Abstract

In this work, for the first time, we present the X-ray diffraction crystal structure and spectral properties of a new, room-temperature polymorph of teriflunomide (TFM), CSD code 1969989. As revealed by DSC, the low-temperature TFM polymorph recently reported by Gunnam et al. undergoes a reversible thermal transition at −40 °C. This reversible process is related to a change in Z’ value, from 2 to 1, as observed by variable-temperature 1H–13C cross-polarization (CP) magic-angle spinning (MAS) solid-state NMR, while the crystallographic system is preserved (triclinic). Two-dimensional 13C–1H and 1H–1H double-quantum MAS NMR spectra are consistent with the new room-temperature structure, including comparison with GIPAW (gauge-including projector augmented waves) calculated NMR chemical shifts. A crystal structure prediction procedure found both experimental teriflunomide polymorphs in the energetic global minimum region. Differences between the polymorphs are seen for the torsional angle describing the orientation of the phenyl ring relative to the planarity of the TFM molecule. In the low-temperature structure, there are two torsion angles of 4.5 and 31.9° for the two Z’ = 2 molecules, while in the room-temperature structure, there is disorder that is modeled with ∼50% occupancy between torsion angles of −7.8 and 28.6°. These observations are consistent with a broad energy minimum as revealed by DFT calculations. PISEMA solid-state NMR experiments show a reduction in the C–H dipolar coupling in comparison to the static limit for the aromatic CH moieties of 75% and 51% at 20 and 40 °C, respectively, that is indicative of ring flips at the higher temperature. Our study shows the power of combining experiments, namely DSC, X-ray diffraction, and MAS NMR, with DFT calculations and CSP to probe and understand the solid-state landscape, and in particular the role of dynamics, for pharmaceutical molecules.
Short abstract
This work presents the X-ray diffraction crystal structure and spectral properties of a new, room-temperature polymorph of teriflunomide that undergoes a reversible thermal transition, and it is related to the change in Z′ value, from 2 to 1. Our study shows the power of combining experiments with DFT calculations and CSP to understand the solid-state landscape for pharmaceutical molecules.
1. Introduction
One particular scientific method is usually not enough to solve a challenging problem relating to solid matter for organic molecules, such as a moderately sized active pharmaceutical ingredient (API) organic molecule. Understanding the complexity of such solid matter which depends on the interplay of a number of subtle, differentiated intermolecular contacts requires the application of different diagnostic tools and should be supported by theoretical methods. For crystalline compounds, X-ray diffraction and solid-state NMR are usually the experimental techniques of choice. In the group of theoretical methods, crystal structure prediction (CSP) developed during the last two decades has become one of the most promising approaches.1−5 CSP methods are based on searching for the most thermodynamically stable crystal structure, making various approximations in evaluating the crystal energy. The most stable (global minimum) structure provides a prediction of an experimental crystal structure. Several successful CSP studies on large but relatively rigid systems, such as organic porous cages, have been reported.6,7 The ab initio random structure search (AIRSS) method that has been used successfully for a number of inorganic solids8−11 has also been applied to a simple organic molecule.12 Organic compounds are more challenging, since they tend to have a considerable conformational flexibility and can crystallize in a variety of conformations.13 In such cases, the CSP search space grows exponentially and the prediction of experimentally observed crystal structures becomes ever more challenging. The support of this process by experimental techniques such as solid-state magic-angle spinning (MAS) NMR spectroscopy14,15 can significantly reduce the complexity of the problem.
Solid-state MAS NMR spectroscopy is a powerful method for characterizing polymorphs, solvates, salts, and cocrystals exhibited by organic molecules.16 The power of this technique is also due to the fact that, using different NMR experiments, subtle structural features as well as dynamic properties that report on the time scale and amplitude of motion can be measured. Solid-state NMR spectroscopy is very sensitive to local molecular disorder (including dynamic disorder) and structural defects. Thus, it is no surprise that this method is frequently used in different branches of science, in particular in the pharmaceutical sciences, to study structural features and to verify the quality and/or homogeneity of the studied material.17−39
As noted above, there is much potential benefit in the complementary application of solid-state NMR spectroscopy and CSP methods. For CSP methods, figuring out the true number of polymorphs that exist and those that have not been found is one of the biggest challenges. Moreover, despite the fact that the CSP methodology explains the range of thermodynamically favored crystal packings very well, even the most robust algorithms still do not consider static or/and dynamic molecular disorder, commonly observed by experimental techniques. These challenges have been discussed, in particular in relation to pharmaceutically important compounds.40,41 On the other hand, solid-state NMR spectroscopy is readily applicable to systems exhibiting conformational flexibility and/or different intermolecular interactions.
One of the simplest and most common applications of solid-state NMR is the use of a one-dimensional 13C cross-polarization (CP) MAS experiment to determine the number of nonequivalent molecules in the asymmetric unit cell: i.e., the crystallographic Z′ value.16 It is a first stage of the strategy called NMR crystallography42−54 where an integral part is the gauge-including projector-augmented wave (GIPAW) method55−57 for the calculation of NMR parameters that is a landmark development in theoretical predictions of NMR parameters for solid materials. This approach can be readily applied to provide experimental verification of CSP solutions; specifically, it is indicative of whether an identified likely solution in the CSP strategy corresponds to an experimental form.12,33,58−60
This paper shows the synergy of experimental solid-state NMR and single crystal X-ray diffraction methods as well as thermal analysis with DFT calculations and the crystal structure prediction technique in the analysis of a system forming polymorphs, undergoing phase transitions and local dynamic processes. Specifically, teriflunomide (TFM) is a moderately sized API (Figure 1) approved for multiple sclerosis treatment in 2012 by the FDA, under the brand name Aubagio.61,62 Teriflunomide has also been recently applied as a noninvasive drug administered to the brain that easily bypasses the blood–brain barrier in glioblastoma treatment.63
Figure 1.

Chemical structure of teriflunomide, including the numbering scheme used here.
Recently Gunnam et al.64 have reported a low-temperature single-crystal X-ray diffraction structure of TFM (CSD reference code: 1885431) as well as TFM cocrystal structures. In this paper, we identify a polymorphic transformation at −40 °C that has not been reported by Gunnam et al. Our paper applies a complementary multitechnique approach, employing 1D and 2D solid-state MAS NMR techniques, low- and room-temperature X-ray diffraction measurements, and differential scanning calorimetry (DSC) as well as the DFT-based GIPAW calculation of NMR chemical shifts and crystal structure prediction (CSP).
2. Results and Discussion
2.1. Variable-Temperature Solid-State NMR study of Teriflunomide
As highlighted in the Introduction, the first step in a solid-state NMR analysis of a crystalline organic sample as part of a structure assignment strategy is to answer the following question: what is the number of independent molecules in the asymmetric part of a crystallographic unit, labeled as Z′? In the simplest case, there is a straightforward correlation between the number of NMR signals in the isotropic part of spectrum and Z′. In the Introduction, we also emphasized that the determination of the exact Z′ value is not always obvious. In some cases, reported in the literature, deducing whether this value is equal to 1 or more is ambiguous.4
In our studies with TFM, we began by carefully selecting a representative material. In order to obtain good-quality samples for the structural investigations, several crystallizations of teriflunomide were carried out by employing various organic solvents. The preliminary solid-state NMR studies proved that, despite numerous efforts and optimization of the crystallization conditions, all obtained samples appeared to be of the same polymorphic form. The results presented in this paper are for TFM crystallized from dichloromethane; a powder X-ray diffraction (PXRD) pattern is shown in Figure S1.
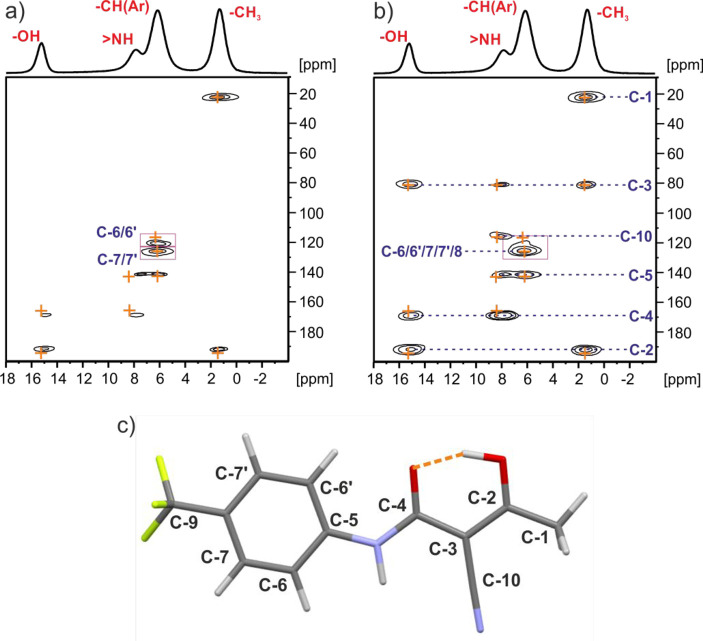
Figure 2a–c shows 1H → 13C, 1H → 15N, and 19F → 13C one-dimensional CP MAS NMR spectra, respectively, and Figure 2d shows a one-pulse 19F MAS NMR spectrum recorded at 28 °C. Note that magnetization transfer from fluorine to 13C (Figure 2c) was performed because the carbon-13 signal from the −CF3 group is missing in classical 1H → 13C CP MAS NMR spectra. In all spectra, narrow resonance lines confirm that the sample is well ordered and is highly crystalline. When the number of resonance lines in the isotropic part of the spectra are accounted for, it is evident that, for the sample under investigation, the Z’ value equals 1. Such information is in conflict with X-ray data reported by Gunnam et al.64 Indeed, our 1H → 13C CP MAS NMR spectrum in Figure 2a is the same as the room-temperature 13C CP MAS spectrum presented by Gunnam et al. in Figure S3b, but they do not comment on the absence of a doubling of 13C resonances.
Figure 2.
1H → 13C (a), 1H → 15N (b), and 19F → 13C (c) CP MAS and one-pulse 19F MAS (d) NMR spectra of TFM recorded at spinning rates of 12 (a, b) and 25 kHz (c, d) at 28 °C at 14 T, corresponding to a 1H Larmor frequency of 600.1 MHz. For a recycle delay of 45 s, 1024, 8192, 2048, and 16 transients were coadded for (a)–(d), respectively. Asterisks indicate spinning sidebands.
In a further analysis of the 1H → 13C CP MAS NMR spectrum, a feature that is ambiguous and surprising is the intensity of signals in the aromatic region. The intensity of the CH carbons (6, 6′, 7, and 7′) is unexpectedly weak, and changing the CP MAS experiment setup, specifically the nutation frequencies and contact time for CP and the relaxation delay, did not change this. However, Figure 3 shows that the signal intensity for these aromatic carbons increases noticeably upon changing the temperature from 40 to 20 °C. This observation is indicative of temperature-dependent phenyl ring dynamics that slow upon a decrease in the temperature (see further discussion below in section 2.4).65
Figure 3.
1H → 13C CP MAS NMR spectra of TFM recorded at 40 °C (a) and 20 °C (b) with a spinning rate of 13 kHz and a 1H Larmor frequency of 600.1 MHz. A total of 256 transients were coadded for a recycle delay of 60 s.
Given the observation of dynamics in the sample, it is instructive to consider Figure 4, which shows a DSC plot for the TFM sample. The heating program, from −150 °C up to 250 °C, shows a strong endothermal peak at around 230 °C, which is the melting point temperature. However, a much more interesting feature is evident from a closer look at the region below room temperature. The enlarged inset around −39 °C clearly indicates a very weak endothermal peak. It is worth mentioning that this thermal effect was repeatedly observed in multiple cooling–heating runs. This suggests a reversible transformation that has only a relatively small effect on the organization of the TFM crystal structure. Although Gunnam et al.64 reported DSC results for TFM, the starting point of their DSC curve (in Figure 8) was 30 °C: i.e., too high to observe this thermal transformation.
Figure 4.

DSC plot for the TFM sample with a heating rate of 5 °C min–1.
With the aim of identifying a correlation between the DSC and solid-state NMR spectroscopy data, we carried out a 13C CP MAS NMR experiment at low temperature, below the phase transition point observed by DSC in Figure 4. Specifically, Figure 5 compares 13C CP MAS NMR spectra recorded at (a) 20 °C and (b) −80 °C.
Figure 5.

1H → 13C CP MAS NMR spectra of TFM recorded at a spinning rate of 12 kHz with an input gas temperature of (a) 20 °C and (b) −80 °C and a 1H Larmor frequency of 850.2 MHz. Totals of 48 and 64 transients were coadded for a recycle delay of 60 s for (a) and (b), respectively. Asterisks indicate spinning sidebands.
From an inspection of the spectra in Figure 5, it is clear that most of the 13C resonances appear as two peaks at the lower temperature. This is highlighted for the −CH3 group, as shown in the insets in Figure 5. This is unambiguous evidence that the low-temperature TFM polymorph has a Z′ value of 2, in agreement with the low-temperature single-crystal X-ray diffraction structure of TFM (CSD reference code 1885431) reported by Gunnam et al. A discussion about the spinning sidebands (denoted by *) seen in Figure 5 can be found in the Supporting Information.
