ABSTRACT
The pathogenic mechanisms underlying severe SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) infection remain largely unelucidated. High-throughput sequencing technologies that capture genome and transcriptome information are key approaches to gain detailed mechanistic insights from infected cells. These techniques readily detect both pathogen- and host-derived sequences, providing a means of studying host-pathogen interactions. Recent studies have reported the presence of host-virus chimeric (HVC) RNA in transcriptome sequencing (RNA-seq) data from SARS-CoV-2-infected cells and interpreted these findings as evidence of viral integration in the human genome as a potential pathogenic mechanism. Since SARS-CoV-2 is a positive-sense RNA virus that replicates in the cytoplasm, it does not have a nuclear phase in its life cycle. Thus, it is biologically unlikely to be in a location where splicing events could result in genome integration. Therefore, we investigated the biological authenticity of HVC events. In contrast to true biological events like mRNA splicing and genome rearrangement events, which generate reproducible chimeric sequencing fragments across different biological isolates, we found that HVC events across >100 RNA-seq libraries from patients with coronavirus disease 2019 (COVID-19) and infected cell lines were highly irreproducible. RNA-seq library preparation is inherently error prone due to random template switching during reverse transcription of RNA to cDNA. By counting chimeric events observed when constructing an RNA-seq library from human RNA and spiked-in RNA from an unrelated species, such as the fruit fly, we estimated that ∼1% of RNA-seq reads are artifactually chimeric. In SARS-CoV-2 RNA-seq, we found that the frequency of HVC events was, in fact, not greater than this background “noise.” Finally, we developed a novel experimental approach to enrich SARS-CoV-2 sequences from bulk RNA of infected cells. This method enriched viral sequences but did not enrich HVC events, suggesting that the majority of HVC events are, in all likelihood, artifacts of library construction. In conclusion, our findings indicate that HVC events observed in RNA-sequencing libraries from SARS-CoV-2-infected cells are extremely rare and are likely artifacts arising from random template switching of reverse transcriptase and/or sequence alignment errors. Therefore, the observed HVC events do not support SARS-CoV-2 fusion to cellular genes and/or integration into human genomes.
IMPORTANCE The pathogenic mechanisms underlying SARS-CoV-2, the virus responsible for COVID-19, are not fully understood. In particular, relatively little is known about the reasons some individuals develop life-threatening or persistent COVID-19. Recent studies identified host-virus chimeric (HVC) reads in RNA-sequencing data from SARS-CoV-2-infected cells and suggested that HVC events support potential “human genome invasion” and “integration” by SARS-CoV-2. This suggestion has fueled concerns about the long-term effects of current mRNA vaccines that incorporate elements of the viral genome. SARS-CoV-2 is a positive-sense, single-stranded RNA virus that does not encode a reverse transcriptase and does not include a nuclear phase in its life cycle, so some doubts have rightfully been expressed regarding the authenticity of HVCs and the role played by endogenous retrotransposons in this phenomenon. Thus, it is important to independently authenticate these HVC events. Here, we provide several lines of evidence suggesting that the observed HVC events are likely artifactual.
KEYWORDS: COVID-19, SARS-CoV-2, RNA sequencing, sequencing reads, chimeric reads, host-virus fusion
INTRODUCTION
Advances in and availability of high-throughput sequencing technologies have enabled the accumulation of detailed molecular-level information from cells, including genome variations, gene transcription, and gene regulation. These technologies are extremely sensitive at capturing nucleic acid sequences regardless of their origin. As such, the data from these techniques contain not only sequences encoded by the cell itself but also sequences encoded by infecting pathogens and/or common contaminating agents (e.g., vectors, plasmids, etc.) (1, 2). In virus-infected cells, captured sequences derived from the host or virus, if appropriately analyzed, represent a powerful tool to study the mechanisms underlying host-pathogen interactions. High-throughput assays are invaluable resources for identifying novel biological events, and they provide exceptionally detailed information about host-virus interactions occurring in vivo. This is exemplified by the discovery of viral integration as a driver of oncogenesis in HPV-associated cancers (3). We have previously deployed these methods to gain mechanistic insights into the pathophysiology of oncogenic viruses like Epstein-Barr virus (EBV), hepatitis B virus (HBV), and human papillomavirus (HPV) (1, 4–6). Even in the absence of infection, detailed analyses of transcriptome sequencing (RNA-seq) data can deliver new insights into cell biology, including, for example, discovery of novel linear or circular genes and isoforms. On the other hand, the technology is extremely sensitive, relies on low-fidelity reverse transcriptases (RTs) during library preparation, and presents many computational challenges during chimeric-sequence alignment. Collectively, these can result in the detection of low-frequency sequencing reads originating from artifactual events or contaminants (including plasmid vectors). Thus, appropriate positive and negative controls during analysis are essential to distinguish real from artifactual events.
RNA-sequencing data from virally infected cells contain reads that map perfectly to either the host genome or the viral genome. However, a significant portion of host sequencing reads can also be aligned to discontiguous sections of the genome and often represent canonical forward-splicing or back-splicing events generated from mRNAs and circular RNAs, respectively (7, 8). In cells infected with DNA viruses that integrate into the host genome (e.g., HPV or HBV), a chimeric read that is partly mapped to the host genome and partly mapped to the virus genome is a signature of transcribed segments of the host genome containing integrated viral DNA (9–11). Similarly, in virus-induced-cancer cells, chimeric reads that are partly mapped to one gene and partly mapped to another gene are the markers of genomic rearrangement and/or gene fusion (12, 13). Thus, chimeric reads can represent real biological events.
The pathogenic mechanisms underlying severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus responsible for pandemic coronavirus disease of 2019 (COVID-19), are under investigation (14–17) but still not fully understood. In particular, relatively little is known about the processes following viral infection and why some individuals develop mild or no symptoms, while others develop life-threatening or persistent (“long”) COVID-19. In this setting, sequencing assays are key methods for uncovering as-yet-undiscovered mechanisms of pathogenesis. Recent studies have identified host-virus chimeric (HVC) reads in RNA-sequencing data from SARS-CoV-2-infected cells and samples from COVID-19 patients (18, 19). Both studies have suggested that HVC events support potential “human genome invasion” and “integration” by SARS-CoV-2. This suggestion has fueled concerns about the long-term effects of current vaccines that incorporate elements of the viral genome (20). SARS-CoV-2 is a positive-sense, single-stranded RNA virus that does not encode a reverse transcriptase and does not include a nuclear phase in its life cycle, so some doubts have rightfully been expressed regarding the authenticity of HVCs and the role played by endogenous retrotransposons in this phenomenon. Thus, it is important to independently authenticate these HVC events.
Therefore, we investigated the presence of HVC events in a large number of currently available RNA-sequencing samples from SARS-CoV-2-infected cells and patients with COVID-19. We also developed and deployed a novel experimental approach that enriched viral sequences from infected cells during RNA-seq library preparation. Collectively, we conclude that current data do not support the authenticity of HVC events in SARS-CoV-2-infected samples.
RESULTS
HVC events are detected in RNA-seq from SARS-CoV-2-infected cells but infrequently in samples from patients with COVID-19.
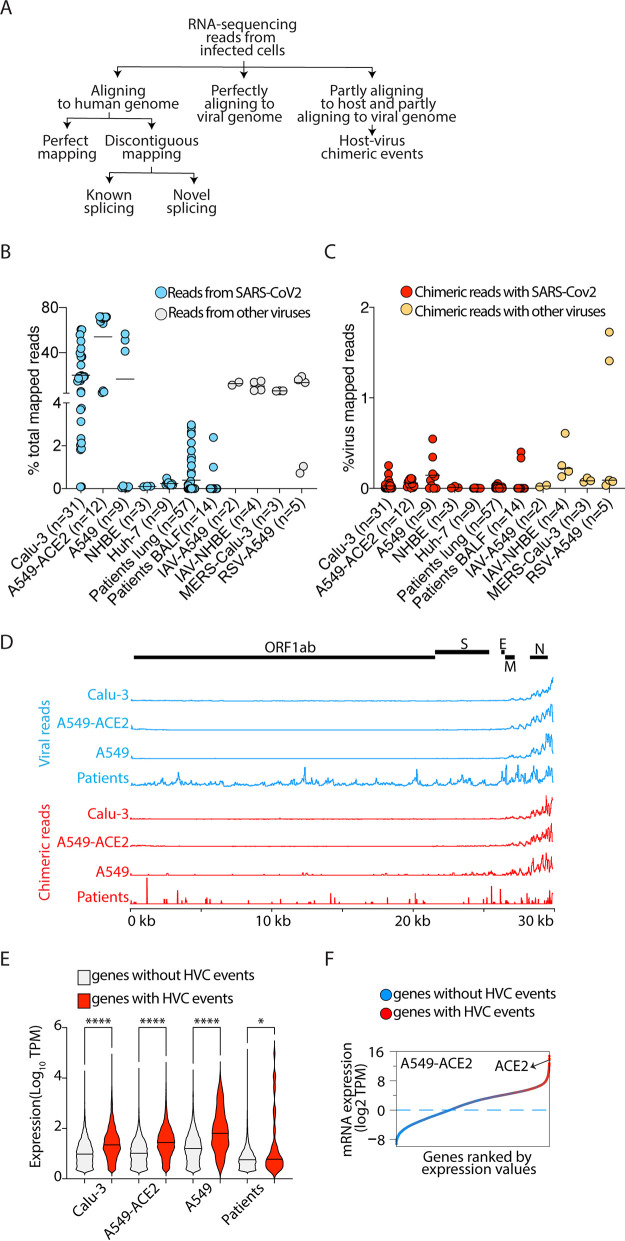
Recent reports (18, 19) about the presence of HVC events in SARS-CoV-2-infected cells have been interpreted as supporting evidence for viral integration into the human genome and as a potential mechanism of viral persistence. To gain insights into the authenticity of these HVC events, we extracted RNA-sequencing reads that partly align to the host genome and partly align to the viral genome. These reads are considered the signature of HVC events in RNA-sequencing data sets. Specifically, we reanalyzed the three available RNA-seq data sets from patients with COVID-19 (n = 57 samples) and in vitro SARS-CoV-2-infected cells (n = 64 samples). We categorized sequencing reads as those that perfectly aligned to the human genome (build hg38) in a contiguous or discontiguous manner (i.e., reads originating from one exon or reads spanning exon-exon junctions), those that perfectly aligned to the SARS-CoV-2 viral genome, and those that partly aligned to both host and viral genomes (potentially representing HVC events) (Fig. 1A and Tables 1 and 2). Viral-genome-mapped reads were detected across several cell lines infected with SARS-CoV-2 (Fig. 1B and Tables 1 and 2). SARS-CoV-2-infected Calu-3 and A549-ACE2 cells had the highest percentages (∼20 to 70%) of viral reads, while other cells, including A549 cells, samples from lung autopsies, or bronchoalveolar lavage fluid (BALF) of patients with COVID-19, had dramatically lower representation of viral reads (Fig. 1B and Tables 1 and 2). This is consistent with the fact that overexpression of angiotensin-converting enzyme 2 (ACE2), a well-characterized SARS-CoV-2 entry receptor, on A549 cells significantly enhances SARS-CoV-2 viral titers. The frequency of viral reads in cells infected in vitro with other viruses, including influenza A virus (IAV), Middle East respiratory syndrome (MERS) coronavirus, and respiratory syncytial virus (RSV), were within the same range (∼10 to 15%) as those of Calu-3 and A549 cells, indicating similar levels of in vitro infectivity of these respiratory viruses (Fig. 1B and Table 3).
FIG 1.
