Abstract
Chikungunya fever (CHIKF) is an arbovirus disease caused by chikungunya virus (CHIKV), an alphavirus of Togaviridae family. Transmission follows a human-mosquito-human cycle starting with a mosquito bite. Subsequently, symptoms develop after 2–6 days of incubation, including high fever and severe arthralgia. The disease is self-limiting and usually resolve within 2 weeks. However, chronic disease can last up to several years with persistent polyarthralgia. Overlapping symptoms and common vector with dengue and malaria present many challenges for diagnosis and treatment of this disease. CHIKF was reported in India in 1963 for the first time. After a period of quiescence lasting up to 32 years, CHIKV re-emerged in India in 2005. Currently, every part of the country has become endemic for the disease with outbreaks resulting in huge economic and productivity losses. Several mutations have been identified in circulating strains of the virus resulting in better adaptations or increased fitness in the vector(s), effective transmission, and disease severity. CHIKV evolution has been a significant driver of epidemics in India, hence, the need to focus on proper surveillance, and implementation of prevention and control measure in the country. Presently, there are no licensed vaccines or antivirals available; however, India has initiated several efforts in this direction including traditional medicines. In this review, we present the current status of CHIKF in India.
Keywords: Chikungunya fever (CHIKF), chikungunya virus (CHIKV), polyarthralgia, epidemiology, disease resolution
Introduction
Chikungunya fever (CHIKF) is an arthropod-borne viral (arbovirus) disease (Mohan, 2006; Mohan and Sharma, 2007) that has become a global health concern following its resurgence since 2006 (Pialoux et al., 2007; Simon et al., 2008). Historically, there have been reports of epidemics of fever along with arthralgia as far back as 1824, similar to present day CHIKF (WHO, 2012; Puntasecca et al., 2021). However, the CHIKF was first officially described in 1952 after an outbreak on the Makonde Plateau, south-eastern Tanzania (Lumsden, 1955; Robinson, 1955). The word “chikungunya” is taken from the verb “kungunyala” in Kimakonde language spoken in the same area which means “to dry up or become contorted” (Lumsden, 1955; Benjamin, 2012). The disease develops the stooped posture due to the rheumatologic manifestations describing the word chikungunya. In Congo region it is known by the name “Buka Buka” meaning “broken-broken” representing incapacitating joint pain (Muyembe-Tamfum et al., 2003).
Classification
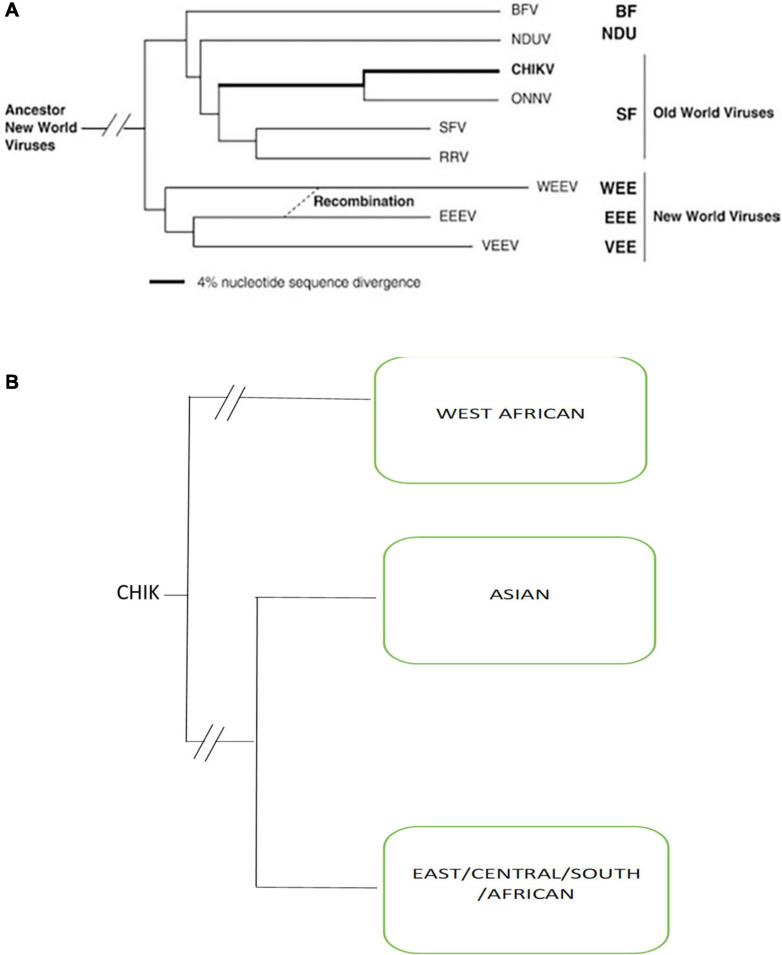
Chikungunya virus (CHIKV) belongs to the genus Alphavirus within the family Togaviridae. Alphaviruses include all viruses of this family that are animal pathogens and have a worldwide geographic distribution based on which they are described classically as either Old World or New World viruses. The New World alphaviruses are distributed across the Americas and cause encephalitis in equines and humans. For example, the eastern equine encephalitis virus, western equine encephalitis virus, and Venezuelan equine encephalitis virus. Whereas Old World alphaviruses to which CHIKV belongs, cause fever, rash, and arthritis in humans, other members of the group include Sindbis virus, o’nyong’nyong virus, Ross River virus, Mayaro virus, Barmah Forest virus, and Semliki Forest virus are present in Asia, Europe, Australia, and parts of Africa (Figure 1). Most alphaviruses are transmitted by arthropod vectors that probably control their geographic dispersal and it is likely that several transoceanic exchanges might have occurred. The alphavirus CHIKV belongs to the Semliki forest virus complex of the Alphaviruses, as seen in Figure 1.
FIGURE 1.
(A) Simplified Phylogenetic tree of all Alphaviruses generated from partial E1 envelope glycoprotein gene sequences by using the neighbor-joining program Adapted from Powers et al. (2001) (BF, Burma Forest virus; CHIKV, chikungunya virus; SF, Semliki Forest; WEE, western equine encephalitis; EEE, eastern equine encephalitis; VEE, Venezuelan equine encephalitis; NDUV, Ndumu virus; ONNV, o’nyong’nyong virus; RRV, Ross River virus). (B) Phylogenetic tree of three genotypes of chikungunya viruses.
Clinical Manifestations
After CHIKV infection most individuals develop symptoms after a silent incubation period that range from 2 to 6 days (can extend up to 12 days) followed by the onset of abrupt symptoms which include high fever and severe arthralgia. The clinical symptoms associated with CHIKV infection are classically divided into acute and chronic phases (Suhrbier, 2019). The acute stage is the febrile phase and can extend until 15 days since the onset of fever. This phase is characterized by common symptoms such as high fever and arthralgias; however, non-classical symptoms, tabulated in Table 1 may also occur during this phase. Fever is usually high and is poorly responsive to antipyretics. Anorexia, myalgias, nausea, and vomiting in adults, and even transient confusion in elderly patient are common with 50–97% of individuals developing these symptoms. Additionally, other less common symptoms like back pain, headache and fatigue have also been reported during this phase (Table 1). Bilateral polyarthralgia is noted as a typical symptom affecting mainly the small joints such as ankles, wrist and phalanges and few large joints such as knees and elbows (WHO, 2009). In about half of the cases, acute phase is also characterized by cutaneous lesions like maculopapular rashes, edematous or itchy skin affecting mainly the face and trunk (Simon et al., 2011). Symptoms such as diarrhoea, abdominal pain, nausea, and vomiting are seen in 15–47% of individuals (Thiberville et al., 2013). Other symptoms are asthenia, erythema, persistent conjunctivitis, conjunctival effusion, and cervical lymphadenopathy. Moderate thrombocytopenia, leukopenia and lymphopenia are also common (Staples et al., 2009; Staples and Fischer, 2014; Madariaga et al., 2016). Severe symptoms like bleeding due to thrombocytopenia, bulbous skin lesions, hepatitis, meningoencephalitis, meningitis have been reported in a few patients (Markoff, 2015; Weaver and Lecuit, 2015; Abeyratne et al., 2018; Saha et al., 2019). Ocular complications like retinitis or uveitis, myocarditis, nephritis, cranial nerve palsies and Guillain-Barre Syndrome have been observed for the first time during the 2006 Indian Ocean outbreak (Wielanek et al., 2007; Lebrun et al., 2009) as referred in Table 1. Renal and neurological complications are also associated with severe chikungunya infection (Weaver and Lecuit, 2015; Mercado et al., 2018). In few cases a very high viral load leads to persistence of the virus in minor joints during acute stage (Thiberville et al., 2013).
TABLE 1.
Classical and non-classical clinical manifestations in chikungunya infection.
| Clinical features of CHIKV infection | |
| Classical features | Complications |
| Fever | Bulbous skin lesions |
| Arthralgia | Fulminant hepatitis |
| Rashes on skin | Meningoencephalitis |
| Headache | Retinitis |
| Back pain | Uveitis |
| Nausea | Myocarditis |
| Vomiting | Nephritis |
| Joint swelling | Convulsions |
| Myalgia | Cranial nerve palsy |
| Lymphadenopathy | Guillain-Barre Syndrome |
| Fatigue | Acute renal failure |
| Restlessness | Respiratory failure |
| Anorexia | Meningitis |
| Abdominal pain | |
| Diarrhea | |
| Leukopenia | |
| Lymphopenia | |
The disease is self-limiting and usually resolve within 2 weeks. However, about 30–40% infected patients can develop chronic stage which is defined as three months after the bulbous skin lesions onset of infection that can last up to several years with persistent polyarthralgia (Schwartz and Albert, 2010).
Chronic stage of the disease is associated with the development of inflammatory joints after the resolution of the acute phase and can last up to several years in some patients. The symptoms of this phase are mainly polyarthralgia and/or polyarthritis, mostly affecting small joints, such as phalanges and wrists, as well as large joints such as shoulders, knees, and ankles affecting the mobility of the affected patients (Hoarau et al., 2010). Inflammation of joints is seen for years, which progresses to chronic inflammatory rheumatism (Borgherini et al., 2008; Rajapakse et al., 2010; Simon et al., 2011; Gupta et al., 2014). Studies in other alphaviruses reveal that there is persistence of viral antigens which causes inflammation of joints (Suhrbier and Mahalingam, 2009; Suhrbier et al., 2012). In case of CHIKV, some studies have shown evidence of prolonged persistence of CHIKV RNA and viral proteins in the synovial tissues (Hoarau et al., 2010). Conversely, other studies have failed to observe viral particles in CHIKV infected joints during the chronic phase and argue that this phase is an outcome of host immune response to the infection (Chang et al., 2018). These contradictory findings suggest that more in-depth studies are required to understand the basis of disease progression to chronic phase in CHIKF.
Clinical Diagnostic Matrix
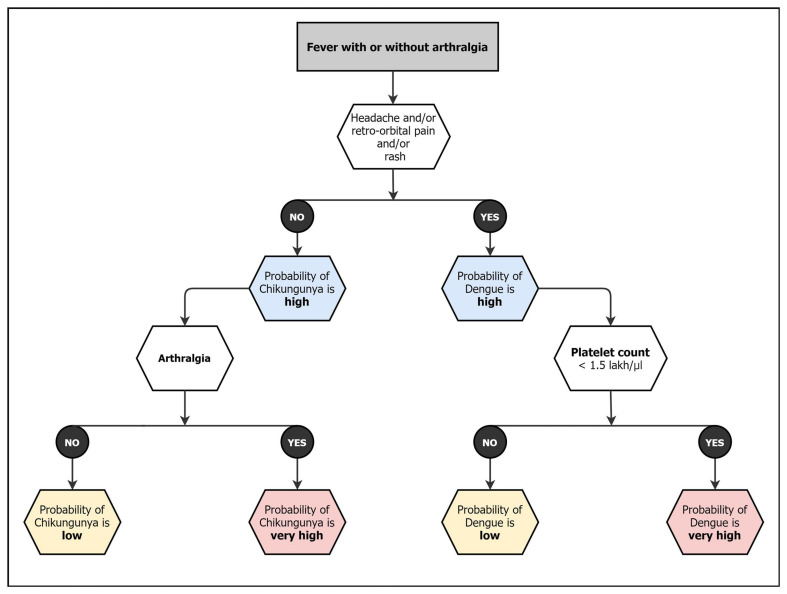
In a country like India, which is an endemic area for chikungunya, Dengue, and malaria, the seasonality of these infections overlaps. Chikungunya and Dengue have a common vector and shows similar clinical presentation; hence coinfections are also reported in several studies (Saswat et al., 2015; Londhey et al., 2016; Jain et al., 2017; Kaur et al., 2017). Despite showing similar symptoms, the outcomes, and management strategies of these two viruses are vastly different. It is important for clinicians to make the differential diagnosis on the basis of various clinical presentations and laboratory methods to start appropriate treatment and avoid complication such as hemorrhages, Acute respiratory distress syndrome (ARDS), renal failure and arthritis (Kaur et al., 2018). It has been shown that simple clinical and laboratory variables can predict these infections at presentation and classify them into symptoms and laboratory tests for appropriate management (Lee et al., 2012). Acute arthritis, shorter duration of fever, rash, myalgia/arthralgia, and conjunctivitis are more prominent in CHIKF, while abdominal pain, leukopenia, neutropenia, and thrombocytopenia are more prominent in dengue fever (DF). Similarly, significant difference is noticed in platelet level cut-offs between DF and CHIKF along with bleeding in patient (Figure 2; Halstead et al., 1963; Nimmannitya et al., 1969; Hochedez et al., 2008; Kularatne et al., 2009; Laoprasopwattana et al., 2012; Lee et al., 2012; Thiberville et al., 2013; Taraphdar et al., 2015).
FIGURE 2.
Diagnostic matrix followed by Indian clinicians for CHIKF and related coinfections (Lee et al., 2012).
There is no specific antivirals or treatment regimen available for chikungunya and symptomatic treatment is the main stay of patient management. Generally, patient is advised for adequate rest and intake of plenty of water to prevent dehydration. Non-steroidal anti-inflammatory drugs are avoided until dengue infection is ruled out. Acetaminophen or paracetamol is given to reduce pain and fever (NVBDCP, 2014-15; CDC, 2018).
Laboratory Diagnosis
In India, the choice of CHIKV test depends on various factors that include the intent of the test, resource availability and time of sample collection (days after onset of illness) (Figure 2). The diagnosis can be achieved by virus isolation, detecting antigen, nucleic acid, and IgM/IgG antibodies. Lab diagnosis is seriously limited by clinician not requesting a PCR test for chikungunya early in infection. The popular test is CHIK IgM, however, some tests show cross reactivity and give false positivity. Generally, the public hospitals receive National Institute of Virology CHIKV IgM kit. Each test has specific advantages, and disadvantages and combination of tests can be performed to increase the credibility of the test result. The laboratory diagnosis of CHIKV can be differentiated into two categories- classical and molecular diagnostics.
Classical Diagnostics
Virus isolation was considered as the gold standard for CHIKV diagnosis for decades but is not commonly used in routine diagnosis now. Virus is detectable in the patient’s serum or plasma samples in high titers between 2 and 6 days following onset of illness. However, the viral load starts decreasing from day five after the onset of illness (Álvarez-Argüelles et al., 2019). Therefore, the usefulness of the test for diagnostic purpose is only limited to the acute phase of the infection. Viral growth and its identification are a time-consuming process and require specialized equipment and skilled staff for performing the procedure. However, the advantage of cell culture is that it allows amplification of virus and identification of viral strain that can be characterized for further epidemiological and research studies (Natrajan et al., 2019).
In the later stages of CHIKV infection (>5 days post-infection), the CHIKV IgM antibody capture ELISA is a dependable testing method for diagnosis (Álvarez-Argüelles et al., 2019). CHIKV specific IgG antibody ELISA can be used for sero-diagnosis by testing in a paired serum samples from the acute and convalescent phase of CHIKV infection; four-fold or more increase in the antibody titer should confirm the presence of recent CHIKV infection (Weaver and Lecuit, 2015). However, the collection of paired serum sample is often not possible, therefore, CHIKV specific IgM antibody in the acute-phase serum sample is generally used for diagnosis (Álvarez-Argüelles et al., 2019). Previous studies have demonstrated the persistence of CHIKV specific IgM antibody up to 10 months post-infection, Chelluboina et al. (2019) which proves that anti-CHIKV IgM antibody ELISA cannot be used as definitive diagnosis method. The cross-reactivity with other arboviruses like o’nyong’nyong virus and Mayaro virus is another disadvantage that affects the sensitivity of the assay (Prat et al., 2014).
