SUMMARY
Although accumulation of DNA damage and genomic instability in resting cells can cause neurodegenerative disorders, our understanding of how transcription produces DNA double-strand breaks (DSBs) is limited. Transcription-blocking topoisomerase I cleavage complexes (TOP1ccs) are frequent events that prime DSB production in non-replicating cells. Here, we report a mechanism of their formation by showing that they arise from two nearby single-strand breaks (SSBs) on opposing DNA strands: one SSB from the removal of transcription-blocking TOP1ccs by the TDP1 pathway and the other from the cleavage of R-loops by endonucleases, including XPF, XPG, and FEN1. Genetic defects in TOP1cc removal (TDP1, PNKP, and XRCC1) or in the resolution of R-loops (SETX) enhance DSB formation and prevent their repair. Such deficiencies cause neurological disorders. Owing to the high frequency of TOP1cc trapping and the widespread distribution of R-loops, these persistent transcriptional DSBs could accumulate over time in neuronal cells, contributing to the neurodegenerative diseases.
INTRODUCTION
In metazoans, most cells are slow- or non-replicating, and the accumulation of DNA damage and genomic instability in such cells can cause several human diseases, including neurodegenerative syndromes (McKinnon, 2017; Rass et al., 2007). Even so, exactly how DNA double-strand breaks (DSBs), the most harmful genomic lesions, are generated in resting cells is unclear. Increasing evidence indicates that transcription can induce DNA damage and genomic instability, particularly through R-loop structures (Aguilera and García-Muse, 2012; Aguilera and Gómez-González, 2017; Sollier and Cimprich, 2015). Furthermore, co-transcriptional R-loops have been associated with neurodegenerative diseases (Groh and Gromak, 2014). R-loops consist of a RNA/DNA hybrid and a displaced single-strand DNA, formed by the reannealing of the nascent transcript with the transcribed DNA strand behind the elongating RNA polymerase II (Pol II). R-loops are widespread dynamic structures (Sanz et al., 2016). They occur naturally during transcription and have important physiological functions, such as gene expression, DNA replication, and repair (Santos-Pereira and Aguilera, 2015; Skourti-Stathaki and Proudfoot, 2014; Sollier and Cimprich, 2015). Several factors regulate R-loop homeostasis and so prevent the accumulation of unscheduled R-loops and DNA damage, including topoisomerase I (TOP1) (Drolet et al., 2003; El Hage et al., 2010; Manzo et al., 2018) and mRNA processing factors (Huertas and Aguilera, 2003; Li and Manley, 2005). Once formed, R-loops can be removed by RNase H enzymes, which degrade the RNA in the RNA/DNA hybrids (Cerritelli and Crouch, 2009), and by RNA/DNA helicases, such as senataxin (SETX) (Groh et al., 2017; Skourti-Stathaki et al., 2011), aquarius (AQR) (Sollier et al., 2014), and DHX9 (Cristini et al., 2018), which unwind the RNA/DNA hybrids.
Deregulation of TOP1 activity is a source of transcription-associated genomic instability, also associated with neurodegenerative disorders (Jiang et al., 2017; Katyal et al., 2014; Pommier et al., 2016; Takashima et al., 2002). TOP1 solves DNA topological problems arising during transcription. It relaxes both positive and negative DNA supercoiling generated ahead and behind the elongating Pol II, respectively, by forming transient TOP1 cleavage complexes (TOP1ccs), which are TOP1linked single-strand breaks (SSBs) (Pommier et al., 2016). After DNA relaxation, TOP1ccs reverse rapidly and TOP1 is released. These transient TOP1ccs are readily stabilized or “trapped” on chromatin under physiological or pathological conditions, giving rise to irreversible TOP1ccs. DNA alterations, including oxidative lesions, base mismatches, modifications and losses, and DNA breaks, can trap TOP1ccs (Huang et al., 2017; Pommier et al., 2016). Genetic defects in TDP1, ATM, and XRCC1 proteins cause neurological disorders and result in long-lived TOP1ccs (Hoch et al., 2017; Jiang et al., 2017; Katyal et al., 2014; Pommier et al., 2016; Takashima et al., 2002). TOP1ccs are selectively trapped by camptothecin (CPT) and its derivatives used as anticancer agents (Pommier, 2006).
Trapped TOP1ccs are potent transcription-blocking DNA lesions (Capranico et al., 2007; Pommier, 2006), and their repair depends primarily on the TDP1 excision pathway (Cristini et al., 2016; El-Khamisy et al., 2005; Miao et al., 2006; Yang et al., 1996). Although TOP1 depletion increases R-loop levels (Drolet et al., 2003; El Hage et al., 2010; Manzo et al., 2018; Tuduri et al., 2009), the effect of TOP1cc trapping on R-loops is less well understood except for specific genes, including snord116 (Powell et al., 2013), frataxin in Friedreich ataxia cells (Groh et al., 2014), and β-actin (Cristini et al., 2018), where R-loops are increased in response to CPT and/or derivatives. A consequence of transcription blocking by TOP1ccs is the production of DSBs, which have been detected in non-replicating cells, such as post-mitotic neurons (Sordet et al., 2009) and quiescent fibroblasts (Cristini et al., 2016). Here, we describe how these DSBs are produced and so provide a molecular mechanism that strictly depends on transcription. We show that these DSBs can arise from two nearby SSBs on opposing DNA strands, both produced during transcription: one SSB results from removal of a transcription-blocking TOP1cc by the TDP1 pathway and the other from cleavage of an R-loop by structure-specific endonucleases. Notably, genetic defects in the transcription-blocking TOP1cc removal pathway or in the resolution of R-loops enhance DSB levels by increasing their production and preventing their repair and lead to cell death. Such defects may underlie neurological diseases. Given the frequency of TOP1ccs and the widespread distribution of R-loops, these persistent transcriptional DSBs will accumulate over time in neuronal cells and so contribute to neurodegenerative diseases.
RESULTS
Transcriptional DSBs Depend on SSB Intermediates Produced during TOP1cc Removal
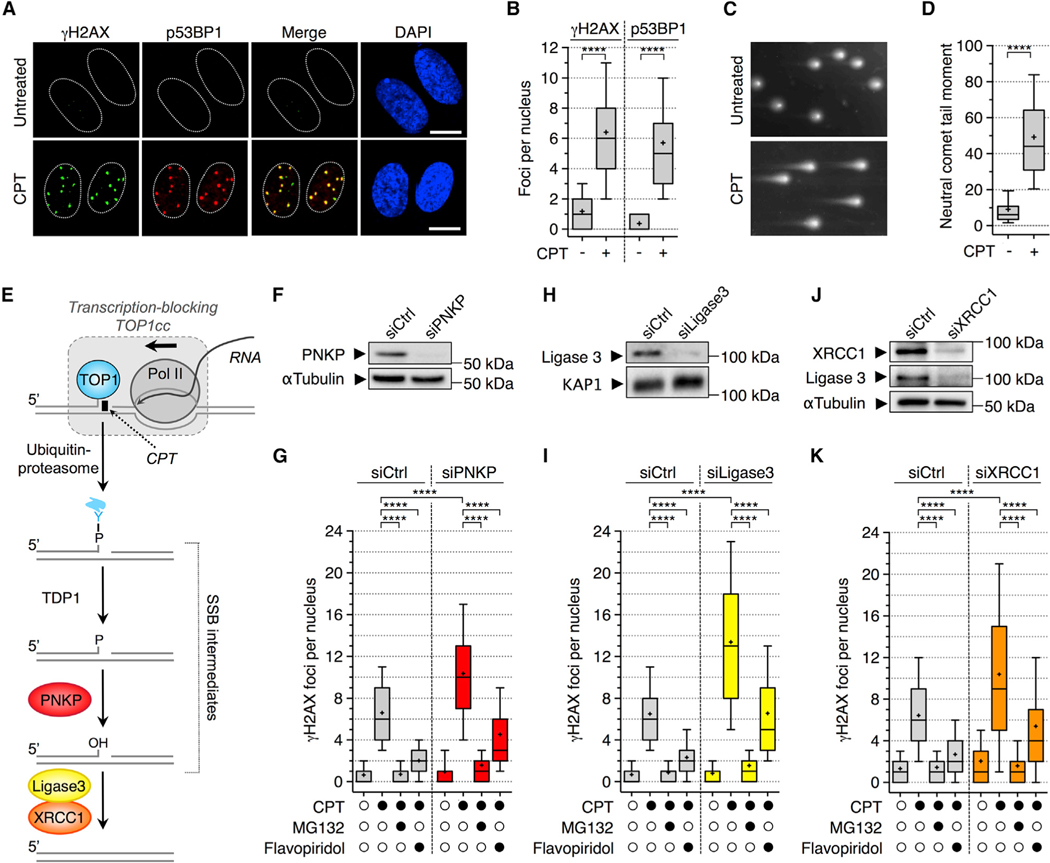
To investigate how DSBs are produced in response to transcription-blocking TOP1ccs, we induced these breaks by exposing quiescent WI38 hTERT fibroblasts to CPT (Cristini et al., 2016). CPT induces high levels of stabilized TOP1ccs (Pommier, 2006). Quiescence was controlled by the lack of 5-ethynyl-2’-deoxyuridine (EdU) incorporation into DNA (Figures S1A and S1B) as reported (Cristini et al., 2016). These transcriptional DSBs can be visualized by microscopy as nuclear foci containing γH2AX (phosphorylated H2AX at S139) and p53BP1 (phosphorylated 53BP1 at S1778; Figures 1A and 1B) and by neutral comet assays as increased comet tail moment (Figures 1C and 1D).
Figure 1. Depletion of PNKP, DNA Ligase 3, or XRCC1 Increases CPT-Induced Transcription- and Proteasome-Dependent DSBs in Quiescent Cells.
(A and B) Quiescent cells were treated with CPT (25 μM; 1 h) and co-stained for γH2AX (green) and p53BP1 (red).
(A) Representative confocal microscopy pictures. Yellow indicates colocalization. Dashed lines indicate nuclei. Scale bars: 10 μm.
(B) Number of γH2AX and p53BP1 foci per nucleus. ****p < 0.0001 (one-way ANOVA).
(C and D) Detection of DSBs by neutral comet assays in quiescent cells treated with CPT (25 μM; 1 h).
(C) Representative pictures of nuclei.
(D) Quantification of neutral comet tail moments. ****p < 0.0001 (two-tailed unpaired t test).
(E) Pathway for the removal of a transcription-blocking TOP1cc. Y: TOP1 catalytic tyrosine.
(F–K) Quiescent cells were transfected with the indicated siRNAs.
(F, H, and J) Western blot probed with PNKP (F), Ligase 3 (H), and XRCC1 and Ligase 3 (J). α-tubulin, KAP1: loading controls.
(G, I, and K) Cells transfected with siRNAs against PNKP (G), Ligase 3 (I), and XRCC1 (K) were treated with MG132 (25 μM) or flavopiridol (1 μM) for 1 h before the addition of CPT (25 μM; 1 h) and co-stained for γH2AX and p53BP1. The number of γH2AX foci per nucleus is shown. ****p < 0.0001 (one-way ANOVA). (B, D, G, I, and K) A representative experiment out of ≥3 is shown.
See also Figure S1.
We previously showed that TOP1 degradation by the ubiquitin-proteasome pathway is required for the production of transcriptional DSBs in CPT-treated quiescent cells (Cristini et al., 2016). TOP1 degradation primes the repair of transcription-blocking TOP1ccs, giving rise to a DNA single-strand break (SSB) intermediate, which is successively processed by TDP1 and PNKP at the 3’ end before DNA re-ligation by the DNA ligase 3-XRCC1 complex (Figure 1E). Here, we asked whether these SSB intermediates contribute to the formation of transcriptional DSBs.
We also reported that TDP1-depleted cells, which accumulate TOP1 peptide-linked SSB intermediates (Figure 1E; El-Khamisy et al., 2005; Interthal et al., 2005; Miao et al., 2006), accumulate transcriptional DSBs in response to CPT (Cristini et al., 2016). To further examine the role of SSB intermediates, we depleted PNKP and the DNA ligase 3-XRCC1 complex to accumulate cellular SSBs with 3’-P and 3’-OH, respectively (Figure 1E; Ashour et al., 2015; Pommier et al., 2014). RNAi-mediated knockdown of PNKP and DNA ligase 3 in quiescent cells increased the number of γH2AX and p53BP1 foci in response to CPT (Figures 1F–1I, S1C, and S1D), which correlated with increased SSBs in alkaline comet assays (Figures S1F and S1G). Inhibition of TOP1 degradation with the proteasome inhibitor MG132, which prevented the generation of these SSB intermediates (Ashour et al., 2015; Pommier et al., 2014), suppressed the induction of γH2AX foci by CPT upon depletion of PNKP and DNA ligase 3 (Figures 1G and 1I). Under these conditions, p53BP1 foci could not be assessed because MG132 prevents the ubiquitin-dependent accumulation of 53BP1 at DSB sites (Mailand et al., 2007). The transcriptional origin of these DSB-associated foci was established by the use of flavopiridol, an inhibitor of Pol II transcription (Figures 1G, 1I, S1C, and S1D). Likewise, depletion of XRCC1, which also decreased the levels of DNA ligase 3 (Figure 1J; Caldecott et al., 1995), induced a MG132- and flavopiridol-sensitive increase of γH2AX and/or p53BP1 foci in response to CPT (Figures 1K and S1E). Together, these results indicate that the SSB intermediates produced during the removal of transcription-blocking TOP1ccs give rise to DSBs in non-replicating cells.
The Formation of Transcriptional DSBs Depends on R-Loops
A possible mechanism for the formation of DSBs in non-replicating cells is that the SSB intermediate produced during the removal of a transcription-blocking TOP1cc would be converted into a DSB by a nearby SSB on the opposing DNA strand. Notably, R-loop structures that form co-transcriptionally in an unscheduled manner are prone to DNA breakage (Aguilera and Gómez-González, 2017; Sollier and Cimprich, 2015). We therefore determined whether R-loops could be linked to the production of a SSB on the opposing DNA strand.
