Abstract
Serine incorporator protein 5 (SERINC5) is the host anti-retroviral factor that reduces HIV-1 infectivity by incorporating into virions and inhibiting the envelope glycoprotein (Env) mediated virus fusion with target cells. We and others have shown that SERINC5 incorporation into virions alters the Env structure and sensitizes the virus to broadly neutralizing antibodies targeting cryptic Env epitopes. We have also found that SERINC5 accelerates the loss of Env function over time compared to control viruses. However, the exact mechanism by which SERINC5 inhibits HIV-1 fusion is not understood. Here, we utilized 2D and 3D super-resolution microscopy to examine the effect of SERINC5 on the distribution of Env glycoproteins on single HIV-1 particles. We find that, in agreement with a previous report, Env glycoproteins form clusters on the surface of mature virions. Importantly, incorporation of SERINC5, but not SERINC2, which lacks antiviral activity, disrupted Env clusters without affecting the overall Env content. We also show that SERINC5 and SERINC2 also form clusters on single virions. Unexpectedly, Env and SERINCs molecules exhibited poor co-distribution on virions, as evidenced by much greater Env-SERINC pairwise distances compare to Env-Env distances. This observation is inconsistent with the previously reported interaction between Env and SERINC5 and suggests an indirect effect of SERINC5 on Env cluster formation. Collectively, our results reveal a multifaceted mechanism of SERINC5-mediated restriction of HIV-1 fusion that, aside from the effects on individual Env trimers, involves disruption of Env clusters, which likely serve as sites of viral fusion with target cells.
Keywords: SERINC5, HIV-1 restriction, envelope glycoprotein clustering, viral fusion, super-resolution microscopy, DBSCAN
SERINC5 (serine incorporator 5) is a host factor that reduces HIV-1 infectivity by incorporating into virions and inhibiting their fusion with a target cell.1-3 SERINC family proteins are conserved across eukaryotes.4 The drosophila SERINC5 structure has been recently solved revealing 10-transmembrane domains arranged in a two sub-domain fold.5 SERINC3 and SERINC4 also reduce HIV-1 infectivity,2, 6 whereas SERINC2 does not exhibit detectable antiviral activity,1, 3, 6 in spite of its efficient incorporation into virions.3, 7 The HIV-1 Nef accessory protein antagonizes SERINCs’ activity by binding these proteins and removing them from the plasma membrane where the viral assembly/budding occurs.1, 2 Nef-mediated internalization targets SERINC5 (hereafter abbreviated SER5) to lysosomal degradation 8 (but see 9 for the opposite result). At least two other retroviral proteins unrelated to Nef antagonize SER5: the Murine Leukemia Virus-encoded glycoGag and Equine Infectious Anemia Virus S2 protein.10-12 These proteins counteract the SER5 activity by a mechanism similar to that employed by Nef.10
Despite the significant antiviral activity of SER5, the mechanism by which this protein restricts HIV-1 is poorly understood. The SER5’s region encompassing the transmembrane domains 5 through 9 appears to be responsible for the antiviral activity.6 A more recent structure-based mutagenesis study revealed a critical role of residues within the extracellular loops and at the sub-domain interface.5 HIV-1’s sensitivity to SER5 restriction varies between virus strains with tier 1, laboratory-adapted HIV-1 strains being generally more sensitive to SER5 restriction than Tier 2/3 strains.1-3, 7, 13 The Env’s resistance to SER5 maps to the gp120 variable loops V1, V2 and V3 2, 7, 13 and appears to correlate with the stability of the “closed” conformation of Env in which the variable loops V1-V3 interact with each other at the apex of Env trimer.2, 14 Consistent with this notion, a recent study reported that incorporation of CD4 into virions promotes an “open” conformation of Env and thereby sensitizes otherwise resistant Env strains to SER5 restriction.7 Moreover, that study has found that Env binds SER5, but not SER2 in the cell membranes and that the extent of Env-SER5 binding correlates with Env’s sensitivity to this restriction factor.7
Consistent with the Env-SER5 interaction, SER5 has been shown to increase the accessibility of the cryptic gp41 domains, membrane-proximal extracellular region and coiled coil region, to neutralizing antibodies 3 and to sensitize viral fusion/infection to neutralizing antibodies and antiviral drugs.3, 6, 13 Hence, SER5 appears to promote Env “opening” and exposure of vulnerable neutralization epitopes. We have also provided evidence that SER5 incorporation accelerates spontaneous loss of Env function over time.3 Interestingly, although we found that SER5 inhibits the formation of a small (~4 nm) fusion pore between viruses and cells,3 HIV-1 fusion is less potently suppressed than infectivity.1-3 This phenotype suggests that SER5 may affect the fusion pore enlargement, which is required for the release of viral core.1, 4
Here, we asked whether, in addition to altering the conformation and promoting spontaneous inactivation of Env,3 SER5 also alters the distribution of Env in the viral membrane, as has been suggested previously.2, 4 A pioneering study using a super-resolution imaging of single HIV-1 particles has shown maturation-dependent clustering of Env on virions and argued that this clustering is essential for viral fusion/infection.15 Indeed, it is generally accepted that the fusion process proceeds through a cooperative action of several glycoproteins forming a functional fusion complex.16-21 Given that there are only a few Env glycoproteins on HIV-1 particles (~14 trimers),22, 23 it is reasonable to assume that Env clustering could create “fusion hotspots” and thereby increase HIV-1 infectivity.
To visualize the Env and SERINC molecule distributions on single HIV-1 particles, we employed 2- and 3-dimensional super-resolution fluorescence microscopy. Fewer Env clusters were observed on mature vs immature virions, in agreement with the previous study.15 Importantly, a comparison of SER5-containing and control SER2-containing particles, which do not exhibit reduction in infectivity,1, 3, 6 revealed that SER5 incorporation into virions disfavored Env clustering compared to control and SER2-containing viruses. Thus, the diminished fraction of Env that forms clusters on mature SER5-containing viruses may be partially responsible for the reduced fusion competence of these particles compared to control viruses. Surprisingly, we did not detect significant co-distribution of Env and SER5 on virions. This finding is inconsistent with Env-SER5 interactions and is indicative of a potential indirect mechanism of SER5-mediated restriction.
Results and Discussion
Single-molecule localization imaging implicates SER5 in disrupting Env clusters on HIV-1 particles.
To determine whether SER5 affects the incorporation and/or distribution of HIV-1 Env glycoproteins on the virus membrane, we produced several independent panels of pseudoviruses bearing SER5-sensitive HXB2 Env by parallel transfections of 293T/17 cells. Each panel consisted of 4 viral preparations: control viruses, viruses containing SER5 or SER2, as well as immature viruses produced in the presence of the HIV-1 protease inhibitor Saquinavir (SQV) to assess the maturation-dependence of Env clustering described previously.15 To visualize single viral particles, all preparations were labeled with GFP-Vpr, which incorporates into the HIV-1 core.24 The quality control for each preparation included the measurements of infectivity, Western blotting for p24 to assess proper virus maturation, immuno-fluorescence staining for Env and SERINCs, as well as single virus imaging to ensure comparable levels of Env, SER5 and SER2 across the viruses in each panel. To detect SER5 and SER2 on single virions, SERINC constructs with an HA-tag inserted into the predicted extra-cellular loop 4 4, 6, 25 were used. Two out of four virus panels passed the quality control and were selected for imaging.
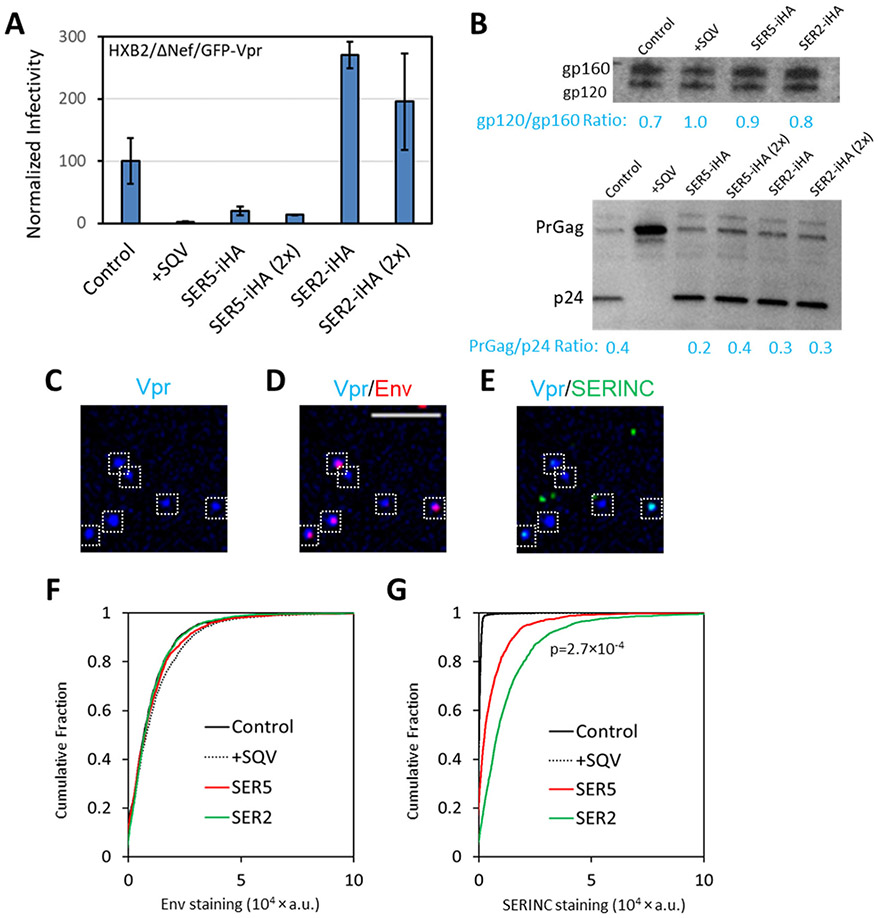
In all virus panels tested (exemplified by the set of viruses designated panel A), infectivity was strongly inhibited by incorporation of SER5, whereas SER2 incorporation did not diminish infectivity (Fig. 1A). In this panel, neither SER5 nor SER2 co-expression affected the release or maturation of HIV-1, as evidenced by comparable p24/PrGag bands on SDS PAGE (Fig. 1B, bottom). As expected, HIV-1 maturation was blocked by SQV treatment. Immuno-fluorescence staining of GFP-Vpr-labeled viruses for Env and SERINCs (Fig. 1C-E) showed that, importantly, all preparations contained comparable levels of Env glycoproteins on viral particles (Fig. 1F). In this panel, the incorporation of SER2 was somewhat more efficient than SER5 (Fig. 1G), in agreement with the previous reports.6, 7 Of note, the levels of Env in virions were not affected by SERINC incorporation, as evidenced by the lack of correlation between the two signals on single particles (Supp. Fig. S1A, B). Similarly, SERINC incorporation did not affect the extent of proteolytic cleavage of Env in virions (Fig. 1B, top). Also, virus aggregation did not noticeably contribute to our analysis, as evidenced by limited correlation between Env and GFP-Vpr signals in all preparations (data not shown).
