Abstract
Segmentation and mutant classification of high-frequency ultrasound (HFU) mouse embryo brain ventricle (BV) and body images can provide valuable information for developmental biologists. However, manual segmentation and identification of BV and body requires substantial time and expertise. This paper proposes an accurate, efficient and explainable deep learning pipeline for automatic segmentation and classification of the BV and body. For segmentation, a two-stage framework is implemented. The first stage produces a low-resolution segmentation map, which is then used to crop a region of interest (ROI) around the target object and serve as the probability map of the auto-context input for the second-stage fine-resolution refinement network. The segmentation then becomes tractable on high-resolution 3D images without time-consuming sliding windows. The proposed segmentation method significantly reduces inference time (102.36 to 0.09 s/volume≈1000x faster) while maintaining high accuracy comparable to previous sliding-window approaches. Based on the BV and body segmentation map, a volumetric convolutional neural network (CNN) is trained to perform a mutant classification task. Through backpropagating the gradients of the predictions to the input BV and body segmentation map, the trained classifier is found to largely focus on the region where the Engrailed-1 (En1) mutation phenotype is known to manifest itself. This suggests that gradient backpropagation of deep learning classifiers may provide a powerful tool for automatically detecting unknown phenotypes associated with a known genetic mutation.
Index Terms—: Image segmentation, classification and visualization, High-frequency ultrasound, Mouse embryo, Deep learning
I. INTRODUCTION
THE mouse is a commonly used animal model in the study of mammalian embryo development due to its high degree of homology with the human genome. Along with complete knowledge of the mouse genome, a wide variety of gene editing tools have enabled the creation of genetic modifications in mice, including many mutations that are lethal in utero [1]. For instance, Engrailed-1 (En1) homozygous mutants exhibit early embryonic deletion of the mid-hindbrain region in the developing central nervous system that leads to a thickening of the brain ventricle (BV) and subsequent death at birth [2]. Observing variations in the shape of the BV and body is an effective way to study how genetic defects, such as the En1 mutation, are manifested during embryonic development [3], [4].
High-throughput, high-frequency ultrasound (HFU) has proven to be an effective imaging modality to generate high-resolution volumetric datasets of mouse embryos in utero over mid-to-late gestational stages [5]. Accurate delineation of anatomical structures from HFU images can provide valuable structural information and enable downstream analysis of complex biomedical image data [6]. As such, accurate and time-efficient BV and body segmentation in HFU data can substantially aid biologists in observing and understanding the development of mouse embryos.
Manual segmentation by imaging experts has long been considered the gold standard in the field of biomedical image analysis. However, manual segmentation of the BV and body (Fig. 1) from 3D HFU volumes is time-consuming, requiring half an hour or more for each volume, which increases considerably with image quality decay. Additionally, the large and ever increasing quantity of HFU images typically used in developmental studies makes manual labelling impractical in the long run. Therefore, it is necessary to develop fully automatic segmentation and classification algorithms to optimize this process [7]. Such an algorithm must overcome five primary challenges related to the image data: (1) extreme imbalance between background and foreground (i.e., the BV makes up only 0.367% of the whole volume, on average, while the body is around 10.6%); (2) differing shapes and locations of the body and BV due to various embryonic stages; (3) large variation in embryo posture and orientation; (4) the presence of missing or ambiguous boundaries (Fig. 1(a)(e)(f)) and motion artifacts (Fig. 1(b)(c)(d)); and (5) large variation in image size, from 150 × 161 × 81 to 210 × 281 × 282 voxels.
Fig. 1.
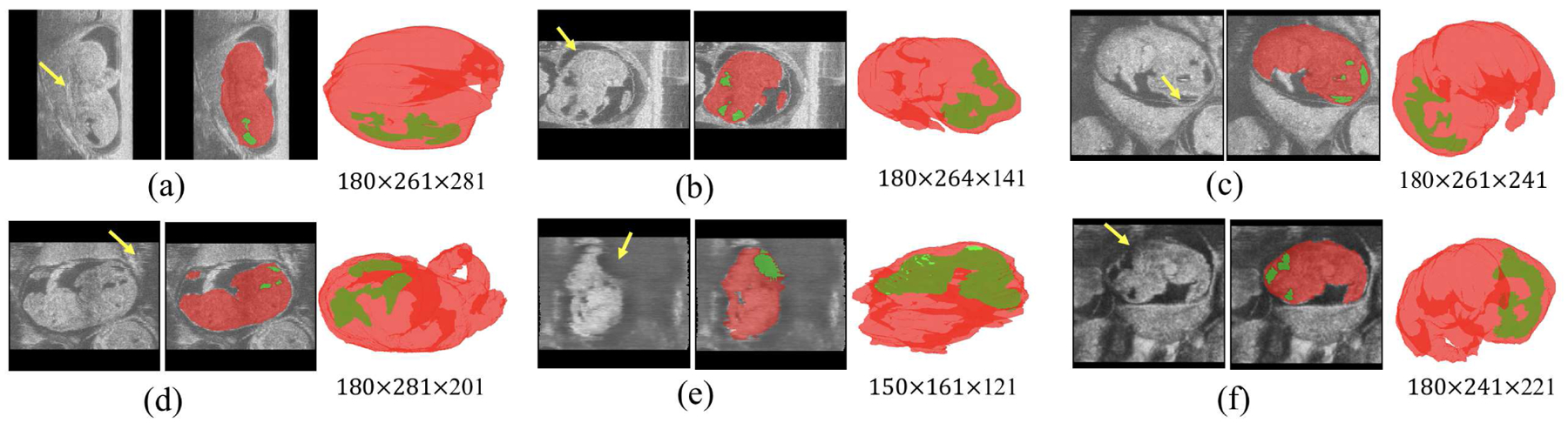
(a–f) 6 embryonic mice HFU volumes are shown with three views each: a B-mode image slice from the 3D volume, a manual BV (green) and body (red) segmentation, and a 3D rendering (visualized in natural orientation relative to the HFU probe). The numbers below each 3D rendering indicate corresponding image size in voxels. The arrow in a) indicates an ambiguous boundary due to contact between the body and uterine wall. The arrows in b), c) and d) indicate motion artifacts because of irregular physiological movements of the anesthetized pregnant mice. The arrows in e) and f) indicate missing head boundaries due to either specular reflections or shadowing from overlaying tissues.
A nested graph cut (NGC) algorithm [8] was first developed to perform segmentation of the BV from the manually selected head portion of HFU of the mouse embryo. NGC relied on the nested structure of the BV, head, uterus and surrounding amniotic fluid and successfully overcame the missing head boundary problem (Fig. 1(e)(f), Challenge 4). This problem is caused by a loss of HFU signal due to either specular reflections or shadowing from overlaying tissues. Subsequent work focused on BV and body segmentation in whole-body images by extending the NGC algorithm to first detect and segment the interior of the uterus and then to detect and segment the BV and body regions [9]. Although this framework performed well on an initial set of 36 embryos [9], it did not generalize well to larger, unseen data sets because the framework was developed based on manually crafted assumptions and several parameters were hand-tuned on the smaller data set.
Given the success of Fully Convolutional Networks (FCN) for semantic segmentation tasks [10], we developed a deep-learning-based framework for BV segmentation [11] that outperformed the NGC-based framework in [9] by a large margin. Because the BV makes up a very small portion (<0.5%) of the whole volume, the algorithm in [11] first applied a volumetric convolutional neural network (CNN) on a 3D sliding window over the entire volume to identify a 3D bounding box containing the whole BV, followed by a FCN to segment the detected bounding box into BV or background. Despite achieving high accuracy of 0.904 Dice Similarity Coefficient (DSC) for BV, this method was inefficient because it required hundreds of thousands of forward passes through a classification network in the first sliding-window-based localization step. The challenges for body segmentation are similar to those for BV segmentation, except that the extreme imbalance between foreground and background is somewhat alleviated (the body makes up 10.6% of the whole volume on average). Hence, the localization step is not necessary for body segmentation. Qiu et al. [12] first applied an FCN to segment each sliding window over the entire volume, and then determined the final body segmentation by merging results from all the sliding windows. However, this sliding-window-segmentation approach suffered from the same inefficiencies as the localization method in [11].
Here, we propose an efficient end-to-end auto-context refinement framework for joint BV and body segmentation from volumetric HFU images. The proposed approach is to: (1) generate a region of interest (ROI) from the original image through one-pass low-resolution segmentation and cropping in order to circumvent the class imbalance problem without the use of a sliding window; and (2) combine the low-resolution map with a cropped, fine-resolution image as an auto-context [13] input so that the fine-resolution segmentation network can utilize valuable global information and produce more accurate results. Specifically, a VNet (VNet I) [14] is first applied to a downsampled HFU 3D image to jointly segment the BV and body (Fig. 2). The resulting low-resolution body segmentation map is then up-sampled to the original resolution, and a bounding box containing the body is generated. Next, the original image and the coarse probability map for the up-sampled body in the bounding box are concatenated as localized auto-context and fed into another VNet (VNet II) to generate the final refined body segmentation map. A parallel process is applied to generate final refined BV segmentation using a third VNet (VNet III). Each VNet is initially trained separately and then fine-tuned in an end-to-end manner.
Fig. 2.
