Abstract
Background
The long noncoding RNA (lncRNA) GAPLINC, or gastric adenocarcinoma predictive long intergenic ncRNA, plays a carcinogenic role in a variety of different tumor types. There is limited information regarding the biological function of GAPLINC in the development of esophageal squamous cell carcinoma (ESCC).
Methods
Surgical tissue samples of 40 patients undergoing ESCC radical surgery were collected, including ESCC tissues and corresponding adjacent normal tissues. Quantitative real-time PCR (qRT-PCR) was used to detect the expression of lncRNA GAPLINC in the human ESCC cell line (TE11). The function role of LncRNA GAPLINC was detected after specific siRNA interference and overexpression in the TE11 cell line. The effects of LncRNA GAPLINC on ESCC cell proliferation, migration and invasion abilities were investigated by flow cytometry, using the Cell Counting Kit-8 (CCK-8), and by Transwell migration assays, respectively.
Results
The expression of lncRNA GAPLINC in ESCC tissues was significantly higher than that in corresponding adjacent normal tissues (P<0.05) and correlated with the degree of tumor differentiation (P<0.05). Compared with human esophageal normal epithelial cell lines, the expression of LncRNA GAPLINC was significantly higher in the human ESCC cell line (P<0.05). CCK-8 assays showed that LncRNA GAPLINC overexpression increased the growth rate of cells (P<0.05). Transwell experiments showed that LncRNA GAPLINC overexpression increased the ability of cell migration and invasion compared to control cells (P<0.05). Annexin V assay revealed that LncRNA GAPLINC silencing increased early stage apoptosis (P<0. 05).
Conclusion
Our results suggest that LncRNA GAPLINC may be used as a biomarker for the diagnosis and monitoring of ESCC, and may play an oncogenic role in ESCC.
Keywords: esophageal squamous cell carcinoma, long non-coding RNA, GAPLINC, biomarker, expression
Background
Esophageal squamous cell carcinoma (ESCC) has become one of the six most frequently diagnosed malignant tumors in the world, with about 572,000 new cases and 509,000 deaths in 2018, ranking seventh and sixth among all tumors in the world, respectively.1 China has the highest incidence and mortality rate of ESCC in the world, accounting for 70%.2 It should be noted that more than 95% of patients with ESCC are in advanced stage at their initial diagnosis, and present poor prognosis with a 5-year survival rate of only about 30%.3,4 The available studies cannot fully explain the pathogenesis of ESCC.5 Numerous factors are related to the occurrence and development of ESCC.6 Thus, it is of great importance for the prevention and treatment of ESCC to better understand its pathogenesis, clarify its biological behavior, identify any susceptible genes and specific biomarkers.
LncRNA is a long non coding RNA of more than 200 nucleotides in length, which is mainly transcribed by RNA Polymerase II and widely distributed in the human genome.7 Numerous lncRNAs have been identified in different malignant tumor. These lncRNAs regulate the expression of oncogenes and tumor suppressor genes in different ways and play an important role in the occurrence, development, metastasis, and recurrence of tumors.8 Furthermore, lncRNAs are stable in circulating body fluids such as plasma and urine; thus, providing a new basis for the diagnosis and treatment of malignant tumors. The search for new lncRNAs as diagnostic biomarkers and therapeutic targets for malignant tumors has become a research hotspot.
GAPLINC is 924 bp in length and is located on the short arm of human chromosome 18; the RNA length after transcription is 643 nt.9 LncRNA GAPLINC was first confirmed to be significantly increased in gastric cancer tissues. GAPLINC expression is strongly correlated with CD44 expression and its role as a proto-oncogene is dependent in part, on CD44.10 At present, few studies have described the functional role of LncRNA GAPLINC. The aim of this study was to explore the biological function of LncRNA GAPLINC in the development of ESCC.
Materials and Methods
Patients and Tumor Specimens
In this study, 40 pairs of specimens were collected, including ESCC cancer tissues and adjacent normal esophageal tissues. These tissues were collected from patients undergoing thoracic surgery as a therapeutic intervention in 2018. The normal tissues adjacent to the cancer required to be ≥5 cm from the edge of the tumor tissue.