Immunofluorescence assay (IFA) is a sensitive test that can be used for diagnosis of CHIKF, by detecting type-specific antibody against CHIKV. It is a labour-intensive method and requires skilled personnel to perform the procedure (Natrajan et al., 2019; Álvarez-Argüelles et al., 2019). This method is usually followed by plaque reduction neutralization test (PRNT) and micro neutralization test (MnT) that shows high sensitivity and specificity (Azami et al., 2016). However, it is not routinely used for diagnostic purpose as it is a tedious process (Álvarez-Argüelles et al., 2019).
Molecular Diagnostics
Molecular diagnosis of acute phase CHIKV by RT-PCR is a commonly used method nowadays. It is a less time consuming and more specific technique. Serum and plasma are the preferred samples, but saliva and urine can also be used as an alternative and in the case of meningitis or encephalitis, CSF can be taken (Musso et al., 2016; Acevedo et al., 2017). Both Real-time RT-PCR and RT-PCR are commonly used and almost all genes of the virus are used for diagnostic purposes (Carletti et al., 2007; Edwards et al., 2007; Chen et al., 2013; Chiam et al., 2013; Parashar et al., 2015; Sarangan et al., 2018). Multiplex assays are also available for the simultaneous detection of CHIKV with other arboviruses (Mishra et al., 2011; Chen et al., 2015). Sensitivity of detecting chikungunya in commercial multiplex kit is a challenge and there is an urgent need to address molecular diagnostics of chikungunya in the country. In addition, commercial immunochromatography antigen detection kits are gaining popularity in the recent past owing to their high sensitivity and specificity with no cross-reactivity with other arboviruses (Okabayashi et al., 2015; Jain et al., 2018a). Other diagnostic initiatives are the use of biosensors using different technologies such as electrochemical impedance spectrometry and optical fibers that have shown promise in CHIKV diagnosis (George et al., 2019, 2021).
Epidemiology of CHIK
Chikungunya outbreaks have been reported in the Indian subcontinent since the early 20th century. Since the earliest outbreaks, even though maximum outbreaks have been reported in India, sporadic have been reported in the neighboring countries. The following section describes the outbreaks that have occurred in the neighboring countries and in India. Summarizing the studies thus far, it appears that India may have been the epicenter of dispersal of CHIKV to neighboring as well as distant countries since the emergence of the IOL, which is significantly endemic to India (Newase et al., 2020).
Epidemiology of CHIKV in Indian Subcontinent and Southeast Asia
As far as neighboring countries are concerned, CHIKV infection has been reported in multiple regions through the period of 2005–2017. During the period 2005–2006 outbreak in South Asia, particularly Myanmar and Sri Lanka, CHIKV positive cases were around 4.6 and 6.1%, of the total suspected cases, respectively; and in Southeast Asia, particularly Indonesia, the Philippines and Vietnam, the same were 27.4, 26.8, and 25.0%, respectively (Ngwe Tun et al., 2016). The Indian Ocean Lineage (IOL), within the ECSA genotype was associated with 2005–2006 outbreak in the Indian subcontinent and the Indian Ocean Islands. The E1-A226V mutation was associated with the reemergence which increased the adaptability of CHIKV to Aedes albopictus mosquitoes (Tsetsarkin et al., 2007) and it has been reported that this species of mosquitoes is predominant in Indian state of Kerala as well as the neighboring island nation, Sri-Lanka (Kumar et al., 2008; Hapuarachchi et al., 2010). In 2006–2007, General Hospital in Peradeniya, Sri Lanka reported an outbreak which was later confirmed as CHIKV infection. Moreover, Bangladesh has had a share of recurrent CHIKV infections during 2008, 2011, and 2017 (Khatun et al., 2015; Rahman et al., 2019). In 2012, CHIKV was reported for the first time in Bhutan (Wangchuk et al., 2013). Further, Nepal reported the upsurge of CHIKV during the period 2013–2015 (Pandey et al., 2017). In Pakistan, although the first detected case was in December 2016, anti-CHIKV antibodies in human sera and rodents have been detected as early as 1980s (Darwish et al., 1983). This virus was introduced into Zhejiang and Tianjin province of China by travelers from Bangladesh to Myanmar in 2017, respectively (Pan et al., 2019; Xie et al., 2020). Again, a study conducted in India through July 2018, involving testing a total of 1549 samples, almost 50% patients tested positive for CHIKV infection (Badar et al., 2019). Although, CHIKV infection dates back to 1950s and has started spreading its roots quite consistently since 2005 yet absence of a nationwide surveillance system for such arboviral diseases and its reported misdiagnosis as Dengue could be one of the reasons for the lack of a detailed prevalence studies. Further, evidence for chikungunya-dengue co-infection has been found in Angola, Gabon, India, Madagascar, Malaysia, Myanmar, Nigeria, Saint Martin, Singapore, Sri Lanka, Tanzania, Thailand, and Yemen which adds to the confusion (Furuya-Kanamori et al., 2016).
Epidemiology of CHIKV in India
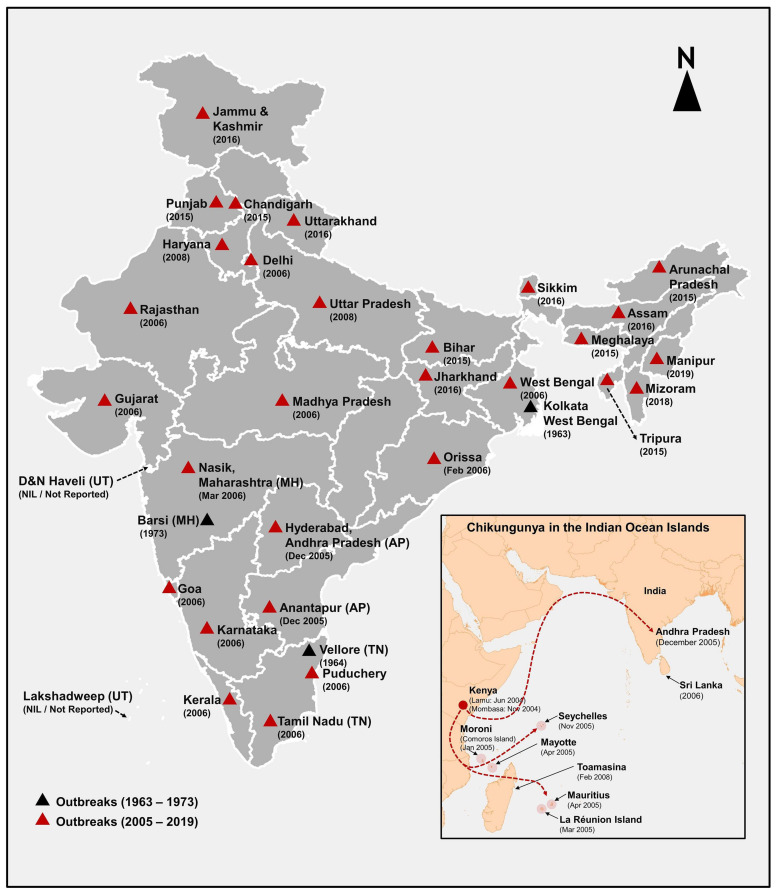
Chikungunya outbreaks have been reported from India during the period 1963–1973 and 2005–2019. The first CHIKF outbreak in India was reported from Kolkata (Calcutta), West Bengal, in 1963 (Sarkar et al., 1964; Shah et al., 1964; Sudeep and Parashar, 2008; Figure 3). However, retrospective serological studies have shown that chikungunya existed in India prior to 1963 (Pavri, 1964; Banerjee, 1965). In 1964, Chennai (Thiruvengadam et al., 1965), Pondicherry, and Vellore reported outbreaks of chikungunya (Jadhav et al., 1965; Naresh Kumar and Sai Gopal, 2010). In 1965, chikungunya outbreaks were reported from Visakhapatnam, Kakinada, Rajahmundry and Nagpur, in 1973 in Barsi in Maharashtra (Rodrigues et al., 1972; Padbidri and Gnaneswar, 1979; Yergolkar et al., 2006) and some authors have opined that lack of surveillance between 1973 and 2005 might have been the reason for lack of CHIKV reports (Pavri, 1986; Ravi, 2006). However, the re-emergence of the virus could be due to several other reasons (Mishra and Ratho, 2006). CHIKV is known to have three genotypes, namely, West African (WA), Asian and East Central South Africa (ECSA) genotypes and the circulating virus in India until the 1970s have been reported to be of the Asian genotype (Kalantri et al., 2006; Naresh Kumar and Sai Gopal, 2010).
FIGURE 3.
Reported chikungunya outbreaks in India and Indian Ocean Islands (IOI) shows the year of onset of chikungunya outbreaks between 1963 and 2019 in India and the change in genotype from Asian (during the 1963–1973) to ECSA (2005–2019) is sketched in black and red triangles. The data source for generating. This figure is from NVBDCP (2014-15); National Health Profile (2019); National Center for Disease Control (2020).
CHIKV in India Post-2005
After the emergence of CHIKV in the Indian Ocean islands in 2005, CHIKV re-emerged in India after 32 years (Lahariya and Pradhan, 2006), affecting Hyderabad and Ananthapur district of Andhra Pradesh in South India and eventually affected 1.4 million people in 13 states (Schuffenecker et al., 2006) with huge economic and productivity loss of 391 million rupees (Krishnamoorthy et al., 2009). Ahmedabad city of Gujarat and Kerala state were the worst affected places (Sudeep and Parashar, 2008). The 2005–2006 outbreak in India was caused by the ECSA genotype (Kalantri et al., 2006; Naresh Kumar and Sai Gopal, 2010). The E1-A226V mutation of the virus and their adaptation to the A. albopictus resulted in increased susceptibility among pediatric population, neurological complications, as well as mortality associated with this outbreak (Naresh Kumar and Sai Gopal, 2010).
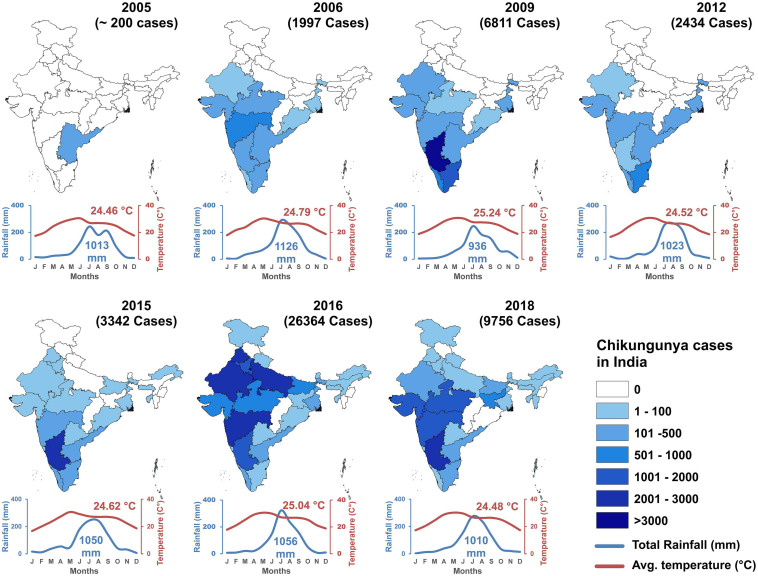
In the 2005–2006 outbreak, 13 states of India reported CHIKF. In 2009, the infection expanded to 15 states in India. The number of states which reported CHIKF increased to 23 by the year 2015, 28 states in 2016, and 30 states/Union territories in the year 2019. Jammu and Kashmir, Mizoram, and Manipur has reported ≤10 laboratory confirmed CHIKF cases until December 2019. Union territories of Dadra and Nagar Haveli and Lakshadweep has not reported CHIKF cases until now. The year-wise prevalence of CHIKF cases in India from 2005 to 2018 along with temperature and rainfall data is shown in Figure 4. Maximum number of laboratory- confirmed cases were reported in the year 2016, followed by 2017, and 2019. Highest confirmed cases were reported in Karnataka, Delhi, and Maharashtra (Supplementary Table 1). Available data on year-wise percentage positivity of CHIKV between 2014 and 2019 show some variation across the different seasons in India. However, there seem to be a general trend of lower percentage positivity values recorded during the summer periods which increases as rainy season sets in and continues to be high through winter (Department of Health and Family Welfare, Government of India, 2018; National Center for Disease Control, 2020) (Supplementary Figure 2). In 2019, a total of 81914 cases were clinically suspected for CHIKV, out of which, 12205 (14.9%) laboratory confirmed chikungunya was reported in 21 Indian states and 3 Union territories in this year. Majority of the CHIKV cases were reported from Karnataka (3664), followed by Maharashtra (1646), Telangana (1358), and least was reported from Uttarakhand (1) (Supplementary Figure 3). In 2020, a total of 18533 cases were clinically suspected for CHIKV and 2812 (15.2%) cases were laboratory confirmed as CHIKF until July 2020 (Department of Health and Family Welfare, Government of India, 2018). Currently, CHIKF is endemic in 24 Indian states and 6 union territories indicating that this as an important health problem in our country (NVBDCP, 2014-15).
FIGURE 4.
Year-wise CHIKF cases in India with annual rainfall and temperature (2005–2018).
Disease Transmission and Vector Biology
Chikungunya virus is transmitted mainly through Aedes mosquitoes. Mosquitoes acquire CHIKV when it bites the infected human with viremia. During this extrinsic incubation period in the mosquito, CHIKV replicates in the midgut, circulates through body cavity, reaches to the salivary gland, establishes infection, and secretes into the saliva. CHIKV reaches to new host when mosquito feeds on their blood. The transmission cycle continues, virus gets amplified in the new host again picked up by mosquito if they feed on the infected person and the transmission cycle goes on.
The most common transmission mechanism of CHIKV is the horizontal transmission through the mosquito saliva. Mosquito injects the saliva carrying virus to a new host when it feeds on their blood. However, a low rate of vertical transmission may also occur through infected eggs (Agarwal et al., 2014). Entomological survey conducted by Jain et al. (2016) has demonstrated the presence of vertical transmission in Aedes aegypti population. Agarwal et al. (2014) confirmed experimental vertical transmission of ECSA genotype of CHIKV in A. aegypti from India. Studies have documented vertical transmission of Indian Ocean Lineage of CHIKV up to F5 and F6 progenies in A. aegypti and A. albopictus (Chompoosri et al., 2016). Likewise, vertical transmission in the host has also been documented in the global scenario. Mother-to-child transmission has been reported in women who developed CHIKF in the last week of gestation prior to delivery (Gerardin et al., 2008; Fritel et al., 2010). One case study from Brazil has documented probable vertical transmission leading to maternal and neonatal death. Hence, clinicians must consider the possibility of neurological complications and find ways to treat this (Bandeira et al., 2016; Cardona-Correa et al., 2017; Oliveira et al., 2018). The risk of transmission appears to be rare before 22 weeks of gestation, however, the pathogenic mechanism is not clear yet (Lenglet et al., 2006). Overall, vertical transmission is linked with poor pregnancy outcomes in some cases (Robillard et al., 2006; Senanayake et al., 2009). In the contrary, vertical transmission of CHIKV has not been documented till date in the Indian scenario (Nigam et al., 2016).
Molecular Epidemiology
Chikungunya virus genome consists of a 11.8 kb linear, single-stranded, positive-sense RNA molecule., organized into two open reading frames (ORFs) encoding non-structural (nsP1-nsP2-nsP3-nsP4) and structural (C-E3-E2-6K-E1) polyproteins at the 5′ and 3′ ends, respectively. These ORFs are flanked by 5′ and 3′ untranslated regions which regulate viral gene expression, replication, translation and host-virus interactions thereby significantly impacting viral evolution, pathogenesis and host range (Hyde et al., 2015). Mutations at these proteins have shown to attribute viral fitness and vector preferences (Tsetsarkin et al., 2007, 2011a,b, 2014; Casal et al., 2015; Agarwal et al., 2016; Jain et al., 2016). Continuous surveillance of phylogeny and mutations of the circulating viral strain is very important for early detection of mutation in the viral genome and evolution of new strain and that can help us to prepare for the prevention and control of future epidemics.
