Abstract.
The presence of intestinal pathogenic Escherichia coli in drinking water is well recognized as a risk for diarrhea. The role of drinking water in extraintestinal infections caused by E. coli—such as urinary tract infections (UTIs)—remains poorly understood. Urinary tract infections are a leading cause of outpatient infections globally, with a lifetime incidence of 50–60% in adult women. We reviewed the scientific literature on the occurrence of uropathogenic E. coli (UPEC) in water supplies to determine whether the waterborne route may be an important, overlooked, source of UPEC. A limited number of studies have assessed whether UPEC isolates are present in drinking water supplies, but no studies have measured whether their presence in water may increase UPEC colonization or the risk of UTIs in humans. Given the prevalence of drinking water supplies contaminated with E. coli across the globe, efforts should be made to characterize UTI-related risks associated with drinking water, as well as other pathways of exposure.
INTRODUCTION
Urinary tract infections (UTIs) are the second most common infection globally, and an estimated 150 million people are diagnosed with UTIs each year, costing more than six billion U.S. dollars in treatment.1,2 In the United States, the CDC estimates that UTIs are responsible for nearly 13,000 deaths every year.3 In addition, uropathogens producing extended spectrum beta-lactamases (ESBLs), and showing resistance to most antimicrobials, are steadily increasing.4,5
Exposure to Escherichia coli—a fecal indicator and a key member of the normal intestinal microflora of humans and other mammals—in drinking water, as well as recreational water, has been linked to an elevated risk of carrying enteric pathogens and diarrhea.6–8 Much less research, however, has been carried out to identify whether the presence of E. coli in water supplies, or the environment where human exposures occur (e.g., recreational water exposures like swimming), may increase the risk for extraintestinal infections, including UTIs. Some highly adapted E. coli strains have acquired specific virulence factors that confer an increased capacity to cause a spectrum of intestinal and extraintestinal diseases.9
Escherichia coli has the potential to cause three broad categories of infection: enteric infections, UTIs, and sepsis/meningitis.9 Infections can be further categorized as intestinal pathogenic E. coli (IPEC), and extraintestinal pathogenic E. coli (ExPEC), which includes uropathogenic E. coli (UPEC) and meningitis-associated E. coli (Figure 1).10,11 Uropathogenic E. coli is the most common etiologic agent of UTIs and causes 68–77% of recurrent UTI infections.12 There are differences in virulence factors between UPEC and commensal E. coli, which make up most of the E. coli that populate the gut.
Figure 1.
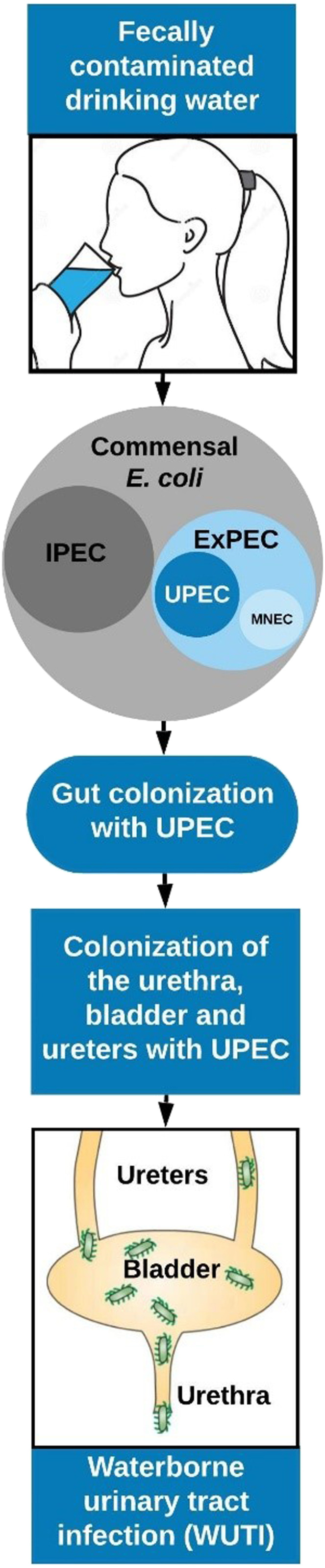
Conceptual illustration of the exposure pathway for waterborne urinary tract infections (WUTIs). E. coli includes commensals, intestinal pathogenic E. coli (IPEC), and extraintestinal pathogenic E. coli (ExPEC), which include meningitis/sepsis-associated E. coli (MNEC) and uropathogenic E. coli (UPEC), the most common infecting agent in the urinary tract. This figure appears in color at www.ajtmh.org.