2.2. TFM X-ray Single-Crystal Structures at 100 and 295 K
In this work, we have obtained single-crystal X-ray structures, for the same crystal kept in the diffractometer, at 100 and 292 K. The crystallographic data for the two polymorphs labeled as TFMLT and TFMRT as well as those determined by Gunnam et al.64 (CSD code 1885431) are presented in Table 1. Both data sets were indexed in the triclinic system with the P1̅ space group, but the a cell length is approximately double in the TFMLT data and the unit cell angles differ (on the frames where a* were visible, reflections were twice as dense). The crystal structures have been deposited in the Cambridge Structural Database under the deposition number 1892916 for TFMLT and 1969989 for TFMRT. The main difference is the number of molecules in the asymmetric unit cell (the Z′ values are equal to 1 and 2 for TFMRT and TFMLT, respectively) in agreement with the solid-state NMR observations.
Table 1. Crystal Structure and Refinement Data for TFM.
| TFMLT | TFM(64) | TFMRT | |
|---|---|---|---|
| CCDC code | 1892916 | 1885431 | 1969989 |
| empirical formula | C12H9F3N2O2 | C12H9F3N2O2 | C12H9F3N2O2 |
| formula wt | 270.21 | 270.21 | 270.21 |
| temp (K) | 100 | 100 | 292 |
| cryst syst | triclinic | triclinic | triclinic |
| space group | P1̅ | P1̅ | P1̅ |
| a (Å) | 9.4030(2) | 9.3951(5) | 4.85221(15) |
| b (Å) | 11.5300(2) | 11.5161(7) | 10.8738(3) |
| c (Å) | 11.9572(3) | 11.9447(8) | 11.7015(3) |
| α (deg) | 95.9600(17) | 95.990(3) | 102.454(2) |
| β (deg) | 105.8499(19) | 105.839(2) | 97.821(2) |
| γ (deg) | 110.763(2) | 110.753(2) | 93.832(2) |
| V (Å3) | 1136.7 | 1133.21(12) | 594.284 |
| Z | 4 | 4 | 2 |
| Z′ | 2 | 2 | 1 |
| density (g cm–3) | 1.579 | 1.579 | 1.510 |
| θ range (deg) | 75.391 | 75.051 | |
| index ranges | –11, +11; −14, +14; −14, +14 | –5, +6; −13, +13; –14, +14 | |
| Nref (I > 2σ(I) | 4638 (4442) | 5004 (3443) | 2413 (2298) |
| R1 (I > 2σ(I)) | 0.0395 (0.0386) | 0.0505 | 0.0448 (0.0438) |
| wR2 (I > 2σ(I)) | 0.1141 (0.1130) | 0.1277 | 0.1286 (0.1275) |
Figure 6 shows a comparison of the asymmetric unit cell of the TFMLT (left, a and b) and TFMRT (right, c and d) polymorphs. In the TFMRT structure, disorder of the phenyl ring (corresponding to a rocking movement) and the trifluoromethyl group (rotation around the C–C bond) was observed. Two orientations of disordered parts were refined with occupancy factors summed to unity and each of them close to 0.5, specifically 0.495 for carbon and hydrogen as well as 0.402 for fluorine atoms for the lower occupancy and 0.505 for carbon and hydrogen as well as 0.598 for fluorine atoms for the higher occupancy. Due to different temperature conditions for both measurements, we observed much larger thermal ellipsoids for TFMRT than for TFMLT. Specifically, the average volumes for the fluorine position was 0.014 and 0.080 Å3 for TFMLT and TFMRT, respectively. The hydrogen atoms connected to carbon atoms and the amide nitrogen were set geometrically and refined as riding with the thermal parameter equal to 1.2 of the thermal vibration of the parent atom. The hydrogen atoms at hydroxyl oxygens were found on the difference Fourier map and refined with geometrical restraints and thermal parameters equal to 1.5 of the thermal vibration of the parent atom. Intermolecular NH···N and intramolecular OH···O hydrogen bonding is indicated by orange dashed lines in Figure 6.
Figure 6.

Asymmetric unit cell of (a, b) the TFMLT and (c, d) the TFMRT polymorphw displayed along the a (a, c) and b (b, d) directions. The atoms with a lower occupancy of disordered fragments in the room-temperature structure are marked with a lighter shade of the corresponding color. The C4–NH–C5–C6 torsional angle is highlighted in orange, and the crystallographic axes are shown in red (a direction), green (b direction), and blue (c direction). Intermolecular NH···N and intramolecular OH···O hydrogen bonds (see Table S3) are indicated by orange dashed lines.
The low-temperature form shows a slight conformational difference of both TFM molecules that is mostly located in the values of the C4–NH–C5–C6 torsional angle (4.5 and 31.9°). Since the TFMRT structure contains one molecule in the asymmetric part of the unit cell (Z′ = 1), a corresponding comparison can be performed between the less and more highly occupied positions, for which the C4–NH–C5–C6 torsional angle values are −7.8 and 28.6°, respectively. Table 2 presents a comparison of C4–NH–C5–C6 torsional angle values for the experimental diffraction structures as well as those after DFT-D (CASTEP) geometry optimization. It is interesting that both TFMRT lower and higher occupancy sites (C4–NH–C5–C6 equal to −7.8 or 28.6°) lead to almost the same final structure with a torsional angle of 4.7 or 5.1° after geometry optimization of all atomic positions with the unit cell parameters fixed to those determined by X-ray diffraction. Optimizing all atomic positions and additionally allowing the unit cell parameters to vary again results in very similar C4–NH–C5–C6 torsional angles of 4.2 and 5.7°. Table S1 in the Supporting Information gives the modified unit cell parameters: the changes in unit cell parameters being similar to those observed previously.43,66Table S2 states the energy differences for the different DFT-D calculations, with DFT having previously been used to evaluate different crystal structures.67−69 Note that for TFMRT, with the less or more highly occupied sites only as starting points, i.e. C4–NH–C5–C6 torsional angle of −7.8 and 28.6° (labeled as A and B), respectively, there remains an energy difference of only 0.8 kJ/mol after optimization of all atomic positions and unit cell parameters.
Table 2. C4–NH–C5–C6 Torsional Angle (deg) Values in TFM for Single-Crystal X-ray Diffraction Structures and Those Obtained after DFT-D (CASTEP) Geometry Optimization.
| C4–NH–C5–C6 torsional angle |
|||
|---|---|---|---|
| structure | X-ray data | DFT-D (opt all)a | DFT-D (opt all + cell)b |
| TFMLT (molecule A) | 4.5 | 3.4 | 4.2 |
| TFMLT (molecule B) | 31.9 | 29.1 | 29.3 |
| TFMRT (lower A occupancy sites)c | –7.8 | 5.1 | 4.2 |
| TFMRT (higher B occupancy sites)c | 28.6 | 4.7 | 5.7 |
DFT-D geometry optimization of all atomic positions with the unit cell parameters fixed to those determined by X-ray diffraction.
DFT-D geometry optimization of all atomic positions and allowing the unit cell parameters to vary (see Table S1 in the Supporting Information)
The DFT calculations for the lower A and higher B occupancy sites of TFMRT were performed for two separate systems where the starting unit cell contains the experimental A or B sites only.
It is to be stressed that the packings of both crystal structures are highly similar to each other. We used the COMPACK algorithm, which is well suited to comparing two crystal structures and determining their metric of similarity.70 The idea is based on the construction and superposition of clusters for comparing crystal forms and the calculation of the difference (root mean square deviation, or RMSD value) among equivalent atomic positions. Figure 7 shows the COMPACK superposition of the clusters TFMLT and TFMRT containing 15 TFM molecules. Note that the COMPACK algorithm comparison procedure presented here considers only the most highly occupied sites.
Figure 7.
Superposition of clusters within the COMPACK algorithm70 for TFMLT (blue) and TFMRT (green) containing 15 TFM molecules (only non-hydrogen atoms are shown). Note that the COMPACK algorithm comparison procedure considers only the most highly occupied sites.
The calculated RMSD15 is equal to 0.396 Å. The value is not large, and a closer inspection of the clusters shows that the main differences concern the orientation of the aromatic ring as well as the fluorine atoms in the −CF3 group. It also cannot be excluded that there is an effect whereby the orientation of the phenyl ring is correlated with the orientation of the −CF3 group.
Since the fluorine atoms in the TFMRT structure are affected by positional disorder, it seems to be justified not to consider fluorine atoms belonging to the −CF3 group during the comparison of both polymorphs. In such a case, the appropriate RMSD15 value (without the disordered −CF3 group and aromatic positions) drops to 0.149 Å. The resulting RMSD15 proved that the same long-range order for both TFMLT and TFMRT structures is preserved. Moreover, on consideration of the intermolecular NH···N and intramolecular OH···O hydrogen bonding, Table S3 in the Supporting Information shows that, after DFT-D geometry optimization of all atomic positions and the unit cell parameters being allowed to vary, the N···N and H···N distances, and the NHN angle and the O···O and H···O distances, and the OHO angle only vary between 2.94 and 2.99 Å, 1.93 and 1.98 Å, and 164 and 166° and between 2.44 and 2.46 Å, 1.45 and 1.47 Å, 154 and 155°, respectively.
We further note that, in the paper by Gunnam et al.,64 it is stated on page 5411 that there is a match of the PXRD experimental line pattern and the calculated line profile from the X-ray crystal structure, even though we have discovered the anomaly in the Z′ value. This observation can be understood, since we observed a great similarity of both simulated PXRD patterns based on TFMRT and TFMLT single-crystal X-ray diffraction structure solutions (see Figure S2): noticeable deviation is only observed for 2θ values above 18°.
2.3. NMR Crystallography of the TFM Room-Temperature Polymorph
The results presented in sections 2.1 and 2.2 show that TFM exhibits temperature-dependent polymorphism. This section extends the solid-state MAS NMR characterization of the room-temperature polymorph (Z′ = 1), specifically also presenting 1H NMR spectra recorded under fast MAS. In particular, this enables the recording of 2D heteronuclear experiments with indirect inverse (inv) observation via 1H such as a 13C–1H invHETCOR MAS NMR experiment that utilizes 1H → 13C and 13C → 1H CP before and after t1 evolution, respectively. Figure 8 shows 13C–1H invHETCOR MAS spectra acquired with a spinning rate, νR = 62.5 kHz, with (a) short (150 μs) and (b) long (1 ms) 13C → 1H CP contact times such that only cross peaks corresponding to short C···H distances, mostly direct C–H bonds, are observed in Figure 8a, while cross peaks due to longer range C···H proximities are seen in Figure 8b.
Figure 8.
13C–1H invHETCOR MAS NMR spectra of TFMRT recorded at 20 °C with a spinning rate of 62.5 kHz at a 1H Larmor frequency of 600.1 MHz with a second (13C → 1H) CP contact time of (a) 150 μs and (b) 1000 μs. A one-pulse 1H MAS spectrum is shown at the top. The orange crosses represent GIPAW calculated NMR correlations for C···H distances up to (a) 2.2 and (b) 3.0 Å. The base contour is at 10% of the maximum peak intensity. (c) Pictographic representation of the OH···H intramolecular hydrogen bond (orange dotted line) in the TFMRT structure.
On consideration of the 13C–1H invHETCOR MAS NMR spectrum in Figure 8a, as well as the peaks for one-bond C–H connectivities, C6/6′, C-7/7′, and the CH3 methyl group, note the strong intensity of correlation peaks between the OH proton and quaternary C2 and C4 carbons that is consistent with an intramolecular hydrogen bonding between the OH group and the carbonyl group, as shown in Figure 8c (see also Figure 6). In Figure 8b, additional strong cross peaks between C3 and the methyl and OH protons are observed as well as between the NH protons and the C3, C4, C5, and C10 carbons, as is consistent with the molecular conformation shown in Figure 8c.
Verification of structures determined by X-ray diffraction (or also CSP) can be achieved by comparing experimental solid-state NMR chemical shifts with GIPAW-calculated chemical shifts.34,42,44,45,66,71 The orange crosses in Figure 8 correspond to the NMR correlations based on GIPAW calculations for the TFMRT crystal structure (after DFT geometry optimization). Excellent agreement is observed between experimental and GIPAW-calculated chemical shifts, as reflected by the small root-mean-squared error (RMSE) values of 0.14 and 2.2 ppm for 1H and 13C, respectively (Table 3).34,42,44,45,66,71 (Note that the GIPAW-calculated shieldings for TFMLT and TFMRT are given in Table S4.) In this context, for organic molecules, Emsley and co-workers have established a threshold root-mean-square error (RMSE) of 0.3 ppm for 1H,72,73 while in further work, they establish RMSEs compared to a machine-learning method of 0.5 ppm for 1H and 4.3 ppm for 13C.74 The RMSE calculation for 13C does not take into account C-9 that is bonded to fluorine atoms and for which there is a very large discrepancy between experimental and GIPAW-calculated values. In this context, Dybowski and co-workers have identified that such discrepancies arise in DFT calculations of 19F magnetic shielding.75
Table 3. Experimental and GIPAWa-Calculated 13C and 1H NMR Chemical Shifts, δ (in ppm), for TFMRT.
| δ(1H) |
δ(13C) |
|||
|---|---|---|---|---|
| structure (position) | exptl | GIPAW | exptl | GIPAW |
| 1 | 1.5 | 1.7b | 22.3 | 23.6 |
| 2 | 191.5 | 194.4 | ||
| 3 | 80.6 | 80.5 | ||
| 4 | 168.6 | 166.2 | ||
| 5 | 141.3 | 141.7 | ||
| 6 | 6.1 | 6.1b | 120.5 | 118.5b |
| 7 | 6.4 | 6.2b | 125.6 | 126.3b |
| 8 | 125.6 | 122.5 | ||
| 9 | 124.8 | 138.3 | ||
| 10 | 116.5 | 120.2 | ||
| –NH– | 8.1 | 8.3 | ||
| –OH | 15.2 | 15.2 | ||
The GIPAW-calculated chemical shifts were determined from the GIPAW-calculated shieldings (see Table S4) using the following equations: δ(1H) = −0.910σ(1H) + 27.0 (ppm) and δ(13C) = −0.969σ(13C) + 167.1 (ppm). Values were obtained after DFT-D geometry optimization, with the unit cell allowed to vary.