Phylogenetic analysis of CHIKV based on whole genome and individual genes grouped the virus into three genotypes: East/Central/South African, West African, and the Asian (Powers et al., 2000). A recent study favored further the division of ECSA into three subgroups: ECSA I, ECSA II, and ECSA III, based on the Bayesian analysis, and E1 gene sequences (Casal et al., 2015).
Starting from 2005, there were reports of the ECSA genotype causing severe epidemics from Kenya to India where over one million cases were recorded and is said to be massively under estimated (Mavalankar et al., 2007). Subsequent studies have also identified the ECSA genotype as the cause of outbreaks in India (Niyas et al., 2010; Agarwal et al., 2019; Tandel et al., 2019; Jain et al., 2020). A recent study revealed the co-circulation of two distinct subclades of ECSA in South India when the circulating strains were characterize from the 2016 outbreak across the country (Harsha et al., 2020). One of these subclades had been earlier reported in 2010 from an outbreak in Delhi (Shrinet et al., 2012). Studies on CHIKV microevolution during the 2005–2006 Indian Ocean epidemic revealed the presence of E1-A226V mutation (Schuffenecker et al., 2006) which was later shown to mediate improved fitness of CHIKV in A. albopictus regarding midgut infectivity, transference to salivary glands and subsequent transmission to the vertebrate host (Tsetsarkin et al., 2007). However, a number of Indian studies have reported the absence of this mutation (Agarwal et al., 2019). Essentially, apart from the Kerala study (Niyas et al., 2010) which reported the E1-A226V strain, many other studies have found this mutation to be absent in their isolates. Consequently, other mutations that aid increased fitness in A. albopictus may be responsible for the dominance of ECSA in Indian outbreaks. Since 2006, A. albopictus has been recognized as the main vector in some parts of India, the Indian Ocean, Thailand, Malaysia, Singapore, Gabon, Sri-Lanka and Italy (Tsetsarkin et al., 2011a). However, a number of mutations in the CHIKV genome were discovered to have epistatic effect (lineage specific) on the E-A226V mutation. For instance, E2-211I in most ECSA strains and E1-98T in all endemic Asian strains tend to block adaptation to A. Albopictus (Tsetsarkin et al., 2009). Whether these mutations that result in epistasis are playing a significant role in controlling the frequency of outbreaks has not been deliberated. It was speculated previously that it would be interesting to evaluate the effect of these mutations with respect to the presence or absence of the E1-A226V mutation (Shrinet et al., 2012). However, it was suspected that the characteristic inter-epidemic quiescence of CHIKV outbreaks might be partly attributed to the effect of epistatic mutations on important adaptive changes in the CHIKV genome (Weaver et al., 2021). In 2010, a novel non-conservative mutation, E2-L210Q was discovered in both human and mosquito samples in Kerala (Niyas et al., 2010). Laboratory investigations revealed that E2-L210Q acts primarily at the level of infection of A. albopictus midgut epithelial cells, thus providing additional selective advantage that may facilitate even more efficient virus circulation and persistence in endemic areas (Tsetsarkin et al., 2011b). Considering the fact that A. albopictus was largely responsible for outbreak in Kerala (Niyas et al., 2010), it is plausible to suggest that the E2-L210Q mutation further enhanced the vector competence, thus driving the epidemic.
Two novel mutations, E1-K211E and E2-V264A were also identified together with molecular signatures that suggest possibility of recent recombination events in Indian CHIKV isolates in an outbreak in Delhi (Shrinet et al., 2012). A study had earlier asserted that homologous recombination did not play a role in the genetic diversity of Asian strains albeit two potential recombination events were found (Cui et al., 2011). However, Casal et al. (2015) proved that recombination was an additional mechanism of CHIKV genetic diversity when it was demonstrated that the recombinant strain KJ679578/2011 (ECSA III) was derived from ECSA III and ECSA I. A more recent study, while highlighting the role of 3′ UTR plasticity in CHIKV evolution, has also proposed that recombination in CHIKV accelerates adaptability (Filomatori et al., 2019; Weaver et al., 2021). Interestingly, these mutations in E1 and E2 (E1-K211E and E2-V264A) have been shown to enhance the fitness of A. aegypti resulting in 13-fold increase in infectivity; 15-fold enhancement in dissemination; and a 62-fold surge in transmission (Agarwal et al., 2016). Notably, A. aegypti is predominant in Delhi, hence the active role of this mutation causing the outbreak within the metropolis is apparent. In the previous study that compared the 2010 and 2016 outbreaks in India, a number of mutations were identified, including the E2-G55R, H73Y protein which was unique to the 2010 outbreak and associated with higher disease severity; and the E1-I317V which was present in all the sequenced 2016 samples (Jain et al., 2020). Some studies have also identified several mutations in CHIKV during epidemics and sporadic infections in India (Table 2). Some of these mutations have been recognized as having high evolutionary potential, higher disease severity (Jain et al., 2020), and capacity to cause epidemics (Singh et al., 2012). Interestingly, some Indian studies have recorded a 100% presence of these mutations including E1-K211E, M269V, and D284E in their sequenced isolates (Mudurangaplar and Peerapur, 2016; Tandel et al., 2019).
TABLE 2.
Few CHIKV mutations reported between 2010 and 2020.
| Location | Period | Protein | Mutation | References |
| Delhi | 2010 | E1 |
K211E M269V D284E V179A S234P R196K R247C |
Agarwal et al. (2016) |
| E2 | V50A C389R A164T I211T V264A S299N T312M A344T S375T V386A |
|||
| G55R H73Y | Jain et al. (2020) | |||
| E3 | V42I P59S | |||
| nsP1 | G230R M314L R85H T215A F391L K128T V172L V452A |
|||
| nsP2 | G641D M290T A256V V639I | |||
| nsP3 | I175V STOP524R | |||
| nsP4 | G85R | |||
| 2016 | C | Q58R | ||
| E1 | I317V N348I S167P T288I | |||
| E2 | K189R L412F | |||
| nsP1 | K224T | |||
| nsP2 |
H130Y E145D T446S P689S H109Q |
|||
| nsP3 | H377R | |||
| nsP4 | S55N | |||
| E1 | R123K T145S V179M K211E P232Q I261D M269V D284E I317V/M V322A N389K |
Kaur et al. (2017) | ||
| 2016 | E1 | K211E M269V D284E T145A N149K G150D G314A |
Tandel et al. (2019) | |
| Central India | 2016–2017 | E1 | K211E M269V D284E I317V V322A |
Agarwal et al. (2019) |
| Southern India | 2011–2014 | E1 | K211E M269V D284E | Mudurangaplar and Peerapur (2016) |
| Tamil Nadu and Andhra Pradesh | 2009–2010 | E1 | K211E P58L G195R | Sumathy and Ella (2012) |
| E2 | V264A K47N G55R K66E | |||
| E3 | D40N | |||
| C | A232V |
Highlighted: 100% of sequenced isolated show this mutation.
Jain et al. (2020) also reported the presence of mutations in nsP1 (G230R) and nsP3 (opal 524R) proteins which were unique to the 2010 outbreak and were associated with higher disease severity. Apart from that, one nsP2 mutation, H130Y, was present in all the sequenced 2016 samples (Jain et al., 2020). It is very important to note that a combination of mutations in the nsP1 protein (G230R and K299E) resulted in CHIKV resistance to 6′-β-Fluoro-Homoaristeromycin and 6′-Fluoro-Homoneplanocin A, which are carbocyclic adenosine analogs and had initially displayed potent antiviral activity on the wild-type virus (Kovacikova et al., 2020). These mutations might have contributed to the difficulties that is currently faced in research toward CHIKV vaccines and therapeutics. A number of other combinations of mutations, some of which have been mentioned in this review, have also been studied and found to increase the fitness of the virus. These include a combination of mutations in the nsP1 and the opal stop codon preceding in the nsP3 protein. Mutations in these regions including nsP3 (opal 524R) - nsP1 (G230R); nsP3 (opal 524R) - nsP1 (V326M); and nsP3 (opal 524R) - nsP1 (G230R) - nsP1 (V326M), have been shown to confer resistance to polyamine depletion in addition to enhanced viral replication (Mounce et al., 2017). Although the authors concede that individual mutations do not induce polyamine depletion resistance, it is still worrisome because some of these combinations have already been reported in Indian outbreaks (Jain et al., 2020).
These reported mutations and further experiments to determine their role in vector range and competence; fitness in vector; disease transmission and severity, have shown that CHIKV evolution has been a significant driving force of epidemics in India. This is evidenced by the current surge in the number of epidemics witnessed in recent years since 2010 despite the under-reporting for obvious reasons. The ability to adapt to a new vector and expand its geographical distribution by a single point mutation as observed in E1-A226V makes CHIKV a dynamic pathogen that has potential to attain a global public health concern.
Antiviral and Vaccine Initiatives
Currently, there is no licensed therapeutics or vaccines for the treatment of CHIKV infections in the market. However, this section describes various efforts at different stages of development, both of therapeutics and vaccines against CHIKV.
Vaccines
Vaccines have a background marked by the beginning in the late 18th century across the globe (Plotkin, 2014) and India was amongst few countries to be involved in these efforts. First CHIKV episode during 1960s prompted the start of research for the development of CHIKV immunization (Halstead et al., 1963). Since then, the quest for CHIKV vaccine candidates that balances the immunogenicity and provides adequate safety to CHIKV pathogenicity continues (Gao et al., 2019).
The very first live attenuated vaccine (LAV) of CHIKV was made in 1986 from Thailand isolate AF15561. LAV was known as 181/clone25 and it was made through 18 plaque passages in Human lung cells (MRC-5). This antibody was profoundly immunogenic in humans, however, it led to arthralgia in few cases (Edelman et al., 2000). Roques et al. (2017) developed vaccine candidates like MVA-CE and DREP-E in non-human primate model that targeted the nsP3 envelope and capsid using ECSA strain of Indian Ocean lineage. They were able to generate neutralizing antibodies against an isolate of the Asian Genotype. The role of nuclear localization sequence (NoLS) in the N-terminal region of capsid protein was characterized for the generation of LAVs. CHIKV infection was effectively neutralized when serum from CHIKV-NoLS-immunized mice was introduced, and the infectious titer of CHIKV-NoLS was found to be insensitive to freeze-thaw cycles, unlike CHIKV-WT, thus demonstrating preclinical safety and stability of CHIKV-NoLS (Taylor et al., 2017; Abeyratne et al., 2018). A few LAVs have been designed based on multiple replacements of synonymous codons thereby leading to decrease in the virus mutational robustness. The approach considerably reduces the risk of reversion because it involves hundreds of synonymous modifications to the genome, making such viruses as promising vaccine candidates (Carrau et al., 2019). Production of LAVs by mutation of non-structural proteins was also investigated by Chan et al. (2019). They found R532H mutation in nsP1 caused reduced infectivity in mouse tail fibroblasts but type-I IFN response was enhanced compared to WT-CHIKV (Chan et al., 2019). Recently, in a novel attempt to generate LAV candidate for CHIKV with complete deficiency of capsid, a single dose of the virus was reported to provide complete protection upon challenge with wild-type CHIKV in mice (Zhang et al., 2019).
Inactivated vaccines are considered to be secured and safe. Initially, researchers attempted to generate CHIKV vaccines by inactivating the virus by the means of heat or chemical treatment. The very first CHIKV vaccine was a formalin inactivated virus prepared from an assortment of cell like chick embryo cells and African green monkey kidney cells way back in 1972 (Eckels et al., 1970; White et al., 1972). Although, the formalin inactivated vaccine induced neutralizing antibody responses successfully and was effective in protecting mice against an intracerebral challenge, but it did not induce potent protective immune response in human volunteers (Harrison et al., 1971). Tiwari et al. (2009) had a trial where inactivated vaccine was produced in Vero cells and the titer of neutralizing antibodies peaked at 6–8 weeks post vaccination.
One of the most successful strategies in the field of vaccine development is the use of virus vectors that involves use of less pathogenic viruses to deliver genetic materials into living cells. In case of CHIKV vaccine development, the CHIKV structural genes are inserted into the genome of the virus vector that initiates the expression of CHIKV structural proteins or contains CHIKV structural proteins embedded in the virion (Ramsauer and Tangy, 2016). A new measles virus vectored vaccine (MV-CHIK) was tested for its efficacy in preventing CHIKV infection in a non-human primate model and found to be protective when challenged with the virulent La Reunion CHIKV strain. These results further documented the immunogenicity and efficacy of such vaccines that exhibited promising results in Phase I-II clinical trials (Reisinger et al., 2019; Rossi et al., 2019).
Another popular technique utilized in CHIKV vaccine development is the use of chimeric vaccine. To defeat the reversion capability of generally attenuated alphavirus vaccine and to address the developing demand to prevent CHIKV infections, chimeric approach has been employed for the development of vaccine. Using the structural genes of CHIKV and either a naturally attenuated strain of eastern equine encephalitis virus (EEEV), Venezuelan equine encephalitis attenuated vaccine strain TC-83, or Sindbis virus, chimeric alphavirus vaccine candidates were developed. The vaccinated mice were found to be fully protected against the disease, characterized by high titers of neutralizing antibodies (Wang et al., 2008; Plante et al., 2011). In another approach, chimeric virus was designed with insect specific alphavirus Eilat virus (EILV), and CHIKV structural proteins. Although, it mimicked the early stages of CHIKV replication in vertebrates, yet, it remained completely defective for viral productive replication, thus, providing a high degree of safety and robust immunity against viremia (Erasmus et al., 2017).
Couderc et al. (2009) purified human polyvalent immunoglobins from plasma samples of patients in the convalescent phase of CHIKV infection and investigated its curative effect in CHIKV mouse models. These immunoglobulins were found to contain anti-CHIKV antibodies and exhibited in vitro neutralizing activity and in vivo prophylactic and therapeutic efficacy. Hence, this approach can provide a safe preventive strategy for CHIKV infected individuals (Couderc et al., 2009). The immunogenic potential of recombinant proteins has been reported with time. Khan et al. (2012) expressed the CHIKV envelope protein in E. coli and found that the recombinant protein not only retained their antigenicity and immunogenicity but also induced production of neutralizing antibodies and cell mediated immune response in immunized mice.
Virus like particles (VLPs) as a vaccine candidate has been evaluated in vitro and in vivo in Balb/C mice and induction of both humoral and cellular immunity were observed. Neonates immunized with anti-CHIKV-VLP antibodies were protected from CHIKV infection, establishing VLP as a promising vaccine candidate (Saraswat et al., 2016).
Many advancements in molecular and biochemical technologies have led to the adoption of various strategies to develop vaccines as summarized in Table 3.
TABLE 3.
