SUMMARY
Zbtb16 encodes the zinc-finger protein PLZF, which is often used as a lineage marker for innate-like T cells and is specifically required for the generation of invariant natural killer T (iNKT) cells in the thymus. Here, we report that not only PLZF expression itself but also the relative abundance of PLZF proteins plays critical roles in iNKT cell development. Utilizing a Zbtb16 hypomorphic allele, PLZFGFPCre, which produces PLZF proteins at only half of the level of the wild-type allele, we show that decreased PLZF expression results in a significant decrease in iNKT cell numbers, which is further associated with profound alterations in iNKT lineage choices and subset composition. These results document that there is a quantitative aspect of PLZF expression in iNKT cells, demonstrating that the availability of PLZF protein is a critical factor for both effective iNKT cell generation and subset differentiation.
INTRODUCTION
The transcription factor PLZF (promyelocytic leukemia zinc finger) plays a critical role in the generation and effector function of invariant natural killer T (iNKT) cells (Kovalovsky et al., 2008; Savage et al., 2008). iNKT cells originate from immature CD4+CD8+ double-positive (DP) thymocytes that are positively selected by T cell receptor (TCR) signaling of glycolipid-loaded major histocompatibility complex (MHC) class I-like CD1d molecules (Cohen et al., 2009). While immature DP thymocytes do not express PLZF, strong agonistic TCR signals are thought to upregulate PLZF expression in DP cells to direct iNKT cell fate and to drive their effector phenotype differentiation (Bedel et al., 2014; Seiler et al., 2012). The precise molecular pathway of PLZF induction and expression remains unmapped. However, both transcriptional mechanisms and posttranscriptional events, such as microRNA (miRNA)-mediated suppression, are known to be involved in this process (D’Cruz et al., 2014; Pobezinsky et al., 2015).
Importantly, PLZF expression in iNKT cells is not unimodal but varies between distinct iNKT subsets. Interleukin-4 (IL-4)-producing iNKT cells are referred to as NKT2 cells, and they express the largest amount of PLZF. Interferon (IFN)γ-secreting iNKT cells, on the other hand, are referred to as NKT1 cells, and they express the smallest amount of PLZF. Lastly, NKT17 cells, which produce IL-17, express PLZF at levels between those of NKT2 and NKT1 cells (Constantinides and Bendelac, 2013; Das et al., 2010; Lee et al., 2013). These observations led us to examine whether the different abundance of PLZF proteins among iNKT subsets would not only correlate with but also direct distinct iNKT lineage fates.
To accurately determine PLZF expression at single-cell resolution, a PLZF reporter mouse strain was recently generated (Constantinides et al., 2014). This gene knockin strain, PLZFGFPcre, was engineered to encode an enhanced GFP-Cre recombinase fusion protein in the 3′ UTR of the endogenous Zbtb16 gene (Constantinides et al., 2014). Thus, PLZF transcription would induce both GFP reporter expression and Cre recombinase activity. Previous analysis demonstrated that the amount of the GFP reporter protein, indeed, correlated with the level of Zbtb16 gene expression, and it further confirmed the successful Cre-mediated recombination in PLZF+ cells with fate mapping experiments (Constantinides et al., 2014).
Using these engineered mice, in this study, we assessed PLZFGFPcre-encoded GFP protein expression to monitor Zbtb16 regulation during thymic iNKT cell differentiation. Notably, we made the serendipitous observation that PLZF protein expression was substantially reduced when the protein was encoded in the PLZFGFPcre knockin allele. Consequently, PLZFGFPcre is a hypomorphic Zbtb16 allele, and this provided us with the opportunity to examine the quantitative facet of PLZF protein expression in thymic iNKT cell generation and subset differentiation.
RESULTS
The PLZFGFPcre Gene Knockin Allele Impairs iNKT Cell Development in the Thymus
PLZF is highly expressed in iNKT cells and in a small fraction of innate-type γδ T cells, but not in conventional αβ T cells (Alonzo and Sant’Angelo, 2011; Constantinides et al., 2014; Kreslavsky et al., 2009; Lynch et al., 2015; Zhang et al., 2015). PLZFGFPcre reporter expression accurately reflected such specificity, because GFP reporter expression was limited to iNKT cells and to some γδ T cells but was not found in conventional αβ T cells (Figure 1A). Moreover, GFP reporter expression in iNKT cells increased with increasing numbers of PLZFGFPcre alleles (Figure 1B), confirming that PLZFGFPcre expression correctly reports Zbtb16 gene transcription and revealing that PLZF is expressed in a biallelic manner in iNKT cells.
Figure 1. The PLZFGFPcre Knockin Encodes a Hypomorphic Zbtb16 Allele.
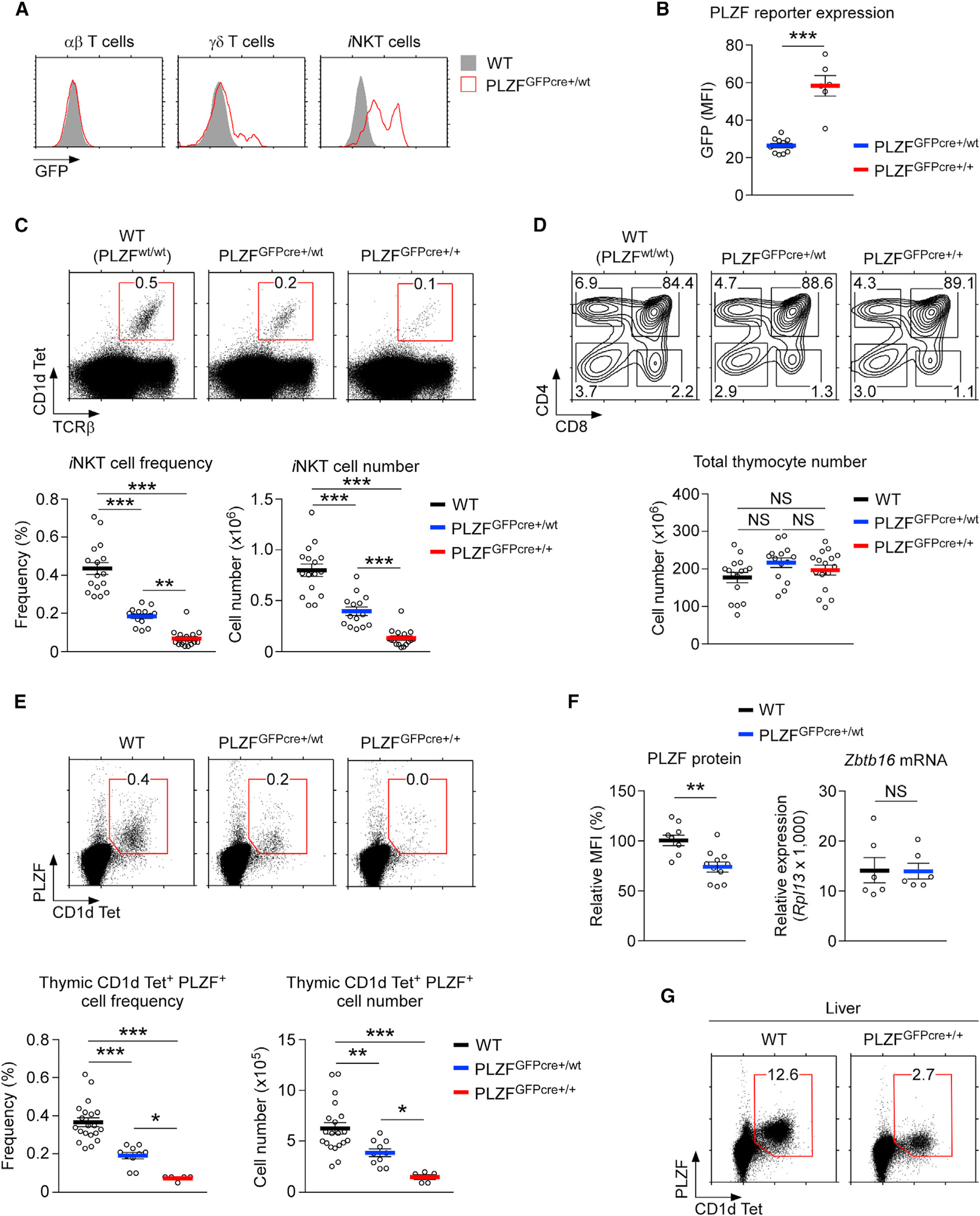
(A) PLZF-GFP reporter expression in thymic αβ T cells (CD1dTet-negative), γδ T cells, and iNKT cells of WT and PLZFGFPcre+/wt mice.
(B) PLZF-GFP reporter expression in thymic iNKT cells of PLZFGFPcre+/wt and PLZFGFPcre+/+ mice. Results show the summary of 4 independent experiments with 11 PLZFGFPcre+/wt and 6 PLZFGFPcre+/+ mice.
(C) Thymic iNKT cells were identified by CD1dTet and anti-TCRβ staining (top), and their frequencies and numbers were determined for the indicated mouse strains (bottom). Results show the summary of 13 independent experiments with 16 WT, 14 PLZFGFPcre+/wt, and 16 PLZFGFPcre+/+ mice.
(D) CD4 versus CD8 profiles (top) and total thymocyte numbers (bottom) of the indicated mice. Results show the summary of 13 independent experiments.
(E) PLZF expression in thymic iNKT cells (top) and their frequencies and numbers (bottom) in WT, PLZFGFPcre+/wt, and PLZFGFPcre+/+ mice. Data are from 5 independent experiments with 20 WT, 10 PLZFGFPcre+/wt, and 5 PLZFGFPcre+/+ mice.