The average of the distinct 1H and 13C chemical shifts is presented.
Further structural information is provided by a 2D homonuclear 1H double-quantum (DQ) single-quantum (SQ) MAS NMR correlation experiment with back to back (BaBa) recoupling sequence that establishes proximities between specific protons, as shown in Figure 9.76−78 Of note is the auto OH DQ peak that is due to an intermolecular OH···HO correlation corresponding to the proximity of TFM molecules in different planes (see Figure 9c). The expected relative intensities of correlation peaks H(i)···H(j) and H(i)···H(k) can be estimated from the proton–proton proximities using the ∼rij6/rik6 formula, where rij and rik are distances between the H(i)···H(j) and H(i)···H(k) coupled protons, respectively.76 For TFMRT, the OH···HO inteplanar distance is 4.87 Å; thus, it is expected that the peak intensity will be only 2.926/4.876 = 0.05 of the CH(aromatic)···HO peak intensity (this is the closest proton to the OH). This is doubled, since there are two such interplanar OH proximities; thus, a 10% relative intensity is consistent with that seen experimentally in Figure 9a.
Figure 9.
(a) 1H–1H DQ-SQ MAS NMR spectrum of TFMRT recorded with one rotor period of BaBa recoupling at ambient temperature and 20 °C with a spinning rate of 62.5 kHz and a 1H Larmor frequency of 600.1 MHz. Blue dotted lines and descriptions indicate observed inter- and intramolecular H–H proximities. The base contour is at 5% of the maximum peak intensity. A one-pulse 1H MAS NMR spectrum is shown at the top. (b, c) Representations of the observed H–H proximities for the OH and NH in the TFMRT crystal structure. The H–H distances in the TFMRT crystal structure after DFT-D geometry optimization are as follows: CH3···HC(Ar), 2.71 Å; CH3···HO, 3.49 Å; (Ar)CH···HO, 2.92 Å; NH···HO, 4.16 Å; OH···HO, 4.87 Å.
2.4. Local Molecular Dynamics in the Crystal Lattice of TFMRT
As noted above, the variable-temperature 13C CP MAS spectra displayed in Figure 3 indicate phenyl ring dynamics in the room-temperature form. Possessing the single-crystal X-ray solution for TFMRT allowed us to investigate the possibility of a dynamic process, by means of theoretical methods. Specifically, following an approach used elsewhere,79−82 we investigate by DFT calculations the effect of changing the C4–NH–C5–C6 torsional angle value which causes aromatic ring rotation around the 1–4 axis. In our procedure, due to the plane symmetry of the aromatic moiety, the torsional angle value was incremented from −90° to +90° (equivalent of −90°) with a 15° interval (omitting the value 0°). Additionally, we added in (green triangles and arrows) values from Table 2 for the TFMRT structure (namely, −7.8 and 28.6° from the diffraction structure as well as 5.1 and 4.7° after DFT-D geometry optimization). Figure 10 shows a plot of the energetics around the equilibrium position upon changing the C4–NH–C5–C6 torsional angle. The red area represents the region where the total relative energy was higher than 35 kJ/mol. Both crystallographically observed torsional angle values are found in the energetic minima; however, the 28.6° torsion angle is more preferred by 2.6 kJ/mol to the −7.8° torsion angle. Note that the DFT-D optimization of the full crystal structure gives the lowest energy point at a torsion angle of 5.1° (see Table 2 and Table S2). The energy change in the wider range between −30 and 45° of the C4–NH–C5–C6 value is lower than 10 kJ/mol, with this value corresponding to an upper limit on typically experimentally accessible rotational barriers, as assessed by literature studies that report on discrete DFT calculations.79−82 Note that such DFT calculations only probe the thermodynamics (i.e., equivalent to 0 K) and temperature-dependent kinetic effects are not considered.
Figure 10.

Energy around the equilibrium position corresponding to the change in DFT-calculated energy for different C4–NH–C5–C6 torsional angles in the full periodic TFMRT structure. The angles observed in the TFMRT crystal structure (namely, −7.8 and 28.6° and the 4.7 and 5.1° values after DFT-D geometry optimization) are indicated by green arrows and green triangles. Energy values of 109.9, 196.4, and 115.9 kJ/mol were calculated for torsional angles of −75, −60, and −45°, respectively; thus, the red area is labeled as restricted. Blue lines linking the symbols are included as a guide to the eye.
In addition to the DFT calculations that explored the energetic barrier corresponding to changing the C4–NH–C5–C6 torsional angle, as presented in Figure 10, this section presents 2D 1H–13C PISEMA MAS NMR spectra by which the changes in the 13C–1H dipolar couplings due to motion of the phenyl rings are quantitatively determined.
Solid-state NMR spectroscopy offers a rich palette of methodological approaches that allows the study of molecular motion on different time scales. Usually, the choice of technique is correlated with the nature of dynamic processes. In our case, we find 2D PISEMA MAS experiments83 to be a useful diagnostic tool. This is a well-established solid-state NMR method to measure 13C–1H dipolar couplings and probe dynamic processes on the microsecond time scale.84,85 The splitting between the singularities in the F1 dimension of 2D PISEMA MAS spectra reflects the dipolar coupling between the specific carbon and closely located protons. According to the equation D = −(μ0ℏ/8π2)(γiγj)/rij3, the dipolar coupling constant for the rigid limit for a 13C–1H distance equal to 1.09 Å is 23.3 kHz. The experimentally measured splitting values are smaller than the calculated coupling values because the observed splitting is reduced by a scaling factor.86 For the PISEMA MAS NMR experiment, the exact Hartmann–Hahn matching condition yields a scaling factor of 0.577 (cos 54.7°), and the expected splitting value is ca. 13.4 kHz (23.3 kHz × 0.577).87 Motional processes can be quantitatively probed by measuring the reduction in the dipolar coupling in comparison to the rigid limit.88−90
Figure 11a,b presents 2D 1H–13C PISEMA MAS NMR spectra for TFMRT carried out at 40 and 20 °C, respectively. For comparison, spectra obtained for l-tyrosine hydrochloride are superimposed, for which previous studies91,92 have shown that the order parameter for the rigid aromatic moiety at these temperatures is close to unity: in Figure 11, the splitting for l-tyrosine hydrochloride is observed to be 13.0 kHz. The observed splittings for the well-separated C-6/6′ aromatic resonances of TFMRT are 6.8 kHz at 40 °C (Figure 11a), increasing to 10.0 kHz as the sample was cooled to 20 °C (Figure 11b): i.e., corresponding to a scaling compared to the rigid limit of 6.8/13.4 = 51% at 40 °C and 10.0/13.4 = 75% at 20 °C. A simple ring flip corresponds to a scaling by ∼60%.88,90 As such, the scaling by 75% at 20 °C can be interpreted as a low-amplitude wobbling of the aromatic ring that is consistent with the broad energy minimum in Figure 10, while the scaling by 51% at 40 °C corresponds to a phenyl flip motion, where there is sufficient energy to overcome the energy barrier in Figure 10. It is interesting to compare the observed values with those in two previous studies by Pawlak et al., specifically, for molecular rotors containing 1,4-diethynylphenylene connected by alkynylene moieties in a steroidal framework93 as well as Tyr-Ala-Phe tripeptides with varying alanine residue stereochemistry (L and D).94 The scalings for dynamic CH aromatic resonances exhibiting ring flips were in the range 31–51% for 1 and 2b in ref (94) and 2A in ref (93), while a scaling of 70% corresponding to small-amplitude wobbling was also observed for 2B in ref (93). Scalings in the range 89–97% were observed for 2a in ref (94) as well as for 1A, 1B, and 1I in ref (93).
Figure 11.
Aromatic region of 2D 1H–13C PISEMA MAS NMR spectra recorded at 40 °C (a) and 20 °C (b) with a spinning rate of 13 kHz and a 1H Larmor frequency of 600.1 MHz for TFM (blue) compared to spectra for l-tyrosine hydrochloride (green). 1H–13C CP MAS spectra recorded with a contact time of 2 ms are shown at the top. The orange rectangles distinguish the relevant C-6/6′ signals of TFM.
2.5. CSP Validation of Experimentally Obtained Structures
On comparison of the crystal lattice energy computed by means of the DFT-D method, the energy differences between the fully optimized TFMLT and TFMRT structures are as much as 2.4 kJ/mol and as small as 0.75 kJ/mol, considering the difference between TFMLT and TFMRT (high-occupancy sites in the crystal structure) after geometry optimization of all atomic positions and unit cell parameters (see Table S2). Discerning structures separated by 2.4 kJ/mol or less is challenging for a CSP strategy and can be considered as a test of method sensitivity and reliability. Another challenge is the high similarity of packing of both crystal structures that was stressed in section 2.2 (see Figure 7). The RMSD values as provided by the COMPACK algorithm for the atomic positions are valuable because they determine the limit when the structure obtained from the CSP step can be accepted as a measure of the equivalence to the specific experimental polymorph.3
After an examination of both TFM crystal structures, the CSP methodology was applied. The procedure included a series of computations using CrystalPredictor as well as CrystalOptimizer algorithms. Further technical details regarding the CSP search are stated in Experimental and Computational Procedures. Extensive calculations with Z′ = 1 and Z′ = 2 among the 20 most common space groups (according to the CSD database)95 were performed.
Figure 12 presents CSP plots of lattice energy versus density for the lowest energy structures within a 10 kJ/mol energy window (see also Tables S5 and S6), noting that a recent analysis of over 1000 experimentally determined crystal structures, including over 500 polymorphs of organic molecules, concluded that, in 95% of the cases, the difference in energy between experimentally observed polymorphs is less than 7.2 kJ/mol.96 Most of the ca. 200 (Z′ = 1) and ca. 150 (Z′ = 2) crystal structures generated within 10 kJ/mol of the global minimum have the same lattice symmetry as the experimental form (P1̅). The Z′ = 2 search dominates the lower energy and higher density region, which shows that this type of crystal is preferred as long as we only consider thermodynamics: i.e., corresponding to 0 K. Moreover, this is consistent with the experimental observation of the conversion of TFMRT into the more stable low-temperature form (TFMLT) when the temperature is reduced. All of the obtained structures were compared to the experimental structures by using the COMPACK algorithm (as introduced above; see Figure 7). There was only one structure (no. 1) from the Z′ = 1 search that we can consider as matching to the TFMRT experimental form on the basis of the RMSD15 criteria described above. Unfortunately, none of the results for the Z′ = 2 search were more similar to the TFMLT than for the similarity obtained between TFMLT and TFMRT forms. The difficulties in this matter are probably because of the fact that both forms are extremely similar to each other, which requires more computationally demanding methods to distinguish them.
Figure 12.
CSP plots for Z′ = 1 (green triangles) and Z′ = 2 (blue circles) searches for TFM for (a) the 10 kJ/mol energy window and (b) an enlarged region corresponding to the 10 lowest energy structures. The orange circles indicate the CSP “parent” structures best matched to the experimental forms after a subsequent CSP-CASTEP step (see Figure 13 and Tables S5 and S6).
As a final evaluation of the CSP-generated structures, DFT-D calculations (implemented in the CASTEP program) were applied—this is referred to here as CSP-CASTEP. The calculations were focused on the optimization of the 10 lowest energy structures obtained from each CSP step (Figure 12b). All atomic coordinates as well as unit cell parameters were allowed to vary. The change in energetic rankings for the five lowest energy CSP-CASTEP structures including the “parent” structures are shown in Figure 13 (see also Tables S5 and S6). The structures obtained after the CSP-CASTEP step were compared to the experimental structures. It was found that the lowest energy structure obtained after CSP-CASTEP for the Z′ = 2 search and the second lowest energy structure for the Z′ = 1 search (CSP no. 1) match very well the experimental forms: after DFT-D (CASTEP) geometry optimization, the RMSD15 values are only 0.276 and 0.222 Å for the TFMRT and TFMLT polymorphs, respectively. These CSP structures are circled in Figure 12b. It is evident that both CSP-CASTEP “parent” structures almost overlay each other in the CSP plot. On the other hand, this is not surprising if we consider the close similarity of the polymorphs TFMLT and TFMRT diffraction structures after DFT-D geometry optimization (see also the above discussion of Table S2).