List of possible CHIKV vaccine candidates.
| Vaccine | Target | Type of vaccine | Status | References |
| Global initiatives | ||||
| T-E CHIKV | Lipid containing envelope of the virus | Inactivated Vaccine; 1st generation | Preclinical | Eckels et al. (1970) |
| USPHSRPB | Structural and envelope proteins | Inactivated Vaccine; 1st generation | Phase I | Harrison et al. (1971) |
| USPHSRPB | Structural and envelope proteins | Inactivated Vaccine; 1st generation | White et al. (1972) | |
| CHIK181/clone 25 | Reversion of attenuating point mutations | LAV; 1st generation | Preclinical | Levitt et al. (1986) |
| TSI-GSD-218 | Inhibition of viral replication | LAV; 1st generation | Phase II | Edelman et al. (2000) |
| 181/c25 | Reversion of attenuating point mutations | Chimeric virus; 3rd generation | Preclinical | Wang et al. (2008) |
| CHIKVIg-01 | Prevention of viral dissemination in tissues | Subunit Vaccine; 2nd generation | Preclinical | Couderc et al. (2009) |
| CMB/R | Envelope proteins | VLP; 2nd generation | Preclinical | Akahata et al. (2010) |
| CHIKV/IRES | Targets viral entry | Recombinant Vaccine; 3rd generation | Preclinical | Plante et al. (2011) |
| VEE/IRES-C/CHIKV | nsP2 envelope and capsid | Chimeric virus; 3rd generation | Preclinical | Wang et al. (2011) |
| pMCE321 | E1 and E2 glycoproteins | DNA vaccine; 3rd generation | Preclinical | Mallilankaraman et al. (2011) |
| dMAb | CHIKV envelope | DNA vaccine; 3rd generation | Preclinical | Muthumani et al. (2016) |
| EILV/CHIK | EILV cDNA clone containing CHIKV structural proteins | Chimeric virus; 3rd generation | Preclinical | Erasmus et al. (2017) |
| MVA-CE | nsP3 envelope and capsid | LAV;1st generation | Preclinical | Roques et al. (2017) |
| DREP-E | nsP3 envelope and capsid | LAV;1st generation | Phase II | Roques et al. (2017) |
| CHIKV-NoLS | N-terminal region of capsid protein | LAV;1st generation | Preclinical | Taylor et al. (2017), Abeyratne et al. (2018) |
| MV-CHIK | Measles vectored CHIKV structural proteins | Vector virus; 3rd generation | Preclinical | Reisinger et al. (2019), Rossi et al. (2019) |
| CHIKV | Multiple synonymous mutations in genome to reduce mutational robustness | LAV; 1st generation | Preclinical | Carrau et al. (2019) |
| SuperStop | Multiple synonymous mutations in genome to reduce mutational robustness | LAV; 1st generation | Preclinical | Carrau et al., 2019) |
| Stop CHIKV | Multiple synonymous mutations in genome to reduce mutational robustness | LAV; 1st generation | Phase I | Carrau et al. (2019) |
| 15nsP3 | E515V-nsp2 | LAV; 1st generation | Preclinical | Chan et al. (2019) |
| 1C-CHIKV | Capsid deletion | LAV; 1st generation | Preclinical | Zhang et al. (2019) |
| RHEV-CHIKV | E515V-nsp2 | LAV; 1st generation | In vitro | Chan et al. (2019) |
| Indian initiatives | ||||
| CK1/2 | Viral replication | Vector virus; 3rd generation | In vitro | Dash et al. (2008) |
| CHIK-FI | Envelope polyprotein | LAV; 1st generation | Preclinical | Tiwari et al. (2009) |
| rCHIKE1/E2 | E1 envelop protein | Subunit; 2nd generation | In vitro | Khan et al. (2012) |
| CHIKV-VLPs | Structural proteins introduced into yeast expression system | VLP; 2nd generation | Preclinical | Saraswat et al. (2016) |
| VSV1G-CHIKV | E3-E2-6K-E1 envelope polyprotein | Vector virus; 3rd generation | Preclinical | Subudhi et al. (2018) |
As a result of the urgent need for vaccines against various viral infections, Government of India started National Immunization Program under which various licensed vaccine manufacturing units were set up in the country. In the beginning of twentieth century, vaccine institutes were setup in India, which eventually moved toward privatization. Bharat Biotech, Indian.
Immunological, Defense Research Development Establishment, National Institute of Virology, Serum Institute of India are few Indian companies/organizations working on CHIKV vaccine dedicatedly.
In the quest to develop an acceptable vaccine against CHIKV, certain considerations must be followed. First, the vaccine titer should be enhanced so that it provides better protection. Second, the future vaccines should provide long term protection. However, short term protection is also required to control local outbreaks. Third, the dose of vaccine would be such that there should not be significant difference in the response to a vaccine owing to the complexity of population (different age groups). Fourth, the vaccine candidates that are effective in animal models should be taken forward for human trials. Fifth, the cost of production of vaccine should be cheap and affordable. Sixth, the vaccines should be thermostable, unlike RNA, so that it could be easy for transport and storage. Lastly, their administration should be convenient so that it does not cause any discomfort or side effects post vaccination. Hence, it can be assumed that with the employment of new strategies and new studies, commercially cost effective CHIKV vaccine with high safety and strong immunogenicity can be developed in the near future.
Antivirals
Albeit different antivirals are accessible for different viral diseases, there are no approved antivirals against CHIKV diseases. Hence, the current therapies mostly involve in management of symptoms using non-salicylate analgesics and non-steroid anti-inflammatory drugs (NSAIDS) (Thiberville et al., 2013). A number of known antiviral agents and other novel compounds have been examined for their activity against CHIKV and many show promises.
Ribavirin and 6-Azauridine show antiviral activity in a concentration dependent manner and found to reduce cytopathic effect and virus titer on infected Vero cells (Briolant et al., 2004). Arbidol a broad-spectrum antiviral drug showed potent inhibitory activity against CHIKV when examined on Vero cells and primary human fibroblasts (IC50 < 10 μg/ml) (Delogu et al., 2011). Furin inhibitors have been reported as potent inhibitors of CHIKV almost comparable to chloroquine (Ozden et al., 2008). CHIKV is highly sensitive to antiviral activity of Type I interferons (IFN-α/β), mediated by 2’,5’-oligoadenylate synthetase (OAS-3) family of proteins (Brehin et al., 2009).
Diterpenoids like prostratin and 12-O-tetradecanoylphorbol 13-acetate (TPA) exhibited antiviral activities in virus-cell-based assays against CHIKV as well as other alphaviruses (Bourjot et al., 2012). Polyinosinic acid (Poly I:C), mimic of double-stranded RNA is recognized by toll like receptor-3 (TLR-3) leading to induction of interferons in many cell types. TLR-3 pathway is suggestive of inducing innate immune responses against many viruses including CHIKV (Li et al., 2012). Since CHIKV infection is usually cleared within 7–8 days post infection before adaptive immune response emerges, therefore innate immune response becomes important for CHIKV clearance.
Several studies have also examined different compounds including viperin; resazurin; human monoclonal antibody C9:also many human and mouse anti-CHIKV monoclonal antibodies have been found protective in vivo; favipiravir; Retinoic acid inducible gene I (RIG-I); Various NSAIDS like mefenamic acid in combination with common antiviral drug like ribavirin; sulfonyl amidines and flavaglines (FL3 and FL23); imipramine; Curcumin; a series of arylalkylidene derivatives of 1, 3-thiazolidin-4-one; chloroquine; and ribavirin (Dash et al., 2008; Ravichandran and Manian, 2008; Padmakumar et al., 2009; Khan et al., 2012; Teng et al., 2012; Cruz et al., 2013; Parashar et al., 2013; Rathore et al., 2013; Selvarajah et al., 2013; Chopra et al., 2014; Delang et al., 2014; Olagnier et al., 2014; Albulescu et al., 2015; Jadav et al., 2015; Wintachai et al., 2015; Rothan et al., 2016; Mounce et al., 2017; Wichit et al., 2017; Subudhi et al., 2018; Jin and Simmons, 2019; Martins et al., 2020; Dong and Dimopoulos, 2021) and reported promising activity against CHIKV as summarized in Table 4. Lack of licensed antivirals or vaccines for CHIKF makes it yearning for the development of potent drugs against it.
TABLE 4.
Showing tested antiviral drugs against CHIKV.
| Antivirals | Target | Assay type | References |
| Global initiatives | |||
| 6-Azauridine | Inhibition of orotidine monophosphate decarboxylse enzyme | In vitro (vero cells) | Briolant et al. (2004) |
| Chloroquine | Inhibition of fusion of the viral E1 protein with endosomal membrane by raising the endosomal pH |
In vitro (vero cells) and in vivo (macaque models) Clinical trials in CHIKV infected Humans |
Roques et al. (2018) |
| Furin inhibitors | Inhibition of virus maturation via inhibition of cellular furins | In vitro (Human muscle satellite cells) | Ozden et al. (2008) |
| Oligoadenylate synthetase (OAS-3) | Inhibits CHIKV replication through RNase L dependant pathway | In vitro (HeLa cells) | Brehin et al. (2009) |
| Arbidol | Inhibits CHIKV replication through single amino acid substitution | In vitro (vero cells) | Delogu et al. (2011) |
| 5,7-Dihydroxyflavones | Unidentified target and mechanism of action | In vitro (BHK replicon cell line) | Pohjala et al. (2011) |
| Prostratin and TPA | Activation of protein kinase C | In vitro (vero cells) | Bourjot et al. (2012) |
| Polyinosinic acid | Stimulation of IFN-α and β and antiviral genes | In vitro (BEAS-2B cells) | Li et al. (2012) |
| Viperin | Targets endoplasmic reticulum | In vivo (monocytes) | Teng et al. (2012) |
| Resazurin | Inhibition of kinases involved in apoptosis | In vitro (HuH-7 cells) | Cruz et al. (2013) |
| Monoclonal antibody C9 | Interaction with CHIKV E2 glycoproteins |
In vitro (vero cells) In vivo (C57/BL6 mice) |
Selvarajah et al. (2013) |
| Favipiravir | Inhibition of viral genome replication |
In vitro (vero cells) In vivo (AG129 mice) |
Delang et al. (2014) |
| RIG-1 agonists | Stimulation of immune response | In vitro (MRC-5 cells) | Olagnier et al. (2014) |
| Suramin | Inhibition of CHIKV RNA synthesis | In vitro (BHK-21, and Vero-E6 cells) | Albulescu et al. (2015) |
| Ribavirin | Inhibition of viral genome replication via GTP pool depletion |
In vitro (vero cells) In vivo (ICR mice) |
Rothan et al. (2015) |
| Flavaglines | Interference with the binding of CHIKV Prohibitin-1 | In vitro (HEK-293T cells) | Wintachai et al. (2015) |
| Benzouracil-Coumarin-Arene conjugates | Unidentified target and mechanism of action | In vitro (vero A cells) | Hwu et al. (2015) |
| Mefenamic acid in combination with ribavirin | Inhibition of viral replication |
In vitro (vero cells) In vivo (ICR mice) |
Rothan et al. (2016) |
| Imipramine | Inhibition of viral replication | In vitro (vero and HFF1 cells) | Wichit et al. (2017) |
| Curcumin | Inhibition of viral binding at the cell surface | In vitro (HeLa, BHK-21, and Vero-E6 cells) | Mounce et al. (2017) |
| Chloroquine | Inhibition of viral replication | In vitro (primary macrophages and fibroblasts cells) in vivo (cynomolgus macaques) | Roques et al. (2018) |
| Indian initiatives | |||
| Ribavirin | Inhibition of viral genome replication | Clinical trials in CHIKV infected Humans | Ravichandran and Manian (2008) |
| Aceclofenac in combination with hydroxychloroquine and prednisolone | inhibition of synthesis of prostaglandins and having chondroprotective effect | Humans | Padmakumar et al. (2009) |
| Chloroquine | Inhibition of fusion of the viral E1 protein with endosomal membrane by raising the endosomal pH | In vitro (vero cells) | Khan et al. (2010), Chopra et al. (2014) |
| Mycophenolic acid (MPA) | Depletion of intracellular guanosine pool | In vitro (vero cells) | Khan et al. (2011) |
| Si-RNAs | Inhibition of protein synthesis by targeting nsp1 and E2 | In vivo (swiss albino and C57/BL6 mice) | Parashar et al. (2013) |
| HSP-90 inhibitors (HS-10 and SNX-2112) | Interaction with CHIKV nsp3 and nsp4 |
In vitro (HEK-293T cells) In vivo (SvA129 mice) |
Rathore et al. (2014) |
| Thiazolidine derivative | Inhibition of CHIKV nsp2 protease activity | In vitro (vero cells) | Jadav et al. (2015) |
Novel Drug Repurposing Initiatives Against Chikungunya
Lots of existing drugs are being repurposed for fighting against CHIKV as they have well established formulations, well known clinical trial safety data. Novel drug discovery requires enormous amount of time, money, and effort. Success rate of drug repurposing or drug repositioning is relatively high because the target drugs have already been tested for their efficiency against other diseases and have been proved to be safe for human use. Also, the existing drugs against CHIKV have achieved only limited success with questionable therapeutic effectiveness (Subudhi et al., 2018). Considering the extensive research in this field, it is worthwhile to mention a list of few drugs available for repurposing and the rest are listed in the table below (Table 5).
TABLE 5.
List of suggested novel antivirals and repurposing initiatives against CHIKV.
| Drugs | Targeted virus | Assay type | References |
| Seco-pregnane steroids | TMV, SINV | In vivo | Li et al. (2007) |
| Naringenin, Apigenin | CHIKV | In vitro | Pohjala et al. (2011) |
| Lupenone, β-amyrone | CHIKV, DENV | In vitro | Bourjot et al. (2012) |
| Baicalin, Quercetagetin | CHIKV | In silico | Seyedi et al. (2016) |
| Hesperetin | CHIKV | In silico | Oo et al. (2016) |
| Doxycycline | CHIKV | In silico | Rothan et al. (2016) |
| Pimozide, Cerulenin, and Tivozanib | CHIKV |
In vitro In vivo |
Karlas et al. (2016) |
| Auranofin | CHIKV |
In vitro In vivo |
Langsjoen et al. (2017) |
| Acrylamides derivative (LQM334) | CHIKV | In silico | Passos et al. (2020) |
| Indian Initiatives | |||
| Harringtonine | CHIKV | In vitro | Kaur et al. (2013) |
| Aplysiatoxin | CHIKV | In vitro | Gupta et al. (2014) |
| CID-5808891 | CHIKV | In silico | Agarwal et al. (2015) |
| Picolinate | CHIKV | In silico | Sharma et al. (2016) |
| Ribostamycin sulfate | CHIKV | In silico | Kumar et al. (2019) |
| Dihydrorugosa flavonoids | CHIKV | In silico | Puranik et al. (2019) |
| Rutin, Moralbanone, and Kaempferol | CHIKV | In silico | Khan et al. (2020) |
Seco-Pregnane Steroids
Seco-pregnane steroid glaucogenin C and its monosugar glycoside cynatratoside A have been shown to be non-toxic to host cells while selectively inhibiting alphavirus-like positive-strand RNA virus (Li et al., 2007).
Baicalin
Compared to other ligands, baicalin has been identified as a potential inhibitor of viral activity by showing a good binding affinity (−9.8 kcal/mol) (Seyedi et al., 2016).
Hesperetin
Hesperetin, a bioflavonoid, exhibits inhibitory effect on multiple viruses in vitro. Results from in silico analysis reveal that hesperetin exhibits drug-like properties which projects its potential as a therapeutic option for CHIKV infection (Oo et al., 2016).
Harringtonine
In an immunofluorescence-based screening platform, harringtonine, a cephalotaxi alkaloid, inhibits early stage of CHIKV replication cycle which occurs after viral entry into cells (Kaur et al., 2013).
Aplysiatoxin
Aplysiatoxin related compound extracted from marine cyanobacteria Trichodesmium erythraeum has also been found to exhibit dose dependent inhibition of CHIKV (Gupta et al., 2014).
Traditional Indian Medical System to Combat CHIKV
Dilip et al. (2010) compared the traditional system of medicines in India like Ayurveda, Unani, Homeopathy. Homeopathy drugs such as, Eupatorium, Pyroginum, Nux Vomica, Gelsemium, Arnica, and Belladonna were used for the treatment and prevention of CHIKV and were found to provide 70–80% relief in symptoms within 5 days. Similarly, Ayurvedic method of medication with Vanatulasi patra, Beruenna oil, Vilwadi gulika, Vettumaran gulika, and Sudarshan tablets, etc., showed promising effects in the treatment and prevention of CHIKV. Hence, it was concluded from the study that all these methods of treatment are equally important for the management of chikungunya (Dilip et al., 2010). Since these studies were performed on a limited scale, hence further investigations are required to evaluate the effectiveness of the aforementioned traditional medicines. A homeopathy therapy in acute phase and post-CHIKV chronic arthritis reported 90% recovery of chronic arthritis patients after an average time of 32.5 days. Initial doses of the homeopathic medication was three times a day gradually being reduced to two and then one with improvement (Wadhwani, 2013). Although, the study was promising yet authors did acknowledge that it had certain experimental limitations. Hence, a revised study overcoming the constraints could indubitably establish its efficacy. The extracts and compounds isolated from Tectona grandis were found to be promising drug candidates to both the Asian and ECSA strains of CHIKV (Sangeetha et al., 2017; Martins et al., 2020). Jain et al. (2018b) tried combinations of herbal formulations, practiced by herbal medicine practitioners according to AYUSH, Govt. of India guidelines, for CHIKV treatment. These herbal formulations like Giloy (Tinospora cordifolia), Spirulina (Spirulina platensis), Ashwagandha (Withania somnifera) and Sonth (Zingiber officinale) are effective against CHIKV and 100% recovery after 10 days of treatment was observed.
