ABSTRACT
The human protein-coding gene ILRUN (inflammation and lipid regulator with UBA-like and NBR1-like domains; previously C6orf106) was identified as a proviral factor for Hendra virus infection and was recently characterized to function as an inhibitor of type I interferon expression. Here, we have utilized transcriptome sequencing (RNA-seq) to define cellular pathways regulated by ILRUN in the context of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection of Caco-2 cells. We find that inhibition of ILRUN expression by RNA interference alters transcription profiles of numerous cellular pathways, including upregulation of the SARS-CoV-2 entry receptor ACE2 and several other members of the renin-angiotensin aldosterone system. In addition, transcripts of the SARS-CoV-2 coreceptors TMPRSS2 and CTSL were also upregulated. Inhibition of ILRUN also resulted in increased SARS-CoV-2 replication, while overexpression of ILRUN had the opposite effect, identifying ILRUN as a novel antiviral factor for SARS-CoV-2 replication. This represents, to our knowledge, the first report of ILRUN as a regulator of the renin-angiotensin-aldosterone system (RAAS).
IMPORTANCE There is no doubt that the current rapid global spread of COVID-19 has had significant and far-reaching impacts on our health and economy and will continue to do so. Research in emerging infectious diseases, such as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is growing rapidly, with new breakthroughs in the understanding of host-virus interactions to assist with the development of innovative and exciting therapeutic strategies. Here, we present the first evidence that modulation of the human protein-coding gene ILRUN functions as an antiviral factor for SARS-CoV-2 infection, likely through its newly identified role in regulating the expression of SARS-CoV-2 entry receptors ACE2, TMPRSS2, and CTSL. These data improve our understanding of biological pathways that regulate host factors critical to SARS-CoV-2 infection, contributing to the development of antiviral strategies to deal with the current SARS-CoV-2 pandemic.
KEYWORDS: COVID-19, ILRUN, RNA virus, SARS-CoV-2, cell biology
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2, family Coronaviridae) is the causative agent of the current coronavirus disease (COVID-19) pandemic, one of the deadliest infectious disease events to have occurred in recent times. Since emerging in Wuhan, Hubei province, China, in December 2019 (1), SARS-CoV-2 has spread across the globe, causing an estimated (as of 9 February 2021) 105.4 million infections and over 2.3 million deaths (2). Comorbidities feature in a high percentage of deaths, with hypertension being the foremost, followed by diabetes and cardiovascular disease (3).
The development of medical countermeasures against COVID-19 is guided by knowledge of cellular factors associated with SARS-CoV-2 infection. Viruses are obligate intracellular pathogens that rely on the hijacking of cellular machinery for successful completion of the infection cycle. SARS-CoV-2 entry requires the type I membrane-bound glycoprotein angiotensin-converting enzyme 2 (ACE2) (4). The biological function of ACE2 is part of the renin-angiotensin-aldosterone system (RAAS), acting as a carboxypeptidase enzyme that converts angiotensin I (Ang-I) and Ang-II into smaller peptides (Ang1-9 and Ang1-7, respectively), counteracting their vasoconstrictive properties and protecting the host from hypertension and severe cardiac dysfunction (5). The ACE2 protein is largely expressed in lung alveolar epithelial cells, consistent with prominent lung pathology in SARS-CoV-2 infection (6). Other permissive cell types predicted by protein-proofed transcript profiling include cardiomyocytes and intestinal enterocytes, consistent with heart injury and the less common intestinal symptoms (7, 8), as well as testis, kidneys, thyroid, and adipose tissues (9). The surface-exposed SARS-CoV-2 spike (S) glycoprotein binds ACE2 to facilitate viral attachment to target cells, while the host serine protease, transmembrane protease serine 2 (TMPRSS2), or lysosomal endopeptidase cathepsin L (CTSL) cleaves the S protein to facilitate receptor recognition and membrane fusion (10, 11). Clinical trials are currently examining RAAS modulators, including the Ang receptor blocker Losartan, in COVID-19 (12), based on the hypothesis that SARS-CoV-2 impairs the regulatory function of ACE2 and leads to unopposed RAAS activation and tissue damage. Such studies highlight how the study of host pathways associated with SARS-CoV-2 pathogenesis can facilitate the rapid clinical testing of novel countermeasures.
The human protein-coding gene ILRUN (inflammation and lipid regulator with UBA-like and NBR1-like domains, previously C6orf106) was identified as a proviral factor for Hendra virus infection from a genome-wide short interfering RNA (siRNA) screen (13) and recently characterized as a novel inhibitor of the transcription regulators p300 and CREB-binding protein (CBP) (14). In the context of virus infection, ILRUN promotes the degradation of p300/CBP to impair the function of the transcription factor interferon (IFN)-regulatory factor 3 (IRF3), thereby acting as a negative regulator of the expression of antiviral (type I IFNs) and proinflammatory (tumor necrosis factor alpha, or TNF-α) cytokines (14). ILRUN is highly phylogenetically conserved and associated with several disease states, including cancer, coronary artery disease, and obesity (15), suggesting it possesses biological functions beyond regulation of the host antiviral response.
To gain further insight into ILRUN function and explore its potential roles in COVID-19 pathogenicity, we performed a transcriptomics study to identify host pathways regulated by ILRUN in the context of SARS-CoV-2 infection using transcriptome sequencing (RNA-seq). From this, we discovered that ILRUN gene knockdown leads to upregulated expression of multiple RAAS pathway members, including ACE2 as well as TMPRSS2 and CTSL. In addition, we show that ILRUN expression negatively correlates with SARS-CoV-2 infection. These data suggest that ILRUN functions as a negative regulator of the RAAS pathway and is antiviral toward SARS-CoV-2. To our knowledge, this study represents the first comprehensive analysis of cellular pathways transcriptionally regulated by ILRUN and suggests that ILRUN is a major checkpoint of both SARS-CoV-2 infection and the RAAS.
RESULTS
ILRUN function and SARS-CoV-2 infection in Caco-2 cells.
To investigate host pathways regulated by ILRUN in the context of SARS-CoV-2 infection, we first established an in vitro system where cells were receptive to both RNA interference (RNAi)-mediated gene silencing and SARS-CoV-2 infection. Previous studies have shown that Caco-2 (human colon epithelial) and Calu-3 (human lung epithelial) cells support SARS-CoV-2 infection (16). Both of these cell types are derived from tissues shown to be productively infected during SARS-CoV-2 infection and contribute to disease symptoms (17, 18). siRNA reagents targeting ILRUN (siILRUN) resulted in a ca. 75% decrease in Caco-2 ILRUN mRNA expression compared to cells transfected with siNEG, a negative-control siRNA (Fig. 1A). To then assess ILRUN function in Caco-2 cells, cells were treated with transfected poly(I·C), a double-stranded RNA mimetic that is recognized by TLR3, retinoic acid-inducible gene I (RIG-I), and melanoma differentiation-associated gene 5 (MDA5) and acts as a viral RNA mimetic that induces a type I IFN response regulated by ILRUN (14). Stimulating Caco-2 cells with poly(I·C) resulted in a robust induction of IFNb, TNF, and IL-6 expression, which was heightened in siILRUN cells (Fig. 1B), thereby validating ILRUN inhibition of the type I IFN response in Caco-2 cells. Despite several rounds of optimization experiments, Calu-3 cells were unreceptive to siRNA or cDNA transfection in our hands (data not shown) and were not investigated further in this study.
FIG 1.