DEFINING UPEC
The features used to classify E. coli as UPEC vary significantly. There are likely diverse, complementary groups of genetic factors that allow E. coli to interact with the host, resulting in UTIs. Progress in determining these mechanisms, however, is being made. Escherichia coli is often identified by serological typing: H (flagellar), O (lipopolysaccharide), and K (capsular) surface antigens. The O serogroup appears to influence pathogenicity, and it has been identified as a causal agent in most UTIs.13 Uropathogenic E. coli isolates are typically found in phylogenetic group B2, and to a lesser extent in group D.14 Although the E. coli isolates that cause UTIs are often clonal, there is no single phenotypic profile that defines UPEC isolates, which are completely distinct for their lack of a defined set of genes that distinguish them from non-UPEC isolates.15,16 Uropathogenic E. coli isolates typically have virulence factors, including adhesins, siderophore systems, toxins, and lipopolysaccharides that enhance their ability to survive outside of the host, colonize humans, and cause infection.17 Most potential UPEC strains carry these virulence genes, but are able to remain as commensals in our gut.16 Johnson et al.15 assessed a diverse set of predictors of virulence in a murine sepsis model and found that various factors—phylogenetic group, clonal complexes, and accessory genes—were important. Studies of putative uropathogenic E. coli (pUPEC) in water have typically classified the isolates based on the presence of specific genes or by their sequence type (ST).18–22
We hypothesize that drinking water represents an important source of UPEC (Figure 1), especially in low- and middle-income countries (LMICs), where fecally contaminated drinking water is more prevalent. The application of advanced genotyping methods in these countries is often more limited, and research has likely missed this important pathway of exposure, in contrast to high- and upper-middle–income countries. A growing body of research, primarily in high-income countries, is rapidly building on foodborne UTIs (FUTIs).23–28
To find relevant documents describing the evidence of pUPEC in waters used as sources for drinking or recreation that may cause UTIs or potentially other extraintestinal infections, we searched PubMed (https://pubmed.ncbi.nlm.nih.gov/) using the following search terms: (“urinary tract infection” OR “Extraintestinal pathogenic E. coli” OR “ExPEC” OR “uropathogenic E. coli” OR “UPEC”) AND “water.” We also searched the resulting reference lists to identify additional articles.
EVIDENCE OF PUTATIVE UPEC IN WATER
We identified 20 studies of 394 search results that assessed the role of water, or wastewater, as a reservoir of ExPEC or UPEC. The studies which used genotyping methods are summarized in Table 1, and they indicate that fecally contaminated water—mainly surface waters and wastewater—are an important source of UPEC.20,29–37 Zhi et al.38,39 identified pUPEC by screening wastewater samples from treatment plants in Alberta, Canada, for E. coli containing at least three of five UPEC virulence genes. When the researchers compared pUPEC with known UPEC from UTIs from international databases, they found > 96% whole genome similarity; one isolate demonstrated 99.5% genome similarity. In a study of 308 E. coli isolates from surface water samples collected from diverse aquatic ecosystems in the United States, researchers used DNA microarray technology and found that most E. coli isolates were putative ExPEC pathotypes and belonged to phylogenetic groups B2 and D.21 Johnson and others18 characterized 280 E. coli isolates from seven surface water sites and found that 5% of isolates were pExPEC strains. In Japan, researchers studied 531 E. coli isolates from the Yamato River and found 58 pExPEC isolates that belonged to lineages of human UPEC (ST95, ST127, ST12, ST14, and ST131).40,41 A study of household drinking water in India applied multi-locus sequence typing (MLST) to identify pUPEC; they found four E. coli STs, ST648, ST92, STc23, and ST58, that are often found to cause UTIs.20 Similarly, a study of surface water in Georgia used MLST to identify ST131, a common ST associated with UTIs globally, in surface waters.22 In Southeast Queensland, Australia, researchers tested 200 E. coli isolates from 22 rainwater tank samples for 20 virulence factors associated with ExPEC; the researchers also classified E. coli by their phylogenetic groups. Putative ExPEC were identified in 15 of the 22 tanks based on the prevalence of ExPEC-associated virulence genes.42 In a study of constructed wetlands in the United States, researchers examined whether crows were carriers of ExPEC and if wetland roost areas contribute to their spread. The study found that 11.2% of the E. coli isolates identified in impacted waters were pExPEC.19 In India, researchers have studied E. coli from coastal estuaries and found that ∼16% were pExPEC, and approximately one-third of isolates contained antimicrobial resistance genes.43,44 In France, researchers evaluated the prevalence of ExPEC in effluents of a municipal wastewater treatment plant receiving wastewater from a slaughterhouse; ExPEC was more prevalent in city wastewater (8.4%) than in slaughterhouse wastewater (1.2%).45 A few studies have used serotyping to identify pUPEC in drinking water and environmental water sources.46–48
Table 1.
