Abstract
Epigenetic landscapes can provide insight into regulation of gene expression and cellular diversity. Here, we examined the transcriptional and epigenetic profiles of seven human blood NK cell populations, including adaptive NK cells. The BCL11B gene, encoding a transcription factor (TF) essential for T cell development and function, was the most extensively regulated, with expression increasing throughout NK cell differentiation. Several Bcl11b-regulated genes associated with T cell-signaling were specifically expressed in adaptive NK cell subsets. Regulatory networks revealed reciprocal regulation at distinct stages of NK cell differentiation, with Bcl11b repressing RUNX2 and ZBTB16 in canonical and adaptive NK cells, respectively. A critical role for Bcl11b in driving NK cell differentiation was corroborated in BCL11B mutated patients and by ectopic Bcl11b expression. Moreover, Bcl11b was required for adaptive NK cell responses in a murine CMV model, supporting expansion of these cells. Together, we define the TF regulatory circuitry of human NK cells and uncover a critical role for Bcl11b in promoting NK cell differentiation and function.
ONE-SENTENCE SUMMARY
Epigenetic and functional analyses of human NK cells identify a role for Bcl11b in canonical and adaptive NK cell differentiation.
INTRODUCTION
Natural killer (NK) cells are ascribed to the family of innate lymphoid cells (ILC) and specialize in killing infected or transformed cells as well as orchestrating immune responses through release of cytokine (1, 2). They express receptors implicated in an evolutionary battle with cytomegalovirus (CMV) (3, 4), a virus which establishes a persistent subclinical infection in the majority of humans but can be pathological in infants and immunocomprised individuals (5, 6).
Mature NK cells found within the blood are phenotypically heterogenous. They can be broadly classified into three subgroups which differ in their proliferative and effector capacity; CD56bright, CD56dimCD57− and CD56dimCD57+ NK cells (7-11). However, additional surface markers, including NKG2A, CD94, killer immunoglobulin-like receptor (KIR) and CD62L, also correlate with differentiation and may define intermediary phenotypes (12, 13). Super-imposed upon differentiation is the incompletely understood process of NK cell education whereby self MHC class I molecule binding of KIR (self-KIR) dictates subsequent NK cell responsiveness to target cells (10, 14).
NK cell differentiation is heavily impacted by CMV infection, as first described as an expansion of NKG2Chi cells (15). Termed adaptive NK cells, they excel in antibody-dependent cytokine responses and mediate potent graft-versus-leukemia effects (16-19). In humans, adaptive NK cells are characterized by low expression of the transcription factor (TF) PLZF, silencing of intracellular signaling molecules, and commonly expressing CD57 (16, 17). Adaptive NK cells exhibit classical features of the adaptive immune system, including clonal expansion, persistence, and recall responses (18, 20-22). These features are best modelled in murine CMV (MCMV), where parallels have been found between the dynamics of chromatin accessibility during the induction of adaptive NK cell responses and T cell memory (23). However, there is still little understanding of the drivers responsible for the generation of human adaptive NK cells and their ontology with canonical NK subsets.
Previous studies have surveyed the epigenetic landscapes of several ILC family members and emphasized the overlap with counterpart T helper cells (24-27). TF axes defining CD56bright compared to CD56dim NK cells have also recently been described (28). However, there is limited understanding of the drivers of peripheral NK cell differentiation as they acquire CD16, NKG2A, KIR and CD57 receptors and accompanying functional compentence. Indeed, in vitro NK cell differentiation models have failed to reliably generate mature canonical NK cells expressing these markers (29).
Here, we dissect the epigenetic cartography spanning human peripheral blood NK cell differentiation by analyzing seven NK cell subsets that map a differentiation trajectory towards adaptive NK cells. Specifically, enhancer analyses identified a set of profoundly regulated TF-encoding genes and unexpectedly indicated a central role for Bcl11b during NK cell development. We provide evidence for a key role of Bcl11b in driving human NK cell differentiation. Furthemore, adaptive NK cell subsets, displaying elevated Bcl11b expression, exhibited additional diversity in the expression of Bcl11b target genes normally restricted to T cells. Finally, knock-out mice demonstrated a requirement for Bcl11b in generation of an adaptive NK cell response to MCMV. Together, we demonstrate that Bcl11b promotes NK cell differentiation.
RESULTS
Delineation of human NK cell subset transcriptional programs
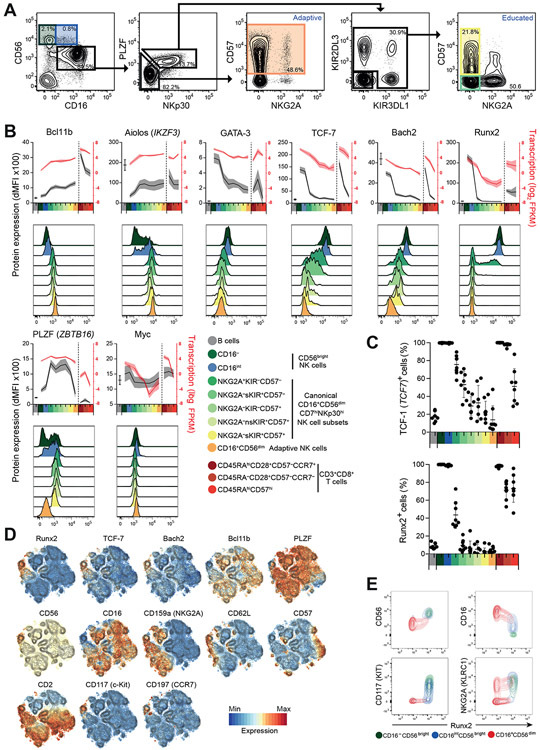
Single cell RNA-seq technology is unravelling immune cell diversity however transcript detection is limited, particularly for lowly abundant TFs and resolution between subsets can be poor compared to bulk RNA-seq analysis (30). In this regard, a comprehensive RNA-seq analysis of phenotypically distinct human NK cell subsets is lacking. We sorted selected NK cell subsets including CD56bright cells; CD56dim cells distinguished by NKG2A, CD57 or KIR-educating phenotypes; adaptive NK cells; and CD8+ T cell subsets for comparison from peripheral blood obtained from CMV seropositive donors (Figure 1A, Figure S1A and S1B, Table 1). An overall low abundance prevented analyses of certain subsets, e.g. canonical CD56dimNKG2A+KIR+ NK cells. Adaptive NK cells are phenotypically heterogenous across CMV seropositive individuals, thus we sorted CD16+CD56dimCD57+NKp30lo adaptive NK cells that either were NKG2Chi or CD7lo (Table 1). These subsets were uniformly PLZFlow and FcεRγlow, but with variable EAT-2 and SYK expression (Figure S1C). Gene expression of selected subset markers validated sorting purity (Figure S1D). Principal component analysis (PCA) of RNA-seq data revealed an NK cell differentiation axis converging towards that of CD8+ T cells (Figure 1B). Both PCA and dendrogram clustering revealed that CD56dimNKG2A− NK cell subsets clustered tightly regardless of KIR or CD57 expression, whereas substantial trancriptome differences were observed between canonical and adaptive CD56dimCD57+ NK cell subsets (Figure 1B, 1C). As such, NK cells could be categorized in to four subgroups, with the canonical CD56dim NK cell subset dissected by NKG2A rather than CD57 expression. Hierarchical clustering was performed on all 2,233 differentially expressed (DE) genes between CD16−CD56bright and canonical CD56dimNKG2A+KIR−CD57− (early) CD56dim NK cells, early CD56dim and canonical CD56dimNKG2A−self-KIR+CD57+ (late) NK cells, or late CD56dim and adaptive NK cells, respectively. Ten clusters reflecting different expression patterns among NK and CD8+ T cell subsets were discerned (Figure 1D). KEGG analysis revealed distinct cellular pathways that were enriched within clusters (Figure 1E).
Figure 1. Transcriptional profiling of human blood NK cell and CD8+ T cell subset.
Ten distinct NK and CD8+ T cell subsets from five healthy adult volunteers were sorted for RNA-seq analyses. (A) Sorting strategy of a representative donor. NK or CD8+ T cells were enriched prior to FACS. Adaptive NK cells were sorted as CD57+NKp30loNKG2C+ (donors 1, 3 and 5) or CD57+NKp30loCD7lo (donors 2 and 4). (B) Principal component analysis and (C) dendrogram of the top 500 highly variable transcripts among all ten cytotoxic lymphocyte subsets. (D-F) 2,233 NK cell differentially expressed (DE genes (FDR < 0.05) were defined based on major NK cell subset comparisons: CD16−D56bright versus CD56dimNKG2A+KIR−CD57− NK cells; CD56dimNKG2A+KIR−CD57− versus CD56dimNKG2A−self-KIR+CD57+ NK cells; or CD56dimNKG2A−self-KIR+CD57+ versus adaptive NK cells. The DE genes were stratified into 10 clusters based on expression profiles across all subsets. (D) Expression profiles and heatmaps of each cluster. (E) KEGG pathway analysis of clusters. (F) Variance in gene expression versus average gene expression levels of each cluster across CD16−CD56bright, canonical early and late CD56dim, and adaptive NK cell subsets. Genes with FDR < 0.01 or < 0.05 in ≥1 comparison are marked with filled or open circles, respectively. TFs are in red.
Table 1. KIR / HLA genotypes and adaptive NK cell phenotypes.
KIR and KIR-ligand genotypes for donors used in RNA-seq and ATAC-seq analysis as determined by multiplex PCR. Adaptive NK cells were sorted as CD3−CD16+CD56dim NKp30loCD57+ and either CD7lo or NKG2Chi, as specified.
| Donor | KIR-Ligands | KIR Genotype | Adaptive phenotype |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| D1 | C1+, Bw4+ | KIR2DL1 | KIR2DL3 | KIR2DL4 | KIR2DP1 | KIR2DS4 | KIR3DL1 | KIR3DL2 | KIR3DL3 | KIR3DP1 | NKp30lo,NKG2C+ |
| D2 | C1+ | KIR2DL1 | KIR2DL3 | KIR2DL4 | KIR2DP1 | KIR2DS4 | KIR3DL1 | KIR3DL2 | KIR3DL3 | KIR3DP1 | NKp30lo,CD7lo |
| D3 | C1+, C2+ | KIR2DL1 | KIR2DL3 | KIR2DL4 | KIR2DP1 | KIR2DS4 | KIR3DL1 | KIR3DL2 | KIR3DL3 | KIR3DP1 | NKp30lo,NKG2C+ |
| D4 | C1+, C2+, Bw4+ | KIR2DL1 | KIR2DL3 | KIR2DL4 | KIR2DP1 | KIR2DS4 | KIR3DL1 | KIR3DL2 | KIR3DL3 | KIR3DP1 | NKp30lo,CD7lo |
| D5 | C1+, C2+, Bw4+ | KIR2DL1 | KIR2DL3 | KIR2DL4 | KIR2DP1 | KIR2DS4 | KIR3DL1 | KIR3DL2 | KIR3DL3 | KIR3DP1 | NKp30lo,NKG2C+ |
| D6 | C1+ | KIR2DL1 | KIR2DL3 | KIR2DL4 | KIR2DP1 | KIR2DS4 | KIR3DL1 | KIR3DL2 | KIR3DL3 | KIR3DP1 | NKp30lo,CD7lo |
To elucidate highly regulated DE genes, gene expression was plotted versus variance within NK cell subsets (Figure 1F). Transcripts in cluster 1 were suppressed upon both NK and CD8+ T cell differentiation and included genes associated with the Notch signaling pathway. Clusters 2 and 3 both contained transcripts that declined upon CD56bright NK cell differentiation, but were distinguished by specifically being down-regulated during NK cell differentiation or also being down-regulated in effector CD8+ T cells, respectively. Genes in cluster 2 were associated with relaxin, phosphoinsitide-3-kinase (PI3K)-Akt signaling, antigen processing, and focal adhesion signaling pathways. Cluster 3 transcripts displayed somewhat higher levels in adaptive NK cells and included the TFs RUNX2, MYC, and TCF7. Cluster 4 grouped genes that were specifically downregulated in adaptive NK cells, including IL12RB2, IL18RAP, and FCER1G as well as TFs SOX4, ZMAT4, MAFF, and ZBTB16 (encoding PLZF). With specific down-regulation in early CD56dimNK cells, cluster 5 included transcripts associated with p53 signaling (i.e. BCL2, SIVA1, SESN3, TNFRSF10B). Clusters 6 and 7 encompassed genes up-regulated throughout NK cell differentiation, but distinguished by being down-regulated or up-regulated during CD8+ T cell differentiation, respectively. This was useful in highlighting genes that potentially are distinctly regulated during NK cell versus T cell differentiation. Cluster 6 was associated with T cell development and MAPK signaling pathways. BCL11B was the most highly regulated TF in this cluster which also included several known Bcl11b target genes (e.g. CD3G, CD3D, CD6, ITGA6) (31, 32). Cluster 7 consisted of genes implicated in cellular cytotoxicity, as well as TFs IKZF3, NFIL3, and ZEB2. Orthologues of these genes are required for mouse NK cell maturation (33-36). Clusters 8-10 had fewer associated genes, nevertheless cluster 9 was a notable profile with genes specifically up-regulated in adaptive NK and effector CD8+ T cells. They included the check-point inhibitory receptor genes LAG3 and PDCD1 (PD-1), in addition to RAB11FIP5 and the TF ZNF683 (Hobit) associated with augmented adaptive NK cell function and tissue-resident memory T cell generation, respectively (37, 38).