Quantitative Real-Time Polymerase Chain Reaction
Fifty mg tissue sample was added to 700 μL of lysate (RNA extraction kit, Shanghai Meixuan Biological Technology Co., Ltd.) for RNA extraction. Next, 20 μL of proteinase K solution and 20 μL magnetic beads to were added to the protocol to obtain purified RNA. After removing, genomic DNA, cDNA was obtained by reverse transcription of RNA (RT Reagent Kit, Takara Co., Ltd.), and Real Time PCR reactions were performed immediately. hGAPLINC (Forward primer sequence: CCACATCCAGGGGCTATCAA; Reverse primer sequence: CACAATCAGGGCTCTTGGACT) and hGAPDH (Forward primer sequence: GCACCGTCAAGGCTGAGAAC; Reverse primer sequence: TGGTGAAGACGCCAGTGGA).
Cell Culture
The human ESCC cell lines KYSE-150, KYSE-450, KYSE510, TE-10, TE-11 and the human-derived normal esophageal epithelial cell line (Het-lA) were obtained from Shanghai Meixuan Biotechnology Co., Ltd.). All cell lines were cultured in complete medium (containing 90% DMEM high glucose plus 10% fetal bovine serum, Hyclone Co., Ltd.) under sterile conditions, and placed in a 37° incubator.
CCK-8 Assay
The proliferative ability of cell lines was detected by CCK-8 assay (CCK-8 Cell Proliferation and Cytotoxicity Detection Kit, Biyuntian Co., Ltd.). A 100 μL cell suspension was added to the 96-well plate and incubated for 72 h. Next, 10 μL CCK-8 solution was added to each well, and the incubation of the plate continued in the incubator for 1 hour. The absorbance at 450 nm was measured using a microplate reader.
Transwell Assay
Approximately 5×104 cells were inoculated in the upper well of the Transwell chamber and 500 μL of fresh medium containing 20% FBS was added in the lower chamber. After incubating for 12 h, the Transwell chamber was removed and cells were fixed with 4% paraformaldehyde for 10 minutes. Crystal violet dyeing solution was added for 5 minutes, the cells were observed under the microscope.
Apoptosis Assay
Cell apoptosis was detected using a flow cytometry assay (Annexin V-Alexa Fluor647/PI Apoptosis Detection Kit, Solarbio Co., Ltd.). Approximately, 50,000–100,000 resuspended cells were centrifuged at 1000 ×g for 5 minutes. Next, 195 μL Annexin V-FITC binding solution, 5 μL Annexin V-Alexa Fluor647, and 10 μL propidium iodide staining solution were added to the cells. After incubating at room temperature in the dark for 20 minutes, the cells were analyzed by flow cytometry.
Results
Expression of LncRNA GAPLINC in ESCC Tissues and Adjacent Tissues
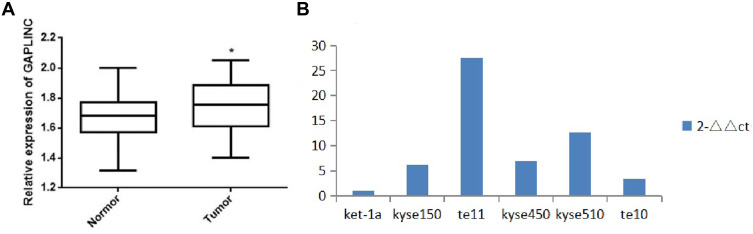
LncRNA GAPLINC expression in ESCC tissue was significantly higher than in normal adjacent tissues (P<0.05) (Figure 1A). The expression of LncRNA GAPLINC in ESCC lines was significantly higher than that of human normal epithelial esophageal cell lines (P<0.05) (Figure 1B). The relative expression of LncRNA GAPLINC was significantly associated with the degree of differentiation of the ESCC (P<0.05), and was not related to the patient’s sex, age, history of tobacco and alcohol consumption, tumor location, esophageal lesion length, infiltration depth, or lymph node metastasis (Table 1).
Figure 1.