Examples of studies that identified putative uropathogenic E. coli (pUPEC) or extraintestinal pathogenic E. coli (pExPEC) in water samples based on multi-locus sequence typing or the presence of virulence genes17
| Reference | Country | Source of samples | Virulence genes and their function used to identify pUPEC or pExPEC | Findings |
|---|---|---|---|---|
| Müller et al.37 | Switzerland | 207 E. coli from surface waters, freshwater fish, fresh vegetables, retail poultry meat, fecal samples of livestock, healthy humans, and primary care patients | Iron uptake (fyuA, chuA, and yfcv), toxin (vat), pathogenicity island (PAI), and protectins/serum resistance (traT) | Overlaps in E. coli genotypes were found for some pUPEC isolates from water and humans |
| Ahmed et al.42 | Australia | 200 E. coli from 22 rainwater tanks used for potable and non-potable purposes | Adhesins (bmaE, papG allele II, papG allele III, papAH, papEF, and focG), toxins (cdtBa and cvaC), invasins (ibeA), siderophores (iutA), capsule synthesis (kpsMT allele III and kpsMT allele K1), pathogenicity island (PAI), and protectins/serum resistance (traT) | Fifteen of 22 rainwater tanks were positive for pExPEC |
| Hamelin et al.21 | United States | 308 E. coli from surface water collected from two large river systems | P pilus-encoding gene (hlyA); Iron uptake (chuA, fepC, cnf1, irp1, irp2, fyuA, iroN, and usp) | E. coli pathotypes were mostly pExPEC and belonged to phylogenetic groups B2 and D |
| Rayasam et al.20 | India | 104 E. coli from 51 drinking water samples collected from elevated storage reservoirs that are piped to households | STs known to cause UTIs in humans | Nineteen of the E. coli STs (18.3%) belonged to known lineages of human UPEC |
| Amato et al.35 | United States | 337 E. coli from streams draining 15 small watersheds of the Chesapeake Bay | Adhesins (papA, papC, and afaC), siderophores (iutA), and capsule synthesis (kpsMII) | Fifty-six isolates (17%) were pExPEC |
| Sen et al.19 | United States | 134 E. coli from wetlands contaminated by corvids | Siderophores (iutA, iroN, and iutA), capsule synthesis (iss, kpsMTII, and traT), adhesins (papEF, pap A/C, papG, sfa/foc, and afa/dra), toxins and hemolysins (cnf1, stx1, stx2, hlyA, and hlyF), and invasion (ibeA) | Fifteen of 134 isolates (11.2%) were pExPEC |
| Cho et al.22 | United States | 34 Antimicrobial-resistant E. coli identified from a mixed-use watershed | STs known to cause UTIs in humans | Three of the 34 isolates were ST131, which are known lineages of human UPEC |
| Divya et al.43 | India | 300 E. coli from tropical estuarine water | Adhesins (papAH, papC, and sfa/foc), capsule synthesis (kpsMT II), and siderophore (iutA) | Forty-nine isolates (16.3%) were pExPEC, and approximately 34.6% of those isolates had antibiotic-resistant genes |
| Johnson et al.18 | United States | 280 E. coli from seven surface water sites | Type 1 fimbriae (fimA), hemolysin (hlyD), P fimbriae (papAH and papC), S and F1C fimbriae (sfa/focDE), Dr-binding adhesins (afa/draBC), group 2 capsule (kpsM II), and aerobactin system (iutA) | Twenty-six isolates (5%) fulfilled the molecular criteria for pExPEC |
| Gomi et al.40 | Japan | 531 E. coli isolates obtained from Yamato River | STs known to cause UTIs in humans | Among 58 pExPEC isolates, several lineages of human UPEC were found (ST95, ST127, ST12, ST14, and ST131) |
| Franz et al.53 | Netherlands | 170 ESBL-producing E. coli from Dutch wastewater (n = 82) and surface water (n = 88) | Afimbrial adhesion (afa), F1C fimbriae (focG), cytolytic protein toxin (hlyD), iron acquisition system (iutA), group 2 polysaccharide capsule (kpsMII), P fimbriae (papA), and S fimbriae (sfaS) | Fifteen of the ESBL-producing E. coli (8.8%) were pExPEC |
| Diallo et al.45 | France | 1,248 E. coli from effluents of a municipal wastewater treatment plant receiving wastewater from a slaughterhouse | S and F1C fimbriae (sfa/focDE), group 2 capsule (kpsMT K10), hemolysin (hlyA), P fimbriae (papEF), adhesins (afa/draBC), toxins, and hemolysins (clbN, f17A, and cnf) | ExPEC was significantly higher in city wastewater (8.4%) than in slaughterhouse wastewater (1.2%) |
| Anastasi et al.36 | Australia | 264 E. coli isolates collected from 129 receiving water sites in a 20-km radius surrounding sewage treatment plants | P fimbriae (papAH, papEF, and papC), siderophore (iroNE.coli), toxins, and hemolysins (cnf1, hlyA, eltA, estII, eaeA, stx1, and stx2) | ExPEC virulence genes were found in 11% of the 15 most common E. coli types identified |
E. coli = Escherichia coli; ESBL = extended spectrum beta-lactamases; ST = sequence type; UTI = urinary tract infections.