Together, analyses broadly categorized NK cells into four subgroups including (i) CD56bright NK cells, (ii-iii) CD56dim NK cells separated by NKG2A rather than CD57 expression and (iv) adaptive NK cells. These subgroups reveal increasing transcription of the TF Bcl11b tracking the NK cell differentiation trajectory. Genetic ablation of Bcl11b in the mouse hematopoietic lineage blocks T cell development and differentiation at numerous stages (39-44), as well as development of innate lymphoid cells type 2 (ILC2) (45-48). However, a role of Bcl11b in NK cell differentiation is unexpected because Bcl11b deficiency in mice does not diminish ILC progenitors and leads to increased numbers of NK-like cells (41).
Chromatin accessibility landscapes of human NK cell subsets
The chromatin architecture of defined peripheral human NK cell subsets, including adaptive NK cells, has not been extensively studied. We performed ATAC-seq on NK cell and T cell subsets to identify chromatin accessibility (49). We catalogued 130,049 ATAC-seq peaks in seven NK cell subsets, of which 52% were common to all (Figure 2A). Clustering analysis of highly variable ATAC-seq peaks largely recapitulated transcriptional relationships with early CD56dim NK cells distinguishable as an intermediary subset and together revealing four NK cell subgroups (Figure 2B), also highlighting four subgroups of NK cells: CD16−CD56bright, canonical early CD56dimNKG2A+ and late CD56dimNKG2A− NK cells, as well as adaptive NK cells. Cumulative changes in chromatin accessibility of NK cells subsets relative to CD56bright NK cells confirmed this trajectory (Figure S2A). However, dendrogram analysis grouped CD56bright NK cells with naïve CD8+ T cells underscoring their less differentiated status (Figure 2C). In mice, the chromatin accessibility of CD27+CD11b− precursor NK cells resemble memory CD8+ T cells which may be true for CD56bright NK cells when compared to other memory CD8+ T cell subsets not analysed here (50).
Figure 2. Epigenetic landscapes of human NK cell subsets.
NK and CD8+ T cell subsets were sorted for ATAC-seq analysis. Adaptive NK cells were sorted as CD57+NKp30loNKG2C+ (donors 1 and 3) or CD57+NKp30loCD7lo (donors 2 and 6). (A) Summary of unique, variable, or common ATAC-seq peaks in NK cell subsets (n=4 donors, average 56,383 peaks per subset). (B) Principal component analysis and (C) dendrogram of the top 500 highly variable ATAC-seq peaks among all nine lymphocyte subsets. (D) Genomic distribution of all NK cell ATAC-seq peaks. (E-G) Genomic classifications of differentially accessible (DA) ATAC-seq peaks (FDR < 0.05) within selected NK cell (E) major (>3000) and (F) minor (<3000) comparisons, and (G) comparisons of adaptive NK cells versus CD8+ T cells. (H-J) Plots show promoter specific fold change (FC) for DA ATAC-seq peaks (FDR < 0.05) versus FC in RNA-seq expression in comparisons of (H) CD16−CD56bright (dark green) versus canonical CD56dimNKG2A+KIR−CD57− (early; light green), (I) early versus canonical CD56dimNKG2A−sKIR−CD57+ (late; yellow), and (J) late versus adaptive CD56dimNKp30lowCD57+ (orange) NK cell subsets. DE genes with FDR < 0.01 or < 0.05 in at least one pairwise comparison are marked with black filled or open circles, respectively. TF-encoding genes are red.
A total of 38,348 differentially accessible (DA) ATAC-seq peaks between CD16−CD56bright and early CD56dim NK cells, early and late NK cells, or late and adaptive NK cells were identified (Figure S2B). While most ATAC-seq peaks were located within gene promoters (Figure 2D), DA ATAC-seq peaks were enriched at intergenic and intronic regions as well (Figure 2E and 2F). Educated NK cells expressing self-KIR have augemented responses towards target cells. However, with the exception of KIR gene loci, no other DA ATAC-seq peaks were identified between self- and non-self KIR-expressing NK cells (Figure 2F), supporting previous data showing little transcriptomic differences between uneducated and educated NK cells (51, 52).
A preferential loss of promoter accessibility with peaks at TFs RUNX2, TCF7, and IRF8 becoming less accessible was observed between early and late CD56dim NK cell subsets (Figure 2F), however not between naïve and effector CD8+ T cells (Figure 2G). By comparison, multiple promoter regions were highly accessible in CD56bright NK cells, including TFs FOXC1, RUNX2, LEF1 and ZEB1 (Figure 2H). A similar number of increasingly accessible promoters were identified in early CD56dim NK cells, exemplified by the TFs BCL11B, GLI3, PRDM1 and IKZF3, with the GLI3 promoter progressively opening throughout CD56dim NK cell differentiation (Figure 2I). Adaptive NK cells displayed restricted accessibility at promoters for TFs ZMAT4, SOX4, and IKZF2, with few TFs epigenetically promoted (Figure 2J).
Thus, chromatin accessibility landscapes confirm four major subgroups of peripheral blood NK cells, but reveal no imprint of education. Many transcriptional differences between NK cell subsets were underpinned by changes in promoter accessibility, as evident for NK cell receptors as well as TFs including RUNX2 and BCL11B.
Active enhancers are associated with regulation of NK cell gene expression
NK cell active enhancers, defined by H3K27 acetylation (H3K27ac) (53), have previously only been broadly analysed using CD56bright and bulk CD56dim human NK cells (28). Thus, to comprehensively define active enhancers (AEs) spanning NK cell differentiation we sorted the seven NK cell subsets and performed H3K27ac chromatin immunoprecipitation (ChIP)-seq. Among the four most distinct NK cell subsets, we identified 4,841 AEs at loci overlapping with DA ATAC-seq peak(s) (Figure 3A), whilst PCA analysis stratified all of the NK cell subsets consistent with RNA-seq clustering (Figure S3A). The majority of AEs with differential chromatin accessibility were identified in the CD16−CD56bright to early CD56dim comparison. Furthermore, 9.1% of AEs with differential chromatin accessibility were identified within these groups and in the early to late CD56dim NK cell comparision (Figure 3A).
Figure 3. Correlation of regulated active enhancers to gene transcription during NK cell differentiation.
(A) Summary of DA ATAC-seq peaks (FDR <0.05) residing within active enhancers (AE). (B-D) SE identified from density of H3K27ac signal versus rank (716-1270 SE identified) detected using ChIP-seq (n=2 donors). (B) Hockey stick plots indicate inflection point and examples of unique SE. (C) Venn diagram depicting common and unique SE. (D) Variation of enhancer rank across all NK cell subsets. Dotted line represents number of in all NK cell subsets, shaded area marking non-SEs, with ranking ≥1300 assigned as 1300. (E-G) AE signals relative to chromatin accessibility and gene expression in (E) CD16−CD56bright versus early, (F) early versus late, and (G) late versus adaptive NK cell subset comparisons. Absolute highest FC DA ATAC-seq peak (FDR < 0.05) assigned to a single gene. Large circles indicate RNA-seq FC > 1.5 log2. (H) Individual DA ATAC-seq peaks (FDR < 0.05) assigned to DE TF genes (FDR < 0.05) with FC > 2 in CD56bright to early, and FC > 1.5 in early to late as well as late to adaptive NK cell transitions. (I) TF motif enrichment within DA ATAC-seq peaks (FDR < 0.05) in AE across NK and T cell subsets normalized to peak length, average NK cell frequency with maximum/minimum score > 2 (n=60) then grouped by similarity according to fixed Euclidean distance (n=26).
Super-enhancers (SE) are broad clusters of active enhancers occupied by TFs and cofactors that maintain expression of genes associated with cell identity (54). Using the ROSE SE pipeline (55), we merged all active enhancers within 12.5kb proximity and ranked them according to overall H3K27ac signal intensity yielding on average 990 SE’s per subset (716-1270 range; Figure 3B). We focused our analysis on the four NK cell subgroups. We identified subset specific SE, with CD56bright NK cells possessing several unique SE, as exemplified by TFs BACH2, AHR and PRMA (Figure 3B). Aggregate distribution of SE revealed the majority were commonly annotated in all NK cell subgroups (Figure 3C). We also analysed SE that were highly variable throughout the NK cell subgroups as exemplified by TCF7 – a key component of the Wnt signaling pathway – and BCL11B, as compared to genes such as BACH2 that was specific to CD56bright NK cells (Figure 3D). Of note, our dataset of SE from CD16−CD56bright NK cells correlated well with SE previously identified in bulk CD56bright NK cells (Figure S3B) (28).
Active enhancers governing gene expression were assessed by correlating transcript levels with proximal DA ATAC-seq peaks overlapping AEs (Figure 3E-3G with colors reflecting fold change in AEs). The transcriptional differences observed in the CD56bright to CD56dim transition were mirrored by the substantial number of differential AE associated peaks (Figure 3E-G).
Many TF-encoding loci possessed multiple DA ATAC-seq peaks within each AE, as exemplified by BCL11B, ZBTB16, BACH2, RUNX2, and TCF7 (Figure S3C), indicating a requirement for several independent TF-binding complexes. Further indicating an unexpected role for BCL11B in NK cell differentiation, our analyses revealed that differentiation from CD16−CD56bright NK cells correlated with increasing accessibility at multiple BCL11B AEs (Figure 3H). By comparison, accessible peaks were lost at TCF7 and RUNX2 loci between CD56bright to early CD56dim NK cellls and were further restricted in late CD56dim NK cells (Figure 3H). Individual DA ATAC-seq peaks were incrementally lost in BACH2 and PRDM8 throughout canonical differentiation and, conversely, stepwise gain of chromatin accessibility in BNC2 and GLI3 correlated positively with differentiation. Notably, BCL11B was unique in possessing DA ATAC-seq peaks at modified AEs in transistions between all four NK cell subgroups, including adaptive NK cells (Figure 3H).