Conclusion and Research Gap
Chikungunya outbreak is a huge public health concern in India. Twenty-four Indian states and six union territories are endemic for CHIKV. Despite causing acute and self-limiting disease with very low mortality, CHIKF is a challenge to the human population because of the painful course of illness and long-term sequelae which affect the quality of life negatively. Therefore, it is important to ensure its prevention and control measures. Year wise increase of the CHIKV epidemics in India is an alarming sign that prompted us to focus on the understanding about virus biology, virus-vector/virus-host interaction, vector distribution and transmission, strain mutation and circulation. Developing extensive clinical and laboratory diagnostic protocols to delineate CHIKV infection from other flaviviruses like dengue, zika and other alphaviruses such as o’nyong’nyong virus, Sindbis virus and malaria for effective diagnosis is the need of the hour. Continuous surveillance has already been advocated by several studies in India and it should be supported for the apparent reasons. This is very important for early detection of adaptive changes in the viral genome, and prompt risk assessment that would encourage implementation of intervention and control measures to moderate the impact of CHIKV outbreaks in India in the future. A number of novel mutations have been reported in Indian studies that might have effect with respect to adaptability of the virus, spread and disease severity. More research is required to describe the significance of many of the observed mutations considering the fact that the E1-A226V is absent in many Indian epidemics. Till now no licensed vaccine is available, however; several potential vaccine candidates have been identified. Even though many drugs have shown effective CHIKV antiviral activities, studies on clinical efficacy to prove the same is still required. Apart from the conventional drug research, India is also focusing on the research on the traditional system of medicines in our country, like Ayurveda, Unani and Homeopathy which is showing promising results. CHIKV eradication is not only a matter of drugs and vaccines but responsible preventive measures are also to be taken. Until such strategies are made available, simple measures like use of mosquito repellents, expulsion of standing water where mosquitos can lay eggs and minimization of skin surface exposed to mosquito bites are the only measures to prevent this disease.
In the wake of the resurgent viral infectious disease including Zika, Nipah, and the covid-19 pandemic there has been tremendous advancement for existing technologies and disease diagnostics such as the CRISPR system, surveillance; therapeutics such as the use of convalescent plasma; and vaccine development such as the mRNA vaccine. The massive employment of bioinformatics and artificial intelligence tools in tracking infectious pathogen mutations and its implication on disease severity and transmission efficiency can also be used in studies of other viral infections like CHIKV. This suggest that developing a bioinformatic infrastructure for an infectious pathogen can be very helpful to track and make a treatment and prevention strategy to control disease spread. The quest for CHIKV vaccines the mRNA vaccine technology offers new hope.
Members of the Translational Research Consortia (TRC) for Chikungunya Virus in India
Anitha Jagadesh, Anup Jayaram, Naren Babu, Piya Paul Mudgal, Robin Sudandiradas, Shahin Sheik, and Ujwal Shetty: Manipal Institute of Virology, Manipal Academy of Higher Education, Manipal, Udupi, Karnataka, India; Dileep Kumar Verma, Shakuntala Mahilkar, Sujatha Sunil*, and Sylvester Agha Ibemgbo: Vector-Borne Diseases Group, International Centre for Genetic Engineering and Biotechnology (ICGEB), New Delhi, India. Prabhudutta Mamidi, Sharad Singh, Soma Chattopadhyay* and Sweta Smita Pani: Infectious Disease Biology, Institute of Life Sciences, (Autonomous Institute of Department of Biotechnology, Government of India), Nalco Square, Bhubaneswar, India. Bijayanthimala Mishra: Department of microbiology, All India Institute of Medical Sciences, (AIIMS), Bhubaneshwar, India; R. K. Ratho: Department of Virology, Post-Graduate Institute of Medical Education and Research, Chandigarh, India; Jayanthi S. Shastri and Sachee Agarwal: Department of Microbiology, TNMC and BYL, Nair Charitable Hospital, Mumbai, Maharashtra, India.
Author Contributions
SSu, SC, AJg, PPM, BM, RR, JS, and SA took part in Conceptualization and reviewing the manuscript. AJy, NB, RS, SSh, US, DK, SI, SM, PM, SSi, and SP drafting, revising, and preparation of the manuscript. SSu, SC, SM, and SI finalized and prepared the manuscript for submission. Further revisions according to the reviewer comments were made by SSu and SM and finalized by SSu.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This research was funded by National Biopharma Mission (NBM), BIRAC, Government of India (BT/NBM0101/02/18).
Contributor Information
The Translational Research Consortia (TRC) for Chikungunya Virus in India:
Anitha Jagadesh, Anup Jayaram, Naren Babu, Piya Paul Mudgal, Robin Sudandiradas, Shahin Sheik, Ujwal Shetty, Dileep Kumar Verma, Shakuntala Mahilkar, Sujatha Sunil*, Sylvester Agha Ibemgbo, Prabhudutta Mamidi, Sharad Singh, Soma Chattopadhyay*, Sweta Smita Pani, Bijayanthimala Mishra, R. K. Ratho, Jayanthi S. Shastri, and Sachee Agarwal
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.695173/full#supplementary-material
References
- Abeyratne E., Freitas J. R., Zaid A., Mahalingam S., Taylor A. (2018). Attenuation and stability of CHIKV-NoLS, a live-attenuated chikungunya virus vaccine candidate. Vaccines 7:2. 10.3390/vaccines7010002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acevedo N., Waggoner J., Rodriguez M., Rivera L., Landivar J., Pinsky B., et al. (2017). Zika virus, chikungunya virus, and dengue virus in cerebrospinal fluid from adults with neurological manifestations, guayaquil, ecuador. Front. Microbiol. 8:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal A., Dash P. K., Singh A. K., Sharma S., Gopalan N., Rao P. V., et al. (2014). Evidence of experimental vertical transmission of emerging novel ECSA genotype of Chikungunya Virus in Aedes aegypti. PLoS Negl. Trop. Dis. 8:e2990. 10.1371/journal.pntd.0002990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal A., Gupta S., Yadav A. K., Nema R. K., Ansari K., Biswas D. (2019). Molecular and phylogenetic analysis of Chikungunya virus in Central India during 2016 and 2017 outbreaks reveal high similarity with recent New Delhi and Bangladesh strains. Infect. Genet. Evol. 75:103940. 10.1016/j.meegid.2019.103940 [DOI] [PubMed] [Google Scholar]
- Agarwal A., Sharma A. K., Sukumaran D., Parida M., Dash P. K. (2016). Two novel epistatic mutations (E1:K211E and E2:V264A) in structural proteins of Chikungunya virus enhance fitness in Aedes aegypti. Virology 497 59–68. 10.1016/j.virol.2016.06.025 [DOI] [PubMed] [Google Scholar]
- Agarwal T., Asthana S., Bissoyi A. (2015). Molecular modeling and docking study to elucidate novel chikungunya virus nsP2 protease inhibitors. Indian J. Pharm. Sci. 77 453–460. 10.4103/0250-474x.164769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akahata W., Yang Z. Y., Andersen H., Sun S., Holdaway H. A., Kong W. P., et al. (2010). A virus-like particle vaccine for epidemic Chikungunya virus protects nonhuman primates against infection. Nat. Med. 16 334–338. 10.1038/nm.2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albulescu I. C., Van Hoolwerff M., Wolters L. A., Bottaro E., Nastruzzi C., Yang S. C., et al. (2015). Suramin inhibits chikungunya virus replication through multiple mechanisms. Antiviral Res. 121 39–46. 10.1016/j.antiviral.2015.06.013 [DOI] [PubMed] [Google Scholar]
- Álvarez-Argüelles M. E., Alba S. R., Pérez M. R., Riveiro J. A. B., García S. M. (2019). “Diagnosis and molecular characterization of chikungunya virus infections,” in Current Topics in Neglected Tropical Diseases ed Rodriguez-Morales A. J. (Germany: BoD – Books on Demand; ). [Google Scholar]
- Azami N. A., Moi M. L., Takasaki T. (2016). Neutralization assay for chikungunya virus infection: plaque reduction neutralization test. Methods Mol. Biol. 1426 273–282. 10.1007/978-1-4939-3618-2_25 [DOI] [PubMed] [Google Scholar]
- Badar N., Salman M., Ansari J., Ikram A., Qazi J., Alam M. M. (2019). Epidemiological trend of chikungunya outbreak in Pakistan: 2016-2018. PLoS Negl. Trop. Dis. 13:e0007118. 10.1371/journal.pntd.0007118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandeira A. C., Campos G. S., Sardi S. I., Rocha V. F., Rocha G. C. (2016). Neonatal encephalitis due to Chikungunya vertical transmission: first report in Brazil. IDCases 5 57–59. 10.1016/j.idcr.2016.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee K. (1965). A note on antibodies to chikungunya virus in human sera collected in Madras state in 1956. Indian J. Med. Res. 53 715–719. [PubMed] [Google Scholar]
- Benjamin M. (2012). “Chikungunya is not a Swahili word, it is from the Makonde language”.). J. Ophthalmic Inflamm. Infect. 3:35. [Google Scholar]
- Borgherini G., Poubeau P., Jossaume A., Gouix A., Cotte L., Michault A., et al. (2008). Persistent arthralgia associated with chikungunya virus: a study of 88 adult patients on reunion island. Clin. Infect. Dis. 47 469–475. 10.1086/590003 [DOI] [PubMed] [Google Scholar]
- Bourjot M., Delang L., Nguyen V. H., Neyts J., Gueritte F., Leyssen P., et al. (2012). Prostratin and 12-O-tetradecanoylphorbol 13-acetate are potent and selective inhibitors of Chikungunya virus replication. J. Nat. Prod. 75 2183–2187. 10.1021/np300637t [DOI] [PubMed] [Google Scholar]
- Brehin A. C., Casademont I., Frenkiel M. P., Julier C., Sakuntabhai A., Despres P. (2009). The large form of human 2’,5’-Oligoadenylate Synthetase (OAS3) exerts antiviral effect against Chikungunya virus. Virology 384 216–222. 10.1016/j.virol.2008.10.021 [DOI] [PubMed] [Google Scholar]
- Briolant S., Garin D., Scaramozzino N., Jouan A., Crance J. M. (2004). In vitro inhibition of Chikungunya and Semliki forest viruses replication by antiviral compounds: synergistic effect of interferon-alpha and ribavirin combination. Antivir. Res. 61 111–117. 10.1016/j.antiviral.2003.09.005 [DOI] [PubMed] [Google Scholar]
- Cardona-Correa S. E., Castano-Jaramillo L. M., Quevedo-Velez A. (2017). Vertical transmission of chikungunya virus infection. case report. Rev. Chil. Pediatr. 88 285–288. [DOI] [PubMed] [Google Scholar]
- Carletti F., Bordi L., Chiappini R., Ippolito G., Sciarrone M. R., Capobianchi M. R., et al. (2007). Rapid detection and quantification of Chikungunya virus by a one-step reverse transcription polymerase chain reaction real-time assay. Am. J. Trop. Med. Hyg. 77 521–524. 10.4269/ajtmh.2007.77.521 [DOI] [PubMed] [Google Scholar]
- Carrau L., Rezelj V. V., Noval M. G., Levi L. I., Megrian D., Blanc H., et al. (2019). Chikungunya virus vaccine candidates with decreased mutational robustness are attenuated in vivo and have compromised transmissibility. J. Virol. 93:e00775-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal P. E., Chouhy D., Bolatti E. M., Perez G. R., Stella E. J., Giri A. A. (2015). Evidence for homologous recombination in Chikungunya Virus. Mol. Phylogenet. Evol. 85 68–75. 10.1016/j.ympev.2015.01.016 [DOI] [PubMed] [Google Scholar]
- CDC (2018). Symptoms, Diagnosis, & Treatment | Chikungunya virus | [Online]. Atlanta, GA: CDC [Google Scholar]
- Chan Y. H., Teo T. H., Utt A., Tan J. J., Amrun S. N., Abu Bakar F., et al. (2019). Mutating chikungunya virus non-structural protein produces potent live-attenuated vaccine candidate. EMBO Mol. Med. 11:e10092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. Y., Encinales L., Porras A., Pacheco N., Reid S. P., Martins K. A. O., et al. (2018). Frequency of chronic joint pain following chikungunya virus infection: a colombian cohort study. Arthritis Rheumatol. 70 578–584. 10.1002/art.40384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelluboina S., Robin S., Aswathyraj S., Arunkumar G. (2019). Persistence of antibody response in chikungunya. Virusdisease 30 469–473. 10.1007/s13337-019-00534-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Wang E., Tsetsarkin K. A., Weaver S. C. (2013). Chikungunya virus 3’ untranslated region: adaptation to mosquitoes and a population bottleneck as major evolutionary forces. PLoS Pathog. 9:e1003591. 10.1371/journal.ppat.1003591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Foo S. S., Taylor A., Lulla A., Merits A., Hueston L., et al. (2015). Bindarit, an inhibitor of monocyte chemotactic protein synthesis, protects against bone loss induced by chikungunya virus infection. J. Virol. 89 581–593. 10.1128/jvi.02034-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiam C. W., Chan Y. F., Loong S. K., Yong S. S., Hooi P. S., Sam I. C. (2013). Real-time polymerase chain reaction for diagnosis and quantitation of negative strand of chikungunya virus. Diagn. Microbiol. Infect. Dis. 77 133–137. 10.1016/j.diagmicrobio.2013.06.018 [DOI] [PubMed] [Google Scholar]
- Chompoosri J., Thavara U., Tawatsin A., Boonserm R., Phumee A., Sangkitporn S., et al. (2016). Vertical transmission of Indian Ocean Lineage of chikungunya virus in Aedes aegypti and Aedes albopictus mosquitoes. Parasit. Vectors 9:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra A., Saluja M., Venugopalan A. (2014). Effectiveness of chloroquine and inflammatory cytokine response in patients with early persistent musculoskeletal pain and arthritis following chikungunya virus infection. Arthritis Rheumatol. 66 319–326. 10.1002/art.38221 [DOI] [PubMed] [Google Scholar]
- Couderc T., Khandoudi N., Grandadam M., Visse C., Gangneux N., Bagot S., et al. (2009). Prophylaxis and therapy for Chikungunya virus infection. J. Infect. Dis. 200 516–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz D. J., Bonotto R. M., Gomes R. G., Da Silva C. T., Taniguchi J. B., No J. H., et al. (2013). Identification of novel compounds inhibiting chikungunya virus-induced cell death by high throughput screening of a kinase inhibitor library. PLoS Negl. Trop. Dis. 7:e2471. 10.1371/journal.pntd.0002471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Gao M., Ren X. (2011). Phylogeny and homologous recombination in Chikungunya viruses. Infect. Genet. Evol. 11 1957–1963. 10.1016/j.meegid.2011.08.026 [DOI] [PubMed] [Google Scholar]
- Darwish M. A., Hoogstraal H., Roberts T. J., Ahmed I. P., Omar F. (1983). A sero-epidemiological survey for certain arboviruses (Togaviridae) in Pakistan. Trans. R. Soc. Trop. Med. Hyg. 77 442–445. 10.1016/0035-9203(83)90106-2 [DOI] [PubMed] [Google Scholar]
- Dash P. K., Parida M., Santhosh S. R., Saxena P., Srivastava A., Neeraja M., et al. (2008). Development and evaluation of a 1-step duplex reverse transcription polymerase chain reaction for differential diagnosis of chikungunya and dengue infection. Diagn. Microbiol. Infect. Dis. 62 52–57. 10.1016/j.diagmicrobio.2008.05.002 [DOI] [PubMed] [Google Scholar]