Sanitation and hygiene likely play important roles in exposure to fecal pathogens such as UPEC. In some contexts, hands have been found to be a more important pathway of exposure to E. coli than water, although no study to date has looked at whether they are potentially UPEC isolates.49 Access to improved sanitation and clean water also impact menstrual hygiene management practices, which may affect the risk of UTIs among women.50,51 Overall, however, there is a paucity of research investigating the impacts of sanitation and hygiene on exposures to UPEC or the risk of UTIs.
ANTIMICAROBIAL RESISTANCE IN UPEC
Antimicrobial-resistant E. coli—many harboring genes that allow them to produce ESBLs and avoid the effects of third-generation cephalosporins—have been identified in different sources of drinking water, including drinking water in high-income countries.30–32,52 In studies of pUPEC in environmental water samples, antimicrobial resistance genes, including ESBL genes, have been identified.38 Rayasam et al.20 found that six of 19 pUPEC were resistant to more than three classes of antimicrobial agents. Hamelin et al.21 found that the river sampling site most impacted by urban municipal wastewater also had a higher prevalence of pUPEC-carrying antimicrobial-resistant genes. A study of ESBL-producing E. coli from Dutch wastewater (n = 82) and surface water (n = 88) found that 8.8% of the 170 isolates studied were pExPEC.53 A study of 22 environmental ESBL-producing E. coli found that six of the isolates were able to colonize bladder cells.54
CONCLUSION
Defining the features that make up UPEC in a local geographic context or for a specific population will be an important step forward. Research that studies a combination of factors (e.g., phylogenetic group, clonal complex, and accessory genes) will be needed, and next-generation sequencing will be essential for assessing the role of fecally contaminated water, or fecally contaminated environments, in cases of human UTIs. Given rapid changes in circulating E. coli clones, human and nonhuman host factors, and resistance phenotypes, our ability to predict what features define UPEC will remain a challenge. As noted earlier, UPEC belongs to a wide variety of serogroups and STs, and may consist of a large variety of virulence factors. Future studies will need to identify the STs, serogroups, phylogenetic groups, and virulence genes associated with UTIs in local contexts to understand the full landscape of UPEC’s defining features. Furthermore, a local set of multiplex panels could be used to understand the relationship between water quality and UTIs.
