Abstract
Despite advances in prostate cancer screening and treatment, available therapy options, particularly in later stages of the disease, remain limited and the treatment-resistant setting represents a serious unmet medical need. Moreover, disease heterogeneity and disparities in patient access to medical advances result in significant variability in outcomes across patients. Disease classification based on genomic sequencing is a promising approach to identify patients whose tumors exhibit actionable targets and make more informed treatment decisions. Here we discuss how we can accelerate precision oncology to inform broader genomically-driven clinical decisions for men with advanced prostate cancer, drug development and ultimately contribute to new treatment paradigms.
Although prostate cancer affects nearly 1.3 million men worldwide each year(1), there is wide variability in outcomes across patients. Prostate cancer can be classified into clinical disease states that help guide therapy, patient counseling, and goals of care (Figure 1). Efforts in the clinically localized setting have focused on minimizing intervention for those patients with indolent prostate cancer that can be safely monitored, and escalating intervention for those with aggressive tumors at highest risk of relapse. Emerging tissue-based molecular and genomic biomarkers and imaging tools have been developed to improve clinical risk stratification and precision medicine in this disease space(2–5), though data on how best to apply these clinically are still accumulating. For those men that develop metastatic disease, genomic sequencing is now more routinely applied to help guide systemic therapy choice including the selection of PARP inhibitor therapy or immunotherapy for those with castration resistant disease(6). In the following sections we review the current treatment landscape and advances in the genomic classification of advanced prostate cancer, with a focus on the challenges in the wider clinical adoption of the latter and potential strategies to overcome these barriers.
Figure 1. The unmet need of precision medicine across different clinical states of prostate cancer.
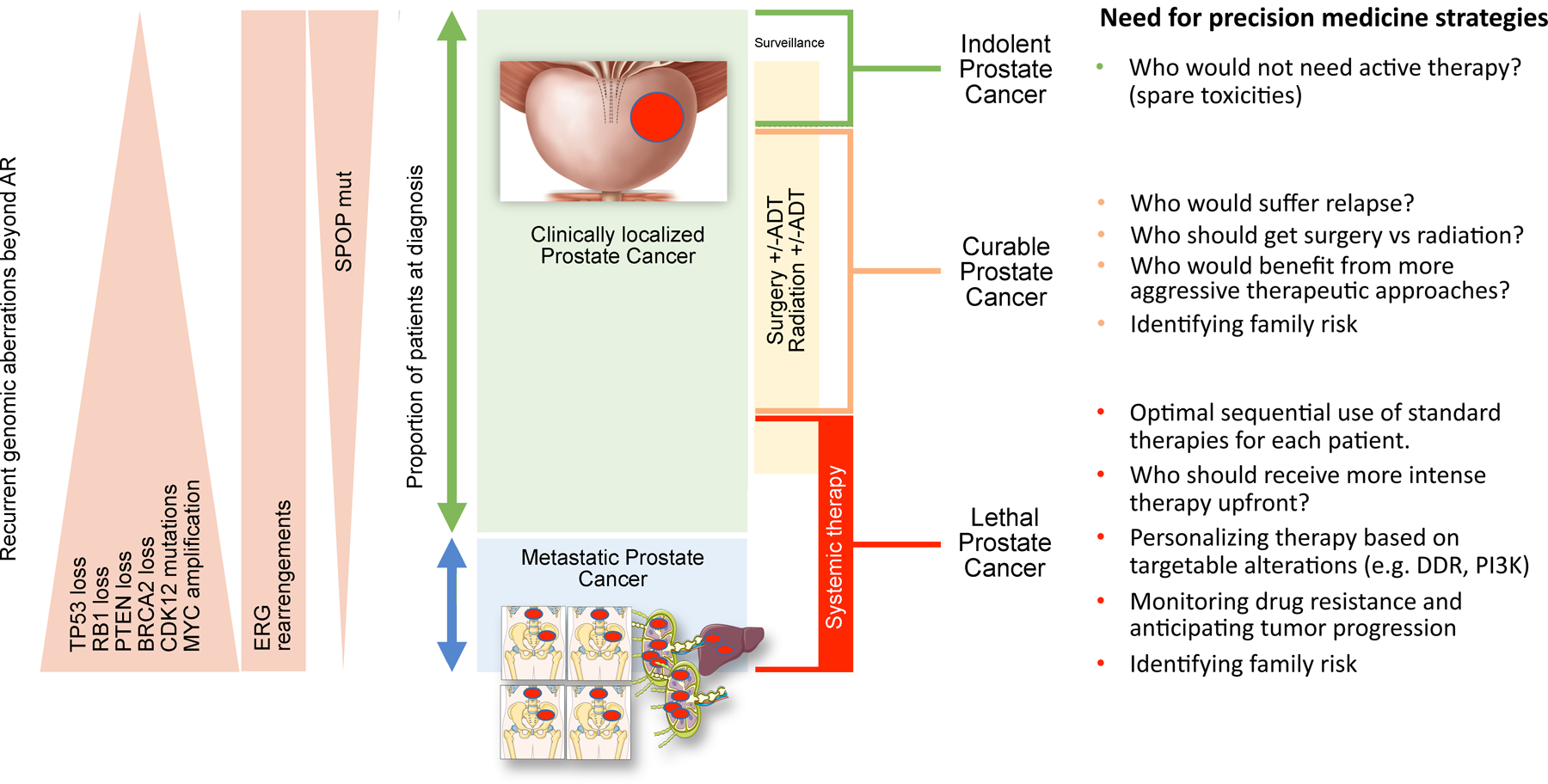
Prostate cancer encompasses a variety of diseases, ranging from indolent to lethal tumors. Localized prostate cancers are managed with active surveillance or treated with surgery, radiation and/or ADT (androgen deprivation therapy). Metastatic prostate cancer is enriched for loss of tumor suppressor genes and MYC amplification, as depicted in the left panel; some of these alterations may be relevant to predict outcome to targeted therapies. The right panel lists key questions that can be addressed by precision medicine studies across the disease spectrum to improve patient outcome.
The clinical landscape of advanced prostate cancer
Metastatic prostate cancer can either develop after local therapy or less commonly, men can present with metastatic disease (Figure 2). Although considered incurable but highly treatable, goals of care for patients with advanced disease have focused on systemic control, prolongation of life and improved quality of life. The most common sites of prostate cancer metastasis are bone and lymph nodes, although some patients do develop visceral disease (e.g, liver, lung) especially in later stages(7). Over 70% of prostate cancer patients develop bone metastases, and this is the sole site of disease in nearly 50%(7, 8). Bone lesions are often sclerotic, identified by bone scan and CT scan, and challenging to evaluate when it comes to response and progression on systemic therapy. Clinical criteria have been developed specifically for prostate cancer that rely on serum prostate specific antigen (PSA) decline and change in size of measurable, non-skeletal, disease, with a definition of progressive disease that includes emergence of new bone lesions observed on bone scans(9). Research incorporating novel imaging techniques may lead to more accurate response and progression assessments(10).
Figure 2. Current therapeutic landscape for different clinical states of advanced forms of prostate cancer.
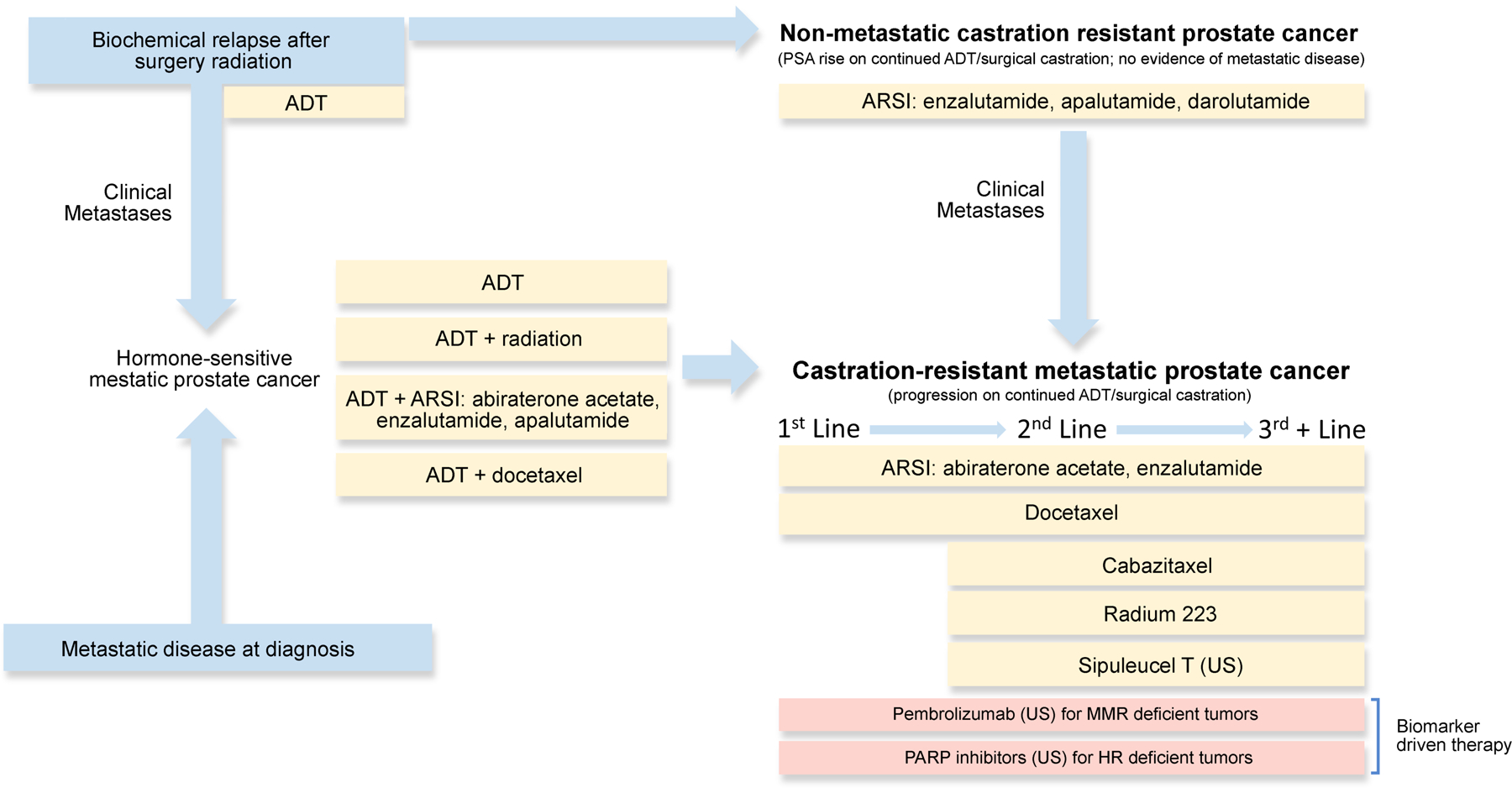
Summary of currently approved therapeutic strategies across advanced prostate cancer. Biochemical relapse after local therapy (top left) can evolve towards emergence of metastasis (left, center) or, alternative, the development of castration-resistant disease in the absence of visible metastatic disease (top right) nonmetastatic castration-resistant prostate cancer). Once metastatic castration-resistant prostate cancer (mCRPC, bottom right) ensues, several therapies are available but there are few tools to prioritize them for each individual patient as subsequent lines of therapy. Some of these approvals refer to the United States only (indicated as “US” in brackets) as of Sept-2020. Yellow boxes indicate drugs approved irrespectively of molecular profiling, whereas pink text boxes show biomarker-driven therapies, approved only for molecularly-defined subpopulations. ADT: androgen deprivation therapy; ARSI: androgen receptor signaling inhibitor.