(F) Intracellular PLZF protein expression in CD1dTet+PLZF+ iNKT cells (left) and the Zbtb16 mRNA contents of mature iNKT cells (right) of WT and PLZFGFPcre+/wt thymocytes. Zbtb16 mRNA was measured in FACS (fluorescence-activated cell sorting)-sorted CD24loCD1dTet+ iNKT cells. Data show the summary of 3 independent experiments.
(G) PLZF expression in liver iNKT cells of WT and PLZFGFPcre+/+ mice. Dot plots are representative of 3 independent experiments.
Data are shown as mean ± SEM. Differences between groups were determined either with two-tailed Mann-Whitney U test or Kruskal-Wallis test to calculate p values, where *p < 0.05; **p < 0.01; ***p < 0.001 were considered statistically significant. NS, not significant.
Surprisingly, we also found that both the frequency and the number of iNKT cells dramatically decreased with increasing gene dosages of PLZFGFPcre (Figure 1C). The detrimental effect of the PLZFGFPcre allele was specific to iNKT cell development because overall thymocyte development was largely unaffected (Figure 1D). Additionally, positive selection of thymocytes, as determined by CD69 and TCRβ expression (Figure S1A; Yamashita et al., 1993), and the generation of Foxp3+CD25+ T regulatory cells (Tregs), which requires strong agonistic selection (Hsieh et al., 2012), was unaffected by the PLZFGFPcre allele (Figure S1B). Collectively, these results validate that PLZFGFPcre is an effective reporter of Zbtb16 gene expression and also document an unexpected and deleterious effect of the PLZFGFPcre allele on thymic iNKT cell generation.
PLZFGFPcre Expression Results in a Paucity of Peripheral iNKT Cells
In the liver of wild-type (WT) mice, iNKT cells constitute a major fraction of hepatic lymphocytes (Crispe, 2009; Lee et al., 2015). However, in the liver of PLZFGFPcre+/+ mice, iNKT cells were conspicuously absent (Figure S1C). This finding suggested that PLZFGFPcre impairs not only the generation but also the survival and homeostasis of iNKT cells. Accordingly, iNKT cell frequencies and numbers in peripheral lymphoid organs were dramatically diminished in PLZFGFPcre+/+ mice, as demonstrated by the loss of spleen iNKT cells, compared to that of WT mice (Figure S1D). Collectively, these results show that not only does PLZFGFPcre impair iNKT cell generation in the thymus, but it is also detrimental for their maintenance in peripheral tissues.
The PLZFGFPcre Allele Attenuates PLZF Protein Expression
PLZF expression is necessary for effective iNKT cell generation (Kovalovsky et al., 2008; Savage et al., 2008). Thus, we examined whether the loss of iNKT cells in PLZFGFPcre mice was associated with decreased PLZF expression. To this end, we assessed PLZF protein expression in WT (PLZFwt/wt), PLZFGFPcre+/wt, and PLZFGFPcre+/+ thymocytes by intranuclear staining (Figure 1E, top). Both the frequency and number of PLZF+ CD1dTet+ iNKT cells dramatically decreased with increasing gene dosages of PLZFGFPcre (Figure 1E, bottom). Heterozygous PLZFGFPcre+/wt mice generated only half the number of PLZF+ iNKT cells, compared to that of the WT mice, and PLZF+ iNKT cells were virtually absent from the thymus of homozygous PLZFGFPcre+/+ mice (Figure 1E, bottom). Notably, we found that not only the number and frequency of PLZF+ cells but also the amount of PLZF proteins was significantly reduced in PLZFGFPcre carriers (Figure 1F), compared to those in WT mice. Accordingly, PLZF protein expression in heterozygous PLZFGFPcre+/wt iNKT cells was reduced to 75% of that in the WT iNKT cells (Figure 1F, left). Curiously, PLZF gene expression, as assessed by Zbtb16 mRNA content, was unaffected in PLZFGFPcre+/wt iNKT cells (Figure 1F, right). These results suggest that production of PLZF protein in PLZFGFPcre carriers is impaired by posttranscriptional mechanisms.
Notably, the diminished PLZF protein expression also affected the homeostasis of peripheral iNKT cells, so there was a significant reduction in the number of PLZF-expressing cells in peripheral tissues of mice carrying the PLZFGFPcre allele, compared to that in the WT mice. The intracellular staining of liver and spleen cells revealed a dramatic reduction in the frequency of PLZF-expressing CD1dTet+ cells (Figure 1G; Figure S1E). Altogether, the genetically engineered PLZFGFPcre allele is hypomorphic regarding its PLZF protein expression, and it reduces the size of the peripheral iNKT cell pool.
To examine whether the detrimental effect of PLZFGFPcre was specific to PLZF+ iNKT cells, we further examined the generation of Vγ1.1Vδ6.3 γδ T cells, which represent another prominent population of PLZF-expressing T cells (Kreslavsky et al., 2009; Figure S1F). We confirmed that PLZF reporter protein was highly and exclusively expressed in the Vγ1.1Vδ6.3 subset among γδ T cells (Figure S1G). Unlike iNKT cells, however, the frequency of Vγ1.1Vδ6.3 γδ T cells in PLZFGFPcre+/wt mice was not different from that in the WT mice (Figure S1H).
PLZF also plays a critical role in limb and axial skeletal patterning (Barna et al., 2000), which is illustrated in the polydactyly of PLZF gene-deficient mice (Figure S1I, middle). Notably, PLZFGFPcre+/+ mice did not show such defects (Figure S1I, right). These results indicate that the quantitative decrease in PLZF proteins in PLZFGFPcre carriers selectively affects the generation and maintenance of iNKT cells but not of other cell types or of cellular processes that rely on PLZF. Thus, iNKT cells are exquisitely sensitive to the abundance of PLZF protein.
PLZF Heterozygosity Impairs iNKT Cell Generation and Homeostasis
To corroborate the effect of PLZF availability, we assessed the development and homeostasis of iNKT cells in Zbtb16 germline-deficient (PLZF−/−) mice as an independent approach to examine the consequences of diminished PLZF availability (Barna et al., 2000). While overall thymocyte differentiation was largely unaffected (Figure S1J), thymic iNKT cell generation and maturation were dramatically impaired in both PLZF−/− and PLZF+/− mice, compared to those in WT mice (Figure 2A). Importantly, losing one allele of PLZF was already sufficient to abolish iNKT cell generation in the thymus (Figure 2A). In fact, PLZF heterozygous (PLZF+/−) mice were indistinguishable from homozygous PLZF-deficient (PLZF−/−) mice regarding their lack of thymic iNKT cells (Figures 2A and 2B). Such a failure to generate iNKT cells further translated into a severe paucity of iNKT cells in peripheral tissues. We found that both the frequencies and numbers of iNKT cells were dramatically diminished in the spleen of both PLZF+/− and PLZF−/− mice, compared to those in WT mice (Figure 2C). This was also the case for iNKT cells in the liver (Figure S1K). Altogether, these results reveal that the ability to produce the full amount of PLZF is critical for effective iNKT cell development.
Figure 2. PLZF Gene Heterozygosity Severely Impairs iNKT Cell Development and Homeostasis.
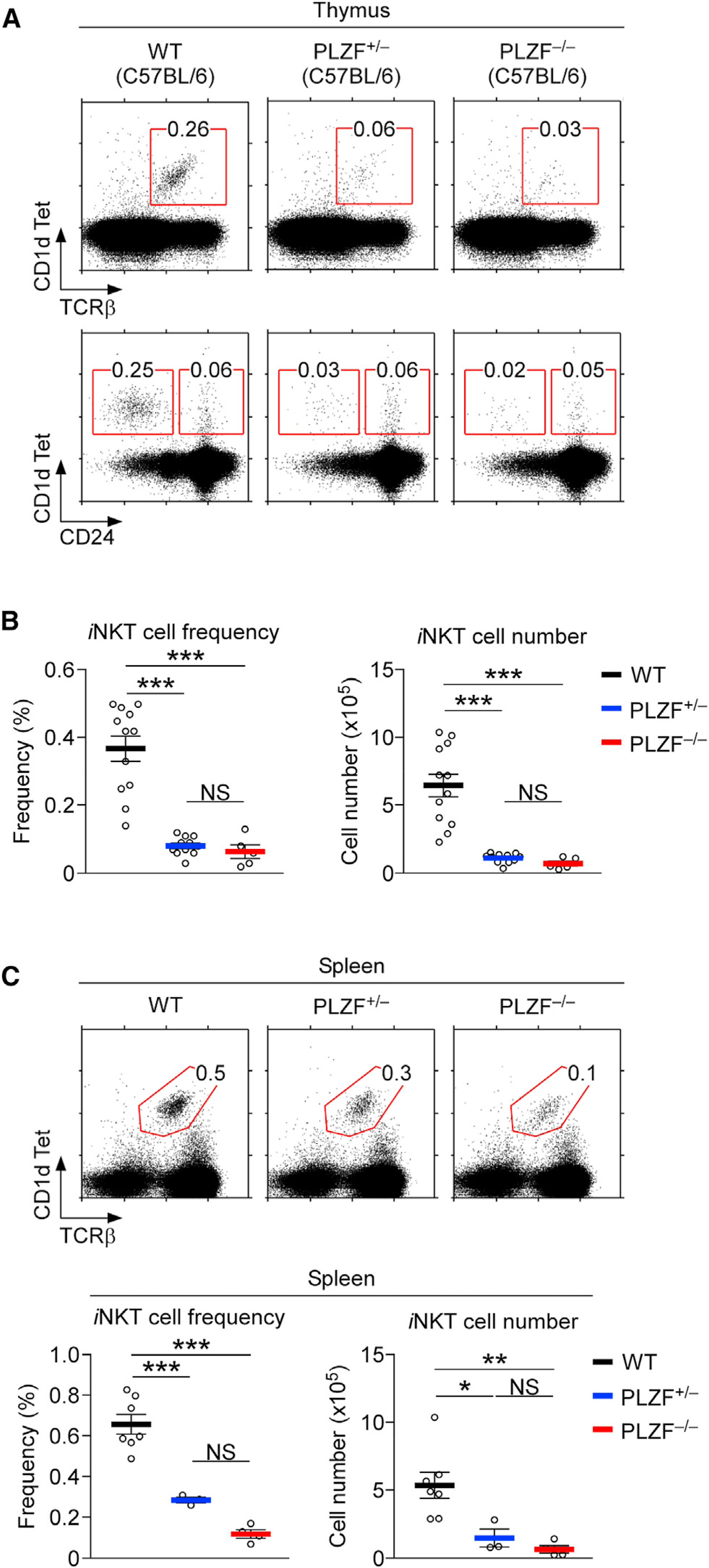
(A) Thymic iNKT cell generation and maturation in PLZF-deficient mice. iNKT cells were identified by CD1dTet and anti-TCRβ antibody staining (top) and assessed for their maturation with CD1dTet versus CD24 staining (bottom) in thymocytes of the indicated mice.
