Abstract
Protein-ligand binding prediction has extensive biological significance. Binding affinity helps in understanding the degree of protein-ligand interactions and is a useful measure in drug design. Protein-ligand docking using virtual screening and molecular dynamic simulations are required to predict the binding affinity of a ligand to its cognate receptor. Performing such analyses to cover the entire chemical space of small molecules requires intense computational power. Recent developments using deep learning have enabled us to make sense of massive amounts of complex data sets where the ability of the model to “learn” intrinsic patterns in a complex plane of data is the strength of the approach. Here, we have incorporated convolutional neural networks to find spatial relationships among data to help us predict affinity of binding of proteins in whole superfamilies toward a diverse set of ligands without the need of a docked pose or complex as user input. The models were trained and validated using a stringent methodology for feature extraction. Our model performs better in comparison to some existing methods used widely and is suitable for predictions on high-resolution protein crystal (⩽2.5 Å) and nonpeptide ligand as individual inputs. Our approach to network construction and training on protein-ligand data set prepared in-house has yielded significant insights. We have also tested DEELIG on few COVID-19 main protease-inhibitor complexes relevant to the current public health scenario. DEELIG-based predictions can be incorporated in existing databases including RSCB PDB, PDBMoad, and PDBbind in filling missing binding affinity data for protein-ligand complexes.
Keywords: Binding affinity, protein-ligand binding, supervised learning, convolutional neural networks, deep learning, PDB, drug discovery
Introduction
Proteins are a diverse class of dynamic macromolecular structures in living organisms and are essential for the biochemistry and physiology of the organism. Proteins may bind to other proteins, peptides, nucleic acids, and other nonpeptide ligands with varying affinities thus fulfilling various functional roles. Determining binding affinity between a protein-ligand complex, typically quantified in terms of inhibition constant (Ki), dissociation constant (Kd), changes in free energy measures (ΔG, ΔH, and IC50), helps in understanding interaction strength, reaction mechanism, and kinetics of the reaction, especially when experimental approaches may not be feasible and has applications in drug development and pharmacology. 1
Although current methods such as flexible docking address several limitations of rigid docking, various problems such including mode of binding, protonation states of charged residues, solvent, and entropic effects still persist. 2 Classical prediction methods to score free binding energies of small ligands to biological macromolecules such as MM/GBSA and MM/PBSA typically rely on molecular dynamic simulations for calculations and aid in-silico docking and virtual screening as well as experimental approaches. However, there is a trade-off between computational resources and accuracy. 3
With a recent shift toward the use of machine-learning and deep-learning based methods in the field of structural biology, making biologically significant predictions using regression and “learning” intrinsic patterns in a complex plane of available data has led to resource-optimal predictions without compromising on accuracy. Deep learning has been known to learn representations and patterns in complex data forms. Our aim was to apply deep learning to predict binding affinity of protein-nonpeptide ligand interaction without the need of a docked pose as input.
Convolutional neural networks (CNN) are deep neural networks that use an input layer, output later as well as convolutional hidden layer(s). The first CNN was incorporated by LeCunn in 1998, 4 the connectivity pattern of which was inspired by the elegant experiments of Hubert and Weisel on the mammalian visual cortex in the 1960s. 5 With the growing technical advancements and massive amounts of data, CNNs have emerged popular in biological fields in the recent decade with various applications. 6
In our study, we have used CNNs to provide a quantitative estimate of protein-ligand binding using various sets of features corresponding to protein and ligand, respectively, by finding spatial relationships among the data without using docked poses as user input. Our approach was validated using ligand-bound complexes from kinases superfamily in the protein data bank (PDB). Kinases belong to a class of enzymes required for substrate-dependent phosphorylation. They are represented across diverse cellular functions like signaling, differentiation, and glycolysis. 7 We have also tested our model on COVID-19 main protease 8 of the novel coronavirus strain complexed with various inhibitors of which binding affinities have not been predicted or experimentally determined so far.
Materials and Methods
Novel data set: raw data
The raw data for our novel database was obtained from the RCSB PDB 9 database, where the following were selected as the query parameters.
Chain type: Protein Chain, No DNA or RNA or DNA/RNA Hybrid.
Binding affinity: Kd or Ki value present.
Chemical components: Has ligand (s)
X-ray crystallography method: Resolution up to 2.5 A.
These criteria resulted in a list of 5464 protein PDB IDs, 2568 complexed ligand(s) and corresponding binding affinity values. The search results include the structures present in PDB, PDBbind 10 , Binding MOAD11-12, and scPDB 13 for its results.
Initial raw data database created contained protein structures in PDB format, protein sequences in FASTA format, ligands in SDF format, and binding affinity values of corresponding protein-ligand pairs for 5464 complexes.