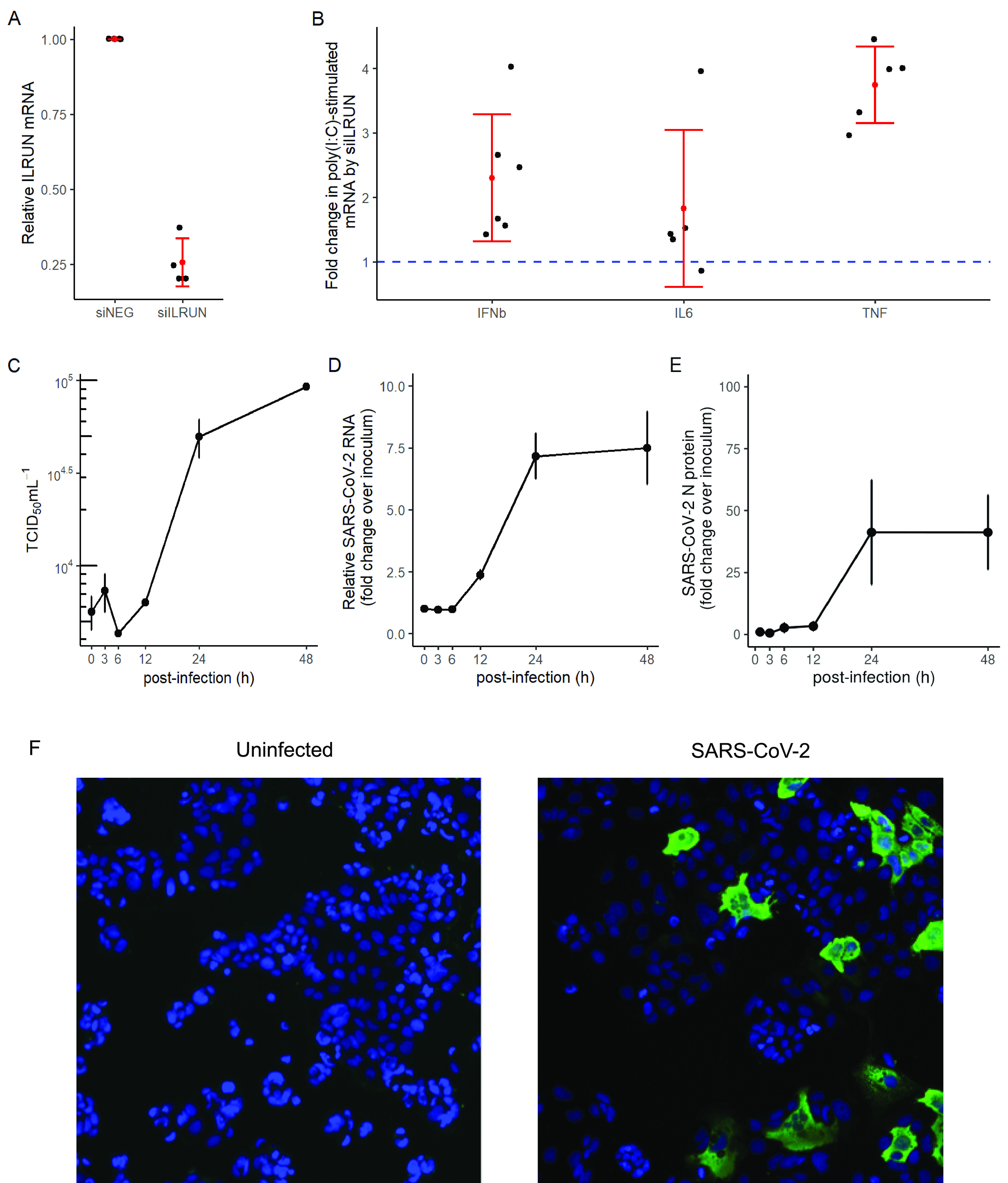
Validation of ILRUN function and SARS-CoV-2 infection in Caco-2 cells. (A) ILRUN mRNA levels (2−ΔΔCT relative to GAPDH) in Caco-2 cells transfected with siRNAs (40 nM, 72 h) targeting ILRUN or a nontargeting control (siNEG). n = 4 biological replicates. (B) Caco-2 cells were transfected with siRNAs (40 nM, 72 h) and then treated with transfected poly(I·C) (10 μg/ml, 6 h). Cells were then harvested and analyzed for mRNA expression of the listed cytokines by qRT-PCR, normalized to GAPDH. n ≥ 5 biological replicates. (C) TCID50 measurements of virus titers. n = 3 biological replicates. (D) qRT-PCR measurements of intracellular viral RNA represented by 2−ΔΔCT normalized first to GAPDH and then to inoculum levels of SARS-CoV-2, set to 1. n = 3 biological replicates. (E) Percent infected cells in Caco-2 cells infected with SARS-CoV-2 compared to inoculum (MOI, 0.1). n = 3 biological replicates. Graphs represent means ± standard deviations (SD). (F) Representative field of view of immunofluorescence microscopy showing SARS-CoV-2 N protein staining (green) in Caco-2 cells infected with SARS-CoV-2 (MOI, 0.1; 24 h). Cell nuclei were stained using DAPI (blue). n = 6 biological replicates.
We next performed a time course experiment to characterize the single-cycle replication kinetics of SARS-CoV-2 in Caco-2 cells. A previous study has shown that increases in SARS-CoV-2 levels in tissue culture supernatant of infected Caco-2 cells are observed within 24 h postinfection (h.p.i.) (16). Consistent with this report, our experiments showed that Caco-2 cells infected with SARS-CoV-2 started producing infectious virions (above inoculum levels) between 12 and 24 h.p.i. (Fig. 1C). This indicates that the length of one cycle of SARS-CoV-2 infection in Caco-2 cells is approximately 12 to 24 h. These results were validated using a quantitative reverse transcription-PCR (qRT-PCR) assay (19) to measure intracellular viral RNA levels at 3, 6, 12, 24, and 48 h.p.i. Intracellular viral RNA levels started increasing above inoculum levels between 6 and 12 h.p.i. (Fig. 1D), which is consistent with the replication kinetics observed with the virus production data (Fig. 1C). When measured by quantitative fluorescence microscopy, intracellular viral nucleoprotein (N) levels (number of infected cells) were also observed to increase between 6 and 12 h, maximizing at 24 h.p.i. (Fig. 1E). We did not observe SARS-CoV-2 infection to induce syncytia or other cytopathic effects in Caco-2 cells (Fig. 1F).
ILRUN regulates multiple cellular pathways, including the renin-angiotensin aldosterone system.
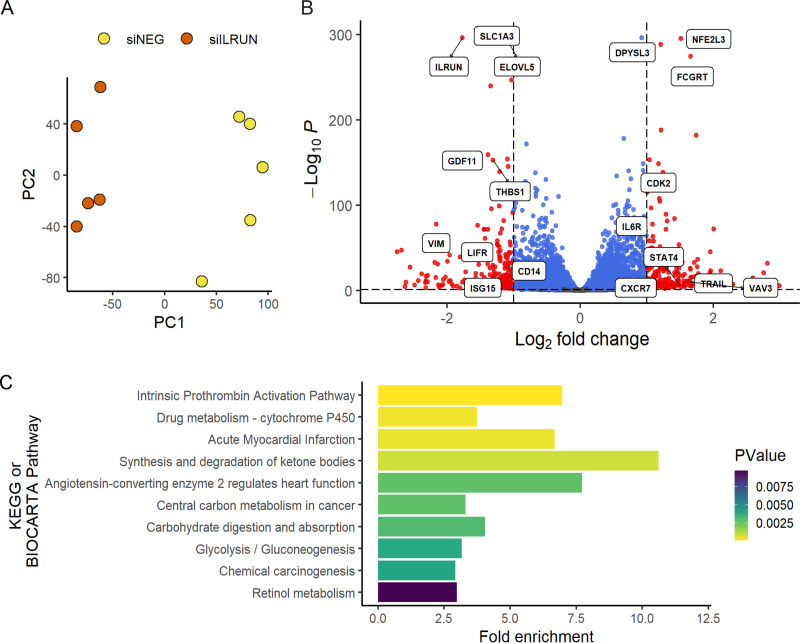
Using Caco-2 cells as a model system, we next utilized RNA-seq to identify genes differentially transcribed as a result of reduced ILRUN expression, including in the context of SARS-CoV-2 infection. RNA was harvested from Caco-2 cells transfected with siILRUN or siNEG (40 nM, 72 h) prior to infection with SARS-CoV-2 for 6 or 24 h. One hundred-base-pair single-end Illumina sequencing reads from two Nova Seq 6000 lanes (technical replicates) were mapped to 33,121 annotated human genes. The DESeq2 R package was then used to normalize read counts for each gene. We first investigated the genes regulated by ILRUN under basal conditions and found that silencing of ILRUN results in a dramatic change in the Caco-2 transcriptome. Unsupervised analysis of variance using principle component analysis (PCA) showed tight clustering of treatment groups (technical replicates not shown for clarity) (Fig. 2A). By comparing siNEG to siILRUN at 96 h posttransfection, we found 457 differentially expressed genes using a conservative cutoff P value of <0.05, log2 fold change (FC) of >1, and baseMean of >5 (Fig. 2B; see also Table S1 in the supplemental material). ILRUN itself was identified as differentially expressed in this analysis (Fig. 2B), validating both siRNA transfection and data analysis protocols. Many of the genes regulated by ILRUN encode proteins with proven roles in innate immunity, including Toll-like receptor 7 (TLR7), interferon-stimulated gene 15 (ISG15), vimentin (VIM), and interleukin-6 receptor (IL6R). Differential expression of some of these genes was validated by qRT-PCR (data not shown).
