ABSTRACT
The International Committee on Taxonomy of Viruses (ICTV) has recently adopted a comprehensive, hierarchical system of virus taxa. The highest ranks in this hierarchy are realms, each of which is considered monophyletic but apparently originated independently of other realms. Here, we announce the creation of a new realm, Adnaviria, which unifies archaeal filamentous viruses with linear A-form double-stranded DNA genomes and characteristic major capsid proteins unrelated to those encoded by other known viruses.
KEYWORDS: A-form DNA, Ligamenvirales, Tokiviricetes, Rudiviridae, Lipothrixviridae, Tristromaviridae, hyperthermophilic archaea, virus evolution, virus classification, virus taxonomy, International Committee on Taxonomy of Viruses (ICTV), major capsid protein, virus structure and assembly
TEXT
In 2018, the International Committee on Taxonomy of Viruses (ICTV) expanded the number of taxonomic ranks available for virus classification from 5 to 15 (1, 2). This development enabled the formal recognition of the evolutionary kinships among distantly related viruses and led to the creation of four realms—virus taxa that are roughly equivalent to the domain rank in cellular taxonomy (3): the realm Riboviria encompasses all RNA viruses and reverse-transcribing viruses that encode homologous RNA-directed RNA polymerases and reverse transcriptases, respectively; the realm Monodnaviria comprises viruses with predominantly single-stranded DNA (ssDNA) genomes and encoding rolling-circle replication initiation endonucleases of the HUH superfamily; the realm Duplodnaviria unifies viruses with double-stranded DNA (dsDNA) genomes that produce virions with icosahedral capsids formed by major capsid proteins (MCPs) with the HK97 fold; and the realm Varidnaviria comprises dsDNA viruses with icosahedral capsids built from MCPs with a double-jelly roll fold (4). However, many virus families remained unassigned to higher taxa due to the lack of demonstrable evolutionary relationships to viruses assigned to the four realms. Here, we announce the creation, and recent official acceptance by the ICTV (https://talk.ictvonline.org/taxonomy/), of a new realm, Adnaviria, which encompasses structurally related archaeal filamentous viruses with dsDNA genomes that adopt the A-form conformation within their virions.
Filamentous viruses infect hosts of all cellular domains, but despite overall similar morphology, virions of eukaryotic, bacterial, and archaeal filamentous viruses are built from capsid proteins with different structural folds (5). Furthermore, the types of the viral genomes that are protected by the corresponding capsid proteins are radically different among viruses infecting hosts from the different domains of life (6–11): all known eukaryotic filamentous viruses have linear ssRNA genomes and are classified in the realm Riboviria (4), all known bacterial filamentous viruses have circular ssDNA genomes and belong to the realm Monodnaviria (4, 12), and all known archaeal filamentous viruses have dsDNA genomes and, until recently, have remained unassigned to taxa ranked higher than order.
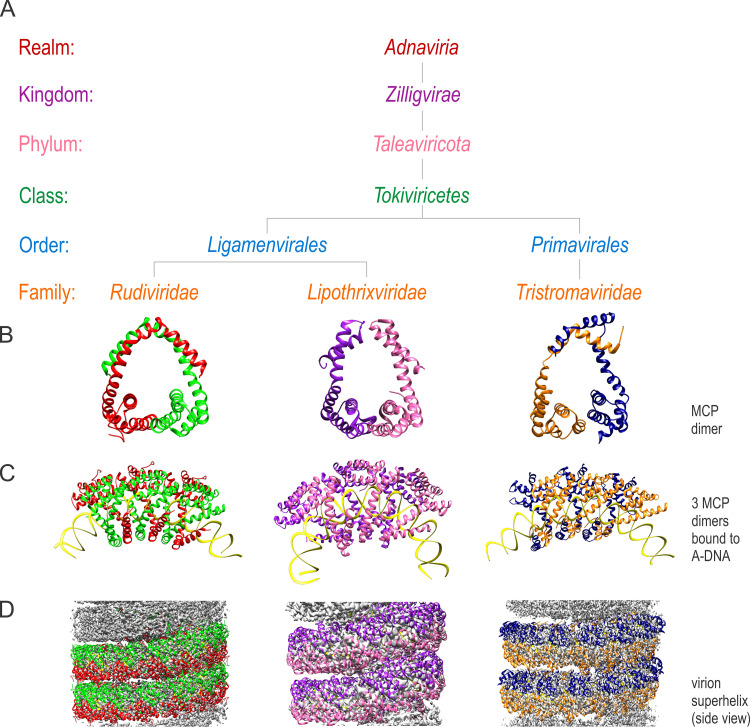
Archaeal viruses that form filamentous virions are currently classified into four families: Clavaviridae, Lipothrixviridae, Rudiviridae, and Tristromaviridae (13). Based on the shared gene content, structural similarity of their virions, and homology of the MCPs, the families Lipothrixviridae and Rudiviridae were included in the order Ligamenvirales (14). In virions of both families, the nucleoprotein helix is composed of asymmetric units containing two MCP molecules, a homodimer in the case of rudivirids and a heterodimer of paralogous MCPs in the case of lipothrixvirids (Fig. 1) (11, 15). The MCPs of ligamenviral particles have a unique α-helical fold first found in the MCP of rudivirid Sulfolobus islandicus rod-shaped virus 2 (SIRV2) (16). Lipothrixvirids and rudivirids share a characteristic feature in that the interaction between the MCP dimer and the linear dsDNA genome maintains the DNA in the A form. Consequently, the entire genome adopts the A form in virions (15–18).
