Abstract
T-cell development is predicated on the successful rearrangement of the T-cell receptor (TCR) gene loci, which encode for antigen-specific receptors. Recombination Activating Gene (RAG) 2 is required for TCR gene rearrangements, which occur during specific stages of T-cell development. Here, we differentiated human pluripotent stem cells with a CRISPR/Cas9-directed deletion of the RAG2 gene (RAG2-KO) to elucidate the requirement for the TCRβ chain in mediating β-selection during human T-cell development. In stark contrast to mice, human RAG2-KO T-lineage progenitors progressed to the CD4+CD8+ double-positive (DP) stage in the absence of TCRβ rearrangements. Nonetheless, RAG2-KO DPs retrovirally-transduced to express a rearranged TCRβ chain showed increased survival and proliferation as compared to control-transduced RAG2-KO DPs. Furthermore, transcriptomic analysis showed that TCRβ- and control-transduced RAG2-KO DPs differed in gene pathways related to survival and proliferation. Our results provide new insights as to the distinct requirement for the TCRβ chain during human T-cell development.
Keywords: RAG2, pre-TCR signaling, β-selection, human T cell development
Introduction
The V(D)J Recombination Activating Genes (RAG) 1 and 2 are two essential DNA processing enzymes required for the rearrangement of the B- and T-cell receptor (BCR or TCR) gene loci (1, 2). RAG1/2 initiate V(D)J recombination by forming a complex that first recognizes and binds to recombination signal sequences (RSS) found adjacent to each V, D, and J gene segment. Upon formation of a synapse with another RSS, the RAG complex induces a double-strand DNA break, which is repaired by non-homologous end-joining process (3, 4). This ultimately results in the imperfect joining of different V, D, and J gene segments to potentially generate millions of different antigen receptors from a few hundred V(D)J segments.
A dysfunction in either RAG1 or RAG2 results in primary immunodeficiencies (PIDs), characterized by a failure to respond to pathogens or properly identify healthy versus malignant cells (5–7). In gene knock-out (KO) mouse studies, either RAG1- or RAG2-KO mice show a complete lack of mature B- or T-cells. In particular, intrathymic T-cell development is arrested at the CD4−CD8− double-negative (DN)3 stage, when RAG1/2-dependent TCRβ rearrangements are completed (8, 9). Successful rearrangement of the TCRβ chain allows for pairing with the surrogate preTα chain to form the preTCR complex, which is required for survival, proliferation, and differentiation to the CD4+CD8+ double-positive (DP) stage (10). Limited access to thymic tissues from individuals with RAG1 or RAG2 gene alterations, which are often hypomorph variants rather than null (11), have led to the assumption that developing human RAG1/2-KO T-cell progenitors would also arrest at the CD7+CD5+CD1a+CD4−CD8− stage (DN3-equivalent in humans) (12). However, we previously reported that induced pluripotent stem cells (iPSCs) derived from severe combined immunodeficient (SCID) patients with RAG1 gene alterations were capable of differentiating well beyond the DN stage and reached the CD7+CD5+CD1a+CD4+CD8+ DP stage in vitro (13). Recently, similar findings were observed with SCID patient-derived CD34+ hematopoietic progenitor cells (14, 15). Nonetheless, this left open the question as to whether preTCR signaling has any function during human T-cell development.
To address the specific roles of the RAG complex and TCRβ chain in human T-cell development, we generated RAG2-deficient (RAG2-KO) human pluripotent stem cell (hPSC) lines, which were differentiated towards the hematopoietic lineage and subsequently induced to become T-cells in vitro. Control wild-type (WT) and RAG2-KO T-cell progenitors both progressed equivalently to the CD4+CD8+ DP stage, but RAG-KO DPs displayed decreased survival and expansion compared to WT DPs. Forced expression of a rearranged TCRβ chain in RAG2-KO DPs resulted in increased survival and expansion, which was further highlighted by the up-regulated expression of survival and proliferation genes. Thus, our results revealed an unexpected stage-specific requirement and function of RAG2-mediated TCRβ rearrangements in developing human T-cells.
Materials and Methods
hESC Maintenance.
Human ESCs (H1; WiCell Research Institute, Madison, WI) were maintained and expanded on plates coated with Matrigel (Corning, NY, USA) in TeSR-E8 medium (STEMCELL Technologies, Vancouver, Canada). Cells were passaged by non-enzymatic dissociation using 0.5 mM EDTA.
Generation of RAG2-KO hESCs.
The pD1321-GFP expression vector, containing cassettes for GFP, Cas9 endonuclease, a CRISPR chimeric cDNA, and the gRNA moiety we designed to target RAG2 [GGTTATGCTTTACATCCAGA], was custom synthesized (DNA2.0). After transfection with Lipofectamine 3000 (Life Technologies, Carlsbad, CA), GFP+ hESCs were sorted using flow cytometry. Individual clones were picked, expanded, and aliquots were collected for purification of genomic DNA using the Genomic DNA kit (Invitrogen). Mutations (indels) were validated by sequencing products of PCR amplification of the regions flanking the targeting sites. The RAG2 KO clone 1 exhibited a 1bp deletion in one allele and 16bp deletion in the other allele, while clone 4 exhibited a 11bp deletion in one allele and 23bp deletion in the other allele.
Western Blot Analysis.