LncRNA GAPLINC expression levels in ESCC tissues, adjacent normal esophagus tissues, and ESCC cell lines. (A) LncRNA GAPLINC expression levels in ESCC tissues and adjacent normal esophagus tissues. (B) LncRNA GAPLINC expression in five human ESCC cell lines. (*p<0.05).
Table 1.
Relationship Between LncRNA GAPLINC and Clinical Characteristics of ESCC Patients
| Clinical Information | No. | % | P value |
|---|---|---|---|
| Gender | P=0.428 | ||
| Male | 34 | 85 | |
| Female | 6 | 15 | |
| Age | P=0.269 | ||
| >60 | 31 | 77.5 | |
| ≤60 | 9 | 22.5 | |
| Smoking | P=0.152 | ||
| Yes | 16 | 40 | |
| No | 24 | 60 | |
| Drinking | P=0.791 | ||
| Yes | 15 | 37.5 | |
| No | 25 | 62.5 | |
| Tumor location | vs Low in chest | ||
| High in chest | 5 | 12.5 | P=0.648 |
| Middle in chest | 14 | 35 | P=0.444 |
| Lower middle in chest | 10 | 25 | P=0.764 |
| Low in chest | 10 | 25 | |
| Unknown | 1 | 2.5 | |
| Tumor size | P=0.629 | ||
| >4cm | 20 | 50 | |
| ≤4cm | 20 | 50 | |
| Differentiation | vs High | ||
| High | 4 | 10 | |
| Higher middle | 6 | 15 | P=0.015 |
| Middle | 13 | 32.5 | P=0.011 |
| Lower middle | 8 | 20 | P=0.017 |
| Low | 9 | 22.5 | P=0.022 |
| Infiltration | vs All | ||
| Mucosa and submucosa | 5 | 12.5 | P=0.700 |
| Superficial muscularis | 2 | 5 | P=0.979 |
| Deep muscularis | 5 | 12.5 | P=0.538 |
| All | 28 | 70 | |
| Lymph node metastasis | |||
| Yes | 15 | 35.5 | P=0.114 |
| No | 25 | 62.5 | |
Note: Chi-square test P<0.05.
Clinical Value of LncRNA GAPLINC in ESCC
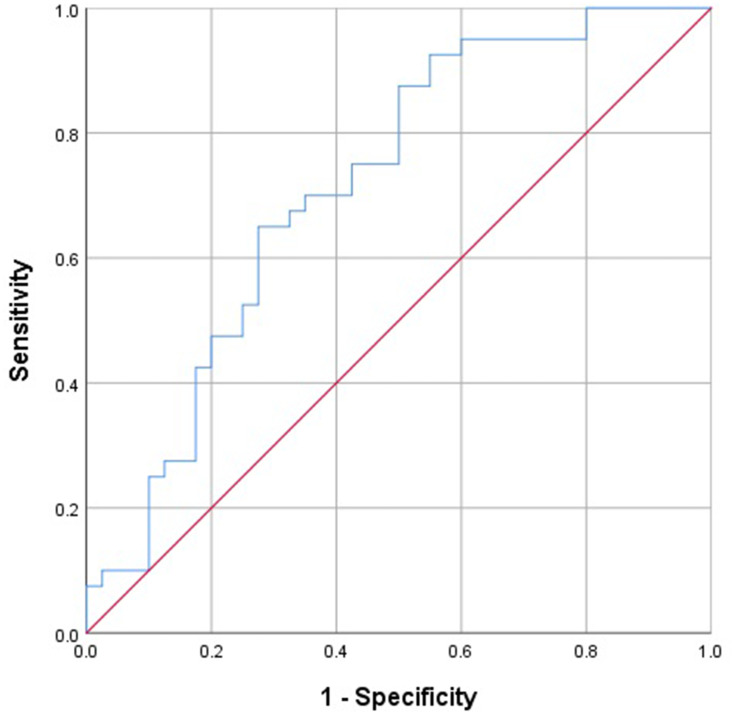
As shown in Figure 2, the area under the curve (AUC) of LncRNA GAPLINC at diagnosis was 0.715, and when the cut-off value for GAPLINC expression was lowered to 0.3865, the specificity and sensitivity of the assay were 72.5% and 65.0%, respectively.