This report highlights a need to investigate the occurrence of pUPEC in drinking water sources to better understand the importance of waterborne UTIs (WUTIs). Given that antimicrobial resistance is predicted to reverse decades of progress in increasing longevity around the world,55 research of WUTIs should test for susceptibility to antimicrobials in assessments of pUPEC. Research has shown the spread of clonal groups of multidrug-resistant E. coli linked to the food supply that cause UTIs in the United States.56 Similar to research that is clarifying FUTIs,23,27,57 we suggest that a similar effort is needed for WUTIs. Drinking water across the globe is commonly contaminated with E. coli, and this pathway deserves more focused investigation. Studies that integrate spatiotemporally matched samples of drinking water and human fecal or urine carriage of pUPEC would provide initial evidence to estimate risks associated with exposures to contaminated drinking water. In addition, case–control studies of UTIs that assess UPEC isolates in both human cases (i.e., urine samples) and pUPEC in household drinking water will clarify the importance of UPEC in drinking water as a risk factor for UTIs in humans.50 This study design has been used to characterize the role of domestic animals in pediatric enteric diseases.58 Advanced molecular methods will be essential for characterizing the genotypic relationships of pUPEC in drinking water and UPEC in humans to elucidate mechanisms of transmission and host invasion pathways. Studies should also consider the effects of sanitation and hygiene on UPEC in drinking water and UTIs. Conducting this research in areas where water supplies are unimproved or poorly managed, such as LMICs, will be important to understand the burden of disease and would likely facilitate gaining a better understanding of this exposure pathway. Even if a small fraction of UTIs or other extraintestinal infections—especially drug-resistant ones—are from contaminated drinking water, the relevance of this exposure to the global burden of disease will likely be substantial, and may generate additional support for efforts that aim to reduce exposures to fecal contamination from drinking water supplies and poor sanitation and hygiene.59
REFERENCES
- 1.Khoshnood S, Heidary M, Mirnejad R, Bahramian A, Sedighi M, Mirzaei H, 2017. Drug-resistant gram-negative uropathogens: a review. Biomed Pharmacother 94: 982–994. [DOI] [PubMed] [Google Scholar]
- 2.Iregbu K, Nwajiobi-Princewill P, 2013. Urinary tract infections in a tertiary hospital in Abuja, Nigeria. Afr J Clin Exp Microbiol 14: 169–173. [Google Scholar]
- 3.Waller TA, Pantin SAL, Yenior AL, Pujalte GG, 2018. Urinary tract infection antibiotic resistance in the United States. Prim Care Clin Off Pract 45: 455–466. [DOI] [PubMed] [Google Scholar]
- 4.Oteo J, Pérez-Vázquez M, Campos J, 2010. Extended-spectrum β-lactamase producing Escherichia coli: changing epidemiology and clinical impact. Curr Opin Infect Dis 23: 320–326. [DOI] [PubMed] [Google Scholar]
- 5.Mazzariol A, Bazaj A, Cornaglia G, 2017. Multi-drug-resistant Gram-negative bacteria causing urinary tract infections: a review. J Chemother 29: 2–9. [DOI] [PubMed] [Google Scholar]
- 6.Loyola S, Sanchez JF, Maguiña E, Canal E, Castillo R, Bernal M, Meza Y, Tilley DH, Oswald WE, Heitzinger K, 2020. Fecal contamination of drinking water was associated with diarrheal pathogen carriage among children younger than 5 years in three Peruvian rural communities. Am J Trop Med Hyg 102: 1279–1285.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhavnani D, Goldstick JE, Cevallos W, Trueba G, Eisenberg JN, 2014. Impact of rainfall on diarrheal disease risk associated with unimproved water and sanitation. Am J Trop Med Hyg 90: 705–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soller JA, Schoen ME, Bartrand T, Ravenscroft JE, Ashbolt NJ, 2010. Estimated human health risks from exposure to recreational waters impacted by human and non-human sources of faecal contamination. Water Res 44: 4674–4691. [DOI] [PubMed] [Google Scholar]
- 9.Kaper J, Nataro J, Mobley H, 2004. Nature reviews. Microbiology. Nat Rev Microbiol 2: 123–140. [DOI] [PubMed] [Google Scholar]