Figure 1. Analysis of Env and SERINC incorporation into pseudoviruses, virus infectivity and maturation.
Four pseudovirus preparations were produced in parallel by transfection of 293T/17 cells and designated as panel A. This panel consisted of control viruses, +SQV (saquinavir treated, immature particles), SER5 and SER2 viruses. For comparison, pseudoviruses were also produced in cells transfected with twice the amount of SER2 or SER5 plasmids. The latter viruses were not used in imaging experiments. (A) Normalized infectivity of pseudoviruses measured by a luciferase reporter assay, using equivalent amounts of p24 to inoculate the cells. (B) Western blotting for HIV-1 gp120 (top) and p24 (bottom) for the virus preparations in the panel. (C-E) SER5-containing viruses were adhered to coverslips, fixed and incubated with anti-gp120 2G12 human (D, F) and anti-HA (SERINC) mouse (E, G) antibodies, followed by staining with anti-human-AF647 and anti-mouse-CF568 antibodies, respectively. White boxes mark particles positive for GFP-Vpr. More than 1,000 single viruses were imaged and analyzed for each preparation of this panel (F, G). SER2 signal was on average twice as strong as SER5 signal with p-value < 0.001, as determined by two-sample Kolmogorov–Smirnov after optimal binning (see Methods). Scale bar: 10 μm.
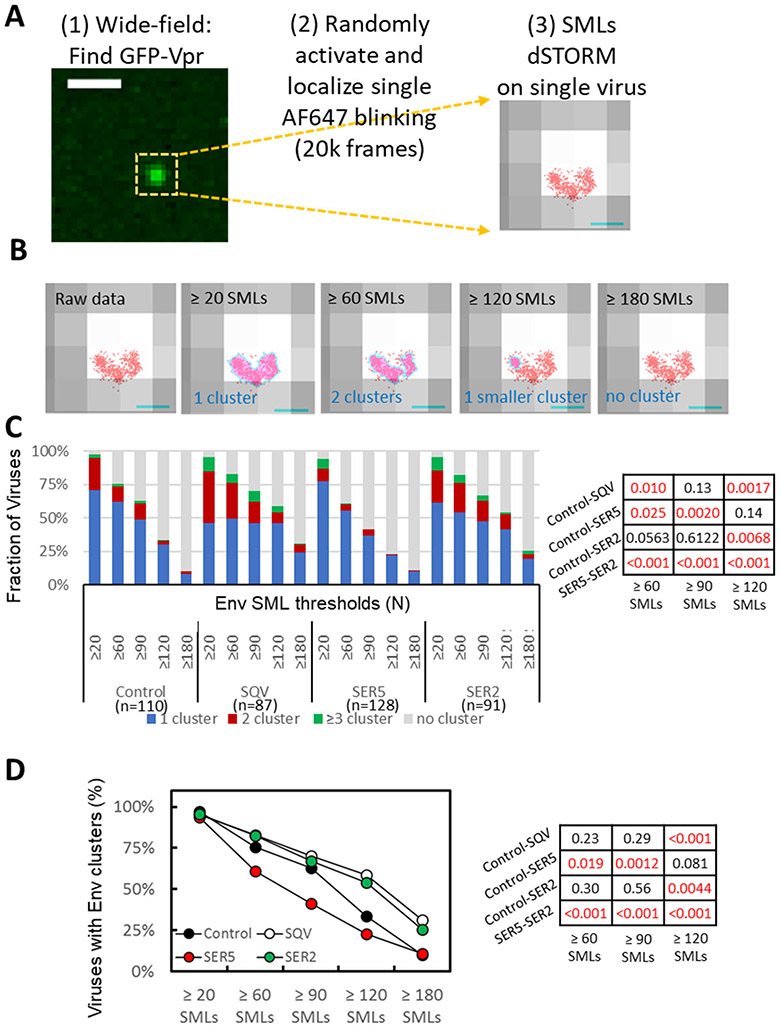
We next visualized the Env distribution on HIV-1 GFP-Vpr labeled pseudoviruses by dSTORM (direct stochastic optical reconstruction microscopy) that enables sub-diffraction resolution imaging down to ~20 nm.26 Given the limited axial resolution of dSTORM (~50 nm),27 we employed this technique to examine 2D-distribution of Env glycoproteins on HIV-1 particles that have a diameter ~120 nm.28-30 Env glycoproteins on the virus surface were visualized by immunostaining, as shown in Figures 1C-E. Briefly, pseudoviruses adhered to coverslips were fixed and incubated with human anti-Env antibodies followed by staining with second antibodies conjugated with AlexaFluor-647 (AF647). dSTORM experiments were carried out on a Vutara 352 system using a 640 nm laser and a low intensity of 405 nm illumination (see Methods) to visualize AF647 blinking associated with each GFP spot (Fig. 2A).
Figure 2. Effect of SERINC incorporation on Env distribution on virions imaged by dSTORM.
(A) Workflow for single virus dSTORM imaging. GFP-Vpr signal was imaged by wide-field fluorescence microscopy to determine the location of single viruses adhered to a coverslip. Samples were then imaged in a dSTORM mode for 20k frames to map single molecule localizations (SMLs, red) associated with single viruses identified by diffraction-limited GFP-Vpr-labeled spot (grey). Scale bar: (Left) 1 μm. (Right) 100 nm. (B) Four panels illustrating the results of DBSCAN analysis of the same virion using different SML thresholds, as indicated, and the search radius 15 nm. Cluster areas are colored light magenta pink with cyan boundaries. Scale bar: 100nm. (C) Distribution of Env clusters as a function of DBSCAN SML threshold categorized into no cluster, 1, 2, and ≥3 clusters per virion. (D) Same as in C but using two virus categories (with/without Env clusters). Statistical comparison for panels C and D is done using Fisher's Exact Test and shown on the right.
dSTORM experiments confirmed comparable levels of Env incorporation across all preparations, as evidenced by similar numbers of single molecule localizations (SMLs) per virion (Supp. Fig. S1C). Visual inspection of Env localizations overlaid onto low-resolution images of single isolated GFP-Vpr spots (Fig. 2A, right) implied that Env glycoproteins were not randomly distributed on HIV-1 particles and appeared to form clusters. To objectively detect and analyze protein clusters on HIV-1 particles, we chose DBSCAN (density-based spatial clustering of applications with noise)31 analysis. DBSCAN allows for an unbiased clustering analysis without assumptions about the shape or size of clusters.31 Here, clusters are defined based upon two userselected parameters – the search radius (R) and the minimal number of single molecule localizations (SMLs) within that radius (N). A cluster encompasses all contiguous points having ≥N SMLs within a search radius around a given localization. The boundaries of a cluster are formed by SMLs that have fewer neighbors than N.
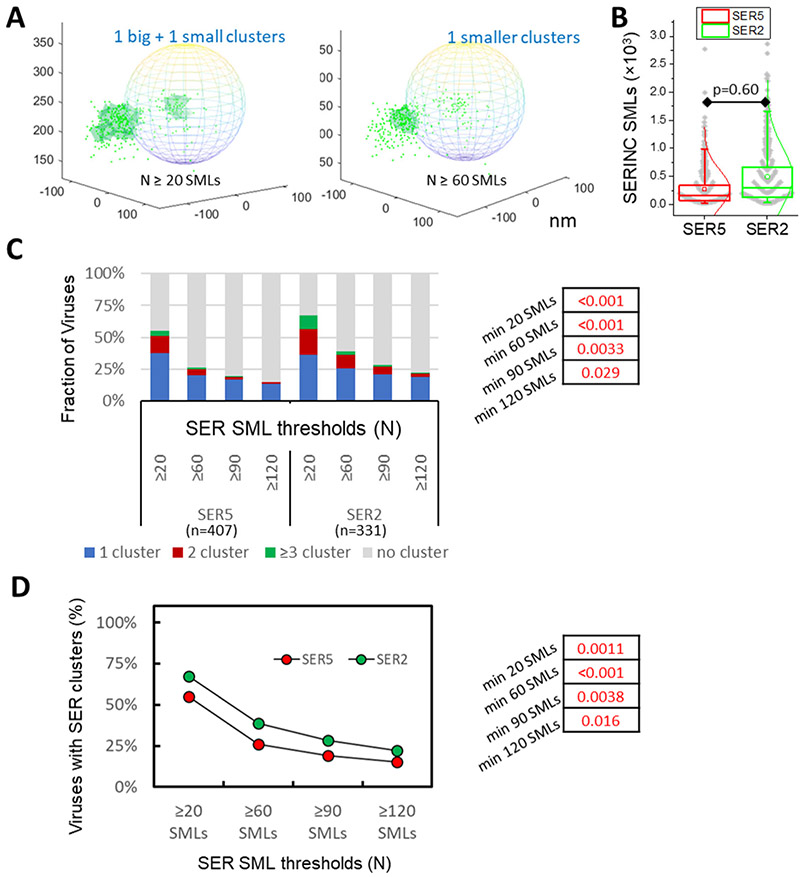
Since the number and size of clusters depend on the selected DBSCAN parameters (R and N), we varied both parameters across a wide range to avoid bias in assessing whether Env clusters differently across the four virus preparations. Varying the SML number threshold N between 20 and 180 for a fixed R=15 nm yielded different number and size of clusters per virus (illustrated in Fig. 2B). Likewise, reducing R from 50 nm to 15 nm (for fixed N=90 SMLs) altered the number and size of Env clusters for all preparations (Supp. Fig. S2A). Overall, the more stringent DBSCAN parameters (smaller R and greater N) selected for smaller and denser clusters. Accordingly, only a small fraction of Env SMLs were found in clusters for the most stringent DBSCAN parameters (Fig. 2B). In order to interpret dSTORM data, we classified the viruses into 4 categories: those containing no clusters, 1 cluster, 2 clusters and 3 or more clusters, and plotted the relative fractions of virions in each category across the range of DBSCAN parameters (Fig. 2C and Supp. Fig. S2B). As expected, the fraction of viruses containing Env clusters, and, especially those that contained ≥3 clusters, decreased as the SML threshold increased or the search radius decreased.
Env cluster analysis revealed that immature viruses contained a greater number of clusters and fewer single clusters per particle than control viruses, in excellent agreement with the previous study that utilized STED imaging to visualize Env clusters.15 Importantly, SER5-containing virions had, on average, significantly fewer clusters than control or SER2-containing viruses across a range of DBSCAN parameters (Fig. 2C). The inhibitory effect of SER5 on Env clustering was also apparent when we analyzed the data while varying the DBSCAN search radius (R) and fixing the SML threshold (N) (Supp. Fig. S2B). We also examined Env’s tendency to cluster using just two categories of viruses – those with and without clusters, irrespective of the number of Env clusters. Similar to the 4-category analysis above, this analysis also revealed a clear tendency of SER5 to inhibit Env clustering compared to control and SER2 viruses (Fig. 2D and Supp. Fig. S2C). These results demonstrate negative regulation of Env clustering on mature viruses by SER5, but not SER2.