Figure 13.
Reordering with respect to energy upon DFT-D (CASTEP) geometry optimization for the (a) Z′ = 1 and (b) Z′ = 2 CSP searches for TFM. The asterisk indicates the CSP "parent" structure best matched to the experimental form in each case. Both graphs have a consistent zero-point energy level.
Additionally, COMPACK results and energy differences for the 10 lowest energy structures after DFT-D geometry optimization for Z′ = 1 and Z′ = 2 are given in Tables S5 and S6, respectively. It is clearly seen that a significant reordering of energetic preferences has occurred. These changes are much larger for the Z′ = 2 search than for the Z′ = 1 search. For the Z′ = 1 search, the structure that is best matched to experiment is finally placed as the second in the CSP-CASTEP energetic ranking. However, the difference is only 0.03 kJ/mol above the most preferred structure, which is well within the accuracy limit of the computational methods.97−99 It is also not meaningless that the Z′ = 1 search was burdened by the fact that the molecular disorder observed at room temperature is not modeled by the applied computational methods. The Z′ = 2 search is much less ambiguous and indicates structure no. 4 as the most energetically stable (considering only thermodynamics: i.e., at 0 K corresponding to the zero-point vibration energy level), which is best matched to the experimental TFMLT structure. Since there are a couple of structures within the threshold of 2 kJ/mol from the lowest energy structure, which is a good estimation of the accuracy of the CSP-CASTEP method,97−99 we cannot eliminate the possibility of formation of other unknown yet experimental polymorphs. It is, however, unlikely if we consider the significant experimental efforts devoted to experimental polymorph screening in this study.
3. Conclusions
In this work for the first time we present an X-ray diffraction structure and characterization by MAS NMR of a new, room-temperature polymorph of TFM (labeled as TFMRT). We observe that the low-temperature TFM polymorph (TFMLT) recently reported by Gunnam et al. undergoes a thermal transition at −40 °C to the TFMRT polymorph. The phase transition is clearly visible on the DSC plot and is reversible. A crystal mounted on the goniometer head can be cooled and warmed without cracking, producing diffraction patterns characteristic for TFMLT and TFMRT lattices, respectively. This reversible process (TFMLT ↔ TFMRT) occurs with a change in Z′ value (from 2 to 1) while the crystallographic system is preserved (triclinic). The two forms have different lattice parameters and show different reflection patterns. Every second h reflection corresponding to lattice direction a in TFMLT disappeared on TFMRT diffraction images. We emphasize that both structures have been determined on the basis of diffraction experiments conducted on the same species of crystal.
Our consideration of differences between TFMLT and TFMRT focuses on changes in the orientation of the aromatic ring associated with the C4–NH–C5–C6 torsional angle. In TFMLT, there are two torsion angles of 4.5 and 31.9° for the two Z′ = 2 molecules, while in the room-temperature structure, there is disorder that is modeled with ∼50% occupancy between torsion angles of −7.8 and 28.6°. These observations are consistent with the broad energy minimum observed by DFT calculations for changes in the C4–NH–C5–C6 torsional angle. PISEMA solid-state NMR experiments show a reduction in the C–H dipolar coupling in comparison to the static limit for the aromatic CH moieties of 75% and 51% at 20 and 40 °C, respectively, that is indicative of ring flips at the higher temperature. The difference in crystal lattice energy computed by means of DFT-D between TFMLT and TFMRT is up to 2.4 kJ/mol. Such a small difference supports the experimentally observed reversible transformation between both forms.
The TFMRT polymorph also exhibits molecular disorder of the CF3 group, and the disorder in the two parts of the molecule hence may be correlated. The transformation from TFMLT and TFMRT appears to be driven by an entropic preference similar to that in a previous study.100 It is curious to compare the case here for TFM, where disorder in TFMRT is associated with a change in comparison to TFMLT in the Z′ value from 2 to 1, with that presented by Szell et al., where disorder leads to a change in Z′ value from 1 to 2.101
It could be thought that TFMLT and TFMRT are not distinct polymorphs but rather the same form with disorder above −40 °C. However, there are clear differences in the calculated PXRD patterns that cannot be explained simply by thermal expansion, and the 13C chemical shifts as observed by solid-state NMR are different for all sites, not just those exhibiting disorder. Moreover, the presented crystal structure prediction procedure that does not consider disorder in an output form described both experimental TFM polymorphs in the energetic global minimum region. In conclusion, our study shows the power of combining experiment, namely DSC, X-ray diffraction, and MAS NMR, with DFT calculation and CSP to probe and understand the solid-state landscape, and in particular the role of dynamics, for pharmaceutical molecules.
4. Experimental and Computational Procedures
4.1. Synthesis and Crystallization of TFM
All investigated compounds were synthesized by the Jinan YSPharma Biotechnology Co. Ltd. (Jinan, China). The purity of the obtained compounds was >98%, as confirmed by HPLC and solution-state NMR (see Figure S3 in the Supporting Information). Crystals of TFM were obtained by crystallization from a dichloromethane solution at 5 °C.
4.2. Single-Crystal X-ray Diffraction of TFM
Single-crystal X-ray diffraction experiments of a single crystal of TFM were carried out at 100 and 292 K using an Oxford SuperNova single-crystal diffractometer with microsource Cu Kα radiation (λ = 1.5418 Å) and a Titan detector Oxford Diffraction (Agilent Technologies, Yarnton, U.K.) equipped with a 800 Cryostream low-temperature unit (Oxford Cryosystems, Oxford, U.K.).
Diffraction data collection, cell refinement, data reduction, and absorption correction were performed using the CrysAlis PRO software (Oxford Diffraction). Structures were solved by the direct method SHELXS102 and then refined using the full-matrix least-squares method SHELXL 2015103 implemented in the OLEX2 package.104 In all of the crystal structures, the non-hydrogen atoms were present in the direct-methods solution.
4.3. Solid-State NMR Spectroscopy of TFM
Cross-polarization magic angle spinning (CP MAS) NMR and one-pulse 1H MAS experiments were performed, except where otherwise stated, on a 600 MHz Avance III spectrometer, operating at 600.13, 564.68, 150.90, and 60.81 MHz for 1H, 19F, 13C, and 15N, respectively, equipped with a HX MAS probe head using 4, 2.5, and 1.3 mm ZrO2 rotors. Except where otherwise stated, a recycle delay of 45 s was used. Additional 1H → 13C CP MAS NMR measurements were performed on a Bruker Neo spectrometer operating at 850.23 and 213.73 MHz for 1H and 13C, respectively, equipped with an HXY probe operating in double-resonance mode using 4 mm ZrO2 rotors. A recycle delay of 60 s was used.
A sample of U-13C,15N-labeled histidine hydrochloride was used to set the Hartmann–Hahn conditions for 13C and 15N. 1H → 13C and 1H → 15N CP MAS experiments on the 600 MHz Avance III spectrometer were performed at a MAS frequency of 12 kHz with a proton 90° pulse length of 4 μs and contact times of 2 ms for 13C and 8 ms for 15N. For cross-polarization, the nutation frequency was 50 kHz for 13C as well as 15N for with a 1H ramp shape from 90% to 100% with a 1H nutation frequency of 62.5 kHz. For 13C and 15N, 3.5k and 2k data points were acquired for spectral widths of 40 and 28 kHz, respectively. 1H → 13C CP MAS experiments on a 850 MHz Bruker Neo spectrometer were performed at a MAS frequency of 12 kHz with a proton 90° pulse length of 3.5 μs and a contact time of 2 ms. For cross-polarization, the nutation frequency was 71 kHz for 13C with a 1H ramp shape from 90% to 100% with a 1H nutation frequency of 83 kHz. A total of 4k data points were acquired for a spectral width of 58.8 kHz. In all cases, a SPINAL-64 decoupling sequence105 with a 1H nutation frequency of 71.4 kHz and pulse length of 7 μs was applied (also for the PISEMA experiment described below). For recording of the 19F →13C CP MAS NMR spectra at a MAS frequency of 28 kHz, a proton 90° pulse length of 2.5 μs, and a contact time of 2 ms with a 1H ramp shape from 90% to 100% with 19F and 13C nutation frequencies of 75 and 47 kHz, respectively, were used. A total of 3.5k data points were recorded for a spectral width of 62.5 kHz. The acquisition data were collected with a decoupling sequence containing one 180° pulse per rotation period.106 In all CP experiments, the following phase cycling was employed: 1H excitation pulse y–y, 1H contact pulse x, 13C contact pulse x, x, −x, −x, y, y, −y, −y, with receiver phase x, −x, −x, x, y, −y, −y, y. A 19F single-pulse MAS spectrum was acquired with a 19F 90° pulse length of 2.5 μs, a spectral width of 250 kHz, and a time domain size of 1k data points. As a sample to setup 19F experiments, polytetrafluoroethylene (PTFE) was used.
The PISEMA MAS experiment83,87,107 was carried out with an 1H nutation frequency of 82.5 kHz in all of the experiments, and the 13C spin-lock field strengths were adjusted to the first-order sideband condition, ω13C = ω1H ± ωr. The spinning frequency was 13 kHz and was regulated to ±3 Hz by a pneumatic control unit. A total of 96 transients were coadded for each of 64 t1 FIDs, corresponding to a total experimental time at 77 h. The 2D PISEMA MAS experiments incremented the SEMA (spin exchange at the magic angle) contact time using a step of 16.28 μs, with a maximum t1 evolution time of approximately 1 ms. The phase cycling was as follows: 1H excitation pulse y–y, 1H magic angle pulse −y, 1H contact pulse −x, 13C contact pulse x, 1H SEMA pulse x, 13C SEMA pulse x, receiver x, −x. Only cosine-modulated data were collected. Thus, a real Fourier transformation was performed on the t1 data that yielded spectra with a symmetrized ω1 dimension and dipolar splitting. Since the t1 time signal increases with increasing SEMA contact time, the ω1 dimension was processed using the baseline correction mode “qfil” in the Bruker TopSpin 3.5 program software,108 which subtracted a constant intensity from the time signals prior to the Fourier transformation and yielded spectra free from the dominant zero-frequency peak that gives the 1H–13C doublet.
Fast MAS spectra were recorded with a spin rate of 62.5 kHz. The 13C–1H invHETCOR (for indirect detection of 13C) experiments were performed using the pulse sequence described by Mao et al.109 The following parameters were used: a proton 90° pulse length of 2.5 μs, a first contact time of 2 ms, a second contact time of 1 ms or 150 μs, both with a 1H ramp shape from 90% to 100%. The 1H and 13C nutation frequencies were 160 and 101 kHz, respectively, for both CP steps. The acquisition data were collected with a SWf-TPPM110,111 decoupling sequence with a 1H nutation frequency of 10 kHz and a pulse length of 50 μs.105 The phase cycling was as follows: 1H excitation pulse x, 1H first contact time pulse y, 13C first contact pulse x, first 13C pulse in z-filter block y, first 1H suppression of 12C magnetization pulse x, second 1H suppression of 12C magnetization pulse y, second 13C pulse in the z-filter block y, – y, 1H second contact pulse x, 13C second contact pulse x, receiver x, −x. The maximum evolution times were t1max = 4.2 ms and t2max = 20 ms, with 8 coadded transients averaged for each of 180 t1 FIDs, corresponding to a total experimental time of 24 h. The 1H–1H DQ-SQ experiment was performed with one rotor period of BaBa recoupling76−78 at a 1H nutation frequency of 160 kHz. A 16-step phase cycle was used to select Δp = ±2 on the DQ excitation block and Δp = −1 on the final 90° pulse, where p is the coherence order. The maximum evolution times were t1max 1.0 ms and t2max 17.2 ms, with 16 coadded transients averaged for each of 128 t1 FIDs, corresponding to a total experimental time of 42 h. The States-TPPI method was employed for sign discrimination.112
Adamantane (resonances at 38.48 and 29.46 ppm) was used as a secondary 13C chemical-shift reference from external tetramethylsilane (TMS) in all experiments.113,114 The 15N chemical shift was referenced indirectly to neat liquid ammonia by using powdered 15N-glycine as an external secondary reference at δ 34.40 ppm.114,115 The 19F chemical shift was referenced indirectly to CCl3F by using PTFE as an external secondary standard at δ −122.7 ppm.114 It is well-known that the real temperature inside the MAS rotor depends on numerous factors, mostly related to frictional effects caused by rotor spinning.116 In this work, Pb(NO3)2 as a commonly accepted solid-state NMR thermometer117 was used for temperature calibration.