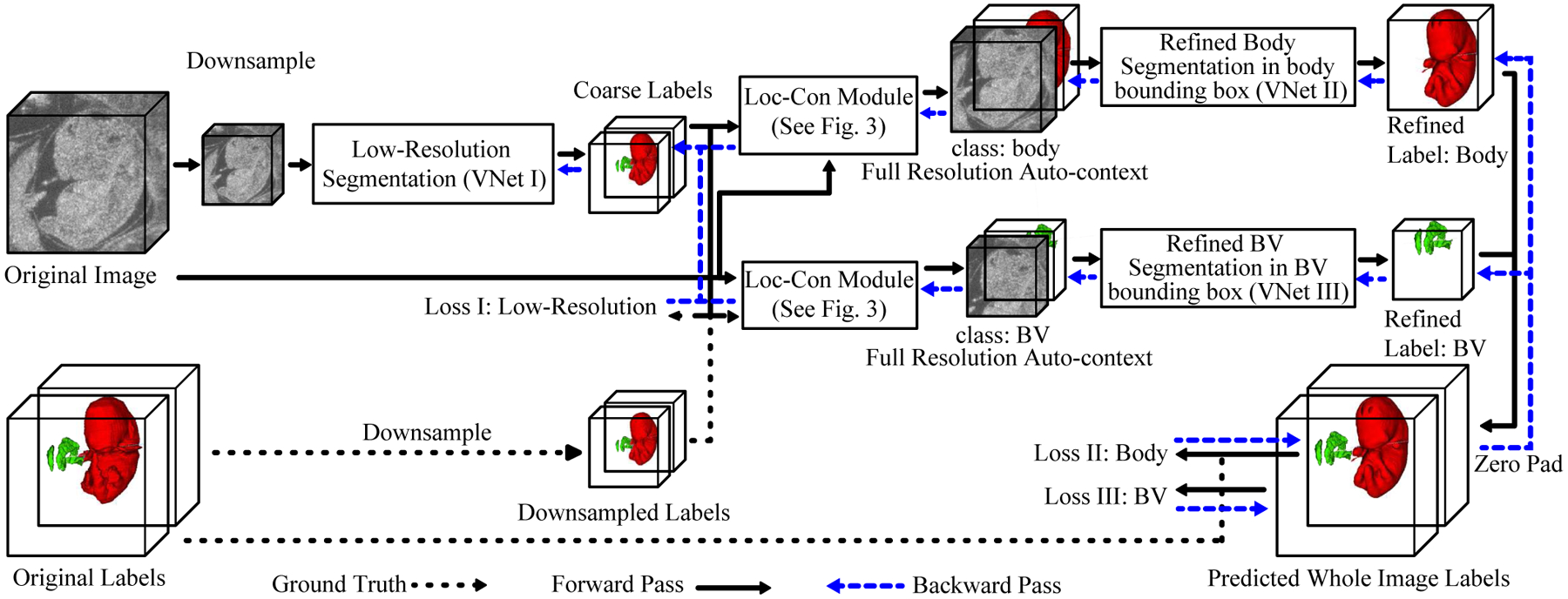
The pipeline of the joint BV and body segmentation from 3D HFU images of mouse embryos.
Compared with previous methods, this segmentation framework has the following advantages:
The class imbalance problem is mitigated by cascading the networks from low resolution of the whole image to fine resolution in localized regions without the need for a time-consuming sliding window.
An auto-context input is created by concatenating the initial blurred low-resolution segmentation map with the high-resolution image (Fig. 3). This auto-context input improves segmentation accuracy by providing a full-resolution refinement network with rich global context information.
The gradient of the refinement networks can flow end-to-end back to the low-resolution segmentation network by combining localization and auto-context modules in a differentiable pipeline which further improves segmentation accuracy.
Fig. 3.
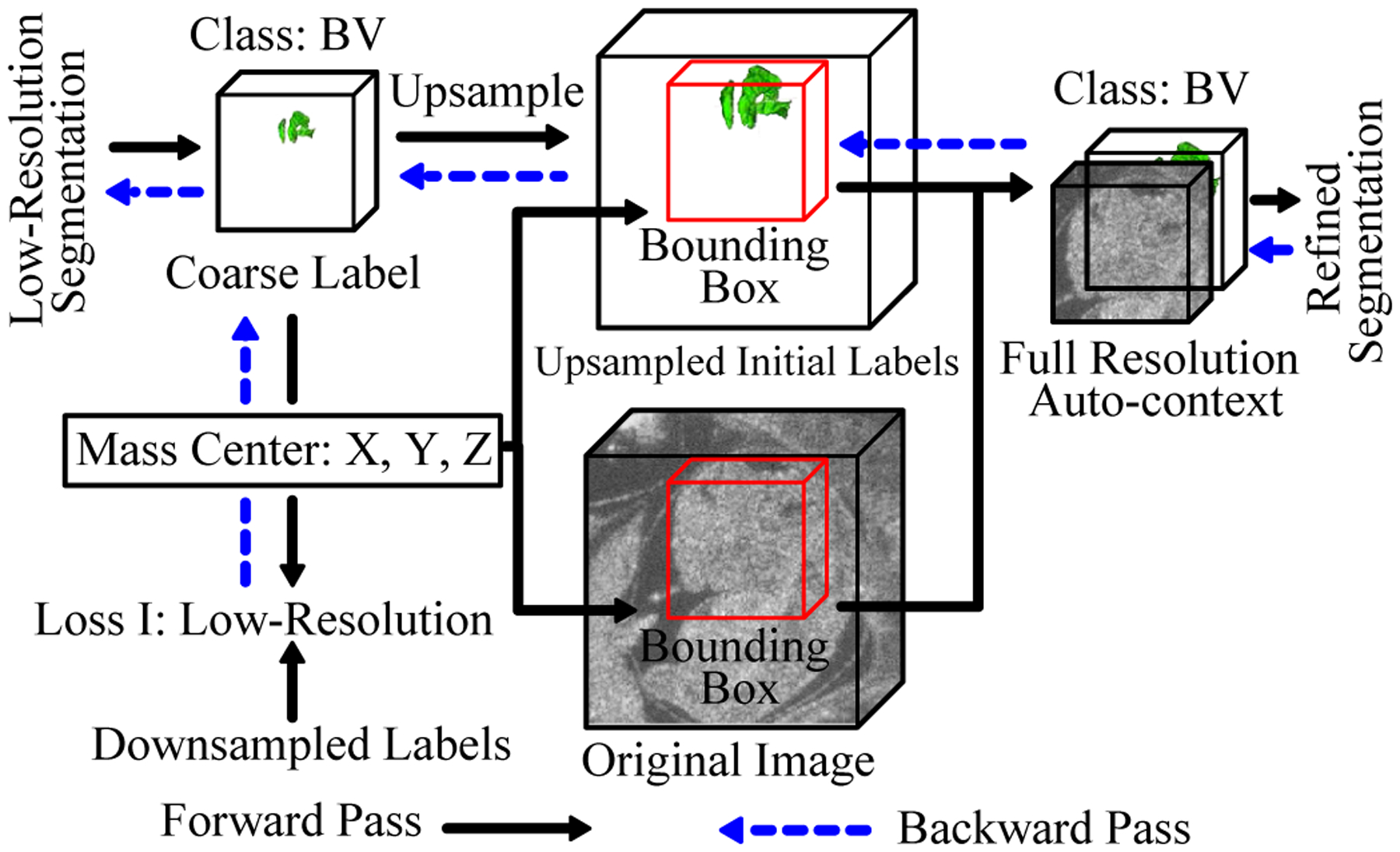
Diagram of localization-auto-context (Loc-Con) module for the BV. A similar configuration is used for the body. The gradient produced by the refinement loss can flow back to the low-resolution segmentation network (blue arrows).
Our proposed segmentation framework allows end-to-end training and efficient, real-time, one-pass inference while achieving comparable segmentation accuracy with the substantially more time consuming sliding-window-based approaches.
The morphology of the BV of a En1 mutant mouse embryos is notably deformed compared to the wildtype phenotype. Moreover, a difference in spine curvature has been reported to exist between En1 mutants and normal mouse embryos [2]. Because visually identifying mutants is time consuming, an automatic classification model for En1 mutants using the BV and body segmentation maps would be advantageous. We therefore extended our BV and body segmentation work to include mutant classification using a volumetric VGG-based CNN approach [15]. The purpose of this work is not simply to classify embryos, but to better understand the underlying morphological changes associated with a mutant phenotype. Therefore, to understand the underlying physiological structures that influence the classification process, the method introduced in [16] is used to visualize the trained network by backpropagating the gradient of the prediction with respect to the input BV and body segmentation map.
In this paper, we first present a real-time and accurate BV and body segmentation algorithm, which is built on our previous efforts [17] with a more thorough exposition of the methods, a more expansive discussion of the results, and a comparison with the prior NGC-based method [9] for the same data set. Then we use the BV and body segmentation to perform mutant classification together with a simple method for automatically rotating the segmentation maps so that the body and BV shapes will all follow the same canonical orientation. A standard 3D image orientation not only helps improve classification results, but also assists better visualization of the 3D volumes. Finally, we leverage gradient-backpropagation-based visualization of the data to understand what features the learnt classifier uses to make its decision. It is worth noting that preliminary mutant classification results based on the BV segmentation only were reported in [12].
II. RELATED WORK
A. Segmentation
Segmentation is a critical component of any pipeline designed to aid in image-based analyses of mouse mutants. Registration-based analysis of magnetic resonance images has been used extensively for studying postnatal brain phenotypes [18], [19] and brain development [20]. This approach makes use of atlases of normal mouse brain anatomy that were derived from image registration and averaging in combination with manual segmentation by experts. Then, individual mouse brains of unknown phenotype are segmented automatically via registration to the atlas. Current pipelines designed for detecting and analyzing mutant mouse embryos have taken a similar approach [1] using ex vivo micro-CT [21] or optical projection tomography (OPT) [22] images. In contrast, HFU is uniquely suited to providing in vivo data on mouse embryonic development [5], but HFU embryo atlases have not been established. In the current study, we investigate deep learning approaches as an alternative to the more conventional, registration-based segmentation methods.
Deep learning has been widely employed in biomedical image analysis tasks [23]–[28]. Milletari et al. [29] applied deep CNNs to localize and segment the midbrain in MRI and ultrasound images in a patch-wise manner with Hough voting. Although this method attempted to implicitly incorporate a shape prior through Hough voting, the patch-wise training strategy ignored the interdependent relationships between neighboring patches during training of the CNN classifiers. Long et al. [10] developed the influential FCN by replacing all the fully connected layers of traditional CNN-based classifiers with a transpose convolutional layer and then Ronneberger et al. [30] improved the FCN model by introducing symmetric skip connections between the encoder and decoder. Liu et al. [31] proposed to use 2D FCN with feature pyramid attention for automatic prostate zonal segmentation in 3D MRI images. Although this 2D-based FCN was shown to outperform UNet, it was still deficient in capturing inter-slice correlation information compared to 3D-based models. Milletari et al. [14] further adapted UNet to VNet for volumetric medical image segmentation and also introduced a Dice-based loss function.
