Abstract
Genetically encoded fluorescent H2O2 probes continue to advance the field of redox biology. Here, we compare the previously established peroxiredoxin-based H2O2 probe roGFP2-Tsa2ΔCR with the newly described OxyR-based H2O2 probe HyPer7, using yeast as the model system. Although not as sensitive as roGFP2-Tsa2ΔCR, HyPer7 is much improved relative to earlier HyPer versions, most notably by ratiometric pH stability. The most striking difference between the two probes is the dynamics of intracellular probe reduction. HyPer7 is rapidly reduced, predominantly by the thioredoxin system, whereas roGFP2-Tsa2ΔCR is reduced more slowly, predominantly by the glutathione system. We discuss the pros and cons of each probe and suggest that future side-by-side measurements with both probes may provide information on the relative activity of the two major cellular reducing systems.
Keywords: yeast, hydrogen peroxide, OxyR, peroxiredoxins, roGFP2, HyPer
Abbreviations: OxD, degree of oxidation; Prx, peroxiredoxin; Trx, thioredoxin
The development of genetically encoded redox probes for the specific sensing of small-molecule oxidants (e.g., H2O2) and redox couples (e.g., GSH/GSSG) has advanced the field of redox biology (1). Such probes provide information on chemically defined redox changes as they occur in living cells, in real time and with subcellular resolution. They have enabled the identification of physiological and pathophysiological redox changes, for example, in development and injury (2).
Two major types of genetically encoded probes are most commonly used for sensing H2O2. Both are fusion proteins in which a fluorescent protein is linked to an H2O2-reactive domain. Both are dynamic, meaning that they reflect the interplay between probe oxidation by H2O2 and probe reduction by intracellular reducing systems. Both are excitation-ratiometric, meaning that dual excitation provides emission ratios indicative of the probe’s redox state.
One type are the roGFP2-thiol peroxidase fusion proteins, specifically roGFP2-Orp1 (3), roGFP2-Tsa2ΔCR (4), roGFP2-Tsa2ΔCPΔCR (4), roGFP2-Tpx1(C169S) (5), roGFP2-Prx2, and roGFP2-Prx2(C172A) (6). In these constructs, selective reactivity toward H2O2 is facilitated by a thiol peroxidase domain, from either the glutathione peroxidase or peroxiredoxin (Prx) family. The peroxidatic cysteine reacts with H2O2 to form a disulfide bond, which is then rearranged by thiol–disulfide exchange to create a disulfide bond on the surface of the attached roGFP2 domain (reviewed in (7)). The roGFP2 disulfide bond causes a small conformational change on the inside of the β-barrel, which in turn alters the protonation state of the chromophore, thus facilitating the ratiometric measurement. The roGFP2 disulfide bond is susceptible to reduction by the glutathione system, but is fully resistant to reduction by thioredoxin (Trx) (8), apparently for steric reasons (9).
The other type are the cpYFP-OxyR fusion proteins, in which a circularly permuted fluorescent protein is inserted into the H2O2-reactive (regulatory) domain of the Escherichia coli transcription factor OxyR, specifically HyPer (10), HyPer2 (11), and HyPer3 (12). Similar to a thiol peroxidase (13), the OxyR regulatory domain harbors an active site that catalyzes the reaction between a cysteine residue and H2O2 (14). The resulting disulfide bond leads to a substantial conformational change (15), which then affects the attached cpYFP domain. E. coli OxyR has been reported to be reducible by glutathione (16), whereas Pseudomonas aeruginosa OxyR has been shown to be reduced by Trx (17), suggesting that the reduction mechanism depends on the specific type of OxyR and/or the host environment. To our knowledge, it has not been clarified how the OxyR domain of HyPer is actually reduced inside cells.
A major problem of the conventional HyPer probes is their pronounced pH sensitivity, which is shared by many other probes based on circularly permuted fluorescent proteins, in particular cpYFP (18). The circular permutation introduces a cleft in the β-barrel that exposes the fluorophore’s pH-sensitive phenoxy group to the ambient environment (19). Therefore, to separate the effects of pH and oxidation, it has been expedient to compare HyPer with a redox-insensitive cysteine mutant, which has been named SypHer (20). In fact, SypHer has since been used as a superior pH probe in its own right (21).
Most recently, a new member of the HyPer family, HyPer7, has been introduced (22). HyPer7 differs from earlier HyPer versions in two important ways. First, it uses the more sensitive regulatory OxyR domain from Neisseria meningitidis. Second, it was subjected to both targeted (Y145F) and random mutagenesis (D135N, G298S, and T379P) to select for favorable properties. Overall, it is reported to be brighter, more sensitive to H2O2, and largely pH insensitive when measured in the ratiometric mode (22).
Because Saccharomyces cerevisiae is a key model eukaryote in redox biology and offers unmatched possibilities for high-throughput genetic screening, we considered it important to test the use of HyPer7 in yeast and compare it to the previously established roGFP2-Tsa2ΔCR probe. In short, we find that HyPer7 is well expressed and brighter than previous probes. As previously reported (22), it does not appear to be influenced by pH in the way that confounded the interpretation of conventional HyPer responses. While exhibiting a larger dynamic range, HyPer7 is not as sensitive to H2O2 as the previously described roGFP2-Tsa2ΔCR probe. The most striking difference between the two probes is the dynamics of intracellular probe reduction. HyPer7 is reduced faster, predominantly by the Trx system, while roGFP2-Tsa2ΔCR is reduced more slowly, by the glutathione system. The more efficient reduction of HyPer7 likely leads to a higher turnover of endogenous H2O2, raising the question if the expression of HyPer7 changes endogenous H2O2 levels and, perhaps, cellular stress resistance and behavior. We conclude that the two probes can be considered complementary in the sense that they provide information on the activity of the two major cellular reducing systems. Future studies using both probes side-by-side may reveal interesting information about the dynamic regulation of the reducing systems.
