Abstract
Campylobacter jejuni is a pathogenic organism that can cause campylobacteriosis in children and adults. Most commonly, campylobacter infection is brought on by consumption of raw or undercooked poultry, unsanitary drinking water, or pet feces. Surrounding the Campylobacter jejuni bacterium is a coat of sugar molecules known as the capsular polysaccharide or CPS. The capsular polysaccharide can be very diverse among the different strains of C. jejuni and this diversity is considered important for evading the host immune system. Modifications to the CPS of C. jejuni NCTC 11168 include O-methylation, phosphoramidylation, and amidation of glucuronate with either serinol or ethanolamine. The enzymes responsible for amidation of glucuronate are currently unknown. In this study, Cj1441, an enzyme expressed from the CPS biosynthetic gene cluster in C. jejuni NCTC 11168, was shown to catalyze the oxidation of UDP-α-d-glucose into UDP-α-d-glucuronic acid with NAD+ as the cofactor. No amide products were found in an attempt to determine whether the putative thioester intermediate formed during the oxidation of UDP-glucose by Cj1441 could be captured in the presence of added amines. The three-dimensional crystal structure of Cj1441 was determined in the presence of NAD+ and UDP-glucose bound in the active site of the enzyme (PDB id: 7KWS). A more thorough bioinformatic analysis of the CPS gene cluster suggests that the amidation activity is localized to the t-terminal half of Cj1438, a bifunctional enzyme that is currently annotated as a sugar transferase.
Graphical Abstract
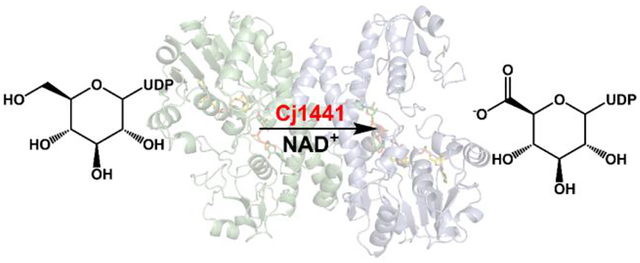
Introduction
Campylobacter jejuni is a zoonotic organism commonly found in the intestinal tracts of poultry, cattle, and dogs. Infection of C. jejuni in humans leads to campylobacteriosis, which is the leading cause of gastroenteritis in the United States, accounting for 1.3 million new cases per year.1 Most commonly, infection by C. jejuni is brought on by consumption of raw or undercooked poultry, unsanitary drinking water, unpasteurized milk, or pet feces.2 C. jejuni is also implicated in Guillain-Barré syndrome (GBS) where ~40% of new GBS cases are preceded by a C. jejuni infection.3, 4 Attached to the exterior of C. jejuni is a polymeric coat of sugar molecules known as the capsular polysaccharide (CPS). The sequence of monosaccharides that comprise the CPS in C. jejuni can be quite diverse among the different strains of this bacterium and this diversity contributes to the evasion of the host immune system and epithelial attachment. More than 60 different strains of C. jejuni are known to exist.5, 6
The CPS from C. jejuni NCTC 11168 is comprised of d-glycero-l-gluco-heptose, d-glucuronic acid, d-N-acetyl-galactose and d-ribose (Figure 1).7, 8 These sugars are further decorated by methylation, phosphoramidylation, and amidylation. The catalytic activities of four enzymes (Cj1415, Cj1416, Cj1417, Cj1418) that are partially responsible for the phosphoramidate modification in C. jejuni NCTC 11168 have been elucidated.9–13 We, and others, have functionally characterized seven other enzymes that are required for the biosynthesis of the d-glycero-l-gluco-heptose moiety (Cj1152, Cj1423, Cj1424, Cj1425, Cj1427, Cj1428, and Cj1430) in C. jejuni NCTC 11168.14–17 Knockout studies have shown that when the gene for Cj1423 from C. jejuni NCTC 11168 is deleted, the heptose sugar is no longer present in the capsular polysaccharide.7, 18, 19 In the same study, a knockout of the gene for Cj1441 resulted in a complete loss of the capsule. The gene cluster that encodes for most, but not all, of the enzymes required for the biosynthesis of the CPS of C. jejuni NCTC 11168 is presented in Figure 2.
Figure 1:
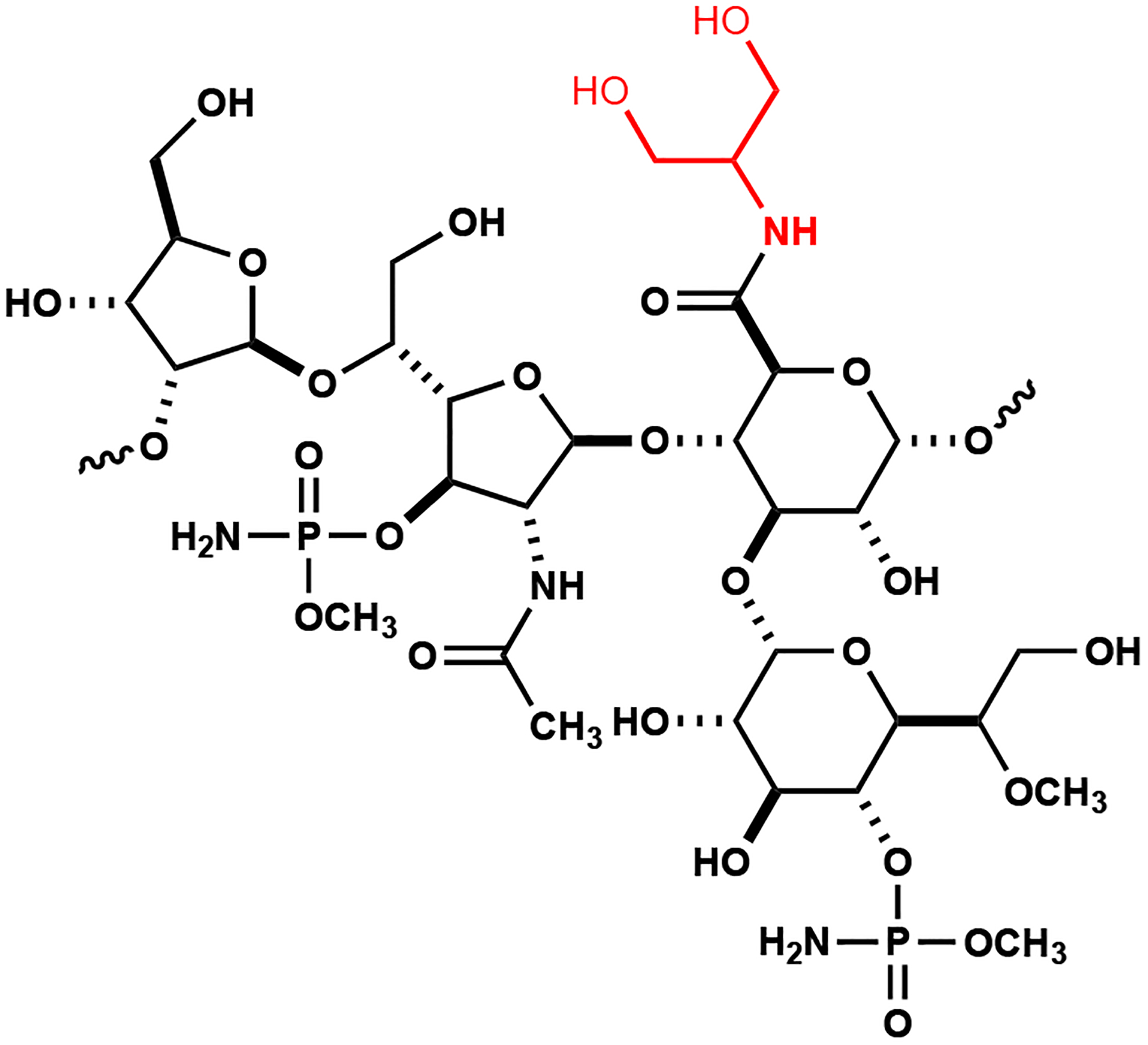
Structure of the repeating unit in the capsular polysaccharide of C. jejuni NCTC 11168. The amide attached to the glucuronamide moiety is highlighted in red.
Figure 2:
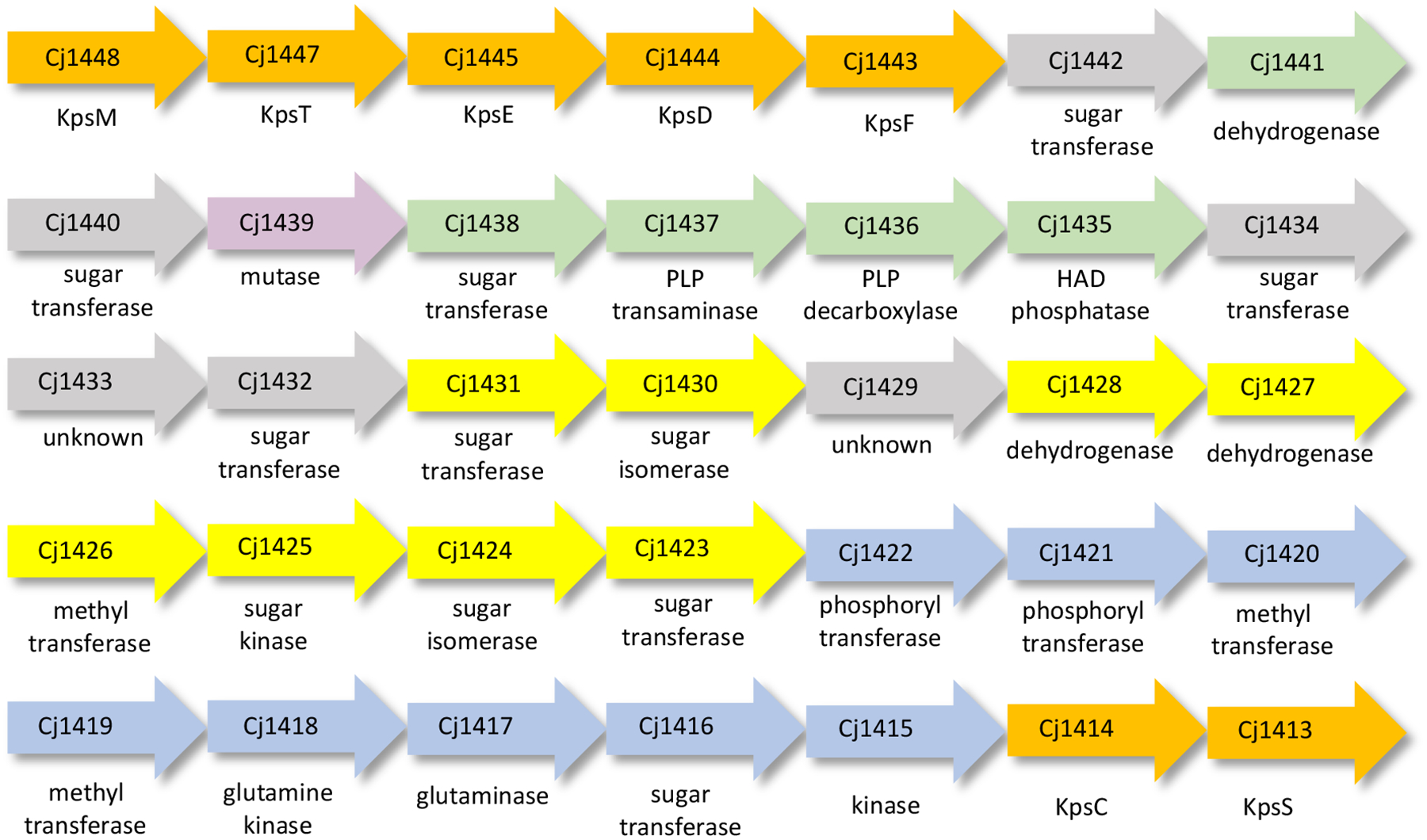
Gene cluster for biosynthesis of the CPS in C. jejuni NCTC 11168. Enzymes of unknown function are colored grey. Enzymes responsible for the biosynthesis of the glucuronamide, heptose, and galactosyl moieties are colored green, yellow, and purple, respectively. Enzymes required for the methyl phosphoramidate modification are colored blue. Enzymes necessary for the export of the capsule are colored orange.