The mainstay of therapy for men with metastatic prostate cancer is androgen deprivation therapy (ADT). Prostate cancer arises as a hormonally-driven disease, relying on activation of the androgen receptor (AR) by endogenous ligands (e.g. testosterone and dihydrotestosterone) for growth, and is initially responsive to ADT, which inhibits testosterone production by the testes(11). Emergence of resistance to ADT leads to castration-resistant prostate cancer (CRPC). AR signaling represents the main driver of prostate cancer progression(12) and understanding its regulation has advanced both therapies that target this signaling axis and our knowledge of the mechanisms that underlie treatment resistance. The majority of castration resistant prostate tumors retain their dependence on the AR cistrome, primarily through genomic amplification or mutation of the AR gene and alterations in androgen synthesis, among other mechanisms(13). More potent next-generation AR-targeting agents now have a dominant role in the treatment of CRPC(14–20) and hormone naive metastatic prostate cancer in combination with ADT(21–23). These drugs act through inhibition of extra-gonadal androgen formation (ie., abiraterone acetate) or through direct inhibition of the AR (ie., enzalutamide, apalutamide, darolutamide). Nonetheless, resistance to these potent agents inevitably occurs, and cross-resistance limits their utility when used sequentially(24, 25). Further, there remains a subset of patients with rapidly progressive, virulent disease that is unresponsive or minimally responsive to manipulation of the AR axis(26,27).
Patients with metastatic CRPC have multiple treatment options with varied mechanisms of action, including additional potent AR targeted agents, taxane chemotherapies(28–31), the bone-targeted radiopharmaceutical radium-223(32), the cell-based immunotherapy sipuleucel-T(33), and more recently biomarker-driven therapy with the immune checkpoint inhibitor pembrolizumab (for those with mismatch repair defects or microsatellite instability) and the PARP inhibitors olaparib and rucaparib (for those with homologous recombination gene deficiency). Despite these advances, the median overall survival from the development of metastatic CRPC remains approximately three years(34) with wide intra-patient variability in response to therapies.
Genomic classification of metastatic prostate cancer
Although the majority of men with potentially lethal disease are not treated based on molecular biomarker selection, genomic observations combined with preclinical studies have pointed to potential predictive and prognostic biomarkers and emerging resistance mechanisms that hold promise for improving selection of existing and developing therapies for patients (Figure 3).
Figure 3. Proposed workflow for implementation of genomic testing in prostate cancer clinical practice, from sample acquisition to clinical-decision making.
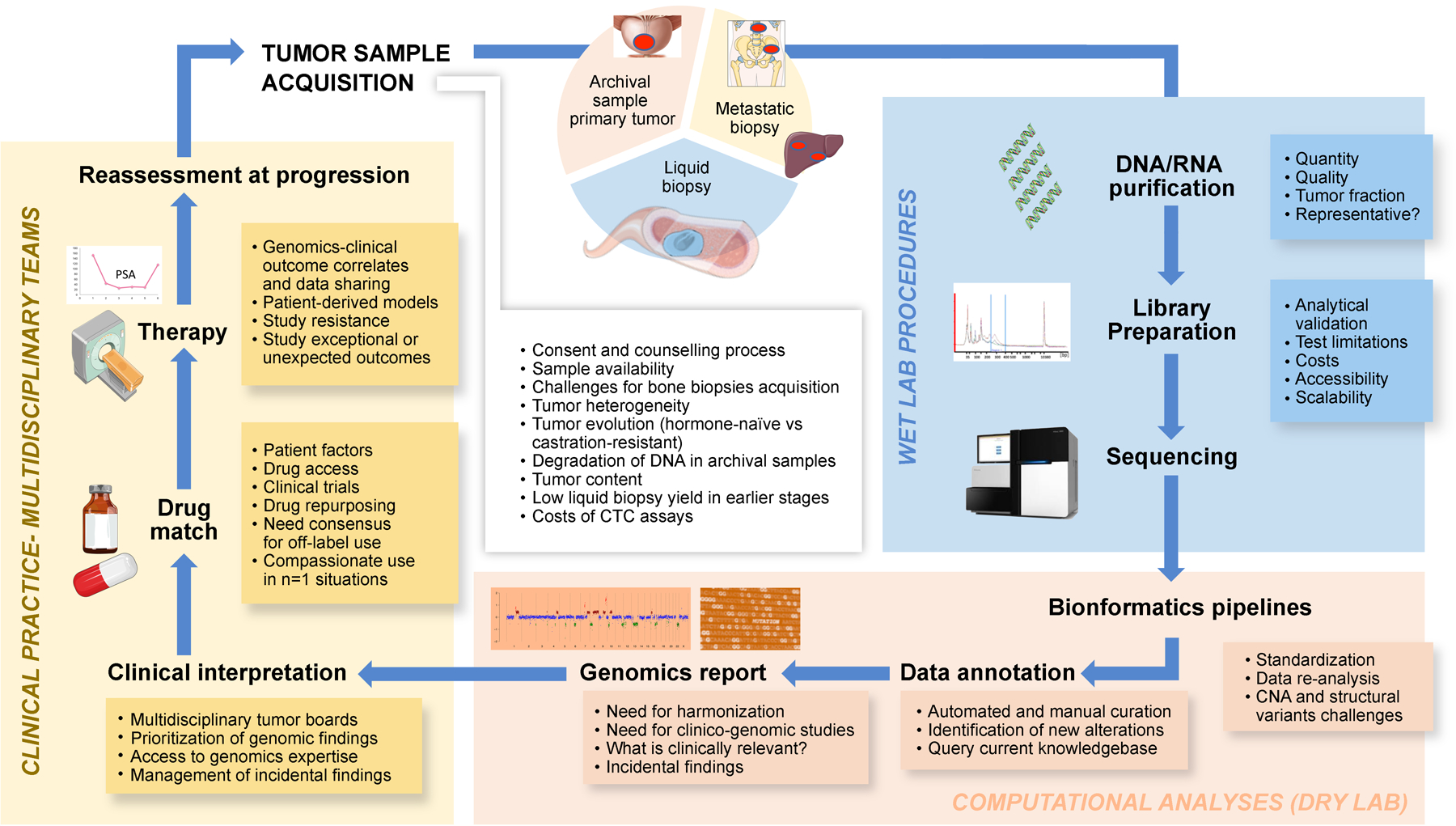
Suitable sources of tumor material for genomic testing include biopsies of the primary or metastatic tumors, or circulating tumor material (top panel). First, tumor DNA/RNA is to be isolated, and sequenced (blue panel); next-generation sequencing (NGS) data is to be processed and reported to physicians (orange panel). These data would then be integrated into the treatment clinical-decision making (yellow panel). The textbox list key concepts to consider at each step of the genomic testing workflow. CTC: circulating tumor cells; CNA: copy-number alterations
Specifically, genomic sequencing studies have identified a high frequency of alterations involving cancer-related genes in metastatic prostate tumors(35–37), including those commonly seen in localized disease and thought to be “early” pathogenesis events such as ETS family gene fusions (approximately 40–50%) (Table 1, Figure 1). Other alterations that are enriched in CRPC include those involving AR (observed in >50% of cases), TP53 (in >40%), PI3K pathway genes such as PTEN (in 45% of cases), BRCA2, BRCA1, ATM and other DNA repair genes (20–25% of cases), CDK12 loss (in 5–7%), RB1 loss (in ~20%), Wnt pathway genes (in ~15%), epigenetic regulator genes (in ~ 20%), and MAP kinase pathway genes (in ~5% of cases). Co-occurrence of alterations is commonly observed between these pathways. Approximately 30% of advanced prostate cancers have been reported to harbor a potentially actionable alteration beyond the AR(38), defined as an alteration which may predict for response to an existing drug at least based on pre-clinical data, although the strength of the data required to define clinical actionability is subject to debate.
Table 1. Prevalence of recurrent genomic alterations across prostate cancer disease states in published cohorts.
Data was extracted from cBioPortal (https://www.cbioportal.org) except for Quigley et al, Cell 2019(46), where data was extracted from the publication. Percentages rounded except if <0.5%.