Briefly, cellular lysates were prepared by incubating the cells in lysis buffer (50 mM Tris-HCl, pH7.5, 150 mM NaCl, 0.5% NP-40, 2 mM EDTA) containing protease inhibitor cocktail (Roche) for 20 min at 4°C, followed by centrifugation at 14000×g for 15 min at 4°C. Proteins were separated by SDS-PAGE, transferred onto PVDF membrane (Millipore, Louis, MO) and probed with anti-RAG2 antibody (Abcam – Ab95955; 1:1000 dilution) overnight at 4°C followed by incubation with secondary antibody. The anti-RAG2 antibody used is a rabbit polyclonal made against a recombinant fragment corresponding to amino acids 271–519 of human RAG2 (Abcam), which are well beyond the gRNA targeting site. Immunoreactive bands were visualized using western blotting Luminol reagent (Thermo). PBMCs from a T-ALL patient were used as positive control (16).
Immunostaining.
Cells were fixed in 4% paraformaldehyde, and permeabilization and blocking were performed in 5% NGS (Abcam, Cambridge, USA) and 1% Triton X-100 (Sigma-Aldrich, Louis, MO) in PBS for 30 min. Cells were stained and analyzed as previously described (17).
Teratoma Formation.
All animal studies were approved by Ethics Committee of Experimental Research of Peking University and were in accordance with the International Animal Care and Use Committee Guidelines. 6–8 week-old non-obese diabetic (NOD)/SCID mice were injected subcutaneously with 1×107 hESCs resuspended in DMED-F12 with 50% Matrigel to allow teratoma formation for 8 weeks. Histological analysis was performed as previously described (17).
hPSC Differentiation and CD34+ Isolation.
hPSCs, H1 embryonic stem cells (ESC) were differentiated as previously described (18). After 8 days of differentiation, embryoid bodies were harvested and dissociated into single-cells using Collagenase type IV and trypsin-EDTA as previously described (18), and positively selected using a MACS column (Miltenyi Biotec) with anti-CD34 PE-conjugated antibody (BD Biosciences) and anti-PE microbeads. The cell yield and purity of the positive selection was assessed pre- and post- MACS by flow cytometry.
OP9-DL4–7FS Co-Culture and Differentiation.
OP9-DL4 cells expressing hIL-7, hFLT3-L, and hSCF (7FS) were generated (manuscript in preparation) and grown in α-MEM containing 10–20% FBS (Gibco), 1% Pen-Strep (Life Technologies), and phospho-ascorbic acid (Sigma-Aldrich) at 37°C, 5% CO2. For co-cultures, OP9-DL4–7FS cells were plated in 6-well plates the previous day. MACS-purified CD34+ cells were counted and seeded at a density of 5–20 ×104 cells per 6-well plate, and differentiated by co-culture with OP9-DL4–7FS cells in OP9 medium at 37°C, 5% CO2, and cell were passaged and co-cultured with fresh OP9-DL4–7FS cells every ~5 days.
Retroviral Transduction.
PG13 cell lines stably expressing empty vector dTomato, TCRα-dTomato, or TCRβ-dTomato were grown towards 70% confluency, at which point the media was switched to α-MEM with 15% FBS and 1% Pen-Strep to condition the supernatant for 48 h before transducing OP9-DL4–7FS/T-cell cultures. On D8+24 of OP9-DL4–7FS/T-cell cultures, cells were transduced with corresponding PG13 supernatants once a day with 1 μl/ml polybrene and centrifuged at 2000xg for 90 min at RT for 4 d (2–3×106 cells per 2 ml of supe). Cells were rested for 3 d and then sorted as dTomato+ CD7+CD5+CD4+CD8+ DPs. Sorted cells were placed in OP9-DL4–7FS co-cultures for up to 10 days.
Flow Cytometry.
Cells were stained for 30 min on ice with the following mouse anti-human antibodies: CD3-BrilliantViolet421, CD4-AlexaFluor700, CD8β-PE, CD31-FITC, CD34-PE, CD45RA-PE/CF594, TCRgd-FITC (BD Biosciences), CD5-PE/Cy7, CD7-AlexaFluor700, CD45-APC/eFluor780, TCRab-APC (eBiosciences), CD8α-PE/Dazzle, CD38-BrilliantViolet421 (BioLegend). Cells were resuspended in flow cytometry buffer containing DAPI, data was collected using LSR Fortessa flow cytometer (BD Biosciences) and analyzed using FlowJo version 9.7.6. For intracellular staining, cells were fixed and permeabilized using Fixation/Permeabilization kit with GolgiPlug™ (BD Biosciences) as per manufacturer’s instructions.
RNA-Seq Analysis of Control and RAG2-KO in vitro Derived CD4+CD8+ cells.
Cells were collected at co-culture day 24–28 and stained with fluorochrome-labelled antibodies to CD45, CD7 (eBioscience), CD5, CD4, CD8 (BD Biosciences), DAPI, and sorted into CD4+CD8+ populations using FACSVantage Diva or FACSAria cell sorters (BD Biosciences). Total RNA was extracted from the sorted cell populations using TRIzol. Purified RNA was subjected to RNA sequencing using Illumina Novaseq 6000. Library preparation was done using Illumina TruSeq Strandard Total RNA Sample Preparation kit. Sequencing was done using 100-cycle paired read protocol and multiplexing to obtain ~40 million reads/sample. Samples were aligned to GRCh38 using HISAT2. Read counts were calculated using HTSeq. Differential gene expression analysis was done using edgeR package in the R platform.
Statistical Analysis.
The data and error bars are presented as mean ± standard error of mean. To determine statistical significance, a one-way ANOVA (comparing three means) was performed using Prism version 6. Statistical significance was determined as *P<0.05 and **P<0.01.
Data Availability.
The data that support the findings of this study are available from the corresponding authors on request. Raw and processed RNA-Seq data are available from the Gene Expression Omnibus under accession number GSE164276 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE164276).
Results and Discussion
Generation and characterization of RAG2-KO hPSC lines.