Data set refinement
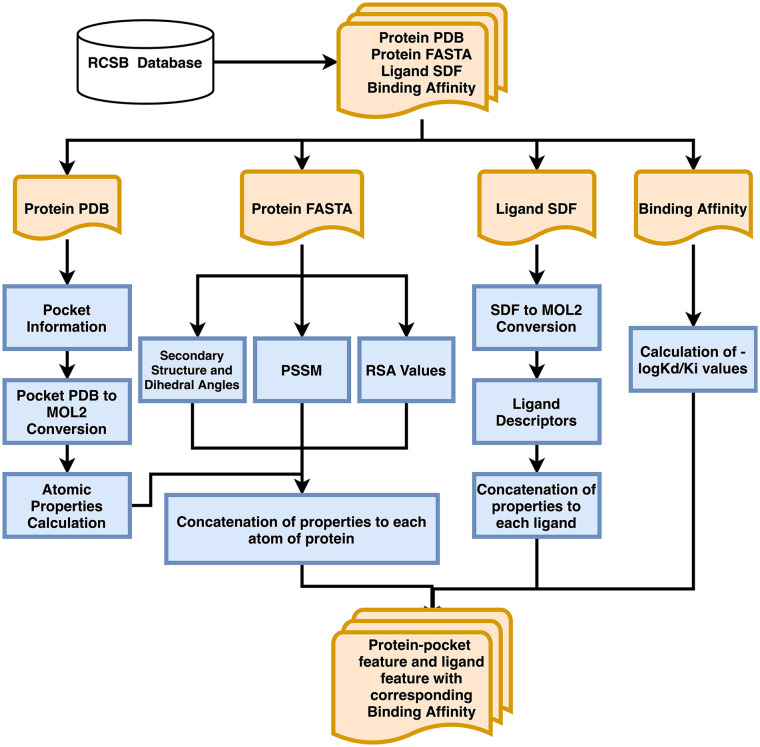
The PDB, FASTA, and SDF files filtered were further processed to refine our novel data set, as shown in Figure 1. Protein-ligand complexes were 5464 in number and corresponded to 29 650 complex unique chain-ligand pairs (SM_File3). Binding affinity values were obtained from the RCSB database and protein chain-ligand pairs with corresponding binding affinity as 0 were discarded to reduce statistical errors. This narrowed down the total complexes to 4750 protein-ligand pairs.
Figure 1.
Feature extraction pipeline.
PSSM indicates Position-Specific Scoring Matrix.
Pocket information was extracted from the protein using Ghecom 14 and converted to MOL2 format using Chimera, 15 which narrowed our dataset from 4699 pocket-ligand pairs to 4286 pocket-ligand pairs.
We discarded other protein-ligand pairs with missing PSSM profiles, secondary structure, or dihedral angle information. It resulted in a total of 4041 pocket-ligand pairs, which corresponds to 7414 pocket-ligand pairs containing unique chains.
Feature extraction
Training the deep learning network on raw information is known to result in a long time for convergence and less accuracy. We followed a conventional methodology for feature extraction and used the deep learning framework to learn the interaction between the protein pocket and ligand for their affinity prediction.
Protein-pocket features
A comprehensive 2-level feature extraction methodology, one at the atomic level and the other at the level of amino acids utilizing structural information and protein sequence respectively.
- Atomic-level (19 bits)
- a. 9 bit 1 hot or all null hot encoding for atom types: B, C, N, O, P, S, Se, halogen, and metal.
- b. 1 integer for hybridization.
- c. 1 integer representing the number of bonds with heavy atoms.
- d. 1 integer representing the number of bonds with hetero atoms.
- e. 5 bits (1 if present) encoding properties defined with SMARTS patterns: hydrophobic, aromatic, acceptor, donor, and ring.
- f. 1 float for partial charges.
- g. 1 integer to distinguish between ligand as −1 and protein as 1.
- Amino acid level (25 bits): we utilized the sequence information of protein to get more features about the protein pocket-ligand interaction.
- a. Position-Specific Scoring Matrix (PSSM): PSSM is a matrix that represents the probability of mutation at each point of the sequence. It gives a 20-bit probability for each amino acid at each location. PSSM profiles were obtained using PSI-BLAST 16 with SwissProt as subject database and E-value threshold as 0.001. Chains with less than 50 amino acids were removed from the input data set.
- b. Relative Solvent Accessibility (RSA): It is encoded by one bit of information for each amino acid that provides whether it is buried or exposed to the solvent. We set a threshold of 25% in RSA values. RSA was obtained using NACCESS. 17
- c. Secondary structure: It is encoded by 1 bit of information about the structure as coil, helix, or plate and was predicted using the DSSP. 18
- d. Dihedral angles: It is encoded by 2 bits of information with phi/psi angles of each of the amino acids and was predicted using DSSP for obtaining dihedral angles.
Ligand features
Standard ligand features were calculated for ligands in our data set using PADEL 19 and fingerprints (1-dimensional [1D], 2-dimensional [2D], and chemical fingerprints), which include hybridization, atom pair interaction, and counts of various functional groups.
We also used QikProp 20 to derive ADMET (Absorption, Distribution, Metabolism, Excretion, and Toxicity) properties, which include the physical properties, solubility, and partition coefficients.
It resulted in a 1D array of 14 716 dimensions containing the various properties of a given ligand. This is used as a feature vector representing the ligand represented in MOL2 format. A detailed list of ligand features has been provided in Supplementary Material (SM_Appendix).
Grid formation
The 3-dimensional (3D) coordinates of atoms were converted into a 3D grid of resolution 10 Å with 1 Å spacing between the 2 axes centered along the centroid of the ligand. Atoms outside each such grid were discarded. The atoms lying inside the grid were rounded up to the nearest coordinate of the grid where features of corresponding atoms that lay in the same coordinates were added up.