4.4. QM Calculations
DFT calculations were performed with periodic boundary conditions using the CASTEP 19.11 code.57 The geometry optimizations were performed until the energy converged to within 10–7 eV using the X-ray diffraction crystal structures as an input file. For all calculations, the generalized density approximation DFT functional PBE with the MBD* dispersion correction scheme (DFT-D method) was applied,118,119 and the maximum plane wave cutoff energy was 630 eV using an ultrasoft pseudopotential.120 A comparison of the average forces remaining on the atoms after geometry optimization was carried out, varying all atoms with the unit cell parameters fixed or varying all atoms and the unit cell parameters (the convergence limit was 0.03 eV/Å). We observed average forces (given as Cartesian components) up to ca. 0.02 eV/Å. The energetic barrier calculations through changes in the C4–NH–C5–C6 torsional angle were carried out by using the DFT-D method for the full periodic system, though with the crystal lattice symmetry reduced to P1. All atomic coordinates were allowed to relax except for the selected torsional angle and unit cell parameters. In all cases, the optimization algorithm was BFSG121 and the Monkhorst–Pack grid122 of minimum sample spacing 0.07 × 2π Å–1 was used to sample the Brillouin zone. The NMR chemical shifts were computed using the gauge including projected augmented wave (GIPAW) method.55,56 The calculated NMR chemical shieldings were transformed to chemical shifts by linear regression between calculated and experimental results.
4.5. CSP Calculations
The molecule was first geometry minimized in the gas phase at the PBEPBE/6-311G(d,p) level of theory using Gaussian16,123 and flexible torsions were determined through second derivatives and finite difference perturbations. We also took into account chemical intuition and the fact that computational time increases significantly when the number of degrees of freedom increases. Local approximation models (LAMs) were therefore constructed for the TFM molecule, treating the two torsional angles describing possible rotation of the aromatic ring as well as the CF3 group as independent degrees of freedom. LAMs were constructed using a uniform grid along the one-dimensional degrees of freedom, at 30° increments. The global search was performed using CrystalPredictor II124 employing a smoothed intramolecular potential,125 with 500000 minimizations in the Z′ = 1 investigation and 1000000 in the Z′ = 2 investigation. Dispersion–repulsion contributions toward the lattice energy were estimated by using a Buckingham exp-6 function with the potential parameters for C, H–C (hydrogen attached to carbon), polar hydrogen, N, O, and F.126−130 Following analysis and clustering, CrystalOptimizer131 was used to refine the 1000 lowest energy structures in each investigation, at the same level of theory, with additional flexibility introduced (angles around torsions treated as flexible in the global search). The lattice energies reported are given per formula unit.
4.6. Other Methods (DSC, Elemental Analysis)
Differential scanning calorimetry (DSC) was recorded using a Mettler-Toledo DSC 3 with a heating/cooling rate of 5 °C min–1.
Elemental analyses of hydrogen, carbon, and nitrogen were performed using CE Instruments.
Acknowledgments
The computational resources were partially provided by the Polish Infrastructure for Supporting Computational Science in the European Research Space (PL-GRID). The project no. 04.04.00-00-4374/17-01 (Homing/2017 4/37) was carried out within the HOMING programme of the Foundation for Polish Science cofinanced by the European Union under the European Regional Development Fund. The UK 850 MHz Solid-state NMR Facility used in this research was funded by the EPSRC and BBSRC, as well as the University of Warwick including via partial funding through Birmingham Science City Advanced Materials Projects 1 and 2 supported by Advantage West Midlands (AWM) and the European Regional Development Fund (ERDF). I.S. acknowledges funding for this research from the EPSRC (EP/J014958/1, EP/J003840/1, EP/P022561/1 and EP/P020194) and the support of Eli Lilly. We thank Professor Kay Saalwächter, Halle, Germany, for helpful discussions about averaged dipolar couplings. The calculated and experimental data for this study are provided as a supporting data set from WRAP, the Warwick Research Archive Portal at http://wrap.warwick.ac.uk/152054/.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.cgd.1c00123.
.magres files for GIPAW calculations (ZIP)
Further discussion of solid-state NMR spectra, experimental X-ray powder pattern calculated diffractograms based on the experimental X-ray single crystal structures, solution-state 1H NMR spectrum, for the geometry optimization, unit cell parameters, densities and energies, and the hydrogen-bonding arrangements, GIPAW-calculated shieldings, crystal structure prediction, and the CASTEP code used during fixed torsion angle calculations (PDF)
Accession Codes
CCDC 1892916 and 1969989 contain the supplementary crystallographic data for this paper. These data can be obtained free of charge via www.ccdc.cam.ac.uk/data_request/cif, or by emailing data_request@ccdc.cam.ac.uk, or by contacting The Cambridge Crystallographic Data Centre, 12 Union Road, Cambridge CB2 1EZ, UK; fax: +44 1223 336033.
The authors declare no competing financial interest.
Supplementary Material
References
- Price S. L. Predicting Crystal Structures of Organic Compounds. Chem. Soc. Rev. 2014, 43 (7), 2098–2111. 10.1039/C3CS60279F. [DOI] [PubMed] [Google Scholar]
- Oganov A. R.Modern Methods of Crystal Structure Prediction; Wiley-VCH: Weinheim, Germany, 2010. [Google Scholar]
- Reilly A. M.; Cooper R. I.; Adjiman C. S.; Bhattacharya S.; Boese A. D.; Brandenburg J. G.; Bygrave P. J.; Bylsma R.; Campbell J. E.; Car R.; Case D. H.; Chadha R.; Cole J. C.; Cosburn K.; Cuppen H. M.; Curtis F.; Day G. M.; DiStasio R. A. Jr; Dzyabchenko A.; van Eijck B. P.; Elking D. M.; van den Ende J. A.; Facelli J. C.; Ferraro M. B.; Fusti-Molnar L.; Gatsiou C.-A.; Gee T. S.; de Gelder R.; Ghiringhelli L. M.; Goto H.; Grimme S.; Guo R.; Hofmann D. W. M.; Hoja J.; Hylton R. K.; Iuzzolino L.; Jankiewicz W.; de Jong D. T.; Kendrick J.; de Klerk N. J. J.; Ko H.-Y.; Kuleshova L. N.; Li X.; Lohani S.; Leusen F. J. J.; Lund A. M.; Lv J.; Ma Y.; Marom N.; Masunov A. E.; McCabe P.; McMahon D. P.; Meekes H.; Metz M. P.; Misquitta A. J.; Mohamed S.; Monserrat B.; Needs R. J.; Neumann M. A.; Nyman J.; Obata S.; Oberhofer H.; Oganov A. R.; Orendt A. M.; Pagola G. I.; Pantelides C. C.; Pickard C. J.; Podeszwa R.; Price L. S.; Price S. L.; Pulido A.; Read M. G.; Reuter K.; Schneider E.; Schober C.; Shields G. P.; Singh P.; Sugden I. J.; Szalewicz K.; Taylor C. R.; Tkatchenko A.; Tuckerman M. E.; Vacarro F.; Vasileiadis M.; Vazquez-Mayagoitia A.; Vogt L.; Wang Y.; Watson R. E.; de Wijs G. A.; Yang J.; Zhu Q.; Groom C. R. Report on the Sixth Blind Test of Organic Crystal Structure Prediction Methods. Acta Crystallogr., Sect. B: Struct. Sci., Cryst. Eng. Mater. 2016, 72 (4), 439–459. 10.1107/S2052520616007447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widdifield C. M.; Nilsson Lili S. O.; Broo A.; Lindkvist M.; Pettersen A.; Ankarberg A. S.; Aldred P.; Schantz S.; Emsley L. Does Z′ Equal 1 or 2? Enhanced Powder NMR Crystallography Verification of a Disordered Room Temperature Crystal Structure of a P38 Inhibitor for Chronic Obstructive Pulmonary Disease. Phys. Chem. Chem. Phys. 2017, 19 (25), 16650–16661. 10.1039/C7CP02349A. [DOI] [PubMed] [Google Scholar]
- Bardwell D. A.; Adjiman C. S.; Arnautova Y. A.; Bartashevich E.; Boerrigter S. X. M.; Braun D. E.; Cruz-Cabeza A. J.; Day G. M.; Della Valle R. G.; Desiraju G. R.; van Eijck B. P.; Facelli J. C.; Ferraro M. B.; Grillo D.; Habgood M.; Hofmann D. W. M.; Hofmann F.; Jose K. V. J.; Karamertzanis P. G.; Kazantsev A. V.; Kendrick J.; Kuleshova L. N.; Leusen F. J. J.; Maleev A. V.; Misquitta A. J.; Mohamed S.; Needs R. J.; Neumann M. A.; Nikylov D.; Orendt A. M.; Pal R.; Pantelides C. C.; Pickard C. J.; Price L. S.; Price S. L.; Scheraga H. A.; van de Streek J.; Thakur T. S.; Tiwari S.; Venuti E.; Zhitkov I. K. Towards Crystal Structure Prediction of Complex Organic Compounds – a Report on the Fifth Blind Test. Acta Crystallogr., Sect. B: Struct. Sci. 2011, 67 (6), 535–551. 10.1107/S0108768111042868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. T. A.; Hasell T.; Wu X.; Bacsa J.; Jelfs K. E.; Schmidtmann M.; Chong S. Y.; Adams D. J.; Trewin A.; Schiffman F.; Cora F.; Slater B.; Steiner A.; Day G. M.; Cooper A. I. Modular and Predictable Assembly of Porous Organic Molecular Crystals. Nature 2011, 474 (7351), 367–371. 10.1038/nature10125. [DOI] [PubMed] [Google Scholar]
- Pyzer-Knapp E. O.; Thompson H. P. G.; Schiffmann F.; Jelfs K. E.; Chong S. Y.; Little M. A.; Cooper A. I.; Day G. M. Predicted Crystal Energy Landscapes of Porous Organic Cages. Chem. Sci. 2014, 5 (6), 2235–2245. 10.1039/C4SC00095A. [DOI] [Google Scholar]
- Strong R. T.; Pickard C. J.; Milman V.; Thimm G.; Winkler B. Systematic Prediction of Crystal Structures: An Application to Sp3-Hybridized Carbon Polymorphs. Phys. Rev. B: Condens. Matter Mater. Phys. 2004, 70 (4), 045101. 10.1103/PhysRevB.70.045101. [DOI] [Google Scholar]
- Pickard C. J.; Needs R. J. Ab Initio Random Structure Searching. J. Phys.: Condens. Matter 2011, 23 (5), 053201. 10.1088/0953-8984/23/5/053201. [DOI] [PubMed] [Google Scholar]
- Martinez-Canales M.; Pickard C. J.; Needs R. J. Thermodynamically Stable Phases of Carbon at Multiterapascal Pressures. Phys. Rev. Lett. 2012, 108 (4), 045704. 10.1103/PhysRevLett.108.045704. [DOI] [PubMed] [Google Scholar]
- Henkelis S. E.; Mazur M.; Rice C. M.; Wheatley P. S.; Ashbrook S. E.; Morris R. E. Kinetics and Mechanism of the Hydrolysis and Rearrangement Processes within the Assembly-Disassembly-Organization-Reassembly Synthesis of Zeolites. J. Am. Chem. Soc. 2019, 141 (10), 4453–4459. 10.1021/jacs.9b00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilka M.; Dudenko D. V.; Hughes C. E.; Williams P. A.; Sturniolo S.; Franks W. T.; Pickard C. J.; Yates J. R.; Harris K. D. M.; Brown S. P. Ab Initio Random Structure Searching of Organic Molecular Solids: Assessment and Validation against Experimental Data. Phys. Chem. Chem. Phys. 2017, 19 (38), 25949–25960. 10.1039/C7CP04186A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantelides C. C.; Adjiman C. S.; Kazantsev A. V.. General Computational Algorithms for Ab Initio Crystal Structure Prediction for Organic Molecules. In Prediction and Calculation of Crystal Structures: Methods and Applications; Atahan-Evrenk S., Aspuru-Guzik A., Eds.; Springer International: Cham, Switzerland, 2014; Topics in Current Chemistry, pp 25–58. 10.1007/128_2013_497. [DOI] [PubMed] [Google Scholar]
- Duer M. J.Introduction to Solid-State NMR Spectroscopy; Blackwell: Oxford, U.K., and Malden, MA, USA, 2004. [Google Scholar]
- Ashbrook S. E.; Griffin J. M.; Johnston K. E. Recent Advances in Solid-State Nuclear Magnetic Resonance Spectroscopy. Annu. Rev. Anal. Chem. 2018, 11 (1), 485–508. 10.1146/annurev-anchem-061417-125852. [DOI] [PubMed] [Google Scholar]
- Harris R. K. NMR Studies of Organic Polymorphs & Solvates. Analyst 2006, 131 (3), 351. 10.1039/b516057j. [DOI] [PubMed] [Google Scholar]
- Pindelska E.; Sokal A.; Kolodziejski W. Pharmaceutical Cocrystals, Salts and Polymorphs: Advanced Characterization Techniques. Adv. Drug Delivery Rev. 2017, 117, 111–146. 10.1016/j.addr.2017.09.014. [DOI] [PubMed] [Google Scholar]
- Virtanen T.; Maunu S. L. Quantitation of a Polymorphic Mixture of an Active Pharmaceutical Ingredient with Solid State 13C CPMAS NMR Spectroscopy. Int. J. Pharm. 2010, 394 (1), 18–25. 10.1016/j.ijpharm.2010.04.017. [DOI] [PubMed] [Google Scholar]
- Zumbulyadis N.; Antalek B.; Windig W.; Scaringe R. P.; Lanzafame A. M.; Blanton T.; Helber M. Elucidation of Polymorph Mixtures Using Solid-State 13C CP/MAS NMR Spectroscopy and Direct Exponential Curve Resolution Algorithm. J. Am. Chem. Soc. 1999, 121 (49), 11554–11557. 10.1021/ja9929786. [DOI] [Google Scholar]
