ABSTRACT
The OC43 coronavirus is a human pathogen that usually causes only the common cold. One of its key enzymes, similar to other coronaviruses, is the 2′-O-RNA methyltransferase (MTase), which is essential for viral RNA stability and expression. Here, we report the crystal structure of the 2′-O-RNA MTase in a complex with the pan-methyltransferase inhibitor sinefungin solved at 2.2-Å resolution. The structure reveals an overall fold consistent with the fold observed in other coronaviral MTases. The major differences are in the conformation of the C terminus of the nsp16 subunit and an additional helix in the N terminus of the nsp10 subunits. The structural analysis also revealed very high conservation of the S-adenosyl methionine (SAM) binding pocket, suggesting that the SAM pocket is a suitable spot for the design of antivirals effective against all human coronaviruses.
IMPORTANCE Some coronaviruses are dangerous pathogens, while some cause only common colds. The reasons are not understood, although the spike proteins probably play an important role. However, to understand the coronaviral biology in sufficient detail, we need to compare the key enzymes from different coronaviruses. We solved the crystal structure of 2′-O-RNA methyltransferase of the OC43 coronavirus, a virus that usually causes mild colds. The structure revealed some differences in the overall fold but also revealed that the SAM binding site is conserved, suggesting that development of antivirals against multiple coronaviruses is feasible.
KEYWORDS: OC43, coronavirus, crystal structure, methyltransferase
INTRODUCTION
Prior to the COVID-19 pandemic, only six other human coronaviruses were known: severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), and four other coronaviruses, OC43 (organ culture 43), 229E, NL63, and HKU1, which are responsible for up to 30% of mild respiratory diseases (1). All of these are members of the subfamily Orthocoronavirinae of the Coronaviridae family. The Orthocoronavirinae subfamily is further divided into four genera: Alpha-, Beta-, Gamma-, and Deltacoronavirus. 229E-CoV and NL63-CoV are members of the genus Alphacoronavirus, while OC43-CoV, HKU1-CoV, SARS-CoV, SARS-CoV-2, and MERS-CoV are all members of the genus Betacoronavirus (2).
The OC43 coronavirus has a 30.7-kbp, positive-sense, single-stranded-RNA (+RNA) genome. That is unusual for a +RNA virus but similar to other coronaviruses (3). It was transmitted to humans recently, in the 19th century, probably from cattle; the bovine CoV is its closet relative. It has also been speculated that OC43-CoV might be responsible for the 1889–1890 pandemic (4), which has usually been attributed to the H2N2 influenza strain (5). Interestingly, while OC43-CoV usually infects the upper respiratory tract and causes respiratory diseases, it is also neurotropic and can be neuroinvasive (6, 7).
The coronaviral genome usually encodes two polyproteins (pp1a and pp1ab), the spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins, and several accessory proteins. OC43, in addition, encodes hemagglutinin esterase (HE), which dramatically increases its infectivity (8). The pp1a and pp1ab polyproteins are autocatalytically cleaved into 16 nonstructural proteins (nsp1 to nsp16) that ensure many enzymatic activities needed for viral replication, most notably the RNA replication machinery (9). nsp7, nsp8, and nsp12 form the viral RNA-dependent RNA polymerase (RdRp), nsp13 functions as a helicase, and nsp10, nsp14, and nsp16 are RNA methyltransferases (MTases). nsp14 is responsible for proofreading during RNA synthesis; it also has an additional enzymatic activity as an exoribonuclease (10).
In this study, we focused on the 2′-O MTase of OC43-CoV. RNA methylation is important for (viral) RNA stability; it protects the RNA from degradation (innate immunity shielding) and facilitates its efficient translation (11–13). Installation of the 5′ cap in coronavirus-infected cells is a four-step process: (i) the 5′ gamma phosphate of the nascent RNA is hydrolyzed, probably by the coronaviral nsp13 helicase; (ii) GMP is transferred to the 5′ end by an unknown guanylyltransferase; (iii) the nsp10:nsp14 protein complex methylates the N-7 position of the newly attached guanosine; and (iv) the nsp10:nsp16 protein complex methylates the 2′-O of the first nucleotide ribose. The goal is to create viral RNA that is stable in human cells, is efficiently translated, and does not induce the innate immune response. Nonmethylated RNA is recognized by the RIG-I (retinoic acid-inducible gene I) pattern recognition receptor and is also recognized by and binds to interferon (IFN)-induced protein with tetratricopeptide repeats 1 and 5 (IFIT1 and IFIT5), which efficiently prevent its translation (14, 15). These facts imply that inhibiting the nsp10:nsp16 MTase is a valid strategy to fight coronaviral infections.
Here, we present the crystal structure of the OC43-CoV 2′-O MTase (nsp10:nsp16 complex) to understand how different coronaviruses evade innate immunity. The structure revealed relatively high conservation of the architecture of the active site between OC43-CoV and SARS-CoV-2 and suggested that the design of MTase inhibitors targeting multiple human coronaviruses is feasible.
RESULTS
Crystallization of the OC43-CoV nsp10:nsp16 MTase.
