Abstract
Objective:
Modeling variable-sized regions of interest (ROIs) in whole slide images using deep convolutional networks is a challenging task, as these networks typically require fixed-sized inputs that should contain sufficient structural and contextual information for classification. We propose a deep feature extraction framework that builds an ROI-level feature representation via weighted aggregation of the representations of variable numbers of fixed-sized patches sampled from nuclei-dense regions in breast histopathology images.
Methods:
First, the initial patch-level feature representations are extracted from both fully-connected layer activations and pixel-level convolutional layer activations of a deep network, and the weights are obtained from the class predictions of the same network trained on patch samples. Then, the final patch-level feature representations are computed by concatenation of weighted instances of the extracted feature activations. Finally, the ROI-level representation is obtained by fusion of the patch-level representations by average pooling.
Results:
Experiments using a well-characterized data set of 240 slides containing 437 ROIs marked by experienced pathologists with variable sizes and shapes result in an accuracy score of 72.65% in classifying ROIs into four diagnostic categories that cover the whole histologic spectrum.
Conclusion:
The results show that the proposed feature representations are superior to existing approaches and provide accuracies that are higher than the average accuracy of another set of pathologists.
Significance:
The proposed generic representation that can be extracted from any type of deep convolutional architecture combines the patch appearance information captured by the network activations and the diagnostic relevance predicted by the class-specific scoring of patches for effective modeling of variable-sized ROIs.
Index Terms—: Digital pathology, breast histopathology, deep feature representation, weakly supervised learning, region of interest classification
I. Introduction
Histopathological image analysis systems aim to provide an accurate modeling of the image content and an objective quantification of the tissue structure. Whole slide imaging has aided these systems via digitization of glass slides into very high resolution images. In addition to the computational challenges due to data sizes, the main semantic challenge is to design an effective representation of the local image details.
For the particular case of breast histopathology, a continuum of histologic features exists in the tissue structures where different types of proliferation have different clinical significance. For example, proliferative changes are considered benign, and do not necessitate additional procedures. However, other diagnoses such as atypical hyperplasia and in situ carcinoma carry different risks of progressing into malignancy and lead to different clinical actions such as surgery, radiation, and hormonal therapy [1], [2]. An automated diagnosis system should involve all intermediate steps that contain both the identification of diagnostically relevant regions and the association of each of these individual regions with a diagnostic category.
Given the high level of uncertainty regarding the correspondence between the diagnostic class of the whole slide and the diverse content in the local details in the image data [3], the main focus of the relevant work has been to perform both training and evaluation tasks on isolated regions of interest (ROI)1 with no ambiguity in their diagnostic labels. Among these works, deep learning-based approaches, in particular convolutional neural networks (CNN), have had the greatest success in recent years [4]. Earlier studies using deep networks focused on the binary (benign vs. malignant) classification problem. For example, Cruz-Roa et al. [5] use a deep network to classify 100×100 pixel patches as benign or invasive for breast histopathology. The BreaKHis data set [6] that consists of 700×460 pixel images has also been popular for benign vs. malignant classification. The common approach is to sample 32×32 or 64×64 pixel patches, classify them by using deep networks, and obtain the image-level diagnoses by combining patch-level outputs using methods such as averaging class probabilities [7] or majority voting [8].
Classifying a tissue as one of multiple cancerous or precancerous lesions as is required in clinical practice holds a higher clinical significance compared to only as benign or malignant. There exist multiple works studying multi-class classification of breast histopathology images with CNNs. For example, the BACH data set [9] that consists of fixed-sized images labeled as normal, benign, in situ, and invasive has been used in several competitions. Uniformly sampling patches over a regular grid, and obtaining the image-level diagnoses via majority voting or averaging patch-level probabilities has been the common choice [10], [11]. Training a separate classifier on the patch-level outputs using logistic regression [12], recurrent neural networks [13], or multiple instance learning [14], [15], [7] are used as alternatives to fixed fusion rules. Besides CNNs, stacked autoencoder-based unsupervised feature representations are also used as patch models [16], [17].
In all of the works reviewed above, typically small, fixed-sized, manually cropped images are used as the final targets in the classification task. However, in a realistic clinical setup, the ROIs often vary significantly in size and in content. In the former scenario where relatively small and isolated ROIs that belong to distinct categories are used, it can be safe to assume that the sampled patches are all similarly relevant for the diagnosis. However, in the latter unconstrained scenario where the ROIs are obtained by manual delineation in free form or by using machine learning-based ROI detectors, typically not all patches are equally informative. Thus, modeling variable-sized ROIs using deep networks remains an open problem. For such ROIs, commonly used transformations such as cropping may lead to loss of important local details, and resizing may result in the loss of important scale information. Furthermore, other popular approaches that involve pooling of pixels into pre-defined grids [18] may also suffer from the aforementioned problems in histopathology images.