Unfortunately, relatively little is known about the biosynthesis of the glucuronamide moiety in the CPS of the NCTC 11168 (HS:2) and NCTC 12517 (HS:19) strains of C. jejuni. In strain NCTC 11168 the glucuronamide moiety is formed from either serinol (2-amino-1,3-propanediol), or ethanolamine,7, 20 whereas in NCTC 12517 the amide bond is only formed with serinol.21 The enzymes most likely involved in the biosynthesis and formation of the glucuronamide moiety to the growing polysaccharide chain include Cj1435, Cj1436, Cj1437, Cj1438, and Cj1441 for HS:2. The genes for these five enzymes are found in the CPS gene cluster of strain NCTC 11168 and close homologues (except for Cj1436) are found in strain NCTC 12517 (HS:19).22 Cj1435 is predicted to be a haloacid dehalogenase (HAD) phosphatase, Cj1436 and Cj1437 are predicted to be pyridoxal phosphate (PLP) containing enzymes, Cj1438 is likely a glycosyltransferase, and Cj1441 is a putative UDP-glucose 6-dehydrogenase.
UDP-glucose 6-dehydrogenase from Streptococcus pyogenes catalyzes the double NAD+-dependent oxidation of UDP-d-glucose to UDP-d-glucuronic acid.23–26 The reaction mechanism has been shown to occur by the initial oxidation of UDP-d-glucose (1) to the corresponding aldehyde intermediate (2) at C6.24 The aldehyde intermediate is tightly bound and not released to solution. The second oxidation step proceeds with the addition of a cysteine thiolate to form a thiohemiacetal intermediate (3) that is followed by a second hydride transfer to form a thioester intermediate (4) that is subsequently hydrolyzed by an activated water molecule to form the ultimate product glucuronic acid (5). This transformation is summarized in Scheme 1.
Scheme 1:
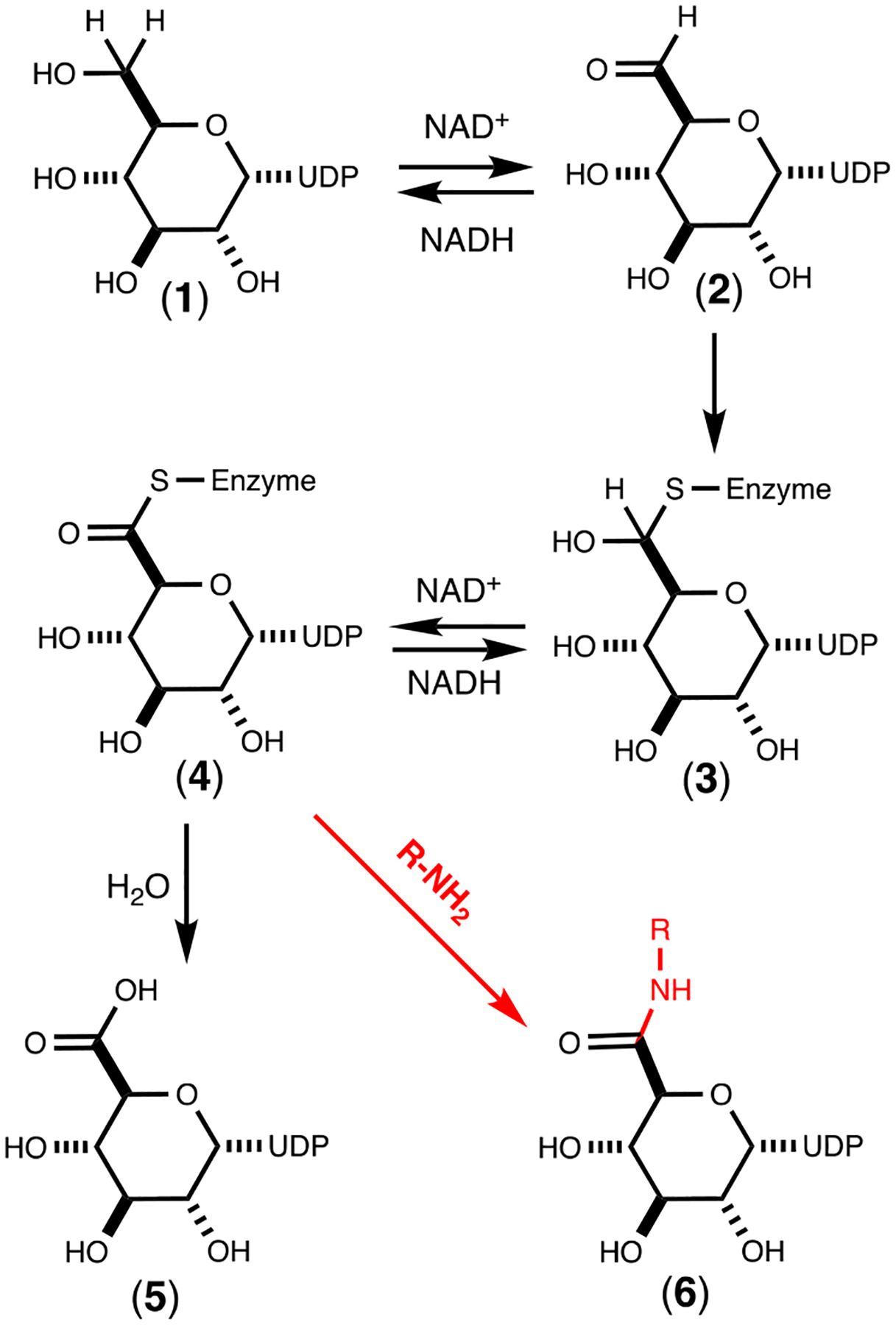
Mechanism for the reaction catalyzed by UDP-glucose 6-dehydrogenase. Shown with the red arrow is a potential mechanism for the formation of the UDP-glucuronamide (6) via nucleophilic attack of an amine with the thioester intermediate.
The source of the primary amine necessary for amide bond formation in the CPS of C. jejuni is likely controlled by the activity of the two PLP containing enzymes (Cj1437 and Cj1436). PLP can be used in a number of reaction types including transamination, decarboxylation, and racemization.27 It is hypothesized here that Cj1436 uses PLP to decarboxylate l-serine or l-serine-phosphate to form ethanolamine or ethanolamine phosphate, while Cj1437 functions to transaminate dihydroxyacetone phosphate (DHAP) to form serinol phosphate.28, 29 In the absence of an obvious amide bond forming enzyme in the putative gene cluster for the biosynthesis of the CPS of C. jejuni, one must look at alternative chemical strategies. One potential scenario is the nucleophilic attack of an amine substrate with the thioester intermediate that is postulated to occur during the reaction catalyzed by UDP-glucose 6-dehydrogenase to form the corresponding UDP-glucuronamide (6) as illustrated in Scheme 1.25, 30 This transformation could occur instead of the “normal” attack by an activated water molecule. This reaction is similar to that catalyzed by glyceraldehyde 3-phosphate dehydrogenases, where a thioester intermediate is attacked by phosphate to form 1,3-diphosphoglycerate. However, the enzyme directly responsible for amide bond formation may occur outside of the gene cluster for formation of the CPS in C. jejuni.
To test our proposal for formation of the glucuronamide moiety of the CPS from C. jejuni we have we have cloned and expressed the gene for Cj1441. The enzyme has been purified, crystallized, and the three-dimensional structure determined. The catalytic properties of the enzyme have been determined in the presence and absence of amine substrates with NAD+ and UDP-d-glucose.
Materials and Methods
Cloning, Expression, and Purification of Cj1441.
The gene for Cj1441 (UniProt id: Q0P8H3) was cloned from the genomic DNA of C. jejuni NCTC 11168 (ATCC-700819D-5). Expression tests of the cloned gene for Cj1441 exhibited low levels of expression, so the gene for Cj1441 was codon optimized and synthesized by GenScript (Piscataway, NJ). The synthesized DNA served as the starting template for PCR using Phusion DNA polymerase (New England Biolabs). For all genes, primers were designed that incorporated NdeI and XhoI restriction sites and the resulting fragments were digested at 37 °C and ligated into a pET31b expression vector, which carried a C-terminal hexahistidine tag.
The pET31b vector was used to transform BL21 Escherichia coli cells (Novagen). The cells harboring the pET31b-Cj1441 plasmid were cultured in lysogeny broth with 100 mg/L ampicillin. The cells were grown at 37 °C with shaking and induced with 1.0 mM isopropyl β-d-1-thiogalactopyranoside when the optical density reached 0.9 at 600 nm. The cells were allowed to express protein at 21 °C for 18 h after induction and then harvested by centrifugation at 15,000 rcf at 4 °C. The cell pellet was then resuspended in loading buffer (50 mM HEPES/K+,300 mM KCl, 20 mM imidazole, pH 8.0) and lysed with sonication. The sonicated cells were centrifuged at 25,000 rcf at 4 °C before the cell lysate was passed through a 0.45 μm filter. The sample was loaded onto a prepacked 5-mL HisTrap (GE Healthcare) nickel affinity column. The protein was eluted from the column using 50 mM HEPES/K+, pH 8.0, 300 mM KCl, and 250 mM imidazole over a gradient of 25 column volumes. The resulting protein was pooled and dialyzed against 10 mM HEPES/K+ pH 8.0, and 200 mM KCl. The protein was concentrated to 22 mg/mL and flash frozen using liquid nitrogen before being stored at −80 °C. Approximately 10 mg of protein was obtained per liter of cell culture.
Mass Spectral Analysis.
Samples of the reaction catalyzed by Cj1441 were tested in 50 mM ammonium bicarbonate and allowed to incubate for 2 h at 25 °C in the presence of dithiothreitol, UDP-glucose and NAD+. The samples were also tested in the presence and absence of ethanolamine phosphate, ethanolamine, serinol-phosphate or serinol. The resulting solution was filtered through an GE Healthcare Vivaspin 500 10kDa filter and the flow through was analyzed using a Thermo Scientific Q Exactive Focus mass spectrometer in ESI (negative) mode.