The exposure of signature binding motifs at AEs indicates potential TF activity. Thus, we determined which known consensus TF binding motifs were enriched at accessible AE loci (Figure 3I). An NFκB signature was prominent in CD16−CD56bright NK cells. Binding partners Runx/Runt and core binding factor (CBF) TF motifs (56), as well as a TCF family motif, correlated with the loss of RUNX2 and TCF7 accessibility at promoters and AEs in canonical NK cells. The AP-1 family of TFs, harbored a higher frequency of target motifs within the early CD56dim subset, suggestive of programs dependent on AP-1 family members. The CTCF insulator motif was also enriched in CD56dim NK cell subsets, where mature NK cell subsets also possessed significantly more ATAC-seq peaks per AE (Figure S3D), together suggesting more compartmentalized and complex enhancer compositions. Bcl11b has low DNA binding affinity and is not included in current motif databases (57).
These data highlight TFs that are extensively epigenetically regulated through NK cell differentiation, some of which imprint strong, differentially regulated signatures. The BCL11B locus acquired SE status in CD56dim NK cells and, further, was the only TF increasingly regulated at distinct sites within all three major NK cell transitions, including adaptive NK cells.
Transcription factor expression in relation to NK cell differentiation
We performed flow cytometry of highly regulated TFs across NK cell differentiation, and included CD16intCD56bright NK cells to enhance the resolution of early differentiation (Figure 4A). Bcl11b and Aiolos were acquired in a subset of CD56bright NK cells and were successively increased throughout differentiation (Figure 4B, S4A). Significant differences were observed in protein levels for these two TFs between intermediary maturing CD56dim NK cells, not seen at the transcript level, while both TCF-7 and Bach2 were gradually lost during differentiation contrasting the sharp reduction of Runx2 and, to a lesser extent, GATA-3, upon transition to the CD56dim NK cell phenotype. Although MYC transcripts were also highly regulated during NK cell differentiation, expression of Myc in unstimulated NK cells was low. In contrast, PLZF appeared more highly regulated at the protein than transcript level. Compared to gradual reduction of TCF-7 (also known as TCF-1) expression throughout NK cell differentiation, Runx2 had a binary expression profile (Figure 4C).
Figure 4. Continuum of transcription factor expression in NK cell differentiation.
(A-E) Intracellular flow cytometric evaluation of selected TFs. (A) Representative gating of NK cell subsets with inclusion of additional CD16intCD56bright cell subset (blue). (B) Graphs depict dMFI values, B-cell values added for comparison, n=8 healthy donors. Histograms from one representative donor. (C) Frequencies of cells expressing specific TFs within B, NK and CD8+ T cell subsets. (D) t-SNE analysis of gated CD3−CD56+ NK cells. (E) Plots depict expression in gated CD16−CD56bright CD16intCD56bright or CD56dim NK cell subsets from a representative donor.
T-distributed stochastic neighborhood embedding (t-SNE) analysis of flow cytometry data including differentiation markers revealed a gradual loss of GATA-3, TCF-7, and Bach2 upon maturation, while Bcl11b expression inversely correlated with TCF-7 and Bach2 (Figure 4D). In contrast, Runx2 was almost exclusively expressed in CD56bright NK cells and a CD56dimCD16+CD62Lhigh subpopulation, correlating closely with CD117 and CD62L and inversely with CD16 expression (Figure 4D, E and S4B). The expression pattern of Aiolos was very similar to Bcl11b across the NK cell subsets (Figure 4B), however t-SNE analysis indicated that the TFs were not strictly co-expressed (Figure S4C).
We find binary regulation pattern of Runx2 expression that was restricted to CD56bright and early CD56dim NK cells, mirroring profound reduction of promoter and AE accessibility. In contast, Bcl11b expression increased throughout differentiation, with highest expression in adaptive NK cells.
Reciprocal transcription factor regulation during NK cell differentiation
To position the TFs within the regulatory network of NK cell differentiation, we conducted ChIP-seq for Runx2, Bach2, GATA-3, Bcl11b, PLZF, and Helios/IKZF2 in peripheral blood NK cells. Helios has previously been shown to be differentially regulated in adaptive NK cells (17). ChIP-seq data quality was high with relatively small peaks discernable from background binding, as shown for exemplar loci NCAM1 (CD56), KLRF1 (NKp80), GZMH, GZMB, KIT, BCL11B and PRF1 (Table S2, Figure S5A). Further, two distinct monoclonal antibodies to Bach2 or Runx2, respectively, displayed high intra-overlap (Figure S5B). To categorize TF binding, we superimposed TF ChIP-seq signals across our transcriptome and open chromatin subset landscapes (Figure S5C). Throughout the NK cell trajectory, Bcl11b preferentially associated with chromatin regions that gained accessibility and transcripts that were up-regulated upon differentiation (Figure S5C), suggesting a role for Bcl11b in promoting human NK cell differentiaton at multiple transitions. Loss of PLZF and gain of Bcl11b expression are associated with NK cell transition to adaptive NK cells. Comparison of DE genes bound by PLZF reveal targets such as MEF2C, IL12BR2 and CD2 at open chromatin loci whilst Bcl11b bound genes including FCER1G, ZBTB16 (PLZF), CCR5, UBASH3A and CD3E (Figure S5D).
Target genes encoding TFs are of particular interest in defining regulatory circuitries. We constructed a de novo TF network of CD16−CD56bright compared to early CD56dim NK cells by integrating ChIP-seq signals with RNA-seq and ATAC-seq datasets to infer TF activity (Figures 5A and S5D). A reciprocal relationship was observed between Runx2 and Bach2 on one hand, and Bcl11b and PLZF on the other. Runx2 bound ZEB1, RFX2, AHR, MYC, PRDM8, and BACH2 (Figure 5A), thus appearing important in maintaining CD56bright NK cell-enriched TFs. Runx2 and Bach2 co-regulated the TF genes TCF7, ZMAT4, and ZNF667. Bcl11b and PLZF bound TF loci including IKZF3, ZEB2, RORA, BNC2, CREM, and PBX4 that were increased in CD56dim NK cells (Figure 5A). The TF network characterizing canonical early to late CD56dim NK cells pointed to TF expression dependence on Runx2, whereby decreased expression of Runx2 correlated with suppression of TFs including TCF7, MAFF, AHR and MYC (Figure 5B). The network also reflected the cementing of less accessible chromatin from CD16−CD56bright to early CD56dim cells whereby chromatin was further constricted upon late CD56dim NK cell differentiation, again particularly at Runx2 target genes such as AHR, ZEB1, TCF4 and PRDM8 (Figure 5B). DA ATAC-seq loci that inversely correlate with lower gene expression can suggest repressive activity. Notably, the majority of PLZF target genes were transcriptionally suppressed in late CD56dim NK cells but correlated with increased chromatin accessibility consistent with the role of PLZF as a transcriptional repressor (Figure 5B). PLZF expression is repressed by Bcl11b in T cells (31). Similarly, multiple Bcl11b peaks targeting ZBTB16 in late CD56dim NK cells and adaptive NK cells relative to early CD56dim NK cells, and late CD56dim NK cells respectively (Figure 5B, 5C, S5E). The adaptive NK cell gene regulation modelled here is mostly shaped by Bcl11b and PLZF. While Bcl11b positively correlated with induction of YBX3, PBX4, and SATB2, it also targeted the down-regulated genes IKZF2 and ZBTB16 (Figure 5C).
Figure 5. Reciprocal transcription factor regulation of NK cell differentiation.
ChIP-seq for TFs GATA-3, Runx2, Bach2, Bcl11b, PLZF, and Helios were performed on isolated human NK cells from individual representative donors. (A-C) Network analysis representing TF downstream targets identified from TF ChIP-seq experiments combined with RNA-seq and ATAC-seq data in (A) CD16−CD56bright versus early, (B) early versus late, and (C) canonical late versus adaptive NK cells. Edge color represents log2 FC in chromatin accessiblity at TF ChIP-seq locus with respect to pairwise subset comparisons (FDR < 0.05) and where multiple ChIP-seq peaks mapped to different DA ATAC-seq peaks, an average value is shown. Edge thickness represents number of ChIP-seq peaks associated with each gene. Node color represents log2 FC of proximally associated genes (< 60 kb) within pairwise subset comparisons (DE RNA-seq FC > 1 and RNA-seq log2 FPKM expression > −11).
Together, our results suggest Runx2 as a major hub in CD56bright cell identity, regulating a number of TF genes together with GATA-3 and Bach2. Bcl11b and PLZF, co-expressed in canonical CD56dim NK cells, appeared to regulate mostly non-overlapping groups of TF genes in early to late canonical CD56dim NK cells. Co-inciding with increased Bcl11b expression in adaptive NK cells, Bcl11b was positioned to potentially antagonize ZBTB16, silencing of which is a defining feature of human adaptive NK cells (17).
Bcl11b promotes canonical NK cell differentiation
Bcl11b displayed an expression profile that correlated with NK cell differentiation. To explore the role of Bcl11b during canonical maturation, we focused on Bcl11b target genes that were successively regulated during the CD16−CD56bright to early CD56dim or early to late CD56dim transitions (Figure S6A). Bcl11b-bound loci strongly correlated with this differentiation trajectory. Binding occurred at repressed chemokine and cytokine receptor loci such as CCR7, CXCR3, CCR1, and IL7R in early CD56dim NK cells, while CXCR1, while CXCR2, CX3CR1 were progressively induced in late CD56dim NK cells (Figure S6A). Likewise, Bcl11b occupied multiple sites within accessible chromatin of RUNX2 and SOX13 in CD16−CD56bright NK cells, whereas MAF, RORA, and ZBTB16 were targeted in open chromatin regions of CD56dim NK cells (Figure S6A). A signature of Bcl11b-regulated genes has previously been derived from a knockout mouse model of T cell development (31), with considerable overlap to genes regulated by Bcl11b in mouse CD4+ and Treg cells (42, 44). We assessed whether orthologous genes were also imprinted during human NK cell differentiation in gene set enrichment analysis (GSEA; Figure S6B). In the CD16−CD56bright to early CD56dim NK cell transition, Bcl11b-repressed target genes including KIT, CD7, and CD160 were significantly depleted in the early CD56dim NK cell subset. Between early to late CD56dim NK cells, where Bcl11b expression is increased, we noted enrichment of Bcl11b-induced genes including TNFRSF1A, NFATC2, SOX13, and CD6 (Figure S6B).
