Abstract
Purpose
To advance fundamental biological and translational research with the bacterium Neisseria gonorrhoeae through the prediction of novel small molecule growth inhibitors via naïve Bayesian modeling methodology.
Methods
Inspection and curation of data from the publicly available ChEMBL web site for small molecule growth inhibition data of the bacterium Neisseria gonorrhoeae resulted in a training set for the construction of machine learning models. A naïve Bayesian model for bacterial growth inhibition was utilized in a workflow to predict novel antibacterial agents against this bacterium of global health relevance from a commercial library of >105 drug-like small molecules. Follow-up efforts involved empirical assessment of the predictions and validation of the hits.
Results
Specifically, two small molecules were found that exhibited promising activity profiles and represent novel chemotypes for agents against N. gonorrrhoeae.
Conclusions
This represents, to the best of our knowledge, the first machine learning approach to successfully predict novel growth inhibitors of this bacterium. To assist the chemical tool and drug discovery fields, we have made our curated training set available as part of the Supplementary Material and the Bayesian model is accessible via the web.
Keywords: Naïve Bayesian classifier, machine learning model, Neisseria gonorrhoeae, diversity
VISUAL ABSTRACT
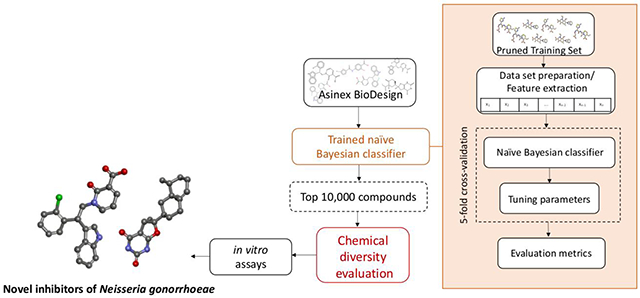
INTRODUCTION
The Gram-negative bacterium Neisseria gonorrhoeae is the etiological agent of the sexually transmitted infection (STI) gonorrhoeae, which if left untreated can cause pelvic inflammatory disease, infertility, ectopic pregnancy, neonatal conjunctivitis resulting in blindness, and increased HIV transmission (1–4).The STI gonorrhoeae is considered by the World Health Organization (WHO) as a major public health challenge because of the high number of infected cases worldwide and the emergence of drug-resistant strains (5). The WHO estimated 78 million new cases of gonorrhea per year among individuals between the ages of 15 and 49 years-old (5–7). The Centers for Disease Control and Prevention reported an increased rate of gonorrhea infections in the United States of 5.0% from 2017 to 2018, which is an increase of over 82.0% since the last historical low in 2009, with the highest rates of reported infection being among adolescents and young adults (8).
The current treatment regimen for gonococcal infection relies on antibacterials due to the lack of an effective vaccine (9). The first-line treatment features a combination of azithromycin and ceftriaxone or a single dose of ceftriaxone, cefixime, or spectinomycin. Drug therapy after first-line treatment failure is based on higher doses of ceftriaxone and azithromycin in combination (5, 8). The rise of antibacterial resistance in different strains of N. gonorrhoeae in recent years to tetracyclines, macrolides (including azithromycin), sulfonamide and trimethoprim combinations, quinolones, and recently third-generation cephalosporins exemplifies a multi-drug resistance global health crisis (7, 8, 10). On a molecular level, this has been traced to mutations in penA that encodes the penicillin-binding protein PBP2 (11), increased efflux of antibacterials via multidrug efflux pumps such as MtrC–MtrE–MtrD and its repressor mtrR (12), and decreased permeability of antibacterials into the bacterium through mutations in genes such as penB, which encode transport proteins(13).
Given the dire need for new therapeutic regimens to treat N. gonorrhoeae infections, our laboratory has explored the potential to extend our machine learning platform from Mycobacterium tuberculosis (14–16) to this Gram-negative bacterium. Specifically, we have explored the application of naïve Bayesian (heretofore referred to as Bayesian) models to learn from publicly available whole-cell phenotypic assay data for N. gonorrhoeae to predict small molecule growth inhibitors of this bacterium. Herein, we report the execution of this strategy to construct and validate the first such models and disclose the identification of new small molecule inhibitors with potential as chemical tools and/or drug discovery entities relevant to N. gonorrhoeae.
MATERIALS AND METHODS
Training Set Construction and Curation:
The model training set was extracted from ChEMBL (17) (https://www.ebi.ac.uk/chembl/) by searching for whole-cell growth inhibition data versus N. gonorrhoeae with the query keywords “Neisseria gonorrhoeae” and “MIC”. The curation process consisted of manually inspecting the dataset to remove duplicates, selecting a conservative MIC value of 8.0 μg/mL, converting as necessary MIC units from μM to μg/mL, as well as removing false positives, which are compounds that were incorrectly deposited in ChEMBL as actives against N. gonorrhoeae. Subsequently, a manual inspection of the 2D structures of all active compounds was performed to detect reactive and Pan Assay INterference compoundS (PAINS) (18, 19), which we then excluded from the training set in a process called “structural pruning” – an extension of the data pruning approach described in our previous work (20, 21). The final dataset contains 282 compounds, of which 160 (56.7%) were labelled as active compounds (Supplemental Information; File: Neisseria training set.sdf).
Neisseria Bayesian Model:
The Neisseria Bayesian model was developed using the Assay Central™ software (https://assaycentral.github.io). It involved the following steps: (1) dataset standardization consisted of removing salts, neutralizing unbalanced charges, and merging duplicate structures. (2) For the model generation phase, Bayesian classifier models were created utilizing Extended-Connectivity Fingerprints (ECFP6; each atom was described by taking into consideration its 6 nearest neighbors) (22–24) as the descriptor, and the model parameters such as the number of bins, bin size, and fingerprint contributions (23–25) were varied and assessed via internal, 5-fold cross-validation. These run parameters for the model were optimized by using the well-known metrics such as recall, precision, specificity, F1-score, Receiver Operating Characteristic (ROC) curve, Cohen’s Kappa (CK) and the Matthews Correlation Coefficient (MCC) (26–28). More detailed descriptions of Assay Central™ have been previously disclosed (23, 24, 29, 30). In conducting this process, two compounds were removed from the initial training set of 282 compounds. This was due to salt differences that when stripped afforded a single compound entry. This pertained to CHEMBL2023883 and CHEMBL2023877 (in this case, the former was considered active and the latter inactive; overall, the parent, neutral molecule was considered active), and to CHEMBL3274638 and CHEMBL3274636.