Reaction Stoichiometry.
To determine the stoichiometry of the Cj1441-catalyzed reaction, solutions consisting of 1.0 μM Cj1441, 2.0 mM NAD+, 1.0 mM DTT, 50 mM triethanolamine pH 8.7, and variable concentrations of UDP-glucose from 10 to 100 μM were incubated for 60 min to enable the reaction to go to completion. The concentration of the NADH was determined from the absorbance at 340 nm.
Determination of Kinetic Constants.
The kinetic constants for Cj1441 were determined by following the reduction of NAD+ to NADH at 340 nm at 25 °C with a Spectramax340 UV-visible spectrophotometer using. Assays were performed in 50 mM triethanolamine/K+, at pH 8.7, 200 mM KCl, and 1.0 mM DTT in 96-well NucC plates. The reaction was initiated with the addition of 150 nM Cj1441 to the reaction well. Kinetic constants were determined with a fixed level of NAD+ (2.0 mM) and variable levels of UDP-glucose (0.01 mM to 2.0 mM) as well as a fixed level of UDP-glucose (5.0 mM) and varying levels of NAD+ (0.05 mM to 4.0 mM). The kinetic parameters were determined by fitting the initial rates to eq. 1 using GraFit 5, where v is the initial velocity of the reaction, Et is the enzyme concentration, kcat is the turnover number, [A] is the substrate concentration, and Km is the Michaelis constant.
| (1) |
Sequence Similarity Network for COG1004.
The identifiers were used to generate a sequence similarity network using the Enzyme Function Initiative – Enzyme similarity tool and Option D.31, 32 The full network was downloaded and used to create a sequence similarity network using Cytoscape 3.7.2.33 The edges of the network were set to 45% sequence identity using the Cytoscape filter function.
Crystallization of Cj1441.
Crystallization conditions were initially surveyed by the hanging drop method of vapor diffusion using a sparse matrix screen from Wizard 3+4 purchased from Molecular Dimensions. The wild-type enzyme was tested in complex with 5.0 mM UDP-glucose and 5.0 mM NAD+ at room temperature. X-ray diffraction quality crystals appeared after two to three days and were subsequently grown by mixing, in a ratio of 1:1, the protein sample at 23 mg/mL and the crystallization buffer. The initial crystallization conditions were 30% pentaerythritol ethoxylate, 50 mM ammonium sulfate, 50 mM BIS-TRIS, at pH 6.5 and 21 °C. These preliminary crystals were tested for diffraction, before being optimized. The crystallization conditions used to grow the samples for data collection were 5.0 mM UDP-glucose, 5.0 mM NAD+, 1.0 mM DTT, 32% pentaerythritol ethoxylate, 50 mM ammonium sulfate, 50 mM BIS-TRIS, pH 6.5, and 200 mM NaCl. The crystals used for data collection were roughly 50 × 50 microns in size.
Diffraction data were collected at the Stanford Synchrotron Radiation Light Source (SSRL), Beamline 14–1, on an Eiger 16M detector at 100K. The data were integrated, scaled, and merged.34 Merged observed intensities were then imported into the Phenix reflection file editor where an Rfree set was assigned using the default settings.35 Relevant X-ray data collection statistics are listed in Table 1. The unit cell was determined to be P21 with cell edges of a = 43.79, b = 148.78, and c = 62.34 and angles of α = 90.0, β = 107.5, and γ = 90.0. The structure was determined using PHASER from the PHENIX suite and using PDB id: 1DLJ as the search model with 49% sequence identity.36, 37 The structure was built using COOT and iterative rounds of refinement using PHENIX refine in the PHENIX suite reduced the Rwork and Rfree to 19.0% and 25.6%, respectively, from 46 to 2.09 Å resolution.35, 38 Refinement statistics are listed in Table 1.
Table 1:
X-ray Data Collection Statistics and Model Refinement Statistics.
| Cj1441 with NAD+ and UDP-glucose | |
|---|---|
| resolution limits | 46.4 – 2.09 (2.14 – 2.09)b |
| Space Group | P21 |
| γ (°) | 90.00 |
| number of independent reflections | 43332 (4336) |
| completeness (%) | 96.6 (97.9) |
| redundancy | 12.8 (4.1) |
| avg I/avg σ(I) | 11.3 (1.43) |
| Rsym (%)a | 4.1 (41.5) |
| resolution limits (Å) | 46.4 – 2.09 |
| cR-factor (overall)%/no. reflections | 19.3/43332 |
| R-factor (working)%/no. reflections | 19.0/ 41328 |
| R-factor (free)%/no. reflections | 25.6/ 2004 |
| number of protein atoms | 6285 |
| number of ligands (NAD or UDP-glucose) | 4 |
| number of water molecules | 180 |
| average B values (Å2) | |
| protein atoms | 48.5 |
| ligand NAD+, UDP-glc | 38.6, 49.1 |
| solvent | 45.7 |
| weighted RMS deviations from ideality | |
| bond lengths (Å) | 0.008 |
| bond angles (°) | 1.04 |
| general planes (°) | 0.006 |
| Ramachandran regions (%) | |
| most favored | 96.5 |
| additionally allowed | 3.3 |
| generously allowed | 0.3 |
Rsym = (∑|I - |/ ∑ I) × 100.
Statistics for the highest resolution bin.
R-factor = (Σ|Fo − Fc|/Σ|Fo|) × 100 where F0 is the observed structure-factor amplitude and Fc is the calculated structure-factor amplitude.
Results
Sequence Similarity Network for UDP-glucose Dehydrogenase.
Cj1441 is a member of COG1004 and the sequence similarity network (SSN) is presented in Figure 3 at a percent identity cutoff of 45%.32 Cj1441 clusters with other known UDP-glucose 6-dehydrogenases, including those from S. pyogenes, and E. coli. The enzymes denoted as UDP-mannose dehydrogenases segregate with one another in a group that is separated from those enzymes that have been shown to catalyze the oxidation of UDP-glucose. Of the 2883 enzymes in COG1004, ~600 of them cluster in the same group as Cj1441. In this group are examples from other pathogenic bacteria, including E. coli O157:H7 and E. coli O6:H1. Cj1441 and the UDP-glucose 6-dehydrogenases from S. pyogenes and E. coli O157:H7exhibit a 47% and 51% sequence identity, respectively.
Figure 3.
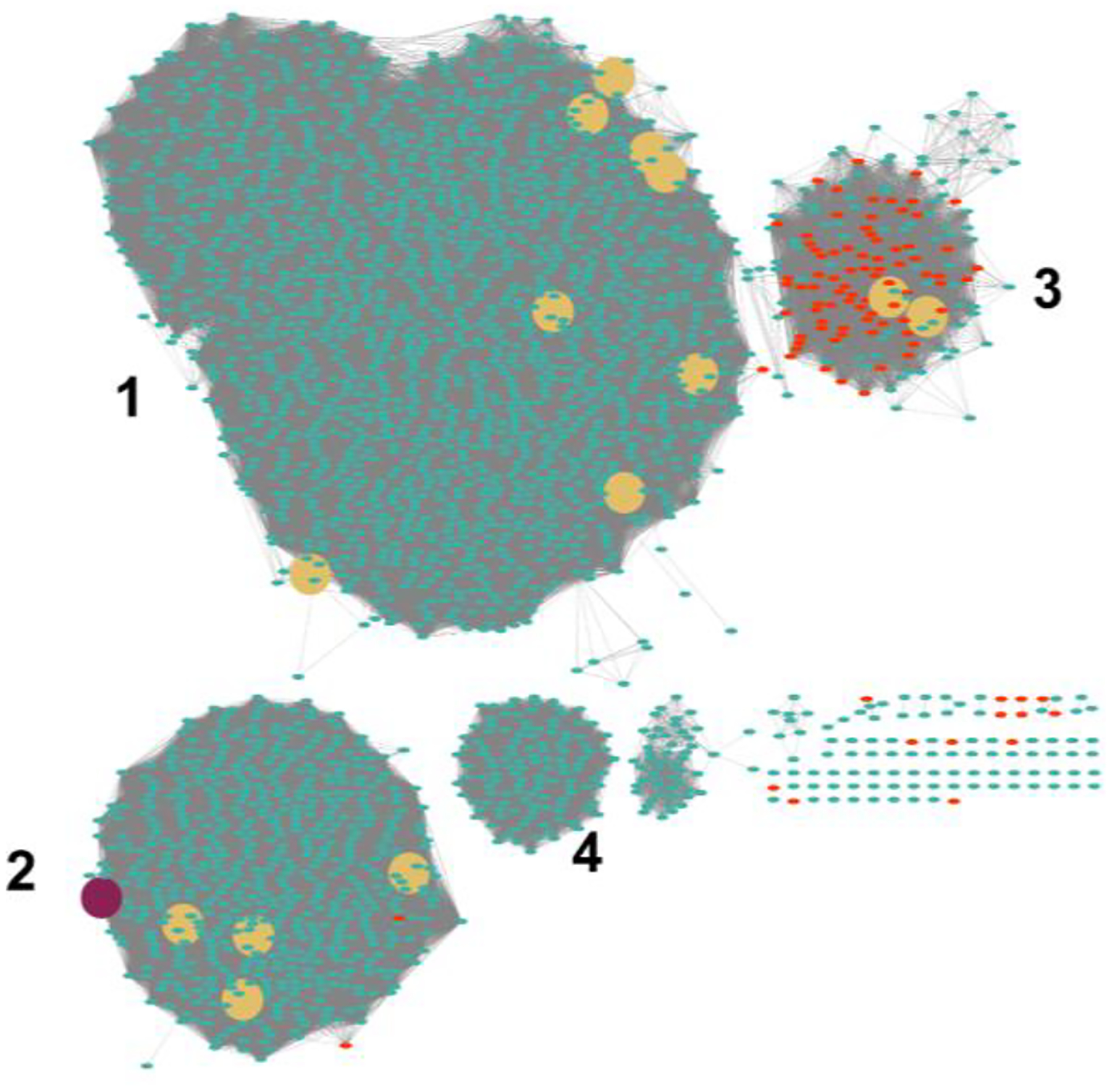
Sequence similarity network for COG1004. Each node in the network represents a single sequence and each edge (depicted as lines) represents the pairwise connection between two sequences at a sequence identify better than 45%. The maroon colored circle represents Cj1441 in Group 2. Group 2 contains the UDP-glucose 6-dehydrogenases from E. coli 0157:H7, E. coli K12, E. coli O6:H1 and S. pyogenes. The red circles are examples annotated as mannose 6-dehydrogenases. The yellow circles are functionally characterized enzymes from Uniprot.org.
Kinetic Analysis of Cj1441.
Cj1441 was shown to catalyze the reduction of NAD+ to NADH in the presence of excess UDP-glucose by following the change in absorbance at 340 nm. To measure the reaction stoichiometry, Cj1441 was mixed with excess NAD+ (2.0 mM) and variable amounts of UDP-glucose (10 – 100 μM). The reaction was followed to completion and the amount of NADH formed was determined from the change in absorbance at 340 nm. On average the ratio of NADH formed at equilibrium with the initial UDP-glucose concentration was determined to be 2:1. This value matches the predicted value from the reaction mechanism presented in Scheme 1.