- 10.Lloyd AL, Rasko DA, Mobley HL, 2007. Defining genomic islands and uropathogen-specific genes in uropathogenic Escherichia coli. J Bacteriol 189: 3532–3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spurbeck RR, Dinh PC, Walk ST, Stapleton AE, Hooton TM, Nolan LK, Kim KS, Johnson JR, Mobley HL, 2012. Escherichia coli isolates that carry vat, fyuA, chuA, and yfcV efficiently colonize the urinary tract. Infect Immun 80: 4115–4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chindre Y, Yellapragada L, Chinthaparthi M, 2015. Antibiotic sensitivity pattern of uropathogens: a comparative study between symptomatic and asymptomaic bacteriuria in pregnant women. Int J Curr Microbiol Appl Sci 4: 689–695. [Google Scholar]
- 13.Paniagua-Contreras GL, Monroy-Pérez E, Rodríguez-Moctezuma JR, Domínguez-Trejo P, Vaca-Paniagua F, Vaca S, 2017. Virulence factors, antibiotic resistance phenotypes and O-serogroups of Escherichia coli strains isolated from community-acquired urinary tract infection patients in Mexico. J Microbiol Immunol Infect 50: 478–485. [DOI] [PubMed] [Google Scholar]
- 14.Johnson JR, Russo TA, 2002. Extraintestinal pathogenic Escherichia coli: “the other bad E. coli”. J Lab Clin Med 139: 155–162. [DOI] [PubMed] [Google Scholar]
- 15.Johnson JR, Johnston BD, Porter S, Thuras P, Aziz M, Price LB, 2019. Accessory traits and phylogenetic background predict Escherichia coli extraintestinal virulence better than does ecological source. J Infect Dis 219: 121–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Köhler C-D, Dobrindt U, 2011. What defines extraintestinal pathogenic Escherichia coli? Int J Med Microbiol 301: 642–647. [DOI] [PubMed] [Google Scholar]
- 17.Sarowska J, Futoma-Koloch B, Jama-Kmiecik A, Frej-Madrzak M, Ksiazczyk M, Bugla-Ploskonska G, Choroszy-Krol I, 2019. Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: recent reports. Gut Pathogens 11: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson JR, Johnston BD, Delavari P, Thuras P, Clabots C, Sadowsky MJ, 2017. Phylogenetic backgrounds and virulence-associated traits of Escherichia coli isolates from surface waters and diverse animals in Minnesota and Wisconsin. Appl Environ Microbiol 83: e01329-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sen K, Shepherd V, Berglund T, Quintana A, Puim S, Tadmori R, Turner RJ, Khalil L, Soares MA, 2020. American crows as carriers of extra intestinal pathogenic E. coli and avian pathogenic-like E. coli and their potential impact on a constructed wetland. Microorganisms 8: 1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rayasam SD, Ray I, Smith KR, Riley LW, 2019. Extraintestinal pathogenic Escherichia coli and antimicrobial drug resistance in a maharashtrian drinking water system. Am J Trop Med Hyg 100: 1101–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamelin K, Bruant G, El-Shaarawi A, Hill S, Edge TA, Fairbrother J, Harel J, Maynard C, Masson L, Brousseau R, 2007. Occurrence of virulence and antimicrobial resistance genes in Escherichia coli isolates from different aquatic ecosystems within the St. Clair River and Detroit River areas. Appl Environ Microbiol 73: 477–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho S, Nguyen HAT, McDonald JM, Woodley TA, Hiott LM, Barrett JB, Jackson CR, Frye JG., 2019. Genetic characterization of antimicrobial-resistant Escherichia coli isolated from a mixed-use watershed in northeast Georgia, USA. Int J Environ Res Public Health 16: 3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu CM, Stegger M, Aziz M, Johnson TJ, Waits K, Nordstrom L, Gauld L, Weaver B, Rolland D, Statham S, 2018. Escherichia coli ST131-H22 as a foodborne uropathogen. MBio 9: e00470-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maluta RP, Logue CM, Casas MRT, Meng T, Guastalli EAL, Rojas TCG, Montelli AC, Sadatsune T, de Carvalho Ramos M, Nolan LK, 2014. Overlapped sequence types (STs) and serogroups of avian pathogenic (APEC) and human extra-intestinal pathogenic (ExPEC) Escherichia coli isolated in Brazil. PloS One 9: e105016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manges AR, Smith SP, Lau BJ, Nuval CJ, Eisenberg JN, Dietrich PS, Riley LW, 2007. Retail meat consumption and the acquisition of antimicrobial resistant Escherichia coli causing urinary tract infections: a case–control study. Foodborne Pathog Dis 4: 419–431. [DOI] [PubMed] [Google Scholar]