HVC events are detectable in RNA-seq from SARS-CoV-2-infected cells but infrequently in samples from COVID-19 patients. (A) Schematic presentation of RNA-sequencing data analysis pipeline. (B) Viral reads in the indicated SARS-CoV-2-infected or other virally infected cells as a proportion of the total reads mapped to the chimeric genome. (C) HVC reads in the indicated SARS-CoV-2-infected or other virally infected cells as a proportion of the total reads mapped to the virus genome. (D) SARS-CoV-2 genome coverage based on reads mapping perfectly to the virus genome (top) or to the viral segments of HVC events (bottom). (E) Violin plots showing the expression of all human genes with or without HVC events in the indicated infected cells. *, P < 0.05, and ****, P < 0.0001, by Kruskal-Wallis and FDR correction. (F) Dot plots showing the expression of all human genes in SARS-CoV-2-infected A549-ACE2 cells ordered by gene expression level. Genes with or without HVC events are highlighted with red and blue, respectively. See Tables 1 and 2 for the sources of data in this figure. TPM, transcripts per million.
TABLE 1.
SARS-CoV-2-infected samples from independent studies used here
| Sample | Accession no. | Reference |
|---|---|---|
| A549-ACE2 | GSE147507 | 25 |
| GSE154613 | 26 | |
| A549 | GSE159191 | 27 |
| GSE147507 | 25 | |
| Calu-3 | GSE147507 | 25 |
| PRJNA665581/SRP285334 | 28 | |
| GSE148729 | 29 | |
| COVID-19 patients | GSE147507 | 25 |
| GSE151803 | 30 | |
| GSE150316 | 31 |
See Table 2 for the complete list of individual samples.
TABLE 2.
Detailed information on RNA-seq libraries from SARS-CoV-2-infected cells used in this studya
| Accession no. | Sample annotation | GEO/SRA data set | Library sizeb | No. of reads mapped to: |
Ratio of reads mapped to virus/total mapped reads (%)c | No. of chimeric reads | Ratio of chimeric reads/reads mapped to virus (%) | No. of unique chimeric reads (HVC events)d | |
|---|---|---|---|---|---|---|---|---|---|
| Human genome | Virus | ||||||||
| GSM4432387 | A549 | GSE147507 | 34,141,057 | 29,554,656 | 11,210 | 0.0379 | 9 | 0.09 | 9 |
| GSM4432388 | A549 | GSE147507 | 29,681,064 | 23,681,134 | 7,589 | 0.03 | 10 | 0.13 | 10 |
| GSM4432389 | A549 | GSE147507 | 20,603,153 | 17,430,125 | 6,252 | 0.04 | 9 | 0.16 | 9 |
| SRR11517677 | A549 | GSE147507 | 22,280,277 | 19,354,844 | 17,481 | 0.09 | 5 | 0.03 | 3 |
| SRR11517678 | A549 | GSE147507 | 24,949,250 | 21,724,729 | 19,129 | 0.09 | 66 | 0.35 | 62 |
| SRR11517679 | A549 | GSE147507 | 44,074,243 | 38,651,231 | 48,607 | 0.13 | 264 | 0.55 | 191 |
| SRR12789544 | A549_12h | GSE159191 | 15,666,722 | 8,422,244 | 5,904,068 | 41.21 | 20 | 0.00 | 17 |
| SRR12789545 | A549_18h | GSE159191 | 12,449,164 | 5,515,627 | 5,770,772 | 51.13 | 14 | 0.00 | 13 |
| SRR12789546 | A549_24h | GSE159191 | 14,774,320 | 5,829,549 | 7,601,829 | 56.60 | 20 | 0.00 | 17 |
| SRR11517741 | A549_ACE2 | GSE147507 | 23,586,566 | 5,470,659 | 12,856,846 | 70.15 | 8,524 | 0.07 | 5,806 |
| SRR11517742 | A549_ACE2 | GSE147507 | 5,720,676 | 1,499,512 | 2,887,501 | 65.82 | 2,800 | 0.11 | 2,476 |
| SRR11517743 | A549_ACE2 | GSE147507 | 4,841,347 | 1,336,788 | 2,815,601 | 67.81 | 1,661 | 0.06 | 1,273 |
| GSM4486160 | A549_ACE2 | GSE147507 | 18,924,539 | 4,717,293 | 11,176,081 | 70.32 | 1,779 | 0.02 | 1,736 |
| GSM4486161 | A549_ACE2 | GSE147507 | 21,401,433 | 5,021,108 | 12,693,264 | 71.66 | 996 | 0.01 | 970 |
| GSM4486162 | A549_ACE2 | GSE147507 | 20,579,555 | 4,940,143 | 12,147,522 | 71.09 | 471 | 0.00 | 456 |
| GSM4486163 | A549_ACE2 | GSE147507 | 29,810,652 | 7,079,255 | 18,102,487 | 71.89 | 8,363 | 0.05 | 8,145 |
| GSM4486164 | A549_ACE2 | GSE147507 | 27,594,875 | 6,637,707 | 16,827,881 | 71.71 | 3,970 | 0.02 | 3,915 |
| GSM4486165 | A549_ACE2 | GSE147507 | 21,641,361 | 5,155,362 | 13,202,056 | 71.92 | 6,721 | 0.05 | 6,579 |
| GSM4675772 | A549_ACE2 | GSE154613 | 17,644,925 | 15,260,808 | 705,436 | 4.42 | 652 | 0.09 | 640 |
| GSM4675773 | A549_ACE2 | GSE154613 | 19,504,193 | 16,548,574 | 1,077,217 | 6.11 | 1,113 | 0.10 | 1,082 |
| GSM4675774 | A549_ACE2 | GSE154613 | 19,491,861 | 16,686,001 | 903,726 | 5.14 | 994 | 0.11 | 975 |
| GSM5097244 | Calu3_HV | GSE167131 | 140,579,755 | 45,289,967 | 65,799,441 | 59.23 | 3,028 | 0.00 | 362 |
| GSM5097245 | Calu3_VH | GSE167131 | 75,242,297 | 30,713,952 | 8,278,786 | 21.23 | 1,496 | 0.02 | 145 |
| GSM5097246 | Calu3_NoEnrich | GSE167131 | 66,883,590 | 21,643,220 | 2,388,074 | 9.94 | 388 | 0.02 | 88 |
| SRR11517747 | Calu-3 | GSE147507 | 23,623,325 | 17,598,926 | 3,152,004 | 15.19 | 4,817 | 0.16 | 4,175 |
| SRR11517748 | Calu-3 | GSE147507 | 13,583,713 | 9,552,755 | 2,332,185 | 19.62 | 1943 | 0.09 | 1,310 |
| SRR11517749 | Calu-3 | GSE147507 | 28,688,015 | 18,300,633 | 4,242,803 | 18.82 | 9,984 | 0.25 | 9,026 |
| GSM4477994 | Calu3_12h | GSE148729 | 231,639,158 | 103,625,245 | 6,038,632 | 5.51 | 1,193 | 0.02 | 980 |
| GSM4477995 | Calu3_12h | GSE148729 | 346,367,074 | 138,220,519 | 6,281,366 | 4.35 | 3,409 | 0.05 | 2,682 |
| GSM4477996 | Calu3_4h | GSE148729 | 447,838,192 | 203,717,693 | 192,681 | 0.09 | 66 | 0.03 | 59 |
| SRR11550087 | Calu3_4h | GSE148729 | 202,773,322 | 91,185,444 | 69,632 | 0.08 | 28 | 0.04 | 26 |
| GSM4477999 | Calu3_8h | GSE148729 | 550,251,760 | 238,772,517 | 9,113,472 | 3.68 | 3,300 | 0.04 | 2,537 |
| SRR11550088 | Calu3_8h | GSE148729 | 216,516,118 | 98,795,456 | 2,149,708 | 2.13 | 580 | 0.03 | 481 |
| SRR11549991 | Calu3_12h | GSE148729 | 11,433,667 | 6,324,394 | 3,621,295 | 36.41 | 66 | 0.00 | 65 |
| SRR11549992 | Calu3_12h | GSE148729 | 8,260,125 | 4,522,110 | 2,589,574 | 36.41 | 42 | 0.00 | 42 |
| SRR11550003 | Calu3_12h | GSE148729 | 27,573,490 | 14,996,384 | 9,865,189 | 39.68 | 199 | 0.00 | 190 |
| SRR11550004 | Calu3_12h | GSE148729 | 23,872,357 | 12,203,177 | 9,462,280 | 43.67 | 220 | 0.00 | 207 |
| SRR11550043 | Calu3_12h | GSE148729 | 31,302,340 | 11,146,014 | 17,077,457 | 60.51 | 270 | 0.00 | 199 |
| SRR11550044 | Calu3_12h | GSE148729 | 31,082,694 | 11,133,581 | 16,977,009 | 60.39 | 352 | 0.00 | 265 |
| SRR11549993 | Calu3_24h | GSE148729 | 7,679,519 | 5,058,810 | 1,193,215 | 19.09 | 49 | 0.00 | 47 |
| SRR11549994 | Calu3_24h | GSE148729 | 9,799,418 | 6,977,556 | 1,199,973 | 14.67 | 34 | 0.00 | 33 |
| SRR11549997 | Calu3_24h | GSE148729 | 3,409,463 | 1,836,844 | 751,057 | 29.02 | 16 | 0.00 | 15 |
| SRR11549998 | Calu3_24h | GSE148729 | 3,710,276 | 2,297,976 | 532,742 | 18.82 | 19 | 0.00 | 17 |
| SRR11550045 | Calu3_24h | GSE148729 | 30,562,938 | 11,731,550 | 14,890,548 | 55.93 | 519 | 0.00 | 394 |
| SRR11550046 | Calu3_24h | GSE148729 | 34,119,394 | 14,646,557 | 14,795,361 | 50.25 | 368 | 0.00 | 278 |
| SRR11549989 | Calu3_4h | GSE148729 | 14,535,594 | 12,050,301 | 249,444 | 2.03 | 2 | 0.00 | 2 |
| SRR11549990 | Calu3_4h | GSE148729 | 17,211,547 | 14,378,232 | 343,858 | 2.34 | 2 | 0.00 | 2 |
| SRR11550009 | Calu3_4h | GSE148729 | 24,407,750 | 20,817,989 | 385,881 | 1.82 | 6 | 0.00 | 5 |
| SRR11550010 | Calu3_4h | GSE148729 | 22,825,048 | 19,402,095 | 367,857 | 1.86 | 5 | 0.00 | 5 |
| SRR11550047 | Calu3_4h | GSE148729 | 32,022,112 | 25,320,980 | 819,284 | 3.13 | 10 | 0.00 | 10 |
| SRR11550048 | Calu3_4h | GSE148729 | 30,837,972 | 23,831,102 | 928,941 | 3.75 | 12 | 0.00 | 11 |
| SRR11550013 | Calu3_8h | GSE148729 | 27,200,369 | 19,692,817 | 4,389,048 | 18.23 | 89 | 0.00 | 86 |
| SRR11550014 | Calu3_8h | GSE148729 | 26,103,647 | 19,011,517 | 3,953,938 | 17.22 | 75 | 0.00 | 71 |
| SRR12709012 | Calu3_1 | SRP285334 | 11,630,824 | 4,701,206 | 1,027,560 | 17.94 | 284 | 0.03 | 243 |