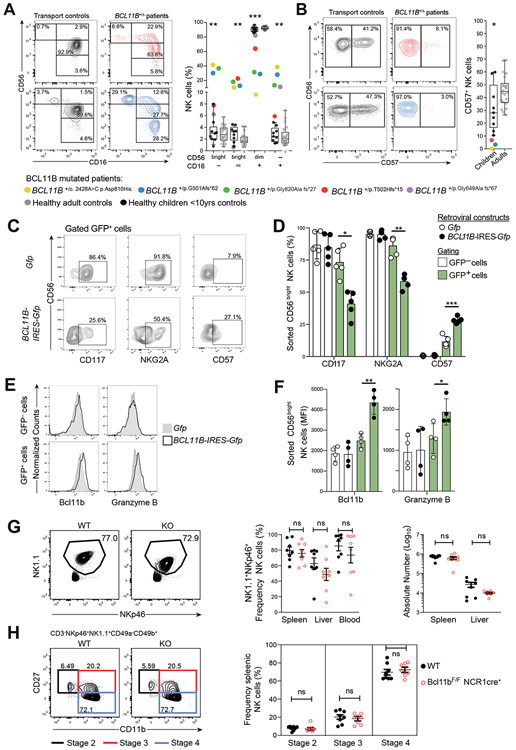
Heterozygous mutations in BCL11B have recently been associated with intellectual disability and varying degrees of immunodeficiency (58, 59). We examined NK cells from five pediatric patients harboring different heterozygous BCL11B loss-of-function mutations (BCL11B+/Δ) to explore the impact of Bcl11b in vivo. One patient had a T cell deficient SCID-like phenotype but the other four patients had relatively normal T cell counts, suggesting a spectrum of disease severity (Table 2). Remarkably, four of the five patients had enrichment of CD16−CD56bright and CD16intCD56bright subsets, with the severe combined immunodeficiency (SCID)-like Patient 1 displaying NK cells devoid of CD16+ NK cells (Figure 6A, S6C). In canonical NK cells Bcl11b expression peaks in CD56dimCD57+ cells. Frequencies of CD57-expressing CD56dim NK cells were also low in BCL11B+/Δ patients relative to age-matched controls (Figure 6B), suggesting a maturation block. Furthermore, retroviral transduction of sorted CD56bright NK cells with a Bcl11b expression construct repressed expression of CD117 and NKG2A, while promoting expression of CD57 and granzyme B in a culture system including IL-15 and feeder cells (Figure 6C-F). These findings were corroborated by selective Bcl11b binding at loci of KIT (CD117), KLRC1 (NKG2A), and B3GAT1 (encoding the glucuronosyltransferase that creates the CD57 epitope) (Figure S6D). Protein expression of Bcl11b itself confirmed significant induction upon Bcl11b-IRES-GFP transduction (Figure 6E-F).
Table 2. Genetic, clinical, and immune cell characteristics of patients with BCL11B mutations.
| Patient | #1 | #2 | #3 | #4 | #5 |
|---|---|---|---|---|---|
| BCL11B mutation | p.Asn810His | p.Gly501Alafs*62 | p.Gly320Afs*27 | p.Thr502Hisfs*15 | p.Gly649Alafs*67 |
| Age (years) | 0.3 | 1.7 | 4 | 4.5 | 15 |
| Descibed in Lessel et al, Brain, 2018 | Novel | Novel | A | D | B |
| Intellectual disability | + | + | + | + | + |
| Speech impairment | + | + | + | + | + |
| Delay in motor development | ++ | + | + | + | + |
| Autistic features | − (too early) | − (too early) | + | − | − |
| Myopathic facial apperance | + | + | + | + | − |
| Hyperteloism | + | + | + | + | − |
| Thin eyebrows | + | + | + | − | + |
| Small palpebral fissures | + | + | + | − | + |
| Hypertension | − | + (steroids) | + | + | − |
| Prominent nose | + | − | + | − | + |
| Long philtrum | + | + | + | + | + |
| Thin upper lip vermilion | + | + | + | + | + |
| Refractive error | ND | + | Hyperopia | − | − |
| Dental anomalies | + (neonatal teeth) | − | + | − | + |
| Feeding difficulties | + | − | − | − | − |
| Immune response | SCID | Refactory autoimmune hemolytic anemia | − | − | Frequent infections |
| T cell numbers /μl | 36 | 1840 | 1080 | 1220 | 3743 |
| NK cell numbers /μl | 1265 | 890 | 107 | 750 | 103 |
Figure 6. Bcl11b regulates canonical NK cell differentiation.
(A) NK cells from Bcl11b+/Δ patients (colored) compared to healthy, age-matched children (grey circles) or adult (black circles) controls gated on CD3− lymphocytes or (B) the frequency of CD16+CD56dim cells expressing CD57. Two-tailed paired Student’s t-test between Bcl11b+/Δ patients and age-matched children. (C-F) Frequency NK cells expressing CD117, NKG2A CD57, Bcl11b and Granzyme B following ectopic expression of Bcl11b. Sorted CD56bright NK cells were transduced with retrovirus encoding either GFP (Gfp) or Bcl11b and GFP (BCL11B-IRES-Gfp), and cultured for 7 days with IL-15 and irradiated EL08-1D2 feeder cells. (C) Protein expression after culture in a representative donor. (D) Summary of frequency of GFP− or GFP+ cells. Bars indicate mean and lines SD, n=5 independent donors. (E) Flow cytometry expression of Bcl11b and Granzyme B from a example donor. (F) Summary of Bcl11b and Granzyme B expression. Two-tailed paired Student’s t-test, (n=4 donors). (G) Frequency and absolute numbers of NK cells from either WT or Bcl11b floxed KO NCR1+ cells assessed in murine spleen, liver and blood. Two-tailed Student’s t test applied, n=8 mice per group. (H) Mouse late stages of NK cell development compared between WT and Bcl11b KO mice. Two-tailed Student’s t test applied, n=8 mice per group.
Human NK cell differentiation has parallels to that of mice, with the mouse CD11b−CD27+ to CD11b+CD27+ NK cell transition resembling human CD56bright to CD56dim NK cell differentiation (60). To determine the role of Bcl11b in murine NK cells, we generated Bcl11bF/F Ncr1cre+ mice to target allele deletion in cells expressing NKp46 (Ncr1), including NK cells. The frequencies and absolute numbers of NK1.1+NKp46+ cells remained unchanged in the absence of Bcl11b in spleen, liver, and blood (Figure 6G), although Bcl11b-deficient NK cells displayed decreased expression of the NK1.1 (Klrb1c) receptor (Figure S6E). Further analysis of Stage 2-4 developmental subsets (CD27+CD11b−, CD27+CD11b+, and CD27−CD11b+) showed no difference between wild-type and Bcl11bF/F Ncr1cre+ mice (Figure 6H). Moreover, distinct from human, RNA-seq analyses of wild-type mouse did not reveal any increase in Bcl11b transcription along the canonical NK cell differentiation trajectory (Figure S6F).
Our results from two distinct lines of investigation provide evidence for a direct role of Bcl11b in promoting canonical human CD56dim NK cell differentiation specifically in suppressing c-KIT and NKG2A whilst promoting CD57 expression. In contrast, Bcl11b ablation in mice did not grossly affect NK cell numbers nor their early development, suggesting differences between human and mouse NK cell maturation pathways.
High Bcl11b expression is associated with diverse adaptive NK cells phenotypes
Our transcriptional and epigenetic profiling pointed to the adaptive NK cell similarity to effector CD8+ T cells, where mouse T cell differentiation requires Bcl11b (32). We examined Bcl11b ChIP-seq target genes commonly regulated between human adaptive NK cells and effector CD8+ T cells relative to canonical late CD56dim and adaptive NK cells respectively using our sorted RNA-seq cell datasets (Figure 7A). Among the shared adaptive NK cell / effector CD8+ T cell Bcl11b-induced target genes included CD3D, CD3E, CD5, THEMIS, IL7R, and LAG3, whilst ZBTB16, FCER1G and SYK were commonly repressed. This suggests that Bcl11b regulates T cell-associated genes in adaptive NK cells whilst repressing genes including ZBTB16. Furthermore, GSEA indicated an imprint of Bcl11b on adaptive NK cell differentiation. Bcl11b-induced genes included CD3 complex genes, CD6, and SOX13 specifically in adaptive NK cells (Figure 7B). Conversely, Bcl11b-repressed genes included phenotypic hallmarks of adaptive NK cells such as ZBTB16, FCER1G, SH2D1B2, CD7, and IL2RB (Figure 7B) (16, 17).
Figure 7. Bcl11b is associated with classical and T-cell associated phenotypes of adaptive NK cells.
(A) Bcl11b-target genes correlating adaptive NK cell and effector CD8+ T cell differentiation. (B) GSEA of Bcl11b target genes over-represented during adaptive NK cell differentiation (FDR < 0.1). Bcl11b-induced or repressed gene sets based on human orthologues of target genes identified in Bcl11b knock-out mice, from Hosokawa et al. (31), and assessed in canonical late CD56dim to adaptive NK cells. Examples listed. (C) Bcl11b ChIP-qPCR of sort purified canonical CD56dimCD57+NKG2C− and adaptive CD56dimCD57+NKG2C+ NK cells. PCR targets reside within indicated DA chromatin loci. Expression normalized to ChIP input. Paired Student’s t-test, n=3. Induction of LAG-3 in NKG2C+ adaptive NK cells after stimulation for 24 hours with plate-bound anti-CD16 or anti-NKG2C antibody. (D-E) Representative flow cytometry plots (D) and summary of LAG-3 induction at 24 hours of stimulation (n=6). (E) NK cells gated as PLZF− and CD56dimCD16+. Paired Student’s t-test. (F) Protein expression of intracellular CD3ε in 196 healthy donors with CMV seropositive donors (filled) and seronegative (open). Positive expression determined from 3x SD of mean CMV seronegative donors (red line). (G) Frequencies of expression in canonical CD56dimPLZFhigh (yellow) and adaptive CD56dimPLZFlow (orange) NK cells. Summary of intracellular CD3ε+ adaptive NK cell populations. Paired Student’s t-test.
Exhaustion markers LAG-3 and PD-1 (PDCD1) are commonly identified within stimulated T cell subsets (61) and represented Bcl11b ChIP-seq target genes in human adaptive NK cells. To confirm preferential binding of Bcl11b at specific target genes FCER1G, ZBTB16, CD3E or LAG3 in adaptive NK cells, we performed BCL11B-ChIP-qPCR in FACS purified conventional CD57+ and adaptive CD57+ NK cells (Figure 7C). Relative to canonical CD56dimCD16+ counterparts, adaptive NK cells exhibited higher expression of LAG-3 after CD16 stimulation, which was synergised by IL-15 (Figure 7D-E). Stimulation with NKG2C and IL-15 for 1 day rapidly induced LAG-3 in adaptive NK cells (defined here as PLZF−), corroborating recently published results at 7 days (62). Our results highlight that adaptive NK cells may be particularly susceptible to early immune checkpoint inhibition.
Adaptive NK cells have heterogenous phenotypes with respect to signaling molecules such as FcεRγ, EAT-2 and SYK (16, 17), but have not previously been associated with T cell-specific proteins. However, transcriptional profiles across all NK cell subsets confirmed highest expression of CD3 complex transcripts CD3D, CD3E, CD3G, CD5, and THEMIS within adaptive NK cells, approximating levels in CD8+ T cells (Figure S7A). Screening 196 healthy donors (65% CMV seropositive) revealed 14 individuals that had NK cells expressing intracellular (i)CD3ε, all of whom were CMV seropositive and displayed adaptive PLZFlow NK cell expansion (Figure 7F). An analysis of protein expression among selected donors revealed that more than 95% of iCD3ε+ NK cells were FcεRγ− (Figure S7B), confirming an adaptive NK cell phenotype. Further, analysis of 12 donors with iCD3ε+ NK cells revealed that expression of iCD3ε, iCD3ẟ, iCD3γ, CD5, and Themis was almost exclusively confined to adaptive PLZFlow NK cells (Figure 7G). Apart from iCD3ẟ and iCD3ε, which were co-expressed (Figure S7C), other T cell-defining markers appeared to be stochastically expressed with t-SNE analysis showing little overlap between cells expressing CD3 isoforms and CD5 or Themis (Figure S7D). Lastly, iCD3ε+ adaptive NK cells expressed greater levels of Bcl11b compared to other adaptive NK cell phenotypes (Figure S7E).
In summary, we identify several unappreciated Bcl11b-regulated, T cell-like phenotypes of adaptive human NK cells that add to the complexity of the adaptive NK cell repertoire. Functional results highlight that adaptive NK cells can be kept in check by LAG-3 while iCD3ε expression correlated with elevated Bcl11b expression.