Results
Expression of HyPer7 in yeast
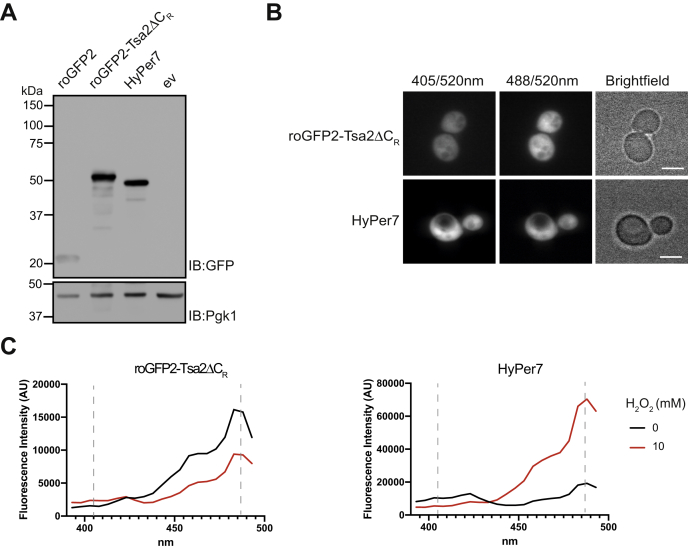
To evaluate the performance of HyPer7 in yeast, and to compare it with roGFP2-Tsa2ΔCR, we expressed corresponding codon-optimized coding sequences under control of the TEF promoter. Both probes were expressed at comparable levels, with only minor signs of degradation (Fig. 1A). Both probes exhibited the expected cytosolic distribution, with HyPer7 fluorescing more strongly than roGFP2-Tsa2ΔCR (Fig. 1B). Excitation spectra recorded from living cells confirmed that both probes respond to an externally applied bolus of H2O2 in the expected manner (Fig. 1C): Oxidation of roGFP2-Tsa2ΔCR (left panel) lowered excitability at 488 nm and increased excitability at 405 nm. In contrast, oxidation of HyPer7 (right panel) increased excitability at 488 nm and decreased excitability at 405 nm. In conclusion, HyPer7 can be expressed in yeast cells in a functional manner.
Figure 1.
Expression of HyPer7 in yeast.A, expression of roGFP2, roGFP2-Tsa2ΔCR, and HyPer7 under the TEF promoter in the WT BY4742 strain. Immunoblotting with anti-GFP and anti-Pgk1 antibodies. B, fluorescence (520 nm) emitted by WT BY4742 yeast cells expressing either roGFP2-Tsa2ΔCR (upper panels) or HyPer7 (lower panels), after excitation at 405 (left panels) and 488 nm (middle panels). The scale bar represents 4 μm. C, fluorescence excitation spectra (emission: 520 nm) of yeast cells expressing roGFP2-Tsa2ΔCR (left panel) or HyPer7 (right panel). Cells were left untreated (black line) or treated with 10 mM H2O2 (red line). Dashed vertical lines indicate the position of the 405- and 488-nm laser lines. Data in this figure are representative of n = 3 independent experiments with n = 3 independent replicates each. ev, empty vector.
For roGFP2-based probes, it is common practice to ‘calibrate’ measurements by treating cells with high amounts of oxidants and reductants to obtain maximal and minimal fluorescence ratios, corresponding to the fully oxidized and full reduced states, respectively. From these reference values, the probe’s degree of oxidation (OxD) can be calculated (9). In principle, such reference measurements and OxD calculations should also be applicable to HyPer7. The fully reduced state is usually obtained by DTT treatment, and oxidized HyPer7 can be reduced by externally added DTT (Fig. S1A). However, DTT unexpectedly increased the baseline 488/405 ratio of HyPer7 (Fig. S1B). Given uncertainty about the proper calibration of HyPer7, we decided to abstain from calculating OxD values and only use fluorescence ratios throughout the study.
Response of cytosolic HyPer7 to externally applied H2O2
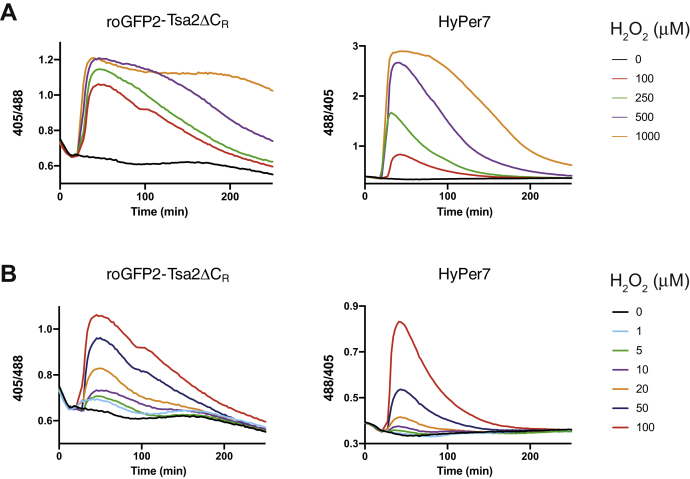
Having confirmed that HyPer7 is expressed and responsive, we performed a titration experiment to determine the minimal amount of exogenous H2O2 that is required to elicit a detectable probe response. In a previous study (4), 10 μM elicited a response of roGFP2-Tsa2ΔCR, while 200 μM was needed to observe a HyPer response. First, we compared the response of both probes to H2O2 concentrations in the range between 100 and 1000 μM (Fig. 2A and Fig. S2A). Interestingly, the two probes showed markedly different dynamic behaviors. Both probes oxidized quickly, but HyPer7 returned more rapidly to the reduced state. HyPer7 clearly responded to a 100 μM H2O2 bolus, thus confirming its higher sensitivity relative to the original HyPer. As described previously (4), the basal roGFP2-Tsa2ΔCR redox state is strongly influenced by changes in oxygen availability, because endogenous H2O2 is generated from oxygen. The continuous lowering of the probe’s baseline ratio reflects ongoing oxygen depletion in the sample. Notably, HyPer7 is not sensitive to the small changes in endogenous peroxide generation related to changes in oxygen pressure. We then compared the response of the two probes to H2O2 bolus concentrations in the range between 1 and 100 μM (Fig. 2B and Fig. S2B). The lowest concentration triggering a detectable response was ≈ 5 μM for roGFP2-Tsa2ΔCR and ≈ 20 μM for HyPer7. In summary, HyPer7 is more sensitive than the original HyPer but not quite as sensitive as the Prx-based probe.