Figure 2.
Receiver operating characteristics (ROC) curves for LncRNA GAPLINC. AUC of LncRNA GAPLINC in the diagnosis of patients was 0.715.
Effect of LncRNA GAPLINC on TE11 Cell Behavior
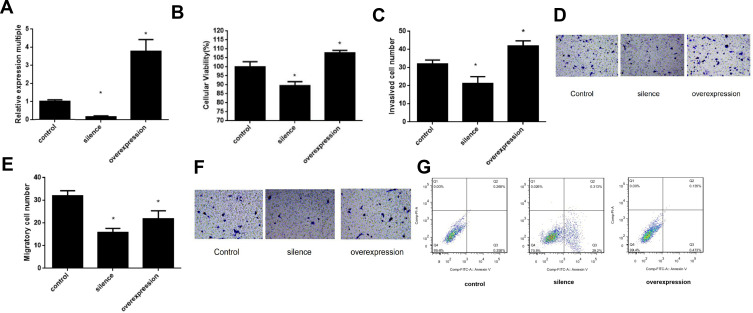
The expression of LncRNA GAPLINC in the control, knockdown, and overexpression groups in TE11 cells are shown in Figure 3A. The relative expression of multiple of Ct values between of the LncRNA GAPLINC silenced group and control group was 18.4%. The knockdown and overexpression of LncRNA GAPLINC in the TE11 cell line were significantly higher than in the control group (P <0.05).
Figure 3.
Effects of LncRNA GAPLINC on TE11 cell behavior. (A) LncRNA GAPLINC expression in control, and cells with LncRNA GAPLINC knockdown and overexpression (B) Cell proliferation and (C) cell migration following lncGAPLINC-knockdown and lncGAPLINC-overexpression. (D) Representative images of the cell migration assay. (E) Cell invasion following lncGAPLINC-knockdown and lncGAPLINC-overexpression. (F) Representative images of the invasion assay. (G) Apoptosis following lncGAPLINC-knockdown and lncGAPLINC-overexpression. (*p<0.05).
CCK-8 Assay
The cellular viability of the TE11 cell line after lncGAPLINC-knockdown and LncGAPLINC-overexpression are shown in Figure 3B. We found that lncGAPLINC-knockdown decreased the cell proliferation rate, and lnGAPLINC-overexpression increased the cell proliferation rate (P<0.05).
Transwell Assay
Transwell cell migration and invasion assays were used to detect TE11 cell migration and invasion ability after lncGAPLINC-knockdown and lncGAPLINC-overexpression. Compared with the control group, the number of migration and invasion decreased after knocking down and increased after following lncGAPLINC overexpression (P<0.05) (Figure 3C–F).
Cell Apoptosis
Cell apoptosis was detected in the LncGAPLINC-knockdown, LncGAPLINC-overexpression, and in control TE11 cell lines. Compared with the control group, knocking down LncRNA GAPLINC lead to an increase in cell apoptosis (35.97±3.58%, P<0.05) (Figure 3G, Table 2). However, there were no significant changes in cell apoptosis between the LncGAPLINC-overexpression cell line and the control cell line.
Table 2.
Apoptosis Rate of LncGAPLINC-Knockdown Cell Line and LncGAPLINC-Overexpression Cell Line
| Control | Silence | Overexpression | |
|---|---|---|---|
| Apoptosis rate (%) | 0.49±0.045 | 35.97±3.58 | 0.70±0.083 |
Discussion
Esophageal cancer is one of the most common malignant tumors. It ranked sixth among the causes of tumor-related mortality worldwide, and morbidity ranked seventh in 2018.1 There are obvious regional differences in esophageal cancer incidence. Northern France, Africa, Central Asia, and Central and South America are the high incidence areas. In China, the high incidence areas include Taihang Mountain, Northern Jiangsu Area, Dabie Mountain, Southern Fujian, Chaoshan of Guangdong, and the Eastern Qinling Mountains.11
The main tissue types of esophageal cancer are esophageal adenocarcinoma (EAC) and ESCC. ESCC is the most common tissue type of esophageal cancer and the main pathological type in China. Recent studies have confirmed that multidisciplinary comprehensive treatment is the main research direction for ESCC treatment. Although the diagnosis and treatment methods of ESCC are continuously being optimized and new drugs are being developed, ESCC is characterized by high invasiveness, treatment resistance, and easy metastasis. Therefore, the overall 5-year survival rate is not obvious. At present, the clinical prognosis of ESCC is mainly defined by the TNM stage. Although the prognosis of patients may vary greatly at the same TNM stage. It is not comprehensive enough to rely on the clinical pathological staging to evaluate prognosis. The inherent molecular biological mechanism is also worth exploring.
