SUMMARY
Bacteria and archaea apply CRISPR-Cas surveillance complexes to defend against foreign invaders. These invading genetic elements are captured and integrated into the CRISPR array as spacer elements, guiding sequence-specific DNA/RNA targeting and cleavage. Recently, in vivo studies have shown that target RNAs with extended complementarity with repeat sequences flanking the target element (tag:anti-tag pairing) can dramatically reduce RNA cleavage by the type VI-A Cas13a system. Here, we report the cryo-EM structure of Leptotrichia shahii LshCas13acrRNA in complex with target RNA harboring tag:anti-tag pairing complementarity, with the observed conformational changes providing a molecular explanation for inactivation of the composite HEPN domain cleavage activity. These structural insights, together with in vitro biochemical and in vivo cell-based assays on key mutants, define the molecular principles underlying Cas13a’s capacity to target and discriminate between self and non-self RNA targets. Our studies illuminate approaches to regulate Cas13a’s cleavage activity, thereby influencing Cas13a-mediated biotechnological applications.
Graphical Abstract
In Brief
Extended complementarity between crRNA and anti-tag target RNA mediates autoimmunity of Cas13a systems. Wang et al. show the structure-function data and reveal the molecular basis for prevention of composite catalytic pocket formation and substrate RNA degradation by anti-tag RNA.
INTRODUCTION
Bacteria and archaea use diverse CRISPR and CRISPR-associated (CRISPR-Cas) systems as a potent adaptive immune defense against foreign nucleic acids such as phages and mobile genetic elements (Dupuis et al., 2013; Hille and Charpentier, 2016; Labrie et al., 2010; Marraffini, 2015; Mohanraju et al., 2016; Nishimasu and Nureki, 2017; Wright et al., 2016). CRISPR-Cas systems use programmable guide RNAs to sequence-specifically target and cleave foreign genetic elements. These systems are broadly categorized into two classes and sub-divided into six types on the basis of the architecture of the interference effector complex: types I, III, and IV in class 1 systems use multi-subunit surveillance ribonucleoprotein complexes, while types II, V, and VI in class 2 systems coordinate single Cas endonucleases (Barrangou et al., 2007; Makarova et al., 2011, 2015; Shmakov et al., 2015). These systems act in three stages (van der Oost et al., 2009; Wright et al., 2016). At the first adaptation stage, a fragment of invading nucleic acid (called protospacer) is recognized, cleaved, and integrated into the CRISPR array to generate a new spacer flanked by repeats on either side. During the second expression stage, the CRISPR array is transcribed into precursor CRISPR RNA (pre-crRNA) and then cut at a specific site in each repeat sequence to generate mature CRISPR RNAs (crRNAs). At the last interference stage, crRNA assembles with related Cas protein(s) and guides sequence-specific targeting of foreign nucleic acids mediated by pairing between the crRNA spacer and the complementary DNA/RNA target segment.
Within class 2, type II Cas9 and type V Cas12 systems target double-stranded DNA, whereas the type VI Cas13 system (formerly named C2c2 in type VI-A systems) targets single-stranded RNA (ssRNA) through pairing with the 20–30 nt spacer in crRNA. Unlike Cas9 and Cas12, which generate site-specific double-stranded breaks (Deveau et al., 2008; Mojica et al., 2009; Zetsche et al., 2015), Cas13 exhibits cis-RNase activities on target RNAs and collateral trans-RNase activities on surrounding non-specific substrate RNAs (Abudayyeh et al., 2016; East-Seletsky et al., 2016, 2017; Konermann et al., 2018; Shmakov et al., 2015; Smargon et al., 2017; Yan et al., 2018). To date, Cas13 has been developed into a rapid detection tool for diagnosis of pathogens, viruses, and genotyping (Freije et al., 2019; Gootenberg et al., 2017, 2018; Myhrvold et al., 2018), as well as for new RNA-targeting tools for RNA knockdown, editing, splicing modulation, and transcript labeling (Abudayyeh et al., 2017; Cox et al., 2017; Konermann et al., 2018).
Cas13 can be further divided into four subtypes, Cas13a–d(types VI A–D), exhibiting diverse primary sequences except for two highly conserved HEPN (higher eukaryotes and prokaryotes nucleotide-binding) domains (Shmakov et al., 2015), which are responsible for both cis- and trans-RNase activities. Previous structural studies revealed that Cas13a (Knott et al., 2017; Liu et al., 2017a, 2017b), Cas13b (Slaymaker et al., 2019; Zhang et al., 2018a), and Cas13d (Zhang et al., 2018b, 2019) adopt a bilobed architecture including recognition (REC) and nuclease (NUC) lobes. Upon target RNA binding, Cas13a and Cas13d undergo large conformational changes, with one consequence being that key residues of the R-X4-H motif within the two HEPN domains are brought into close proximity to generate a composite pocket, indicating the existence of an auto-inhibited conformation that is released on target RNA loading (Liu et al., 2017a; Zhang et al., 2018b).
The sequences flanking the target region are essential for target recognition and discrimination between self and non-self targets. In types I, II, and V DNA-targeting systems, a short nucleotide sequence named protospacer-adjacent motif (PAM) is required for target recognition and discrimination (Deveau et al., 2008; Gasiunas et al., 2012; Jinek et al., 2012; Semenova et al., 2011; Zetsche et al., 2015). In type III systems, complementarity between target flanking and repeat regions of crRNA inhibits DNase activity of effectors, thereby distinguishing between self and non-self targets (Elmore et al., 2016; Estrella et al., 2016; Kazlauskiene et al., 2016; Samai et al., 2015). In type VI systems, target RNA discrimination and substrate RNA cleavage follow different rules. A single nucleotide called protospacer flanking site (PFS) is required for target cleavage in type VI-A Leptotrichia shahii LshCas13a (Abudayyeh et al., 2016, 2017) and type VI-B Cas13b (Cox et al., 2017; Smargon et al., 2017) systems, while PFS is not essential for type VI-A Leptotrichia buccalis LbuCas13a system (East-Seletsky et al., 2016). PFS requirement is absent in type VI-D Cas13d systems (Konermann et al., 2018). Recent in vivo functional studies demonstrated that type VI-A systems use a similar strategy as that of type III systems for discrimination between self and non-self targets (Meeske and Marraffini, 2018). Extending complementarity between 3′-flank of crRNA (designated as tag) and target RNA beyond the guide:target RNA duplex, called tag:anti-tag RNA pairing (the segment in target RNA pairing with the crRNA tag is designated as anti-tag), abolishes substrate RNA degradation in Listeria seeligeri LseCas13a and LbuCas13a systems, thereby providing a potent inhibitor for Cas13-based applications (Meeske and Marraffini, 2018).
Here, we took a structural approach to investigate the inhibition mechanism associated with pairing between the anti-tag segment of target RNA and the tag segment of crRNA in Cas13a systems. We observed that target RNA harboring an anti-tag sequence on pairing with the tag segment of crRNA alters the conformation of the Cas13a-crRNA complex. To better understand the molecular mechanism, we determined the cryo-electron microscopy (cryo-EM) structure of LshCas13a-crRNA complex bound to anti-tag target RNA. The tag:anti-tag pairing changes the conformation of the tag region and induces domain movements in the NUC lobe, thereby blocking formation of a composite HEPN catalytic pocket required for substrate RNA cleavage activity. Our structural studies complemented by in vitro and in vivo mutational analyses on the LshCas13a surveillance complex both illustrate and highlight the inhibition mechanism associated with anti-tag RNA and provide guidelines for development of Cas13-based diagnostics and RNA-targeting applications, as well as the development of applications using anti-tag as an inhibitor of RNA substrate cleavage.
RESULTS
Anti-tag inhibits cis- and trans-cleavage activity of LshCas13a and LbuCas13a
Previous in vivo functional studies reported that the 5–8 nt complementary anti-tag (henceforth simplified as anti-tag) segments in target RNA prevents nuclease activation of LseCas13a, while shorter anti-tag segments provide partial protection against cleavage (Meeske and Marraffini, 2018). In addition, target RNA containing an 8 nt anti-tag inhibits both cis- and trans-RNase activity of LbuCas13a with a 21 nt spacer in vitro (Meeske and Marraffini, 2018). Sequence alignment (East-Seletsky et al., 2016, 2017) and structural studies suggested that the repeat region of crRNAs in type VI-A present common features, including a 5–6 bp stem, a 7–9 nt hairpin loop, and an embedded 2 nt flipped-out bulge (Figure S1A) (Knott et al., 2017; Liu et al., 2017a, 2017b). The 8 nt crRNA tag segment under consideration (labeled −1 to −8) covers the 3′-flank and the 3′-bulge segments. Specially, LshCas13a, LbuCas13a, and LseCas13a, which belong to uridine (U)-cleaving subfamily members, contain identical sequences in the tag region (Figure S1A) (East-Seletsky et al., 2017), implying that anti-tag-based inhibition could be a common feature in these Cas13a systems.
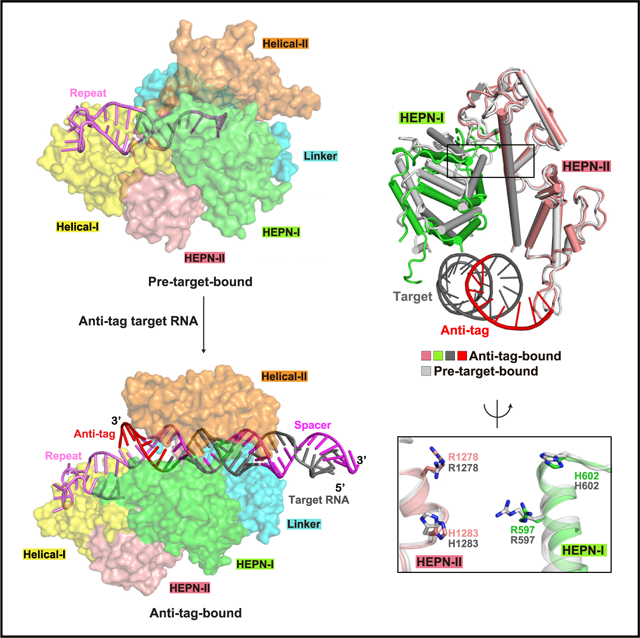
To investigate such a commonality concept, we tested the effects of anti-tag complementarity using both LshCas13a and LbuCas13a with a 28 nt spacer (Figures 1A–1D, S1B, and S1C). For cis-RNase activity, we designed three target RNAs containing different lengths of anti-tag (Figure 1A). RNA1, a 68 nt ssRNA without anti-tag, contains a 28 nt target segment for pairing with spacer. Another two target RNAs, RNA2 and RNA3, are derived from RNA1 by inserting 5 or 8 nt anti-tag segments into the 3′ end of the target segment. After incubating with Cas13a-crRNA complex, cleavage of target RNA was either reduced or eliminated in the presence of 5 or 8 nt anti-tag sequences, respectively (Figure 1B).
Figure 1. Extended tag:anti-tag pairing prevents RNA cleavage by Cas13a.
(A and C) Schematic of target RNAs designed for in vitro cis-RNase and trans-RNase cleavage assays. The spacer and repeat of crRNA are shown in magenta and pink, respectively. The target segment is shown in gray, with anti-tag indicated in red.
(B and D) In vitro cleavage assays by LshCas13a and LbuCas13a monitoring substrate RNA degradation on formation of target RNA (B) and non-target substrate RNA (D) ternary complexes in the presence or absence of anti-tag. Two reported dead mutations, LshCas13a-R1278A and LbuCas13a-R472A/H477A, are used as negative controls and labeled as dCas13a. The sequence and schematic of crRNA and target RNAs are shown in (A), (C), and Figures S1B and S1C.
(E and G) Quantification of EYFP mRNA knockdown by LshCas13a (E) and LbuCas13a (G) in E. coli cells. EYFP mRNA contains a 28 nt segment complementary to spacer followed by an 8 nt segment with or without anti-tag. Transcription of EYFP mRNA was induced by tetracycline. Error bars represent standard error of mean (SEM) of three biological replicates.
(F and H) Effect on growth rate of E. coli cells upon EYFP mRNA interference by LshCas13a (F) and LbuCas13a (H). Error bars represent SEM of three biological replicates.
(I and J) Elution profiles run from a Superdex 200 10/300 GL size exclusion column in the presence or absence of anti-tag, showing that anti-tag has no effect on target RNA loading and formation of LshCas13a-crRNA-target RNA (I) and LbuCas13a-crRNA-target RNA (J) ternary complexes.
See also Figure S1.