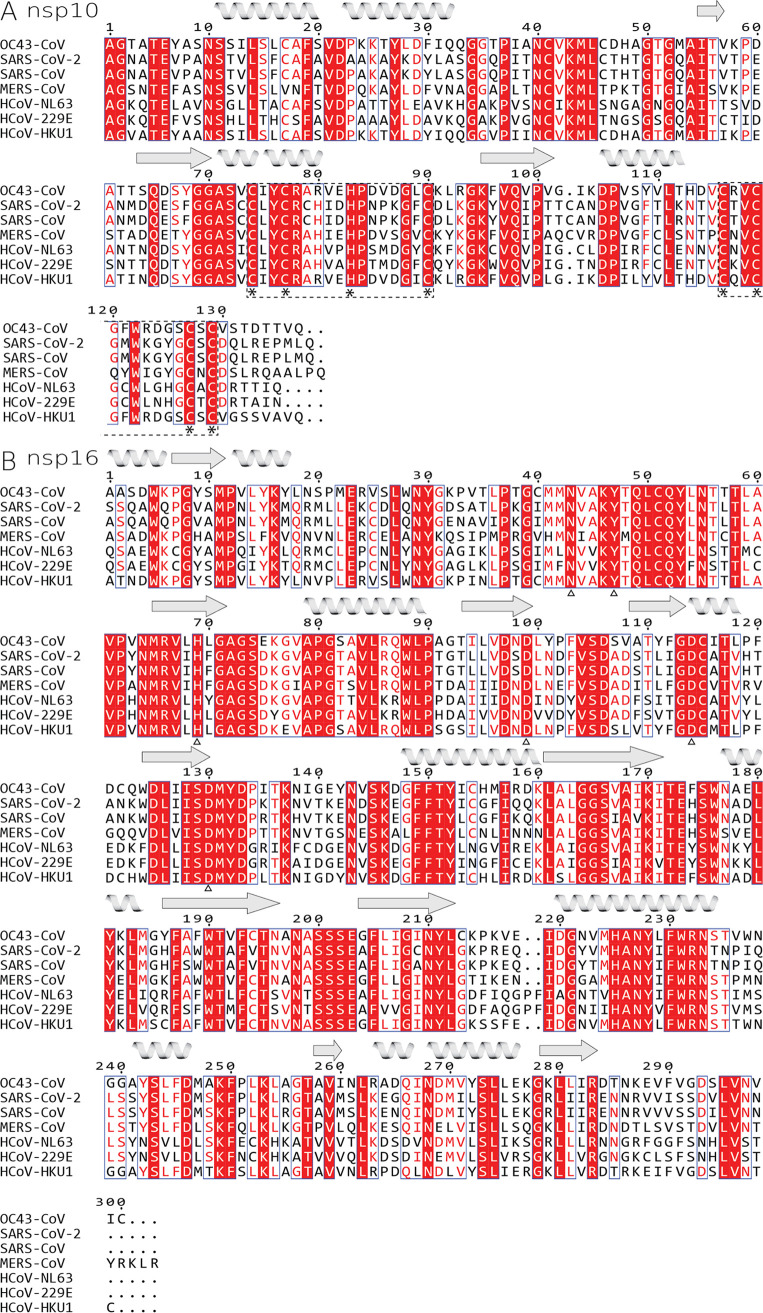
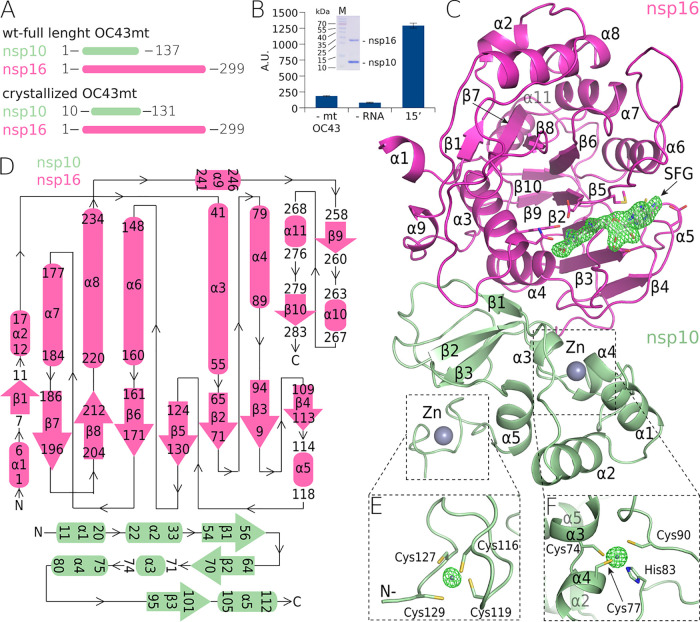
We analyzed the sequences of all human coronaviral nsp10 and nsp16 proteins (Fig. 1). The OC43 nsp10 has 53% sequence identity and 68% sequence similarity to the nsp10 of SARS-CoV-2, and similarly, the OC43 nsp16 displays 66% sequence identity and 76% sequence similarity to the nsp16 of SARS-CoV-2, which are not high degrees of identity and similarity for viruses of the same genus. We aimed to solve the crystal structure of the nsp10:nsp16 MTase of OC43 to learn the degree of conservation of the three-dimensional (3-D) structure of an essential enzyme between these two coronaviruses. We used full-length nsp16 and slightly truncated nsp10 (residues 10 to 131) for crystallization trials (Fig. 2A). Both proteins were expressed recombinantly and purified to homogeneity, and the enzymatic activity of the recombinant nsp10:nsp16 protein complex was verified (Fig. 2B). Next, we started the crystallization trials. Eventually, we obtained crystals that diffracted to 2.2 Å. We solved the structure by molecular replacement and refined it to good R factors and geometry (details in Table 1 and Materials and Methods).
