Abstract
Background:
A coding variant in MTARC1 (rs2642438; p.Ala165Thr) was recently associated with protection from cirrhosis in European individuals. However, its impact on overall and cause-specific mortality remained elusive.
Methods:
Using a genome-first approach, we explored a range of metabolic phenotypes and outcomes associated with MTARC1 p.Ala165Thr in the UKBiobank and the Penn-Medicine BioBank.
Findings:
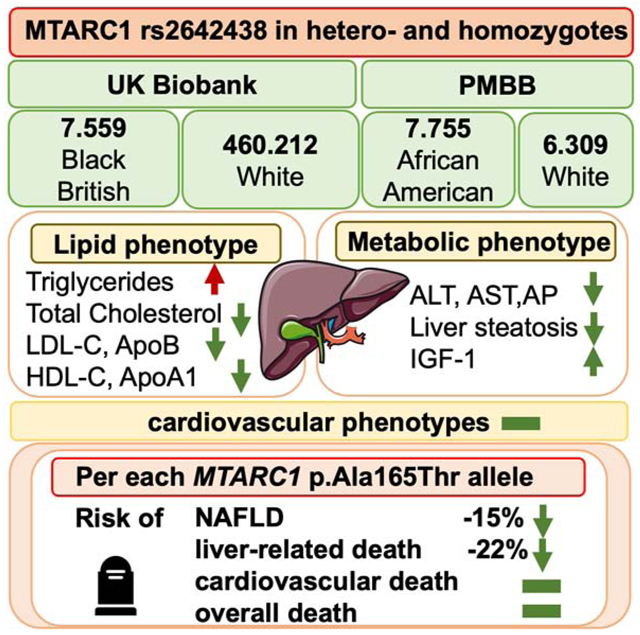
MTARC1 p.Ala165Thr was significantly associated with higher triglycerides, lower total cholesterol, lower LDL-C, lower ApoB, lower HDL-C, lower ApoA-I and higher IGF-1. Per each minor allele, the risk of NAFLD was reduced by ~15%. The ALT-lowering and NAFLD-protective effect of MTARC1 p.Ala165Thr was amplified by obesity, diabetes mellitus and presence of PNPLA3 rs738409:G. In African-American and Black-British individuals, the allele frequency of MTARC1 p.Ala165Thr was lower, but carriers showed the same distinctive lipid phenotype. Importantly, MTARC1 p.Ala165Thr carriers did not show higher cardiovascular disease burden as evidenced by cardiac MRI and carotid ultrasound. In prospective analyses, the homozygous minor allele was associated with up to 39% lower rates of liver-related mortality, while no risk of increased overall or cardiovascular death could be observed. Strikingly, liver-related mortality was more than 50% reduced in diabetic participants or carriers of PNPLA3 rs738409:G.
Conclusions:
Together these data highlight MTARC1 as an important liver disease modifier that influences plasma lipids in an allele-dose-dependent manner without increasing cardiovascular outcomes. Our results point toward potential mechanisms and reveal a remarkable association with liver-related mortality calling for future studies exploring its therapeutic potential.
Funding:
This study was funded by the German Research Foundation (DFG).
Keywords: MTARC1, Liver disease, NAFLD, SNP, Survival, Liver-related death
Graphical Abstract
Introduction
The non-alcoholic fatty liver disease (NAFLD) pandemic affects nearly 2 billion people worldwide1. It is well known that obesity is the most important risk factor for the development of NAFLD2. NAFLD is also strongly associated with type 2 diabetes mellitus, dyslipidemia, and atherosclerotic cardiovascular disease 1. In addition, several genomic loci were revealed to be associated with increased risk of NAFLD (e.g. PNPLA3 rs738409:G)3.
Recently, a coding variant (MTARC1 p.Ala165Thr) was reported to be associated with protection against NAFLD and its progression,4-6 as well as protection from alcohol-related and all-cause cirrhosis7,8. Other reports indicated that NAFLD severity was reduced in MTARC1 p.Ala165Thr carriers9,10. MTARC1, the Mitochondrial Amidoxime Reducing Component 1 gene, encodes the mitochondrial amidoxime-reducing component, which is involved in metabolic processes in the liver (e.g. activating N-hydroxylated prodrugs or reducing nitrite to produce nitric oxide)11. The exact function of the MTARC1 protein is unknown and the mechanism by which MTARC1 p.Ala165Thr may protect against liver damage remains elusive5. It is unclear whether the variant exerts an effect through modulation of specific risk factors (e.g. obesity), or if it exercises a direct hepatoprotective effect. As the published studies were focused on case-control cohorts5,7, it remains unknown whether MTARC1 p.Ala165Thr affects liver- related morbidity and cardiometabolic risk factors in the general population. In addition, the effects of MTARC1 p.Ala165Thr have so far only been described in cohorts of predominantly European ancestry and its impact on overall and cause-specific mortality have not been studied yet.
In this study, we performed deeper phenotyping of MTARC1 p.Ala165Thr carriers in unselected populations, both in Europeans, African-Americans and Black British participants, focusing on liver and cardiometabolic phenotypes as well as survival. After demonstrating an increased protective effect of MTARC1 p.Ala165Thr on NAFLD development in individuals who are obese or diabetic, we looked at overall and liver-related survival in MTARC1 p.Ala165Thr carriers. We found that MTARC1 p.Ala165Thr carriers have significantly higher triglycerides (TG), lower HDL-C and apoA-I levels, but also have lower LDL-C and apoB levels, while no difference in cardiovascular disease risk was observed. While MTARC1 p.Ala165Thr is less common in individuals of African American or Black British ancestry, it also has similar protective effects on liver outcomes. We are the first to show, that liver-related survival is improved in MTARC1 p.Ala165Thr carriers, especially in participants prone to liver disease development. This reduction in liver-related survival does not increase the risk of cardiovascular-related death. Together, our study reveals a deeper phenotype of MTARC1 p.Ala165Thr carriers, its relationship with clinical liver disease, and hints toward potential mechanisms by which MTARC1 could protect from NAFLD.