Figure 2.
Response of cytosolic HyPer7 to externally applied H2O2.A, response of BY4742 cells expressing roGFP2-Tsa2ΔCR (left panel) or HyPer7 (right panel) to exogenously applied H2O2 boli in the concentration range of 100 to 1000 μM. B, response of BY4742 cells expressing roGFP2-Tsa2ΔCR (left panel) or HyPer7 (right panel) to exogenously applied H2O2 boli in the concentration range 1 to 100 μM. Data in this figure are representative of n = 3 independent experiments with n = 3 independent replicates.
pH sensitivity of HyPer7 in yeast
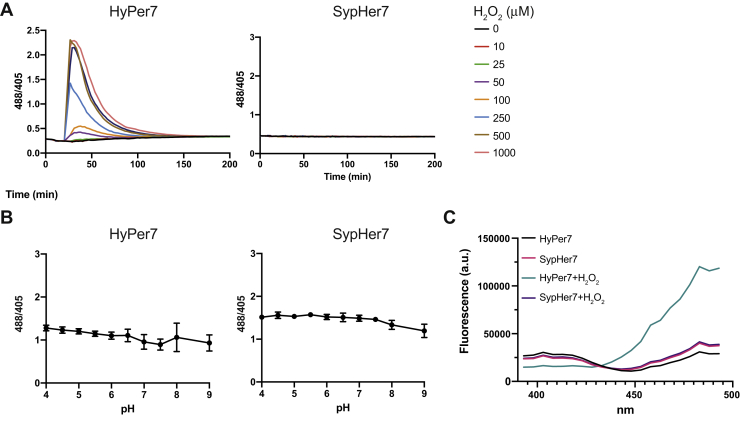
The central problem of previous HyPer probes (HyPer1-3) is their marked pH sensitivity. To deconvolute the influence of H2O2 and pH, we generated and expressed the H2O2-insensitive mutant HyPer7(C121S), which we named SypHer7 in analogy to the original HyPer–SypHer pair of probes (23, 24). SypHer7 did not respond to any of the H2O2 concentrations tested (Fig. 3A). Equilibration of probe-expressing yeast cells in different pH buffers (pH 4–9) had a relatively minor influence on the steady-state fluorescence ratio (Fig. 3B). In conclusion, the ratiometric H2O2 response of HyPer7 in yeast does not seem to be confounded by pH effects. The excitation spectrum of SypHer7 was completely unaffected by H2O2 treatment (Fig. 3C); however, we also noticed something unexpected: SypHer7 should behave like a fully reduced HyPer7 (as it cannot form the disulfide bond that links the two OxyR half domains) and therefore should never exhibit a higher 488/405 fluorescence ratio than HyPer7, yet its excitation spectrum (and the corresponding ratio) indicated it to be in a (seemingly) more oxidized state than HyPer7 in untreated cells (Fig. 3C). This baseline ratio difference is also evident in Figure 3B. This observation suggests that the C121S mutation not only abolishes redox sensitivity but also influences the protonation state of the cpYFP fluorophore.
Figure 3.
pH sensitivity of HyPer7 in yeast.A, response of BY4742 cells expressing HyPer7 (left panel) or SypHer7 (right panel) to exogenously applied H2O2 boli in the concentration range of 10 to 1000 μM. B, response of BY4742 cells expressing HyPer7 (left panel) or SypHer7 (right panel) to exogenously applied pH buffers in the range between 4 and 9. C, fluorescence excitation spectra (emission: 520 nm) of yeast cells expressing HyPer7 or SypHer7, collected at pH 6. Cells were left untreated or treated with 10 mM H2O2. Data in this figure are representative of n = 3 independent experiments with n = 3 independent replicates each.
