Abstract
Recent advances in CRISPR present attractive genome-editing toolsets to implement therapeutic strategies at the genetic level. Here, we report a liposome-coated mesoporous silica nanoparticle (lipoMSN) as an effective CRISPR delivery system for multiplex gene-editing in the liver. The use of MSN provides a large surface area for efficient loading of the large Cas9 plasmid as well as Cas9 protein/guide RNA ribonucleoprotein complex (RNP), while liposome coating offers improved serum stability and enhanced cell uptake. Hypothesizing that a loss-of-function mutation in the lipid metabolism-related pcsk9, apoc3, and angptl3 genes would improve cardiovascular health by lowering blood cholesterol and triglycerides, we used this lipoMSN platform to deliver a combination of RNPs targeting three genes.[1] When targeting a single gene, the lipoMSN achieved a 54% gene editing efficiency, higher than the state-of-art Lipofectamine CRISPRMax. In the multiplex scenario, the lipoMSN maintained significant gene editing at each gene target despite reduced dosage of target-specific RNP. By delivering combinations of targeting RNPs in the same nanoparticle, synergistic effects on lipid metabolism were observed both in vitro and in vivo. These effects, such as a 50% decrease in serum cholesterol 4-weeks post-treatment with lipoMSN carrying both pcsk9- and angptl3- targeted RNPs, could not be reached with a single gene-editing approach. Taken together, this lipoMSN represents a versatile platform for the development of efficient, combinatorial gene editing therapeutics.
Keywords: multiplex gene editing, CRISPR/Cas9, nanoparticle, cardiovascular disease, gene therapy
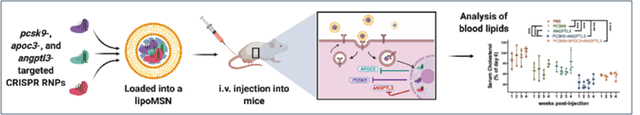
Graphical Abstract
Liposome-coated mesoporous silica nanoparticle enables multiplex gene editing to understand potential therapeutic targets in liver lipid metabolism. Using this novel nonviral platform to deliver CRISPR/Cas9 ribonucleoprotein, we demonstrated gene editing at the three potential therapeutic targets (pcsk9, apoc3, angptl3) for cardioprotection in vitro and in vivo.
Cardiovascular disease remains a leading cause of death globally, with high plasma low-density lipoprotein-cholesterol (LDL-C) level, or hypercholesterolemia, and high plasma triglyceride level, or hyperlipidemia, as major determinants of risk.[2] Reduction of cholesterol is an attractive therapeutic objective, with 30–40% reduction in LDL-C correlating with paralleled reduction in cardiovascular disease risk.[3] Statins, the current standard-of-care, neglect 10–20% of the high-risk patient-population due to intolerance and adverse effects with increased dosage, which motivates a genetic approach to find alternatives.[3–4] The first gene target for cardioprotection was discovered when a gain-of-function mutation in proprotein convertase subtilisin/kexin type 9 (PCSK9) was identified as the cause of autosomal dominant hypercholesterolemia, driving patients into high levels of LDL-C and early coronary heart disease (CHD).[5] Loss-of-function sequence variations of PCSK9 lead to significant (40%) reduction in the LDL-C level and 88% reduction in CHD.[6] PCSK9 is an LDL receptor (LDLR) antagonist expressed in the liver, such that overexpression leads to less LDL receptors and a decrease in LDL-C removal from the plasma.[7] Monoclonal antibodies targeting PCSK9 were considered the potential solution for the significant unmet need unfulfilled by statin drugs.[8] However, PCSK9 antibodies such as alirocumab showed adverse effects including injection site reactions, neurocognitive events, ophthalmologic events and anti-drug antibody production in clinical trials.[9] Small interfering RNAs (siRNAs), e.g., inclisiran, have been developed to provide a similar cardioprotective effect as the antibody therapies.[10] While these siRNAs enable significant down-regulation of PCSK9, high off-target effects associated with this modality of gene manipulation remain a concern. CRISPR/Cas9-mediated gene disruption offers an alternative for higher-precision, lower frequency treatment.[11]
Derived from the prokaryotic immune system, Cas9 endonuclease allows for precisely controllable gene targeting in mammalian cells when complexed with a specific guide RNA (gRNA), thereby generating a specifically-localized double-stranded break at the target site.[12] During the DNA repair process, the dominant pathway, nonhomologous end-joining of the break, often leads to frameshift errors and results in knockout of the gene.[13] This provides a simple mechanism to explore cause-and-effect in the context of lipid metabolism pathways, as specific genetic perturbations can be made and blood lipid profile changes can be measured. Similar to PCSK9, naturally occurring heterozygous mutations in ANGPTL3 and APOC3 yield cardioprotective effects through their impact in lipid metabolism in the liver.[6, 14] Leveraging the CRISPR/Cas9 system to explore these three gene targets separately and in various combinations could provide valuable information for the development of cardioprotective therapeutics.