Tu [13] first proposed the auto-context concept for high-level vision tasks, such as image segmentation. Specifically, the idea behind auto-context [13] is to iterate in order to approach the reference segmentation through a sequence of models, where the input and output of the previous model are concatenated to form the input for the next model such that the next model can make use of richer context information from the output of the previous model. It is possible to cascade two or more segmentation networks for the purpose of either localization or auto-context. Roth et al. [32] focused on abdominal CT image segmentation. They applied two cascaded 3D FCNs using the initial segmentation results to localize the foreground organs, which were then input into the second FCN. The initial segmentation was used only for localization and was not concatenated with the raw image as auto-context input to the second FCN. Tang et al. [33] cascaded four UNets and trained them in an end-to-end manner for skin lesion segmentation. However, this framework did not use the segmentation output of a previous UNet to reduce the spatial region to the next UNet, and was restricted to 2D binary segmentation. Chen et al. [34] cascaded two residual FCNs for volumetric MRI brain segmentation in order to use the first FCN’s output as context information for the second FCN. This framework also did not use the initial segmentation for localization of desired structures to reduce spatial input into the second network. In contrast to these efforts, the cascading networks we propose not only serve to localize the ROI, but also function as an auto-context module for multi-class volumetric image segmentation. The Mask-RCNN [35] has been well-known for accurate object detection and instance segmentation. It is less appropriate in our application, because each 3D HFU image has only a single embryo (as opposed to imaging multiple embryos at once) and Mask-RCNN was designed to detect and segment multiple objects in an image.
B. Classification and Visualization
Increasingly powerful neural network architectures (e.g. AlexNet [36], VGGNet [15], ResNet [37], DenseNet [38] and SENet [39]) have led to successful breakthroughs in a variety of classification tasks. For example, Wang et al. [40] demonstrated that a VGG16 model can outperform a radiomics-based method for thyroid nodules classification in ultrasound images. Moreover, numerous works have focused on interpreting the decision making process of these neural networks. Simonyan et al. [16] proposed visualizing the image-specific class saliency map by backpropagating the gradient from the top-1 class prediction unit to the input image. Springenberg et al. [41] adapted [16] to only backpropagate the positive gradient to reduce noise in the saliency map. Zhou et al. [42] proposed using global average pooling to replace fully connected layers such that the predicted class score can be mapped back to the previous convolutional layer to generate the class activation maps (CAMs). The CAM can highlight the class-specific discriminative regions. Selvaraju et al. [43] generalized CAM to any CNN-based architecture by using gradient backpropogation and also combined guided backpropagation [41] for better visualization.
In biomedical applications, it is essential that classification results are accurate and interpretable. Wang et al. [44] embedded Grad-CAM [43] as an attention branch into a classification network (ResNet-152 model) for 14 thorax diseases diagnosed in chest X-ray images. The embedded Grad-CAM branch enabled the learned feature maps from the classification branch to be converted into an attention map that highlighted the locations of disease-specific regions under the supervision of image-level class labels.
Given a small data set, we adopted a shallow volumetric VGG-like network with 9 layers to perform the mutant classification. We developed an automatic procedure to rotate the BV and body segmentation maps to a canonical orientation. This greatly reduced the orientation variance among the training samples and between the training and testing samples, leading to significant improvement in the classification accuracy. Then, the technique in [16] was utilized to visualize the saliency map by backpropagating the gradient from the top-1 class prediction unit to the input. The pipeline was simple and fully automatic and proved to be highly effective for our classification and visualization tasks.
III. DATA SET
The data set used for developing our segmentation framework consists of 231 embryonic mouse HFU volumes which were acquired in utero and in vivo from pregnant mice (10–14.5 days after mating) using a 5-element 40-MHz annular array [5], [45]. The dimensions of the HFU volumes varied from 150 × 161 × 81 to 210 × 281 × 282 voxels and the size of each voxel is 50 × 50 × 50 μm. For each of the 231 volumes, manual BV and body segmentations were conducted by trained research assistants using commercial software (Amira, FEI, Hillsboro, Oregon, USA). It is worth noting that the BV could be segmented accurately because BV is a relatively small and dark region in the brain and can be segmented using the region growing function in Amira. Then, our trained research assistants would refine the BV boundaries. For BV, the labeling process took around 10 minutes for each image volume. Because the body is much larger than the BV and has more variations, the manual body segmentation was achieved by labeling every ≈ 10 2D slices in a volume and then using the label interpolation function in Amira to complete the 3D segmentation. Then, the interpolated slices were examined and slices with large errors were corrected. The number of 2D slices manually segmented in each 3D volume varied depending on the image quality and the number of slices of the image. The body labeling process took around 30 minutes for each 3D image. Any interpolation artifacts from this approach had minimal impact on the analyses. The data set containing 231 images was randomly split into 139 cases for training, 46 cases for validation and 46 cases for testing. The validation set was used to determine the stopping criterion during training. The data used in this paper will be provided upon request.
Among the 231 data sets with manual BV and body segmentation, there were only 35 mutant images, which was not sufficient to train and test a mutant classification algorithm. In addition, we also had 336 unlabeled HFU embryo data sets. Hence, the developed segmentation algorithm was applied to these 336 unlabeled HFU images. After auto segmentation of the unlabeled data sets, we manually reviewed the results and selected 321 sets with visually satisfactory segmentation. This process resulted in a total of 552 (231 manual + 321 automatic) data sets with BV and body segmentation containing 102 mutant and 440 normal. Because 102 mutant images were a small data set, six-fold cross validation was employed to develop and evaluate the mutant classification algorithm.
IV. METHODS
A. Segmentation Framework
An overview of our proposed end-to-end BV and body segmentation framework is shown in Fig. 2. The pipeline consists of an coarse segmentation stage and a segmentation refinement stage. The initial coarse segmentation produces low-resolution segmentation maps for BV and body, simultaneously. Next, the original data and the low-resolution label for each object are passed to a Loc-Con module (Fig. 3), which first generates a bounding box for the object using the centroid of the up-sampled predicted probability map. Then, for the corresponding object, the Loc-Con module concatenates the full-resolution original image and the initial up-sampled predicted probability map in the bounding box as auto-context input for the refinement network. Next, the refinement network generates a full resolution segmentation map within the bounding box. This segmentation is then projected back to the entire image volume through zero padding. We first train the initial coarse segmentation network and then separately train the BV and body refinement networks using output from the trained initial segmentation network. Finally, we fine-tune all three networks (VNet I, VNet II, VNet III) in an end-to-end manner. These three networks follow the exact VNet structure [14] and only the first and last layers are changed based on how many channels the input has and how many classes are predicted.
1). Initial Segmentation on Low-Resolution Volumes:
Because of memory constraints and large variations in image sizes, all the images were padded and downsampled (3rd order spline interpolation) to a low resolution volume of 1603 voxels. A VNet [14] (VNet I) was trained to perform BV and body segmentation, simultaneously, at low resolution. The output of the VNet had 3 channels, representing the background, BV, and body. The Dice loss [14] for each class was summed and used as the training loss (loss I in Fig. 2).
2). Localization-auto-context Module:
To better utilize the information obtained from the low-resolution segmentation result for each structure, the Loc-Con module was introduced to produce the localized auto-context input for refinement. For each foreground object (BV or body), the Loc-Con module steps are as follows (Fig. 3):
Up-sample (trilinear interpolation) the initial coarse segmentation map to original resolution.
Generate a fixed-pixel size bounding box (1443 for BV and 2243 for body) located at the corresponding centroid of the up-sampled predicted probability map for each class. Images were zero-padded when smaller than the bounding box.
Concatenate the original resolution image and the initial predicted probability map (after up-sampling) in the bounding box to create the auto-context input for the refinement network.
Going beyond previous works [32]–[34], our Loc-Con module served as a hard attention mechanism by leveraging the low-resolution, rough segmentation to crop an ROI (the bounding box) at the original resolution. It made use of a conventional auto-context strategy [13] by employing the initial predicted probability map, in conjunction with the original image, as an additional input channel. The initial coarse segmentation map obtained from the whole image at low resolution can provide global context information, which improved the results of the subsequent segmentation networks (VNet II and III). Because trilinear up-sampling was used, the gradient from the subsequent refinement networks (VNet II and III) was able to flow back to the initial segmentation network (VNet I), which made end-to-end fine-tuning feasible (Fig. 2).
3). Pre-training of Fine-Resolution Refinement Network:
Two refinement networks (VNet II and III) were trained for body and BV segmentation, respectively. For each object, the fine-resolution raw image and the up-sampled initial segmentation probability map in the localized bounding box were concatenated and used as the input. The structure of the refinement network was exactly the same as the initial segmentation, except that it required 2 channels as input (original cropped image and corresponding initial segmentation probability map), and produced a single channel as output (indicating the probability of being part of the body or BV at each pixel). Using the detected centroid information on the object, the output was zero-padded back to original image size.
4). End-to-end Refinement on Fine-Resolution:
During the pretraining of the refinement stage, the parameters of the initial segmentation network (VNet I) were frozen until the refinement network for each object (VNet II and III) converged. After that, all three networks were jointly optimized to minimize the sum of Dice losses measured on the fine-resolution image (loss II and III in Fig. 2). The gradient backpropagation path for this end-to-end refinement is indicated in Fig. 2.
B. Mutant Classification and Visualization Pipeline
Subtle structural differences in the BV and body shapes between mutant and normal mouse embryos have been reported [2]. Hence, it is possible to use the BV and body segmentation map (the output of our proposed segmentation algorithm in Sec. IV-A) to train a volumetric CNN for mutant vs. normal binary classification. Due to the small number of training images, each with varying orientation, it was important to first rotate all images into a canonical orientation in order to reduce the input variance prior to feeding it into a classifier. This approach is more efficient for discovering subtle structural differences between the normal and mutant images, and for improving classification performance.
