Abstract
PURPOSE
Genetic testing is important for breast and ovarian cancer risk reduction and treatment, yet little is known about its evolving use.
METHODS
SEER records of women of age ≥ 20 years diagnosed with breast or ovarian cancer from 2013 to 2017 in California or Georgia were linked to the results of clinical germline testing through 2019. We measured testing trends, rates of variants of uncertain significance (VUS), and pathogenic variants (PVs).
RESULTS
One quarter (25.2%) of 187,535 patients with breast cancer and one third (34.3%) of 14,689 patients with ovarian cancer were tested; annually, testing increased by 2%, whereas the number of genes tested increased by 28%. The prevalence of test results by gene category for breast cancer cases in 2017 were BRCA1/2, PVs 5.2%, and VUS 0.8%; breast cancer–associated genes or ovarian cancer–associated genes (ATM, BARD1, BRIP1, CDH1, CHEK2, EPCAM, MLH1, MSH2, MSH6, NBN, NF1, PALB2, PMS2, PTEN, RAD51C, RAD51D, STK11, and TP53), PVs 3.7%, and VUS 12.0%; other actionable genes (APC, BMPR1A, MEN1, MUTYH, NF2, RB1, RET, SDHAF2, SDHB, SDHC, SDHD, SMAD4, TSC1, TSC2, and VHL) PVs 0.6%, and VUS 0.5%; and other genes, PVs 0.3%, and VUS 2.6%. For ovarian cancer cases in 2017, the prevalence of test results were BRCA1/2, PVs 11.0%, and VUS 0.9%; breast or ovarian genes, PVs 4.0%, and VUS 12.6%; other actionable genes, PVs 0.7%, and VUS 0.4%; and other genes, PVs 0.3%, and VUS 0.6%. VUS rates doubled over time (2013 diagnoses: 11.2%; 2017 diagnoses: 26.8%), particularly for racial or ethnic minorities (47.8% Asian and 46.0% Black, v 24.6% non-Hispanic White patients; P < .001).
CONCLUSION
A testing gap persists for patients with ovarian cancer (34.3% tested v nearly all recommended), whereas adding more genes widened a racial or ethnic gap in VUS results. Most PVs were in 20 breast cancer–associated genes or ovarian cancer–associated genes; testing other genes yielded mostly VUS. Quality improvement should focus on testing indicated patients rather than adding more genes.
INTRODUCTION
Genetic testing is increasingly valuable for cancer risk assessment, screening, risk reduction, and treatment. Innovation in sequencing technology and declining costs have facilitated the progress of genetic testing into the mainstream of cancer care.1 Epidemiologic studies are defining cancer risks associated with many genes, and guidelines offer screening and prevention protocols for pathogenic variants (PVs) in more than 40 genes.2-4 There is growing evidence for using germline results to guide therapy, with poly(ADP-ribose) polymerase (PARP) inhibitors approved to treat breast, pancreatic, prostate, and ovarian cancers in carriers of PVs in BRCA1 and BRCA2 (BRCA1/2).5-8
CONTEXT
Key Objective
To understand how the use and results of germline genetic testing are evolving over time among women diagnosed with breast cancer or ovarian cancer.
Knowledge Generated
Most pathogenic variant results were found in 20 breast cancer–associated genes and/or ovarian cancer–associated genes. There is persistent underuse of genetic testing among patients with ovarian cancer, whereas testing more genes per patient is associated with a growing racial or ethnic disparity in uncertain results.
Relevance
Focused testing of a limited, clinically relevant subset of genes may optimize the yield of genetic testing for women diagnosed with breast cancer or ovarian cancer, particularly among racial or ethnic minorities.
The expanding role of genetic testing in cancer care motivates research about how testing is deployed in the community and the implications of results for patients and their families.9 In particular, there is concern about gaps and disparities in testing and how results influence cancer risk reduction and treatment decisions.10-14 However, most prior studies have been limited to small, clinic-based samples or predate the modern era of multiple-gene panels (MGPs). The Georgia-California SEER Genetic Testing Linkage Demonstration Project enabled a population-based assessment of genetic testing for cancer risk. In prior work, we reported on patterns and correlates of genetic testing among women diagnosed with breast or ovarian cancer in 2013 and 2014 and found underutilization of testing after diagnosis of ovarian cancer (only 31% tested v nearly 100% recommended) and racial or ethnic differences in receipt of variants of uncertain significance (VUS).15,16
In this article, we examine trends in germline testing over 7 years among all women diagnosed with breast cancer or ovarian cancer in Georgia or California from 2013 to 2017, followed for testing and result interpretation from 2012 to 2019. As interest in genetic risk evaluation has surged, our hypotheses were that (1) MGP would entirely replace testing BRCA1/2 only, (2) testing underutilization in patients with ovarian cancer would improve, (3) more patients would be tested at lower levels of pretest risk for PVs, (4) sociodemographic differences in testing trends would not be observed, (5) detection of both PVs and VUS would increase, and (6) racial or ethnic disparities in rates of VUS would diminish.