Three-dimensional super-resolution imaging of SER5-mediated disruption of Env clusters on HIV-1 particles.
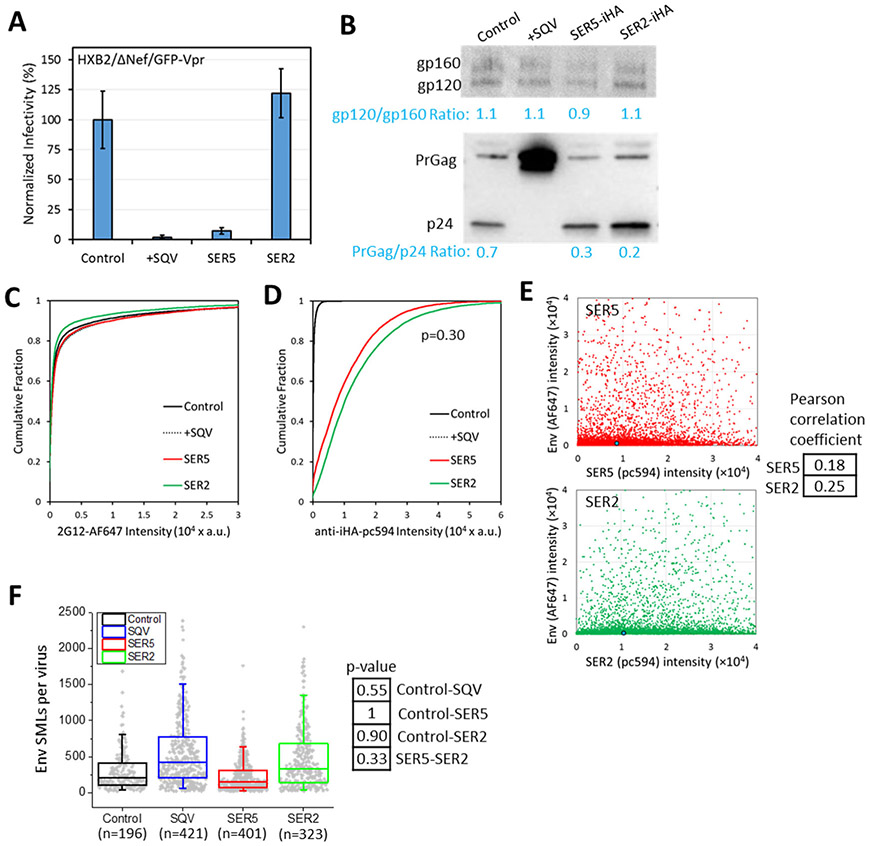
Since projection of SMLs distributed over a 3-dimensional object onto a single plane should increase their apparent clustering, we further validated the results of 2D dSTORM imaging using a 3D super-resolution microscopy technique referred to as iPALM (interferometric photo-activation localization microscopy), which achieves a nearly isotropic resolution of ~15 nm in all three dimensions (https://www.aicjanelia.org/ipalm).32 Using two-color iPALM, we visualized 3D distributions of Env and SERINCs on an independently produced panel of pseudoviruses (designated panel B) consisting of control, immature (SQV), SER5 and SER2 particles. As with the first virus panel, the incorporation of SER5, but not SER2, strongly inhibited specific infectivity (Fig. 3A). All preparations, except SQV-treated viruses, properly matured (Fig. 3B, bottom). The efficiency of SER5 and SER2 incorporation into virions was similar and this incorporation did not affect the levels or proteolytic cleavage of viral Env, as measured by Western blotting (Fig. 3B, top) and immuno-fluorescence imaging of single particles (Fig. 3C, D), as well as by single-molecule localization microscopy (Fig. 3F). Encouragingly, these preparations did not exhibit a noticeable correlation between Env and SERINC incorporation (Fig. 3E).
Figure 3. Analysis of Env and SERINC incorporation into pseudoviruses, virus infectivity and maturation.
Four pseudovirus preparations were produced in parallel by transfection of 293T/17 cells and designated panel B, which consisted of control viruses, +SQV (saquinavir treated, immature particles), SER5 and SER2 (viruses containing SERINC5 or SERINC2, respectively). (A) Normalized infectivity of pseudoviruses of the panel measured by fluorescence microscopy as the number of cells expressing the mKate2-reporter gene. (B) Western blot for HIV-1 gp120 (top) and p24 (bottom) of the viruses in the panel. (C, D) Viruses were adhered to coverslips, fixed and incubated with anti-gp120 2G12 human (C) or anti-HA (SERINC) mouse (D) antibodies, followed by staining with AF647- with pc594-conjugated second anti-human and anti-mouse antibodies, respectively. More than 5,000 single viruses were imaged and analyzed for each preparation of this panel. No significant differences between SER5 and SER2 incorporation. (E) No significant correlation is observed between Env and SERINC incorporation (Pearson correlation < 0.3). Blue dot is median (50%) of the intensity. (F) Single molecule localization (SML) distributions per virion for this panel obtained by iPALM. No significant differences were observed between control, SQV, SER5 and SER2 pseudoviruses (p>0.05), using a two-sample Kolmogorov–Smirnov test after optimal binning of data (see Methods). Box plot includes 1st, 2nd, and 3rd quantiles. Whiskers are 5% and 95% values.
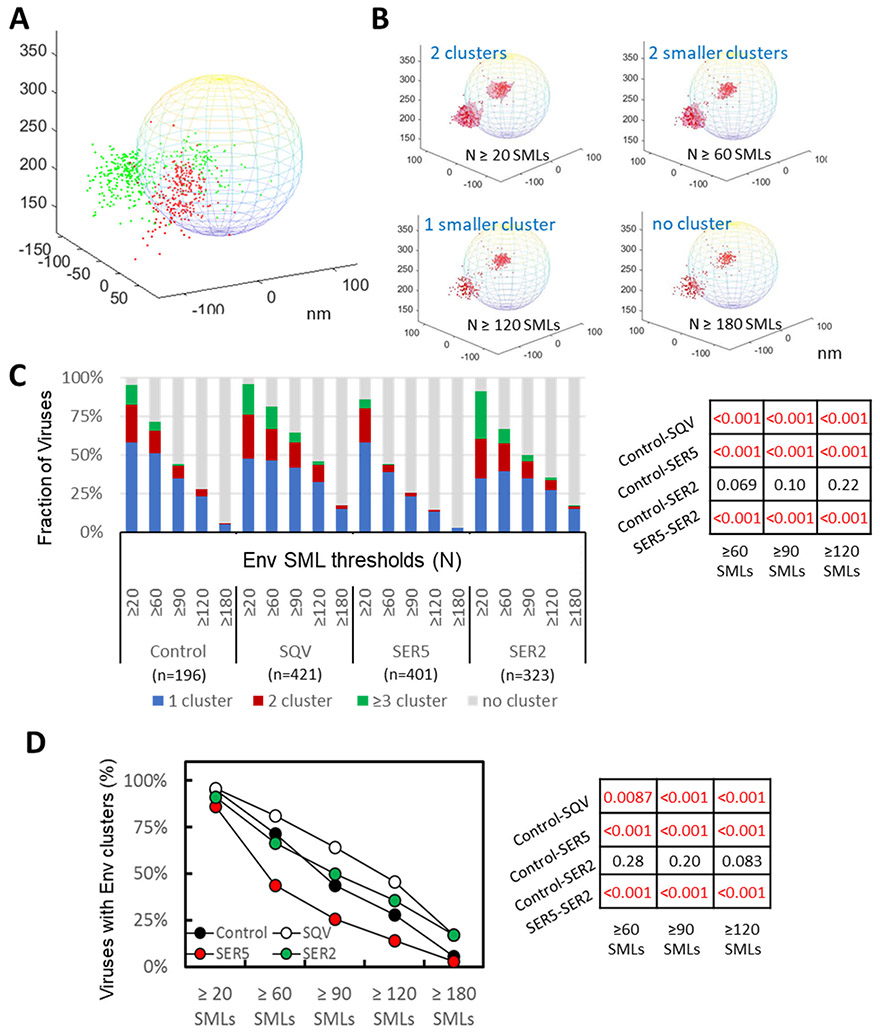
Coverslip-adhered viruses were fixed, washed and incubated with human anti-Env antibody and mouse anti-HA antibody (to visualize SERINCs), followed by staining with respective AF647- and pc594-conjugated second antibodies. The obtained 3D SML coordinates were rendered onto a virus-sized sphere with the center corresponding to the center of a diffraction-limited GFP-Vpr spot (Fig. 4A). Visual inspection of Env and SERINC distribution on single virions suggested nonrandom distributions of these proteins on the virus surface. We then performed 3D DBSCAN analysis of Env clustering on single virions by varying the parameters R and N. As was the case for 2D analysis, more stringent DBSCAN parameters selected for smaller and denser Env clusters, to the point where only a small fraction of particles had clusters, as illustrated in Fig. 4B. We plotted the fraction of viruses with no clusters, 1, 2 and ≥3 clusters as a function of the minimum number of SMLs (N, Fig. 4C) or the search radius (R, Supp. Fig. S3A, B). Across the selected ranges of DBSCAN parameters, a larger fraction of SER5-containing viruses lacked Env clusters than control and SER2-containing particles (Fig. 4C and Supp. Fig. S3B). The same analysis performed using just two categories of viruses (with or without clusters) confirmed that SER5 incorporation markedly disfavored Env clustering on mature virions. Similar to the dSTORM analysis above, Env clusters were more abundant on immature particles (SQV) compared to control virions, irrespective of DBSCAN parameters.
Figure 4. 3D iPALM imaging and analysis of HIV-1 Env distribution on virions.
(A) Illustration of 2-color 3D iPALM imaging on single Env molecule localizations (SMLs, red dots) and SER5 SMLs (green dots) projected onto a 200 nm-diameter sphere representing a GFP-Vpr labeled viral particle. (B) Illustration of Env SMLs (red dots) in 3D overlaid onto an idealized viral particle shown as a sphere with diameter of 200 nm. Dependence of Env clustering (light red area) on the DBSCAN SML threshold N of 20, 60, 120, 180 localizations, and the same searching distance R of 20 nm. X, Y, Z axes are in nm. (C) Analysis of Env clustering for a panel of HIV-1 pseudoviruses consisting of control and immature (SQV) particles and virions containing SER2 and SER5, using different DBSCAN SML threshold parameters, as indicated. The results are categorized into no cluster, 1, 2, and ≥3 clusters per virion. The same R=20 nm for DBSCAN analysis with N=20, 60, 90, 120, 180 localizations. (D) Fraction of pseudoviruses containing or lacking Env clusters (2 categories) for the same DBSCAN parameters as in B. Statistical analyses of data in panel B and C using Fisher's Exact Test are shown on the right.