As shown in Fig. 4, we made use of the structural characteristics and relative positions of the BV and body to rotate the BV and body segmentation map into a canonical space. Specifically, we rotated the shape images so that the first Principal Component (PC) of the BV shape was aligned with the X-axis. We then rotated the images so that the first PC of the body shape was in the X–Y plane. Then the centroid positions of the BV and body were used to flip the images to make sure they all have the same up-right orientation along the Y axis. Finally, we made use of the fact that the front BV is wider than the back BV to flip the images along the X axis such that all the images had the same orientation. With all the BV and body shapes in the same orientation, a 256 × 192 × 160 bounding box was cropped at the centroid of the body to remove unrelated regions. This bounding box was then downsampled by 2 to reduce input size. Finally, a 9-layer volumetric VGG-like CNN (Fig. 5) was trained on the rotated, cropped, and downsampled segmentation map to perform mutant vs. normal binary classification. Note that the input segmentation map has only one channel with different values indicating the BV, body and background.
Fig. 4.
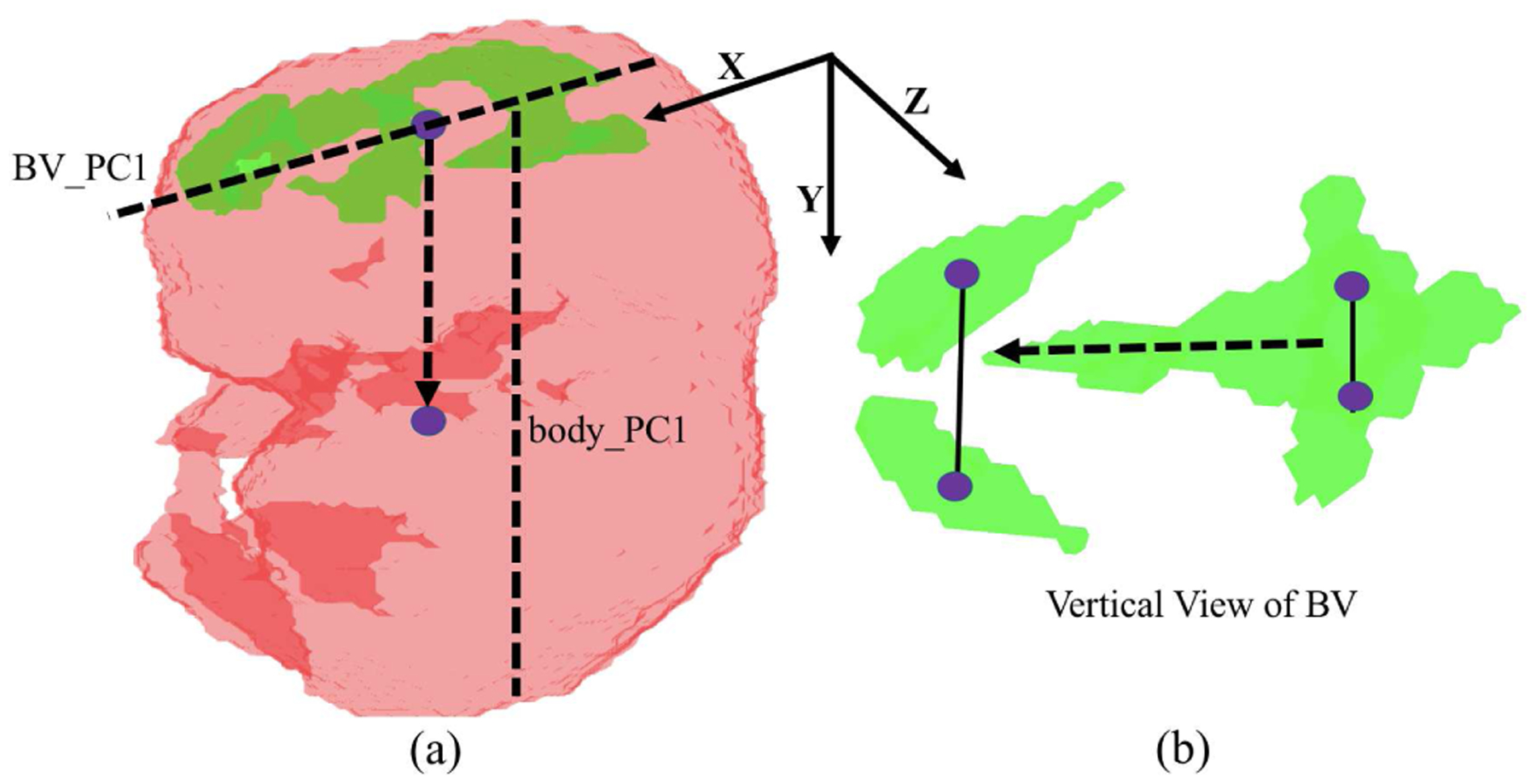
Procedure for rotating each BV and body segmentation map into a canonical orientation. The first Principle Component (PC) of BV and the first PC of body are used (indicated as dash line in (a)). The up-down direction (dash arrow in (a)) is determined by the comparative centroid positions of BV and body. The front-back direction (dash arrow in (b)) is determined by the structural characteristic of BV because the front BV region is wider than the back BV region (solid line in (b)).
Fig. 5.
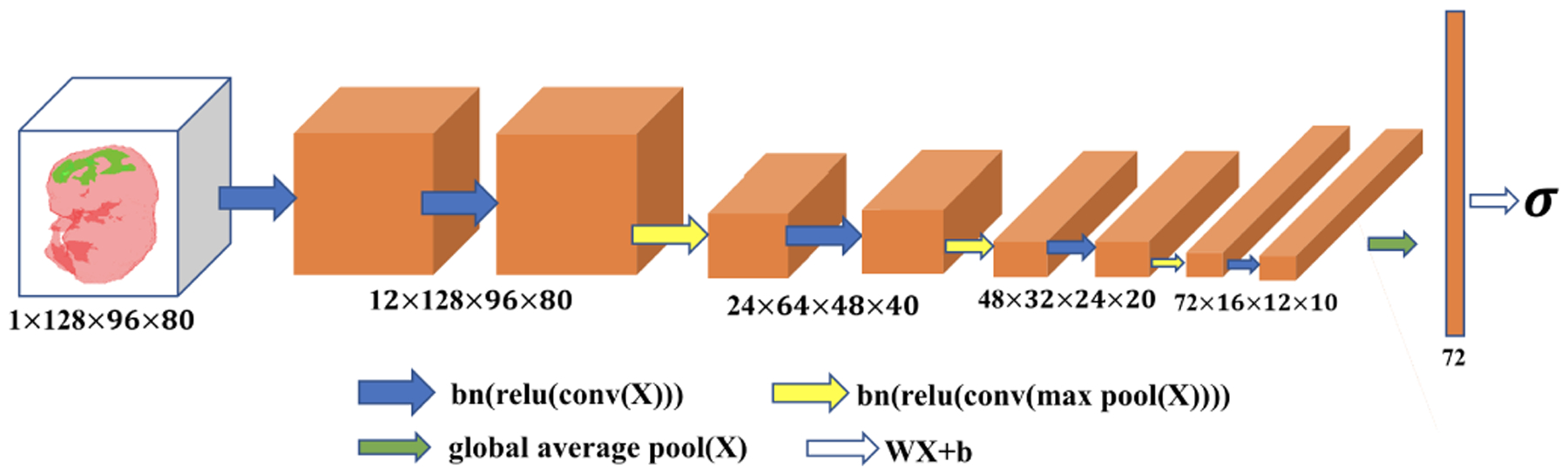
Pictorial representation of the mutant classification network. The numbers below each box indicate channel, depth, height and width. “bn”, “relu”, “conv”, “max pool”, “global average pool” and “WX+b” indicate batch normalization, rectified linear unit, convolution, max pooling, global average pooling and fully connected layer operations, respectively.
In order to understand the decisions made by the trained network, we visualized the trained network through the gradient of the prediction with respect to the input segmentation map [16]. 20% of the maximum gradient value was used as the threshold to obtain a binary saliency image. Guided gradient backpropagation [41] was also implemented, and similar visualization results were obtained. These saliency images served as the explanations of the classifier’s decisions between mutant and normal mice.
C. Implementation Details
All the codes were written in Python 3.6.3. All neural network models were implemented in PyTorch 1.2 [46], with CentOS 7.4, CUDA 9.1 using 2 NVIDIA Tesla P40 graphic-processing-units (NVIDIA Corp., Santa Clara, CA, USA) with 2 × 24 GB of memory.
To compensate for the limited amount of training data for the segmentation networks, data augmentation was employed. Available volumes were randomly rotated from −180° to 180° along each of the three axes, then randomly translated −30 to 30 voxels and, finally, randomly flipped. During the initial pretraining step (VNet I), the Dice loss (loss I) between the predicted segmentations and the downsampled manual labels were averaged across the three classes (background, body and BV). During the pre-training refinement stage (VNet II & III) and end-to-end refinement stage (VNet I, II and III), the Dice loss for body and BV (loss II and III) were used to train the networks. All networks were trained with the Adam optimizer [47] using a learning rate of 10−2 for the initial segmentation and pre-training refinement stages. A learning rate of 10−3 was used for the end-to-end refinement stage. The batchsize was 8 for the first stage segmentation; 2 for the second stage and refinement stage. The training data size was 139 and we applied a drop-last data-loader. Thus, for the first stage segmentation, 17 updates were performed per epoch; for the second stage and refinement segmentation, 69 updates were performed per epoch. An independent validation data set was used to determine the stop criterion. The first stage took 803 epochs; the second stage took 349 and 270 epoch for BV and body, respectively; and the refinement stage took 117 epochs. The models were chosen to minimize validation loss.
In this work, ITK-SNAP [48] was used to visualize some initial and end-to-end segmentation results of our proposed framework.