FIG 1.
Taxonomic organization of the realm Adnaviria. (A) Taxonomic ranks from family to realm rendered with different colors. (B to D) Cryogenic electron microscopy showing structures of the major capsid protein (MCP) homodimer (characteristic of rudivirids) and heterodimer (characteristic of lipothrixvirids and tristromavirids) (B), MCPs bound to the viral double-stranded DNA (dsDNA) genome in the A-form conformation (C), and side views of the viral nucleocapsid for each constituent family (D). Rudiviridae, Lipothrixviridae, and Tristromaviridae are represented by structures of Sulfolobus islandicus rod-shaped virus 2 (PDB accession no. 3J9X), Acidianus filamentous virus 1 (PDB accession no. 5W7G), and Pyrobaculum filamentous virus 2 (PDB accession no. 6V7B), respectively.
Clavavirids (19, 20) and tristromavirids (21) do not encode proteins with recognizable sequence similarity to proteins of ligamenvirals (22). Hence, the two families were not included in Ligamenvirales. Recently, the structure of particles produced by tristromavirid Pyrobaculum filamentous virus 2 (PFV2) has been characterized by cryogenic electron microscopy (cryo-EM) (23). Unexpectedly, the two nucleocapsid proteins of tristromavirids were found to be structurally related to those of ligamenvirals, and the virion organizations were discovered to be remarkably similar, including the A-form conformation of the genomic dsDNA (23) (Fig. 1). Similar to lipothrixvirids, virions of tristromavirids are enveloped by a lipid membrane (24). Based on the structural similarity between members of the Ligamenvirales and Tristromaviridae, we inferred that they share an origin (23). To formalize this evolutionary relationship, the family Tristromaviridae was assigned to a new order, Primavirales, which, along with Ligamenvirales, was included in a new class, Tokiviricetes.
Members of the Tokiviricetes represent one of the most commonly detected virus groups in terrestrial hot springs and are globally distributed (25–27). Rudivirids have been extensively studied in particular, and many aspects of their life cycles are understood in considerable detail (28). Notably, the mechanisms of genome replication do not appear to be conserved among tokiviricetes. It has been proposed that the rudivirus SIRV2 uses a combination of strand displacement, rolling-circle, and strand-coupled genome replication mechanisms, which generate multimeric, highly branched, “brush-like” intermediates reaching lengths of >1,200 kb (∼34 genome units) (29). The latter are then processed into unit-length, linear genomes with covalently linked hairpin ends. The genome replication of the lipothrixvirid Acidianus filamentous virus 1 has been suggested to start by the formation of a D loop and to progress by the strand displacement replication mechanism, whereas termination relies on recombination events through the formation of terminal loop-like structures, although the genes involved in this unique mechanism of replication remain unknown (30). Similar to rudivirid genomes, tristromavirid genomes have terminal inverted repeats (21), but the replication mechanism of these viruses has not been investigated.