Virtual screening:
The Neisseria Bayesian model was used to virtually screen the Asinex BioDesign Library, a commercial library of over 190,000 small molecules, described as synthetic molecules with features in common with, or inspired by, natural product architectures and approved drugs (http://www.asinex.com/libraries_biodesign-html/). The screening library was downloaded from the Asinex website in .sdf format. Subsequently, the library file was processed to remove salts, neutralize unbalanced charges, and merge duplicate structures. The output of the virtual screen with the Neisseria Bayesian model was a ranked list of compounds, where each molecule’s score scaled with the likelihood of it meeting the activity criterion of an MIC ≤ 8.0 μg/mL. The 10,000 top-ranked compounds (Supplemental Information; File: Asinex top 10000 by Bayesian.sdf) then underwent a structural diversity evaluation, which consisted of selecting the most mutually diverse structures from the top scoring compounds by using ECFP6 fingerprints and Tanimoto similarity. The top-scoring 100 most diverse compounds (Supplemental Information; File: Asinex top 10000 by Bayesian then top 100 by diversity.sdf) were visually inspected to triage chemically reactive compounds(31) and nitroheterocycles given our previous work with them (32, 33). The filtered top-scoring 20 compounds were then commercially sourced, verified as to their purity by HPLC (typically ≥95% at 250 nm) and identity by MS (typically observing the M+H ion). Overall, these 20 compounds displayed an HPLC purity ≥ 95%, except for the compounds JSF-4437 (BDE 32402339) and JSF-4438 (BDE 33873286) which each exhibited a purity of 81%.
In vitro assays:
For the MIC assay, seven representative strain isolates (FA1090 (ATCC 700825), FA19 (ATCC BAA-1838), F62 (ATCC 33084), FA6140 (https://www.ncbi.nlm.nih.gov/genome/?term=Neisseria%20gonorrhoeae%20FA6140), F89 (34), 35/02 (https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=528346), and 59/03 (35)) were sub-cultured from frozen stocks onto GC medium base agar supplemented with hemoglobin powder (Sigma) and IsoVitaleX Enrichment (BD Biosciences) at 36 – 37 °C with 5% CO2 for 24 or 48 h. The MIC values were determined by the broth microdilution method using fastidious broth (FB; Hardy Diagnostics) with hematin solution (Sigma), Tween 80 (Sigma), and IsoVitaleX Enrichment (BD Biosciences) (36, 37). Overnight cultures of the strain being tested were diluted 1:10 with FB to approximately 5 × 105 CFU/mL (5 × 104 CFU/well), and 100 μL of the suspension were inoculated into a 96-well plate with serial dilutions; the last column of each plate contained only bacteria and served as a no-drug control. The plates were incubated for 24 or 48 h at 36 – 37 °C in a moist atmosphere containing 5% CO2. The MIC was defined as the lowest concentration of the compound tested that inhibited visible growth of the bacterium under the defined conditions. A subset of ESKAPE bacteria were utilized in a similar protocol to assess the MIC of a compound.
The MBC for select compounds was quantified using the strain FA1090 by culturing 20 μL of the MIC well and that from the wells above and below it (2x change in compound concentration in each direction from the MIC) onto GC agar plates. The number of colonies were counted after the inoculated GC agar plates were incubated for 24 h at 36 – 37 °C with 5% CO2 (36, 38). The MBC was defined as the lowest concentration of the compound to kill 99% of the initial bacterial inoculum tested.
Mouse liver microsome stability and kinetic aqueous solubility assays: These assays were performed by BioDuro, Inc. (https://bioduro.com) and were as according to literature procedures (39).
Principal Component Analyses and Tanimoto similarity calculations:
Principal Analysis (PCA) was used to compare the chemical space between the training set and the Asinex library that was screened. PCA was also used to compare the chemical property space between the set of 20 filtered, top-scoring compounds and the 160 known N. gonorrhoeae inhibitors in the training set. In both cases, the datasets to be compared were converted to .sdf files and then combined into a single .sdf. The single .sdf file set was utilized in a PCA through Discovery Studio Version 2017 R2 (40) and Pipeline Pilot Version 2017 R2 (41), using the following eight interpretable descriptors: LogP, molecular weight, number of rotatable bonds, number of rings, number of aromatic rings, number of hydrogen-bond acceptors, number of hydrogen-bond donors, and molecular fractional polar surface area. The pairwise Tanimoto similarity (42) was calculated between the three most promising compounds (JSF-4439, JSF-4444, and JSF-4447) from the top 20 compounds selected and the 160 known N. gonorrhoeae inhibitors available in the training set. To calculate the pairwise Tanimoto similarity we used the “find similar molecules by fingerprints” protocol in Discovery Studio, which consists of measuring the similarity by comparing a selected fingerprint property (ECFP6).
Synthetic Chemistry:
All reagents were obtained from commercial vendors and used as received unless noted otherwise. Whenever necessary, reactions were performed under a nitrogen atmosphere and anhydrous solvents were used. NMR spectra of the synthesized compounds were recorded on Avance III 500 MHz spectrometers from the Bruker Corporation (Billerica, MA, USA). Reverse-phase high performance liquid chromatography (HPLC) and electrospray ionization (ESI) mass spectra were obtained on an Agilent 6120 single quadrupole LC/MS system using a reverse-phase EMD Millipore Chromolith SpeedRod RP-18e column (50 × 4.6 mm). In general, a 10 - 100% gradient of acetonitrile/water containing 0.1% formic acid was used for the analysis of the samples. All compounds were purified to >95% peak area (i.e., purity) via an HPLC UV trace at 220 nm or 250 nm with observation of a low-resolution MS m/z consistent with each compound. High-resolution mass spectral data were acquired with an Agilent 6230B Accurate Mass TOF spectrometer. Purification of samples by flash chromatography was performed on a Teledyne ISCO CombiFlash Rf+ system using a Teledyne RediSep normal phase silica gel column. For TLC, aluminum plates coated by silica gel 60 with F254 fluorescent indicator from EMD Millipore were used. Preparative reverse-phase HPLC was performed on a Varian (now Agilent) SD-1 preparative HPLC system equipped with an Agilent Pursuit (10 μm, 250 x 21.2 mm) C-18 column with detection UV wavelength set at 220 nm or 250 nm. A gradient of acetonitrile in water at a flow rate of 20 mL/min was used for the separation.