| SRR12709013 | Calu3_2 | SRP285334 | 10,195,438 | 4,191,265 | 855,638 | 16.95 | 232 | 0.03 | 207 |
| SRR12340058 | Patient_10_lung1 | GSE150316 | 16,714,994 | 8,901,706 | 26 | 0.00 | 0 | 0.00 | 0 |
| SRR12340059 | Patient_10_lung2 | GSE150316 | 19,964,550 | 10,250,204 | 114 | 0.00 | 0 | 0.00 | 0 |
| SRR12340060 | Patient_10_lung3 | GSE150316 | 13,569,562 | 8,228,681 | 22 | 0.00 | 0 | 0.00 | 0 |
| SRR12340063 | Patient_11_lung1 | GSE150316 | 16,853,822 | 10,671,637 | 71,977 | 0.67 | 12 | 0.02 | 4 |
| SRR12340064 | Patient_11_lung2 | GSE150316 | 15,393,954 | 9,324,512 | 17,258 | 0.18 | 3 | 0.02 | 1 |
| SRR12340065 | Patient_11_lung3 | GSE150316 | 21,366,972 | 10,264,261 | 1,316 | 0.01 | 0 | 0.00 | 0 |
| SRR11772358 | Patient_1_lung1 | GSE150316 | 51,535,310 | 37,786,103 | 514,304 | 1.34 | 5 | 0.00 | 3 |
| SRR11772359 | Patient_1_lung2 | GSE150316 | 7,028,852 | 3,686,027 | 54,566 | 1.46 | 0 | 0.00 | 0 |
| SRR11772360 | Patient_1_lung3 | GSE150316 | 5,572,502 | 2,722,176 | 20,742 | 0.76 | 0 | 0.00 | 0 |
| SRR11772361 | Patient_1_lung4 | GSE150316 | 3,122,594 | 1,700,851 | 37,261 | 2.14 | 0 | 0.00 | 0 |
| SRR11772363 | Patient_2_lung1 | GSE150316 | 7,395,980 | 5,445,877 | 473 | 0.01 | 0 | 0.00 | 0 |
| SRR11772364 | Patient_2_lung2 | GSE150316 | 8,383,638 | 5,574,204 | 42 | 0.00 | 0 | 0.00 | 0 |
| SRR11772366 | Patient_2_lung3 | GSE150316 | 8,219,320 | 5,868,544 | 10 | 0.00 | 0 | 0.00 | 0 |
| SRR11772368 | Patient_3_lung1 | GSE150316 | 49,775,452 | 33,568,539 | 12 | 0.00 | 0 | 0.00 | 0 |
| SRR11772370 | Patient_3_lung2 | GSE150316 | 15,191,288 | 7,497,674 | 4 | 0.00 | 0 | 0.00 | 0 |
| SRR11772371 | Patient_4_lung1 | GSE150316 | 13,642,316 | 3,581,379 | 0 | 0.00 | 0 | 0.00 | 0 |
| SRR11772374 | Patient_4_lung2 | GSE150316 | 12,003,794 | 2,689,156 | 12 | 0.00 | 0 | 0.00 | 0 |
| SRR11772378 | Patient_5_lung1 | GSE150316 | 13,837,028 | 4,865,695 | 217 | 0.00 | 0 | 0.00 | 0 |
| SRR11772379 | Patient_5_lung2 | GSE150316 | 11,400,420 | 2,221,320 | 37 | 0.00 | 0 | 0.00 | 0 |
| SRR11772380 | Patient_5_lung3 | GSE150316 | 14,612,006 | 9,892,592 | 654 | 0.01 | 0 | 0.00 | 0 |
| SRR11772381 | Patient_5_lung4 | GSE150316 | 16,156,442 | 10,006,048 | 513 | 0.01 | 0 | 0.00 | 0 |
| SRR11772383 | Patient_5_lung5 | GSE150316 | 15,833,086 | 11,097,238 | 730 | 0.01 | 0 | 0.00 | 0 |
| SRR12340068 | Patient_6_lung1 | GSE150316 | 19,929,798 | 9,933,705 | 14 | 0.00 | 0 | 0.00 | 0 |
| SRR12340069 | Patient_6_lung2 | GSE150316 | 17,246,168 | 11,487,143 | 0 | 0.00 | 0 | 0.00 | 0 |
| SRR12340070 | Patient_6_lung3 | GSE150316 | 19,233,156 | 15,468,962 | 0 | 0.00 | 0 | 0.00 | 0 |
| SRR12340071 | Patient_6_lung4 | GSE150316 | 17,960,546 | 14,997,315 | 4 | 0.00 | 0 | 0.00 | 0 |
| SRR12340072 | Patient_6_lung5 | GSE150316 | 21,122,880 | 10,277,008 | 0 | 0.00 | 0 | 0.00 | 0 |
| SRR12340073 | Patient_7_lung1 | GSE150316 | 24,700,670 | 20,563,657 | 16 | 0.00 | 0 | 0.00 | 0 |
| SRR12340074 | Patient_7_lung2 | GSE150316 | 16,504,676 | 12,820,340 | 156 | 0.00 | 0 | 0.00 | 0 |
| SRR12340075 | Patient_7_lung3 | GSE150316 | 15,469,042 | 13,034,225 | 25 | 0.00 | 0 | 0.00 | 0 |
| SRR12340076 | Patient_7_lung4 | GSE150316 | 14,943,496 | 11,926,324 | 0 | 0.00 | 0 | 0.00 | 0 |
| SRR12340077 | Patient_7_lung5 | GSE150316 | 16,729,928 | 11,800,435 | 1,276 | 0.01 | 0 | 0.00 | 0 |
| SRR12340081 | Patient_8_lung1 | GSE150316 | 19,548,066 | 11,455,200 | 3,184 | 0.03 | 0 | 0.00 | 0 |
| SRR12340082 | Patient_8_lung2 | GSE150316 | 16,898,698 | 8,361,934 | 200 | 0.00 | 0 | 0.00 | 0 |
| SRR12340083 | Patient_8_lung3 | GSE150316 | 20,488,048 | 15,322,949 | 24,592 | 0.16 | 13 | 0.05 | 2 |
| SRR12340084 | Patient_8_lung4 | GSE150316 | 15,908,804 | 12,422,143 | 5,799 | 0.05 | 0 | 0.00 | 0 |
| SRR12340085 | Patient_8_lung5 | GSE150316 | 15,876,038 | 12,108,990 | 103 | 0.00 | 0 | 0.00 | 0 |
| SRR12340086 | Patient_9_lung1 | GSE150316 | 18,607,658 | 13,330,919 | 45,654 | 0.34 | 8 | 0.02 | 2 |
| SRR12340087 | Patient_9_lung2 | GSE150316 | 15,029,230 | 11,513,925 | 153,942 | 1.32 | 36 | 0.03 | 16 |
| SRR12340088 | Patient_9_lung3 | GSE150316 | 16,505,706 | 12,800,471 | 138,501 | 1.07 | 5 | 0.00 | 4 |
| SRR12340089 | Patient_9_lung4 | GSE150316 | 17,251,878 | 13,254,898 | 361,225 | 2.65 | 82 | 0.02 | 32 |
| SRR12340090 | Patient_9_lung5 | GSE150316 | 15,101,984 | 9,965,292 | 179,437 | 1.77 | 28 | 0.02 | 3 |
| SRR12340091 | Patient_A_lung | GSE150316 | 15,766,426 | 867,123 | 1,201 | 0.14 | 0 | 0.00 | 0 |
| SRR12340092 | Patient_B_lung | GSE150316 | 16,187,020 | 10,895,560 | 6 | 0.00 | 0 | 0.00 | 0 |
| SRR12340093 | Patient_C_lung | GSE150316 | 17,034,088 | 5,697,554 | 168,623 | 2.87 | 4 | 0.00 | 3 |
| SRR12340094 | Patient_D_lung | GSE150316 | 16,500,666 | 1,665,413 | 43,552 | 2.55 | 0 | 0.00 | 0 |
| SRR12340095 | Patient_E_lung | GSE150316 | 12,535,746 | 4,278,988 | 131,460 | 2.98 | 6 | 0.00 | 4 |
| SRR12340096 | Patient_F_lung | GSE150316 | 9,457,786 | 6,054,890 | 14 | 0.00 | 0 | 0.00 | 0 |
| SRR12340097 | Patient_G_lung | GSE150316 | 9,834,674 | 6,149,855 | 20 | 0.00 | 0 | 0.00 | 0 |
| SRR12340098 | Patient_H_lung | GSE150316 | 7,720,090 | 5,612,412 | 0 | 0.00 | 0 | 0.00 | 0 |
| SRR12340099 | Patient_I_lung | GSE150316 | 9,091,456 | 2,874,051 | 0 | 0.00 | 0 | 0.00 | 0 |
| SRR12340100 | Patient_J_lung | GSE150316 | 10,431,476 | 5,824,824 | 7 | 0.00 | 0 | 0.00 | 0 |
| SRR11924416 | Patient_Lung1 | GSE151803 | 3,904,878 | 2,614,576 | 3 | 0.00 | 0 | 0.00 | 0 |
| SRR11924417 | Patient_Lung2 | GSE151803 | 20,136,650 | 16,918,888 | 4 | 0.00 | 0 | 0.00 | 0 |
| SRR11924418 | Patient_Lung3 | GSE151803 | 16,330,983 | 13,934,664 | 73 | 0.00 | 0 | 0.00 | 0 |
| GSM4462415 | Patient_lung | GSE147507 | 10,561,476 | 107,344 | 6 | 0.01 | 0 | 0.00 | 0 |
| GSM4462416 | Patient_lung | GSE147507 | 9,514,219 | 1,414,534 | 189 | 0.01 | 0 | 0.00 | 0 |
| GSM4432381 | NHBE | GSE147507 | 15,032,096 | 13,800,150 | 15,156 | 0.11 | 1 | 0.01 | 1 |
| GSM4432382 | NHBE | GSE147507 | 15,108,090 | 13,892,225 | 12,670 | 0.09 | 3 | 0.02 | 3 |
| GSM4432383 | NHBE | GSE147507 | 44,210,735 | 40,537,603 | 45,524 | 0.11 | 1 | 0.00 | 1 |
| HRR057161 | Patient_BALF | HRA000143 | 2,935,311 | 2,492,465 | 2 | 0.00 | 0 | 0.00 | 0 |
| HRR057162 | Patient_BALF | HRA000144 | 19,906,482 | 17,269,935 | 0 | 0.00 | 0 | 0.00 | 0 |
| HRR057163 | Patient_BALF | HRA000145 | 9,322,656 | 7,784,478 | 0 | 0.00 | 0 | 0.00 | 0 |
| HRR057164 | Patient_BALF | HRA000146 | 27,544,048 | 24,398,263 | 0 | 0.00 | 0 | 0.00 | 0 |
| HRR057165 | Patient_BALF | HRA000147 | 3,974,104 | 3,403,134 | 0 | 0.00 | 0 | 0.00 | 0 |
| HRR057166 | Patient_BALF | HRA000148 | 31,721,102 | 28,454,823 | 0 | 0.00 | 0 | 0.00 | 0 |
| HRR057167 | Patient_BALF | HRA000149 | 12,941,494 | 11,526,537 | 0 | 0.00 | 0 | 0.00 | 0 |
| HRR057168 | Patient_BALF | HRA000150 | 38,612,800 | 35,749,762 | 0 | 0.00 | 0 | 0.00 | 0 |
| HRR057169 | Patient_BALF | HRA000151 | 13,353,298 | 12,032,341 | 0 | 0.00 | 0 | 0.00 | 0 |
| HRR057170 | Patient_BALF | HRA000152 | 31,382,014 | 28,622,595 | 0 | 0.00 | 0 | 0.00 | 0 |
| HRR057171 | Patient_BALF | HRA000153 | 1,059,967 | 1,007,836 | 0 | 0.00 | 0 | 0.00 | 0 |
| HRR057172 | Patient_BALF | HRA000154 | 6,873,885 | 6,269,049 | 0 | 0.00 | 0 | 0.00 | 0 |
| CRR119894-5 | Patient_BALF | CRA002390 | 24,259,658 | 16,626,357 | 407,354 | 2.39 | 1,632 | 0.40 | 1,628 |
| CRR119896-7 | Patient_BALF | CRA002390 | 14,052,564 | 9,620,062 | 97,457 | 1.00 | 326 | 0.33 | 326 |
The library size and the total number of reads mapped to the human genome, SARS-CoV-2 genome, or chimeric reads are reported.