To more directly compare the results of 2D and 3D SML analyses, we projected the 3D iPALM point coordinates onto a single plane and analyzed Env clustering by DBSCAN. As illustrated in Supp. Fig. S4A, for the same DBSCAN parameters, projecting 3D SML coordinates onto a single plane altered the apparent extent of Env clustering due to an overlap between clusters separated in Z-direction. Because of this, 2D projection of iPALM data resulted in: (i) reduction in the apparent number of clusters and (ii) increase in the apparent density of clusters in 2D-projections. However, in spite of the changes in the relative cluster abundance in 2D projections, DBSCAN analysis of 2D-projected iPALM data confirmed that SER5 virions contained fewer clusters compared to the control and SER2 preparations (Supp. Fig. S4B, C), supporting the validity of 2D dSTORM imaging for analysis of Env clusters on single virions (Fig. 2).
Virus-incorporated SER2 is more prone to form clusters than SER5
We next measured the propensity of SER5 and SER2 to cluster on HIV-1 particles by analyzing 2-color iPALM images (Fig. 4A). Selection of more stringent DBSCAN parameters diminished the size and the number of SERINC clusters per virion (illustrated in Fig. 5A). Since protein clustering is likely affected by the surface density, we examined iPALM data to determine whether SER5 and SER2 incorporated into virions at comparable levels. Although SER2 tended to more efficiently incorporate into virions, the difference between distributions of SER5 and SER2 SMLs/virus was not significant (Fig. 5B). We therefore proceeded with SERINC clustering analysis as a function of the SML number threshold and found that a larger fraction of SER2 molecules on virions formed clusters compared to SER5 (Fig. 5C, D).
Figure 5. SERINC clustering analysis.
(A) Illustration of single molecule localizations (SMLs) of SER5 (green dots) in 3D overlaid onto an idealized viral particle shown as sphere and dependence of SERINC clustering (light green area) on the DBSCAN SML threshold parameter (N=20 and 60 SMLs and R=20 nm). X, Y, Z axes labels are in nm. (B) Distributions of SMLs per virion are similar for SER5 and SER2. (C) DBSCAN analysis of SER5 and SER2 clustering using a fixed distance parameter (20 nm) and varied SML thresholds, as indicated. Pseudoviruses with less than 20 SER localizations were excluded from analysis. (D) Fractions of pseudoviruses containing or lacking SER clusters (2 categories) as a function of SER SMLs thresholds. Fisher's Exact Test was used for statistical analysis in panels C and D (shown on the right).
SERINC molecules do not self-aggregate in cell membrane.
In order to assess whether the observed Env and SERINC foci on single virions result from protein self-aggregation, we used an image correlation technique known as Number and Brightness (N&B) analysis.33 N&B assesses oligomerization of fluorescently tagged molecules based upon the molecular brightness (ε) of diffusing species, which is derived from the ratio of pixel fluctuation variance over the mean. We were unable to examine HIV-1 Env oligomerization by this technique due to a quick clearance of Env from the plasma membrane mediated by the Y712SPL motif in its cytoplasmic tail.34 By contrast, wild-type and GFP-tagged SER5 and SER2 proteins are predominantly localized to the plasma membrane (1, 2 and Supp. Fig. S5) and are thus amenable to N&B analysis. In control experiments, N&B analysis of the GPI-anchored GFP and a tandem GFP construct (1xGFP-GPI and 2xGFP-GPI, respectively, Supp. Fig. S5B and C) showed a ~2-fold difference in ε, as expected (Supp. Fig. S5F). Analysis of cells expressing SER5-GFP or SER2-GFP revealed a low molecular brightness for both proteins (Supp. Fig. S5D, E and G) that was indistinguishable from 1xGFP-GPI and was significantly lower that 2xGFP-GPI (Supp. Fig. S5H). This result implies that SER2 and SER5 are predominantly expressed in the plasma membrane as monomers and that, therefore, clustering of these proteins on virions observed by single molecule localization microscopy is not driven by pre-oligomerization of these proteins in the plasma membrane.
Env and SERINCs do not co-cluster on HIV-1 particles
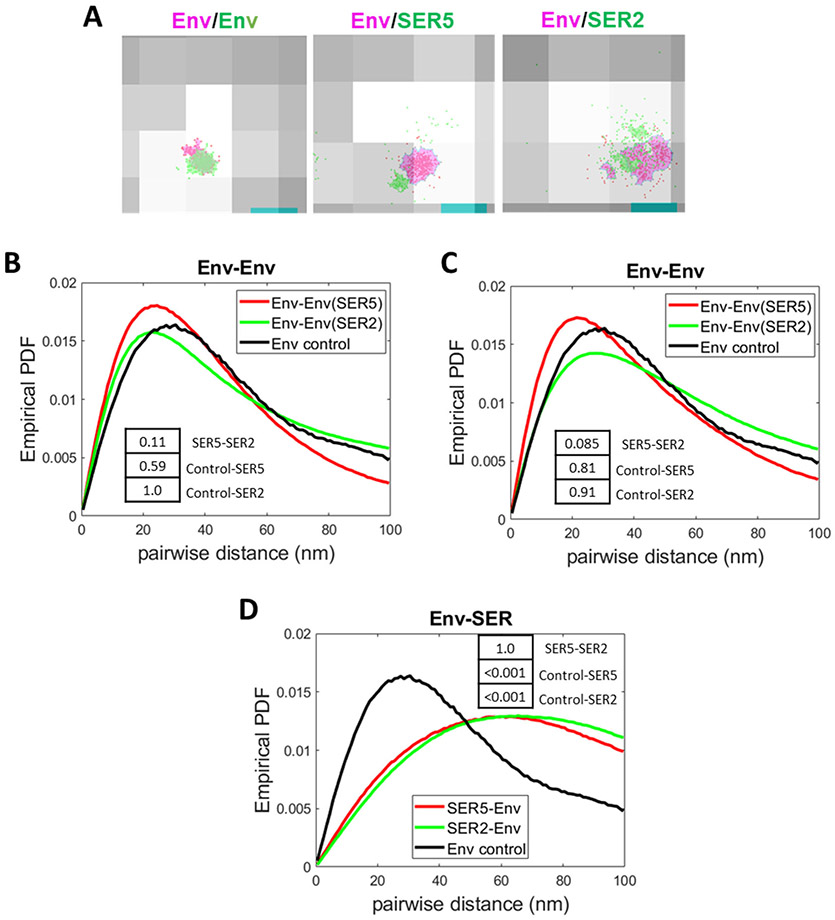
We next examined 2-color iPALM data (for representative images, see Fig. 4A and Supp. Fig. 6A) to assess the degree of Env and SERINC co-distribution on virions. For this purpose, we analyzed the distributions of pairwise distances between SMLs within single particles (see Methods). Before examining the Env-SERINC distance distributions, we analyzed the Env-Env distances in order to establish a baseline for a “self-clustering” distribution. As a positive control for co-distribution, we incubated viruses with anti-Env 2G12 antibody, co-stained with a mixture of two second antibodies conjugated with AF647 and CF568 and imaged by 2color dSTORM (referred to as co-staining control, Fig. 6A, left). The obtained pairwise distribution for the co-staining control was indistinguishable from the distributions of inter-Env distances derived from iPALM data for SER5- and SER2-containing viruses (virus panel B) after projecting the point coordinates onto 2D (Fig. 6B) or from Env-Env distances obtained by dSTORM (virus panel A, Fig. 6C). The overlapping distributions of Env-Env distances and distances for an Env co-staining control (Fig. 6B, C) support the notion that Env trimers form tight clusters on virions.
Figure 6. Analysis of Env-SERINC co-distribution on HIV-1 by 2-color iPALM and dSTORM.
(A) Representative images of Env co-stained with both anti-human AF647 (red) and anti-human CF568 (green, left image). The cluster areas are shaded in magenta (AF647) and light green (CF568). Middle and right panels show examples of Env SMLs (red) with a magenta-colored cluster and SER5 SMLs (middle) or SER2 SMLs (right) with a green-colored cluster. Env, SER, and Env co-stained clusters were defined by DBSCAN, R=15 nm and N=20 SMLs. (B) Comparison of probability density function (PDF) of Env-Env pairwise distances between SER5- (red) and SER2-containing viruses (from panel B) imaged by iPALM and projected on a single plane. A pairwise distance distribution for Env control co-stained with two second antibodies is shown for comparison. (C) Comparison of PDF of Env-Env pairwise distances between SER5- (red) and SER2- (green) containing viruses (from panel A) and control Env co-staining obtained by dSTORM. (D) Comparison of PDF of Env-SERINC pairwise distances between SER5- (red) and SER2- (green) containing viruses imaged by iPALM (2D projection). Env-Env distance distribution for control viruses co-stained with a mixture of two second antibodies is shown for comparison. Insets: p-values obtained statistical analysis of the PDF of pairwise distance distributions using two-sample Kolmogorov–Smirnov test after optimal binning (see Methods).
We next compared the Env-SER5 and Env-SER2 pairwise distance distributions and found that the distributions of Env-SER5 and Env-SER2 pairwise distances were indistinguishable (Fig. 6D). Note that 3D analysis of pairwise distances also did not reveal any difference in Env-SERINC or Env-Env distributions on SERINC-containing or control viruses (Supp. Fig. S6). Importantly, Env-SERINC distances were much longer than the Env-Env distances in control co-stained samples (Fig. 6D). Thus, Env-SERINC distance distributions are not as tight as Env-Env distance distribution, implying that these two molecules do not co-distribute or co-cluster on virions.
Here, we employed single-molecule localization microscopy to visualize the distribution of Env glycoproteins on the surface of HIV-1 particles and to examine the effect of SER5 on Env distribution. Specifically, we asked if SER5 could disrupt the formation of Env clusters that had been proposed to be essential for the virus’ ability to fuse with a target cell.15 Two- and threedimensional super-resolution imaging of single virions containing or lacking SERINCs lead to the following principal findings. First, SER5 interferes with Env clustering on mature virions, whereas SER2 is without effect (no significant differences were observed across DBSCAN parameters). Thus, disruption of Env clusters may contribute to the HIV-1 fusion-inhibitory activity of SER5. Second, in spite of SER5’s effect on Env clustering (this paper), on the antigenic properties of Env,3, 6, 13 and on the rate of loss of HIV-1 fusion-competence,3 we observed poor co-distribution of Env and SERINCs on virions. This finding is inconsistent with the reports of specific interaction between Env and SER5.7
A correlation between the lower fraction of Env in clusters on SER5-containing viruses and impaired infectivity, as compared to control or SER2-containing viruses, supports the significance of Env clustering for the virus’ fusion-competence. Indeed, viral protein-mediated membrane fusion is thought to proceed through a cooperative action of several glycoproteins assembled into a fusion complex 16-21 (but see 35 for the opposite conclusion). A concerted action of several glycoproteins would more readily overcome the energetic barriers for creating and expanding a fusion pore.36-38 The minimal number of fusion proteins in a functional complex is referred to as “fusion stoichiometry”.16, 35 Increasing the local surface density of Env through clustering (15, 39 and this study) is expected to favor the formation of Env complexes and thereby facilitate fusion with a target cell.