- Zencirci N.; Griesser U. J.; Gelbrich T.; Apperley D. C.; Harris R. K. Crystal Polymorphs of Barbital: News about a Classic Polymorphic System. Mol. Pharmaceutics 2014, 11 (1), 338–350. 10.1021/mp400515f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corlett E. K.; Blade H.; Hughes L. P.; Sidebottom P. J.; Walker D.; Walton R. I.; Brown S. P. Investigating Discrepancies between Experimental Solid-State NMR and GIPAW Calculation: NC–N 13C and OH···O 1H Chemical Shifts in Pyridinium Fumarates and Their Cocrystals. Solid State Nucl. Magn. Reson. 2020, 108, 101662. 10.1016/j.ssnmr.2020.101662. [DOI] [PubMed] [Google Scholar]
- Corlett E. K.; Blade H.; Hughes L. P.; Sidebottom P. J.; Walker D.; Walton R. I.; Brown S. P. 5-Amino-2-Methylpyridinium Hydrogen Fumarate: An XRD and NMR Crystallography Analysis. Magn. Reson. Chem. 2020, 58 (11), 1026–1035. 10.1002/mrc.5021. [DOI] [PubMed] [Google Scholar]
- Corlett E. K.; Blade H.; Hughes L. P.; Sidebottom P. J.; Walker D.; Walton R. I.; Brown S. P. An XRD and NMR Crystallographic Investigation of the Structure of 2,6-Lutidinium Hydrogen Fumarate. CrystEngComm 2019, 21 (22), 3502–3516. 10.1039/C9CE00633H. [DOI] [Google Scholar]
- Mann S. K.; Pham T. N.; McQueen L. L.; Lewandowski J. R.; Brown S. P. Revealing Intermolecular Hydrogen Bonding Structure and Dynamics in a Deep Eutectic Pharmaceutical by Magic-Angle Spinning NMR Spectroscopy. Mol. Pharmaceutics 2020, 17 (2), 622–631. 10.1021/acs.molpharmaceut.9b01075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhari S. R.; Griffin J. M.; Broch K.; Lesage A.; Lemaur V.; Dudenko D.; Olivier Y.; Sirringhaus H.; Emsley L.; Grey C. P. Donor–Acceptor Stacking Arrangements in Bulk and Thin-Film High-Mobility Conjugated Polymers Characterized Using Molecular Modelling and MAS and Surface-Enhanced Solid-State NMR Spectroscopy. Chem. Sci. 2017, 8 (4), 3126–3136. 10.1039/C7SC00053G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsh D. A.; Wijesekara A. V.; Carnahan S. L.; Hung I.; Lubach J. W.; Nagapudi K.; Rossini A. J. Rapid Characterization of Formulated Pharmaceuticals Using Fast MAS 1H Solid-State NMR Spectroscopy. Mol. Pharmaceutics 2019, 16 (7), 3121–3132. 10.1021/acs.molpharmaceut.9b00343. [DOI] [PubMed] [Google Scholar]
- Harris R. K.; Hodgkinson P.; Larsson T.; Muruganantham A. Quantification of Bambuterol Hydrochloride in a Formulated Product Using Solid-State NMR. J. Pharm. Biomed. Anal. 2005, 38 (5), 858–864. 10.1016/j.jpba.2005.01.052. [DOI] [PubMed] [Google Scholar]
- Brus J.; Urbanova M.; Sedenkova I.; Brusova H. New Perspectives of 19F MAS NMR in the Characterization of Amorphous Forms of Atorvastatin in Dosage Formulations. Int. J. Pharm. 2011, 409 (1–2), 62–74. 10.1016/j.ijpharm.2011.02.030. [DOI] [PubMed] [Google Scholar]
- Yarava J. R.; Nishiyama Y.; Raghothama S.; Ramanathan K. V. Conformational Investigation of Peptides Using Solid-state NMR Spectroscopy—A Study of Polymorphism of β-turn Peptides Containing Diprolines. Chem. Biol. Drug Des. 2020, 95 (3), 394–407. 10.1111/cbdd.13649. [DOI] [PubMed] [Google Scholar]
- Hong Y.; Manjunatha Reddy G. N.; Nishiyama Y. Selective Detection of Active Pharmaceutical Ingredients in Tablet Formulations Using Solid-State NMR Spectroscopy. Solid State Nucl. Magn. Reson. 2020, 106, 101651. 10.1016/j.ssnmr.2020.101651. [DOI] [PubMed] [Google Scholar]
- Hirsh D. A.; Su Y.; Nie H.; Xu W.; Stueber D.; Variankaval N.; Schurko R. W. Quantifying Disproportionation in Pharmaceutical Formulations with 35Cl Solid-State NMR. Mol. Pharmaceutics 2018, 15 (9), 4038–4048. 10.1021/acs.molpharmaceut.8b00470. [DOI] [PubMed] [Google Scholar]
- Hildebrand M.; Hamaed H.; Namespetra A. M.; Donohue J. M.; Fu R.; Hung I.; Gan Z.; Schurko R. W. 35Cl Solid-State NMR of HCl Salts of Active Pharmaceutical Ingredients: Structural Prediction, Spectral Fingerprinting and Polymorph Recognition. CrystEngComm 2014, 16 (31), 7334–7356. 10.1039/C4CE00544A. [DOI] [Google Scholar]
- Baias M.; Dumez J.-N.; Svensson P. H.; Schantz S.; Day G. M.; Emsley L. De Novo Determination of the Crystal Structure of a Large Drug Molecule by Crystal Structure Prediction-Based Powder NMR Crystallography. J. Am. Chem. Soc. 2013, 135 (46), 17501–17507. 10.1021/ja4088874. [DOI] [PubMed] [Google Scholar]
- Pinon A. C.; Rossini A. J.; Widdifield C. M.; Gajan D.; Emsley L. Polymorphs of Theophylline Characterized by DNP Enhanced Solid-State NMR. Mol. Pharmaceutics 2015, 12 (11), 4146–4153. 10.1021/acs.molpharmaceut.5b00610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caulkins B. G.; Young R. P.; Kudla R. A.; Yang C.; Bittbauer T. J.; Bastin B.; Hilario E.; Fan L.; Marsella M. J.; Dunn M. F.; Mueller L. J. NMR Crystallography of a Carbanionic Intermediate in Tryptophan Synthase: Chemical Structure, Tautomerization, and Reaction Specificity. J. Am. Chem. Soc. 2016, 138 (46), 15214–15226. 10.1021/jacs.6b08937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukami T.; Ishii T.; Io T.; Suzuki N.; Suzuki T.; Yamamoto K.; Xu J.; Ramamoorthy A.; Tomono K. Nanoparticle Processing in the Solid State Dramatically Increases the Cell Membrane Permeation of a Cholesterol-Lowering Drug, Probucol. Mol. Pharmaceutics 2009, 6 (3), 1029–1035. 10.1021/mp9000487. [DOI] [PubMed] [Google Scholar]
- Damron J. T.; Kersten K. M.; Pandey M. K.; Nishiyama Y.; Matzger A.; Ramamoorthy A. Role of Anomalous Water Constraints in the Efficacy of Pharmaceuticals Probed by 1H Solid-State NMR. ChemistrySelect 2017, 2 (23), 6797–6800. 10.1002/slct.201701547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Q. Z.; Yang F.; Can T. V.; Sergeyev I. V.; D’Addio S. M.; Jawla S. K.; Li Y.; Lipert M. P.; Xu W.; Williamson R. T.; Leone A.; Griffin R. G.; Su Y. In Situ Characterization of Pharmaceutical Formulations by Dynamic Nuclear Polarization Enhanced MAS NMR. J. Phys. Chem. B 2017, 121 (34), 8132–8141. 10.1021/acs.jpcb.7b07213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.; Xu W.; Su Y. Solid-State NMR Spectroscopy in Pharmaceutical Sciences. TrAC, Trends Anal. Chem. 2021, 135, 116152. 10.1016/j.trac.2020.116152. [DOI] [Google Scholar]
- Woollam G. R.; Neumann M. A.; Wagner T.; Davey R. J. The Importance of Configurational Disorder in Crystal Structure Prediction: The Case of Loratadine. Faraday Discuss. 2018, 211, 209–234. 10.1039/C8FD00072G. [DOI] [PubMed] [Google Scholar]
- Price S. L.; Braun D. E.; Reutzel-Edens S. M. Can Computed Crystal Energy Landscapes Help Understand Pharmaceutical Solids?. Chem. Commun. 2016, 52 (44), 7065–7077. 10.1039/C6CC00721J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlak T.; Potrzebowski M. J. Fine Refinement of Solid-State Molecular Structures of Leu- and Met-Enkephalins by NMR Crystallography. J. Phys. Chem. B 2014, 118 (12), 3298–3309. 10.1021/jp500379e. [DOI] [PubMed] [Google Scholar]
- Ashbrook S. E.; McKay D. Combining Solid-State NMR Spectroscopy with First-Principles Calculations – a Guide to NMR Crystallography. Chem. Commun. 2016, 52 (45), 7186–7204. 10.1039/C6CC02542K. [DOI] [PubMed] [Google Scholar]
- Webber A. L.; Emsley L.; Claramunt R. M.; Brown S. P. NMR Crystallography of Campho[2,3-c]Pyrazole (Z ′ = 6): Combining High-Resolution 1H-13C Solid-State MAS NMR Spectroscopy and GIPAW Chemical-Shift Calculations. J. Phys. Chem. A 2010, 114 (38), 10435–10442. 10.1021/jp104901j. [DOI] [PubMed] [Google Scholar]
- Kerr H. E.; Mason H. E.; Sparkes H. A.; Hodgkinson P. Testing the Limits of NMR Crystallography: The Case of Caffeine–Citric Acid Hydrate. CrystEngComm 2016, 18 (35), 6700–6707. 10.1039/C6CE01453D. [DOI] [Google Scholar]
- Fernandes J. A.; Sardo M.; Mafra L.; Choquesillo-Lazarte D.; Masciocchi N. X-Ray and NMR Crystallography Studies of Novel Theophylline Cocrystals Prepared by Liquid Assisted Grinding. Cryst. Growth Des. 2015, 15 (8), 3674–3683. 10.1021/acs.cgd.5b00279. [DOI] [Google Scholar]
- Hodgkinson P. NMR Crystallography of Molecular Organics. Prog. Nucl. Magn. Reson. Spectrosc. 2020, 118–119, 10–53. 10.1016/j.pnmrs.2020.03.001. [DOI] [PubMed] [Google Scholar]
- Harris R. K. NMR Crystallography: The Use of Chemical Shifts. Solid State Sci. 2004, 6 (10), 1025–1037. 10.1016/j.solidstatesciences.2004.03.040. [DOI] [Google Scholar]
- Elena B.; Pintacuda G.; Mifsud N.; Emsley L. Molecular Structure Determination in Powders by NMR Crystallography from Proton Spin Diffusion. J. Am. Chem. Soc. 2006, 128 (29), 9555–9560. 10.1021/ja062353p. [DOI] [PubMed] [Google Scholar]
- Taulelle F.Fundamental Principles of NMR Crystallography. In Encyclopedia of Magnetic Resonance; Harris R. K., Ed.; Wiley: Chichester, U.K., 2009. 10.1002/9780470034590.emrstm1003. [DOI] [Google Scholar]
- Salager E.; Stein R. S.; Pickard C. J.; Elena B.; Emsley L. Powder NMR Crystallography of Thymol. Phys. Chem. Chem. Phys. 2009, 11 (15), 2610. 10.1039/b821018g. [DOI] [PubMed] [Google Scholar]
- Bonhomme C.; Gervais C.; Babonneau F.; Coelho C.; Pourpoint F.; Azaïs T.; Ashbrook S. E.; Griffin J. M.; Yates J. R.; Mauri F.; Pickard C. J. First-Principles Calculation of NMR Parameters Using the Gauge Including Projector Augmented Wave Method: A Chemist’s Point of View. Chem. Rev. 2012, 112 (11), 5733–5779. 10.1021/cr300108a. [DOI] [PubMed] [Google Scholar]
- Brus J.; Czernek J.; Kobera L.; Urbanova M.; Abbrent S.; Husak M. Predicting the Crystal Structure of Decitabine by Powder NMR Crystallography: Influence of Long-Range Molecular Packing Symmetry on NMR Parameters. Cryst. Growth Des. 2016, 16 (12), 7102–7111. 10.1021/acs.cgd.6b01341. [DOI] [Google Scholar]
- Thureau P.; Sturniolo S.; Zilka M.; Ziarelli F.; Viel S.; Yates J. R.; Mollica G. Reducing the Computational Cost of NMR Crystallography of Organic Powders at Natural Isotopic Abundance with the Help of 13C-13C Dipolar Couplings. Magn. Reson. Chem. 2019, 57 (5), 256–264. 10.1002/mrc.4848. [DOI] [PubMed] [Google Scholar]
- Pickard C. J.; Mauri F. All-Electron Magnetic Response with Pseudopotentials: NMR Chemical Shifts. Phys. Rev. B: Condens. Matter Mater. Phys. 2001, 63 (24), 245101. 10.1103/PhysRevB.63.245101. [DOI] [Google Scholar]
- Yates J. R.; Pickard C. J.; Mauri F. Calculation of NMR Chemical Shifts for Extended Systems Using Ultrasoft Pseudopotentials. Phys. Rev. B: Condens. Matter Mater. Phys. 2007, 76 (2), 024401. 10.1103/PhysRevB.76.024401. [DOI] [Google Scholar]
- Clark S. J.; Segall M. D.; Pickard C. J.; Hasnip P. J.; Probert M. J.; Refson K.; Payne M. C. First Principles Methods Using CASTEP. Z. Kristallogr. - Cryst. Mater. 2005, 220, 567–570. 10.1524/zkri.220.5.567.65075. [DOI] [Google Scholar]
- Price S. L.; Reutzel-Edens S. M. The Potential of Computed Crystal Energy Landscapes to Aid Solid-Form Development. Drug Discovery Today 2016, 21 (6), 912–923. 10.1016/j.drudis.2016.01.014. [DOI] [PubMed] [Google Scholar]
- Li X.; Neumann M. A.; van de Streek J. The Application of Tailor-Made Force Fields and Molecular Dynamics for NMR Crystallography: A Case Study of Free Base Cocaine. IUCrJ 2017, 4 (2), 175–184. 10.1107/S2052252517001415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman J. D.; Monaco S.; Schatschneider B.; Beran G. J. O. Fragment-Based 13C Nuclear Magnetic Resonance Chemical Shift Predictions in Molecular Crystals: An Alternative to Planewave Methods. J. Chem. Phys. 2015, 143 (10), 102809. 10.1063/1.4922649. [DOI] [PubMed] [Google Scholar]
- Klotz L.; Eschborn M.; Lindner M.; Liebmann M.; Herold M.; Janoschka C.; Garrido B. T.; Schulte-Mecklenbeck A.; Gross C. C.; Breuer J.; Hundehege P.; Posevitz V.; Pignolet B.; Nebel G.; Glander S.; Freise N.; Austermann J.; Wirth T.; Campbell G. R.; Schneider-Hohendorf T.; Eveslage M.; Brassat D.; Schwab N.; Loser K.; Roth J.; Busch K. B.; Stoll M.; Mahad D. J.; Meuth S. G.; Turner T.; Bar-Or A.; Wiendl H. Teriflunomide Treatment for Multiple Sclerosis Modulates T Cell Mitochondrial Respiration with Affinity-Dependent Effects. Sci. Transl. Med. 2019, 11 (490), eaao5563. 10.1126/scitranslmed.aao5563. [DOI] [PubMed] [Google Scholar]
- Commissioner of the U.S. Food and Drug Administration; http://www.fda.gov/home (accessed Dec 2, 2019).