Delivery mechanisms for CRISPR/Cas9 rely heavily on viral machinery, most popularly adeno-associated virus (AAV).[15] While AAV have lower immunogenicity compared with lentivirus or adenovirus, AAV has the lowest packaging capacity of approximately 5 kb.[16] This makes transduction of both the Cas9 and gRNA difficult and increasingly so for multiplexing. Further, cloning is required for each gene target, which contributes to slower workflow when screening potential gRNA designs.[17] Nanoparticle delivery of CRISPR/Cas9 elements has become a viable alternative, providing transient delivery of various forms of the CRISPR/Cas9 cassette, ranging from plasmid encoding the Cas9 endonuclease and gRNA, Cas9 mRNA with gRNA, to the Cas9/gRNA ribonucleoprotein (RNP) complex.[18] Liposomes and lipid-based nanoparticles have been investigated extensively as nonviral carriers, enabling effective drug/gene delivery with limited risk of immunogenicity and allowing for tunable surface properties via lipid composition.[19] However, many commercially available lipid-based carriers have limitations in vivo, and often rely on electrostatic self-assembly with cargo, providing relatively low loading efficiency of low solubility or charge density cargos.[20] Hypothesizing that integrating liposome with a core capable of loading diverse therapeutic cargo may resolve the aforementioned limitation and preserve liposome’s favorable cell entry, we designed a mesoporous silica nanoparticle (MSN) core to provide a liposome-coated MSN (lipoMSN) system for delivery of CRISPR/Cas9 elements.[21] MSN provides high surface area for the electrostatic loading of lower charge density Cas9/gRNA RNP cargo and additionally shelters its gRNA component susceptible to the degradative extracellular and endosomal environments.[22]
While lipoMSNs have proven their efficacy in the delivery of a variety of cargos, from small molecule drugs, peptides, to nucleic acids (siRNA and plasmid), they have not yet been successfully employed in the context of multiplex gene editing using multiple Cas9/gRNA RNPs.[23] In this work, we demonstrate that our lipoMSN delivery system is versatile in its ability to deliver Cas9/gRNA RNP as well as Cas9 plasmid with gRNA through electrostatic loading. Further, we apply this system to target three different cardioprotective genes simultaneously in order to study the potential synergistic effects that arise from multiple pathway manipulation. Since previous multiplex gene editing relied on in vivo transcription of the CRISPR/Cas9 components via plasmid or RNA delivery, or delivery in separate vehicles, our work provides a unique opportunity to study the effects of multiple Cas9/gRNA RNPs co-loaded into a singular delivery vehicle.[24] Chadwick et al., used separated adenoviruses to deliver CRISPR base editors targeting angptl3 and pcsk9 but could not detect synergistic effect.[6] This could be due to the un-synchronized editing of any given cell. With our proposed system, we are able to reduce the potential compensation mechanisms of these nonredundent pathways of lipid metabolism, which can provide insight of potential synergistic effects.[25]
We first tested whether our lipoMSN system could deliver various CRISPR/Cas9 editing elements, including low charge density Cas9 protein (~160 kDa), short gRNA (~100 nucleotides), and large Cas9 plasmid (~10 kb).[26] While gRNAs and plasmids have been previously loaded into various nanoparticle platforms,[27] loading of non-uniform, weakly-charged Cas9/gRNA RNP (−1.4 mV) presents a challenge, compared with the loading of uniformly negative-charged Cas9 plasmid (−16 mV) or gRNA (−14 mV; Figure 1A).[28] Screening of two different MSN cores—functionalized with carboxyl (-COOH) or amino (-NH2) group—showed that while all cores were comparable (Figures S1 and S2, Supporting Information), amine-functionalization on MSN, yielding a positive charge between 30–40 mV, resulted in efficient loading of Cas9/gRNA RNP as well as the Cas9 plasmid and gRNA at an MSN-to-cargo ratio of 20 to 1 (w/w) (Figure S3, Supporting Information).
Figure 1. In vitro design and optimization of the lipoMSN for hepatic CRISPR delivery.