METHODS
Study Cohort and Data Set
We linked all female patients with breast cancer or ovarian cancer diagnosed from January 1, 2013, to December 31, 2017, in Georgia and California and reported to one of the four SEER registries that provided statewide coverage (in Georgia, the Georgia Cancer Registry and in California, the Los Angeles Cancer Surveillance Program, the Greater Bay Area Cancer Registry, and the Cancer Registry of Greater California) to germline genetic testing results from four laboratories (Ambry Genetics, Aliso Viejo, CA; GeneDx, Gaithersburg, MD; Invitae, San Francisco, CA; and Myriad Genetics, Salt Lake City, UT) that performed the majority of clinical testing in the regions. We implemented probabilistic methods to maximize ascertainment and accuracy of the linkage. The analytic data set combined laboratory testing data (genetic tests and results from all laboratories) with variables from SEER registries. We obtained the genetic test use and results from reports dated 2012 through the first quarter of 2019. We excluded patients below 20 years old, missing race or ethnicity information, having more than one primary tumor, and diagnosed from death certificate only (Data Supplement, online only). For a patient who was tested more than once (12.7% of tested patients), we kept only the most recent result for each gene. Thus, the final status for each patient included any additional testing or reclassification of a prior finding reported by the laboratory over 2012-2019. The analytic file containing both registry and laboratory information was stripped of protected health information (as defined by the Health Information Portability and Accountability Act Privacy Rule17), and some variables (age, race, marital status, poverty, insurance, histology, and test result) were collapsed to minimize reidentification risk. This study was approved by institutional review boards associated with the SEER registries.
Test Results From Laboratories
Laboratories provided the results at the gene level, including the interpretation according to American College of Medical Genetics (ACMG) criteria18 that were sent to the ordering clinician: PV or likely PV (combined for analysis as PV), VUS, and benign or likely benign (combined for analysis as normal). We combined the results from all laboratories to ensure anonymity of laboratories; gene-specific results were retained in the analytic data set only for genes tested by two or more laboratories (n = 86). We grouped results for the remaining 252 genes tested by only one laboratory: PVs in these genes were rare (1.4% for ovarian and 0.5% for breast cancer), but VUS were more common (6.4% for ovarian and 5.4% for breast cancer).
Measures
We categorized a test as MGP if it included other genes in addition to BRCA1/2. We grouped PVs according to the level of evidence that supported clinical testing, as follows: BRCA1/2; genes designated by National Comprehensive Cancer Network guidelines as associated with breast and/or ovarian cancer (breast or ovarian genes: ATM, BARD1, BRIP1, CDH1, CHEK2, EPCAM, MLH1, MSH2, MSH6, NBN, NF1, PALB2, PMS2, PTEN, RAD51C, RAD51D, STK11, and TP53)2; genes designated by ACMG guidelines as medically actionable, for which reporting is advised if a PV result is detected incidentally (other actionable genes: APC, BMPR1A, MEN1, MUTYH, NF2, RB1, RET, SDHAF2, SDHB, SDHC, SDHD, SMAD4, TSC1, TSC2, and VHL)19; and any other tested genes (other genes). Patients with VUS in any gene but no PVs were categorized as VUS. Patients with multiple VUS were categorized by the most clinically significantly affected gene, as follows: BRCA1/2, breast or ovarian, other actionable, and other genes. SEER measures included diagnosis age, summary stage, bilateral disease (breast cancer only), triple-negative biologic subtype (breast cancer only), percent poverty at the census level (< 10%, 10%-19%, and ≥ 20%), and race or ethnicity (non-Hispanic White, Black, Hispanic, Asian, and Others).
Analysis Plan
We evaluated clinical and sociodemographic correlates of testing over time for breast cancer and ovarian cancer separately. We tested for differences in trends in receipt of any test and in type of test using logistic regression, with race, age, and geographic site as covariates. Among testers, we described trends in the number of genes tested and used multivariate Poisson models to examine the association between sociodemographic factors and the number of genes tested. We then examined trends in test results (grouped by clinical testing evidence as defined above) among testers and whether these trends differed by race or ethnicity. To display trends in the patterns and results of testing, we used a nonparametric technique of local weighted regression, choosing a smoothing parameter using the Akaike Information Criterion.
RESULTS
Study Population
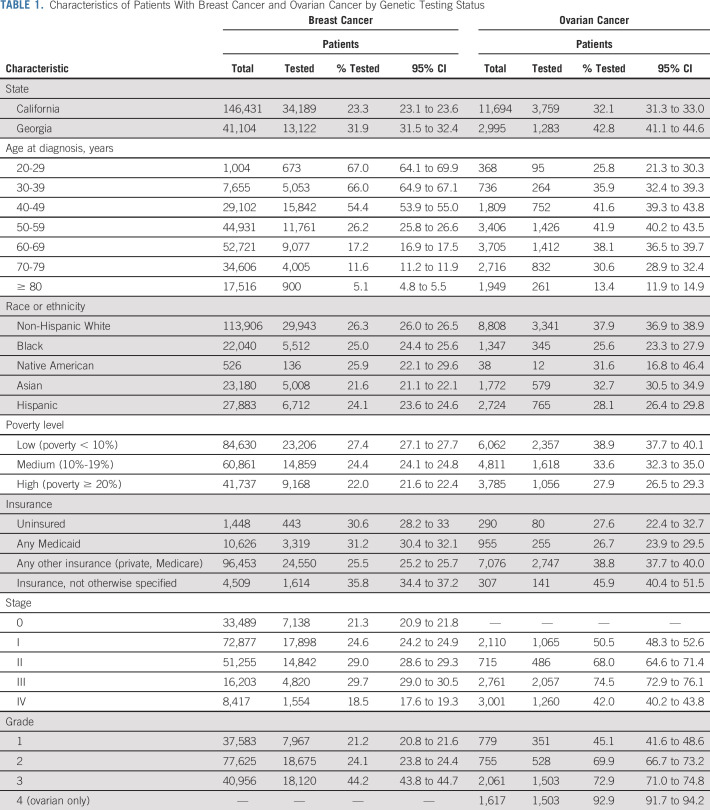
The study cohort included 187,535 patients with breast cancer and 14,689 patients with ovarian cancer, who were diagnosed in Georgia or California in 2013-2017. The Data Supplement shows the flow of patients into the study. California represented 78.1% and 79.6% of the breast and ovarian cancer cases, respectively (Table 1).