This resulted in projecting ligand-interacting residues into a 3D cube with features representing the atomic as well as protein-based properties of each atom of the protein pocket.
Strategies
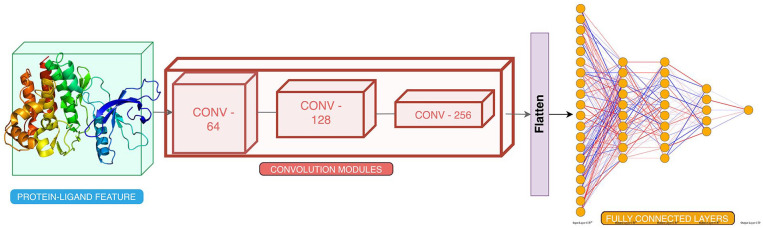
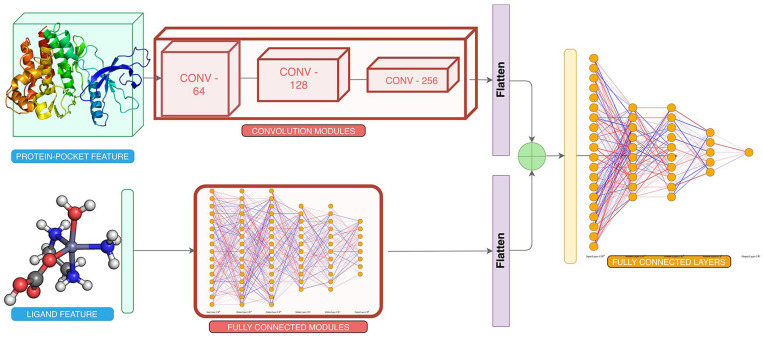
Detailed and complete block diagrams with inputs are provided in Figures 2 and 3 as well as in Supplementary Material.
Figure 2.
Training framework for atomic model. The framework is trained on 19 bits features each for protein pocket and ligand together as input.
Figure 3.
Training framework for composite model. The framework is trained on 44 bits features of protein pocket and 14 716 bits of ligand as separate inputs.
Atomic model
Preprocessing
Features were calculated at the atomic level (Methods and Materials - Feature Extraction) corresponding to each atom of an amino acid and ligand. A 19-bit vector was calculated that uniquely identified each of the atoms in the 3D coordinates of a given protein pocket and ligand complex. A 4-dimensional (4D) tensor each of size m × m × m × 19, that is, the 3 coordinates (x, y, and z) and the features, where m represents the number of atoms present in a complex, was constructed as the feature vector representing the given protein pocket-ligand. The 4D vector contains the protein-pocket features and was converted to a 3D grid using grid featurization (section “Feature Extraction”). The 3D-featurized grid is essentially a 4D tensor, where the coordinates are approximated to the points on the grid. The data set is converted to vectors and is divided into training:validation: test sets in the ratio of 80:10:10.
Architecture
Convolutional neural networks have been used to capture spatial features in an image. 21 We use CNNs to capture the interaction between ligand and protein atoms in 3D space. A network was constructed (Figure 2) with a 3D CNN of varying channel sizes of [64, 128, 256] with nonlinear activation ReLU after each layer, each 3D CNN had a filter of 5 Å cube which was used to perform convolution operations. MaxPool 22 layer acts in 3 dimensions to lower the dimension with a pool size of 2 Å cube, and batch normalization 23 layer is added after each CNN layer, which in turn decreases the training time and helps in faster convergence.
The latent features learned from the above CNN layers were then flattened and used for calculating the binding affinity of the protein pocket-ligand pair. The CNN derives the relation among the 3D coordinates and their features, which would correspond well to the binding affinities of complexes.
The features from the last CNN layer are then flattened out and passed through a fully connected neural network having the number of neurons as [5000, 2000, 500, 200] with ReLU as nonlinearity after each layer. 24 Dropout 24 is added after each layer to prevent overfitting by forcing the neural network to learn various other pathways by randomly assigning neurons to 0, 0.50 as the dropout threshold. A dense network predicts a regressive value of binding affinity, corresponding to a single neuron output. The training framework is shown in Figure 2 and a detailed layer network (Appendix_FigureA1) and network parameters have been provided in Supplementary Material (SM_Appendix).
Training
The featurized protein-pocket grid formed was rotated to all 24 combinations possible, such that the network is able to learn in an orientation invariant form. The network was trained by taking the mean square error between the predicted and actual values as a loss function. The network was optimized using Adam 25 as the optimizer with a learning rate of 1e−5 and weight decay of 0.001 for 20 epochs. The network was trained on an Nvidia Pascal GPU using Pytorch 26 as the framework.
Composite model
Preprocessing
Features were calculated at the amino acid level (Methods and Materials – Feature Extraction) and were concatenated alongside the atomic-level features (section 4.1.1) to each atom of amino acid. It results in a 44-bit vector uniquely identifying each of the atoms in the 3D coordinates of a given protein. A 4-dimensional (4D) tensor each of sizes m × m × m × 44, that is, the 3 coordinates (x, y, z) and the features, where m represents the number of atoms present in a complex, is constructed as the feature vector of the protein pocket. Here, the grid size of the binding pocket was 10*10*10*44, that is, 44 000 bits.