The apparent kinetic constants for the oxidation of UDP-glucose by Cj1441 were determined by variation of the UDP-glucose concentration at a fixed concentration of NAD+ (5.0 mM) and then variation of the NAD+ concentration at a fixed concentration of UDP-glucose (5.0 mM). The plot for the variation of UDP-glucose is presented in Figure 4. The apparent kcat values are 0.80 to 1.1 s−1, with kcat/Km values of 4.2 × 103 M−1 s−1 and 4.8 × 103 M−1 s−1 for the variation of UDP-glucose and NAD+, respectively. To determine whether or not the addition of potential amine substrates to the reaction mixtures could alter the rate of NAD+ reduction, UDP-glucose was varied at a fixed concentration of NAD+ and the apparent kinetic constants determined for the oxidation of UDP-glucose. Within experimental error the addition of either ethanolamine, ethanolamine phosphate, serinol, or serinol phosphate had no measurable effect on the catalytic properties of Cj1441. The apparent kinetic constants are presented in Table 2.
Figure 4.
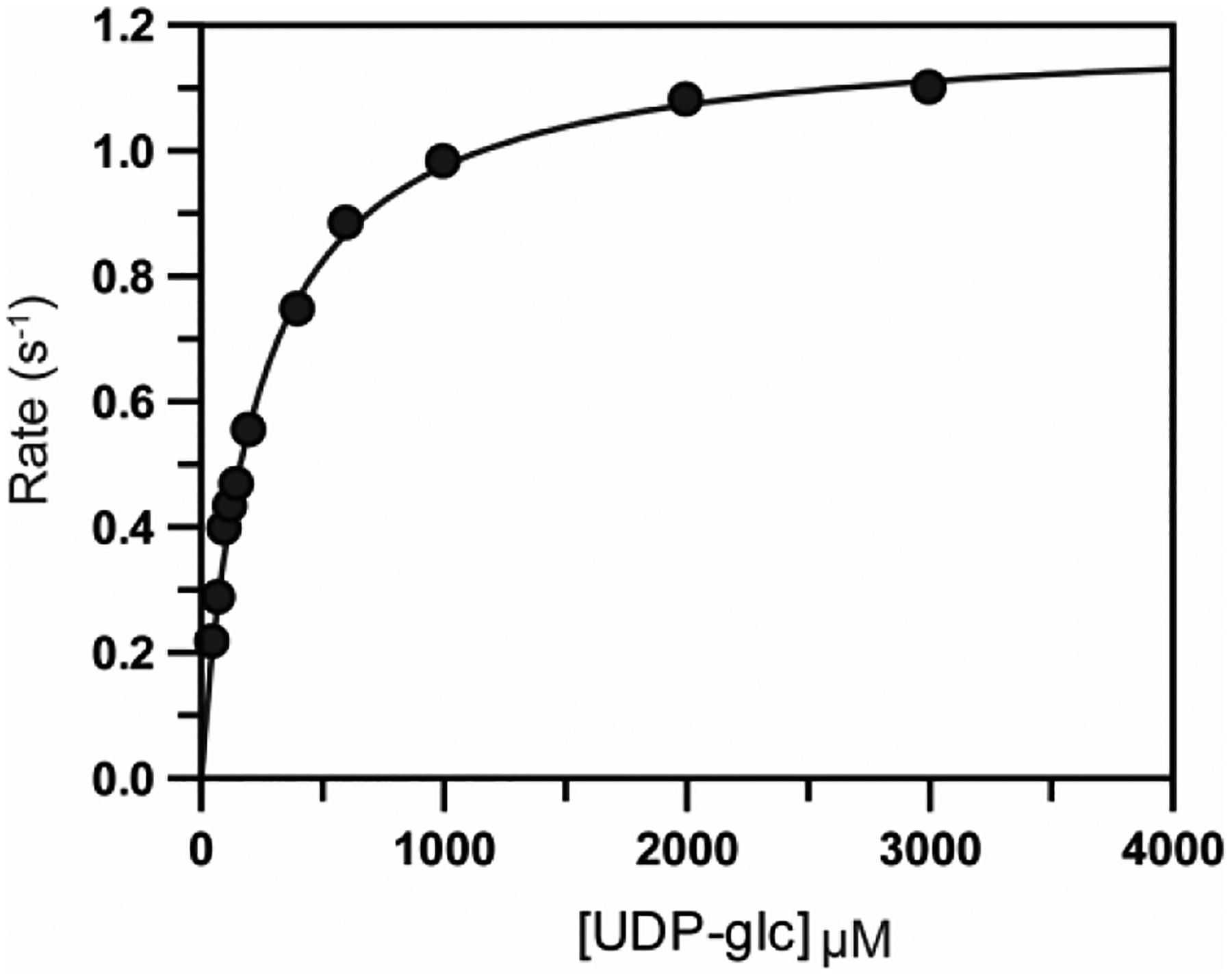
Michaelis-Menten plot for the oxidation of UDP-glucose catalyzed by Cj1441 at 25 °C. The reaction mixture contained saturating conditions of NAD+ (2.0 mM) and 1.0 mM DTT. The solid line represents the fit of the data to eq 1.
Table 2.
Steady State Kinetic Constants for Cj1441a
| fixed substrate | variable substrate (mM) |
kcat
(s−1) |
Km (μM) |
kcat/Km
(M−1 s−1) |
|---|---|---|---|---|
| NAD+
(5.0 mM) |
UDP-glucose (0.05 – 3.0) |
1.10 ± 0.02 | 210 ± 8.0 | 5200 ± 180 |
| UDP-glucose (5.0 mM) | NAD+ (0.05 – 3.0) |
0.80 ± 0.02 | 190 ± 12 | 4200 ± 290 |
| ethanolamine (2.0 mM) NAD+ (5.0 mM) |
UDP-glucose (0.05 – 3.0) |
0.90 ± 0.04 | 300 ± 14 | 3000 ± 190 |
| ethanolamine-P (2.0 mM) NAD+ (5.0 mM) |
UDP-glucose (0.05 – 3.0) |
0.80 ± 0.03 | 210 ± 11 | 3800 ± 210 |
| serinol (2.0 mM) NAD+ (5.0 mM) |
UDP-glucose (0.05 – 3.0) |
0.80 ± 0.03 | 230 ± 15 | 3500 ± 240 |
| serinol phosphate (2.0 mM) NAD+ (5.0 mM) |
UDP-glucose (0.05 – 3.0) |
0.80 ± 0.03 | 200 ± 11 | 4000 ± 220 |
pH 8.7, 25 °C
Verification of Reaction Products.
The products of the reaction catalyzed by Cj1441 were analyzed by negative ion ESI mass spectrometry. As a control, commercial UDP-glucuronic acid was shown to have an m/z value of 579.02, identical to the calculated value of 579.02. Cj1441 was subsequently incubated with NAD+, DTT, and UDP-glucose and the products were analyzed similarly with m/z values of 565.04 (UDP-glucose), 579.02 (UDP-glucuronate) and 664.11 (NADH). Amidated products were not observed in any of the mass spectra when ammonia, ethanolamine, serinol, ethanolamine phosphate, or serinol phosphate were added to the reaction mixtures (Figures S1 – S6).
Crystallization and Structure Determination of Cj1441.
Cj1441 crystallized as a dimer in the asymmetric unit and was solved by molecular replacement using UDP-glucose 6-dehydrogenase from S. pyogenes (PDB id: 1DLJ) as the search model. Cj1441 co-crystallized with UDP-glucose and NAD+ in the active site and the structure was determined to a resolution of 2.09 Å. The Cj1441 monomer contains 393 amino acids and the dimeric form of the enzyme is presented in Figure 5. The enzyme consists of two major domains, each containing an α/β fold. The N-terminal domain contains a Rossmann fold bearing the conserved GXGXXG motif between residues 7 and 12, in addition to six parallel β-strands arranged in the canonical order (3, 2, 1, 4, 5, 6). The two subunits were aligned using Pymol and they superimpose with a root mean square deviation of 0.3 Å.39 The overall buried surface area between the dimeric interface was calculated using PISA to be 2504 Å2 (http://pdbe.org/pisa). The two α/β domains of the protein are linked to one another by a 48 Å long central α-helix from residues Asp191 through Asn223. The N-terminal domain (residues 1–191) is responsible for binding the NAD+ cofactor. The C-terminal domain (residues 223–393) binds UDP-glucose using a similar α/β fold. The defining difference between the two domains of the protein is the lack of the Ω loop from residues 301–317. A structural comparison of Cj1441 with the UDP-glucose 6-dehydrogenase from S. pyogenes (PDB id: 1DLJ) is shown in Figure S7.
Figure 5.
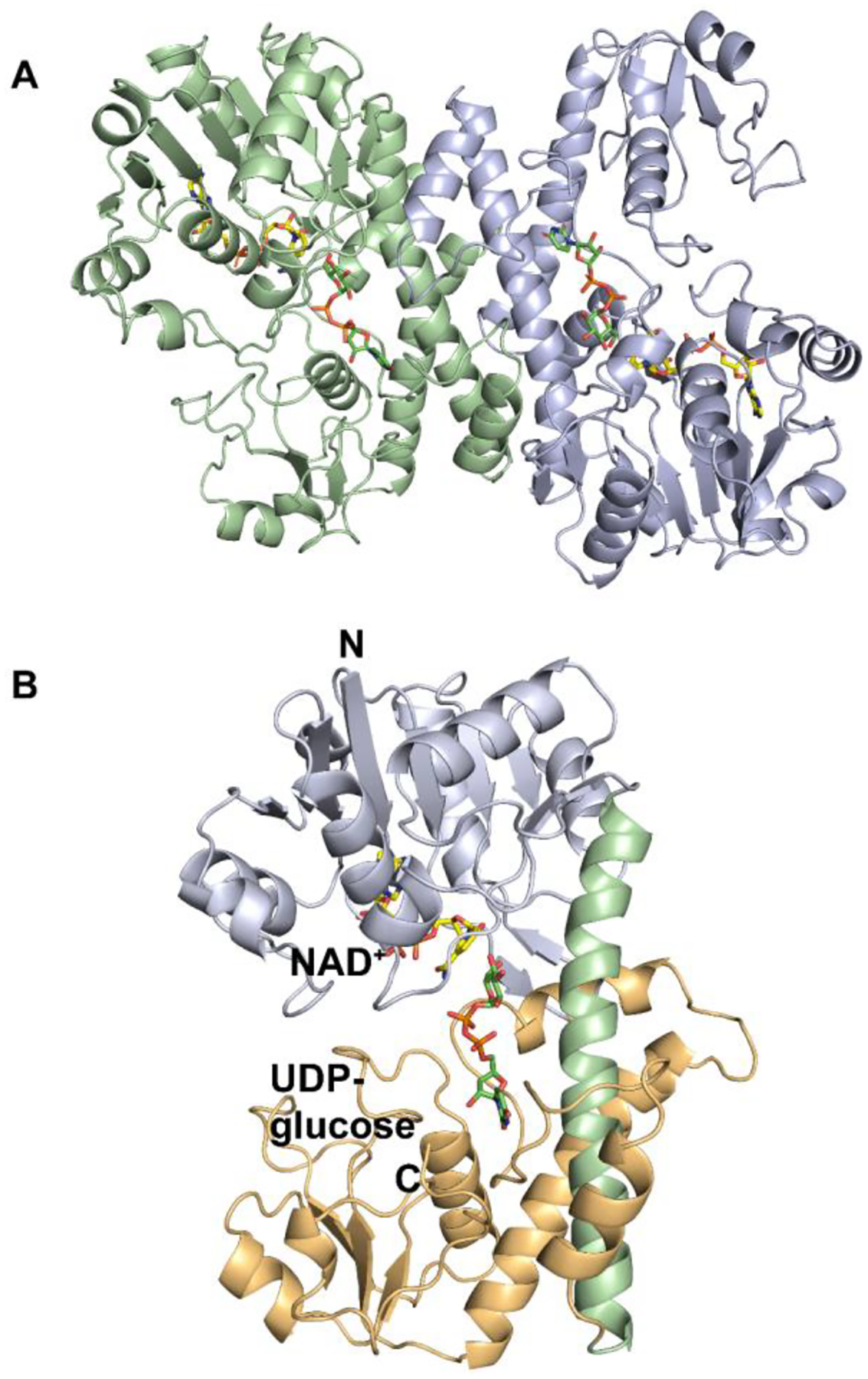
(A) Ribbon diagram of Cj1441 homodimer. Chain A is shown in pale green, and Chain B is shown in pale blue. UDP-glucose is depicted in green and NAD+ is displayed in yellow. (B) Ribbon diagram of Cj1441 monomer where the N-terminal domain is pale blue and the C-terminal domain is shown in tan. The alpha helix at the dimer interface is shown in light green.