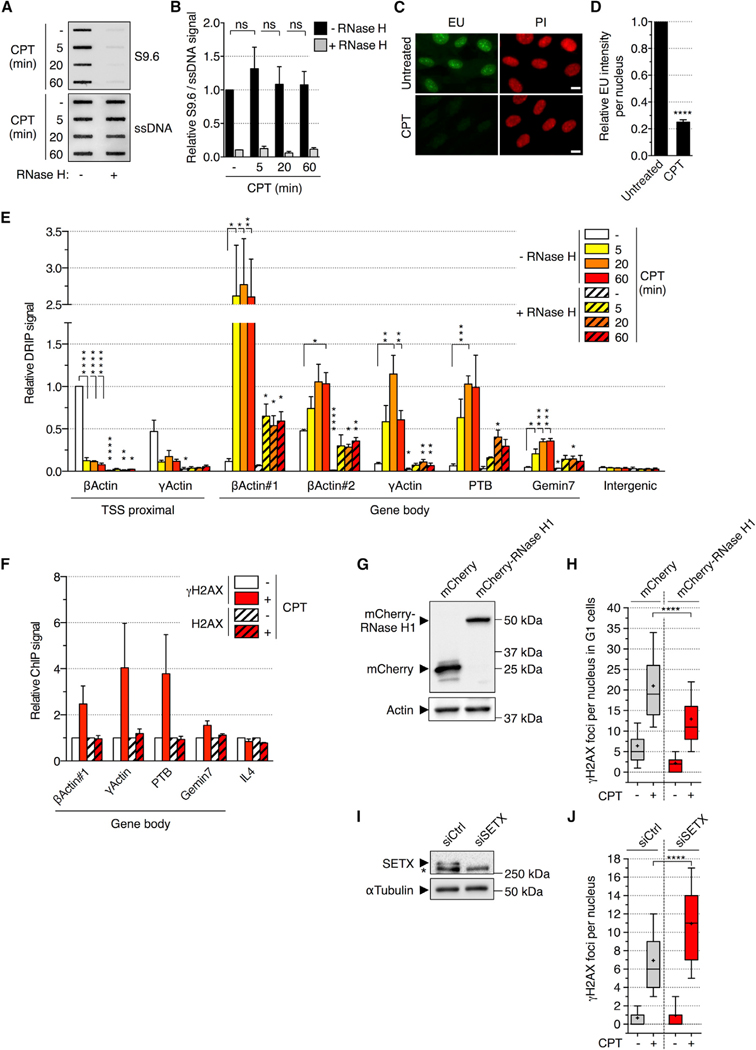
Although TOP1 depletion is known to increase R-loop levels (Drolet et al., 2003; El Hage et al., 2010; Manzo et al., 2018; Tuduri et al., 2009), the effect of TOP1cc trapping on R-loops is still unclear. To address this, quiescent cells were exposed to CPT and R-loop levels were assessed by a DNA slot blot assay employing S9.6 antibody (Boguslawski et al., 1986). CPT did not significantly affect S9.6 signal, albeit a small and transient increase was observed after 5 min, consistent with previous reports (Figures 2A and 2B; Marinello et al., 2013, 2016). RNase H treatment suppressed S9.6 signal, indicating its specificity for RNA/DNA hybrids (Figures 2A and 2B). These results indicate that TOP1cc trapping does not significantly increase R-loop levels. We therefore hypothesized that TOP1cc trapping could instead cause a genomic redistribution of R-loops. In support to this possibility, CPT readily inhibited transcription (Figures 2C and 2D; Cristini et al., 2016; Desai et al., 2003) and therefore could decrease the levels of physiological R-loops that have a rapid turnover (10–20 min) and are enriched 1 to 2 kb downstream of transcription start site (TSS) of expressed genes (Sanz et al., 2016). Conversely, CPT could increase R-loops in the body of genes where it primarily traps TOP1ccs and blocks transcription (Baranello et al., 2016; Solier et al., 2013; Sordet et al., 2008).
Figure 2. CPT Induces R-Loop-Dependent DSBs in the Absence of DNA Replication.
(A and B) RNA/DNA hybrid slot blot of genomic DNA ± RNase H from quiescent cells treated with 25 μM CPT.
(A) Representative slot blot. ssDNA: loading control.
(B) Values are normalized to ssDNA (means ± SEM; n ≥ 2). not significant (ns; two-tailed unpaired t test).
(C and D) Immunofluorescence (IF) analysis of 5-ethynyl uridine (EU) incorporation in quiescent cells treated with CPT (25 μM; 1 h) and incubated with 1 mM EU for the last 30 min of CPT treatment.
(C) Representative pictures. EU (green); propidium iodide (PI; red); scale bars: 10 μm.
(D) Values are normalized to untreated cells (means ± SEM; n = 3). ****p < 0.0001 (two-tailed unpaired t test).
(E) DRIP analysis in quiescent cells treated with 25 μM CPT. Values are normalized to β-actin “TSS proximal” amplicon of the “RNase H” sample from untreated cells (means ± SEM; n = 3). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 (two-tailed unpaired t test). Intergenic: negative control.
(F) γH2AX and H2AX ChIP analysis in quiescent cells treated with CPT (25 μM; 1 h). Values are normalized to untreated cells (means ± SEM; n ≥ 2). Interleukin-4 (IL-4): negative control.
(G and H) U2OS cells were treated with doxycycline to express mCherry or mCherry-RNase H1.
(G) Western blot of mCherry. Actin: loading control.
(H) Cells were incubated with 10 μM EdU for 30 min before treatment with CPT (25 μM; 1 h) and stained for γH2AX and Hoechst 33342 (DNA). The number of γH2AX foci per G1 nucleus (EdU-negative and low Hoechst 33342) is shown.
(I and J) Quiescent cells were transfected with siCtrl or siSETX.
(I) Western blot of SETX. *non-specific band. α-tubulin: loading control.
(J) siRNA-transfected cells were treated with CPT (25 μM; 1 h) and co-stained for γH2AX and p53BP1. The number of γH2AX foci per nucleus is shown.
A representative experiment out of 2 (H) or 3 (J) is shown. ****p < 0.0001 (two-tailed unpaired t test).
See also Figure S2.
Therefore, we examined R-loops at the TSS-proximal and gene body regions using RNA/DNA hybrid immunoprecipitation (DRIP) with the S9.6 antibody. Consistent with the prevalent localization of physiological R-loops at promoter-proximal regions (Sanz et al., 2016; Skourti-Stathaki et al., 2011), RNase H-sensitive DRIP signals were detected at TSS-proximal regions of β-actin and γ-actin genes in untreated quiescent cells (Figure 2E, white bars). CPT induced a rapid drop of DRIP signals at TSS-proximal regions together with an increase in the body of these genes (Figure 2E). Such increase of DRIP signals was also observed over PTB and Gemin7 genes (Figure 2E). These results indicate that TOP1cc trapping by CPT treatment increases R-loop levels over gene bodies.
To assess the role of R-loops in TOP1cc-dependent transcriptional DSBs, we investigated whether R-loops form at DSB sites. Chromatin immunoprecipitation (ChIP) analysis showed an enrichment of γH2AX after CPT exposure on β-actin, γ-actin, PTB, and Gemin7 genes (Figure 2F), coinciding with positions of R-loop enrichment (Figure 2E). To assess the direct consequences of R-loops, we tested whether modulating their cellular levels could impact DSB formation. First, we decreased R-loops by overexpressing RNase H1. We took advantage of U2OS cells inducible for mCherry or mCherry-RNase H1 fusion protein (Figure 2G; Britton et al., 2014). These cells cannot enter quiescence; hence, we analyzed DSBs in G1, prior to DNA replication. CPT-induced DSBs in G1 cells were also dependent on transcription and TOP1 degradation (Figure S2A), indicating that these DSBs are produced by a mechanism similar to quiescent cells (Figures 1 and S1). RNase H1 overexpression decreased the number of γH2AX foci induced by CPT in G1 cells (Figure 2H). Second, we increased R-loops by depleting SETX or AQR (Groh et al., 2017; Skourti-Stathaki et al., 2011; Sollier et al., 2014). Small interfering RNA (siRNA)-mediated knockdown of SETX or AQR increased the number of γH2AX and p53BP1 foci in CPT-treated quiescent cells (Figures 2I, 2J, and S2B– S2D). SETX depletion also increased DRIP signals over β-actin, PTB, and Gemin7 genes (Figure S2E). Together, these data indicate that R-loops contribute to the formation of TOP1cc-induced transcriptional DSBs in non-replicating cells.
Structure-Specific Endonucleases Cleave R-Loops to Produce Transcriptional DSBs
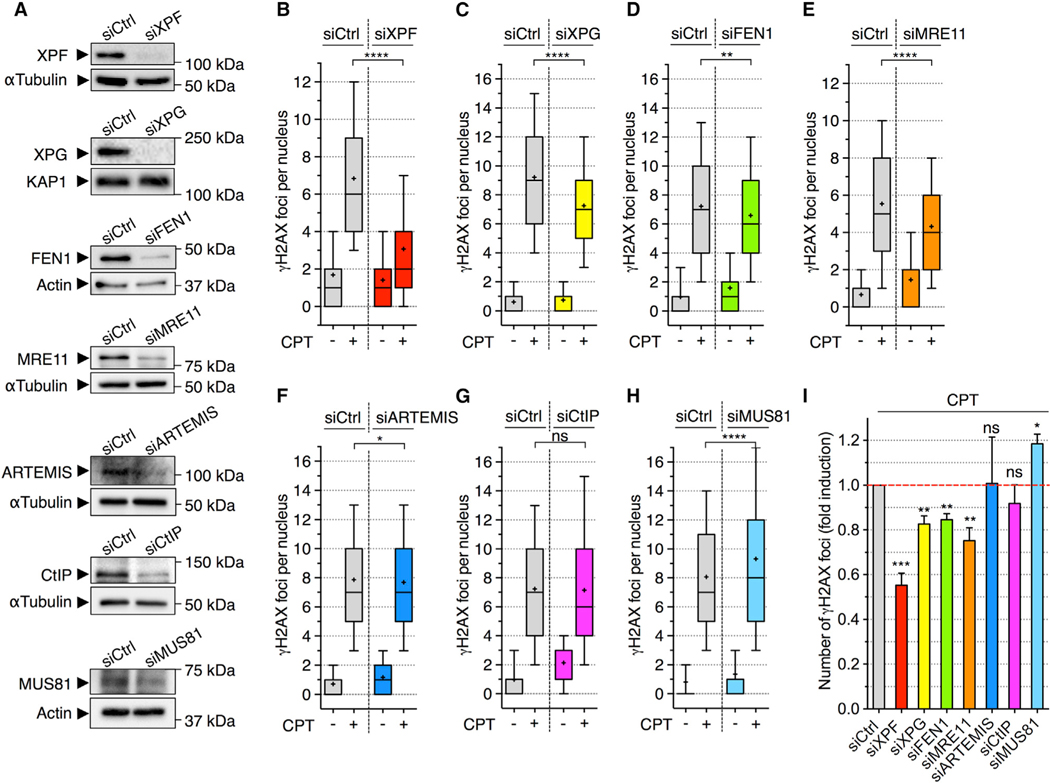
Next, we asked whether the SSBs in R-loops could contribute to DSB production. Because R-loops form single-stranded DNA structures with flap extremities, we tested whether the inhibition of structure-specific endonucleases that can cleave related structures can prevent the induction of DSBs in CPT-treated quiescent cells. XPF, XPG, FEN1, MRE11, ARTEMIS, CtIP, and MUS81 all possess flap endonuclease activity (Dehé and Gaillard, 2017; Regairaz et al., 2011). RNAi-mediated depletion of XPF, XPG, FEN1, or MRE11, but not of ARTEMIS, CtIP, or MUS81, reduced the induction of γH2AX and p53BP1 foci by CPT (Figures 3 and S3). These results support a role for R-loop cleavage in the production of TOP1cc-mediated transcriptional DSBs and further suggest that these breaks can be mediated by several endonucleases, including XPF, XPG, FEN1, and MRE11. To further examine the mechanism of endonuclease-mediated R-loop cleavage in the production of these DSBs, we initially focused on XPF because its depletion had the most severe defect in DSB induction (Figures 3 and S3).
Figure 3. Depletion of the Nucleases XPF, XPG, FEN1, and MRE11 Reduces the Induction of DSBs in CPT-Treated Quiescent Cells.
Quiescent cells were transfected with the indicated siRNAs.
(A) Western blot probed with the indicated antibodies. Actin, α-tubulin, and KAP1: loading controls.
(B–H) Cells transfected with siRNAs against XPF (B), XPG (C), FEN1 (D), MRE11 (E), ARTEMIS (F), CtIP (G), and MUS81 (H) were treated with CPT (25 μM; 1 h) and co-stained for γH2AX and p53BP1. The number of γH2AX foci per nucleus is shown. A representative experiment out of ≥3 is shown. The data with siCtrl cells in (D) and (G) are from the same experiment. ns, *p < 0.05, **p <0.01, ****p < 0.0001 (two-tailed unpaired t test).
(I) The fold induction of γH2AX was calculated by subtracting the number of foci of untreated cells from that of CPT-treated cells and normalized to siCtrl cells treated with CPT (means ± SEM; n ≥ 3). ns, *p < 0.05, **p < 0.01, ***p < 0.001 (two-tailed unpaired t test).
See also Figure S3.