Library size is the total number of reads in the RNA-seq library.
(No. of reads mapped to virus/total no. of mapped reads) × 100.
(No. of chimeric reads/no. of reads mapped to virus) × 100.
TABLE 3.
Detailed information on RNA-seq libraries from IAV-, RSV-, or MERS-infected cells used in this studya
| Accession no. | Sample annotation | GEO/SRA data set | Library sizeb | No. of reads mapped to: |
Ratio of reads mapped to virus/total mapped reads (%)c | No. of chimeric reads | Ratio of chimeric reads/reads mapped to virus (%)d | |
|---|---|---|---|---|---|---|---|---|
| Human genome | virus | |||||||
| GSM4432396 | A549_IAV | GSE147507 | 13,418,279 | 9,503,073 | 1,517,013 | 13.77 | 235 | 0.02 |
| GSM4432397 | A549_IAV | GSE147507 | 4,464,185 | 2,318,324 | 291,465 | 11.17 | 102 | 0.03 |
| GSM4432392 | A549_RSV | GSE147507 | 11,230,884 | 9,155,717 | 95,783 | 1.04 | 1,349 | 1.41 |
| GSM4432393 | A549_RSV | GSE147507 | 6,420,293 | 5,326,409 | 38,429 | 0.72 | 663 | 1.73 |
| GSM4462357 | A549_RSV | GSE147507 | 18,265,188 | 13,991,448 | 2,173,813 | 13.45 | 719 | 0.03 |
| GSM4462358 | A549_RSV | GSE147507 | 10,113,566 | 7,168,172 | 1,664,353 | 18.84 | 1,487 | 0.09 |
| GSM4462359 | A549_RSV | GSE147507 | 17,024,730 | 12,665,503 | 2,395,363 | 15.90 | 1,811 | 0.08 |
| SRR10357369 | Calu3_MERS | GSE139516 | 41,727,622 | 35,420,732 | 2,318,768 | 6.14 | 1,599 | 0.07 |
| SRR10357370 | Calu3_MERS | GSE139516 | 39,671,252 | 33,878,259 | 2,167,867 | 6.01 | 2,491 | 0.11 |
| SRR10357371 | Calu3_MERS | GSE139516 | 44,724,750 | 38,097,990 | 2,381,305 | 5.88 | 1,998 | 0.08 |
| GSM4462367 | NHBE_IAV | GSE147507 | 43,108,363 | 29,007,597 | 2,143,637 | 6.88 | 2,763 | 0.13 |
| GSM4462368 | NHBE_IAV | GSE147507 | 10,822,990 | 6,785,007 | 458,523 | 6.33 | 864 | 0.19 |
| GSM4462369 | NHBE_IAV | GSE147507 | 9,991,901 | 7,432,860 | 1,178,928 | 13.69 | 7,165 | 0.61 |
| GSM4462370 | NHBE_IAV | GSE147507 | 5,174,748 | 3,790,576 | 655,191 | 14.74 | 1,649 | 0.25 |
The library size and the total number of reads mapped to the human genome, virus genome, or human-virus chimeric reads are reported.
Library size is the total number of reads in the RNA-seq library.
(No. of reads mapped to virus/total no. of mapped reads) × 100.
(No. of chimeric reads/no. of reads mapped to virus) × 100.
We next quantified the reads that partly mapped to the human genome and partly mapped to the SARS-CoV-2 genome (see Materials and Methods). We found that nearly 0.05 to 1% of all viral reads are formed of hybrid sequences between host and virus RNAs, a frequency consistent with those recently reported by others (Fig. 1C) (18, 19). Infected A549-ACE2 and Calu-3 cells had the highest percentages of chimeric reads, while others, including normal human bronchial epithelial cells (NHBE) and lung autopsy samples or BALF of patients with COVID-19, had ∼1.5 to 2 orders of magnitude fewer chimeric reads (Fig. 1C). Similar percentages of chimeric reads were observed in cells infected with other viruses (Fig. 1C).
To test whether there are regions of the viral genome that more frequently participate in chimeric events, we separately aligned the viral reads and the viral fragments of the chimeric reads to the SARS-CoV-2 genome (Fig. 1D). Consistent with previous studies, we found higher coverage of the 3′ end of the SARS-CoV-2 genome in sequencing libraries across different cells (Fig. 1D, top). This portion of the virus encodes the viral N protein. Similarly, we observed that viral fragments from chimeric reads are also biased toward the 3′ end of the SARS-CoV-2 genome (Fig. 1D, bottom). This is consistent with a stochastic model in which chimeric events are dependent on the availability of template RNA, i.e., the more viral-RNA fragments present, the higher the chance of participation in chimeric events. Based on this model, we hypothesized that host fragments participating in chimeric reads will also be overrepresented in genes that are more highly expressed. Indeed, we observed that human genes with HVC events are more highly expressed than those without HVC events across all SARS-CoV-2-infected cells (Fig. 1E). This is exemplified by A549-ACE2 cells, which support high level of virus replication. In these cells, ACE2 was one of the top loci participating in chimeric events (Fig. 1F).
HVC events are not reproducible and have frequencies comparable to those of artifactual chimeric events.
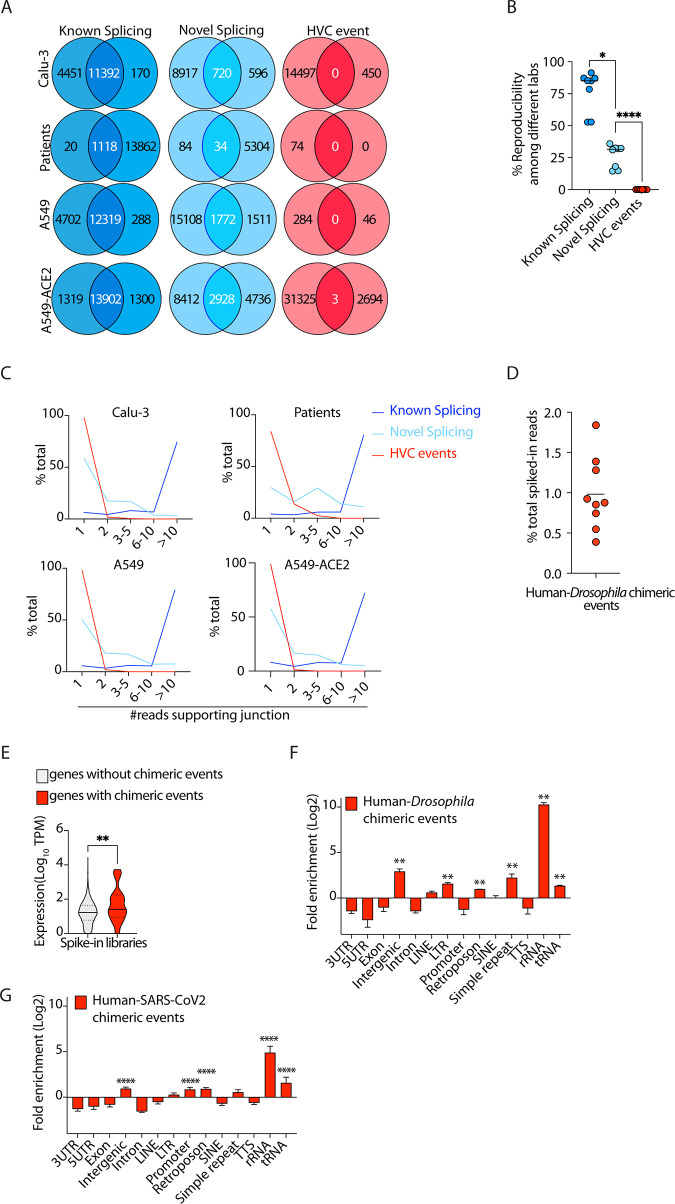
A precise and reproducible junction between host and viral fragments of a chimeric event would be evidence of authentic HVC events occurring as part of the natural life cycle of the virus. To determine whether the junctions of HVC events are precise and reproducible, we compared RNA-seq data from independent studies (Tables 1 and 2) and looked for reads that spanned known or novel exon-exon splicing junctions, as well as HVCs (Fig. 1A and B). For each cell type, we specifically sourced two or more RNA-seq libraries from independent studies (Table 1). As expected, ∼90% of known splicing events sourced from the RefSeq database were reproducible between independent studies (Fig. 2A and B). We also found that nearly one-third of novel (i.e., unannotated) splicing events could also be independently replicated between different studies (Fig. 2A and B). Conversely, almost none of the exact HVC events were reproducible in independent data sets (Fig. 2A and B). Of the three overlapping HVC events found only in A549-ACE2 cells, two mapped to the mitochondrial chromosome and one to a region of chromosome 12 annotated as an rRNA repeat.
FIG 2.
HVC events are not reproducible and have frequencies comparable to those of artifactual chimeric events. (A, B) Representative Venn diagrams (A) and cumulative data (B) comparing known splicing, novel splicing, and HVC events across independent studies (see Table 1 for the list of independent studies used here). The accession numbers of data from representative studies used in panel A are GSE147507 and PRJNA665581/SRP285334 for Calu-3 cells, GSE147507 and GSE151803 for patient samples, GSE147507 and GSE159191 for A549 cells, and GSE147507 and GSE154613 for A549-ACE2 cells. (C) Histograms showing the numbers of reads spanning junctions of the indicated events. (D) The fractions of spiked-in Drosophila RNA detected to be chimeras with human RNA. Data are from the data set with accession number PRJNA311567. (E) Violin plots showing expression of all human genes with or without human-Drosophila chimeric events. TPM, transcripts per million. (F) Distribution of genomic features in the human segment of human-Drosophila chimeric events. (G) Distribution of genomic features in the host segment of human–SARS-CoV-2 HVC events. *, P < 0.05, **, P < 0.01, and ****, P < 0.0001, by Wilcoxon test (B, E) and FDR correction (F and G).
Another way to determine whether specific HVCs are reproducible is to identify the proportion of unique reads in a given RNA-seq data set that span the HVC junction. The higher the number of reads, the more likely it is that the HVC is not a stochastic event. We therefore examined the number of unique reads spanning known splicing, novel splicing, and HVC junctions in each RNA-seq data set (Fig. 2C). We found that only 2 to 15% of HVC events had more than one read spanning their junctions. This is in clear contrast to 90 to 95% and 40 to 70% of known and novel splicing events, respectively, that have more than one supporting read (Fig. 2C).