FIG 1.
Sequence alignment of all human coronaviral nsp10 and nsp16 proteins. (A) nsp10 and (B) nsp16. Identical residues are highlighted in red fields, while conserved residues are in red font. Important residues involved in coordination of zinc are marked by asterisks below the alignments, and whole zinc-binding regions are in dashed-line boxes, while residues coordinating sinefungin are marked by empty triangles. Secondary-structure features of OC43-CoV are symbolized by helices and arrows for beta sheets.
FIG 2.
The crystal structure of the methyltransferase complex (nsp10:nsp16) from OC43-CoV. (A) Schematic representation of the crystallized and full-length nsp10:nsp16 proteins. (B) SDS-PAGE gel illustrating the purity of the recombinant nsp10:nsp16 complex and a graph illustrating its enzymatic activity. Values are for the reaction performed with all components except the enzyme (1st bar), with all components except for the RNA (2nd bar), and with all components (3rd bar). Error bars show standard deviations. A.U., arbitrary units. (C) Overall crystal structure of the nsp10:nsp16 complex (ribbon) with a sinefungin molecule (white) bound in the active site (SFG) and a “kick-out” omit map (Fo − Fc) of electron density (green) at 3.5 sigma with SFG excluded from the calculation. (D) Topological representation of the secondary-structure features of the complex. (E and F) Zn2+ coordination centers in nsp10 in kick-out omit maps (Fo − Fc) of electron density (green) at 10 sigma with Zn2+ excluded from the calculation.
TABLE 1.
Crystallographic data collection and refinement statistics
| Parametera | Value(s) for OC43 nsp10:nsp16b |
|---|---|
| PDB accession code | 7NH7 |
| Data collection and processing | |
| Space group | P31 |
| Cell dimensions—a, b, c (Å) | 61.9, 61.9, 109.7 |
| X-ray source | Home source |
| Wavelength (Å) | 1.5419 |
| Resolution range (Å) | 36.57–2.2 (2.28–2.2) |
| No. of unique reflections | 23,828 (2,385) |
| Completeness (%) | 99.74 (99.13) |
| Multiplicity | 5.1 (4.7) |
| Mean I/σ〈I〉 | 11.66 (2.48) |
| CC1/2 | 0.981 (0.687) |
| CC* | 0.995 (0.902) |
| Rmerge (%) | 13.05 (62.57) |
| Structure solution and refinement | |
| Rwork (%) | 16.41 (21.51) |
| Rfree (%) | 21.01 (29.29) |
| RMSD bond length (Å)/bond angle (°) | 0.005/0.86 |
| Avg B factors (Å2) | 28.24 |
| Clashscore | 7.84 |
| Ramachandran plot—favored regions/outlier regions (%) | 97.32/0 |
CC1/2, correlation of one half of the observations with the other half; CC*, correlation of the observed data with the unknown “true” intensities.
Numbers in parentheses refer to the highest-resolution shell.
The overall fold of OC43-CoV nsp10:nsp16 MTase.
The overall fold is consistent with the fold of previously analyzed coronaviral nsp10:nsp16 MTases from SARS-CoV-2, SARS-CoV, and MERS-CoV (16–19). We could trace the entire protein chain except for the very last five residues of nsp16 and the last residue of nsp10. The nsp16 subunit is composed of 10 β-sheets and 11 helices (Fig. 2). It exhibits a Rossmann fold where the β-sheets are arranged in a central β-motif in the shape of a letter “J” that is surrounded by α-helices (Fig. 2C and D). The small nsp10 subunit is composed only of three small β-sheets and five α-helices (Fig. 2C and D). The nsp10 fold is stabilized by two zinc fingers. The first zinc finger stabilizes the central part of the nsp10 molecule and is formed by Cys74, Cys77, His83, and Cys90. The second zinc finger stabilizes the very C terminus and is formed by Cys166, Cys119, Cys127, and Cys129 (Fig. 2E and F). Notably, all the residues forming both zinc fingers are absolutely conserved among human coronaviruses, highlighting their importance for the function of the nsp10 protein.
S-Adenosyl methionine (SAM) binding pocket.