(B) Frequency and number of thymic iNKT cells were determined for the indicated mouse strains. Results are the summary of 8 independent experiments with 12 WT, 11 PLZF+/−, and 5 PLZF−/− mice.
(C) Spleen iNKT cells in PLZF-deficient mice. iNKT cells were identified by CD1dTet and anti-TCRβ staining (top). Frequency and number of splenic iNKT cells were determined in the indicated mouse strains (bottom). Results are the summary of 8 independent experiments with 12 WT, 11 PLZF+/−, and 5 PLZF−/− mice.
Data are shown as mean ± SEM. Differences between groups were determined either with two-tailed Mann-Whitney U test or Kruskal-Wallis test to calculate p values, where *p < 0.05; **p < 0.01; ***p < 0.001 were considered statistically significant. NS, not significant.
PLZFGFPcre Impairs iNKT Cell Generation in BALB/c Thymocytes
To further demonstrate that the loss of PLZF is intrinsic to the PLZFGFPcre knockin gene, we explanted the PLZFGFPcre allele onto a different genetic background by backcrossing it from the C57BL/6 background to BALB/cAnNCrl mice that we obtained from the Charles River Laboratories (Frederick, MD, USA). Differences in genetic background can result in distinct outcomes of thymic iNKT cell development (Lee et al., 2013, 2015). Thus, we considered it important to examine whether the hypomorphic effect of PLZFGFPcre could be replicated independently of the mouse strain. To our satisfaction, the PLZFGFPcre allele significantly blunted iNKT cell generation also in mice of BALB/c background (Figure 3A). Both the frequencies and numbers of thymic iNKT cells were substantially reduced in heterozygous PLZFGFPcre+/wt and homozygous PLZFGFPcre+/+ BALB/c mice, compared with those in WT (PLZFwt/wt) BALB/c mice (Figure 3B). The generation of CD24lo mature iNKT cells was also significantly blunted in carriers of the PLZFGFPcre allele, compared to that in controls (Figure S2A). These results indicated that the PLZFGFPcre allele directly impairs iNKT cell generation, which was presumably caused by the attenuation of PLZF protein expression when produced from the PLZFGFPcre allele (Figure 3C). Indeed, the abundance of PLZF protein was significantly diminished in PLZFGFPcre carriers, as assessed by intracellular staining for PLZF proteins in iNKT cells (Figure S2B).
Figure 3. The PLZFGFPcre Knockin Allele Suppresses iNKT Cell Generation in BALB/c Thymocytes.
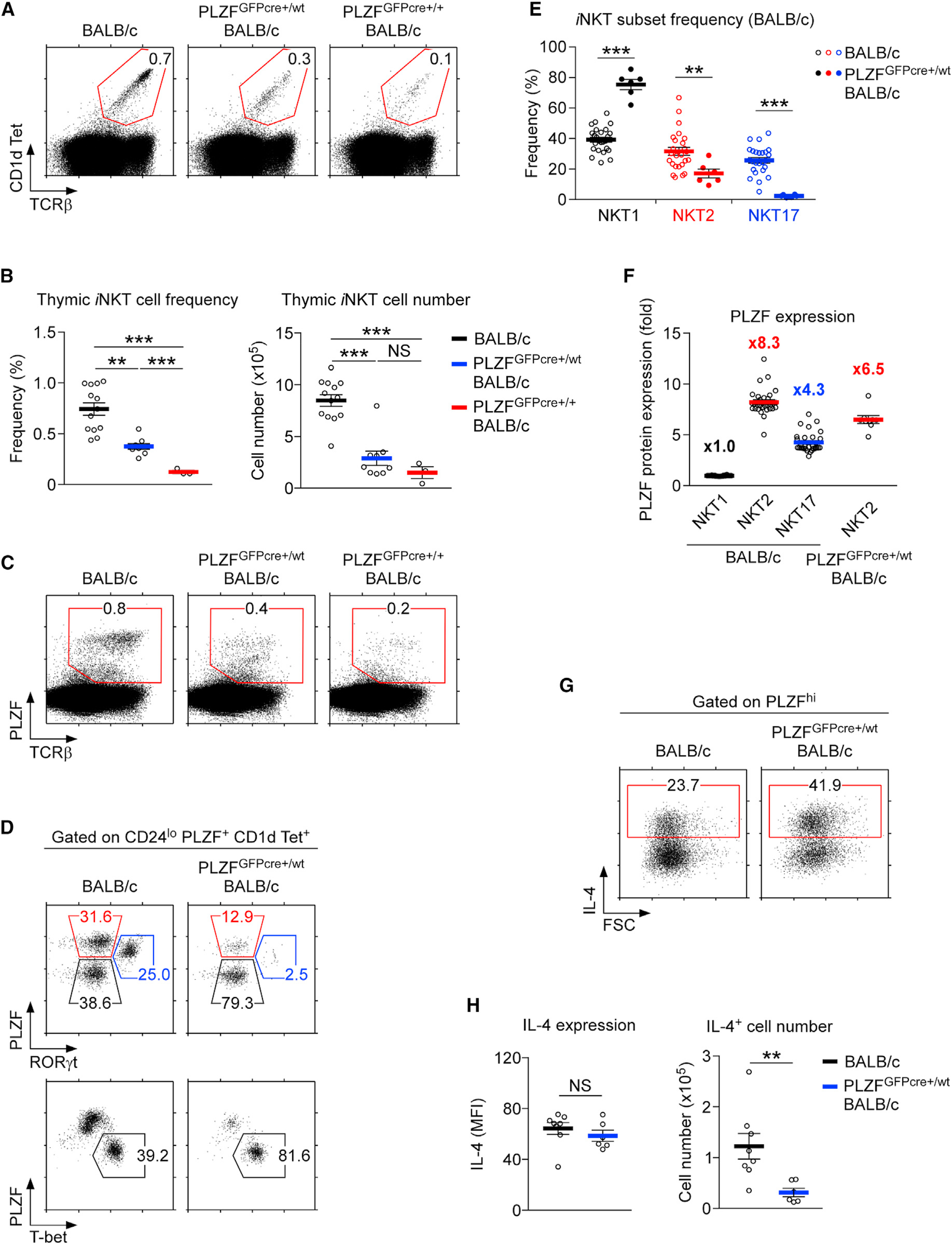
(A) Frequencies of thymic iNKT cells in BALB/c, PLZFGFPcre+/wt BALB/c, and PLZFGFPcre+/+ BALB/c mice. Dot plots are representative of 3 independent experiments.
(B) Frequencies (left) and numbers (right) of thymic iNKT cells in the indicated mouse strains. Results show the summary of 7 independent experiments with 13 BALB/c, 9 PLZFGFPcre+/wt BALB/c, and 3 PLZFGFPcre+/+ BALB/c mice.
(C) PLZF expression in thymic iNKT cells of indicated mice. Dot plots are representative of 3 independent experiments.
(D) iNKT subset distribution in BALB/c and PLZFGFPcre+/wt BALB/c mice. NKT1, NKT2, and NKT17 subsets were identified with PLZF versus RORγt (top) and PLZF versus T-bet staining (bottom). Results are representative of 6 independent experiments.
(E) Frequency of iNKT subsets in BALB/c and PLZFGFPcre+/wt BALB/c thymocytes. Results show the summary of 6 independent experiments with 25 BALB/c and 6 PLZFGFPcre+/wt BALB/c mice.
(F) Relative PLZF expression among iNKT subsets in BALB/c thymocytes, which was calculated as the fold difference compared to PLZF expression in NKT1 cells. Results show the summary of 6 independent experiments with 25 BALB/c and 6 PLZFGFPcre+/wt BALB/c mice.
(G) IL-4 expression was assessed in PLZFhi cells of PMA + ionomycin-stimulated BALB/c and PLZFGFPcre+/wt BALB/c thymocytes. Results are representative of 4 independent experiments.
(H) Amounts of IL-4 expression were quantified on a per-cell basis by mean fluorescence intensity (MFI; left). Numbers of IL-4-producing cells were assessed (right) from thymocytes of the indicated mice. Results indicate the summary of 4 independent experiments with 8 BALB/c and 6 PLZFGFPcre+/wt BALB/c mice.