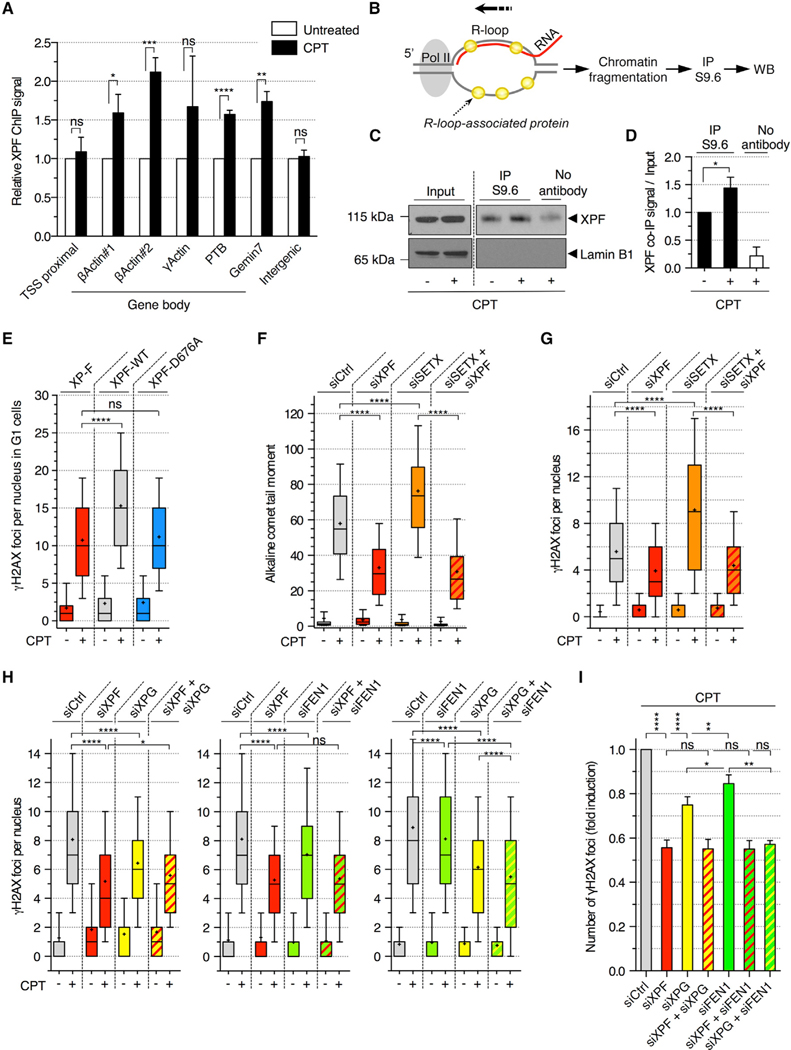
We first asked whether XPF is recruited to R-loops in response to CPT. ChIP analysis in quiescent cells showed an enrichment of XPF after CPT treatment over β-actin, γ-actin, PTB, and Gemin7 genes (Figure 4A), coinciding with R-loop accumulation (Figure 2E). To test whether XPF binds to R-loops, we used the R-loop immunoprecipitation (IP) method we recently developed, which allows the identification of RNA/DNA hybrid-interacting proteins in cells (Figure 4B; Cristini et al., 2018). XPF was enriched in RNA/DNA hybrid IP samples compared to the nuclear lamin B protein and “no antibody” IP samples in untreated quiescent cells (Figure 4C), indicating that XPF binds to R-loops under physiological conditions. Cellular exposure to CPT resulted in a further enrichment of XPF in the RNA/DNA hybrid IP (Figures 4C and 4D), and the quantity of RNA/DNA hybrid IPs was comparable (Figure S4A). The interaction of XPF with RNA/DNA hybrids is specific as it was reduced by RNase H treatment and prevented by synthetic RNA/DNA hybrid competitors added into IP reaction (Figures S4B–S4D). These results indicate that XPF is enriched on gene body R-loops in response to CPT in non-replicating cells.
Figure 4. R-Loop-Dependent DSBs Are Mediated by XPF, XPG, and FEN1 in Non-replicating Cells Treated with CPT.
(A) ChIP analysis of XPF in quiescent cells treated with CPT (25 μM; 20 min). Values are normalized to untreated cells (means ± SEM; n = 4). ns, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 (two-tailed unpaired t test). Intergenic: negative control.
(B) R-loop IP method for the detection of R-loop-associated proteins.
(C and D) R-loop IP was carried out in quiescent cells treated with CPT (25 μM; 20 min). Input and IP fractions were probed with XPF or lamin B1 antibodies.
(C) Representative immunoblots.
(D) Quantification of R-loop IP. Values were normalized to input and DMSO sample (means ± SEM; n = 3). *p < 0.05 (two-tailed unpaired t test).
(E) XPF-deficient patient cell line (XP-F) complemented with a wild-type (XPF-WT) or a nuclease-dead (XPF-D676A) XPF were incubated with 10 μM EdU for 30 min before treatment with CPT (25 μM; 1 h) and stained for γH2AX and Hoechst 33342 (DNA). The number of γH2AX foci per G1 nucleus (EdU-negative and low Hoechst 33342) is shown. ns, ****p < 0.0001 (two-tailed unpaired t test).
(F–I) Quiescent cells were transfected with the indicated siRNAs and treated with CPT (25 μM; 1 h).
(F) Detection of SSBs by alkaline comet assays. The quantification of comet tail moments is shown. ****p < 0.0001 (one-way ANOVA).
(G) Number of γH2AX foci per nucleus. ****p < 0.0001 (one-way ANOVA).
(H) Number of γH2AX foci per nucleus. ns, *p < 0.05, ****p < 0.0001 (one-way ANOVA).
(I) The fold induction of γH2AX was calculated by subtracting the number of foci of untreated cells from that of CPT-treated cells and normalized to siCtrl cells treated with CPT in each experiment (means ± SEM; n ≥ 3). ns, *p < 0.05, **p < 0.01, ****p < 0.0001 (two-tailed unpaired t test).
A representative experiment out of 2 (E and G) or R3 (F and H) is shown.
See also Figure S4.
To determine whether XPF cleaves R-loops to produce DSBs, we tested whether the endonuclease activity of XPF is required for this process. We analyzed γH2AX induction by CPT in the G1 population of XPF-deficient cells (XP-F) complemented with either a wild-type (XPF-WT) or a nuclease-dead (XPF-D676A) XPF (Staresincic et al., 2009). CPT increased the number of γH2AX foci in XPF-WT cells compared to XPF-deficient cells, and this increase was lost in XPF-nuclease-dead cells (Figure 4E). Thus, the nuclease activity of XPF is required for the production of TOP1cc-dependent transcriptional DSBs. To further test whether XPF cleaves R-loops, we analyzed the effects of XPF depletion on R-loop-dependent DNA breaks. By using alkaline comet assays to detect SSBs, we showed that increasing R-loop levels upon RNAi-mediated depletion of SETX resulted in an increase of SSBs in CPT-treated quiescent cells (Figure 4F). Such increase was suppressed by XPF depletion (Figure 4F), suggesting that XPF induces SSBs within R-loops. To evaluate the contribution of XPF-mediated R-loop cleavage to the formation of DSBs, we carried out the same experiments but analyzed DSBs instead of SSBs. XPF depletion also suppressed the increase of γH2AX foci induced by CPT upon SETX depletion (Figure 4G). Similar results were obtained by increasing R-loops upon AQR depletion (Figure S4E). Conversely, decreasing R-loops by RNase H1 overexpression did not further decrease the induction of γH2AX by CPT upon XPF depletion in the G1 population of U2OS cells (Figure S4F). Taken together, these results indicate that XPF can cleave R-loops formed upon TOP1cc trapping and that such cleavage can generate DSBs independently of replication.
To assess whether XPG and FEN1 act on the same R-loops as XPF to induce DSB formation, we depleted these endonucleases in combination in quiescent cells. Co-depletion of XPF with XPG or FEN1 did not further decrease the number of γH2AX foci induced by CPT than the depletion of XPF alone (Figures 4H, left and middle panels, 4I, and S4G), suggesting that XPF and XPG, as well as XPF and FEN1, act on the same R-loops. The FEN1 nuclease inhibitor myricetin (Ma et al., 2019) decreased the induction of γH2AX and p53BP1 foci by CPT (Figures S4H and S4I), further supporting the role of FEN1 in the cleavage of R-loops. Because XPF possesses a 3’-flap endonuclease activity and XPG and FEN1 a 5’-flap endonuclease activity (Dehé and Gaillard, 2017), these results suggest that R-loops are cleaved at both 3’- and 5’-extremities to induce DSB formation in CPT-treated quiescent cells. Then, the two 5’-flap endonucleases, XPG and FEN1, were depleted together to assess whether they act on the same R-loops. Co-depletion of XPG and FEN1 showed an additive effect in decreasing the number of γH2AX foci induced by CPT (Figures 4H, right panel, and 4I), suggesting that XPG and FEN1 act on different R-loops. Notably, co-depletion of XPG and FEN1 decreased the number of γH2AX to the same extent as the depletion of XPF alone (Figure 4I), further suggesting that XPG and FEN1 are the main 5’-flap endonucleases that act with the 3’-flap endonuclease XPF on the R-loops. Taken together, these results suggest that R-loops are cleaved at both extremities by a dual incision mediated by XPG and XPF or FEN1 and XPF to induce the formation of co-transcriptional DSB in CPT-treated quiescent cells.
The Formation of Transcriptional DSBs Requires the Dual Processing of TOP1ccs and R-Loops
Our data are consistent with the possibility that, in non-replicating cells, the SSB intermediates produced during the removal of transcription-blocking TOP1ccs are converted into DSBs by the presence of nearby SSBs on opposing DNA strand generated by the cleavage of R-loops. However, another possibility is that the endonucleases cleave R-loops on both strands, generating DSBs independently of TOP1cc-dependent SSB intermediates.
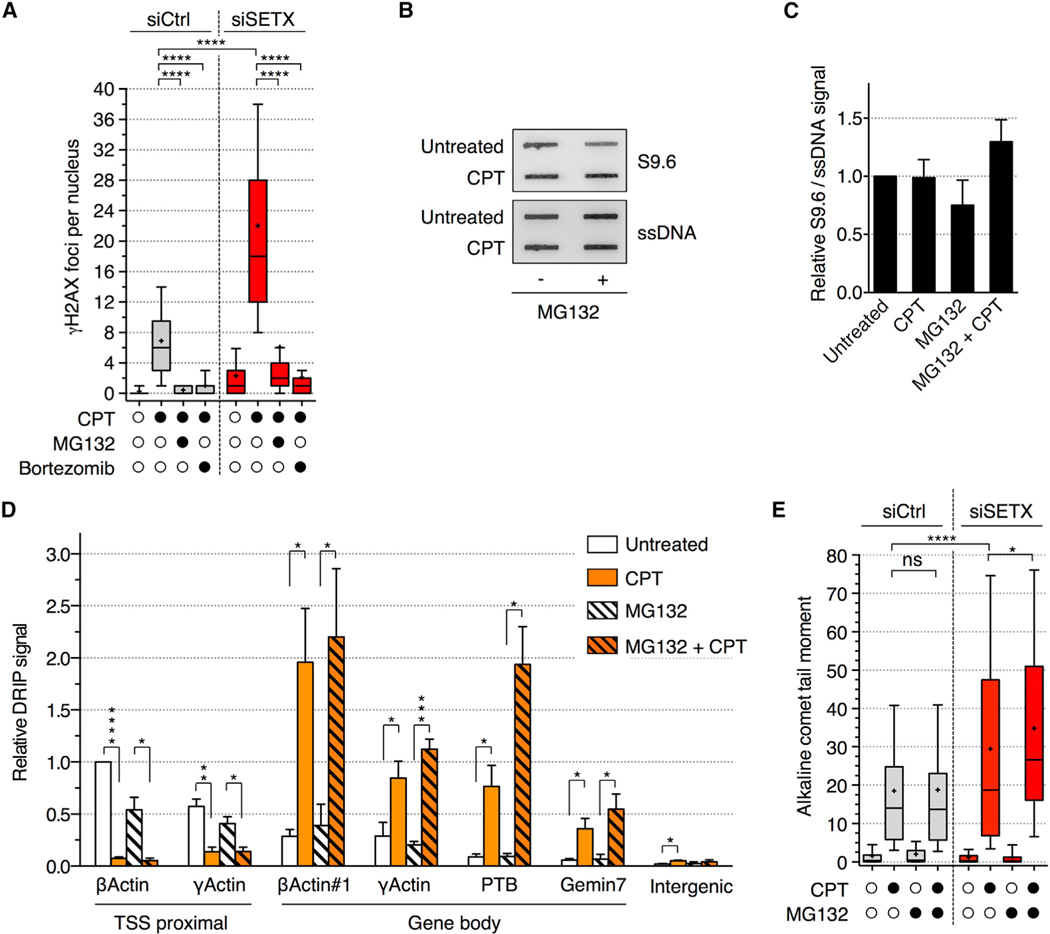
To distinguish between these two possibilities, we tested whether R-loop-dependent DSBs were also dependent on SSBs generated during TOP1cc removal. We therefore induced R-loop-dependent DSBs by depleting SETX in CPT-treated quiescent cells while inhibiting TOP1 proteolysis with proteasome inhibitors to prevent the formation of TOP1cc-induced SSB intermediates (Figure 1E; Ashour et al., 2015; Pommier et al., 2014). Proteasome inhibitors MG132 and bortezomib suppressed the induction of γH2AX foci by CPT upon SETX depletion (Figure 5A). Although SETX can be recruited at DSBs (Cohen et al., 2018), ChIP analysis showed that MG132 did not prevent SETX recruitment at the β-actin, γ-actin, and PTB genes (Figure S5), indicating that SETX recruitment to these loci is not due to DSBs but rather due to the presence of R-loops. Consistent with that, MG132 also did not reduce R-loop levels measured by DNA slot blot (Figures 5B and 5C) and by DRIP (Figure 5D), excluding the possibility that fewer R-loops account for the lack of DSB induction. In addition, alkaline comet assays showed that MG132 did not prevent the induction of SSBs within the R-loops (Figure 5E). These results indicate that, in non-replicating cells, the cleavage of R-loops alone is insufficient to generate DSBs and that it requires the formation of SSB intermediates produced during the removal of TOP1ccs. Altogether, these results support a model where, in non-replicating cells, a transcription-blocking TOP1cc is converted into a DSB by the presence of two SSBs on opposing DNA strand: one SSB resulting from TOP1cc removal and the other from cleavage of an R-loop by endonucleases.
Figure 5. Proteasome Inhibition Prevents R-Loop-Dependent DSBs in CPT-Treated Quiescent Cells.