- 26.Mora A, Viso S, López C, Alonso MP, García-Garrote F, Dabhi G, Mamani R, Herrera A, Marzoa J, Blanco M, 2013. Poultry as reservoir for extraintestinal pathogenic Escherichia coli O45: K1: H7-B2-ST95 in humans. Vet Microbiol 167: 506–512. [DOI] [PubMed] [Google Scholar]
- 27.Nordstrom L, Liu CM, Price LB, 2013. Foodborne urinary tract infections: a new paradigm for antimicrobial-resistant foodborne illness. Front Microbiol 4: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamaji R, Friedman CR, Rubin J, Suh J, Thys E, McDermott P, Hung-Fan M, Riley LW, 2018. A population-based surveillance study of shared genotypes of Escherichia coli isolates from retail meat and suspected cases of urinary tract infections. mSphere 3: e00179-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Zhou Y, Guo S, Chang W, 2015. Prevalence and characteristics of extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae isolated from rural well water in Taian, China, 2014. Environ Sci Pollut Res 22: 11488–11492. [DOI] [PubMed] [Google Scholar]
- 30.De Boeck H, Miwanda B, Lunguya-Metila O, Muyembe-Tamfum JJ, Stobberingh E, Glupczynski Y, Jacobs J, 2012. ESBL-positive enterobacteria isolates in drinking water. Emerg Infect Dis 18: 1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madec JY, Haenni M, Ponsin C, Kieffer N, Rion E, Gassilloud B, 2016. Sequence type 48 Escherichia coli carrying the blaCTX-M-1 IncI1/ST3 plasmid in drinking water in France. Antimicrob Agents Chemoth 60: 6430–6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanner WD, VanDerslice JA, Goel RK, Leecaster MK, Fisher MA, Olstadt J, Gurley CM, Morris AG, Seely KA, Chapman L, 2019. Multi-state study of Enterobacteriaceae harboring extended-spectrum beta-lactamase and carbapenemase genes in US drinking water. Sci Rep 9: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walsh TR, Weeks J, Livermore DM, Toleman MA, 2011. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis 11: 355–362. [DOI] [PubMed] [Google Scholar]
- 34.Coleman B, Salvadori M, McGeer A, Sibley K, Neumann N, Bondy S, Gutmanis I, McEwen S, Lavoie M, Strong D, 2012. The role of drinking water in the transmission of antimicrobial-resistant E. coli. Epidemiol Infect 140: 633–642. [DOI] [PubMed] [Google Scholar]
- 35.Amato HK, Wong NM, Pelc C, Taylor K, Price LB, Altabet M, Jordan TE, Graham JP, 2020. Effects of concentrated poultry operations and cropland manure application on antibiotic resistant Escherichia coli and nutrient pollution in Chesapeake Bay watersheds. Sci Total Environ 735: 139401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anastasi EM, Matthews B, Stratton HM, Katouli M, 2012. Pathogenic Escherichia coli found in sewage treatment plants and environmental waters. Appl Environ Microbiol 78: 5536–5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Müller A, Stephan R, Nüesch-Inderbinen M, 2016. Distribution of virulence factors in ESBL-producing Escherichia coli isolated from the environment, livestock, food and humans. Sci Total Environ 541: 667–672. [DOI] [PubMed] [Google Scholar]
- 38.Zhi S, Stothard P, Banting G, Scott C, Huntley K, Ryu K, Otto S, Ashbolt N, Checkley S, Dong T, 2020. Characterization of water treatment-resistant and multidrug-resistant urinary pathogenic Escherichia coli in treated wastewater. Water Res 182: 115827. [DOI] [PubMed] [Google Scholar]
- 39.Zhi S, Banting G, Stothard P, Ashbolt NJ, Checkley S, Meyer K, Otto S, Neumann NF, 2019. Evidence for the evolution, clonal expansion and global dissemination of water treatment-resistant naturalized strains of Escherichia coli in wastewater. Water Res 156: 208–222. [DOI] [PubMed] [Google Scholar]
- 40.Gomi R, Matsuda T, Matsumura Y, Yamamoto M, Tanaka M, Ichiyama S, Yoneda M, 2017. Whole-genome analysis of antimicrobial-resistant and extraintestinal pathogenic Escherichia coli in river water. Appl Environ Microbiol 83: 115827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gomi R, Matsuda T, Fujimori Y, Harada H, Matsui Y, Yoneda M, 2015. Characterization of pathogenic Escherichia coli in river water by simultaneous detection and sequencing of 14 virulence genes. Environ Sci Technol 49: 6800–6807. [DOI] [PubMed] [Google Scholar]