To evaluate the role of RAG2 in human T-cell development, we used CRISPR-Cas9 gene editing to target exon 3 of the RAG2 gene (Supplemental Fig. 1A). hPSCs were transfected with a plasmid encoding the RAG2-targeting guide RNA, the Cas9 enzyme, and green fluorescent protein (GFP). Transfected GFP+ hPSCs were single-cell sorted and cultured. After expanding individual clones, we identified two clones that contained unique insertion-deletions with bi-allelic mutations (KO-1 and KO-4) (Supplemental Fig. 1A). To evaluate the impact of the RAG2 mutations on protein expression, we performed Western blot analysis, which confirmed the absence of detectable RAG2 protein in both KO-1 and −4 derived T-lineage cells (Supplemental Fig. 1B).
To assess whether RAG2-KO hPSCs maintained pluripotency, we evaluated the expression of key markers and teratoma formation. Immunofluorescence staining showed that RAG2-KO hPSCs expressed OCT4, NANOG, SOX2, and SSEA-4 (Supplemental Fig. 1C). To functionally test pluripotency, we injected RAG2-KO hPSCs into immunodeficient mice, and histological analysis revealed RAG2-KO teratoma formation with all three germ layers (Supplemental Fig. 1D), indicating that RAG2-KO hPSCs retained key features of pluripotency. To determine the capacity of RAG2-KO hESCs to generate hematopoietic progenitors, we analyzed CD34 expression after 8 days of embryoid-body differentiation cultures (18). Control WT, RAG2-KO-1 and −4 hPSCs gave rise to similar frequencies of hemogenic endothelial CD34+ cells, which could be further enriched by magnetic-assisted cell sorting (MACS) (Supplemental Fig. 1E).
T-cell development from RAG2-KO hPSCs.
CD34+ hemogenic endothelial cells were MACS-enriched and cultured with OP9-DL4 cells, expressing human IL-7, FLT3-ligand and stem cell factor (7FS), to induce T-cell differentiation. After 10 days of culture, Control WT, RAG2-KO-1 and −4 cells proceeded along the T-cell lineage, as marked by expression of both CD7 and CD5 (Fig. 1A). All three groups reached the CD4+ intermediate single positive (ISP) stage by day 15 and displayed intracellular CD3 expression by day 20 (Fig. 1A). After 24 days of culture, the majority of cells from Control WT, RAG2-KO-1 and −4 groups were CD7+CD5+ (Fig. 1A). Of note, by 29–34 days of culture, Control WT, RAG2-KO-1 and −4 cells all reached the CD4+CD8+ DP stage (Fig. 1A). However, as expected, only Control WT cells displayed intracellular TCRβ expression, and cell surface CD3/TCRβ expression (Fig. 1A). These results indicate that RAG2-deficient human T-cell progenitors can differentiate up to the CD4+CD8+ DP stage.
Figure 1. T-cell development from Control WT and RAG2-KO hESC lines.
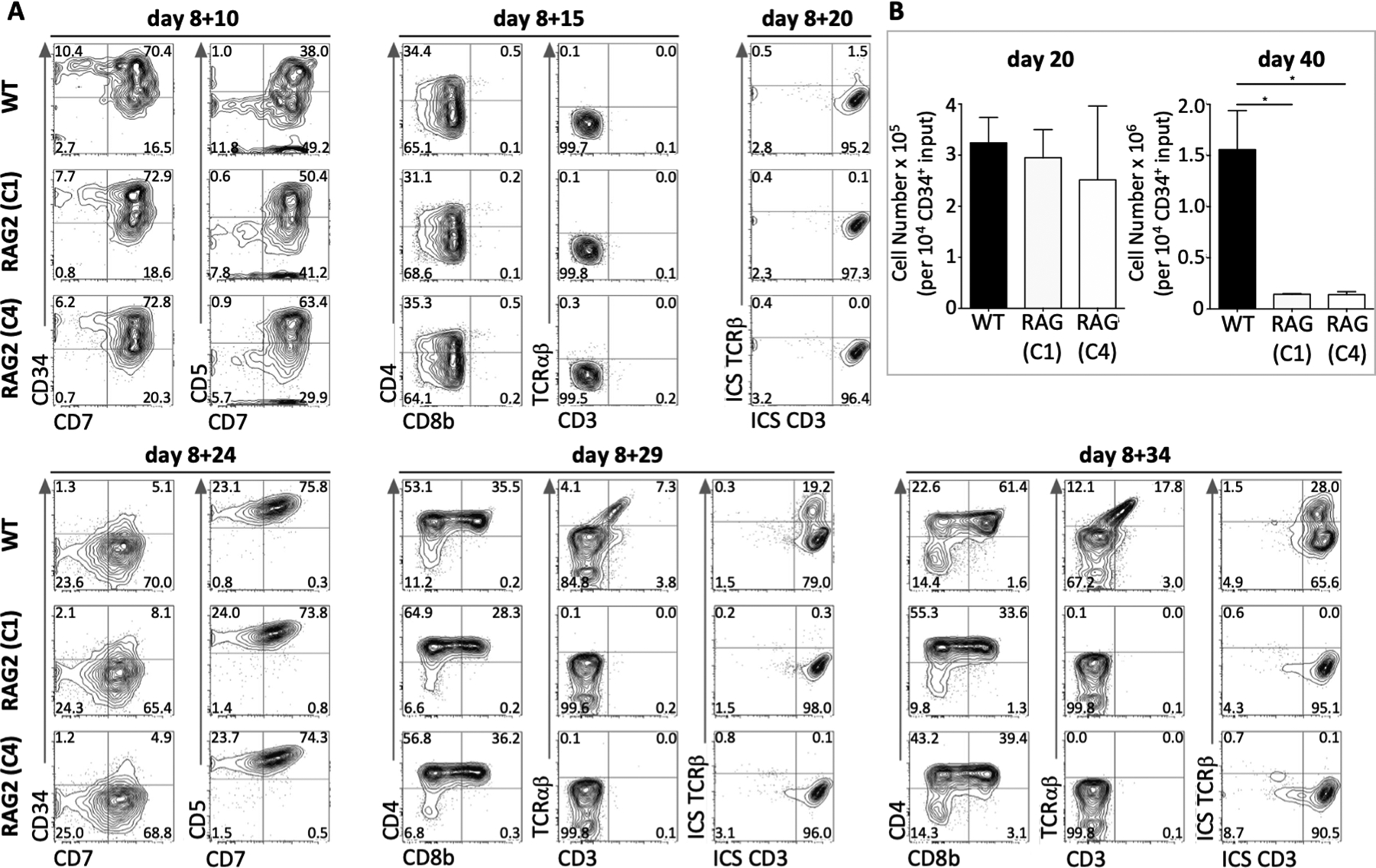
(A) Representative flow cytometry analysis of Control WT and RAG2-KO (clones 1 and 4) hPSC-derived T-lineage cells from d8 EBs + 10–34d OP9-DL4–7FS co-cultures, as indicated. Cells were pre-gated for DAPI−CD45+. (n=5 of five independent experiments). (B) Cell numbers of Control WT and RAG2-KO (clones 1 and 4) hPSC-derived T-lineage cells per 10,000 input of CD34+ cells after indicated number of days of culture on OP9-DL4–7FS. (n=3 of three independent experiments).