Data are shown as mean ± SEM. Differences between groups were determined either with two-tailed Mann-Whitney U test or Kruskal-Wallis test to calculate p values, where **p < 0.01; ***p < 0.001 were considered statistically significant. NS, not significant.
PLZF expression differs among iNKT subsets. IL-4-producing NKT2 cells express the largest amount of PLZF, while IFNγ-producing NKT1 cells express the smallest amount of PLZF. To examine whether the failure to express the full amount of the PLZF protein would interfere with iNKT subset differentiation, we assessed iNKT subset distribution in WT and PLZFGFPcre+/wt heterozygous BALB/c mice. Compared to WT mice, in PLZFGFPcre+/wt BALB/c mice, we observed a dramatic decrease in the frequency of PLZFhi NKT2 cells (Figures 3D and 3E), which was accompanied by a relative increase in PLZFlow NKT1 cells (Figures 3D and 3E). Along these lines, we also documented a dramatic decrease in NKT2 cell numbers in PLZFGFPcre+/wt BALB/c thymocytes (Figure S2C). Of note, we did not examine the subset distribution in PLZFGFPcre+/+ homozygous BALB/c mice, because they lacked any meaningful number of iNKT cells.
To determine whether increased cell death or decreased cell generation were the reason for diminished NKT2 cell numbers, we examined intracellular expression of the pro-survival factors Bcl-xL and Bcl-2 in individual iNKT subsets. While Bcl-xL was abundant in all iNKT cells, the amount of Bcl-2 significantly differed among subsets (Figure S2D), suggesting that Bcl-2 is potentially a contributing factor to the differential survival of iNKT subsets. Next, we assessed intracellular Bcl-2 contents in WT versus PLZFGFPcre+/wt NKT2 cells. Notably, Bcl-2 expression was not decreased by the presence of PLZFGFPcre. In fact, Bcl-2 levels were increased in PLZFGFPcre NKT2 cells (Figure S2E), which effectively excluded increased cell death as the molecular basis of diminished NKT2 cell numbers in the thymus of PLZFGFPcre+/wt mice. Altogether, these results favor a model in which the PLZFGFPcre allele interferes with the differentiation of iNKT cells but not with their survival.
Along these lines, we found that PLZF expression significantly differed in NKT2 cells of WT and PLZFGFPcre+/wt mice (Figure 3F). In WT BALB/c mice, NKT2 cells expressed 8.3-fold more PLZF protein than that in NKT1 cells (Figure 3F; Figure S2F). NKT2 cells in PLZFGFPcre+/wt BALB/c thymocytes, on the other hand, only expressed 6.5-fold more PLZF protein than that in WT NKT1 cells (Figure 3F). Thus, PLZF levels in PLZFGFPcre+/wt NKT2 cells were approximately 25% lower than those in WT BALB/c NKT2 cells (Figure 3F). Notably, this was also the case for NKT1 and NKT17 cells of PLZFGFPcre+/wt mice, which contained smaller amounts of PLZF protein than those in their WT counterparts (Figure S2G). Despite their lower PLZF expression, however, PLZFGFPcre+/wt BALB/c NKT2 cells remained functional, as they expressed copious amounts of IL-4 upon Phorbol 12-myristate 13-acetate (PMA) and ionomycin stimulation (Figure 3G), which were comparable to those amounts in WT NKT2 cells on a per-cell basis (Figure 3H, left). Thus, the decreased amount of PLZF did not affect the function of PLZFGFPcre+/wt NKT2 cells. However, the decrease in PLZF significantly reduced the number of IL-4-producing NKT2 cells that were generated (Figure 3H, right). In contrast to PLZF, the abundance of other subset-specific transcription factors, such as T-bet for NKT1 and RORγt for NKT17 (Lee et al., 2013, 2015), remained unaffected (Figure S2H). Altogether, these results revealed that there was a quantitative aspect of PLZF expression in iNKT lineage choice, in which the high abundance of PLZF is associated with NKT2 lineage commitment that is separate from the effector function of NKT2 cells.
To directly demonstrate the quantitative effect of PLZF protein expression on iNKT subsets, and specifically on NKT2 cell differentiation, we bred the Zbtb16 knockout allele (PLZF−) into BALB/c mice. Unlike C57BL/6 mice, BALB/c mice contain large numbers of NKT2 cells (Lee et al., 2013, 2015), so it was important to assess the effect of PLZF haploinsufficiency in mice on a BALB/c background. PLZF+/− BALB/c mice can produce only half the amount of PLZF proteins compared to that in WT BALB/c mice. Therefore, PLZF+/− BALB/c mice are comparable to PLZFGFPcre+/+ mice regarding their PLZF protein contents, and both mice only express 50% of PLZF compared to WT BALB/c mice. Strikingly, losing one copy of Zbtb16 was already enough to virtually abolish iNKT cell generation in the thymus, and we could not detect any consequential numbers of thymic iNKT cells in PLZF+/− BALB/c mice (Figure S3A). Because the frequency and number of peripheral iNKT cells were also significantly reduced in PLZF+/− BALB/c mice (Figure S3B), these results agree with the results from our PLZFGFPcre+/wt BALB/c analysis (Figures 3A and 3B), which showed that the ability to produce the full amount of PLZF is critical for effective iNKT cell generation and maintenance. Moreover, assessing the subset composition of splenic iNKT cells revealed a substantial decrease in NKT2 cell frequencies that was concomitant with a dramatic increase in NKT1 abundance (Figure S3C). These results agree with and validate our findings that the quantity of the PLZF protein plays a critical role in determining iNKT lineage fate.
The PLZFGFPcre Allele Attenuates Innate CD8 T Cell Development
A major function of IL-4-producing NKT2 cells is the generation of innate CD8 T cells in the thymus (Lee et al., 2013; Weinreich et al., 2010). In contrast to conventional CD8 T cells, innate CD8 T cells are thymus-generated effector T cells that are antigen inexperienced but that express large amounts of IFNγ (Jameson et al., 2015). Generation of innate CD8 T cells requires IL-4 signaling, and NKT2 cells constitute a major source of intrathymic IL-4 (Lee et al., 2013; Yoshimoto and Paul, 1994). BALB/c thymocytes harbor a large population of innate CD8 T cells because they contain a large fraction of IL-4-producing NKT2 cells (Lai et al., 2011; Lee et al., 2013). Notably, introducing the PLZFGFPcre allele into BALB/c mice caused a significant decrease in the frequency of CD8 single-positive (SP) thymocytes (Figure 4A; Figure S4A). The loss of CD8SP thymocytes was further associated with decreased frequencies and numbers of CD44+ IL-2Rβ+ memory phenotype CD8 cells (Figures 4B and 4C). CD8SP cells in PLZFGFPcre+/wt BALB/c thymocytes were further marked by the downregulation of IL-4Rα and the upregulation of CD24 expression (Figure 4D), demonstrating that the loss of NKT2 cells halted the development of innate CD8 T cells in the thymus. In agreement, expression of the transcription factor eomesodermin (Eomes) (Figure 4E) and PMA and ionomycin-induced IFNγ expression, both of which are markers of innate CD8 T cells (Jameson et al., 2015), were dramatically reduced among CD8SP cells in PLZFGFPcre+/wt BALB/c mice compared to those in WT mice (Figure 4F). Interestingly, the frequency and numbers of thymic Vγ1.1Vδ6.3 γδ T cells remained unaffected in these mice (Figures S4B–S4D), indicating that quantitative differences in PLZF protein expression specifically affect iNKT and innate CD8 T cells. Collectively, these data demonstrated that the quantity of PLZF proteins controls not only the generation but also the subset composition of thymic iNKT cells, resulting in the attenuation of innate CD8 T cell generation.
Figure 4. PLZFGFPcre Knockin BALB/c Mice Fail to Generate Innate CD8 T Cells in the Thymus.