In order to compensate for the imbalance between the amount of mutant and normal images (102 mutant and 440 normal), weighted cross entropy loss was used to train the classification networks with weights 3.5 and 1.0 for the mutant and normal classes, respectively. No data augmentation was used for the training of the classification network because all of the images were rotated into a canonical orientation before feeding into the network. The network was trained using the SGD optimizer with momentum 0.9 and weight decay 10−5. The learning rate was set to 10−2 for the first 70 epochs and decreased to 5 × 10−3 for the remaining 30 epochs. Approximately, each epoch would have 57 updates.
V. EXPERIMENTAL RESULTS AND DISCUSSION
A. Segmentation Results
In this work, we evaluated the performance of the proposed framework using the DSC score with standard deviation, which is widely employed to evaluate segmentation performance in biomedical imaging. Given reference segmentation G and predicted segmentation P, the DSC can be computed as: . As mentioned in Sec. III, we used 139 image volumes for training, 46 image volumes for validation and 46 image volumes for testing. We used further data augmentation as described in Sec. IV-C during the training. Note that because each voxel can be treated as a training sample, the relatively small number of image volumes for training was sufficient to generalize well to the test image volumes.
As shown in Table I, the initial results of our proposed segmentation framework achieved a satisfactory average DSC score of 0.924 for the body and lower DSC score of 0.887 for the BV. This was expected because the BV was much smaller than the body. Hence, it was necessary to localize the ROI and refine the segmentation. The refinement using the raw image as well as the initial predicted probability map improved the average DSC to 0.898 for the BV. In order to determine the efficacy of the auto-context approach, only the raw image cropped from the bounding box found in the localization step was fed to the refinement network. Compared to jointly using the initial segmentation and the raw image (auto-context input [13]), this refinement-without-auto-context approach yielded a lower DSC for the BV (between 0.893 and 0.898), which indicates context information is important to BV segmentation. In our application, jointly using the initial segmentation and the raw image (auto-context input) was similar to stacking two VNet together with the first one being frozen. In this way, the second VNet (the refinement network) had larger receptive field and was able to utilize more context information to improve the final segmentation results. Note that for the body segmentation, the gain from the refinement compared to the initial coarse segmentation was limited, because the body boundary was fairly smooth and did not suffer from a downsampled representation. Finally, end-to-end refinement improved BV DSC to 0.899 and body DSC to 0.934. As shown in Fig. 6, the initial segmentation produced reasonable BV and body segmentation results. After end-to-end refinement, the segmentation accuracy was improved along with better boundaries, and sometimes eliminated structural segmentation errors in the first stage. This implies that the fine-resolution refinement was more beneficial than suggested by the small improvement in the DSC metric.
TABLE I.
DSC with standard deviation and inference time per volume averaged over 46 test volumes
| Methods╲Results | BV DSC | Body DSC | Inference Time* |
|---|---|---|---|
| NGC-based Framework [9] | 0.762±0.254 | 0.775±0.183 | 699.3 s |
| Sliding-window Benchmark | 0.904±0.050 [11] | 0.924±0.023 [12] | 102.4 s |
| Coarse Segmentation | 0.887± 0.055 | 0.924 ± 0.023 | 6 ms |
| Refinement Without Auto-Context Input | 0.893±0.057 | 0.918±0.059 | 80 ms |
| Refinement With Auto-Context Input | 0.898±0.052 | 0.927±0.026 | 90 ms |
| Refinement End-to-end | 0.899±0.056 | 0.934±0.015 | 90 ms |
Fig. 6.
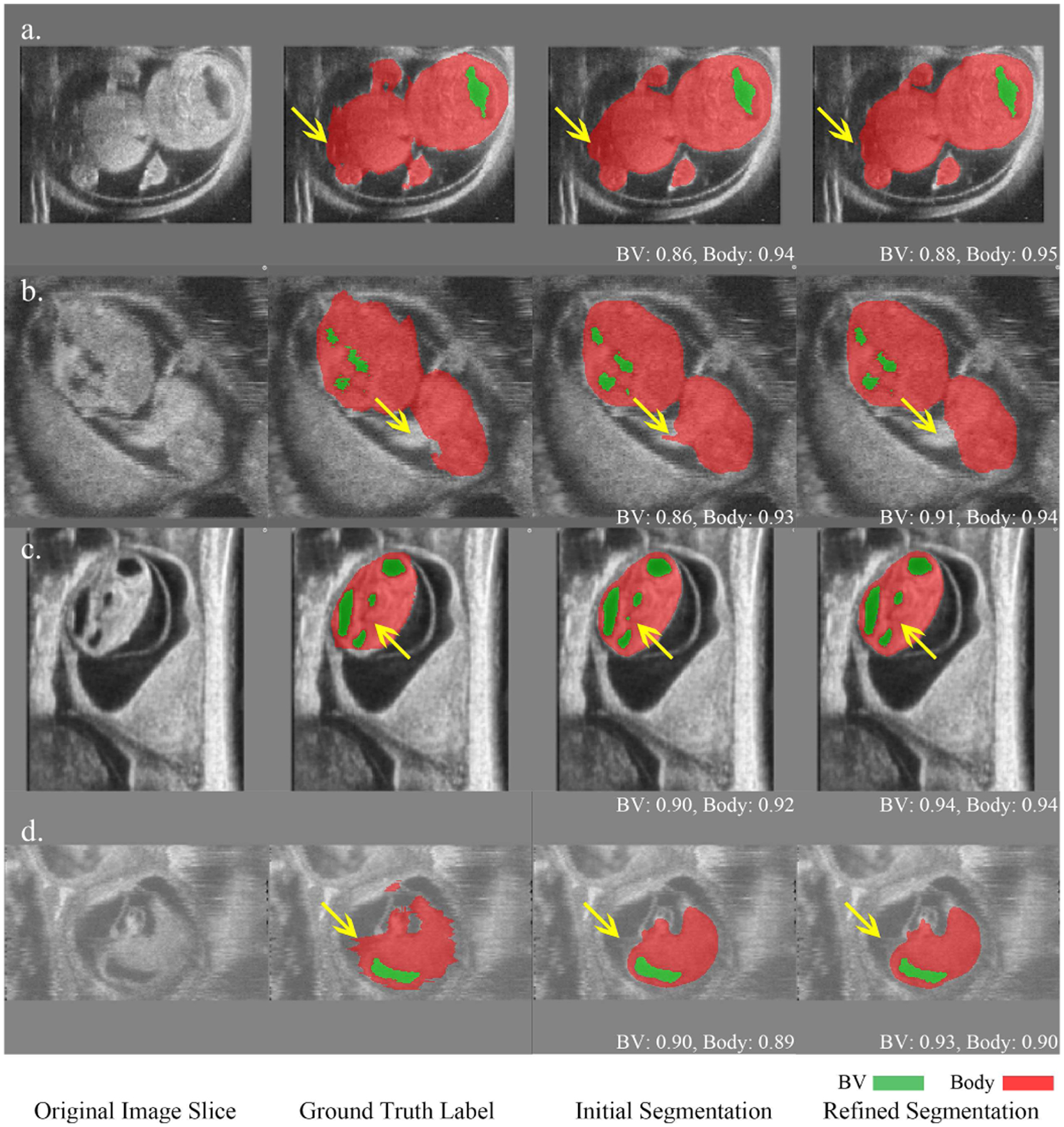
Comparison of initial coarse segmentation and refined segmentation for four HFU volumes. Green indicates BV, red indicates body and the numbers below the predicted segmentation correspond to DSC. In a), b) and c), yellow arrows indicate that the refinement improved the segmentation in terms of boundary and structure. d) represents an image with motion artifacts where the manual segmentation was noisy in the body background boundary while the refinement network produced a smooth boundary which was closer to the true physical structure. The refined BV segmentation in d) is also more accurate than the initial BV segmentation.
Compared with other existing methods, our proposed segmentation framework outperforms the rule-based segmentation NGC-based framework (Tab. I) [9] by a large margin. Moreover, the NGC-based framework was not robust with 8 failure cases in BV and 3 in body (DSC < 0.6) while our proposed framework and the sliding-window-based methods [11], [12] did not have any failure cases. Although the performance was comparable to the sliding-window-based methods, our proposed method achieved a 1000 fold inference time reduction from 102.36 to 0.09 seconds per volume, enabling real-time segmentation. For fair comparison, the networks from [11] and [12] were retrained using the same training set described here and evaluated on the same testing set. Therefore, the numbers reported here are slightly different from those reported in [11], [12].
Qualitatively, the proposed segmentation pipeline and sliding-window-based methods performed consistently well when challenged with ambiguous or missing boundaries, motion artifacts, and differing image contrast (Fig. 7). In contrast, the NGC-based framework performed worse and failed to produce correct segmentation when boundaries were missing or ambiguous. Our manual body segmentation was achieved by labeling every few images in a volume and then using a label interpolation function to complete the 3D segmentation. Although we determined that the interpolated manual segmentation was reliable for algorithm development and other down-stream analyses, this protocol can potentially lead to interpolation artifacts (e.g. Fig. 7(b)(c)). Advantageously, these artifacts were mitigated by our deep-learning-based framework with segmentation results closer to the true physical structure. For similar interpolated 2D slices across different 3D images, some were slightly under-segmented on the boundaries while others were slightly over-segmented. Moreover, there are still sufficient number of manually labeled slices which are accurate on the boundaries. When we trained our deep neural networks with over a hundred such 3D images, the network learned the true boundaries so that the interpolation artifacts were mitigated. This is similar to small random noise being added to the manual label, but the trained network can still generalize well [49].
Fig. 7.