In recent years, studies have found that genetic abnormalities are one of the main causes of ESCC, indicating that ESCC occurrence might be related to environmental factors and genetic factors. In order to achieve an accurate early diagnosis of ESCC and better prognosis evaluation, we need to further explore its pathogenesis at the genomic level. Previous studies have confirmed that the genesis and development of esophageal cancer involved many genes, such as C-MYC, EGFR, INT-2, and P53.
LncRNAs are more than 200 nucleotides long and do not encode protein. According to their position, LncRNAs can be divided into 5 types: sense LncRNA, antisense LncRNA, bidirectional LncRNA, intron LncRNA, and intergenic LncRNA.12 Studies on lncRNA in esophageal cancer are not rare, but it is still a hot spot in current tumor research. Compared with miRNA, lncRNA is much more abundant in terms of function and mechanism, and it may be a core factors in RNA regulation. LncRNAs control biological events by numerous ways.13 An increasing number of studies have shown that LncRNA is differentially expressed in tumors.14 Several studies have shown that lncRNA has a relatively stable secondary structure and can be detected in plasma or other body fluids.15
LncRNA may become a new class of tumor markers and potential therapeutic targets.16 An important function of LncRNA is to regulate the expression of neighboring protein-coding genes. The molecular mechanisms include histone modification, DNA methylation, splicing regulation, and protein ubiquitination. LncRNA can be used as a signal or guide the recruitment of chromatin modification complexes to induce transcription, and can even be used as a bait that binds transcription factors to prevent them from binding to target gene promoter regions, and thereby inhibiting transcription.17 In addition, lncRNA can hybridize with the mRNA precursor, block the recognition of the splice site through the spliceosome, and regulate the alternative splicing of the mRNA precursor to produce alternate transcripts.18 Another biological function of lncRNA is to act as a “microRNA sponge” by interacting with microRNA, inactivating these small regulatory RNAs, thereby increasing the expression of microRNA target genes.19,20 Finally, lncRNA may be involved in the regulation of protein localization, activity, and function.21 Thus, identifying lncRNA expression in ESCC may further expand our understanding of ESCC and open up a new strategies for the prevention and treatment of esophageal cancer.
LncRNA GAPLINC was first confirmed to be highly expressed in gastric cancer tissues, where it promotes cell proliferation by acting as a molecular sponge of miR-378.22 GAPLINC up-regulates the expression of CD44 by competing with miR-211-3p.23 In colorectal cancer, GAPLINC inhibits cell proliferation and invasion of cancer cells by combining with SNAI2 and the up-regulation of GAPLINC expression has been associated with late clinical stage and overall survival of colorectal cancer patients.24 In glioma, GAPLINC acts as an oncogene by regulating NRP2 and ERBB-2 in combination with miR-331-3p.25 In non-small cell lung cancer, GAPLINC is located downstream of TGF-β signaling and is up-regulated by TGF-β1. GAPLINC may act as a mediator of TGF-β and its downstream effectors to promote the migration and invasion of NSCLC cells.26 Overall, the above studies show that LncRNA GAPLINC plays a role as an oncogene in gastric cancer, colorectal cancer, glioma, and non-small cell lung cancer. The function and mechanism of LncRNA GAPLINC in ESCC is unknown.