Results:
MTARC1 and liver disease, liver enzymes, and liver imaging
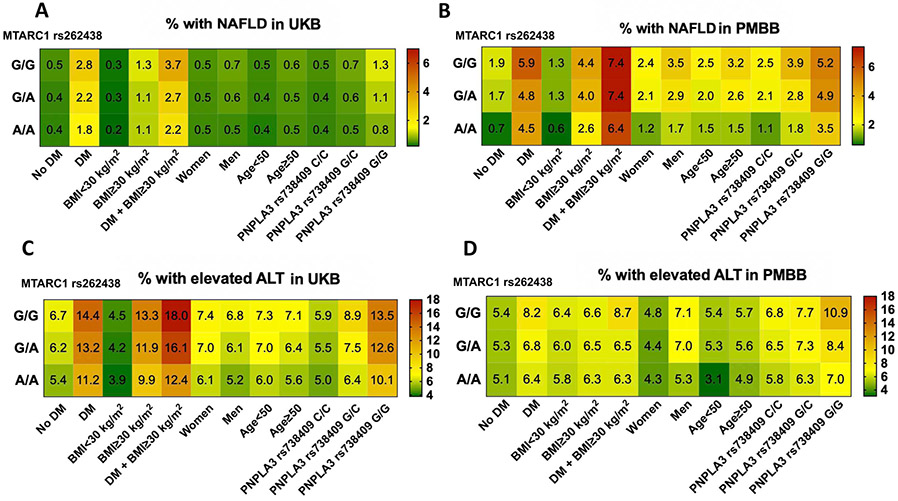
Of 460,212 White UK Biobank participants, 9% were homozygous for MTARC1 rs2642438 and 42% were heterozygous (Table 1). We analyzed the association between rs2642438 and diagnoses of liver disease in White UKB participants. For both homo-and heterozygous carriers of rs2642438, the risk of physician diagnosed NAFLD (ICD-10) was significantly reduced (A/A vs G/G: aOR=0.78[0.67-0.91]; and G/A vs G/G: aOR=0.86[0.79-0.94], Table 2). For each rs2642438 minor allele, the risk of NAFLD development was reduced by up to 14%. Regarding liver enzymes, mean alanine aminotransferase (ALT) and aspartate aminotransferase (AST) values were significantly lower in individuals with MTARC1 rs2642438 than in non-carriers (all p<0.0001; Table 1). A subgroup of UKB participants received a liver MRI. Heterozygous MTARC1 rs2642438 carriers had significantly lower proton density fat fraction, a measurement of liver steatosis (p=0.013, Table S1). We also determined the association between MTARC1 rs2642438 and clinically relevant diseases such as type 2 diabetes and obesity (Figure 2). Among the analyzed parameters, the protective association of MTARC1 rs2642438 on NAFLD and ALT elevation were significantly more pronounced in the presence of diabetes, obesity or presence of PNPLA3 rs738409:G (p<.03, Figure S1). For each rs2642438 minor allele, the risk of NAFLD development in diabetic individuals was reduced by 21% and in obese and diabetic individuals it was reduced by 27%.
Table 1:
Characteristics of rs2642438 homozygous or heterozygous Carriers compared with non-carriers in UKB.
| Non carriers (G/G) n=227 685 |
Heterozygotes (G/A) n=191 936 |
Homozygotes (A/A) n=40 591 |
p-Value G/G vs G/A |
p-Value G/G vs A/A |
|
|---|---|---|---|---|---|
| Characteristics | Univ. | Univ. | |||
| Age (years) | 56.6±8.1 | 56.8±8.0 | 56.8±8.0 | .11 | .089 |
| Women (%) | 55 | 54 | 54 | .18 | .11 |
| BMI (kg/m2) | 27.4±4.8 | 27.4±4.8 | 27.4±4.8 | .24 | .55 |
| Alcohol (g/d) | 8.9±10.1 | 9.0±10.2 | 9.2±10.1 | .038 | .00002 |
| Diabetes mellitus (%) | 5 | 5 | 5 | .59 | .085 |
| Ethnicity (% white) | 100 | 100 | 100 | ||
| Frequency of well-known NAFLD influencing genes* | |||||
| HSD17B13 rs72621367:TA | 0.55±0.63 | 0.55±0.63 | 0.55±0.63 | .10 | .18 |
| PNPLA3 rs738409:G | 0.43±0.58 | 0.43±0.58 | 0.43±0.58 | .34 | .26 |
| Liver status | Multiv, | Multiv. | |||
| ALT (U/l) | 23.7±14.5 | 23.3±13.9 | 22.8±13.4 | 1.8*10-25 | 7.2*10-38 |
| AST (U/l) | 26.2±10.8 | 26.0±10.2 | 25.9±9.9 | .0001 | 3.3*10-9 |
| GGT (U/l) | 37.1±41.9 | 36.8±41.0 | 36.8±37.5 | .009 | .11 |
| Bilirubin (mg/dl) | 0.54±0.26 | 0.53±0.26 | 0.53±0.26 | .000009 | .000009 |
| AP (U/l) | 83.9±26.3 | 83.3±26.0 | 82.6±26.8 | 2.8*10-16 | 3.7*10-22 |
| Lipid metabolism | |||||
| Triglycerides (mg/dl) | 154±89 | 156±91 | 157±91 | .000008 | 1.1*10-9 |
| HDL cholesterol (mg/dl) | 57±15 | 56±15 | 55±15 | 9.2*10-18 | 1.6*10-26 |
| LDL cholesterol (mg/dl) | 138±34 | 137±34 | 136±34 | 9.8*10-15 | 5.3*10-20 |
| Cholesterol (mg/dl) | 221±44 | 219±44 | 220±44 | 3.7*10-19 | 2.4*10-25 |
| Apolipoprotein A1 (g/l) | 1.54±0.27 | 1.54±0.27 | 1.53±0.27 | 2.5*10-19 | 1.1*10-29 |
| Apolipoprotein B (g/l) | 1.04±0.24 | 1.03±0.24 | 1.03±0.24 | 5.9*10-7 | 1.7*10-8 |
| Lipoprotein A (nmol/l) | 44.05±48.9 | 44.04±49.4 | 44.08±49.4 | .99 | .90 |
| Additional serum parameters | |||||
| Urate (umol/L) | 308.2±80.0 | 309.0±80.4 | 310.2±80.4 | .011 | .00001 |
| Vitamin D (nmol/L) | 49.2±20.9 | 49.5±21.0 | 49.8±21.2 | .000005 | 4.7*10-8 |
| IGF-1 (nmol/l) | 21.3±5.8 | 21.5±5.6 | 21.6±5.6 | 3.2*10-14 | 2.2*10-16 |
Quantitative measures are expressed as means and standard deviations or as relative frequencies (%). All multivariable analyses were adjusted for age, sex, BMI, and PC1-4.
(0=non carrier, 1=heterozygous, 2=homozygous). A FDR adjusted significance level of p <= 0.03 was used.
Table 2:
Characteristics of rs2642438 homozygous or heterozygous carriers compared with non-carriers in UKB.