(A) Quiescent cells were transfected with siCtrl or siSETX, treated with MG132 (25 μM; 1 h) or bortezomib (1 μM; 4 h) before the addition of CPT (25 μM; 1 h), and stained for γH2AX. The number of γH2AX foci per nucleus is shown.
(B and C) RNA/DNA hybrid slot blot of genomic DNA from quiescent cells treated with MG132 (10 μM; 1 h) before the addition of CPT (25 μM; 1 h), probed with S9.6 and ssDNA antibodies.
(B) Representative slot blot. ssDNA: loading control.
(C) Quantification of RNA/DNA hybrid slot blot. Values are normalized to ssDNA (means ± SEM; n = 4).
(D) DRIP analysis from quiescent cells treated as in (B) and (C). Values are normalized to β-actin TSS proximal from untreated cells (means ± SEM; n = 3). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 (two-tailed unpaired t test).
(E) Detection of SSBs by alkaline comet assays in cells transfected and treated as in (A). The quantification of comet tail moments is shown.
A representative experiment out of 2 is shown in (A) and (E); ns, *p < 0.05, ****p < 0.0001 (one-way ANOVA).
See also Figure S5.
Removal of TOP1ccs and R-Loops Allows the Repair of Transcriptional DSBs
Based on this model, TDP1-deficient cells should have DSBs with 3’-DNA broken strands attached to TOP1 peptides (i.e., TOP1 partially proteolyzed; see Figure 1E), which is likely to prevent DSB repair. Indeed, the DNA end must be cleansed to allow DSB repair by non-homologous end joining (NHEJ) (Kawale et al., 2018; Menon and Povirk, 2016), which is the main repair pathway in G0/G1 cells (Hendrickson, 1997). Similarly, cells with deficiencies in R-loop resolution factors, such as SETX, will have persistent RNA/DNA hybrids between the two SSBs, which would need to be resolved to allow DSB repair. Our next question was therefore whether cells deficient for one of these pathways, in addition to forming higher levels of transcriptional DSBs, would also be unable to repair them. We therefore analyzed the effects of TDP1 and SETX depletion on the repair of CPT-induced transcriptional DSBs.
To determine the kinetics of DSB repair, we analyzed the reversal kinetics of DSB-associated foci (Löbrich et al., 2010). Cells were exposed to CPT, washed free of drug, and γH2AX/p53BP1 foci were monitored post-CPT treatment (Figure 6A) as reported (Mamouni et al., 2014). In quiescent cells, the number of γH2AX and p53BP1 foci decreased rapidly after CPT removal. However, reversal kinetic was faster for p53BP1 foci than for γH2AX foci (Figures S6A and S6B). Neutral comet assays showed a reversal of about 80%, 1 h after CPT removal (Figures S6C and S6D), which correlates with p53BP1 reversal kinetics. Therefore, we chose p53BP1 to study DSB repair.
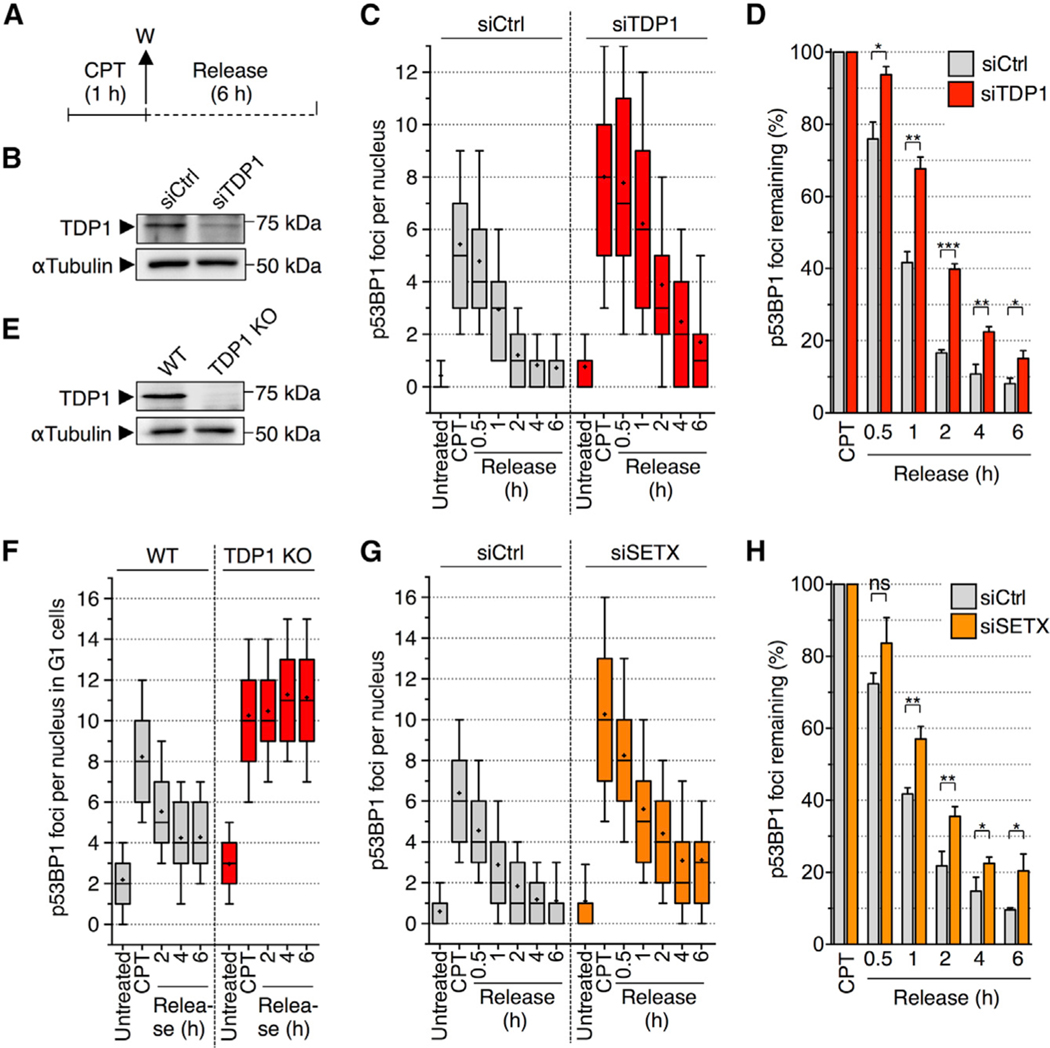
Figure 6. TDP1- and SETX-Deficient Cells Are Defective for the Repair of CPT-Induced Non-replicating DSBs.
(A) Protocol to study p53BP1 foci reversal following CPT removal used in (C), (D), and (F)–(H). Cells were treated with CPT (25 μM; 1 h), washed (W), and cultured in CPT-free medium for up to 6 h (release).
(B–D) Quiescent cells were transfected with siCtrl or siTDP1.
(B) Western blot of TDP1. α-tubulin: loading control.
(C) Number of p53BP1 foci per nucleus.
(D) The percentages of p53BP1 foci remaining following CPT removal were calculated by subtracting the number of foci of untreated cells from that of treated cells and normalized to cells treated with CPT (means ± SEM; n = 4). *p < 0.05, **p < 0.01, ***p < 0.001 (two-tailed unpaired t test). (E) Western blot of TDP1 in TDP1 WT and TDP1 KO HCT116 cells.
(F) p53BP1 foci in G1 nuclei (EdU-negative and low Hoechst 33342) of TDP1 WT and TDP1 KO HCT116 cells.
(G) Similar experiment as in (C) in quiescent cells transfected with siSETX.
(H) Similar experiment as in (D) in quiescent cells transfected with siSETX. Means ± SEM; n = 3. ns, *p < 0.05, **p < 0.01 (two-tailed unpaired t test). A representative experiment out of ≥2 is shown in (C), (F), and (G).
See also Figure S6.
RNAi-mediated depletion of TDP1 led to persistent p53BP1 foci after CPT removal in quiescent cells (Figures 6B–6D). Such a defect in p53BP1 reversal kinetics was also observed in the G1 population of HCT116 cells knockout (KO) for TDP1 (Figures 6E and 6F). Similarly, RNAi-mediated depletion of SETX led to persistent p53BP1 foci after CPT removal in quiescent cells (Figures 6G and 6H). These results indicate that cells deficient for TOP1cc removal or R-loop resolution form high levels of transcriptional DSBs due to their defective repair.
Besides NHEJ, DSBs can be repaired in G0/G1 cells by the single-strand annealing (SSA) pathway. SSA consists of the annealing of homologous repeat sequences and requires the RAD52 protein (Ceccaldi et al., 2016). RNAi-mediated depletion of RAD52 in quiescent cells did not affect the reversal kinetics of p53BP1 foci upon CPT removal (Figures S6E and S6F). These results suggest that, in quiescent cells, transcriptional DSBs are not repaired by the SSA pathway, thus supporting repair by NHEJ.
TOP1cc- and R-Loop-Dependent Transcriptional DSBs Accumulate under Physiological Conditions
CPT selectively traps TOP1ccs (Nitiss and Wang, 1988; Pommier, 2006), and hence, it is a precise tool to dissect the production mechanism of TOP1cc-dependent transcriptional DSBs. Besides CPT, a wide range of DNA alterations can trap TOP1ccs, and many of them are frequent (Pommier et al., 2016). To determine whether transcriptional DSBs could form under physiological conditions by a mechanism that relies on TOP1ccs and R-loop processing, we analyzed DSBs in quiescent cells under normal cultured conditions without CPT.
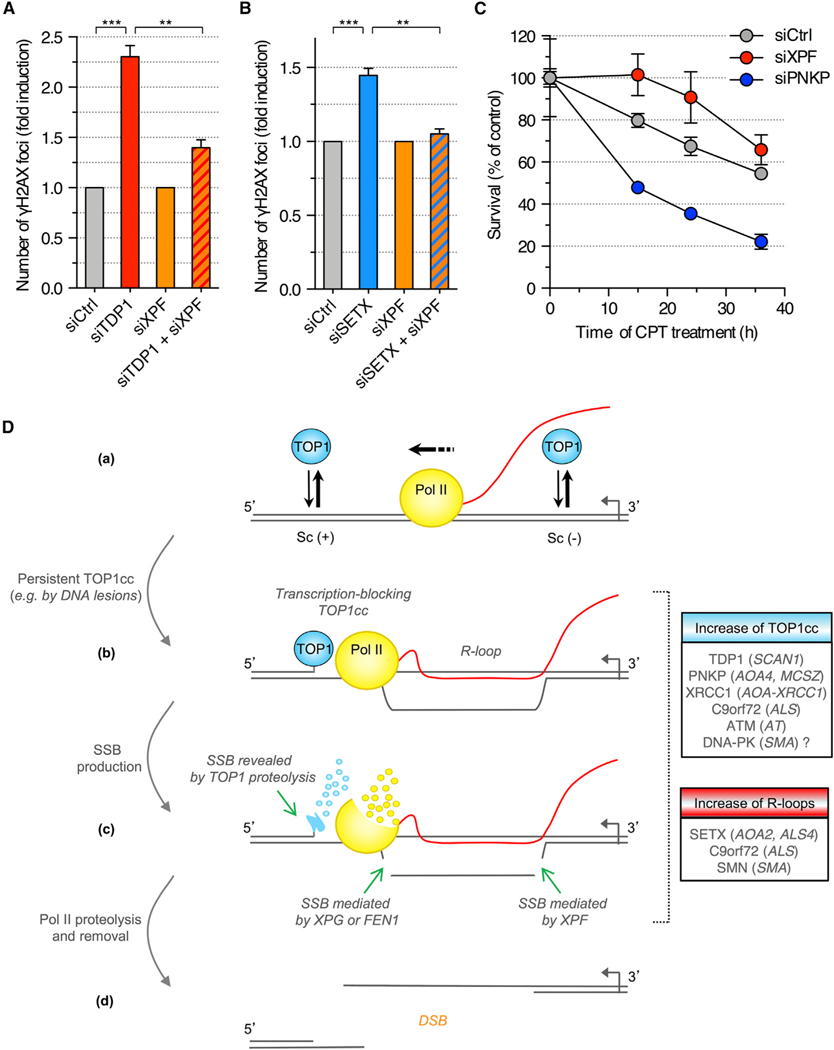
RNAi-mediated depletion of TDP1 (Figure 6B) increased the number of γH2AX and p53BP1 foci (Figures 7A, S7A, and S7C), indicating that SSB intermediates produced during TOP1cc removal promote DSB formation. Concurrent depletion of XPF suppressed the induction of γH2AX and p53BP1 foci upon TDP1 knockdown (Figures 7A, S7A, and S7C), suggesting that DSBs formed upon depletion of TDP1 depend on SSBs within the R-loops. To control that XPF cleaves R-loops to form DSBs, we increased R-loop levels and prevented their cleavage by depletion of SETX and XPF, respectively. The increase of γH2AX and p53BP1 foci upon depletion of SETX was abrogated by the concurrent depletion of XPF (Figures 7B, S7B, and S7C). Together, these results led us to conclude that TOP1cc-dependent transcriptional DSBs form under physiological conditions and involve the dual processing of TOP1cc and R-loops.
Figure 7. Depletion of XPF Prevents the Induction of DSBs upon Depletion of TDP1 in Quiescent Cells.
(A and B) Quiescent cells were transfected with the indicated siRNAs and co-stained for γH2AX and p53BP1. To compare XPF-proficient (siCtrl) and XPF-deficient (siXPF) cells upon depletion of TDP1 (A) or SETX (B), γH2AX induction was normalized to 1 in both siCtrl and siXPF cells (means ± SEM; n = 3). **p < 0.01, ***p < 0.001 (two-tailed unpaired t test).
(C) Viability of siRNA-transfected quiescent cells treated with 25 μM CPT (means ± SEM of triplicates).