| TCGA, Cell 2015 | Abida et al, JCO-PO 2017 | Abida et al, PNAS 2019 | Quigley et al, Cell 2019 | |||||
|---|---|---|---|---|---|---|---|---|
| n | 333 | 200 | 140 | 164 | 444* | 101 | ||
| NGS assay | WES | Panel (MSK-IMPACT) | Panel (MSK-IMPACT) | Panel (MSK-IMPACT) | WES | WGS | ||
| Population | Locoregional PC | Locoregional PC | Hormone-sensitive metastatic PC | mCRPC | mCRPC | mCRPC | ||
| Gene | Alterations | Percentage altered | ||||||
| AR and ERG | AR | Mut, Amp | 1 | 2 | 4 | 51 | 59 | 70 |
| ERF | Mut, HomDel | 2 | 0 | 0 | 0 | 5 | N/A | |
| ERG | Fusion | 46 | N/A | N/A | N/A | 41 | 43 | |
| ETV1 | Fusion | 9 | N/A | N/A | N/A | 6 | 10 | |
| ETV4 | Fusion | 5 | N/A | N/A | N/A | 4 | 5 | |
| ETV5 | Fusion | 0 | N/A | N/A | N/A | 0,4 | 2 | |
| FOXA1 | Mut | 4 | 15 | 10 | 10 | 9 | 19 | |
| Cell Cycle | CDK1NB | Mut | 2 | 1 | 2 | 7 | 5 | 7 |
| CDKN2A | Mut | 2 | 2 | 2 | 2 | 3 | N/A | |
| RB1 | Mut, HomDel | 1 | 2 | 7 | 18 | 13 | 12 | |
| TP53 | Mut, HomDel | 8 | 27 | 30 | 48 | 40 | 57 | |
| DNA repair | ATM | Mut, HomDel | 6 | 2 | 2 | 11 | 7 | 6 |
| BRCA1 | Mut, HomDel | 1 | 1 | 1 | 2 | 2 | 1 | |
| BRCA2 | Mut, HomDel | 3 | 6 | 7 | 10 | 11 | 10 | |
| CDK12 | Mut, HomDel | 2 | 4 | 6 | 10 | 7 | 3 | |
| FANCA | Mut, HomDel | 8 | 1 | 3 | 7 | 1 | N/A | |
| MLH1 | Mut, HomDel | 0,3 | 1 | 2 | 1 | 1 | 1 | |
| MSH2 | Mut, HomDel | 1 | 0 | 2 | 3 | 2 | 2 | |
| MSH6 | Mut, HomDel | 1 | 0 | 1 | 1 | 2 | 1 | |
| PALB2 | Mut, HomDel | 0,3 | 1 | 0 | 3 | 1 | N/A | |
| PI3K | AKT1 | Mut | 1 | 0 | 2 | 1 | 1 | 2 |
| PIK3CA | Mut | 2 | 3 | 4 | 3 | 3 | 1 | |
| PIK3CB | Mut | 1 | 0 | 1 | 3 | 2 | N/A | |
| PTEN | Mut, HomDel | 17 | 11 | 17 | 27 | 32 | 45 | |
| WNT | APC | Mut, HomDel | 5 | 3 | 13 | 14 | 8 | 9 |
| CTNNB1 | Mut | 2 | 2 | 6 | 3 | 4 | 6 | |
| Other | ARID1A | Mut | 1 | 1 | 1 | 2 | 3 | N/A |
| BRAF | Mut | 4 | 2 | 2 | 3 | 7 | 4 | |
| CHD1 | Mut, HomDel | 7 | N/A | N/A | N/A | 7 | 9 | |
| MYC | Amp | 7 | 2 | 6 | 10 | 23 | 33 | |
| SPOP | Mut | 11 | 12 | 11 | 5 | 6 | 5 | |
The denominator for assessment of ERG, ETV1, ETV4 and ETV5 fusions is 266, corresponding only to patients with both WES and RNAseq data available.
Approximately 3–5% of prostate cancers harbor evidence of DNA mismatch repair deficiency (dMMR), hyper-mutation, or increased microsatellite instability (MSI-high tumors) that can be identified through DNA sequencing and/or immunohistochemistry detecting loss of the MMR protein(39). These patients may benefit from treatment with pembrolizumab, a PD-1-targeting antibody that is FDA-approved for all dMMR, MSI-high, or tumor mutational burden (TMB)-high solid tumors, and some have shown durable responses to this or other immune checkpoint inhibitors(40). Despite the availability of an FDA-approved therapy, it is not clear how often patients with metastatic CRPC undergo testing for dMMR or MSI, if/when they are also being tested for germline aberrations such as Lynch syndrome, and what to expect regarding degree and duration of response to immunotherapy.
In addition, ~20% of men with metastatic CRPC (mCRPC) harbor germline or somatic alterations involving homologous recombination mediated DNA repair genes, such as BRCA2, which may predict for response to PARP inhibitors and platinum chemotherapy. The Phase 3 PROfound clinical trial identified a significant radiographic progression free survival and overall survival benefit for olaparib in patients with DNA repair aberrations previously treated with a potent AR-pathway inhibitor for CRPC(41). This was the first phase 3 biomarker-based trial in mCRPC, and these data provide a clear rationale for molecular testing in patients with advanced disease. Although responses were observed across patients with different alterations, the benefit was especially evident for men with BRCA2 alterations. The Phase 2 TRITON2 trial of the PARP inhibitor rucaparib has also demonstrated antitumor activity for patients with germline or somatic BRCA alterations(42). Based on these studies, olaparib and rucaparib were recently FDA approved for patients with mCRPC and certain homologous recombination deficiencies. Exceptional responders to platinum chemotherapy have also been identified in patients with BRCA2-mutated prostate cancer(43,44). There is still much to learn regarding the impact of less common DNA repair genes on responsiveness to PARP inhibitors and other potential mediators of sensitivity and resistance to PARP inhibitors and platinum chemotherapy. The use of other DNA damage response targeting agents such as ATR inhibitors may be particularly relevant for ATM-mutant prostate cancer(45,46). Alterations CDK12, proposed to be involved in DNA repair and characterized by focal tandem duplications, high neoantigen burden, and poor prognosis, have been associated with sensitization to immune checkpoint blockade and clinical trials are ongoing(47–50).
Approximately 45% of mCRPC tumors harbor alterations within the PI3K pathway. Rarely these are PIK3CA hotspot mutations, which may be relevant given the FDA approval of alpelisib, a PI3K alpha inhibitor, for the treatment of PIK3CA-mutated breast cancer(51). However, in most cases they occur as loss of function of the PTEN gene. Single agent drugs targeting this pathway have demonstrated limited efficacy for PTEN-deficient tumors(52), although preclinical data suggest benefit from concurrent inhibition of AR(53). A phase 2 study showed benefit for the combination of abiraterone with the AKT inhibitor ipatasertib in PTEN-deficient mCRPC(54), and the Phase 3 IPATential150 trial of ipatasertib plus abiraterone versus abiraterone in first line metastatic CRPC recently met its co-primary endpoint of radiographic progression free survival in those with PTEN loss (de Bono et al, ESMO 2020). Rare hotspot alterations (~1%) in AKT1 and its homologs may be potentially targetable with AKT inhibitors, which have shown benefit in AKT-mutated breast cancer and other solid tumors(55). Other less common hotspot alterations (<5%) in CRPC occur in BRAF, MAP2K1 and KRAS.
In addition, a subset of CRPCs with unusually aggressive clinical behavior, include those that develop histologic features of small cell neuroendocrine carcinoma (30)(26,56,57) and/or harbor molecular characteristics typified by loss of the tumor suppressors RB1 and TP53 (31). These are particularly evident in later stages of the disease, partly as a result of tumor evolution and treatment-driven selective pressure(58). RB1 loss in particular has been shown to associate strongly with poor clinical outcomes in advanced prostate cancer(40,59,60). Strategies to selectively target tumors with aggressive or atypical histologic, clinical, or molecular findings include the use of platinum chemotherapy(57, 61, 62) or other targeted approaches(26,63,64). Insights into how these alterations might be further leveraged are particularly needed.
Challenges for Precision Medicine in Prostate Cancer
With an increased understanding of the molecular landscape of advanced prostate cancer, there has been a rise in the clinical use of genomic sequencing to identify actionable targets. However, the optimal use of genomics to guide clinical decision-making in prostate cancer is not well defined, and there are no current guidelines to inform the timing, type of tissue, or optimal set of clinically-validated laboratory tests. With a growing number of agents being used off-label, reported data have mostly been non-systematic. There remain significant barriers for the implementation of precision oncology for patients (Table 2) and the development of strategies for overcoming these challenges is critical.
Table 2:
Priorities to accelerate precision medicine in metastatic prostate cancer
| Challenges | Short Term Goals | Longer Term Goals |
|---|---|---|
| Challenge 1: Access to tumor tissue for molecular profiling | Standardization of protocols for the collection and processing of specimens for genomic profiling Partnership between oncologists, radiologists, and pathologists regarding optimal techniques and team education Incorporation of sample processing protocols for future molecular tests (eg., RNA, protein) |
Functional imaging to improve diagnosis and biopsy acquisition Generation of patient-derived prostate cancer models for precision medicine ex-vivo clinical trials. |
| Challenge 2: Addressing tumor heterogeneity | Expanded correlation of liquid biopsies and molecular imaging with single site tumor biopsies at different stages of the disease and under distinct therapeutic contexts Optimization of ctDNA, CTC, and other liquid biopsy approaches to improve sensitivity to detect common prostate cancer alterations in the circulation including copy number aberrations Implementation of approaches to capture tumor microenvironment, including immune infiltrates, at different disease states |
Integration of multi-omic approaches (eg., genomics with transcriptomics and protein-based functional assays) to provide additional readouts of pathway activation/suppression. Utilization of functional imaging and liquid biopsies to identify early acquired resistance Improved understanding of the intersection between tumor genomics and tumor-immune system regulation |
| Challenge 3: Detection of complex drivers | Clinical characterization of not only single gene aberrations, including rare long tail events, but also how co-occurring alterations modulate the predictive value for a biomarker-drug matches Functional characterization of rare /uncommon molecular alterations and co-occurring lesions in preclinical models |
Understanding mechanisms of response and resistance to therapies Drug discovery for targeting rare aberrations Clinical-grade tests beyond targeted genomic panels, such as WGS, transcriptomics, methylation Precision oncology trials focused on gene expression signatures, metabolism, epigenetics, or other non-genomic features |
| Challenge 4: Clinical and genomic integration | Streamline of consent process to facilitate broader sharing of de-identified data Generation of a prostate cancer specific N=1 database collecting and linking genomic and clinical data from academic and community partners Institutional or supra-institutional protocols collecting tumor and liquid biopsies for later evaluation of exceptional responders and exceptional non-responders as well as acquired resistance |
Harmonization of genomics data coming from different tests for uniform /comparable annotations, reporting, and visualization Machine-learning approaches to integrate emerging data for genomics-clinical outcome correlations, validation of complex algorithms to prioritize choices of therapies or clinical trials Developing paradigms for precision oncology based on specific alterations |
| Challenge 5: Understanding the impact of genomics in diverse patient populations. | Partnership with patients, community providers, local health departments, the Veterans administration, and other stakeholders Improving access to somatic and germline sequencing for all men with advanced prostate cancer, including underserved populations Point of care educational resources for physicians and patients New technologies and social media platforms to facilitate partnerships with patients and advocacy groups throughout the process Provide access of N=1 database to patients/providers beyond academic institutions |
Genomic landscape studies focused on previously underrepresented populations Improved understanding regarding how germline genetics impacts tumor somatic genomic profiles and tumor phenotypes Elucidation of the interaction between genomic, environmental and lifestyle factors for determining tumor progression and response to therapy Long term impact on reducing disparities |
| Challenge 6: Access to matched therapies and clinical trials | Decision support and education materials for practitioners and patients Live and virtual prostate cancer molecular tumor boards to support clinical decision making Matching of patients with molecularly-defined clinical trials and therapies (eg., via MatchMiner) |
Development of a prostate cancer umbrella protocol for research testing of a platform of agents in molecularly selected populations Early phase trials testing novel molecularly targeted and immune based therapeutic targets and drugs Repurposing of available drugs for molecularly selected populations (“off-label clinical trials”) Development of consensus guidelines for stakeholders (insurance companies, health systems) on off-label use Streamlining N=1 Investigational New Drug processes for compassionate use of unapproved agents |
Challenge 1: Access to tumor tissue for molecular profiling
Procurement of tumor material is a vital step for molecular characterization. Over 70% of metastatic prostate cancer patients develop bone metastases, with nearly half having bone-predominant disease(7, 8). Despite imaging-guidance, biopsy of osteoblastic bone metastases can be technically challenging and often yields insufficient tumor tissue. Moreover, sample decalcification may impact nucleic acid quantity and quality. Even in high-volume academic centers that have tissue acquisition protocols in place, the viable tumor tissue yield from bone biopsies for genomic assays is relatively low. In a whole-exome sequencing study, 43/76(57%) of bone biopsies were successfully sequenced(65). A second study obtained adequate tissue from 76/110 image-guided bone biopsies (67%)(66). In both series, careful patient selection led to increased yields. Lymph node biopsies of have higher success rates; however, accessibility to pelvic and retroperitoneal lymph nodes may not be feasible in many patients.