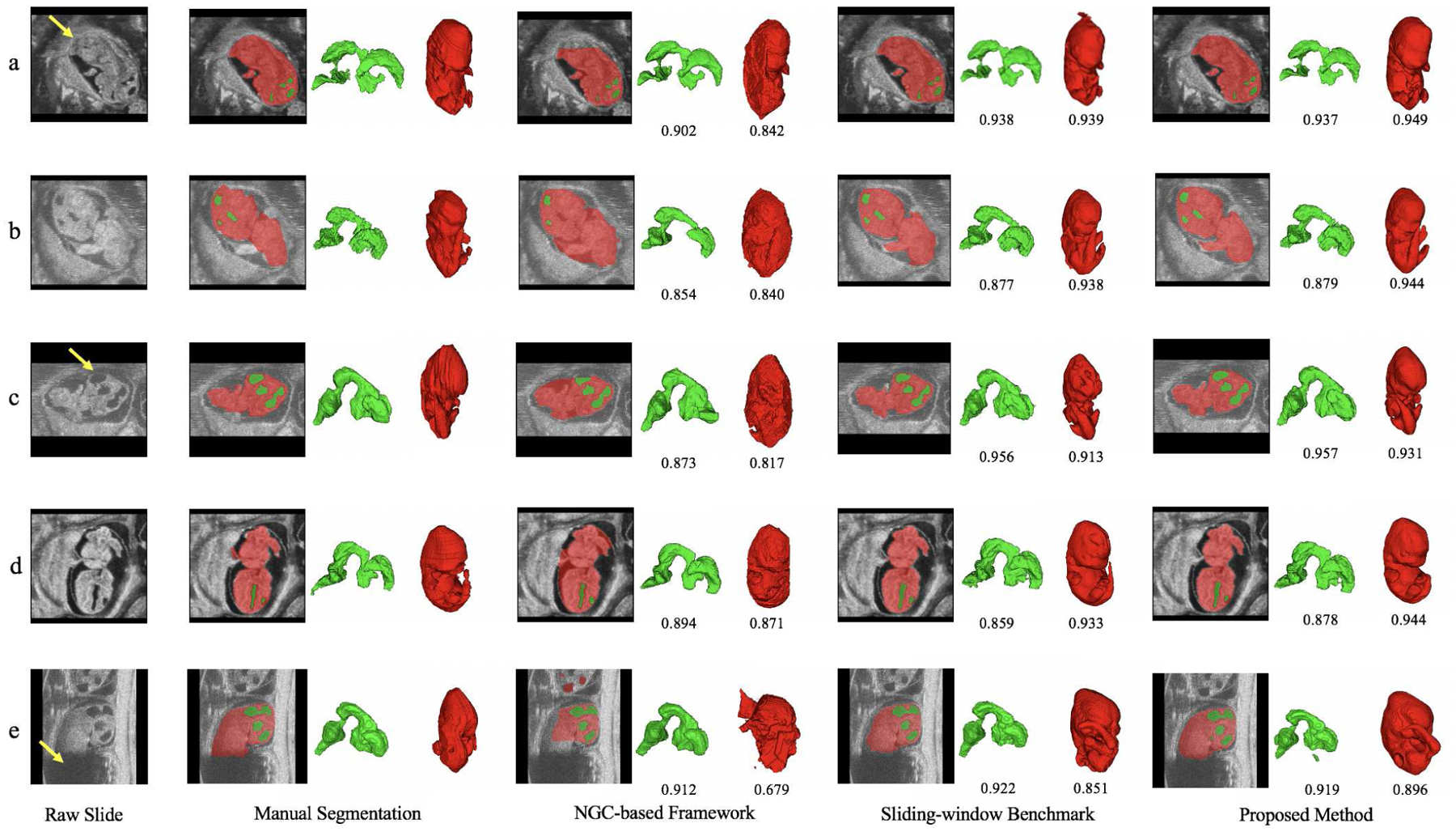
Comparison of qualitative segmentation results among different methods for five HFU images. Green indicates BV, red indicates body and the numbers below the predicted segmentation are corresponding DSC. Yellow arrow in a) indicates ambiguous boundary due to the deep touching of the body and uterine wall. Image b) has severe motion artifacts. Yellow arrow in c) indicates missing head boundary. Image d) has different contrast with image b) and c). Yellow arrow in e) indicates severe missing signal of body, which leads to unsatisfactory automatic body segmentation results across different methods.
B. Classification and Visualization Results
Using the rotated BV and body segmentation maps, a volumetric CNN (Fig. 5) was trained to perform mutant vs. normal binary classification. Due to the limited number of mutant images (102 mutant vs. 440 normal), we conducted a six-fold cross validation, where each fold had the same mutant vs. non-mutant ratio. In each run, one fold was used for validation while the other five folds were used for training. The average classification accuracy among validation samples are shown in Table II. We also show the Receiver Operating Characteristic (ROC) curve in Fig. 8 (red curve), which was obtained by using different thresholds on the predicted probability for the mutant class. The average classification accuracy was 0.969, and the area under the ROC curve (AUC) was 0.9893.
TABLE II.
Confusion matrix of mutant classification results summed over validation samples with six-fold cross validation. The threshold value was set to 0.5 and the average accuracy is 0.969.
| True╲Predict | Mutant | Normal |
|---|---|---|
| Mutant | 96 | 6 |
| Normal | 11 | 429 |
Fig. 8.
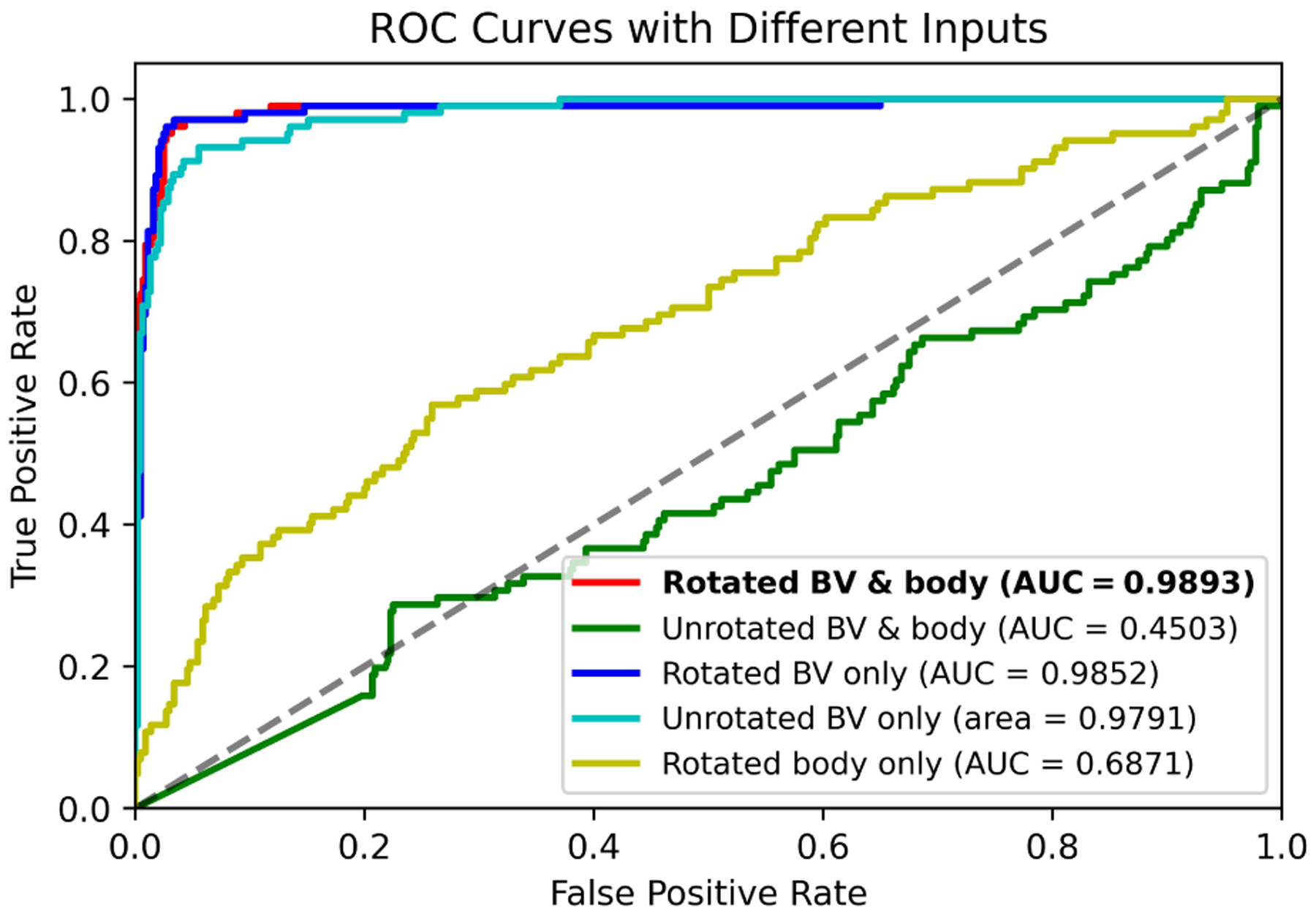
ROC curves and AUC scores of the mutant classification results with different input combinations (each obtained with six-fold cross validation).
In order to verify the utility of rotating the shape images into a canonical space, we also used unrotated BV and body segmentation maps as input to train the same classification network. Without pre-processing rotation, the BV and body were in a wide variety of orientations in the segmentation maps. Because of the limited amount of training data, additional segmentation maps were generated by using existing data but with random combinations of 90-, 180-, or 270-degree rotations around each axis or image flipping along each axis. Using this data, the network still failed to converge to good results (AUC of 0.4503, Fig. 8 green curve). The reason for the divergence of the network could be that the subtle structural differences between normal and mutant mouse embryos (Fig. 9 blue arrows) were overwhelmed by the large variations of embryo orientation. Hence, for our somewhat limited data set it was critical to perform the rotation into a canonical space such that the embryo orientations were aligned. A standard 3D image orientation also assists better visualization of the 3D volumes.
Fig. 9.