(D) Model for the induction of DSBs during transcription. (a) TOP1 removes both positive and negative DNA supercoiling (Sc) generated during transcription. (b) Stabilization of a TOP1cc on the template strand blocks Pol II transcription and promotes R-loop formation. (c) The transcription-blocking TOP1cc is partially proteolyzed by the ubiquitin-proteasome system, which reveals the SSB. The R-loop is cleaved by a dual incision mediated by XPG and XPF or FEN1 and XPF (legend continued on next page) generating a SSB on the opposing DNA strand. Stalled Pol II is degraded and/or removed from the chromatin. (d) The two DNA strands separate forming a DSB. Boxes indicate human disorders associated with increase of TOP1cc (blue) or R-loop levels (red). Genes mutated in these disorders are indicated. ALS, amyotrophic lateral sclerosis; AOA, ataxia with oculomotor apraxia; AT, ataxia telangiectasia; SCAN, spinocerebellar ataxia with axonal neuropathy; SMA, spinal muscular atrophy; SMN, survival motor neuron.
See also Figure S7.
To assess whether these transcriptional DSBs affect the viability of non-replicative cells, we tested the effect of CPT-dependent DSB formation on the survival of quiescent cells. Increasing DSBs by depleting PNKP with siRNA (Figures 1F and 1G) decreased cell survival in response to CPT (Figure 7C), whereas decreasing DSBs by depleting XPF with siRNA (Figures 3A and 3B) increased cell survival (Figure 7C). These results highlight the physiological relevance of transcription-dependent DSBs induced by the dual processing of TOP1ccs and R-loops in non-replicating cells.
DISCUSSION
Genomic instability in resting cells is an emerging cause of neurodegenerative syndromes. Even so, our understanding of how DSBs are produced in non-replicating cells is limited. Here, we report a mechanism of DSB formation that strictly depends on transcription. DSBs are generated by two nearby SSBs on opposing DNA strands, both produced during transcription. One SSB arises from the repair of a transcription-blocking TOP1cc by the TDP1 pathway and the other from cleavage of an R-loop by endonucleases. Our data can be explained by the model in Figure 7D, where the SSB resulting from TOP1cc removal is on the transcribed DNA strand ahead of Pol II and the SSB in the R-loop is behind Pol II on the non-transcribed DNA strand displaced by the RNA/DNA hybrid. This polarity is in agreement with previous work showing that a TOP1cc located specifically on the transcribed strand blocks transcription (Bendixen et al., 1990; Wu and Liu, 1997) and is converted into an irreversible TOP1cc (Wu and Liu, 1997), the removal of which depends primarily on the TDP1 pathway (Cristini et al., 2016; El-Khamisy et al., 2005; Miao et al., 2006; Yang et al., 1996). In addition, R-loops form behind the elongating Pol II (Huertas and Aguilera, 2003), and the single-stranded nature of the displaced DNA strand within the R-loop makes it more vulnerable to DNA damage (Sollier and Cimprich, 2015).
An alternative model could be that the SSB generated during TOP1cc removal is located on the RNA/DNA hybrid within the R-loop or a few base pairs away from it. This is plausible as TOP1 removes negative DNA supercoiling behind the elongating Pol II (Pommier et al., 2016) and TOP1 co-immunoprecipitates with RNA/DNA hybrids (Cristini et al., 2018). However, it has not been reported that CPT can trap TOP1ccs in RNA/DNA hybrids and, if so, whether this would lead to irreversible TOP1ccs, the removal of which would depend on TDP1. Nevertheless, our results showing that the production of SSBs at R-loops is suppressed by XPF depletion (Figure 4F) argue against the possibility that TOP1ccs generate SSBs in RNA/DNA hybrids in our cell model.
Our data support that CPT increases R-loops in gene bodies, which is consistent with previous work showing that this is where TOP1ccs are primarily trapped (Baranello et al., 2016; Solier et al., 2013). Persistent TOP1ccs represent a barrier to the elongating Pol II (Bendixen et al., 1990; Sordet et al., 2008; Wu and Liu, 1997), which could promote R-loop formation by displacing spliceosomes (Tresini et al., 2015). Also, TOP1ccs could favor R-loop formation by increasing non-coding RNAs, such as antisense RNAs (Graf et al., 2017; Marinello et al., 2013, 2016; Nojima et al., 2018). A recent genome-wide analysis following TOP1 depletion shows R-loop gains in gene bodies (Manzo et al., 2018), suggesting that the decrease in TOP1 activity could also account for the effects of CPT on R-loops. Such a decrease in TOP1 activity could promote R-loop formation as a result of negative supercoiling accumulating behind the elongating Pol II (Drolet et al., 2003) and/or inhibition of the serine and argininerich (SR)-kinase activity of TOP1 interfering with splicing (Li and Manley, 2005; Tuduri et al., 2009).
Unscheduled R-loops can induce DSBs and genomic instability (Aguilera and García-Muse, 2012; Sollier and Cimprich, 2015). Our analysis in non-replicating cells suggests that a DSB does not form directly in the R-loop by the cleavage of both strands. Instead, a SSB is produced on the displaced DNA strand within the R-loop and the DSB is created by a second SSB next to the R-loop on the opposing DNA strand. In replicating cells, it is also plausible that a SSB within the R-loop could be converted into a DSB as the DNA replicates. Our data show that the SSB in the R-loop is mediated by the nucleases XPF, XPG, and FEN1. Consistently, XPF and XPG have been reported to cleave R-loops in vitro (Tian and Alt, 2000) and in cells (Sollier et al., 2014; Yasuhara et al., 2018), and FEN1 contributes to R-loop resolution during telomere replication (Teasley et al., 2015). XPF has a 3’-flap endonuclease activity, whereas XPG and FEN1 have a 5’-flap activity (Dehé and Gaillard, 2017). Our results from nuclease co-depletion experiments support a model in which R-loops are cleaved at both 3’- and 5’-extremities to induce DSBs by a dual incision mediated by XPF and XPG or XPF and FEN1. Although the cleavage at both ends of the R-loops might occur independently of each other, it is possible that it is more coordinated, e.g., that the cleavage at one end primes the cleavage at the other end. Although MRE11 is primarily involved in DSB repair by homologous recombination (Ceccaldi et al., 2016), it is unlikely that it contributes to the repair of transcriptional DSBs in quiescent cells due to the absence of sister chromatids for recombination. MRE11 has both an endonuclease and an exonuclease activity. It may therefore promote DSB formation by cleaving R-loops and/or by resecting DNA after a break is produced. Nuclease-independent functions of MRE11 may also contribute, including recruitment and activation of ATM (Buis et al., 2008), ultimately resulting in TOP1 degradation (Katyal et al., 2014) and further induction of transcriptional DSBs (this study; Cristini et al., 2016).
Although the two SSBs on opposing DNA strands can be distant from each other, it is plausible that they give rise to a DSB because the DNA strands in between, i.e., in the R-loop and the transcription bubble, are not annealed. Previous work also reported that Pol II arrested by TOP1ccs is released from chromatin (Sordet et al., 2008) and degraded by the proteasome (Desai et al., 2003), which would enable strand separation. DNA strand separation may be further facilitated by the removal of single-stranded DNA (ssDNA) fragments of R-loops that we report here. Lastly, although the DNA strands between TOP1ccs and stalled Pol II may be annealed, their length has been estimated in vitro to be 10 bp (Bendixen et al., 1990), and melting of the DNA duplex can readily occur over such a short length (Ran et al., 2013).
Non-replicative DSBs likely occur spontaneously in cells becauseTOP1ccscanbetrappedunderphysiologicalconditions, including oxidative base damage, alkylation and nicks (Pommier et al., 2014), and ribonucleotide misincorporation (Huang et al., 2015, 2017; Kim et al., 2011; Sparks and Burgers, 2015). Consistently, our data indicate that DSBs dependent on the processing of both TOP1ccs and R-loops are produced in non-replicating cells under physiological conditions (Figures 7A and 7B). Genetic defects in the transcription-blocking TOP1cc repair pathway (TDP1, PNKP, and XRCC1) and in the resolution of R-loops (SETX) are associated with genomic instability and cause neurological diseases, primarily cerebellar ataxia and amyotrophic lateral sclerosis (see Figure 7D; Dumitrache and McKinnon, 2017; Groh et al., 2017; Hoch et al., 2017; Takashima et al., 2002). This study reveals that such genetic defects enhance transcriptional DSB sin CPT-treated quiescent cells due to anincrease in their production and a defect in repair. This raises the possibility that these persistent DSBs would accumulate over time, contributing to the neurodegenerative phenotype. Consistently, this study (Figure 7C) and previous work (Cristini et al., 2016) showed that such DSBs and their modulation by factors controlling their production can kill non-replicating cells. A further connection between TOP1ccs, R-loops, and neurological disease has recently been reported. Cells bearing the C9orf72 expansion repeat, the most common genetic cause of amyotrophic lateral sclerosis, accumulate both TOP1ccsand R-loops, resulting in DSBs (Walker et al., 2017). In addition, deficiency of ATM or DNA-PK, which elevates TOP1cc levels (Cristini et al., 2016; Katyal et al., 2014), can also cause neurological diseases (Kannan et al., 2018; Savitsky et al., 1995). Neurons may be prone to produce transcriptional DSBs as a result of high rates of oxygen consumption, which produces reactive oxygen species that can stabilize TOP1ccs (Daroui et al., 2004; Pommier, 2006; Pommier et al., 2014). We also note that oxidative stress has been implicated in several neurodegenerative diseases (Barnham et al., 2004). Furthermore, DSBs may be particularly deleterious in neurons due to their reduced DNA repair capability as compared to proliferating cells (Rass et al., 2007).
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Olivier Sordet (olivier.sordet@inserm.fr). This study did not generate new unique reagents.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cells and Treatments
Primary human lung embryonic WI38 fibroblasts (from a 3-month-gestation aborted female fetus) immortalized with hTERT were obtained from Carl Mann (CEA, Gif-sur-Yvette, France) (Jeanblanc et al., 2012). Cells were cultured at 37°C in modified Eagle’s medium (MEM) supplemented with 10% (v/v) fetal bovine serum, 1 mM sodium pyruvate, 2 mM glutamine and 0.1 mM non-essential amino acids. To induce quiescence, cells were washed twice with serum-free medium and cultured for 72 h in medium supplemented as described above but with 0.2% (v/v) serum. To control the induction of quiescence in Figures S1A and S1B, cells were incubated with 10 μM EdU for 30 min and the incorporated EdU into DNA was detected using the Click-iT EdU Alexa Fluor 647 imaging kit (ThermoFisher Scientific) according to the manufacturer’s protocol. SV40-transformed human fibroblasts XP2Y0 (XPF-deficient, GM08437, from a 64-year-old female), XP2Y0 + XPF WT, and XP2Y0 + XPF-D676A were gifts from Orlando Schärer (Stony Brook University, NY, USA) (Staresincic et al., 2009) and cultured at 37°C in Dulbecco’s Modified Eagle’s Medium (DMEM) high glucose supplemented with 1 mM sodium pyruvate and 10% (v/v) fetal bovine serum. Human colon carcinoma HCT116 cells (from an adult male) parental and TDP1 KO (Al Abo et al., 2017) and human osteosarcoma U2OS cells (from a 15-year-old female) inducible for mCherry or mCherry-RNH1 (Britton et al., 2014) were cultured at 37°C in DMEM supplemented with 10% (v/v) fetal bovine serum. To express mCherry or mCherry-RNase H1, U2OS cells were treated with 2 μg/ml doxycycline for 24 h. Drugs and chemicals used for treatments are CPT, FLV, MG132, myricetin and doxycycline from Sigma-Aldrich, and bortezomib from Selleckchem. All these agents were dissolved in DMSO except for doxycycline, which was in water. In all the experiments, mock samples were treated with the vehicle only.
METHOD DETAILS
Immunofluorescence Microscopy
Cells were seeded in poly-L-lysine (Sigma-Aldrich)-coated 96-well plates (CellCarrier; PerkinElmer). After treatment, cells were washed twice with PBS and fixed with 3.7% paraformaldehyde for 15 min. After two washes with PBS, cells were permeabilized with 0.5% Triton X-100 for 15 min and washed twice with PBS. Cells were incubated with 8% bovine serum albumin (BSA) in PBS for 1 h before incubation with a mouse anti-γH2AX antibody (05–636; Millipore) and/or a rabbit anti-p53BP1 antibody (#2675; Cell Signaling Technology) diluted at 1/500 in 1% BSA in PBS for 2 h. Cells were washes three times with PBS and incubated with the appropriate secondary antibody coupled to Alexa Fluor 488, 594 or 647 (ThermoFisher Scientific) diluted at 1/500 in 1% BSA in PBS for 1 h. After three washes with PBS, nuclei were stained with 1 μg/ml Hoechst 33342 for 15 min, washed twice with PBS and stored at 4°C until analysis. To identify G1 nuclei, cells were incubated with 10 μM EdU for 30 min before treatment with CPT. The incorporated EdU into DNA was detected using the Click-iT EdU Alexa Fluor 647 Imaging Kit (ThermoFisher Scientific) according to the manufacturer’s protocol. After the Click-iT reaction, cells were processed for immunolabelling as described above starting at the 8% BSA step.
96-well plates were scanned with a 20X objective using an Operetta High-Content Imaging System or an Operetta CLS High-Content Imaging System (PerkinElmer) with Harmony software (version 4.1 or 4.8). After data acquisition, subsequent analyses were performed with Columbus software (version 2.5.0 or 2.8.2). γH2AX and p53BP1 foci were detected with the “C” method. Variations of the number of foci between independent experiments can be related to experimental variabilities, which include the automatic counting of foci with Columbus software that depends on the ratio between the signal intensity of foci and background noise. To compare independent experiments, the number of γH2AX/p53BP1 foci were normalized in each individual experiment as indicated in the figure legends. For graphical representation of foci distribution, we used box-and-whisker plots with GraphPad Prism 6 software with the following settings: boxes: 25–75 percentile range; whiskers: 10–90 percentile range; horizontal bars: median number of foci; “+”: mean number of foci.
In Figures 1A and 5A, cells were seeded in poly-L-lysine-coated Lab-Tek™ RS chamber slides (NalgeNunc), labeled as described above and slides were mounted using Mowiol 4–88 (Millipore) containing 4’,6-diamidino-2-phenylindole (DAPI). Slides were visualized with an inverted confocal microscope (LSM 780; ZEISS) with the objective Plan-Apochromat 63x / 1.4 Oil DIC III. In Figure 5A, γH2AX foci were counted with ImageJ (version 1.48v) as described previously (Cristini et al., 2016).