The use of primary prostate tumor, such as biopsy cores or radical prostatectomy tissue, for molecular stratification of advanced prostate cancer could overcome some of the challenges in acquiring metastatic biopsies for certain genomic alterations such as DNA repair genes. Indeed, some driver alterations show high concordance between primary and metastatic tumors(67). However, prostate biopsy cores may often contain insufficient tumor content, or the DNA obtained may be too low in yield or degraded for genomic testing. In the phase 3 PROfound trial, quality control failures accounted for 31% of all samples prospectively analyzed(41). Primary localized prostate tumors are commonly multifocal(68, 69), and therefore the obtained sample may not always represent the clone driving metastatic spread. Furthermore, treatment-naive biopsies do not capture acquired alterations that may develop after progression on systemic therapies reflecting treatment resistance and/or clonal selection.
Challenge 2: Tumor heterogeneity
Multifocal prostate cancer consists of spatially and clonally distinct tumors within the primary prostate gland(69). Most studies to date support a monoclonal origin of metastatic prostate cancer, with metastatic lesions traceable back to one founding clone within the primary tumor(70, 71). Tumors subsequently acquire alterations with disease progression and treatment resistance, and polyclonal metastasis-to-metastasis seeding can further lead to intra-patient heterogeneity(72, 73). Although autopsy studies have supported limited intra-individual heterogeneity across metastases when looking at alterations involving common oncogenes and tumor suppressors in late-stage disease(74), more data are needed to understand how faithfully single site metastatic biopsies represent the overall metastatic tumor burden in prostate cancer patients, and how tumor heterogeneity is impacted by selective pressures from exposure to specific therapies. This is particularly relevant in the setting of oligo-progression, or cases where differential responses are observed across sites of metastases, with some lesions progressing and others maintaining good responses due to therapy response and resistance mechanisms differing between anatomic sites. A further understanding the molecular mechanisms that promote heterogeneity and resistance is critical. For instance, certain genomic or epigenomic alterations may promote plasticity or transcriptional dysregulation under therapy pressure(58), and in other cases subclonal genomic alterations emerge and/or cooperate to drive treatment resistance(75).
Challenge 3: Detection of loss of function events and complex drivers
An additional layer of complexity for prostate cancer precision medicine is intrinsic to the genomic make-up of the disease. Beyond alterations involving the AR gene, which a major driver of treatment resistance, many of the genomic alterations enriched in metastatic prostate cancer involve loss of tumor suppressor genes (such as PTEN, TP53, RB1) rather than activating events (such as hotspot mutations in BRAF, PIK3CA, AKT). Loss of tumor suppressor genes can be the result of truncating mutations (which can occur anywhere along the gene), gene deletions (complete or partial) or complex gene rearrangements. Assessing the presence of loss-of-function events in a given gene is challenging compared with activating hotspot mutations identified through multiplexed panels. Further, whereas loss of PTEN or BRCA2 may be predictive for tumors more susceptible to AKT or PARP inhibition respectively, deletion of other genes, such as RB1, may have been shown to be prognostic(36, 59) and their value as predictive biomarkers to guide therapy selection is not yet supported by strong data. Co-occurrence of BRCA2 and RB1 deletion is also frequent, given these genes’ physical proximity(76). Loss of RB1 may have a different role in facilitating lineage plasticity in the context of concurrent TP53 loss or other features(76–78).
There is a long list of less common alterations in metastatic CRPC for whose predictive or prognostic role or functional significance little is known(79). This long tail, representing alterations occurring in <5% of patients, may provide unique insights in identifying rare subsets of patients with distinct biology or therapeutic responses, alone or in combination with other events. However, traditional study designs are not geared towards investigating the clinical value of low-prevalence events.
Most clinical assays for precision medicine rely on targeted exome sequencing, either tumor-only sequencing or paired tumor and germline testing. Integration of tumor with germline sequencing offers the additional benefit of not requiring separate tests and the ability to assess loss of heterozygosity and the clonal hematopoiesis. There is also a wealth of data in non-coding regions. Whole genome sequencing, while not clinically used today, is capable of detecting structural alterations and rearrangements that lead to dysregulation of cancer-related genes and resistance-causing alterations(49, 80). An additional layer of information may be retrieved through tumor mRNA analysis to identify gene fusions and gene expression profiles, protein expression, and DNA methylation to capture epigenetic alterations, although none of these approaches are currently considered clinically actionable. Despite the potential to exploit emerging molecular findings to learn about disease biology and discover new therapeutic targets, the gap between what is theoretically possible and what is practically feasible, especially in the clinic, remains substantial.
Challenge 4: Clinical and genomic integration
Prostate cancer is unique compared with other cancer types due to its long natural history, clinical use of PSA as a routine biomarker and tendency to metastasize to bone, making measurements of response and resistance complex. Therefore, the relevant clinical data elements and endpoints in prostate cancer differ from other cancers, which is particularly pertinent when it comes to defining “exceptional responder” and “non-responder” classifiers that are not always based on standard RECIST criteria and may be influenced by patterns of metastases (eg., bone versus other sites), PSA dynamics, and disease state. An ‘exceptional’ responder likely requires a definition that goes beyond the traditional complete response, radiographic or PSA, but should also encompass those patients achieving durable benefit (eg., lack of need to restart or switch systemic therapy for ≥24 months).
In addition to integrating clinical features with genomic and other molecular alterations to identify clinically meaningful biomarkers, decision support for clinicians based on existing information is also needed. More specifically, accurate interpretation of genomic findings requires a harmonized use of terminologies. Most laboratory providers offer a clinician-oriented report as the final product from a DNA sequencing test. However, beyond well validated biomarkers for approved drugs, such reports often highlight gene aberrations based on their functional impact rather than on the clinical actionability of the detected event, thereby making the linking of patients with the most appropriate treatments or clinical trials challenging. As patients go on to receive biomarker-driven therapies, collection of specimens to learn from responders and non-responders and those who develop acquired resistance can help catalyze future mechanistic studies.
Challenge 5: Understanding the impact of genomics in diverse patient populations
Our knowledge of the extent to which molecular and genomic mechanisms contribute to prostate cancer progression and treatment resistance in diverse patient populations is limited. Most published genomic studies were conducted at select academic institutions and were limited to patients of primarily European ancestry(65, 81). ETS gene fusions are less common in prostate tumors of men of Asian ancestry compared with those of European origin, and other alterations such as CHD1 loss are more common(82, 83). MYC amplifications have been reported to be more frequent in tumors from African American men compared with those of European origin, and PTEN deletion and ETS gene fusion less common(84). The question of how to address prostate cancer disparities is of particular relevance, given a higher incidence and mortality rate in African American men(85). However, there exist reports that black men have similar or better outcomes than white men in certain settings and when access to care settings was equal(86)(87, 88). There is also a need to expand access to genomic and genetic testing and to better understand how specific data from academic centers translate to the broader community where the majority of the population receives care. Otherwise, there is a risk of genomic testing contributing to health disparities, if data from select population groups are incorrectly attributed to the wider society.
Challenge 6: Access to matched therapies and clinical trials
Advancements in understanding prostate tumor biology, accelerated development of new targeted drugs, and acknowledgement of often suboptimal drug approval timelines using traditional approaches has resulted in novel clinical trial designs, including basket, platform and umbrella trials, intended to accelerate drug development and approval (89). Umbrella trials enroll patients with the same histologic cancer type and assign them to different treatment cohorts based on the presence of a specific biomarker. Platform trials are an extension of umbrella trials: patients are randomized to different cohorts and by following statistical algorithms, new therapies are adapted or existing therapies dropped from an ongoing study (90). Basket trials group patients based on a specific biomarker, regardless of their tumor type, which is particularly helpful for biomarkers that are present at low frequencies. Biomarker-driven strategies require validated, reproducible, and scalable biomarker tests. Several multi-institutional efforts combine the umbrella and basket concepts by enrolling patients with multiple cancer types, including the NCI-MATCH Trial (Molecular Analysis for Therapy Choice), ECOG-ACRIN ComboMATCH, a successor to NCI-MATCH that focuses on drug combinations, ASCO’s TAPUR Trial, and the iMATCH trial in the United Kingdom. Despite available biomarker trials, including those dedicated to prostate cancer patients, trial accessibility and effective matching to patients remain challenging especially outside of academic institutions. The underlying factors include trial slot limitations, patient proximity to clinical trial centers, exclusion from pan-cancer studies due to the need for concurrent androgen deprivation therapy or requirement of measurable non-bony disease, and insufficient familiarity of clinical practitioners with genomic biomarkers and trial matching tools.
Strategies for Overcoming Barriers
In this section we propose strategies to address the limitations listed above and to facilitate the translation of precision oncology to the broader population of prostate cancer patients.
Strategy 1: Improving tumor tissue acquisition.
Considering the challenges of obtaining biopsies from metastatic sites of prostate cancer, there is clearly a need for engagement of radiologists, pathologists and oncologists to establish guidelines that can improve biopsy yield and can be widely implemented in clinical practice. Different groups have reported their experience in optimizing bone biopsy procedures and pathology processing protocols to 1) maximize the quality of the obtained material(91, 92); 2) validate clinical algorithms to stratify patients based on the likelihood of successful biopsy (93); and 3) develop imaging assays to guide selection of target bone lesions(10). Training, experience and feedback are key to improving the skills of clinicians and biomedical professionals, thus cross-institutional educational initiatives, such as teaching videos showing current best practices and emerging techniques for bone biopsy and procurement, may be useful. Although protocols are currently not standardized, our review of current studies that evaluate biopsies from metastatic prostate cancer sites have identified a number of parameters to consider when optimizing tumor yield for successful molecular sequencing (Box 1). The advent of next-generation imaging may also offer the opportunity to incorporate higher resolution and functional assessments of the tumor. These techniques, including multiparametric MRI with diffusion-weighted imaging and PET-CT or PET-MRI fusions could help optimize biopsy acquisition(10). Moreover, the use of primary diagnostic samples may facilitate implementation of genomic testing beyond academic institutions. Diagnostic guidelines should account for collecting FFPE blocks specifically for future genomic testing when planning biopsy acquisition and handling procedures.