FIG 2.
Host pathways regulated by ILRUN in Caco-2 cells. (A) Unsupervised variance analysis of 5 biological replicate samples of Caco-2 cells transfected with 40 nM siNEG or siILRUN for 96 h and analyzed by RNA-seq. Samples plotted based on principle component 1 (PC1) and PC2 dimensions. (B) Volcano plot showing global transcriptional changes of samples described in panel A. A total of 457 transcripts (shown in red) were differentially expressed by ILRUN knockdown based on the cutoff P value of <0.05, log2 fold change (FC) of >1, and baseMean of >5. Genes that did not exceed the fold change threshold are shown in blue. Genes of interest related to antiviral immunity are labeled. (C) Functional annotation clustering of the 457 differentially expressed genes in host pathways (BIOCARTA and KEGG) are plotted against the fold enrichment and ordered based on P value.
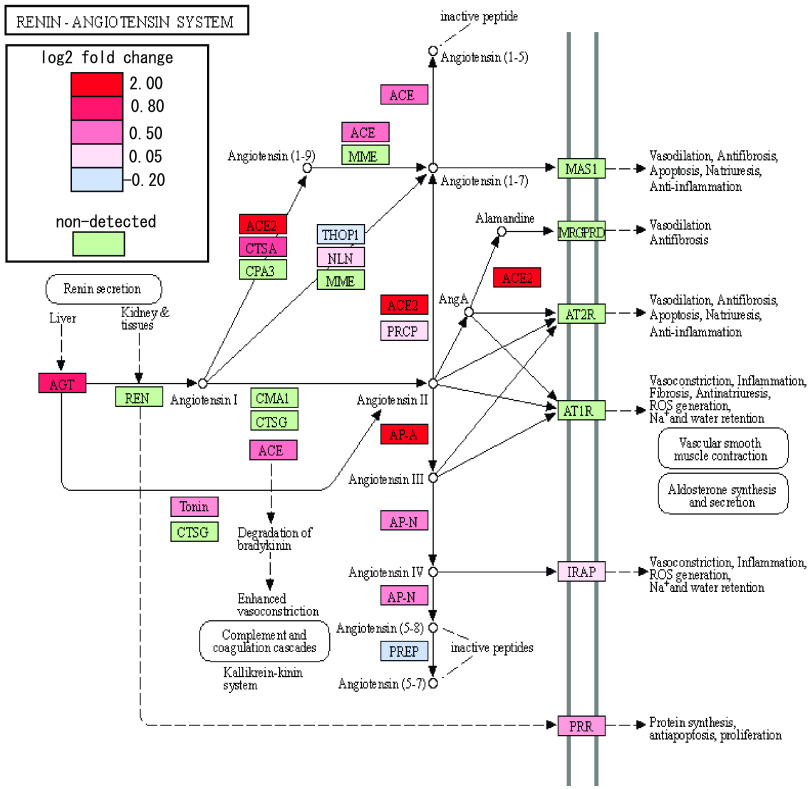
Next, functional annotation clustering was performed to determine whether particular molecular pathways were overrepresented by the set of ILRUN-regulated genes (Fig. 2C). The significantly enriched pathways were largely associated with various metabolic pathways, some of which may be associated with the function(s) of ILRUN in lipoprotein metabolism (20), such as synthesis and degradation of ketone bodies and glycolysis/gluconeogenesis. Other pathways identified may involve the role of ILRUN in regulating inflammation, albeit distinct from its established function as an inhibitor of the type I IFN pathway (14). Of particular interest was the enrichment of the “ACE2 regulates heart function” pathway, which largely consists of the RAAS and includes the SARS-COV-2 entry receptor, ACE2. Our transcriptomics revealed that along with ACE2, ACE, AGT, AP-A, and AP-N were upregulated following ILRUN knockdown (Fig. 3). Thus, our analysis has revealed that ILRUN participates in numerous biological pathways with diverse roles in cell biology, including as a novel regulator of the RAAS.
FIG 3.
ILRUN regulates expression of key RAAS genes. The impact on siILRUN (40 nM, 96 h) on the RAAS was investigated by quantitatively coloring the “Renin-Angiotensin System” KEGG pathway map (https://www.genome.jp/kegg/) using a diverging color palette to show fold change effects across the entire pathway based on our transcriptomics analysis. Shades of red indicate upregulation, while shades of blue indicate downregulation. Genes that were not detected in this study are shown in green.
ILRUN suppresses SARS-CoV-2 growth by regulating the expression of enzymes critical to cell entry.
We next examined the SARS-CoV-2-infected samples to assess the impact of modulating ILRUN expression on infection by mapping RNA-seq reads to the SARS-CoV-2 genome using Bowtie2. After normalizing for library size, we detected elevated reads mapping to SARS-CoV-2 in cells depleted of ILRUN at both the 6-h and 24-h time points (Fig. 4A); this was confirmed via qRT-PCR (Fig. 4B). Overexpression of ILRUN was also found to have the opposing effect, modestly decreasing viral replication (Fig. 4B). Virus infectivity assays also confirmed that siILRUN treatment increased SARS-CoV-2 production in Caco-2 cells, while ILRUN overexpression inhibited production (Fig. 4C).
FIG 4.