Chromatin Immunoprecipitation (ChIP)
Chromatin immunoprecipitation was performed as described previously (Cristini et al., 2018; Groh et al., 2014). Briefly, cells were crosslinked with 1% formaldehyde at 37C for 15 min and the reaction was stopped by adding 0.125 M glycine for 5 min. Pelleted cells were first subjected to lysis with cell lysis buffer (5 mM PIPES pH 8.0, 85 mM KCl, 0.5% NP-40 supplemented with 0.5 mM PMSF and 1X Complete EDTA-free protease inhibitors, Sigma-Aldrich) to isolate nuclei. Pelleted nuclei were then incubated in nuclear lysis buffer (50 mM Tris-HCl pH 8.0, 5 mM EDTA, 1% SDS supplemented with 0.5 mM PMSF and 1X Complete EDTA-free protease inhibitors, Sigma-Aldrich) and sonicated (Diagenode Bioruptor). After removal of insoluble material by centrifugation, samples were diluted with ChIP IP buffer (16.7 mM Tris-HCl pH 8.0, 1.2 mM EDTA pH 8.0, 167 mM NaCl, 0.01% SDS, 1.1% Triton X-100 supplemented with 0.5 mM PMSF and 1X Complete EDTA-free protease inhibitors, Sigma-Aldrich) and precleared by adding protein A agarose beads (16–157; Millipore). Chromatin was then incubated overnight at 4C with anti-γH2AX (07–164; Millipore), anti-H2AX (07–627; Millipore), anti-SETX (NB100–57542; Novus Biologicals), anti-XPF (ab76948; Abcam Lot#GR172751) antibodies or no antibody. Immunocomplexes were retrieved by incubation with protein A agarose beads (16–157; Millipore). Beads were then washed once with buffer A (20 mM Tris-HCl pH 8.0, 2 mM EDTA, 0.1% SDS, 1% Triton X-100 and 0.15 M NaCl), once with buffer B (20 mM Tris-HCl pH 8.0, 2 mM EDTA, 0.1% SDS, 1% Triton X-100 and 0.5 M NaCl), once with buffer C (10 mM Tris-HCl pH 8.0, 1 mM EDTA, 1% NP-40, 1% Sodium Deoxycholate and 0.25 M LiCl) and then twice with buffer D (10 mM Tris-HCl pH 8.0 and 1 mM EDTA). Beads were eluted in 1% SDS and 0.1 M NaHCO3 and samples decross-linked by addition of RNase A and NaCl (0.3 M) at 65C for at least 4 h. After 2 h proteinase K (Sigma-Aldrich) digestion at 45°C, DNA was purified with QIAquick PCR purification kit (QIAGEN), and analyzed by qPCR with Rotor-Gene Q (QIAGEN). The amount of immunoprecipitated material at a particular gene region was calculated as the percentage of input after subtracting the background signal (no antibody control). Unless otherwise stated, data were normalized to the DMSO-treated sample in each experiment, which was set equal to 1. The primers used for ChIP are listed in Table S1.
RNA/DNA Immunoprecipitation (DRIP)
RNA/DNA immunoprecipitation was carried out as described in (Cristini et al., 2018; Skourti-Stathaki et al., 2011) with the S9.6 antibody (Boguslawski et al., 1986). Briefly, isolated non-crosslinked nuclei were subjected to nuclear lysis (50 mM Tris-HCl pH 8.0, 5 mM EDTA, 1% SDS) and digestion with Proteinase K (Sigma-Aldrich) for 3 h at 55°C. Genomic DNA was precipitated, resuspended in IP dilution buffer (16.7 mM Tris-HCl pH 8.0, 1.2 mM EDTA, 167 mM NaCl, 0.01% SDS, 1.1% Triton X-100) and sonicated (Diagenode Bioruptor) to obtain DNA fragments of about 500 bp. Sonicated genomic DNA was then precleared with protein A agarose beads (16157; Millipore) in presence of protease inhibitors (0.5 mM PMSF, 0.8 μg/ml pepstatin A, 1 μg/ml leupeptin). 10 μg of genomic DNA were incubated overnight at 4°C with 15 μl of S9.6 antibody or no antibody control. Addition of beads, washes, and elution steps were carried out following the same procedure described for ChIP. RNase H sensitivity was performed by adding 1.7 U RNase H (M0297; NEB) per microgram of genomic DNA for 2.5 h at 37°C before IP step. The amount of immunoprecipitated material at a particular gene region was calculated as the percentage of input after subtracting the background signal (no antibody control). Unless otherwise stated, data were normalized to the β-actin TSS proximal probe of the DMSO-treated sample in each experiment, which was set equal to 1. The primers used for DRIP are listed in Table S1.
RNA/DNA Hybrid Slot Blot
Slot blot experiments were carried out as described (Cristini et al., 2018) using the S9.6 antibody (Boguslawski et al., 1986). Genomic DNA preparation and RNase H digestion were performed following the procedure described for DRIP. For loading control, 350 ng of genomic DNA were heated at 95°C for 10 min, loaded on the slot blot and the membrane was denatured for 10 min in 0.5 M NaOH, 1.5 M NaCl, and neutralized for 2 min in 0.5 M Tris-HCl pH 7.2, 1.5 M NaCl. The membrane was probed with an anti-ssDNA antibody (MAB3034; Millipore) after UV crosslinking and saturating. Images were acquired with LAS-4000 (Fujifilm). S9.6 and ssDNA signals were quantified using Image Studio Lite software (Li-COR Biosciences).
RNA/DNA Hybrid Co-immunoprecipitation
RNA/DNA hybrid co-IP was performed as described (Cristini et al., 2018) using the S9.6 antibody (Boguslawski et al., 1986). Briefly, non-crosslinked nuclei were incubated in RSB buffer (10 mM Tris-HCl pH 7.5, 200 mM NaCl, 2.5 mM MgCl2) with 0.2% sodium deoxycholate, 0.1% SDS, 0.05% sodium lauroyl sarcosinate and 0.5% Triton X-100, and sonicated for 10 min (Diagenode Bioruptor). Samples were then diluted 4 times in RSB with 0.5% Triton X-100 (RSB + T) before IP with the S9.6 antibody, bound to protein A dynabeads (Invitrogen), and preblocked with 0.5% BSA/PBS for 2 h. IP was performed in presence of 0.1 ng of RNase A (PureLink, Invitrogen) per microgram of genomic DNA. Beads were then washed 4 times with RSB + T and 2 times with RSB. IPs were eluted in 1x LDS (Invitrogen), 100 mM DTT for 10 min at 70°C for SDS-PAGE, and in 1% SDS and 0.1 M NaHCO3 for 30 min at room temperature for RNA/DNA hybrid slot blot. Where indicated, RNA/DNA hybrid competitors were added during the IP step at 1.3 mM concentration (Phillips et al., 2013). RNA/DNA hybrids were prepared as described in (Cristini et al., 2018; Phillips et al., 2013) (ssDNA- CGGTGTGAATCAGAC; ssRNA-GUCUGAUUCACACCG). RNA/DNA hybrid IP with RNase H treatment was carried out as described previously (Cristini et al., 2018). Proteins were separated on 4%–12% Bis-Tris or 3%–8% Tris-Acetate gels by SDS-PAGE (Invitrogen) and immunoblotted with anti-XPF (ab76948; Abcam Lot#GR172751) and anti-lamin B1 (ab16048; Abcam) antibodies. XPF signal was quantified by using ImageJ (version 1.52h). XPF IP signal was normalized to XPF input signal and the DMSO-treated sample was set to 1.
Cell Extracts and Immunoblotting
Cell extracts were obtained by lysing cells for 15 min at 4C in buffer containing 50 mM Tris-HCl (pH 8.0), 300 mM NaCl, 0.4% NP-40, 10 mM MgCl2 and 5 mM DTT, supplemented with protease and phosphatase inhibitors (Halt Protease & Phosphatase Inhibitor Cocktail; ThermoFisher Scientific). After centrifugation (10,000 x g, 20 min), supernatants were diluted (v/v) in 50 mM Tris-HCl (pH 8.0), 0.4% NP-40 and 5 mM DTT. Proteins were separated by SDS-PAGE and immunoblotted with the following antibodies at dilutions recommended by the manufacturer: anti-pan-actin (MAB1501; Millipore), anti-ARTEMIS (#13381; Cell Signaling Technology), antimCherry (GTX128509; GeneTex), anti-CtIP (A300–488A; Bethyl), anti-FEN1 (A300–255A; Bethyl), anti-KAP1 (A300–274A; Bethyl), anti-DNA ligase3 (A301–636A; Bethyl), anti-MRE11 (A303–998A; Bethyl), anti-MUS81 (ab14387; Abcam), anti-PNKP (A300–257A; Bethyl), anti-SETX (A301–104A; Bethyl), anti-TDP1 (H00055775-A01; Abnova Corporation), anti-atubulin (T5168; Sigma-Aldrich), anti-XPF (A301–315A; Bethyl), anti-XPG (A301–484A; Bethyl), and anti-XRCC1 (A300–065A; Bethyl). Immunoblotting was revealed by chemiluminescence using a ChemiDoc MP Imaging System (Bio-Rad).
Comet Assays
Neutral and alkaline comet assays were performed according to the manufacturer’s instructions (Trevigen), except that electrophoresis was performed at 4C. Slides were scanned by using a AxioObserver Z1 fluorescence microscope (ZEISS) with the objective EC Plan-Neofluar 10X / 0.3 Ph1. Comet tail moments were measured with ImageJ software (version 1.51n) with the plugin OpenComet (http://www.cometbio.org).
siRNA Transfection
Cells were transfected with siRNA duplexes using Dharmafect 4 transfection reagent (Dharmacon) for 24 h before inducing quiescence for 72 h. siRNAs used are pools of 4 siRNAs per gene from Dharmacon [CtIP: M-011376–00-0005; ARTEMIS (DCLRE1C): M-004269–02; DNA ligase 3: M-009227–02; FEN1: M-010344–01; MRE11: M-009271–01; MUS81: L-016143–01; PNKP: M-006783–02; RAD52: M-011760–01; TDP1: M-016112–01; XPF: M-019946–00; XPG: M-006626–01; XRCC1: M-009394–01], or are individual siRNAs from Eurofins Genomics (SETX: 5’-UUGGAGUAGUUGAUACCCGAAdTdT-3’), Dharmacon (AQR: D-022214–03; LUC control sequence: D-001400–01), or Eurogentec (nontargeting control sequence: SR-CL000–005).
Quantification of Global RNA Transcription
Global RNA transcription was detected in cells using the Click-iT RNA Alexa Fluor 488 Imaging Kit (ThermoFisher Scientific). Cells were seeded in poly-L-lysine-coated Lab-Tek™ RS chamber slides (NalgeNunc) and incubated with 1 mM EU for 30 min to label newly synthesized RNA, which was detected according to the manufacturer’s instructions. Slides were mounted using Mowiol 4–88 (Millipore) containing PI and scanned by using an AxioObserver Z1 fluorescence microscope (ZEISS) with the objective Plan-Apochromat 20X / 0.8. Pictures were analyzed with ImageJ (version 1.48v).
Cell Viability Assays
CellTiter-Blue cell viability assays were performed into 96-well microplates (CellCarrier; PerkinElmer) according to the manufacturer’s instructions (Promega) and fluorescence was measured at 545–20 nm Ex/600–40 nm Em using a CLARIOstar microplate reader (BMG Labtech). Viability of untreated cells was set to 100%.