Text Box 1. Parameters to consider for optimum tumor yield for molecular profiling.
Use of larger needle or drill when safe during biopsy procedure(91).
Discussion between clinicians and radiologists about selection of lesions for biopsy based on radiographic parameters such as size, location, evidence of progression, degree of sclerosis(93), proximity to the skin, and corresponding activity on technetium-99 bone scan, PET imaging(130) or multiparametric MRI(131, 132). A clear understanding of the number of cores required for genomic and other molecular assays should be established in advance with selection of soft tissue over bone when possible and avoidance of areas with extensive necrosis on imaging or of previously irradiated bone lesions. Aiming the needle at the edge of an osseous metastatic lesion also increases yield.
Communication between clinicians and pathologists to ensure appropriate bone decalcification methods to maximize nucleic acid preservation (133). Discussion of tissue procurement needs in advance (e.g., formalin fixed versus fresh/frozen, use of cytology techniques, inclusion of blood clots).
Continued training and feedback between oncology and the interventional radiology and pathology teams as additional methods are developed that improve yield.
Strategy 2: Accounting for tumor heterogeneity in clinical practice.
Advanced imaging assays could also help select lesions with resistance to systemic agents for re-biopsy. Molecular imaging, such as prostate specific membrane antigen (PSMA)-PET/CT, provides insights into tumor heterogeneity across and within lesions. Nearly all hormone-naive and castration-resistant prostate cancer cells express PSMA. PSMA-PET imaging is a highly sensitive and specific imaging modality, and PSMA-directed theranostics are in clinical development. However, in later stages of prostate cancer, some castration resistant prostate tumors lose PSMA expression and/or demonstrate PSMA-PET versus FDG-PET discordance, which has been associated with AR-independent disease and aggressive clinical behavior(94, 95). Therefore PSMA-imaging may help guide patient selection for PSMA-directed therapies and/or targeted biopsies to look for AR-independent disease or other molecular targets.
Isolation of tumor material from blood, such as circulating tumor DNA (ctDNA) or circulating tumor cells (CTCs), also offers opportunities to non-invasively characterize molecular features of metastatic tumors in a manner tailored to the patient. Liquid biopsies also capture intra-patient tumor heterogeneity by identifying tumor alterations across metastases whose cells have entered the bloodstream and can address temporal evolution of alterations, as repeated samples may be obtained over time without major discomfort for patients(96–99). ctDNA tumor content increases with disease progression, making most assays most sensitive in the metastatic disease setting(100). The detection of AR gene alterations (eg., mutations, amplification) in ctDNA have been associated with inferior outcomes in men with CRPC treated with AR pathway inhibitors(101,102). Targeted and whole exome ctDNA sequencing has shown high concordance with matched tumor biopsies in capturing clinically relevant and recurrent prostate cancer aberrations(103, 104). Nevertheless, cell free DNA fragments are usually small, rendering copy-number estimation challenging. This is progressively being addressed by technological improvements and advanced computational pipelines (101,105,106)
CTC enumeration in patients with CRPC are prognostic and changes in CTC numbers on therapy have been identified as surrogates of overall survival at a single-patient level, offering a promising intermediate endpoint for accelerating development of new agents in phase II clinical trials(107). CTC assessment of the AR splice variant, AR-V7, expression has been associated with inferior response to AR-pathway inhibitors in men with CRPC(108). Single cell CTC genomics can also capture tumor heterogeneity (109,110). CTCs are amenable to different types of analyses, including DNA, mRNA and protein tests, as well morphological assessment in terms of shape, size, clustering, to identify specific resistance phenotypes(111–113). However, CTC counts are variable across patients and not all patients are amenable to all these analyses at once; their interpretation is influenced by tumor burden and tumor heterogeneity. It is important to also recognize that while both ctDNA and CTC approaches may be useful tools for the non-invasive detection of certain alterations, important features of the tumor (such as tumor morphology and cell surface proteins) as well as the microenvironment, are crucially missing, and tumor biopsies will likely remain standard when selecting patients for drugs targeting this type of non-genomic features.
Strategy 3: Improving detection of clinically relevant genotypes and phenotypes.
There is a pressing need for the clinical characterization of not only single gene aberrations, including long tail events, but also of combined/co-occurring alterations. Understanding their incidence, context, and clinical significance will require detailed data capture (Figure 4). Functional characterization of less common alterations is also needed. Understanding their biologic significance and incorporating preclinical approaches such as genome wide library screening may point to therapeutic vulnerabilities and provide new opportunities for target discovery (eg., synthetic lethal approaches).
Figure 4. Learning from exceptional responders.
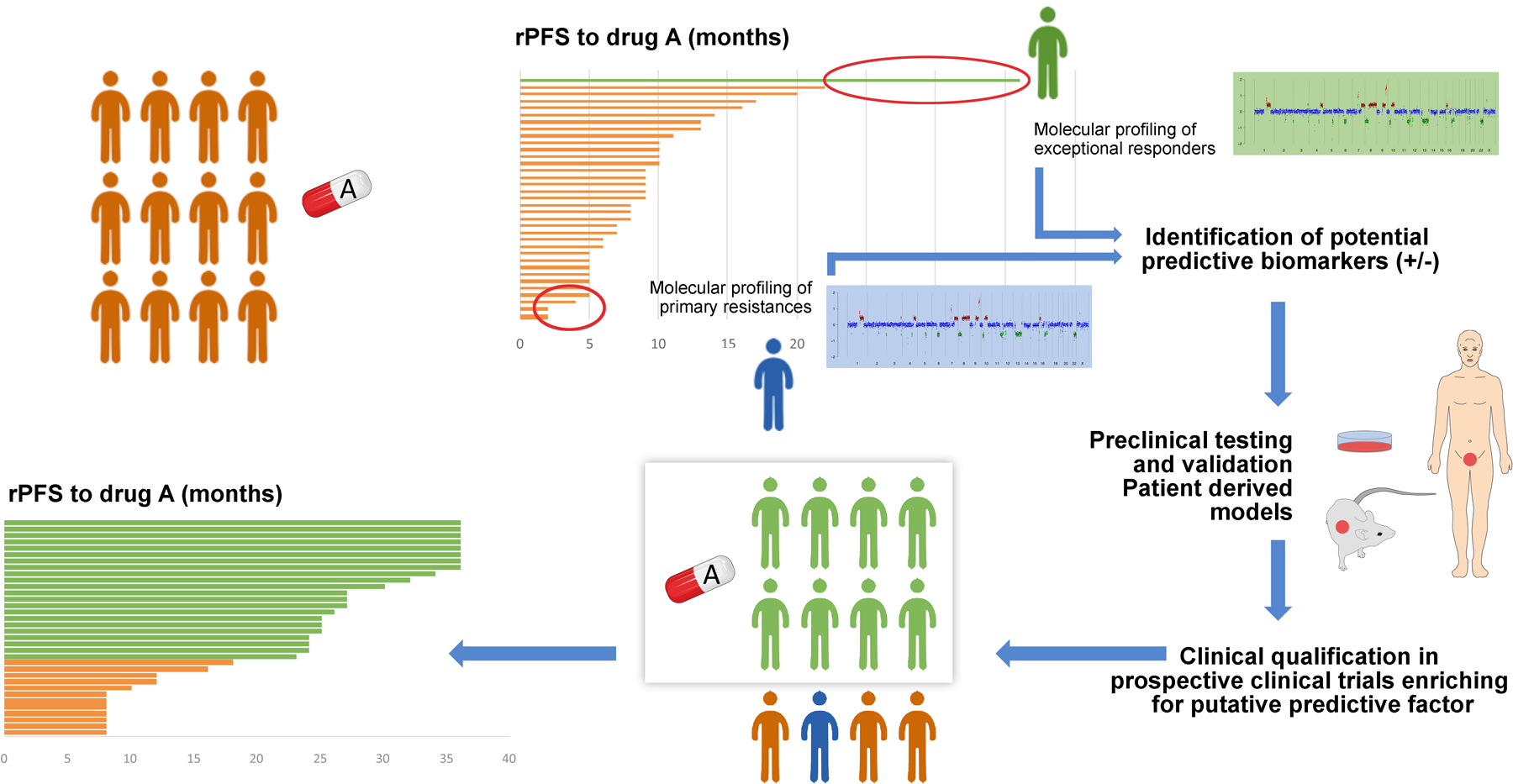
The diagram represents how the study of exceptional responders can lead to advances in prostate cancer treatment. In the top left, a group of patients with prostate cancer receive treatment “A”. The swimmers plot shows how only some patients (in green) achieve a long-lasting response, whereas other patients (in blue) are primarily resistant to drug “A”. Comparing the molecular profiles of sensitive (green) vs resistant (blue) patients may lead to identification of putative relevant predictive biomarkers of response and resistance, to be validated in functional laboratory studies (right panel). Biologically validated biomarkers would then be tested back in clinical trials for clinical qualification, ideally enriched for those patients presenting the biomarker of interest (“green” patients in the figure). If qualified, this enrichment would lead to improved outcome on treatment “A” for patients with the putative biomarker (waterfall plot in the bottom left panel). rPFS: radiographic progression-free survival
Although whole genome sequencing and non-genomic assays are currently mostly limited to research studies, these broader platforms will likely enter the clinic in the future and these data should be systematically assessed similar to targeted panels. Beyond genomically-selected trials, some therapeutic trials are targeted to non-genomic biomarkers and require specific biomarker tests available only as part of trial screening. Examples include PSMA-targeted therapies that require PSMA imaging for eligibility, AKT inhibitor studies that require immunohistochemical detection of PTEN protein loss, and CDK4-targeted therapy that requires immunohistochemical assessment of Rb. Although the use of the majority of these assays is considered investigational in CRPC, integration of these data with genomics may inform the optimal genotype and phenotype for patient selection in the future. Transcriptome analyses may be useful to identify pathway alterations such as PI3K/AKT activation, AR activity, neuroendocrine programs, or glucocorticoid receptor-driven programs, which may be useful clinically for selection of existing or emerging drugs. Developing ways to adopt these emerging assays in clinical practice by understanding how to standardize, validate, and implement them on a routine basis will be critical.
Strategy 4: Integrating clinical and genomic landscapes in routine practice
Building platforms that link genomic and clinical outcome data can help uncover the significance of an individual mutation or spectrum of alterations, especially for infrequent genomic events which are unlikely to be appreciated in small data series. This framework has the potential to also inform the design of biomarker-driven clinical trials. National genomic-medicine programs provide the opportunity to promote best practices in data sharing by structuring consent processes, harmonizing clinical and genomic data collection, organizing data access, and committing to global data exchange.