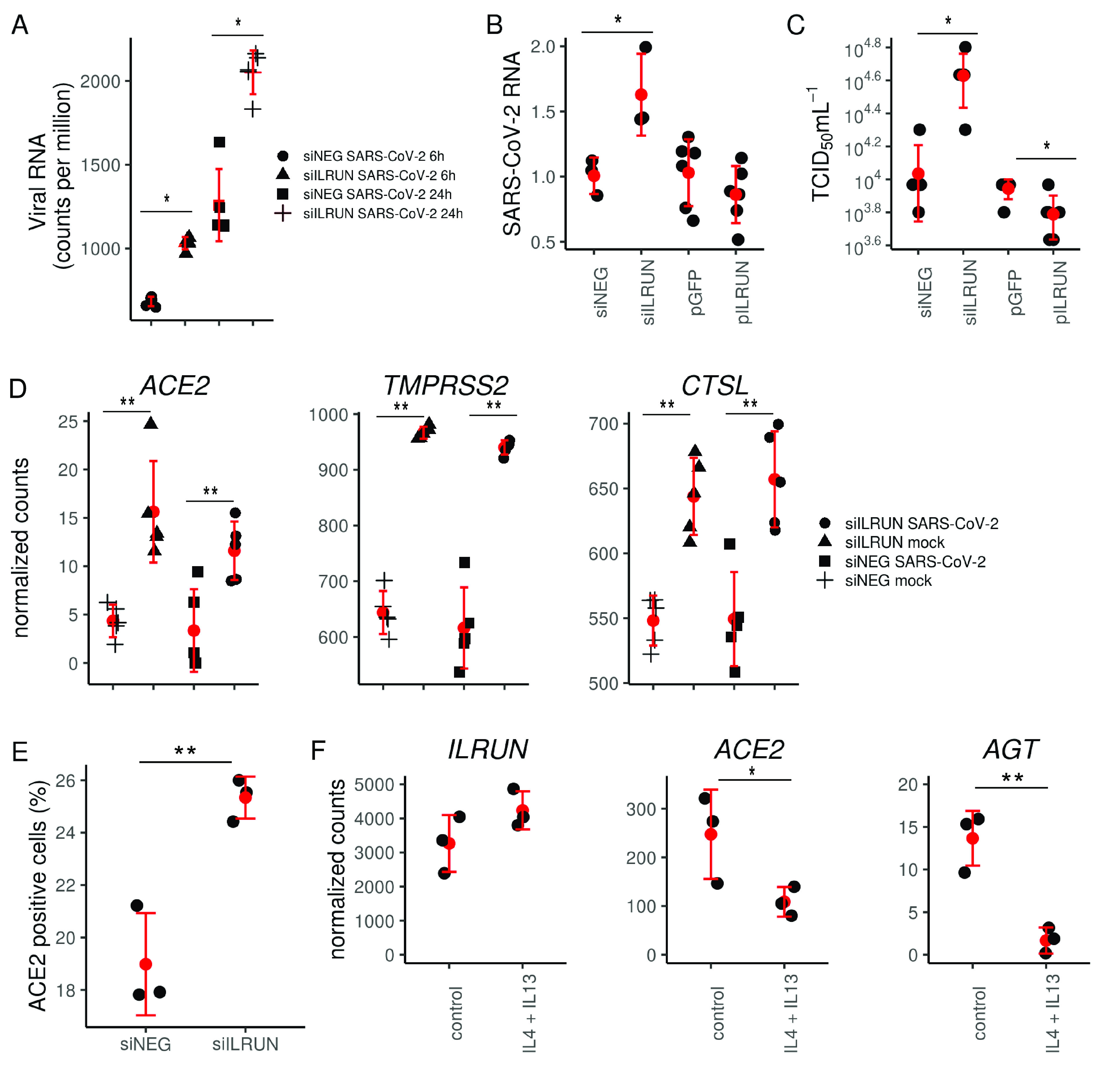
ILRUN suppresses SARS-CoV-2 infection and downregulates host genes essential for SARS-CoV-2 entry. (A) RNA profile of SARS-CoV-2 in Caco-2 cells transfected with 40 nM siNEG or siILRUN for 72 h at 6 h and 24 h postinfection (MOI, 0.1) and analyzed by RNA-seq. n = 5 biological replicates. (B) Caco-2 cells were transfected with indicated siRNAs (40 nM, 72 h) or expression plasmids (500 ng, 48 h) and infected with SARS-CoV-2 (24 h; MOI, 0.1). Cells were then harvested and analyzed for viral RNA by qRT-PCR, normalized to GAPDH. n ≥ 3 biological replicates. (C) SARS-CoV-2 titers of supernatants from Caco-2 cells infected with SARS-CoV-2 (24 h; MOI, 0.3) posttransfection with indicated siRNAs (40 nM, 72 h) or expression plasmids (100 ng, 48 h). n = 5 biological replicates. (D) Normalized RNA-seq counts measuring expression of ACE2, TMPRSS2, and CTSL in Caco-2 cells transfected with indicated siRNAs (40 nM, 72 h) with or without infection with SARS-CoV-2 (24 h; MOI, 0.1). n = 5 biological replicates. (E) Percent ACE2-positive Caco-2 cells following transfection with indicated siRNAs (40 nM, 72 h) and staining with anti-ACE2 antibody and Alexa488-tagged secondary antibody and analyzed by flow cytometry. n = 3 biological replicates. (F) ILRUN, ACE2, and AGT expression in primary human bronchial epithelial cells stimulated with Th2 cytokines (30 ng/ml of IL-4 and IL-13) for 24 h (data set from publicly available Gene Expression Omnibus series GSE113185 [21]). n = 3 biological replicates. *, P < 0.05; **, P < 0.01. Graphs represent means ± SD.
These antiviral properties of ILRUN against SARS-CoV-2 infection prompted further analysis of ILRUN-regulated host genes linked to SARS-CoV-2 infection. Due to its involvement in the RAAS, we investigated whether ILRUN impacts the expression of ACE2 in infected cells. Our RNA-seq data revealed that siILRUN also increased ACE2 gene expression in infected cells and to a similar degree to that observed in uninfected cells (Fig. 4D), indicating that the differences are due to the effects of ILRUN knockdown and not viral infection. This upregulation of ACE2 was confirmed via qRT-PCR (data not shown), and an increase in plasma membrane-bound ACE2 protein was also observed in siILRUN-treated cells compared to siNEG controls, indicated by an increase in ACE2-postive cells detected by flow cytometry (Fig. 4E). Thus, our transcript-level data correlate with levels of protein at the cell surface, suggesting that increased levels of plasma membrane-bound ACE2 are responsible for the increased infection and replication of SARS-CoV-2 in the absence of ILRUN. Indeed, we found that overexpression of ACE2 in Caco-2 cells increases their susceptibility to SARS-CoV-2 infection (data not shown), further supporting this hypothesis.
Although not part of the RAAS, we also investigated whether ILRUN affects the expression of the SARS-CoV-2 coreceptors CTSL and TMPRSS2. Transcripts of both these genes were upregulated in both infected and uninfected cells treated with siILRUN (Fig. 4D), similar to ACE2. Taken together, these data indicate that ILRUN has antiviral function toward SARS-CoV-2 and that this is likely through regulating the expression of key SARS-CoV-2 entry and fusion molecules ACE2, TMPRSS2, and CTSL.
Finally, to investigate the relationship between ILRUN and RAAS gene expression in primary cells, we accessed publicly available RNA-seq data measuring gene expression in normal primary human bronchial epithelial cells from a single donor grown in triplicate (21). In this data set, once fully differentiated, cells were stimulated with interleukin-4 (IL-4) and IL-13 to elicit a Th2-type inflammatory response (22). This resulted in higher levels of ILRUN and a concomitant decrease in ACE2 and AGT expression (Fig. 4F), consistent with our data in Caco-2 cells. This observation provides additional physiological relevance to the role of ILRUN in the expression of the RAAS gene and its potential contribution to the pathology of COVID-19 in the primary site of SARS-CoV-2 infection in the lungs.
DISCUSSION
Here, we present the first evidence that ILRUN modulates SARS-CoV-2 infection as an antiviral factor. Intriguingly, this contrasts with its initially identified function as a proviral factor for Hendra virus (13). This difference is likely due to our new data that show ILRUN participates in multiple cellular processes and, therefore, has the potential to impact virus infection at different stages of their life cycles. Here, we have identified a novel role for ILRUN in regulating the expression of key elements of the RAAS, the body’s mechanism for regulating blood pressure and fluid homeostasis (23). In particular, we found that ILRUN modulates expression of the SARS-CoV-2 (in addition to the other human coronaviruses, HCoV-NL63 [24] and SARS-CoV [25]) entry receptor, ACE2, a constituent of the RAAS (4, 26).
Our data suggest that ILRUN inhibits SARS-CoV-2 infection by blocking the early stages of the virus infection cycle. First, inhibition of ILRUN results in increases in intracellular viral RNA levels at 6 h postinfection (Fig. 4A), a time point that precedes virus replication in Caco-2 cells (Fig. 1D). Second, we have shown ILRUN negatively regulates expression of three host proteins critical for coronavirus virus entry, ACE2, TMPRSS2, and CTSL (Fig. 4D). While the coronavirus S protein mediates target cell attachment via ACE2 (4, 26), the virus-host membrane fusion reaction requires S protein subunits S1 and S2 to undergo a series of conformational changes induced by proteolysis, which is mediated at the plasma membrane by TMPRSS2 (26) and within endosomal compartments by cathepsin L (27). In addition to facilitating virus entry, TMPRSS2 also mediates cell-cell fusion of infected cells (28). Pharmacological inhibitors of both TMPRSS2 (26) and CTSL (27) potently inhibit coronavirus infection in vitro, highlighting the potential of targeting these proteases as therapeutic countermeasures against SARS-CoV-2.