To determine cell survival and expansion, total cellularity was quantified from Control WT, RAG2-KO-1 and −4 developing T-cells. After 20 days of culture, all three samples showed similar cell numbers (Fig. 1B). However, after 40 days of culture, Control WT T-lineage cells further increased their survival and expansion as opposed to RAG2-KO-1 and −4 T-lineage cells, with approximately 10-fold greater cellularity by this time-point (Fig. 1B). This suggests that RAG2-dependent TCRβ rearrangement outcomes are limited to mediating the survival and proliferation of human DP T-cells.
Forced expression of a TCRβ chain in RAG2-KO CD4+CD8+ DPs.
RAG2-KO hPSC-derived CD34+ cells were cultured on OP9-DL4–7FS cells for 24 days, and DP cells were retrovirally-transduced with an empty vector (dTomato), a rearranged TCRα chain (TRA 1383i), or a rearranged TCRβ chain (TRB 1383i) (Fig. 2A). Transduced RAG-KO cells were sorted for CD7+CD5+CD4+CD8+ DP cells, and placed back on OP9-DL4–7FS cells for an additional 10 days to assess for cell survival and expansion (Fig. 2A). Both dTomato- and TCRα-transduced DPs showed similar cell numbers after 10 days of culture (Fig. 2B). However, TCRβ-transduced DPs displayed significantly higher cell numbers after 10 days of culture compared to dTomato- and TCRα-transduced DPs (Fig. 2B). This suggests that, in contrast to a TCRα chain, expression of a rearranged TCRβ chain promotes cell survival and/or proliferation of developing human T-cells at the DP stage.
Figure 2. Forced expression of a rearranged TCRβ chain in RAG2-KO CD4+CD8+ DP cells results in cell expansion.
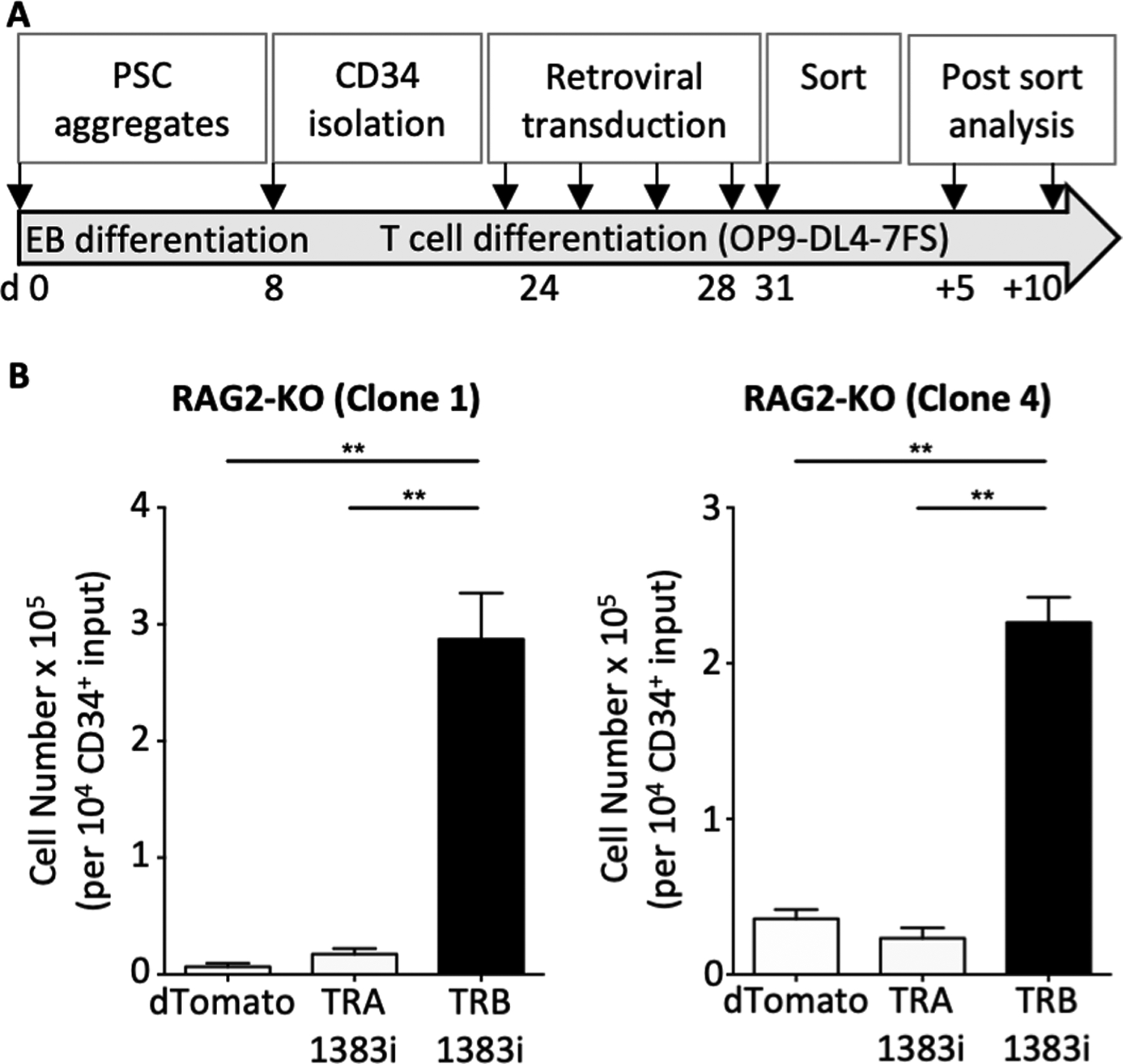
(A) Schematic of the experimental approach. (B) Cell counts of RAG2-KO DPs (clones 1 and 4) retrovirally-transduced with empty vector (dTomato), TCR alpha chain (TRA 1383i), or TCR beta chain (TRB 1383i) per 10,000 input of CD34+ cells. Transduced T-lineage cells (dTomato+) were first sorted for CD7+CD5+CD4+CD8+ double positive cells and then re-cultured on OP9-DL4–7FS for 10 days. (n=3 of three independent experiments).
Transcriptomic analysis of Control WT, RAG2-KO, and RAG2-KO TCRβ-transduced CD4+CD8+ DP cells.