TABLE 1.
Characteristics of Patients With Breast Cancer and Ovarian Cancer by Genetic Testing Status
Testing Receipt
Overall, 25.2% of patients with breast cancer and 34.3% of patients with ovarian cancer linked to one or more tests. There were differences in testing rates between states: Georgia linked 31.9% and 42.8% versus California 23.4% and 32.1% for breast and ovarian cancer, respectively (P < .001). Among tested patients, 87.3% had one test, 10.7% had two, and 2.0% three or more. Most patients (71.8%) were first tested within 6 months after diagnosis, 24.2% more than 6 months after diagnosis, and 4.0% before diagnosis.
Trends in Testing Receipt
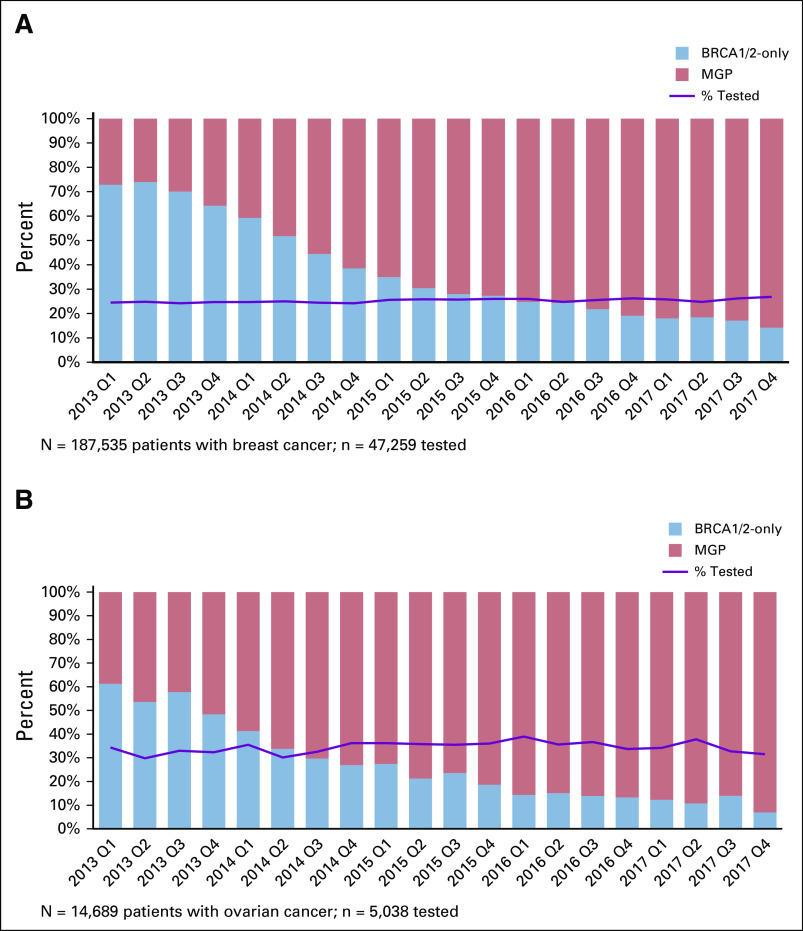
Figure 1 shows that rates of receiving any genetic test increased little over time. On average, testing rates increased 2% per year. By contrast, there was a marked increase in the number of genes tested: about one quarter of tested patients with breast cancer diagnosed in early 2013 received MGP versus > 80% of those diagnosed in late 2017. Most patients diagnosed in late 2017 who had BRCA1/2-only testing were tested before cancer diagnosis. The trend for ovarian cancer was similar: about 40% of patients diagnosed in early 2013 received MGP versus > 90% diagnosed in late 2017. These findings were similar between states. The Data Supplement shows testing trends by age: older patients were more likely to be tested in later years. In patients of age > 60 years (who accounted for > 50% of both cancer cohorts), testing rates increased from 11.1% in 2013 to 14.9% for breast cancer and 25.3% to 31.4% for ovarian cancer. By contrast, patients of age < 45 years (< 15% of the sample) had lower testing rates over time. We did not observe substantial changes in testing rates by other sociodemographic or clinical variables over time.
FIG 1.
(A) Trends in test rates and test type for patients with breast cancer and (B) patients with ovarian cancer. The solid line indicates the proportion of patients who received any genetic test over the observation period by quarter (Q) and year of diagnosis. The bars indicate the percent of testers in each time period who received BRCA1/2-only testing (blue) or MGP testing (orange). MGP, multiple-gene panel.
The Data Supplement shows trends in the number of genes tested: from 1 to 82 for breast cancer (mean, 19) and 1 to 81 for ovarian cancer (mean, 21). There was a consistent upward trend in gene number for patients with both breast and ovarian cancer, from approximately 10 to 35 genes. The increase in tested gene number for patients with breast cancer was 28.0% (95% CI, 27.5 to 28.8) annually, after adjusting for geographic site, race, and age.