The SER5-induced disruption of Env clusters may account for the observed differences in the SER5-sensitivity of different HIV-1 strains, which has been mapped to the gp120 variable loops V1-V3.2, 7, 13 We surmise that SER5-sensitivity may correlate with the stoichiometry of fusion complexes formed by different HIV-1 Envs. Indeed, there is evidence that a smaller number of Env trimers from some primary HIV-1 isolates, like SER5-resistant JRFL Env, is involved in a functional fusion complex, as compared to SER5-sensitive laboratory adapted strains, like NL4-3 17, 18 and HXB2. Thus, HIV-1 strains capable of forming functional fusion complexes with fewer Envs should be less sensitive to SER5-mediated disruption of Env clusters, in agreement with the greater SER5 resistance of the low-stoichiometry JRFL Env compared to the higher-stoichiometry NL4-3 Env.17
Although STED imaging revealed that single Env focus formation is disfavored in immature particles,15, 39 the block of Env function through the Gag-Env cytoplasmic tail interactions in immature virions 40 does not allow one to assess the relevance of single Env cluster formation to viral fusion. Whereas our findings are in general agreement with STED data,15, 39 we interpret our results as supporting the functional relevance of dense Env clusters, instead of only a single Env cluster per virion proposed previously.15 In order to match the STED imaging-based cluster analysis presented in that study, we replotted the results in Figures 2 and 4 after excluding virions without clusters (Supp. Fig. S7). While this analysis confirmed that cluster-containing immature particles had a significantly greater number of Env foci than mature virions, as reported in,15 a much greater fraction of SER5 viruses contained a single Env cluster compared to SER2 viruses (Supp. Fig. S7). Thus, the prevalence of single Env clusters in SER5- vs SER2-containing virions appears to inversely correlate with infectivity. The relative fractions of viruses with single vs. multiple clusters was not consistently different between SER5 and control viruses (Supp. Fig. S7). By contrast, the fraction of Env in clusters was markedly lower on SER5-containing particles (Figs. 2 and 4). Thus, the increase in fraction of SER5-containing virions with no clusters correlates well with reduced infectivity.
The functional relevance of multiple Env clusters is further supported by our previous data showing that HIV-mediated cell-cell fusion (also known as fusion-from-without), which proceeds through the formation of two fusion pores connecting a single virion to two adjacent cells, is markedly augmented by deletion of the Env cytoplasmic tail.41 Enhancement of fusion-from-without upon deletion of the Env cytoplasmic tail is likely due to the increased mobility of truncated Env compared to the full-length Env, which likely facilitates the formation of at least two clusters at the sites of virus-cell contact.15 We therefore propose that it is not the formation of a single (presumably large) Env cluster per virion, but the formation of any number of dense clusters exceeding the fusion stoichiometry of a given Env, that increases the likelihood of fusion with a target cell.
N&B analysis revealed a lack of considerable oligomerization of SER5 or SER2 in the plasma membrane (Supp. Fig. S5). This finding is consistent with our previous fluorescence recovery after photobleaching (FRAP) results in cells expressing SER5-GFP and CCR5-GFP, which yielded similar diffusion coefficients for these two proteins in the plasma membrane.3 The lack of selfoligomerization argues against the possibility that SERINC clustering on HIV-1 virions is driven by strong interactions between monomers. We surmise that the proximity of SERINC SMLs in virions is due to the protein sequestration into membrane domains, possibly into lipid rafts formed by cholesterol and sphingomyelin, which are enriched in the HIV-1 membrane compared to the plasma membrane.42, 43 Similarly, in the absence of known Env-Env interactions, Env clustering in virions is likely driven by the interaction between its cytoplasmic tail and the Gag lattice in immature/assembling virions 44-46 or the cytoplasmic tail and the matrix protein lattice in mature virions.15
A recent study using co-immunoprecipitation and bi-molecular fluorescence complementation (BiFC) assays reported the Env and SER5 interaction in cell membranes.7 We surmise that the reason for the apparent specific interaction between Env and SER5, but not SER2, observed in 7 could be due to a marked difference in the amounts of respective plasmids used to transfect the cells in an attempt to match the surface expressions of these two proteins. This would result in a lower fraction of SER2-positive cells and thus a smaller fraction of cells that could possibly generate a BiFC signal with co-expressed Env compared to SER5-transfected cells. Our super-resolution experiments reveal poor co-distribution of these two molecules in virions (Fig. 6). The pairwise Env-SERINC distance distributions was markedly longer compared to Env-Env distances (Fig. 6 and Supp. Fig. S6), implying the lack of co-clustering of the two proteins. These results are inconsistent with the interaction between Env and SER5 or SER2. Thus, SER5 appears to segregate into clusters that are distinct from those formed by Env and is thus unlikely to disrupt Env clusters by insinuating itself between the Env trimers.
The lack of Env and SER5 co-distribution on virions observed in our experiments supports an indirect mechanism of inhibition of Env function that does not involve a direct interaction between these proteins. However, we cannot rule out the possibility that Env-SER5 interactions occur transiently, perhaps early upon virus assembly/budding, and that this restriction factor no longer interacts with inactivated Env on mature cell-free virions. It is also possible that virus adhesion to glass coverslips and/or fixation can adversely affect Env and SERINC distribution. We note, however, that, under these experimental conditions, SER5, but not SER2, selectively disrupts Env clustering. It is also worth noting that, although we did not probe the effects of SERINCs on infectious HIV-1, pseudoviruses used in this study faithfully recapitulate SER5-mediated inhibition of viral fusion.
It is currently unclear how SER5 can alter the Env structure 3, 6, 13 or accelerate loss of its function 3 without binding to Env. A previous report concluded that SERINCs function as facilitators of phospholipid synthesis.47 One could thus speculate that SER5 inhibits viral fusion by altering the lipid composition of virions and perhaps stiffening the viral membrane. However, more recent studies found no changes in cell or virus lipidome upon expression/incorporation of SER5.48, 49 In spite of the lack of SER5 effect on the virus’ lipid composition, it is possible that this protein inhibits viral fusion by segregating lipids required for the Env’s stability and/or function. For instance, HIV-1 fusion is inhibited by cholesterol depletion 50-53 or mutations in the Env’s CRAC-like cholesterol binding motif located in the gp41 membrane-proximal extracellular region.54, 55 It is therefore possible that cholesterol sequestration by SER5, which has been shown to bind cholesterol and phosphatidylserine,5 can compromise the stability and/or fusion-competence of Envs. It is conceivable that, by virtue of their greater Env stability, SER5-resistant viruses could better tolerate changes in the virus lipid composition than the less stable SER5-sensitive Envs. Future studies addressing this and other models of inhibition of HIV-1 infectivity will further delineate the mechanism of SER5 restriction.
Conclusions
Our 2D and 3D single-virus super-resolution imaging results reveal that HIV-1 Env glycoproteins form clusters in the membrane of mature virions and that SER5, but not SER2 disrupts Env clusters. Since isolated Env glycoproteins that do not form clusters are less likely to promote viral fusion, we conclude that SER5 restricts HIV-1, in part, by dispersing the Env clusters. The surprisingly low colocalization of Env and SER5 molecules observed in our experiments argues against their interaction in the viral membrane and suggests an indirect mechanism of SER5-mediated inhibition of HIV-1 fusion.
Methods
Cell lines, reagents, and plasmids.
HeLa and HEK293T/17 cells were acquired from ATCC (Manassas, VA). HeLa-derived TZM-bl cells were obtained from AIDS Research and Reference Reagent Program, National Institutes of Health (NIH-ARP). Both cell lines were grown in highglucose DMEM supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Atlanta Biologicals, Flowery Branch, GA) and 100 units/ml penicillin/streptomycin (Gemini Bio-Products, Sacramento, CA). The growth medium for HEK293T/17 cells was supplemented with 0.5 mg/ml G418 (Cellgro, Mediatech, Manassas, VA).
Bright-Glo luciferase kit was purchased from Promega (Madison, WI). Poly-L-lysine and poly-D-lysine were from Sigma (St. Louis, MO). The viral protease inhibitor Saquinavir (cat # 4658), human 2G12 antibody (cat #1476), and human HIV immunoglobulin (HIV IG) (cat # 3957) were obtained from the NIH AIDS Reagent Program. The DMEM without phenol red was obtained from Life Technologies (Grand Island, NY). The anti-human AlexaFluor-647 (cat #A21445), and the mouse anti-HA.11 (cat #901501) were purchased from Invitrogen (Waltham, MA) and BioLegend (San Diego, CA), respectively. The anti-mouse CF568-conjugated (cat #20800), anti-human CF568-conjugated (cat #20097), and goat anti-human HRP-conjugated (cat # 31412) antibodies were acquired from Biotium (Fremont, CA) and Thermo Scientific (Waltham, MA), respectively. The 16% formaldehyde stock (cat #28908) was purchased from Thermo Scientific.
The pCAGGS plasmid encoding HIV-1 HXB2 envelope glycoprotein, the pR9ΔEnvΔNef HIV-1-based packaging vector, pcRev, GFP-Vpr, pBJ5-SER2-GFP and pBJ5-SER5-GFP expression vectors have been described previously.3 The GFP-Vpr plasmid was a gift from Dr. T. Hope (Northwestern University). The psPAX2 lentiviral packaging vector was from NIH-ARP. The CMV-SERINC2-iHA and CMV-SERINC5-iHA expression vectors were kindly provided by Dr. M. Pizzato (University of Trento, Italy). The pVPX-mKate2 lentiviral vector was a gift from Dr. A. Brass (University of Massachusetts). pCR3-GFP-GPI (here denoted as 1xGFP-GPI) and pBunny encoding the dimer GFP were gifts from Dr. C. Bron (University of Konstanz, Germany), and Dr. S. Padilla-Parra (Oxford, UK), respectively.
To obtain pCR3-2xGFP-GPI (2xGFP-GPI), the GFP sequence from pCR3-GFP-GPI plasmid flanked by XhoI and BamHI unique restriction sites was replaced with the dimer GFP. The dimer GFP fragment was amplified by PCR using TaqDNA high fidelity polymerase (Invitrogen), pBunny as plasmid template, and the following forward and reverse primers, containing XhoI and BamHI restriction sites, respectively: 5’-GGGCTCGAGGTGAGCAAGGGCGAGGAGCTG-3’ and 5’- GGGGGATCCCCTTGTACAGCTCGTCCATGC-3’. The PCR fragment was purified on a 1% agarose gel, digested with XhoI and BamHI, and ligated with pCR3-GFP-GPI digested and purified in a similar manner.