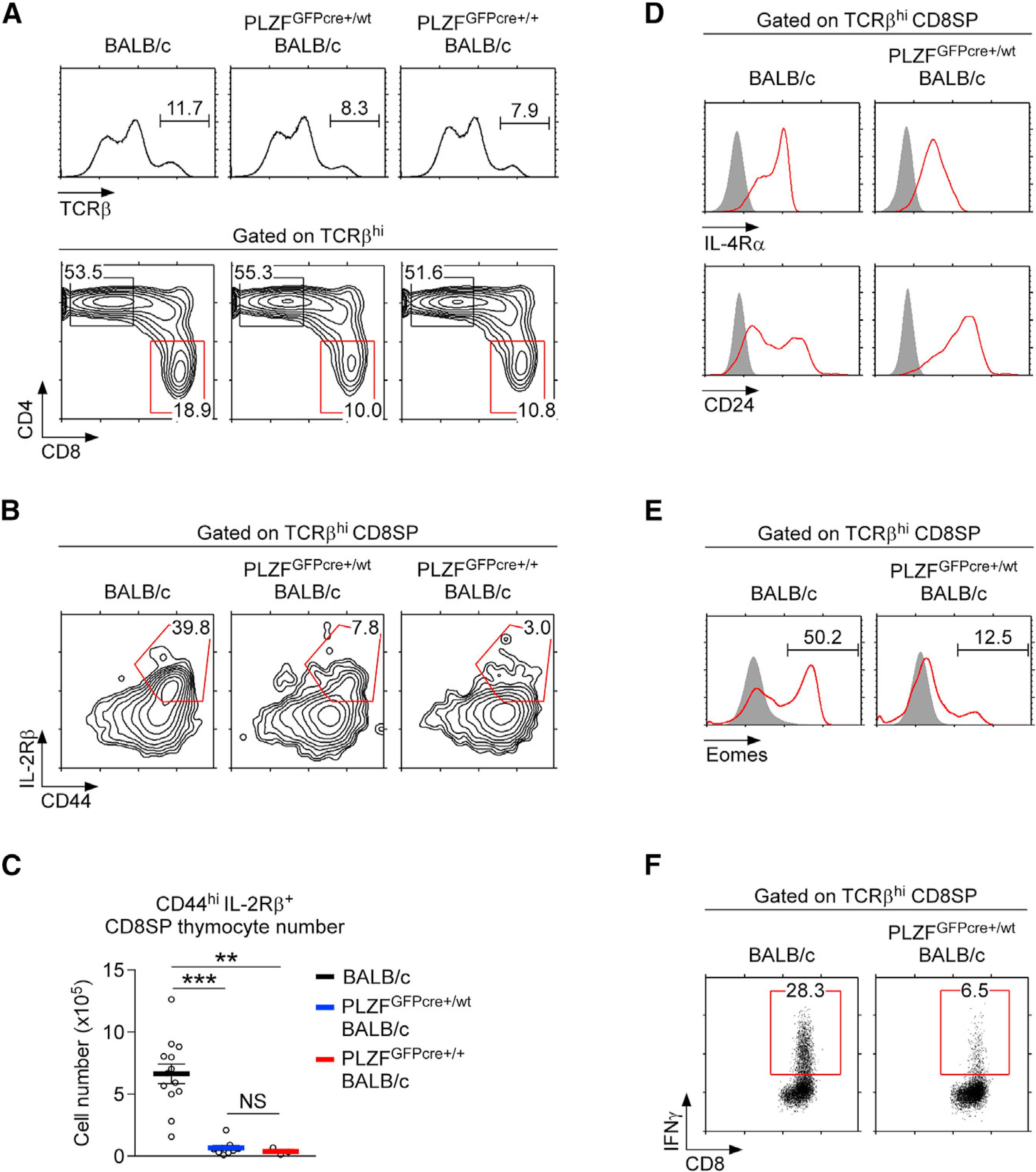
(A) CD4 versus CD8 profiles of TCRβhi mature thymocytes in BALB/c, PLZFGFPcre+/wt BALB/c, and PLZFGFPcre+/+ BALB/c mice. Results are representative of 3 independent experiments with 13 BALB/c, 9 PLZFGFPcre+/wt BALB/c, and 3 PLZFGFPcre+/+ BALB/c mice.
(B and C) CD44 and IL-2Rβ expression profiles (B) and numbers of CD44hi IL-2Rβ+ cells (C) were assessed in TCRβhi CD8SP thymocytes of BALB/c, PLZFGFPcre+/wt BALB/c, and PLZFGFPcre+/+ BALB/c mice. Results are representative or the summary of 3 independent experiments with 13 BALB/c, 9 PLZFGFPcre+/wt BALB/c, and 3 PLZFGFPcre+/+ BALB/c mice.
(D and E) Surface IL-4Rα and CD24 expression (D) and intracellular eomesodermin expression (E) were assessed on TCRβhi CD8SP thymocytes of BALB/c and PLZFGFPcre+/wt BALB/c mice.
Results are representative of 3 independent experiments.
(F) IFNγ expression in PMA + ionomycin-stimulated TCRβhi CD8SP thymocytes of BALB/c and PLZFGFPcre+/wt BALB/c mice. Results are representative of 4 independent experiments. Results are representative of 3 independent experiments Data are shown as mean ± SEM. Differences between groups were determined either with two-tailed Mann-Whitney U test or Kruskal-Wallis test to calculate p values, where **p < 0.01; ***p < 0.001 were considered statistically significant. NS, not significant.
DISCUSSION
Mounting evidence suggests that quantitative differences in transcription factors can play critical roles in the lineage choice and differentiation of lymphocytes (David-Fung et al., 2009; Scripture-Adams et al., 2014; Sigvardsson, 2012). Whether distinct amounts of PLZF protein can control iNKT subset fate is an interesting question that remains unresolved (Buechel et al., 2015). In the present study, we characterized a hypomorphic allele of Zbtb16, PLZFGFPcre, which was originally designed with the aim of reporting Zbtb16 gene transcription (Constantinides et al., 2014). For reasons that remain unknown to us, we found that the EGFP-Cre reporter gene insertion impaired the expression of the PLZF protein from this allele. Consequently, carriers of the PLZFGFPcre allele produced significantly decreased amounts of PLZF proteins in iNKT cells, compared to that of WT mice. Notably, an ~25% reduction in PLZF abundance, as observed in PLZFGFPcre+/wt heterozygous mice, was sufficient to markedly impair iNKT cell generation in the thymus, compared to that in WT mice.
Surprisingly, we also observed that the subset composition of PLZFGFPcre+/wt thymic iNKT cells was substantially altered. In PLZFGFPcre+/wt mice, the iNKT lineage choice was heavily skewed toward NKT1 cells, and we found that NKT2 cell frequencies and numbers were significantly diminished, compared to those in WT mice. Because the generation of IFNγ-producing innate CD8 T cells depends on NKT2 cells (Jameson et al., 2015; Lee et al., 2013), PLZFGFPcre carriers further failed to produce innate CD8 T cells when this allele was introduced to the BALB/c background. Collectively, these results directly illustrate that not only PLZF expression itself but also the abundance of the PLZF protein is a controlling factor of iNKT cell development.
While NKT2 cells reportedly contain the largest amount of PLZF among iNKT subsets (Constantinides and Bendelac, 2013; Lee et al., 2013), it was not clear how much more PLZF protein they express compared to that in NKT1 and NKT17 cells. Here, we quantified the amount of PLZF protein for each iNKT subset, and we found that NKT2 cells contained approximately 8-fold more and that NKT17 cells contained approximately 4-fold more PLZF protein than that in NKT1 cells. Because we also demonstrated that Zbtb16 is transcribed in a biallelic manner, these results suggest that NKT2 cells express 4 times more PLZF protein than NKT1 cells per Zbtb16 allele. Along these lines, NKT17 cells express 2 times more PLZF proteins per Zbtb16 allele than NKT1 cells do.
The notion that the abundance of PLZF differed by a series of 2-fold differences between the iNKT subsets was intriguing to us. Conventionally, the common precursor of all iNKT subsets is thought to express high levels of PLZF that are comparable to the levels expressed in mature NKT2 cells (Kovalovsky et al., 2008; Lee et al., 2013; Pobezinsky et al., 2015). In this scenario, the amount of PLZF in postselection iNKT cells would be set either by maintaining or degrading pre-existing PLZF proteins (Pobezinsky et al., 2015). Because we found that PLZF expression was diminished in a 2-fold manner, we considered the possibility that the decrease in PLZF expression could be a result of cell proliferation upon thymic positive selection. Accordingly, pre-existing PLZF protein would be diluted by binary cell division so that NKT1 cells, which are the progenies of PLZFhigh immature iNKT precursors (Gapin, 2016; Lee et al., 2013; Pobezinsky et al., 2015), would have undergone three cell divisions (23, 8-fold dilution), NKT17 cells would have undergone one cell division (21, 2-fold dilution), and NKT2 cells would not have divided during the lineage choice. Whether such dilution of pre-existing PLZF protein is sufficient to explain the distinct PLZF levels among iNKT cells remains to be assessed.
As an alternative model, we also considered the possibility that all iNKT subsets branch out from a PLZF-negative precursor population and that lineage-specific signals would induce upregulation of PLZF protein expression to specific levels. In this scenario, qualitative or quantitative differences in TCR or cytokine receptor signaling would induce distinct amounts to PLZF that would result in different amounts of PLZF expression, directing distinct lineage fates. In this model, however, it is difficult to explain the mechanism and to identify the signals that would induce an 8-fold versus a 4-fold increase in PLZF expression, depending on the iNKT subset. A better understanding of the upstream regulatory signals of PLZF expression is necessary to answer these questions.
Recent chromatin immunoprecipitation sequencing (ChIP-seq) analysis of total thymocytes revealed that more than 60% of PLZF-occupied sites contained consensus sequences for ETS, E proteins, and RUNX family factors (Mao et al., 2016). These data also raised the possibility that PLZF does not directly bind to DNA but rather indirectly associates with DNA and utilizes other prominent transcription factors to control gene expression. These results agree with other reports that highlighted a critical function of PLZF as an epigenetic regulator involved in chromosome remodeling that indirectly facilitated the expression of target genes (Barna et al., 2002; Mathew et al., 2012). Based on these reports, we consider the possibility that discrete abundance of PLZF expression in distinct iNKT subsets may result in differences in the coverage of PLZF binding sites in the genome. Variations in PLZF availability would then generate a different epigenomic landscape, depending on the amount of PLZF protein. Accordingly, NKT2 cells that express the largest amount of PLZF proteins would have almost every PLZF target site occupied, and such saturated promoter or enhancer occupancy of target genes would induce expression of a full spectrum of PLZF-controlled genes, which are necessary to impose NKT2 lineage fate. In this scenario, the decreased abundance of PLZF proteins, as found in PLZFGFPcre+/− and PLZF+/− mice, would result in the diminished or redistributed occupancy of PLZF-controlled gene regulatory sites, thus altering iNKT subset fate. NKT1 cells express the lowest amount of PLZF among iNKT subsets, and the low abundance of PLZF protein would be insufficient to saturate all PLZF-target loci. Consequently, in NKT1 cells, PLZF would be preferentially recruited to high-affinity PLZF-binding sites, thus activating a distinct transcriptional profile that is unique to the NKT1 subset. In this regard, the loss of NKT2 and the increased frequency of NKT1 cells in PLZFGFPcre+/− and PLZF+/− mice agree with there being a quantitative aspect of PLZF expression in iNKT subset differentiation.