- Gadhave D.; Gorain B.; Tagalpallewar A.; Kokare C. Intranasal Teriflunomide Microemulsion: An Improved Chemotherapeutic Approach in Glioblastoma. J. Drug Delivery Sci. Technol. 2019, 51, 276–289. 10.1016/j.jddst.2019.02.013. [DOI] [Google Scholar]
- Gunnam A.; Nangia A. K. High-Solubility Salts of the Multiple Sclerosis Drug Teriflunomide. Cryst. Growth Des. 2019, 19 (9), 5407–5417. 10.1021/acs.cgd.9b00914. [DOI] [Google Scholar]
- Apperley D.; Harris R.; Hodgkinson P.. Solid-State NMR: Basic Principles & Practice, 1st ed.; Momentum Press: 2012. 10.5643/9781606503522. [DOI] [Google Scholar]
- Dudenko D. V.; Yates J. R.; Harris K. D. M.; Brown S. P. An NMR Crystallography DFT-D Approach to Analyse the Role of Intermolecular Hydrogen Bonding and π–π Interactions in Driving Cocrystallisation of Indomethacin and Nicotinamide. CrystEngComm 2013, 15 (43), 8797. 10.1039/c3ce41240g. [DOI] [Google Scholar]
- van de Streek J.; Neumann M. A. Validation of Molecular Crystal Structures from Powder Diffraction Data with Dispersion-Corrected Density Functional Theory (DFT-D). Acta Crystallogr., Sect. B: Struct. Sci., Cryst. Eng. Mater. 2014, 70 (6), 1020–1032. 10.1107/S2052520614022902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Streek J.; Neumann M. A. Validation of Experimental Molecular Crystal Structures with Dispersion-Corrected Density Functional Theory Calculations. Acta Crystallogr., Sect. B: Struct. Sci. 2010, 66 (5), 544–558. 10.1107/S0108768110031873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widdifield C. M.; Farrell J. D.; Cole J. C.; Howard J. A. K.; Hodgkinson P. Resolving Alternative Organic Crystal Structures Using Density Functional Theory and NMR Chemical Shifts. Chem. Sci. 2020, 11 (11), 2987–2992. 10.1039/C9SC04964A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm J. A.; Motherwell S. COMPACK: A Program for Identifying Crystal Structure Similarity Using Distances. J. Appl. Crystallogr. 2005, 38 (1), 228–231. 10.1107/S0021889804027074. [DOI] [Google Scholar]
- Tatton A. S.; Blade H.; Brown S. P.; Hodgkinson P.; Hughes L. P.; Lill S. O. N.; Yates J. R. Improving Confidence in Crystal Structure Solutions Using NMR Crystallography: The Case of β-Piroxicam. Cryst. Growth Des. 2018, 18 (6), 3339–3351. 10.1021/acs.cgd.8b00022. [DOI] [Google Scholar]
- Salager E.; Day G. M.; Stein R. S.; Pickard C. J.; Elena B.; Emsley L. Powder Crystallography by Combined Crystal Structure Prediction and High-Resolution 1H Solid-State NMR Spectroscopy. J. Am. Chem. Soc. 2010, 132 (8), 2564–2566. 10.1021/ja909449k. [DOI] [PubMed] [Google Scholar]
- Hofstetter A.; Balodis M.; Paruzzo F. M.; Widdifield C. M.; Stevanato G.; Pinon A. C.; Bygrave P. J.; Day G. M.; Emsley L. Rapid Structure Determination of Molecular Solids Using Chemical Shifts Directed by Unambiguous Prior Constraints. J. Am. Chem. Soc. 2019, 141 (42), 16624–16634. 10.1021/jacs.9b03908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paruzzo F. M.; Hofstetter A.; Musil F.; De S.; Ceriotti M.; Emsley L. Chemical Shifts in Molecular Solids by Machine Learning. Nat. Commun. 2018, 9 (1), 4501. 10.1038/s41467-018-06972-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkan F.; Holmes S. T.; Dybowski C. Role of Exact Exchange and Relativistic Approximations in Calculating 19F Magnetic Shielding in Solids Using a Cluster Ansatz. J. Chem. Theory Comput. 2017, 13 (10), 4741–4752. 10.1021/acs.jctc.7b00555. [DOI] [PubMed] [Google Scholar]
- Bradley J. P.; Tripon C.; Filip C.; Brown S. P. Determining Relative Proton–Proton Proximities from the Build-up of Two-Dimensional Correlation Peaks in 1H Double-Quantum MAS NMR: Insight from Multi-Spin Density-Matrix Simulations. Phys. Chem. Chem. Phys. 2009, 11 (32), 6941–6952. 10.1039/b906400a. [DOI] [PubMed] [Google Scholar]
- Brown S. P. Probing Proton–Proton Proximities in the Solid State. Prog. Nucl. Magn. Reson. Spectrosc. 2007, 50 (4), 199–251. 10.1016/j.pnmrs.2006.10.002. [DOI] [Google Scholar]
- Brown S. P. Applications of High-Resolution 1H Solid-State NMR. Solid State Nucl. Magn. Reson. 2012, 41, 1–27. 10.1016/j.ssnmr.2011.11.006. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Fortea A.; Canadell E.; Wzietek P.; Lemouchi C.; Allain M.; Zorina L.; Batail P. Nanoscale Rotational Dynamics of Four Independent Rotators Confined in Crowded Crystalline Layers. Nanoscale 2020, 12 (15), 8294–8302. 10.1039/D0NR00858C. [DOI] [PubMed] [Google Scholar]
- Simonov S.; Zorina L.; Wzietek P.; Rodríguez-Fortea A.; Canadell E.; Mézière C.; Bastien G.; Lemouchi C.; Garcia-Garibay M. A.; Batail P. Static Modulation Wave of Arrays of Halogen Interactions Transduced to a Hierarchy of Nanoscale Change Stimuli of Crystalline Rotors Dynamics. Nano Lett. 2018, 18 (6), 3780–3784. 10.1021/acs.nanolett.8b00956. [DOI] [PubMed] [Google Scholar]
- Lemouchi C.; Yamamoto H. M.; Kato R.; Simonov S.; Zorina L.; Rodríguez-Fortea A.; Canadell E.; Wzietek P.; Iliopoulos K.; Gindre D.; Chrysos M.; Batail P. Reversible Control of Crystalline Rotors by Squeezing Their Hydrogen Bond Cloud Across a Halogen Bond-Mediated Phase Transition. Cryst. Growth Des. 2014, 14 (7), 3375–3383. 10.1021/cg5002978. [DOI] [Google Scholar]
- Lemouchi C.; Mézière C.; Zorina L.; Simonov S.; Rodríguez-Fortea A.; Canadell E.; Wzietek P.; Auban-Senzier P.; Pasquier C.; Giamarchi T.; Garcia-Garibay M. A.; Batail P. Design and Evaluation of a Crystalline Hybrid of Molecular Conductors and Molecular Rotors. J. Am. Chem. Soc. 2012, 134 (18), 7880–7891. 10.1021/ja301484b. [DOI] [PubMed] [Google Scholar]
- Ramamoorthy A.; Opella S. J. Two-Dimensional Chemical Shift/Heteronuclear Dipolar Coupling Spectra Obtained with Polarization Inversion Spin Exchange at the Magic Angle and Magic-Angle Sample Spinning (PISEMAMAS). Solid State Nucl. Magn. Reson. 1995, 4 (6), 387–392. 10.1016/0926-2040(95)00054-T. [DOI] [PubMed] [Google Scholar]
- Krushelnitsky A.; Reichert D. Solid-State NMR and Protein Dynamics. Prog. Nucl. Magn. Reson. Spectrosc. 2005, 47 (1–2), 1–25. 10.1016/j.pnmrs.2005.04.001. [DOI] [Google Scholar]
- Schanda P.; Ernst M. Studying Dynamics by Magic-Angle Spinning Solid-State NMR Spectroscopy: Principles and Applications to Biomolecules. Prog. Nucl. Magn. Reson. Spectrosc. 2016, 96, 1–46. 10.1016/j.pnmrs.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu R.; Tian C.; Kim H.; Smith S. A.; Cross T. A. The Effect of Hartmann–Hahn Mismatching on Polarization Inversion Spin Exchange at the Magic Angle. J. Magn. Reson. 2002, 159 (2), 167–174. 10.1016/S1090-7807(02)00036-8. [DOI] [PubMed] [Google Scholar]
- Dvinskikh S. V.; Zimmermann H.; Maliniak A.; Sandström D. Heteronuclear Dipolar Recoupling in Liquid Crystals and Solids by PISEMA-Type Pulse Sequences. J. Magn. Reson. 2003, 164 (1), 165–170. 10.1016/S1090-7807(03)00180-0. [DOI] [PubMed] [Google Scholar]
- Hong M.; Yao X.; Jakes K.; Huster D. Investigation of Molecular Motions by Lee-Goldburg Cross-Polarization NMR Spectroscopy. J. Phys. Chem. B 2002, 106 (29), 7355–7364. 10.1021/jp0156064. [DOI] [Google Scholar]
- Lorieau J.; McDermott A. E. Order Parameters Based On 13C1H, 13C1H2 and 13C1H3 Heteronuclear Dipolar Powder Patterns: A Comparison of MAS-Based Solid-State NMR Sequences. Magn. Reson. Chem. 2006, 44 (3), 334–347. 10.1002/mrc.1773. [DOI] [PubMed] [Google Scholar]
- Achilles A.; Bärenwald R.; Lechner B.-D.; Werner S.; Ebert H.; Tschierske C.; Blume A.; Bacia K.; Saalwächter K. Self-Assembly of X-Shaped Bolapolyphiles in Lipid Membranes: Solid-State NMR Investigations. Langmuir 2016, 32 (3), 673–682. 10.1021/acs.langmuir.5b03712. [DOI] [PubMed] [Google Scholar]
- Ladizhansky V.; Vinogradov E.; van Rossum B.-J.; de Groot H. J. M.; Vega S. Multiple-Spin Effects in Fast Magic Angle Spinning Lee–Goldburg Cross-Polarization Experiments in Uniformly Labeled Compounds. J. Chem. Phys. 2003, 118 (12), 5547–5557. 10.1063/1.1517299. [DOI] [Google Scholar]
- van Rossum B.-J.; de Groot C. P.; Ladizhansky V.; Vega S.; de Groot H. J. M. A Method for Measuring Heteronuclear (1H–13C) Distances in High Speed MAS NMR. J. Am. Chem. Soc. 2000, 122 (14), 3465–3472. 10.1021/ja992714j. [DOI] [Google Scholar]
- Pawlak T.; Czajkowska-Szczykowska D.; Jastrzebska I.; Santillan R.; Seroka B.; Maj J.; Morzycki J. W.; Labra-Vázquez P.; Farfán N.; Bujacz G. D.; Potrzebowski M. J. Influence of Hydrogen/Fluorine Substitution on Structure, Thermal Phase Transitions, and Internal Molecular Motion of Aromatic Residues in the Crystal Lattice of Steroidal Rotors. Cryst. Growth Des. 2020, 20 (4), 2202–2216. 10.1021/acs.cgd.9b01179. [DOI] [Google Scholar]
- Pawlak T.; Trzeciak-Karlikowska K.; Czernek J.; Ciesielski W.; Potrzebowski M. J. Computed and Experimental Chemical Shift Parameters for Rigid and Flexible YAF Peptides in the Solid State. J. Phys. Chem. B 2012, 116 (6), 1974–1983. 10.1021/jp2111567. [DOI] [PubMed] [Google Scholar]