Control WT, RAG2-KO control-transduced, and RAG2-KO TCRβ-transduced DP cells were sorted for RNA sequencing (RNA-Seq) analysis. As expected, Control WT DP cells expressed a large set of TCRα, TCRβ, TCRγ, and TCRδ genes that were absent in RAG2-KO control-transduced and RAG-KO TCRβ-transduced DP cells, with the notable exception of some TCR genes, including the 1383i TCRβ used in the transduction and a few TCRα genes, likely the result of germline transcripts induced by the β-selection signals, as seen in mice (19) (Supplemental Table 1, and Supplemental Fig. 2A). Furthermore, we compared the expression of genes from RAG2-KO control- and TCRβ-transduced DP cells with that of umbilical cord-blood (UCB)-hematopoietic stem cell-derived DP cells for a set of DP-associated signature genes (20) (Supplemental Table 1). This analysis revealed that TCRβ-transduced RAG2-KO DP cells gained the expression of a suite of genes that were present in the UCB-derived DP cells, including RORC, which is induced following β-selection in mice (21) (Supplemental Fig. 2B).
Differentially expressed gene analysis was performed to determine genes that were up-regulated in Control WT compared to RAG2-KO (KO-1 and KO-4) DP cells, and vice versa. Genes significantly up-regulated in Control WT compared to both RAG2-KO-1 and RAG2-KO-4 include CCDC152, GPR183, IL32, and MAL (Supplemental Table 1 and Fig. 3A). Genes significantly up-regulated (p<0.05) in Control WT compared to RAG2-KO-1 or −4 included ADAMTS17, IL1RL1, PLXNA2, and TEAD1, or CTSW, IKBKG, IL21R, and IL4R, respectively (Supplemental Table 1 and Fig. 3A). Of note, many of these genes (such as TEAD1, IKBKG, and IL32) were also highly expressed in TCRβ-transduced RAG2-KO DP cells similar to Control WT DP cells (Fig. 3A). Interestingly, the expression of a subset of genes (such as MAL) were not rescued with forced expression of a TCRβ chain (Fig. 3A). Genes significantly up-regulated in both RAG2-KO-1 and RAG2-KO-4 compared to Control WT include HBG1, HBG2, LOC100240735, and LOC339975 (Supplemental Table 1 and Fig. 3B). Genes significantly up-regulated in RAG2-KO-1 or −4 compared to Control WT include CCDC8, CPA4, EPHA4, and ID1, or ANKRD1, CMTM8, MET, and RHOU, respectively (Supplemental Table 1 and Fig. 3B). Of note, many of these genes (such as ID1, RHOU, and HBG1) also showed low expression in TCRβ-transduced RAG2-KO DPs similar to Control WT DP cells (Fig. 3B). Interestingly, the expression of a subset of genes (such as LOC100240735) were not reduced with forced expression of a TCRβ chain (Fig. 3B). Phylogenetic tree analysis again showed that TCRβ-transduced RAG2-KO DP cells were more similar to Control WT DP cells than to control-transduced RAG2-KO DP cells (Fig. 3).
Figure 3. Transcriptomic analysis of Control WT, RAG2-KO, and RAG2-KO TCRβ-transduced CD4+CD8+ DP cells.