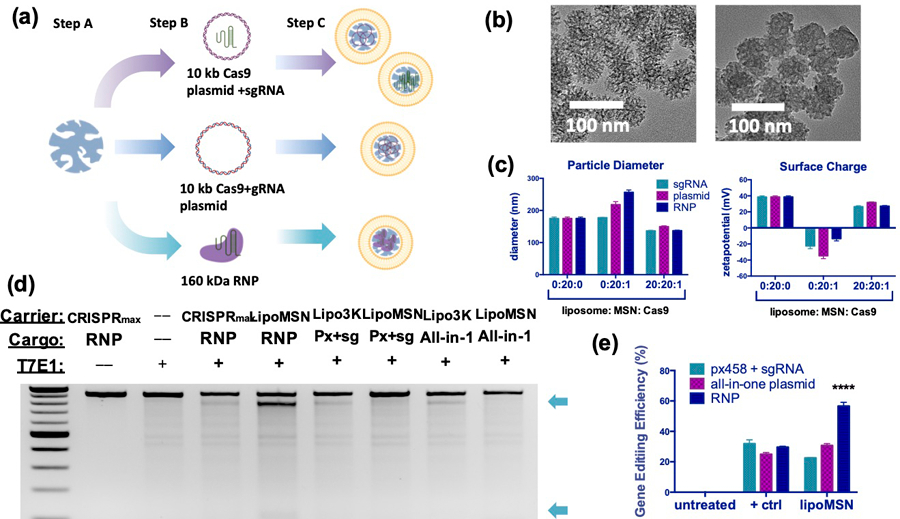
(A) Schematic illustration of lipoMSN loading capabilities spanning small gRNA and larger RNP composed of both Cas9 protein and gRNA. Step A: MSN preparation; step B: loading of CRISPR/Cas9 elements (Cas9 plasmid + gRNA separately, all-in-one Cas9/gRNA plasmid, or Cas9/gRNA RNP); step C: liposome coating to produce a consistent delivery system for various CRIPSR/Cas9 elements. (B) Representative TEM images of MSN (left) and Cas9/gRNA-loaded lipoMSN (right) (C) Size and surface charge characterization of the lipoMSN. (D) Representative gel images obtained from the Surveyor assay for comparison of gene editing efficiency. (E) Semi-quantitative analysis of the gene editing efficiency obtained from the Surveyor assay using Image J software. Results are presented as average ± standard errors of mean (SEM, n = 4). CRISPR/Cas9 elements were delivered in a 1 ug/mL dosage for (D-E). Significance was determined using one-way ANOVA with Tukey’s posthoc test, and represented as **** p < 0.0001.
Because liver was the initial therapeutic target, we then optimized the formulations in the primary mouse hepatic cell line (AML-12). Despite the loading capabilities, MSN alone provided low uptake and poor serum stability (Figures S1A and S4, Supporting Information). In contrast, coating with liposome— confirmed via transmission electron microscopy (TEM) (Figure 1B and Figure S5, Supporting Information)— improved both cellular uptake and transfection efficiency of MSN (Figures S1A and S1B, Supporting Information). Under the optimized liposome/MSN/cargo ratio (20/20/1), gRNA-loaded and Cas9-T2A-EGFP plasmid (px458)- lipoMSNs showed improved uptake (98% at 4h) and transfection (25% at 24h), respectively. Seeing positive results given by liposome-coating, an iterative optimization process on lipid composition of the liposome was subsequently carried out (Figure S6, Supporting Information), yielding the best composition with 65% DOTAP, 30% cholesterol, 3.75% DOPE and 1.25% DSPE-PEG (Figure S7, Supporting Information).
With the optimized liposome composition and liposome/MSN/cargo ratio, the lipoMSN showed relatively uniform physical characteristics in size and surface charge despite the varied cargos (Figure 1C). To date, there is no single platform allowing direct comparison of gene editing efficacy between different formats of CRISPR/Cas9 elements, although some attempts have been made.[29] As the first delivery system capable of delivering CRISPR elements in three different formats (Cas9 plasmid + gRNA, all-in-one plasmid encoding both Cas9 and gRNA, Cas9/gRNA RNP), we carried out a head-to-head comparison between these formats. When delivered by the lipoMSN, the Cas9/gRNA RNP gave the highest gene editing (Figures 1D and 1E), similar to reported results obtained through electroporation.[30] Notably, our lipoMSN outperformed the current gold standard for Cas9/gRNA RNP delivery, Lipofectamine CRISPRMax. Surveyor assays showed Cas9/gRNA-loaded lipoMSN produced a gene disruption efficiency of 54%, which was superior to Lipofectamine CRISPRMax with Cas9/gRNA RNP (30%) as well as Lipofectamine 3000 with the all-in-one Cas9/gRNA plasmid (33%). The use of Cas9/gRNA RNP has increased in popularity in the field because of its high editing fidelity.[29–30] This in conjunction with our maximized editing efficiency led us to continue our work using our lipoMSN system with Cas9/gRNA RNP.
After seeing effective gene editing with Cas9/gRNA RNP-loaded lipoMSN, we next explored multiplex gene editing with our system to disrupt the three genes (pcsk9, apoc3 and angptl3) in disparate pathways involved in LDL metabolism. As shown in Figure 2A, Pcsk9 inhibits LDLR recycling, and Apoc3 inhibits lipoprotein lipase activity, while Angptl3 inhibits the expression of LDLR as well as lipoprotein lipase.[31] Simultaneous disruption of these three genes may show synergy on lowering LDL-C level, thereby boosting cardioprotection efficacy; yet, multiplex gene editing provided the next challenge of maintaining significant gene editing for these three different gene targets while keeping the total Cas9/gRNA RNP dose constant. Editing efficiency at the pcsk9 target site remains consistent despite dosage of pcsk9-targeting Cas9/gRNA RNP being a half or a third of that in the single-targeting group. Similar results were obtained at the apoc3 and angptl3 loci as well (Figures 2B–2E). Our results imply that the limiting factor for effective gene editing lies with the delivery of CRISPR/Cas9 elements into the cell, not the quantity delivered per cell, which is also supported by previous reports with Cas9 plasmid and mRNA.[32] It may be that the number of RNP only needs to meet a threshold to provide targeted gene editing, such that when the combination of three different RNP are delivered into one cell, they provide similar amounts of gene editing to all three gene targets as a monogenic Cas9/gRNA RNP.