| Non carriers (G/G) n=227 685 |
Heterozygotes (G/A) n = 191 936 |
Homozygotes (A/A) n=40 591 |
p-Value G/G vs G/A |
p-Value G/G vs A/A |
aOR G/G vs G/A |
aOR G/G vs A/A |
|
|---|---|---|---|---|---|---|---|
| Liver status | |||||||
| ALT ≥ULN, N(%) | 15 531 (6.8) | 11 999 (6.3) | 2 211 (5.4) | 1.3*10-13 | 2.7*10-25 | 0.91 [0.89-0.93] | 0.78[0.74-0.82] |
| AST ≥ULN, N(%) | 10 317 (4.5) | 8 357 (4.4) | 1 664 (4.1) | .008 | .00001 | 0.96[0.93-0.99] | 0.90[0.85-0.95] |
| ICD10 coded diagnoses | |||||||
| Overall liver disease (K70-K76), N(%) | 3 609 (1.59) | 2 874 (1.50) | 580 (1.43) | .015 | .010 | 0.94[0.89-0.99] | 0.89[0.81-0.97] |
| Alcoholic liver disease (K70), N(%) | 516 (0.23) | 397 (0.21) | 78 (0.19) | .17 | .16 | ||
| Toxic liver disease (K71), N(%) | 40 (0.02) | 39 (0.02) | 6 (0.01) | .45 | .48 | ||
| Hepatic failure (K72), N(%) | 258 (0.11) | 177 (0.09) | 41 (0.10) | .050 | .90 | ||
| Chronic Hepatitis (K73), N(%) | 78 (0.03) | 68 (0.04) | 14 (0.03) | .80 | .99 | ||
| Fibrosis and cirrhosis (K74), N(%) | 575 (0.25) | 467 (0.24) | 73 (0.18) | .59 | .007 | ||
| Inflammatory liver diseases (K75), N(%) | 481 (0.25) | 404 (0.24) | 80 (0.24) | .85 | .52 | ||
| NASH (K75.8), N(%) | 170 (0.07) | 129 (0.07) | 19 (0.05) | .32 | .044 | ||
| Other liver diseases (K76), N(%) | 2 650 (1.16) | 2 071 (1.08) | 403 (0.99) | .006 | .001 | 0.92[0.87-0.98] | 0.84[0.75-0.93] |
| NAFLD (K76.0), N(%) | 1 347 (0.59) | 991 (0.52) | 191 (0.47) | .0004 | .002 | 0.86[0.79-0.94] | 0.78[0.67-0.91] |
| Malignant neoplasm of the liver and/or bile duct (C22), N(%) | 268 (0.12) | 200 (0.10) | 35 (0.09) | .17 | .072 | ||
| Survival | |||||||
| All-cause mortality, N(%) | 14 045 (6) | 12 061 (6) | 2501 (6) | .71 | .35 | ||
| Liver related death, N(%) | 474 (0.21) | 345 (0.18) | 52 (0.13) | .030 | .001 | 0.86[0.74-0.99] | 0.61[0.46-0.81] |
| Cardiovascular death, N(%) | 2 734 (1.2) | 2 338 (1.2) | 544 (1.3) | .63 | .35 |
Quantitative measures are expressed as number of participants (N) and as relative frequencies (%). All multivariable analyses were adjusted for age, sex, BMI, and PC1-4). A FDR adjusted significance level of p <= 0.03 was used.
Figure 2: Frequency of NAFLD or elevated ALT for each MTARC1 genotype among various MTARC1 genotypes in the UKB (A,C) and PMBB cohorts (B,D).
Relative frequencies (%) are shown and visualized by a color coding (right panel). Abbreviations: ALT, alanine aminotransferase; BMI, body mass index (kg/m2); DM, diabetes mellitus.
Next, we confirmed the association between rs2642438 and metabolic phenotypes in the PMBB, a biobank which contrasts UKB in its higher percentage of diseased participants as well as its multiethnicity. Of 6,309 White PMBB participants, 8% were homozygous and 42% were heterozygous for rs2642438, similar to UKB. In contrast, out of 7,755 African American PMBB participants, 1% were homozygous and 16% were heterozygous (Table 3; Table S2). In all PMBB participants, the risk of physician diagnosed NAFLD (ICD-9/10) was reduced in homozygous and heterozygous carriers of rs2642438 (A/A vs. G/G: aOR=0.70[0.50-0.98]; and G/A vs G/G: aOR=0.85[0.69-0.96], Table 3). For each rs2642438 minor allele, the risk of NAFLD development was reduced by 15% (Table 3).
Table 3:
Baseline characteristics of homozygous (G/G) or heterozygous rs2642438 carriers (G/A) compared with non-carriers (A/A) in PMBB.
| Non carriers (G/G) n=9 581 |
Heterozygotes (G/A) n=3 889 |
Homozygotes (A/A) n=594 |
p-Value G/G vs G/A |
p-Value G/G vs A/A |
|
|---|---|---|---|---|---|
| Characteristics | Univ. | Univ. | |||
| Age (years) | 60.6±17.3 | 65.9±13.9 | 69.5±15.4 | 1.7*10-64 | 4.1*10-13 |
| Women (%) | 57 | 48 | 41 | 2.0*10-18 | 1.9*10-37 |
| BMI (kg/m2) | 29.0±5.9 | 30.0±6.1 | 28.4±5.3 | 3.5*10-21 | 6.8*10-21 |
| Diabetes mellitus (%) | 26 | 24 | 22 | .048 | .040 |
| Ethnicity (% Whites) | 33 | 68 | 88 | 4.6*10-306 | 5.0*10-185 |
| Frequency of well-known NAFLD influencing genes* | |||||
| HSD17B13 rs72621367:TA | 0.55±0.63 | 0.56±0.63 | 0.57±0.63 | .51 | .95 |
| PNPLA3 rs738409:G | 0.35±0.58 | 0.41±0.58 | 0.46±0.58 | .47 | .67 |
| Liver status | Multiv. | Multiv. | |||
| ALT (U/l) | 24.4±36.1 | 23.6±17.8 | 23.9±17.0 | .001 | .034 |
| ALT ≥ULN (%) | 7 | 7 | 6 | .22 | .064 |
| AST (U/l) | 25.3±44.8 | 24.0±15.7 | 24.0±17.9 | .006 | .12 |
| AST ≥ULN (%) | 6 | 6 | 6 | .021a | .86 |
| Lipid metabolism | |||||
| Triglycerides (mg/dl) | 112±78 | 119±69 | 128±69 | .041 | .029 |
| HDL cholesterol (mg/dl) | 50±15 | 50±17 | 48±16 | .37 | .062 |
| LDL cholesterol (mg/dl) | 100±33 | 95±32 | 90±34 | .057 | .052 |
| Cholesterol (mg/dl) | 174±39 | 169±39 | 163±38 | .035 | .011 |
| ICD10 coded diagnoses | |||||
| Toxic liver disease (K71) | 0.7 | 0.6 | 0.5 | .20 | .22 |
| Hepatic failure (K72) | 1.0 | 0.9 | 1.0 | .99 | .77 |
| Chronic Hepatitis (K73) | 0.7 | 0.05 | 0.0 | .59 | .99 |
| Fibrosis and cirrhosis (K74) | 0.4 | 0.4 | 0.2 | .90 | .23 |
| Inflammatory liver diseases (K75) | 2.2 | 1.8 | 1.3 | .019b | .10 |
| NASH (K75.8) | 2.2 | 2.0 | 1.9 | .065 | .28 |
| Other liver diseases (K76) | 4.0 | 3.5 | 2.2 | .14 | .052 |
| NAFLD (K76.0) | 3.0 | 2.5 | 1.5 | .021c | .030d |
Quantitative measures are expressed as means and standard deviations or as relative frequencies (%). All multivariable analyses were adjusted for age, sex, BMI, and PC1-4.
(0=non carrier, 1=heterozygous, 2=homozygous);
aOR=0.81[0.67-0.97]
aOR=0.71 [0.54-0.94]
aOR=0.85[0.69-0.96]
aOR=0.70[0.50-0.98]. A FDR adjusted significance level of p <= 0.03 was used.