An alternative is to design a representation that can capture the variations in local details of variable-sized ROIs. For example, Mehta et al. [19] propose the Y-Net framework that is jointly trained for segmentation and classification where the classification output is used with a threshold to obtain a tissue-level discriminative segmentation mask. Then, the frequencies of the selected tissue components are used with a multi-layer perceptron to obtain the ROI-level diagnosis. Mercan et al. [20] use superpixels to aggregate the pixel-level tissue segmentation, estimate the duct locations from the epithelium regions, and compute histograms of the tissue types within layers of superpixels both inside and outside of these ductal components as structure features for ROI-level classification.
Our whole slide analysis pipeline involves three stages illustrated in Figure 1: 1) detection of ROIs; 2) modeling of these ROIs using a variable number of fixed-sized patches; 3) modeling of slides using these ROIs. This model can be considered within the weakly supervised learning paradigm where ROI-level class labels are missing when only slide-level diagnoses are available, and the contributions of individual patches to an ROI are also not known. We proposed both traditional [21] and deep learning-based [22] solutions to the first stage. We also proposed a multi-instance multi-label learning formulation [3] for the third stage. In this paper, we focus on the second stage of modeling the individual ROIs by designing an ROI-level representation via weighted aggregation of patch-level representations. The proposed approach can be applied to both manually and automatically identified ROIs. The weights can be considered as confidence scores that quantify the importance and informativeness of the patches for the ROI-level diagnosis. We have shown that such weighted combinations of patch-level representations are quite powerful for simultaneous learning of attention and classification models when only image-level labels are available during weakly supervised learning [23]. Here, the patch-level feature representations are obtained from both fully-connected layer activations and pixel-level convolutional layer activations, and the weights are obtained from the class predictions. Our main contributions include a new patch-level representation based on convolutional activation maps, a generic representation for modeling variable-sized ROIs that is illustrated by using two different deep network architectures, and extensive evaluation using a challenging multi-class breast histopathology data set that covers the whole histologic spectrum. We compare this representation to alternative representations as well as to operations such as cropping, resizing, and pooling. A preliminary version of this work was presented in [24].
Fig. 1.
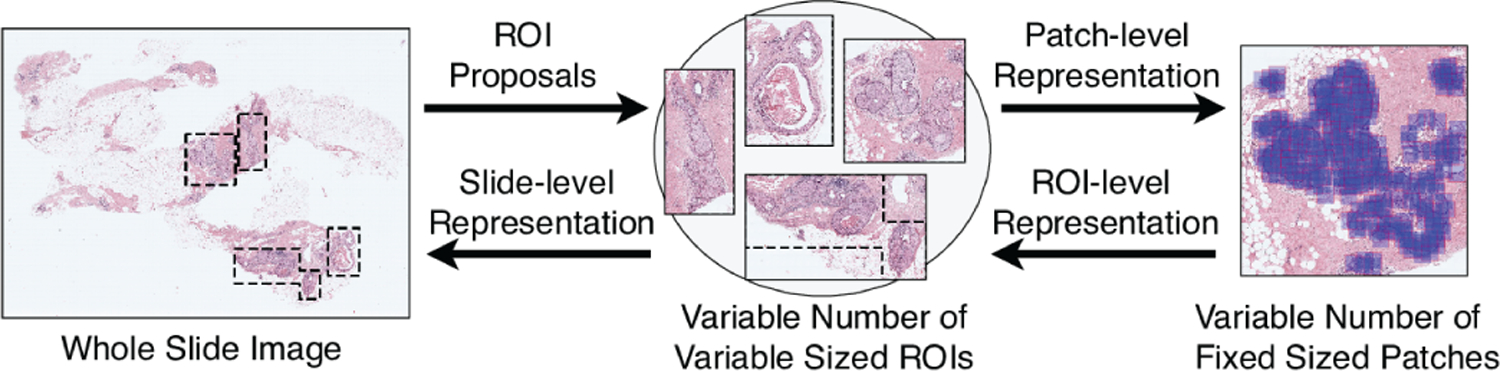
Modeling of a WSI in terms of ROIs and patches.
The paper is organized as follows. Section II introduces the data set. Section III describes the deep feature representation methodology. Section IV presents how these representations can be used for classification. Section V provides the experimental results. Finally, Section VI gives the conclusions.
II. Data Set
We use a data set of 240 breast biopsies that was developed as part of an NIH-sponsored project to study variability in the interpretation of breast histopathology [1]. The haematoxylin and eosin (H&E) stained slides that belonged to independent cases from different patients were selected from cancer registries associated with the Breast Cancer Surveillance Consortium by stratified sampling to cover the full range of diagnostic categories from benign to cancer. The study was approved by the institutional review boards at Bilkent University, University of Washington, and University of Vermont.