- 42.Ahmed W, Hodgers L, Masters N, Sidhu J, Katouli M, Toze S, 2011. Occurrence of intestinal and extraintestinal virulence genes in Escherichia coli isolates from rainwater tanks in Southeast Queensland, Australia. Appl Environ Microbiol 77: 7394–7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Divya SP, Hatha AAM, 2019. Screening of tropical estuarine water in south-west coast of India reveals emergence of ARGs-harboring hypervirulent Escherichia coli of global significance. Int J Hyg Environ Health 222: 235–248. [DOI] [PubMed] [Google Scholar]
- 44.Sukumaran D, Mohamed Hatha AA, 2015. Antibiotic resistance and virulence genes of extraintestinal pathogenic Escherichia coli from tropical estuary, South India. J Infect Dev Ctries 9: 496–504. [DOI] [PubMed] [Google Scholar]
- 45.Diallo AA, Brugère H, Kérourédan M, Dupouy V, Toutain PL, Bousquet-Mélou A, Oswald E, Bibbal D, 2013. Persistence and prevalence of pathogenic and extended-spectrum beta-lactamase-producing Escherichia coli in municipal wastewater treatment plant receiving slaughterhouse wastewater. Water Res 47: 4719–4729. [DOI] [PubMed] [Google Scholar]
- 46.Chandran A, Hatha AAM, Varghese S, Sheeja KM, 2008. Prevalence of multiple drug resistant Escherichia coli serotypes in a tropical estuary, India. Microbes Environ 23: 153–158. [DOI] [PubMed] [Google Scholar]
- 47.Ramteke PW, Tewari S, 2007. Serogroups of Escherichia coli from drinking water. Environ Monit Assess 130: 215–220. [DOI] [PubMed] [Google Scholar]
- 48.Verma T, Ramteke PW, Garg SK, 2008. Quality assessment of treated tannery wastewater with special emphasis on pathogenic E. coli detection through serotyping. Environ Monit Assess 145: 243–249. [DOI] [PubMed] [Google Scholar]
- 49.Mattioli MCM, Davis J, Boehm AB, 2015. Hand-to-mouth contacts result in greater ingestion of feces than dietary water consumption in Tanzania: a quantitative fecal exposure assessment model. Environ Science Technol 49: 1912–1920. [DOI] [PubMed] [Google Scholar]
- 50.Das P, Baker KK, Dutta A, Swain T, Sahoo S, Das BS, Panda B, Nayak A, Bara M, Bilung B, 2015. Menstrual hygiene practices, WASH access and the risk of urogenital infection in women from Odisha, India. PLoS One 10: e0130777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Janoowalla H, Keppler H, Asanti D, Xie X, Negassa A, Benfield N, Rulisa S, Nathan LM., 2020. The impact of menstrual hygiene management on adolescent health: the effect of Go! pads on rate of urinary tract infection in adolescent females in Kibogora, Rwanda. Int J Gynecol Obstet 148: 87–95. [DOI] [PubMed] [Google Scholar]
- 52.McKeon DM, Calabrese JP, Bissonnette GK, 1995. Antibiotic resistant gram-negative bacteria in rural groundwater supplies. Water Res 29: 1902–1908. [Google Scholar]
- 53.Franz E, Veenman C, van Hoek AHAM, de Roda Husman A, Blaak H, 2015. Pathogenic Escherichia coli producing extended-spectrum β-Lactamases isolated from surface water and wastewater. Sci Rep 5: 14372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hong H, Chun J, Lee Y, 2004. Detection of extended-spectrum beta-lactamase-producing, multidrug-resistant environmental isolates of Escherichia coli that bind to human bladder cells. Microb Drug Resist 10: 184–189. [DOI] [PubMed] [Google Scholar]
- 55.World Health Organization , 2017. Antimicrobial Resistance: Global Report on Surveillance. Geneva, Switzerland: WHO. [Google Scholar]
- 56.Manges AR, Johnson JR, Foxman B, O’Bryan TT, Fullerton KE, Riley LW, 2001. Widespread distribution of urinary tract infections caused by a multidrug-resistant Escherichia coli clonal group. New Engl J Med 345: 1007–1013. [DOI] [PubMed] [Google Scholar]
- 57.Manges A, 2016. Escherichia coli and urinary tract infections: the role of poultry-meat. Clin Microbiol Infect 22: 122–129. [DOI] [PubMed] [Google Scholar]
- 58.Conan A, O’Reilly CE, Ogola E, Ochieng JB, Blackstock AJ, Omore R, Ochieng L, Moke F, Parsons MB, Xiao L, 2017. Animal-related factors associated with moderate-to-severe diarrhea in children younger than five years in western Kenya: a matched case-control study. PLoS Negl Trop Dis 11: e0005795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pickering AJ, Crider Y, Sultana S, Swarthout J, Goddard FG, Islam SA, Sen S, Ayyagari R, Luby SP, 2019. Effect of in-line drinking water chlorination at the point of collection on child diarrhoea in urban Bangladesh: a double-blind, cluster-randomised controlled trial. Lancet Glob Health 7: e1247–e1256. [DOI] [PubMed] [Google Scholar]