As 55% of PMBB participants were self-reported African American, we analyzed the association between rs2642438 and liver disease in the African American subpopulation (Figure 1C). Due to a reduced frequency of MTARC1 p.Ala165Thr in African Americans, the number of homozygous carriers was low (1% vs. 8% in Whites). Moreover, PNPLA3 rs738409:G, which is itself highly associated with NAFLD/NASH development12, was more frequent in MTARC1 rs2642438 carriers (Table S2). Still, homozygous MTARC1 rs2642438 carriage was associated with a significant reduction in NASH (aOR=0.70[0.50-0.98], Table S2). Heterozygous MTARC1 rs2642438 carriers had a marked negative association with NAFLD frequency (aOR=0.75[0.59-0.96], Table S2). Thus, MTARC1 rS264238 is associated with reduced NAFLD in individuals of both Europeans and African Americans.
Figure 1: Overview of the analyzed cohorts.
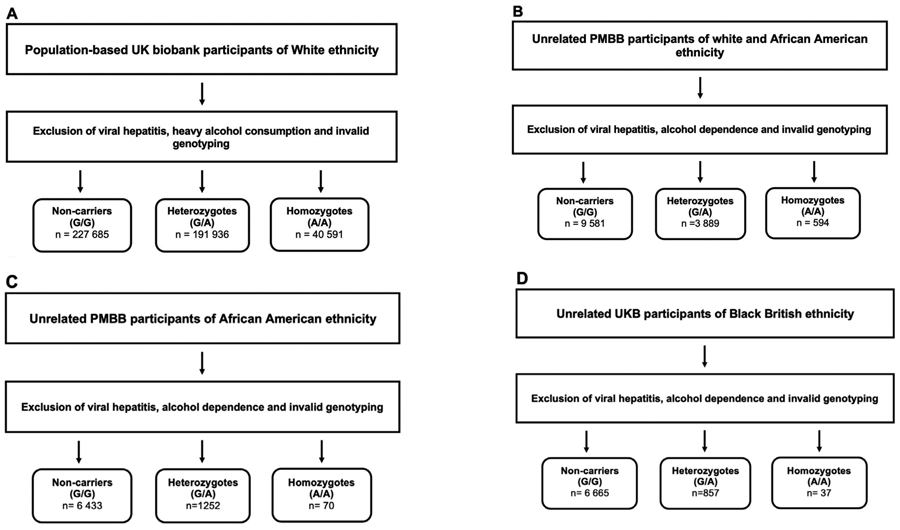
A) UK Biobank participants of European ancestry aged 37 to 73 years
B) Penn medicine BioBank (PMBB) participants of 55% African American ancestry aged 25 to 105 years
C) African American Penn medicine BioBank (PMBB) participants
D) Black British UK Biobank participants
Plasma lipids and apolipoproteins
Given the strong association between NAFLD and dyslipidemia, we analyzed the relationship between MTARC1 rs2642438, plasma lipids and apolipoproteins. In White UKB participants, both homo- and heterozygous MTARC1 rs2642438 carriers showed significantly higher plasma TG than non-carriers (p<0.0001, Table 1). The TG elevating effect was enhanced with higher allele-dose of MTARC1 rs2642438. Consistent with the higher TGs, rs2642438 was significantly associated with lower levels of HDL cholesterol (HDL-C) and ApoA-I in an allele-dose specific manner (Table 1). On the other hand, both homo- and heterozygous MTARC1 rs2642438 carriers had reduced levels of total cholesterol, LDL cholesterol (LDL-C) and ApoB (Table 1). No difference was observed in plasma levels of lipoprotein(a) [Lp(a)].
Next, we confirmed this association in Black British UKB participants, which had been excluded from the previous UKB analyses. Here, out of 7,559 Black British UKB participants, only 0.4% were homozygous and 12% were heterozygous for MTARC1 rs2642438 (Table S3). Still, heterozygous MTARC1 rs2642438 carriage was associated with a significant reduction of cholesterol and LDL-C (Table S3). Due to the reduced frequency of MTARC1 p.Ala165Thr NAFLD and NASH trended to be reduced in carriers, but did not reach statistical significance (Table S3).
In all PMBB patients, we confirmed the association of rs2642438 with significantly elevated TGs (p<0.05, Table 3). Here, for each rs2642438 minor allele, TGs were elevated by 8 mg/dl. Moreover, MTARC1 rs264238 was associated with an allele-dose-dependent reduction of total cholesterol (Table 3). In African American participants in PMBB, homozygous MTARC1 rs2642438 carriers had significantly higher TGs and significantly reduced levels of total cholesterol (p<0.03, Table S2). Thus, MTARC1 rs264238 is associated with a distinctive lipid profile in European, Black British and African American individuals.
Additional cardiometabolic phenotypes
To enhance our knowledge of metabolic changes associated with MTARC1 rs2642438, we analyzed additional metabolic serum markers in the UKB. MTARC1 p.Ala165Thr carriers had elevated serum urate and vitamin D levels (p<0.01, Table 1). In addition, individuals with MTARC1 rs2642438 were also more likely to have elevated levels of IGF-1 (p<0.0001, Table 1). A subset of UKB participants had cardiac MRI and carotid ultrasound performed. MTARC1 rs2642438 carriers had no impairment in cardiac output, ejection fraction or intima-media thickness compared with non-carriers (Table S4,S5).
MTARC1 rs2642438 is associated with reduced liver-related mortality without elevating the cardiovascular risk
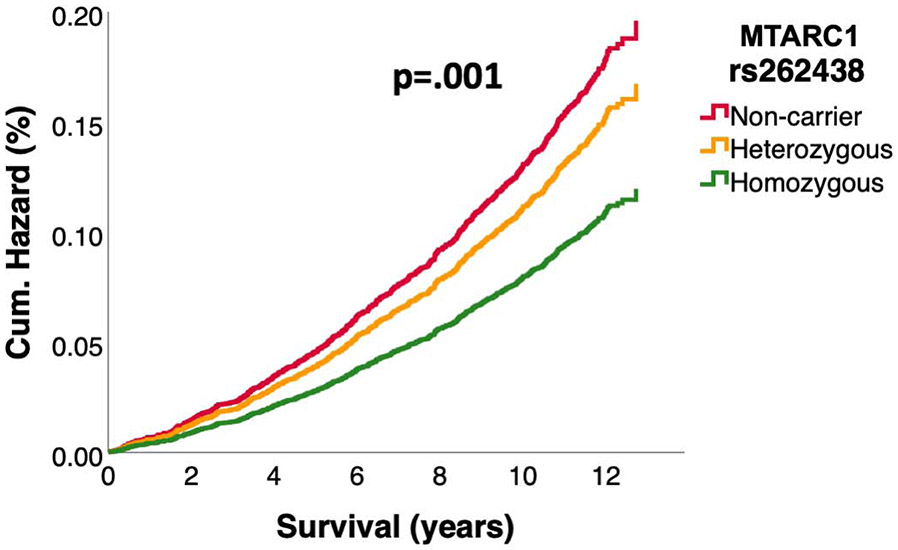
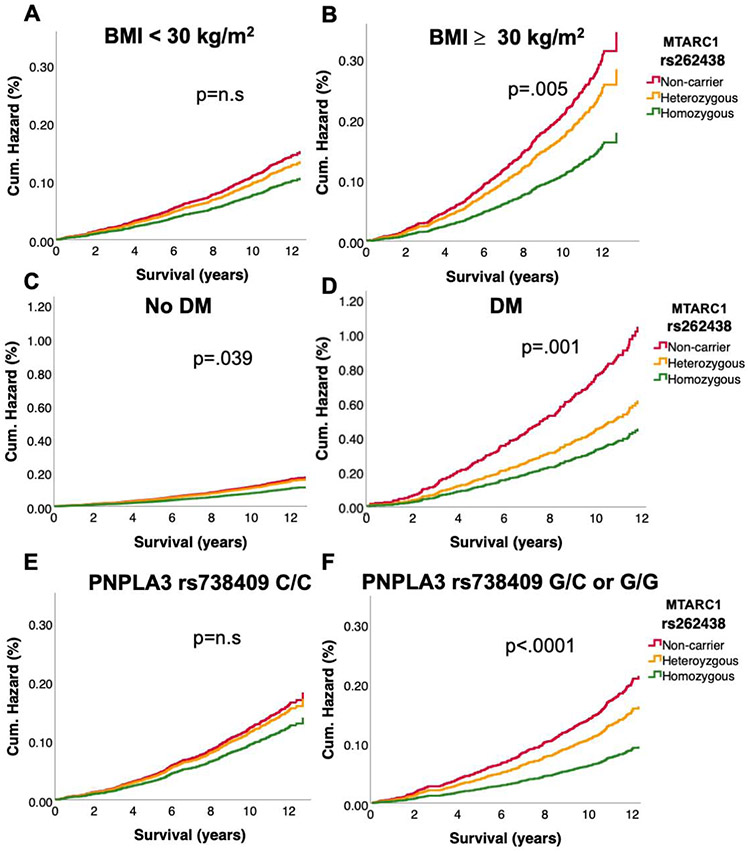
In up to 14 years of follow up 28,607 (6%) of the UKB participants and 4170 (30%) of the PMBB Participants died (Table 2,S6). We tested whether MTARC1 rs2642438 was associated with liver-related, cardiovascular and all-cause mortality. Compared with non-carriers, in the UKB the hazard ratios for liver-related mortality in MTARC1 rs2642438 homozygotes were 0.61[0.46-0.81] and 0.85[0.74-0.98] for heterozygotes (Figure 3), while a similar trend could be observed in all PMBB participants, in the African-American PMBB subgroup and the Black British UKB subgroup (Figure S2,S3). Interestingly, the effect of reduced liver-related mortality in homozygotes in white UKB participants was even more pronounced in obese (aHR=0.52[0.34-0.80]) and diabetic (aHR=0.44[0.22-0.86]) MTARC1 