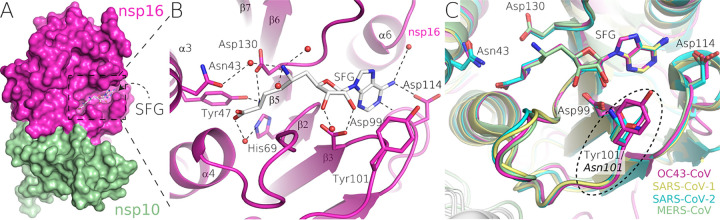
Prior to crystallization trials, the nsp10:nsp16 2′-O MTase was supplemented with 1 mM sinefungin, an adenosine derivative originally isolated from Streptomyces griseolus by Eli Lilly and Co. as a potential antibiotic (20) that is a pan-methyltransferase inhibitor. The electron density for sinefungin is clearly visible (Fig. 2C). Detailed analysis revealed residues that are responsible for sinefungin binding. Asp114 binds the adenine amino group, while Asp99 forms hydrogen bonds with both ribosyl hydroxyl groups. The amino acid moiety of sinefungin is coordinated by hydrogen bonds to Asp130 and Tyr47, while Asn43 and His69 contribute to ligand binding via water bridges (Fig. 3). All of these residues are absolutely conserved in human coronaviruses (Fig. 1), even Asn43 and His69, which contribute to ligand binding indirectly.
FIG 3.
Detail of sinefungin binding to nsp16. (A) Overall surface representation of the nsp10:nsp16 complex of OC43-CoV. (B) Detailed view of the sinefungin interaction with the active site of the enzyme. The amino acids involved in the interaction are shown as sticks, the waters are shown as red spheres, and the hydrogen bonds are depicted as dashed lines. (C) Structural comparison of OC43-CoV, SARS-CoV, SARS-CoV-2, and MERS-CoV nsp16 sinefungin binding sites.
Comparison to SARS-CoV-2 nsp10:nsp16 MTase.
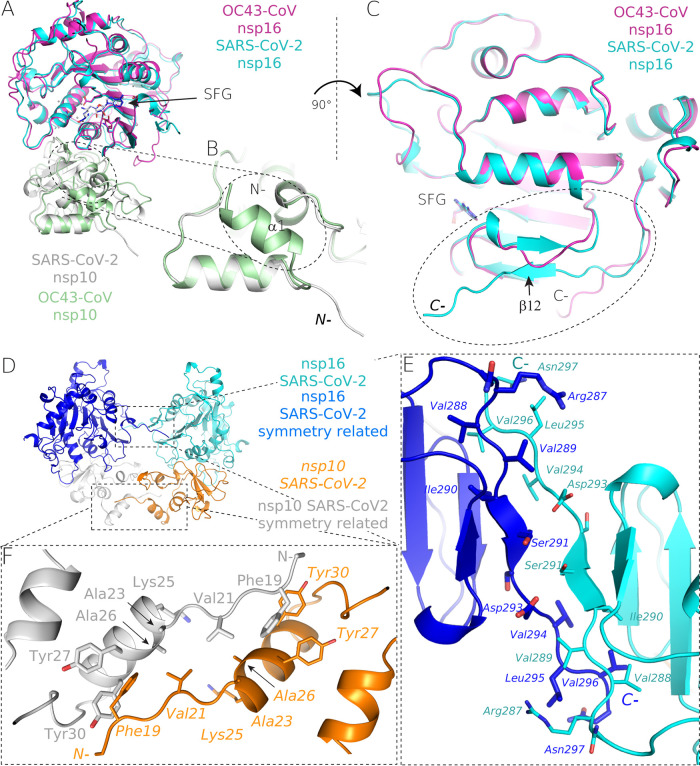
We compared the structures of OC43 and SARS-CoV-2 nsp10:nsp16 MTases to gain insight into the structural conservation of this enzyme within human coronaviruses. The overall fold remains conserved, with root mean square deviations (RMSD) of 0.99 Å2 for the nsp16 and 1.03 Å2 for the nsp10 protein (Fig. 4A), as does the sinefungin binding pocket, with the exception of Tyr101, where most human CoVs (SARS-CoV, SARS-CoV-2, MERS-CoV, NL63-CoV, and HKU1-CoV) have an Asn residue instead (Fig. 1 and 3C).
FIG 4.
Structural alignment of the OC43-CoV and SARS-CoV-2 2′-O-RNA MTases. (A) The overall fold of structurally aligned SARS-Cov-2 and OC43-CoV MTases. (B) The N-terminal region of nsp10 of OC43-CoV (green) contains an additional α-helix. (C to F) The C terminus of nsp16 of OC43-CoV (magenta) lacks the final β-sheet of nsp16 SARS-CoV-2 (cyan), and it is driven toward nsp10, where it is a part of the nsp10:nsp16 interface (C), while the SARS-CoV-2 C terminus of nsp16 is involved in crystal contacts just as the SARS-CoV-2 N terminus is (D to F).
However, the structural alignment also revealed significant differences. The C terminus of OC43-CoV nsp16 is located at the nsp10:nsp16 interface and contributes to nsp10:nsp16 binding. While the SARS-CoV-2 nsp16 C terminus is folded away from the interface and actually forms crystal contacts, which the SARS-CoV-2 nsp16 N terminus does as well (Fig. 4D to F). SARS-CoV-2 nsp16 also has a final β-sheet (β-12) that is antiparallel to β-5. Surprisingly, this β-sheet (β-12) is missing on the OC43 nsp16. On the other hand, the OC43 nsp10 has an additional α-helix (residues 11 to 20) that is not present in the SARS-CoV-2 nsp10 (Fig. 4B).