Next, we detected Cas13a trans-RNase activity on substrate RNA when using different target RNAs in the presence or absence of anti-tag complementarity. We designed three target RNAs of the same lengths, labeled RNA4 to RNA6 (Figures 1C, S1B, and S1C). Target RNA4 consists of a 28 nt target segment pairing with spacer and an 8 nt extended non-specific sequence following the 3′ end of the target segment. RNA5 and RNA6 are derived from RNA4 by replacing the non-specific sequence with 5 and 8 nt anti-tag segments, respectively. Notably, anti-tag containing target RNAs block the degradation of substrate RNA, whereas target RNA without anti-tag retain cleavage capacity on substrate RNA (Figure 1D).
We further performed in vivo RNA interference assays to investigate the effect of anti-tag in LshCas13 and LbuCas13 in E. coli cells (Figures 1E–1H, S1D, and S1E). We co-transformed E. coli with a plasmid encoding Cas13a and a CRISPR array targeting the mRNA of enhanced yellow fluorescent protein (EYFP), as well as a tetracycline-inducible plasmid expressing EYFP mRNA in the presence or absence of anti-tag. Plasmids lacking the CRISPR array were used as controls. The experimental data include quantification of EYFP mRNA knockdown by LshCas13a (Figure 1E) or LbuCas13a (Figure 1G) in E. coli cells and the effect on the growth rate of E. coli cells upon EYFP mRNA interference by LshCas13a (Figures 1F and S1D) and LbuCas13a (Figures 1H and S1E).
After adding tetracycline, we observed significant decrease in EYFP fluorescence in the absence of anti-tag, indicating that Cas13a mediates EYFP mRNA knockdown. Upon transcription of EYFP mRNA carrying anti-tag, no decrease or partial decrease in EYFP fluorescence was observed, indicating failure of RNA targeting by Cas13a (Figures 1E and 1G). Previous data show that RNA targeting by LshCas13a induces growth restriction phenotype via trans-RNase activity on cellular mRNAs (Abudayyeh et al., 2016). We then examined the effect on growth of cells mediated by anti-tag (Figures 1F, 1H, S1D, and S1E). Upon induction of EYFP transcription, cells showed substantial growth suppression in the absence of anti-tag. In contrast, we observed partial or no growth restriction on cells with transcribed anti-tag containing EYFP mRNA. In our in vitro and in vivo assays for LbuCas13a, 5 nt anti-tag provides partial protection of RNA targets and reduced EYFP fluorescence and growth rate, indicative of incomplete inhibition (Figures 1B, 1G, 1H, and S1E). To digest the same amount of RNA targets, higher concentrations and much longer reaction times were required for LshCas13a in our in vitro cleavage assays (see details in STAR methods), indicating that LbuCas13a exhibits much higher cis- and trans-enzymatic activities than LshCas13a under our reaction conditions. A higher reaction rate of LbuCas13a might explain the observation of incomplete protection by 5 nt anti-tag.
Collectively, anti-tag segments inhibit the cis- and trans-RNase activity of both LshCas13a and LbuCas13a in vitro and in vivo. In addition, the 8 nt anti-tag segment provides full protection against Cas13a-generated degradation, similar to the effect of mutations in dCas13a (deactivated Cas13a), such that our in vitro data are consistent with previously reported in vivo data (Meeske and Marraffini, 2018).
Previous biochemical data showed that addition of anti-tag had no impact on the integrity of the crRNA-bound Cas13a complex (Meeske and Marraffini, 2018). We next addressed whether RNA harboring anti-tag directly interacts with Cas13a in solution using size exclusion chromatography (SEC). We used RNA4, RNA5, and RNA6 to eliminate the effect of different RNA lengths, as well as dCas13a to avoid the potential cleavage of target RNA. After incubation with Cas13a-crRNA binary complex, target RNAs formed stable complexes with both LshCas13a and LbuCas13a in solution regardless of the presence or absence of anti-tag (Figures 1I and 1J). We noted minor shifts in elution volumes of RNA5 and RNA6 compared with RNA4, indicating the potential for conformational changes of Cas13a induced by pairing of the anti-tag segment. To rule out the effect of crRNA spacer length, we also investigated whether the conformational changes can be induced with a 21 nt spacer. The shift of elution volume was observed with the addition of target RNA carrying anti-tag segment, suggestive of potential conformational changes induced by tag:anti-tag pairing (Figures S1F and S1G). Collectively, extended tag:anti-tag duplex formation blocks RNA degradation and the blockage may potentially be indicative of conformational changes of Cas13a.
Overall structure of LshCas13a-crRNA-target RNA complex harboring an anti-tag segment
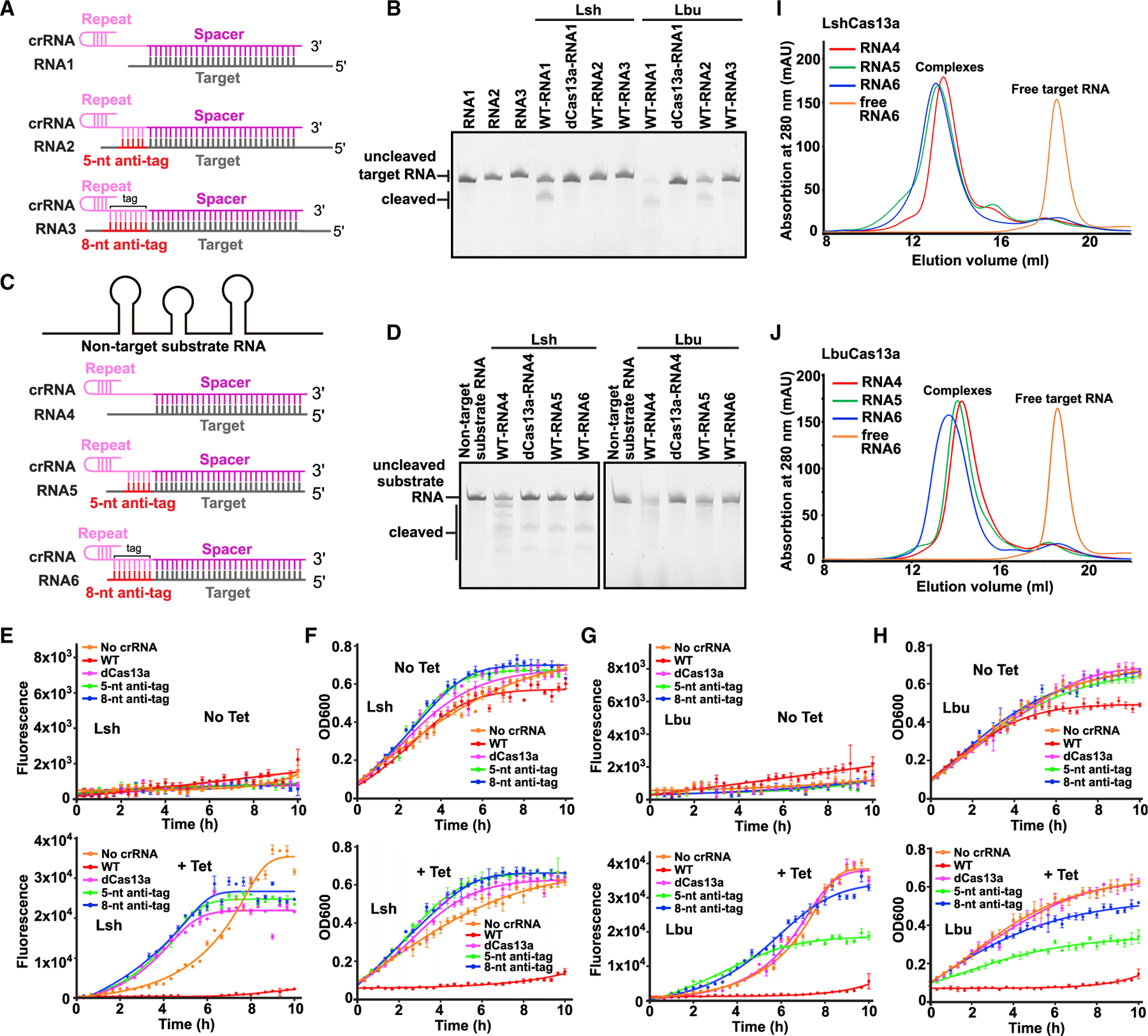
The domain architecture of LshCas13a is depicted in Figure 2A. To understand the inhibition mechanism of anti-tag harboring RNA, we determined the cryo-EM structure of LshCas13a-crRNA-target RNA ternary complex containing an extended 8 nt anti-tag at 3.06 Å resolution (Figures 2B–2D and S2; Table S1). We used the 37 nt RNA6 for sample preparation, which contains a 28 nt target segment followed by an 8 nt anti-tag complementary to the 3′-flank of the repeat tag element (Figure S1B, lower panel). LshCas13a assumes a bilobal architecture with the REC and NUC lobes organized around the guide:target duplex, similar to previously reported crystallographic structures of complexes lacking the anti-tag segment (Liu et al., 2017a, 2017b). The REC lobe, which is composed of the N-terminal domain (NTD) and Helical-I domains, binds to the crRNA repeat, while the NUC lobe, composed of the Helical-II, Linker, HEPN-I, and HEPN-II domains, binds to the guide:target duplex. All but one of the Cas13a domains and the guide:target RNA duplex could be reliably modeled, but no clear electron density was observed for the NTD domain (Figures S3A and S3B). In addition, the first 5 nt of the 5′-handle of crRNA and the terminal 2 bp of the RNA duplex extending outside of the bound protein have no clear electron density (Figures 2D and S3B).
Figure 2. Overall structure of LshCas13a-crRNA-anti-tag RNA complex.
(A) Domain organization of LshCas13a. The NTD domain of LshCas13a has no clear density and is indicated by the dashed box.
(B) Ribbon representation of LshCas13a-crRNA-anti-tag RNA6 ternary complex. Color codes of RNA and Cas13a are defined as in Figure 1C and (A), respectively.
(C and D) Ribbon representation and schematic of crRNA:target RNA duplex. The anti-tag is complementary with 3′-flank of crRNA repeat and forms an extended A-form RNA duplex beyond the guide:target duplex. Nucleotides not observed in the structures are colored gray in (D).
(E) Schematic representation of the conformational changes occurring in crRNA upon anti-tag RNA or target RNA loading. Nucleotides not observed in the structures are indicated by the dashed box. The RNA duplexes inside the binding channel are indicated by yellow boxes.
See also Figures S1–S3 and Table S1.
In previous structures of Cas13a-crRNA complexes in the presence or absence of target RNA lacking an anti-tag segment, the intact stem-loop in the repeat region (four C-G pairs and one A-U pair flanking a 2 nt bulge) (schematic in Figure 2E, middle and bottom panels) is sandwiched by the NTD and Helical-I domains (PDB: 5WTK, 5W1H, and 5XWY for LshCas13a, LbaCas13a, and LbuCas13a in complex with crRNA, respectively; PDB: 5XWP for LbuCas13a-crRNA-target RNA complex) (Knott et al., 2017; Liu et al., 2017a, 2017b). When bound to anti-tag RNA as observed in our structure of the complex, the 8 nt repeat tag segment immediately upstream of the spacer region, which is complementary with the anti-tag segment, forms a continuous A-form RNA duplex (schematic in Figure 2E, top panel), resulting in the disruption of A(−26):U(−6) base pair and base pair formation between U(−6) of crRNA and A(−6)of target RNA anti-tagregion (schematic in Figure 2D and structure in Figure 2C). The remaining four C:G pairs of the stem-loop retain their helical alignment and interact with the Helical-I domain (Figure 2B), while interaction with the NTD domain remains unclear because of poor density in this region.
Together, tag:anti-tag pairing involving residues −1 to −8 of the crRNA tag element disrupts the lower segment of the stem-loop (Figures 2E, top panel) such that the tag:anti-tag duplex is collinearly stacked with the guide:target duplex but not with the residual stem-loop duplex composed of four C:G base pairs (Figure 2C).