Preparation of 1-(2-(2-chlorophenyl)-2-(1H-indol-3-yl)ethyl)-2-oxo-1,2-dihydropyridine-3-carboxylic acid (JSF-4439): Methyl 2-oxo-2H-pyran-3-carboxylate (15 mg, 0.097 mmol) was dissolved in 1 mL DMF. The solution was cooled to 0 °C, and a solution of 2-(2-chlorophenyl)-2-(1H-indol-3-yl)ethan-1-amine (26 mg, 0.097 mmol) in 1 mL DMF was added dropwise. The mixture was warmed gradually to rt and was stirred for 7 - 9 h. After confirming the formation of an intermediate (Michael addition) product via LC-MS, the mixture was cooled back to 0 °C, after which DMAP (2.0 mg, 0.019 mmol) and EDC•HCl (22 mg, 0.11 mmol) were added sequentially, and the mixture was warmed gradually to rt and stirred for an additional 12 h. The reaction mixture was diluted with water and the organic layer was extracted with ethyl acetate. The combined organic layers were washed successively with water, and dried over anhydrous sodium sulfate. The solvent was removed in vacuo and the crude product was dissolved in 2 mL of 3:1 THF/MeOH. Then, LiOH•H2O (18 mg, 0.29 mmol) was added and the mixture was stirred for 3 - 4 h at rt. The mixture was cooled to 0 °C, and a few drops of 1 N HCl were added until reaching pH ~ 2. The product was then extracted with ethyl acetate. The combined organic layers were dried over anhydrous sodium sulfate. The solvent was removed in vacuo, and the crude product was purified via HPLC (H2O/CH3CN 100 to 20% gradient for 15 min; seven runs). Pure fractions, as judged by LC-MS, were combined and lyophilized for 18 h to afford the desired product as a pale yellow product (7.0 mg, 18% overall yield): 1H NMR (500 MHz, CDCl3) δ 8.43 (d, J = 6.7 Hz, 1), 8.18 (s, 1), 7.41 (d, J = 7.9 Hz, 1), 7.36 (m, 3), 7.19 (m, 4), 7.05 (t, J = 7.4 Hz, 1), 6.29 (t, J = 6.8 Hz, 1), 5.42 (t, J = 7.7 Hz, 1), 5.19 (dd, J = 12.7, 7.1 Hz, 1), 4.28 (dd, J = 12.5, 8.5 Hz, 1). Two hydrogens (presumably the COOH, and NH) were unaccounted for. 13C NMR (126 MHz, CDCl3) δ 166.2, 164.6, 145.9, 142.4, 137.9, 136.6, 134.6, 130.1, 129.6, 128.9, 127.7, 126.7, 123.0, 122.0, 120.2, 119.2, 118.0, 114.3, 111.5, 108.0, 53.9, 38.5. Calculated for C22H18ClN2O3 (M+H)+ = 393.1008; Observed 393.1001.
Preparation of 6-((8S,8aR)-8,8a-dimethyl-1,2,3,4,6,7,8,8a-octahydronaphthalen-2-yl)-6-methyl-5,6-dihydrofuro[2,3-d]pyrimidine-2,4(1H,3H)-dione (JSF-4444): To a stirred solution of valencene (2.55 g, 12.5 mmol) and barbituric acid (0.64 g, 5.0 mmol) in acetonitrile at −10 °C was added ceric ammonium nitrate (5.48 g, 10.0 mmol) in portions over several minutes. The reaction mixture was stirred at −10 °C for 2 h. The reaction mixture was quenched with saturated aqueous ammonium chloride solution, and the solvent was removed in vacuo. The reaction mixture was extracted with diethyl ether and the combined organics were washed with saturated aqueous brine solution, dried over anhydrous sodium sulfate, and the volatiles were removed in vacuo to provide a yellow oil. The crude reaction product was purified by flash chromatography on silica gel, eluting with dichloromethane, to afford the desired product as a white solid (12 mg, 0.73%): 1H NMR (500 MHz, CDCl3) δ 9.42 (br s, 1), 8.16 (s, 1), 5.36 (d, J = 2.2 Hz, 1), 2.95 (dd, J = 13.7, 5.3 Hz, 1), 2.61 (t, J = 13.5 Hz, 1), 2.27 (t, J = 15.0 Hz, 1), 2.12 (d, J = 11.3 Hz, 1), 2.00-1.92 (m, 4), 1.88-1.84 (m, 1), 1.80-1.72 (m, 2), 1.44(s, 2), 1.42 (s, 2), 1.16-0.96 (m, 2), 0.92 (d, J = 3.9 Hz, 2), 0.88 (dd, J = 10.5, 5.8 Hz, 3), 0.85-0.81 (m, 1). 13C NMR (125 MHz, DMSO) δ 162.3, 161.9*, 151.5, 142.5, 120.5, 97.9*, 85.2*, 67.2, 46.6, 42.4*, 40.6, 37.5*, 34.9, 34.6, 31.9*, 28.3*, 27.1, 25.7, 23.9*, 18.7*, 16.1*. *Corresponded to carbons in 1:1 diastereomeric ratio. Calculated for C19H26N2O3 (M+H)+ = 331.2022; Observed 331.2016.
RESULTS
The dataset used to train the Neisseria Bayesian model was extracted from ChEMBL (17) (https://www.ebi.ac.uk/chembl/) with the query keywords “Neisseria gonorrhoeae” and “MIC”. The results were manually curated to remove false positive compounds, via comparison of tabulated data on the web site versus the published data, duplicates, and Pan Assay INterference compoundS (PAINS) (18, 19) or colloidal aggregators (43). Ultimately, the curated training set (Supplemental Information; File: Neisseria training set.sdf) contained 282 molecules, of which 160 compounds (56.7%) were deemed as “active,” given a practical minimum inhibitory concentration (MIC; typically the minimal concentration to result in a no-growth phenotype) cutoff of ≤ 8.0 μg/mL given our experience. A Principal Component Analysis (PCA) was performed to visualize the placement of active and inactive compounds in chemical space (Fig. S1). Graphically, the majority of the actives and inactives in the training set are clustered together.
Given our previously published work disclosing the utilization of naive Bayesian (heretofore Bayesian) models to discover novel antitubercular agents (44), we began with utilization of the Assay Central™ software and its Bayesian approach (45). Further curation of the training set included removal of molecular charges and duplicates. The run parameters were varied and optimized to produce the final Bayesian model. This was accomplished with ECFP6 fingerprints (22–24) and the use of 5-fold cross-validation with typical model evaluation metrics (recall, precision, specificity, F1-score, ROC, CK, and MCC as defined in the Materials and Methods section). The optimized Neisseria Bayesian model displayed the following internal statistics: AUC ROC = 0.8470, recall = 0.7736, precision = 0.8200, specificity = 0.7769, F1 score = 0.7961, CK = 0.5456, and MCC = 0.5467 (Fig. 1). The Neisseria Bayesian model was subsequently used to virtually screen over 190,000 small molecules from the Asinex BioDesign Library, characterized as containing chemical features present in approved drugs and natural products in synthetically accessible molecules. This library is shown (Fig. S2) to occupy a similar chemical space as does the training set. The top 10,000 compounds ranked by the Neisseria Bayesian model (Supplemental Information; File: Asinex top 10000 by Bayesian.sdf) were subjected to chemical diversity evaluation using the Assay Central™ workflow. The evaluation consisted of selecting the 100 most diverse compounds by using ECFP6 fingerprints as the descriptor and the Tanimoto score to calculate the dissimilarity index and subsequently rank this set using the Neisseria Bayesian score. These 100 compounds (Supplemental Information; File: Asinex top 10000 by Bayesian then top 100 by diversity.sdf) were visually inspected to remove chemically reactive compounds(31) and nitroheterocycles given our desire to explore chemotypes different from those being studied in the laboratory at the time (32, 33). The resulting top-scoring 20 compounds were purchased from Asinex and assayed for purity by HPLC and identity by MS.
Figure 1. Receiver Operator Characteristic (ROC) curve for the Neisseria Bayesian model.