DISCUSSION
Before the appearance of SARS and MERS, the coronaviruses were rather an insignificant group that did not attract that much attention (21), despite some of the most important discoveries, such as proteolytical processing of the spike protein (at that time called the E2 glycoprotein) or that the leader sequence of subgenomic RNAs comes from the 5′ end of the genome, having already been made (22, 23). Upon the appearance of MERS and SARS, the coronaviruses attracted significant scientific interest that led to the molecular description of their life cycle (reviewed in Snijder et al. [9]), including a structural understanding of their key enzymes (19, 24–30). However, the four human coronaviruses that generally do not cause any serious illness (OC43-, 229E-, NL63-, and HKU1-CoVs) are still understudied. Nonetheless, it would be interesting and important to understand why certain coronaviruses cause deadly diseases and others just mild colds. Certainly, the spike protein is the primary determinant of pathogenicity: both the SARS-CoV and the SARS-CoV-2 spike proteins recognize angiotensin-converting enzyme 2 (ACE2) as the receptor (31, 32), while MERS-CoV uses dipeptidyl peptidase 4 (33), OC43- and HKU1-CoV use 9-O-acetylated sialic acids (34), and E229-CoV uses aminopeptidase N (35). However, NL63-CoV also uses ACE2 as its receptor (36, 37) and causes only common colds, suggesting that other determinants of coronaviral perilousness also exist.
Here, we report the structure of the human OC43-CoV 2′-O MTase, a complex of two nonstructural proteins, nsp10 and nsp16. The main function of this complex is to create the cap structure on the 5′ end of viral RNA. The structure revealed that despite significant sequence differences between OC43 and other human coronaviruses, the overall fold is well preserved: the biggest differences are an additional helix (α1) in the nsp10 subunit that is not observed in the SARS-CoV and SARS-CoV-2 coronaviruses and a different conformation of the OC43 nsp16 C terminus. The C terminus is also missing a β-sheet compared to its SARS-CoV-2 counterpart.
The development of inhibitors of coronaviral MTases (both nsp14 and nsp16) has just begun. The compounds discovered so far are potent in vitro; however, most of them suffer from low membrane permeability, hindering their direct testing in antiviral assays (38–40). Several compounds inhibiting the flaviviral MTase (the N-terminal domain of the NS5 protein) are known, and some of them are potent in cell-based antiviral assays; however, none of these inhibitors is approved as a drug (41–44).
Importantly, the structure revealed high conservation of the SAM binding pocket. All residues that make direct contact with the ligand (sinefungin) are absolutely conserved among human coronaviruses, suggesting that mutations of these residues are not tolerated. This is definitely good news for drug design, because compounds that target the SAM pocket will likely be effective against all human coronaviruses and the evolution of resistance against them is unlikely.
MATERIALS AND METHODS
Protein expression and purification.
The genes encoding OC43 nsp10 and nsp16 proteins were codon optimized for Escherichia coli and commercially synthesized (Thermo Fisher Scientific). The genes were cloned in a home-made pSUMO-Kan plasmid (a pRSFD-derived vector already encoding a His8x-SUMO solubility and purification tag). The proteins were purified using our established protocols for viral enzymes (45, 46). Briefly, E. coli BL21 cells were transformed with nps10 and nsp16 expression vectors and cultivated at 37°C in LB medium with the addition of ZnSO4 (10 μM) and kanamycin (50 μg/ml). The production of nsp16 and nsp10 proteins was induced by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) (300 μM) at the late exponential growth phase (optical density [OD] = 0.6 to 0.8), and the temperature was lowered to 18°C for 18 h. The bacterial cells were collected by centrifugation, resuspended in lysis buffer (50 mM Tris, pH 8, 300 mM NaCl, 20 mM imidazole, 5 mM MgSO4, 3 mM β-mercaptoethanol, 10% glycerol) and lysed by sonication. The lysate was centrifuged, the supernatant was incubated with Ni2+ agarose (Macherey-Nagel) and washed with lysis buffer, and the recombinant proteins were eluted by lysis buffer supplemented with 300 mM imidazole. The His8x-SUMO tag was cleaved by the Ulp1 protease during dialysis against lysis buffer. After dialysis, the His8x-SUMO tag was removed by a second round of affinity chromatography. The proteins were further purified by gel filtration on a HiLoad 16/600 Superdex 75 column (GE Healthcare) in size exclusion buffer {50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM TCEP [Tris(2-carboxyethyl)phosphine hydrochloride], 5% glycerol}. The purified proteins were mixed in a 1:1 molar ratio, concentrated to 5 mg/ml, and immediately used for crystallographic trials. After purification, proteins were concentrated to 5 mg/ml and stored at −80°C.
Crystallization and structural analysis.