Bcl11b is required for the adaptive NK cell response in mice
Kinetics of adaptive NK cell expansion and persistence can be readily modelled upon MCMV infection in mice (23). Upon infection, Bcl11b transcription was induced and durably maintained in Ly49H+ NK cells responding to MCMV (Figure 8A). We assessed the contribution of Bcl11b to the generation of adaptive Ly49H+ NK cells in response to MCMV. Wildtype (WT; CD45.1+CD45.2+) and Bcl11b KO (CD45.1−CD45.2+) NK cells were adoptively co-transferred in equal numbers into Ly49H-deficient (Klra8−/−) recipient mice, followed by infection with MCMV and tracking of the transferred NK cell response. Bcl11b-deficient Ly49H+ NK cells were significantly reduced at 7 days post infection (DPI) in comparison to WT NK cells in blood, spleen, and liver (Figure 8B-C). Both Ly49H− and Ly49H+ NK cells proliferate upon MCMV infection (63). Notably, splenic Ly49H− donor Bcl11b KO NK cells were reduced at day 7 following MCMV infection (Figure S8A), indicating that cytokine-driven NK cell proliferation is Bcl11b-dependent. On 28 DPI, Bcl11b-deficient memory NK cells were further diminished compared to WT counterparts, suggesting that Bcl11b is essential for optimal accumulation of adaptive NK cells during MCMV infection. Of the Cd3 genes, only Cd3g was transcribed in adaptive mouse NK cells and was not regulated by Bcl11b. Furthermore, Cd7, Kit, Fcer1g, Themis, Il7r, and Lag3 and Zbtb16 were not differentialy regulated in Bcl11b-deficient in NK cells from infected mice.
Figure 8. Bcl11b is critical to adaptive NK cell responses to MCMV.
(A) Relative transcripts of Bcl11b following RNA-seq performed on mouse Ly49H+ NK cell population throughout the course of murine CMV infection. (B) WT Ncr1+Cre+ CD45.1+CD45.2+ or KO NK cells from Bcl11bF/F Ncr1+Cre+ CD45.1−CD45.2+ were isolated from mouse spleen and adoptively co-transferred in equal numbers into Ly49H-deficient (Klra8−/−) recipient mice. Ly49H+ NK cells were compared at 7 and 28 days post-infection (DPI), n=13 and n=4 mice respectively, Representative flow cytometry plot shown from blood harvested NK cells. (C) Comparison of Ly49H+ transferred cells in blood, spleen and liver. Two-tailed paired Student’s t-test. (D-G) RNA-seq of WT and KO NK cells isolated after adoptively transferred into Ly49H− mice infected with MCMV. Splenic Ly49H+ WT and KO NK cells were sorted for RNA-seq 7 days post infection, n=3 mice. (D) Heatmap of the top 100 DEGs. (E) Gene set enrichment analysis showing enrichment of cell cycle associated genes in WT NK cells. (F) Heatmap of selected NK receptors or effector genes (G) that were dependent on Bcl11b. FDR-adjusted P values were calculated using the Benjamini-Hochberg method, significance cutoff adjusted to p < 0.10.
To investigate Bcl11b regulated pathways that control the murine adaptive NK cell response, we conducted RNA-seq on sort-purified WT and Bcl11b-deficient NK cells from the spleens of mice co-transferred and MCMV-infected (Figure 8D). The majority DE genes (96%; FDR < 0.1) were downregulated in the absence of Bcl11b, suggesting that Bcl11b predominantly acts as a transcriptional activator upon murine NK cell expansion. Gene set enrichment analysis (GSEA) revealed enrichment of pathways associated with cell cycle in WT NK cells (Figure 8E, S8B). These results suggest that Bcl11b may be essential for NK cell proliferation during MCMV infection. In addition, several NK receptors were down-regulated in the absence of Bcl11b, including Klrb1 (NK1.1), Klrb1b, Klrc1 (NKG2A), Klra10 (Ly49J), Klra1 (Ly49B), Klri2 and Klra3 (Ly49C) (Figure 8F). This was surprising because removal of Bcl11b in multiple T cell populations represses several of these genes, including in mature T cells (42, 44). Furthermore, genes indicative of NK cell differentiation and effector function, including Klrg1, Ly6c1, Gzma, Gzmb, and Prf1 (64), were significantly down-regulated in the absence of Bcl11b (Figure 8G). These data suggest that Bcl11b sustains a transcriptional program that promotes NK cell receptor expression, effector function and expansion in response to viral infection.
Together, we find that loss of Bcl11b substantially impacts the generation and durablity of mouse adaptive NK cells, suggesting an important role of Bcl11b in promoting adaptive NK cell differentiation.
DISCUSSION
NK cells share many common features with T cells (17, 65), where Bcl11b is critical for T cell development and differentiation including effector CD8+ T cell expansion, memory formation and cytotoxic function (32, 66). However, Bcl11b has not previously been implicated in NK cell differentiation. By assessing distinct human NK cell subsets, we identified a gradient of BCL11B transcription throughout differentiation. This was reflected in pronounced epigenetic modification of the BCL11B locus as well as regulation of Bcl11b target genes. We found that Bcl11b directly promotes human canonical CD56dim NK cell differentiation and correlates with suppression of markers that define adaptive NK cells while inducing T cell associated proteins. A critical requirement for Bcl11b was also found in mouse adaptive NK cell responses, revealing a conserved role of Bcl11b in cytotoxic lymphocytes’ adaptive responses.
Bcl11b is a Kruppel-like C2H2-type zinc finger TF (67). Knock-out mice, which die shortly after birth, uncovered an essential role for Bcl11b in the development of αβ T cells and neuronal corticospinal motor neurons (68, 69). Bcl11b is also active in ILC precursors committed to the ILC2 lineage and is critical for ILC2 development (45-48, 70) as well as iNKT cell development (71, 72). In pro-T cells, Bcl11b represses Id2 to suppress ILC fate. However, Bcl11b can promote ILC2 commitment by occupying different cell type-specific binding motifs (73). Our data indicate that Bcl11b also promotes human NK cell differentiation, as patients with heterozygous BCL11B mutations displayed impaired early NK cell maturation and abrogated CD57 expression. The heterozygous BCL11B mutations were associated with a spectrum of clinical phenotypes of varying severity, with one SCID-like patient lacking both T cells and CD16+ NK cells, resembling the phenotype of Bcl11b−/− mice. At the other end of the spectrum, one patient exhibited relatively normal T and NK cell composition. Overall, the majority (3/5) of patients with BCL11B mutation had approximately 1000 T cells/μl but highly elevated CD56bright NK cell frequencies, much more so than Omenn syndrome or atypical SCID patients that retain similar T cell numbers (74). Furthermore, ectopic expression of Bcl11b in CD56bright NK cells drove NK cell maturation by suppressing NKG2A and inducing CD57 and granzyme B expression (32). Given the difficulty in inducing mature canonical NK cells in vitro (75), ectopic expression of Bcl11b appears to be a pivotal factor in directing human NK cell differentiation. Augmenting Bcl11b expression may be beneficial in designing durable NK cell-mediated immunotherapeutic strategies.
While several lines of data support an important role for BCL11B in early human NK cell differentiation, experiments did not reveal a requirement for Bcl11b in differentiation of canonical CD11b-expressing mouse NK cells. Although transient expression of Bcl11b in immature mouse NK cells has been noted (41), Bcl11b transcription did not appreciably increase with mature mouse NK cell differentiation, in agreement with early NK cell development appearing intact in Bcl11b-deficient mice (41). This suggests regulatory differences between human and mouse canonical NK cell differentiation. Supporting this, comparison of regulatory regions shared between human and mouse NK cells revealed high conservation of CD56bright and murine CD27+CD11b− cells, but far less overlap between CD56dim and CD27−CD11b+ cells (28). Similarly, scRNA-seq of human and mouse NK cells indicates the CD56bright / CD56dim distinction resembles separation of CD27+CD11b− versus CD27−CD11b+ cells in mice, however overall there are relatively few common orthologues (76). These differences may reflect the pathogen-defined environment of laboratory mice attenuating inflammatory priming cues required for NK cell development and Bcl11b activity resembling that found in humans. Alternatively, different Bcl11b binding partners account for human and mouse differences in canonical NK cell differentiation, as previously observed in cell-type specific binding patterns in early T-lineage and ILC2 cells (73).
While canonical NK cell differentiation between humans and mice appears divergent, Bcl11b appeared critical in generating adaptive NK cell responses to CMV. In both humans and mice, BCL11B was most highly transcribed in adaptive NK cells. Adoptive transfer experiments in mice demonstrated a requirement for Bcl11b in adaptive NK cell expansion. Analyses indicated that Bcl11b acts as a transcriptional activator promoting proliferation and differentiation. Notably, Bcl11b regulated mouse NK cell expression of lectin-like receptors, encoded by the natural killer complex (NKC), which is suppressed by Bcl11b during T cell development and in mature T cells (41, 42, 44). As ectopic expression of Bcl11b in human CD56bright NK cells suppressed NKG2A expression, these data suggest that Bcl11b may play an important role in orchestrating dynamic regulation of the NKC locus during NK cell differentiation. Adaptive human NK cells are defined by silencing of ZBTB16 and exhibit stochastic loss of multiple NK cell signaling molecules including FCER1G, SH2D1B2 (EAT-2) and SYK. In a mouse T cell development model Zbtb16 (PLZF), Fcer1g and Sh2d1b2 were suppressed by Bcl11b (31). Moreover, our ChIP-seq experiments identified Bcl11b-targeted genes ZBTB16, FCER1G and SYK that suggests Bcl11b may directly drive human adaptive NK cell phenotypes. We further noted preferential Bcl11b binding at these loci in adaptive relative to conventional NK cells. These genes were not repressed during formation of murine adaptive NK cells, consistent with previous observations (16). Fetal NK cells express iCD3ε, iCD3ẟ, and iCD3γ, whereas adult NK cells can have low levels of iCD3ε that is increased upon activation (77, 78). However, the preferential acquisition of T cell signaling molecules in adaptive NK cells has not been reported. We identified subsets of adaptive NK cells expressing Bcl11b-regulated T cell signaling components, most frequently iCD3ε. We find interesting parallels between the function of Bcl11b in T cells and NK cells. In mice, Bcl11b is required for the antigen-driven expansion and function of mature CD8+ T cells (32, 66). Likewise, we find deficient adaptive NK cell responses in our murine model at both day 7 and day 28, suggesting defective proliferation at the early phase of the MCMV response. As such, our data suggest a conserved requirement for Bcl11b in memory responses by cytotoxic lymphocytes.
A blueprint of NK cell development has been suggested as progressing from CD56brightCD16−CD57− (Stage 4b), to CD56dimCD16+CD57− (Stage 5), to CD56dimCD16+CD57+ (Stage 6) (7). However, rather than CD57, our data highlight a profound transcriptional and epigenetic distinction between CD56dimCD16+ NK cells in terms of NKG2A expression. Differentiation of early CD56dimNKG2A+ NK cells coincided with loss of promoter accessibility and more complex AE compositions, possibly reflecting greater TF occupancy. As such, NKG2A expression defines a continuum of transcriptionally intermediate canonical CD56dim NK cells. Furthermore, although self-KIR expression enhances NK cell function, we found no epigenetic imprint of educating KIR.