rs2642438 homozygotes (Figure 4). In participants, who are carriers of the known risk variant PNPLA3 rs738409, MTARC1 rs2642438 homozygotes showed the strongest reduction in liver-related death (HR: 0.43 [0.27-0.71], Figure 4). MTARC1 rs2642438 was not associated with all-cause and cardiovascular mortality neither in UKB nor in PMBB (Figure S4). To further corroborate the lower risk of liver-related mortality without elevated cardiovascular mortality, we constructed multivariable cox regression models accounting for several potential metabolic confounders (Table S7). In all models, MTARC1 rs2642438 carriers had significantly lower odds for overall liver-related mortality, while no risk for elevated cardiovascular mortality could be observed (Table S7).
Figure 3: Prospective liver-related and cardiovascular mortality as a function of MTARC1 rs2642438 genotype in the UKB.
Liver-related mortality was defined as death due to liver diseases or hepatocellular carcinoma. (A) Liver-related mortality in the UKB population as a function of MTARC1 genotype. Participants were followed prospectively from the time of study entry until death or end of follow-up. (B) Cardiovascular mortality in the UKB population as a function of MTARC1 genotype. Hazard ratios were calculated by Cox regression, adjusted for age, sex, BMI, and PC1-4. Hazard ratios: 3A) HR homozygotes vs. non-carrier: 0.61[0.46-0.81]; HR heterozygotes vs. non-carrier: 0.85 [0.74-0.98].
Figure 4: Liver disease related hazard in UKB cohort stratified by MTARC1 rs264238 and presence of diabetes, obesity and presence of PNPLA3 rs738409:G.
Liver-related mortality was defined as death due to liver diseases or hepatocellular carcinoma. A) Liver-related mortality in non-obese participants as a function of MTARC1 genotype. B) Liver-related mortality in obese participants as a function of MTARC1 genotype. C) Liver-related mortality in non-diabetic participants as a function of MTARC1 genotype. D) Liver-related mortality in diabetic participants as a function of MTARC1 genotype. E) Liver-related mortality in PNPLA3 rs738409:G non-carriers (genotype C/C) as a function of MTARC1 genotype. D) Liver-related mortality in PNPLA3 rs738409:G-carriers (genotype G/C and G/G) as a function of MTARC1 genotype. Participants were followed prospectively from the time of study entry until death or end of follow-up. Hazard ratios were calculated by Cox regression, adjusted for age, sex, BMI, and PC1-4. HR homozygous vs. non-carriers: A) HR: 0.70 [0.48-1.02], B) HR: 0.52 [0.34-0.80], C) HR: 0.67 [0.49-0.91], D) HR: 0.44 [0.22-0.86], F) HR: 0.43 [0.27-0.71]. HR heterozygous vs. non-carriers: D) HR: 0.59 [0.35-0.81], F) HR:0.76 [0.62-0.94].
Analyzing LoF variants in MTARC1
Lastly, we investigated whether loss of MTARC1 function might be responsible for the distinctive lipid phenotype and the protective effect on several metabolic phenotypes. In UKB, we leveraged a rare stop gain mutation in the MTARC1 gene (rs139321832, p.Arg200Ter), which truncates the enzymatic domain of MTARC1 before the catalytic site 13, in 83 carriers and in PMBB we found 10 carriers the same MTARC1 LoF variant. In UKB the carriers of MTARC1 p.R200Ter showed significantly reduced HDL-C (Table S9), compared to non-carriers and in PMBB the carriers of the rare MTARC1 LoF variant showed significantly elevated Triglycerides (Table S8). MTARC1 LoF variants in both cohorts showed the same directionality as MTARC1 rs2642438 for the other analyzed metabolic phenotypes (Table S8,9). Thus, MTARC1 rs2642438 shows the same directionality of changes in metabolic phenotypes as a MTARC1 LoF variant.
Discussion
Here we expand the range of phenotypes associated with MTARC1 rs2642438, a genetic variant encoding a missense variant p.Ala165Thr that was previously reported to be associated with protection from liver disease. Using analyses in White participants in the UKB, we found that carriers of MTARC1 rs2642438 had reduced risk of NAFLD, NASH, and liver-related mortality and that the effect is even more pronounced in participants with obesity, type 2 diabetes or in presence of the known risk variant PNPLA3 rs738409:G. We found that MTARC1 rs2642438 is associated with an allele- dose dependent distinctive plasma lipid phenotype of elevated TGs and lower HDL-C as well as reduced LDL-C and apoB levels. We replicated these findings in the PMBB and extended these findings to African Americans and Black British individuals. Our results extend previous observations for this variant to heterozygotes, establishing a clear allele-dose dependent effect. Thus, MTARC1 rs2642438 is associated with a range of metabolic phenotypes that may provide insight into putative protective mechanisms against NAFLD and its consequences.