Trends in Test Results
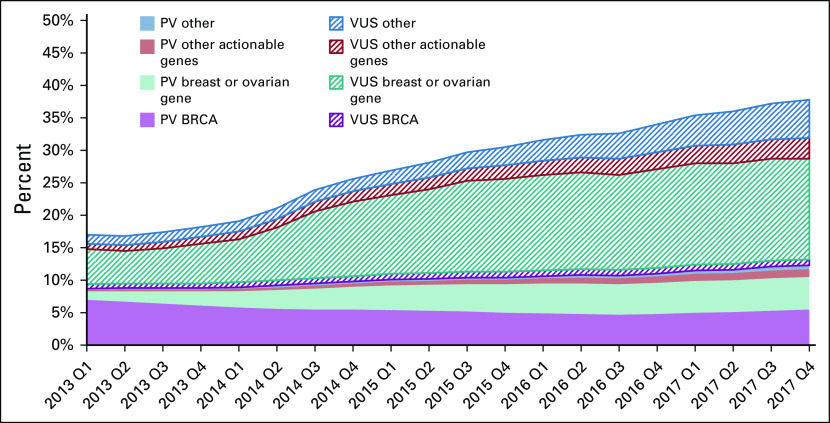
Figure 2 shows results of trends for patients with breast cancer by clinical categories of genes. In early 2013, 18.3% of testers had a PV or VUS-only result, which increased to 37.2% in late 2017. The proportion of tested patients with breast cancer with PVs in BRCA1/2 decreased from 7.5% to 5.0% (P < .001), whereas PV yield for the two other clinically salient categories (breast or ovarian and other actionable genes) increased: in any breast or ovarian gene from 1.3% to 4.6% and in any other actionable gene from 0.3% to 1.3%. Breast or ovarian genes in which PVs were found included ATM, BARD1, BRIP1, CDH1, CHEK2, EPCAM, MLH1, MSH2, MSH6, NBN, NF1, PMS2, PALB2, PTEN, RAD51C, RAD51D, STK11, and TP53; other actionable genes in which PVs were found were APC, MUTYH, RB1, RET, SDHB, SDHC, SDHD, SMAD4, and VHL. PVs in any of the other 51 genes were generally < 1% per gene (Data Supplement). In contrast to PVs, VUS-only rates increased markedly: from 8.5% in patients diagnosed in early 2013 to 22.4% in patients diagnosed in late 2017.
FIG 2.
Trends by diagnosis year and quarter (Q) in PV and VUS results for patients with breast cancer by gene categories: BRCA1/2 (n = 2); breast cancer–associated genes or ovarian cancer–associated genes: ATM, BARD1, BRIP1, CDH1, CHEK2, EPCAM, MLH1, MSH2, MSH6, NBN, NF1, PALB2, PMS2, PTEN, RAD51C, RAD51D, STK11, and TP53 (n = 18); other actionable genes: APC, BMPR1A, MEN1, MUTYH, NF2, RB1, RET, SDHAF2, SDHB, SDHC, SDHD, SMAD4, TSC1, TSC2, and VHL (n = 15); and other genes (n = 51). See the Data Supplement for the list of genes. PV, pathogenic variant; VUS, variant of unknown significance.
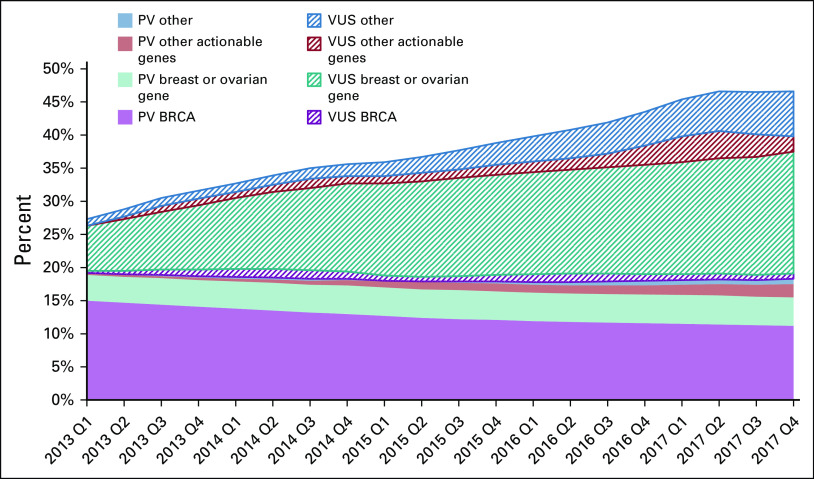
Figure 3 shows results of trends for patients with ovarian cancer by clinical categories of genes. In early 2013, 30.8% of testers had a PV or VUS-only result, which increased to 43.0% in late 2017: this was entirely due to the increase in VUS-only rates. The yield of PVs in BRCA1/2 decreased from 15.7% to 12.4% (P < .001), whereas the PV yield for breast or ovarian genes changed from 3.9% to 4.3% and for other actionable genes from 0.3% to 2.0% (Data Supplement). Breast or ovarian genes in which PVs were found were ATM, BARD1, BRIP1, CHEK2, EPCAM, MLH1, MSH2, MSH6, NBN, NF1, PALB2, PMS2, PTEN, RAD51C, RAD51D, STK11, and TP53; other actionable genes in which PVs were found were APC, MUTYH, and SDHC. VUS-only rates increased markedly, from 8.1% in patients diagnosed in early 2013 to 28.3% in patients diagnosed in late 2017.
FIG 3.
Trends by diagnosis year and quarter (Q) in PV and VUS results for patients with ovarian cancer by gene categories: BRCA1/2 (n = 2); breast cancer–associated genes or ovarian cancer–associated genes: ATM, BARD1, BRIP1, CDH1, CHEK2, EPCAM, MLH1, MSH2, MSH6, NBN, NF1, PALB2, PMS2, PTEN, RAD51C, RAD51D, STK11, and TP53 (n = 18); other actionable genes: APC, BMPR1A, MEN1, MUTYH, NF2, RB1, RET, SDHAF2, SDHB, SDHC, SDHD, SMAD4, TSC1, TSC2, and VHL (n = 15); and other genes (n = 51). See the Data Supplement for the list of genes. PV, pathogenic variant; VUS, variant of unknown significance only.