Recognition of extended duplex formed by crRNA and anti-tag target RNA
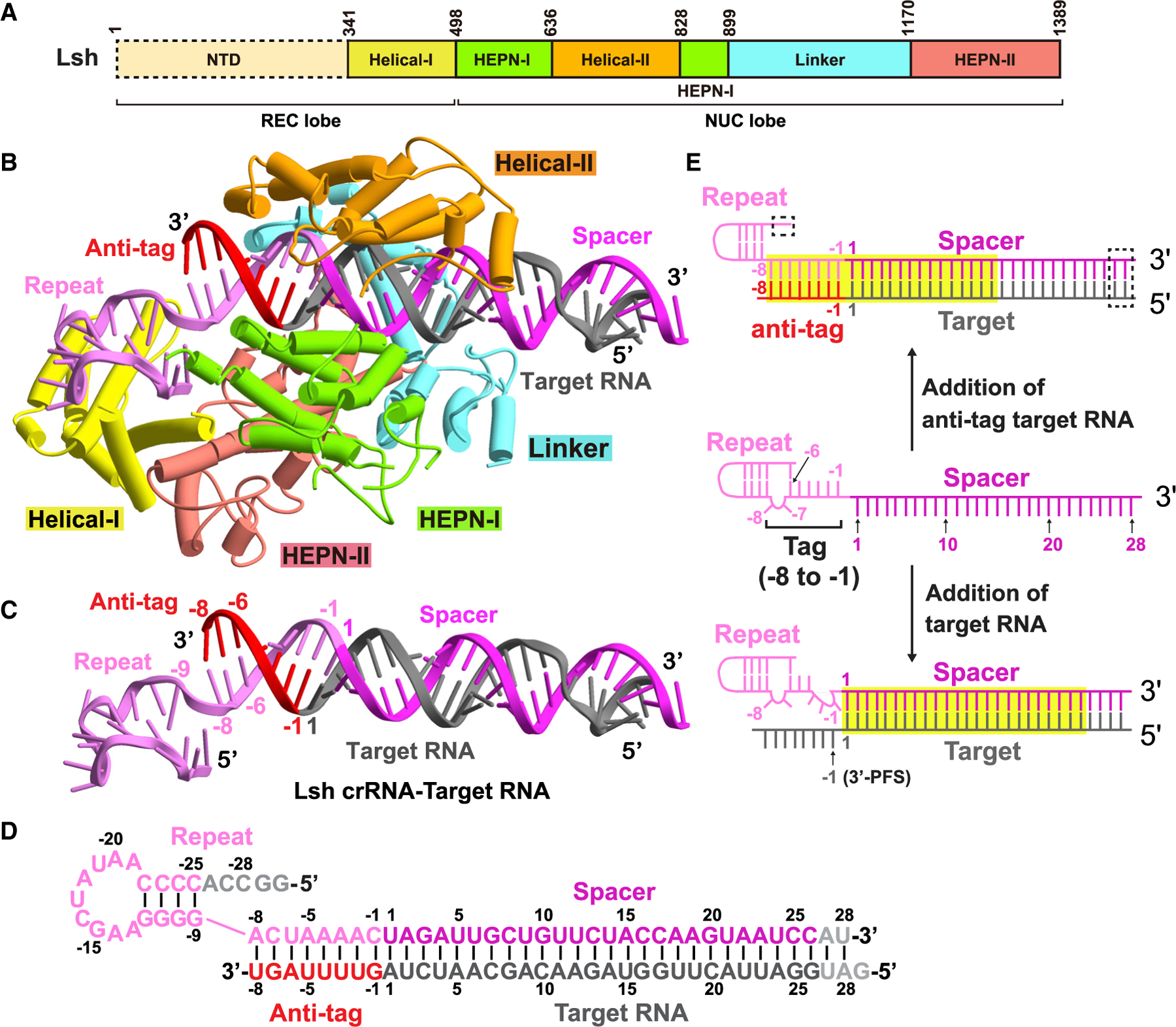
The recognition of crRNA:anti-tag target RNA extended duplex by LshCas13a NUC lobe comprises two parts: the tag:anti-tag duplex and the guide:target duplex (Figures 2B and 3A). In the reported RNA target-bound LbuCas13a structure, the guide:target duplex binding channel accommodates around 24 bp, while the tag region of crRNA is bound by the REC lobe (Figure 2E, lower panel) (Liu et al., 2017a). In our structure of the ternary complex, which contains an anti-tag element, the A(−8)-C(−1) tag segment of the crRNA repeat region is complementary with its anti-tag counterparts and forms a 8 bp RNA duplex (Figure 2E, top panel) that protrudes into the RNA binding channel (Figures 2B and 3A). The recognition of the tag:anti-tag duplex is contributed mainly by the Helical-II and Linker domains in a non-sequence-specific manner (Figure 3B). In detail, the phosphates of A(−4) to C(−1) of the tag in the repeat region hydrogen-bond to Ser672 in the Helical-II domain and Asn1119 and Gly1120 in the Linker domain. The riboses of A(−4) to A(−5) in the anti-tag region are stabilized by the side chains of Ser763 and Gln759 in the Helical-II domain. The 3 bp downstream of the stem-loop in the repeat region have no contact with Cas13a and are exposed to solvent.
Figure 3. Detailed interactions of LshCas13a with crRNA:anti-tag RNA duplex.
(A) Surface views of the interfaces between Cas13a and duplex formed by crRNA and anti-tag RNA.
(B) Recognition of tag:anti-tag duplex by the Linker and Helical-II domains.
(C and D) Interactions between Cas13a and base pairs 2–6 (C) and 7–15 (D) in the guide:target duplex.
(E) Surface views of pre-target-bound LshCas13a-crRNA complex, with interactions between crRNA tag region and LshCas13a shown in the zoomed-in panel.
See also Figure S3.
The 28 bp guide:target duplex immediately downstream of tag:anti-tag duplex extends along the channel, with the terminal 13 bp located beyond the channel (Figures 2B and 3A). Given that the tag:anti-tag duplex occupies a part of the binding channel, only the remaining segment of the channel becomes available for accommodation of guide:target duplex. This explains why anti-tag prevents catalytic activity with either 21 or 28 nt spacers and implies a common inhibition mechanism for 21- to 28-nt-length spacers.
In the anti-tag-proximal guide:target duplex region (positions 2–6), the sugar-phosphate backbones of the G3 to U6 segment of crRNA interacts with the HEPN-I and HEPN-II domains, while the A6 to U3 segment of target RNA hydrogen bonds with the Linker, Helical-II, and HEPN-II domains (Figure 3C). The anti-tag-distal guide:target duplex (positions 7–15) is recognized by hydrophilic interactions with backbones coordinated mainly by the Linker and Helical-II domains in a sequence-independent manner (Figure 3D).
lshCas13a presents a catalytically inactive HEPN domain architecture with anti-tag RNA
We next compared the conformations of the two HEPN domains of LshCas13a in the pre-target-bound and anti-tag RNA-bound states with the structures aligned relative to their HEPN-II domains. Despite observing conformational changes for the two HEPN-I domains between the two states (Figure 4A), the four conserved Arg597, His602, Arg1278, and His1283 catalytic residues in the two HEPN domains in the anti-tag-bound state share nearly identical main-chain position and side-chain direction to those in pre-target-bound state (Figure 4A, expanded panel). Collectively, these results definitively demonstrate that anti-tag RNA loading locks Cas13a into a catalytically inactive architecture as defined from the relative positioning of key catalytic residues in the HEPN-I and HEPN-II domains.
Figure 4. Comparison of HEPN pocket alignments and global folds between ternary complexes involving bound anti-tag (LshCas13) and target (LbuCas13) RNAs.
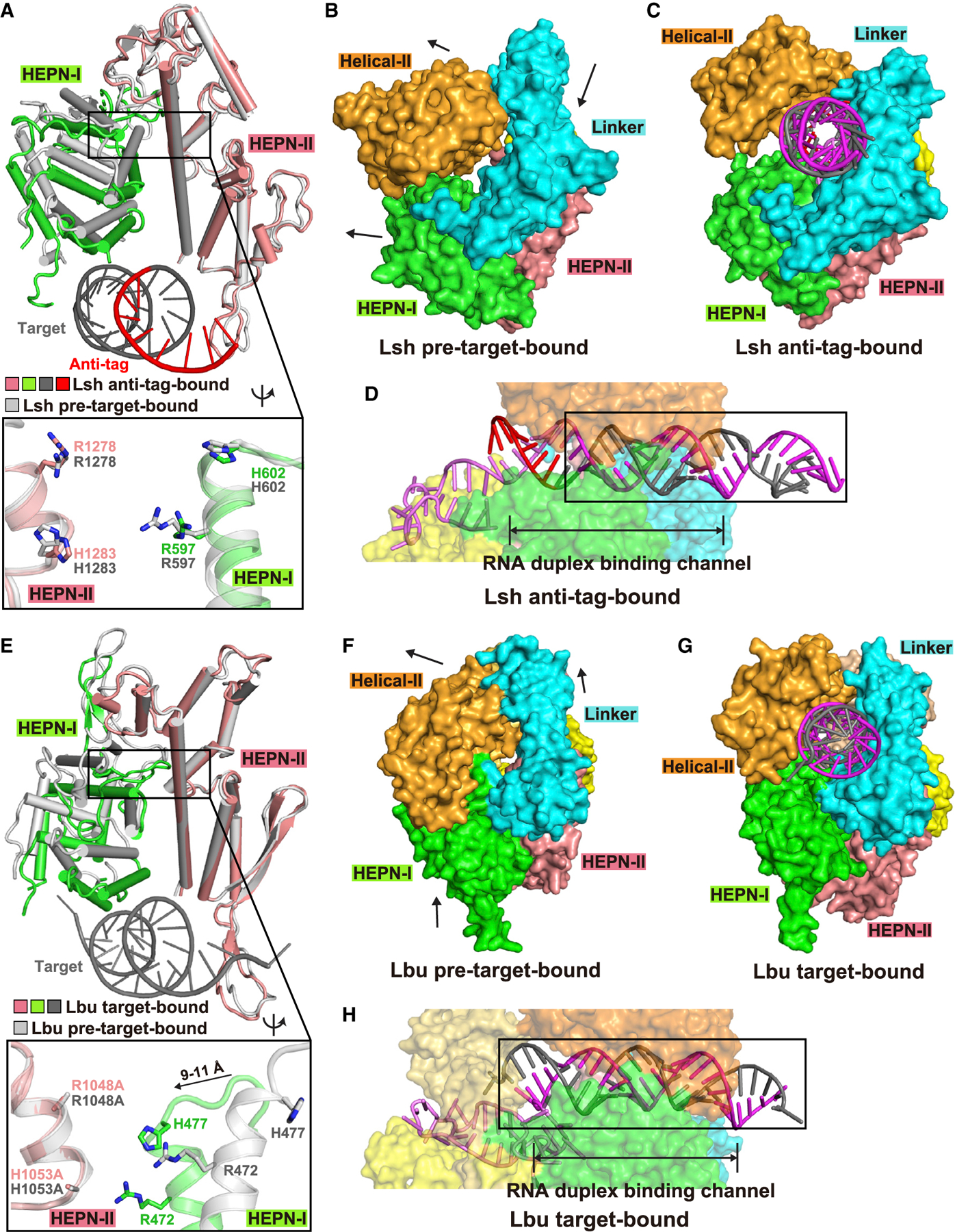
(A) Structural comparisons of two HEPN domains of LshCas13a in anti-tag-bound (HEPN-I in green, HEPN-II in salmon, target segment in gray, and anti-tag in red) and pre-target-bound (in silver) states by superposing the HEPN-II domains. For simplicity, only target RNA is shown. Comparison of the positioning of the four catalytic residues from the pair of HEPN domains is shown in the zoomed-in segment (inset).
(B and C) Surface of LshCas13a showing domain rearrangements to generate crRNA:target RNA duplex binding channel from pre-target-bound (B) to anti-tag-bound (C) states. Black arrows in (B) show the directions of domain movements on ternary complex formation with anti-tag RNA.
(D) Surface views of the interfaces between crRNA-bound LshCas13a and duplex formed by bound anti-tag RNA. The guide:target duplex segment is boxed.
(E) Structural comparisons of two HEPN domains of LbuCas13a in target-bound (HEPN-I in green, HEPN-II in salmon, target segment in gray) and pre-target-bound (in silver) states by superposing the HEPN-II domains (Liu et al., 2017a). For simplicity, only target RNA is shown. Double mutation R1048A/H1053A was used for structural studies of target RNA-bound LbuCas13a. Comparison of the positioning of the four catalytic residues from the pair of HEPN domains is shown in the zoomed-in segment (inset).
(F and G) Surface of LbuCas13a showing domain rearrangements to generate crRNA:target RNA duplex binding channel from pre-target-bound (F) to target-bound (G) states. Black arrows in (F) show the directions of domain movements on ternary complex formation with target RNA.
(H) Surface views of the interfaces between crRNA-bound LbuCas13a and duplex formed by bound target RNA. The guide-target duplex segment is boxed.
See also Figure S4.
Surface views of the pre-target-bound and anti-target-bound structures of LshCas13 are shown in Figures 4B and 4C, with the alignment of the guide-target duplex in the binding channel shown in Figure 4D (boxed segment).
Domain rearrangement of LshCas13a on complex formation with anti-tag target RNA
A structural comparison between the pre-target-bound (Liu et al., 2017b) and anti-tag-bound (this study) LshCas13a reveals a minor domain shift in the REC lobe and a large “rigid body” shift in the NUC lobe, with the two lobes moving apart on complex formation (Figure 5A). The Helical-I domain that interacts with the stem-loop of the repeat region aligns well in the two states and exhibits small conformational changes (Figure 5B). The HEPN-I and HEPN-II domains move away from the REC lobe and present modest domain shift rotations (larger rotation and shift for HEPN-I relative to HEPN-II, with relatively minor changes for positions of catalytic residues) in a similar direction (Figures 4A and 5B) to form a part of the RNA duplex binding channel. By contrast, the Helical-II and Linker domains show large conformational changes and move away from the NUC lobe in opposite directions (Figure 5B). All these domain movements generate a binding channel to accommodate the guide:target heteroduplex (compare Figures 4B and 4C and Figures 5C and 5D).