MTARC1 rs2642438 was recently shown to protect from all-cause cirrhosis and was confirmed to protect from alcoholic cirrhosis4,5,7. Moreover, MTARC1 rs2642438 carriers displayed less severe NAFLD biopsy histology9,10. MTARC1 is expressed in the liver as well as in subcutaneous adipose tissue14, and the MTARC1 protein is located in the outer mitochondrial membrane11,14. Murine data suggest that MTARC1 may be involved in hepatic drug and lipid metabolism15. In addition, MTARC1 is involved in the regulation of nitric oxide (NO) production, which is a vasodilator that can alter intrahepatic vascular resistance11.
The protective effect of MTARC1 rs2642438 on liver disease was most pronounced among those at the highest risk of NAFLD, namely those with obesity, type 2 diabetes mellitus and carriers of PNPLA3 rs738409:G. The same pattern has been described for another recently discovered liver-protective variant HSD17B13 rs72613567:TA4. In our study, A-homozygotes had a 39% lower risk of dying from liver disease compared to non-carriers. Among diabetic patients and PNPLA3 rs738409:G carriers, the corresponding associated risk reduction was more than 50%. These results are comparable with the effects of the HSD17B13 rs72613567:TA16 variant and underline the significance of the MTARC1 rs2642438 mutation. Mechanistically, these findings are in accordance with data showing that activity of the N-reductive enzyme systems, such as MTARC1 are affected by fasting (decreased MTARC1 activity) and high-fat-diet (increased MTARC1 activity) in mice and upregulated under adipogenic conditions17. A loss of function in MTARC1 as in the rs2642438 variant could therefore be particularly beneficial in those at highest risk of liver disease. While the functional effect of the p.Ala165Thr variant on MTARC1 protein function is uncertain, the direction of effect on NAFLD and all other metabolic phenotypes tested in this study is similar to that of true loss-of-function variant in MTARC1, suggesting that it is a reduced function allele5,18.
The plasma lipid phenotype associated with MTARC1 rs2642438 is interesting and may hint at a possible physiological mechanism by which MTARC1 could protect from NAFLD. In an allele-dose dependent manner, the liver protective MTARC1 rs2642438 is associated with increased plasma TGs and decreased LDL-C and apoB. Interestingly, reduced apoB levels have been linked to less NAFLD19. This pattern is consistent with the possibility of reduced MTARC1 activity leading to increased hepatic secretion of a TG-enriched VLDL without an accompanying increase or even possibly a decrease in secretion of apoB. This could result from increased loading of hepatocyte TG onto TG-enriched VLDL particles, which would be predicted to decrease hepatic fat. A recent study showed, that MTARC1 rs2642438 carriers had higher concentrations of hepatic polyunsaturated phosphatidylcholines10 and higher sphingomyelin levels20 compared to non-carriers. This hepatic lipid profile was remarkably similar to that in carriers of HSD17B13 rs72613567:TA21 and, interestingly, opposite to that of PNPLA3 rs738409:G carriers22. Increased phosphatidylcholine is known to be associated with less NAFLD23 and is also strongly related to the synthesis of VLDL24. Phosphatidylcholine is a required component of the VLDL envelope and regulates the size and dynamics of lipid droplets24.
We analyzed the interaction between MTARC1 and IGF-1 as IGF-1 might be involved in mechanisms of hepatocyte TG secretion25,26. Consistently, we show increased IGF-1 levels in MTARC1 p.Ala165Thr carriers, which were associated with higher serum TGs. On the other hand, the fact that MTARC1 p.Ala165Thr carriers showed elevated levels of urate is surprising, especially given the fact that xanthine oxidase, the leading enzyme in urate production, belongs to the same ancient group of molybdenum containing enzymes as the MTARC1 complex15. Moreover, we analyzed Vitamin D in MTARC1 p.Ala165Thr carriers as Vitamin D is associated with lower risk of NAFLD27 Here, it is unclear whether MTARC1 leads to higher vitamin D levels due to its liver protective nature, or if MTARC1 might even be involved in vitamin D synthesis. Together, these data underline that MTARC1 is an important metabolic orchestrator.
Previous studies of MTARC1 were mostly performed in European participants5. Analysis of trans-ethnic cohorts may help to clarify whether variation in MTARC1 rs2642438 confers NAFLD protection among non-European individuals. Although we found a reduced minor allele frequency in our African American and Black British subcohorts, African American MTARC1 p.Ala165Thr carriers showed significantly reduced NAFLD/NASH and both subgroups showed the distinctive lipid profile. Further studies are warranted to confirm these associations and to explore the effect of MTARC1 rs2642438 in other ethnic groups.
Limitations of study
While the UK Biobank was utilized as part of the initial report of the association between MTARC1 rs2642438 and cirrhotic liver disease,5 our genotype-first approach revealed several new phenotypes that have not been reported, including the favorable effect of MTARC1 rs2642438 on liver-related survival. We also used the PMBB to confirm our findings in an unrelated bi-ethnic medical biobank. A limitation is that outcomes based on ICD codes are likely to suffer from some degree of misclassification or underdiagnosis. In particular, we found the numbers of NAFLD classified participants in the UKB to be quite low compared to other cohorts28,29. This might lead to an underestimation of the effect of MTARC1 in the general population. However, we were able to show a robust association between MTARC1 and obesity, diabetes, steatogenic risk, plasma ALT and NAFLD diagnosis in two independent cohorts. Strengths of our study include the long follow-up time for a median of 10-12 years and the large number of participants from two unrelated cohorts.
In summary, MTARC1 rs2642438 is associated in an allele-dose dependent manner with reduced NAFLD, NASH, and liver-related mortality, especially in subpopulations prone to NAFLD development. Furthermore, it is associated with an interesting plasma lipid phenotype characterized by elevated TGs but reduced LDL-C and apoB, suggestive of a potential physiological mechanism by which MTARC1 influences NAFLD and its progression. MTARC1 presents an interesting therapeutic target for NAFLD and its consequences without raising the risk of cardiovascular outcomes. Inhibition of MTARC1 may be expected to increase plasma triglyceride levels even as it reduces hepatic fat.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests should be directed to and will be fulfilled by the Lead Contact Dr. med. Carolin V. Schneider (carolin.schneider@pennmedicine.upenn.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The datasets used in the current study have not been deposited in a public repository, but are available after approval of a reasonable application at https://www.ukbiobank.ac.uk for UKB and for PMBB under https://www.itmat.upenn.edu/biobank/researchers.html. For further information, please contact the corresponding author.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
The ‘UK biobank’ (UKB) is a population-based cohort study conducted in the United Kingdom from 2006 to 2010, which recruited 502,505 volunteers aged 37 to 73 years at baseline (Figure 1A). All participants were registered with the UK National Health Service and attended an initial examination, which is followed by a long-term follow-up taking place continuously. On all analyzed visits, blood samples were taken and physical measures were performed. All participants gave informed consent for genotyping and data linkage to medical reports. Cardiac MRI and liver MRI were performed in subsets of participants (Table S1,S4,S5). Genotyping of the MTARC1 variant rs2642438 was conducted in a total of 488,377 White subjects. We excluded participants with chronic hepatitis B or C (n=850) or heavy alcohol consumption (>60g alcohol/day for men or >40g alcohol/day for women, n=4490) and participants with ethnicity listed as missing/other (n=847).