The 4D vector contains the protein-pocket features: it was converted to a 3D grid using grid featurization (section “Feature Extraction”). The 3D-featurized grid is essentially a 4D tensor, where the coordinates are approximated to the points on the grid. The ligands were separately featurized by calculating the ligand properties (section “Data set Refinement”), which results in a 1D tensor. The data set is converted to vectors and is divided into training:validation: test sets in ratio 80:10:10.
Architecture
A multi-input network was constructed with a 3D CNN of varying channel sizes of [64, 128, 256] with nonlinear activation ReLU after each layer, each 3D CNN had a filter of 5 Å cube which was used to perform convolution operations. We also added a MaxPool layer that acts three-dimensionally to lower dimensionality while retraining features learned after each CNN layer. A filter size of 2 Å cube was used. A batch normalization layer was added after each CNN module for faster convergence.
The ligand features were passed through the dense layers of sizes [7000, 5000, 2000] with ReLU as nonlinearity after each layer, and we also perform dropout operations after each dense layer to prevent it from overfitting. 23 This results in a latent vector representing the relevant features for each ligand.
The latent output from the CNN layers is flattened and concatenated with the latent feature vector of ligand, to create one single-feature vector of protein pocket-ligand interactions. This vector is passed through a densely connected neural network having the number of neurons as [7000, 2000, 500, 200] with ReLU as nonlinearity after each layer, and we used dropout after each layer also to prevent overfitting forcing the neural network to learn various other pathways by randomly assigning weights of neurons to zero, with 0.50 as dropout threshold. This dense network finally predicts a regressive value of binding affinity, corresponding to a single-neuron output.
The training framework is shown in Figure 3, a detailed layer network (Appendix_FigureA1), network parameters, and hyperparameters for the Composite Model are shown in the Appendix file under the Supplementary Material section (SM_Appendix).
Training
The featurized protein-pocket grid formed was rotated to all 24 combinations possible, such that the network is able to learn in an orientation invariant form.
The featurized protein pocket-ligand pair of the training set was passed through corresponding the network and trained by taking mean square error between the predicted and actual values as a loss function. The network was optimized using Adam as the optimizer with a learning rate of 1e−5 and a weight decay of 0.001. The network was trained on an Nvidia Pascal GPU using Pytorch as the framework.
Calculation of binding affinity
The predicted value of our regression-based approach is the negative natural logarithmic value of Kd or Ki. This is then converted to its antilog to obtain Kd or Ki value in nanoMolar quantity.
Performance
The accuracy is measured in terms of scores mean absolute error (MAE), root mean squared error (RMSE), and standard deviation (SD).
Time complexity
For the purpose of training and testing models, one NVIDIA Tesla P100 GPU cluster was used. Computational time taken for featurization of data set, training, and testing were 52 hr, 22 hr, and 8 min, respectively.
Additional case studies of specific protein families
Recently deposited complexes of COVID-19 main protease with various inhibitors deposited in the PDB were used for the purpose of our study (Table 4). The crystal structure complexes (PDB IDs: 5R7Y, 5R7Z, 5R82, 5R84) of the COVID-19 main protease with inhibitors ((Z45617795: N-[(5-methylisoxazol-3-yl)carbonyl]alanyl-L-valyl-N~1~-((1R,2Z)-4-(benzyloxy)-4-oxo-1-{[(3R)-2-oxopyrrolidin-3-yl]methyl)but-2-enyl)-L-leucinamide); Z1220452176: (~{N)-[2-(5-fluoranyl-1~{H)-indol-3-yl)ethyl]ethanamide); Z219104216: 6-(ethylamino)pyridine-3-carbonitrile; Z31792168: 2-cyclohexyl-~{N)-pyridin-3-yl-ethanamide)), respectively, has been recently deposited in PDB (2020; unpublished).
Table 4.
Predictions of binding affinity on COVID-19 main protease-ligand complexes.
| PDB | Ligand | −Log (Kd/Ki) | [Kd] or [Ki] (nM) |
|---|---|---|---|
| 5R7Y | Z45617795 | 11.96 | 6.39e+3 |
| 5R7Z | Z1220452176 | 7.69 | 4.57e+5 |
| 5R82 | Z219104216 | 6.12 | 2.18e+6 |
| 5R84 | Z31792168 | 8.32 | 2.43e+5 |
| 6W63 | X77 | 15.34 | 2.17e+2 |
| 7BQY | N3 | 10.38 | 3.10e+4 |
Another study has deposited the complex of the COVID-19 main protease with a broad-spectrum inhibitor X77 (N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl) ethyl]-1H-imidazole-4-carboxamide) (2020; unpublished).
To compare affinity of deoxycholate with homologous proteins of the periplasmic C-type cytochrome (Table 5), Ppc homologs PpcA (PDB: 1OS6), PpcB (PDB: 3BXU), PpcC (PDB: 3H33), PpcD (PDB: 3H4N), and PpcE (PDB: 3H34) and ligand deoxycholic acid (Pubchem CID: 222528) were gathered. DEELIG was used to predict the binding affinity of each homolog with the ligand.
Table 5.