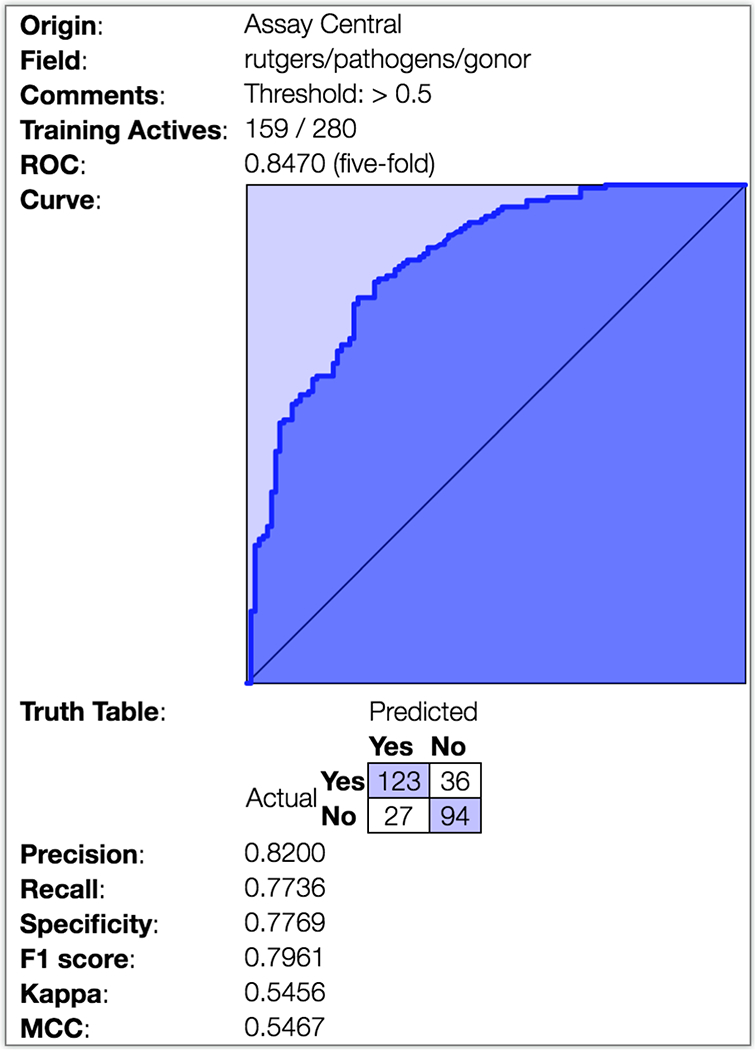
The Assay Central workflow was utilized in conjunction with the training set to produce an optimize a Bayesian model where the ROC curve, generated through 5-fold cross-validation, is depicted along with key model parameters.
The top 20 compounds selected by the Neisseria Bayesian model were then assayed for in vitro growth inhibition activity (Table 1 and Table S1) against three representative strains of N. gonorrhoeae most commonly used as laboratory strains (FA1090 (ATCC 700825), FA19 (ATCC BAA-1838), and F62 (ATCC 33084)), demonstrated to be susceptible to cefixime, ceftriaxone, ciprofloxacin, doxycycline, rifampicin, and azithromycin (Table S2) (46). Two compounds may be considered active compounds or hits (2/20 = 10% hit rate). JSF-4439 (BDE 33956561) and JSF-4444 (BDG 33899218), were active against all three strains. JSF-4439 and JSF-4444 exhibited the same MIC value of 7.0 μg/mL (18 μM for JSF-4439 and 21 μM for JSF-4444) versus FA1090 and 8.0 μg/mL (20 μM for JSF-4439 and 24 μM for JSF-4444) against F62, while displaying values of 6.2 μg/mL (16 μM) and 12 μg/mL (36 μM), respectively, versus the strains FA19 strain. JSF-4447 (LAS 13556723) did not meet the activity criteria of MIC ≤ 8.0 μg/mL versus any of these laboratory strains, demonstrating MIC values of 25, 50, and 50 μg/mL (77, 150, and 150 μM) versus the FA1090, FA19, and F62 strains. The remaining 17 tested compounds were inactive against all three laboratory strains (MIC > 50 μg/mL; Table S1). Finally, all 20 compounds were also inactive against four multi-drug resistant N. gonorrhoeae strains: FA6140 (https://www.ncbi.nlm.nih.gov/genome/?term=Neisseria%20gonorrhoeae%20FA6140), F89 (34), 35/02 (https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=528346), and 59/03 (35) (Tables S2 and S3). JSF-4439 and JSF-4444 exhibited minimum bactericidal concentrations (MBC; minimum concentration to afford a 2 log10 reduction in bacterial colony-forming units) of 3.2 – 6.3 and 12 – 25 μg/mL, respectively, versus the FA1090 strain (Fig. S3).
Table 1. In vitro growth inhibition profile for hit compounds versus drug-susceptible N. gonorrhoeae strains.
The MIC values tabulated are representative of experiments conducted in at least duplicate.
| Compound Structure | Compound Name (Asinex) |
MIC in μg/mL versus the N. gonorrhoeae strain | ||
|---|---|---|---|---|
| FA1090 | FA19 | F62 | ||
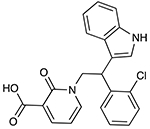 |
JSF-4439 (BDE 33956561) |
7.0 | 6.2 | 8.0 |
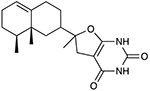 |
JSF-4444 (BDG 33899218) |
7.0 | 12 | 8.0 |
JSF-4439 and JSF-4444 were next assayed against a subset of ESKAPE bacteria to query for broad-spectrum antibacterial activity. The ESKAPE panel consisted of Enterococcus faecium (NCTC 7171), Staphylococcus aureus (ATCC 43300), Klebsiella pneumoniae (BAA 2146), Acinetobacter baumannii (ATCC 19606), Pseudomonas aeruginosa (HER 1018), and Enterobacter cloacae (ATCC 13047) strains. JSF-4439 and JSF-4444 exhibited selective inhibition of N. gonorrhoeae growth, as each demonstrated an MIC of >50 μg/mL versus all six strains.
JSF-4439 was sourced through synthesis via adaptation of a route disclosed in a Takeda Pharmaceutical Company patent (47) (Fig. 2A). Commercially available 2-(2-chlorophenyl)-2-(1H-indol-3-yl)ethan-1-amine and methyl 2-oxo-2H-pyran-3-carboxylate were subjected to the addition of N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride in the presence of 4-dimethylaminopyridine in N,N-dimethylformamide to afford methyl 1-(2-(2-chlorophenyl)-2-(1H-indol-3-yl)ethyl)-2-oxo-1,2-dihydropyridine-3-carboxylate (1). Saponification of the methyl ester in 1 afforded synthetic JSF-4439 in 18% overall yield. JSF-4444 was prepared in one low-yielding step, via a literature route (48), involving the oxidative coupling of commercial (+)-valencene and barbituric acid (Fig. 2B). In both cases, the LC-MS, 1H NMR, and high-resolution mass spectrum of the synthetic materials matched those of the commercial materials, and their assays profiles were similar.