Despite being highly evolutionarily conserved, with orthologues expressed in nearly all metazoans (15), ILRUN remains a relatively poorly characterized protein. Our previous studies show that ILRUN inhibits IRF3-dependent antiviral cytokine transcription, while other studies have identified ILRUN single-nucleotide polymorphisms (SNPs) associated with increased risk of obesity (29, 30), coronary artery disease (CAD) (31), and altered timing of pubescent growth spurts (32, 33). Our current hypothesis is that ILRUN is linked to these processes and the RAAS, through its regulation of the histone acetyltransferases p300 and CBP. p300/CBP are considered master regulators of gene transcription that control proliferation, differentiation, infection, and apoptosis, among other processes (34). AGT expression is tightly regulated by p300/CBP (35), while the ACE2 promoter region also contains a p300/CBP binding site (36), nominating a potential mechanism by which ILRUN regulates expression of these genes. Notably, the RAAS is a key pathway associated with CAD, with inhibitors standardly prescribed for treatment (37). RAAS genes are also found to be upregulated in the dendritic cells of CAD patients, potentially contributing to pathogenesis. Similarly, the RAAS is involved in lipid metabolism and upregulated in adipocytes as a result of obesity (38, 39). Thus, the role of ILRUN in CAD and obesity may be via its transcription regulation of RAAS components.
IFNs are key to antiviral immunity and viral recognition elicits IFN production, which triggers the transcription of IFN-stimulated genes (ISGs), which engage in various antiviral activities. Nevertheless, type I IFNs are widely expressed and can result in immunopathology during viral infections. An intriguing finding from this study is that while ILRUN regulates the type I IFN induction pathway, SARS-CoV-2 did not induce a noticeable type I IFN response in Caco-2 cells (no ISGs were observed to be upregulated following during infection; Table S1), indicating this is independent of the ILRUN-induced IFN expression. Another study has similarly observed a lack of type I IFN induction in Caco-2 cells infected with SARS-CoV-2, including at a high multiplicity of infection (MOI) (40). While one explanation for this result is that Caco-2 cells do not mount robust innate immune responses to viral infection, previous studies have shown infection of Caco-2 cells with Sendai virus and Newcastle disease virus induces a strong type I IFN response, while SARS-CoV-1 suppresses these cytokines (41). Our own data also show that Caco-2 cells respond to the viral RNA mimetic poly(I·C) (Fig. 1B). In Calu-3 cells (which support SARS-CoV-2 replication to higher titers than Caco-2 cells [16]), SARS-CoV-2 induces a type I IFN response that is markedly delayed compared to those of other viral infections, such as influenza A virus (42, 43) and Sendai virus (44). SARS-CoV-2 appears to be particularly adept at immune evasion, encoding seven proteins that inhibit IFN-β promoter activation (45), among them the ORF6 protein that blocks both type I IFN induction and signaling pathways. Several groups have hypothesized that the lack of timely and robust antiviral responses to SARS-CoV-2 infection contributes to COVID-19 pathogenesis (45, 46).
Hadjadj et al. (47) showed that peripheral blood immune cells from severe and critical COVID-19 patients show enhanced inflammatory responses. In our analysis, of note were the inflammation-associated cytokines, IL-6R and IL-1β, which were differentially regulated upon suppression of ILRUN. Inflammation is known to play a crucial role in the pathogenesis of severe infections and ARDS, and evidence is emerging that the IL-1/IL-6 pathway is highly upregulated in patients with severe disease. Both IL-6R and IL-1β have been shown to be significantly upregulated in severe COVID infection (47). Importantly, controlling excessive immune activation is critical, as these cytokines can drive inflammasome activation and, in an autoinflammatory loop, lead to uncontrolled inflammation.
Our data also reveal that ILRUN promotes basal transcription of ISG15, a host member of the ubiquitin family that is targeted by SARS-CoV-2 as an evasion mechanism against antiviral immune responses (48). ISG15 plays diverse roles in antiviral immunity as both a promoter and negative feedback inhibitor of IFN signaling, in addition to direct antiviral activity (49). These functions are carried out via conjugation onto both cellular and viral proteins via an enzymatic cascade (termed ISGlyation) or secreted as an unconjugated protein that acts as a cytokine (49). Coronaviruses, including SARS-CoV-2, encode papain-like protease (PLpro) that, in addition to processing viral proteins to activate the virus replicase complex, cleave posttranslational modifications on host proteins to deactivate immune signaling cascades (49). While the PLpro of SARS-CoV-1 predominantly targets ubiquitin chains, SARS-CoV-2 PLpro preferentially cleaves ISG15, including that from IRF3, resulting in attenuated type I IFN signaling during infection (48). GRL-0617, a chemical compound targeting SARS-CoV-2 PLpro, restores ISG15-mediated activation of IRF3 during SARS-CoV-2 infection and inhibits virus replication (48). Beyond antiviral signaling, extracellular signal-related kinase-1 (ERK1) has been shown to be ISGylated, although the functional relevance of this modification has yet to be elucidated (49). ERK1 participates in the Ras-Raf-MEK-ERK mitogen-activated protein (MAP) kinase signaling pathway, which regulates the cell cycle in response to extracellular signals (50). Notably, ILRUN has been shown to activate a member of this pathway, 90-kDa ribosomal r6 kinase (p90RSK), a target of ERK1 (51). The presence of a UBA-like domain in ILRUN (15), which generally facilitates binding to ubiquitin and ubiquitin-like protein, also offers the intriguing possibility that ILRUN directly interacts with ISG15 to modulate its function. Thus, future studies to elucidate the functions and mode of action of ILRUN may additionally further our understanding of the complex roles of ISG15 in cellular biology, including during infection with pathogenic viruses such as SARS-CoV-2.
In summary, this study sheds further light on the regulatory function of ILRUN, a recently characterized inhibitor of p300/CBP with links to infectious disease, cancer, obesity, and cardiovascular disease. The study reveals a novel role for ILRUN in the regulation of the RAAS and, remarkably, three key genes (ACE2, TMPRSS2, and CTSL) required for infection of human cells by SARS-CoV-2 and other coronaviruses. Future work will aim to understand the molecular mechanisms underlying ILRUN function and progress this molecule as a potential drug target for a number of diseases of significant concern to human health.
MATERIALS AND METHODS
Cell lines.
Vero E6 cells (ATCC CRL-1586) were maintained in Gibco Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (vol/vol) fetal calf serum (FCS), 100 U/ml penicillin, and 100 μg/ml streptomycin (Life Technologies). Caco-2 cells were maintained in Gibco Eagle’s minimum essential medium (EMEM) supplemented with 20% (vol/vol) FCS, 10 mM HEPES, 0.1 mM nonessential amino acids, 2 mM glutamine, 1 mM sodium pyruvate, 100 U/ml penicillin, and 100 μg/ml streptomycin (Life Technologies). All cells were kept at 37°C in a humidified incubator (5% CO2).
Virus.
All virology work was conducted at the CSIRO Australian Centre for Disease Preparedness at physical containment 4 (PC-4). The isolate of SARS-CoV-2 (BetaCoV/Australia/VIC01/2020) was received from the Victorian Infectious Disease Reference Laboratory (VIDRL; Melbourne, Australia) and passaged in Vero E6 cells for isolation, followed by passaging in Vero E6 cells for stock generation. All virus stocks were aliquoted and stored at −80°C for inoculations.
Transfections.