- Delang L., Segura Guerrero N., Tas A., Querat G., Pastorino B., Froeyen M., et al. (2014). Mutations in the chikungunya virus non-structural proteins cause resistance to favipiravir (T-705), a broad-spectrum antiviral. J. Antimicrob. Chemother. 69 2770–2784. 10.1093/jac/dku209 [DOI] [PubMed] [Google Scholar]
- Delogu I., Pastorino B., Baronti C., Nougairede A., Bonnet E., De Lamballerie X. (2011). In vitro antiviral activity of arbidol against Chikungunya virus and characteristics of a selected resistant mutant. Antivir. Res. 90 99–107. 10.1016/j.antiviral.2011.03.182 [DOI] [PubMed] [Google Scholar]
- Department of Health and Family Welfare, Government of India (2018). “Chapter 5: disease control program,” in Annual Report 2017-2018, Department of Health and Family Welfare, Government of India. Available online at: https://main.mohfw.gov.in/sites/default/files/05Chapter.pdf [Google Scholar]
- Dilip C., Saraswathi R., Krishnan P., Azeem A. (2010). Comparitive evaluation of different systems of medicines and the present scenario of chikungunya in Kerala. Asian Pac. J. Trop. Med. 3 443–447. 10.1016/s1995-7645(10)60106-x [DOI] [Google Scholar]
- Dong S., Dimopoulos G. (2021). Antiviral compounds for blocking arboviral transmission in mosquitoes. Viruses 13:108. 10.3390/v13010108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckels K. H., Harrison V. R., Hetrick F. M. (1970). Chikungunya virus vaccine prepared by Tween-ether extraction. Appl. Microbiol. 19 321–325. 10.1128/aem.19.2.321-325.1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman R., Tacket C. O., Wasserman S. S., Bodison S. A., Perry J. G., Mangiafico J. A. (2000). Phase II safety and immunogenicity study of live chikungunya virus vaccine TSI-GSD-218. Am. J. Trop. Med. Hyg. 62 681–685. 10.4269/ajtmh.2000.62.681 [DOI] [PubMed] [Google Scholar]
- Edwards C. J., Welch S. R., Chamberlain J., Hewson R., Tolley H., Cane P. A., et al. (2007). Molecular diagnosis and analysis of Chikungunya virus. J. Clin. Virol. 39 271–275. 10.1016/j.jcv.2007.05.008 [DOI] [PubMed] [Google Scholar]
- Erasmus J. H., Auguste A. J., Kaelber J. T., Luo H., Rossi S. L., Fenton K., et al. (2017). A chikungunya fever vaccine utilizing an insect-specific virus platform. Nat. Med. 23 192–199. 10.1038/nm.4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filomatori C. V., Bardossy E. S., Merwaiss F., Suzuki Y., Henrion A., Saleh M. C., et al. (2019). RNA recombination at Chikungunya virus 3’UTR as an evolutionary mechanism that provides adaptability. PLoS Pathog. 15:e1007706. 10.1371/journal.ppat.1007706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritel X., Rollot O., Gerardin P., Gauzere B. A., Bideault J., Lagarde L., et al. (2010). Chikungunya virus infection during pregnancy. reunion, France, 2006. Emerg. Infect. Dis. 16 418–425. 10.3201/eid1604.091403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya-Kanamori L., Liang S., Milinovich G., Soares Magalhaes R. J., Clements A. C. A., Hu W., et al. (2016). Co-distribution and co-infection of chikungunya and dengue viruses. BMC Infect. Dis. 16:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S., Song S., Zhang L. (2019). Recent progress in vaccine development against Chikungunya Virus. Front. Microbiol. 10:2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George A., Amrutha M. S., Srivastava P., Sunil S., Sai V. V. R., Srinivasan R. (2021). Development of a U-bent plastic optical fiber biosensor with plasmonic labels for the detection of chikungunya non-structural protein 3. Analyst 146 244–252. 10.1039/d0an01603a [DOI] [PubMed] [Google Scholar]
- George A., Amrutha M., Srivastava P., Sai V., Sunil S., Srinivasan R. (2019). Label-free detection of Chikungunya non-structural protein 3 using electrochemical impedance spectroscopy. J. Electrochem. Soc. 166:B1356. [Google Scholar]
- Gerardin P., Barau G., Michault A., Bintner M., Randrianaivo H., Choker G., et al. (2008). Multidisciplinary prospective study of mother-to-child chikungunya virus infections on the island of La Reunion. PLoS Med. 5:e60. 10.1371/journal.pmed.0050060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta D. K., Kaur P., Leong S. T., Tan L. T., Prinsep M. R., Chu J. J. (2014). Anti-Chikungunya viral activities of aplysiatoxin-related compounds from the marine cyanobacterium Trichodesmium erythraeum. Mar. Drugs 12 115–127. 10.3390/md12010115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead S., Yamarat C., Scanlon J. (1963). The Thai hemorrhagic fever epidemic of 1962: a preliminary report. J. Med. Assoc. Thai. 46 449–462. [Google Scholar]
- Hapuarachchi H. C., Bandara K. B., Sumanadasa S. D., Hapugoda M. D., Lai Y. L., Lee K. S., et al. (2010). Re-emergence of Chikungunya virus in South-east Asia: virological evidence from Sri Lanka and Singapore. J. Gen. Virol. 91 1067–1076. 10.1099/vir.0.015743-0 [DOI] [PubMed] [Google Scholar]
- Harrison V. R., Eckels K. H., Bartelloni P. J., Hampton C. (1971). Production and evaluation of a formalin-killed Chikungunya vaccine. J. Immunol. 107 643–647. [PubMed] [Google Scholar]
- Harsha P. K., Reddy V., Rao D., Pattabiraman C., Mani R. S. (2020). Continual circulation of ECSA genotype and identification of a novel mutation I317V in the E1 gene of Chikungunya viral strains in southern India during 2015-2016. J. Med. Virol. 92 1007–1012. 10.1002/jmv.25662 [DOI] [PubMed] [Google Scholar]
- Hoarau J. J., Jaffar Bandjee M. C., Krejbich Trotot P., Das T., Li-Pat-Yuen G., Dassa B., et al. (2010). Persistent chronic inflammation and infection by Chikungunya arthritogenic alphavirus in spite of a robust host immune response. J. Immunol. 184 5914–5927. 10.4049/jimmunol.0900255 [DOI] [PubMed] [Google Scholar]
- Hochedez P., Canestri A., Guihot A., Brichler S., Bricaire F., Caumes E. (2008). Management of travelers with fever and exanthema, notably dengue and chikungunya infections. Am. J. Trop. Med. Hyg. 78 710–713. 10.4269/ajtmh.2008.78.710 [DOI] [PubMed] [Google Scholar]
- Hwu J. R., Kapoor M., Tsay S. C., Lin C. C., Hwang K. C., Horng J. C., et al. (2015). Benzouracil-coumarin-arene conjugates as inhibiting agents for chikungunya virus. Antivir. Res. 118 103–109. 10.1016/j.antiviral.2015.03.013 [DOI] [PubMed] [Google Scholar]
- Hyde J. L., Chen R., Trobaugh D. W., Diamond M. S., Weaver S. C., Klimstra W. B., et al. (2015). The 5’ and 3’ ends of alphavirus RNAs–Non-coding is not non-functional. Virus Res. 206 99–107. 10.1016/j.virusres.2015.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadav S. S., Sinha B. N., Hilgenfeld R., Pastorino B., De Lamballerie X., Jayaprakash V. (2015). Thiazolidone derivatives as inhibitors of chikungunya virus. Eur. J. Med. Chem. 89 172–178. 10.1016/j.ejmech.2014.10.042 [DOI] [PubMed] [Google Scholar]
- Jadhav M., Namboodripad M., Carman R. H., Carey D. E., Myers R. M. (1965). Chikungunya disease in infants and children in Vellore: a report of clinical and haematological features of virologically proved cases. Indian J. Med. Res. 53 764–776. [PubMed] [Google Scholar]
- Jain J., Dubey S. K., Shrinet J., Sunil S. (2017). Dengue Chikungunya co-infection: a live-in relationship? Biochem. Biophys. Res. Commun. 492 608–616. 10.1016/j.bbrc.2017.02.008 [DOI] [PubMed] [Google Scholar]
- Jain J., Kaur N., Haller S. L., Kumar A., Rossi S. L., Narayanan V., et al. (2020). Chikungunya outbreaks in India: a prospective study comparing neutralization and sequelae during two outbreaks in 2010 and 2016. Am. J. Trop. Med. Hyg. 102 857–868. 10.4269/ajtmh.19-0481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain J., Kushwah R. B. S., Singh S. S., Sharma A., Adak T., Singh O. P., et al. (2016). Evidence for natural vertical transmission of chikungunya viruses in field populations of Aedes aegypti in Delhi and Haryana states in India-a preliminary report. Acta Trop. 162 46–55. 10.1016/j.actatropica.2016.06.004 [DOI] [PubMed] [Google Scholar]
- Jain J., Okabayashi T., Kaur N., Nakayama E., Shioda T., Gaind R., et al. (2018a). Evaluation of an immunochromatography rapid diagnosis kit for detection of chikungunya virus antigen in India, a dengue-endemic country. Virol. J. 15:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain J., Pai S., Sunil S. (2018b). Standardization of in vitro assays to evaluate the activity of polyherbal siddha formulations against Chikungunya virus infection. Virusdisease 29 32–39. 10.1007/s13337-018-0421-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Simmons G. (2019). Antiviral functions of monoclonal antibodies against Chikungunya Virus. Viruses 11:305. 10.3390/v11040305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantri S. P., Joshi R., Riley L. W. (2006). Chikungunya epidemic: an Indian perspective. Natl. Med. J. India 19 315–322. [PubMed] [Google Scholar]
- Karlas A., Berre S., Couderc T., Varjak M., Braun P., Meyer M., et al. (2016). A human genome-wide loss-of-function screen identifies effective chikungunya antiviral drugs. Nat. Commun. 7:11320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur M., Singh K., Sidhu S. K., Devi P., Kaur M., Soneja S., et al. (2018). Coinfection of chikungunya and dengue viruses: a serological study from North Western region of Punjab. India. J. Lab. Physicians 10 443–447. 10.4103/jlp.jlp_13_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur N., Jain J., Kumar A., Narang M., Zakaria M. K., Marcello A., et al. (2017). Chikungunya outbreak in Delhi, India, 2016: report on coinfection status and comorbid conditions in patients. New Microbes New Infect. 20 39–42. 10.1016/j.nmni.2017.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur P., Thiruchelvan M., Lee R. C., Chen H., Chen K. C., Ng M. L., et al. (2013). Inhibition of chikungunya virus replication by harringtonine, a novel antiviral that suppresses viral protein expression. Antimicrob. Agents Chemother. 57 155–167. 10.1128/aac.01467-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M., Dhanwani R., Patro I., Rao P., Parida M. (2011). Cellular IMPDH enzyme activity is a potential target for the inhibition of Chikungunya virus replication and virus induced apoptosis in cultured mammalian cells. Antivir. Res. 89 1–8. 10.1016/j.antiviral.2010.10.009 [DOI] [PubMed] [Google Scholar]
- Khan M., Dhanwani R., Rao P. V., Parida M. (2012). Subunit vaccine formulations based on recombinant envelope proteins of Chikungunya virus elicit balanced Th1/Th2 response and virus-neutralizing antibodies in mice. Virus Res. 167 236–246. 10.1016/j.virusres.2012.05.004 [DOI] [PubMed] [Google Scholar]
- Khan M., Santhosh S. R., Tiwari M., Lakshmana Rao P. V., Parida M. (2010). Assessment of in vitro prophylactic and therapeutic efficacy of chloroquine against Chikungunya virus in vero cells. J. Med. Virol. 82 817–824. 10.1002/jmv.21663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan N., Bhat R., Jain V., Raghuvendara B. S., Patel A. K., Ray P. (2020). Epidemiology and molecular characterization of Chikungunya virus from human cases in North India, 2016. Microbiol. Immunol. 21:12869. 10.1111/1348-0421.12869 [DOI] [PubMed] [Google Scholar]
- Khatun S., Chakraborty A., Rahman M., Nasreen Banu N., Rahman M. M., Hasan S. M., et al. (2015). An outbreak of Chikungunya in Rural Bangladesh, 2011. PLoS Negl. Trop. Dis. 9:e0003907. 10.1371/journal.pntd.0003907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacikova K., Morren B. M., Tas A., Albulescu I. C., Van Rijswijk R., Jarhad D. B., et al. (2020). 6’-beta-fluoro-homoaristeromycin and 6’-fluoro-homoneplanocin a are potent inhibitors of Chikungunya virus replication through their direct effect on viral nonstructural protein 1. Antimicrob. Agents Chemother. 64:e02532-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamoorthy K., Harichandrakumar K. T., Krishna Kumari A., Das L. K. (2009). Burden of chikungunya in India: estimates of disability adjusted life years (DALY) lost in 2006 epidemic. J. Vector Borne Dis. 46 26–35. [PubMed] [Google Scholar]
- Kularatne S. A., Gihan M. C., Weerasinghe S. C., Gunasena S. (2009). Concurrent outbreaks of Chikungunya and Dengue fever in Kandy, Sri Lanka, 2006-07: a comparative analysis of clinical and laboratory features. Postgrad. Med. J. 85 342–346. 10.1136/pgmj.2007.066746 [DOI] [PubMed] [Google Scholar]
- Kumar N. P., Joseph R., Kamaraj T., Jambulingam P. (2008). A226V mutation in virus during the 2007 chikungunya outbreak in Kerala, India. J. Gen. Virol. 89 1945–1948. 10.1099/vir.0.83628-0 [DOI] [PubMed] [Google Scholar]
- Kumar P., Kumar D., Giri R. (2019). Targeting the nsp2 cysteine protease of chikungunya virus using FDA approved library and selected cysteine protease inhibitors. Pathogens 8:128. 10.3390/pathogens8030128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahariya C., Pradhan S. K. (2006). Emergence of chikungunya virus in Indian subcontinent after 32 years: a review. J. Vector Borne Dis. 43 151–160. [PubMed] [Google Scholar]
- Langsjoen R. M., Auguste A. J., Rossi S. L., Roundy C. M., Penate H. N., Kastis M., et al. (2017). Host oxidative folding pathways offer novel anti-chikungunya virus drug targets with broad spectrum potential. Antivir. Res. 143 246–251. 10.1016/j.antiviral.2017.04.014 [DOI] [PubMed] [Google Scholar]
- Laoprasopwattana K., Kaewjungwad L., Jarumanokul R., Geater A. (2012). Differential diagnosis of Chikungunya, dengue viral infection and other acute febrile illnesses in children. Pediatr. Infect. Dis. J. 31 459–463. 10.1097/inf.0b013e31824bb06d [DOI] [PubMed] [Google Scholar]
- Lebrun G., Chadda K., Reboux A. H., Martinet O., Gauzere B. A. (2009). Guillain-Barre syndrome after chikungunya infection. Emerg. Infect. Dis. 15 495–496. 10.3201/eid1503.071482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee V. J., Chow A., Zheng X., Carrasco L. R., Cook A. R., Lye D. C., et al. (2012). Simple clinical and laboratory predictors of Chikungunya versus dengue infections in adults. PLoS Negl. Trop. Dis. 6:e1786. 10.1371/journal.pntd.0001786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenglet Y., Barau G., Robillard P. Y., Randrianaivo H., Michault A., Bouveret A., et al. (2006). [Chikungunya infection in pregnancy: evidence for intrauterine infection in pregnant women and vertical transmission in the parturient. survey of the Reunion Island outbreak]. J. Gynecol. Obstet. Biol. Reprod. 35 578–583. [DOI] [PubMed] [Google Scholar]
- Levitt N. H., Ramsburg H. H., Hasty S. E., Repik P. M., Cole F. E., et al. (1986). Development of an attenuated strain of chikungunya virus for use in vaccine production. Vaccine 4 157–162. 10.1016/0264-410x(86)90003-4 [DOI] [PubMed] [Google Scholar]
- Li Y. G., Siripanyaphinyo U., Tumkosit U., Noranate N. A., Pan Y. (2012). Poly (I:C), an agonist of toll-like receptor-3, inhibits replication of the Chikungunya virus in BEAS-2B cells. Virol. J. 9:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang L., Li S., Chen X., Shen Y., Zhang Z., et al. (2007). Seco-pregnane steroids target the subgenomic RNA of alphavirus-like RNA viruses. Proc. Natl. Acad. Sci. U. S. A. 104 8083–8088. 10.1073/pnas.0702398104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londhey V., Agrawal S., Vaidya N., Kini S., Shastri J., Sunil S. (2016). Dengue and Chikungunya virus co-infections: the inside story. J. Assoc. Physicians India 64 36–40. [PubMed] [Google Scholar]