Binding Site for NAD+.
The N-terminal domain of the protein is primarily responsible for binding the NAD+ cofactor. The binding site for NAD+ is marked by the β1-α1 turn of the Rossmann fold that directly interacts with the adenine ribose. With the exception of a small portion of the adenine ring, which is buried in the interior of the protein, the vast majority of the cofactor is exposed to solvent. This may enable the cofactor to be readily exchangeable, which is consistent with the two-fold oxidation mechanism.40 Additionally, the nicotinamide ring is in the syn conformation, as determined previously for the enzyme from S. pyogenes (PDB id: 1DLI) and Klebsiella pneumoniae (PDB id: 3PLR).37, 40 The residues directly responsible for binding the coenzyme are Val11, Gly12, Asp31, Lys36, Ala59, Thr85, Thr120, and Arg316. Additionally, four ordered waters interact with the cofactor to help orient and bind the cofactor to the protein surface. UDP-glucose points directly toward the si face of the nicotinamide ring. There are six residues that are apparently conserved across all UDP-glucose 6-dehydrogenases involved in NAD+ binding.41 These include the three glycine residues from the Rossmann fold, a Thr83/Pro84 pair which interacts with the adenine ring, and Arg316, which forms a salt bridge with the pyrophosphate of the nucleotide (Figure 6a).
Figure 6.
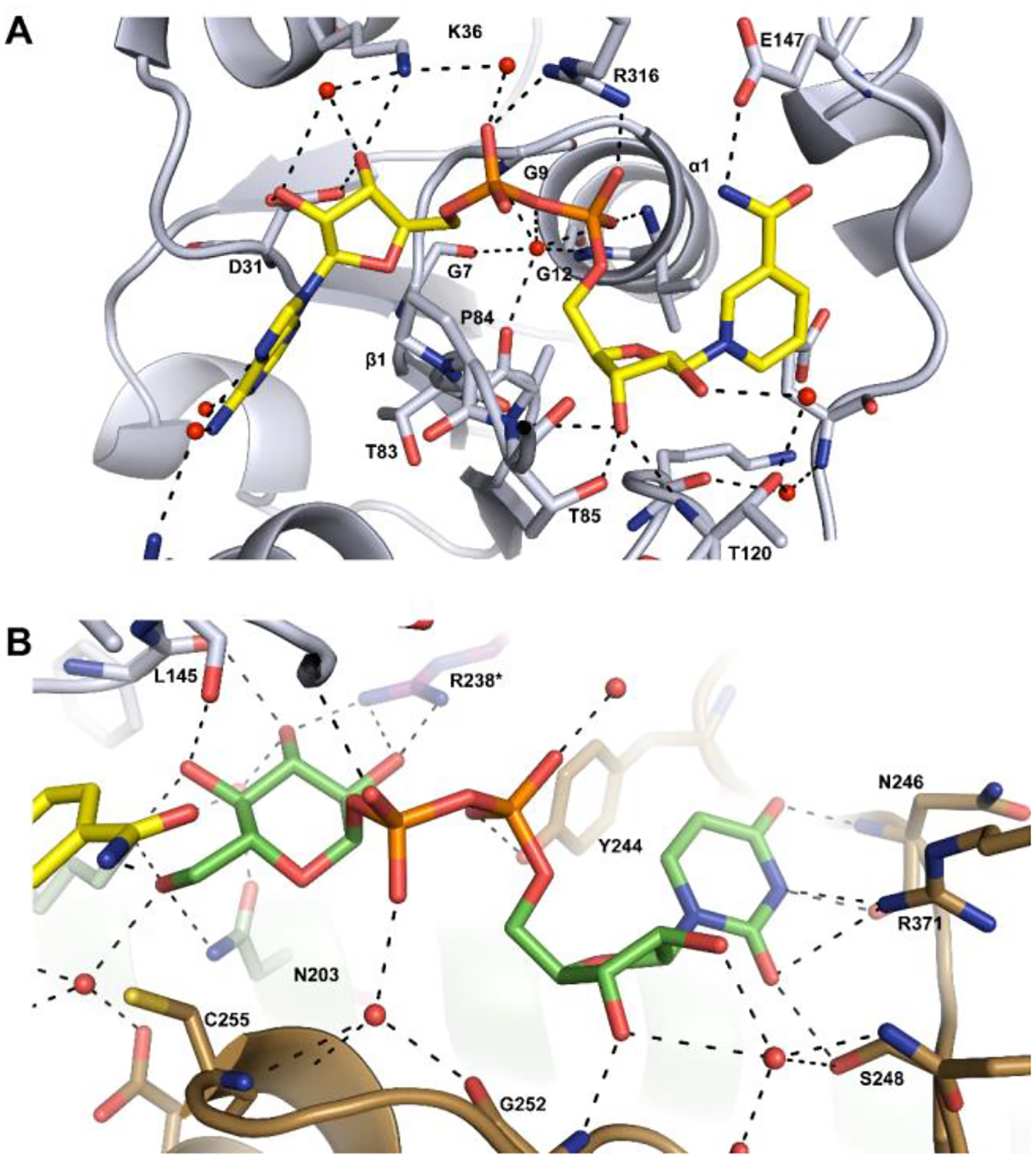
Cartoon representations of the binding sites for NAD+ and UDP-glucose. Potential hydrogen bonding interactions (<3.2 Å) are shown in black. (A) Binding site for NAD+ (colored yellow) in the N-terminal domain of the protein. (B) Binding site for UDP-glucose (colored green) in the C-terminal domain of the protein. Additional details are provided in the text.
Binding of UDP-glucose.
UDP-glucose primarily interacts with the C-terminal domain of Cj1441. The protein crystallized at pH 6.5 which explains, in part, why UDP-glucose, rather than UDP-glucuronate, is present in the active site since the reaction equilibrium favors the substrates at low pH (Figure S8). Cys255 is located 3.6 Å from C6 of UDP-glucose, which is sufficient to facilitate nucleophilic attack with the aldehyde intermediate and formation of a thioester intermediate during the catalytic cycle (Scheme 1).25 Additionally, C4 of the nicotinamide ring is positioned 3.2 Å from C6 of UDP-glucose to facilitate hydride transfer (Figure 7). UDP-glucose is held in place by interactions with Asn203, Leu145, Lys199, Glu147, Tyr244, Gly252, Asn246, Ser248, and Arg371. Arg238 from the adjacent subunit is found on a flexible loop near the dimeric interface and is capable of interacting with the C2’ and C3’ hydroxyl groups of the UDP-glucose (Figure 6b).
Figure 7.
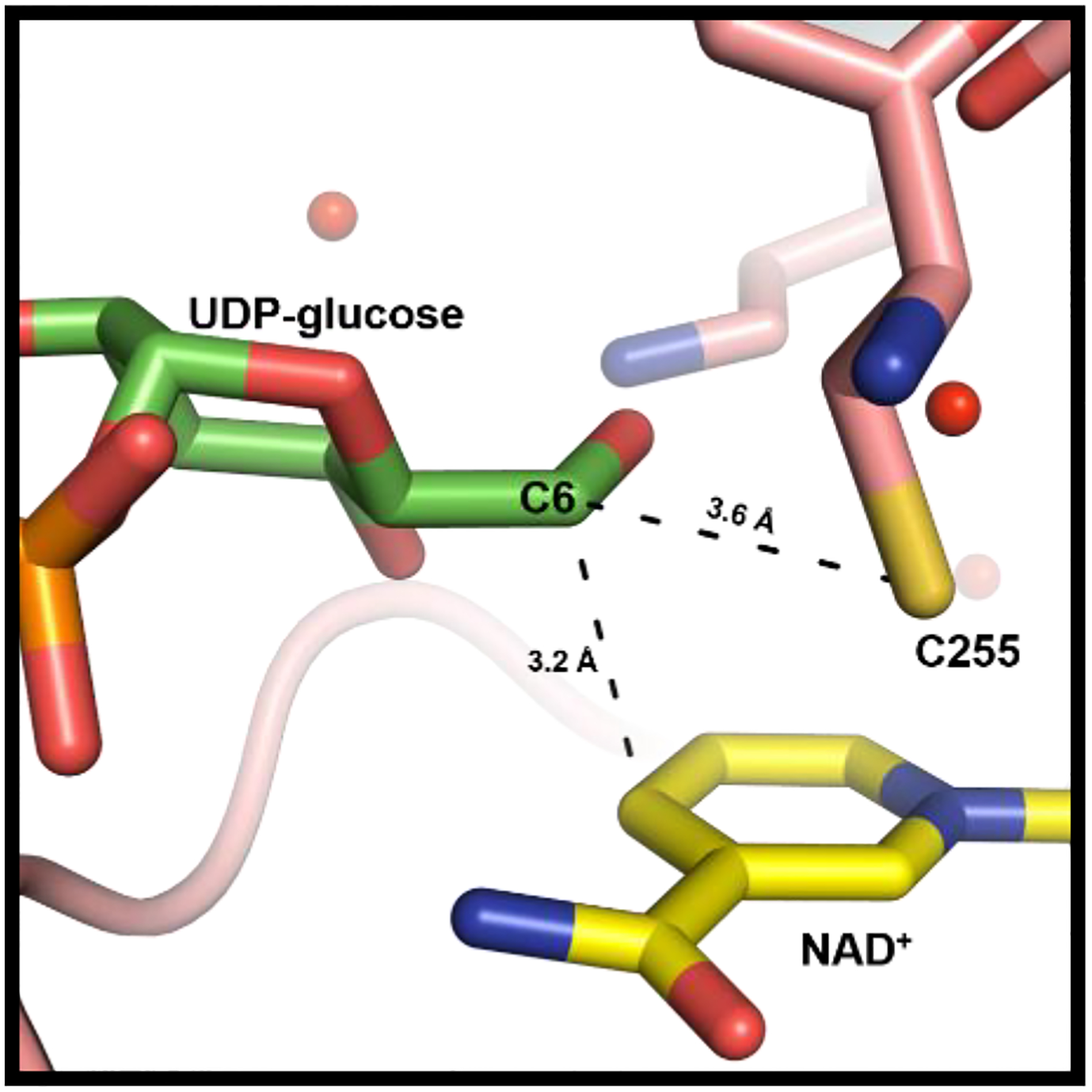
Ligand interactions between NAD+ and UDP-glucose in the active site of Cj1441. The nicotinamide portion of NAD+ is presented in yellow and the glucose portion of UDP-glucose is shown in green. The catalytic cysteine is 3.6 Å from C6 of the glucose moiety. NAD+ is 3.2 Å from C6 of the glucose moiety of the substrate.
