Abstract
The complete mitochondrial genome of Ostrinia kasmirica (Moore, 1888) was sequenced in this study. The circular mitogenome is 15,214 bp in length, containing 37 typical encoded genes and a non-coding control region. The gene organization and nucleotide composition are similar to those of most other sequenced Ostrinia species. All protein-coding genes (PCGs) initiate with ATN and terminate with TAN, except cox1 starts with CGA and cox1, cox2, nad5 terminate with an incomplete codon T. The control region of 308 bp contains three conserved features including the motif ‘TTAGA’ preceded a poly-T stretch, a microsatellite-like (TA)n element, and a poly-A stretch upstream of trnM. Phylogenetic analysis based on mitogenome sequences revealed that the O. kasmirica (the second species group) was more closely related to the third species group of the genus and the first species group was not at the basal position of this genus as that Mutuura and Munroe indicated.
Keywords: Mitochondrial genome, Crambidae, Ostrinia, phylogenetic analysis
The genus Ostrinia Hübner, 1825 (Crambidae: Pyraustinae) includes 23 species, with a few well-known destructive pests that caused huge economic losses (Mutuura and Munroe 1970). Mutuura and Munroe (1970) recognized three species groups of the genus based on the morphology of male genitalia. The first species group (Group I) only contains a single species Ostrinia penitalis (Grote, 1876) with unarmed sacculus and one-lobed uncus which is regarded as the ‘primitive’ species of this genus. The second species group (Group II) includes nine species with simple or weakly bifid uncus in the male genitalia. The third species group (Group III) comprises 10 species such as Ostrinia furnacalis (Guenée, 1854) and Ostrinia nubilalis (Hübner, 1796) . The remaining species including Ostrinia maysalis Leraut, 2012, Ostrinia ovalipennis Ohno, 2003, and Ostrinia avarialis Amsel, 1970 not classified into the above three species groups.
The Ostrinia kasmirica (Moore, 1888) is commonly found in the northeast India, southeast Russia, and Siberia (Mutuura and Munroe 1970). It was first recorded by Li and Tang in 1981 in China (Li and Tang 1985). The larva of this species mainly feeds on the thistle Cnicus wallichi. Mutuura and Munroe (1970) purported that the O. kasmirica, having bell-shaped and weakly bifid uncus, is most likely to be closely related with Ostrinia latipennis (Warren, 1892) and Ostrinia palustralis (Hübner,1796) based on the male genitalia within the second species group. However, the O. kasmirica is similar to O. nubilalis and Ostrinia scapulalis (Walker, 1859) in appearance (Bidzilya and Budashkin 2017), implying that O. kasmirica is closely related to the members of the third species group. Due to the lack of the molecular data of O. kasmirica its phylogenetic position is still unclear. To address this question, we sequenced the complete mitogenome sequence of O. kasmirica and inferred the phylogenetic relationships of this genus based on this mitogenome and other six published mitogenome sequences of the genus Ostrinia .
The adult specimens of O. kamirica were collected by light trap in July 2018 from Jalai Nur District (49°31'N, 117°43'E), Hulunbuir Prefecture, Inner Mongolia Autonomous Region, China. Specimens were preserved in a freezer in pure ethanol at −20 °C. The voucher sample was deposited at the Entomological Museum (Voucher number ZLNE3), Northwest A&F University, Yangling, Shaanxi, China. Genomic DNA was extracted from thoracic muscle. Illumina HiSeq platform (Illumina, San Diego, CA) was used for sequencing and generated 2 × 150 bp paired-end reads. Mitochondrial gene annotation was performed by MitoZ version 2.4 (Meng et al. 2019) under all module which includes four steps: raw data pretreatment, de novo assembly with SOAPdenvo-Trans, mitogenome sequence identification, and mitogenome annotation . Before data analyses, the annotated mitogenome was rechecked with previously published mitogenomes of Ostrinia species.