This study collected samples from 40 cases of ESCC tissues and adjacent normal tissue. The results of qRT-PCR assay showed that lncRNA GAPLINC had a significantly higher expression in ESCC. The study revealed a correlation between LncRNA expression level and tumor differentiation degree, indicating the potential of LncRNA GAPLINC as a biomarker for diagnosis in ESCC. In order to further explore the function of lncRNA GAPLINC in esophageal tumors, we used qRT-PCR technology to detect lncRNA GAPLINC expression in five human ESCC lines (KYSE-450, KYSE-510, KYSE-150, TE-10, and TE- 11). Compared with the normal esophageal epithelial cell line (HET-1A), lncRNA GAPLINC expression was up-regulated in these five ESCC lines, which was similar to that observed in ESCC tissue. We selected the TE-11 cell line to knock down and overexpress lncRNA GAPLINC. The Annexin V apoptosis assay showed that knocking down of GAPLINC lead to a significant difference in cell apoptosis rates, while overexpression of GAPLINC had no effect on apoptosis. Knocking down of lncRNA GAPLINC lead to a decrease in the rates of cell proliferation, migration and invasion. Conversely, overexpressing lncRNA GAPLINC led to an increase on cell proliferation, migration and invasion. Thus, downregulation of LncRNA GAPLINC could significantly inhibit the proliferation, migration, and invasion ability of ESCC cells in vitro. Altogether our findings show that LncRNA GAPLINC acts as an oncogene in the development of ESCC, and may represent a potential treatment target for ESCC.
Conclusion
Our results revealed that LncRNA GAPLINC could acts as an oncogene in the occurrence and development of ESCC and may be an effective biomarker for ESCC diagnosis and a potential therapeutic target.
Acknowledgments
This manuscript has been released as a pre-print at Research Square, Peng Wang et al.
Funding Statement
There is no funding to report.
Abbreviations
CCK-8, Cell counting kit-8; EAC, esophageal adenocarcinoma; ESCC, esophageal squamous cell carcinoma; GAPLINC, Gastric adenocarcinoma predictive long intergenic noncoding RNA; lncRNA, Long non-coding RNA; qRT-PCR, Quantitative real-time PCR.
Data Sharing Statement
The raw data supporting the conclusions of this article will be made available by the author (Peng Wang, 5655818@qq.com), without undue reservation.
Ethics Approval and Consent to Participate
Human samples experimented in this study were performed referring to the Helsinki Declaration and approved by the ethical committee of Jiangsu Taizhou People’s Hospital. Participants were in accordance with the ethical standards of the institutional research committee. Informed consent was obtained from all individual participants included in the study.
Consent for Publication
All patients included were required to offer written informed consent. All authors agree to submit the article for publication.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that the research was conducted in the absence of any commercial or financial relationships.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.DeSantis CE, Lin CC, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64(4):252–271. doi: 10.3322/caac.21235 [DOI] [PubMed] [Google Scholar]
- 3.Minashi K, Nihei K, Mizusawa J, et al. Efficacy of endoscopic resection and selective chemoradiotherapy for stage I esophageal squamous cell carcinoma. Gastroenterology. 2019;157(2):382–390. doi: 10.1053/j.gastro.2019.04.017 [DOI] [PubMed] [Google Scholar]
- 4.Zeng HM, Chen WQ, Zheng RS, et al. Changing cancer survival in China during 2003–15: a pooled analysis of 17 population-based cancer registries. Lancet Glob Health. 2018;6(5):e555–e567. doi: 10.1016/S2214-109X(18)30127-X [DOI] [PubMed] [Google Scholar]
- 5.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349(23):2241–2252. doi: 10.1056/NEJMra035010 [DOI] [PubMed] [Google Scholar]
- 6.Matejcic M, Parker MI. Gene-environment interactions in esophageal cancer. Crit Rev Clin Lab Sci. 2015;52(5):21. doi: 10.3109/10408363.2015.1020358 [DOI] [PubMed] [Google Scholar]