Figure 2. Surveyor Assay confirmation of multiplex gene targeting using lipoMSN.
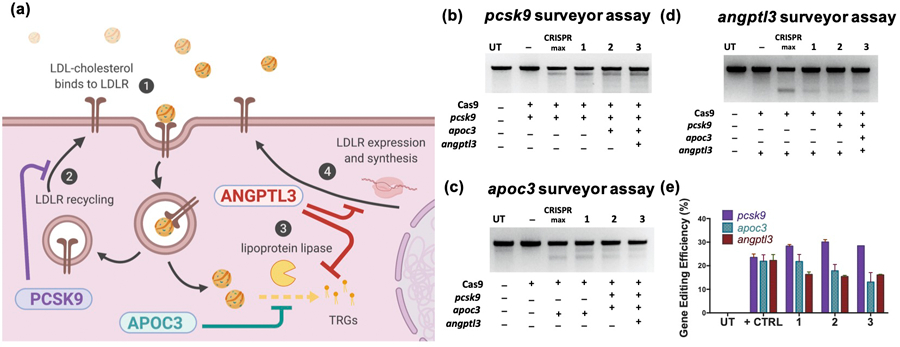
(A) Schematic illustrating the roles of Pcsk9, Apoc3 and Angptl3 on lipid metabolism. Surveyor assays showing in vitro editing of (B) pcsk9, (C) apoc3, and (D) and angptl3 was durable despite lowered doses of target gRNA. (E) Gene editing efficiency quantification using Image J. Results are presented as average ± SEM (n = 4). Positive control of Lipofectamine CRISPRMax delivering targeted RNP noted by ‘+’, while negative control of Cas9 protein only noted with ‘-’. Control and singular target (1) RNP was delivered at 2 ug/mL, while target-specific RNP for dual-targeting (2) was at 1 ug/mL each (2 ug/mL total) and triple-targeting (3) was at 0.67 ug/mL (2 ug/mL total).
To validate that our gene editing resulted in reduced expression of the three targets, pcsk9, angptl3, and apoc3, we first performed reverse-transcription quantitative PCR (RT-qPCR) on the treated AML-12 cells. Interestingly, in addition to the expected results of reduced expression of the gene target with treatment by the respective Cas9/gRNA RNP, we found collateral effects of the gene editing. For example, pcsk9 expression was significantly upregulated by 50% and 25% with editing of apoc3 and angptl3, respectively. In contrast, when treated with a combination of all three Cas9/gRNA RNPs, the expression levels of pcsk9, apoc3 and angptl3 were most significantly reduced by 50%, 80% and 85%, respectively (Figures 3A–3C). This suggests the potential for Pcsk9, Angptl3 or Apoc3 to compensate for each other through uncharacterized feedback loops as they all show effects on lipid uptake and metabolism in the liver.[33] Our singular lipoMSN delivery approach targeting all three genes may take advantage of the overlap of these pathways by removal of these compensation mechanisms. We looked at increased ldlr expression as a result of the different RNP treatments in order to predict most effective synergistic gene editing combinations for further exploration. Disruption of all three target genes led to up-regulation of ldlr expression by 5-fold at 24h post-treatment (Figure 3D), whereas the ldlr level was significantly increased at 48h post-treatment in the single- and dual- (pcsk9 + angptl3) disruption groups (Figure 3E). ELISA assay also confirmed the Ldlr upregulation after Cas9-mediated pcsk9 disruption using our lipoMSN (Figure 3F).
Figure 3. qPCR measurement of gene regulation after CRISPR/Cas9 disruption of singular or multiple genes.