Pseudovirus production and characterization.
The HIV-1 HXB2 Env pseudotyped viruses were produced by transfecting HEK293T/17 cells with JetPRIME transfection reagent (Polyplus-transfection, Illkirch-Graffenstaden, France). To produce viruses for iPALM imaging (panel B), HEK293T/17 cells seeded in 6-well tissue culture plates were transfected with 0.4 μg of HXB2 Env, 0.33 μg of pcRev, 1 μg of psPAX2, 0.13 μg of GFP-Vpr, 0.2 μg of either CMV-SERINC2-iHA, CMV-SERINC5-iHA, or empty pcDNA3.1 vector, and 1 μg of pVPX-mKate2. To generate viruses for dSTORM imaging (panel A), 0.64 μg of HXB2 Env, 0.28 μg of pcRev, 0.85 μg of pR9ΔEnvΔNef, 0.14 μg of GFP-Vpr, and 0.09 μg of either CMV-SERINC2-iHA, CMV-SERINC5-iHA, or empty pcDNA3.1. The DNA transfection mix and HEK293T/17 cells were incubated overnight at 37 °C, 5% CO2, at which point the medium was removed, and DMEM without phenol red was added. Where indicated, DMEM contained 300 nM Saquinavir to inhibit HIV-1 protease. Forty-eight hours post-transfection, the supernatants were collected, passed through 0.45 μm pore filters, aliquoted and stored at −80 °C.
The p24 content of viral preparations was determined by ELISA, as previously described.41 For Western blotting, equal amounts of p24 were loaded onto 4–15% polyacrylamide gel (Bio-Rad, Hercules, CA), and transferred on a nitrocellulose membrane. The membrane was incubated overnight at 4 °C with either HIV IG (1:2000 dilution) or goat anti-gp120 (Fitzgerald, Acton, MA) (1:700 dilution), washed and incubated for 1 h at room temperature with either with goat anti-human HRP-conjugated IgG (1:5000 dilution) or donkey anti-goat HRP-conjugated (Santa Cruz, Dallas, TX) (1:700 dilution). The chemiluminescence signal was measured on ChemiDoc XR+ (Bio-Rad). The densitometry was performed using Image Lab software (Bio-Rad).
For infectivity assays, TZM-bl cells seeded in black-clear 96-well plates (Corning) at 0.2x105 cells/well were infected with viruses by centrifugation at 4 °C for 30 min at 1550 × g. Forty-eight hours later, infected cells were either lysed and incubated with Bright-Glo luciferase substrate for 5 min at room temperature or immediately analyzed for the presence of fluorescent cells. The luciferase signal was measured using a TopCount NXT reader (PerkinElmer Life Sciences). For virus panel B, infectivity was measured by visualizing mKate2-expressing cells 48 hours postinfection with psPAX2/pVPX-mKate2 pseudoviruses, using a Zeiss LSM880 microscope. The results were normalized to the p24 content.
iPALM sample preparation, acquisition and processing.
For iPALM measurements, 25 mm #1.5 glass coverslips containing fiducial markers were prepared, as described previously.32 Prior to the experiment, the coverslips were washed in 1M KOH for 30 min, washed, coated with 0.1 mg/ml poly-L-lysine for 30 min and air-dried for 10 min. To bind the viral particles, 100 μl of viral suspension containing equivalent amounts of GFP-Vpr viral particles was spotted onto a 70% ethanol-sterilized parafilm sheet, covered with a coverslip, and incubated for 30 min at room temperature in the biosafety cabinet. The coverslips were then washed 3 times, with 10 min intervals, by immerging in 3 ml of PBS containing calcium and magnesium (PBS++). The coverslip-bound viruses were fixed with 2% paraformaldehyde for 30 min at room temperature. Paraformaldehyde was quenched by washing two times, with 10 min intervals, using a 1% glycine solution, and once with PBS++. Fixed samples were blocked with PBS++/15% FBS for 2 h, and incubated at 4 °C for ~12 h with 20 μg/ml 2G12 (NIH-ARP, cat #1476) for Env staining or 20 μg/ml mouse anti-HA.11 (BioLegend, cat #901501) for SERINC staining. The coverslips were washed 3 times with PBS++/15% FBS, with 10 min intervals, and incubated for 1 h at room temperature with 20 μg/ml of anti-human AlexaFluor-647 for Env staining or goat anti-mouse antibody conjugated to a photochromic self-blinking derivative of AlexaFluor-594 (pc594, kindly provided by Luke Lavis at Janelia Research Campus) for SERINC staining. The coverslips were washed 3 times, with 10 min intervals between solution changes, with large excess of PBS++. Samples were then immersed in dSTORM imaging buffer described in,56 and a second 18 mm diameter coverslip was affixed to the bottom of a 25 mm coverslip and sealed to prevent buffer evaporation and oxygen infiltration into the sample.
Samples were mounted on the iPALM system and illuminated with 3-7 kW/cm2 laser intensity (647 and 561 nm for the AlexaFluor-647 and pc594 dyes, respectively). Images were acquired through a pair of Nikon 60x Plan Apo 1.49 NA TIRF objectives, combined and allowed to interfere in a custom made three-way beam splitter.32 The resulting signal was detected via three electron multiplying charge couple devices (EMCCD) cameras (iXon DU-897, Andor). We acquired 20,000 frames for AF647 and 30,000 frames for pc594 with 50 ms exposure time per frame. Single molecule blinks were localized in 3D using the PeakSelector software (Janelia Research Campus). Corrections were made for drift in all directions, as well as for any residual tilt in the specimen plane (typically less than 50 nm). Two-color images were registered with respect to each other via the gold fiducial markers embedded in the coverslip.
dSTORM sample preparation and image acquisition.
Eight-chambered glass coverslips (#1.5, Lab-Tek, Nalge Nunc International, Penfield, NY) were washed with 70% ethanol twice and incubated with 0.1 mg/ml poly-D-lysine in water for 30 min and solution aspirated. Treated chambered coverslips were next incubated with pre-sonicated 100 nm nanoparticles (Cytodiagnostics, G-100-20) diluted 5-fold in water for 30 min, washed with water and blocked with 10% FBS in water for 30 min. After blocking with FBS, the chamber slide was washed with water and then incubated with poly-D-Lysine 0.1 mg/ml for 30 min again and solution aspirated. The chamber was washed with water and stored at 4 °C for several months.
Chambered coverslips pretreated as above were incubated with pseudovirus stocks diluted (4- to 10-fold) in PBS++ for 30 min at room temperature, washed and fixed with 2% paraformaldehyde for 30 min at room temperature. Paraformaldehyde was quenched by washing 5x with 20 mM TRIS/PBS++. Each washing step was done without completely removing the solution in order to avoid sample drying. Samples were then blocked with 15% FBS/ PBS++ for 2 hr at room temperature and incubated with primary antibodies (5 μg/ml 2G12 for Env staining and mouse anti-HA.11 for SERINC staining) at 4 °C overnight and washed 9 times with 15% FBS/ PBS++. Next, samples were incubated with second antibodies, 2 μg/ml anti-human AlexaFluor-647 (for Env) or 4 μg/ml anti-mouse CF568 (for SERINC). After 9 washes with 15% FBS/ PBS++, the samples were used for immunofluorescence imaging and dSTORM.
Image drift correction was performed using more than two fiducial markers (gold nanoparticles (SRX, Vutara software, Bruker, Billerica, MA). After drift correction, the full width at half maximum of the distribution of localizations from >90% gold nanoparticles should be less than 20 nm in XY, or the images were discarded. GFP-Vpr-positive particles with fewer than 20 SMLs were excluded from analyses, reasoning that even non-specific binding of a single AF647-labeled antibody can produce around 20 blinking events under our experimental conditions.
Owing to the uncertainty of localization of diffraction-limited GFP-Vpr spots, we assigned all Env SMLs in dSTORM and iPALM experiments falling within a 200 nm distance from the center of the GFP signal to that particle.
Wide-field fluorescence and dSTORM imaging.
Prior to dSTORM imaging, fixed/stained viruses (panel A) were imaged in a single z-plane using an Elite DeltaVision microscope (Applied Precision, GE, Pittsburgh, PA), using an UPlanFluo 40x/1.3 NA oil objective (Olympus, Tokyo, Japan) and a FITC/TRITC/Cy5 filter set (Chroma, Bellows Falls, VT). Prior to iPALM imaging, samples (panel B) were imaged using a Nikon Eclipse Ti microscope equipped with a 100x Plan Apo 1.4NA objective, a white light LED excitation source (Sola, Lumencore), FITC and TRITC filter sets (Semrock), and EMCCD detector (DU-885, Andor). For dSTORM, imaging buffer was made based on the Nikon STORM protocol sample preparation. Buffer A: 10 mM Tris (pH 8.0) with 50 mM NaCl in PBS++. Buffer B: 50 mM Tris (pH 8.0), 10 mM NaCl, and 10% glucose in PBS++. GLOX solution: 28 mg glucose oxidase (from Aspergillus niger, ≥100,000 units/g, Sigma, cat #G2133-50KU), 100 μl of Catalase (17 mg/mL) (prepared from Catalase lyophilized powder, ≥10,000 units/mg, Sigma, cat #C40-100MG) with 400 μl Buffer A, vortexed to dissolve and stored up to two weeks. STORM imaging buffer contained 100 mM MEA (cysteamine hydrochloride, Sigma M6500-25G) with 1% GLOX in Buffer B. Chambered coverslips filled with the dSTORM imaging buffer were covered with parafilm to seal the wells and limit oxygen access. Freshly prepared dSTORM imaging buffer was used every 2 hours to avoid buffer acidification.
dSTORM imaging of Env was performed on a Vutara 352 microscope (Bruker, Billerica, MA) using 3% 640 nm laser power with 50 ms frame rate for a total 20,000 frames. The power of a 405 nm laser was adjusted (0.1-0.3%) to increase the blinking rate. 2-color dSTORM imaging of Env co-staining control was done using the same condition as above for AF647 staining. For the second color, CF568, 20% 561 nm laser power with 50 ms frame rate for a total 30,000 frames. The power of a 405 nm laser was turned on at 0.5% after 10,000 frames of acquisition to increase the blinking rate.
Number and Brightness analysis.
HeLa cells were seeded on collagen-coated 8-well chamber slides ((#1.5, Lab-Tek) the day before transfection. At 60-80% confluency, cells were transfected with JetPRIME transfection reagent (Polyplus-transfection) using 0.1-0.2 μg SER5-GFP, SER2-GFP, 1xGFP-GPI, or 2xGFP-GPI expression vectors. Cells were imaged 26-40 hours after transfection on a Zeiss LSM880 confocal microscope (Carl Zeiss Microimaging, Germany).