Predictions of binding affinity on homologs of periplasmic C-type cytochrome (Ppc) family.
| Homolog | PDB ID | Prediction Kd or Ki (uM) |
|---|---|---|
| PpcA | 1OS6 | 4.512 |
| PpcB | 3BXU | 416.042 |
| PpcC | 3H33 | 835.232 |
| PpcD | 3H4N | 483.678 |
| PpcE | 3H34 | 187.157 |
Results
We have created a refined data set that represents a diverse set of ligands by having strict filtering criteria. We created a composite model and pipeline for featurization that takes into account the protein-level properties along with atomic-level properties for proteins. The deep–learning-based model predicts the binding affinity by considering protein and ligand in separate networks.
We have trained 2 models to predict the binding affinity between protein and ligand in a given complex. The first model was trained using a small set of features for protein and ligand, which were represented together in a 3D grid space. This approach has also been part of a previous study. 27 However, the previous study uses a restricted ligand set that does not involve larger ligands. Here, we have used a diverse set of ligands as one of our inputs (SM_File1). While we have used ~68% of the PDBbind core set in our training data set, this subset of the PDBbind core set incorporated constitutes only ~2.9% of our entire training data set.
The performance of the models was quantified using MAE and RMSE. It was tested on validation and testing sets which were initially divided from our data set as mentioned in the training section. Lower error corresponds to better learning capacity of the model. Standard deviation among the real and predicted values was also calculated.
The MAE, RMSE, and SD values are shown in Table 1. Training of atomic model for 35 epochs achieved MAE score of 2.84 (Table 1). In addition, the composite model achieved mean squared error (MSE) score of less than 2 by 20 epochs itself (SM_Appendix).
Table 1.
Predictions accuracy on test set of our novel data set.
| Method | MAE | RMSE | SD | PCC |
|---|---|---|---|---|
| Atomic model | 2.84 | 3.93 | 2.62 | 0.758 |
| Composite model | 2.27 | 3.07 | 2.06 | 0.794 |
Abbreviations: MAE, mean absolute error; RMSE, root mean squared error; SD, standard deviation; PCC, percent correct classification.
For the purpose of training and testing models, one NVIDIA Tesla P100 GPU cluster was used. Computational time taken to featurize our data set, training, and testing were 52 hours, 22 hours, and 8 min, respectively. This timeline involves generating features used for atomic- and protein-level properties, training the models, and testing on CASF sets.
We constructed another model that enabled us to improve on the ligand- and protein-based information. To this purpose, we used an increased feature vector size which amounted to 14 716 bits in size for ligand and 44 bits for each atom of protein.
In the case of the atomic model, training for 35 epochs yielded an MAE score of 2.84, whereas training for 20 epochs in case of the composite model yielded MAE score of 2.27 (Table 1), the training and validation curve for the process is attached in Supplementary Material (SM_Appendix). The composite model was chosen further based on input richness and relative performance. The novel framework (composite model) was further tested on the PDBbind core set and with a percent correct classification (PCC) score of 0.89, it outperformed Autodock Vina 28 (core set v13; Table 2) and other methods29-32 tested on PDBbind (core sets v2013 and 2016) (Table 2).
Table 2.
Pearson correlations coefficient on PDBbind core set.
The performance of the composite model was further evaluated using ligand-bound complexes from the kinase superfamily from PDB (SM_Appendix). The composite model outperformed the atomic model significantly and with a lower standard deviation (Table 3).
Table 3.
Predictions accuracy on kinases.
| Method | MAE | RMSE | SD | PCC |
|---|---|---|---|---|
| Atomic model | 2.48 | 3.24 | 3.11 | 0.73 |
| Composite model | 2.24 | 2.71 | 2.67 | 0.77 |
Abbreviations: MAE, mean absolute error; RMSE, root mean squared error; SD, standard deviation.
In light of the ongoing coronavirus pandemic, we tested protein-ligand complexes from the coronavirus (CoV) family. The COVID-19 main protease is a key enzyme for the novel strain of coronavirus, that is, being implicated in the pandemic. A recent study involved testing of in-vitro binding efficacy of coronavirus COVID-19 virus main protease (Mpro) with a potent irreversible synthetic inhibitor, N3. 8 However, the highly potent inhibition by N3 rendered the experimental determination of binding affinity not achievable. Using the structure of Mpro at high resolution (7BQY: 1.7 Angstrom), we have been able to predict the binding affinity of N3 to 3.1e+4 nanoMolar (Table 4). This value agrees with the observed high affinity in the course of recent experiments. 8
We used complexes of COVID-19 main protease with various inhibitors (Materials and Methods; Table 4) to predict their respective binding affinities as their experimental values have not been made available. Based on our model-based predictions, broad-spectrum inhibitor X77 scores for highest affinity followed by ligands Z45617795, N3, Z31792168, Z1220452176, and Z219104216 in the order of decreasing binding affinity (Table 4) strengthening the suitability of X77 as a potential candidate against COVID-19 virus protease.
A triheme cytochrome from the sulfur-, metal-, and radionuclide-reducing bacteria, Geobacter sulfurreducens, named PpcA crystallizes only in the presence of anionic deoxycholate 33 facilitated by its excessively positively charged binding cavity not observed in its paralogs. 34 However, its triheme paralogous counterparts PpcB, PpcC, PpcD, and PpcE do not require deoxycholate to crystallize.34,35 Our results also predict that ligand deoxycholate binds with high affinity to periplasmic C-type cytochrome A (PpcA) but not to its homologs PpcB, PpcC, PpcD, and PpcE (Table 5).