QUANTIFICATION AND STATISTICAL ANALYSIS
Information on biological replicates (n) is indicated in the figure legends. For immunofluorescence microscopy experiments, at least 500 nuclei were analyzed when using the Operetta/Columbus system and at least 150 nuclei when using counting with ImageJ in Figure 5A. For comet assays, at least 150 nuclei were analyzed. Experimental differences were tested for significance with one-way ANOVA or two-tailed unpaired t test by using GraphPad Prism 6 software. Ns indicates not significant differences. p < 0.05 is considered significant. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
DATA AND CODE AVAILABILITY
This study did not generate/analyze datasets/code.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| p53BP1 (phospho-Ser1778) rabbit polyclonal antibody (used for: IF) | Cell Signaling Technology | Cat# 2675; RRID:AB_490917 |
| Actin mouse monoclonal antibody (clone C4) (used for: WB) | Millipore | Cat# MAB1501; RRID:AB_2223041 |
| ARTEMIS (DCLRE1C) rabbit monoclonal antibody (clone D708V) (used for: WB) | Cell Signaling Technology | Cat# 13381; RRID:AB_2798197 |
| mCherry rabbit polyclonal antibody (used for: WB) | GeneTex | Cat# GTX128509 |
| CtIP (RBBP8) rabbit polyclonal antibody (used for: WB) | Bethyl | Cat# A300-488A; RRID:AB_2175262 |
| FEN1 rabbit polyclonal antibody (used for: WB) | Bethyl | Cat# A300-255A; RRID:AB_185550 |
| H2AX rabbit polyclonal antibody (used for: ChIP) | Millipore | Cat# 07-627; RRID:AB_2233033 |
| γH2AX (phospho-Ser139) mouse monoclonal antibody (clone JBW301) (used for: IF) | Millipore | Cat# 05-636; RRID:AB_309864 |
| γH2AX (phospho-Ser139) rabbit polyclonal antibody (used for: ChIP) | Millipore | Cat# 07-164; RRID:AB_310406 |
| KAP-1 (TRIM28) rabbit polyclonal antibody (used for: WB) | Bethyl | Cat# A300-274A; RRID:AB_185559 |
| Lamin B1 rabbit polyclonal antibody (used for: WB) | Abcam | Cat# ab16048; RRID:AB_443298 |
| DNA ligase 3 (LIG3, DNL3) rabbit polyclonal antibody (used for: WB) | Bethyl | Cat# A301-636A; RRID:AB_1210932 |
| MRE11 rabbit polyclonal antibody (used for: WB) | Bethyl | Cat# A303-998A; RRID:AB_2620347 |
| MUS81 mouse monoclonal antibody (clone MTA30 2G10/3 (used for: WB) | Abcam | Cat# ab14387; RRID:AB_301167 |
| PNKP (PNK1) rabbit polyclonal antibody (used for: WB) | Bethyl | Cat# A300-257A; RRID:AB_263356 |
| S9.6 mouse monoclonal antibody (used for: DRIP, Slot-blot) | Gromak Lab | N/A; RRID:AB_2810829 |
| ssDNA (single stranded DNA) mouse monoclonal antibody (clone 16-19) (used for: Slot-blot) | Millipore | Cat# MAB3034; RRID:AB_94645 |
| SETX (Senataxin) rabbit polyclonal antibody (used for: WB) | Bethyl | Cat# A301-104A; RRID:AB_873128 |
| SETX (Senataxin) rabbit polyclonal antibody (used for: ChIP) | Novus | NB100-57542; RRID:AB_922411 |
| TDP1 mouse polyclonal antibody (used for: WB) | Abnova Corporation | Cat# H00055775-A01; RRID:AB_461513 |
| aTubulin mouse monoclonal antibody (clone B-5-1-2) (used for: WB) | Sigma-Aldrich | Cat# T5168; RRID:AB_477579 |
| XPF (ERCC4) rabbit polyclonal antibody (used for: WB) | Bethyl | Cat# A301-315A; RRID:AB_938089 |
| XPF (ERCC4) rabbit polyclonal antibody (used for: ChIP and WB) | Abcam | Cat# ab76948; RRID:AB_1524575; Lot#GR172751 |
| XPG (ERCC5) rabbit polyclonal antibody (used for: WB) | Bethyl | Cat# A301-484A; RRID:AB_999684 |
| XRCC1 rabbit polyclonal antibody (used for: WB) | Bethyl | Cat# A300-065A; RRID:AB_2218477 |
| Donkey anti-mouse secondary antibody, Alexa Fluor 488 (used for: IF) | ThermoFisher Scientific | Cat# A-21202; RRID:AB_141607 |
| Goat anti-rabbit secondary antibody, Alexa Fluor 594 (used for: IF) | ThermoFisher Scientific | Cat# A-11037; RRID:AB_2534095 |
| Goat anti-mouse secondary antibody, Alexa Fluor 594 (used for: IF) | ThermoFisher Scientific | Cat# A-11032; RRID:AB_2534091 |
| Goat anti-rabbit secondary antibody, Alexa Fluor 647 (used for: IF) | ThermoFisher Scientific | Cat# A-21245; RRID:AB_2535813 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Bortezomib (PS-341) | Selleckchem | Cat# S1013; CAS: 179324-69-7 |
| CPT [(S)-(+)-Camptothecin] | Sigma-Aldrich | Cat# C9911; CAS: 7689-03-4 |
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
| Dharmafect 4 transfection reagent | Dharmacon | Cat# T-2004 |
| Doxycycline hyclate | Sigma-Aldrich | Cat# D9891; CAS 24390-14-5 |
| FLV (Flavopiridol hydrochloride hydrate) | Sigma-Aldrich | Cat# F3055 |
| Halt Protease & Phosphatase Inhibitor Cocktail | ThermoFisher Scientific | Cat# 1861281 |
| MG132 (Z-Leu-Leu-Leu-al) | Sigma-Aldrich | Cat# C9911; CAS: 133407-82-6 |
| Myricetin | Sigma-Aldrich | Cat# M6760; CAS: 529-44-2 |
| Poly-L-lysine solution | Sigma-Aldrich | Cat# P4832 |
| Proteinase K | Sigma-Aldrich | Cat# 03115828001 |
| RNase H | NEB | Cat# M0297 |
| Critical Commercial Assays | ||
| CellTiter-Blue cell viability assay | Promega | Cat# G8080 |
| Click-iT EdU Alexa Fluor 647 Imaging Kit | ThermoFisher Scientific | Cat# C10340 |
| Click-iT RNA Alexa Fluor 488 Imaging Kit | ThermoFisher Scientific | Cat# C10329 |
| CometAssay Kit | Trevigen | Cat# 4250-050-K |
| Experimental Models: Cell Lines | ||
| Human: WI38 hTERT cells | Carl Mann laboratory (Jeanblanc et al., 2012) | N/A |
| Human: XP2Y0 (XPF-deficient) cells | Orlando Schärer laboratory (Staresincic et al., 2009) | N/A |
| Human: XP2Y0 + XPF WT cells | Orlando Schärer laboratory (Staresincic et al., 2009) | N/A |
| Human: XP2Y0 + XPF-D676A (nuclease-dead) cells | Orlando Schärer laboratory (Staresincic et al., 2009) | N/A |
| Human: HCT116 cells | Yves Pommier laboratory (Al Abo et al., 2017) | N/A |
| Human: HCT116 TDP1 KO cells | Yves Pommier laboratory (Al Abo et al., 2017) | N/A |
| Human: U2OS cells inducible for mCherry | Patrick Calsou laboratory (Britton et al., 2014) | N/A |
| Human: U2OS cells inducible for mCherry-RNH1 | Patrick Calsou laboratory (Britton et al., 2014) | N/A |
| Oligonucleotides | ||
| AQR (Aquarius) (human) individual siRNA | Dharmacon | Cat# D-022214-03 |
| ARTEMIS (DCLRE1C) (human) SMARTpool siRNA | Dharmacon | Cat# M-004269-02 |
| CtIP (RBBP8) (human) SMARTpool siRNA | Dharmacon | Cat# M-011376-00 |
| DNA ligase 3 (LIG3, DNL3) (human) SMARTpool siRNA | Dharmacon | Cat# M-009227-02 |
| FEN1 (human) SMARTpool siRNA | Dharmacon | Cat# M-010344-01 |
| LUC (Luciferase GL3 Duplex) siRNA | Dharmacon | Cat# D-001400-01 |
| MRE11 (human) SMARTpool siRNA | Dharmacon | Cat# M-009271-01 |
| MUS81 (human) SMARTpool siRNA | Dharmacon | Cat# L-016143-01 |
| PNKP (human) SMARTpool siRNA | Dharmacon | Cat# M-006783-02 |
| RAD52 (human) SMARTpool siRNA | Dharmacon | Cat# M-011760-01 |
| SETX (Senataxin) (human) siRNA: 5’-UUGGAGUAGUU GAUACCCGAAdTdT-3’ | This paper | N/A |
| TDP1 (human) SMARTpool siRNA | Dharmacon | Cat# M-016112-01 |
| XPF (ERCC4) (human) SMARTpool siRNA | Dharmacon | Cat# M-019946-00 |
| XPG (ERCC5) (human) SMARTpool siRNA | Dharmacon | Cat# M-006626-01 |
| XRCC1 (human) SMARTpool siRNA | Dharmacon | Cat# M-009394-01 |
| Nontargeting control siRNA | Eurogentec | Cat# SR-CL000-005 |
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
| ssDNA: CGGTGTGAATCAGAC (used for DNA/RNA hybrid competition) | Sigma-Aldrich | N/A |
| ssRNA: GUCUGAUUCACACCG (used for DNA/RNA hybrid competition) | Dharmacon | N/A |
| See Table S1 for primers used in this study. | ||
| Software and Algorithms | ||
| Columbus software (versions 2.5.0 and 2.8.2) | PerkinElmer | N/A |
| GraphPad Prism 6.0 | GraphPad Prism | https://www.graphpad.com/; RRID:SCR_002798 |
| Harmony software (versions 4.1 and 4.8) | PerkinElmer | N/A |
| ImageJ software (versions 1.48, 1.51, and 1.52) | National Institutes of Health | https://imagej.nih.gov/ij/; RRID:SCR_003070 |
| OpenComet (version 1.3.1) | OpenComet Software | http://www.cometbio.org |
| Other | ||
| AxioObserver Z1 fluorescence microscope | ZEISS | https://www.zeiss.com/corporate/int/home.html |
| LSM 780 confocal microscope | ZEISS | https://www.zeiss.com/corporate/int/home.html |
| CLARIOstar microplate reader | BMG Labtech | https://www.bmglabtech.com |
| ChemiDoc MP Imaging System | Bio-Rad | http://www.bio-rad.com |
| Operetta High-Content Imaging System | PerkinElmer | https://www.perkinelmer.com |
| Operetta CLS High-Content Imaging System | PerkinElmer | https://www.perkinelmer.com |
| CellCarrier 96-well | PerkinElmer | Cat# 6005550 |
| Lab-Tek RS chamber slides | ThermoFisher Scientific | Cat# 154526K |
ACKNOWLEDGMENTS
We thank Carl Mann for WI38 hTERT cells; Orlando Schärer for XP2Y0, XP2Y0 + XPF-WT, and XP2Y0 + XPF-D676A cells; and the Toulouse Regional Imaging-Light Imaging Toulouse CBI Platform, as well as the CRCT platform, for technical assistance in high-content microscopy. We also thank Nicholas Proudfoot for editing the manuscript. This work was supported by funds from the Ligue Nationale Contre le Cancer (LNCC) Comité Départemental 31 and the Fondation pour la Recherche Médicale (FRM) (Equipe labellisée FRM [DEQ20170839117]). N.G. lab is supported by the Royal Society University Research fellowship (UF 100243, UF 150656) and an MRC Grant (MR/ J007870/1) to N.G. Y.P. and S.N.H. are supported by the Intramural Program of the National Cancer Institute, NIH (Z01-BC-006161). The G.C. lab is supported by Associazione Italiana per la Ricerca sul Cancro, Milan (IG15886). S.S. is supported by a PhD fellowship under the French-Italian University VINCI Program.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.08.041.
REFERENCES
- Aguilera A, and García-Muse T. (2012). R loops: from transcription byproducts to threats to genome stability. Mol. Cell 46, 115–124. [DOI] [PubMed] [Google Scholar]
- Aguilera A, and Gómez-González B. (2017). DNA-RNA hybrids: the risks of DNA breakage during transcription. Nat. Struct. Mol. Biol. 24, 439–443. [DOI] [PubMed] [Google Scholar]
- Al Abo M, Sasanuma H, Liu X, Rajapakse VN, Huang SY, Kiselev E, Takeda S, Plunkett W, and Pommier Y. (2017). TDP1 is critical for the repair of DNA breaks induced by sapacitabine, a nucleoside also targeting ATM- and BRCA-deficient tumors. Mol. Cancer Ther. 16, 2543–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashour ME, Atteya R, and El-Khamisy SF (2015). Topoisomerase-mediated chromosomal break repair: an emerging player in many games. Nat. Rev. Cancer 15, 137–151. [DOI] [PubMed] [Google Scholar]
- Baranello L, Wojtowicz D, Cui K, Devaiah BN, Chung HJ, Chan-Salis KY, Guha R, Wilson K, Zhang X, Zhang H, et al. (2016). RNA polymerase II regulates topoisomerase 1 activity to favor efficient transcription. Cell 165, 357–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnham KJ, Masters CL, and Bush AI (2004). Neurodegenerative diseases and oxidative stress. Nat. Rev. Drug Discov. 3, 205–214. [DOI] [PubMed] [Google Scholar]
- Bendixen C, Thomsen B, Alsner J, and Westergaard O. (1990). Campto-thecin-stabilized topoisomerase I-DNA adducts cause premature termination of transcription. Biochemistry 29, 5613–5619. [DOI] [PubMed] [Google Scholar]
- Boguslawski SJ, Smith DE, Michalak MA, Mickelson KE, Yehle CO, Patterson WL, and Carrico RJ (1986). Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. J. Immunol. Methods 89, 123–130. [DOI] [PubMed] [Google Scholar]
- Britton S, Dernoncourt E, Delteil C, Froment C, Schiltz O, Salles B, Frit P, and Calsou P. (2014). DNA damage triggers SAF-A and RNA biogenesis factors exclusion from chromatin coupled to R-loops removal. Nucleic Acids Res. 42, 9047–9062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S, and Ferguson DO (2008). Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell 135, 85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldecott KW, Tucker JD, Stanker LH, and Thompson LH (1995). Characterization of the XRCC1-DNA ligase III complex in vitro and its absence from mutant hamster cells. Nucleic Acids Res. 23, 4836–4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capranico G, Ferri F, Fogli MV, Russo A, Lotito L, and Baranello L. (2007). The effects of camptothecin on RNA polymerase II transcription: roles of DNA topoisomerase I. Biochimie 89, 482–489. [DOI] [PubMed] [Google Scholar]
- Ceccaldi R, Rondinelli B, and D’Andrea AD (2016). Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 26, 52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerritelli SM, and Crouch RJ (2009). Ribonuclease H: the enzymes in eukaryotes. FEBS J. 276, 1494–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Puget N, Lin YL, Clouaire T, Aguirrebengoa M, Rocher V, Pasero P, Canitrot Y, and Legube G. (2018). Senataxin resolves RNA:DNA hybrids forming at DNA double-strand breaks to prevent translocations. Nat. Commun. 9, 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristini A, Park JH, Capranico G, Legube G, Favre G, and Sordet O. (2016). DNA-PK triggers histone ubiquitination and signaling in response to DNA double-strand breaks produced during the repair of transcription-blocking topoisomerase I lesions. Nucleic Acids Res. 44, 1161–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristini A, Groh M, Kristiansen MS, and Gromak N. (2018). RNA/DNA hybrid interactome identifies DXH9 as a molecular player in transcriptional termination and R-loop-associated DNA damage. Cell Rep. 23, 1891–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daroui P, Desai SD, Li TK, Liu AA, and Liu LF (2004). Hydrogen peroxide induces topoisomerase I-mediated DNA damage and cell death. J. Biol. Chem. 279, 14587–14594. [DOI] [PubMed] [Google Scholar]
- Dehé PM, and Gaillard PH (2017). Control of structure-specific endonucleases to maintain genome stability. Nat. Rev. Mol. Cell Biol. 18, 315–330. [DOI] [PubMed] [Google Scholar]
- Desai SD, Zhang H, Rodriguez-Bauman A, Yang JM, Wu X, Gounder MK, Rubin EH, and Liu LF (2003). Transcription-dependent degradation of topoisomerase I-DNA covalent complexes. Mol. Cell. Biol. 23, 2341–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drolet M, Broccoli S, Rallu F, Hraiky C, Fortin C, Masse E, and Baaklini I. (2003). The problem of hypernegative supercoiling and R-loop formation in transcription. Front. Biosci. 8, d210–d221. [DOI] [PubMed] [Google Scholar]