High-resolution structures are now available for virions of six distinct tokiviricetes (11, 15). Like many structurally related viruses in the two other realms of dsDNA viruses (Duplodnaviria and Varidnaviria), there is no detectable sequence similarity among the capsid proteins of viruses from different tokiviricete families, suggesting a vast undescribed diversity of viruses in this part of the virosphere. Indeed, it has been suggested that tokiviricetes were present in the last archaeal common ancestor and possibly even in the last universal cellular ancestor (LUCA) (31). Regardless, the available data unequivocally show that archaeal filamentous viruses do not fall into any of the four established realms. Thus, given the lack of a detectable relationship with other viruses and officially acknowledging the uniqueness of the three families of archaeal filamentous viruses that contain A-form DNA in their virions, a new taxon of the highest rank, the realm Adnaviria, was created for their classification. To bridge the gap between the class and realm taxa, intermediate kingdom and phylum taxa, named Zilligvirae and Taleaviricota, respectively, were established.
In contrast, structural characterization of clavavirid Aeropyrum pernix bacilliform virus 1 (APBV1) particles by cryo-EM confirmed that the fold of its MCP and its overall virion organization are unrelated to those of ligamenviral, tristromavirid, or other characterized virus particles (32). Therefore, notwithstanding their filamentous virions, clavavirids are not included in the realm Adnaviria and likely represent another realm to be established in the future.
ETYMOLOGY OF NEW TAXA
Adnaviria, from A-form DNA characteristic of viruses in this realm and the suffix -viria for realm taxa.
Zilligvirae, after Wolfram Zillig (1925 to 2005), a pioneer of research on hyperthermophilic archaeal viruses, and the suffix -virae for kingdom taxa.
Taleaviricota, from Latin talea, meaning “rod” (referring to the virion morphology), and the suffix -viricota for phylum taxa.
Tokiviricetes, from Georgian თოკი (toki), meaning “thread,” and the suffix -viricetes for class taxa.
Primavirales, from Latin prima, meaning “first,” referring to the fact that Thermoproteus tenax virus 1, classified in this order, was the first hyperthermophilic archaeal virus to be isolated in 1983 (33).
ACKNOWLEDGMENTS
We are grateful to Anya Crane (National Institutes of Health [NIH] National Institute of Allergy and Infectious Diseases [NIAID] Division of Clinical Research [DCR] Integrated Research Facility at Fort Detrick [IRF-Frederick], Frederick, MD, USA) for editing the manuscript.
M.K. was supported by l’Agence Nationale de la Recherche (grant ANR-20-CE20-0009-02) and Emergence(s) project MEMREMA from the Ville de Paris. E.V.K. is supported by the Intramural Research Program of the U.S. National Institutes of Health (National Library of Medicine). E.H.E. was supported by NIH grant R35GM122510. This work was supported in part through a Laulima Government Solutions, LLC, prime contract with the U.S. NIAID under contract no. HHSN272201800013C. J.H.K. performed this work as an employee of Tunnell Government Services (TGS), a subcontractor of Laulima Government Solutions, LLC, under contract no. HHSN272201800013C.
The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the U.S. Department of Health and Human Services or of the institutions and companies affiliated with the authors.
The views expressed in this article do not necessarily reflect the views of the journal or of ASM.
Contributor Information
Mart Krupovic, Email: mart.krupovic@pasteur.fr.
Rozanne M. Sandri-Goldin, University of California, Irvine
REFERENCES
- 1.International Committee on Taxonomy of Viruses Executive Committee. 2020. The new scope of virus taxonomy: partitioning the virosphere into 15 hierarchical ranks. Nat Microbiol 5:668–674. 10.1038/s41564-020-0709-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siddell SG, Walker PJ, Lefkowitz EJ, Mushegian AR, Adams MJ, Dutilh BE, Gorbalenya AE, Harrach B, Harrison RL, Junglen S, Knowles NJ, Kropinski AM, Krupovic M, Kuhn JH, Nibert M, Rubino L, Sabanadzovic S, Sanfaçon H, Simmonds P, Varsani A, Zerbini FM, Davison AJ. 2019. Additional changes to taxonomy ratified in a special vote by the International Committee on Taxonomy of Viruses (October 2018). Arch Virol 164:943–946. 10.1007/s00705-018-04136-2. [DOI] [PubMed] [Google Scholar]