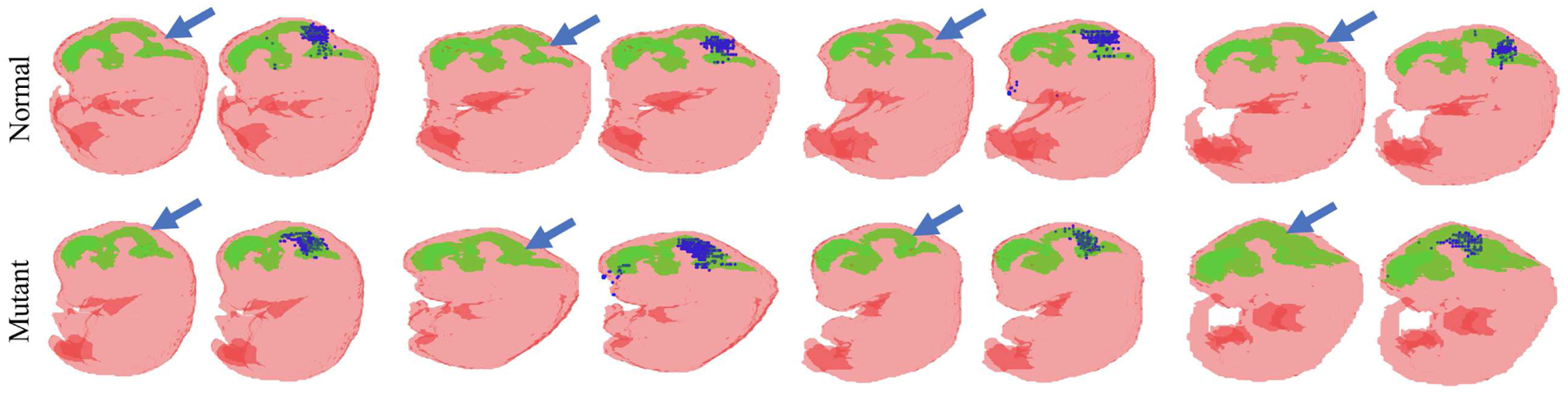
Saliency images of the trained mutant classification neural network. The first row is the normal mouse embryo BV (green) and body (red) segmentation while the second row is mutant. Two images are presented for each sample. The blue arrow in the first image indicates the known structural differences between En1 mutant and normal BVs while the blue dots in the second image (salient points) indicate where the trained network focused when making the prediction.
To investigate which shape information (BV, body or both) was important for final classification, only the rotated BV or body segmentation maps were used to train the same classifier. This approach achieved an AUC of 0.9852 for BV and 0.6871 for body (Fig. 8 blue and yellow ROC curves), respectively. It is worth noting that the AUC of 0.9852 obtained based on just the BV segmentation is similar to the AUC of 0.9893 obtained using BV and body segmentation maps. These AUC values indicate that the BV contributes the most to successful mutant classification. This is consistent with the saliency images (Fig. 9), where most salient points were located around the known structural differences between En1 mutant and normal BVs. We then defined a tight bounding box around unrotated BV data and trained the same classifier with the same data augmentation as the above unrotated BV and body segmentation maps (Fig. 8 green curve). Using this approach, we achieved an AUC of 0.9791 (Fig. 8 cyan curve), which indicates that unrotated BV (with data augmentation) is sufficient to train an accurate mutant classification network. These results also explain why, even with sufficient data augmentation, unrotated BV and body data (Fig. 8 green curve) fails, because the large bounding box includes the BV and body. The large bounding box makes the BV too small relative to the input image size such that the subtle BV differences between En1 mutant and normal embryos were easily overwhelmed by the variations of embryo orientation. Our rational for using the BV and body together to train a classifier is that a difference in spine curvature exists between En1 mutant and normal mouse embryos [2]. Using the BV and body together, the visualization in (Fig. 9) would have the potential to highlight differences in spine curvature. Unfortunately, our trained classifier does not seem to make use of the spine curvature in its decision. The reason might be the spine curvature difference is not as consistent and conspicuous as the BV difference between mutant and normal mouse embryos.
More importantly, as shown in Fig. 9, the visualization of the trained classifier (using rotated BV and body segmentation maps) demonstrated that the trained network focused on regions where En1 mutation is known to cause the loss of brain tissue and thickening of the BV (Fig. 9 blue arrows). This BV region is the main ROI when performing manual segmentation for mutant vs. normal classification En1. If had not known a priori where to detect the difference in BV between normal and En1 mutant mouse embryos beforehand, the visualization results of the trained network would have highlighted these relevant regions. This observation indicates that gradient backpropagation of trained, deep-learning classifiers has the potential to automatically detect unknown phenotypes associated with a known genetic mutation.
C. Limitations
Our study had a few limitations. First, the proposed two-stage segmentation framework only provided limited improvement over the first stage initial segmentation. If less-accurate automatic segmentation quality for some down-stream analyses is acceptable, the initial segmentation would be enough, which will further reduce the inference time by another factor of 15 (from 90 ms to 6 ms per image). Second, the reference manual segmentation for the 3D images was obtained by labeling every few frames and then using a label interpolation function, which inevitably introduced some interpolation artifacts. Manual segmentation of every image slice could be labeled in future studies. Finally, although a difference in spine curvature has been reported to exist between En1 mutant and normal mouse embryos [2], our experiments did not seem to use this difference to perform the classification (Fig. 9). Further work is necessary to understand why our methods did not detect these differences.
VI. CONCLUSION
An end-to-end two-stage segmentation framework was proposed for accurate and real-time segmentation of the BV and body in 3D, in vivo and in utero HFU images of mouse embryos. The initial coarse segmentation stage acted as an ROI localization module and provided global context information for the second-stage, fine-resolution refinement network. The results demonstrated the efficacy of this two-stage structure. The proposed method achieved high DSC scores of 0.899 for BV and 0.934 for body segmentation, comparable to the previous benchmark (i.e. sliding-window-based methods), and was approximately one thousand times faster in inference time. A deep-learning-based method was also developed for mutant vs. normal classification using the BV and body segmentation maps. To overcome the limited data problem, a fully automatic method was developed to rotate the raw segmentation maps such that the BV and body shapes were in a canonical orientation, thus, removing uninformative input variations. Using this pre-processing approach, the model achieved a high average accuracy of 0.969 and AUC of 0.9893 over six cross validation folds. The trained classification model was shown to differentiate between mutant and normal mouse embryos by focusing on the BV region where the phenotype associated with the En1 mutation typically manifests. The proposed pipeline has the potential to uncover unknown phenotypes manifested as shape changes associated with different gene mutations. Our segmentation and mutant classification algorithms may be applicable and invaluable in streamlining developmental biology studies.
Acknowledgments
The research described in this paper was supported in part by NIH grant EB022950 and HD097485.
Contributor Information
Jack Langerman, Department of Computer Science and Engineering, New York University, New York, 11201, USA.
William Das, Hunter College High School, New York, 10128, USA.
Chuiyu Wang, School of Electronic and Information Engineering, Beihang University, Beijing, 100191, China.
Nitin Nair, Department of Electrical and Computer Engineering, New York University, New York, 11201, USA.
Orlando Aristizábal, Skirball Institute of Biomolecular Medicine, New York University, New York, 10016, USA.
Jonathan Mamou, F. L. Lizzi Center for Biomedical Engineering, Riverside Research, New York, 10038, USA.
Daniel H. Turnbull, Skirball Institute of Biomolecular Medicine, New York University, New York, 10016, USA
Jeffrey A. Ketterling, F. L. Lizzi Center for Biomedical Engineering, Riverside Research, New York, 10038, USA
Yao Wang, Department of Electrical and Computer Engineering, New York University, New York, 11201, USA.