The complete mitogenome of O. kasmirica (GenBank accession number. MT978075) is circular molecular structure of 15,214 bp in length, containing 37 typical genes (13 PCGs, 22 transfer RNAs and 2 ribosomal RNAs) and a non-coding control region. The gene arrangement and content are identical to most of the mitogenomes in Lepidoptera. Nucleotide compositions of 41.8% A, 39.2% T, 11.3% C, and 7.7% G indicated that the O. kasmirica mitogenome is biased toward higher A + T content. All PCGs initiate with standard start codon ATN except cox1 with the special start codon CGA. Most PCGs terminate with TAN, whereas cox1, cox2, and nad5 stop with an incomplete codon T. All tRNAs exhibit cloverleaf secondary structure except trnS1(AGN) which was missing the dihydrouridine (DHU) arm. The length of 12S rRNA and 16S rRNA are 777 and 1,339 bp, respectively. The control region of the O. kasmirica is 308 bp in length with several conserved blocks, including a ‘TTAGA’ motif proceeding a 19-bp poly-T stretch, a microsatellite-like (TA)n element, and an ‘A-rich’ upstream of trnM, which are commonly found in other published mitogenomes of the genus Ostrinia (Zhou et al. 2020).
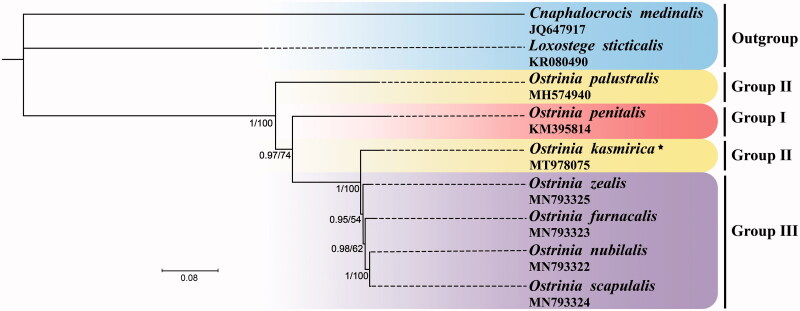
Phylogeny of the genus Ostrinia was inferred based on the concatenated nucleotide sequences of 13 PCGs with all codon positions using both maximum likelihood (ML) and Bayesian inference (BI) methods. Best substitution model for ML and BI was selected based on PartitionFinder version 2.1.1, respectively (Lanfear et al. 2016). There are seven partition schemes for the ML analysis (Subset 1: GTR + I, atp6_pos1, cox1_pos1, cox2_pos1, cox3_pos1, cytb_pos1; Subset 2: HKY + I, atp6_pos2, cox1_pos2, cox2_pos2, cox3_pos2, cytb_pos2; Subset 3: GTR + I+G, atp6_pos3, atp8_pos3, cox1_pos3, cox2_pos3, cox3_pos3, cytb_pos3, nad2_pos3, nad3_pos3, nad6_pos3; Subset 4: GTR + I, atp8_pos1, atp8_pos2, nad2_pos1, nad3_pos1, nad6_pos1; Subset 5: HKY + I, nad1_pos1, nad4_pos1, nad4L_pos1 nad5_pos1; Subset 6: HKY + I, nad1_pos2, nad2_pos2, nad3_pos2, nad4_pos2, nad4L_pos2, nad5_pos2, nad6_pos2; Subset 7: GTR + G, nad1_pos3, nad4_pos3, nad4L_pos3, nad5_pos3) and seven partition schemes for the BI analysis (Subset 1: TRN + I, atp6_pos1, cox1_pos1, cox2_pos1, cox3_pos1, cytb_pos1; Subset 2: HKY + I, atp6_pos2, cox2_pos2, cytb_pos2, nad1_pos2, nad2_pos2, nad3_pos2, nad4_pos2, nad4L_pos2, nad5_pos2, nad6_pos2; Subset 3: TIM + I+G, atp6_pos3, atp8_pos3, cox1_pos3, cox2_pos3, cox3_pos3, cytb_pos3, nad2_pos3, nad3_pos3, nad6_pos3; Subset 4: TIM + I, atp8_pos1, nad2_pos1, nad3_pos1, nad6_pos1; Subset 5: TRN + I+G, atp8_pos2, nad1_pos1, nad4_pos1, nad4L_pos1, nad5_pos1; Subset 6: F81 + I, cox1_pos2, cox3_pos2; Subset 7: GTR + G, nad1_pos3, nad4_pos3, nad4L_pos3, nad5_pos3). Six species within the genus Ostrinia were selected as ingroups (Coates and Abel 2016; Hwang et al. 2019; Zhou et al. 2020). Loxostege sticticalis (Linnaeus, 1761) (Crambidae: Pyraustinae) and Cnaphalocrocis medinalis (Guenée, 1854) (Crambidae: Spilomelinae) were selected as outgroups (Wan et al. 2013; Ma et al. 2016). The ML analyses were performed using IQ-TREE version 1.6.8 (Nguyen et al. 2015) and BI analyses were conducted with MrBayes version 3.2.6 (Ronquist et al. 2012 ). The phylogenetic topologies of the ML and BI analyses agreed with each other (Figure 1). The O. kasmirica (Group II) and four species including O. zealis, O. furnacalis, O. nubilalis, and O. scapulalis of the third species group were clustered into a clade with high nodal support values, indicating O. kasmirica is likely to be more closely related to the trifid-uncus species group other than O. palustralis (Group II). Our results confirmed that the O. penitalis (Group I) is not the primitive species among the genus Ostirnia, which is consistent with the previous study (Zhou et al. 2020).