Our data described above indicated that observed HVCs likely represent nonbiological artifacts. However, how these artifacts are generated remained unclear. Reverse transcriptase enzymes (RTs) are error prone and susceptible to a process called random template switching (21). In this process, RTs synthesizing cDNA infrequently dissociate from their template RNA and associate with a secondary template RNA, resulting in the creation of an artifactual fusion cDNA containing both the original template and the secondary RNA. Reverse transcription is one of the main steps in most commonly used RNA-sequencing methods, and thus, it is conceivable that some of the HVC events are artifacts of reverse transcription. To test this, we took advantage of control spike-in libraries that are typically utilized for internal calibration and normalization. In those libraries, a small quantity of RNA from an unrelated species is spiked into the RNA of interest, followed by RNA-sequencing-library preparation. We sourced existing human RNA-sequencing libraries that harbored spiked-in Drosophila melanogaster RNA and that were prepared using a common library preparation kit from Illumina. We mapped these libraries to the human-Drosophila chimeric genome, using the exact same method that we employed when analyzing the host-virus chimeric genome (see Materials and Methods and Table 4). Nearly 5% of all reads were mapped to the Drosophila genome. We then identified the fraction of Drosophila-mapped RNAs that were human-Drosophila chimeric. Since there is no actual possibility of biological fusion events between host and spiked-in RNAs, we considered any chimeric reads identified as artifactual. This could therefore determine the expected background level (“noise”) of chimeric events created as artifacts of reverse transcription and/or alignment errors. We observed ∼1% of all Drosophila-mapped reads to participate in chimeric events (Fig. 2D). Interestingly, in all analyzed libraries from SARS-CoV-2-infected cells, the observed fractions of HVC reads were lower than 1%, indicating that the frequency of HVC events in SARS-CoV-2-infected libraries was comparable to the expected background “noise” of chimeric events created as artifacts of reverse transcription and/or alignment errors.
TABLE 4.
Detailed information on Drosophila spike-in RNA-seq libraries used in this studya
| Accession no. | Sample annotation | GEO/SRA data set | Library sizeb | No. of reads mapped to: |
No. of chimeric reads | Ratio of chimeric reads/reads mapped to Drosophila (%)c | |
|---|---|---|---|---|---|---|---|
| Human genome | Drosophila chr4 | ||||||
| SRR4934910 | P493_FlyS2_A | SRP075325 | 19,333,486 | 14,866,599 | 25,994 | 114 | 0.44 |
| SRR4934934 | P493_FlyS2_A | SRP075325 | 20,972,522 | 15,808,054 | 33,233 | 105 | 0.32 |
| SRR4934935 | P493_FlyS2_A | SRP075325 | 20,579,126 | 15,663,884 | 28,772 | 197 | 0.68 |
| SRR4934311 | P493_FlyS2_B | SRP075325 | 23,229,139 | 18,482,501 | 18,498 | 138 | 0.75 |
| SRR4934799 | P493_FlyS2_B | SRP075325 | 20,416,688 | 16,460,316 | 13,049 | 134 | 1.03 |
| SRR4934936 | P493_FlyS2_B | SRP075325 | 21,016,231 | 17,054,532 | 15,935 | 112 | 0.71 |
| SRR4934495 | P493_FlyS2_C | SRP075325 | 26,464,903 | 21,675,460 | 10,466 | 155 | 1.48 |
| SRR4934687 | P493_FlyS2_C | SRP075325 | 21,191,865 | 17,584,755 | 8,330 | 93 | 1.12 |
| SRR4934937 | P493_FlyS2_C | SRP075325 | 17,256,607 | 14,298,342 | 5,587 | 34 | 0.61 |
The library size and the total number of reads mapped to the human genome, chr4 of the Drosophila genome, or human-Drosophila (chr4) chimeric reads are reported.
Library size is the total number of reads in the RNA-seq library.
(No. of chimeric reads/no. of reads mapped to chr4 of Drosophila) × 100.
We next examined the expression of human genes with and without Drosophila chimeras. Similar to what we had observed in SARS-CoV-2-infected cells (Fig. 1E), human genes with chimeric events were more highly expressed than those without such events (Fig. 2E). This was consistent with a stochastic model in which chimeric events are dependent on the availability of template RNA and driven by random RT template switching. Repeat sequences of RNA are known substrates for RT template switching (21). To test this, we examined the genomic distribution of the host segments of chimeric events between human and spiked-in Drosophila RNA and found that these artifactual chimeric events were, indeed, enriched in RNAs with highly repetitive structures, including rRNAs and tRNAs (Fig. 2F). We next sought to determine whether the same observation holds true in virally infected cells. In RNA-seq libraries of SARS-CoV-2-infected cells, we found that HVCs were similarly enriched in RNAs with repetitive motifs, including rRNAs and tRNAs, compared to the total transcriptome (see Materials and Methods and Fig. 2G). Thus, the frequency and the genomic distribution of HVC events were comparable to those from artifactual chimeric events generated by RT template switching, and host RNAs partaking in chimera formation were enriched in structures conducive to template switching.
Experimental enrichment of viral RNA during RNA-seq library construction does not enrich HVC events.
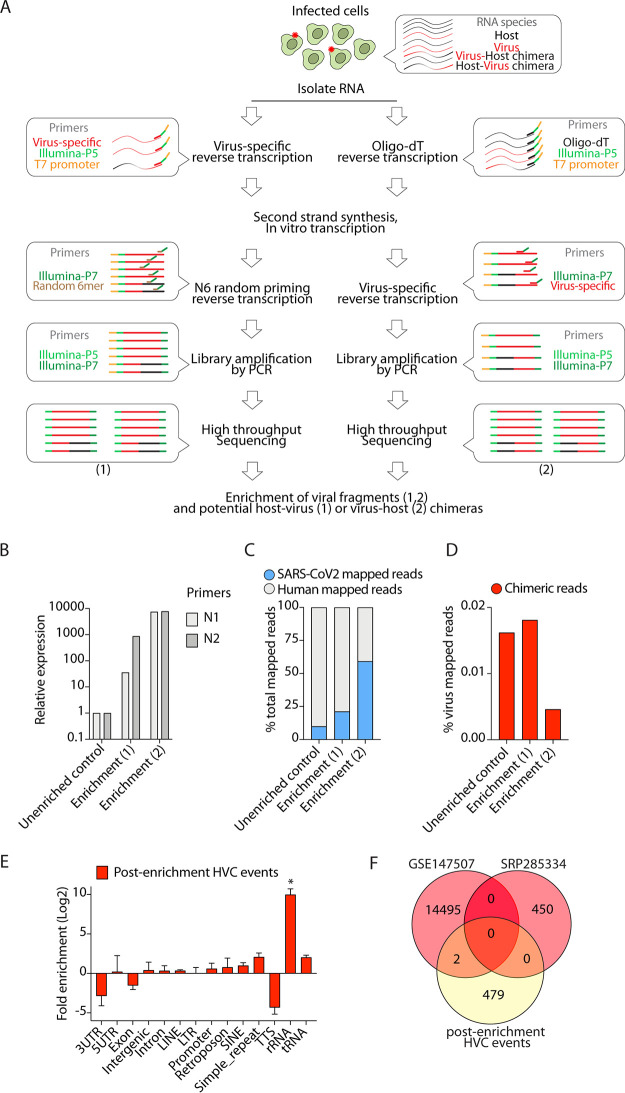
Although viral reads in most infected RNA-sequencing libraries were readily detectable, the fraction of viral reads to total mapped reads was low (Fig. 1B), presumably due to heterogeneous infectivity rates within cell cultures or patient samples. Thus, it is possible that detection of HVC events and junctional reads is too infrequent to allow robust detection of identical species across different samples. Therefore, we developed a technique to experimentally enrich viral RNAs during RNA-seq library preparation that would also enrich any bona fide HVC events as well. To this end, we designed a pool of 30 specific oligonucleotides that spanned the entire SARS-CoV-2 genome (Table 5). Using these oligonucleotides, we developed a novel methodology to specifically amplify viral RNAs from SARS-CoV-2-infected cells and constructed sequencing libraries (see Materials and Methods and schematic in Fig. 3A). Two types of chimeric events are possible, 5′-to-3′ host-virus chimeras and 5′-to-3′ virus-host chimeras. To enrich viral sequences and ensure “capture” of both types of chimeras, we used two approaches (enrichment methods 1 and 2, respectively, in Fig. 3A). To enrich viral sequences that also contain 5′-to-3′ host-virus chimeras (enrichment method 1 in Fig. 3A), we carried out virus-specific reverse transcription to construct cDNA incorporating an Illumina P5 adaptor sequence and T7 RNA polymerase promoter, followed by second-strand DNA synthesis and in vitro RNA transcription using T7 RNA polymerase. Reverse transcription primed with a random hexamer was then carried out to incorporate an Illumina P7 adaptor sequence before library amplification by PCR and high-throughput Illumina sequencing. To also enrich viral sequences that included 5′-to-3′ virus-host chimeras (enrichment method 2 in Fig. 3A), we performed oligo(dT)-primed reverse transcription to construct cDNA incorporating an Illumina P5 adaptor sequence and T7 RNA polymerase promoter, followed by second-strand DNA synthesis and in vitro RNA transcription using T7 RNA polymerase. Reverse transcription primed with virus-specific primers was then carried out to incorporate an Illumina P7 adaptor sequence before library amplification by PCR and high-throughput Illumina sequencing. Any RNA amplified using this technique would be enriched in viral sequences, including those mapping solely to the viral genome and those mapping partially to host as well as viral genomes (HVCs). For comparison, we also prepared cDNAs from RNAs of infected cells without any enrichment (unenriched control) (see Materials and Methods).
TABLE 5.