REFERENCES
- [1].Dickinson ME, Flenniken AM, Ji X,et al. “High-throughput discovery of novel developmental phenotypes,” in Nature, vol. 537, pp. 508–514, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wurst W, Auerbach AB, Joyner AL, “Multiple developmental defects in Engrailed-1 mutant mice: an early mid-hindbrain deletion and patterning defects in forelimbs and sternum,” in Development, vol. 120, no. 7, pp. 2065–75, 1994. [DOI] [PubMed] [Google Scholar]
- [3].Kuo JW, Wang Y, Aristiz’abal O, Turnbull DH, Ketterling J, and Mamou J, “Automatic mouse embryo brain ventricle segmentation, gestation stage estimation, and mutant detection from 3D 40-MHz ultrasound data,” in Proc. IEEE Int. Ultrasonics Symp, 2015, pp. 1–4. [Google Scholar]
- [4].Martínez-Martínez MA, Pacheco-Torres J, Borrell V, Canals S, “Phenotyping the central nervous system of the embryonic mouse by magnetic resonance microscopy”, NeuroImage, volume 97, pp. 95–106, 2014. [DOI] [PubMed] [Google Scholar]
- [5].Aristiz’abal O, Mamou J, Ketterling JA, and Turnbull DH, “High-throughput, high-frequency 3-D ultrasound for in utero analysis of embryonic mouse brain development,” Ultrasound Med. Biol, vol. 39, no. 12, pp. 2321–2332, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Mayhew TM, Lucocq JM, “From gross anatomy to the nanomor-phome: stereological tools provide a paradigm for advancing research in quantitative morphomics”, Journal of Anatomy, vol. 25, no. 4, pp. 309–321, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Henkelman RM, “Systems biology through mouse imaging centers: experience and new directions,” Annu. Rev. Biomed. Eng, vol. 12, pp. 143–166, 2010. [DOI] [PubMed] [Google Scholar]
- [8].Kuo JW, Mamou J, Aristiz’abal O, Zhao X, Ketterling JA, and Wang Y, “Nested graph cut for automatic segmentation of high-frequency ultrasound images of the mouse embryo,” IEEE Trans. Med. Imag, vol. 35, no. 2, pp. 427–441, 2015. [DOI] [PubMed] [Google Scholar]
- [9].Kuo JW, Qiu Z, Aristiz’abal O, Mamou J, Turnbull DH, Ketterling J, and Wang Y, “Automatic body localization and brain ventricle segmentation in 3D high frequency ultrasound images of mouse embryos,” in Proc. IEEE Int. Symp. Biomed. Imag, 2018, pp. 635–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Long J, Shelhamer E, and Darrell T, “Fully convolutional networks for semantic segmentation,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit, 2015, pp. 3431–3440. [DOI] [PubMed] [Google Scholar]
- [11].Qiu Z, Langerman J, Nair N, Aristiz’abal O, Mamou J, Turnbull DH, Ketterling J, and Wang Y, “Deep BV: A fully automated system for brain ventricle localization and segmentation in 3D ultrasound images of embryonic mice,” in Proc. IEEE Signal Process. Medicine and Biol. Symp, 2018, pp. 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Qiu Z, Nair N, Langerman J, Aristiz’abal O, Mamou J, Turnbull DH, Ketterling JA, and Wang Y, “Automatic mouse embryo brain ventricle & body segmentation and mutant classification from ultrasound data using deep learning,” in Proc. IEEE Int. Ultrasonics Symp, 2019, pp. 12–15. [Google Scholar]
- [13].Tu Z, “Auto-context and its application to high-level vision tasks,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit, 2008, pp. 1–8. [Google Scholar]
- [14].Milletari F, Navab N, and Ahmadi SA, “V-net: Fully convolutional neural networks for volumetric medical image segmentation,” in Proc. 4th Int. Conf. 3D Vision, 2016, pp. 565–571. [Google Scholar]
- [15].Simonyan K, and Zisserman A, “Very deep convolutional networks for large-scale image recognition,” 2014, arXiv:1409.1556. [Google Scholar]
- [16].Simonyan K, Vedaldi A, and Zisserman A, “Deep inside convolutional networks: Visualising image classification models and saliency maps,” 2013, arXiv:1312.6034. [Google Scholar]
- [17].Xu T, Qiu Z, Das W, Wang C, Langerman J, Nair N, Aristiz’abal O, Mamou J, Turnbull DH, Ketterling JA, Wang Y, “Deep Mouse: An End-to-End Auto-Context Refinement Framework for Brain Ventricle & Body Segmentation in Embryonic Mice Ultrasound Volumes,” in Proc. IEEE Int. Symp. Biomed. Imag, 2020, pp. 122–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dorr AE, Lerch JP, Spring S, Kabani N and Henkelman RM, “High resolution three-dimensional brain atlas using an average magnetic resonance image of 40 adult C57Bl/6J mice,” NeuroImage, vol. 42, pp. 60–69, 2008. [DOI] [PubMed] [Google Scholar]
- [19].Nieman BJ, van Eede MC, Spring S, Dazai J, Henkelman RM and Lerch JP, “MRI to assess neurological function,” Current protocols in mouse biology, vol. 8, pp. 44, 2018. [DOI] [PubMed] [Google Scholar]
- [20].Szulc KU, Lerch JP, Nieman BJ, Bartelle BB, Friedel M, Suero-Abreu GA, Watson C, Joyner AL and Turnbull DH, “4D MEMRI atlas of neonatal FVB/N mouse brain development,” Neuroimage, vol. 118, pp. 49–62, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wong MD, Maezawa Y, Lerch JP and Henkelman RM, “Automated pipeline for anatomical phenotyping of mouse embryos using micro-CT,” Development, vol. 141, pp. 2533–2541, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wong MD, van Eede MC, Spring S, Jevtic S, Boughner JC, Lerch JP and Henkelman RM, “4D atlas of the mouse embryo for precise morphological staging,” Development, vol. 142, pp. 3583–3591, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Shen D, Wu G, and Suk HI, “Deep learning in medical image analysis,” Annu. Rev. Biomed. Eng, vol. 19, pp. 221–248, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, Van Der Laak JA, Van Ginneken B, and S’anchez CI, “A survey on deep learning in medical image analysis,” Med. Image Anal, vol. 42, pp. 60–88, 2017. [DOI] [PubMed] [Google Scholar]
- [25].Li M, Wang C, Zhang H, and Yang G, “MV-RAN: Multiview recurrent aggregation network for echocardiographic sequences segmentation and full cardiac cycle analysis,” Comput. in Biol. and Med, vol. 120, pp. 103728, 2020. [DOI] [PubMed] [Google Scholar]
- [26].Yang G, Chen J, Gao Z, Li S, Ni H, Angelini E, Wong T et al. , “Simultaneous left atrium anatomy and scar segmentations via deep learning in multiview information with attention,” Future Generation Comput. Syst, vol. 107, pp. 215–228, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Aristizabal O, Turnbull DH, Ketterling JA, Wang Y, Qiu Z, Xu T, Goldman H and Mamou J, “Scanner Independent Deep Learning-Based Segmentation Framework Applied to Mouse Embryos,” in Proc. IEEE Int. Ultrasonics Symp, 2020, pp. 1–4. [Google Scholar]
- [28].Zhu W, Liao H, Li W, Li W and Luo J, “Alleviating the Incompatibility Between Cross Entropy Loss and Episode Training for Few-Shot Skin Disease Classification,” in Int. Conf. on Med. Image Comput. and Computer-Assisted Intervention 2020, pp. 330–339. [Google Scholar]
- [29].Milletari F, Ahmadi SA, Kroll C, Plate A, Rozanski V, Maiostre J, Levin J, Dietrich O, Ertl-Wagner B, Bötzel K, and Navab N, “Hough-CNN: deep learning for segmentation of deep brain regions in MRI and ultrasound,” Comput. Vision and Image Understanding, vol. 164, pp. 92–102, 2017. [Google Scholar]
- [30].Ronneberger O, Fischer P, and Brox T, “U-net: Convolutional networks for biomedical image segmentation,” in Proc. Int. Conf. Medical image comput. computer-assisted intervention, 2015, pp. 234–241. [Google Scholar]
- [31].Liu Y, Yang G, Mirak SA, Hosseiny M, Azadikhah A, Zhong X, Reiter RE, Lee Y, Raman SS, and Sung K, “Automatic prostate zonal segmentation using fully convolutional network with feature pyramid attention,” IEEE Access, vol. 7, pp. 163626–163632, 2019. [Google Scholar]
- [32].Roth HR, Oda H, Zhou X, Shimizu N, Yang Y, Hayashi Y, Oda M, Fujiwara M, Misawa K, and Mori K, “An application of cascaded 3D fully convolutional networks for medical image segmentation,” Computerized Med. Imag. Graph, vol. 66, pp. 90–99, 2018. [DOI] [PubMed] [Google Scholar]
- [33].Tang Y, Yang F, and Yuan S, “A multi-stage framework with context information fusion structure for skin lesion segmentation,” in Proc. IEEE Int. Symp. Biomed. Imag, 2019, pp. 1407–1410. [Google Scholar]
- [34].Chen H, Dou Q, Yu L, and Heng PA, “Voxresnet: Deep voxelwise residual networks for volumetric brain segmentation,” 2016, arXiv:1608.05895. [DOI] [PubMed] [Google Scholar]
- [35].He K, Gkioxari G, Doll’ar P and Girshick R, “Mask r-cnn,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit, 2017, pp. 2961–2969. [Google Scholar]
- [36].Krizhevsky A, Sutskever I, and Hinton GE, “Imagenet classification with deep convolutional neural networks,” Advances Neural Inf. Process. Syst, vol. 25, pp. 1097–1105, 2012. [Google Scholar]
- [37].He K, Zhang X, Ren S, and Sun J, “Deep residual learning for image recognition,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit, 2016, pp. 770–778. [Google Scholar]
- [38].Huang G, Liu Z, Van Der Maaten L, and Weinberger KQ, “Densely connected convolutional networks,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit, 2017, pp. 4700–4708. [Google Scholar]
- [39].Hu J, Shen L, and Sun G, “Squeeze-and-excitation networks,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit, 2018, pp. 7132–7141. [Google Scholar]
- [40].Wang Y, Yue W, Li X, Liu S, Guo L, Xu H, Zhang H, and Yang G, “Comparison Study of Radiomics and Deep Learning-Based Methods for Thyroid Nodules Classification Using Ultrasound Images,” IEEE Access, vol. 8, pp.52010–52017, 2020. [Google Scholar]
- [41].Springenberg JT, Dosovitskiy A, Brox T, and Riedmiller M, “Striving for simplicity: The all convolutional net,” 2014, arXiv:1412.6806. [Google Scholar]
- [42].Zhou B, Khosla A, Lapedriza A, Oliva A, and Torralba A, “Learning deep features for discriminative localization,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit, 2016, pp. 2921–2929. [Google Scholar]
- [43].Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, and Batra D, “Grad-cam: Visual explanations from deep networks via gradient-based localization,” in Proc. IEEE Int. Conf. Comput. Vis, 2017, pp. 618–626. [Google Scholar]
- [44].Wang H, Jia H, Lu L, and Xia Y, “Thorax-Net: An Attention Regularized Deep Neural Network for Classification of Thoracic Diseases on Chest Radiography,” IEEE J. Biomed. Health Inform, vol. 24, no. 2, pp. 475–485, 2019. [DOI] [PubMed] [Google Scholar]
- [45].Ketterling JA, Aristizábal O, Turnbull DH, and Lizzi FL, “Design and fabrication of a 40-MHz annular array transducer” IEEE Trans. Ultrason. Ferroelect. Freq. Contr, vol. 52, no. 4, pp. 672–681, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Paszke A, Gross S, Chintala S, Chanan G, Yang E, DeVito Z, Lin Z, Desmaison A, Antiga L, and Lerer A, “Automatic differentiation in PyTorch,” 2017.
- [47].Kingma DP, and Ba J, “Adam: A method for stochastic optimization,” 2014, arXiv:1412.6980. [Google Scholar]
- [48].Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, and Gerig G, “User-guided 3D Active Contour Segmentation of Anatomical Structures: Significantly Improved Efficiency and Reliability,” Neuroimage, vol. 31, no. 3, pp. 1116–1128, 2006. [DOI] [PubMed] [Google Scholar]
- [49].An G, “The effects of adding noise during backpropagation training on a generalization performance,” Neural Computation, vol. 8, pp. 643–674, 1996. [Google Scholar]