Recently, Colonna and colleagues epigenetically assessed ILC subsets including peripheral blood CD56bright as well as two CD56dim NK cell subsets. These results implicated a TCF-7-LEF1-Myc TF axis in maintaining CD56bright NK cells, with Bach2 suppression of PRDM1 and ZEB2 defining CD56dim NK cells (28). In addition, enrichment of Runx2 expression was noted for CD56bright NK cells, although to what extent Runx2 integrated with other TFs remained undefined. Our data display concordance with this data with respect to CD56bright NK cells and confirm apical Runx2 protein expression in CD56bright but also in a small subset of early CD56dim NK cells that were CD62Lhi, representing a transitional population. We found that loss of Runx2 expression coincided with suppression of MYC, BACH2, and TCF7 promoter and enhancer accessibility, with ChIP-seq confirming these as Runx2 target genes as well as LEF1, ZEB1, and AHR. This was further illustrated through de novo TF networks, which positioned RUNX2 as an instrumental factor in CD56bright cell regulation. Taken together, Runx2 may represent a major hub in maintaining CD56bright NK cell identity.
In summary, our data delineate changes in chromatin accessibility throughout human peripheral blood NK cell differentiation, uncovering networks of TF regulation. Several lines of evidence revealed a crucial role for Bcl11b in NK cell differentiation, with a gradient of expression defining this trajectory from CD56bright through to adaptive NK cells.
MATERIALS & METHODS
Study design
The purpose of this study was to determine epigenetic and transcriptional landscapes of peripheral human NK cell subsets with reference to cytotoxic CD8+ T cells and identify TFs that direct NK cell differentiation. Validation of TF expression was performed by flow cytometry and functional assessment of Bcl11b used multiple lines of evidence including Bcl11b ChIP-seq, in vitro transduction experiments and knock-out mouse models. The same donors were used for RNA-seq, ATAC-seq and AE ChIP-seq interrogation as indicated in Figures 1-2. Healthy human donors were selected with haplotype A KIR and adaptive NK cell populations. Data was not excluded from the study. The study was designed to have high statistical confidence in calling transcriptional and chromatin accessibility differences using 4-5 donors in each experiment. Active enhancer analysis required larger input cell numbers and was therefore limited to 2 donors, hence calling differential epigenetic elements typically combined chromatin accessibility and AE datasets. Experimental study numbers were otherwise not predetermined. Replicates and statistical tests are provided in the figure legends. Mouse MCMV transfer experiments used well-described previously published endpoints. Experimenters were not blinded to BCL11B patient or mouse genotype status upon phenotyping analysis.
Cells and patients
Studies were approved by The Regional Ethical Review Boards in Stockholm, Ethics Commission of Medical Association in Hamburg (Ärztekammer Hamburg), and the The Cincinnati Children's Hospital Institutional Review Board. Healthy blood donors were recruited from Karolinska University Hospital blood banks. Patients were recruited with informed written consent. Blood samples were screened using flow cytometry for adaptive NK cell populations. Peripheral blood mononuclear cells (PBMC) were isolated from selected donor buffy coats by density gradient centrifugation and cryopreserved in liquid nitrogen. Donors were genotyped for KIR and KIR-ligands using genotyping KIR SSO Luminex (One Lambda). Heterozygous BCL11B mutated patients are described in Table 2.
Fluorescent activated cell sorting
Cryopreserved PBMC were thawed, washed in RPMI with 10% fetal bovine serum (FBS) and enriched for either NK cells or CD8+ T cells using negative magnetic selection (NK cell depletion kit or CD8+ T cell depletion kit; Miltenyi Biotech). Purified NK cells were fluorescent activated cell sorted (FACS) on basis of CD14−CD19−DCM−CD3−CD56+ lymphocytes (DCM: dead cell marker). T cells were labeled independently and gated as CD14−CD19−DCM−CD3+CD8+ lymphocytes. Fluorochrome-conjugated antibodies (Table S3) were incubated for 30 mins on ice, washed twice in PBS with 2 mM EDTA and 1% bovine serum albumin and sorted using an ARIA II SORP instrument (BD Biosciences). The purity of specific cell subsets was >98%, as determined by flow cytometry.
Flow cytometry
Antibodies and reagents are specified in Table S3. For phenotypic analyses, PBMCs were surface-stained with indicated antibodies and a fixable dead cell stain (ThermoFisher) in FACS buffer (PBS supplemented with 2% FBS and 2 mM EDTA). Cells were thereafter fixed in 2% formaldehyde (Pierce) in PBS, permeabilized in 0.05% Triton X-100 (Sigma-Aldrich) in PBS and stained intracellularly in FACS buffer. For quantification of LAG-3 induction, cryopreserved PBMCs were thawed, cultured overnight in complete medium (RPMI1640 with Glutamax [Gibco] and 10% fetal bovine serum), then stimulated with plate-bound monoclonal antibodies for 24 hours in the presence of 10 ng/ml IL-15 (Peprotech). Coating of antibodies was performed for 24 hours at 4°C in with 5 μg/ml antibody in PBS with 0.1 M NaHCO3 (Sigma-Aldrich), followed by two washes with PBS and a blocking step at 37°C for 20 minutes using complete medium. Samples were acquired on an LSRFortessa or FACSymphony A5 instrument (BD Biosciences) and analyzed with FlowJo software (v9.9.6 or v10.2, BD Biosciences).
RNA-seq
Approximately 2×104 cells were sorted directly into 300 μl Trizol LS reagent (ThermoFisher Scientific) and purified with Direct-zol columns (Zymo) with on-column DNase I treatment. Ribosomal RNA was removed using RiboMinus (ThermoFisher Scientific) diluted 10-fold, followed by repurification of RNA on concentrator columns (Zymo). Libraries were prepared from ribosomal depleted RNA using CAT-seq total RNA kit (Diagenode). Samples were purified with nucleotide-binding beads (AMPure, Beckman Coulter) and assessed on a bioanalyzer with high-sensitivity chips and quantified using Qubit fluorescence. Pooled libraries were run on a NextSeq instrument (Illumina) acquiring 40 bp paired-end reads.
ATAC-seq
Between 20,000-40,000 cells were FACS sorted into PBS with 2% bovine serum albumin and subjected to OmniATAC-seq as previously described (49). Briefly, cells were pelleted and permeabilized on ice for 3 minutes in resuspension buffer (RSB: 0.1% Tween-20, 10 mM Tris-HCl, 10 mM NaCl, 3 mM MgCl2) supplemented with 0.1% NP-40 (Sigma-Aldrich) and 0.01% Digitonin (Sigma-Aldrich) detergents. Cells were washed in cold RSB and then transposed using Nextera Tn5 kit (Illumina) supplemented with 0.1% NP-40, and 0.01% Digitonin for 30 minutes at 37°C. DNA was purified on columns (Zymo), amplified using barcoded PCR primers for 9-11 cycles and purified 2-3 times using AMPure beads. DNA libraries were assessed on a bioanalyzer and Qubit spectrometer. Pooled libraries were run on a NextSeq instrument (Illumina) acquiring 40 bp paired-end reads.
Statistical Analysis
Statistics were calculated using either GraphPad Prism (v7.0) or in R base and R package edgeR (v 3.22.3). Statistical methods are given in figure legends and raw data tables. P-values are denoted by *, **, and *** representing p < 0.05, p < 0.01, and p < 0.001, respectively.
Supplementary Material
Figure S1. Sorting strategies and phenotype validation
Figure S2. DA ATAC-seq peak distribution and clustering
Figure S3. DB H3K27ac peaks and trace profiles
Figure S4. Flow cytometric TF staining in NK cells
Figure S5. TF ChIP-seq peak traces and ATAC-seq overlap
Figure S6. Bcl11b regulates canonical NK cell differentiation
Figure S7. T-cell associated phenotypes of adaptive NK cells
Figure S8. GSEA in Bcl11b−/− mice following MCMV infection
Table S2. Summary of TF ChIP-seq peak intensities
Table S3. List of antibodies and fluorochromes
Table S1. Raw data
ACKNOWLEDGEMENTS
We thank Dr. Davor Lessel (Human Genetics, UKE) for recruiting patients with BCL11B mutations and the MedH Flow Cytometry core facility (Karolinska Institutet) for cell analysis and sorting.
FUNDING
This work was supported by the Swedish Research Council, Swedish Foundation for Strategic Research, Swedish Cancer Foundation, Swedish Children's Cancer Foundation, Knut and Alice Wallenberg Foundation, the Karolinska Institute Research Foundation, the Frontiers in Biomedical Research Fellowship to Y.T.B and NIH grants K99/R00 HL123638 to F.C., NIH R01AI067846 to D.A., NIH 2T32DK074367 to T.T.D.
NIH grant K99/R00 HL123638 to F.C.
Footnotes
Competing interests
H.S. and Y.T.B. are inventors on patent WO2017048809 held by the University of Minnesota that covers preparation and clinical use of adapative NK cells.