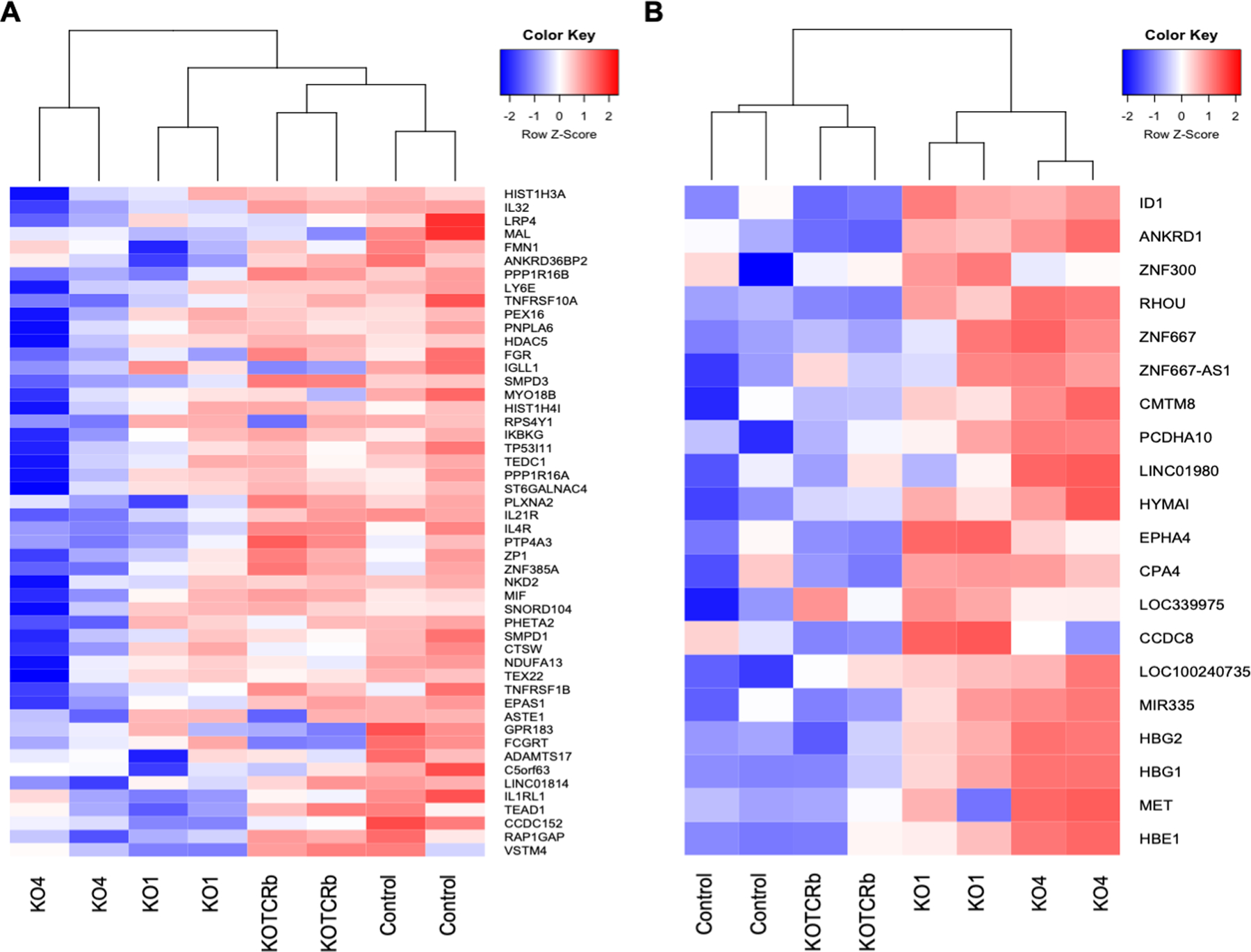
RNA-seq analysis of Control WT, RAG2-KO1/4, and TCRβ+ RAG2-KO1/4 CD4+CD8+ DPs. Shown are genes that are differentially highly expressed in Control WT DPs compared to RAG2-KO1/4 DPs (A) and differentially highly expressed in RAG2-KO1/4 DPs compared to Control WT DPs (B). (n=2 for Control WT, RAG2-KO-1, and RAG2-KO-4, and n=1 for RAG2-KO-1 TCRβ-transduced and RAG2-KO-4 TCRβ-transduced of one independent experiment).
Analysis of cell cycle regulators, survival and differentiation genes, which are known to be involved in mouse β-selection (22–24), revealed a set of genes, such as RORC, CD27, ERG and CCDN3, that are also regulated following TCRβ expression in RAG2-KO DPs (Supplemental Fig. 3A). A gene ontology analysis to determine biological pathways that involve genes up-regulated in Control WT compared to RAG2-KO DP cells revealed a genetic program involved in “negative regulation of intrinsic apoptotic signaling pathway” (Supplemental Fig. 3B). Furthermore, a gene set pathway pertaining to leukocyte regulation, included genes involved in “positive regulation of lymphocyte activation” and “regulation of lymphocyte proliferation” (Supplemental Fig. 3B). Thus, these data provide additional evidence that RAG2-dependent TCRβ expression in developing human T-cells supports their survival and/or proliferation.
Conclusions.
Here, we generated RAG2-KO hPSCs to assess the role of TCRβ during human T-cell development. In contrast to the effects seen with Rag1- or Rag2-deficient mice, which show a complete lack of DP thymocytes, developing human T-cells were able to reach the CD4+CD8+ DP stage in the absence of RAG2 expression. Lack of RAG1/2 in mice results in a definitive block at the CD44−CD25+ DN3 stage, as it is well documented that RAG1/2-mediated TCRβ rearrangement controls the transition from the DN3 to the CD44−CD25− DN4 and DP stages (25, 26). However, the precise developmental block in the absence of RAG1/2 expression or TCRβ rearrangement during human T-cell development was largely unresolved (27, 28).