- Lumsden W. H. (1955). An epidemic of virus disease in Southern Province, Tanganyika territory, in 1952-53. II. general description and epidemiology. Trans. R. Soc. Trop. Med. Hyg. 49 33–57. 10.1016/0035-9203(55)90081-x [DOI] [PubMed] [Google Scholar]
- Madariaga M., Ticona E., Resurrecion C. (2016). Chikungunya: bending over the Americas and the rest of the world. Braz. J. Infect. Dis. 20 91–98. 10.1016/j.bjid.2015.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallilankaraman K., Shedlock D. J., Bao H., Kawalekar O. U., Fagone P., Ramanathan A. A., et al. (2011). A DNA vaccine against chikungunya virus is protective in mice and induces neutralizing antibodies in mice and nonhuman primates. PLoS Negl. Trop. Dis. 5:e928. 10.1371/journal.pntd.0000928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markoff L. (2015). “Alphaviruses,” in Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases, 8th Edn, eds John R. D., Bennett E., Blaser M. J.. Hoboken, NJ: Wiley [Google Scholar]
- Martins D. O. S., Santos I. A., De Oliveira D. M., Grosche V. R., Jardim A. C. G. (2020). Antivirals against Chikungunya virus: is the solution in nature? Viruses 12:272. 10.3390/v12030272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavalankar D., Shastri P., Raman P. (2007). Chikungunya epidemic in India: a major public-health disaster. Lancet Infect. Dis. 7 306–307. 10.1016/s1473-3099(07)70091-9 [DOI] [PubMed] [Google Scholar]
- Mercado M., Acosta-Reyes J., Parra E., Guzmán L., Beltrán M., Gasque P., et al. (2018). Renal involvement in fatal cases of chikungunya virus infection. J. Clin. Virol. 103 16–18. 10.1016/j.jcv.2018.03.009 [DOI] [PubMed] [Google Scholar]
- Ministry of Health and Family Welfare, Government of India (2019). Ministry of Health and Family Welfare National Health Profile -2019, Central Bureau of Health Intelligence, Directorate General of Health Services. Ministry of Health and Family Welfare, Government of India. Available online at: http://www.cbhidghs.nic.in/showfile.php?lid=1147 [Google Scholar]
- Ministry of Health and Family Welfare, Government of India (2020). Chikungunya situation in India. Epidemiological Profile of Chikungunya Fever in the Country since 2015. Directorate of National Vector Borne Disease Control Program. Ministry of Health and Family Welfare, Government of India. Available online at: https://nvbdcp.gov.in/index4.php?lang=1&level=0&linkid=486&lid=3765 (accessed July 24, 2020). [Google Scholar]
- Mishra B., Ratho R. K. (2006). Chikungunya re-emergence: possible mechanisms. Lancet 368:918. 10.1016/s0140-6736(06)69370-7 [DOI] [PubMed] [Google Scholar]
- Mishra B., Sharma M., Pujhari S. K., Ratho R. K., Gopal D. S., Kumar C. N., et al. (2011). Utility of multiplex reverse transcriptase-polymerase chain reaction for diagnosis and serotypic characterization of dengue and chikungunya viruses in clinical samples. Diagn. Microbiol. Infect. Dis. 71 118–125. 10.1016/j.diagmicrobio.2011.06.020 [DOI] [PubMed] [Google Scholar]
- Mohan A. (2006). Chikungunya fever: clinical manifestations & management. Indian J. Med. Res. 124 471–474. [PubMed] [Google Scholar]
- Mohan A., Sharma S. K. (2007). “Chikungunya fever,” in Medicine Update, ed. Singal R. K. (Mumbai: Association of Physicians of India; ). [Google Scholar]
- Mounce B. C., Cesaro T., Vlajnic L., Vidina A., Vallet T., Weger-Lucarelli J., et al. (2017). Chikungunya Virus overcomes polyamine depletion by mutation of nsP1 and the Opal stop codon to confer enhanced replication and fitness. J. Virol. 91:e00344-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudurangaplar B., Peerapur B. V. (2016). Molecular characterisation of clinical isolates of Chikungunya virus: a study from tertiary care hospitals in Southern India. J. Clin. Diagn. Res. 10 DC14–DC17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musso D., Teissier A., Rouault E., Teururai S., De Pina J. J., Nhan T. X. (2016). Detection of chikungunya virus in saliva and urine. Virol. J. 13:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthumani K., Block P., Flingai S., Muruganantham N., Chaaithanya I. K., Tingey C., et al. (2016). Rapid and long-term immunity elicited by DNA-encoded antibody Prophylaxis and DNA vaccination against Chikungunya virus. J. Infect. Dis. 214 369–378. 10.1093/infdis/jiw111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyembe-Tamfum J. J., Peyrefitte C. N., Yogolelo R., Mathina Basisya E., Koyange D., Pukuta E., et al. (2003). [Epidemic of Chikungunya virus in 1999 and 200 in the Democratic Republic of the Congo]. Med. Trop. 63 637–638. [PubMed] [Google Scholar]
- Naresh Kumar C. V., Sai Gopal D. V. (2010). Reemergence of Chikungunya virus in Indian subcontinent. Indian J. Virol. 21 8–17. 10.1007/s13337-010-0012-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Center for Disease Control (2020). Disease Alert. Integrated Disease Surveillance Programme (IDSP). Ministry of Health and Family Welfare. Government of India. National Center for Disease Control. Available online at: https://idsp.nic.in/index1.php?lang=1&level=1&sublinkid=5922&lid=3885 (accessed July 24, 2020). [Google Scholar]
- National Health Profile (2019). Central Bureau of Health Intelligence, Directorate General of Health Services, Ministry of Health and Family Welfare, Government of India. Available online at: http://www.cbhidghs.nic.in/showfile.php?lid=1147 [Google Scholar]
- Natrajan M. S., Rojas A., Waggoner J. J. (2019). Beyond fever and pain: diagnostic methods for Chikungunya virus. J. Clin. Microbiol. 57:e00350-19. 10.1128/JCM.00350-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newase P., More A., Patil J., Patil P., Jadhav S., Alagarasu K., et al. (2020). Chikungunya phylogeography reveals persistent global transmissions of the Indian Ocean lineage from India in association with mutational fitness. Infect. Genet. Evol. 82:104289. 10.1016/j.meegid.2020.104289 [DOI] [PubMed] [Google Scholar]
- Ngwe Tun M. M., Inoue S., Thant K. Z., Talemaitoga N., Aryati A., Dimaano E. M., et al. (2016). Retrospective seroepidemiological study of chikungunya infection in South Asia, Southeast Asia and the Pacific region. Epidemiol. Infect. 144 2268–2275. 10.1017/s095026881600056x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigam A., Sharma S., Jain A., Gupta A., Prakash A. (2016). Vertical transmission of Chikungunya manifesting as foetal pericardial effusion. J. Assoc. Physicians India 64 76–79. [PubMed] [Google Scholar]
- Nimmannitya S., Halstead S. B., Cohen S. N., Margiotta M. R. (1969). Dengue and chikungunya virus infection in man in Thailand, 1962-1964. I. observations on hospitalized patients with hemorrhagic fever. Am. J. Trop. Med. Hyg. 18 954–971. 10.4269/ajtmh.1969.18.954 [DOI] [PubMed] [Google Scholar]
- Niyas K. P., Abraham R., Unnikrishnan R. N., Mathew T., Nair S., Manakkadan A., et al. (2010). Molecular characterization of Chikungunya virus isolates from clinical samples and adult Aedes albopictus mosquitoes emerged from larvae from Kerala. South India. Virol. J. 7:189. 10.1186/1743-422x-7-189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- NVBDCP (2014–15). Chikungunya Situation in India. Epidemiological Profile of Chikungunya Fever in the Country since 2015. [Google Scholar]
- Okabayashi T., Sasaki T., Masrinoul P., Chantawat N., Yoksan S., Nitatpattana N., et al. (2015). Detection of chikungunya virus antigen by a novel rapid immunochromatographic test. J. Clin. Microbiol. 53 382–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olagnier D., Scholte F. E., Chiang C., Albulescu I. C., Nichols C., He Z., et al. (2014). Inhibition of dengue and chikungunya virus infections by RIG-I-mediated type I interferon-independent stimulation of the innate antiviral response. J. Virol. 88 4180–4194. 10.1128/jvi.03114-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira R., Barreto F. K. A., Maia A., Gomes I. P., Simiao A. R., Barbosa R. B., et al. (2018). Maternal and infant death after probable vertical transmission of chikungunya virus in Brazil - case report. BMC Infect. Dis. 18:333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oo A., Hassandarvish P., Chin S. P., Lee V. S., Abu Bakar S., Zandi K. (2016). In silico study on anti-Chikungunya virus activity of hesperetin. PeerJ 4:e2602. 10.7717/peerj.2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozden S., Lucas-Hourani M., Ceccaldi P. E., Basak A., Valentine M., Benjannet S., et al. (2008). Inhibition of Chikungunya virus infection in cultured human muscle cells by furin inhibitors: impairment of the maturation of the E2 surface glycoprotein. J. Biol. Chem. 283 21899–21908. 10.1074/jbc.m802444200 [DOI] [PubMed] [Google Scholar]
- Padbidri V. S., Gnaneswar T. T. (1979). Epidemiological investigations of chikungunya epidemic at Barsi, Maharashtra state, India. J. Hyg. Epidemiol. Microbiol. Immunol. 23 445–451. [PubMed] [Google Scholar]
- Padmakumar B., Jayan J. B., Menon R. M., Krishnankutty B., Payippallil R., Nisha R. (2009). Comparative evaluation of four therapeutic regimes in chikungunya arthritis: a prospective randomized parallel-group study. Indian J. Rheumatol. 4 94–101. 10.1016/s0973-3698(10)60189-6 [DOI] [Google Scholar]
- Pan J., Fang C., Yan J., Yan H., Zhan B., Sun Y., et al. (2019). Chikungunya fever outbreak, zhejiang province, China, 2017. Emerg. Infect. Dis. 25 1589–1591. 10.3201/eid2508.181212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey K., Pandey B. D., Chaurasiya R. R., Thakur M., Neupane B., Shah Y., et al. (2017). Evidence of Chikungunya virus circulation in the Terai region of Nepal in 2014 and 2015. Trans. R. Soc. Trop. Med. Hyg. 111 294–299. 10.1093/trstmh/trx059 [DOI] [PubMed] [Google Scholar]
- Parashar D., Amdekar S., More A., Patil P., More R., Babu V. R. (2015). Chikungunya fever outbreak in Guntur, Andhra Pradesh, India. Indian J. Med. Res. 142(Suppl.) S111–S115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parashar D., Paingankar M. S., Kumar S., Gokhale M. D., Sudeep A. B., Shinde S. B., et al. (2013). Administration of E2 and NS1 siRNAs inhibit chikungunya virus replication in vitro and protects mice infected with the virus. PLoS Negl. Trop. Dis. 7:e2405. 10.1371/journal.pntd.0002405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passos G. F. S., Gomes M. G. M., Aquino T. M., Araujo-Junior J. X., Souza S. J. M., Cavalcante J. P. M., et al. (2020). Computer-aided design, synthesis, and antiviral evaluation of Novel Acrylamides as potential Inhibitors of E3-E2-E1 glycoproteins complex from Chikungunya Virus. Pharmaceuticals 13:141. 10.3390/ph13070141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri K. (1986). Disappearance of Chikungunya virus from India and South East Asia. Trans. R. Soc. Trop. Med. Hyg. 80:491. 10.1016/0035-9203(86)90358-5 [DOI] [PubMed] [Google Scholar]
- Pavri K. M. (1964). Presence of Chikungunya antibodies in human sera collected from calcutta and Jamshedpur before 1963. Indian J. Med. Res. 52 698–702. [PubMed] [Google Scholar]
- Pialoux G., Gauzere B. A., Jaureguiberry S., Strobel M. (2007). Chikungunya, an epidemic arbovirosis. Lancet Infect. Dis. 7 319–327. 10.1016/s1473-3099(07)70107-x [DOI] [PubMed] [Google Scholar]
- Plante K., Wang E., Partidos C. D., Weger J., Gorchakov R., Tsetsarkin K., et al. (2011). Novel chikungunya vaccine candidate with an IRES-based attenuation and host range alteration mechanism. PLoS Pathog. 7:e1002142. 10.1371/journal.ppat.1002142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin S. (2014). History of vaccination. Proc. Natl. Acad. Sci. U. S. A. 111 12283–12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohjala L., Utt A., Varjak M., Lulla A., Merits A., Ahola T., et al. (2011). Inhibitors of alphavirus entry and replication identified with a stable Chikungunya replicon cell line and virus-based assays. PLoS One 6:e28923. 10.1371/journal.pone.0028923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers A. M., Brault A. C., Shirako Y., Strauss E. G., Kang W., Strauss J. H., et al. (2001). Evolutionary relationships and systematics of the alphaviruses. J. Virol. 75 10118–10131. 10.1128/jvi.75.21.10118-10131.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers A. M., Brault A. C., Tesh R. B., Weaver S. C. (2000). Re-emergence of Chikungunya and O’nyong-nyong viruses: evidence for distinct geographical lineages and distant evolutionary relationships. J. Gen. Virol. 81 471–479. 10.1099/0022-1317-81-2-471 [DOI] [PubMed] [Google Scholar]
- Prat C. M., Flusin O., Panella A., Tenebray B., Lanciotti R., Leparc-Goffart I. (2014). Evaluation of commercially available serologic diagnostic tests for chikungunya virus. Emerg. Infect. Dis. 20 2129–2132. 10.3201/eid2012.141269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puntasecca C. J., King C. H., Labeaud A. D. (2021). Measuring the global burden of chikungunya and Zika viruses: a systematic review. PLoS Negl. Trop. Dis. 15:e0009055. 10.1371/journal.pntd.0009055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranik N. V., Rani R., Singh V. A., Tomar S., Puntambekar H. M., Srivastava P. (2019). Evaluation of the antiviral potential of Halogenated Dihydrorugosaflavonoids and molecular modeling with nsP3 protein of Chikungunya Virus (CHIKV). ACS Omega 4 20335–20345. 10.1021/acsomega.9b02900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M., Yamagishi J., Rahim R., Hasan A., Sobhan A. (2019). East/central/South African genotype in a chikungunya outbreak. Dhaka, Bangladesh, 2017. Emerg. Infect. Dis. 25 370–372. 10.3201/eid2502.180188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajapakse S., Rodrigo C., Rajapakse A. (2010). Atypical manifestations of chikungunya infection. Trans. R. Soc. Trop. Med. Hyg. 104 89–96. 10.1016/j.trstmh.2009.07.031 [DOI] [PubMed] [Google Scholar]
- Ramsauer K., Tangy F. (2016). Chikungunya virus vaccines: viral vector-based approaches. J. Infect. Dis. 214 S500–S505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathore A. P., Haystead T., Das P. K., Merits A., Ng M.-L., Vasudevan S. G. (2014). Chikungunya virus nsP3 & nsP4 interacts with HSP-90 to promote virus replication: HSP-90 inhibitors reduce CHIKV infection and inflammation in vivo. Antivir. Res. 103 7–16. [DOI] [PubMed] [Google Scholar]
- Rathore A. P., Ng M. L., Vasudevan S. G. (2013). Differential unfolded protein response during Chikungunya and Sindbis virus infection: CHIKV nsP4 suppresses eIF2alpha phosphorylation. Virol. J. 10:36. 10.1186/1743-422x-10-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi V. (2006). Re-emergence of chikungunya virus in India. Indian J. Med. Microbiol. 24 83–84. 10.1016/s0255-0857(21)02403-8 [DOI] [PubMed] [Google Scholar]
- Ravichandran R., Manian M. (2008). Ribavirin therapy for Chikungunya arthritis. J. Infect. Dev. Ctries. 2 140–142. 10.3855/t2.2.140 [DOI] [PubMed] [Google Scholar]