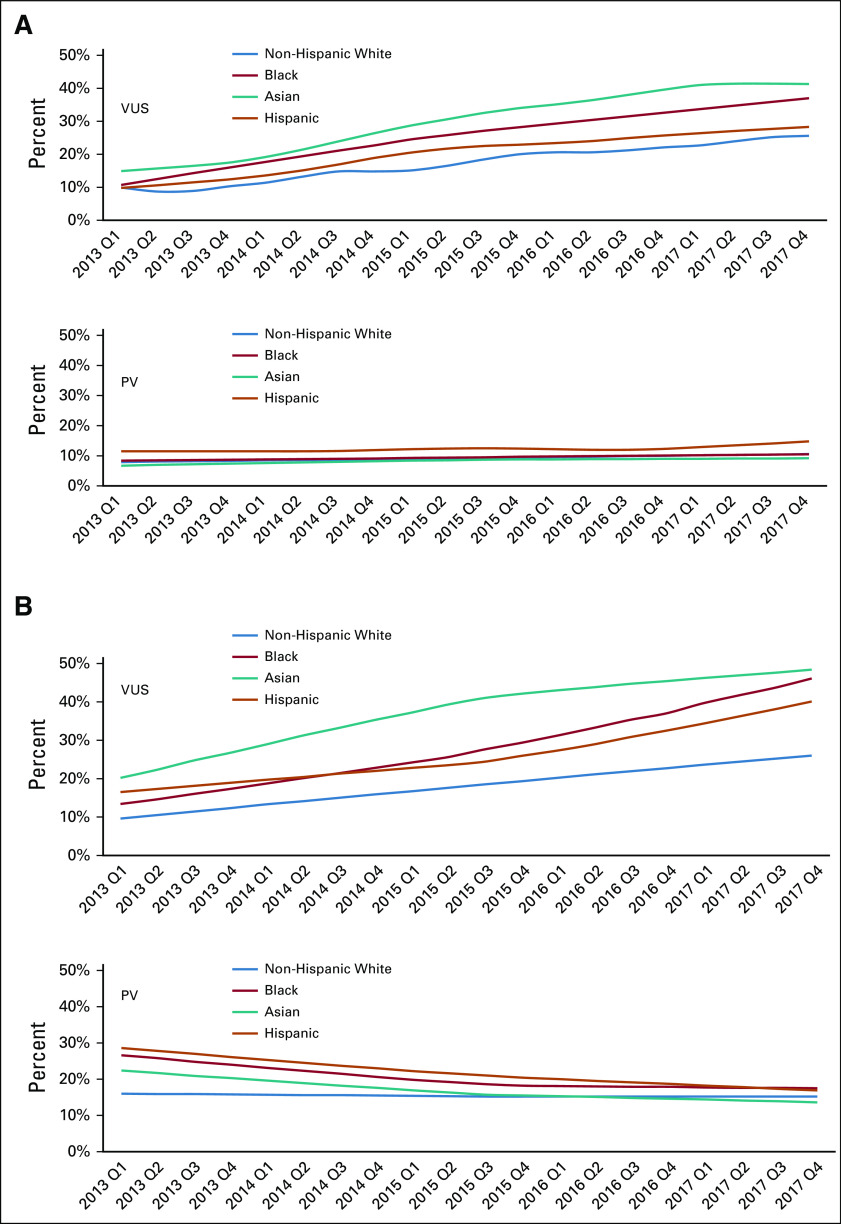
Figure 4 shows trends in PV and VUS-only rates among testers with breast cancer and with ovarian cancer by race or ethnicity. Among patients with breast cancer, racial or ethnic differences in PV rates were small and did not change over time. However, large differences in VUS-only rates across race or ethnicity persisted: in 2017, VUS-only rates were substantially higher in Asian (42.4%), Black (36.6%), and Hispanic (27.7%) than non-Hispanic White patients with breast cancer (24.5%, P < .001). Among patients with ovarian cancer, PV rates across racial or ethnic groups diminished over time. By contrast, large racial or ethnic differences in VUS-only rates persisted throughout the study period: in 2017, VUS-only rates were substantially higher in Asian (47.8%), Black (46.0%), and Hispanic (36.8%) than non-Hispanic White patients with ovarian cancer (24.6%, P < .001).
FIG 4.
Trends by diagnosis year and quarter (Q) in the percent of VUS-only and PV results in any gene among testers by selected racial or ethnic groups with (A) breast cancer and (B) ovarian cancer. PV, pathogenic variant; VUS, variant of unknown significance only.
Multivariable logistic regressions were performed separately for tested patients with breast cancer and ovarian cancer, regressing the VUS-only result against date of diagnosis, race or ethnicity, age, stage, and geographic site. In both models, date of diagnosis and race or ethnicity were significant predictors of VUS-only results (all P < .001). There was no significant interaction between race or ethnicity and date, confirming no significant change in racial or ethnic differences across the study period (Data Supplement).
DISCUSSION
We studied receipt of genetic testing and outcomes for nearly 200,000 patients with breast cancer and 15,000 patients with ovarian cancer diagnosed from 2013 to 2017. We examined test trends over time that integrated information from repeat testing and reinterpretation of VUS results in individual patients through 2019. Consistent with our hypotheses, we observed marked expansion in the number of genes sequenced; a modest trend in selection toward patients with lower pretest risk; no sociodemographic differences in testing trends; a small increase in PV rates; and a substantial increase in VUS-only rates. Contrary to our hypotheses, we observed a major, sustained deficit in testing of patients with ovarian cancer (only 34.3% v nearly 100% recommended) and no evidence of reduction in the racial or ethnic disparity in VUS: these persistent gaps are key targets for intervention.