- Groom C. R.; Bruno I. J.; Lightfoot M. P.; Ward S. C. The Cambridge Structural Database. Acta Crystallogr., Sect. B: Struct. Sci., Cryst. Eng. Mater. 2016, 72 (2), 171–179. 10.1107/S2052520616003954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyman J.; Day G. M. Static and Lattice Vibrational Energy Differences between Polymorphs. CrystEngComm 2015, 17 (28), 5154–5165. 10.1039/C5CE00045A. [DOI] [Google Scholar]
- Brandenburg J. G.; Grimme S.. Dispersion Corrected Hartree–Fock and Density Functional Theory for Organic Crystal Structure Prediction. In Prediction and Calculation of Crystal Structures; Atahan-Evrenk S., Aspuru-Guzik A., Eds.; Springer International: Cham, Switzerland, 2013; Vol. 345, pp 1–23. 10.1007/128_2013_488. [DOI] [PubMed] [Google Scholar]
- Goerigk L.; Kruse H.; Grimme S. Benchmarking Density Functional Methods against the S66 and S66 × 8 Datasets for Non-Covalent Interactions. ChemPhysChem 2011, 12 (17), 3421–3433. 10.1002/cphc.201100826. [DOI] [PubMed] [Google Scholar]
- Grimme S.; Hansen A.; Brandenburg J. G.; Bannwarth C. Dispersion-Corrected Mean-Field Electronic Structure Methods. Chem. Rev. 2016, 116 (9), 5105–5154. 10.1021/acs.chemrev.5b00533. [DOI] [PubMed] [Google Scholar]
- Be̅rziņš A.; Hodgkinson P. Solid-State NMR and Computational Investigation of Solvent Molecule Arrangement and Dynamics in Isostructural Solvates of Droperidol. Solid State Nucl. Magn. Reson. 2015, 65, 12–20. 10.1016/j.ssnmr.2014.09.001. [DOI] [PubMed] [Google Scholar]
- Szell P. M. J.; Brown S. P.; Hughes L. P.; Blade H.; Nilsson Lill S. O. A Curious Case of Dynamic Disorder in Pyrrolidine Rings Elucidated by NMR Crystallography. Chem. Commun. 2020, 56 (90), 14039–14042. 10.1039/D0CC05236A. [DOI] [PubMed] [Google Scholar]
- Sheldrick G. M. A Short History of SHELX. Acta Crystallogr., Sect. A: Found. Crystallogr. 2008, 64 (1), 112–122. 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- Sheldrick G. M. Crystal Structure Refinement with SHELXL. Acta Crystallogr., Sect. C: Struct. Chem. 2015, 71 (1), 3–8. 10.1107/S2053229614024218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolomanov O. V.; Bourhis L. J.; Gildea R. J.; Howard J. a. K.; Puschmann H. OLEX2: A Complete Structure Solution, Refinement and Analysis Program. J. Appl. Crystallogr. 2009, 42 (2), 339–341. 10.1107/S0021889808042726. [DOI] [Google Scholar]
- Fung B. M.; Khitrin A. K.; Ermolaev K. An Improved Broadband Decoupling Sequence for Liquid Crystals and Solids. J. Magn. Reson. 2000, 142 (1), 97–101. 10.1006/jmre.1999.1896. [DOI] [PubMed] [Google Scholar]
- Liu S.-F.; Schmidt-Rohr K. High-Resolution Solid-State 13C NMR of Fluoropolymers. Macromolecules 2001, 34 (24), 8416–8418. 10.1021/ma0113576. [DOI] [Google Scholar]
- Dvinskikh S. V.; Sandström D. Frequency Offset Refocused PISEMA-Type Sequences. J. Magn. Reson. 2005, 175 (1), 163–169. 10.1016/j.jmr.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Topspin, Ver. 3.5; Bruker Biospin: Karlsruhe, Germany.
- Mao K.; Wiench J. W.; Lin V. S.-Y.; Pruski M. Indirectly Detected Through-Bond Chemical Shift Correlation NMR Spectroscopy in Solids under Fast MAS: Studies of Organic–Inorganic Hybrid Materials. J. Magn. Reson. 2009, 196 (1), 92–95. 10.1016/j.jmr.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Thakur R. S.; Kurur N. D.; Madhu P. K. Swept-Frequency Two-Pulse Phase Modulation for Heteronuclear Dipolar Decoupling in Solid-State NMR. Chem. Phys. Lett. 2006, 426 (4–6), 459–463. 10.1016/j.cplett.2006.06.007. [DOI] [Google Scholar]
- Vinod Chandran C.; Madhu P. K.; Kurur N. D.; Bräuniger T. Swept-Frequency Two-Pulse Phase Modulation (SWf-TPPM) Sequences with Linear Sweep Profile for Heteronuclear Decoupling in Solid-State NMR. Magn. Reson. Chem. 2008, 46 (10), 943–947. 10.1002/mrc.2285. [DOI] [PubMed] [Google Scholar]
- Marion D.; Ikura M.; Tschudin R.; Bax A. Rapid Recording of 2D NMR Spectra without Phase Cycling. Application to the Study of Hydrogen Exchange in Proteins. J. Magn. Reson. 1989, 85 (2), 393–399. 10.1016/0022-2364(89)90152-2. [DOI] [Google Scholar]
- Morcombe C. R.; Zilm K. W. Chemical Shift Referencing in MAS Solid State NMR. J. Magn. Reson. 2003, 162 (2), 479–486. 10.1016/S1090-7807(03)00082-X. [DOI] [PubMed] [Google Scholar]
- Harris R. K.; Becker E. D.; Cabral de Menezes S. M.; Goodfellow R.; Granger P. NMR Nomenclature. Nuclear Spin Properties and Conventions for Chemical Shifts(IUPAC Recommendations 2001). Pure Appl. Chem. 2001, 73 (11), 1795–1818. 10.1351/pac200173111795. [DOI] [PubMed] [Google Scholar]
- Bertani P.; Raya J.; Bechinger B. 15N Chemical Shift Referencing in Solid State NMR. Solid State Nucl. Magn. Reson. 2014, 61–62, 15–18. 10.1016/j.ssnmr.2014.03.003. [DOI] [PubMed] [Google Scholar]
- Bielecki A.; Burum D. P. Temperature Dependence of 207Pb MAS Spectra of Solid Lead Nitrate. An Accurate, Sensitive Thermometer for Variable-Temperature MAS. J. Magn. Reson., Ser. A 1995, 116 (2), 215–220. 10.1006/jmra.1995.0010. [DOI] [Google Scholar]
- Hronský V. Measurement of Sample Temperatures and Temperature Gradients in Magic-Angle Spinning NMR. Acta Electrotech. Inform. 2013, 13 (1), 95. 10.2478/aeei-2013-0021. [DOI] [Google Scholar]
- McNellis E. R.; Meyer J.; Reuter K. Azobenzene at Coinage Metal Surfaces: Role of Dispersive van Der Waals Interactions. Phys. Rev. B: Condens. Matter Mater. Phys. 2009, 80, 205414. 10.1103/PhysRevB.80.205414. [DOI] [Google Scholar]
- Ambrosetti A.; Reilly A. M.; DiStasio R. A.; Tkatchenko A. Long-Range Correlation Energy Calculated from Coupled Atomic Response Functions. J. Chem. Phys. 2014, 140, 18A508. 10.1063/1.4865104. [DOI] [PubMed] [Google Scholar]
- Vanderbilt D. Soft Self-Consistent Pseudopotentials in a Generalized Eigenvalue Formalism. Phys. Rev. B: Condens. Matter Mater. Phys. 1990, 41 (11), 7892–7895. 10.1103/PhysRevB.41.7892. [DOI] [PubMed] [Google Scholar]
- Nocedal J.; Wright S. J.. Numerical Optimization; Springer Science+Business Media: New York, NY, 2006. [Google Scholar]
- Monkhorst H. J.; Pack J. D. Special Points for Brillouin-Zone Integrations. Phys. Rev. B 1976, 13 (12), 5188–5192. 10.1103/PhysRevB.13.5188. [DOI] [Google Scholar]
- Frisch M. J.; Trucks G. W.; Schlegel H. B.; Scuseria G. E.; Robb M. A.; Cheeseman J. R.; Scalmani G.; Barone V.; Petersson G. A.; Nakatsuji H.; Li X.; Caricato M.; Marenich A. V.; Bloino J.; Janesko B. G.; Gomperts R.; Mennucci B.; Hratchian H. P.; Ortiz J. V.; Izmaylov A. F.; Sonnenberg J. L.; Williams; Ding F.; Lipparini F.; Egidi F.; Goings J.; Peng B.; Petrone A.; Henderson T.; Ranasinghe D.; Zakrzewski V. G.; Gao J.; Rega N.; Zheng G.; Liang W.; Hada M.; Ehara M.; Toyota K.; Fukuda R.; Hasegawa J.; Ishida M.; Nakajima T.; Honda Y.; Kitao O.; Nakai H.; Vreven T.; Throssell K.; Montgomery J. A. Jr.; Peralta J. E.; Ogliaro F.; Bearpark M. J.; Heyd J. J.; Brothers E. N.; Kudin K. N.; Staroverov V. N.; Keith T. A.; Kobayashi R.; Normand J.; Raghavachari K.; Rendell A. P.; Burant J. C.; Iyengar S. S.; Tomasi J.; Cossi M.; Millam J. M.; Klene M.; Adamo C.; Cammi R.; Ochterski J. W.; Martin R. L.; Morokuma K.; Farkas O.; Foresman J. B.; Fox D. J.. Gaussian 16 Rev. A.01; Gaussian Inc.: Wallingford, CT, 2016.
- Habgood M.; Sugden I. J.; Kazantsev A. V.; Adjiman C. S.; Pantelides C. C. Efficient Handling of Molecular Flexibility in Ab Initio Generation of Crystal Structures. J. Chem. Theory Comput. 2015, 11 (4), 1957–1969. 10.1021/ct500621v. [DOI] [PubMed] [Google Scholar]
- Sugden I. J.; Adjiman C. S.; Pantelides C. C. Accurate and Efficient Representation of Intramolecular Energy in Ab Initio Generation of Crystal Structures. II. Smoothed Intramolecular Potentials. Acta Crystallogr., Sect. B: Struct. Sci., Cryst. Eng. Mater. 2019, 75 (3), 423–433. 10.1107/S2052520619005778. [DOI] [PubMed] [Google Scholar]
- Williams D. E.; Cox S. R. Nonbonded Potentials for Azahydrocarbons: The Importance of the Coulombic Interaction. Acta Crystallogr., Sect. B: Struct. Sci. 1984, 40 (4), 404–417. 10.1107/S010876818400238X. [DOI] [Google Scholar]
- Coombes D. S.; Price S. L.; Willock D. J.; Leslie M. Role of Electrostatic Interactions in Determining the Crystal Structures of Polar Organic Molecules. A Distributed Multipole Study. J. Phys. Chem. 1996, 100 (18), 7352–7360. 10.1021/jp960333b. [DOI] [Google Scholar]
- Beyer T.; Price S. L. Dimer or Catemer? Low-Energy Crystal Packings for Small Carboxylic Acids. J. Phys. Chem. B 2000, 104 (12), 2647–2655. 10.1021/jp9941413. [DOI] [Google Scholar]
- Cox S. R.; Hsu L.-Y.; Williams D. E. Nonbonded Potential Function Models for Crystalline Oxohydrocarbons. Acta Crystallogr., Sect. A: Cryst. Phys., Diffr., Theor. Gen. Crystallogr. 1981, 37 (3), 293–301. 10.1107/S0567739481000703. [DOI] [Google Scholar]
- Williams D. E.; Houpt D. J. Fluorine Nonbonded Potential Parameters Derived from Crystalline Perfluorocarbons. Acta Crystallogr., Sect. B: Struct. Sci. 1986, 42 (3), 286–295. 10.1107/S010876818609821X. [DOI] [Google Scholar]
- Kazantsev A. V.; Karamertzanis P. G.; Adjiman C. S.; Pantelides C. C. Efficient Handling of Molecular Flexibility in Lattice Energy Minimization of Organic Crystals. J. Chem. Theory Comput. 2011, 7 (6), 1998–2016. 10.1021/ct100597e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.