Influence of thiol-reducing systems on cytosolic HyPer7
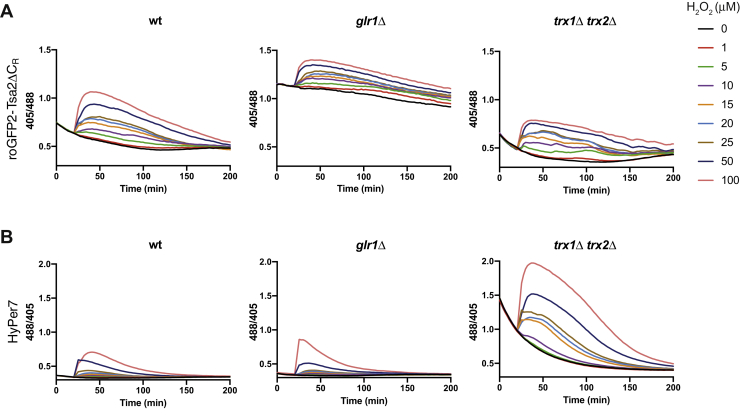
roGFP2-Tsa2ΔCR is known to be predominantly reduced by the glutathione system (4). Having observed pronounced differences in the reduction kinetics of the two probes (Fig. 2, A and B), we wondered which thiol-reducing system is mainly responsible for the reduction of HyPer7 in the cytosol of S. cerevisiae. To this end, we measured probe responses in strains lacking either glutathione reductase (Glr1) or the two cytosolic Trxs (Trx1 and Trx2). In the absence of Glr1, but not in the absence of Trx1/2, the basal redox state of roGFP2-Tsa2ΔCR shifted to a higher fluorescence ratio (Fig. 4A). In contrast, the HyPer7 response was only slightly altered in the absence of Glr1 (Fig. 4B, left and middle panel). Instead, it was enhanced and prolonged in the absence of Trx1/2. Moreover, in the absence of Trx1/2, HyPer7 showed a higher basal fluorescence ratio followed by increased reduction over time (Fig. 4B, right panel), suggesting that it has become sensitive to oxygen availability, similar to the Prx-based probe. In conclusion, HyPer7 differs from roGFP2-Tsa2ΔCR in that its reduction is mostly dependent on the Trx system.
Figure 4.
Influence of thiol-reducing systems on cytosolic HyPer7.A, response of WT (left panel), Glr1-deficient (middle panel), and Trx1/Trx2-deficient (right panel) BY4742 cells expressing roGFP2-Tsa2ΔCR to a range of different H2O2 concentrations (0–100 μM). B, response of WT (left panel), Glr1-deficient (middle panel), and Trx1/Trx2-deficient (right panel) BY4742 cells expressing HyPer7 to a range of different H2O2 concentrations (0–100 μM). Data in this figure are representative of n = 3 independent experiments with n = 3 independent replicates. Trx, thioredoxins.
Influence of HyPer7 expression on the cell’s H2O2 removal capacity
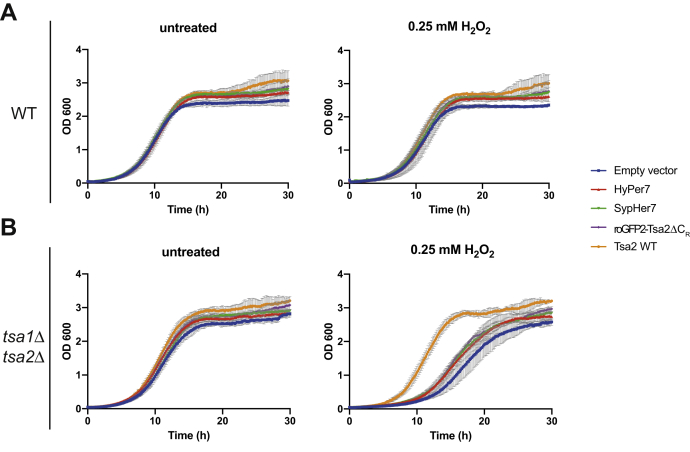
Both HyPer7 and roGFP2-Tsa2ΔCR react with and therefore eliminate H2O2 from the cell. As the probes are ectopically expressed at relatively high levels, we asked to which extent probe expression contributes to the cell’s overall capacity to remove H2O2. Previous experiments have shown that roGFP2-Tsa2ΔCR makes a negligible contribution to overall peroxidase activity (4). To make a broad assessment also for HyPer7, we asked if probe expression can rescue a growth defect that is caused by the lack of the two endogenous cytosolic Prxs Tsa1 and Tsa2. As expected, ectopic re-expression of WT Tsa2 efficiently rescued growth of a tsa1Δ tsa2Δ strain under H2O2 exposure (Fig. 5). Neither roGFP2-Tsa2ΔCR nor HyPer7 rescued to a significant extent, and HyPer7 behaved mostly like SypHer7 (Fig. 5B, right panel).
Figure 5.
Influence of the probes on cell growth and endogenous H2O2scavenging.A, growth curve of a WT BY4742 strain transformed with an empty vector, Tsa2 WT, roGFP2-Tsa2ΔCR, HyPer7, or SypHer7. The cells were exposed to 0.25 mM of H2O2 2 h after the start of the experiment. B, growth curve of a tsa1Δ tsa2Δ strain transformed with an empty vector, Tsa2 wt, roGFP2-Tsa2ΔCR, HyPer7, or SypHer7. The cells were exposed to 0.25 mM of H2O2 2 h after the start of the experiment.
Discussion
In this study, we investigated the utility of HyPer7 in S. cerevisiae and directly compared it with the previously established roGFP2-Tsa2ΔCR probe. We found that HyPer7 is suitable for expression in yeast and highly preferable over earlier HyPer versions in terms of brightness and H2O2 sensitivity. Importantly, ratiometric HyPer7 responses do not seem to be confounded by ambient pH, at least not within the specific context of our experiments. It seems to be an interesting open question which of the mutations specific to HyPer7 renders the probe ratiometrically pH stable and why.