The nsp10:16 protein complex was supplemented by 1 mM sinefungin. Crystals of the nsp10:nsp16 protein complex grew in 3 days in a sitting drop that was created by mixing 300 nl of protein solution and 300 nl of mother liquor (0.1 M MES [morpholineethanesulfonic acid], pH 6.5, 15% [vol/vol] polyethylene glycol monomethyl ether 500 [PEG 500 MME]). They were cryoprotected in mother liquor supplemented with 20% glycerol and flash frozen in liquid nitrogen. The crystals diffracted to 2.2-Å resolution and belonged to the trigonal P31 space group. Data were collected using a home source (Rigaku) equipped with a Pilatus detector (Dectris). The data were indexed, scaled, and integrated using XDS (47). The structure was solved by molecular replacement using the structure of the SARS-CoV-2 nsp10:nsp16 (PDB accession code 6YZ1) as the search model in Phaser (48) and was refined in Phenix (49), together with manual building in Coot (50). The structure was refined to good R factors (Rwork = 16.41% and Rfree = 21.01%) and good geometry as summarized in Table 1. RMSDs were also calculated in Coot using the secondary-structure-matching algorithm.
RNA preparation.
m7GpppA was prepared chemically according to a published protocol (51). A 35-mer m7GpppA-capped RNA was prepared by in vitro transcription using an DNA template (5′-CAGTAATACGACTCACTATAGGGGAAGCGGGCATGCGGCCAGCCATAGCCGATCA-3′) and the TranscriptAid T7 high-yield transcription kit (Thermo Scientific). The reaction was performed in a 50-μl mixture containing 1× TranscriptAid reaction buffer, 7.5 mM nucleoside triphosphates [NTPs], 6 mM cap analog, 1 μg template DNA, and 1× TranscriptAid enzyme mix. The mixture was incubated for 8 h at 37°C. Next, the RNA was purified using RNA Clean & Concentrator-5 from Zymo Research. DNase I treatment was performed directly on the column according to the purification protocol for 15 min at room temperature.
Analysis of the nsp10:nsp16 enzymatic activity.
The reaction was performed in a total volume of 15 μl. The starting reaction mixture contained 10 μM SAM (S-adenosyl methionine), 5 μM m7GpppA-capped RNA, and 1 μM nsp10 in the MTase reaction buffer (5 mM Tris, pH 8.0, 1 mM MgCl2, 3 mM dithiothreitol [DTT]). The reaction was initiated by adding 500 nM nsp16 and incubated at 30°C while shaking at 350 rpm. After 15 min of incubation, 15 μl of 40% methanol was added and the samples were analyzed on an Echo mass spectrometry system coupled with a Sciex 6500 triple-quadrupole mass spectrometer operating with an electrospray ionization source. The spectrometer was run in the multiple-reaction-monitoring (MRM) mode with the interface heated to 350°C. The declustering potential was 20 V, the entrance potential 10 V, and the collision energy 28 eV. Ten nanoliters was injected in the mobile phase (flow rate of 0.46 ml/min; 70% acetonitrile with 0.1% formic acid). The characteristic product ion of S-adenosylhomocysteine (SAH), m/z 385.1 > 134.1, was used for quantification.
ACKNOWLEDGMENTS
The project was supported by the Czech Science Foundation (grant number 21-25280S); the support of the Academy of Sciences of the Czech Republic (RVO 61388963) is also acknowledged. We are grateful to Mike Downey for critical reading of the manuscript.
P.D., P.K., J.K., D.C., K.C., and E.B. performed experiments, J.S., K.C., and E.B. analyzed data, and E.B. designed and supervised the project and wrote the manuscript.
We declare no conflicts of interest.
Contributor Information
Evzen Boura, Email: boura@uochb.cas.cz.
Rebecca Ellis Dutch, University of Kentucky College of Medicine.
REFERENCES
- 1.Paules CI, Marston HD, Fauci AS. 2020. Coronavirus infections—more than just the common cold. JAMA 323:707–708. 10.1001/jama.2020.0757. [DOI] [PubMed] [Google Scholar]
- 2.Lefkowitz EJ, Dempsey DM, Hendrickson RC, Orton RJ, Siddell SG, Smith DB. 2018. Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res 46:D708–D717. 10.1093/nar/gkx932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res 117:17–37. 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vijgen L, Keyaerts E, MoëS E, Thoelen I, Wollants E, Lemey P, Vandamme A-M, Van Ranst M. 2005. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J Virol 79:1595–1604. 10.1128/JVI.79.3.1595-1604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mulder J, Masurel N. 1958. Pre-epidemic antibody against 1957 strain of Asiatic influenza in serum of older people living in the Netherlands. Lancet 271:810–814. 10.1016/S0140-6736(58)91738-0. [DOI] [PubMed] [Google Scholar]