Binding of the uridine nucleotide moiety of UDP-glucose is facilitated by the C-terminal domain through backbone interactions with residues 244–252. Of the 6 hydrogen bonds formed with the UMP portion of the substrate, only two of them are from amino acid side chains (Ser248, and Tyr244). The rest of the interactions are made through hydrogen bond interactions with the carbonyl oxygens and amide nitrogens of the protein backbone. The glucose 1-phosphate binding region is rather compact compared to the UMP binding region, consisting of only 4 residues (Phe144-Glu147). There is an additional hydrogen bond provided by Asn203 on the central helix.
Other Amide Bond Forming Enzymes.
A bioinformatic search was conducted in an attempt to find additional candidates for the amide bond forming enzyme within the CPS of C. jejuni. The initial probe focused on the LOS region of the C. jejuni genome. Specifically, we searched for regions of genes that might contain a ligase domain that would be capable of catalyzing amide bond formation. Generally, these bonds are formed through the use of an ATP-grasp domain, so our investigation focused on finding genes that contained such a region. It was subsequently determined that the LOS of E. coli L19 and Shigella boydii type 8 contained a glucuronamide moiety with serinol.42–44 The appropriate gene clusters within these organisms contained a phosphatase (WfdQ), a PLP-dependent transaminase (WfdP), and a putative transferase with an ATP-grasp domain (WfdR). This discovery led to the interrogation of the sugar transferases of unknown specificities in the capsular polysaccharide cluster of C. jejuni. We identified a putative glycosyltransferase (Cj1438) in the gene cluster of C. jejuni NCTC 11168 that also contains an ATP-grasp domain at the C-terminal half of the protein. This enzyme has all of the necessary residues to bind ATP when compared to other ATP-grasp enzymes, including those homologs in E. coli L19 and S. boydii type 8.
DISCUSSION
The putative UDP-glucose 6-dehydrogenase (Cj1441) from C. jejuni NCTC 11168 was successfully purified to homogeneity after recombinant expression in E. coli. We demonstrated that the enzyme requires two equivalents of NAD+ and that the ultimate product is UDP-d-glucuronate (5). The enzyme was crystallized in the presence of UDP-d-glucose and NAD+, and the three-dimensional structure subsequently determined to a resolution of 2.09 Å. However, attempts to demonstrate amide bond formation through nucleophilic attack of added amines on the proposed thioester intermediate (4) were unsuccessful (Scheme 1). Therefore, it is highly unlikely that this enzyme is directly responsible for formation of the glucuronamide moiety found in the CPS of C. jejuni NCTC 11168 (Figure 1). However, it remains possible that amines other than the ones tested are required for amide bond formation.
How then is the glucuronamide moiety actually synthesized for the HS:2 capsule? A cursory look at the annotations of the enzymes derived from the C. jejuni NCTC 11168 gene cluster (Figure 2) for the biosynthesis of the CPS does not identify any obvious candidates. However, we have previously demonstrated that Cj1152 can be used for the biosynthesis of GDP-d-glycero-d-manno-heptose in C. jejuni NCTC 11168 and thus genes outside of this specific gene cluster are required for the biosynthesis of the CPS.16 In this specific case, Cj1152 catalyzes the hydrolysis of phosphate from d-glycero-α-d-manno-heptose-1,7-bisphosphate for the biosynthesis of lipooligosaccharides (LOS) in C. jejuni.45 However, no amide bonds are formed in the LOS of C. jejuni and thus the missing enzyme is not to be found there.
The aminoglycerol modification within the CPS of C. jejuni NCTC 11168 (HS:2) is, however, also found in other structurally elucidated polysaccharide capsules including those from C. jejuni NCTC 12517 (HS:19),46, 47 V. cholerae H1122,48 E. coli O143, S. boydii type 8,42, 43 and E. coli L19.44 The capsule from C. jejuni NCTC 12517 is presented in Figure 8. Comparison of the gene clusters for the biosynthesis of the capsular polysaccharides in C. jejuni NCTC 11168 (HS:2) and C. jejuni NCTC 12517 (HS:19) highlights the production of enzymes of nearly identical function (Table 3). These proteins include the eight enzymes known to be responsible for the O-methyl phosphoramidate decoration of the capsule [Cj1415 through Cj1421 in C. jejuni NCTC 11168 (HS:2) and HS19.01 through HS19.07 in C. jejuni NCTC 12517 (HS:19)].9–13, 49, 50 Except for HS19.07 and Cj1421/Cj1422, the sequence identities for these proteins are ≥97%. The other relevant homologous enzymes include Cj1435 (HAD phosphatase), Cj1437 (PLP-dependent amino transferase) and Cj1441 (UDP-glucose 6-dehydrogenase) where the amino acid sequence identity is >55%. The two putative sugar transferases (Cj1438 and Cj1434) are less similar to their apparent counterparts for assembly of the HS19 capsule (34 and 21% identical to HS19.11 and HS19.08, respectively). This is quite reasonable since the acceptor/donor pairs are not identical in the two capsules.
Figure 8:
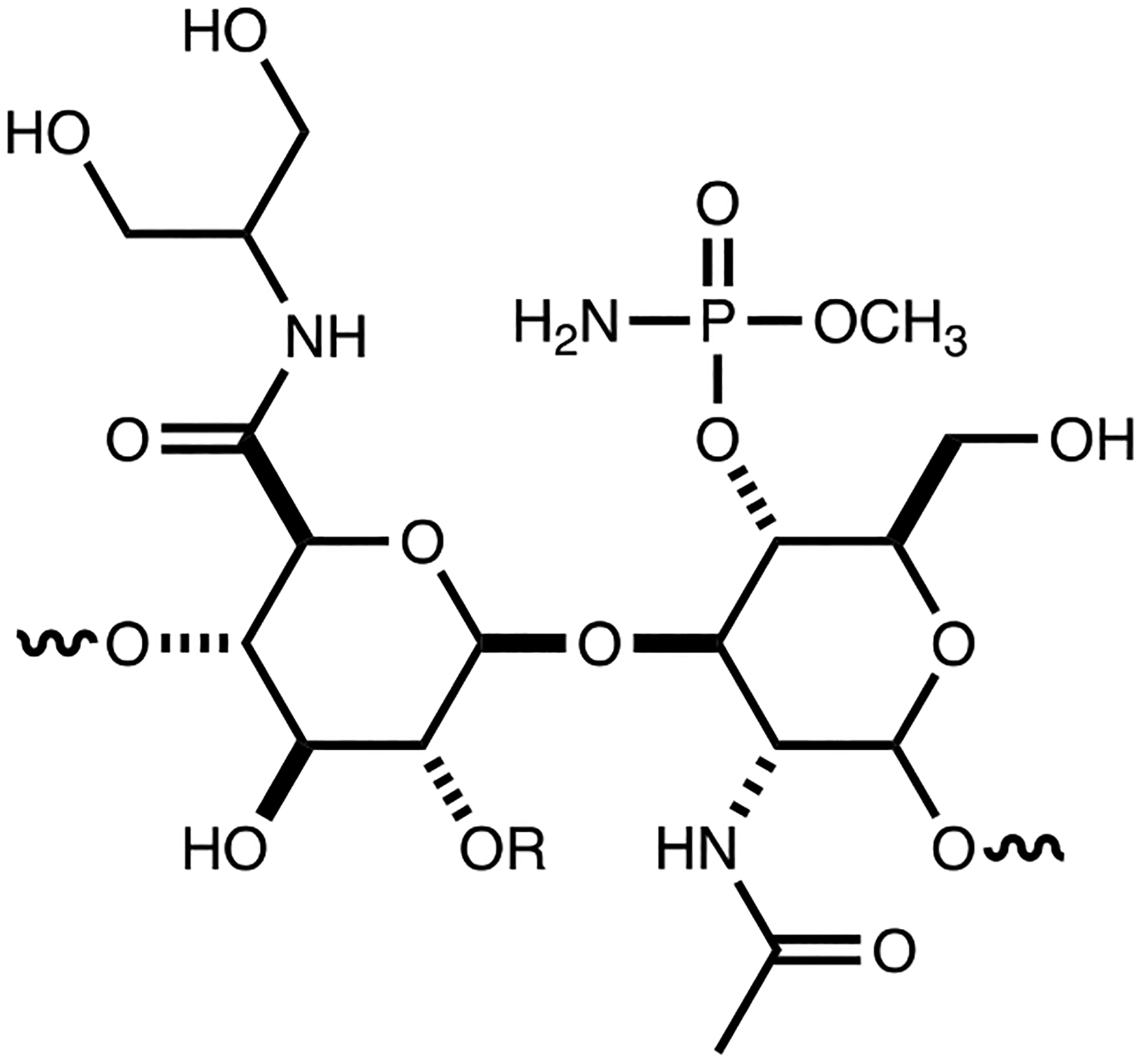
Structure of the repeating unit for the capsular polysaccharide from C. jejuni NCTC 12517 (HS:19). C2 of the glucuronamide moiety can be derivatized with l-sorbose.