Additionally, a second group of 7,559 Black British UKB participants without chronic hepatitis C and heavy alcohol consumption and with available MTARC1 rs2642438 genotyping was analyzed (Table S3).
For both groups, ongoing inpatient hospital records beginning in 1996 until 2018 were used to identify diagnoses according to the International Classification of Diseases, Tenth Revision (ICD-10) codes. The presence of the following primary ICD10 codes was evaluated: Overall liver disease (K70-K76), Alcoholic liver disease (K70), Toxic liver disease (K71), Hepatic failure (K72), Chronic Hepatitis (K73), Fibrosis and cirrhosis (K74), Inflammatory liver diseases (K75), NASH (K75.8), Other liver diseases (K76), NAFLD (K76.0) and Malignant neoplasm of the liver and/or intrahepatic bile duct (C22). The UK Biobank receives death notifications (age at death and primary ICD diagnosis that led to death) through linkage to national death registries. End of follow-up was defined as death or end of hospital inpatient data collection in June 2020. Specific causes of death included cardiovascular diseases (I00–I99) and liver diseases (K70–K77 and C22). The study has been approved by the UKB Access Committee (Project #59657).
Participants in the Penn Medicine BioBank (PMBB) were recruited from clinical practice sites throughout the University of Pennsylvania Health System. Participants consented for access to EHR data. Whole exome sequences were generated from DNA extracted from stored buffy coats by the Regeneron Genetics Center and mapped to Genome Reference Consortium Build 38 (GRCh38). For subsequent phenotypic analyses, we removed samples with low exome sequencing coverage (i.e., less than 75% of targeted bases achieving 20×coverage; n=46), high missingness (i.e., greater than 5% of targeted bases; n=14), high heterozygosity (n=97), dissimilar reported and genetically determined sex (n=104), genetic evidence of sample duplication (n=89), and cryptic relatedness (i.e., closer than third-degree relatives; n=145) with overlap among categories, leading to a total of 455 removed from our database. For the PMBB cohort, ICD-9 and ICD-10 diagnosis codes were extracted. In total, the MTARC1 variant rs2642438 was analyzed in a total of 15,859 unrelated subjects (Figure 1B). We excluded participants with chronic hepatitis B or C (n=593) or subjects with alcohol-related conditions or dependence (n=301), such as alcoholic liver disease (571.0, K70.0), alcoholic hepatitis (571.1, K70.1), alcoholic fibrosis and sclerosis of the liver (571.2, K70.3), alcoholic cirrhosis of liver and/or ascites (571.2, K70.2), alcoholic hepatic failure, coma, and unspecified alcoholic liver disease (571.3, K70.4, K70.40, K70.41, K70.9), and alcohol dependence (303.0, 303.9, F10.229, F10.20) and participants with ethnicity listed as other or missing ethnicity (n=901). Daily alcohol consumption data was not available in PMBB. Self-reported Ethnicity was assessed at baseline using questionnaires and EHR data. Ongoing inpatient and outpatient records between the baseline examination and July 2020 were used to identify diagnoses according to ICD-10 codes. The same ICD10-diagnoses as in the UKB cohort were used. In addition, the following ICD-9 codes were used and translated into ICD-10 codes: Hepatic failure (570), chronic hepatitis (571.4), fibrosis and cirrhosis (571.5) and NAFLD (571.81/571.89). Additionally, serum parameters were extracted for participants from the time of enrollment in the PMBB until June 1, 2020; if multiple measurements were available the median value was chosen. Measurements above the 20xULN were excluded. In addition, values for age, sex, BMI and diabetes mellitus were also extracted from the EHR. The PMBB receives death notifications (age at death and primary ICD diagnosis that led to death) through linkage to the EHR. End of follow-up was defined as death or end of hospital inpatient data collection at the end of May 2020. Specific causes of death included cardiovascular diseases (I00–I99) and liver diseases (K70–K77 and C22).
METHOD DETAILS
To examine whether MTARC1 deficiency may also protect against the analyzed phenotypes, we leveraged a rare stop codon in MTARC1 (Arg200Ter, rs139321832) in UKB and PMBB (Table S8,9). This stop codon truncates the enzymatic domain of MTARC1 before the catalytic site 13 The same exclusion criteria as mentioned above were used. 83 carriers were analyzed in UKB and 10 carriers in PMBB.
QUANTIFICATION AND STATISTICAL ANALYSIS
All continuous variables were analyzed by unpaired, two-tailed t-tests or Mann-Whitney U test, and by an appropriate multivariable model. The results are presented as mean±standard deviation (normal distribution). All categorical variables were displayed as relative (%) frequencies and the corresponding contingency tables were analyzed using the Chi-square test. Odds/hazard ratios (ORs/HRs) were presented with their corresponding 95% confidence intervals(CI) given in brackets. HRs were calculated using Cox proportional hazard regression models. Multivariable logistic regression was performed to test for independent associations. All multivariable analyses were adjusted for age, sex, BMI and principal components of ancestry 1-4. The reference groups in this association study for analyzing any specific ICD10 code are participants without the specific ICD10 code. A FDR adjusted significance level of p <= 0.03 was calculated using the Benjamini-Hochberg method30.