Figure 5. Conformational changes in LshCas13 upon anti-tag RNA loading.
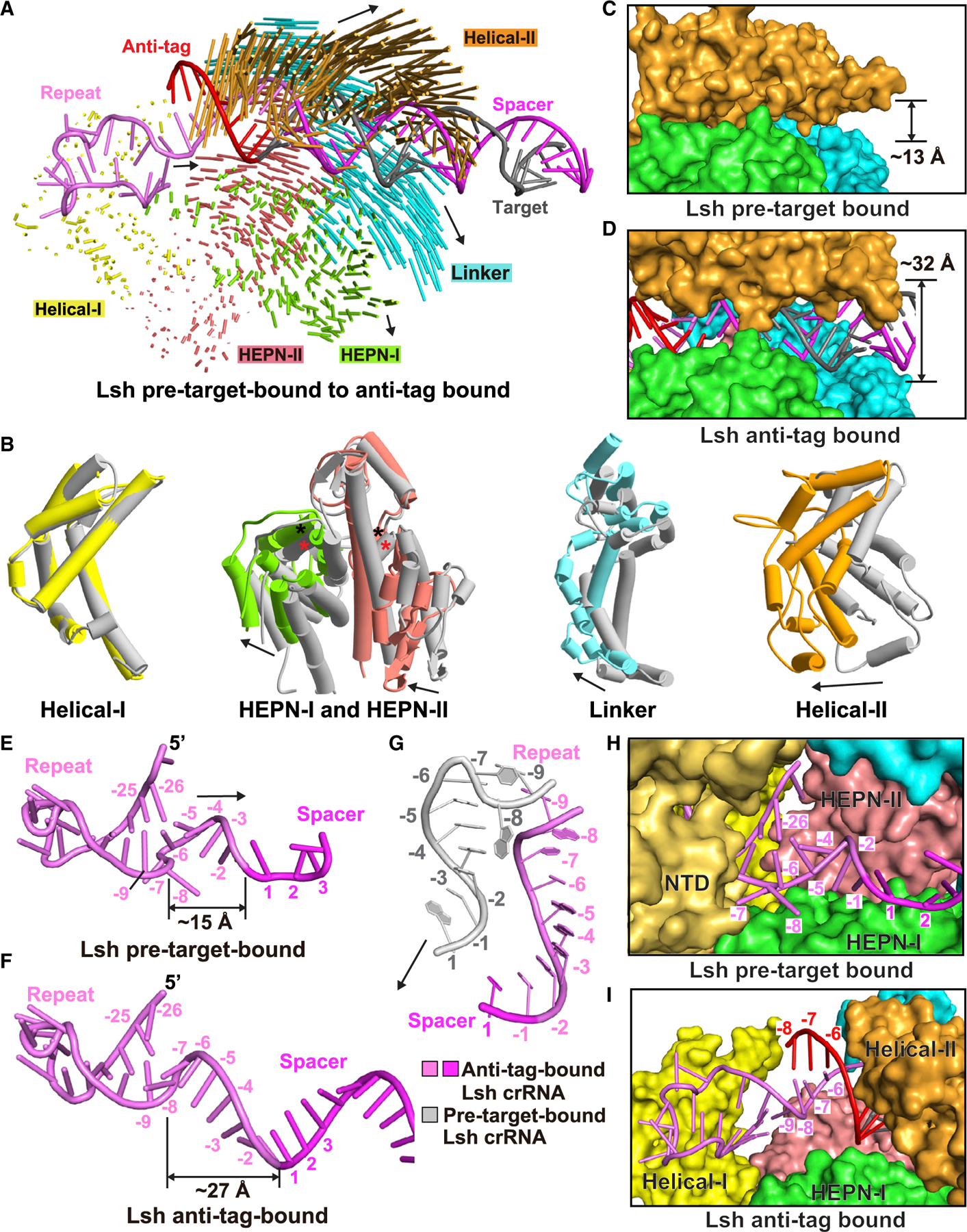
(A) Structural comparison between LshCas13a-crRNA binary complex and LshCas13a-crRNA-anti-tag RNA ternary complex. Vector lengths correlate with the domain motion scales. Arrows show the directions of domain movement from pre-target-bound to anti-tag-bound states.
(B) Structural comparison of the Helical-I, HEPN-I, HEPN-II, Linker, and Helical-II domains between LshCas13a-crRNA binary (in silver) and LshCas13a-crRNA-anti-tag RNA ternary (in color) complexes. Arrows indicate the domain movements. The key catalytic residues from the pair of HEPN domains are indicated by black (anti-tag-bound) and red (pre-target-bound) asterisks, respectively.
(C and D) Binding with target RNA harboring anti-tag widens the guide:target duplex-binding channel on proceeding from LshCas13a-crRNA binary complex (C) to the LshCas13a-crRNA-anti-tag RNA ternary complex (D).
(E–G) Architectures of crRNA in LshCas13a-crRNA binary (E) and LshCas13a-crRNA-anti-tag RNA ternary (F) complexes. The details of tag region are shown in (G), with anti-tag-bound crRNA in color and pre-target-bound crRNA in silver.
(H and I) Comparisons of the tag region of crRNA in LshCas13a-crRNA binary (H) and LshCas13a-crRNA-anti-tag RNA ternary (I) complexes. The Helical-II domain, which interacts with and covers the tag region in LshCas13a-crRNA complex, is hidden in (H) to show the position of tag region.
See also Figure S4.
It should be noted that the tag region (positions −8 to −1) undergoes significant conformational changes upon binding anti-tag RNA, while the upper segment of the crRNA stem-loop (positions −26 to −9) keeps a similar architecture (Figures 5E–5G). In the reported LshCas13a-crRNA complex, A(−8) and C(−7) flip out and form a 2 nt bulge between base pairs of C(−25):G(−9) and A(−26):U(−6) (Figure 5E) (Liu et al., 2017b). U(−6) stacks with A(−5) and the remaining A(−5) to C(−1) segment of the tag region, together with the following nucleotides 1–4 of the spacer region, forms a nearly A-form conformation (Figure 5E). After anti-tag RNA binding, region A(−5) to C(−1) remains in a similar conformation and pairs with anti-tag, forming an extended A-form RNA duplex (Figure 5F). In contrast, the A(−8) to U(−6) region exhibits dramatically conformational changes. The A(−26):U(−6) base pair is disrupted, and nucleotides A(−8) and C(−7) flip back and stack with U(−6) to exhibit a continuous A-form conformation with the following 3′-part of the tag region (Figure 5F), forcing the tag region to move apart from the NTD and HEPN-I domains and positioning it within the nucleic acid-binding channel (Figures 5G–5I).
Furthermore, the interactions between the tag region and Cas13 are broken and rebuilt on proceeding from the binary to anti-tag containing ternary complex. Gln759 and Ser763 in the Helical-II domain hydrogen bond to the base of A(−8) in the pre-target-bound state (Figure 3E), whereas these two residues hydrogen bond to A(−4) and A(−5) of anti-tag segment in the anti-tag-bound state (Figure 3B). Moreover, residues of the HEPN-I domain involved in the stabilization of A(−8) and C(−7) in the pre-target-bound state (Figures 5H and 3E) have no contact with the RNA duplex on anti-tag ternary complex formation (Figure 5I).
Collectively, the 5′-part of the tag region (positions −8 to −6) undergoes significant conformational changes upon binding anti-tag RNA, while the 3′-part of tag region (positions −5 to −1) retains a similar architecture. The tag:anti-tag pairing results in large conformational changes in the Helical-II and Linker domains necessary for positioning of the tag:anti-tag duplex in the binding channel on proceeding from the pre-target-bound to anti-tag-bound states of LshCas13a. Nevertheless, both states retain the separated alignments of HEPN-I and HEPN-II domains (Figure 4A, expanded panel), preventing formation of a composite catalytic pocket necessary for RNA cleavage.
LbuCas13a presents a catalytically active HEPN domain architecture with target RNA
Previous structural studies of LbuCas13a on ternary complex formation with target RNA established that formation of guide:target duplex results in a conformational change in the HEPN domains (Figure 4E) that facilitates the catalytic residues in the two HEPN domains to shift closer together to form a composite pocket (Figure 4E, expanded panel), resulting in competent RNase activity (Knott et al., 2017; Liu et al., 2017a, 2017b; Slaymaker et al., 2019; Zhang et al., 2018a, 2018b, 2019). Surface views of the pre-target-bound and target-bound structures of LbuCas13 are shown in Figures 4F and 4G, with the alignment of the guide-target duplex in the binding channel shown in Figure 4H.
Notably, there is a register shift in the positioning of the guide-target duplex (boxed segment) on proceeding from the target-bound LbuCas13a ternary complex (Figure 4H) to its anti-tag-bound LshCas13a ternary complex counterpart (Figure 4D). The collinearly stacked tag:anti-tag duplex and guide:target duplex (boxed segment in Figure 4D) are both accommodated within the nucleic acid-binding channel in the ternary complex with anti-tag target RNA (this study), in contrast to the sole accommodation of the guide:target duplex in the ternary complex with target RNA (boxed segment in Figure 4H) (Liu et al., 2017a).
Domain rearrangement of LbuCas13a on complex formation with target RNA
We briefly summarize the previously reported conformational changes on proceeding from the LbuCas13-crRNA binary complex to the ternary complex with added bound-target RNA (Liu et al., 2017a). Upon target RNA loading, similar domain movements in the NUC lobe are observed during the transition from pre-target-bound to target-bound states (Figures S4A and S4B) to form the guide:target duplex binding channel (Figure S4C), except for the movement of the HEPN domains in different directions (Figure S4B), thereby positioning them spatially in close proximity to generate a catalytically competent composite binding pocket (Figure 4E, expanded panel). Furthermore, pre-ordered nucleotides A(−3) to C(−1) in the 3′-part of the tag region flip out and form a U-shaped kink upon target RNA loading (Figures S4E and S4F), with C(−1) stacking with A(−8) and the following spacer region moving toward the REC lobe to position the guide:target duplex in the binding channel (Figure S4F). Numerous interactions are formed between the bases of flip-out nucleotides A(−3) to C(−1) and the HEPN-I domain during the transition from binary to ternary complex (Figures S4G and S4H).
Collectively, the addition of target RNA induces large conformational changes of the 3′-part of tag region (positions −3 to −1), while the 5′-part of tag region (positions −8 to −4) retains a similar conformation, with significant domain movements of LbuCas13a for accommodation of guide:target duplex and formation of a composite catalytic pocket for RNA hydrolysis (Liu et al., 2017a).
Attempts at cryo-EM structure determination of LshCas13a-crRNA-target RNA complex
We attempted a multitude of approaches toward cryo-EM structural characterization of target RNA-bound LshCas13a ternary complex for direct comparison with its anti-tag counterpart. We observed a large amount of target RNA disassociation for the target RNA-bound LshCas13a ternary complex during cryo-EM specimen preparation. We tried different sample preparation conditions, including blotting, variations in pH and salt conditions, cross-linking, and different target RNAs. None of these five separate attempts were successful for high-resolution structure determination. As an example, we collected a large cryo-EM dataset that yielded after three-dimensional (3D) classification a limited number of particles of target RNA-bound ternary Cas13a complex with strong preferred orientation (Figure S5). These data yielded crRNA-bound binary LshCas13a complex at 3.7 Å resolution, but target RNA-bound ternary LshCas13a could only be solved at low 4.0 Å resolution (Figure S5). However, the resolution of the reconstructed map of target-bound LshCas13a ternary complex is overestimated because of the preferred orientation issue. The map was severely elongated along one direction because of preferred orientation and may contain artifactual densities, which prevents the accurate model building of target-bound LshCas13a ternary complex (Figures S5D and S5E).
As mentioned in an earlier section, a structure is available for the LbuCas13a-crRNA-target RNA complex (Liu et al., 2017a), and hence we used this structure as a surrogate for its unavailable Lsh counterpart. LshCas13a and LbuCas13a share 22.1% identity and 36.9% similarity (calculated using Clustal Omega; Madeira et al., 2019), with the sequence alignment shown in Figure S6. The key residues (catalytic residues R597, H602, R1278, and H1283 in LshCas13a and R472, H477, R1048, and H1053 in LbuCas13a, as well as most residues contributing to RNase activities as discussed below) and secondary structures are conserved between the two species (Figure S6). Besides, the Lsh-crRNA tag region (positions −8 to −1) in pre-target-bound LshCas13a-crRNA complex shares nearly identical architecture to that of Lbu-crRNA tag region (Figure S7).