Caco-2 cells were transfected with 40 nM small interfering RNA (siRNA) (GE Life Sciences) and 2 μl (24-well plates) or 0.6 μl (96-well plates) Dharmafect-1 (GE Life Sciences) in serum-free EMEM. For DNA transfections, plasmids encoding ILRUN-FLAG (pILRUN) and green fluorescent protein (pGFP) are described in reference 14. DNA was incubated with FugeneHD (Promega) in serum-free EMEM at a ratio of 1:4 (micrograms of DNA to microliters of FugeneHD). Cells were stimulated with transfected high-molecular-weight poly(I·C) (Invivogen) (10 μg/ml with 3 μl FugeneHD) for 6 h.
RNA-seq.
Total RNA was isolated from cells in TRIzol using the RNeasy kit (Qiagen) per the manufacturer’s instructions. The quality and quantity of RNA was assessed for all samples using a Bioanalyzer (Agilent, Santa Carla, CA). RNA-seq was performed by the Australian Genome Research Facility (AGRF). Illumina TruSeq stranded mRNA libraries were prepared, followed by sequencing on an Illumina Novaseq-6000 for generation of 100-bp single-end reads on two lanes (technical replicates). Between ∼15 and 21 million raw single-end reads were obtained per sample. Raw data were assessed for overall quality using fastqc v0.11.8 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
Bioinformatic analysis of RNA-seq data.
Quality and adapter trimming was performed using TrimGalore v0.6.4 (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) with default settings for automatic adapter detection. Trimmed reads were mapped to the National Centre for Biotechnology Information (NCBI) human reference genome (GRCh38), downloaded from Illumina iGenomes (http://igenomes.illumina.com.s3-website-us-east-1.amazonaws.com/Homo_sapiens/NCBI/GRCh38/Homo_sapiens_NCBI_GRCh38.tar.gz), using Tophat v2.1.1 (52). The number of reads overlapping each gene in the NCBI annotated reference genome (GRCh38) was counted using htseq-count v0.11.2 within Python v3.7.2 (53), using the intersection-nonempty mode to handle reads overlapping more than one feature. A custom python script was used to compile counts from individual samples into a single-count matrix file. The Bioconductor package DESeq2 was used to test for differential expression between different experimental groups (54). Volcano plots were prepared in R using (https://github.com/kevinblighe/EnhancedVolcano). Gene lists were submitted to DAVID bioinformatics resources (https://david.ncifcrf.gov/summary.jsp), and functional annotation clustering was performed using default settings to retrieve GO terms in each annotation cluster. For Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway mapping (55), custom python scripts were used to prepare input lists for conversion of gene symbols to Entrez gene IDs using bioDBnet’s db2db tool (https://biodbnet-abcc.ncifcrf.gov/db/db2db.php) and generation of hex codes based on fold change for color mapping of KEGG pathways (https://www.genome.jp/kegg/tool/map_pathway2.html). Trimmed reads were mapped to a SARS-CoV-2 reference genome of the virus used in experiments (MT007544; SARS-CoV-2/human/AUS/VIC01/2020) using bowtie v2.3.4 (56). Mapped viral reads were counted using SAMtools v1.10.0 (57) and coverview v1.4.4 (58) and then normalized to library read content. Normalized counts from viral reads and DESeq2 (host genes) were plotted using the geom_boxplot function in the ggplot2 R package using the 25th and 75th percentiles to form box and whiskers no larger than 1.5 times the interquartile range. Data points beyond the whiskers are outliers.
Analysis of publicly available data.
The NCBI Gene Expression Omnibus was searched for suitable RNA-seq data sets and GSE accession number entered into the GEO RNA-seq Experiments Interactive Navigator (http://www.ilincs.org/apps/grein/) to retrieve metadata and normalized counts. Normalized counts were plotted for each gene using ggplot2 package in R.
Quantitative real-time PCR.
Five hundred nanograms of RNA was reverse transcribed to DNA using SensiFast reverse transcriptase (Bioline) first-strand cDNA synthesis protocols. qRT-PCR was performed on a Quant Studio 3 thermocycler (Applied Biosystems) with TaqMan probes for SARS-CoV-2 E gene, ILRUN (Hs00256056_s1), IFNβ (Hs01077958_s1), TNF-α (Hs00174128_m1), IL-6 (Hs00174131_m1), ISG15 (Hs01921423_s1), IL6R (Hs01075664_m1), VIM (Hs00958111_m1), and glyceraldeyde-3-phosphate dehydrogenase (GAPDH) (Hs02758991_g1) purchased from ThermoFisher Scientific. ACE2 mRNA expression was measured using SYBR green dye and primers (forward, GGAGTTGTGATGGGAGTGAT; reverse, GATGGAGGCATAAGGATTTT) as described previously (55). PCR cycling for gene detection was at 95°C for 10 min followed by 45 cycles of 95°C for 15 s and 60°C for 1 min. RNA transcription data were analyzed using the 2−ΔΔCT method and were normalized to GAPDH expression.
TCID50 analysis.
The infectious titers of SARS-CoV-2 stocks were determined by 50% tissue culture infective doses (TCID50) assays performed as described previously (59). Samples were titrated in quadruplicate in 96-well plates, cocultured with Vero E6 cells for 4 days, and monitored for development of cytopathic effects (CPE). Infectious titers were calculated by the method of Reed and Muench (60).
Immunofluorescence and quantification of relative antigen staining.
Caco-2 cells were fixed for 30 min in 4% paraformaldehyde (PFA) and stained with a polyclonal antibody targeting the SARS-CoV-2 nucleocapsid (N) protein (catalog number 40588-T62; used at 1/2,000; Sino Biological) for 1 h. Cells were subsequently stained with a 1/1,000 dilution of an anti-rabbit AF488 antibody (catalogue number A11008; Invitrogen). Nuclei were counterstained with diamidino-2-phenylindole (DAPI). Caco-2 cells were imaged using the CellInsight quantitative fluorescence microscope (Thermo Fisher Scientific) at a magnification of ×10, 49 fields/well, capturing the entire well. The relative viral antigen staining was quantified using the Compartmental analysis bioapplication of the Cellomics Scan software.
Flow cytometry.
Caco-2 cells transfected with siRNA targeting ILRUN or nontargeting control for 72 h were washed and stained in phosphate-buffered saline containing 0.1%, wt/vol, bovine serum albumin with goat-anti-hACE IgG polyclonal antibody (AF933; R&D Systems), followed by secondary staining with donkey-anti-goat IgG-AF488 (A11055; Thermofisher). Cells were then fixed in 4% paraformaldehyde, run on an LSRII (BD Immunocytometry Systems) flow cytometer, and analyzed with FLOWLOGIC software (Miltenyi Biotech).
Statistics.
The difference between two groups was analyzed in GraphPad Prism by a two-tailed Student's t test and between multiple groups by one-way analysis of variance (ANOVA). A P value of <0.05 was considered significant.
ACKNOWLEDGMENTS
We are grateful for support from our colleagues at the Australian Centre for Disease Preparedness (https://www.grid.ac/institutes/grid.413322.5) for providing the facility used in the completion of this work.
This work was funded by the CSIRO. L.T., M.R.A., A.M.B., and K.M. are the recipients of CSIRO CERC postdoctoral fellowships. M.M. is the recipient of an Australian Postgraduate Award.
We have no completing interests to declare.
Footnotes
Supplemental material is available online only.
Contributor Information
Cameron R. Stewart, Email: cameron.stewart@csiro.au.
Tom Gallagher, Loyola University Chicago.