It was previously suggested that the requirement for TCRβ-induced survival/proliferation, or β-selection, occurs at the CD4+ ISP stage, in which 5% of cells express cell-surface TCRβ protein (29). In another study, it was suggested that β-selection instead happens later, at the CD4+CD8α+CD8β− early double positive (EDP) stage in which 25% of cells express intracellular TCRβ protein (30). Thus, based on these two studies, it was proposed that β-selection begins at the CD4+ ISP stage and continues until the EDP stage. Here, we detected robust development past the ISP and EDP to the DP stage, as CD4, CD8α, and CD8β expression was observed in RAG2-KO developing T-cells. While the requirement for the TCRβ chain in the phenotypic differentiation to the DP stage differs between mice and humans, the TCRβ chain similarly promotes cellular expansion (survival/proliferation) in both mouse and human developing T-cells, as enforced TCRβ expression rescued cellular expansion.
The requirement for TCRβ, and thus the preTCR, during, rather than prior to, the DP stage during human T-cell development may reflect differential expression of cell cycle and survival genes. In mice, expression of a preTCR in DN3 cells leads to expression of Bmi-1, which represses the expression of the cell cycle inhibitor Cdkn2a. Repression of Cdkn2a is required for preTCR-induced cell proliferation and the DN3-DP transition (31). Future studies could investigate whether preTCR-mediated regulation of Bmi-1, Cdkn2a, Cdkn1a (32), as well as other factors involved in cell cycling, are initially acting at the DP stage during human T-cell development. In summary, this study reveals the unexpected timing required for TCRβ-mediated β-selection in developing human T-cells.
Supplementary Material
Key points.
Human RAG2-deficient developing T-cells progress up to the CD4+CD8+ stage
TCRβ-selection of human CD4+CD8+ cells allows for their survival/proliferation.
Acknowledgements
We thank Dr. Geneve Awong and Mr. Paul Oleynik (Centre for Flow Cytometry & Microscopy, Sunnybrook Research Institute, Toronto, Ontario, Canada) for their cell sorting support.
This work was supported by grants from the Canadian Institutes of Health Research (CHIR, FND-154332), National Institutes of Health (NIH-1P01AI102853-01), Innovation to Impact grant from the Canadian Cancer Society (705960), Medicine by Design: A Canada First Research Excellence Fund Program at the University of Toronto, and The Krembil Foundation. Y.L. is supported by the National Natural Science Foundation of China (31571517) and the Beijing Municipal Natural Science Foundation (7192091). J.C.Z.P. is supported by a Canada Research Chair in Developmental Immunology.
References
- 1.Schatz DG, Oettinger MA, and Baltimore D. 1989. The V(D)J recombination activating gene, RAG-1. Cell 59: 1035–1048. [DOI] [PubMed] [Google Scholar]
- 2.Oettinger MA, Schatz DG, Gorka C, and Baltimore D. 1990. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science 248: 1517–1523. [DOI] [PubMed] [Google Scholar]
- 3.Jones JM, and Gellert M. 2004. The taming of a transposon: V(D)J recombination and the immune system. Immunol Rev 200: 233–248. [DOI] [PubMed] [Google Scholar]
- 4.Smith AL, Scott JNF, and Boyes J. 2019. The ESC: The Dangerous By-Product of V(D)J Recombination. Front Immunol 10: 1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Herz W, Bousfiha A, Casanova JL, Chatila T, Conley ME, Cunningham-Rundles C, Etzioni A, Franco JL, Gaspar HB, Holland SM, Klein C, Nonoyama S, Ochs HD, Oksenhendler E, Picard C, Puck JM, Sullivan K, and Tang ML. 2014. Primary immunodeficiency diseases: an update on the classification from the international union of immunological societies expert committee for primary immunodeficiency. Front Immunol 5: 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delmonte OM, Schuetz C, and Notarangelo LD. 2018. RAG Deficiency: Two Genes, Many Diseases. J Clin Immunol 38: 646–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gennery A 2019. Recent advances in understanding RAG deficiencies. F1000Res 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, and Papaioannou VE. 1992. RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68: 869–877. [DOI] [PubMed] [Google Scholar]
- 9.Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall AM, and et al. 1992. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell 68: 855–867. [DOI] [PubMed] [Google Scholar]
- 10.Shah DK, and Zuniga-Pflucker JC. 2014. An overview of the intrathymic intricacies of T cell development. J Immunol 192: 4017–4023. [DOI] [PubMed] [Google Scholar]
- 11.Notarangelo LD, Kim MS, Walter JE, and Lee YN. 2016. Human RAG mutations: biochemistry and clinical implications. Nat Rev Immunol 16: 234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taghon T, and Rothenberg EV. 2008. Molecular mechanisms that control mouse and human TCR-alphabeta and TCR-gammadelta T cell development. Semin Immunopathol 30: 383–398. [DOI] [PubMed] [Google Scholar]