- 3.Walker PJ, Siddell SG, Lefkowitz EJ, Mushegian AR, Dempsey DM, Dutilh BE, Harrach B, Harrison RL, Hendrickson RC, Junglen S, Knowles NJ, Kropinski AM, Krupovic M, Kuhn JH, Nibert M, Rubino L, Sabanadzovic S, Simmonds P, Varsani A, Zerbini FM, Davison AJ. 2019. Changes to virus taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2019). Arch Virol 164:2417–2429. 10.1007/s00705-019-04306-w. [DOI] [PubMed] [Google Scholar]
- 4.Koonin EV, Dolja VV, Krupovic M, Varsani A, Wolf YI, Yutin N, Zerbini FM, Kuhn JH. 2020. Global organization and proposed megataxonomy of the virus world. Microbiol Mol Biol Rev 84:e00061-19. 10.1128/MMBR.00061-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krupovic M, Koonin EV. 2017. Multiple origins of viral capsid proteins from cellular ancestors. Proc Natl Acad Sci U S A 114:E2401–E2410. 10.1073/pnas.1621061114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DiMaio F, Chen C-C, Yu X, Frenz B, Hsu Y-H, Lin N-S, Egelman EH. 2015. The molecular basis for flexibility in the flexible filamentous plant viruses. Nat Struct Mol Biol 22:642–644. 10.1038/nsmb.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klug A. 1999. The tobacco mosaic virus particle: structure and assembly. Philos Trans R Soc Lond B Biol Sci 354:531–535. 10.1098/rstb.1999.0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stubbs G, Kendall A. 2012. Helical viruses. Adv Exp Med Biol 726:631–658. 10.1007/978-1-4614-0980-9_28. [DOI] [PubMed] [Google Scholar]
- 9.Tarafder AK, von Kügelgen A, Mellul AJ, Schulze U, Aarts DGAL, Bharat TAM. 2020. Phage liquid crystalline droplets form occlusive sheaths that encapsulate and protect infectious rod-shaped bacteria. Proc Natl Acad Sci U S A 117:4724–4731. 10.1073/pnas.1917726117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zamora M, Méndez-López E, Agirrezabala X, Cuesta R, Lavín JL, Sánchez-Pina MA, Aranda MA, Valle M. 2017. Potyvirus virion structure shows conserved protein fold and RNA binding site in ssRNA viruses. Sci Adv 3:eaao2182. 10.1126/sciadv.aao2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baquero DP, Liu Y, Wang F, Egelman EH, Prangishvili D, Krupovic M. 2020. Structure and assembly of archaeal viruses. Adv Virus Res 108:127–164. 10.1016/bs.aivir.2020.09.004. [DOI] [PubMed] [Google Scholar]
- 12.Roux S, Krupovic M, Daly RA, Borges AL, Nayfach S, Schulz F, Sharrar A, Matheus Carnevali PB, Cheng J-F, Ivanova NN, Bondy-Denomy J, Wrighton KC, Woyke T, Visel A, Kyrpides NC, Eloe-Fadrosh EA. 2019. Cryptic inoviruses revealed as pervasive in bacteria and archaea across Earth’s biomes. Nat Microbiol 4:1895–1906. 10.1038/s41564-019-0510-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prangishvili D, Bamford DH, Forterre P, Iranzo J, Koonin EV, Krupovic M. 2017. The enigmatic archaeal virosphere. Nat Rev Microbiol 15:724–739. 10.1038/nrmicro.2017.125. [DOI] [PubMed] [Google Scholar]
- 14.Prangishvili D, Krupovic M. 2012. A new proposed taxon for double-stranded DNA viruses, the order “Ligamenvirales.” Arch Virol 157:791–795. 10.1007/s00705-012-1229-7. [DOI] [PubMed] [Google Scholar]
- 15.Wang F, Baquero DP, Beltran LC, Su Z, Osinski T, Zheng W, Prangishvili D, Krupovic M, Egelman EH. 2020. Structures of filamentous viruses infecting hyperthermophilic archaea explain DNA stabilization in extreme environments. Proc Natl Acad Sci U S A 117:19643–19652. 10.1073/pnas.2011125117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DiMaio F, Yu X, Rensen E, Krupovic M, Prangishvili D, Egelman EH. 2015. A virus that infects a hyperthermophile encapsidates A-form DNA. Science 348:914–917. 10.1126/science.aaa4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasson P, DiMaio F, Yu X, Lucas-Staat S, Krupovic M, Schouten S, Prangishvili D, Egelman EH. 2017. Model for a novel membrane envelope in a filamentous hyperthermophilic virus. Elife 6:e26268. 10.7554/eLife.26268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Osinski T, Wang F, Krupovic M, Schouten S, Kasson P, Prangishvili D, Egelman EH. 2018. Structural conservation in a membrane-enveloped filamentous virus infecting a hyperthermophilic acidophile. Nat Commun 9:3360. 10.1038/s41467-018-05684-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prangishvili D, Mochizuki T, Liu Y, Krupovic M, ICTV Report Consortium. 2019. ICTV virus taxonomy profile: Clavaviridae. J Gen Virol 100:1267–1268. 10.1099/jgv.0.001295. [DOI] [PubMed] [Google Scholar]