We found that the trend toward sequencing more genes, which we observed in 2013-2014,15 progressed to near-total replacement of BRCA1/2-only with MGP testing by 2019. The mean number of genes tested per patient increased over time but did not vary meaningfully by race or ethnicity and other demographic or clinical factors. Receipt of testing increased in older versus younger patients, which suggested a decreasing pretest probability of PV carriage over time among those tested, potentially supported by emerging studies of PV prevalence and clinical utility.2,7,20-22 Recent studies that found little evidence for patient worry or other psychological harms of testing might have offered reassurance about sequencing more genes in lower-risk patients.23-25
Ovarian is the cancer for which genetic testing has been most unequivocally indicated for more than 10 years. The prevalence of BRCA1/2 PVs is higher in ovarian than in breast, prostate, pancreatic, or other cancers26,27; germline-targeted therapy with a PARP inhibitor was first approved for ovarian cancer28; guidelines have advised, and most insurance has covered testing all patients with high-grade, serous epithelial carcinoma of the ovary, fallopian tube, and/or peritoneum since 2009.2,29 Despite these very strong indications, we previously identified substantial undertesting of patients with ovarian cancer diagnosed in 2013-2014,16 but we anticipated that mounting evidence for PARP inhibitors30-33 and declining test costs would quickly close this gap. Strikingly and concerningly, however, follow-up through early 2019 reveals virtually no improvement in testing rates, which remain at only 34.3% of patients with ovarian cancer. We previously identified characteristics associated with less testing, including Black race, greater poverty, and less insurance.16 There is urgent need to further define the patient, clinician, and healthcare system factors that limit testing of patients with ovarian cancer and to develop interventions that surmount these barriers.
The yield of PVs changed over time. For patients with breast cancer, the proportion of all PVs that were in BRCA1/2 fell substantially as adoption of MGP testing doubled the probability of detecting a PV in any tested gene. A lower pretest probability of BRCA1/2 PV carriage might have also contributed. Most of the PV increase was in genes with an established breast or ovarian cancer association as noted by National Comprehensive Cancer Network guidelines2; fewer PVs were found in other genes designated by ACMG guidelines as actionable.19 By contrast, very few tested patients had a PV in other tested genes. Our findings suggest that, based on current understanding of breast and ovarian cancer genetics, testing a panel of 20 breast cancer–associated and/or ovarian cancer–associated genes may optimize the signal-to-noise ratio of PVs to VUS.
We found that the increase in VUS-only results outpaced the increase in PVs, although VUS reclassification has accelerated over time.34 Notably, these data incorporate VUS reclassification by laboratories through early 2019, so our findings offer a contemporary measure of the uncertainty that accompanies MGP testing. Our findings imply that population-based testing, which would include those with lower pretest PV probability than the clinically selected patients in this study, would exacerbate the proliferation of VUS over PVs.
We and others have previously reported that racial or ethnic minorities are more likely to receive a VUS-only result, much greater with MGP versus BRCA1/2-only testing.15,35-37 This VUS gap arises from the substantially larger volume of clinical genetic testing data from non-Hispanic White patients, such that the spectrum of normal variation is less well-defined in other racial or ethnic groups and they more often receive VUS results. Our findings now show that the racial or ethnic VUS gap did not close but rather widened over time, with Black and Asian patients having nearly twofold more VUS although they were not tested for more genes than non-Hispanic White patients. Questions remain about the impact of VUS on patients. Although some studies found little evidence of overtreatment because of VUS misinterpretation as PV, another study recently reported unwarranted risk–reducing salpingo-oophorectomy with VUS results.38,39 Moreover, VUS may exacerbate the challenge of family result communication and cascade genetic testing of relatives. It is essential to accelerate approaches to VUS reclassification, particularly as VUS results are increasing more markedly in racial or ethnic minorities than non-Hispanic White patients.
Aspects of our study warrant comment. Strengths include a very large, diverse, contemporary population-based sample that is relevant to clinically tested patients with breast and ovarian cancer, with testing and result classification updated through early 2019. Limitations include the lack of detailed genetic sequence data, because of privacy concerns, and the lack of patient and physician-reported data about test selection and results management. We previously confirmed by survey of patients from the participating SEER registries and clinicians practicing in these regions that the four participating laboratories performed the great majority of tests in the regions and years under study15,16; however, it is possible that some tests from other laboratories were missed. We studied two states only, which may not represent the whole United States.
Our findings suggest that a more delimited panel composition could improve the clinical validity and utility of genetic testing for women with breast cancer or ovarian cancer. Most PVs were found in 20 genes among patients with breast and ovarian cancer (ATM, BARD1, BRCA1, BRCA2, BRIP1, CDH1, CHEK2, EPCAM, MLH1, MSH2, MSH6, NBN, NF1, PMS2, PALB2, PTEN, RAD51C, RAD51D, STK11, and TP53); testing only these genes could maximize clinically relevant PV yield while minimizing the VUS results, particularly for racial or ethnic minority patients. Since 2012, genetic testing rates have increased annually by 2%, whereas the number of tested genes has increased annually by 28%. Quality improvement efforts should focus on closing the genetic testing gap in indicated patients, notably those with ovarian cancer, rather than adding more genes per test.
ACKNOWLEDGMENT
We thank Lynne S. Penberthy, MD and Valentina I. Petkov, MD, at the National Cancer Institute; Nicola Schussler at Information Management Services; Jill S. Dolinsky, MS, Lily Hoang, BA, and Amal Yussuf, BS, at Ambry Genetics; Delores Bowman, RN and Rachel Klein, MS, CGC, at Bioreference/GeneDx; Edward Esplin, MD, PhD, and Stephen Lincoln, PhD, at Invitae; and Krystal Brown, PhD, Jerry Lanchbury, PhD, Thomas P. Slavin, MD, and Brian Dechairo, PhD, at Myriad Genetics for their collaboration on the genetic test data linkage to SEER data. Written permission was obtained to include the names of all acknowledged individuals.