Finally, we want to point out that the PLZFGFPcre allele is also used to recombine floxed genes for their conditional deletion in PLZF-expressing cells (Constantinides et al., 2014; Thapa et al., 2017). However, because PLZFGFPcre significantly impairs the generation of PLZF+ iNKT cells, it needs to be cautioned that the effect of gene deletions by PLZFGFPcre should be interpreted in the context of diminished production of PLZF-expressing cells. Thus, if the PLZFGFPcre allele is utilized only for the purpose of expressing the Cre recombinase under the control of Zbtb16 regulatory elements, it would be advisable to utilize mice that express PLZF-driven Cre without interfering with the endogenous Zbtb16 gene, as recently described for a PLZF-Cre BAC transgene (Zhang et al., 2015).
Altogether, the PLZFGFPcre reporter mice provided us with a tool to examine a quantitative aspect of PLZF expression during iNKT cell development, and here, we demonstrated that iNKT lineage choice is exquisitely sensitive to the abundance of PLZF protein. Thus, these results demonstrate that the quantitative regulation of PLZF protein is a previously unappreciated control mechanism of cell-fate decision during thymic T cell development.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jung-Hyun Park (parkhy@mail.nih.gov).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
C57BL/6 and BALB/c mice were obtained from the Charles River Laboratories. PLZFGFPcre (B6(SJL)-Zbtb16 tm1.1(EGFP/cre)Aben/J) mice were previously described (Constantinides et al., 2014), and purchased from the Jackson Laboratory. PLZFGFPcre BALB/c mice were generated in house by backcrossing PLZFGFPcre mice onto BALB/c background for > 8 generations. PLZF-deficient mice (PLZF−/−) were kindly provided by Dr. P.P. Pandolfi (Harvard Medical School, Boston, MA) (Barna et al., 2000). All experimental mice were analyzed between 6–12 weeks of age, using age- and sex-matched controls. Animal experiments were approved by the NCI Animal Care and Use Committee. All mice were cared for in accordance with NIH guidelines. Genotypes of mice were determined by PCR analysis using tail DNA. The PLZF-knock-out allele was identified by genomic PCR using the following primer pairs: Genotype Zbtb16 WT (Forward: 5′-GCAGATCTGGGACCACCATC-3′; Reverse: 5′-GCTGCATACAGCAGGTCATC-3′), Genotype Zbtb16 KO (Forward: 5′-GCAGATCTGGGACCACCATC-3′; Reverse: 5′-GCTTCCTCGTGCTTTACGGTATC-3′). The PCR reactions were set up as follows: one cycle of denaturation for 5 min at 95°C, followed by 35 cycles of 30 s at 95°C, 30 s at 62°C, and 30 s at 72°C, followed by a final extension cycle of 10 min at 72°C. The expected PCR product size of the wild-type allele is 523 bp and the knock-out allele is 530 bp. The PLZFGFPcre allele was screened using the following primer pairs; WT Forward: 5′-CTCCTCCATGCAGAAACACA-3′, Rep Forward: 5′-CCCCAGGAAATAATCCAAGG-3′, Reverse: 5′-TAGTGAAACAGGGGCAATGG-3′). The PCR reactions were set up as follows: one cycle of denaturation for 5 min at 95°C, followed by 40 cycles of 25 s at 95°C, 20 s at 62°C, and 30 s at 72°C, followed by a final extension cycle of 6 min at 72°C. The expected PCR product size of the wild-type allele is 281 bp and the PLZFGFPcre allele is 380 bp.
METHOD DETAILS
Flow cytometry
Lymphocyte suspensions were prepared from thymus, spleen, and liver, and subjected to flow cytometry using specific gating strategies as previously described (Park et al., 2016). In brief, live cells were gated using the forward scatter exclusion of dead cells stained with propidium iodide. Flow cytometric data were acquired on LSR Fortessa or LSRII flow cytometers (BD Biosciences) and were analyzed with either a software (Active Control version 4.2) designed by the Division of Computer Research and Technology, NIH, or with FlowJo (FlowJo). Antibodies with the following antigen specificities were used for staining: CD8 (clone 53–6.7), CD4 (clone GK1.5), TCRβ (clone H57–597), CD44 (clone IM7), CD25 (clone PC61.5), CD69 (clone H1.2F3), and CD45 (clone 30-F11) from eBioscience Thermo Fisher; CD122 (clone TM-β1), CD124 (clone mIL4R-M1), Vδ6.3/2 (clone 8F4H7B7), and TCRγδ (clone GL3) from BD Biosciences; TCRβ (clone H57–597), Vγ1.1 (clone 2.11) and CD24 (clone M1/69) from BioLegend. Fluorochrome-conjugated CD1d tetramers loaded with PBS-57 and unloaded controls were obtained from the NIH tetramer facility (Emory University, Atlanta, GA).
Lymphocyte isolation from lymphoid tissues and the liver
Single cell suspensions were prepared from thymus and spleen by gently teasing the tissues with forceps, resuspending the processed tissues in ice-cold harvest media (10% FCS in RPMI-1640 medium), and filtering the cell suspension through a 70 μm Nylon filter mesh (Millipore Sigma). Cells were washed once in harvest media by centrifugation for 7 min at 1,500 rpm, and resuspended in FACS buffer (0.5% BSA, 0.1% sodium azide in PBS) for staining. Liver mononuclear cells (MNC) were prepared as previously described with minor modifications (Watarai et al., 2008, Zhang et al., 2005). In brief, liver tissues were pressed through a 70 μm cell strainer (BD Biosciences) and resuspended in ice-cold PBS. Cell suspensions were centrifuged at 100 g for 3 min to remove tissue debris, and supernatants were collected, spun down, and washed again with ice-cold PBS. The cell pellet was resuspended in 10 mL of 40% Percoll in PBS (GE Life Sciences) and layered on 10 mL of 70% Percoll in PBS. The Percoll gradient was centrifuged at room temperature for 30 min at 1,250 rpm. Lymphocytes at the interphase were harvested and transferred into a new 50 mL conical tube. Collected interphase lymphocytes were then washed twice with harvest media (10% FCS in RPMI-1640 medium) and resuspended in harvest media before further analysis. MNCs were identified by expression of CD45.
Reverse transcription real-time PCR
iNKT cells were electronically sorted from pooled thymocytes of wild-type or PLZFGFPcre+/wt mice using a FACSAria (BD Biosciences). Total RNA was isolated from sorted cells using the RNeasy Plus Micro kit (QIAGEN) and reverse transcribed into cDNA by oligo (dT) priming with the QuantiTect reverse transcription kit (QIAGEN). Quantitative RT-PCR (qRT-PCR) was performed with the QuantiTect SYBR green detection system (QIAGEN) and analyzed on an ABI PRISM 7900HT (Life technologies). The following primers were used for detecting gene expression: Zbtb16 (Forward: 5′-TTGGGACGGACCCACACCCA-3′; Reverse: 5′-GGCGTGGCCGGTGAATAGGG-3′), Rpl13 (Forward: 5′-CGAGGCATGCTGCCCCACAA-3′; Reverse: 5′-AGCAGGGACCACCATCCGCT-3′).
iNKT subset staining
In brief, iNKT cells were stained with PBS-57-loaded mouse CD1d tetramers followed by staining for other surface markers, as previously described (Park et al., 2016). Specifically, for each analysis, 10 million thymocytes were stained with fluorochrome-conjugated CD1d tetramers in FACS buffer (0.5% BSA, 0.1% sodium azide in Ca2+ and Mg2+-free HBSS) for 20 min at 4°C. Without removing the tetramer reagents, antibodies against surface proteins were then added, and cells were incubated for an additional 30 min at 4°C. Excess reagents were then washed out with FACS buffer by centrifugation for 7 min at 1,500 rpm. Pelleted cells were then resuspended in 150 μL of 1:3 mixture of concentrate/diluent working solution of the Foxp3 Transcription Factor Staining Buffer kit (eBioscience Thermo Fisher) and 100 μL of FACS buffer and incubated at room temperature for 20 minutes. Cells were then washed twice with 1x permeabilization buffer (eBioscience Thermo Fisher), before adding antibodies for transcription factors, such as PLZF, RORγt and T-bet. Cells were incubated at room temperature for 1 hour, before washing out excess reagents with FACS buffer and analysis. Antibodies with the following antigen specificities were used for iNKT subset staining: Eomes (clone Dan11mag), RORγt (clone AFKJS-9) and T-bet (clone eBio4B10) from eBioscience Thermo Fisher; PLZF (clone R17–809) and RORγt (clone Q31–378) from BD Biosciences; PLZF (clone 9E12) from BioLegend.
Intracellular cytokine staining
Thymocytes were stimulated for 4 hours with PMA (25 ng/ml) and ionomycin (1 μM) (both from Sigma) in the presence of 3.0 μg/ml brefeldin A (eBioscience Thermo Fisher) for intracellular cytokine staining. Stimulation was terminated by washing cells in ice-cold FACS buffer (0.5% BSA 0.1% sodium azide in HBSS). Dead cells were counter-stained with 1 ul of Aqua Live/Dead solution per staining (eBioscience Thermo Fisher) for 30 min at 4°C, and excess reagents were washed out with ice-cold FACS buffer. Surface proteins were stained for 20 min at 4°C, and excess reagents were washed out with FACS buffer by centrifugation for 7 min at 1,500 rpm. Pelleted cells were then resuspended in 100 μL of FACS buffer, to which 150 μL of a 1:3 mixture of concentrate/diluent working solution of the Foxp3 Transcription Factor Staining Buffer kit (eBioscience Thermo Fisher) was added. Cells were then incubated at room temperature for 20 minutes. Cells were washed twice with 1x permeabilization buffer (eBioscience Thermo Fisher) before adding antibodies for transcription factors and cytokines, such as PLZF, IL-4, IL-17, and IFNγ. Excess reagents were washed out with FACS buffer before analysis. Antibodies with the following antigen specificities were used; IL-4 (clone 11B11) and IL-17 (clone eBio17B7) from eBioscience Thermo Fisher; IFNγ (clone XMG1.2) from BioLegend; PLZF (clone R17–809) from BD Biosciences.