Structural differences between target-bound LbuCas13 and anti-tag-bound LshCas13 ternary complexes
As the LshCas13a-crRNA-target RNA ternary complex has not been determined yet, we used the corresponding LbuCas13a ternary complex with bound target RNA (Liu et al., 2017a) for structural comparison. This allowed us to investigate the structural difference between anti-tag-bound LshCas13a (this study) and target-bound LbuCas13a (Liu et al., 2017a) and found large conformational differences between these two ternary complexes (Figure 6A), with the domain shift difference occurring mainly in the NUC lobe. It is worth noting that nearly all domains in the NUC lobe move in the opposite direction during the anti-tag-bound to target-bound transition, compared with the transition from pre-target-bound to anti-tag-bound state (compare Figures 5A and 6A).
Figure 6. Structural comparison between target-bound (LbuCas13a) and anti-tag-bound (LshCas13a) ternary complexes.
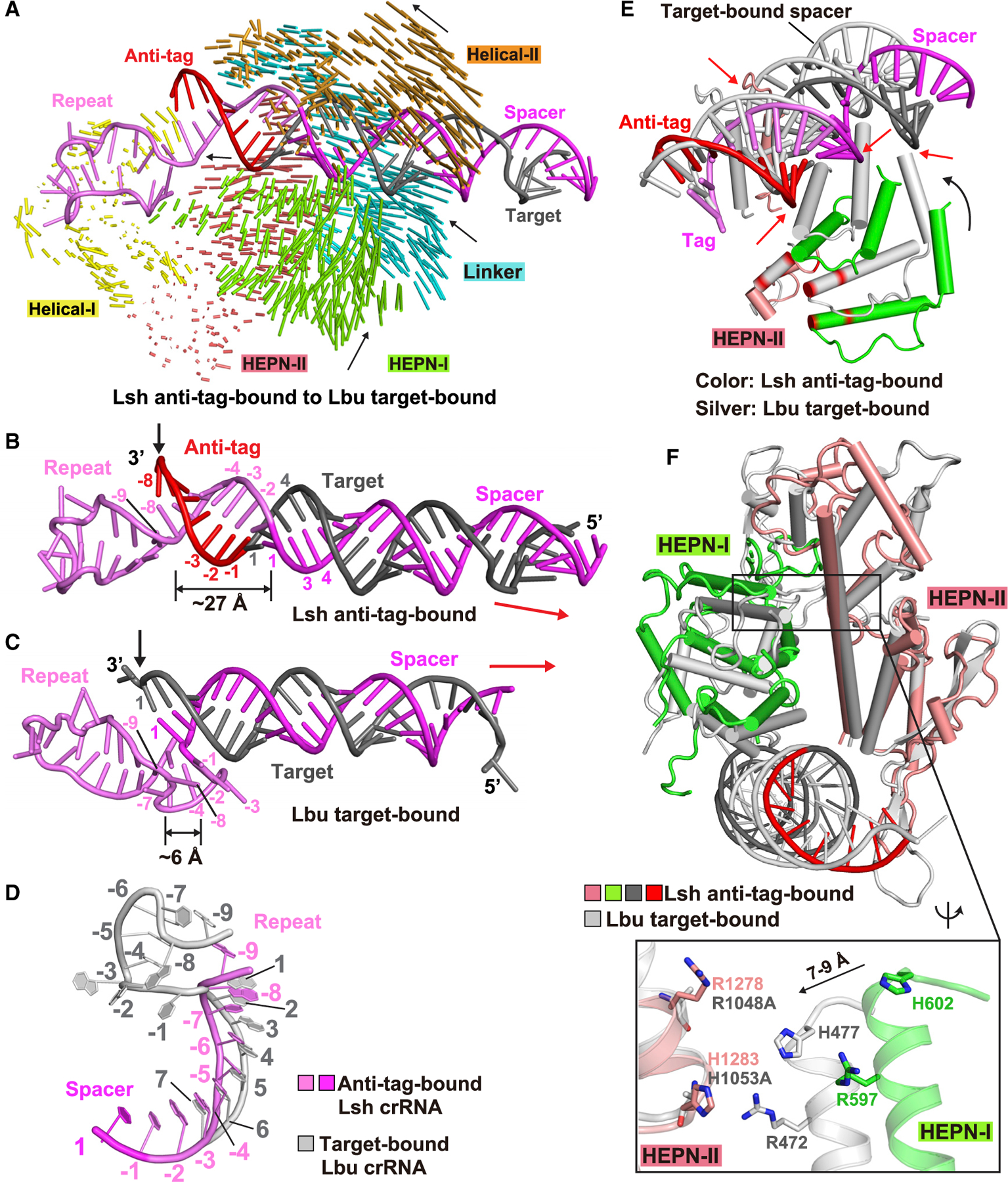
(A) Structural comparison between LshCas13a-crRNA-anti-tag RNA (this study) and LbuCas13a-crRNA-target RNA (Liu et al., 2017a) ternary complexes. Vector lengths correlate with the domain motion scales. Arrows show the directions of domain movement from anti-tag-bound to target-bound states.
(B–D) Architectures of crRNA in LshCas13a-crRNA-anti-tag RNA (B) and LbuCas13a-crRNA-target RNA (C) ternary complexes. The positions of 3′ end of target RNAs are indicated by black arrows. The extension directions of RNA duplexes are indicated by red arrows. The details conformational changes of the tag region are shown in (D), with anti-tag bound crRNA in color and target-bound crRNA in silver.
(E) Superposition of LshCas13a-crRNA-anti-tag RNA (in color) and LbuCas13a-crRNA-target RNA (in silver) ternary complexes with the focus on HEPN-I and HEPN-II domains. The key catalytic residues in HEPN domains are highlighted in red. The black arrow indicates the movements of HEPN-I domain toward the HEPN-II domain from anti-tag-bound to target-bound states. The red arrows indicates the steric clashes between crRNA:anti-tag RNA duplex and the HEPN domains in target-bound state, indicating that the formation of tag:anti-tag RNA duplex prevents the movements of HEPN domains to generate a competent composite catalytic pocket.
(F) Structural comparisons of two HEPN domains LshCas13a in anti-tag-bound (HEPN-I in green, HEPN-II in salmon, target segment in gray, and anti-tag in red) and LbuCas13a in target-bound bound (in silver) states by superposing the HEPN-II domains. For simplicity, only target RNA is shown. Comparison of the positioning of the four catalytic residues from the pair of HEPN domains is shown in the zoomed-in segment (inset).
See also Figure S4.
The nucleotides A(−8) to A(−4) in the 5′-part of the tag region retain a similar conformation during the transition from pre-target-bound to target-bound (Figure S4D) states of LbuCas13a (Figure S4E), while the A(−8) to U(−6) segment undergoes large conformational changes during the transition from pre-target-bound (Figure 5E) to anti-tag-bound (Figure 5F) states of LshCas13a (Figure 5G). These differences in the tag region of the two ternary complexes results in differences in the position and extension direction of guide:target duplex.
The formation of U-shaped kink in the tag region upon target RNA loading shortens the distance between nucleotides A(−8) and A(−1) (compare Figure 6C for target-bound versus Figure 6B for anti-tag-bound ternary complexes), possibly driving the guide:target duplex to move toward to the REC lobe. Furthermore, A(−1) stacks with the following spacer region upon anti-tag RNA binding (Figure 5F), whereas A(−1) flips and stacks with A(−8) and forms a kink with the following spacer region upon target RNA binding (Figures S4E and S4F), with a consequence that the RNA duplex extends in different direction (Figures 6B–6D). The position of tag:anti-tag duplex prevents the movement of the HEPN-I domain toward to the HEPN-II domain, thereby forcing the separation apart of the catalytic residues (Figures 6E and 6F).
Also note that the spacer region in the target-bound state (Figure 4H) is located at a relatively similar position to that of tag A(−8) to C(−1) segment in the anti-tag-bound state (Figure 4D) in the duplex binding channel.
Collectively, distinctly different conformational transitions are observed on proceeding from the pre-target-bound binary complex to target-bound (Liu et al., 2017a) versus anti-tag bound (this study) ternary complexes. Formation of the target-bound complex does not disrupt the base pair and the bulge of the tag segment but instead the conformational transitions of stacked nucleotides A(−3) to C(−1) (Figures S4E and S4F), resulting in bringing the two HEPN domains into close proximity to generate a catalytically competent composite pocket (Figure 4E, expanded panel) (Liu et al., 2017a). In contrast, formation of the anti-tag-bound ternary complex disrupts and realigns the tag segment as a result of tag:anti-tag pairing (Figure 5F) but retains separation of the catalytic residues of HEPN-I and HEPN-II domains (Figure 4A, expanded panel), thereby suppressing RNA cleavage activity.
Key residues contributing to Cas13a activities
Upon anti-tag RNA binding, the conformational changes of A(−8) to U(−6) facilitates the relocation of the RNA duplex and formation of new Cas13-RNA contacts, thereby preventing enzymatic activity. In efforts to monitor the cellular consequences (Figure 7) associated with the above conformational change, we generated RNA and protein mutants and undertook in vitro RNA cleavage assays (Figures 7A–7D) and in vivo RNA interference assays in indicated E. coli strains to evaluate the quantities of RNA knockdown levels by various LshCas13a (Figure 7E) and LbuCas13a (Figure 7G) mutants, as well as growth defects in liquid and solid media for LshCas13a (Figures 7F and 7I) and LbuCas13a (Figures 7H and 7J) mutants upon Cas13a-dependent RNA interference.
Figure 7. Inhibition of RNA cleavage by blockage of conformational changes in tag segment.
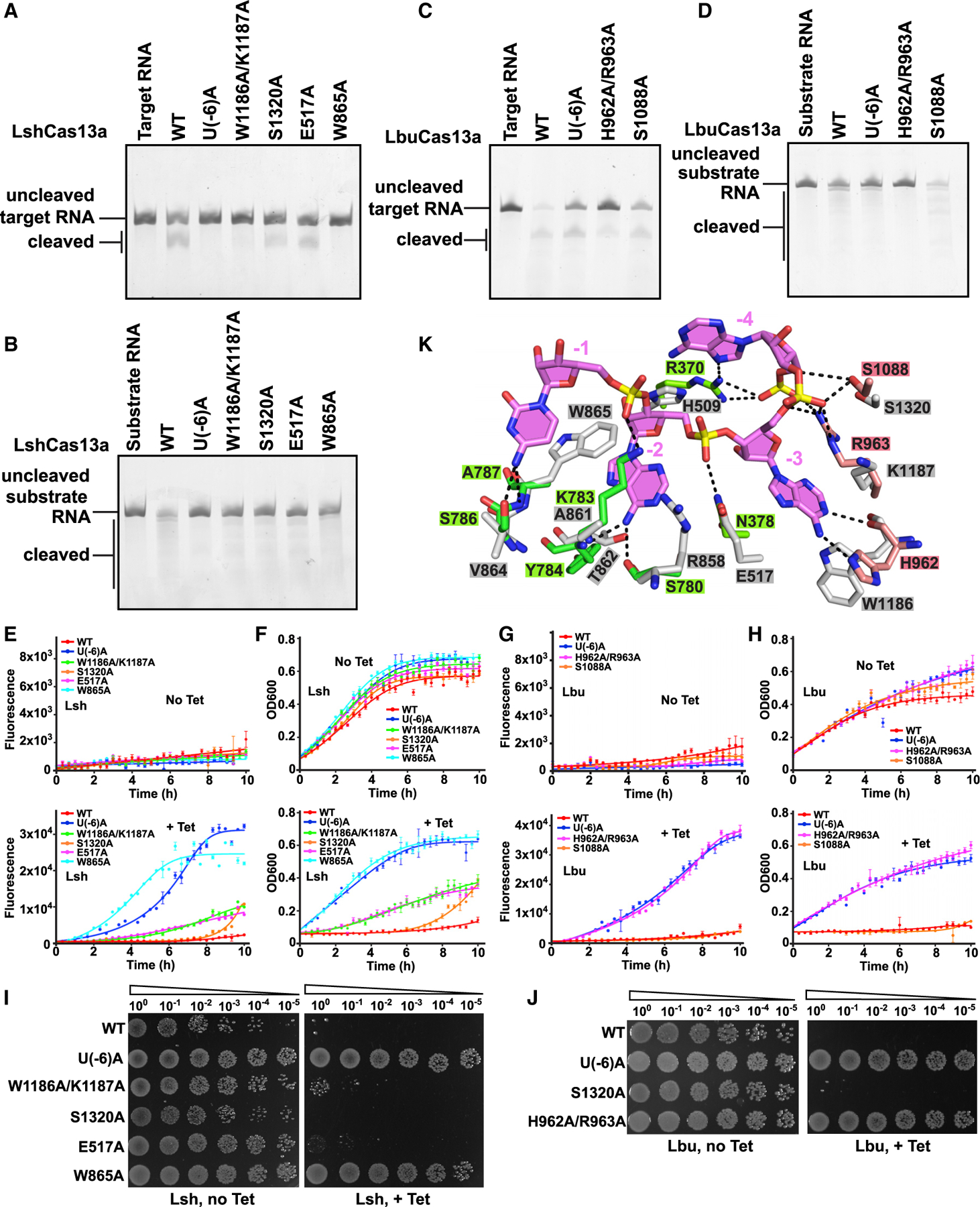
(A and B) Cleavage assays monitoring degradation on target RNA (A) and substrate RNA (B) with Lsh-crRNA or LshCas13a mutants.