Allison W. Kurian
Research Funding: Myriad Genetics
Other Relationship: Ambry Genetics, Color Genomics, GeneDx/BioReference, InVitae, Genentech
Monica Morrow
Honoraria: Genomic Health, Roche/Genentech
Travel, Accommodations, Expenses: Genomic Health, Roche/Genentech
Jonathan S. Berek
Leadership: Oncoquest
Research Funding: Tesaro, Karyopharm Therapeutics
No other potential conflicts of interest were reported.
DISCLAIMER
The ideas and opinions expressed herein are those of the author(s) and do not necessarily reflect the opinions of the State of California, Department of Public Health, the National Cancer Institute, and the Centers for Disease Control and Prevention or their Contractors and Subcontractors.
PRIOR PRESENTATION
Presented in part at the 2020 ASCO Virtual Quality Care Symposium, October 9-10, 2020, and the 2020 Virtual San Antonio Breast Cancer Symposium, December 8-12, 2020.
SUPPORT
Supported by the National Cancer Institute (NCI) of the National Institutes of Health under award numbers P01 CA163233 and P30 CA046592 to the University of Michigan and R01 CA225697 to Stanford University. The collection of cancer incidence data in Georgia was supported by contract HHSN261201800003I, Task Order HHSN26100001 from the NCI, and cooperative agreement 5NU58DP006352-03-00 from the CDC. The collection of cancer incidence data used in this study was supported by the California Department of Public Health pursuant to California Health and Safety Code Section; 103885 Centers for Disease Control and Prevention's (CDC) National Program of Cancer Registries, under cooperative agreement 5NU58DP006344; and the National Cancer Institute's SEER Program under contract HHSN261201800032I awarded to the University of California, San Francisco, contract HHSN261201800015I awarded to the University of Southern California, and contract HHSN261201800009I awarded to the Public Health Institute, Cancer Registry of Greater California. Research funding to her institution for an unrelated project was provided to Allison Kurian, MD, MSc, by Myriad Genetics.
AUTHOR CONTRIBUTIONS
Conception and design: Allison W. Kurian, Kevin C. Ward, Paul Abrahamse, Dennis Deapen, Jonathan S. Berek, Timothy P. Hofer, Steven J. Katz
Financial support: Allison W. Kurian, Steven J. Katz
Administrative support: Steven J. Katz
Provision of study materials or patients: Allison W. Kurian, Kevin C. Ward, Ann S. Hamilton, Steven J. Katz
Collection and assembly of data: Allison W. Kurian, Kevin C. Ward, Paul Abrahamse, Ann S. Hamilton, Dennis Deapen, Steven J. Katz
Data analysis and interpretation: Allison W. Kurian, Kevin C. Ward, Paul Abrahamse, Irina Bondarenko, Dennis Deapen, Monica Morrow, Jonathan S. Berek, Timothy P. Hofer, Steven J. Katz
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS’ DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Time Trends in Receipt of Germline Genetic Testing and Results for Women Diagnosed With Breast Cancer or Ovarian Cancer, 2012-2019
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Allison W. Kurian
Research Funding: Myriad Genetics
Other Relationship: Ambry Genetics, Color Genomics, GeneDx/BioReference, InVitae, Genentech
Monica Morrow
Honoraria: Genomic Health, Roche/Genentech
Travel, Accommodations, Expenses: Genomic Health, Roche/Genentech
Jonathan S. Berek
Leadership: Oncoquest
Research Funding: Tesaro, Karyopharm Therapeutics
No other potential conflicts of interest were reported.
REFERENCES
- 1.Katz SJ, Kurian AW, Morrow M.Treatment decision making and genetic testing for breast cancer: Mainstreaming mutations JAMA 314997–9982015 [DOI] [PubMed] [Google Scholar]
- 2.Daly MB, Pilarski R, Yurgelun MB, et al. NCCN guidelines insights: Genetic/familial high-risk assessment: Breast, ovarian, and pancreatic, version 1.2020 J Natl Compr Canc Netw 18380–3912020 [DOI] [PubMed] [Google Scholar]
- 3.Gupta S, Provenzale D, Regenbogen SE, et al. NCCN guidelines insights: Genetic/familial high-risk assessment: Colorectal, version 3.2017 J Natl Compr Canc Netw 151465–14752017 [DOI] [PubMed] [Google Scholar]
- 4.Couch FJ, Shimelis H, Hu C, et al. Associations between cancer predisposition testing panel genes and breast cancer JAMA Oncol 31190–11962017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Bono J, Mateo J, Fizazi K, et al. Olaparib for metastatic castration-resistant prostate cancer N Engl J Med 3822091–21022020 [DOI] [PubMed] [Google Scholar]
- 6.Golan T, Hammel P, Reni M, et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer N Engl J Med 381317–3272019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robson M, Im SA, Senkus E, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation N Engl J Med 377523–5332017 [DOI] [PubMed] [Google Scholar]
- 8.Ledermann J, Harter P, Gourley C, et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer N Engl J Med 3661382–13922012 [DOI] [PubMed] [Google Scholar]
- 9.Palmer JR, Polley EC, Hu C, et al. Contribution of germline predisposition gene mutations to breast cancer risk in African American women J Natl Cancer Inst 1121213–12212020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robson M, Domchek S. Broad application of multigene panel testing for breast cancer susceptibility—Pandora's Box is opening wider. JAMA Oncol. [epub ahead of print on October 3, 2019] [DOI] [PubMed]
- 11.Kilbride MK, Domchek SM, Bradbury AR.Ethical implications of direct-to-consumer hereditary cancer tests JAMA Oncol 41327–13282018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robson ME, Bradbury AR, Arun B, et al. American Society of Clinical Oncology policy statement update: Genetic and genomic testing for cancer susceptibility J Clin Oncol 333660–36672015 [DOI] [PubMed] [Google Scholar]