Intracellular Bcl-2 and Bcl-xL staining of iNKT cells
iNKT cells were first identified by PBS-57-loaded mouse CD1d tetramers followed by staining for other surface markers such as TCRβ and CD24. Excess reagents were then washed out with FACS buffer by centrifugation for 7 min at 1,500 rpm. Pelleted cells were resuspended in 150 μL of 1:3 mixture of concentrate/diluent working solution of the Foxp3 Transcription Factor Staining Buffer kit (eBioscience Thermo Fisher) and 100 μL of FACS buffer and incubated at room temperature for 20 minutes. Cells were washed twice with 1x permeabilization buffer before adding antibodies for transcription factors, such as PLZF, RORγt and T-bet, and antibodies for cytosolic Bcl-2 or Bcl-xL proteins. Cells were incubated with the antibodies at room temperature for 1 hour, before analysis. Antibodies for Bcl-xL (clone 54H6) and isotype control (clone DA1E) were purchased from Cell Signaling Technology. Antibodies for Bcl-2 (clone 3F11) and isotype control (clone A19–3) were purchased from BD Biosciences.
Intranuclear staining for Foxp3 proteins
Single cell suspensions of thymocytes were first stained with saturating amounts of anti-CD4, CD8 and CD25 antibodies for 30 min on ice. Excess reagents were washed out with FACS buffer (0.5% BSA, 0.1% sodium azide in PBS) by centrifugation for 7 min at 1,500 rpm. Pelleted cells were then resuspended in 200 μL of 1:3 mixture of concentrate/diluent working solution of the Foxp3 Transcription Factor Staining Buffer kit (eBioscience Thermo Fisher) at room temperature for 20 minutes. Cells were washed twice with 1x permeabilization buffer and stained with anti-Foxp3 antibodies (clone FJK16s; eBioscience Thermo Fisher) for 1 hour, before washing out excess reagents with FACS buffer for analysis.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis
Data are shown as means ± s.e.m. Differences between groups were determined either with two-tailed Mann-Whitney U test or Kruskal-Wallis test to calculate P-values, where *, p < 0.05; **, p < 0.01; ***, p < 0.001 were considered statistically significant. NS, not significant. Number of experimental repeats is indicated in each figure legend. Samples were not randomized, and investigators were not blinded to sample identities. All statistical data analyses were performed using GraphPad Prism 7 software (GraphPad software).
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| APC-eFluor 780 anti-CD8 (clone 53-6.7) | eBioscience Thermo Fisher | Cat#: 47-0081-82; RRID:AB_1272185 |
| PE-Cy7 anti-CD4 (clone GK1.5) | eBioscience Thermo Fisher | Cat#: 25-0041-82; RRID: AB_ 469575 |
| FITC anti-TCRβ (clone H57-597) | eBioscience Thermo Fisher | Cat#: 11-5961-82; RRID: AB_ 465322 |
| PE-Cy7 anti-TCRβ (clone H57-597) | eBioscience Thermo Fisher | Cat#: 25-5961-82; RRID:AB_ 2573506 |
| APC anti-CD44 (clone IM7) | eBioscience Thermo Fisher | Cat#:17-0441-81; RRID:AB_469389 |
| eFluor 660 anti-Eomes (clone Dan11mag) | eBioscience Thermo Fisher | Cat#: 50-4875-82; RRID:AB_ 2574226 |
| APC anti-RORγt (clone AFKJS-9) | eBioscience Thermo Fisher | Cat#: 17-6988-82; RRID:AB_ 1633425 |
| APC anti-IL-4 (clone 11B11) | eBioscience Thermo Fisher | Cat#: 17-7041-82; RRID:AB_469493 |
| APC anti-IL-17 (clone eBio17B7) | eBioscience Thermo Fisher | Cat#: 17-7177-81; RRID:AB_ 763580 |
| eFluor 660 anti-T-bet (clone eBio4B10) | eBioscience Thermo Fisher | Cat#: 50-5825-82; RRID:AB_ 10596655 |
| eFluor 660 anti-Foxp3 (clone FJK-16 s) | eBioscience Thermo Fisher | Cat#: 50-5773-82; RRID:AB_ 11220082 |
| PE anti-CD25 (clone PC61.5) | eBioscience Thermo Fisher | Cat#: 12-0251-82; RRID:AB_ 465606 |
| PE-Cy7 anti-CD25 (clone PC61.5) | eBioscience Thermo Fisher | Cat#: 25-0251-82; RRID:AB_ 469607 |
| PE anti-CD69 (clone H1.2F3) | eBioscience Thermo Fisher | Cat#: 12-0691-82; RRID:AB_ 465731 |
| APC-eFluor 780 anti-CD45 (clone 30-F11) | eBioscience Thermo Fisher | Cat#: 47-0451-82; RRID:AB_ 1548790 |
| PE anti-Bcl-xL (clone 54H6) | Cell Signaling Technology | Cat#: 13835S; RRID:AB_ 2228008 |
| PE isotype control (clone DA1E) | Cell Signaling Technology | Cat#: 5742S; RRID:AB_1550038 |
| Alexa Fluor 647 anti-PLZF (clone R17-809) | BD Biosciences | Cat#: 563490; RRID:AB_2738238 |
| FITC anti-CD122 (clone TM-β1) | BD Biosciences | Cat#: 553361; RRID:AB_ 394808 |
| PE anti-CD124 (clone mIL4R-M1) | BD Biosciences | Cat#: 552509; RRID:AB_ 394407 |
| Alexa Fluor 647 anti-RORγt (clone Q31-378) | BD Biosciences | Cat#: 562682; RRID:AB_2687546 |
| PE anti-Bcl-2 (clone 3F11) | BD Biosciences | Cat#: 556537; RRID:AB_396457 |
| PE Bcl-2 isotype control (clone A19-3) | BD Biosciences | Cat#: 556537; RRID:AB_470073 |
| PE anti-Vδ 6.3/2 (clone 8F4H7B7) | BD Biosciences | Cat#: 555321; RRID:AB_395729 |
| PE anti-TCRγδ (clone GL3) | BD Biosciences | Cat#: 553178; RRID:AB_394689 |
| Alexa 647 anti-TCRβ (clone H57-597) | BioLegend | Cat#: 109218; RRID:AB_ 493346 |
| APC anti-IFNγ (clone XMG1.2) | BioLegend | Cat#: 505810; RRID:AB_ 315404 |
| PE anti-PLZF (clone 9E12) | BioLegend | Cat#: 145804; RRID:AB_ 2561973 |
| PE-Cy7 anti-CD24 (clone M1/69) | BioLegend | Cat#: 101822; RRID:AB_ 756048 |
| APC anti-Vγ1.1 (clone 2.11) | BioLegend | Cat#: 141108; RRID:AB_ 10901177 |
| CD1d tetramers (PBS-57 loaded) | NIH tetramer facility | Emory University, Atlanta, Georgia |
| Chemicals, Peptides, and Recombinant Proteins | ||
| LIVE/DEAD Fixable Aqua Dead Cell Stain Kit | eBioscience Thermo Fisher | Cat#: L34957 |
| PMA | Sigma Aldrich | Cat#: P8139 |
| Ionomycin | Sigma Aldrich | Cat#: I0634 |
| Brefeldin A | eBioscience Thermo Fisher | Cat#: 00-4506-51 |
| QuantiTect SYBR® Green PCR Kits | QIAGEN | Cat#: 204145 |
| RNeasy Plus Micro kit | QIAGEN | Cat#: 74034 |
| QuantiTect reverse transcription kit | QIAGEN | Cat#: 205313 |
| Percoll | GE Life Sciences | Cat#: 17089101 |
| Critical Commercial Assays | ||
| eBioscience Fixation/Perm diluents | eBioscience Thermo Fisher | Cat#:00-5223-56 |
| eBioscience Fixation/Perm concentrate | eBioscience Thermo Fisher | Cat#:00-5213-43 |
| Permeabilization Buffer 10x | eBioscience Thermo Fisher | Cat#: 00-8333-56 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6 (C57BL/6NCrl) | Charles River Laboratories | Stock#: 24107773, 24107757 |
| Mouse: BALB/c (BALB/cAnNCrl) | Charles River Laboratories | Stock#: 24107700, 24107684 |
| Mouse: PLZFGFPcre (B6(SJL)-Zbtb16 tm1.1(EGFP/cre)Aben/J) | (Constantinides et al., 2014) The Jackson Laboratory | Stock#: 024529 |
| Mouse: PLZF-deficient mice (PLZF−/−) | Barna et al., 2000 | Provided by Dr. P.P. Pandolfi (Harvard Medical School, Boston, MA) |
| Oligonucleotides | ||
| RT-PCR Zbtb16-Forward: 5′-TTGGGACGGACCCACACCCA-3′ | This paper | N/A |
| RT-PCR Zbtb16-Reverse: 5′-GGCGTGGCCGGTGAATAGGG-3′ | This paper | N/A |
| RT-PCR Rpl13-Forward: 5′-CGAGGCATGCTGCCCCACAA-3′ | This paper | N/A |
| RT-PCR Rpl13-Reverse: 5′-AGCAGGGACCACCATCCGCT-3′ | This paper | N/A |
| Genotype Zbtb16 WT Forward: 5′-GCAGATCTGGGACCACCATC-3′ | This paper | N/A |
| Genotype Zbtb16 WT Reverse: 5′-GCTGCATACAGCAGGTCATC-3′ | This paper | N/A |
| Genotype Zbtb16 KO Forward: 5′- GGTGCCAATGTCACTTATAGAACC-3′ | This paper | N/A |
| Genotype Zbtb16 KO Reverse: 5′- GCTTCCTCGTGCTTTACGGTATC-3′ | This paper | N/A |
| Genotype PLZFGFP/CRE WT Forward: 5′-CTCCTCCATGCAGAAACACA-3′ | This paper | N/A |
| Genotype PLZFGFP/CRE Reverse: 5′-TAGTGAAACAGGGGCAATGG-3′ | This paper | N/A |
| Genotype PLZFGFP/CRE Rep Forward: 5′-CCCCAGGAAATAATCCAAGG-3′ | This paper | N/A |
| Software and Algorithms | ||
| GraphPad Prism 7 | GraphPad software | N/A |
| FlowJo software version 10.2 | FlowJo | N/A |
| Active Control 4.2.0.7 | Division of Computer Research and Technology, NIH | N/A |
| Other | ||
| Canvas X | Canvas GFX | N/A |
| BD FACS LSRII | BD Biosciences | N/A |
| BD FACS LSRFortessa | BD Biosciences | N/A |
ACKNOWLEDGMENTS
We thank Drs. D. Kovalovsky and A. Singer for the critical review of this manuscript. We also thank the members of the Park lab for technical assistance and discussion. This work was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.05.012.