The development of tools to accumulate and visualize clinical-genomic datasets is also important. The cBioPortal for Cancer Genomics is an open source software system that allows visualization and analysis of large-scale cancer genomic datasets(114,115). Although the portal facilitates the exploration of multidimensional cancer genomics, at present it has limited information on treatment-specific variables, including treatment types, duration, and response. Utilizing cBioPortal as a strategic partner, the AACR Genomics Evidence Neoplasia Information Exchange (GENIE) is an international pan-cancer registry of real-world data assembled through data sharing between academic cancer centers contributing clinical-grade genomic and clinical outcomes data(116,117). Leveraging these existing platforms may help fast-track research in prostate cancer through data sharing between a wider scope of academic centers and serve as a framework for future expansion of similar efforts to community practices.
Integration of genomic data into the Electronic Medical Record (EMR), which was initially designed as an ordering, billing, and clinical documentation system, could also help facilitate clinical decision making and act as a scientific discovery platform. A significant amount of longitudinal clinical data is accessible within the EMR and having this information linked to genomic data could help unveil previously unrecognized correlations and patterns. Alternatives to human curation for large sets of clinical variables, such as machine learning and artificial intelligence strategies for integration of pathology and radiology with genomic data, will be critical features to ensure scalability(118). The formulation of consistent standards for the transfer of genomic results and defining the optimal legal and ethical framework with regard to access and use of human data is also critical.
Platforms that integrate various levels of evidence including mutation and cancer type, and therapeutic targets can help facilitate treatment decisions. Several pan-cancer resources, such as OncoKB(119), PMKB(120) and other knowledgebases(121), and the ESMO Scale for Clinical Actionability of Molecular Targets (ESCAT) (122), utilize levels of evidence systems to assign clinical actionability to an individual genomic event with the goal of supporting optimal treatment decisions and providing a shared vocabulary to facilitate communication between the relevant stakeholders, including patients, physicians, healthcare systems, pharmaceutical industry and regulatory bodies. Harmonized interpretation of these knowledgebases can improve patient matching(121). As the treatment of cancer becomes increasingly driven by genomic data, molecular tumor boards or other decision support systems are essential, similar to the current model of disease-specific tumor boards today(123,124). Point-of-care access to decision support through prostate cancer specific virtual tumor boards or electronic applications could also help provide real-time guidance to practicing clinicians. Additionally, data capture for tracking the frequency and reasons driving clinical decisions is also needed, particularly with regard to uncommon yet potentially actionable alterations. Understanding molecularly targeted responses is not only important for patients with confirmed MSI-high or BRCA-mutated prostate cancer but should also be explored in the context of other molecularly-defined tumor subsets.
Strategy 5: Incorporating diverse patient populations into genomic analyses to enable clinical decisions reflective of the real-world situation.
Partnering with patients and advocacy groups is an essential step for raising awareness about personalized cancer care and ensuring that the diverse patient population is appropriately represented in available datasets and subsequently developed analytical approaches. To that end, internet-based programs, such as the Count Me In Project (https://mpcproject.org), and social media platforms can enhance visibility, engage patients and caregivers in research efforts and provide educational tools. The GENTleMEN study (NCT03053097) is an example of a germline genetic testing study that is enrolling geographically distributed, diverse prostate cancer patient populations through internet consent and with testing kits delivered by mail. Engaging patients and other stakeholders will be important as platform costs and insurance reimbursements are considered as precision oncology expands beyond academic centers, and these considerations may differ based on geography, health delivery models, and other factors.
A better understanding of variation in prostate cancer treatment and care across diverse populations is needed, as is being addressed by the IRONMAN global population-based registry (NCT03151629). Given that disparities in health care are magnified in clinical research, focused efforts are also needed to engage diverse patient populations and different ancestries using culturally sensitive methods. Studies that do not offer treatment, such as those focused on clinical data and specimen collection for genomic evaluation, should try to deliver substantive benefits to patients(125). Discussions with patients should be comprehensive, honest, culturally sensitive and should highlight unique aspects relevant to large scale initiatives, such as genomic data collection and sample sharing to advance discovery. Additionally, safeguards need to be in place to protect patient information and genomic data privacy during the process.
Partnering with community providers, local health departments, and other stakeholders will be important for expanding access to genomic testing and precision medicine to diverse community patient populations. Programs such as IMPACT (IMProving Access, Counseling & Treatment For Californians with Prostate Cancer), which has a mission to provide high quality prostate cancer treatment to Californian men with little or no health insurance, are models for expanding access to care for underserved patients with prostate cancer. The Prostate Cancer Foundation has developed a partnership with the Veterans Administration to increase research and precision medicine studies offered to veterans with prostate cancer within the VA healthcare system (https://www.pcf.org/va-partnership/).
Strategy 6: Matching patients to appropriate therapies and clinical trials.
Critical to precision medicine is an infrastructure that allows patients to identify and receive matched therapies, either drugs approved for a particular indication or in the context of therapeutic trials. Several tools are available to this end, including the NIH clinicaltrials.gov database, local molecular tumor boards at larger academic institutions, and increasingly detailed annotation of commercially-available molecular testing reports that offer ease of access to ordering clinicians, including in non-academic settings. Matchminer (https://matchminer.org) is an open source computational platform that aims to improve matching of patient-specific genomic profiles with locally available trials based on both genomics and curated sets of inclusion criteria. Establishment of a matching platform for prostate cancer along with a collaborative prostate cancer tumor board across institutions may help facilitate the identification of available trials for patients. In addition to recruitment to existing trials and ensuring that prostate cancer patients are eligible for studies, the development of new and accessible biomarker-driven trials for men with prostate cancer is also needed. This will hopefully limit the use of off-label drugs with preliminary efficacy and safety data in prostate cancer(125), though reporting responses and tracking outcomes of these specific off-label situations will still be critical.
A Call for Action
Translational studies regarding the utility of next generation sequencing technologies in the broader population and their impact are largely lacking. “Exceptional responders” (N=1) to drugs based on genomic testing have been reported in prostate cancer, including responses to both targeted therapies and immunotherapies, with little known about how often this occurs or details regarding those patients that do not respond(63,126–128). In addition, there is a long list of less common alterations in metastatic prostate cancer with little data assessing their predictive or prognostic roles or functional significance. Identifying biomarker-specific trials for these individuals remains a significant barrier to clinical practice. By leveraging existing infrastructures including those developed by pan-cancer efforts, to collect and share N=1 biomarker-driven precision medicine experiences regardless of their level of success, we can accelerate precision oncology for prostate cancer patients. Sharing such experiences can be used to inform broader genomically-driven clinical decision making and also facilitate drug development.
An N=1 effort should be accessible to both academic and community practitioners and potentially other partners and would establish needed data regarding the clinical relevance of less prevalent but actionable alterations through pooled analyses. In addition to clinical actions, data regarding specific mutations or data from patients treated with off label and single patient IND use of specific drugs could also be captured. Future directions may encompass streamlined central consent, data entry, and specimen collection. By providing decision support and facilitating drug access, this would accelerate the accessibility of precision medicine for prostate cancer patients. Select patients with rare targets or unexpected responses could be referred for expanded molecular assessment and clinical trial enrollment.
Conclusions
A number of opportunities and challenges lie ahead for precision medicine in advanced prostate cancer. We outline some of them here with specific short- and longer- term collaborative strategies (Table 2). Genomic testing (both germline and somatic) is increasingly being performed and is indicated for patients with metastatic castration resistant prostate cancer, with a subset of men harboring actionable targets (eg. in DNA repair genes) that have been confirmed through clinical trial data. Improving access and implementation of genomic testing across diverse populations will be critical to optimizing access to better therapies. More widespread testing, based on updated guidelines(129), is also now identifying alterations in patients at earlier stages of the disease, for instance patients with high-risk localized or metastatic hormone-naive prostate cancer.
If we aim for precision medicine to make a difference for patients with prostate cancer, it is our duty as clinicians and researchers to define the optimal timing and type of genomic testing and application of test results for the treatment of patients. This will require first capturing data across diverse populations and understanding how these tests are being or could be used. By understanding practice patterns, we envision a collaborative and global network to learn from both responders and non-responders and understand the clinical impact of rare biologically significant molecular alterations and response and resistance to therapies. Capturing the decision-making process and potentially expanding it to collect tissues for a focused prostate cancer group across a wide network is a unique endeavor not currently addressed by pan-cancer efforts and should be prioritized. It is also important to consider, that when it comes to therapy response, it is not solely tumor-related contributing factors that should be considered, but also barriers, including patient-specific considerations (e.g. alternative options, urgency of therapy, comorbidities, patient preference, toxicities, financial burden), assay limitations, and drug access limitations. Addressing these challenges will help establish a framework for future genomic and non-genomic biomarkers, and shape future guidelines for molecular profiling in prostate cancer, including who and when should perform it and how patients should be biopsied/liquid biopsied to identify targets for precision oncology therapy.
Footnotes
Competing Interests Statement. J.M. reports advisory board participation for Amgen, AstraZeneca, Roche, Janssen, MSD and Clovis Oncology; research funding from AstraZeneca and Pfizer Oncology; W.A. reports consulting /advisory for Clovis, Janssen, More Health, ORIC, Daiichi Sankyo; research funding from AstraZeneca, Zenith Epigenetics, Clovis, GlaxoSmithKline, ORIC, Epizyme; travel from GlaxoSmithKline, Clovis, ORIC; and honoraria from CARET. R.R.M. received research funding from Bayer, Pfizer, Tempus; serves on Advisory Board for Bayer, Bristol Myers Squib, Exelixis, Janssen, Novartis, Pfizer, Sanofi, Tempus; is a consultant for Dendreon, Vividion. R.A. reports advisory board participation and research funding from Merck, AstraZeneca, and Janssen; B.M. reports research funding from AstraZeneca, Janssen, Clovis, Astellas, Beigene; M.R. reports consulting: Amgen, Ambryx, Constellation; educational writing and consulting: Plexus; speaking: Bayer, Janssen; funding and clinical research support: Novartis, Astellas, Medivation, Merck; D.B.S. has consulted for/received honoraria from Pfizer, Loxo Oncology, Lilly Oncology, BioBridge, Vivideon Therapeutics, and Illumina; E.V. reports advisory/consulting: Tango Therapeutics, Genome Medical, Invitae, Enara Bio, Janssen, Manifold Bio, Monte Rosa; research support: Novartis, BMS; Equity: Tango Therapeutics, Genome Medical, Syapse, Enara Bio, Manifold Bio, Microsoft, Monte Rosa; travel reimbursement: Roche/Genentech; institutional patents on chromatin mutations and immunotherapy response, and methods for clinical interpretation; D.V. reports honoraria from Clovis Oncology. H.B. reports advisory/consulting from Janssen, Amgen, Astra Zeneca, Pfizer, Astellas, Sanofi Genzyme and research funding from Janssen, Abbvie Stemcentryx, Eli Lilly, Millenium, Astellas. J.V. is employed by the Prostate Cancer Clinical Trials Consortium. H.R.S., J.W.S., and A.K.M. are employed by the Prostate Cancer Foundation.