Primers and oligonucleotides used in this study
| Pool for enrichment (method) or primera | Sequence (5′→3′)b |
|---|---|
| T7-P5-VSP pool (1) | TAATACGACTCACTATAGGGATACCACCATGGCTCTTTCCCTACACGACGCTCTTCCGATCTcactgctatgtttagtgttc |
| TAATACGACTCACTATAGGGATACCACCATGGCTCTTTCCCTACACGACGCTCTTCCGATCTcaacataagagaacacacag | |
| TAATACGACTCACTATAGGGATACCACCATGGCTCTTTCCCTACACGACGCTCTTCCGATCTtgcttttcactcttcatttc | |
| TAATACGACTCACTATAGGGATACCACCATGGCTCTTTCCCTACACGACGCTCTTCCGATCTaaacctagatgtgctgatg | |
| TAATACGACTCACTATAGGGATACCACCATGGCTCTTTCCCTACACGACGCTCTTCCGATCTgtgtgaaggtattgtttgtt | |
| TAATACGACTCACTATAGGGATACCACCATGGCTCTTTCCCTACACGACGCTCTTCCGATCTtttttgtcctttttaggctc | |
| TAATACGACTCACTATAGGGATACCACCATGGCTCTTTCCCTACACGACGCTCTTCCGATCTctttaccagacattttgctc | |
| TAATACGACTCACTATAGGGATACCACCATGGCTCTTTCCCTACACGACGCTCTTCCGATCTatctttcattttaccgtcac | |
| TAATACGACTCACTATAGGGATACCACCATGGCTCTTTCCCTACACGACGCTCTTCCGATCTgttctccattctggttactg | |
| TAATACGACTCACTATAGGGATACCACCATGGCTCTTTCCCTACACGACGCTCTTCCGATCTgtgctatgtagttacgagaa | |
| TAATACGACTCACTATAGGGATACCACCATGGCTCTTTCCCTACACGACGCTCTTCCGATCTatagaagtgaataggacacg | |
| TAATACGACTCACTATAGGGATACCACCATGGCTCTTTCCCTACACGACGCTCTTCCGATCTtcaagtcctccctaatgtt | |
| TAATACGACTCACTATAGGGATACCACCATGGCTCTTTCCCTACACGACGCTCTTCCGATCTattggttgctcttcatctaa | |
| TAATACGACTCACTATAGGGATACCACCATGGCTCTTTCCCTACACGACGCTCTTCCGATCTgtctgaaagaagcaatgaag | |
| TAATACGACTCACTATAGGGATACCACCATGGCTCTTTCCCTACACGACGCTCTTCCGATCTagaggatgaaatggtgaatt | |
| TAATACGACTCACTATAGGGATACCACCATGGCTCTTTCCCTACACGACGCTCTTCCGATCTcaagtgagaaccaaaagataa | |
| TAATACGACTCACTATAGGGATACCACCATGGCTCTTTCCCTACACGACGCTCTTCCGATCTgagtaaacgtaaaaagaaggt | |
| TAATACGACTCACTATAGGGATACCACCATGGCTCTTTCCCTACACGACGCTCTTCCGATCTaagccaaagcctcattatta | |
| TAATACGACTCACTATAGGGATACCACCATGGCTCTTTCCCTACACGACGCTCTTCCGATCTaatctactgatgtcttggtc | |
| TAATACGACTCACTATAGGGATACCACCATGGCTCTTTCCCTACACGACGCTCTTCCGATCTcctttccacaaaaatcaact | |
| TAATACGACTCACTATAGGGATACCACCATGGCTCTTTCCCTACACGACGCTCTTCCGATCTtaatcagcaatctttccagt | |
| TAATACGACTCACTATAGGGATACCACCATGGCTCTTTCCCTACACGACGCTCTTCCGATCTacagaaagtagtgaaaccat | |
| TAATACGACTCACTATAGGGATACCACCATGGCTCTTTCCCTACACGACGCTCTTCCGATCTcaaaataggcatacaccatc | |
| TAATACGACTCACTATAGGGATACCACCATGGCTCTTTCCCTACACGACGCTCTTCCGATCTctttccatccaacttttgtt | |
| TAATACGACTCACTATAGGGATACCACCATGGCTCTTTCCCTACACGACGCTCTTCCGATCTttctgtgtaactccaatacc | |
| TAATACGACTCACTATAGGGATACCACCATGGCTCTTTCCCTACACGACGCTCTTCCGATCTaaacccacttctcttgttat | |
| TAATACGACTCACTATAGGGATACCACCATGGCTCTTTCCCTACACGACGCTCTTCCGATCTtgggtggtttatgtgattta | |
| TAATACGACTCACTATAGGGATACCACCATGGCTCTTTCCCTACACGACGCTCTTCCGATCTggtcaaggttaatataggca | |
| TAATACGACTCACTATAGGGATACCACCATGGCTCTTTCCCTACACGACGCTCTTCCGATCTtacgtccattcataccattt | |
| P7-N6 | GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNNN |
| P7-VSP pool (2) | GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTgaacactaaacatagcagtg |
| GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTctgtgtgttctcttatgttg | |
| GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTgaaatgaagagtgaaaagca | |
| GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTcatcagcacatctaggttt | |
| GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTaacaaacaataccttcacac | |
| GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTgagcctaaaaaggacaaaaa | |
| GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTgagcaaaatgtctggtaaag | |
| GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTgtgacggtaaaatgaaagat | |
| GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTcagtaaccagaatggagaac | |
| GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTtctcgtaactacatagcac | |
| GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTcgtgtcctattcacttctat | |
| GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTaacattagggaggacttga | |
| GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTttagatgaagagcaaccaat | |
| GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTcttcattgcttctttcagac | |
| GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTaattcaccatttcatcctct | |
| GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTttatcttttggttctcacttg | |
| GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTaccttctttttacgtttactc | |
| GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTtaataatgaggctttggctt | |
| GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTgaccaagacatcagtagatt | |
| GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTagttgatttttgtggaaagg | |
| GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTactggaaagattgctgatta | |
| GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTatggtttcactactttctgt | |
| GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTgatggtgtatgcctattttg | |
| GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTaacaaaagttggatggaaag | |
| GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTggtattggagttacacagaa | |
| GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTataacaagagaagtgggttt | |
| GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTtaaatcacataaaccaccca | |
| GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTtgcctatattaaccttgacc | |
| GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTaaatggtatgaatggacgta | |
| T7-P5-dT | TAATACGACTCACTATAGGGATACCACCATGGCTCTTTCCCTACACGACGCTCTTCCGATCTtttttttttttttttttt |
Oligonucleotides used in viral-fragment enrichment method 1 and 2, respectively. P7, Illumina primer; VSP, SARS-CoV-2-specific primer; P5, Illumina primer; T7, T7 promoter for in vitro transcription; N6, random hexamer; dT, oligo(dT).
Lowercase letters represent virus specific primers/regions.
FIG 3.
Experimental enrichment for viral-RNA-containing fragments does not enrich HVC events. (A) Schematic presentation of viral-RNA enrichment from infected host cells. Cellular RNA from infected cells comprises host RNA, viral RNA, and presumably, any fusion RNA between virus and host. A pool of oligonucleotide probes that are specific to SARS-CoV-2 were used in a series of reverse transcription, in vitro transcription, and PCR amplification steps to amplify viral RNAs and potential host-virus (1) or virus-host (2) chimeras (see Materials and Methods). (B) Expression of N protein in control and virus-enriched (1 or 2) samples using N1 and N2 qPCR probes recommended by the CDC. (C) Viral reads in the indicated libraries from SARS-CoV-2-infected Calu-3 cells as a proportion of the total reads mapped to the chimeric genome. (D) HVC reads in the indicated libraries from SARS-CoV-2-infected Calu-3 cells as a proportion of the total reads mapped to the SARS-CoV-2 genome. (E) Distribution of genomic features in the human segment of HVC events detected after enrichment for viral-RNA-containing transcripts. *, P < 0.05, by Wilcoxon test. (F) Venn diagram comparing HVC events in Calu-3 cells from the data shown in Fig. 2A with postenrichment HVC events.
To validate this approach, we performed quantitative PCR (qPCR) on sequencing libraries for the SARS-CoV-2 N gene using CDC-recommended primer sets. We specifically chose the N gene since it is the most highly expressed gene and the site of most HVC events. We observed dramatic enrichment (more than 30-fold) of viral N gene mRNA in enriched libraries compared to the level in the control (Fig. 3B). We next performed high-throughput sequencing on all libraries and their corresponding controls and performed the same analysis presented in Fig. 1. Consistent with our qPCR data, we found that the total number of reads mapped to the virus genome was much higher in enriched libraries than in the control (Fig. 3C), indicating mean enrichment for viral reads of 2- to 6-fold. We then compared the total number of HVC events before and after the enrichment. Despite the significant enrichment of viral reads, HVC events were not enriched and their frequency remained at <0.05% (Fig. 3D), comparable to the expected level from background “noise” denoted previously (Fig. 2D). Moreover, the genomic distribution of the host portion of these HVC events (Fig. 3E) was similar to those observed from artifactual chimeric events. In addition, postenrichment HVC events did not overlap significantly with HVC events from existing RNA-seq data (Fig. 3F). There were only two HVC events that overlapped with one of the data sets, both from a region of the human genome annotated as rRNA repetitive elements (Fig. 3F). These data indicate that even after enrichment of transcripts containing viral sequences, HVC events remained at the level of noise expected from random RT template switching.
DISCUSSION
Here, we found several lines of evidence that indicate that the observed HVC events between SARS-CoV-2 and human genetic material in sequencing libraries are most likely artifactual. We identified HVC events in RNA-seq from SARS-CoV-2-infected cells. These events were very rare in samples from patients with COVID-19. The precise locations and the nucleic acid sequences of HVC events are not reproducible across different libraries prepared by different laboratories, suggesting that they are either stochastic or artifactual. In addition, these events were mostly supported by only one read. The lack of reproducibility of the exact HVC event does not on its own rule out the possibility of stochastic integration events. However, if an integration had occurred and was being transcribed, then the junction between host and virus DNA would be expected to be evident multiple times across an RNA-sequencing data set (i.e., independent sequencing reads in the experiment would show the same host-virus junction). Consistent with previous reports, we also find the viral part of HVC events to be enriched in sequences from the 3′ end of SARS-CoV-2 virus. This is the portion of the virus that contains the most highly expressed gene, encoding the N protein (22). Likewise, we also observed that chimeric events incorporated the more highly expressed host genes. Additionally, A549-ACE2 cells express the entry receptor for SARS-CoV-2 and, thus, have higher viral entry. Consequently, they have much higher SARS-CoV-2 RNA levels than other cells, resulting in higher availability of template to form HVC events. Thus, there are more HVC events observed in A549-ACE2 cells than in A549 and other cells (Fig. 2A). A model consistent with these observations is that HVCs are likely the result of stochastic events occurring at the RNA level that incorporate components of more highly expressed transcripts (templates) from both the host and virus.
One of the potential mechanisms that could generate artifactual HVC events is random template switching by RTs used during RNA-seq library preparation to convert RNA to cDNA. RTs are known to occasionally switch from one template to another, thus creating artifactual fusion cDNA. Using spiked-in control RNA, we estimated that errors in in vitro reverse transcription result in ∼1% of RNA-seq reads being artifactually chimeric, approximately the same frequency as for HVCs observed in SARS-CoV-2-infected cells. These artifacts can be explained by random template switching during library preparation. Such analysis provides an expected level of artifactual chimeric events for the RTs used in common RNA-sequencing library preparation kits (e.g., SuperScript II). We found that the frequency of HVC events from all SARS-CoV-2-infected samples was below 1%, indicating that these events are likely to be artifacts of RTs. Although the mechanistic details of random template switching are not fully understood, repeat sequences of RNA are known substrates for template switching (21). Not surprisingly, we found that the host part of HVC events was enriched in RNAs with highly repetitive structures, including rRNAs and tRNAs (Fig. 2G). This further supports undesired template switching by RTs as the origin of observed HVC events.
Finally, we developed a novel method to enrich viral-RNA fragments from infected cells during RNA-seq library preparation. Deploying this method, we found that, although we could enrich viral transcripts by more than 30-fold, the rate of HVCs remained unchanged and at or below the expected level of noise introduced by in vitro RTs. A benefit of our technique is that it is a general method that can easily be used for enrichment of any RNA and its chimeric partners, as long as sequences for oligonucleotide design are known (e.g., a genome build is available). This is particularly useful because cellular RNAs in infected cells are typically dominant over RNA derived from infecting pathogens, especially when infection rates and/or viral titers are low. One example for the utility of this method is to help identify “cap-snatching” and “start-snatching” events. In IAV-infected cells, for example, viral transcripts form chimeras with the 5′ portion of host transcripts containing 5′ caps in order to stabilize viral transcripts and create bona fide fusion proteins (23, 24). Although there are computational challenges in aligning sequencing reads if very short fragments (<18 bp) are “snatched” from the host, one would anticipate seeing enrichment of host 5′ untranslated region (UTR) elements in HVC events if similar cap-snatching mechanisms were utilized by SARS-CoV-2. However, we observed quite the opposite, if anything (Fig. 2G). In fact, the overall conclusion on successfully enriching viral-RNA reads but observing no enrichment of HVC events above the background level is that the majority of HVCs are the result of artifacts generated by reverse transcription errors during library preparation.
Collectively, our data analyses and experimental findings indicate that currently observed and widely reported HVC events are infrequent, not reproducible, and likely to be artifacts of reverse transcription during RNA-seq library preparation. As anticipated from the cytoplasmic replication stage of positive-strand RNA viruses, viral integration is not expected to be a major pathological factor for SARS-CoV-2 and, by extension, not a cause for concern in the use of SARS-CoV-2 mRNA vaccines. In summary, current data do not support the authenticity of HVC events in SARS-CoV-2-infected samples.