REFERENCES
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team . 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. 2021. COVID-19 weekly epidemiological update 9 February 2021. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 3.Sanyaolu A, Okorie C, Marinkovic A, Patidar R, Younis K, Desai P, Hosein Z, Padda I, Mangat J, Altaf M. 2020. Comorbidity and its impact on patients with COVID-19. SN Compr Clin Med 2020:1–8. 10.1007/s42399-020-00363-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crackower MA, Sarao R, Oudit GY, Yagil C, Kozieradzki I, Scanga SE, Oliveira-dos-Santos AJ, da Costa J, Zhang L, Pei Y, Scholey J, Ferrario CM, Manoukian AS, Chappell MC, Backx PH, Yagil Y, Penninger JM. 2002. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature 417:822–828. 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- 6.Hamming I, Timens W, Bulthuis MLC, Lely AT, Navis GJ, van Goor H. 2004. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol 203:631–637. 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mao R, Qiu Y, He J-S, Tan J-Y, Li X-H, Liang J, Shen J, Zhu L-R, Chen Y, Iacucci M, Ng SC, Ghosh S, Chen M-H. 2020. Manifestations and prognosis of gastrointestinal and liver involvement in patients with COVID-19: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol 5:667–678. 10.1016/S2468-1253(20)30126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han H, Xie L, Liu R, Yang J, Liu F, Wu K, Chen L, Hou W, Feng Y, Zhu C. 2020. Analysis of heart injury laboratory parameters in 273 COVID-19 patients in one hospital in Wuhan, China. J Med Virol 92:819–823. 10.1002/jmv.25809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li M-Y, Li L, Zhang Y, Wang X-S. 2020. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect Dis Poverty 9:45. 10.1186/s40249-020-00662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L, Zhang Y, Zhang S. 2020. Cardiovascular impairment in COVID-19: learning from current options for cardiovascular anti-inflammatory therapy. Front Cardiovasc Med 7:78. 10.3389/fcvm.2020.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, Li F. 2020. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci U S A 117:11727–11734. 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vaduganathan M, Vardeny O, Michel T, McMurray JJV, Pfeffer MA, Solomon SD. 2020. Renin-angiotensin-aldosterone system inhibitors in patients with Covid-19. N Engl J Med 382:1653–1659. 10.1056/NEJMsr2005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deffrasnes C, Marsh GA, Foo CH, Rootes CL, Gould CM, Grusovin J, Monaghan P, Lo MK, Tompkins SM, Adams TE, Lowenthal JW, Simpson KJ, Stewart CR, Bean AGD, Wang L-FF. 2016. Genome-wide siRNA screening at biosafety level 4 reveals a crucial role for fibrillarin in henipavirus infection. PLoS Pathog 12:e1005478. 10.1371/journal.ppat.1005478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ambrose RL, Liu YC, Adams TE, Bean AGD, Stewart CR. 2018. C6orf106 is a novel inhibitor of the interferon-regulatory factor 3-dependent innate antiviral response. J Biol Chem 293:10561–10573. 10.1074/jbc.RA117.001491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ambrose RL, Brice AM, Caputo AT, Alexander MR, Tribolet L, Liu YC, Adams TE, Bean AGDD, Stewart CR. 2020. Molecular characterisation of ILRUN, a novel inhibitor of proinflammatory and antimicrobial cytokines. Heliyon 6:e04115. 10.1016/j.heliyon.2020.e04115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chu H, Chan JF-W, Yuen TT-T, Shuai H, Yuan S, Wang Y, Hu B, Yip CC-Y, Tsang JO-L, Huang X, Chai Y, Yang D, Hou Y, Chik KK-H, Zhang X, Fung AY-F, Tsoi H-W, Cai J-P, Chan W-M, Ip JD, Chu AW-H, Zhou J, Lung DC, Kok K-H, To KK-W, Tsang OT-Y, Chan K-H, Yuen K-Y. 2020. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe 1:e14–e23. 10.1016/S2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leung WK, To K-F, Chan PKS, Chan HLY, Wu AKL, Lee N, Yuen KY, Sung JJY. 2003. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology 125:1011–1017. 10.1016/S0016-5085(03)01215-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiersinga WJ, Rhodes A, Cheng AC, Peacock SJ, Prescott HC. 2020. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA 324:782–793. 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 19.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, Brünink S, Schneider J, Schmidt ML, Mulders DG, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette J-L, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MP, Drosten C. 2020. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 25:2000045. 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bi X, Kuwano T, Lee PC, Millar JS, Li L, Shen Y, Soccio RE, Hand NJ, Rader DJ. 2020. ILRUN, a human plasma lipid GWAS locus, regulates lipoprotein metabolism in mice. Circ Res 127:1347–1361. 10.1161/CIRCRESAHA.120.317175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jevnikar Z, Östling J, Ax E, Calvén J, Thörn K, Israelsson E, Öberg L, Singhania A, Lau LCK, Wilson SJ, Ward JA, Chauhan A, Sousa AR, De Meulder B, Loza MJ, Baribaud F, Sterk PJ, Chung KF, Sun K, Guo Y, Adcock IM, Payne D, Dahlen B, Chanez P, Shaw DE, Krug N, Hohlfeld JM, Sandström T, Djukanovic R, James A, Hinks TSC, Howarth PH, Vaarala O, van Geest M, Olsson H, Unbiased Biomarkers in Prediction of Respiratory Disease Outcomes Study Group . 2019. Epithelial IL-6 trans-signaling defines a new asthma phenotype with increased airway inflammation. J Allergy Clin Immunol 143:577–590. 10.1016/j.jaci.2018.05.026. [DOI] [PubMed] [Google Scholar]
- 22.Choy DF, Modrek B, Abbas AR, Kummerfeld S, Clark HF, Wu LC, Fedorowicz G, Modrusan Z, Fahy JV, Woodruff PG, Arron JR. 2011. Gene expression patterns of Th2 inflammation and intercellular communication in asthmatic airways. J Immunol 186:1861–1869. 10.4049/jimmunol.1002568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reid IA, Morris BJ, Ganong WF. 1978. The renin-angiotensin system. Annu Rev Physiol 40:377–410. 10.1146/annurev.ph.40.030178.002113. [DOI] [PubMed] [Google Scholar]
- 24.Hofmann H, Pyrc K, van der Hoek L, Geier M, Berkhout B, Pöhlmann S. 2005. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc Natl Acad Sci U S A 102:7988–7993. 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450–454. 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu N-H, Nitsche A, Müller MA, Drosten C, Pöhlmann S. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simmons G, Gosalia DN, Rennekamp AJ, Reeves JD, Diamond SL, Bates P. 2005. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci U S A 102:11876–11881. 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glowacka I, Bertram S, Müller MA, Allen P, Soilleux E, Pfefferle S, Steffen I, Tsegaye TS, He Y, Gnirss K, Niemeyer D, Schneider H, Drosten C, Pöhlmann S. 2011. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol 85:4122–4134. 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahmad S, Poveda A, Shungin D, Barroso I, Hallmans G, Renström F, Franks PW. 2016. Established BMI-associated genetic variants and their prospective associations with BMI and other cardiometabolic traits: the GLACIER Study. Int J Obes 40:1346–1352. 10.1038/ijo.2016.72. [DOI] [PubMed] [Google Scholar]