References:
- 1.Bray F et al. , Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68, 394–424 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Eggener SE et al. , Molecular Biomarkers in Localized Prostate Cancer: ASCO Guideline. J Clin Oncol 38, 1474–1494 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Sonni I et al. , Impact of. J Nucl Med, (2020). [Google Scholar]
- 4.Klotz L et al. , Active Surveillance Magnetic Resonance Imaging Study (ASIST): Results of a Randomized Multicenter Prospective Trial. Eur Urol 75, 300–309 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Kasivisvanathan V et al. , MRI-Targeted or Standard Biopsy for Prostate-Cancer Diagnosis. N Engl J Med 378, 1767–1777 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ku SY, Gleave ME, Beltran H, Towards precision oncology in advanced prostate cancer. Nat Rev Urol 16, 645–654 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gandaglia G et al. , Distribution of metastatic sites in patients with prostate cancer: A population-based analysis. Prostate 74, 210–216 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Halabi S et al. , Meta-Analysis Evaluating the Impact of Site of Metastasis on Overall Survival in Men With Castration-Resistant Prostate Cancer. J Clin Oncol 34, 1652–1659 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scher HI et al. , Trial Design and Objectives for Castration-Resistant Prostate Cancer: Updated Recommendations From the Prostate Cancer Clinical Trials Working Group 3. J Clin Oncol 34, 1402–1418 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perez-Lopez R et al. , Multiparametric Magnetic Resonance Imaging of Prostate Cancer Bone Disease: Correlation With Bone Biopsy Histological and Molecular Features. Invest Radiol 53, 96–102 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huggins C, Hodges CV, Studies on prostatic cancer. I. The effect of castration, of estrogen and androgen injection on serum phosphatases in metastatic carcinoma of the prostate. CA Cancer J Clin 22, 232–240 (1972). [DOI] [PubMed] [Google Scholar]
- 12.Chen CD et al. , Molecular determinants of resistance to antiandrogen therapy. Nat Med 10, 33–39 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Watson PA, Arora VK, Sawyers CL, Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer 15, 701–711 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ryan CJ et al. , Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med 368, 138–148 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Bono JS et al. , Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med 364, 1995–2005 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scher HI et al. , Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med 367, 1187–1197 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Beer TM, Tombal B, Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med 371, 1755–1756 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Hussain M et al. , Enzalutamide in Men with Nonmetastatic, Castration-Resistant Prostate Cancer. N Engl J Med 378, 2465–2474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fizazi K et al. , Darolutamide in Nonmetastatic, Castration-Resistant Prostate Cancer. N Engl J Med 380, 1235–1246 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Chi KN et al. , Apalutamide for Metastatic, Castration-Sensitive Prostate Cancer. N Engl J Med 381, 13–24 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Fizazi K et al. , Abiraterone plus Prednisone in Metastatic, Castration-Sensitive Prostate Cancer. N Engl J Med 377, 352–360 (2017). [DOI] [PubMed] [Google Scholar]
- 22.James ND et al. , Abiraterone for Prostate Cancer Not Previously Treated with Hormone Therapy. N Engl J Med 377, 338–351 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis ID et al. , Enzalutamide with Standard First-Line Therapy in Metastatic Prostate Cancer. N Engl J Med 381, 121–131 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Attard G et al. , Abiraterone Alone or in Combination With Enzalutamide in Metastatic Castration-Resistant Prostate Cancer With Rising Prostate-Specific Antigen During Enzalutamide Treatment. J Clin Oncol 36, 2639–2646 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rathkopf DE et al. , Safety and Antitumor Activity of Apalutamide (ARN-509) in Metastatic Castration-Resistant Prostate Cancer with and without Prior Abiraterone Acetate and Prednisone. Clin Cancer Res 23, 3544–3551 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beltran H et al. , The role of lineage plasticity in prostate cancer therapy resistance. Clin Cancer Res, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beltran H et al. , Aggressive variants of castration-resistant prostate cancer. Clin Cancer Res 20, 2846–2850 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tannock IF et al. , Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med 351, 1502–1512 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Petrylak DP et al. , Docetaxel and estramustine compared with mitoxantrone and prednisone for advanced refractory prostate cancer. N Engl J Med 351, 1513–1520 (2004). [DOI] [PubMed] [Google Scholar]
- 30.de Wit R et al. , Cabazitaxel versus Abiraterone or Enzalutamide in Metastatic Prostate Cancer. N Engl J Med 381, 2506–2518 (2019). [DOI] [PubMed] [Google Scholar]
- 31.de Bono JS et al. , Prednisone plus cabazitaxel or mitoxantrone for metastatic castration-resistant prostate cancer progressing after docetaxel treatment: a randomised open-label trial. Lancet 376, 1147–1154 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Parker C et al. , Alpha emitter radium-223 and survival in metastatic prostate cancer. N Engl J Med 369, 213–223 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Kantoff PW et al. , Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N Engl J Med 363, 411–422 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Beer TM et al. , Enzalutamide in Men with Chemotherapy-naïve Metastatic Castration-resistant Prostate Cancer: Extended Analysis of the Phase 3 PREVAIL Study. Eur Urol 71, 151–154 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson D et al. , Integrative clinical genomics of advanced prostate cancer. Cell 161, 1215–1228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abida W et al. , Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci U S A 116, 11428–11436 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stopsack KH et al. , Oncogenic genomic alterations, clinical phenotypes, and outcomes in metastatic castration-sensitive prostate cancer. Clin Cancer Res, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abida W et al. , Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci U S A, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nava Rodrigues D et al. , Immunogenomic analyses associate immunological alterations with mismatch repair defects in prostate cancer. J Clin Invest 128, 4441–4453 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abida W et al. , Analysis of the Prevalence of Microsatellite Instability in Prostate Cancer and Response to Immune Checkpoint Blockade. JAMA Oncol 5, 471–478 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Bono J et al. , Olaparib for Metastatic Castration-Resistant Prostate Cancer. N Engl J Med, (2020). [DOI] [PubMed] [Google Scholar]
- 42.Abida W et al. , Rucaparib in Men With Metastatic Castration-Resistant Prostate Cancer Harboring a. J Clin Oncol, JCO2001035 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng HH, Pritchard CC, Boyd T, Nelson PS, Montgomery B, Biallelic Inactivation of BRCA2 in Platinum-sensitive Metastatic Castration-resistant Prostate Cancer. Eur Urol 69, 992–995 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zafeiriou Z et al. , Genomic Analysis of Three Metastatic Prostate Cancer Patients with Exceptional Responses to Carboplatin Indicating Different Types of DNA Repair Deficiency. Eur Urol 75, 184–192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rafiei S et al. , ATM Loss Confers Greater Sensitivity to ATR Inhibition than PARP Inhibition in Prostate Cancer. Cancer Res, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yap TA et al. , First-in-Human Trial of the Oral Ataxia Telangiectasia and Rad3-Related Inhibitor BAY 1895344 in Patients with Advanced Solid Tumors. Cancer Discov, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu YM et al. , Inactivation of CDK12 Delineates a Distinct Immunogenic Class of Advanced Prostate Cancer. Cell 173, 1770–1782.e1714 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Viswanathan SR et al. , Structural Alterations Driving Castration-Resistant Prostate Cancer Revealed by Linked-Read Genome Sequencing. Cell 174, 433–447.e419 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quigley DA et al. , Genomic Hallmarks and Structural Variation in Metastatic Prostate Cancer. Cell 175, 889 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Nguyen B et al. , Pan-cancer Analysis of CDK12 Alterations Identifies a Subset of Prostate Cancers with Distinct Genomic and Clinical Characteristics. Eur Urol, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.André F et al. , Alpelisib for. N Engl J Med 380, 1929–1940 (2019). [DOI] [PubMed] [Google Scholar]
- 52.George DJ et al. , Phase 2 clinical trial of TORC1 inhibition with everolimus in men with metastatic castration-resistant prostate cancer. Urol Oncol, (2019). [DOI] [PubMed] [Google Scholar]
- 53.Schwartz S et al. , Feedback suppression of PI3Kα signaling in PTEN-mutated tumors is relieved by selective inhibition of PI3Kβ. Cancer Cell 27, 109–122 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Bono JS et al. , Randomized Phase II Study Evaluating Akt Blockade with Ipatasertib, in Combination with Abiraterone, in Patients with Metastatic Prostate Cancer with and without PTEN Loss. Clin Cancer Res 25, 928–936 (2019). [DOI] [PubMed] [Google Scholar]
- 55.Hyman DM et al. , AKT Inhibition in Solid Tumors With AKT1 Mutations. J Clin Oncol 35, 2251–2259 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aggarwal RR et al. , Whole Genome and Transcriptional Analysis of Treatment-Emergent Small Cell Neuroendocrine Prostate Cancer Demonstrates Intra-Class Heterogeneity. Mol Cancer Res, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aparicio AM et al. , Combined Tumor Suppressor Defects Characterize Clinically Defined Aggressive Variant Prostate Cancers. Clin Cancer Res 22, 1520–1530 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beltran H et al. , Circulating tumor DNA profile recognizes transformation to castration-resistant neuroendocrine prostate cancer. J Clin Invest, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen WS et al. , Genomic Drivers of Poor Prognosis and Enzalutamide Resistance in Metastatic Castration-resistant Prostate Cancer. Eur Urol 76, 562–571 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hamid AA et al. , Compound Genomic Alterations of TP53, PTEN, and RB1 Tumor Suppressors in Localized and Metastatic Prostate Cancer. Eur Urol 76, 89–97 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Corn PG et al. , Cabazitaxel plus carboplatin for the treatment of men with metastatic castration-resistant prostate cancers: a randomised, open-label, phase 1–2 trial. Lancet Oncol 20, 1432–1443 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aparicio AM et al. , Platinum-based chemotherapy for variant castrate-resistant prostate cancer. Clin Cancer Res 19, 3621–3630 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beltran H et al. , A phase II trial of the aurora kinase A inhibitor alisertib for patients with castration resistant and neuroendocrine prostate cancer: efficacy and biomarkers. Clin Cancer Res, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Puca L et al. , Delta-like protein 3 expression and therapeutic targeting in neuroendocrine prostate cancer. Sci Transl Med 11, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Robinson D et al. , Integrative Clinical Genomics of Advanced Prostate Cancer. Cell 162, 454 (2015). [DOI] [PubMed] [Google Scholar]