Table 3:
Sequence identities in similar proteins for assembly of CPS in HS:2 and HS:19.
| C. jejuni NCTC 11168 (HS:2) | C. jejuni NCTC 12517 (HS:19) | ||||
|---|---|---|---|---|---|
| Protein | Uniprot | function | Protein | Uniprot | sequence identity |
| Cj1415 | Q0P8J9 | kinase | HS19.01 | Q6EF72 | 98 |
| Cj1416 | Q0P8J8 | transferase | HS19.02 | Q5M6N1 | 98 |
| Cj1417 | Q0P8J7 | hydrolase | HS19.03 | Q5M6N0 | 97 |
| Cj1418 | Q0P8J6 | glutamine kinase | HS19.04 | Q5M6M9 | 98 |
| Cj1419 | Q0P8J5 | methyl transferase | HS19.05 | Q5M6M8 | 98 |
| Cj1420 | Q0P8J4 | methyl transferase | HS19.06 | Q5M6M7 | 98 |
| Cj1421 | Q0P8J3 | phosphotransferase | HS19.07 | Q5M6M6 | 47 |
| Cj1422 | Q0P8J2 | phosphotransferase | HS19.07 | Q5M6M6 | 49 |
| Cj1434 | Q0P8I0 | sugar transferase | HS19.08 | Q5M6M5 | 21 |
| Cj1435 | Q0P8H9 | phosphatase | HS19.09 | Q5M6M4 | 65 |
| Cj1437 | Q0P8H7 | aminotransferase | HS19.10 | Q5M6M3 | 55 |
| Cj1438 | Q0P8H6 | sugar transferase | HS19.11 | Q5M6M2 | 36 |
| Cj1441 | Q0P8H3 | dehydrogenase | HS19.12 | Q5M6M1 | 56 |
On further inspection the putative sugar transferase (Cj1438) is particularly interesting because the N-terminal half of this protein is homologous to a large number of other functionally characterized GT2 family of sugar transferases,51–54 whereas the C-terminal half of the enzyme shows significant similarity to TupA.55 TupA is an enzyme from the ATP-grasp family that is important for amide bond formation during the synthesis of the teichuronopeptide where an amide bond is formed between glucuronate and polyglutamate.56 This observation suggests that the C-terminal half of Cj1438 harbors the catalytic machinery for amide bond formation with glucuronate. This conclusion is also supported by the high sequence identity between the C-terminal half of Cj1438 (residues 420 – 776) with the C-terminal half of HS19.11 (residues 471 – 832) and the TupA-like sequence found in the gene cluster for capsule formation in E. coli O143 (WfdR). The residues responsible for the binding of ATP are clearly conserved (Figure S9).
Our proposal for the assembly of the glucuronamide moiety of the HS2 capsule in C. jejuni NCTC 11168 utilizes the catalytic function of Cj1437 as a PLP-dependent aminotransferase to convert dihydroxyacetone phosphate to serinol phosphate. Cj1435 functions to hydrolyze the phosphorylated product to serinol, and then Cj1438 catalyzes the ATP-dependent amide bond formation between UDP-glucuronate and serinol. However, the exact order of these chemical transformations is clearly not known at this time. It is quite possible that amide bond formation does not occur until after the glucuronate has been incorporated into the growing polysaccharide and the phosphatase does not have to function until after amide bond formation. One possibility is highlighted in Scheme 2.
Scheme 2:
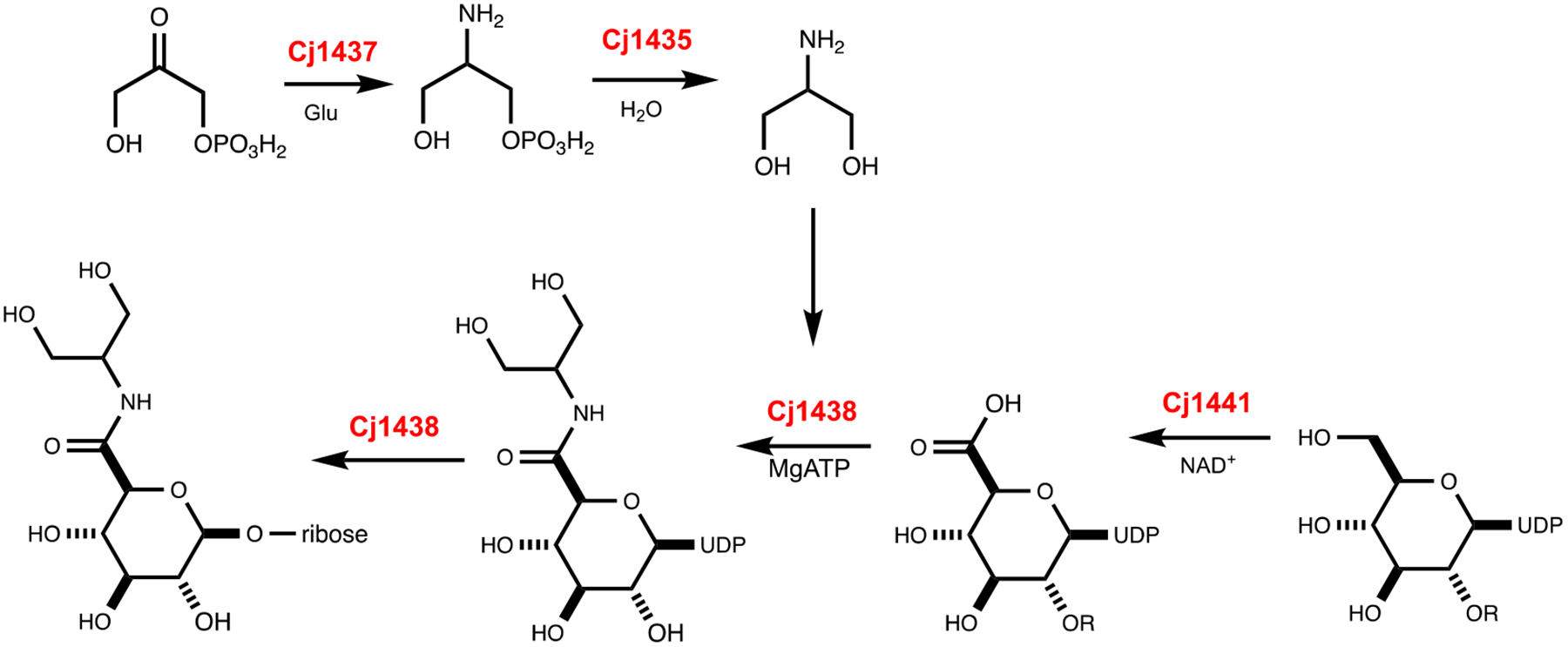
Proposed reactions for the catalytic activities of Cj1435, Cj1437, Cj1438, and Cj1441.
CONCLUSIONS
The putative UDP-glucose 6-dehydrogenase (Cj1441) from C. jejuni NCTC 11168 was purified to homogeneity and shown to catalyze the oxidation of UDP-d-glucose to UDP-d-glucuronate in the presence of NAD+. The enzyme does not facilitate the nucleophilic attack of added amines with the proposed thioester intermediate and thus this enzyme is not responsible for formation of the glucuronamide moiety in the HS:2 capsular polysaccharide. The enzyme was crystallized in the presence of NAD+ and UDP-glucose, and the three-dimensional structure determined to a resolution of 2.09 Å. The positioning of the hydroxyl group attached to C6 of UDP-glucose is ideal for hydride transfer to C4 of the bound NAD+. A bioinformatic analysis of the respective gene clusters for the biosynthesis of the HS:2 and HS:19 capsular polysaccharides in C. jejuni suggests that the catalytic activity for amide bond formation for assembly of the glucuronamide moiety resides in the C-terminal domain of Cj1438 from C. jejuni NCTC 11168.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Seth Cory and the Barondeau laboratory at Texas A&M for their help with data processing issues. Use of the Stanford Synchrotron Radiation Lightsource (SSRL), SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC-2-76SF00515.
Funding
This work was supported in part by grants from the National Institutes of Health (GM 122825) and the Robert A. Welch Foundation (A-840).
Footnotes
The authors declare no competing financial interests.
Supporting Information
Mass spectra of reaction products; structural comparison of Cj1441 with UDP-glucose 6-dehydrogenase from S. pyogenes; electron density for UDP-glucose in active site of Cj1441; and sequence alignment between Cj1438 and WfdR from E. coli O143.
Accession Codes
Cj1441, UniProt id: Q0PH3
PDB id: 7KWS
References
- (1).Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, and Griffin PM (2011). Foodborne illness acquired in the United States--major pathogens. Emerg. Infect. Dis 17, 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Burnham PM, and Hendrixson DR (2018). Campylobacter jejuni: collective components promoting a successful enteric lifestyle. Nat. Rev. Microbiol 16, 551–565. [DOI] [PubMed] [Google Scholar]
- (3).Nachamkin I, Allos BM, and Ho T (1998) Campylobacter species and Guillain-Barré syndrome. Clinical Microbiology Reviews 11, 555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Yuki N (1997) Molecular mimicry between gangliosides and lipopolysaccharides of Campylobacter jejuni isolated from patients with Guillain-Barré Syndrome and Miller Fisher Syndrome. The Journal of Infectious Diseases 176, S150–S153. [DOI] [PubMed] [Google Scholar]
- (5).van Alphen LB, Wenzel CQ, Richards MR, Fodor C, Ashmus RA, Stahl M, Karlyshev AV, Wren BW, Stintzi A, Miller WG, Lowary TL, and Szymanski CM (2014) Biological roles of the O-methyl phosphoramidate capsule modification in Campylobacter jejuni. PLoS One 9, e87051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).McNally DJ, Lamoureux MP, Karlyshev AV, Fiori LM, Li J, Thacker G, Coleman RA, Khieu NH, Wren BW, Brisson J-R, Jarrell HC, and Szymanski CM (2007) Commonality and biosynthesis of the O-methyl phosphoramidate capsule modification in Campylobacter jejuni. J. Biol. Chem 282, 28566-28576. [DOI] [PubMed] [Google Scholar]
- (7).St Michael F, Szymanski CM, Li J, Chan KH, Khieu NH, Larocque S, Wakarchuk WW, Brisson JR, and Monteiro MA (2002) The structures of the lipooligosaccharide and capsule polysaccharide of Campylobacter jejuni genome sequenced strain NCTC 11168. European Journal of Biochemistry 269, 5119–5136. [DOI] [PubMed] [Google Scholar]
- (8).Young KT, Davis LM, and Dirita VJ (2007) Campylobacter jejuni: molecular biology and pathogenesis. Nature Reviews. Microbiology 5, 665–679. [DOI] [PubMed] [Google Scholar]
- (9).Taylor ZW, Brown HA, Holden HM, and Raushel FM (2017) Biosynthesis of nucleoside diphosphoramidates in Campylobacter jejuni. Biochemistry 56, 6079–6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Taylor ZW, Chamberlain AR, and Raushel FM (2018) Substrate specificity and chemical mechanism for the reaction catalyzed by glutamine kinase. Biochemistry 57, 5447–5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Taylor ZW, and Raushel FM (2019) Manganese-induced substrate promiscuity in the reaction catalyzed by phosphoglutamine cytidylyltransferase from Campylobacter jejuni. Biochemistry 58, 2144–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Taylor ZW, and Raushel FM (2018) Cytidine diphosphoramidate kinase: an enzyme required for the biosynthesis of the O-methyl phosphoramidate modification in the capsular polysaccharides of Campylobacter jejuni. Biochemistry 57, 2238–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Taylor ZW, Brown HA, Narindoshvili T, Wenzel CQ, Szymanski CM, Holden HM, and Raushel FM (2017) Discovery of a glutamine kinase required for the biosynthesis of the O-methyl phosphoramidate modifications found in the capsular polysaccharides of Campylobacter jejuni. J. Am. Chem. Soc, 139, 9463–9466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Huddleston JP, Anderson TK, Spencer KD, Thoden JB, Raushel FM, and Holden HM (2020) Structural analysis of Cj1427, an essential NAD-dependent dehydrogenase for the biosynthesis of the heptose residues in the capsular polysaccharides of Campylobacter jejuni. Biochemistry, 59, 1314–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Huddleston JP, and Raushel FM (2020) Functional characterization of Cj1427, a unique ping-pong dehydrogenase responsible for the oxidation of GDP-d-glycero-α-d-manno-heptose in Campylobacter jejuni. Biochemistry, 59, 1328–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Huddleston JP, and Raushel FM (2019) Biosynthesis of GDP-d-glycero-α-d-manno-heptose for the capsular polysaccharide of Campylobacter jejuni. Biochemistry, 58, 3893–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).McCallum M, Shaw GS, and Creuzenet C (2013) Comparison of predicted epimerases and reductases of the Campylobacter jejuni d-altro- and l-gluco-heptose synthesis pathways. J. Biol. Chem 288, 19569–19580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Karlyshev AV, Linton D, Gregson NA, Lastovica AJ, and Wren BW (2000) Genetic and biochemical evidence of a Campylobacter jejuni capsular polysaccharide that accounts for Penner serotype specificity. Mol. Micro 35, 529–541. [DOI] [PubMed] [Google Scholar]