(C and D) Cleavage assays by LbuCas13a monitoring degradation on target RNA (C) and substrate RNA (D).
(E and G) In vivo RNA knockdown assays of indicated E. coli strains expressing various LshCas13a (E) and LbuCas13a (G). Error bars represent SEM of three biological replicates.
(F and H) Liquid growth assays for LshCas13a (F) and LbuCas13a (H) illustrating growth defects upon Cas13a-dependent RNA interference. Error bars represent SEM of three biological replicates.
(I and J) Plasmids interference assays for E. coli strains co-transformed with various LshCas13a (I) or LbuCas13a (J) plasmids as well as a tet-induced EYFP plasmid. Ten-fold serial dilution of cells as indicated were spotted onto plates.
(K) Recognition of crRNA 3′-flank (positions −3 to −1) by LbuCas13a (HEPN-I in green, HEPN-II in salmon, and crRNA tag segment in violet) and modeled LshCas13a (in silver).
See also Figure S4.
We generated mutation U(−6)A in LshCas13a and LbuCas13a crRNAs to prohibit formation of A(−26):U(−6) base pair. As expected, the mutant crRNAs incapacitated cis- and trans-RNase cleavage by Cas13a (Figures 7A–7D). We further validated RNA interference in vivo and observed that U(−6)A mutation had no capacity to reduce EYFP mRNA, as well as the growth rate (Figures 7E–7J).
We propose that LshCas13a undergoes similar conformational changes as LbuCas13a upon binding target RNA. Addition of anti-tag RNA induces different conformational changes and thereby inhibits the activation of Cas13a. As LshCas13a-crRNA-target RNA ternary complex has not been determined yet, we modeled a LshCas13a target-RNA-bound ternary complex on the basis of the reported structure of the LbuCas13a complex. We found that the residues in LbuCas13a hydrogen bonding with A(−3) to C(−1) tag segment share similar positions with their corresponding counterparts in LshCas13a (Figure 7K). We generated Ala mutations for residues (Glu517, Trp865, Trp1186, Lys1187, and Ser1320 in LshCas13a and His962, Arg963, and Ser1088 in LbuCas13a) interacting with the A(−3) to C(−1) tag segment through their side chains to impair the conformational changes of the tag segment. As we expected, all mutations show reduced or eliminated both cis- and trans-enzymatic activities, except for the LbuCas13a S1088A mutation, which showed higher trans-RNase activity (Figures 7A–7D), which is consistent with the reported reduced cis-RNase activities with mutations of H509A, R858A, and W865A in LshCas13a (Liu et al., 2017b) and H962A, R964A, and K783A in LbuCas13a (Liu et al., 2017a). We also investigated the effect of these mutations on in vivo RNA interference in E. coli cells (Figures 7E–7J). Similar to our cleavage assays, LbuCas13a S1088A mutation exhibited comparable capacity in RNA knockdown to wild-type protein (Figures 7H and 7J). Besides, the LshCas13a S1320A mutation retained comparable activities, while LshCas13a W1186A/K1187A and E517A mutations retained partial activities (Figures 7E, 7F, and 7I).
Collectively, the unique conformation of the crRNA tag region A(−3) to C(−1) is essential for Cas13a RNase activity. On addition with anti-tag RNA, formation of tag:anti-tag base-pairing in A(−3) to C(−1) tag segment blocks the conformational changes of tag segment and thus inhibits the catalytic activity.
DISCUSSION
One constraint of the present study has been our inability despite repeated attempts to solve the cryo-EM structure of LshCas13a-crRNA bound to target RNA at high resolution for direct comparison with LshCas13a-crRNA bound to anti-tag target RNA. This required us to instead make comparisons involving the published structure of LbuCas13a-crRNA bound to target RNA (Liu et al., 2017a) with LshCas13a-crRNA bound to anti-tag target RNA (this study), with its inherent limitations when comparing results from different Cas13a species (Figures S6 and S7).
Nevertheless, we would like to emphasize that the key comparison in the paper is the transition from pre-target-bound to anti-tag target-bound states, and such a comparison has been done on the same LshCas13 system (Figures 4A–4D and 5). Furthermore, the literature does provide information on the pre-target-bound to target-bound states for the same LbuCas13 system (Figures 4E–4H and S4) (Liu et al., 2017a), providing added confidence in our conclusions.
Anti-tag inhibition of cleavage activity in Cas13a systems
As reported previously, crRNA-bound Cas13a exhibits a catalytic inactive conformation before target RNA loading (Knott et al., 2017; Liu et al., 2017a, 2017b). The tag region (positions −8 to −1) exhibits a twisted 2 nt bulge (positions −8 to −7) followed by a 6 nt nearly A-form single-stranded conformation. In this state, the catalytic residues of the pair of HEPN domains are kept apart, reflective of an inactive state.
Upon the addition of target RNA lacking an anti-tag element, the flip-out of A(−3) to C(−1) tag segment in LbuCas13a undergoes large conformational changes and rebuilds the interactions with surrounding residues (Figures S4G and S4H), thereby changing the location of the spacer (Figures S4E and 6C). The unique conformation of the tag region is accommodated and stabilized mainly by the HEPN domains, facilitating the movements of two HEPN domains to form a composite pocket in the stabilized active state (Figure 4E, expanded panel) (Liu et al., 2017a, 2017b). We anticipate that LshCas13a shares a similar activation mechanism. Mutations in the key residues contributing to the maintenance of unique tag conformation impairs or eliminates the Cas13a activity in vitro and in vivo (Figures 7A–7J).
Addition of the anti-tag RNA results in the relocation of 2 nt bulge and the tag region (positions −8 to −1) is pulled out of its binding pocket because of tag:anti-tag duplex formation, disrupting the interactions between tag region and HEPN domains (Figures 3B and 3E). The tag:anti-tag duplex in turn collinearly stacks with the guide:target duplex and protrudes into and is encapsulated by the binding channel in the NUC lobe (Figure 3A). The rigid A-form tag:anti-tag duplex fixes the direction and position of the collinearly stacked guide:target duplex, which in turn occupies the location of the active state positioning of the HEPN-I domain. Such occupancy forces the HEPN domains apart from each other, thereby locking Cas13a in an inactive conformation (Figure 4A).
Previous in vivo studies reported that at least a 6 nt anti-tag was required for full inhibition of LseCas13a activity, while a shorter anti-tag exhibited some protective effect (Meeske and Marraffini, 2018), which is consistent with our in vitro and in vivo results for LshCas13a and LbuCas13a (Figures 1B, 1D–1H, S1D, and S1E). Combined with our structural data, we anticipate that the formation of a 5 bp tag:anti-tag could lock the 3′-part of the tag region (positions −5 to −1) and then further block the movement of guide:target duplex. The shorter tag:anti-tag duplex leaves open the possibility that the guide:target duplex moves away to allow the two HEPN domains to get closer together. The longer anti-tag breaks the base pair A(−26):U(−6) and relocates nucleotides −8 to −7, resulting in full protection.
Non-G PFS requirement for RNA cleavage in LshCas13a
Previous studies reported that 3′-G at position −1 of target RNA repressed cis- and trans-RNA cleavage activity of LshCas13a in E. coli, which implies a non-G PFS requirement for cleavage (Abudayyeh et al., 2016). Together with our biochemical and structural data, we believe that 3′-G at position −1 of target RNA could easily pair with C(−1) of tag segment, which limits the conformational changes at this region and then further reduces the enzymatic activity. In contrast, unpaired 3′-PFS of target RNA flips out and allows the movement of C(−1) of tag segment.
Comparison with tag:anti-tag inhibition in type III systems
The self/non-self discrimination mediated by tag:anti-tag pairing is also found in type III CRISPR-Cas systems (Elmore et al., 2016; Estrella et al., 2016; Kazlauskiene et al., 2016; Niewoehner and Jinek, 2016; Samai et al., 2015). The extended tag:anti-tag duplex specifically induces minimal conformational changes of the Csm complex but prohibits non-specific ssDNA cleavage activity and cyclase activity of Cas10 and thereby prohibits non-specific ssRNA cleavage activity of Csm6, with no effect on cleavage of target RNA by Csm3/Cmr4 subunit (Jia et al., 2019b; You et al., 2019). Furthermore, the enzymatic activity of HEPN-domain-containing Csm6 is regulated indirectly by the secondary messenger cyclic oligoadenylates (cOAs) synthesized by Cas10 and involves minor conformational changes (Jia et al., 2019a; Kazlauskiene et al., 2017; Niewoehner et al., 2017). Unlike type III systems, which contain multiple subunits for RNA degradation and an indirect regulation mechanism for RNase activity, type VI-A systems apply a single Cas13a protein to achieve both target RNA and substrate RNA degradation and a direct regulation mechanism for RNase activity. The tag:anti-tag duplex directly interacts and induces large conformational changes of Cas13a to inhibit cis- and trans-RNase activities, which involves domain movements of the two HEPN domains.
The different inhibition mechanisms determine the time frame for shutdown of the whole system, as well as the design of regulators or inhibitors for related applications. For Cas13a-based tools, it may be convenient to use anti-tag RNA as an inhibitor to turn off activated Cas13a rather than eliminate crRNA or Cas13a protein. The inhibition could also be recovered after removal of anti-tag RNA. Small molecules might be designed on the basis of the anti-tag-bound Cas13a structure to inactivate Cas13a cleavage activity without an effect on target recognition and loading.
Summary
Overall, our structure-function data elucidate the molecular principles underlying anti-tag-mediated inhibition in the Cas13a system. We observe NUC lobe-mediated conformational changes on proceeding from the pre-target-bound inactive binary complex to the anti-tag-bound and also inactive ternary complex. Tag:anti-tag duplex formation alters the pairing alignments in the repeat 3′-flank region, with the tag:anti-tag duplex collinearly stacking with the guide:target duplex, and together the collinearly aligned duplexes are positioned within the NUC lobe binding channel. These conformational changes prevent the two HEPN domains from getting closer together to generate a composite HEPN catalytic pocket, thereby suppressing the cleavage activity for degradation of substrate RNA.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for reagents could be directed to and will be fulfilled by the lead contact, Hui Yang (yanghui@sibcb.ac.cn).
Materials availability
Plasmids generated in this study will be made available on request.
Data and code availability
The atomic coordinates and cryo-EM density map of the Cas13a anti-tag RNA ternary complex have been deposited in the Protein Data Bank (PDB) with accession number 7DMQ and Electron Microscopy Data Bank (EMDB) with accession number EMD-30767. Original experimental data are available from Mendeley Data at https://dx.doi.org/10.17632/6zxkdbckfp.12.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacterial strains
Proteins were obtained through recombinant expression in Escherichia coli BL21 (DE3) (Novagen). E. coli Turbo competent cells (Weidi) were used for in vivo RNA interefence studies.
METHOD DETAILS
Protein expression and purification
The full-length Leptotrichia shahii cas13a and Leptotrichia buccalis cas13a genes were synthesized and inserted into a modified pCDF-Duet-1 and pRSF-Duet-1 vector (Novagen) with N-terminal His6-SUMO tag following a ubiquitin-like protease (ULP1) cleavage site, respectively. Recombinant Cas13 protein was overexpressed in E. coli BL21 (DE3) strain in Lysogeny broth (LB) medium (Sangon Biotech). The cells were grown at 37°C until OD600 reached 0.8 and then induced with 0.2 mM isopropyl β-D-1-thiogalac-topyranoside (IPTG, Sangon Biotech) at 18°C for 20 hr. Cell pellets were resuspended in buffer A (20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 5% glycerol, 20 mM imidazole, 5 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride), lysed by the high-pressure cell disrupter, and centrifuged at 18,000 rpm for 1 hr at 4°C. The supernatant containing Cas13 protein was loaded to 5 mL HisTrap Fast-flow column (GE Healthcare) pre-equilibrated in buffer A and eluted with buffer A supplemented with 480 mM imidazole. The His6-SUMO tag was removed by ULP1 and during dialysis against buffer A and then separated by re-loading to HisTrap column. The flow-through fraction was further dialyzed against buffer B (20 mM Tris-HCl, pH 7.5, 350 mM NaCl, 5% glycerol, 5 mM β-mercaptoethanol) and loaded on 5 mL HiTrap Heparin HP Sepharose column (GE Healthcare) pre-equilibrated in buffer B and eluted by a linear gradient from 350 mM to 1 M NaCl in 20 column volumes. Fractions containing recombinant Cas13 protein were concentrated in 30 kDa molecular mass cut-off concentrators (Amicon) and further purified by Superdex 200 16/60 GL column (GE Healthcare) in buffer C (20 mM HEPES, pH 7.2, 350 mM NaCl, 2 mM MgCl2, 5 mM DTT). Different mutations were generated based on PCR-based method. The mutants were purified by the same method as described above.