Figure 2. Synthetic schemes to provide the two hit compounds (A) JSF-4439 and (B) JSF-4444.
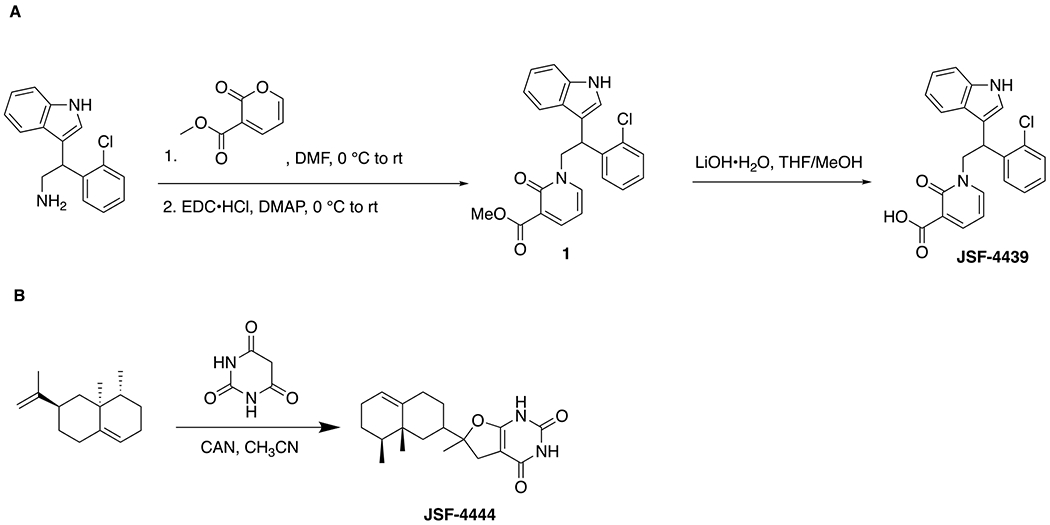
JSF-4439 was prepared in two steps from commercial materials, while JSF-4444 was attained in one reaction from commercial materials.
JSF-4439 and JSF-4444 were compared to the curated actives in the training set via calculation of their pairwise Tanimoto scores using ECFP6 fingerprints. With this Tanimoto calculation, the score ranges from 0 to 1.0 (for identical compounds). JSF-4439 and JSF-4444 presented maximum Tanimoto similarities to the training set actives of 0.31 and 0.39, respectively, with minimum Tanimoto similarities of 0.10 (Table S4). Visually, the two hits are chemically distinct from their closest actives in the training set. The PCA (Fig. S4) indicates that JSF-4439 and JSF-4444 have predicted chemical properties similar to those of known N. gonorrhoeae inhibitors, but they are generally not within the main clusters of these previously characterized inhibitors. Additionally, searches of SciFinder (http://scifinder.cas.org) and PubChem (https://pubchem.ncbi.nlm.nih.gov failed to uncover previous disclosure of these two compounds as growth inhibitors of N. gonorrhoeae or as antibacterial agents, in general. Representing novel hits against N. gonorrhoeae, JSF-4439 and JSF-4444 were profiled for mouse liver microsome (MLM) stability and kinetic aqueous solubility (S) in pH 7.4 phosphate-buffered saline to inform future optimization campaigns. JSF-4439 exhibited an MLM half-life (t1/2) of 15.6 min and a S = 202.0 μM, while JSF-4444 was characterized by MLM t1/2 = 19.6 min and S = 301.5 μM.
DISCUSSION
Although an urgent need exists for new drug treatments for Neisseria gonorrhoeae infection, the previously reported machine leaning efforts pertaining to this STI have not been directly relevant to drug discovery. Machine learning methods for N. gonorrhoeae have been limited to: 1) phenotype prediction by using Multiple Instance Learning models (49); 2) antibacterial-resistant DNA identification by using Random Forest, naïve Bayesian, and Support Vector Machine (50) approaches; and 3) bacteria imaging identification through a combination of Deep Neural Networks and Support Vector Machine (51, 52) methods. To the best of our knowledge, we present the first application of machine learning, a Bayesian model, for successfully predicting novel compounds with in vitro antibacterial activity against N. gonorrhoeae. Significantly, this modeling effort produced a training set, curated from the ChEMBL database, which we provide as an .sdf file in the Supplemental Information so that it may serve as a training set for the efforts of other laboratories, using other modeling approaches. In addition, this Bayesian model, upon publication, will be made publicly available at www.assaycentral.org.
The Neisseria Bayesian model workflow identified two promising hit compounds (JSF-4439 and JSF-4444) out of a library of over 190,000 candidates. JSF-4439 and JSF-4444 were initially ranked by the Neisseria Bayesian model at positions 83 and 791, respectively (Supplemental Information; File: Asinex top 10000 by Bayesian.sdf). After the diversity filter was applied, and compounds were sorted by the Neisseria Bayesian model score, JSF-4439 and JSF-4444 ascended to the 2nd and 4th positions, respectively (Supplemental Information; File: Asinex top 10000 by Bayesian then top 100 by diversity.sdf). Without the diversity filter, we would not have inspected JSF-4444; consequently, it would not have been assayed. However, the diversity filter alone was not sufficient to identify the hit compounds against N. gonorrhoeae. It is interesting to note that the two hits were not amongst the top 100 most diverse molecules chosen from the Asinex library of over 190,000 compounds (Supplemental Information; File: Asinex top 100 by diversity.sdf), but both were amongst the top 1000 most diverse molecules (Supplemental Information; File: Asinex top 1000 by diversity.sdf). Therefore, combining a diversity filter with the Neisseria Bayesian model improved the performance of the workflow for this screening campaign.
Diversity has occupied an important role in machine learning with regard to decreasing the redundancy between data and model, providing informative data, and generating representative models (53). Diversity as a strategy is mainly applied to improve the representativeness of the training set (54, 55), to select informative parameters (56, 57), and to combine distinct models (ensemble models) to reduce the generalization error of the prediction (58) (59). The chemical diversity of compounds has been intensively scrutinized to design virtual screening libraries (60), to improve the training data (61), and to select parameters and/or models (62, 63).
In this work, we present an alternative use of chemical diversity for improving machine learning models by decreasing the redundancy amongst the model’s top-ranked compounds. The Neisseria Bayesian model selected as active (score > 0.5) over 10,000 compounds, before the diversity filter (Supplemental Information; File: Asinex top 10000 by Bayesian.sdf). In the top 100 compounds (Supplementary Information; File: Asinex top 100 by Bayesian.sdf; score refers to column entitled Ngonor_Prediction), the difference in score between the 1st scored compound (score 0.678) and the 100th scored compound (0.633) was only 0.044, and the difference between the 56th (0.640) and 100th (0.633) was even smaller (0.007). Often, the top compounds ranked by the Neisseria Bayesian model (without the diversity filter) presented an overlapping of chemical space. This behavior might be related to the over-representation of features among the training compounds labeled as active, which directly reflects on the top ranked compounds. The top compounds with similar chemical structure will likely share the same over-represented characteristics. A similar assumption was presented by the Lounkine group (63), who used feature selection to improve an active learning model. We assert that by using the chemical diversity filter we have reduced the redundancy of the model’s top-ranked compounds.