- 30.Riveros-McKay F, Mistry V, Bounds R, Hendricks A, Keogh JM, Thomas H, Henning E, Corbin LJ, O’Rahilly S, Zeggini E, Wheeler E, Barroso I, Farooqi IS, O’Rahilly S, Zeggini E, Wheeler E, Barroso I, Farooqi IS, Understanding Society Scientific Group . 2019. Genetic architecture of human thinness compared to severe obesity. PLoS Genet 15:e1007603. 10.1371/journal.pgen.1007603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LeBlanc M, Zuber V, Andreassen BK, Witoelar A, Zeng L, Bettella F, Wang Y, McEvoy LK, Thompson WK, Schork AJ, Reppe S, Barrett-Connor E, Ligthart S, Dehghan A, Gautvik KM, Nelson CP, Schunkert H, Samani NJ, Consortium C, Ridker PM, Chasman DI, Aukrust P, Djurovic S, Frigessi A, Desikan RS, Dale AM, Andreassen OA, Ridker PM, Chasman DI, Aukrust P, Djurovic S, Frigessi A, Desikan RS, Dale AM, Andreassen OA, CARDIoGRAM Consortium . 2016. Identifying novel gene variants in coronary artery disease and shared genes with several cardiovascular risk factors. Circ Res 118:83–94. 10.1161/CIRCRESAHA.115.306629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JJ, Essers JB, Kugathasan S, Escher JC, Lettre G, Butler JL, Stephens MC, Ramoni MF, Grand RJ, Hirschhorn J. 2010. Association of linear growth impairment in pediatric Crohn’s disease and a known height locus: a pilot study. Ann Hum Genet 74:489–497. 10.1111/j.1469-1809.2010.00606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sovio U, Bennett AJ, Millwood LY, Molitor JJ, O’Reilly PF, Timpson N, Kaakinen M, Laitinen J, Haukka J, Pillas D, Tzoulaki I, Molitor JJ, Hoggart C, Coin LJMM, Whittaker J, Pouta A, Hartikainen A-LL, Freimer NB, Widen E, Peltonen L, Elliott PI, McCarthy M, Jarvelin M-RR, Millwood IY, Molitor JJ, O’Reilly PF, Timpson NJ, Kaakinen M, Laitinen J, Haukka J, Pillas D, Tzoulaki I, Molitor JJ, Hoggart C, Coin LJMM, Whittaker J, Pouta A, Hartikainen A-LL, Freimer NB, Widen E, Peltonen L, Elliott P, McCarthy MI, Jarvelin M-RR. 2009. Genetic determinants of height growth assessed longitudinally from infancy to adulthood in the northern Finland birth cohort 1966. PLoS Genet 5:e1000409. 10.1371/journal.pgen.1000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan HM, La Thangue NB. 2001. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J Cell Sci 114:2363–2373. 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- 35.Ray S, Sherman CT, Lu M, Brasier AR. 2002. Angiotensinogen gene expression is dependent on signal transducer and activator of transcription 3-mediated p300/cAMP response element binding protein-binding protein coactivator recruitment and histone acetyltransferase activity. Mol Endocrinol 16:824–836. 10.1210/mend.16.4.0811. [DOI] [PubMed] [Google Scholar]
- 36.Clarke NE, Belyaev ND, Lambert DW, Turner AJ. 2014. Epigenetic regulation of angiotensin-converting enzyme 2 (ACE2) by SIRT1 under conditions of cell energy stress. Clin Sci 126:507–516. 10.1042/CS20130291. [DOI] [PubMed] [Google Scholar]
- 37.Sheppard RJ, Schiffrin EL. 2013. Inhibition of the renin-angiotensin system for lowering coronary artery disease risk. Curr Opin Pharmacol 13:274–279. 10.1016/j.coph.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 38.Frigolet ME, Torres N, Tovar AR. 2013. The renin-angiotensin system in adipose tissue and its metabolic consequences during obesity. J Nutr Biochem 24:2003–2015. 10.1016/j.jnutbio.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 39.Favre GA, Esnault VLM, Van Obberghen E. 2015. Modulation of glucose metabolism by the renin-angiotensin-aldosterone system. Am J Physiol Endocrinol Metab 308:E435–E449. 10.1152/ajpendo.00391.2014. [DOI] [PubMed] [Google Scholar]
- 40.Shuai H, Chu H, Hou Y, Yang D, Wang Y, Hu B, Huang X, Zhang X, Chai Y, Cai J-P, Chan JF-W, Yuen K-Y. 2020. Differential immune activation profile of SARS-CoV-2 and SARS-CoV infection in human lung and intestinal cells: implications for treatment with IFN-β and IFN inducer. J Infect 81:e1–e10. 10.1016/j.jinf.2020.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spiegel M, Weber F. 2006. Inhibition of cytokine gene expression and induction of chemokine genes in non-lymphatic cells infected with SARS coronavirus. Virol J 3:17. 10.1186/1743-422X-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, Oberley-Deegan R, Wang S, Nikrad M, Funk CJ, Hartshorn KL, Mason RJ. 2009. Differentiated human alveolar type II cells secrete antiviral IL-29 (IFN-lambda 1) in response to influenza A infection. J Immunol 182:1296–1304. 10.4049/jimmunol.182.3.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khaitov MR, Laza-Stanca V, Edwards MR, Walton RP, Rohde G, Contoli M, Papi A, Stanciu LA, Kotenko SV, Johnston SL. 2009. Respiratory virus induction of alpha-, beta- and lambda-interferons in bronchial epithelial cells and peripheral blood mononuclear cells. Allergy 64:375–386. 10.1111/j.1398-9995.2008.01826.x. [DOI] [PubMed] [Google Scholar]
- 44.Monson EA, Crosse KM, Das M, Helbig KJ. 2018. Lipid droplet density alters the early innate immune response to viral infection. PLoS One 13:e0190597. 10.1371/journal.pone.0190597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lei X, Dong X, Ma R, Wang W, Xiao X, Tian Z, Wang C, Wang Y, Li L, Ren L, Guo F, Zhao Z, Zhou Z, Xiang Z, Wang J. 2020. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat Commun 11:3810. 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shuai L, Feng N, Wang X, Ge J, Wen Z, Chen W, Qin L, Xia X, Bu Z. 2015. Genetically modified rabies virus ERA strain is safe and induces long-lasting protective immune response in dogs after oral vaccination. Antiviral Res 121:9–15. 10.1016/j.antiviral.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 47.Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, Péré H, Charbit B, Bondet V, Chenevier-Gobeaux C, Breillat P, Carlier N, Gauzit R, Morbieu C, Pène F, Marin N, Roche N, Szwebel T-A, Merkling SH, Treluyer J-M, Veyer D, Mouthon L, Blanc C, Tharaux P-L, Rozenberg F, Fischer A, Duffy D, Rieux-Laucat F, Kernéis S, Terrier B. 2020. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 369:718–724. 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shin D, Mukherjee R, Grewe D, Bojkova D, Baek K, Bhattacharya A, Schulz L, Widera M, Mehdipour AR, Tascher G, Geurink PP, Wilhelm A, van der Heden van Noort GJ, Ovaa H, Müller S, Knobeloch K-P, Rajalingam K, Schulman BA, Cinatl J, Hummer G, Ciesek S, Dikic I. 2020. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature 587:657–662. 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perng Y-C, Lenschow DJ. 2018. ISG15 in antiviral immunity and beyond. Nat Rev Microbiol 16:423–439. 10.1038/s41579-018-0020-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pelech SL, Charest DL. 1995. MAP kinase-dependent pathways in cell cycle control. Prog Cell Cycle Res 1:33–52. 10.1007/978-1-4615-1809-9_4. [DOI] [PubMed] [Google Scholar]
- 51.Li X, Dong M, Zhou J, Zhu D, Zhao J, Sheng W. 2019. C6orf106 accelerates pancreatic cancer cell invasion and proliferation via activating ERK signaling pathway. Mol Cell Biochem 454:87–95. 10.1007/s11010-018-3455-0. [DOI] [PubMed] [Google Scholar]
- 52.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anders S, Pyl PT, Huber W. 2015. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44:D457–D462. 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Münz M, Mahamdallie S, Yost S, Rimmer A, Poyastro-Pearson E, Strydom A, Seal S, Ruark E, Rahman N. 2018. CoverView: a sequence quality evaluation tool for next generation sequencing data. Wellcome Open Res 3:36. 10.12688/wellcomeopenres.14306.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stewart CR, Marsh GA, Jenkins KA, Gantier MP, Tizard ML, Middleton D, Lowenthal JW, Haining J, Izzard L, Gough TJ, Deffrasnes C, Stambas J, Robinson R, Heine HG, Pallister JA, Foord AJ, Bean AG, Wang L-FF. 2013. Promotion of Hendra virus replication by microRNA 146a. J Virol 87:3782–3791. 10.1128/JVI.01342-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reed L, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am J Hyg 27:493–497. 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download JVI.00327-21-s0001.xlsx, XLSX file, 219 KB (218KB, xlsx)
Table S2. Download JVI.00327-21-s0002.xlsx, XLSX file, 22 KB (21.7KB, xlsx)
Supplemental Table Captions. Download JVI.00327-21-s0003.pdf, PDF file, 6 KB (5.8KB, pdf)