Data and materials availability
Human RNA-seq, ATAC-seq, and ChIP-seq data are deposited under EGA EGAS00001005025. Murine RNA-seq data is deposited under GEO Accession number GSE162472. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
REFERENCES
- 1.Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, Yokoyama WM, Ugolini S, Innate or adaptive immunity? The example of natural killer cells. Science 331, 44–49 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie AN, Mebius RE, Powrie F, Vivier E, Innate lymphoid cells--a proposal for uniform nomenclature. Nature reviews. Immunology 13, 145–149 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Lanier LL, Evolutionary struggles between NK cells and viruses. Nat Rev Immunol 8, 259–268 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parham P, Moffett A, Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nature reviews. Immunology 13, 133–144 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cannon MJ, Congenital cytomegalovirus (CMV) epidemiology and awareness. J Clin Virol 46 Suppl 4, S6–10 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Cannon MJ, Schmid DS, Hyde TB, Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev Med Virol 20, 202–213 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Freud AG, Mundy-Bosse BL, Yu J, Caligiuri MA, The Broad Spectrum of Human Natural Killer Cell Diversity. Immunity 47, 820–833 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baume DM, Robertson MJ, Levine H, Manley TJ, Schow PW, Ritz J, Differential responses to interleukin 2 define functionally distinct subsets of human natural killer cells. Eur J Immunol 22, 1–6 (1992). [DOI] [PubMed] [Google Scholar]
- 9.Cooper MA, Fehniger TA, Turner SC, Chen KS, Ghaheri BA, Ghayur T, Carson WE, Caligiuri MA, Human natural killer cells: a unique innate immunoregulatory role for the CD56(bright) subset. Blood 97, 3146–3151 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Bjorkstrom NK, Riese P, Heuts F, Andersson S, Fauriat C, Ivarsson MA, Bjorklund AT, Flodstrom-Tullberg M, Michaelsson J, Rottenberg ME, Guzman CA, Ljunggren HG, Malmberg KJ, Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood 116, 3853–3864 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Lopez-Verges S, Milush JM, Pandey S, York VA, Arakawa-Hoyt J, Pircher H, Norris PJ, Nixon DF, Lanier LL, CD57 defines a functionally distinct population of mature NK cells in the human CD56dimCD16+ NK-cell subset. Blood 116, 3865–3874 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Juelke K, Killig M, Luetke-Eversloh M, Parente E, Gruen J, Morandi B, Ferlazzo G, Thiel A, Schmitt-Knosalla I, Romagnani C, CD62L expression identifies a unique subset of polyfunctional CD56dim NK cells. Blood 116, 1299–1307 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Yu J, Mao HC, Wei M, Hughes T, Zhang J, Park IK, Liu S, McClory S, Marcucci G, Trotta R, Caligiuri MA, CD94 surface density identifies a functional intermediary between the CD56bright and CD56dim human NK-cell subsets. Blood 115, 274–281 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anfossi N, Andre P, Guia S, Falk CS, Roetynck S, Stewart CA, Breso V, Frassati C, Reviron D, Middleton D, Romagne F, Ugolini S, Vivier E, Human NK cell education by inhibitory receptors for MHC class I. Immunity 25, 331–342 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Guma M, Angulo A, Vilches C, Gomez-Lozano N, Malats N, Lopez-Botet M, Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood 104, 3664–3671 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Lee J, Zhang T, Hwang I, Kim A, Nitschke L, Kim M, Scott JM, Kamimura Y, Lanier LL, Kim S, Epigenetic Modification and Antibody-Dependent Expansion of Memory-like NK Cells in Human Cytomegalovirus-Infected Individuals. Immunity 42, 431–442 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schlums H, Cichocki F, Tesi B, Theorell J, Beziat V, Holmes TD, Han H, Chiang SC, Foley B, Mattsson K, Larsson S, Schaffer M, Malmberg KJ, Ljunggren HG, Miller JS, Bryceson YT, Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity 42, 443–456 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun JC, Beilke JN, Lanier LL, Adaptive immune features of natural killer cells. Nature 457, 557–561 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cichocki F, Cooley S, Davis Z, DeFor TE, Schlums H, Zhang B, Brunstein CG, Blazar BR, Wagner J, Diamond DJ, Verneris MR, Bryceson YT, Weisdorf DJ, Miller JS, CD56(dim)CD57(+)NKG2C(+) NK cell expansion is associated with reduced leukemia relapse after reduced intensity HCT. Leukemia 30, 456–463 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cichocki F, Wu CY, Zhang B, Felices M, Tesi B, Tuininga K, Dougherty P, Taras E, Hinderlie P, Blazar BR, Bryceson YT, Miller JS, ARID5B regulates metabolic programming in human adaptive NK cells. J Exp Med 215, 2379–2395 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlums H, Jung M, Han H, Theorell J, Bigley V, Chiang SC, Allan DS, Davidson-Moncada JK, Dickinson RE, Holmes TD, Hsu AP, Townsley D, Winkler T, Wang W, Aukrust P, Nordoy I, Calvo KR, Holland SM, Collin M, Dunbar CE, Bryceson YT, Adaptive NK cells can persist in patients with GATA2 mutation depleted of stem and progenitor cells. Blood 129, 1927–1939 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corat MA, Schlums H, Wu C, Theorell J, Espinoza DA, Sellers SE, Townsley DM, Young NS, Bryceson YT, Dunbar CE, Winkler T, Acquired somatic mutations in PNH reveal long-term maintenance of adaptive NK cells independent of HSPCs. Blood 129, 1940–1946 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lau CM, Adams NM, Geary CD, Weizman OE, Rapp M, Pritykin Y, Leslie CS, Sun JC, Epigenetic control of innate and adaptive immune memory. Nat Immunol 19, 963–972 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gury-BenAri M, Thaiss CA, Serafini N, Winter DR, Giladi A, Lara-Astiaso D, Levy M, Salame TM, Weiner A, David E, Shapiro H, Dori-Bachash M, Pevsner-Fischer M, Lorenzo-Vivas E, Keren-Shaul H, Paul F, Harmelin A, Eberl G, Itzkovitz S, Tanay A, Di Santo JP, Elinav E, Amit I, The Spectrum and Regulatory Landscape of Intestinal Innate Lymphoid Cells Are Shaped by the Microbiome. Cell 166, 1231–1246 e1213 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Koues OI, Collins PL, Cella M, Robinette ML, Porter SI, Pyfrom SC, Payton JE, Colonna M, Oltz EM, Distinct Gene Regulatory Pathways for Human Innate versus Adaptive Lymphoid Cells. Cell 165, 1134–1146 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shih HY, Sciume G, Mikami Y, Guo L, Sun HW, Brooks SR, Urban JF Jr., Davis FP, Kanno Y, O'Shea JJ, Developmental Acquisition of Regulomes Underlies Innate Lymphoid Cell Functionality. Cell 165, 1120–1133 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pokrovskii M, Hall JA, Ochayon DE, Yi R, Chaimowitz NS, Seelamneni H, Carriero N, Watters A, Waggoner SN, Littman DR, Bonneau R, Miraldi ER, Characterization of Transcriptional Regulatory Networks that Promote and Restrict Identities and Functions of Intestinal Innate Lymphoid Cells. Immunity 51, 185–197 e186 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collins PL, Cella M, Porter SI, Li S, Gurewitz GL, Hong HS, Johnson RP, Oltz EM, Colonna M, Gene Regulatory Programs Conferring Phenotypic Identities to Human NK Cells. Cell 176, 348–360 e312 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cichocki F, Grzywacz B, Miller JS, Human NK Cell Development: One Road or Many? Front Immunol 10, 2078 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saliba AE, Westermann AJ, Gorski SA, Vogel J, Single-cell RNA-seq: advances and future challenges. Nucleic Acids Res 42, 8845–8860 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hosokawa H, Romero-Wolf M, Yui MA, Ungerback J, Quiloan MLG, Matsumoto M, Nakayama KI, Tanaka T, Rothenberg EV, Bcl11b sets pro-T cell fate by site-specific cofactor recruitment and by repressing Id2 and Zbtb16. Nat Immunol 19, 1427–1440 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang S, Rozell M, Verma RK, Albu DI, Califano D, VanValkenburgh J, Merchant A, Rangel-Moreno J, Randall TD, Jenkins NA, Copeland NG, Liu P, Avram D, Antigen-specific clonal expansion and cytolytic effector function of CD8+ T lymphocytes depend on the transcription factor Bcl11b. J Exp Med 207, 1687–1699 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Helden MJ, Goossens S, Daussy C, Mathieu AL, Faure F, Marcais A, Vandamme N, Farla N, Mayol K, Viel S, Degouve S, Debien E, Seuntjens E, Conidi A, Chaix J, Mangeot P, de Bernard S, Buffat L, Haigh JJ, Huylebroeck D, Lambrecht BN, Berx G, Walzer T, Terminal NK cell maturation is controlled by concerted actions of T-bet and Zeb2 and is essential for melanoma rejection. J Exp Med 212, 2015–2025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holmes ML, Huntington ND, Thong RP, Brady J, Hayakawa Y, Andoniou CE, Fleming P, Shi W, Smyth GK, Degli-Esposti MA, Belz GT, Kallies A, Carotta S, Smyth MJ, Nutt SL, Peripheral natural killer cell maturation depends on the transcription factor Aiolos. EMBO J 33, 2721–2734 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gascoyne DM, Long E, Veiga-Fernandes H, de Boer J, Williams O, Seddon B, Coles M, Kioussis D, Brady HJ, The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat Immunol 10, 1118–1124 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Kamizono S, Duncan GS, Seidel MG, Morimoto A, Hamada K, Grosveld G, Akashi K, Lind EF, Haight JP, Ohashi PS, Look AT, Mak TW, Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. The Journal of experimental medicine 206, 2977–2986 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mackay LK, Minnich M, Kragten NA, Liao Y, Nota B, Seillet C, Zaid A, Man K, Preston S, Freestone D, Braun A, Wynne-Jones E, Behr FM, Stark R, Pellicci DG, Godfrey DI, Belz GT, Pellegrini M, Gebhardt T, Busslinger M, Shi W, Carbone FR, van Lier RA, Kallies A, van Gisbergen KP, Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 352, 459–463 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Bradley T, Peppa D, Pedroza-Pacheco I, Li D, Cain DW, Henao R, Venkat V, Hora B, Chen Y, Vandergrift NA, Overman RG, Edwards RW, Woods CW, Tomaras GD, Ferrari G, Ginsburg GS, Connors M, Cohen MS, Moody MA, Borrow P, Haynes BF, RAB11FIP5 Expression and Altered Natural Killer Cell Function Are Associated with Induction of HIV Broadly Neutralizing Antibody Responses. Cell 175, 387–399 e317 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ikawa T, Hirose S, Masuda K, Kakugawa K, Satoh R, Shibano-Satoh A, Kominami R, Katsura Y, Kawamoto H, An essential developmental checkpoint for production of the T cell lineage. Science 329, 93–96 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Li L, Leid M, Rothenberg EV, An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science 329, 89–93 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li P, Burke S, Wang J, Chen X, Ortiz M, Lee SC, Lu D, Campos L, Goulding D, Ng BL, Dougan G, Huntly B, Gottgens B, Jenkins NA, Copeland NG, Colucci F, Liu P, Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science 329, 85–89 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drashansky TT, Helm E, Huo Z, Curkovic N, Kumar P, Luo X, Parthasarathy U, Zuniga A, Cho JJ, Lorentsen KJ, Xu Z, Uddin M, Moshkani S, Zhou L, Avram D, Bcl11b prevents fatal autoimmunity by promoting Treg cell program and constraining innate lineages in Treg cells. Sci Adv 5, eaaw0480 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Avram D, Califano D, The multifaceted roles of Bcl11b in thymic and peripheral T cells: impact on immune diseases. J Immunol 193, 2059–2065 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lorentsen KJ, Cho JJ, Luo X, Zuniga AN, Urban JF Jr., Zhou L, Gharaibeh R, Jobin C, Kladde MP, Avram D, Bcl11b is essential for licensing Th2 differentiation during helminth infection and allergic asthma. Nat Commun 9, 1679 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Califano D, Cho JJ, Uddin MN, Lorentsen KJ, Yang Q, Bhandoola A, Li H, Avram D, Transcription Factor Bcl11b Controls Identity and Function of Mature Type 2 Innate Lymphoid Cells. Immunity 43, 354–368 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walker JA, Oliphant CJ, Englezakis A, Yu Y, Clare S, Rodewald HR, Belz G, Liu P, Fallon PG, McKenzie AN, Bcl11b is essential for group 2 innate lymphoid cell development. J Exp Med 212, 875–882 