Derived from this Bayesian workflow leveraging compound diversity, JSF-4439 and JSF-4444 represent novel hit chemotypes that will require mechanism of action studies and compound optimization to achieve early lead compound status (39, 64). We envision mechanistic studies would involve the generation of drug-resistant mutants by established methods which upon validation of the mutation/s would help clarify biological target/s (65, 66). With regard to compound evolution, limited molecular profiling demonstrated the need to improve the in vitro profile of both hits. While an MIC ≤ 8.0 μg/mL is reasonable for a hit in our experience, we would prefer to see an MIC ≤ 1.0 μM (or ~0.3 μg/mL for a compound of 300 g/mol molecular weight) for both compounds against a range of both N. gonorrhoeae drug-susceptible and drug-resistant strains. To this end, the previously stated future goal of probing the mechanism should provide insight into potential issues due to cross-resistance with existing antibacterials. However, given the novel chemotypes of these two hits, we believe the likelihood of them sharing a target with existing antibacterials is relatively low. In addition, the identification of molecular target/s may be coupled with X-ray crystallography to enable a structure-based design optimization (39). The MLM stability of both compounds will need to be improved as the respective values are demonstrative of metabolic instability. Typically, we would prefer an early lead to have MLM t1/2 ≥ 60 min (64). The aqueous solubility of both compounds significantly exceeded the goal value of 100 μM (64). We anticipate that an optimization of these two hits will rely on medicinal chemistry heuristics, the Neisseria Bayesian model for growth inhibition and specifically its ability to identify which atoms within these compounds are contributing to, or detracting from, whole-cell activity (Table S5), our models for MLM t1/2 (67, 68), and our concise syntheses of both compounds (Fig. 2). An acceptable early lead compound profile would enable mouse pharmacokinetic studies to assess the plasma concentration versus time relationship and set the stage for approaching in vivo efficacy studies to pharmacologically validate both the chemotype and the elucidated mechanism of action.
CONCLUSIONS
The Bayesian model approach presented herein has supplied what we believe to be the first report of a machine learning workflow to predict novel small molecule growth inhibitors of N. gonorrhoeae. A key element to the strategy appears to be a diversity filter to perhaps better spread the “risk” amongst a wider range of chemotypes when scoring a very large library of candidate molecules. In so doing, we disclose JSF-4439 and JSF-4444, which represent new hit molecules. Further evolution of their properties should afford valuable chemical tools and/or drug discovery molecules. We also anticipate that application of this machine learning approach to other bacteria of global health relevance should prove fruitful.
Supplementary Material
Acknowledgments
J.S.F. was supported by award number U19AI109713 NIH/NIAID for the “Center to develop therapeutic countermeasures to high-threat bacterial agents,” from the National Institutes of Health: Centers of Excellence for Translational Research (CETR). S.E. kindly acknowledges NIH/NIGMS for R44GM122196 which funded Assay Central™ and Dr. Alex Clark (Molecular Materials Informatics) for assistance with Assay Central™. We thank BIOVIA for providing J.S.F. and S.E. with Discovery Studio and Pipeline Pilot.
ABBREVIATIONS
- STI
sexually transmitted infection
- WHO
World Health Organization
- PAINS
Pan Assay INterference compoundS
- ROC
Receiver Operating Characteristic
- CK
Cohen’s Kappa
- MCC
Matthews Correlation Coefficient
- MIC
minimum inhibitory concentration
- MBC
minimum bactericidal concentration
- PCA
Principal Component Analysis
- MLM
mouse liver microsome
- t1/2
half-life
- S
solubility
Footnotes
Supplementary Material Available: document with supplemental figures and tables; and relevant training set and prediction files.
Conflicts of Interest: S.E. is the founder and CEO of Collaborations Pharmaceuticals, Inc. and K.Z. is an employee of Collaborations Pharmaceuticals, Inc.
REFERENCES
- 1.Quillin SJ, Seifert HS. Neisseria gonorrhoeae host adaptation and pathogenesis. Nature Rev Microbiol. 2018;16(4):226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wi T, Lahra MM, Ndowa F, Bala M, Dillon J-AR, Ramon-Pardo P, Eremin SR, Bolan G, Unemo M. Antimicrobial resistance in Neisseria gonorrhoeae: Global surveillance and a call for international collaborative action. PLoS Med. 2017;14(7):e1002344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ison CA, Dillon J-AR, Tapsall JW. The epidemiology of global antibiotic resistance among Neisseria gonorrhoeae and Haemophilus ducreyi. Lancet. 1998;351:S8–S11. [DOI] [PubMed] [Google Scholar]
- 4.Cehovin A, Harrison OB, Lewis SB, Ward PN, Ngetsa C, Graham SM, Sanders EJ, Maiden MC, Tang CM. Identification of Novel Neisseria gonorrhoeae Lineages Harboring Resistance Plasmids in Coastal Kenya. J Infect Dis. 2018;218(5):801–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO. WHO guidelines for the treatment of Neisseria gonorrhoeae. 2016. [PubMed] [Google Scholar]
- 6.WHO. Global action plan to control the spread and impact of antimicrobial resistance in Neisseria gonorrhoeae. 2012. [Google Scholar]
- 7.Tanaka M, Furuya R, Kobayashi I, Kanesaka I, Ohno A, Katsuse AK. Antimicrobial resistance and molecular characterisation of Neisseria gonorrhoeae isolates in Fukuoka, Japan, 1996–2016. J Glob Antimicrob Resist. 2019;17:3–7. [DOI] [PubMed] [Google Scholar]
- 8.CDC. Sexually Transmitted Disease Surveillance 2018. 2019. October 09. Available from: https://www.cdc.gov/std/stats18/Gonorrhea.htm.