MATERIALS AND METHODS
Cell culture and viral infections.
Human adenocarcinomic lung epithelial (Calu-3) cells (HTB-55; ATCC) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; GIBCO) supplemented with 10% fetal bovine serum (FBS; Corning), HEPES, nonessential amino acids, l-glutamine, and 1× antibiotic-antimycotic solution (Gibco). All cells were maintained at 37°C and 5% CO2. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) isolate USA-WA1/2020 (NR-52281) was obtained from Bei resources and was propagated in Vero E6 cells in DMEM supplemented with 2% FBS, 4.5 g/liter d-glucose, 4 mM l-glutamine, 10 mM nonessential amino acids, 1 mM sodium pyruvate, and 10 mM HEPES. Infectious titers of SARS-CoV-2 were determined using the 50% tissue culture infective dose (TCID50) method of Reed and Muench. Fifty thousand Calu-3 cells were seeded in 48-well plates and allowed to form 80% confluent monolayers. SARS-CoV-2 virus was pretreated with porcine trypsin (10 μg/ml) for 15 min at 37 degrees. Cells were then infected with the pretreated virus preparation at a multiplicity of infection (MOI) of 10 for 1 h in culture medium (the final concentration of trypsin on cells was 2 μg/ml). After absorption, the virus inoculum was removed and replaced with fresh culture medium. Forty-eight hours postinfection, cells were harvested and RNA was isolated. Briefly, infected cells were lysed in TRIzol (Invitrogen) and RNA was extracted using the Direct-zol RNA miniprep kit (Zymo Research) according to the manufacturer’s instructions. Experiments using SARS-CoV-2 were performed at the University of Michigan under biosafety level 3 (BSL3) protocols in compliance with containment procedures in laboratories approved for use by the University of Michigan Institutional Biosafety Committee (IBC) and Environment, Health & Safety (EHS) Department.
Library preparation and virus enrichment assay.
To experimentally enrich viral RNAs from total RNA of SARS-CoV-2-infected cells prior to RNA-seq library preparation, we developed a series of in vitro amplification steps using SARS-CoV-2-specific primers (VSPs) as follows. The VSP pool contained ∼30 oligonucleotides that span all SARS-CoV-2 genes (at least one oligonucleotide per gene and nearly 1 oligonucleotide per 1 kb of the genome). Given our goal to additionally enrich potential HVC events, we used two approaches, enrichment method 1 (5′-to-3′ host-virus chimeras) and enrichment method 2 (5′-to-3′ virus-host chimeras) (Fig. 3A).
First-strand cDNA synthesis reaction.
To capture and enrich virus, virus-host, or host-virus transcripts, we first set up a 20-μl reverse transcription reaction mixture using 100 ng of mRNA isolated from SARS-CoV-2-infected cells using SuperScript III reverse transcriptase. We used 2 pmol T7-P5-VSP oligonucleotide pool (for enrichment method 1) or 50 pmol T7-P5-oligo(dT) (for enrichment method 2) as the “gene-specific primer” for the reverse transcription reaction (Table 5). We also incorporated a T7 promoter and Illumina P5 sequence at the 5′ end of every oligonucleotide, as shown in the schematic in Fig. 3A. After combining all the components according to the recommended protocol (catalog numbers 18080044 and 18064014; Thermo Fisher), we incubated the entire reaction mixture at 25°C for 15 min, followed by 50°C for 30 min for SuperScript III. Next, we inactivated the reaction mixture by heating at 70°C for 15 min. To remove RNA/cRNA to the cDNA, we added 1 μl of PureLink RNase A (20 mg/ml) (catalog number 12091021; Invitrogen) and 1 μl (5 units) of RNase H (catalog number E018; Applied Biological Materials) and incubated at 37°C for 1 h. We then purified cDNA using 1× Mag-Bind total pure next-generation sequencing (NGS) beads (catalog number M1378-01; Omega Bio-tek) according to the manufacturer’s instructions and eluted the cDNA in 15 μl of sterile water. To further remove the excess single-stranded short oligonucleotides, we treated the purified reverse transcription reaction mixture with 1 μl of exonuclease I (catalog number M0293L; NEB) at 37°C for 30 min. Next, we added excess sterile water to the sample to get a total volume of 40 μl. The reaction mixture was then purified with 1× Mag-Bind total pure NGS beads and eluted in 15 μl of sterile water.
Second-strand cDNA synthesis and in vitro transcription.
Following this, we performed second-strand synthesis using the NEBNext ultra II nondirectional RNA second-strand synthesis module according to the suggested protocol (catalog number E6111L; NEB). The synthesized DNA was purified via 1× Mag-Bind total pure NGS beads and eluted in ∼12 μl of sterile water. Ten microliters of this was then used as an input for T7 polymerase-mediated in vitro transcription using the NEB HiScribe T7 high-yield RNA synthesis kit (catalog number E2040S; NEB). Briefly, all the components were mixed as mentioned in the kit protocol and incubated at 37°C (lid at 50°C) for 16 h. The reaction mixture was eluted in 20 μl of sterile water after a round of 1× Mag-Bind total pure NGS bead cleanup. This newly transcribed RNA was quantified using a NanoDrop, and to improve the hybridization kinetics and enhance the signal, 500 ng of the amplified RNA was fragmented using RNA fragmentation reagent in a total reaction mixture volume of 10 μl according to specifications (catalog number AM8740; Thermo Fisher).
Final reverse transcription and PCR enrichment of the library.
Next, to generate final enriched libraries, we performed reverse transcription of the fragmented RNA with 50 pmol of P7-N6 for enrichment method 1 and 2 pmol of the P7-VSP primer pool for enrichment method 2 (Table 5), using SuperScript III reverse transcriptase according to the steps mentioned above. After the reverse transcription, the reaction mixture was purified using 1× Mag-Bind total pure NGS beads and eluted in 20 μl of sterile water. Five microliters of this reverse transcription reaction mixture was saved for running on a Bioanalyzer and to perform a real-time quantitative PCR validation assay. The remaining 15 μl was used to PCR amplify the library by using high-fidelity Q5 DNA polymerase (catalog number M0491L; NEB) for 16 cycles using universal primer and unique indices (catalog numbers E7335L and E7500L; NEB) in a total reaction mixture volume of 50 μl. Finally, the amplified and enriched library was purified using the 0.8× Mag-Bind total pure NGS beads, quantified by using the Bioanalyzer/Tape station and then sequenced using Illumina platform.
Real-time quantitative PCR validation assay.
The enrichment of viral genes was determined by performing a real-time quantitative PCR assay on the libraries generated. Briefly, the cDNA generated by reverse transcription, prior to library amplification by Q5-PCR, was diluted 10- to 20-fold and used to amplify target gene N of SARS-CoV-2 using CDC-recommended primers 2019-nCoV_N1-F (5′-GAC CCC AAA ATC AGC GAA AT-3′), 2019-nCoV_N1-R (5′ TCT GGT TAC TGC CAG TTG AAT CTG-3′), 2019-nCoV_N2-F (5′ TTA CAA ACA TTG GCC GCA AA-3′), and 2019-nCoV_N2-R (5′ GCG CGA CAT TCC GAA GAA3′). The UBC gene as the housekeeping gene was amplified using primers UBC-F (5′-CCT GGA GGA GAA GAG GAA AGA GA-3′) and UBC-R (5′-TTG AGG ACC TCT GTG TAT TTG TCA A-3′). The real-time quantitative PCR was performed on the Bio-Rad CFX connect system. All experiments were performed in independent triplicates in total reaction mixture volumes of 15 μl using PowerUp SYBR green master mix (catalog number A25778; Applied Biosystems). The expression level was calculated by the cycle threshold (2−ΔCT) method and normalized to that of the indicated housekeeping gene in the same sample.
Host-virus chimeric read analysis.
The raw sequencing files were downloaded from the Sequence Read Archive (SRA) as shown in Tables 1 and 2. Fastqc (version 0.11.7) was used for data quality control. Sequencing reads were aligned as single end to the chimeric genome of human (hg38) and SARS-CoV-2 (accession number NC_045512.2) using STAR aligner (version 2.7.7a). For the analyses of the other viruses, the influenza A virus (IAV) genome (A/Puerto Rico/8/1934 [H1N1], accession number GCA_000865725.1), Middle East respiratory syndrome (MERS) coronavirus genome (accession number NC_019843.3), and respiratory syncytial virus (RSV) genome (A2 strain, accession number M11486) were all sourced from NCBI.
To estimate the background level of chimeric reads in RNA-seq libraries, a fruit fly RNA spike-in control library (accession number PRJNA311567) was used. Briefly, a chimeric genome between human (hg38) and fruit fly chr4 (dm6) was constructed and the sequencing reads were aligned by the STAR aligner using parameters –outFilterMultimapNmax 1 –outFilterMismatchNmax 3 –chimSegmentMin 30 –chimOutType Junctions SeparateSAMold WithinBAM SoftClip –chimJunctionOverhangMin 30 –chimScoreMin 1 –chimScoreDropMax 30 –chimScoreJunctionNonGTAG 0 –chimScoreSeparation 1 –alignSJstitchMismatchNmax -1 -1 -1 -1 –chimSegmentReadGapMax 3.
The known annotated and novel unannotated splicing junctions were extracted from the STAR output as positive controls. The chimeric junctions for human-virus and human-Drosophila were extracted from the STAR chimeric output. The unique chimeric junctions were considered our chimeric events. To estimate the reproducibility for each independent study and each cell type, the numbers of unique junctions were extracted. For every pair of independent studies in each cell type, the number of overlapping junctions was calculated and was divided to the number of junctions in each study. The average of the two values was then recorded as the reproducibility between that pair.
To examine the genomic features of the HVC reads, HOMER (version 4.11) annotatePeaks.pl was used to annotate the HVC junctions and the corresponding RNA-seq library. In brief, reads in each RNA-seq library were converted to genomic regions by bamTobed (bedtools, version 2.30.0) and the unique regions were kept using the command “sort -k1,1 -k2,2n | uniq.” The reported “Log2 Ratio (obs/exp)” for each annotation (e.g, tRNA or long terminal repeat [LTR]) was compared between HVC junctions and the corresponding RNA-seq library. Mann-Whitney’s U test was used for statistical analysis.
Data availability.
Raw data are available from the Gene Expression Omnibus under accession no. GSE167131.
ACKNOWLEDGMENTS
This research was financed by the National Institute of General Medical Sciences of the NIH (grant number R35GM138283 to M.K.) and supported in part by the Intramural Research Program of the NIH, the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (project number ZIA/DK075149 to B.A.), and the National Institute of Allergy and Infectious Diseases (NIAID) (project number ZIA/AI001175 to M.S.L.). D.C. and C.M. are supported by an NIH Office of Dietary Supplements research scholar award (awarded to D.C.), a Marie-Slodowska Curie individual fellowship (GA–841247 awarded to C.M.), and the MICHR postdoctoral translational scholars program (grant number UL1TR002240 awarded to C.M.).
B.Y., S.C., L.W., B.A., and M.K. analyzed data and wrote the manuscript. C.M., S.C., D.C., D.K., and C.E.W. performed experiments and analyzed data. J.L.T.-O., D.C., D.K., M.S.L., M.R.O., C.E.W., B.A., and M.K. provided intellectual input and wrote the manuscript. C.E.W., B.A., and M.K. conceived and supervised the work.
Contributor Information
Christiane E. Wobus, Email: cwobus@umich.edu.
Behdad Afzali, Email: behdad.afzali@nih.gov.
Majid Kazemian, Email: kazemian@purdue.edu.