To assemble the Cas13-crRNA-ssRNA ternary complex, the purified Cas13 protein was mixed with crRNA and ssRNA target at a molar ratio of 1:1.2:1.2, and incubated on ice for 60 min. The reconstituted ternary complex was purified by gel filtration chromatography using Superdex 200 10/300 GL column pre-equilibrated in buffer D (20 mM HEPES, pH 7.2, 150 mM NaCl, 2 mM MgCl2, 5 mM DTT).
In vitro transcription and purification of crRNA and ssRNA targets
The crRNA, target RNAs 1–3, and 102-nt non-target RNA were transcribed in vitro using home-made T7 RNA polymerase with a linearized plasmid DNA as template. Transcription reactions were performed at 37°C for 4–5 hr in buffer containing 100 mM Tris-HCl, pH 7.9, 30 mM DTT, 15 mM MgCl2, 2 mM spermidine, 4 mM each NTP, 100 μg/ml DNA template, 100 μg/ml T7 RNA polymerase. The transcribed RNAs were purified by 15% denaturing TBE-urea PAGE, extracted from gel by electroelution using Elutrap (GE Healthcare). The RNAs were resuspended in DEPC (diethylpyrocarbonate)-treated water, denatured at 95°C for 5 min, and then slowly cooling to room temperature.
Cryo-EM sample preparation and imaging
Freshly purified LshCas13a-crRNA-RNA6 ternary complex (anti-tag bound) and LshCas13a-crRNA-RNA4 ternary complex (target-bound) were spotted onto freshly glow-discharged UltrAufoil R1.2/1.3 300 mesh grids (Quantifoil) at a concentration of ~0.35 mg/ml or 0.2 mg/ml, respectively. Excess samples were blotted for 1.5–2.0 s at 100% humidity using the Vitrobot Mark IV (FEI). The images were recorded on a 300 kV FEI Titan Krios transmission electron microscope (FEI) and equipped with a K2 Summit direct detector (Gatan), using super-resolution mode with a 0.86 Å (for anti-tag-bound) and 1.014 Å (for target-bound) physical pixel size. Movies were collected using SerialEM (Mastronarde, 2005) with a dose rate of 8 e−/pixel/s with a total exposure time of 6 s, with 0.13 s for each frame, generating 46 frames per micrograph. Defocus values range was set between −0.8 μm and −2.2 μm.
Image processing
All processing procedures were performed using RELION 3.0 (Scheres, 2012). Motion correction was performed by MotionCor2 (Zheng et al., 2017) and the contrast transfer function (CTF) was estimated by CTFFIND4 (Rohou and Grigorieff, 2015) using the summed image without dose-weighting. Initial 2D templates were generated from ~5,000 manually picked particles for automatic particle picking. For anti-tag-bound LshCas13a, approximately 829,579 particles were picked from 2,536 images and processed by reference free 2D classification for 2 rounds. 446,585 particles were selected for further 3D classification using initial model generated by RELION. After two rounds of 3D classification, best classes containing 212,861 particles was selected for 3D auto-refinement and further for particle polishing, resulting in a 3.06 Å density map. Local resolution measurements were carried out with the program ResMap blocres (Kucukelbir et al., 2014) using unfiltered half maps. Additional processing details specific to each of the datasets are summarized in Figure S2 and Table S1. For target-bound LshCas13a, a large amount of target RNA disassociation occurs during specimen preparation and the LshCas13a particles exhibit a strong preferred orientation, resulting in resolution limitation of 3D reconstruction. Briefly, 1,046,285 particles were selected from 12,446 images after 3 rounds of 2D classification. After 2 rounds of 3D classification, 117,621 particles were selected for 3D auto-refinement and further for Bayesian polishing to yield a low 4.0 Å density map. It is worth mentioning that the resolution of the reconstructed map of target-bound LshCas13a ternary complex is overestimated due to the preferred orientation issue. The map was severely elongated along one direction due to the preferred orientation, and this may produce artifactual densities. We cannot fit the model of target-bound LshCas13a ternary complex into map accurately. All details are summarized in Figure S5.
Atomic model building and refinement
Initial model for LshCas13a-crRNA-RNA4 ternary complex was generated by per-chain rigid-body fitting into electron density maps, using the model of reported crystal structure of LshCas13-crRNA binary complex (PDB code: 5WTK) (Liu et al., 2017b) and then manually rebuilt in COOT (Emsley et al., 2010). All models were refined against summed maps using phenix.real_space_refine with geometric and secondary structure restraints (Adams et al., 2010). No clear electron density was observed for the NTD domain (res 1–340), the first five nucleotides of the 5′-handle of crRNA, and the terminal two base pairs of the RNA duplex extending outside. Poor electron density was observed for the C-terminal region of HEPN-I domain (res 886–890) and N-terminal region of Linker domain res 914–1012. All figures were generated by UCSF Chimera (Pettersen et al., 2004), PyMol (https://pymol.org/2/) and CueMol (http://www.cuemol.org).
In vitro RNA cleavage assay
The cleavage reactions were performed in cleavage buffer (20 mM HEPES, pH 7.0, 50 mM KCl, 2 mM MgCl2, 5 mM DTT, 5% glycerol) with 2 μg RNA targets as reported previously with some modification (Liu et al., 2017a, 2017b; Meeske and Marraffini, 2018). Briefly, crRNA and Cas13a proteins at a molar ratio of 1:1 were incubated at room temperature for 30 min in cleavage buffer ahead of reactions. For target RNA cleavage assays, 2 μg target RNA was added into Cas13a-crRNA mixture at a molar ratio of 5000:1:1 for LbuCas13a and 500:1:1 for LshCas13a, respectively. For non-target RNA cleavage assays, 2 μg non-target RNA and various target RNA were added into Cas13a-crRNA mixture at a molar ratio of 3000:1:1:1 for LbuCas13a and 10:1:1:1 for LshCas13a, respectively. All reactions were performed at 37°C for 3 min (LbuCas13a) or 50 min (LshCas13a), and then terminated by adding 2 × loading buffer and quenched at 75°C for 5 min. Samples were analyzed by 10% TBE Urea gels.
In vivo RNA targeting assay
The in vivo RNA interference assays were performed as previously described with some modification (Abudayyeh et al., 2016). Lshcas13a gene and the CRISPR array targeting EYFP mRNA were inserted into the two MCS of a modified pCDF-Duet-1 vector with constitutive express promoter, respectively. We subcloned Lbucas13a gene and the CRISPR array into a modified pRSF-Duet-1 vector in the same manner. Eyfp gene containing 8-nt non-specific sequence, 5-nt anti-tag, or 8-nt anti-tag were cloned into a modified tetracycline-inducible pBR322 vector, respectively. 50 ng plasmids carrying cas13a and eypf were transformed into 10 μL E. coli Turbo competent cells (Weidi) and spread them on antibiotic selective plates. Colonies were isolated from plates and grown in LB at 37°C overnight with double selection to maintain both plasmids. Cells were diluted to an OD600 of 0.1 and tested by liquid and solid cultures, respectively. For liquid culture, the cells were further grown at 37°C for 1 hr. EYFP expression was then induced by 15 μg/ml tetracycline (Solarbio) at 37°C for 10 hr. Fluorescence and OD600 were measured every 20 min by Synergy NEO microplate reader (BioTek, Winooski, VT). PRISM 7.0 was used for all statistical analysis. For solid culture, the cells were diluted to 100-fold, and then 3 μL serial 10-fold dilutions were spotted to plates and incubated at 37°C overnight.
QUANTIFICATION AND STATISTICAL ANALYSIS
In Figure S2, resolution estimations of cryo-EM density maps are based on the 0.143 Fourier Shell Correlation (FSC) criterion (Chen et al., 2013; Rosenthal and Henderson, 2003).
In Figures 1E–1H and 7E–7H, statistical analyses of the fluorescence and OD600 were performed in GraphPad PRISM 7.0, with error bars representing the standard deviation across three replicates.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| E.coli: BL21(DE3) Competent cells | Novagen | Cat#69450–3 |
| E.coli: DH5α Competent cells | ThermoFisher | Cat#18265017 |
| E.coli: Turbo Competent cells | Weidi | Cat#DL1081 |
| Chemicals, peptides, and recombinant proteins | ||
| ULP1 | Home-made | N/A |
| T7 RNA polymerase | Home-made | N/A |
| Urea | Sangon Biotech | CAS: 57-13-6 Cat#A610148-0500 |
| Diethyl pyrocarbonate | Sangon Biotech | CAS: 1609-47-8 Cat#B600154-0100 |
| 40% acrylamide and bis-acrylamide solution, 29:1 | Sangon Biotech | Cat#B546013–0500 |
| Critical commercial assays | ||
| PureLink HiPure Plasmid Megaprep Kit | ThermoFisher | Cat#K210008 |
| Deposited data | ||
| Coordinate of LshCas13a-crRNA-anti-tag RNA ternary complex | This paper | PDB: 7DMQ |
| Density map of LshCas13a-crRNA-anti-tag RNA ternary complex | This paper | EMDB: EMD-30767 |
| Recombinant DNA | ||
| pRSF-Duet-1-His6-SUMO-LshCas13a, wild-type and various mutants | This paper | N/A |
| pRSF-Duet-1-His6-SUMO-LbuCas13a, wild-type and various mutants | This paper | N/A |
| pUT7-LshCas13a crRNA, wild-type and mutant | This paper | N/A |
| pUT7-LbuCas13a crRNA, wild-type and mutant | This paper | N/A |
| pUT7-target RNA (RNA1–6, 21-spacer RNA) | This paper | N/A |
| pUT7-non-target RNA | This paper | N/A |
| pCDF-Duet-1-LshCas13a-CRISPR, wild-type and various mutants | This paper | N/A |
| pRSF-Duet-1-LbuCas13a-CRISPR, wild-type and various mutants | This paper | N/A |
| pBR322-EYFP, with or without anti-tag sequence | This paper | N/A |
| Oligonucleotides | ||
| 56-nt LshCas13a crRNA: GGCCACCCCAAUAUCGAAGGGGACUAAAACUAGAUUGCUGUUCUACCAAGUAAUCCAU | This paper | N/A |
| 57-nt LbuCas13a crRNA: GGACCACCCCAAAAAUGAAGGGGACUAAAACUAGAUUGCUGUUCUACCAAGUAAUCCAU | This paper | N/A |
| 50-nt LbuCas13a crRNA: GGACCACCCCAAAAAUGAAGGGGACUAAAACUAGAUUGCUGUUCUACCAAGU | This paper | N/A |
| Target RNA1: GGGAAGUUCAACUCUUUAUGUAUUGAUCUUCCAAAAUauggauuacuugguagaacagcaaucuaCUC | This paper | N/A |
| Target RNA2: GGGAAGUUCAACUCUUUAUGUAUUGAUCUUCCAAAAUauggauuacuugguagaacagcaaucuaGUUUUCUC | This paper | N/A |
| Target RNA3: GGGAAGUUCAACUCUUUAUGUAUUGAUCUUCCAAAAUauggauuacuugguagaacagcaaucuaGUUUUAGUCUC | This paper | N/A |
| Target RNA4: GauggauuacuugguagaacagcaaucuaCAAAAUCA | IDT | N/A |
| Target RNA5: GauggauuacuugguagaacagcaaucuaGUUUUUCA | IDT | N/A |
| Target RNA6: GauggauuacuugguagaacagcaaucuaGUUUUAGU | IDT | N/A |
| 21-nt Target RNA without anti-tag: GacuugguagaacagcaaucuaCAAAAUCA | IDT | N/A |
| 21-nt Target RNA with anti-tag: GacuugguagaacagcaaucuaGUUUUAGU | IDT | N/A |
| 102-nt non-target RNA: GGAUAACUCAAUUUGUAAAAAAGUUUUAGAGCUAGAAAUAGCAAGUUAAAAUAAGGCUAGUCCGUUAUCAACUUGAAAAAGUGGCACCGAGUCGGUGCUUUC | This paper | N/A |
| Software and algorithms | ||
| Coot | Emsley et al., 2010 |
https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot; RRID:SCR_014222 |
| Phenix | Adams et al., 2010 |
https://www.phenix-online.org; RRID: SCR_014224 |
| SerialEM data collection software | Mastronarde, 2005 |
https://bio3d.colorado.edu/SerialEM/; RRID: SCR_017293 |
| MotionCor2 | Zheng et al., 2017 |
https://msg.ucsf.edu/em/software/motioncor2.html; RRID: SCR_016499 |
| CTFFIND4 | Rohou and Grigorieff, 2015 |
https://grigoriefflab.janelia.org/ctffind4/; RRID: SCR_016732 |
| RELION | Scheres, 2012 |
https://www3.mrc-lmb.cam.ac.uk/relion//index.php?title=Main_Page%2F; RRID: SCR_016274 |
| ResMap | Kucukelbir et al., 2014 | http://resmap.sourceforge.net/ |
| USCF Chimera | Pettersen et al., 2004 |
https://www.cgl.ucsf.edu/chimera/; RRID: SCR_004097 |
| PyMOL | The PyMOL Molecular Graphics System, Version 2.2.3, Schrodinger, LLC |
https://pymol.org/2/; RRID: SCR_000305 |
| CueMol | Version 2.2.3.434 |
http://www.cuemol.org; RRID: SCR_019052 |
| Clustal Omega | Madeira et al., 2019 |
https://www.ebi.ac.uk/Tools/msa/clustalo/; RRID: SCR_001591 |
| PRISM | Version 7.0 |
https://www.graphpad.com/scientific-software/prism/; RRID: SCR_002798 |
| Other | ||
| Amicon concentrators (30K) | Millipore | Cat#UFC903024 |
| HisTrap FF (5 ml) | GE Healthcare | Cat#17-5255-01 |
| HiTrap Heparin HP (5 ml) | GE Healthcare | Cat#17-0407-03 |
| HiLoad 16/600 Superdex 200 pg | GE Healthcare | Cat#28989335 |
| Superdex 200 10/300 GL | GE Healthcare | Cat#17517501 |
| UltrAufoil R1.2/1.3 300 mesh grids | Quantifoil | Cat# BQRU1.2/1.3–3A-B |
| Whatman Elutrap electroelution system | Sigma-Aldrich | Cat#WHA10447700 |
Highlights.