Differences were considered to be statistically significant when p<0.05. The data were analyzed using R version 4.0.2 (R Foundation for Statistical Computing; Vienna, Austria), SPSS Statistics version 26 (IBM; Armonk, NY, USA) and Prism version 8 (GraphPad, LaJolla, CA, USA).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Bacterial and virus strains | ||
| Biological samples | ||
| Chemicals, peptides, and recombinant proteins | ||
| Critical commercial assays | ||
| Deposited data | ||
| Experimental models: cell lines | ||
| Experimental models: organisms/strains | ||
| Oligonucleotides | ||
| Recombinant DNA | ||
| Software and algorithms | ||
| SPSS IBM; Armonk, NY, USA | SPSS 26 | IBM; Armonk, NY, USA |
| Prism version 8, GraphPad, LaJolla, CA, USA | Prism | GraphPad, LaJolla, CA, USA |
| R version 4.0.2- | R | R Foundation for Statistical Computing; Vienna, Austria |
| Other | ||
Acknowledgments:
This research has been conducted using the UK Biobank Resource under Application Number 59657. C.V.S is supported by Walter-Benjamin Fellowship from German Research Foundation (SCHN- 1640/1-1). K.M.S is supported by the German Research Foundation (DFG) consortium (SCHN 1626/1-1). P.S. is supported by the German Research Foundation (DFG) consortium SFB 1382 “Gut-liver axis” and DFG grant STR 1095/6-1 (Heisenberg professorship). Dr. Carr has received research support from Intercept Pharmaceuticals and NIH 1R01AA026302. The Penn Medicine BioBank is funded by the Perelman School of Medicine at the University of Pennsylvania and a gift from the Smilow family. C.V.S analyzed the data overseen by K.M.S. C.V.S and D.J.R had unrestricted access to all the data. C.V.S, D.J.R and K.M.S drafted the first version of the manuscript and D.M.C., J.P, M.V, I.Z, Y.K, C.T, R.M.C and P.S. reviewed and edited it. All authors agreed to submit the manuscript, read and approved the final draft and take full responsibility of its content, including the accuracy of the data and its statistical analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest: The authors declare no competing interests
References
- 1.Younossi ZM, Koenig AB, Abdelatif D, et al. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016;64:73–84. [DOI] [PubMed] [Google Scholar]
- 2.Hardy T, Oakley F, Anstee QM, et al. Nonalcoholic Fatty Liver Disease: Pathogenesis and Disease Spectrum. Annu Rev Pathol 2016;11:451–496. [DOI] [PubMed] [Google Scholar]
- 3.Buch S, Stickel F, Trepo E, et al. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat Genet 2015;47:1443–1448. [DOI] [PubMed] [Google Scholar]
- 4.Abul-Husn NS, Cheng X, Li AH, et al. A protein-truncating HSD17B13 variant and protection from chronic liver disease. N Engl J Med 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emdin C, Haas M, Khera A, et al. A missense variant in Mitochondrial Amidoxime Reducing Component 1 gene and protection against liver disease. PLOS Genet 2020;16:e1008629. Available at: http://biorxiv.org/content/early/2019/03/31/594523.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anstee QM, Darlay R, Cockell S, et al. Genome-wide association study of nonalcoholic fatty liver and steatohepatitis in a histologically characterised cohort(☆). J Hepatol 2020;73:505–515. [DOI] [PubMed] [Google Scholar]
- 7.Innes H, Buch S, Hutchinson S, et al. Genome-wide Association Study for Alcohol-related Cirrhosis Identifies Risk Loci in MARC1 and HNRNPUL1. Gastroenterology 2020. [DOI] [PubMed] [Google Scholar]
- 8.Schwantes-An T-H, Darlay R, Mathurin P, et al. Genome-wide association study and meta-analysis on alcohol-related liver cirrhosis identifies novel genetic risk factors. Hepatology 2020. [DOI] [PubMed] [Google Scholar]
- 9.Alisi A, Anstee Q. Variants in MARC1 and HSD17B13 reduce severity of NAFLD in children , perturb phospholipid metabolism , and suppress fibrotic pathways. 2020. [Google Scholar]
- 10.Luukkonen PK, Juuti A, Sammalkorpi H, et al. MARC1 variant rs2642438 increases hepatic phosphatidylcholines and decreases severity of non-alcoholic fatty liver disease in humans. J Hepatol 2020;73:725–726. [DOI] [PubMed] [Google Scholar]
- 11.Sparacino-Watkins CE, Tejero J, Sun B, et al. Nitrite reductase and nitric-oxide synthase activity of the mitochondrial molybdopterin enzymes mARC1 and mARC2. J Biol Chem 17205BC;289:10345,10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai G, Liu P, Li X, et al. Association between PNPLA3 rs738409 polymorphism and nonalcoholic fatty liver disease (NAFLD) susceptibility and severity: A meta-analysis. Medicine (Baltimore) 2019;98:e14324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kubitza C, Bittner F, Ginsel C, et al. Crystal structure of human mARC1 reveals its exceptional position among eukaryotic molybdenum enzymes. Proc Natl Acad Sci 2018;115:11958–11963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neve EPA, Köfeler H, Hendriks DFG, et al. Expression and Function of mARC: Roles in Lipogenesis and Metabolic Activation of Ximelagatran. PLoS One 2015;10:e0138487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ott G, Havemeyer A, Clement B. The mammalian molybdenum enzymes of mARC. 2015:265–275. [DOI] [PubMed] [Google Scholar]
- 16.Gellert-Kristensen H, Nordestgaard BG, Tybjaerg-Hansen A, et al. High Risk of Fatty Liver Disease Amplifies the Alanine Transaminase–Lowering Effect of a HSD17B13 Variant. Hepatology 2020;71:56–66. Available at: 10.1002/hep.30799. [DOI] [PubMed] [Google Scholar]
- 17.Jakobs HH, Mikula M, Havemeyer A, et al. The N-Reductive System Composed of Mitochondrial Amidoxime Reducing The N-Reductive System Composed of Mitochondrial Amidoxime Reducing Component ( mARC ), Cytochrome b5 ( CYB5B ) and Cytochrome b5 Reductase ( CYB5R ) Is Regulated by Fasting and High Fat . 2014;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hudert CA, Alisi A, Anstee QM, et al. reduce severity of NAFLD in children , perturb phospholipid metabolism , and suppress fibrotic pathways. 2020. [Google Scholar]
- 19.Wang J, Zhu W, Huang S, et al. Serum apoB levels independently predict the development of non-alcoholic fatty liver disease: A 7-year prospective study. Liver Int Off J Int Assoc Study Liver 2017;37:1202–1208. [DOI] [PubMed] [Google Scholar]
- 20.Mann JP, Pietzner M, Wittemans LB, et al. Insights into genetic variants associated with NASH-fibrosis from metabolite profiling. Hum Mol Genet 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luukkonen PK, Tukiainen T, Juuti A, et al. Hydroxysteroid 17-β dehydrogenase 13 variant increases phospholipids and protects against fibrosis in nonalcoholic fatty liver disease. JCI insight 2020;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luukkonen PK, Nick A, Hölttä-Vuori M, et al. Human PNPLA3-I148M variant increases hepatic retention of polyunsaturated fatty acids. JCI insight 2019;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tiwari-Heckler S, Gan-Schreier H, Stremmel W, et al. Circulating Phospholipid Patterns in NAFLD Patients Associated with a Combination of Metabolic Risk Factors. Nutrients 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veen JN van der, Kennelly JP, Wan S, et al. The critical role of phosphatidylcholine and phosphatidylethanolamine metabolism in health and disease. Biochim Biophys acta Biomembr 2017;1859:1558–1572. [DOI] [PubMed] [Google Scholar]
- 25.Arturi F, Succurro E, Procopio C, et al. Nonalcoholic Fatty Liver Disease Is Associated with Low Circulating Levels of Insulin-Like Growth Factor-I. J Clin Endocrinol Metab 2011;96:E1640–E1644. Available at: 10.1210/jc.2011-1227. [DOI] [PubMed] [Google Scholar]
- 26.Adamek A, Kasprzak A. Insulin-Like Growth Factor (IGF) System in Liver Diseases. Int J Mol Sci 2018;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z, Thorne JL, Moore JB. Vitamin D and nonalcoholic fatty liver disease. Curr Opin Clin Nutr Metab Care 2019;22:449–458. [DOI] [PubMed] [Google Scholar]
- 28.Dunn W, Xu R, Wingard DL, et al. Suspected nonalcoholic fatty liver disease and mortality risk in a population-based cohort study. Am J Gastroenterol 2008;103:2263–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cleveland ER, Ning H, Vos MB, et al. Low Awareness of Nonalcoholic Fatty Liver Disease in a Population-Based Cohort Sample: the CARDIA Study. J Gen Intern Med 2019;34:2772–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benjamini Y, Drai D, Elmer G, et al. Controlling the false discovery rate in behavior genetics research. Behav Brain Res 2001;125:279–284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used in the current study have not been deposited in a public repository, but are available after approval of a reasonable application at https://www.ukbiobank.ac.uk for UKB and for PMBB under https://www.itmat.upenn.edu/biobank/researchers.html. For further information, please contact the corresponding author.