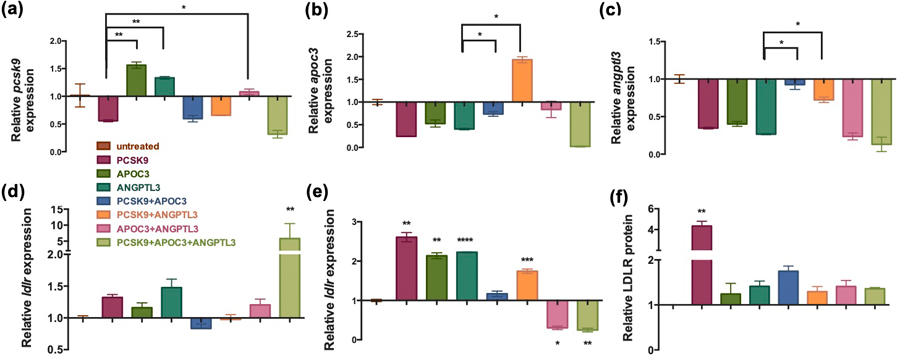
RT-qPCR validation on the (A) pcsk9, (B) apoc3 and (C) angptl3 expression levels in the AML-12 cells at 48h post-treatement of the lipoMSN with Cas9/gRNA RNP relative to the untreated control after normalization to GAPDH expression. RT-qPCR calculated expression of ldlr at (D) 24 h and (E) 48 h. (F) Ldlr protein amount is quantified at 48h post-treatment measured by ELISA. Results are presented as average ± SEM (n = 4). (A-E) qPCR data is relative to the untreated control after normalization to GAPDH expression. Significance was determined using one-way ANOVA with Tukey’s posthoc test, and represented as * p < 0.05 and ** p < 0.01.
To determine the lipoMSN delivery system’s efficacy at RNP multiplex gene editing in vivo, six groups of 5-week-old C57BL/6J female mice were treated in various combinations (Figure 4A) These combinations (pcsk9, angptl3, pcsk9 + angptl3, pcsk9 + apoc3 + angptl3, Cas9 protein alone, PBS control) were designed to validate the synergistic effects between pcsk9- and angptl3-targeting. LipoMSN was given twice through intravenous administration, for a total dose of 10 mg/kg of Cas9/gRNA RNP. Blood of each mouse was drawn weekly beginning one week before treatment in order to determine treatment effects on triglycerides and cholesterol. We also monitored the changes in weight, blood HDL-cholesterol (HDL-C) and alanine transaminase (ALT) levels to determine potential toxicity of our lipoMSN. Serum triglycerides showed a significant lasting effect in the single angptl3-targted group, with a 25% decrease observed even at week 4 post-treatment (Figure 4B). The unexpected finding in the PBS control group, which showed an observable drop in serum triglycerides from week 1 to week 2, may have been due to variations in time of blood collection, which has previously been shown to have an impact on blood lipids.[34] Serum cholesterol measurements showed a significant effect for all the treated groups. At week 4 post-treatment, single gene disruption lowered the cholesterol level by ~30% (31.7% and 28.2% for the pcsk9- and angptl3- targeted groups, respectively), while dual- (pcsk9 + angptl3) and triple- (pcsk9+angptl3+apoc3) gene disruption gave more substantial reduction (56.5% and 43.18%, respectively; Figure 4C). The results of mouse weight, HDL-C and ALT measurements indicated that our lipoMSN did not cause any significant adverse effects, as no significant difference was observed in each indicator between groups (Figure 4D–4F). Further, collection and H&E staining of the heart, liver, lung, kidney and spleen yielded no observable damage in any treatment groups (Figure S11, Supporting Information). Dual disruption of both pcsk9 and angptl3 was more effective on lowering serum cholesterol than any singular disruption. This finding was different from that by the Musunuru group in their adenovirus-based gene editing where no synergy between the two targets was observed.[6] The main reason could be due to the difference in delivery approach; the previous approach, where two individual adenoviruses were applied to target pcsk9 and angptl3, did not ensure a high probability that any given cell would receive both viral vectors.[35] Plausibly, as the editing kinetics of Cas9/gRNA RNP is faster, similar effect might also take a longer period to show when using viral machinery for gene targeting.[36] Nevertheless, to put our lipoMSN efficacy into context, alirocumab, an anti-PCSK9 drug in clinical trials, provides human patients with approximately a 61% reduction in LDL-C with a biweekly dosing, which in preclinical studies have shown an approximately 50% reduction in total cholesterol in mice.[37]
Figure 4. Multiplex liver gene editing by lipoMSN in vivo yields significant effect on blood lipids.
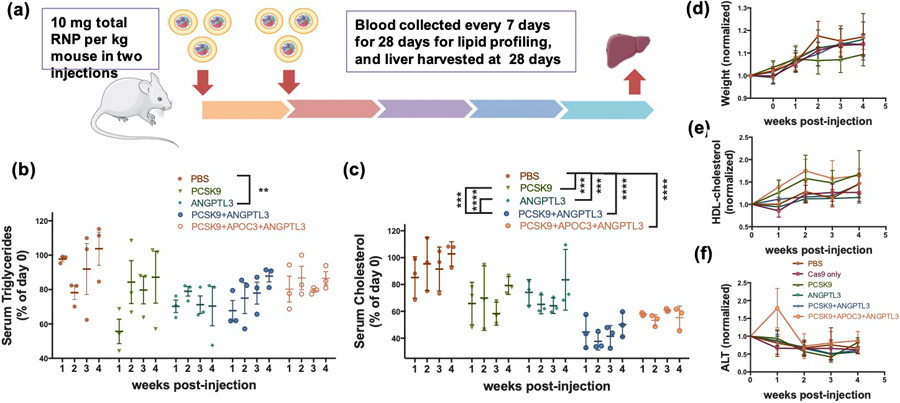
(A) Scheme of the mouse study workflow. Serum (B) triglycerides and (C) cholesterol profiles of treated groups. The changes of (D) weight, (E) HDL-C and (F) ALT of each group post-treatment. Results are presented as average ± SEM (n = 4). Significance was determined using two-way ANOVA with Tukey’s posthoc test, and represented as ** p < 0.01, *** p< 0.001, **** p < 0.0001.