- 9.Edwards JL, Jennings MP, Seib KL. Neisseria gonorrhoeae vaccine development: hope on the horizon? Current Opin Infect Dis. 2018;31(3):246–250. [DOI] [PubMed] [Google Scholar]
- 10.WHO. Emergence of multi-drug resistant Neisseria gonorrhoeae: Threat of global rise in untreatable sexually transmitted infections. 2011. [Google Scholar]
- 11.Zhao S, Duncan M, Tomberg J, Davies C, Unemo M, Nicholas RA. Genetics of chromosomally mediated intermediate resistance to ceftriaxone and cefixime in Neisseria gonorrhoeae. Antimicrob Agents Chemother. 2009;53(9):3744–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagman KE, Pan W, Spratt BG, Balthazar JT, Judd RC, Shafer WM. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology. 1995;141(3):611–622. [DOI] [PubMed] [Google Scholar]
- 13.Cristillo AD, Bristow CC, Torrone E, Dillon J-A, Kirkcaldy RD, Dong H, Grad YH, Nicholas RA, Rice PA, Lawrence K. Antimicrobial Resistance in Neisseria gonorrhoeae: Proceedings of the STAR Sexually Transmitted Infection-Clinical Trial Group Programmatic Meeting. Sexually transmitted diseases. 2019;46(3):e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ekins S, Freundlich JS, Reynolds RC. Fusing dual-event data sets for Mycobacterium tuberculosis machine learning models and their evaluation. J Chem Inf Model. 2013;53(11):3054–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ekins S, Reynolds RC, Franzblau SG, Wan B, Freundlich JS, Bunin BA. Enhancing hit identification in Mycobacterium tuberculosis drug discovery using validated dual-event Bayesian models. PLoS One. 2013;8(5):e63240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ekins S, Reynolds RC, Kim H, Koo M-S, Ekonomidis M, Talaue M, Paget SD, Woolhiser LK, Lenaerts AJ, Bunin BA. Bayesian models leveraging bioactivity and cytotoxicity information for drug discovery. Chem Biol. 2013;20(3):370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D, Al-Lazikani B. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2011;40(D1):D1100–D1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mendgen T, Steuer C, Klein CD. Privileged scaffolds or promiscuous binders: a comparative study on rhodanines and related heterocycles in medicinal chemistry. J Med Chem. 2012;55(2):743–753. [DOI] [PubMed] [Google Scholar]
- 19.Zinglé C, Tritsch D, Grosdemange-Billiard C, Rohmer M. Catechol–rhodanine derivatives: Specific and promiscuous inhibitors of Escherichia coli deoxyxylulose phosphate reductoisomerase (DXR). Bioorg Med Chem. 2014;22(14):3713–3719. [DOI] [PubMed] [Google Scholar]
- 20.Perryman AL, Stratton TP, Ekins S, Freundlich JS. Predicting mouse liver microsomal stability with “pruned” machine learning models and public data. Pharm Res. 2016;33(2):433–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perryman AL, Patel JS, Russo R, Singleton E, Connell N, Ekins S, Freundlich JS. Naive Bayesian models for vero cell cytotoxicity. Pharm Res. 2018;35(9):170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogers D, Hahn M. Extended-connectivity fingerprints. J Chem Inf Model. 2010;50(5):742–754. [DOI] [PubMed] [Google Scholar]
- 23.Clark AM, Dole K, Coulon-Spektor A, McNutt A, Grass G, Freundlich JS, Reynolds RC, Ekins S. Open source Bayesian models. 1. Application to ADME/Tox and drug discovery datasets. J Chem Inf Model. 2015;55(6):1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clark AM, Ekins S. Open source Bayesian models. 2. Mining a “big dataset” to create and validate models with ChEMBL. J Chem Inf Model. 2015;55(6):1246–1260. [DOI] [PubMed] [Google Scholar]
- 25.Clark AM, Dole K, Ekins S. Open source bayesian models. 3. Composite models for prediction of binned responses. J Chem Inf Model. 2016;56(2):275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carletta J Assessing agreement on classification tasks: the kappa statistic. Comp Ling. 1996;22(2):249–254. [Google Scholar]
- 27.Cohen J A coefficient of agreement for nominal scales. Ed Psychol Meas. 1960;20(1):37–46. [Google Scholar]
- 28.Matthews BW. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim Biophys Acta. 1975;405(2):442–451. [DOI] [PubMed] [Google Scholar]
- 29.Korotcov A, Tkachenko V, Russo DP, Ekins S. Comparison of deep learning with multiple machine learning methods and metrics using diverse drug discovery data sets. Mol Pharm. 2017;14(12):4462–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lane T, Russo DP, Zorn KM, Clark AM, Korotcov A, Tkachenko V, Reynolds RC, Perryman AL, Freundlich JS, Ekins S. Comparing and validating machine learning models for Mycobacterium tuberculosis drug discovery. Mol Pharm. 2018;15(10):4346–4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walters WP, Stahl MT, Murcko MA. Virtual screening - an overview. Drug Discov Today. 1998;3:160–178. [Google Scholar]
- 32.Wang X, Inoyama D, Russo R, Li SG, Jadhav R, Stratton TP, Mittal N, Bilotta JA, Singleton E, Kim T, Paget SD, Pottorf RS, Ahn YM, Davila-Pagan A, Kandasamy S, Grady C, Hussain S, Soteropoulos P, Zimmerman MD, Ho HP, Park S, Dartois V, Ekins S, Connell N, Kumar P, Freundlich JS. Antitubercular Triazines: Optimization and Intrabacterial Metabolism. Cell Chem Biol. 2020;27(2):172–185 e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Perryman AL, Li SG, Paget SD, Stratton TP, Lemenze A, Olson AJ, Ekins S, Kumar P, Freundlich JS. Intrabacterial Metabolism Obscures the Successful Prediction of an InhA Inhibitor of Mycobacterium tuberculosis. ACS Infect Dis. 2019;5(12):2148–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goire N, Lahra MM, Ohnishi M, Hogan T, Liminios AE, Nissen MD, Sloots TP, Whiley DM. Polymerase chain reaction-based screening for the ceftriaxone-resistant Neisseria gonorrhoeae F89 strain. Euro Surveill. 2013;18(14):20444. [DOI] [PubMed] [Google Scholar]
- 35.John CM, Feng D, Jarvis GA. Treatment of human challenge and MDR strains of Neisseria gonorrhoeae with LpxC inhibitors. J Antimicrob Chemother. 2018;73(8):2064–2071. [DOI] [PubMed] [Google Scholar]
- 36.Takei M, Yamaguchi Y, Fukuda H, Yasuda M, Deguchi T. Cultivation of Neisseria gonorrhoeae in liquid media and determination of its in vitro susceptibilities to quinolones. J Clin Micro. 2005;43(9):4321–4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiegand I, Hilpert K, Hancock RE. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nature Prot. 2008;3(2):163. [DOI] [PubMed] [Google Scholar]