- 13.Brauer PM, Pessach IM, Clarke E, Rowe JH, Ott de Bruin L, Lee YN, Dominguez-Brauer C, Comeau AM, Awong G, Felgentreff K, Zhang YH, Bredemeyer A, Al-Herz W, Du L, Ververs F, Kennedy M, Giliani S, Keller G, Sleckman BP, Schatz DG, Bushman FD, Notarangelo LD, and Zuniga-Pflucker JC. 2016. Modeling altered T-cell development with induced pluripotent stem cells from patients with RAG1-dependent immune deficiencies. Blood 128: 783–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bifsha P, Leiding JW, Pai SY, Colamartino ABL, Hartog N, Church JA, Oshrine BR, Puck JM, Markert ML, and Haddad E. 2020. Diagnostic assay to assist clinical decisions for unclassified severe combined immune deficiency. Blood Adv 4: 2606–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bosticardo M, Pala F, Calzoni E, Delmonte OM, Dobbs K, Gardner CL, Sacchetti N, Kawai T, Garabedian EK, Draper D, Bergerson JRE, DeRavin SS, Freeman AF, Gungor T, Hartog N, Holland SM, Kohn DB, Malech HL, Markert ML, Weinacht KG, Villa A, Seet CS, Montel-Hagen A, Crooks GM, and Notarangelo LD. 2020. Artificial thymic organoids represent a reliable tool to study T-cell differentiation in patients with severe T-cell lymphopenia. Blood Adv 4: 2611–2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bories JC, Cayuela JM, Loiseau P, and Sigaux F. 1991. Expression of human recombination activating genes (RAG1 and RAG2) in neoplastic lymphoid cells: correlation with cell differentiation and antigen receptor expression. Blood 78: 2053–2061. [PubMed] [Google Scholar]
- 17.Li Y, Brauer PM, Singh J, Xhiku S, Yoganathan K, Zuniga-Pflucker JC, and Anderson MK. 2017. Targeted Disruption of TCF12 Reveals HEB as Essential in Human Mesodermal Specification and Hematopoiesis. Stem Cell Reports 9: 779–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kennedy M, Awong G, Sturgeon CM, Ditadi A, LaMotte-Mohs R, Zuniga-Pflucker JC, and Keller G. 2012. T lymphocyte potential marks the emergence of definitive hematopoietic progenitors in human pluripotent stem cell differentiation cultures. Cell Rep 2: 1722–1735. [DOI] [PubMed] [Google Scholar]
- 19.Villey I, Quartier P, Selz F, and de Villartay JP. 1997. Germ-line transcription and methylation status of the TCR-J alpha locus in its accessible configuration. Eur J Immunol 27: 1619–1625. [DOI] [PubMed] [Google Scholar]
- 20.Casero D, Sandoval S, Seet CS, Scholes J, Zhu Y, Ha VL, Luong A, Parekh C, and Crooks GM. 2015. Long non-coding RNA profiling of human lymphoid progenitor cells reveals transcriptional divergence of B cell and T cell lineages. Nat Immunol 16: 1282–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He YW 2000. The role of orphan nuclear receptor in thymocyte differentiation and lymphoid organ development. Immunol Res 22: 71–82. [DOI] [PubMed] [Google Scholar]
- 22.Lefebvre JM, Haks MC, Carleton MO, Rhodes M, Sinnathamby G, Simon MC, Eisenlohr LC, Garrett-Sinha LA, and Wiest DL. 2005. Enforced expression of Spi-B reverses T lineage commitment and blocks beta-selection. J Immunol 174: 6184–6194. [DOI] [PubMed] [Google Scholar]
- 23.Klein F, Mitrovic M, Roux J, Engdahl C, von Muenchow L, Alberti-Servera L, Fehling HJ, Pelczar P, Rolink A, and Tsapogas P. 2019. The transcription factor Duxbl mediates elimination of pre-T cells that fail beta-selection. J Exp Med 216: 638–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sicinska E, Aifantis I, Le Cam L, Swat W, Borowski C, Yu Q, Ferrando AA, Levin SD, Geng Y, von Boehmer H, and Sicinski P. 2003. Requirement for cyclin D3 in lymphocyte development and T cell leukemias. Cancer Cell 4: 451–461. [DOI] [PubMed] [Google Scholar]
- 25.von Boehmer H, Aifantis I, Feinberg J, Lechner O, Saint-Ruf C, Walter U, Buer J, and Azogui O. 1999. Pleiotropic changes controlled by the pre-T-cell receptor. Curr Opin Immunol 11: 135–142. [DOI] [PubMed] [Google Scholar]
- 26.Michie AM, and Zuniga-Pflucker JC. 2002. Regulation of thymocyte differentiation: pre-TCR signals and beta-selection. Semin Immunol 14: 311–323. [DOI] [PubMed] [Google Scholar]
- 27.Rothenberg EV, and Taghon T. 2005. Molecular genetics of T cell development. Annu Rev Immunol 23: 601–649. [DOI] [PubMed] [Google Scholar]
- 28.Carrasco YR, Navarro MN, de Yebenes VG, Ramiro AR, and Toribio ML. 2002. Regulation of surface expression of the human pre-T cell receptor complex. Semin Immunol 14: 325–334. [DOI] [PubMed] [Google Scholar]
- 29.Blom B, Verschuren MC, Heemskerk MH, Bakker AQ, van Gastel-Mol EJ, Wolvers-Tettero IL, van Dongen JJ, and Spits H. 1999. TCR gene rearrangements and expression of the pre-T cell receptor complex during human T-cell differentiation. Blood 93: 3033–3043. [PubMed] [Google Scholar]
- 30.Carrasco YR, Trigueros C, Ramiro AR, de Yebenes VG, and Toribio ML. 1999. Beta-selection is associated with the onset of CD8beta chain expression on CD4(+)CD8alphaalpha(+) pre-T cells during human intrathymic development. Blood 94: 3491–3498. [PubMed] [Google Scholar]
- 31.Miyazaki M, Miyazaki K, Itoi M, Katoh Y, Guo Y, Kanno R, Katoh-Fukui Y, Honda H, Amagai T, van Lohuizen M, Kawamoto H, and Kanno M. 2008. Thymocyte proliferation induced by pre-T cell receptor signaling is maintained through polycomb gene product Bmi-1-mediated Cdkn2a repression. Immunity 28: 231–245. [DOI] [PubMed] [Google Scholar]
- 32.Zhao B, Yoganathan K, Li L, Lee JY, Zuniga-Pflucker JC, and Love PE. 2019. Notch and the pre-TCR coordinate thymocyte proliferation by induction of the SCF subunits Fbxl1 and Fbxl12. Nat Immunol 20: 1381–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors on request. Raw and processed RNA-Seq data are available from the Gene Expression Omnibus under accession number GSE164276 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE164276).