REFERENCES
- Alonzo ES, and Sant’Angelo DB (2011). Development of PLZF-expressing innate T cells. Curr. Opin. Immunol. 23, 220–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barna M, Hawe N, Niswander L, and Pandolfi PP (2000). Plzf regulates limb and axial skeletal patterning. Nat. Genet. 25, 166–172. [DOI] [PubMed] [Google Scholar]
- Barna M, Merghoub T, Costoya JA, Ruggero D, Branford M, Bergia A, Samori B, and Pandolfi PP (2002). Plzf mediates transcriptional repression of HoxD gene expression through chromatin remodeling. Dev. Cell 3, 499–510. [DOI] [PubMed] [Google Scholar]
- Bedel R, Berry R, Mallevaey T, Matsuda JL, Zhang J, Godfrey DI, Rossjohn J, Kappler JW, Marrack P, and Gapin L (2014). Effective functional maturation of invariant natural killer T cells is constrained by negative selection and T-cell antigen receptor affinity. Proc. Natl. Acad. Sci. USA 111, E119–E128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buechel HM, Stradner MH, and D’Cruz LM (2015). Stages versus subsets: Invariant natural killer T cell lineage differentiation. Cytokine 72, 204–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen NR, Garg S, and Brenner MB (2009). Antigen presentation by CD1 lipids, T cells, and NKT cells in microbial immunity. Adv. Immunol. 102, 1–94. [DOI] [PubMed] [Google Scholar]
- Constantinides MG, and Bendelac A (2013). Transcriptional regulation of the NKT cell lineage. Curr. Opin. Immunol. 25, 161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinides MG, McDonald BD, Verhoef PA, and Bendelac A (2014). A committed precursor to innate lymphoid cells. Nature 508, 397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispe IN (2009). The liver as a lymphoid organ. Annu. Rev. Immunol. 27, 147–163. [DOI] [PubMed] [Google Scholar]
- D’Cruz LM, Stradner MH, Yang CY, and Goldrath AW (2014). E and Id proteins influence invariant NKT cell sublineage differentiation and proliferation. J. Immunol. 192, 2227–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Sant’Angelo DB, and Nichols KE (2010). Transcriptional control of invariant NKT cell development. Immunol. Rev. 238, 195–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David-Fung ES, Butler R, Buzi G, Yui MA, Diamond RA, Anderson MK, Rowen L, and Rothenberg EV (2009). Transcription factor expression dynamics of early T-lymphocyte specification and commitment. Dev. Biol. 325, 444–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapin L (2016). Development of invariant natural killer T cells. Curr. Opin. Immunol. 39, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh CS, Lee HM, and Lio CW (2012). Selection of regulatory T cells in the thymus. Nat. Rev. Immunol. 12, 157–167. [DOI] [PubMed] [Google Scholar]
- Jameson SC, Lee YJ, and Hogquist KA (2015). Innate memory T cells. Adv. Immunol. 126, 173–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, Chua K, Eidson M, Kim HJ, Im JS, et al. (2008). The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat. Immunol. 9, 1055–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreslavsky T, Savage AK, Hobbs R, Gounari F, Bronson R, Pereira P, Pandolfi PP, Bendelac A, and von Boehmer H (2009). TCR-inducible PLZF transcription factor required for innate phenotype of a subset of γδ T cells with restricted TCR diversity. Proc. Natl. Acad. Sci. USA 106, 12453–12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai D, Zhu J, Wang T, Hu-Li J, Terabe M, Berzofsky JA, Clayberger C, and Krensky AM (2011). KLF13 sustains thymicmemory-like CD8(+) T cells in BALB/c mice by regulating IL-4-generating invariant natural killer T cells. J. Exp. Med. 208, 1093–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Holzapfel KL, Zhu J, Jameson SC, and Hogquist KA (2013). Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat. Immunol. 14, 1146–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Wang H, Starrett GJ, Phuong V, Jameson SC, and Hogquist KA (2015). Tissue-specific distribution of iNKT cells impacts their cytokine response. Immunity 43, 566–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch L, Michelet X, Zhang S, Brennan PJ, Moseman A, Lester C, Besra G, Vomhof-Dekrey EE, Tighe M, Koay HF, et al. (2015). Regulatory iNKT cells lack expression of the transcription factor PLZF and control the homeostasis of T(reg) cells and macrophages in adipose tissue. Nat. Immunol. 16, 85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao AP, Constantinides MG, Mathew R, Zuo Z, Chen X, Weirauch MT, and Bendelac A (2016). Multiple layers of transcriptional regulation by PLZF in NKT-cell development. Proc. Natl. Acad. Sci. USA 113, 7602–7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Seiler MP, Scanlon ST, Mao AP, Constantinides MG, Bertozzi-Villa C, Singer JD, and Bendelac A (2012). BTB-ZF factors recruit the E3 ligase cullin 3 to regulate lymphoid effector programs. Nature 491, 618–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, Jo Y, Ko E, Luckey MA, Park YK, Park SH, Park JH, and Hong C (2016). Soluble γc cytokine receptor suppresses IL-15 signaling and impairs iNKT cell development in the thymus. Sci. Rep. 6, 36962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pobezinsky LA, Etzensperger R, Jeurling S, Alag A, Kadakia T, McCaughtry TM, Kimura MY, Sharrow SO, Guinter TI, Feigenbaum L, and Singer A (2015). Let-7 microRNAs target the lineage-specific transcription factor PLZF to regulate terminal NKT cell differentiation and effector function. Nat. Immunol. 16, 517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, Lantz O, and Bendelac A (2008). The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity 29, 391–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scripture-Adams DD, Damle SS, Li L, Elihu KJ, Qin S, Arias AM, Butler RR 3rd, Champhekar A, Zhang JA, and Rothenberg EV (2014). GATA-3 dose-dependent checkpoints in early T cell commitment. J. Immunol. 193, 3470–3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler MP, Mathew R, Liszewski MK, Spooner CJ, Barr K, Meng F, Singh H, and Bendelac A (2012). Elevated and sustained expression of the transcription factors Egr1 and Egr2 controls NKT lineage differentiation in response to TCR signaling. Nat. Immunol. 13, 264–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigvardsson M (2012). Transcription factor dose links development to disease. Blood 120, 3630–3631. [DOI] [PubMed] [Google Scholar]
- Thapa P, Manso B, Chung JY, Romera Arocha S, Xue HH, Angelo DBS, and Shapiro VS (2017). The differentiation of ROR-γt expressing iNKT17 cells is orchestrated by Runx1. Sci. Rep. 7, 7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watarai H, Nakagawa R, Omori-Miyake M, Dashtsoodol N, and Taniguchi M (2008). Methods for detection, isolation and culture of mouse and human invariant NKT cells. Nat. Protoc. 3, 70–78. [DOI] [PubMed] [Google Scholar]
- Weinreich MA, Odumade OA, Jameson SC, and Hogquist KA (2010). T cells expressing the transcription factor PLZF regulate the development of memory-like CD8+ T cells. Nat. Immunol. 11, 709–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita I, Nagata T, Tada T, and Nakayama T (1993). CD69 cell surface expression identifies developing thymocytes which audition for T cell antigen receptor-mediated positive selection. Int. Immunol. 5, 1139–1150. [DOI] [PubMed] [Google Scholar]
- Yoshimoto T, and Paul WE (1994). CD4pos, NK1.1pos T cells promptly produce interleukin 4 in response to in vivo challenge with anti-CD3. J. Exp. Med. 179, 1285–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Dong Z, Zhou R, Luo D, Wei H, and Tian Z (2005). Isolation of lymphocytes and their innate immune characterizations from liver, intestine, lung and uterus. Cell. Mol. Immunol. 2, 271–280. [PubMed] [Google Scholar]
- Zhang S, Laouar A, Denzin LK, and Sant’Angelo DB (2015). Zbtb16 (PLZF) is stably suppressed and not inducible in non-innate T cells via T cell receptor-mediated signaling. Sci. Rep. 5, 12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