- 13.Bradbury AR, Patrick-Miller L, Domchek S.Multiplex genetic testing: Reconsidering utility and informed consent in the era of next-generation sequencing Genet Med 1797–982015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kilbride MK, Bradbury AR.The need to improve the clinical utility of direct-to-consumer genetic tests: Either too narrow or too broad JAMA 3231443–14442020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurian AW, Ward KC, Hamilton AS, et al. Uptake, results, and outcomes of germline multiple-gene sequencing after diagnosis of breast cancer JAMA Oncol 41066–10722018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurian AW, Ward KC, Howlader N, et al. Genetic testing and results in a population-based cohort of breast cancer patients and ovarian cancer patients J Clin Oncol 371305–13152019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.United States Department of Health and Human Services Guidance regarding methods for de-identification of protected health information in accordance with the Health Insurance Portability and Accountability Act (HIPAA) privacy rule. https://www.hhs.gov/hipaa/for-professionals/privacy/special-topics/de-identification/index.html#safeharborguidance
- 18.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology Genet Med 17405–4242015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalia SS, Adelman K, Bale SJ, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): A policy statement of the American College of Medical Genetics and Genomics Genet Med 19249–2552017 [DOI] [PubMed] [Google Scholar]
- 20.Desmond A, Kurian AW, Gabree M, et al. Clinical actionability of multigene panel testing for hereditary breast and ovarian cancer risk assessment JAMA Oncol 1943–9512015 [DOI] [PubMed] [Google Scholar]
- 21.Tung N, Lin NU, Kidd J, et al. Frequency of germline mutations in 25 cancer susceptibility genes in a sequential series of patients with breast cancer J Clin Oncol 341460–14682016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Litton JK, Rugo HS, Ettl J, et al. Talazoparib in patients with advanced breast cancer and a germline BRCA mutation N Engl J Med 379753–7632018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bradbury AR, Patrick-Miller LJ, Egleston BL, et al. Patient feedback and early outcome data with a novel tiered-binned model for multiplex breast cancer susceptibility testing Genet Med 1825–332016 [DOI] [PubMed] [Google Scholar]
- 24.Bradbury AR, Patrick-Miller LJ, Egleston BL, et al. Randomized noninferiority trial of telephone vs in-person disclosure of germline cancer genetic test results J Natl Cancer Inst 110985–9932018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katz SJ, Ward KC, Hamilton AS, et al. Association of germline genetic test type and results with patient cancer worry after diagnosis of breast cancer JCO Precis Oncol 2018PO.18.002252018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Norquist BM, Harrell MI, Brady MF, et al. Inherited mutations in women with ovarian carcinoma JAMA Oncol 2482–4902016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lilyquist J, LaDuca H, Polley E, et al. Frequency of mutations in a large series of clinically ascertained ovarian cancer cases tested on multi-gene panels compared to reference controls Gynecol Oncol 147375–3802017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim G, Ison G, McKee AE, et al. FDA approval summary: Olaparib monotherapy in patients with deleterious germline BRCA-mutated advanced ovarian cancer treated with three or more lines of chemotherapy Clin Cancer Res 214257–42612015 [DOI] [PubMed] [Google Scholar]
- 29.American College of Obstetricians and Gynecologists ACOG Practice Bulletin No. 103: Hereditary breast and ovarian cancer syndrome Obstet Gynecol 113957–9662009 [DOI] [PubMed] [Google Scholar]
- 30.Pujade-Lauraine E, Ledermann JA, Selle F, et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): A double-blind, randomised, placebo-controlled, phase 3 trial Lancet Oncol 181274–12842017 [DOI] [PubMed] [Google Scholar]
- 31.Matulonis UA, Penson RT, Domchek SM, et al. Olaparib monotherapy in patients with advanced relapsed ovarian cancer and a germline BRCA1/2 mutation: A multistudy analysis of response rates and safety Ann Oncol 271013–10192016 [DOI] [PubMed] [Google Scholar]
- 32.Swisher EM, Lin KK, Oza AM, et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): An international, multicentre, open-label, phase 2 trial Lancet Oncol 1875–872017 [DOI] [PubMed] [Google Scholar]
- 33.Mirza MR, Monk BJ, Herrstedt J, et al. Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer N Engl J Med 3752154–21642016 [DOI] [PubMed] [Google Scholar]
- 34.Mersch J, Brown N, Pirzadeh-Miller S, et al. Prevalence of variant reclassification following hereditary cancer genetic testing JAMA 3201266–12742018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hall MJ, Reid JE, Burbidge LA, et al. BRCA1 and BRCA2 mutations in women of different ethnicities undergoing testing for hereditary breast-ovarian cancer Cancer 1152222–22332009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manrai AK, Funke BH, Rehm HL, et al. Genetic misdiagnoses and the potential for health disparities N Engl J Med 375655–6652016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caswell-Jin JL, Gupta T, Hall E, et al. Racial/ethnic differences in multiple-gene sequencing results for hereditary cancer risk Genet Med 20234–2392018 [DOI] [PubMed] [Google Scholar]
- 38.Kurian AW, Ward KC, Abrahamse P, et al. Association of germline genetic testing results with locoregional and systemic therapy in patients with breast cancer. JAMA Oncol. 2020;6:e196400. doi: 10.1001/jamaoncol.2019.6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Domchek SM, Brower J, Symecko H, et al. Uptake of oophorectomy in women with findings on multigene panel testing: Results from the Prospective Registry of Multiplex Testing (PROMPT) J Clin Oncol38, 2020 (suppl; abstr 1508) [Google Scholar]