- 7.Ernst C, Morton CC. Identification and function of long non-coding RNA. Front Cell Neurosci. 2013;7:168. doi: 10.3389/fncel.2013.00168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camacho CV, Choudhari R, Gadad SS. Long noncoding RNAs and cancer, an overview. Steroids. 2018;133:1–12. doi: 10.1016/j.steroids.2017.12.012 [DOI] [PubMed] [Google Scholar]
- 9.Li T, Mo X, Fu L, et al. Molecular mechanisms of long noncoding RNAs on gastric cancer. Oncotarget. 2016;7(8):8601–8612. doi: 10.18632/oncotarget.6926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu Y, Wang J, Qian J, et al. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer Res. 2014;74(23):6890–6902. doi: 10.1158/0008-5472.CAN-14-0686 [DOI] [PubMed] [Google Scholar]
- 11.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516 [DOI] [PubMed] [Google Scholar]
- 12.Huang JL, Zheng L, Hu YW, et al. Characteristics of long non-coding RNA and its relation to hepatocellular carcinoma. Carcinogenesis. 2014;35(3):507–514. doi: 10.1093/carcin/bgt405 [DOI] [PubMed] [Google Scholar]
- 13.Talebi A, Masoodi M, Mirzaei A, et al. Biological and clinical relevance of metastasis‐associated long noncoding RNAs in esophageal squamous cell carcinoma: a systematic review. J Cell Physiol. 2020;235(2):848–868. doi: 10.1002/jcp.29083 [DOI] [PubMed] [Google Scholar]
- 14.Huang W, Huang F, Lei Z, Luo H. LncRNA SNHG11 promotes proliferation, migration, apoptosis, and autophagy by regulating hsa-miR-184/AGO2 in HCC. Onco Targets Ther. 2020;13:413–421. doi: 10.2147/OTT.S237161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitra SA, Mitra AP, Triche TJ. A central role for long non-coding RNA in cancer. Front Genet. 2011;3:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reis EM, Sergio VA. Perspectives of long non-coding RNAs in cancer diagnostics. Front Genet. 2012;3:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palazzo AF, Lee ES. Non-coding RNA: what is functional and what is junk? Front Genet. 2015;6:2. doi: 10.3389/fgene.2015.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He Y, Meng XM, Huang C, et al. Long noncoding RNAs: novel insights into hepatocelluar carcinoma. Cancer Lett. 2014;344(1):20–27. doi: 10.1016/j.canlet.2013.10.021 [DOI] [PubMed] [Google Scholar]
- 19.Bartonicek N, Maag J, Dinger ME. Long noncoding RNAs in cancer: mechanisms of action and technological advancements. Mol Cancer. 2016;15(1):43. doi: 10.1186/s12943-016-0530-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Das S, Ghosal S, Sen R, et al. lnCeDB: database of human long noncoding RNA acting as competing endogenous RNA. PLoS One. 2014;9(6):e98965. doi: 10.1371/journal.pone.0098965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi: 10.1016/j.ccell.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diao L, Wang S, Sun Z. Long noncoding RNA GAPLINC promotes gastric cancer cell proliferation by acting as a molecular sponge of miR-378 to modulate MAPK1 expression. Onco Targets Ther. 2018;11:2797–2804. doi: 10.2147/OTT.S165147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He Y, Wu Z, Qiu C, et al. Long non‐coding RNA GAPLINC promotes angiogenesis by regulating miR‐211 under hypoxia in human umbilical vein endothelial cells. J Cell Mol Med. 2019;23(12):8090–8100. doi: 10.1111/jcmm.14678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang P, Chen T, Xu Z, et al. Long noncoding RNA GAPLINC promotes invasion in colorectal cancer by targeting SNAI2 through binding with PSF and NONO. Oncotarget. 2016;7(27):42183–42194. doi: 10.18632/oncotarget.9741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen HH, Zong J, Wang SJ. LncRNA GAPLINC promotes the growth and metastasis of glioblastoma by sponging miR-331-3p. Eur Rev Med Pharmacol Sci. 2019;23(1):262–270. doi: 10.26355/eurrev_201901_16772 [DOI] [PubMed] [Google Scholar]
- 26.Zhao J, Wang C, Liu S, et al. TGF-β1 mediates lncRNA GAPLINC expression to promote the migration and invasion of non-small cell lung cancer. Onco Targets Ther. 2019;12:6175–6180. doi: 10.2147/OTT.S207079 [DOI] [PMC free article] [PubMed] [Google Scholar]