Structure of LshCas13a ternary complex with extended repeat:target pairing
Architecture of crRNA tag segment is critical for Cas13a enzymatic activity
Extended tag:anti-tag complementarity triggers domain rearrangements of LshCas13
Mechanistic insights into substrate RNA cleavage inhibition by tag:anti-tag pairing
ACKNOWLEDGMENTS
We thank the staff of the BL18U and BL19U1 beamlines of National Center for Protein Science in Shanghai (NCPSS) and BL17U of Shanghai Synchrotron Radiation Facility (SSRF). We thank the staff of the Electron Microscopy facility of NCPSS, Cryo-EM Facility Center of Southern University of Science and Technology (Shenzhen), Cryo-EM Facility of Memorial Sloan Kettering Cancer Center, Bio-Electron Microscopy Facility of ShanghaiTech University, and Center of Cryo-Electron Microscopy of Zhejiang University for technical assistance in data collection. This research was supported by grants from the National Natural Science Foundation of China (grant 31971135 to H.Y.), the Shanghai Rising-Star Program (grant 20QA1410700 to H.Y.), the Thousand Talents Plan-Youth to H.Y., the NIH (GM129430) and the Geoffrey Beene Cancer Research Center to D.J.P., and a Memorial Sloan Kettering Cancer Center Core Grant (P30CA008748).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.molcel.2020.12.033.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB, Shmakov S, Makarova KS, Semenova E, Minakhin L, et al. (2016). C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, et al. (2017). RNA targeting with CRISPR-Cas13. Nature 550, 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, and Horvath P (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. [DOI] [PubMed] [Google Scholar]
- Chen S, McMullan G, Faruqi AR, Murshudov GN, Short JM, Scheres SH, and Henderson R (2013). High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, and Zhang F (2017). RNA editing with CRISPR-Cas13. Science 358, 1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, Labonté J, Fremaux C, Boyaval P, Romero DA, Horvath P, and Moineau S (2008). Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol 190, 1390–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis ME, Villion M, Magadán AH, and Moineau S (2013). CRISPR-Cas and restriction-modification systems are compatible and increase phage resistance. Nat. Commun 4, 2087. [DOI] [PubMed] [Google Scholar]
- East-Seletsky A, O’Connell MR, Knight SC, Burstein D, Cate JH, Tjian R, and Doudna JA (2016). Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 538, 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East-Seletsky A, O’Connell MR, Burstein D, Knott GJ, and Doudna JA (2017). RNA targeting by functionally orthogonal type VI-A CRISPR-Cas enzymes. Mol. Cell 66, 373–383.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore JR, Sheppard NF, Ramia N, Deighan T, Li H, Terns RM, and Terns MP (2016). Bipartite recognition of target RNAs activates DNA cleavage by the type III-B CRISPR-Cas system. Genes Dev. 30, 447–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrella MA, Kuo FT, and Bailey S (2016). RNA-activated DNA cleavage by the Type III-B CRISPR-Cas effector complex. Genes Dev. 30, 460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freije CA, Myhrvold C, Boehm CK, Lin AE, Welch NL, Carter A, Metsky HC, Luo CY, Abudayyeh OO, Gootenberg JS, et al. (2019). Programmable inhibition and detection of RNA viruses using Cas13. Mol. Cell 76, 826–837.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, and Siksnys V (2012). Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U S A 109, E2579–E2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer NM, Freije CA, et al. (2017). Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356, 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, and Zhang F (2018). Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 360, 439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille F, and Charpentier E (2016). CRISPR-Cas: biology, mechanisms and relevance. Philos. Trans. R. Soc. Lond. B Biol. Sci 371, 20150496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia N, Jones R, Yang G, Ouerfelli O, and Patel DJ (2019a). CRISPR-Cas III-A Csm6 CARF domain is a ring nuclease triggering stepwise cA4 cleavage with ApA>p formation terminating RNase activity. Mol. Cell 75, 944–956.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia N, Mo CY, Wang C, Eng ET, Marraffini LA, and Patel DJ (2019b). Type III-A CRISPR-Cas Csm complexes: assembly, periodic RNA cleavage, DNase activity regulation, and autoimmunity. Mol. Cell 73, 264–277.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, and Charpentier E (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskiene M, Tamulaitis G, Kostiuk G, Venclovas Č, and Siksnys V (2016). Spatiotemporal control of type III-A CRISPR-Cas immunity: coupling DNA degradation with the target RNA recognition. Mol. Cell 62, 295–306. [DOI] [PubMed] [Google Scholar]
- Kazlauskiene M, Kostiuk G, Venclovas Č, Tamulaitis G, and Siksnys V (2017). A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science 357, 605–609. [DOI] [PubMed] [Google Scholar]
- Knott GJ, East-Seletsky A, Cofsky JC, Holton JM, Charles E, O’Connell MR, and Doudna JA (2017). Guide-bound structures of an RNA-targeting A-cleaving CRISPR-Cas13a enzyme. Nat. Struct. Mol. Biol 24, 825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Lotfy P, Brideau NJ, Oki J, Shokhirev MN, and Hsu PD (2018). Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell 173, 665–676.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukelbir A, Sigworth FJ, and Tagare HD (2014). Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrie SJ, Samson JE, and Moineau S (2010). Bacteriophage resistance mechanisms. Nat. Rev. Microbiol 8, 317–327. [DOI] [PubMed] [Google Scholar]
- Liu L, Li X, Ma J, Li Z, You L, Wang J, Wang M, Zhang X, and Wang Y (2017a). The Molecular architecture for RNA-guided RNA cleavage by Cas13a. Cell 170, 714–726.e10. [DOI] [PubMed] [Google Scholar]
- Liu L, Li X, Wang J, Wang M, Chen P, Yin M, Li J, Sheng G, and Wang Y (2017b). Two distant catalytic sites are responsible for C2c2 RNase activities. Cell 168, 121–134.e12. [DOI] [PubMed] [Google Scholar]
- Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, and Lopez R (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47 (W1), W636–W641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. (2011). Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol 9, 467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, et al. (2015). An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol 13, 722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA (2015). CRISPR-Cas immunity in prokaryotes. Nature 526, 55–61. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol 152, 36–51. [DOI] [PubMed] [Google Scholar]
- Meeske AJ, and Marraffini LA (2018). RNA guide complementarity prevents self-targeting in type VI CRISPR systems. Mol. Cell 71, 791–801.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanraju P, Makarova KS, Zetsche B, Zhang F, Koonin EV, and van der Oost J (2016). Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science 353, aad5147. [DOI] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, and Almendros C (2009). Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155, 733–740. [DOI] [PubMed] [Google Scholar]
- Myhrvold C, Freije CA, Gootenberg JS, Abudayyeh OO, Metsky HC, Durbin AF, Kellner MJ, Tan AL, Paul LM, Parham LA, et al. (2018). Field-deployable viral diagnostics using CRISPR-Cas13. Science 360, 444–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewoehner O, Garcia-Doval C, Rostøl JT, Berk C, Schwede F, Bigler L, Hall J, Marraffini LA, and Jinek M (2017). Type III CRISPR–Cas systems produce cyclic oligoadenylate second messengers. Nature 548, 543–548. [DOI] [PubMed] [Google Scholar]
- Niewoehner O, and Jinek M (2016). Structural basis for the endoribonu-clease activity of the type III-A CRISPR-associated protein Csm6. RNA 22, 318–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, and Nureki O (2017). Structures and mechanisms of CRISPR RNA-guided effector nucleases. Curr. Opin. Struct. Biol 43, 68–78. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Rohou A, and Grigorieff N (2015). CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol 192, 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal PB, and Henderson R (2003). Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol 333, 721–745. [DOI] [PubMed] [Google Scholar]
- Samai P, Pyenson N, Jiang W, Goldberg GW, Hatoum-Aslan A, and Marraffini LA (2015). Co-transcriptional DNA and RNA cleavage during type III CRISPR-Cas immunity. Cell 161, 1164–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH (2012). RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, van der Oost J, Brouns SJ, and Severinov K (2011). Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. U S A 108, 10098–10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, et al. (2015). Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol. Cell 60, 385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker IM, Mesa P, Kellner MJ, Kannan S, Brignole E, Koob J, Feliciano PR, Stella S, Abudayyeh OO, Gootenberg JS, et al. (2019). High-resolution structure of Cas13b and biochemical characterization of RNA targeting and cleavage. Cell Rep. 26, 3741–3751.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smargon AA, Cox DBT, Pyzocha NK, Zheng K, Slaymaker IM, Gootenberg JS, Abudayyeh OA, Essletzbichler P, Shmakov S, Makarova KS, et al. (2017). Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol. Cell 65, 618–630.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Oost J, Jore MM, Westra ER, Lundgren M, and Brouns SJ (2009). CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem. Sci 34, 401–407. [DOI] [PubMed] [Google Scholar]
- Wright AV, Nuñez JK, and Doudna JA (2016). Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell 164, 29–44. [DOI] [PubMed] [Google Scholar]
- Yan WX, Chong S, Zhang H, Makarova KS, Koonin EV, Cheng DR, and Scott DA (2018). Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol. Cell 70, 327–339.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You L, Ma J, Wang J, Artamonova D, Wang M, Liu L, Xiang H, Severinov K, Zhang X, and Wang Y (2019). Structure studies of the CRISPR-Csm complex reveal mechanism of co-transcriptional interference. Cell 176, 239–253.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, et al. (2015). Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Ye W, Ye Y, Zhou H, Saeed AFUH, Chen J, Lin J, Perčulija V, Chen Q, Chen CJ, et al. (2018a). Structural insights into Cas13b-guided CRISPR RNA maturation and recognition. Cell Res. 28, 1198–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Konermann S, Brideau NJ, Lotfy P, Wu X, Novick SJ, Strutzenberg T, Griffin PR, Hsu PD, and Lyumkis D (2018b). Structural basis for the RNA-guided ribonuclease activity of CRISPR-Cas13d. Cell 175, 212–223.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Ye Y, Ye W, Perčulija V, Jiang H, Chen Y, Li Y, Chen J, Lin J, Wang S, et al. (2019). Two HEPN domains dictate CRISPR RNA maturation and target cleavage in Cas13d. Nat. Commun 10, 2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and cryo-EM density map of the Cas13a anti-tag RNA ternary complex have been deposited in the Protein Data Bank (PDB) with accession number 7DMQ and Electron Microscopy Data Bank (EMDB) with accession number EMD-30767. Original experimental data are available from Mendeley Data at https://dx.doi.org/10.17632/6zxkdbckfp.12.