Although HyPer7 is much more sensitive than earlier HyPer probes, it is not quite as sensitive as the Prx-based roGFP2-Tsa2ΔCR probe. In our bolus response experiments, HyPer7 detected ≈ 20 μM of exogenously added H2O2, whereas roGFP2-Tsa2ΔCR detected ≈ 5 μM. It should be noted, as discussed previously (4, 25), that these exogenous concentrations translate to endogenous concentrations in the low nanomolar range. The higher sensitivity of the Prx-based probe is also evident in two other ways: First, roGFP2-Tsa2ΔCR records the drop in H2O2 steady-state levels that follows the drop in oxygen availability, while HyPer7 is insensitive to these changes. Second, roGFP2-Tsa2ΔCR becomes maximally oxidized at bolus concentrations around 750 μM H2O2, while HyPer can resolve boli in a higher concentration range.
A clear advantage of HyPer7 over roGFP2-Tsa2ΔCR is its larger dynamic range, that is, the fold change in ratio between the fully reduced and fully oxidized states, which is mostly due to the larger intensity change in the 488 nm excitation channel. However, for reasons that remain unclear to us, we found it difficult to reliably “calibrate” HyPer7 inside cells, in particular to determine the exact ratio for the fully reduced state. This is why we decided to present intensity ratios only. It seems that HyPer7 can be slightly modulated by factors unrelated to the thiol-disulfide state of its OxyR domain, and this is potentially also reflected by our observation that SypHer7 shows a higher fluorescence ratio than reduced HyPer7. However, this may not be of practical relevance for most applications.
A key finding of this study is that the two probes differ significantly in how they are reduced in the cytosol of S. cerevisiae. Although both probes are oxidized rapidly, HyPer7 is much more quickly reduced than roGFP2-Tsa2ΔCR. We found that HyPer7 is predominantly reduced by the Trx system, whereas roGFP2-Tsa2ΔCR is predominantly reduced by the glutathione system, as engineered by design (4). The predominant coupling of the two probes to different reducing systems leads to differences in the probe response dynamics (Fig. 2). There is also a difference in how the probes respond to the absence of their respective primary reducing system: For roGFP2-Tsa2ΔCR, the main effect is an upward shift of the baseline fluorescence ratio, which is expected because the probe is already sensitive to baseline H2O2 fluctuations in WT cells and hence responds with a substantial upward baseline shift when the predominant reducing system is deleted. Because of the shifted baseline, all bolus-induced changes are compressed into a smaller ratio window, for which resolution is much more limited. In contrast, HyPer7 is not sensitive enough to “see” baseline changes in WT cells, but the deletion of the relevant reducing system makes it sensitive to baseline fluctuations, as evidenced by the appearance of ‘baseline decay’ (caused by ongoing oxygen depletion) in Trx1/2-deficient cells.
It is important to recall that both probes, although commonly perceived only as H2O2 probes, are actually reporting on the dynamic interplay between H2O2-dependent thiol oxidation and NADPH-dependent thiol reduction. This means that they are sensitive not only to changes in H2O2 availability but also to changes in thiol-reducing capacity. Hence, the response depends on the interplay between oxidation and reduction kinetics in a given situation and environment, as pointed out earlier (1).
It is difficult to say which of the probes is oxidized more rapidly. The observed overall oxidation rate should approximately correspond to the rate of formation of the relevant disulfide bond, that is, the one that alters the fluorescence properties of the attached fluorescent protein. It seems quite likely that the rate of the initial reaction with H2O2 (leading to sulfenic acid formation) is faster for the Prx-based probe than for the OxyR-based probe. However, the Prx probe requires two additional steps (sulfenic acid–thiol condensation followed by thiol–disulfide exchange) to transform the oxidizing equivalent into the roGFP2 disulfide bond. HyPer7 forms the conformation-altering disulfide bond more directly, in one condensation step from the sulfenic acid. Indeed, under the bolus response conditions tested, a major difference in the initial rate of probe oxidation does not seem to exist (Fig. 2).
Obviously, in a situation of sudden H2O2 exposure, it is the kinetics of probe reduction that determines if (and how fast) probe oxidation (i.e., accumulation of probes in the disulfide state) can build up to rise above the detection level. An increase of H2O2 concentration will not translate into increased probe oxidation as long as the probe reduction system can keep up; only the flux of oxidizing equivalents through the probe system will increase. The roGFP2-Tsa2ΔCR probe has been deliberately designed to limit reduction by the Trx system: the removal of the resolving cysteine in the Prx domain prevents the formation of Trx-reducible Prx–Prx intersubunit disulfides, and the roGFP2 disulfide bond is not sensitive to reduction by Trx (8), for steric reasons (9). Indeed, previous experiments have demonstrated that the sensitivity of the roGFP2-Tsa2(wt) fusion protein is limited by the action of the Trx system on the probe (4). Our data suggest that the Trx system also limits the sensitivity of HyPer7 because both basal oxidation levels and the H2O2 response are increased in a Trx1/2 KO background (Fig. 4). Thus, it seems that for HyPer7 slow and small changes in H2O2 availability are “ironed out” by fast and efficient reduction. This makes the probe less sensitive but has the potential advantage of allowing for better temporal resolution under conditions of rapidly changing H2O2 levels. Overall, it seems impossible to say which probe is “more authentic,” given that the two probes are coupled to different reducing systems and therefore reflect different aspects of redox homoeostasis.
We suggest that roGFP2-Tsa2ΔCR and HyPer7 can be considered complementary probes, in the sense that they couple to different reducing systems, at least in the yeast cytosol. A side-by-side comparison of the two probes may be of interest to uncover changes in one reducing system relative to the other. However, it must be kept in mind that the reducing system acting on HyPer7 may, to some extent, depend on the host environment. It is possible that in a different subcellular compartment, or organism, or under different nutrient conditions, the relative contribution of the two major reducing systems is changing. It seems expedient that the interpretation of probe responses considers changes or differences in reduction, not just in oxidation.