Cell imaging was performed at room temperature in a Live Cell Imaging Buffer (Invitrogen, Carlsbad, CA) supplemented with 2% FBS using a Plan-Apochromat 63x/1.4 oil objective after the medium was changed to. N&B imaging parameters imaging was performed at 8.19 μs/pixel dwell time with total of 256x256 pixels (1.26 s per frame).33, 57 Fifty consecutive image frames were analyzed for single variance (σ2) and average value (<k>) after detrending the images (time constant, 20 frames) on Zen Black (Zen 2.3 SP1, Carl Zeiss AG, Oberkochen, Germany) to compensate for slow photobleaching. Molecular brightness (ε) of each pixel was calculated using a custom Matlab code with formula:
For western-blotting analysis of 1xGFP-GPI and 2xGFP-GPI, 70% confluent HeLa cells in 6-well plate were transfected with JetPRIME reagent using 1 μg of either 1xGFP-GPI or 2xGFP-GPI expression vectors. Thirty-six to forty hours post-transfection, the cells were lysed in RIPA buffer (Sigma) supplemented with protease inhibitor cocktail (Complete Mini, Roche, Mannheim, Germany), and the total protein was determined using Micro BCA protein assay kit (Themo Scientific). Equal amounts of total protein were loaded onto 4–15% polyacrylamide gel (Bio-Rad) and transferred onto nitrocellulose membranes. The membranes were incubated overnight at 4 °C with either mouse anti-GFP antibody (Clontech) diluted 1:1000 or mouse-anti-tubulin (Sigma) diluted 1:3000. The membranes were washed in PBS/0.1% Tween20 and incubated for 1h at room temperature with rabbit anti-mouse HRP-conjugated (EMD Millipore, Burlington, MA) at 1:3000 dilution. The chemiluminescence signal was measured on ChemiDoc XR+ (Bio-Rad).
DBSCAN and statistical analyses.
Single virus particles were identified by the GFP-Vpr fluorescence signal. The coordinates of single-molecule localizations (SMLs) were assigned to a virus, using a search distance of ±200 nm from the center of a low-resolution GFP spot. Single virus particles with less than 20 Env localizations were excluded from analysis. Clustering analysis was performed by density-based spatial clustering of applications with noise (DBSCAN) described by S.M.K. Heris in “Implementation of DBSCAN Clustering in MATLAB” (https://yarpiz.com/255/ypml110-dbscan-clustering). This algorithm uses two user-selectable parameters, the search radius (R) and the minimal number (N) of SMLs within that radius, to identify densely located SMLs, regardless of the cluster shape. DBSCAN finds the neighbors within R for each SML. If more than N neighbors within the search radius are found, this location is considered a cluster. For any neighboring localization, the cluster grows if a neighboring SML has more than N neighbors within the radius R. The cluster stops growing whenever a neighbor has less than N neighbors. That is, DBSCAN identifies clusters when SMLs are dense enough to satisfy the selected parameters, N and R. Unless stated otherwise, we used R=20 nm in 3D iPALM images, and R=15 nm in 2D iPALM or 2D dSTORM images to adjust for an increase in SML density upon projecting 3D localizations on a single plane.
Statistical analysis of categorized clustering data was performed by Fisher's Exact Test using R program, while statistical analysis of continuous distributions was done using two-sample Kolmogorov-Smirnov test in Matlab. Since a very large sample size yields smaller p-value and thus greater statistical significance, we used an optimal binning method for nonparametric density estimation58 to represent each sample population without the influence of a large sample size. We applied optimal binning, W (bin width) = 2·(3rd quantile −1st quantile)·N−1/3, for n>100 prior to running the Kolmogorov–Smirnov test.
Pairwise distance distribution analyses.
Pairwise distance analysis of SMLs on single viruses was calculated for each pair of one-color Env-Env SMLs using the pdist() function in Matlab; for two-color Env-SER SML pairwise distances were calculated using pdist2() function in Matlab. The pairwise distance was statistically compared by two-sample Kolmogorov–Smirnov test in Matlab after optimal binning58 (see above).
Supplementary Material
ACKNOWLEDGMENT
iPALM imaging was conducted in collaboration with the Advanced Imaging Center at Janelia Research Campus, a facility jointly supported by the Gordon and Betty Moore Foundation, and the Howard Hughes Medical Institute. pc594-labeled second antibodies were kindly provided by Luke Lavis (Janelia Research Campus) and SER5-iHA and SER2-iHA expression vectors by Massimo Pizzato (University of Trento). We are grateful to the NIH AIDS Reagent Program for providing saquinavir, 2G12 antibody, human HIV immunoglobulin and TZM-bl cells. We thank Dr. Aaron Lifland (GT) for the help with Vutara imaging system, Dr. Li-Hsiang Lin (School of Industrial Engineering, GT) for the assistance with statistical analysis, and Dr. Alexa Mattheyses for helpful discussions of dSTORM data, and Matthew Prellberg for technical help with manuscript. This work was supported by the NIH R01 grant GM054787 to GBM and NIH P41-GM103540 to EG.
Footnotes
Supporting Information. This document is available at https://pubs.acs.org/journal/ancac3 and contains seven Supplemental Figures and figure legends. Suppl. Fig. S1 presents analysis of correlation of Env and SERINC intensities associated with single virions and dSTORM-based analysis of number of Env SMLs per virion. Suppl. Fig. S2 shows DBSCAN analysis of Env clusters using varied search distance R. Suppl. Fig. S3 includes DBSCAN analysis of Env distribution obtained by 3D iPALM. Suppl. Fig. S4 presents HIV-1 Env cluster analysis of 2D-projected iPALM data. Suppl. Fig. S5 shows the lack of significant dimerization of SER5-GFP or SER2-GFP in the plasma membrane. Suppl. Fig. S6 includes pairwise distance analysis for Env-Env, SER-SER, and Env-SER localizations by 2-color iPALM. Suppl. Fig. S7 presents analysis of Env clustering after excluding particles that did not contain clusters.
REFERENCES
- 1.Rosa A; Chande A; Ziglio S; De Sanctis V; Bertorelli R; Goh SL; McCauley SM; Nowosielska A; Antonarakis SE; Luban J; Santoni FA; Pizzato M, HIV-1 Nef Promotes Infection by Excluding Serinc5 from Virion Incorporation. Nature 2015, 526, 212–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Usami Y; Wu Y; Gottlinger HG, Serinc3 and Serinc5 Restrict HIV-1 Infectivity and Are Counteracted by Nef. Nature 2015, 526, 218–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sood C; Marin M; Chande A; Pizzato M; Melikyan GB, Serinc5 Protein Inhibits HIV-1 Fusion Pore Formation by Promoting Functional Inactivation of Envelope Glycoproteins. J Biol Chem 2017, 292, 6014–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Firrito C; Bertelli C; Vanzo T; Chande A; Pizzato M, Serinc5 as a New Restriction Factor for Human Immunodeficiency Virus and Murine Leukemia Virus. Annu Rev Virol 2018, 323–340. [DOI] [PubMed] [Google Scholar]
- 5.Pye VE; Rosa A; Bertelli C; Struwe WB; Maslen SL; Corey R; Liko I; Hassall M; Mattiuzzo G; Ballandras-Colas A; Nans A; Takeuchi Y; Stansfeld PJ; Skehel JM; Robinson CV; Pizzato M; Cherepanov P, A Bipartite Structural Organization Defines the Serinc Family of HIV-1 Restriction Factors. Nat Struct Mol Biol 2020, 27, 78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schulte B; Selyutina A; Opp S; Herschhorn A; Sodroski JG; Pizzato M; Diaz-Griffero F, Localization to Detergent-Resistant Membranes and HIV-1 Core Entry Inhibition Correlate with HIV-1 Restriction by Serinc5. Virology 2018, 515, 52–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X; Shi J; Qiu X; Chai Q; Frabutt DA; Schwartz RC; Zheng YH, Cd4 Expression and Env Conformation Are Critical for HIV-1 Restriction by Serinc5. J Virol 2019, 93, e00544–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi J; Xiong R; Zhou T; Su P; Zhang X; Qiu X; Li H; Li S; Yu C; Wang B; Ding C; Smithgall TE; Zheng YH, HIV-1 Nef Antagonizes Serinc5 Restriction by Downregulation of Serinc5 Via the Endosome/Lysosome System. J Virol 2018, 92, e00196–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trautz B; Pierini V; Wombacher R; Stolp B; Chase AJ; Pizzato M; Fackler OT, The Antagonism of HIV-1 Nef to Serinc5 Particle Infectivity Restriction Involves the Counteraction of Virion-Associated Pools of the Restriction Factor. J Virol 2016, 90, 10915–10927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahmad I; Li S; Li R; Chai Q; Zhang L; Wang B; Yu C; Zheng YH, The Retroviral Accessory Proteins S2, Nef, and Glycoma Use Similar Mechanisms for Antagonizing the Host Restriction Factor Serinc5. J Biol Chem 2019, 294, 7013–7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chande A; Cuccurullo EC; Rosa A; Ziglio S; Carpenter S; Pizzato M, S2 from Equine Infectious Anemia Virus Is an Infectivity Factor Which Counteracts the Retroviral Inhibitors Serinc5 and Serinc3. Proc Natl Acad Sci U S A 2016, 113, 13197–13202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li S; Ahmad I; Shi J; Wang B; Yu C; Zhang L; Zheng YH, Murine Leukemia Virus Glycosylated Gag Reduces Murine Serinc5 Protein Expression at Steady-State Levels Via the Endosome/Lysosome Pathway to Counteract Serinc5 Antiretroviral Activity. J Virol 2019, 93, e01651–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beitari S; Ding S; Pan Q; Finzi A; Liang C, Effect of HIV-1 Env on Serinc5 Antagonism. J Virol 2017, 91, e02214–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Usami Y; Gottlinger H, HIV-1 Nef Responsiveness Is Determined by Env Variable Regions Involved in Trimer Association and Correlates with Neutralization Sensitivity. Cell Rep 2013, 5, 802–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chojnacki J; Staudt T; Glass B; Bingen P; Engelhardt J; Anders M; Schneider J; Muller B; Hell SW; Krausslich HG, Maturation-Dependent HIV-1 Surface Protein Redistribution Revealed by Fluorescence Nanoscopy. Science 2012, 338, 524–8. [DOI] [PubMed] [Google Scholar]