- 20.Mochizuki T, Yoshida T, Tanaka R, Forterre P, Sako Y, Prangishvili D. 2010. Diversity of viruses of the hyperthermophilic archaeal genus Aeropyrum, and isolation of the Aeropyrum pernix bacilliform virus 1, APBV1, the first representative of the family Clavaviridae. Virology 402:347–354. 10.1016/j.virol.2010.03.046. [DOI] [PubMed] [Google Scholar]
- 21.Prangishvili D, Rensen E, Mochizuki T, Krupovic M, ICTV Report Consortium. 2019. ICTV virus taxonomy profile: Tristromaviridae. J Gen Virol 100:135–136. 10.1099/jgv.0.001190. [DOI] [PubMed] [Google Scholar]
- 22.Iranzo J, Koonin EV, Prangishvili D, Krupovic M. 2016. Bipartite network analysis of the archaeal virosphere: evolutionary connections between viruses and capsidless mobile elements. J Virol 90:11043–11055. 10.1128/JVI.01622-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang F, Baquero DP, Su Z, Osinski T, Prangishvili D, Egelman EH, Krupovic M. 2020. Structure of a filamentous virus uncovers familial ties within the archaeal virosphere. Virus Evol 6:veaa023. 10.1093/ve/veaa023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rensen EI, Mochizuki T, Quemin E, Schouten S, Krupovic M, Prangishvili D. 2016. A virus of hyperthermophilic archaea with a unique architecture among DNA viruses. Proc Natl Acad Sci U S A 113:2478–2483. 10.1073/pnas.1518929113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baquero DP, Contursi P, Piochi M, Bartolucci S, Liu Y, Cvirkaite-Krupovic V, Prangishvili D, Krupovic M. 2020. New virus isolates from Italian hydrothermal environments underscore the biogeographic pattern in archaeal virus communities. ISME J 14:1821–1833. 10.1038/s41396-020-0653-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bautista MA, Black JA, Youngblut ND, Whitaker RJ. 2017. Differentiation and structure in Sulfolobus islandicus rod-shaped virus populations. Viruses 9:120. 10.3390/v9050120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prangishvili D, Arnold HP, Gotz D, Ziese U, Holz I, Kristjansson JK, Zillig W. 1999. A novel virus family, the Rudiviridae: structure, virus-host interactions and genome variability of the sulfolobus viruses SIRV1 and SIRV2. Genetics 152:1387–1396. 10.1093/genetics/152.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prangishvili D, Koonin EV, Krupovic M. 2013. Genomics and biology of rudiviruses, a model for the study of virus-host interactions in Archaea. Biochem Soc Trans 41:443–450. 10.1042/BST20120313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martínez-Alvarez L, Bell SD, Peng X. 2016. Multiple consecutive initiation of replication producing novel brush-like intermediates at the termini of linear viral dsDNA genomes with hairpin ends. Nucleic Acids Res 44:8799–8809. 10.1093/nar/gkw636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pina M, Basta T, Quax TEF, Joubert A, Baconnais S, Cortez D, Lambert S, Le Cam E, Bell SD, Forterre P, Prangishvili D. 2014. Unique genome replication mechanism of the archaeal virus AFV1. Mol Microbiol 92:1313–1325. 10.1111/mmi.12630. [DOI] [PubMed] [Google Scholar]
- 31.Krupovic M, Dolja VV, Koonin EV. 2020. The LUCA and its complex virome. Nat Rev Microbiol 18:661–670. 10.1038/s41579-020-0408-x. [DOI] [PubMed] [Google Scholar]
- 32.Ptchelkine D, Gillum A, Mochizuki T, Lucas-Staat S, Liu Y, Krupovic M, Phillips SEV, Prangishvili D, Huiskonen JT. 2017. Unique architecture of thermophilic archaeal virus APBV1 and its genome packaging. Nat Commun 8:1436. 10.1038/s41467-017-01668-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janekovic D, Wunderl S, Holz I, Zillig W, Gierl A, Neumann H. 1983. TTV1, TTV2 and TTV3, a family of viruses of the extremely thermophilic, anaerobic, sulfur reducing archaebacterium Thermoproteus tenax. Mol Gen Genet 192:39–45. 10.1007/BF00327644. [DOI] [Google Scholar]