The probes we are discussing are facilitating the reduction of H2O2 to water, and there is always the question if the ectopic (over)expression of such probes can alter endogenous H2O2 levels or transients. In other words, the concern is that an H2O2 probe may potentially perturb its own measurement. The rapid reduction of HyPer7 (Fig. 2) by the Trx system (Fig. 4) suggests that it turns over oxidizing equivalents rapidly, potentially consuming more H2O2 than roGFP2-Tsa2ΔCR. To make at least a rough assessment, we tested the ability of HyPer7 to compensate for the lack of endogenous peroxidase activity and found no significant influence, suggesting that the impact of HyPer7 expression on the overall cellular H2O2 reduction capacity is rather limited.
There are some additional aspects that may be counted as pros and cons for one or the other probe. Prx-based probes offer an opportunity that is not available to the HyPer-type probes. The double mutant roGFP2-Tsa2ΔCPΔCR lacking both peroxidatic and resolving cysteines is almost as sensitive as the roGFP2-Tsa2ΔCR probe (4). This probe is catalytically inactive; it does not consume H2O2 on its own, yet it receives oxidizing equivalents from endogenous Prx with which it forms mixed oligomers. This probe makes it possible to observe H2O2 responses without enhancing the cell’s intrinsic H2O2 consumption capacity. However, the oligomeric nature of roGFP2-Tsa2ΔCPΔCR and roGFP2-Tsa2ΔCR (or similar probes) may also be a disadvantage for certain kinds of applications, especially when fusing the probe to other proteins. In such applications, the use of HyPer7 is likely to be an advantage, as it does not have to form dimers to be functional. It is also conceivable that Prx-based probes to some extent interfere with the sensing and signaling functions of endogenous Prxs (by oligomerizing with them). In this respect, HyPer7 can be seen as advantageous because it is completely foreign to the yeast cell and therefore less likely to interact with endogenous systems.
On a topical note, a very recent study has compared HyPer7 with the roGFP2-Tpx1(C169S) probe, which is the corresponding Prx-based probe in Schizosaccharomyces pombe (26). In agreement with our study, it finds that the Prx-based probe is more sensitive than HyPer7, and that the latter is predominantly reduced by the Trx system. Another new study evaluated the use of HyPer7 in plants (27).
In conclusion, HyPer7 is a welcome addition to the yeast redox biology toolbox. Future studies using different types of probes side-by-side are likely to reveal interesting information about the dynamic regulation of not only H2O2 levels but also reducing activities.
Experimental procedures
Yeast strains and plasmids
All experiments used the BY4742 strain (MATα his3Δ1 leu2Δ0 lys2Δ0 MET15 ura3Δ0). Single-gene deletion strains were taken from the deletion collection (Euroscarf) (28), generated using the KANMX marker. Strains lacking Trx1 and Trx2 (trx1Δ trx2Δ) or Tsa1 and Tsa2 (tsa1Δ tsa2Δ) have been described previously (9). All plasmids used in this study are based on the p415 vector backbone and expressed under control of the TEF promoter (29, 30). Plasmids expressing roGFP2, Tsa2(wt), and roGFP2-Tsa2ΔCR (in which the resolving cysteine of Tsa2 (C171) is mutated to alanine) were published previously (9, 17). The yeast codon optimized coding sequence of HyPer7 was synthesized (Life Technologies) and cloned into p415TEF. SypHer7, that is, HyPer7(C121S), was generated by site-directed mutagenesis. All constructs were verified by DNA sequencing (Eurofins).
Growth conditions
Yeast strains were grown in synthetic complete dextrose medium (0.17% w/v yeast nitrogen base without amino acids, with 0.5% w/v ammonium sulfate (Difco, BD), 2% w/v glucose (Sigma), Kaiser complete amino acid mix with or without Leu (Formedium)) at 30 °C with shaking (180 rpm). Media were solidified by the addition of 2% (w/v) agar (Sigma). For H2O2 bolus treatment of liquid cultures, hydrogen peroxide (Sigma) was added at the indicated concentrations.
Fluorescence measurements
Plate reader–based fluorescence measurements were performed as described previously (9, 18). Briefly, yeast strains were grown to the mid-exponential phase and harvested by centrifugation. Cells were resuspended in MES-Tris buffer (pH 6) and 1.5 A600 units were dispensed (per well) into black 96-well flat bottom plates (Falcon). Plates were centrifuged briefly (30 rpm, 2 min) and then measured in a PHERAstar FSX or CLARIOstar plate reader (BMG Labtech) at 30 °C. Excitation/emission wavelengths were 405/520 nm and 488/520 nm with a bandwidth of 10 nm. In experiments investigating the influence of pH, cells were resuspended in a buffer (50 mM NaH2PO4, 150 mM NaCl, 1 mM EDTA) adjusted to different pHs between 4 and 9, incubated for 15 min, and then measured. Hydrogen peroxide (Sigma) was added at the indicated concentrations. For reduction experiments, 25 mM DTT (AppliChem) was added as indicated.
Cell lysis, SDS-PAGE, and immunoblotting
Yeast cells were lysed by bead beating in PBS containing cOmplete Protease Inhibitor Cocktail (Roche). Lysates were run on 12% BIS-Tris acrylamide gels. Probe expression was visualized by immunoblotting with a polyclonal anti-GFP antibody (PABG1, Chromotek) recognizing a broad range of GFP derivatives, including roGFP2 and cpYFP. Detection of phosphoglycerate kinase with an anti-Pgk1 antibody (Life Technologies) served as a loading control.