- Reisinger E. C., Tschismarov R., Beubler E., Wiedermann U., Firbas C., Loebermann M., et al. (2019). Immunogenicity, safety, and tolerability of the measles-vectored chikungunya virus vaccine MV-CHIK: a double-blind, randomised, placebo-controlled and active-controlled phase 2 trial. Lancet 392 2718–2727. 10.1016/s0140-6736(18)32488-7 [DOI] [PubMed] [Google Scholar]
- Robillard P. Y., Boumahni B., Gerardin P., Michault A., Fourmaintraux A., Schuffenecker I., et al. (2006). [Vertical maternal fetal transmission of the chikungunya virus. Ten cases among 84 pregnant women]. Presse Med. 35 785–788. [DOI] [PubMed] [Google Scholar]
- Robinson M. C. (1955). An epidemic of virus disease in Southern Province, Tanganyika Territory, in 1952-53. i. clinical features. Trans. R. Soc. Trop. Med. Hyg. 49 28–32. 10.1016/0035-9203(55)90080-8 [DOI] [PubMed] [Google Scholar]
- Rodrigues F. M., Patankar M. R., Banerjee K., Bhatt P. N., Goverdhan M. K., Pavri K. M., et al. (1972). Etiology of the 1965 epidemic of febrile illness in Nagpur city, Maharashtra State, India. Bull. World Health Organ. 46 173–179. [PMC free article] [PubMed] [Google Scholar]
- Roques P., Ljungberg K., Kummerer B. M., Gosse L., Dereuddre-Bosquet N., Tchitchek N., et al. (2017). Attenuated and vectored vaccines protect nonhuman primates against Chikungunya virus. JCI Insight 2:e83527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roques P., Thiberville S.-D., Dupuis-Maguiraga L., Lum F.-M., Labadie K., Martinon F., et al. (2018). Paradoxical effect of chloroquine treatment in enhancing chikungunya virus infection. Viruses 10:268. 10.3390/v10050268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi S. L., Comer J. E., Wang E., Azar S. R., Lawrence W. S., Plante J. A., et al. (2019). Immunogenicity and efficacy of a measles virus-vectored Chikungunya vaccine in Nonhuman primates. J. Infect. Dis. 220 735–742. 10.1093/infdis/jiz202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothan H. A., Bahrani H., Abdulrahman A. Y., Mohamed Z., Teoh T. C., Othman S., et al. (2016). Mefenamic acid in combination with ribavirin shows significant effects in reducing chikungunya virus infection in vitro and in vivo. Antivir. Res. 127 50–56. 10.1016/j.antiviral.2016.01.006 [DOI] [PubMed] [Google Scholar]
- Rothan H. A., Bahrani H., Mohamed Z., Teoh T. C., Shankar E. M., Rahman N. A., et al. (2015). A combination of doxycycline and ribavirin alleviated chikungunya infection. PLoS One 10:e0126360. 10.1371/journal.pone.0126360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S., Ramesh A., Kalantar K., Malaker R., Hasanuzzaman M., Khan L. M., et al. (2019). Unbiased metagenomic sequencing for pediatric meningitis in Bangladesh reveals neuroinvasive Chikungunya virus outbreak and other unrealized pathogens. mBio 10:e002877-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangeetha K., Purushothaman I., Rajarajan R. (2017). Spectral characterisation, antiviral activities, in silico ADMET and molecular docking of the compounds isolated from Tectona grandis to chikungunya virus. Biomed. Pharmacother. 87 302–310. 10.1016/j.biopha.2016.12.069 [DOI] [PubMed] [Google Scholar]
- Sarangan G., Nayar S. A., Mani M., Sundarrajan S., Sankar S., Palani G., et al. (2018). Genetic characterization of Chikungunya virus 2009 isolates from South India. Bioinformation 14 106–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraswat S., Athmaram T. N., Parida M., Agarwal A., Saha A., Dash P. K. (2016). Expression and characterization of yeast derived Chikungunya virus like particles (CHIK-VLPs) and its evaluation as a potential vaccine candidate. PLoS. Trop. Dis. 10:e0004782. 10.1371/journal.pntd.0004782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar J. K., Pavri K. M., Chatterjee S. N., Chakravarty S. K., Anderson C. R. (1964). Virological and serological studies of cases of haemorrhagic fever in calcutta. material collected by the calcutta school of tropical medicine. Indian J. Med. Res. 52 684–691. [PubMed] [Google Scholar]
- Saswat T., Kumar A., Kumar S., Mamidi P., Muduli S., Debata N. K., et al. (2015). High rates of co-infection of Dengue and Chikungunya virus in Odisha and Maharashtra, India during 2013. Infect. Genet. Evol. 35 134–141. 10.1016/j.meegid.2015.08.006 [DOI] [PubMed] [Google Scholar]
- Schuffenecker I., Iteman I., Michault A., Murri S., Frangeul L., Vaney M. C., et al. (2006). Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med. 3:e263. 10.1371/journal.pmed.0030263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz O., Albert M. L. (2010). Biology and pathogenesis of chikungunya virus. Nat. Rev. Microbiol. 8 491–500. 10.1038/nrmicro2368 [DOI] [PubMed] [Google Scholar]
- Selvarajah S., Sexton N. R., Kahle K. M., Fong R. H., Mattia K. A., Gardner J., et al. (2013). A neutralizing monoclonal antibody targeting the acid-sensitive region in chikungunya virus E2 protects from disease. PLoS Negl. Trop. Dis. 7:e2423. 10.1371/journal.pntd.0002423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senanayake M. P., Senanayake S. M., Vidanage K. K., Gunasena S., Lamabadusuriya S. P. (2009). Vertical transmission in chikungunya infection. Ceylon Med. J. 54 47–50. 10.4038/cmj.v54i2.865 [DOI] [PubMed] [Google Scholar]
- Seyedi S. S., Shukri M., Hassandarvish P., Oo A., Shankar E. M., Abubakar S., et al. (2016). Computational approach towards exploring potential Anti-Chikungunya activity of selected flavonoids. Sci. Rep. 6:24027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah K. V., Gibbs C. J., Banerjee G. (1964). Virological investigation of the epidemic of Haemorrhagic fever in calcutta: isolation of three strains of Chikungunya virus. Indian J. Med. Res. 52 676–683. [PubMed] [Google Scholar]
- Sharma R., Fatma B., Saha A., Bajpai S., Sistla S., Dash P. K., et al. (2016). Inhibition of chikungunya virus by picolinate that targets viral capsid protein. Virology 498 265–276. 10.1016/j.virol.2016.08.029 [DOI] [PubMed] [Google Scholar]
- Shrinet J., Jain S., Sharma A., Singh S. S., Mathur K., Rana V., et al. (2012). Genetic characterization of Chikungunya virus from New Delhi reveal emergence of a new molecular signature in Indian isolates. Virol. J. 9:100. 10.1186/1743-422x-9-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon F., Javelle E., Oliver M., Leparc-Goffart I., Marimoutou C. (2011). Chikungunya virus infection. Curr. Infect. Dis. Rep. 13 218–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon F., Savini H., Parola P. (2008). Chikungunya: a paradigm of emergence and globalization of vector-borne diseases. Med. Clin. North Am. 92 1323–1343, ix. [DOI] [PubMed] [Google Scholar]
- Singh P., Mittal V., Rizvi M., Chhabra M., Sharma P., Rawat D., et al. (2012). The first dominant co-circulation of both dengue and chikungunya viruses during the post-monsoon period of 2010 in Delhi, India. Epidemiol. Infect. 140 1337–1342. 10.1017/s0950268811001671 [DOI] [PubMed] [Google Scholar]
- Staples J. E., Fischer M. (2014). Chikungunya virus in the Americas–what a vectorborne pathogen can do. N. Engl. J. Med. 371 887–889. 10.1056/nejmp1407698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staples J. E., Breiman R. F., Powers A. M. (2009). Chikungunya fever: an epidemiological review of a re-emerging infectious disease. Clin. Infect. Dis. 49 942–948. 10.1086/605496 [DOI] [PubMed] [Google Scholar]
- Subudhi B. B., Chattopadhyay S., Mishra P., Kumar A. (2018). Current strategies for inhibition of Chikungunya infection. Viruses 10:235. 10.3390/v10050235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudeep A. B., Parashar D. (2008). Chikungunya: an overview. J. Biosci. 33 443–449. 10.1007/s12038-008-0063-2 [DOI] [PubMed] [Google Scholar]
- Suhrbier A. (2019). Rheumatic manifestations of chikungunya: emerging concepts and interventions. Nat. Rev. Rheumatol. 15 597–611. 10.1038/s41584-019-0276-9 [DOI] [PubMed] [Google Scholar]
- Suhrbier A., Mahalingam S. (2009). The immunobiology of viral arthritides. Pharmacol. Ther. 124 301–308. 10.1016/j.pharmthera.2009.09.005 [DOI] [PubMed] [Google Scholar]
- Suhrbier A., Jaffar-Bandjee M. C., Gasque P. (2012). Arthritogenic alphaviruses–an overview. Nat. Rev. Rheumatol. 8 420–429. 10.1038/nrrheum.2012.64 [DOI] [PubMed] [Google Scholar]
- Sumathy K., Ella K. M. (2012). Genetic diversity of Chikungunya virus, India 2006-2010: evolutionary dynamics and serotype analyses. J. Med. Virol. 84 462–470. 10.1002/jmv.23187 [DOI] [PubMed] [Google Scholar]
- Tandel K., Kumar M., Shergill S. P. S., Sahai K., Gupta R. M. (2019). Molecular characterization and phylogenetic analysis of Chikungunya virus from Delhi, India. Med. J. Armed Forces India 75 266–273. 10.1016/j.mjafi.2018.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraphdar D., Roy B. K., Chatterjee S. (2015). Chikungunya virus infection amongst the acute encephalitis syndrome cases in West Bengal, India. Indian J. Med. Microbiol. 33(Suppl.) 153–156. [DOI] [PubMed] [Google Scholar]
- Taylor A., Liu X., Zaid A., Goh L. Y., Hobson-Peters J., Hall R. A., et al. (2017). Mutation of the N-terminal region of Chikungunya virus capsid protein: implications for vaccine design. mBio 8 e01970–16. 10.1128/mBio.01970-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng T. S., Foo S. S., Simamarta D., Lum F. M., Teo T. H., Lulla A., et al. (2012). Viperin restricts chikungunya virus replication and pathology. J. Clin. Invest. 122 4447–4460. 10.1172/jci63120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiberville S. D., Moyen N., Dupuis-Maguiraga L., Nougairede A., Gould E. A., Roques P., et al. (2013). Chikungunya fever: epidemiology, clinical syndrome, pathogenesis and therapy. Antivir. Res. 99 345–370. 10.1016/j.antiviral.2013.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiruvengadam K. V., Kalyanasundaram V., Rajgopal J. (1965). Clinical and pathological studies on chikungunya fever in Madras city. Indian J. Med. Res. 53 729–744. [PubMed] [Google Scholar]
- Tiwari M., Parida M., Santhosh S. R., Khan M., Dash P. K., Rao P. V. (2009). Assessment of immunogenic potential of vero adapted formalin inactivated vaccine derived from novel ECSA genotype of Chikungunya virus. Vaccine 27 2513–2522. 10.1016/j.vaccine.2009.02.062 [DOI] [PubMed] [Google Scholar]
- Tsetsarkin K. A., Chen R., Leal G., Forrester N., Higgs S., Huang J., et al. (2011a). Chikungunya virus emergence is constrained in Asia by lineage-specific adaptive landscapes. Proc. Natl. Acad. Sci. U. S. A. 108 7872–7877. 10.1073/pnas.1018344108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsetsarkin K. A., Chen R., Sherman M. B., Weaver S. C. (2011b). Chikungunya virus: evolution and genetic determinants of emergence. Curr. Opin. Virol. 1 310–317. 10.1016/j.coviro.2011.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsetsarkin K. A., Chen R., Yun R., Rossi S. L., Plante K. S., Guerbois M., et al. (2014). Multi-peaked adaptive landscape for chikungunya virus evolution predicts continued fitness optimization in Aedes albopictus mosquitoes. Nat. Commun. 5:4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsetsarkin K. A., Mcgee C. E., Volk S. M., Vanlandingham D. L., Weaver S. C., Higgs S. (2009). Epistatic roles of E2 glycoprotein mutations in adaption of chikungunya virus to Aedes albopictus and Ae. Aegypti mosquitoes. PLoS One 4:e6835. 10.1371/journal.pone.0006835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsetsarkin K. A., Vanlandingham D. L., Mcgee C. E., Higgs S. (2007). A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 3:e201. 10.1371/journal.ppat.0030201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhwani G. G. (2013). Homeopathic drug therapy. homeopathy in Chikungunya fever and post-chikungunya chronic arthritis: an observational study. Homeopathy 102 193–198. 10.1016/j.homp.2013.02.001 [DOI] [PubMed] [Google Scholar]
- Wang D., Suhrbier A., Penn-Nicholson A., Woraratanadharm J., Gardner J., Luo M., et al. (2011). A complex adenovirus vaccine against chikungunya virus provides complete protection against viraemia and arthritis. Vaccine 29 2803–2809. 10.1016/j.vaccine.2011.01.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E., Volkova E., Adams A. P., Forrester N., Xiao S. Y., Frolov I., et al. (2008). Chimeric alphavirus vaccine candidates for chikungunya. Vaccine 26 5030–5039. 10.1016/j.vaccine.2008.07.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangchuk S., Chinnawirotpisan P., Dorji T., Tobgay T., Dorji T., Yoon I. K., et al. (2013). Chikungunya fever outbreak, Bhutan, 2012. Emerg. Infect. Dis. 19 1681–1684. 10.3201/eid1910.130453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver S. C., Lecuit M. (2015). Chikungunya virus and the global spread of a mosquito-borne disease. N. Engl. J. Med. 372 1231–1239. 10.1056/nejmra1406035 [DOI] [PubMed] [Google Scholar]
- Weaver S. C., Forrester N. L., Liu J., Vasilakis N. (2021). Population bottlenecks and founder effects: implications for mosquito-borne arboviral emergence. Nat. Rev. Microbiol. 19 184–195. 10.1038/s41579-020-00482-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White A., Berman S., Lowenthal J. P. (1972). Comparative immunogenicities of Chikungunya vaccines propagated in monkey kidney monolayers and chick embryo suspension cultures. Appl. Microbiol. 23 951–952. 10.1128/am.23.5.951-952.1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2009). Guidelines for Prevention and Control of Chikungunya Fever [Online]. Available Online at: https://apps.who.int/iris/bitstream/handle/10665/205166/B4289.pdf?sequence=1&isAllowed=y (accessed March 15, 2021). [Google Scholar]
- WHO (2012). Chikungunya Fever, a Re-Emerging Disease in Asia [Online]. Available Online at: http://www.searo.who.int/en/Section10/Section2246.htm (accessed March 15, 2021). [Google Scholar]
- Wichit S., Hamel R., Bernard E., Talignani L., Diop F., Ferraris P., et al. (2017). Imipramine inhibits Chikungunya Virus Replication in human skin fibroblasts through interference with intracellular cholesterol trafficking. Sci. Rep. 7:3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielanek A. C., Monredon J. D., Amrani M. E., Roger J. C., Serveaux J. P. (2007). Guillain-Barre syndrome complicating a Chikungunya virus infection. Neurology 69 2105–2107. 10.1212/01.wnl.0000277267.07220.88 [DOI] [PubMed] [Google Scholar]
- Wintachai P., Kaur P., Lee R. C., Ramphan S., Kuadkitkan A., Wikan N., et al. (2015). Activity of andrographolide against chikungunya virus infection. Sci. Rep. 5:14179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T., Lyu L. K., Tan Z. L., Li L., Lyu J., Li X. Y. (2020). [Genotyping on one case with Chikungunya infection introduced into Tianjin in China from Myanmar]. Zhonghua Liu Xing Bing Xue Za Zhi 41 2131–2134. [DOI] [PubMed] [Google Scholar]
- Yergolkar P. N., Tandale B. V., Arankalle V. A., Sathe P. S., Sudeep A. B., Gandhe S. S., et al. (2006). Chikungunya outbreaks caused by African genotype, India. Emerg. Infect. Dis. 12 1580–1583. 10.3201/eid1210.060529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. N., Deng C. L., Li J. Q., Li N., Zhang Q. Y., Ye H. Q., et al. (2019). Infectious Chikungunya Virus (CHIKV) with a complete capsid deletion: a new approach for a CHIKV vaccine. J. Virol. 93 e504–e519. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.