- 6.Desforges M, Le Coupanec A, Brison E, Meessen-Pinard M, Talbot PJ. 2014. Neuroinvasive and neurotropic human respiratory coronaviruses: potential neurovirulent agents in humans. Adv Exp Med Biol 807:75–96. 10.1007/978-81-322-1777-0_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arbour N, Day R, Newcombe J, Talbot PJ. 2000. Neuroinvasion by human respiratory coronaviruses. J Virol 74:8913–8921. 10.1128/JVI.74.19.8913-8921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desforges M, Desjardins J, Zhang C, Talbot PJ. 2013. The acetyl-esterase activity of the hemagglutinin-esterase protein of human coronavirus OC43 strongly enhances the production of infectious virus. J Virol 87:3097–3107. 10.1128/JVI.02699-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snijder EJ, Decroly E, Ziebuhr J. 2016. The nonstructural proteins directing coronavirus RNA synthesis and processing. Adv Virus Res 96:59–126. 10.1016/bs.aivir.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.V’kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. 2021. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol 19:155–170. 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bradrick SS. 2017. Causes and consequences of flavivirus RNA methylation. Front Microbiol 8:2374. 10.3389/fmicb.2017.02374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzales-van Horn SR, Sarnow P. 2017. Making the mark: the role of adenosine modifications in the life cycle of RNA viruses. Cell Host Microbe 21:661–669. 10.1016/j.chom.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cross ST, Michalski D, Miller MR, Wilusz J. 2019. RNA regulatory processes in RNA virus biology. Wiley Interdiscip Rev RNA 10:e1536. 10.1002/wrna.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Decroly E, Canard B. 2017. Biochemical principles and inhibitors to interfere with viral capping pathways. Curr Opin Virol 24:87–96. 10.1016/j.coviro.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin TY, Schneller S, Zust R, Dong H, Thiel V, Sen GC, Fensterl V, Klimstra WB, Pierson TC, Buller RM, Gale M, Jr, Shi PY, Diamond MS. 2010. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468:452–456. 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krafcikova P, Silhan J, Nencka R, Boura E. 2020. Structural analysis of the SARS-CoV-2 methyltransferase complex involved in RNA cap creation bound to sinefungin. Nat Commun 11:3717. 10.1038/s41467-020-17495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosas-Lemus M, Minasov G, Shuvalova L, Inniss NL, Kiryukhina O, Brunzelle J, Satchell KJF. 2020. High-resolution structures of the SARS-CoV-2 2′-O-methyltransferase reveal strategies for structure-based inhibitor design. Sci Signal 13:eabe1202. 10.1126/scisignal.abe1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Viswanathan T, Arya S, Chan SH, Qi S, Dai N, Misra A, Park JG, Oladunni F, Kovalskyy D, Hromas RA, Martinez-Sobrido L, Gupta YK. 2020. Structural basis of RNA cap modification by SARS-CoV-2. Nat Commun 11:3718. 10.1038/s41467-020-17496-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Decroly E, Debarnot C, Ferron F, Bouvet M, Coutard B, Imbert I, Gluais L, Papageorgiou N, Sharff A, Bricogne G, Ortiz-Lombardia M, Lescar J, Canard B. 2011. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog 7:e1002059. 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamil RL, Hoehn MM. 1973. A9145, a new adenine-containing antifungal antibiotic. I. Discovery and isolation. J Antibiot 26:463–465. 10.7164/antibiotics.26.463. [DOI] [PubMed] [Google Scholar]
- 21.Weiss SR. 2020. Forty years with coronaviruses. J Exp Med 217:e20200537. 10.1084/jem.20200537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai MMC, Patton CD, Stohlman SA. 1982. Further characterization of messenger-RNAs of mouse hepatitis-virus—presence of common 5'-end nucleotides. J Virol 41:557–565. 10.1128/JVI.41.2.557-565.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frana MF, Behnke JN, Sturman LS, Holmes KV. 1985. Proteolytic cleavage of the E2-glycoprotein of murine coronavirus—host-dependent differences in proteolytic cleavage and cell-fusion. J Virol 56:912–920. 10.1128/JVI.56.3.912-920.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anand K, Ziebuhr J, Wadhwani P, Mesters JR, Hilgenfeld R. 2003. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 300:1763–1767. 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 25.Jia Z, Yan L, Ren Z, Wu L, Wang J, Guo J, Zheng L, Ming Z, Zhang L, Lou Z, Rao Z. 2019. Delicate structural coordination of the severe acute respiratory syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res 47:6538–6550. 10.1093/nar/gkz409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao Y, Ma Q, Restle T, Shang W, Svergun DI, Ponnusamy R, Sczakiel G, Hilgenfeld R. 2012. Nonstructural proteins 7 and 8 of feline coronavirus form a 2:1 heterotrimer that exhibits primer-independent RNA polymerase activity. J Virol 86:4444–4454. 10.1128/JVI.06635-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dinesh DC, Chalupska D, Silhan J, Koutna E, Nencka R, Veverka V, Boura E. 2020. Structural basis of RNA recognition by the SARS-CoV-2 nucleocapsid phosphoprotein. PLoS Pathog 16:e1009100. 10.1371/journal.ppat.1009100. [DOI] [PMC free article] [PubMed]