To validate that the blood lipid profile was a result of the lipoMSN-RNP treatment, ELISA assays were used to measure the decrease in Pcsk9 and Angptl3 after the treatment. Reduced circulating Pcsk9 was observed in the pcsk9-targeting group’s serum samples at both weeks 1 and 4 post-treatment (Figure S8B and S8C, Supporting Information). Circulating Angptl3 showed no significant differences, but this could be due to the decreased editing efficiency at angptl3, which was supported by our sequencing results. Further, clinical studies measuring circulating Angptl3 in patients with homozygous and heterozygous loss-of-function mutations show that heterozygous mutations do not provide a statistically significant decrease in Angptl3 compared to a healthy control, implying that significant gene disruption may be required to provide measurable decreases in Angptl3.[38] At our end-point (week 4 post-administration), we were able to detect an indel rate of 24.8% at the target pcsk9 locus, but only 7.2% at the angptl3 site (Figure S10, Supporting Information). Similar disparities in gene editing were observed in our in vitro validation as well (Figure 2). The gene disruption efficiency of angptl3 was lower than that of pcsk9, which could be due to differences in gRNA’s targeting capability or Cas9 affinity, and a further optimization on gRNA design may resolve this issue.
In conclusion, we designed and tested this lipoMSN platform for effective Cas9/gRNA delivery for multiplex gene editing, which enabled exploration of three cardioprotective gene targets in the liver. This easy-to-assemble delivery system leverages on the MSN core to load variable cargos from small gRNA, plasmid, to large protein, while the liposome coating provides consistent and predictable physical characteristics despite the cargo. Gene editing efficiency when delivering a single gRNA reached 54% in vitro and 24.8% in vivo at week 4 post-treatment. The efficiency was not significantly compromised with co-delivery of three different Cas9/gRNA RNP, which allowed synergistic effects to be detected when a singular vehicle was used to disrupt the three cardioprotective genes, pcsk9, apoc3 and angptl3. The in vivo gene disruption of these genes provides encouraging evidence that the multiplex lipoMSN platform offers significant improvement over single-target therapy. Collectively, this study suggests an effective approach of discovering synergistic therapeutic targets using multiplexed nonviral gene editing.
Supplementary Material
Acknowledgements
The authors would like to acknowledge the technical support from Flow Cytometry Core Facility (Columbia Center for Translational Immunology) and Columbia Medical Center Molecular Pathology Core Facility. This work is supported by NSF Graduate Research Fellowships Program (J.G., DGE 1644869), NIH (UG3-NS115598, UH3-TR002151, UH3-TR002142), DARPA (HR00111920009). The animal study was approved and supervised by the Institutional Animal Care and Use Committees at Columbia University.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Jing Gong, Department of Biomedical Engineering, Columbia University, New York, NY 10027, USA.
Hong-Xia Wang, Department of Biomedical Engineering, Columbia University, New York, NY 10027, USA.
Yeh-Hsing Lao, Department of Biomedical Engineering, Columbia University, New York, NY 10027, USA.
Hanze Hu, Department of Biomedical Engineering, Columbia University, New York, NY 10027, USA.
Naazanene Vatan, Department of Biomedical Engineering, Columbia University, New York, NY 10027, USA.
Jonathan Guo, Department of Biomedical Engineering, Columbia University, New York, NY 10027, USA.
Tzu-Chieh Ho, Department of Biomedical Engineering, Columbia University, New York, NY 10027, USA.
Dantong Huang, Department of Biomedical Engineering, Columbia University, New York, NY 10027, USA.
Mingqiang Li, Department of Biomedical Engineering, Columbia University, New York, NY 10027, USA.
Dan Shao, Department of Biomedical Engineering, Columbia University, New York, NY 10027, USA; Institute of Life Sciences, School of Biomedical Science and Engineering and National Engineering Research Center for Tissue Restoration and Reconstruction, South China University of Technology, Guangzhou, Guangdong 510006, China.
