et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Santos A et al. Comprehensive comparison of large-scale tissue expression datasets. Peer J. 3, e1054 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mi H, Muruganujan A & Thomas PD PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41, 377–386 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kanehisa M, Furumichi M, Tanabe M, Sato Y & Morishima K KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tani H & Akimitsu N Genome-wide technology for determining RNA stability in mammalian cells: historical perspective and recent advantages based on modified nucleotide labeling. RNA Biol. 9, 37–41 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dölken L High resolution gene expression profiling of RNA synthesis, processing, and decay by metabolic labeling of newly transcribed RNA using 4-thiouridine. Methods Mol. Biol 1064, 91–100 (2013). [DOI] [PubMed] [Google Scholar]
- 70.Trapnell C et al. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol 28, 516–520 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim D et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Imamachi N et al. BRIC-seq: a genome-wide approach for determining RNA stability in mammalian cells. Methods 67, 55–63 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing datasets (FASTQ files), including RNA-seq and RIP-seq, have been deposited in the DNA Data Bank of Japan under the accession number DRA005644. Proteomics datasets have been deposited in the ProteomeXchange Consortium under the accession number PXD014901. Source data are provided with this paper. All other data supporting the findings of this study are available from the corresponding author on reasonable request.