Discussion
Deep-learning-based approaches have been implemented for the prediction of binding affinity. Previously, a study used atomic-level features of complexes in a CNN-based framework for binding affinity prediction, 36 while another study used protein sequence level features in a CNN-based framework for prediction. 37 Another approach used has been to use feature learning along with gradient boosting algorithms to predict binding affinity. 38 Here, we provide a composite model that incorporates tripartite structural, sequence, and atomic-level features with those of the atomic and other chemical features of the ligand to predict the binding affinity of a putative complex.
We propose a deep-learning-based approach to predict ligand (eg, drug)—target-binding affinity using only structures of target protein (PDB format) and ligand (SDF format) as inputs. Convolutional neural networks were used to learn representations from the features extracted from these inputs and the hidden layers in the affinity prediction task. We used 2 approaches for feature extraction (Table 1; SM_Appendix)—atomic level as well as the composite level and compared their performance using the same network. We have trained on complexes from PDB across all taxa filtered as per a few starting criteria including crystal quality. Our results are validated and reflected in the performance scores. The baseline to the results of our approach is the study by Stepniewska-Dziubinska et al, 27 2018 the performance of which our study has exceeded (Results).
Our algorithm relies on certain inputs including sensitive binding cavity detection by the Ghecom algorithm (Kawabata, 2010) that uses mathematical morphology to find both deep and shallow pockets (if any) in a given protein. The coordinates of the predicted binding cavity of the protein (grid) are rotated to various combinations and are placed around the centroid of the ligand and the resultant 4D tensor is processed further for features along with the CNN (Materials and Method). Hence, ligand-bound poses are not used as input. Our data set has ~5k + complexes (SM_File2 and SM_File3) and also includes complexes that were not part of PDBbind (which is usually used to benchmark and is derived from PDB). The ligand set we have used also represents a diverse set (SM_Fi1e1) and is one of the highlights of our approach. The predictions from DEELIG can in fact help existing databases like RSCB PDB, PDBMoad, and PDBbind in filling missing binding affinity data for complexes.
We have constructed a novel data set that represents a diverse set of ligands and using a novel deep-learning-based approach, we have achieved significant improvement in the prediction of the binding affinity of protein-ligand complexes. To counter filtering and noise reduction in our data set, our data set constructed is smaller than PDBbind,39-40 but we have overcome constraints on ligand selection part of a previous study. 27 Although our data set contains 5464 complexes compared with 16 151 complexes found in PDBbind, the ligands used as part of our training include 452 unique ligands absent in PDBbind. This helps in achieving ligand diversity (SM_File3) while training the CNN model. The similarity matrix constructed from the binary fingerprints of ligands used in the data set supports our claim of improved ligand diversity in our data set.
As our data set has been derived directly from PDB, our data set also contains complexes that were not part of PDBbind, which updates itself from PDB itself. With increasingly refined methods of biophysical techniques being used to determine complex structures with attention to resolution, we have aimed to minimize noise due to technical constraints in training by filtering our data using few criteria.
We have highlighted a few examples such as complexes of kinases and viral drug targets only to reinforce the broader applicability of our approach (Tables 3 and 4). Our predictions are in line with crystallography-based experimental observations33-35 that deoxycholate is required to crystallize PpcA cytochrome but not to obtain crystals of homologs PpcB—E cytochrome (Table 5).
We have also eliminated the need to provide ligands in a complex form with protein. Thus a given protein pocket may be tested for the degree of binding for any given ligand. This can be extended to predicting potential binding partners for proteins in other superfamilies as well. It is also important to consider that docking score and pose may not reliably correlate with MM/GBSA poses. 1 DEELIG can be used to predict binding affinity for a member of any protein superfamily and a nonpeptide ligand, the docking pose of which may or may not be known. Binding affinity predictions through DEELIG can be extended to protein-ligand complexes of protein superfamilies where the affinity is quantitatively unknown due to experimental limitations or where the potential for binding is yet to be explored in vitro.
A webserver to implement DEELIG for easy online access would be useful for the general scientific community and this is in the pipeline. A later version of DEELIG which is trained on peptide ligand data set will also be worked on.
Supplemental Material
Supplemental material, sj-pdf-1-bbi-10.1177_11779322211030364 for DEELIG: A Deep Learning Approach to Predict Protein-Ligand Binding Affinity by Asad Ahmed, Bhavika Mam and Ramanathan Sowdhamini in Bioinformatics and Biology Insights
Supplemental material, sj-pdf-2-bbi-10.1177_11779322211030364 for DEELIG: A Deep Learning Approach to Predict Protein-Ligand Binding Affinity by Asad Ahmed, Bhavika Mam and Ramanathan Sowdhamini in Bioinformatics and Biology Insights
Supplemental material, sj-pdf-3-bbi-10.1177_11779322211030364 for DEELIG: A Deep Learning Approach to Predict Protein-Ligand Binding Affinity by Asad Ahmed, Bhavika Mam and Ramanathan Sowdhamini in Bioinformatics and Biology Insights
Supplemental material, sj-pdf-4-bbi-10.1177_11779322211030364 for DEELIG: A Deep Learning Approach to Predict Protein-Ligand Binding Affinity by Asad Ahmed, Bhavika Mam and Ramanathan Sowdhamini in Bioinformatics and Biology Insights
Acknowledgments
All authors acknowledge NCBS for infrastructural support.