Kam W. Leong, Department of Biomedical Engineering, Columbia University, New York, NY 10027, USA; Department of Systems Biology, Columbia University Medical Center, New York, NY 10032, USA
References
- [1].a) Zhang L, Wang L, Xie Y, Wang P, Deng S, Qin A, Zhang J, Yu X, Zheng W, Jiang X, Angewandte Chemie International Edition 2019, 58, 12404; [DOI] [PubMed] [Google Scholar]; b) Sun W, Wang J, Hu Q, Zhou X, Khademhosseini A, Gu Z, Science Advances 2020, 6, eaba2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].a) Sistino JJ, Perfusion 2003, 18, 73; [DOI] [PubMed] [Google Scholar]; b) Nordestgaard BG, Varbo A, Lancet 2014, 384, 626. [DOI] [PubMed] [Google Scholar]
- [3].Steinberg D, Witztum JL, Proc Natl Acad Sci U S A 2009, 106, 9546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Krähenbühl S, Pavik-Mezzour I, von Eckardstein A, Drugs 2016, 76, 1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Cohen JC, Boerwinkle E, Mosley TH Jr, Hobbs HH, New England Journal of Medicine 2006, 354, 1264. [DOI] [PubMed] [Google Scholar]
- [6].Chadwick AC, Evitt NH, Lv W, Musunuru K, Circulation 2018, 137, 975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].a) Maxwell KN, Breslow JL, Proc Natl Acad Sci U S A 2004, 101, 7100; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Cohen JC, Boerwinkle E, Mosley TH Jr., Hobbs HH, N Engl J Med 2006, 354, 1264. [DOI] [PubMed] [Google Scholar]
- [8].Athyros VG, Katsiki N, Dimakopoulou A, Patoulias D, Alataki S, Doumas M, Current pharmaceutical design 2018, 24, 3638. [DOI] [PubMed] [Google Scholar]
- [9].Robinson JG, Farnier M, Krempf M, Bergeron J, Luc G, Averna M, Stroes ES, Langslet G, Raal FJ, El Shahawy M, New England Journal of Medicine 2015, 372, 1489. [Google Scholar]
- [10].Ray KK, Landmesser U, Leiter LA, Kallend D, Dufour R, Karakas M, Hall T, Troquay RP, Turner T, Visseren FL, New England Journal of Medicine 2017, 376, 1430. [DOI] [PubMed] [Google Scholar]
- [11].Stojic L, Lun ATL, Mangei J, Mascalchi P, Quarantotti V, Barr AR, Bakal C, Marioni JC, Gergely F, Odom DT, Nucleic acids research 2018, 46, 5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Xiao-Jie L, Hui-Ying X, Zun-Ping K, Jin-Lian C, Li-Juan J, J Med Genet 2015, 52, 289. [DOI] [PubMed] [Google Scholar]
- [13].Ding QR, Strong A, Patel KM, Ng SL, Gosis BS, Regan SN, Cowan CA, Rader DJ, Musunuru K, Circ Res 2014, 115, 488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lai CQ, Parnell LD, Ordovas JM, Curr Opin Lipidol 2005, 16, 153. [DOI] [PubMed] [Google Scholar]
- [15].Senís E, Fatouros C, Große S, Wiedtke E, Niopek D, Mueller AK, Börner K, Grimm D, Biotechnology journal 2014, 9, 1402. [DOI] [PubMed] [Google Scholar]
- [16].Gong J, Tang D, Leong K, Current Opinion in Biomedical Engineering 2018, 7, 9. [Google Scholar]
- [17].Meghrous J, Aucoin MG, Jacob D, Chahal PS, Arcand N, Kamen AA, Biotechnology progress 2005, 21, 154. [DOI] [PubMed] [Google Scholar]
- [18].a) Wang H-X, Li M, Lee CM, Chakraborty S, Kim H-W, Bao G, Leong KW, Chem Rev 2017, 117, 9874; [DOI] [PubMed] [Google Scholar]; b) Lee K, Conboy M, Park HM, Jiang F, Kim HJ, Dewitt MA, Mackley VA, Chang K, Rao A, Skinner C, Nature biomedical engineering 2017, 1, 889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].a) Nelson CE, Gersbach CA, Annu Rev Chem Biomol Eng 2016, 7, 637; [DOI] [PubMed] [Google Scholar]; b) Torchilin VP, Nature reviews Drug discovery 2005, 4, 145; [DOI] [PubMed] [Google Scholar]; c) Suk JS, Xu Q, Kim N, Hanes J, Ensign LM, Advanced drug delivery reviews 2016, 99, 28; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Veronese FM, Mero A, BioDrugs 2008, 22, 315; [DOI] [PubMed] [Google Scholar]; e) Milla P, Dosio F, Cattel L, Current drug metabolism 2012, 13, 105. [DOI] [PubMed] [Google Scholar]
- [20].a) Allen TM, Cullis PR, Adv Drug Deliv Rev 2013, 65, 36; [DOI] [PubMed] [Google Scholar]; b) Gong Y, Tian S, Zhang S, Available at SSRN 3401354 2019.