Figure 1.
Phylogenetic trees using ML analyses based on 13 PCGs of nine mitogenomes. BI analyses show the same topology (not shown). The numbers under the branches are Bayesian posterior probabilities (PP) and bootstrap support values (BS). Alphanumeric terms indicate the GenBank accession numbers.
Funding Statement
This research was supported by the National Natural Science Foundation of China [31772508] and National Key Research and Development Program of China [2016YFC0501502].
Disclosure statement
No potential conflict of interest was reported by the authors. Conceptualization, Nan Zhou and Zhaofu Yang; Specimen collection and identification, Nan Zhou and Zhaofu Yang; Data analysis, Qiuyu Luo and Nan Zhou; Writing-Original Draft Preparation, Qiuyu Luo and Nan Zhou; Writing-Review and Editing, Qiuyu Luo, Nan Zhou and Zhaofu Yang.
Data availability statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov, reference number MT978075. The associated BioProject accession number, SRA data, and BioSample accession number are PRJNA716609, SRR14044899, and SAMN18439942 respectively.
References
- Bidzilya OV, Budashkin YI.. 2017. New records of Lepidoptera from Ukraine and description of a new species of Caloptilia Hübner, 1825 (Lepidoptera, Gracillariidae) from the mountains of Crimea. Nota Lepidopterol. 40(2):145–161. [Google Scholar]
- Coates BS, Abel CA.. 2016. The mitochondrial genome of the American lotus borer, Ostrinia penitalis (Lepidoptera: Crambidae). Mitochondrial DNA Part A. 27(3):1938–1939. [DOI] [PubMed] [Google Scholar]
- Hwang EJ, Kim MJ, Kim SS, Kim I.. 2019. Complete mitochondrial genome of Ostrinia palustralis memnialis Walker, 1859 (Lepidoptera: Crambidae). Mitochondrial DNA Part B. 4(1):1364–1366. [Google Scholar]
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B.. 2016. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34(3):772–773. [DOI] [PubMed] [Google Scholar]
- Li WD, Tang DZ.. 1985. Ostrinia kasmirica (Moore) – a new record from China. Entomotaxonomia. 7(1):24. [Google Scholar]
- Ma HF, Zheng XX, Peng MH, Bian HX, Chen MM, Liu YQ, Jiang X, Qin L.. 2016. Complete mitochondrial genome of the meadow moth, Loxostege sticticalis (Lepidoptera: Pyraloidea: Crambidae), compared to other Pyraloidea moths. J Asia-Pac Entomol. 19(3):697–706. [Google Scholar]
- Meng GL, Li YY, Yang CT, Liu SL.. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63–e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutuura A, Munroe E.. 1970. Taxonomy and distribution of the European corn borer and allied species: genus Ostrinia (Lepidoptera: Pyralidae). Mem Entomol Soc Can. 102(S71):1–112. [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, P Van Der M, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP.. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan XL, Kim MJ, Kim I.. 2013. Description of new mitochondrial genomes (Spodoptera litura, Noctuoidea and Cnaphalocrocis medinalis, Pyraloidea) and phylogenetic reconstruction of Lepidoptera with the comment on optimization schemes. Mol Biol Rep. 40(11):6333–6349. [DOI] [PubMed] [Google Scholar]
- Zhou N, Dong YL, Qiao PP, Yang ZF.. 2020. Complete mitogenomic structure and phylogenetic implications of the genus Ostrinia (Lepidoptera: Crambidae). Insects. 11(4):232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov, reference number MT978075. The associated BioProject accession number, SRA data, and BioSample accession number are PRJNA716609, SRR14044899, and SAMN18439942 respectively.