Footnotes
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: A.A. acknowledges funding awarded by the Indian Academy of Sciences, Bangalore (2019). B.M. acknowledges support from the Tata Education and Development Trust (2018-2019). R.S. would like to acknowledge her JC Bose Fellowship (JC Bose fellowship (SB/S2/JC-071/2015) from the Science and Engineering Research Board, Bioinformatics Center Grant funded by the Department of Biotechnology, India (BT/PR40187/BTIS/137/9/2021) and NCBS (TIFR) for infrastructural facilities.
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: Conceptualization, B.M. and A.A.; methodology, R.S., B.M., and A.A.; software, A.A. and B.M.; validation, A.A. and B.M.; formal analysis, A.A. and B.M.; investigation, A.A. and B.M.; resources, R.S.; data curation, A.A. and B.M.; writing—original draft preparation, A.A. and B.M.; writing—review and editing, R.S.; visualization, A.A.; supervision, R.S.; project administration, R.S.; funding acquisition, R.S. All authors have read and agreed to the published version of the manuscript.
Availability: The code for the article is openly available at https://github.com/asadahmedtech/DEELIG.
ORCID iDs: Asad Ahmed  https://orcid.org/0000-0003-3775-9320
https://orcid.org/0000-0003-3775-9320
Bhavika Mam  https://orcid.org/0000-0002-3130-0925
https://orcid.org/0000-0002-3130-0925
Ramanathan Sowdhamini  https://orcid.org/0000-0002-6642-2367
https://orcid.org/0000-0002-6642-2367
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Pantsar T, Poso A. Binding affinity via docking: fact and fiction. Molecules. 2018;23:1899. doi: 10.3390/molecules23081899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pagadala N, Syed K, Tuszynski J. Software for molecular docking: a review. Biophys Rev. 2017;9:91-102. doi: 10.1007/s12551-016-0247-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Genheden S, Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Discov. 2015;10:449-461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. LeCun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proc IEEE. 1998;86:2278-2324. doi: 10.1109/5.726791. [DOI] [Google Scholar]
- 5. Hubel DH, Wiesel TN. Receptive fields, binocular interaction and functional architecture in the cat’s visual cortex. J Physiol. 1962;160:106-154. doi: 10.1113/jphysiol.1962.sp006837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Min S, Lee B, Yoon S. Deep learning in bioinformatics. Brief Bioinform. 2017;18:851-869. doi: 10.1093/bib/bbw068. [DOI] [PubMed] [Google Scholar]
- 7. Stone J, Walker J. Plant protein kinase families and signal transduction. Plant Physiol. 1995;108:451-457. doi: 10.1104/pp.108.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jin Z, Du X, Xu Y, et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289-293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 9. Rose PW, Beran B, Bi C, et al. The RCSB protein data bank: redesigned website and web services. Nucleic Acids Res. 2010;39:D392-D401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu Z, Li Y, Han L, et al. PDB-wide collection of binding data: current status of the PDBbind database. Bioinformatics. 2014;31:405-412. doi: 10.1093/bioinformatics/btu626. [DOI] [PubMed] [Google Scholar]
- 11. Hu L, Benson ML, Smith RD, Lerner MG, Carlson HA. Binding MOAD (Mother Of All Databases). Proteins. 2005;60:333-340. doi: 10.1002/prot.20512. [DOI] [PubMed] [Google Scholar]
- 12. Benson ML, Smith RD, Khazanov NA, et al. Binding MOAD, a high-quality protein-ligand database. Nucleic Acids Res. 2008;36:D674-D678. doi: 10.1093/nar/gkm911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Desaphy J, Bret G, Rognan D, Kellenberger E. sc-PDB: a 3D-database of ligandable binding sites – 10 years on. Nucleic Acids Res. 2015;43:D399-D404. doi: 10.1093/nar/gku928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kawabata T. Detection of multiscale pockets on protein surfaces using mathematical morphology. Proteins. 2010;78:1195-1211. doi: 10.1002/prot.22639. [DOI] [PubMed] [Google Scholar]
- 15. Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605-1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 16. Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389-3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hubbard SJ, Thornton JM. “NACCESS,” Computer Program. London, England: Department of Biochemistry and Molecular Biology, University College, 1993. [Google Scholar]
- 18. Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577-2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 19. Yap CW. PaDEL-descriptor: an open source software to calculate molecular descriptors and fingerprints. J Comput Chem. 2011;32:1466-1474. doi: 10.1002/jcc.21707. [DOI] [PubMed] [Google Scholar]
- 20. Schrödinger Release 2020-3: QikProp . New York, NY: Schrödinger, LLC, 2020. [Google Scholar]
- 21. Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Commun ACM. 2017;60:1097-1105. doi: 10.1145/3065386. [DOI] [Google Scholar]
- 22. Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv. 2014, preprint arXiv:1409.1556. https://arxiv.org/pdf/1409.1556.pdf.
- 23. Ioffe S, Szegedy C. Batch normalization: accelerating deep network training by reducing internal covariate shift. arXiv. 2015, preprint arXiv:1502.03167. https://arxiv.org/pdf/1502.03167.pdf.