Colin R. Parrish, Cornell University
REFERENCES
- 1.Kazemian M, Ren M, Lin JX, Liao W, Spolski R, Leonard WJ. 2015. Possible human papillomavirus 38 contamination of endometrial cancer RNA sequencing samples in the Cancer Genome Atlas Database. J Virol 89:8967–8973. 10.1128/JVI.00822-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kazemian M, Ren M, Lin JX, Liao W, Spolski R, Leonard WJ. 2015. Comprehensive assembly of novel transcripts from unmapped human RNA-Seq data and their association with cancer. Mol Syst Biol 11:826. 10.15252/msb.156172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McBride AA, Warburton A. 2017. The role of integration in oncogenic progression of HPV-associated cancers. PLoS Pathog 13:e1006211. 10.1371/journal.ppat.1006211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mani SKK, Yan B, Cui Z, Sun J, Utturkar S, Foca A, Fares N, Durantel D, Lanman N, Merle P, Kazemian M, Andrisani O. 2020. Restoration of RNA helicase DDX5 suppresses hepatitis B virus (HBV) biosynthesis and Wnt signaling in HBV-related hepatocellular carcinoma. Theranostics 10:10957–10972. 10.7150/thno.49629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L, Laing J, Yan B, Zhou H, Ke L, Wang C, Narita Y, Zhang Z, Olson M, Afzali B, Zhao B, Kazemian M. 2020. Epstein-Barr virus episome physically interacts with active regions of the host genome in lymphoblastoid cells. J Virol 94:e01390-20. 10.1128/JVI.01390-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakravorty S, Yan B, Wang C, Wang L, Quaid JT, Lin CF, Briggs SD, Majumder J, Canaria DA, Chauss D, Chopra G, Olson MR, Zhao B, Afzali B, Kazemian M. 2019. Integrated pan-cancer map of EBV-associated neoplasms reveals functional host-virus interactions. Cancer Res 79:6010–6023. 10.1158/0008-5472.CAN-19-0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nellore A, Jaffe AE, Fortin JP, Alquicira-Hernandez J, Collado-Torres L, Wang S, Phillips RA, III, Karbhari N, Hansen KD, Langmead B, Leek JT. 2016. Human splicing diversity and the extent of unannotated splice junctions across human RNA-seq samples on the Sequence Read Archive. Genome Biol 17:266. 10.1186/s13059-016-1118-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J, Chen LL, Yang L. 2016. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res 26:1277–1287. 10.1101/gr.202895.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brant AC, Menezes AN, Felix SP, de Almeida LM, Sammeth M, Moreira MAM. 2019. Characterization of HPV integration, viral gene expression and E6E7 alternative transcripts by RNA-Seq: a descriptive study in invasive cervical cancer. Genomics 111:1853–1861. 10.1016/j.ygeno.2018.12.008. [DOI] [PubMed] [Google Scholar]
- 10.Sung WK, Zheng H, Li S, Chen R, Liu X, Li Y, Lee NP, Lee WH, Ariyaratne PN, Tennakoon C, Mulawadi FH, Wong KF, Liu AM, Poon RT, Fan ST, Chan KL, Gong Z, Hu Y, Lin Z, Wang G, Zhang Q, Barber TD, Chou WC, Aggarwal A, Hao K, Zhou W, Zhang C, Hardwick J, Buser C, Xu J, Kan Z, Dai H, Mao M, Reinhard C, Wang J, Luk JM. 2012. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet 44:765–769. 10.1038/ng.2295. [DOI] [PubMed] [Google Scholar]
- 11.Zhao LH, Liu X, Yan HX, Li WY, Zeng X, Yang Y, Zhao J, Liu SP, Zhuang XH, Lin C, Qin CJ, Zhao Y, Pan ZY, Huang G, Liu H, Zhang J, Wang RY, Yang Y, Wen W, Lv GS, Zhang HL, Wu H, Huang S, Wang MD, Tang L, Cao HZ, Wang L, Lee TL, Jiang H, Tan YX, Yuan SX, Hou GJ, Tao QF, Xu QG, Zhang XQ, Wu MC, Xu X, Wang J, Yang HM, Zhou WP, Wang HY. 2016. Genomic and oncogenic preference of HBV integration in hepatocellular carcinoma. Nat Commun 7:12992. 10.1038/ncomms13591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palacios-Flores K, Castillo A, Uribe C, Garcia Sotelo J, Boege M, Davila G, Flores M, Palacios R, Morales L. 2019. Prediction and identification of recurrent genomic rearrangements that generate chimeric chromosomes in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 116:8445–8450. 10.1073/pnas.1819585116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kannan K, Wang L, Wang J, Ittmann MM, Li W, Yen L. 2011. Recurrent chimeric RNAs enriched in human prostate cancer identified by deep sequencing. Proc Natl Acad Sci U S A 108:9172–9177. 10.1073/pnas.1100489108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGregor R, Chauss D, Freiwald T, Yan B, Wang L, Nova-Lamperti E, Zhang Z, Teague H, West EE, Bibby J, Kelly A, Malik A, Freeman AF, Schwartz D, Portilla D, John S, Lavender P, Lionakis MS, Mehta NN, Kemper C, Cooper N, Lombardi G, Laurence A, Kazemian M, Afzali B. 2020. An autocrine vitamin D-driven Th1 shutdown program can be exploited for COVID-19. bioRxiv 10.1101/2020.07.18.210161. [DOI]
- 15.Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, Liu S, Zhao P, Liu H, Zhu L, Tai Y, Bai C, Gao T, Song J, Xia P, Dong J, Zhao J, Wang FS. 2020. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med 8:420–422. 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fung TS, Liu DX. 2019. Human coronavirus: host-pathogen interaction. Annu Rev Microbiol 73:529–557. 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- 17.Yan B, Freiwald T, Chauss D, Wang L, West E, Mirabelli C, Zhang CJ, Nichols EM, Malik N, Gregory R, Bantscheff M, Ghidelli-Disse S, Kolev M, Frum T, Spence JR, Sexton JZ, Alysandratos KD, Kotton DN, Pittaluga S, Bibby J, Niyonzima N, Olson MR, Kordasti S, Portilla D, Wobus CE, Laurence A, Lionakis MS, Kemper C, Afzali B, Kazemian M. 2021. SARS-CoV-2 drives JAK1/2-dependent local complement hyperactivation. Sci Immunol 6:eabg0833. 10.1126/sciimmunol.abg0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L, Richards A, Khalil A, Wogram E, Ma H, Young RA, Jaenisch R. 2020. SARS-CoV-2 RNA reverse-transcribed and integrated into the human genome. bioRxiv 10.1101/2020.12.12.422516. [DOI]
- 19.Yin Y, Liu X-Z, He X, Zhou L-Q. 2021. Exogenous coronavirus interacts with endogenous retrotransposon in human cells. Front Cell Infect Microbiol 10.3389/fcimb.2021.609160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen J. 2020. The coronavirus may sometimes slip its genetic material into human chromosomes. Science 10.1126/science.abg2000. [DOI] [Google Scholar]
- 21.Cocquet J, Chong A, Zhang G, Veitia RA. 2006. Reverse transcriptase template switching and false alternative transcripts. Genomics 88:127–131. 10.1016/j.ygeno.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 22.Kim D, Lee JY, Yang JS, Kim JW, Kim VN, Chang H. 2020. The architecture of SARS-CoV-2 transcriptome. Cell 181:914–921.e10. 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sikora D, Rocheleau L, Brown EG, Pelchat M. 2017. Influenza A virus cap-snatches host RNAs based on their abundance early after infection. Virology 509:167–177. 10.1016/j.virol.2017.06.020. [DOI] [PubMed] [Google Scholar]
- 24.Ho JSY, Angel M, Ma Y, Sloan E, Wang G, Martinez-Romero C, Alenquer M, Roudko V, Chung L, Zheng S, Chang M, Fstkchyan Y, Clohisey S, Dinan AM, Gibbs J, Gifford R, Shen R, Gu Q, Irigoyen N, Campisi L, Huang C, Zhao N, Jones JD, van Knippenberg I, Zhu Z, Moshkina N, Meyer L, Noel J, Peralta Z, Rezelj V, Kaake R, Rosenberg B, Wang B, Wei J, Paessler S, Wise HM, Johnson J, Vannini A, Amorim MJ, Baillie JK, Miraldi ER, Benner C, Brierley I, Digard P, Luksza M, Firth AE, Krogan N, Greenbaum BD, MacLeod MK, van Bakel H, et al. 2020. Hybrid gene origination creates human-virus chimeric proteins during infection. Cell 181:1502–1517.e23. 10.1016/j.cell.2020.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blanco-Melo D, Nilsson-Payant BE, Liu WC, Uhl S, et al. 2020. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 181(5):1036–1045.e9. 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoagland DA, Clarke DJB, Møller R, Han Y, Yang L, Wojciechowicz ML, Lachmann A, Oguntuyo KY, Stevens C, Lee B, Chen S, Ma'ayan A, tenOever BR. 2020. bioRxiv 10.1101/2020.07.12.199687. [DOI] [Google Scholar]
- 27.Weingarten-Gabbay S, Klaeger S, Sarkizova S, Pearlman LR, Chen D-Y, Bauer MR, Taylor HB, Conway HL, Tomkins-Tinch CH, Finkel Y, Nachshon A, Gentili M, Rivera KD, Keskin DB, Rice CM, Clauser KR, Hacohen N, Carr SA, Abelin JG, Saeed M, Sabeti PC. 2020. SARS-CoV-2 infected cells present HLA-I peptides from canonical and out-of-frame ORFs. bioRxiv 10.1101/2020.10.02.324145. [DOI] [Google Scholar]
- 28.Banerjee AK, Blanco MR, Bruce EA, Honson DD, Chen LM, Chow A, Bhat P, Ollikainen N, Quinodoz SA, Loney C, Thai J, Miller ZD, Lin AE, Schmidt MM, Stewart DG, Goldfarb D, De Lorenzo G, Rihn SJ, Voorhees RM, Botten JW, Majumdar D, Guttman M. 2020. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell 183(5):1325–1339.e21. 10.1016/j.cell.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wyler E, Mösbauer K, Franke V, Diag A, Gottula LT, Arsie R, Klironomos F, Koppstein D, Ayoub S, Buccitelli C, Richter A, Legnini I, Ivanov A, Mari T, Del Giudice S, Papies JP, Praktiknjo S, Müller MA, Niemeyer D, Selbach M, Akalin A, Rajewsky N, Drosten C, Landthaler M. 2021. Transcriptomic profiling of SARS-CoV-2 infected human cell lines identifies HSP90 as target for COVID-19 therapy. iScience 24(3):102151. 10.1016/j.isci.2021.102151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han Y, Duan X, Yang L, Nilsson-Payant BE, Wang P, Duan F, Tang X, Yaron TM, Zhang T, Uhl S, Bram Y, Richardson C, Zhu J, Zhao Z, Redmond D, Houghton S, Nguyen DT, Xu D, Wang X, Jessurun J, Borczuk A, Huang Y, Johnson JL, Liu Y, Xiang J, Wang H, Cantley LC, tenOever BR, Ho DD, Pan FC, Evans T, Chen HJ, Schwartz RE, Chen S. 2021. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 589(7841):270–275. 10.1038/s41586-020-2901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Desai N, Neyaz A, Szabolcs A, Shih AR, et al. 2020. Temporal and spatial heterogeneity of host response to SARS-CoV-2 pulmonary infection. Nat Commun 11:6319. 10.1038/s41467-020-20139-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw data are available from the Gene Expression Omnibus under accession no. GSE167131.