Fluorescence microscopy
For fluorescence microscopy, an Olympus IX81 inverted microscope was used with a PL APO 100×/1.45 oil differential interference contrast objective and a Hamamatsu ORCA-R2 camera. Olympus xCellence software was used for image acquisition. Images were further analyzed with FIJI software (31).
Growth rate measurements
To assess the influence of exogenously applied H2O2 on the cellular growth rate, growth curves were measured as reported previously (4). Briefly, yeast cells were grown until reaching the stationary phase and then diluted to A600 = 0.1. The suspension was dispensed in triplicates into a 96-well plate (200 μl/well) and the A600 monitored with a plate reader (OMEGA, BMG Labtech) at 30 °C, measuring every 15 min after 10 s of shaking. The H2O2 bolus (0.25 mM) was added to individual wells 2 h after the start of the experiment as indicated.
Data availability
All data presented in this study are contained within this article.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Valeriy Pak and Seva Belousov (Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Moscow) for providing the HyPer7 coding sequence. We thank Daria Ezeriņa (Vrije Universiteit Brussel, Brussels) for helpful discussions. We thank Holger Lorenz (Bioimaging core facility, ZMBH, Heidelberg) for microscopy support.
Author contributions
P. K. and T. P. D. conceptualization; P. K. and T. P. D. supervision; P. K. validation; P. K. and T. K. S. investigation; P. K. and T. K. S. visualization; P. K. and T. P. D. writing–original draft; P. K. and T. P. D. writing–review and editing; T. P. D. funding acquisition.
Funding and additional information
P. K. acknowledges support by the CNV Foundation. This work has been supported by the European Commission (to T. P. D.).
Edited by Ursula Jakob
Supporting information
References
- 1.Schwarzlander M., Dick T.P., Meyer A.J., Morgan B. Dissecting redox biology using fluorescent protein sensors. Antioxid. Redox Signal. 2016;24:680–712. doi: 10.1089/ars.2015.6266. [DOI] [PubMed] [Google Scholar]
- 2.Kostyuk A.I., Panova A.S., Kokova A.D., Kotova D.A., Maltsev D.I., Podgorny O.V., Belousov V.V., Bilan D.S. In vivo imaging with genetically encoded redox biosensors. Int. J. Mol. Sci. 2020;21:8164. doi: 10.3390/ijms21218164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gutscher M., Sobotta M.C., Wabnitz G.H., Ballikaya S., Meyer A.J., Samstag Y., Dick T.P. Proximity-based protein thiol oxidation by H2O2-scavenging peroxidases. J. Biol. Chem. 2009;284:31532–31540. doi: 10.1074/jbc.M109.059246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morgan B., Van Laer K., Owusu T.N., Ezerina D., Pastor-Flores D., Amponsah P.S., Tursch A., Dick T.P. Real-time monitoring of basal H2O2 levels with peroxiredoxin-based probes. Nat. Chem. Biol. 2016;12:437–443. doi: 10.1038/nchembio.2067. [DOI] [PubMed] [Google Scholar]
- 5.Carmona M., de Cubas L., Bautista E., Moral-Blanch M., Medrano-Fernandez I., Sitia R., Boronat S., Ayte J., Hidalgo E. Monitoring cytosolic H2O2 fluctuations arising from altered plasma membrane gradients or from mitochondrial activity. Nat. Commun. 2019;10:4526. doi: 10.1038/s41467-019-12475-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pastor-Flores D., Talwar D., Pedre B., Dick T.P. Real-time monitoring of peroxiredoxin oligomerization dynamics in living cells. Proc. Natl. Acad. Sci. U. S. A. 2020;117:16313–16323. doi: 10.1073/pnas.1915275117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roma L.P., Deponte M., Riemer J., Morgan B. Mechanisms and applications of redox-sensitive green fluorescent protein-based hydrogen peroxide probes. Antioxid. Redox Signal. 2018;29:552–568. doi: 10.1089/ars.2017.7449. [DOI] [PubMed] [Google Scholar]
- 8.Gutscher M., Pauleau A.L., Marty L., Brach T., Wabnitz G.H., Samstag Y., Meyer A.J., Dick T.P. Real-time imaging of the intracellular glutathione redox potential. Nat. Methods. 2008;5:553–559. doi: 10.1038/nmeth.1212. [DOI] [PubMed] [Google Scholar]
- 9.Meyer A.J., Dick T.P. Fluorescent protein-based redox probes. Antioxid. Redox Signal. 2010;13:621–650. doi: 10.1089/ars.2009.2948. [DOI] [PubMed] [Google Scholar]
- 10.Belousov V.V., Fradkov A.F., Lukyanov K.A., Staroverov D.B., Shakhbazov K.S., Terskikh A.V., Lukyanov S. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat. Methods. 2006;3:281–286. doi: 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- 11.Markvicheva K.N., Bilan D.S., Mishina N.M., Gorokhovatsky A.Y., Vinokurov L.M., Lukyanov S., Belousov V.V. A genetically encoded sensor for H2O2 with expanded dynamic range. Bioorg. Med. Chem. 2011;19:1079–1084. doi: 10.1016/j.bmc.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 12.Bilan D.S., Pase L., Joosen L., Gorokhovatsky A.Y., Ermakova Y.G., Gadella T.W., Grabher C., Schultz C., Lukyanov S., Belousov V.V. HyPer-3: A genetically encoded H(2)O(2) probe with improved performance for ratiometric and fluorescence lifetime imaging. ACS Chem. Biol. 2013;8:535–542. doi: 10.1021/cb300625g. [DOI] [PubMed] [Google Scholar]