- 66.Aggarwal R et al. , Clinical and Genomic Characterization of Treatment-Emergent Small-Cell Neuroendocrine Prostate Cancer: A Multi-institutional Prospective Study. J Clin Oncol, JCO2017776880 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mateo J et al. , Genomics of lethal prostate cancer at diagnosis and castration resistance. J Clin Invest 130, 1743–1751 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lovf M et al. , Multifocal Primary Prostate Cancer Exhibits High Degree of Genomic Heterogeneity. Eur Urol, (2018). [DOI] [PubMed] [Google Scholar]
- 69.Boutros PC et al. , Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat Genet 47, 736–745 (2015). [DOI] [PubMed] [Google Scholar]
- 70.Haffner MC et al. , Tracking the clonal origin of lethal prostate cancer. J Clin Invest 123, 4918–4922 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu W et al. , Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med 15, 559–565 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gundem G et al. , The evolutionary history of lethal metastatic prostate cancer. Nature 520, 353–357 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hong MK et al. , Tracking the origins and drivers of subclonal metastatic expansion in prostate cancer. Nat Commun 6, 6605 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kumar A et al. , Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med 22, 369–378 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nava Rodrigues D et al. , RB1 Heterogeneity in Advanced Metastatic Castration-Resistant Prostate Cancer. Clin Cancer Res 25, 687–697 (2019). [DOI] [PubMed] [Google Scholar]
- 76.Chakraborty G et al. , Significance of. Clin Cancer Res 26, 2047–2064 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ku SY et al. , Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 355, 78–83 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mu P et al. , SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science 355, 84–88 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Armenia J et al. , The long tail of oncogenic drivers in prostate cancer. Nat Genet 50, 645–651 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Takeda DY et al. , A Somatically Acquired Enhancer of the Androgen Receptor Is a Noncoding Driver in Advanced Prostate Cancer. Cell 174, 422–432.e413 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Network CGAR, The Molecular Taxonomy of Primary Prostate Cancer. Cell 163, 1011–1025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ren S et al. , Whole-genome and Transcriptome Sequencing of Prostate Cancer Identify New Genetic Alterations Driving Disease Progression. Eur Urol, (2017). [DOI] [PubMed] [Google Scholar]
- 83.Li J et al. , A genomic and epigenomic atlas of prostate cancer in Asian populations. Nature 580, 93–99 (2020). [DOI] [PubMed] [Google Scholar]
- 84.Koga Y et al. , Genomic Profiling of Prostate Cancers from Men with African and European Ancestry. Clin Cancer Res 26, 4651–4660 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.DeSantis CE et al. , Cancer statistics for African Americans, 2016: Progress and opportunities in reducing racial disparities. CA Cancer J Clin 66, 290–308 (2016). [DOI] [PubMed] [Google Scholar]
- 86.Dess RT et al. , Association of Black Race With Prostate Cancer-Specific and Other-Cause Mortality. JAMA Oncol 5, 975–983 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Halabi S et al. , Overall Survival of Black and White Men With Metastatic Castration-Resistant Prostate Cancer Treated With Docetaxel. J Clin Oncol 37, 403–410 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ramalingam S et al. , Prostate-specific antigen response in black and white patients treated with abiraterone acetate for metastatic castrate-resistant prostate cancer. Urol Oncol 35, 418–424 (2017). [DOI] [PubMed] [Google Scholar]
- 89.Cecchini M et al. , Challenges with Novel Clinical Trial Designs: Master Protocols. Clin Cancer Res 25, 2049–2057 (2019). [DOI] [PubMed] [Google Scholar]
- 90.Berry SM, Connor JT, Lewis RJ, The platform trial: an efficient strategy for evaluating multiple treatments. JAMA 313, 1619–1620 (2015). [DOI] [PubMed] [Google Scholar]
- 91.Sailer V et al. , Bone biopsy protocol for advanced prostate cancer in the era of precision medicine. Cancer 124, 1008–1015 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Holmes MG et al. , CT-Guided Bone Biopsies in Metastatic Castration-Resistant Prostate Cancer: Factors Predictive of Maximum Tumor Yield. J Vasc Interv Radiol 28, 1073–1081.e1071 (2017). [DOI] [PubMed] [Google Scholar]
- 93.Lorente D et al. , Castration-Resistant Prostate Cancer Tissue Acquisition From Bone Metastases for Molecular Analyses. Clin Genitourin Cancer 14, 485–493 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Thang b. Sue Ping a et al. (2018).
- 95.Tosoian JJ et al. , Correlation of PSMA-Targeted. Clin Genitourin Cancer 15, e65–e68 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lambros MB et al. , Single-Cell Analyses of Prostate Cancer Liquid Biopsies Acquired by Apheresis. Clin Cancer Res 24, 5635–5644 (2018). [DOI] [PubMed] [Google Scholar]
- 97.Goodall J et al. , Circulating Cell-Free DNA to Guide Prostate Cancer Treatment with PARP Inhibition. Cancer Discov 7, 1006–1017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.De Laere B et al. , Comprehensive Profiling of the Androgen Receptor in Liquid Biopsies from Castration-resistant Prostate Cancer Reveals Novel Intra-AR Structural Variation and Splice Variant Expression Patterns. Eur Urol 72, 192–200 (2017). [DOI] [PubMed] [Google Scholar]
- 99.Quigley D et al. , Analysis of Circulating Cell-Free DNA Identifies Multiclonal Heterogeneity of BRCA2 Reversion Mutations Associated with Resistance to PARP Inhibitors. Cancer Discov 7, 999–1005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Conteduca V et al. , Plasma tumour DNA as an early indicator of treatment response in metastatic castration-resistant prostate cancer. Br J Cancer 123, 982–987 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Romanel A et al. , Plasma AR and abiraterone-resistant prostate cancer. Sci Transl Med 7, 312re310 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Azad AA et al. , Androgen Receptor Gene Aberrations in Circulating Cell-Free DNA: Biomarkers of Therapeutic Resistance in Castration-Resistant Prostate Cancer. Clin Cancer Res 21, 2315–2324 (2015). [DOI] [PubMed] [Google Scholar]
- 103.Wyatt AW et al. , Concordance of Circulating Tumor DNA and Matched Metastatic Tissue Biopsy in Prostate Cancer. J Natl Cancer Inst 109, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Adalsteinsson VA et al. , Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun 8, 1324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Carreira S et al. , Tumor clone dynamics in lethal prostate cancer. Sci Transl Med 6, 254ra125 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Annala M et al. , Circulating Tumor DNA Genomics Correlate with Resistance to Abiraterone and Enzalutamide in Prostate Cancer. Cancer Discov 8, 444–457 (2018). [DOI] [PubMed] [Google Scholar]
- 107.Scher HI et al. , Circulating tumor cell biomarker panel as an individual-level surrogate for survival in metastatic castration-resistant prostate cancer. J Clin Oncol 33, 1348–1355 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Antonarakis ES et al. , AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med 371, 1028–1038 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lambros MB et al. , Single-Cell Analyses of Prostate Cancer Liquid Biopsies Acquired by Apheresis. Clin Cancer Res 24, 5635–5644 (2018). [DOI] [PubMed] [Google Scholar]
- 110.Scher HI et al. , Phenotypic Heterogeneity of Circulating Tumor Cells Informs Clinical Decisions between AR Signaling Inhibitors and Taxanes in Metastatic Prostate Cancer. Cancer Res 77, 5687–5698 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Keomanee-Dizon K, Shishido SN, Kuhn P, Circulating Tumor Cells: High-Throughput Imaging of CTCs and Bioinformatic Analysis. Recent Results Cancer Res 215, 89–104 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Beltran H et al. , The Initial Detection and Partial Characterization of Circulating Tumor Cells in Neuroendocrine Prostate Cancer. Clin Cancer Res 22, 1510–1519 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Miyamoto DT et al. , An RNA-Based Digital Circulating Tumor Cell Signature Is Predictive of Drug Response and Early Dissemination in Prostate Cancer. Cancer Discov 8, 288–303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cerami E et al. , The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gao J et al. , Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Consortium APG, AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov 7, 818–831 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Printz C, AACR releases large cancer genomic data set from project GENIE. Cancer 123, 1685 (2017). [DOI] [PubMed] [Google Scholar]
- 118.Kehl KL et al. , Assessment of Deep Natural Language Processing in Ascertaining Oncologic Outcomes From Radiology Reports. JAMA Oncol, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chakravarty D et al. , OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol 2017, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Huang L et al. , The cancer precision medicine knowledge base for structured clinical-grade mutations and interpretations. J Am Med Inform Assoc 24, 513–519 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wagner AH et al. , A harmonized meta-knowledgebase of clinical interpretations of somatic genomic variants in cancer. Nat Genet 52, 448–457 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mateo J et al. , A framework to rank genomic alterations as targets for cancer precision medicine: the ESMO Scale for Clinical Actionability of molecular Targets (ESCAT). Ann Oncol 29, 1895–1902 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Knepper TC et al. , Key Lessons Learned from Moffitt’s Molecular Tumor Board: The Clinical Genomics Action Committee Experience. Oncologist 22, 144–151 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tafe LJ et al. , Implementation of a Molecular Tumor Board: The Impact on Treatment Decisions for 35 Patients Evaluated at Dartmouth-Hitchcock Medical Center. Oncologist 20, 1011–1018 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pfister DG, Off-label use of oncology drugs: the need for more data and then some. J Clin Oncol 30, 584–586 (2012). [DOI] [PubMed] [Google Scholar]
- 126.Nickols NG et al. , MEK-ERK signaling is a therapeutic target in metastatic castration resistant prostate cancer. Prostate Cancer Prostatic Dis 22, 531–538 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Subudhi SK et al. , Neoantigen responses, immune correlates, and favorable outcomes after ipilimumab treatment of patients with prostate cancer. Sci Transl Med 12, (2020). [DOI] [PubMed] [Google Scholar]
- 128.Vlachostergios PJ et al. , Exceptional Response to Pembrolizumab in a Patient With Castration-Resistant Prostate Cancer With Pancytopenia From Myelophthisis. J Oncol Pract 15, 343–345 (2019). [DOI] [PubMed] [Google Scholar]
- 129.Mohler JL et al. , Prostate Cancer, Version 2.2019, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 17, 479–505 (2019). [DOI] [PubMed] [Google Scholar]
- 130.McKay RR et al. , Imaging, procedural and clinical variables associated with tumor yield on bone biopsy in metastatic castration-resistant prostate cancer. Prostate Cancer Prostatic Dis 17, 325–331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Houlahan KE et al. , Molecular Hallmarks of Multiparametric Magnetic Resonance Imaging Visibility in Prostate Cancer. Eur Urol, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kasivisvanathan V et al. , MRI-Targeted or Standard Biopsy for Prostate-Cancer Diagnosis. N Engl J Med, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Choi SE, Hong SW, Yoon SO, Proposal of an appropriate decalcification method of bone marrow biopsy specimens in the era of expanding genetic molecular study. J Pathol Transl Med 49, 236–242 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]