- 16.Brandenberg OF; Magnus C; Regoes RR; Trkola A, The HIV-1 Entry Process: A Stoichiometric View. Trends Microbiol 2015, 23, 763–74. [DOI] [PubMed] [Google Scholar]
- 17.Brandenberg OF; Magnus C; Rusert P; Regoes RR; Trkola A, Different Infectivity of HIV-1 Strains Is Linked to Number of Envelope Trimers Required for Entry. PLoS Pathog 2015, 11, e1004595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magnus C; Rusert P; Bonhoeffer S; Trkola A; Regoes RR, Estimating the Stoichiometry of HIV Entry. J Virol 2009, 83, 1523–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blijleven JS; Boonstra S; Onck PR; van der Giessen E; van Oijen AM, Mechanisms of Influenza Viral Membrane Fusion. Semin Cell Dev Biol 2016, 60, 78–88. [DOI] [PubMed] [Google Scholar]
- 20.Ivanovic T; Choi JL; Whelan SP; van Oijen AM; Harrison SC, Influenza-Virus Membrane Fusion by Cooperative Fold-Back of Stochastically Induced Hemagglutinin Intermediates. Elife 2013, 2, e00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Markovic I; Leikina E; Zhukovsky M; Zimmerberg J; Chernomordik LV, Synchronized Activation and Refolding of Influenza Hemagglutinin in Multimeric Fusion Machines. J Cell Biol 2001, 155, 833–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu P; Winkler H; Chertova E; Taylor KA; Roux KH, Cryoelectron Tomography of HIV-1 Envelope Spikes: Further Evidence for Tripod-Like Legs. PLoS Pathog 2008, 4, e1000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stano A; Leaman DP; Kim AS; Zhang L; Autin L; Ingale J; Gift SK; Truong J; Wyatt RT; Olson AJ; Zwick MB, Dense Array of Spikes on HIV-1 Virion Particles. J Virol 2017, 91, e00415–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDonald D; Vodicka MA; Lucero G; Svitkina TM; Borisy GG; Emerman M; Hope TJ, Visualization of the Intracellular Behavior of HIV in Living Cells. J Cell Biol 2002, 159, 441–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai W; Usami Y; Wu Y; Gottlinger H, A Long Cytoplasmic Loop Governs the Sensitivity of the Anti-Viral Host Protein Serinc5 to HIV-1 Nef. Cell Rep 2018, 22, 869–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rust MJ; Bates M; Zhuang X, Sub-Diffraction-Limit Imaging by Stochastic Optical Reconstruction Microscopy (Storm). Nat Methods 2006, 3, 793–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang B; Wang W; Bates M; Zhuang X, Three-Dimensional Super-Resolution Imaging by Stochastic Optical Reconstruction Microscopy. Science 2008, 319, 810–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Briggs JA; Wilk T; Welker R; Krausslich HG; Fuller SD, Structural Organization of Authentic, Mature HIV-1 Virions and Cores. Embo J 2003, 22, 1707–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pham S; Tabarin T; Garvey M; Pade C; Rossy J; Monaghan P; Hyatt A; Bocking T; Leis A; Gaus K; Mak J, Cryo-Electron Microscopy and Single Molecule Fluorescent Microscopy Detect Cd4 Receptor Induced HIV Size Expansion Prior to Cell Entry. Virology 2015, 486, 121–33. [DOI] [PubMed] [Google Scholar]
- 30.Sougrat R; Bartesaghi A; Lifson JD; Bennett AE; Bess JW; Zabransky DJ; Subramaniam S, Electron Tomography of the Contact between T Cells and SIV/HIV-1: Implications for Viral Entry. PLoS Pathog 2007, 3, e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ester MK, H.-P., Sander J, Xu X, A Density-Based Algorithm for Discovering Clusters a Density-Based Algorithm for Discovering Clusters in Large Spatial Databases with Noise. KDD-96 Proceedings 1996, AAAI Press: Portland, Oregon, 226–231. [Google Scholar]
- 32.Shtengel G; Wang Y; Zhang Z; Goh WI; Hess HF; Kanchanawong P, Imaging Cellular Ultrastructure by Palm, Ipalm, and Correlative Ipalm-Em. Methods Cell Biol 2014, 123, 273–94. [DOI] [PubMed] [Google Scholar]
- 33.Digman MA; Dalal R; Horwitz AF; Gratton E, Mapping the Number of Molecules and Brightness in the Laser Scanning Microscope. Biophys J 2008, 94, 2320–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boge M; Wyss S; Bonifacino JS; Thali M, A Membrane-Proximal Tyrosine-Based Signal Mediates Internalization of the HIV-1 Envelope Glycoprotein Via Interaction with the Ap-2 Clathrin Adaptor. J Biol Chem 1998, 273, 15773–8. [DOI] [PubMed] [Google Scholar]
- 35.Yang X; Kurteva S; Ren X; Lee S; Sodroski J, Stoichiometry of Envelope Glycoprotein Trimers in the Entry of Human Immunodeficiency Virus Type 1. J Virol 2005, 79, 12132–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chernomordik LV; Kozlov MM, Mechanics of Membrane Fusion. Nat Struct Mol Biol 2008, 15, 675–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozlovsky Y; Chernomordik LV; Kozlov MM, Lipid Intermediates in Membrane Fusion: Formation, Structure, and Decay of Hemifusion Diaphragm. Biophys J 2002, 83, 2634–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen FS; Melikyan GB, The Energetics of Membrane Fusion from Binding, through Hemifusion, Pore Formation, and Pore Enlargement. J Membr Biol 2004, 199, 1–14. [DOI] [PubMed] [Google Scholar]
- 39.Carravilla P; Chojnacki J; Rujas E; Insausti S; Largo E; Waithe D; Apellaniz B; Sicard T; Julien JP; Eggeling C; Nieva JL, Molecular Recognition of the Native HIV-1 Mper Revealed by Sted Microscopy of Single Virions. Nat Commun 2019, 10, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murakami T; Ablan S; Freed EO; Tanaka Y, Regulation of Human Immunodeficiency Virus Type 1 Env-Mediated Membrane Fusion by Viral Protease Activity. J Virol 2004, 78, 1026–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kondo N; Marin M; Kim JH; Desai TM; Melikyan GB, Distinct Requirements for HIV-Cell Fusion and HIV-Mediated Cell-Cell Fusion. J Biol Chem 2015, 290, 6558–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brugger B; Glass B; Haberkant P; Leibrecht I; Wieland FT; Krausslich HG, The HIV Lipidome: A Raft with an Unusual Composition. Proc Natl Acad Sci U S A 2006, 103, 2641–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lorizate M; Sachsenheimer T; Glass B; Habermann A; Gerl MJ; Krausslich HG; Brugger B, Comparative Lipidomics Analysis of HIV-1 Particles and Their Producer Cell Membrane in Different Cell Lines. Cell Microbiol 2013, 15, 292–304. [DOI] [PubMed] [Google Scholar]
- 44.Muranyi W; Malkusch S; Muller B; Heilemann M; Krausslich HG, Super-Resolution Microscopy Reveals Specific Recruitment of HIV-1 Envelope Proteins to Viral Assembly Sites Dependent on the Envelope C-Terminal Tail. PLoS Pathog 2013, 9, e1003198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roy NH; Chan J; Lambele M; Thali M, Clustering and Mobility of Hiv-1 Env at Viral Assembly Sites Predict Its Propensity to Induce Cell-Cell Fusion. J Virol 2013, 87, 7516–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pezeshkian N; Groves NS; van Engelenburg SB, Single-Molecule Imaging of HIV-1 Envelope Glycoprotein Dynamics and Gag Lattice Association Exposes Determinants Responsible for Virus Incorporation. Proc Natl Acad Sci U S A 2019, 116, 25269–25277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inuzuka M; Hayakawa M; Ingi T, Serinc, an Activity-Regulated Protein Family, Incorporates Serine into Membrane Lipid Synthesis. J Biol Chem 2005, 280, 35776–83. [DOI] [PubMed] [Google Scholar]
- 48.Trautz B; Wiedemann H; Luchtenborg C; Pierini V; Kranich J; Glass B; Krausslich HG; Brocker T; Pizzato M; Ruggieri A; Brugger B; Fackler OT, The Host-Cell Restriction Factor Serinc5 Restricts HIV-1 Infectivity without Altering the Lipid Composition and Organization of Viral Particles. J Biol Chem 2017, 292, 13702–13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chu EP; Elso CM; Pollock AH; Alsayb MA; Mackin L; Thomas HE; Kay TW; Silveira PA; Mansell AS; Gaus K; Brodnicki TC, Disruption of Serinc1, Which Facilitates Serine-Derived Lipid Synthesis, Fails to Alter Macrophage Function, Lymphocyte Proliferation or Autoimmune Disease Susceptibility. Mol Immunol 2017, 82, 19–33. [DOI] [PubMed] [Google Scholar]
- 50.Campbell SM; Crowe SM; Mak J, Virion-Associated Cholesterol Is Critical for the Maintenance of HIV-1 Structure and Infectivity. AIDS 2002, 16, 2253–61. [DOI] [PubMed] [Google Scholar]
- 51.Viard M; Parolini I; Sargiacomo M; Fecchi K; Ramoni C; Ablan S; Ruscetti FW; Wang JM; Blumenthal R, Role of Cholesterol in Human Immunodeficiency Virus Type 1 Envelope Protein-Mediated Fusion with Host Cells. J Virol 2002, 76, 11584–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rawat SS; Viard M; Gallo SA; Rein A; Blumenthal R; Puri A, Modulation of Entry of Enveloped Viruses by Cholesterol and Sphingolipids (Review). Mol Membr Biol 2003, 20, 243–54. [DOI] [PubMed] [Google Scholar]
- 53.Graham DR; Chertova E; Hilburn JM; Arthur LO; Hildreth JE, Cholesterol Depletion of Human Immunodeficiency Virus Type 1 and Simian Immunodeficiency Virus with Beta-Cyclodextrin Inactivates and Permeabilizes the Virions: Evidence for Virion-Associated Lipid Rafts. J Virol 2003, 77, 8237–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vishwanathan SA; Thomas A; Brasseur R; Epand RF; Hunter E; Epand RM, Large Changes in the Crac Segment of Gp41 of HIV Do Not Destroy Fusion Activity If the Segment Interacts with Cholesterol. Biochemistry 2008, 47, 11869–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vishwanathan SA; Hunter E, Importance of the Membrane-Perturbing Properties of the Membrane-Proximal External Region of Human Immunodeficiency Virus Type 1 Gp41 to Viral Fusion. J Virol 2008, 82, 5118–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dempsey GT; Vaughan JC; Chen KH; Bates M; Zhuang X, Evaluation of Fluorophores for Optimal Performance in Localization-Based Super-Resolution Imaging. Nat Methods 2011, 8, 1027–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nolan R; Iliopoulou M; Alvarez L; Padilla-Parra S, Detecting Protein Aggregation and Interaction in Live Cells: A Guide to Number and Brightness. Methods 2018, 140-141, 172–177. [DOI] [PubMed] [Google Scholar]
- 58.Izenman AJ, Recent Developments in Nonparametric Density Estimation. J Am Stat Assoc 1991, 86, 205–224. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.