- 13.Dalla Tiezza M., Bickelhaupt F.M., Flohe L., Maiorino M., Ursini F., Orian L. A dual attack on the peroxide bond. The common principle of peroxidatic cysteine or selenocysteine residues. Redox Biol. 2020;34:101540. doi: 10.1016/j.redox.2020.101540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pedre B., Young D., Charlier D., Mourenza A., Rosado L.A., Marcos-Pascual L., Wahni K., Martens E., G de la Rubia A., Belousov V.V., Mateos L.M., Messens J. Structural snapshots of OxyR reveal the peroxidatic mechanism of H2O2 sensing. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E11623–E11632. doi: 10.1073/pnas.1807954115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee C., Lee S.M., Mukhopadhyay P., Kim S.J., Lee S.C., Ahn W.S., Yu M.H., Storz G., Ryu S.E. Redox regulation of OxyR requires specific disulfide bond formation involving a rapid kinetic reaction path. Nat. Struct. Mol. Biol. 2004;11:1179–1185. doi: 10.1038/nsmb856. [DOI] [PubMed] [Google Scholar]
- 16.Zheng M., Aslund F., Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]
- 17.Wei Q., Minh P.N., Dotsch A., Hildebrand F., Panmanee W., Elfarash A., Schulz S., Plaisance S., Charlier D., Hassett D., Haussler S., Cornelis P. Global regulation of gene expression by OxyR in an important human opportunistic pathogen. Nucleic Acids Res. 2012;40:4320–4333. doi: 10.1093/nar/gks017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ezerina D., Morgan B., Dick T.P. Imaging dynamic redox processes with genetically encoded probes. J. Mol. Cell. Cardiol. 2014;73:43–49. doi: 10.1016/j.yjmcc.2013.12.023. [DOI] [PubMed] [Google Scholar]
- 19.Schwarzlander M., Wagner S., Ermakova Y.G., Belousov V.V., Radi R., Beckman J.S., Buettner G.R., Demaurex N., Duchen M.R., Forman H.J., Fricker M.D., Gems D., Halestrap A.P., Halliwell B., Jakob U. The 'mitoflash' probe cpYFP does not respond to superoxide. Nature. 2014;514:E12–14. doi: 10.1038/nature13858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poburko D., Santo-Domingo J., Demaurex N. Dynamic regulation of the mitochondrial proton gradient during cytosolic calcium elevations. J. Biol. Chem. 2011;286:11672–11684. doi: 10.1074/jbc.M110.159962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breckwoldt M.O., Armoundas A.A., Aon M.A., Bendszus M., O'Rourke B., Schwarzlander M., Dick T.P., Kurz F.T. Mitochondrial redox and pH signaling occurs in axonal and synaptic organelle clusters. Sci. Rep. 2016;6:23251. doi: 10.1038/srep23251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pak V.V., Ezerina D., Lyublinskaya O.G., Pedre B., Tyurin-Kuzmin P.A., Mishina N.M., Thauvin M., Young D., Wahni K., Martinez Gache S.A., Demidovich A.D., Ermakova Y.G., Maslova Y.D., Shokhina A.G., Eroglu E. Ultrasensitive genetically encoded indicator for hydrogen peroxide identifies roles for the oxidant in cell migration and mitochondrial function. Cell Metab. 2020;31:642–653.e646. doi: 10.1016/j.cmet.2020.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Demaurex N., Poburko D., Frieden M. Regulation of plasma membrane calcium fluxes by mitochondria. Biochim. Biophys. Acta. 2009;1787:1383–1394. doi: 10.1016/j.bbabio.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 24.Matlashov M.E., Bogdanova Y.A., Ermakova G.V., Mishina N.M., Ermakova Y.G., Nikitin E.S., Balaban P.M., Okabe S., Lukyanov S., Enikolopov G., Zaraisky A.G., Belousov V.V. Fluorescent ratiometric pH indicator SypHer2: Applications in neuroscience and regenerative biology. Biochim. Biophys. Acta. 2015;1850:2318–2328. doi: 10.1016/j.bbagen.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Domenech A., Ayte J., Antunes F., Hidalgo E. Using in vivo oxidation status of one- and two-component redox relays to determine H2O2 levels linked to signaling and toxicity. BMC Biol. 2018;16:61. doi: 10.1186/s12915-018-0523-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Cubas L., Pak V.V., Belousov V.V., Ayté J., Hidalgo E. The mitochondria-to-cytosol H2O2 gradient Is caused by peroxiredoxin-dependent cytosolic scavenging. Antioxidants (Basel) 2021;10:731. doi: 10.3390/antiox10050731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ugalde J.M., Schlößer M., Dongois A., Martinière A., Andreas J. Meyer A.J. The latest HyPe(r) in plant H2O2 biosensing. Plant Physiology. 2021 doi: 10.1093/plphys/kiab306. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Entian K.D., Schuster T., Hegemann J.H., Becher D., Feldmann H., Guldener U., Gotz R., Hansen M., Hollenberg C.P., Jansen G., Kramer W., Klein S., Kotter P., Kricke J., Launhardt H. Functional analysis of 150 deletion mutants in Saccharomyces cerevisiae by a systematic approach. Mol. Gen. Genet. 1999;262:683–702. doi: 10.1007/pl00013817. [DOI] [PubMed] [Google Scholar]
- 29.Mumberg D., Muller R., Funk M. Regulatable promoters of Saccharomyces cerevisiae: Comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mumberg D., Muller R., Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 31.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.Y., White D.J., Hartenstein V., Eliceiri K., Tomancak P. Fiji: An open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data presented in this study are contained within this article.