- Dumitrache LC, and McKinnon PJ (2017). Polynucleotide kinase-phosphatase (PNKP) mutations and neurologic disease. Mech. Ageing Dev. 161 (Pt A), 121–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hage A, French SL, Beyer AL, and Tollervey D. (2010). Loss of topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 24, 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Khamisy SF, Saifi GM, Weinfeld M, Johansson F, Helleday T, Lupski JR, and Caldecott KW (2005). Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature 434, 108–113. [DOI] [PubMed] [Google Scholar]
- Graf M, Bonetti D, Lockhart A, Serhal K, Kellner V, Maicher A, Jolivet P, Teixeira MT, and Luke B. (2017). Telomere length determines TERRA and R-loop regulation through the cell cycle. Cell 170, 72–85.e14. [DOI] [PubMed] [Google Scholar]
- Groh M, and Gromak N. (2014). Out of balance: R-loops in human disease. PLoS Genet. 10, e1004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groh M, Lufino MM, Wade-Martins R, and Gromak N. (2014). R-loops associated with triplet repeat expansions promote gene silencing in Friedreich ataxia and fragile X syndrome. PLoS Genet. 10, e1004318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groh M, Albulescu LO, Cristini A, and Gromak N. (2017). Senataxin: genome guardian at the interface of transcription and neurodegeneration. J. Mol. Biol. 429, 3181–3195. [DOI] [PubMed] [Google Scholar]
- Hendrickson EA (1997). Cell-cycle regulation of mammalian DNA double-strand-break repair. Am. J. Hum. Genet. 61, 795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch NC, Hanzlikova H, Rulten SL, Tétreault M, Komulainen E, Ju L, Hornyak P, Zeng Z, Gittens W, Rey SA, et al. ; Care4Rare Canada Con-sortium (2017). XRCC1 mutation is associated with PARP1 hyperactivation and cerebellar ataxia. Nature 541, 87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SY, Ghosh S, and Pommier Y. (2015). Topoisomerase I alone is sufficient to produce short DNA deletions and can also reverse nicks at ribonucleotide sites. J. Biol. Chem. 290, 14068–14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SN, Williams JS, Arana ME, Kunkel TA, and Pommier Y. (2017). Topoisomerase I-mediated cleavage at unrepaired ribonucleotides generates DNA double-strand breaks. EMBO J. 36, 361–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, and Aguilera A. (2003). Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell 12, 711–721. [DOI] [PubMed] [Google Scholar]
- Interthal H, Chen HJ, Kehl-Fie TE, Zotzmann J, Leppard JB, and Champoux JJ (2005). SCAN1 mutant Tdp1 accumulates the enzyme–DNA intermediate and causes camptothecin hypersensitivity. EMBO J. 24, 2224–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanblanc M, Ragu S, Gey C, Contrepois K, Courbeyrette R, Thuret JY, and Mann C. (2012). Parallel pathways in RAF-induced senescence and conditions for its reversion. Oncogene 31, 3072–3085. [DOI] [PubMed] [Google Scholar]
- Jiang B, Glover JN, and Weinfeld M. (2017). Neurological disorders associated with DNA strand-break processing enzymes. Mech. Ageing Dev. 161 (Pt A), 130–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan A, Bhatia K, Branzei D, and Gangwani L. (2018). Combined deficiency of Senataxin and DNA-PKcs causes DNA damage accumulation and neurodegeneration in spinal muscular atrophy. Nucleic Acids Res. 46, 8326–8346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katyal S, Lee Y, Nitiss KC, Downing SM, Li Y, Shimada M, Zhao J, Russell HR, Petrini JH, Nitiss JL, and McKinnon PJ (2014). Aberrant topoisomerase-1 DNA lesions are pathogenic in neurodegenerative genome instability syndromes. Nat. Neurosci. 17, 813–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawale AS, Akopiants K, Valerie K, Ruis B, Hendrickson EA, Huang SN, Pommier Y, and Povirk LF (2018). TDP1 suppresses mis-joining of radiomimetic DNA double-strand breaks and cooperates with Artemis to promote optimal nonhomologous end joining. Nucleic Acids Res. 46, 8926–8939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Huang SN, Williams JS, Li YC, Clark AB, Cho JE, Kunkel TA, Pommier Y, and Jinks-Robertson S. (2011). Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science 332, 1561–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, and Manley JL (2005). Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 122, 365–378. [DOI] [PubMed] [Google Scholar]
- Löbrich M, Shibata A, Beucher A, Fisher A, Ensminger M, Goodarzi AA, Barton O, and Jeggo PA (2010). gammaH2AX foci analysis for monitoring DNA double-strand break repair: strengths, limitations and optimization. Cell Cycle 9, 662–669. [DOI] [PubMed] [Google Scholar]
- Ma L, Cao X, Wang H, Lu K, Wang Y, Tu C, Dai Y, Meng Y, Li Y, Yu P, et al. (2019). Discovery of myricetin as a potent inhibitor of human flap endonuclease 1, which potentially can be used as sensitizing agent against HT-29 human colon cancer cells. J. Agric. Food Chem. 67, 1656–1665. [DOI] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, and Lukas J. (2007). RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131, 887–900. [DOI] [PubMed] [Google Scholar]
- Mamouni K, Cristini A, Guirouilh-Barbat J, Monferran S, Lemarié A, Faye JC, Lopez BS, Favre G, and Sordet O. (2014). RhoB promotes γH2AX dephosphorylation and DNA double-strand break repair. Mol. Cell.Biol. 34, 3144–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzo SG, Hartono SR, Sanz LA, Marinello J, De Biasi S, Cossarizza A, Capranico G, and Chedin F. (2018). DNA topoisomerase I differentially modulates R-loops across the human genome. Genome Biol. 19, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinello J, Chillemi G, Bueno S, Manzo SG, and Capranico G. (2013). Antisense transcripts enhanced by camptothecin at divergent CpG-island promoters associated with bursts of topoisomerase I-DNA cleavage complex and R-loop formation. Nucleic Acids Res. 41, 10110–10123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinello J, Bertoncini S, Aloisi I, Cristini A, Malagoli Tagliazucchi G, Forcato M, Sordet O, and Capranico G. (2016). Dynamic effects of topoisomerase I inhibition on R-loops and short transcripts at active promoters. PLoS ONE 11, e0147053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon PJ (2017). Genome integrity and disease prevention in the nervous system. Genes Dev. 31, 1180–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon V, and Povirk LF (2016). End-processing nucleases and phosphodi-esterases: An elite supporting cast for the non-homologous end joining pathway of DNA double-strand break repair. DNA Repair (Amst.) 43, 57–68. [DOI] [PubMed] [Google Scholar]
- Miao ZH, Agama K, Sordet O, Povirk L, Kohn KW, and Pommier Y. (2006). Hereditary ataxia SCAN1 cells are defective for the repair of transcription-dependent topoisomerase I cleavage complexes. DNA Repair (Amst.) 5, 1489–1494. [DOI] [PubMed] [Google Scholar]
- Nitiss J, and Wang JC (1988). DNA topoisomerase-targeting antitumor drugs can be studied in yeast. Proc. Natl. Acad. Sci. USA 85, 7501–7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima T, Tellier M, Foxwell J, Ribeiro de Almeida C, Tan-Wong SM, Dhir S, Dujardin G, Dhir A, Murphy S, and Proudfoot NJ (2018). Deregulated expression of mammalian lncRNA through loss of SPT6 induces R-loop formation, replication stress, and cellular senescence. Mol. Cell 72, 970– 984.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips DD, Garboczi DN, Singh K, Hu Z, Leppla SH, and Leysath CE (2013). The sub-nanomolar binding of DNA-RNA hybrids by the single-chain Fv fragment of antibody S9.6. J. Mol. Recognit. 26, 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y. (2006). Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer 6, 789–802. [DOI] [PubMed] [Google Scholar]
- Pommier Y, Huang SY, Gao R, Das BB, Murai J, and Marchand C. (2014). Tyrosyl-DNA-phosphodiesterases (TDP1 and TDP2). DNA Repair (Amst.) 19, 114–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y, Sun Y, Huang SN, and Nitiss JL (2016). Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol. 17, 703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell WT, Coulson RL, Gonzales ML, Crary FK, Wong SS, Adams S, Ach RA, Tsang P, Yamada NA, Yasui DH, et al. (2013). R-loop formation at Snord116 mediates topotecan inhibition of Ube3a-antisense and allele-specific chromatin decondensation. Proc. Natl. Acad. Sci. USA 110, 13938–13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, and Zhang F. (2013). Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rass U, Ahel I, and West SC (2007). Defective DNA repair and neurodegenerative disease. Cell 130, 991–1004. [DOI] [PubMed] [Google Scholar]
- Regairaz M, Zhang YW, Fu H, Agama KK, Tata N, Agrawal S, Aladjem MI, and Pommier Y. (2011). Mus81-mediated DNA cleavage resolves replication forks stalled by topoisomerase I-DNA complexes. J. Cell Biol. 195, 739–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Pereira JM, and Aguilera A. (2015). R loops: new modulators of genome dynamics and function. Nat. Rev. Genet. 16, 583–597. [DOI] [PubMed] [Google Scholar]
- Sanz LA, Hartono SR, Lim YW, Steyaert S, Rajpurkar A, Ginno PA, Xu X, and Ché din, F. (2016). Prevalent, dynamic, and conserved R-loop structures associate with specific epigenomic signatures in mammals. Mol. Cell 63, 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle DA, Smith S, Uziel T, Sfez S, et al. (1995). A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science 268, 1749–1753. [DOI] [PubMed] [Google Scholar]
- Skourti-Stathaki K, and Proudfoot NJ (2014). A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 28, 1384–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Proudfoot NJ, and Gromak N. (2011). Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell 42, 794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solier S, Ryan MC, Martin SE, Varma S, Kohn KW, Liu H, Zeeberg BR, and Pommier Y. (2013). Transcription poisoning by topoisomerase I is controlled by gene length, splice sites, and miR-142–3p. Cancer Res. 73, 4830–4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollier J, and Cimprich KA (2015). Breaking bad: R-loops and genome integrity. Trends Cell Biol. 25, 514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollier J, Stork CT, García-Rubio ML, Paulsen RD, Aguilera A, and Cimprich KA (2014). Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol. Cell 56, 777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sordet O, Larochelle S, Nicolas E, Stevens EV, Zhang C, Shokat KM, Fisher RP, and Pommier Y. (2008). Hyperphosphorylation of RNA polymerase II in response to topoisomerase I cleavage complexes and its association with transcription- and BRCA1-dependent degradation of topoisomerase I. J. Mol. Biol. 381, 540–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sordet O, Redon CE, Guirouilh-Barbat J, Smith S, Solier S, Douarre C, Conti C, Nakamura AJ, Das BB, Nicolas E, et al. (2009). Ataxia telangiectasia mutated activation by transcription- and topoisomerase I-induced DNA double-strand breaks. EMBO Rep. 10, 887–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks JL, and Burgers PM (2015). Error-free and mutagenic processing of topoisomerase 1-provoked damage at genomic ribonucleotides. EMBO J. 34, 1259–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staresincic L, Fagbemi AF, Enzlin JH, Gourdin AM, Wijgers N, Dunand-Sauthier I, Giglia-Mari G, Clarkson SG, Vermeulen W, and Schärer OD (2009). Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO J. 28, 1111–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima H, Boerkoel CF, John J, Saifi GM, Salih MA, Armstrong D, Mao Y, Quiocho FA, Roa BB, Nakagawa M, et al. (2002). Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat. Genet. 32, 267–272. [DOI] [PubMed] [Google Scholar]
- Teasley DC, Parajuli S, Nguyen M, Moore HR, Alspach E, Lock YJ, Honaker Y, Saharia A, Piwnica-Worms H, and Stewart SA (2015). Flap endonuclease 1 limits telomere fragility on the leading strand. J. Biol. Chem. 290, 15133–15145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, and Alt FW (2000). Transcription-induced cleavage of immunoglobulin switch regions by nucleotide excision repair nucleases in vitro. J. Biol. Chem. 275, 24163–24172. [DOI] [PubMed] [Google Scholar]
- Tresini M, Warmerdam DO, Kolovos P, Snijder L, Vrouwe MG, Demmers JA, van IJcken WF, Grosveld FG, Medema RH, Hoeijmakers JH, et al. (2015). The core spliceosome as target and effector of non-canonical ATM signalling. Nature 523, 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuduri S, Crabbe L, Conti C, Tourrière H, Holtgreve-Grez H, Jauch A, Pantesco V, De Vos J, Thomas A, Theillet C, et al. (2009). Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat. Cell Biol. 11, 1315–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker C, Herranz-Martin S, Karyka E, Liao C, Lewis K, Elsayed W, Lukashchuk V, Chiang SC, Ray S, Mulcahy PJ, et al. (2017). C9orf72 expansion disrupts ATM-mediated chromosomal break repair. Nat. Neurosci. 20, 1225–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, and Liu LF (1997). Processing of topoisomerase I cleavable complexes into DNA damage by transcription. Nucleic Acids Res. 25, 4181–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SW, Burgin AB Jr., Huizenga BN, Robertson CA, Yao KC, and Nash HA (1996). A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases. Proc. Natl. Acad. Sci. USA 93, 11534–11539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuhara T, Kato R, Hagiwara Y, Shiotani B, Yamauchi M, Nakada S, Shibata A, and Miyagawa K. (2018). Human Rad52 promotes XPG-mediated R-loop processing to initiate transcription-associated homologous recombination repair. Cell 175, 558–570.e11. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate/analyze datasets/code.