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu Y, Wang C, Clare S, Wang J, Lee SC, Brandt C, Burke S, Lu L, He D, Jenkins NA, Copeland NG, Dougan G, Liu P, The transcription factor Bcl11b is specifically expressed in group 2 innate lymphoid cells and is essential for their development. J Exp Med 212, 865–874 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu Y, Tsang JC, Wang C, Clare S, Wang J, Chen X, Brandt C, Kane L, Campos LS, Lu L, Belz GT, McKenzie AN, Teichmann SA, Dougan G, Liu P, Single-cell RNA-seq identifies a PD-1(hi) ILC progenitor and defines its development pathway. Nature 539, 102–106 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, Satpathy AT, Rubin AJ, Montine KS, Wu B, Kathiria A, Cho SW, Mumbach MR, Carter AC, Kasowski M, Orloff LA, Risca VI, Kundaje A, Khavari PA, Montine TJ, Greenleaf WJ, Chang HY, An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods 14, 959–962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zook EC, Li ZY, Xu Y, de Pooter RF, Verykokakis M, Beaulieu A, Lasorella A, Maienschein-Cline M, Sun JC, Sigvardsson M, Kee BL, Transcription factor ID2 prevents E proteins from enforcing a naive T lymphocyte gene program during NK cell development. Sci Immunol 3, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guia S, Jaeger BN, Piatek S, Mailfert S, Trombik T, Fenis A, Chevrier N, Walzer T, Kerdiles YM, Marguet D, Vivier E, Ugolini S, Confinement of activating receptors at the plasma membrane controls natural killer cell tolerance. Science signaling 4, ra21 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Goodridge JP, Jacobs B, Saetersmoen ML, Clement D, Hammer Q, Clancy T, Skarpen E, Brech A, Landskron J, Grimm C, Pfefferle A, Meza-Zepeda L, Lorenz S, Wiiger MT, Louch WE, Ask EH, Liu LL, Oei VYS, Kjallquist U, Linnarsson S, Patel S, Tasken K, Stenmark H, Malmberg KJ, Remodeling of secretory lysosomes during education tunes functional potential in NK cells. Nat Commun 10, 514 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B, Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA, Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA, Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Golling G, Li L, Pepling M, Stebbins M, Gergen JP, Drosophila homologs of the proto-oncogene product PEBP2/CBF beta regulate the DNA-binding properties of Runt. Mol Cell Biol 16, 932–942 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Avram D, Fields A, Senawong T, Topark-Ngarm A, Leid M, COUP-TF (chicken ovalbumin upstream promoter transcription factor)-interacting protein 1 (CTIP1) is a sequence-specific DNA binding protein. Biochem J 368, 555–563 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Punwani D, Zhang Y, Yu J, Cowan MJ, Rana S, Kwan A, Adhikari AN, Lizama CO, Mendelsohn BA, Fahl SP, Chellappan A, Srinivasan R, Brenner SE, Wiest DL, Puck JM, Multisystem Anomalies in Severe Combined Immunodeficiency with Mutant BCL11B. N Engl J Med 375, 2165–2176 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lessel D, Gehbauer C, Bramswig NC, Schluth-Bolard C, Venkataramanappa S, van Gassen KLI, Hempel M, Haack TB, Baresic A, Genetti CA, Funari MFA, Lessel I, Kuhlmann L, Simon R, Liu P, Denecke J, Kuechler A, de Kruijff I, Shoukier M, Lek M, Mullen T, Ludecke HJ, Lerario AM, Kobbe R, Krieger T, Demeer B, Lebrun M, Keren B, Nava C, Buratti J, Afenjar A, Shinawi M, Guillen Sacoto MJ, Gauthier J, Hamdan FF, Laberge AM, Campeau PM, Louie RJ, Cathey SS, Prinz I, Jorge AAL, Terhal PA, Lenhard B, Wieczorek D, Strom TM, Agrawal PB, Britsch S, Tolosa E, Kubisch C, BCL11B mutations in patients affected by a neurodevelopmental disorder with reduced type 2 innate lymphoid cells. Brain 141, 2299–2311 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Crinier A, Milpied P, Escaliere B, Piperoglou C, Galluso J, Balsamo A, Spinelli L, Cervera-Marzal I, Ebbo M, Girard-Madoux M, Jaeger S, Bollon E, Hamed S, Hardwigsen J, Ugolini S, Vely F, Narni-Mancinelli E, Vivier E, High-Dimensional Single-Cell Analysis Identifies Organ-Specific Signatures and Conserved NK Cell Subsets in Humans and Mice. Immunity 49, 971–986 e975 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Woo SR, Turnis ME, Goldberg MV, Bankoti J, Selby M, Nirschl CJ, Bettini ML, Gravano DM, Vogel P, Liu CL, Tangsombatvisit S, Grosso JF, Netto G, Smeltzer MP, Chaux A, Utz PJ, Workman CJ, Pardoll DM, Korman AJ, Drake CG, Vignali DA, Immune inhibitory molecules LAG-3 and PD-1 synergistically regulate T-cell function to promote tumoral immune escape. Cancer Res 72, 917–927 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Merino A, Zhang B, Dougherty P, Luo X, Wang J, Blazar BR, Miller JS, Cichocki F, Chronic stimulation drives human NK cell dysfunction and epigenetic reprograming. J Clin Invest 130, 3770–3785 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dokun AO, Kim S, Smith HR, Kang HS, Chu DT, Yokoyama WM, Specific and nonspecific NK cell activation during virus infection. Nature immunology 2, 951–956 (2001). [DOI] [PubMed] [Google Scholar]
- 64.Bezman NA, Kim CC, Sun JC, Min-Oo G, Hendricks DW, Kamimura Y, Best JA, Goldrath AW, Lanier LL, ImmGen Report: Molecular definition of Natural Killer cell identity and activation. Nat Immunol 13, 1000–1009 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bezman NA, Kim CC, Sun JC, Min-Oo G, Hendricks DW, Kamimura Y, Best JA, Goldrath AW, Lanier LL, Gautier EL, Jakubzick C, Randolph GJ, Best AJ, Knell J, Goldrath A, Miller J, Brown B, Merad M, Jojic V, Koller D, Cohen N, Brennan P, Brenner M, Shay T, Regev A, Fletcher A, Elpek K, Bellemare-Pelletier A, Malhotra D, Turley S, Jianu R, Laidlaw D, Collins JJ, Narayan K, Sylvia K, Kang J, Gazit R, Rossi DJ, Kim F, Rao TN, Wagers A, Shinton SA, Hardy RR, Monach P, Heng T, Kreslavsky T, Painter M, Ericson J, Davis S, Mathis D, Benoist C, Molecular definition of the identity and activation of natural killer cells. Nature immunology 13, 1000–1009 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abboud G, Stanfield J, Tahiliani V, Desai P, Hutchinson TE, Lorentsen KJ, Cho JJ, Avram D, Salek-Ardakani S, Transcription Factor Bcl11b Controls Effector and Memory CD8 T cell Fate Decision and Function during Poxvirus Infection. Front Immunol 7, 425 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Avram D, Fields A, Pretty On Top K, Nevrivy DJ, Ishmael JE, Leid M, Isolation of a novel family of C(2)H(2) zinc finger proteins implicated in transcriptional repression mediated by chicken ovalbumin upstream promoter transcription factor (COUP-TF) orphan nuclear receptors. J Biol Chem 275, 10315–10322 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wakabayashi Y, Watanabe H, Inoue J, Takeda N, Sakata J, Mishima Y, Hitomi J, Yamamoto T, Utsuyama M, Niwa O, Aizawa S, Kominami R, Bcl11b is required for differentiation and survival of alphabeta T lymphocytes. Nat Immunol 4, 533–539 (2003). [DOI] [PubMed] [Google Scholar]
- 69.Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD, Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron 45, 207–221 (2005). [DOI] [PubMed] [Google Scholar]
- 70.Harly C, Cam M, Kaye J, Bhandoola A, Development and differentiation of early innate lymphoid progenitors. J Exp Med 215, 249–262 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Albu DI, VanValkenburgh J, Morin N, Califano D, Jenkins NA, Copeland NG, Liu P, Avram D, Transcription factor Bcl11b controls selection of invariant natural killer T-cells by regulating glycolipid presentation in double-positive thymocytes. Proc Natl Acad Sci U S A 108, 6211–6216 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Uddin MN, Sultana DA, Lorentsen KJ, Cho JJ, Kirst ME, Brantly ML, Califano D, Sant’Angelo DB, Avram D, Transcription factor Bcl11b sustains iNKT1 and iNKT2 cell programs, restricts iNKT17 cell program, and governs iNKT cell survival. Proc Natl Acad Sci U S A 113, 7608–7613 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hosokawa H, Romero-Wolf M, Yang Q, Motomura Y, Levanon D, Groner Y, Moro K, Tanaka T, Rothenberg EV, Cell type-specific actions of Bcl11b in early T-lineage and group 2 innate lymphoid cells. J Exp Med 217, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dobbs K, Tabellini G, Calzoni E, Patrizi O, Martinez P, Giliani SC, Moratto D, Al-Herz W, Cancrini C, Cowan M, Bleesing J, Booth C, Buchbinder D, Burns SO, Chatila TA, Chou J, Daza-Cajigal V, Ott de Bruin LM, de la Morena M, Di Matteo G, Finocchi A, Geha R, Goyal RK, Hayward A, Holland S, Huang CH, Kanariou MG, King A, Kaplan B, Kleva A, Kuijpers TW, Lee BW, Lougaris V, Massaad M, Meyts I, Morsheimer M, Neven B, Pai SY, Parvaneh N, Plebani A, Prockop S, Reisli I, Soh JY, Somech R, Torgerson TR, Kim YJ, Walter JE, Gennery AR, Keles S, Manis JP, Marcenaro E, Moretta A, Parolini S, Notarangelo LD, Natural Killer Cells from Patients with Recombinase-Activating Gene and Non-Homologous End Joining Gene Defects Comprise a Higher Frequency of CD56(bright) NKG2A(+++) Cells, and Yet Display Increased Degranulation and Higher Perforin Content. Front Immunol 8, 798 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cichocki F, Miller JS, In vitro development of human Killer-Immunoglobulin Receptor-positive NK cells. Methods Mol Biol 612, 15–26 (2010). [DOI] [PubMed] [Google Scholar]
- 76.Yang C, Siebert JR, Burns R, Gerbec ZJ, Bonacci B, Rymaszewski A, Rau M, Riese MJ, Rao S, Carlson KS, Routes JM, Verbsky JW, Thakar MS, Malarkannan S, Heterogeneity of human bone marrow and blood natural killer cells defined by single-cell transcriptome. Nat Commun 10, 3931 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Phillips JH, Hori T, Nagler A, Bhat N, Spits H, Lanier LL, Ontogeny of human natural killer (NK) cells: fetal NK cells mediate cytolytic function and express cytoplasmic CD3 epsilon,delta proteins. The Journal of experimental medicine 175, 1055–1066 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lanier LL, Chang C, Spits H, Phillips JH, Expression of cytoplasmic CD3 epsilon proteins in activated human adult natural killer (NK) cells and CD3 gamma, delta, epsilon complexes in fetal NK cells. Implications for the relationship of NK and T lymphocytes. Journal of immunology 149, 1876–1880 (1992). [PubMed] [Google Scholar]
- 79.Narni-Mancinelli E, Chaix J, Fenis A, Kerdiles YM, Yessaad N, Reynders A, Gregoire C, Luche H, Ugolini S, Tomasello E, Walzer T, Vivier E, Fate mapping analysis of lymphoid cells expressing the NKp46 cell surface receptor. Proceedings of the National Academy of Sciences of the United States of America 108, 18324–18329 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vanvalkenburgh J, Albu DI, Bapanpally C, Casanova S, Califano D, Jones DM, Ignatowicz L, Kawamoto S, Fagarasan S, Jenkins NA, Copeland NG, Liu P, Avram D, Critical role of Bcl11b in suppressor function of T regulatory cells and prevention of inflammatory bowel disease. J Exp Med 208, 2069–2081 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gomez-Sanchez D, Schlotterer C, ReadTools: A universal toolkit for handling sequence data from different sequencing platforms. Mol Ecol Resour 18, 676–680 (2018). [DOI] [PubMed] [Google Scholar]
- 82.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome S Project Data Processing, The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Robinson MD, McCarthy DJ, Smyth GK, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR, Weirauch MT, The Human Transcription Factors. Cell 172, 650–665 (2018). [DOI] [PubMed] [Google Scholar]
- 86.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK, Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.E. P. Consortium, An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gupta R, Wikramasinghe P, Bhattacharyya A, Perez FA, Pal S, Davuluri RV, Annotation of gene promoters by integrative data-mining of ChIP-seq Pol-II enrichment data. BMC Bioinformatics 11 Suppl 1, S65 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Sorting strategies and phenotype validation
Figure S2. DA ATAC-seq peak distribution and clustering
Figure S3. DB H3K27ac peaks and trace profiles
Figure S4. Flow cytometric TF staining in NK cells
Figure S5. TF ChIP-seq peak traces and ATAC-seq overlap
Figure S6. Bcl11b regulates canonical NK cell differentiation
Figure S7. T-cell associated phenotypes of adaptive NK cells
Figure S8. GSEA in Bcl11b−/− mice following MCMV infection
Table S2. Summary of TF ChIP-seq peak intensities
Table S3. List of antibodies and fluorochromes
Table S1. Raw data