- 24. Dahl GE, Sainath TN, Hinton GE. Improving deep neural networks for LVCSR using rectified linear units and dropout. Paper presented at: IEEE International Conference on Acoustics, Speech and Signal Processing, 26-31 May 2013:8609-8613; Vancouver, BC, Canada. doi: 10.1109/ICASSP.2013.6639346. [DOI] [Google Scholar]
- 25. Kingma DP, Ba J. Adam: a method for stochastic optimization. arXiv. 2014, preprint arXiv:1412.6980. https://arxiv.org/pdf/1412.6980.pdf.
- 26. Paszke A, Gross S, Massa F, et al. PyTorch: an imperative style, high-performance deep learning library. Adv Neural Inf Process Syst. 2019;32:8024-8035. [Google Scholar]
- 27. Stepniewska-Dziubinska MM, Zielenkiewicz P, Siedlecki P. Development and evaluation of a deep learning model for protein–ligand binding affinity prediction. Bioinformatics. 2018;34:3666-3674. doi: 10.1093/bioinformatics/bty374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen P, Ke Y, Lu Y, et al. DLIGAND2: an improved knowledge-based energy function for protein–ligand interactions using the distance-scaled, finite, ideal-gas reference state. J Cheminform. 2019;11:52. doi: 10.1186/s13321-019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li H, Leung K-S, Wong M-H, Ballester PJ. Low-quality structural and interaction data improves binding affinity prediction via random forest. Molecules. 2015;20:10947-10962. doi: 10.3390/molecules200610947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang C, Zhang Y. Improving scoring-docking-screening powers of protein−ligand scoring functions using random forest. J Comput Chem. 2017;38:169-177. doi: 10.1002/jcc.24667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cang ZX, Mu L, Wei GW. Representability of algebraic topology for biomolecules in machine learning based scoring and virtual screening. PLoS Comput Biol. 2018;14:e1005929. doi: 10.1371/journal.pcbi.1005929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nguyen DD, Wei GW. AGL-Score: algebraic graph learning score for protein–ligand binding scoring, ranking, docking, and screening. J Chem Inf Model. 2019;59:3291-3304. doi: 10.1021/acs.jcim.9b00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pokkuluri PR, Londer YY, Duke NE, Long C, Schiffer M. Family of cytochrome c7-type proteins from geobacter sulfurreducens: structure of one cytochrome c7 at 1.45 Å resolution. Biochemistry. 2004;43:849-859. doi: 10.1021/bi0301439. [DOI] [PubMed] [Google Scholar]
- 34. Pokkuluri PR, Londer YY, Yang X, et al. Structural characterization of a family of cytochromes c(7) involved in Fe(III) respiration by Geobacter sulfurreducens. Biochim Biophys Acta. 2010;1797:222-232. doi: 10.1016/j.bbabio.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 35. Pokkuluri PR, Londer YY, Duke NE, et al. Structure of a novel dodecaheme cytochrome c from Geobacter sulfurreducens reveals an extended 12 nm protein with interacting hemes. J Struct Biol. 2011;174:223-233. doi: 10.1016/j.jsb.2010.11.022. [DOI] [PubMed] [Google Scholar]
- 36. Li Y, Rezaei MA, Li C, Li X, Wu D. DeepAtom: a framework for protein-ligand binding affinity prediction. arXiv. 2019, preprintarXiv:1912.00318. https://arxiv.org/pdf/1912.00318.pdf.
- 37. Öztürk H, Özgür A, Ozkirimli E. DeepDTA: deep drug–target binding affinity prediction. Bioinformatics. 2018;34:i821-i829. doi: 10.1093/bioinformatics/bty593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. He T, Heidemeyer M, Ban F, Cherkasov A, Ester M. Simboost: a read-across approach for predicting drug–target binding affinities using gradient boosting machines. J Cheminform. 2017;9:24. doi: 10.1186/s13321-017-0209-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang R, Fang X, Lu Y, Yang C-Y, Wang S. The PDBbind database: methodologies and updates. J Med Chem. 2005;48:4111-4119. doi: 10.1021/jm048957q. [DOI] [PubMed] [Google Scholar]
- 40. Wang R, Fang X, Lu Y, Wang S. The PDBbind database: collection of binding affinities for protein-ligand complexes with known three-dimensional structures. J Med Chem. 2004;47:2977-2980. doi: 10.1021/jm030580l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-bbi-10.1177_11779322211030364 for DEELIG: A Deep Learning Approach to Predict Protein-Ligand Binding Affinity by Asad Ahmed, Bhavika Mam and Ramanathan Sowdhamini in Bioinformatics and Biology Insights
Supplemental material, sj-pdf-2-bbi-10.1177_11779322211030364 for DEELIG: A Deep Learning Approach to Predict Protein-Ligand Binding Affinity by Asad Ahmed, Bhavika Mam and Ramanathan Sowdhamini in Bioinformatics and Biology Insights
Supplemental material, sj-pdf-3-bbi-10.1177_11779322211030364 for DEELIG: A Deep Learning Approach to Predict Protein-Ligand Binding Affinity by Asad Ahmed, Bhavika Mam and Ramanathan Sowdhamini in Bioinformatics and Biology Insights
Supplemental material, sj-pdf-4-bbi-10.1177_11779322211030364 for DEELIG: A Deep Learning Approach to Predict Protein-Ligand Binding Affinity by Asad Ahmed, Bhavika Mam and Ramanathan Sowdhamini in Bioinformatics and Biology Insights