- 28.Hillen HS, Kokic G, Farnung L, Dienemann C, Tegunov D, Cramer P. 2020. Structure of replicating SARS-CoV-2 polymerase. Nature 584:154–156. 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- 29.Konkolova E, Klima M, Nencka R, Boura E. 2020. Structural analysis of the putative SARS-CoV-2 primase complex. J Struct Biol 211:107548. 10.1016/j.jsb.2020.107548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, Becker S, Rox K, Hilgenfeld R. 2020. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science 368:409–412. 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li WH, Moore MJ, Vasilieva N, Sui JH, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450–454. 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shang J, Wan YS, Luo CM, Ye G, Geng QB, Auerbach A, Li F. 2020. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci U S A 117:11727–11734. 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raj VS, Mou HH, Smits SL, Dekkers DHW, Muller MA, Dijkman R, Muth D, Demmers JAA, Zaki A, Fouchier RAM, Thiel V, Drosten C, Rottier PJM, Osterhaus ADME, Bosch BJ, Haagmans BL. 2013. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495:251–254. 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang XC, Dong WJ, Milewska A, Golda A, Qi YH, Zhu QK, Marasco WA, Baric RS, Sims AC, Pyrc K, Li WH, Sui JH. 2015. Human coronavirus HKU1 spike protein uses O-acetylated sialic acid as an attachment receptor determinant and employs hemagglutinin-esterase protein as a receptor-destroying enzyme. J Virol 89:7202–7213. 10.1128/JVI.00854-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeager CL, Ashmun RA, Williams RK, Cardellichio CB, Shapiro LH, Look AT, Holmes KV. 1992. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 357:420–422. 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu K, Li W, Peng G, Li F. 2009. Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc Natl Acad Sci U S A 106:19970–19974. 10.1073/pnas.0908837106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hofmann H, Pyrc K, van der Hoek L, Geier M, Berkhout B, Pohlmann S. 2005. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc Natl Acad Sci U S A 102:7988–7993. 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomáš O, Michal Š, Fengling L, Jindřich F, Kanchan D, Paknoosh P, Pavel H, Masoud V, Evzen B, Radim N. 2021. The structure-based design of SARS-CoV-2 Nsp14 methyltransferase ligands yields nanomolar inhibitors. ChemRxiv 10.26434/chemrxiv.14075408.v1. [DOI] [PubMed] [Google Scholar]
- 39.Devkota K, Schapira M, Perveen S, Yazdi AK, Li F, Chau I, Ghiabi P, Hajian T, Loppnau P, Bolotokova A, Satchell KJF, Wang K, Li D, Liu J, Smil D, Luo M, Jin J, Fish PV, Brown PJ, Vedadi M. 2021. Probing the SAM binding site of SARS-CoV-2 nsp14 in vitro using SAM competitive inhibitors guides developing selective bi-substrate inhibitors. bioRxiv 10.1101/2021.02.19.424337. [DOI] [PMC free article] [PubMed]
- 40.Yazdi AK, Li F, Devkota K, Perveen S, Ghiabi P, Hajian T, Bolotokova A, Vedadi M. 2021. A high-throughput radioactivity-based assay for screening SARS-CoV-2 nsp10-nsp16 complex. bioRxiv 10.1101/2021.02.03.429625. [DOI] [PMC free article] [PubMed]
- 41.Brecher M, Chen H, Li Z, Banavali NK, Jones SA, Zhang J, Kramer LD, Li HM. 2015. Identification and characterization of novel broad-spectrum inhibitors of the flavivirus methyltransferase. ACS Infect Dis 1:340–349. 10.1021/acsinfecdis.5b00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lim SP, Sonntag LS, Noble C, Nilar SH, Ng RH, Zou G, Monaghan P, Chung KY, Dong HP, Liu BP, Bodenreider C, Lee G, Ding M, Chan WL, Wang G, Jian YL, Chao AT, Lescar J, Yin Z, Vedananda TR, Keller TH, Shi PY. 2011. Small molecule inhibitors that selectively block dengue virus methyltransferase. J Biol Chem 286:6233–6240. 10.1074/jbc.M110.179184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coutard B, Barral K, Lichiere J, Selisko B, Martin B, Aouadi W, Lombardia MO, Debart F, Vasseur JJ, Guillemot JC, Canard B, Decroly E. 2017. Zika virus methyltransferase: structure and functions for drug design perspectives. J Virol 91:e02202-16. 10.1128/JVI.02202-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hercik K, Brynda J, Nencka R, Boura E. 2017. Structural basis of Zika virus methyltransferase inhibition by sinefungin. Arch Virol 162:2091–2096. 10.1007/s00705-017-3345-x. [DOI] [PubMed] [Google Scholar]
- 45.Klima M, Chalupska D, Różycki B, Humpolickova J, Rezabkova L, Silhan J, Baumlova A, Dubankova A, Boura E. 2017. Kobuviral non-structural 3A proteins act as molecular harnesses to hijack the host ACBD3 protein. Structure 25:219–230. 10.1016/j.str.2016.11.021. [DOI] [PubMed] [Google Scholar]
- 46.Sebera J, Dubankova A, Sychrovsky V, Ruzek D, Boura E, Nencka R. 2018. The structural model of Zika virus RNA-dependent RNA polymerase in complex with RNA for rational design of novel nucleotide inhibitors. Sci Rep 8:11132. 10.1038/s41598-018-29459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kabsch W. 2010. XDS. Acta Crystallogr D Biol Crystallogr 66:125–132. 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baranowski MR, Nowicka A, Rydzik AM, Warminski M, Kasprzyk R, Wojtczak BA, Wojcik J, Claridge TD, Kowalska J, Jemielity J. 2015. Synthesis of fluorophosphate nucleotide analogues and their characterization as tools for (1)(9)F NMR studies. J Org Chem 80:3982–3997. 10.1021/acs.joc.5b00337. [DOI] [PubMed] [Google Scholar]