- (19).Sternberg MJ, Tamaddoni-Nezhad A, Lesk VI, Kay E, Hitchen PG, Cootes A, van Alphen LB, Lamoureux MP, Jarrell HC, Rawlings CJ, Soo EC, Szymanski CM, Dell A, Wren BW, and Muggleton SH (2013) Gene function hypotheses for the Campylobacter jejuni glycome generated by a logic-based approach. J. Mol. Biol 425, 186–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Szymanski CM, Michael FS, Jarrell HC, Li J, Gilbert M, Larocque S, Vinogradov E, and Brisson JR (2003) Detection of conserved N-linked glycans and phase-variable lipooligosaccharides and capsules from campylobacter cells by mass spectrometry and high-resolution magic angle spinning NMR spectroscopy. J. Biol. Chem 278, 24509–24520. [DOI] [PubMed] [Google Scholar]
- (21).McNally DJ, Jarrell HC , Khieu NH, Li J, Vinogradov E, Whitfield DM, Szymanski CM, and Brisson JR (2006) The HS:19 serostrain of Campylobacter jejuni has a hyaluronic acid-type capsular polysaccharide with a nonstoichiometric sorbose branch and O-methyl phosphoramidate group. FEBS J. 273, 3975–3989. [DOI] [PubMed] [Google Scholar]
- (22).Parker CT, Huynh S, and Heikema AP (2016) Complete genomic sequence of Campylobacter jejuni subsp. jejuni HS:19 Strain RM1285 isolated from packaged chicken. Genome Announcements, 4, e01100–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Campbell RE, Sala RF, van de Rijn I, and Tanner ME (1997) Properties and kinetic analysis of UDP-glucose dehydrogenase from group A streptococci. Irreversible inhibition by UDP-chloroacetol. J. Biol. Chem 272, 3416–3422. [DOI] [PubMed] [Google Scholar]
- (24).Campbell RE and Tanner ME (1997) Uridine diphospho-α-d-gluco-hexodialdose: Synthesis and kinetic competence in the reaction catalyzed by UDP-glucose dehydrogenase. Angew. Chem. Int. Ed. Engl 36, 1520–1522. [Google Scholar]
- (25).Ge X, Campbell RE, van de Rijn I, and Tanner ME (1998) Covalent adduct formation with a mutated enzyme: Evidence for a thioester intermediate in the reaction catalyzed by UDP-glucose dehydrogenase. J. Am. Chem. Soc 120, 6613–6614. [Google Scholar]
- (26).Campbell RE, and Tanner ME (1999). UDP-glucose analogues as inhibitors and mechanistic probes of UDP-glucose dehydrogenase. J. Org. Chem 64, 9487–9492. [Google Scholar]
- (27).Eliot AC, and Kirsch JF (2004) Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations. Annual Review of Biochemistry 73, 383–415. [DOI] [PubMed] [Google Scholar]
- (28).Keller S, Wetterhorn KM, Vecellio A, Seeger M, Rayment I, & Schubert T (2019). Structural and functional analysis of an l-serine O-phosphate decarboxylase involved in norcobamide biosynthesis. FEBS Letters, 593, 3040–3053. [DOI] [PubMed] [Google Scholar]
- (29).Andreeßen Björn, and Steinbüchel A (2011) Serinol: small molecule – big impact. AMB Express 1:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Egger S, Chaikuad A, Klimacek M, Kavanagh KL, and Oppermann U (2012). Structural and kinetic evidence that catalytic reaction of human UDP-glucose 6-dehydrogenase involves covalent thiohemiacetal and thioester enzyme intermediates. J. Biol. Chem, 287, 2119–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Gerlt JA, Bouvier JT, Davidson DB, Imker HJ, Sadkhin B, Slater DR, and Whalen KL (2015) Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): A web tool for generating protein sequence similarity networks. Biochim. Biophys. Acta, Proteins Proteomics 1854, 1019-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Atkinson HJ, Morris JH, Ferrin TE, and Babbitt PC (2009) Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS One 4, e4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, and Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Battye TG, Kontogiannis L, Johnson O, Powell HR, and Leslie AG (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta crystallographica. Section D, Biological crystallography 67, 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, and Zwart PH (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. Section D, Biological crystallography, 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007) Phaser crystallographic software. Journal of Applied Crystallography, 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Campbell RE, Mosimann SC, van De Rijn I, Tanner ME, and Strynadka NC (2000) The first structure of UDP-glucose dehydrogenase reveals the catalytic residues necessary for the two-fold oxidation. Biochemistry, 39, 7012–7023. [PubMed] [Google Scholar]
- (38).Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010) Features and development of Coot. Acta crystallographica. Section D, Biological Ccrystallography, 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).DeLano WL (2002) The PyMOL Molecular Graphics System. DeLano Scientific, San Carlos, CA, USA, The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA, USA. [Google Scholar]
- (40).Chen YY, Ko TP, Lin CH, Chen WH, and Wang AH (2011). Conformational change upon product binding to Klebsiella pneumoniae UDP-glucose dehydrogenase: a possible inhibition mechanism for the key enzyme in polymyxin resistance. Journal of Structural Biology 175, 300–310. [DOI] [PubMed] [Google Scholar]
- (41).Egger S, Chaikuad A, Kavanagh KL, Oppermann U, and Nidetzky B (2011). Structure and mechanism of human UDP-glucose 6-dehydrogenase. J. Biol. Chem, 286, 23877–23887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).L’vov VL, Tokhtamysheva NV, Shashkov AS, Dmitriev BA and Kochetkov NK (1983) Bacterial antigenic polysaccharides. 12. Structure and 13C NMR spectrum of the polysaccharide chain of Shigella boydii type 8 lipopolysaccharide. Bioorg. Khim 9, 60–73. [PubMed] [Google Scholar]
- (43).Landersjö C, Weintraub A, and Widmalm G (1996) Structure determination of the O-antigen polysaccharide from the enteroinvasive Escherichia coli (EIEC) O143 by component analysis and NMR spectroscopy. Carbohydrate Research 291, 209–216. [DOI] [PubMed] [Google Scholar]
- (44).Zdorovenko EL, Varbanets LD, Liu B, Valueva OA, Wang Q, Shashkov AS, Garkavaya EG, Brovarskaya OS, Wang L, and Knirel YA (2014). Structure and gene cluster of the O antigen of Escherichia coli L-19, a candidate for a new O-serogroup. Microbiology (Reading, England), 160, 2102–2107. [DOI] [PubMed] [Google Scholar]
- (45).Gilbert M, Brisson JR, Karwaski MF, Michniewicz J, Cunningham AM, Wu Y, Young NM, and Wakarchuk WW (2000). Biosynthesis of ganglioside mimics in Campylobacter jejuni OH4384. Identification of the glycosyltransferase genes, enzymatic synthesis of model compounds, and characterization of nanomole amounts by 600-Mhz 1H and 13C NMR analysis. J. Biol. Chem, 275, 3896–3906. [DOI] [PubMed] [Google Scholar]
- (46).McNally DJ, Jarrell HC, Khieu NH, Li J, Vinogradov E, Whitfield DM, Szymanski CM, and Brisson JR (2006). The HS:19 serostrain of Campylobacter jejuni has a hyaluronic acid-type capsular polysaccharide with a nonstoichiometric sorbose branch and O-methyl phosphoramidate group. The FEBS Journal, 273, 3975–3989. [DOI] [PubMed] [Google Scholar]
- (47).Aspinall GO, McDonald AG, Pang H, Kurjanczyk LA and Penner JL (1994) Lipopolysaccharides of Campylobacter jejuni serotype O:19: structures of core oligosaccharide regions from the serostrain and two bacterial isolates from patients with the Guillain–Barré syndrome. Biochemistry 33, 241–249. [DOI] [PubMed] [Google Scholar]
- (48).Vinogradov EV, Holst O, Thomas-Oates JE, Broady KW and Brade H (1992) The structure of the O-antigenic polysaccharide from lipopolysaccharide of Vibrio cholerae strain H11 (non-O1). Eur. J. Biochem 210, 491–498. [DOI] [PubMed] [Google Scholar]
- (49).Parker CT, Huynh S, & Heikema AP (2016) Complete Genomic Sequence of Campylobacter jejuni subsp. jejuni HS:19 Strain RM1285 Isolated from Packaged Chicken. Genome Announcements, 4, e01100–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S, Jagels K, Karlyshev AV, Moule S, Pallen MJ, Penn CW, Quail MA, Rajandream MA, Rutherford KM, van Vliet AH, Whitehead S, and Barrell BG (2000) The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature, 403, 665–668. [DOI] [PubMed] [Google Scholar]
- (51).Vrielink A, Rüger W, Driessen HP, and Freemont PS (1994). Crystal structure of the DNA modifying enzyme beta-glucosyltransferase in the presence and absence of the substrate uridine diphosphoglucose. EMBO J, 13, 3413–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Tarbouriech N, Charnock SJ, and Davies GJ (2001). Three-dimensional structures of the Mn and MgdTDP complexes of the family GT-2 glycosyltransferase SpsA: a comparison with related NDP-sugar glycosyltransferases. Journal of Molecular Biology, 314, 655–661. [DOI] [PubMed] [Google Scholar]
- (53).Breton C, Snajdrová L, Jeanneau C, Koca J, and Imberty A (2006). Structures and mechanisms of glycosyltransferases. Glycobiology, 16, 29R–37R. [DOI] [PubMed] [Google Scholar]
- (54).Lairson LL, Henrissat B, Davies GJ, and Withers SG (2008). Glycosyltransferases: structures, functions, and mechanisms. Annual Review of Biochemistry, 77, 521–555. [DOI] [PubMed] [Google Scholar]
- (55).Aono R, Ito M, and Machida T (1999). Contribution of the cell wall component teichuronopeptide to pH homeostasis and alkaliphily in the alkaliphile Bacillus lentus C-125. Journal of Bacteriology, 181 6600–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Iyer LM, Abhiman S, Maxwell Burroughs A, & Aravind L (2009). Amidoligases with ATP-grasp, glutamine synthetase-like and acetyltransferase-like domains: synthesis of novel metabolites and peptide modifications of proteins. Molecular BioSystems, 5, 1636–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