- 38.Hauser C, Hirzberger L, Unemo M, Furrer H, Endimiani A. In vitro activity of fosfomycin alone and in combination with ceftriaxone or azithromycin against clinical Neisseria gonorrhoeae isolates. Antimicrob Agents Chemother. 2015;59(3):1605–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inoyama D, Awasthi D, Capodagli GC, Tsotetsi K, Sukheja P, Zimmerman M, Li SG, Jadhav R, Russo R, Wang X, Grady C, Richmann T, Shrestha R, Li L, Ahn YM, Ho Liang HP, Mina M, Park S, Perlin DS, Connell N, Dartois V, Alland D, Neiditch MB, Kumar P, Freundlich JS. A Preclinical Candidate Targeting Mycobacterium tuberculosis KasA. Cell Chem Biol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.BIOVIA DS. Discovery Studio Modeling Environment. San Diego: Dassault Systèmes; Release 2017. [Google Scholar]
- 41.BIOVIA DS. Pipeline Pilot. San Diego: Dassault Systèmes; Release 2017. [Google Scholar]
- 42.Willett P Similarity-based virtual screening using 2D fingerprints. Drug Discov Today. 2006;11(23-24):1046–1053. [DOI] [PubMed] [Google Scholar]
- 43.McGovern SL, Caselli E, Grigorieff N, Shoichet BK. A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J Med Chem. 2002;45(8):1712–1722. [DOI] [PubMed] [Google Scholar]
- 44.Ekins S, Reynolds RC, Kim H, Koo M-S, Ekonomidis M, Talaue M, Paget SD, Woolhiser LK, Lenaerts A, Bunin BA, Connell N, Freundlich JS. Bayesian models leveraging bioactivity and cytotoxicity information for drug discovery. Chem Biol. 2013;20:370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ekins S, Freundlich JS. Computational models for tuberculosis drug discovery. Methods Mol Bio. 2013;993:245–262. [DOI] [PubMed] [Google Scholar]
- 46.Jordan PW, Snyder LA, Saunders NJ. Strain-specific differences in Neisseria gonorrhoeae associated with the phase variable gene repertoire. BMC Microbiol. 2005;5(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miyamoto N, Matsumoto S, Imamura S. Fused heterocyclic derivatives and use thereof. WO 2009/136663. Takeda Pharmaceutical Company Limited; 2009. [Google Scholar]
- 48.Kobayashi K, Tanaka H, Tanaka K, Yoneda K, Morikawa O, Konishi H. One-step synthesis of furo[2,3-d]pyrimidine-2,4(1H,3H)-diones using the CAN-mediated furan ring formation. Synth Commun. 2000;30:4277–4291. [Google Scholar]
- 49.Aridhi S, Sghaier H, Maddouri M, Nguifo EM. Computational phenotype prediction of ionizing-radiation-resistant bacteria with a multiple-instance learning model. Proceedings of the 12th International Workshop on Data Mining in Bioinformatics: ACM; 2013. p. 18–24. [Google Scholar]
- 50.Lingle JI, Santerre J. Using Machine Learning for Antimicrobial Resistant DNA Identification. SMU Data Science Rev. 2019;2(2):12. [Google Scholar]
- 51.Wahid MF, Ahmed T, Habib MA. Classification of Microscopic Images of Bacteria Using Deep Convolutional Neural Network. 2018 10th International Conference on Electrical and Computer Engineering (ICECE): IEEE; 2018. p. 217–220. [Google Scholar]
- 52.Ahmed T, Wahid MF, Hasan MJ. Combining Deep Convolutional Neural Network with Support Vector Machine to Classify Microscopic Bacteria Images. 2019 International Conference on Electrical, Computer and Communication Engineering (ECCE): IEEE; 2019. p. 1–5. [Google Scholar]
- 53.Gong Z, Zhong P, Hu W. Diversity in Machine Learning. IEEE Access. 2019;7:64323–64350. [Google Scholar]
- 54.Prudent R, Moucadel V, López-Ramos M, Aci S, Laudet B, Mouawad L, Barette C, Einhorn J, Einhorn C, Denis J-N. Expanding the chemical diversity of CK2 inhibitors. Mol Cellular Biochem. 2008;316(1-2):71–85. [DOI] [PubMed] [Google Scholar]
- 55.Xu J A new approach to finding natural chemical structure classes. J Med Chem. 2002;45(24):5311–5320. [DOI] [PubMed] [Google Scholar]
- 56.Zhong P, Gong Z, Li S, Schönlieb C-B. Learning to diversify deep belief networks for hyperspectral image classification. IEEE Trans Geosci Remote Sensing. 2017;55(6):3516–3530. [Google Scholar]
- 57.Brinker K Incorporating diversity in active learning with support vector machines. Proceedings of the 20th international conference on machine learning (ICML-03); 2003. p. 59–66. [Google Scholar]
- 58.Melville P, Mooney RJ. Creating diversity in ensembles using artificial data. Information Fusion. 2005;6(1):99–111. [Google Scholar]
- 59.Shoham R, Permuter H. Amended Cross-Entropy Cost: An Approach for Encouraging Diversity in Classification Ensemble (Brief Announcement). In.International Symposium on Cyber Security Cryptography and Machine Learning: Springer; 2019. p. 202–207. [Google Scholar]
- 60.Nikitina N, Ivashko E, Tchernykh A. Congestion game scheduling for virtual drug screening optimization. J Comput Aided Mol Des. 2018;32(2):363–374. [DOI] [PubMed] [Google Scholar]
- 61.Cheng F, Shen J, Yu Y, Li W, Liu G, Lee PW, Tang Y. In silico prediction of Tetrahymena pyriformis toxicity for diverse industrial chemicals with substructure pattern recognition and machine learning methods. Chemosphere. 2011;82(11):1636–1643. [DOI] [PubMed] [Google Scholar]
- 62.Afolabi LT, Saeed F, Hashim H, Petinrin OO. Ensemble learning method for the prediction of new bioactive molecules. PLoS One. 2018;13(1):e0189538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maciejewski M, Wassermann AM, Glick M, Lounkine E. Experimental design strategy: weak reinforcement leads to increased hit rates and enhanced chemical diversity. J Chem Inf Model. 2015;55(5):956–962. [DOI] [PubMed] [Google Scholar]
- 64.Inoyama D, Paget SD, Russo R, Kandasamy S, Kumar P, Singleton E, Occi J, Tuckman M, Zimmerman MD, Ho HP, Perryman AL, Dartois V, Connell N, Freundlich JS. Novel Pyrimidines as Antitubercular Agents. Antimicrob Agents Chemother. 2018;62(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chu J, Vila-Farres X, Inoyama D, Ternei M, Cohen LJ, Gordon EA, Reddy BV, Charlop-Powers Z, Zebroski HA, Gallardo-Macias R, Jaskowski M, Satish S, Park S, Perlin DS, Freundlich JS, Brady SF. Discovery of MRSA active antibiotics using primary sequence from the human microbiome. Nat Chem Biol. 2016;12(12):1004–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kumar P, Capodagli GC, Awasthi D, Shrestha R, Maharaja K, Sukheja P, Li SG, Inoyama D, Zimmerman M, Ho Liang HP, Sarathy J, Mina M, Rasic G, Russo R, Perryman AL, Richmann T, Gupta A, Singleton E, Verma S, Husain S, Soteropoulos P, Wang Z, Morris R, Porter G, Agnihotri G, Salgame P, Ekins S, Rhee KY, Connell N, Dartois V, Neiditch MB, Freundlich JS, Alland D. Synergistic Lethality of a Binary Inhibitor of Mycobacterium tuberculosis KasA. MBio. 2018;9(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perryman AL, Stratton TP, Ekins S, Freundlich JS. Predicting Mouse Liver Microsomal Stability with “Pruned” Machine Learning Models and Public Data. Pharm Res. 2016;33(2):433–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stratton TP, Perryman AL, Vilcheze C, Russo R, Li SG, Patel JS, Singleton E, Ekins S, Connell N, Jacobs WR Jr., Freundlich JS. Addressing the Metabolic Stability of Antituberculars through Machine Learning. ACS Med Chem Lett. 2017;8(10):1099–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