- [21].Tarn D, Ashley CE, Xue M, Carnes EC, Zink JI, Brinker CJ, Accounts of chemical research 2013, 46, 792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].a) Chen G, Abdeen AA, Wang Y, Shahi PK, Robertson S, Xie R, Suzuki M, Pattnaik BR, Saha K, Gong S, Nature nanotechnology 2019, 14, 974; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Möller K, Müller K, Engelke H, Bräuchle C, Wagner E, Bein T, Nanoscale 2016, 8, 4007. [DOI] [PubMed] [Google Scholar]
- [23].a) Singh RK, Patel KD, Leong KW, Kim H-W, ACS applied materials & interfaces 2017, 9, 10309; [DOI] [PubMed] [Google Scholar]; b) Chen Y, Chen H, Shi J, Advanced Materials 2013, 25, 3144; [DOI] [PubMed] [Google Scholar]; c) Brinker CJ, Sandia National Lab(SNL-NM; ), Albuquerque, NM (United States), 2018. [Google Scholar]
- [24].a) Sakuma T, Nishikawa A, Kume S, Chayama K, Yamamoto T, Scientific reports 2014, 4, 1; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Nissim L, Perli SD, Fridkin A, Perez-Pinera P, Lu TK, Molecular cell 2014, 54, 698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Shen JP, Zhao D, Sasik R, Luebeck J, Birmingham A, Bojorquez-Gomez A, Licon K, Klepper K, Pekin D, Beckett AN, Nature methods 2017, 14, 573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Mout R, Ray M, Lee Y-W, Scaletti F, Rotello VM, Bioconjugate Chem 2017, 28, 880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tong S, Moyo B, Lee CM, Leong K, Bao G, Nat Rev Mater 2019, 4, 726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wang L, Zheng W, Liu S, Li B, Jiang X, ChemBioChem 2019, 20, 634. [DOI] [PubMed] [Google Scholar]
- [29].a) Liang XQ, Potter J, Kumar S, Zou YF, Quintanilla R, Sridharan M, Carte J, Chen W, Roark N, Ranganathan S, Ravinder N, Chesnut JD, J Biotechnol 2015, 208, 44; [DOI] [PubMed] [Google Scholar]; b) Yu X, Liang XQ, Xie HM, Kumar S, Ravinder N, Potter J, du Jeu XD, Chesnut JD, Biotechnol Lett 2016, 38, 919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wu YX, Zeng J, Roscoe BP, Liu PP, Yao QM, Lazzarotto CR, Clement K, Cole MA, Luk K, Baricordi C, Shen AH, Ren CY, Esrick EB, Manis JP, Dorfman DM, Williams DA, Biffi A, Brugnara C, Biasco L, Brendel C, Pinello L, Tsai SQ, Wolfe SA, Bauer DE, Nature Medicine 2019, 25, 776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Dewey FE, Gusarova V, Dunbar RL, O’Dushlaine C, Schurmann C, Gottesman O, McCarthy S, Van Hout CV, Bruse S, Dansky HM, New England Journal of Medicine 2017, 377, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].a) Finn JD, Smith AR, Patel MC, Shaw L, Youniss MR, van Heteren J, Dirstine T, Ciullo C, Lescarbeau R, Seitzer J, Cell reports 2018, 22, 2227; [DOI] [PubMed] [Google Scholar]; b) Lao YH, Li MQ, Gao MA, Shao D, Chi CW, Huang DT, Chakraborty S, Ho TC, Jiang WQ, Wang HX, Wang SH, Leong KW, Adv Sci 2018, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Nordestgaard BG, Nicholls SJ, Langsted A, Ray KK, Tybjærg-Hansen A, Nature Reviews Cardiology 2018, 15, 261. [DOI] [PubMed] [Google Scholar]
- [34].a) Rivera-Coll A, Fuentes-Arderiu X, Díez-Noguera A, Clinical chemistry 1994, 40, 1549; [PubMed] [Google Scholar]; b) Pocock S, Ashby D, Shaper A, Walker M, Broughton P, Journal of clinical pathology 1989, 42, 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Xie S, Duan J, Li B, Zhou P, Hon GC, Molecular cell 2017, 66, 285. [DOI] [PubMed] [Google Scholar]
- [36].DeWitt MA, Corn JE, Carroll D, Methods 2017, 121, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].a) Roth EM, Moriarty PM, Bergeron J, Langslet G, Manvelian G, Zhao J, Baccara-Dinet MT, Rader DJ, Atherosclerosis 2016, 254, 254; [DOI] [PubMed] [Google Scholar]; b) Kühnast S, van der Hoorn JW, Pieterman EJ, van den Hoek AM, Sasiela WJ, Gusarova V, Peyman A, Schäfer H-L, Schwahn U, Jukema JW, Journal of lipid research 2014, 55, 2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Robciuc MR, Maranghi M, Lahikainen A, Rader D, Bensadoun A, Öörni K, Metso J, Minicocci I, Ciociola E, Ceci F, Arteriosclerosis, thrombosis, and vascular biology 2013, 33, 1706. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