The slides were scanned by the same iScan Coreo Au digital slide scanner (Roche). The cases were independently interpreted by three experienced pathologists who then met in consensus meetings to define a single consensus diagnosis for each case. At the end, each case was classified into one of the following 4 classes with example diagnostic terms: class I benign without atypia (Benign), class II atypical ductal hyperplasia (ADH), class III ductal carcinoma in situ (DCIS), and class IV invasive cancer (INV). The benign class includes samples that contain non-proliferative changes, fibroadenoma, intraductal papilloma without atypia, usual ductal hyperplasia, columnar cell hyperplasia, sclerosing adenosis, complex sclerosing lesion, and flat epithelial atypia. The ADH class includes atypical ductal and lobular hyperplasia, and intraductal papilloma with atypia. The DCIS class includes both ductal and lobular carcinoma in situ. The difficulty of this multi-class problem can also be confirmed from the evaluations in [1], [25] where a large set of pathologists’ concordance rates compared with the consensus diagnoses were 82% for Benign, 43% for ADH, 79% for DCIS, and 93% for INV. The study in [9] also reported that the benign and in situ classes were the most difficult to classify. Our data set contains a very diverse mix of sub-categories considered benign. It also includes the challenging and clinically significant ADH class that was not present in any of the work (except [3], [19], [20], [22] that used the same data set) reviewed in Section I.
We divided the data set equally into two as training and test sets so that the slide-level class distribution between the two sets are kept as close as possible while each subset has slides from different patients. The ROI-level analysis studied in this paper uses the ROIs marked by the pathologists as one or more representative regions in each slide to support the corresponding diagnosis for that slide. In total, there are 437 consensus ROIs, having the same diagnostic labels as the slide-level consensus labels. The class distribution of slides and ROIs are shown in Table I. The ROIs have considerable amount of variability in size as shown in Table II. Note that all diagnostic categories exhibit this variability that further supports the need for developing new feature representations for multi-class modeling of variable-sized ROIs.
TABLE I.
Class distribution of slides and ROIs in the data set.
| Benign | ADH | DCIS | INV | Total | ||
|---|---|---|---|---|---|---|
| Slide | Training set | 34 | 35 | 41 | 10 | 120 |
| Test set | 22 | 48 | 38 | 12 | 120 | |
| ROI | Training set | 60 | 58 | 85 | 17 | 220 |
| Test set | 37 | 81 | 80 | 19 | 217 | |
TABLE II.
Statistics of ROI bounding box sizes (number of pixels at 40×).
| Benign | ADH | DCIS | INV | |
|---|---|---|---|---|
| Min. | 1,400 × 1,200 | 1,320 × 1,120 | 1,041 × 1,400 | 1,708 × 2,987 |
| Max. | 41,652 × 39,617 | 39,585 × 28,975 | 73,612 × 64,843 | 72,442 × 55,151 |
| Mean | 10,495 × 8,943 | 7,075 × 6,206 | 11,063 × 9,812 | 25,238 × 22,899 |
| Std.dev. | 8,877 × 7,081 | 5,455 × 4,347 | 11,051 × 8,266 | 17,514 × 14,994 |
III. Deep Feature Representation
We introduce a deep feature extraction method for variable-sized ROIs, while preserving patch-level local information and their relative contribution for ROI-level diagnosis. First, a CNN is trained on the patches sampled from the ROIs. Then, a patch-level feature representation is obtained by concatenation of weighted instances of feature activations computed for the patch by the network. The weights are also obtained from the network as the class probabilities for the corresponding patch. Finally, the ROI-level representation is obtained via aggregation of the patch-level representations by average pooling. This generic representation that can be extracted from any type of deep convolutional architecture aims to combine the patch appearance information modeled by the network activations and the diagnostic relevance modeled by the class-specific scoring of the patches. The details of each step of this representation are described below.
A. Patch-level Deep Network Training
State-of-the-art deep convolutional architectures that aim to produce image-level class probability scores face a challenge for ROIs with significantly different shapes and sizes. Our proposed solution is to model each ROI as a combination of variable number of potentially salient fixed-sized patches.
1). Identification of Patches from ROI:
The first step involves identification of informative and diverse set of patches to represent the structural and contextual information in the ROI. According to pathologists, appearance of the cell nuclei and their spatial distribution within the ducts are important indicators for the diagnosis [17]. Eye tracking studies also show high correlation between the regions viewed by the pathologists and the computer vision-based saliency detector outputs that highly overlap with epithelium-rich regions [26]. In this paper, informativeness is achieved by sampling the patches from nuclei-dense areas in the ROI, and diversity is attained by enforcing a constraint that two patches should not overlap by more than a margin. We show in Section V-C that this is empirically an effective choice as well.
An efficient way of locating nuclei-dense regions as potential locations for ductal structures is to use the haematoxylin channel estimated from the RGB image. We use the built-in stain vectors in the ImageJ implementation (https://imagej.net/Colour_Deconvolution) of the color deconvolution algorithm in [27]. After obtaining the haematoxylin value at each pixel, we compute a non-parametric Parzen density estimate [28], and apply a threshold to this estimate to eliminate the regions with little to no nuclei. The remaining regions are used to sample the center pixels of patches on a uniform grid to enforce a limit on the patch overlap. The image magnification used is determined in coordination with the patch size required by the network architecture as described in Section V-A. The patches should not be too large to risk simultaneous inclusion of details from irrelevant proliferations and too small to contain insufficient context. The patch selection process is illustrated in Figure 2.
Fig. 2.
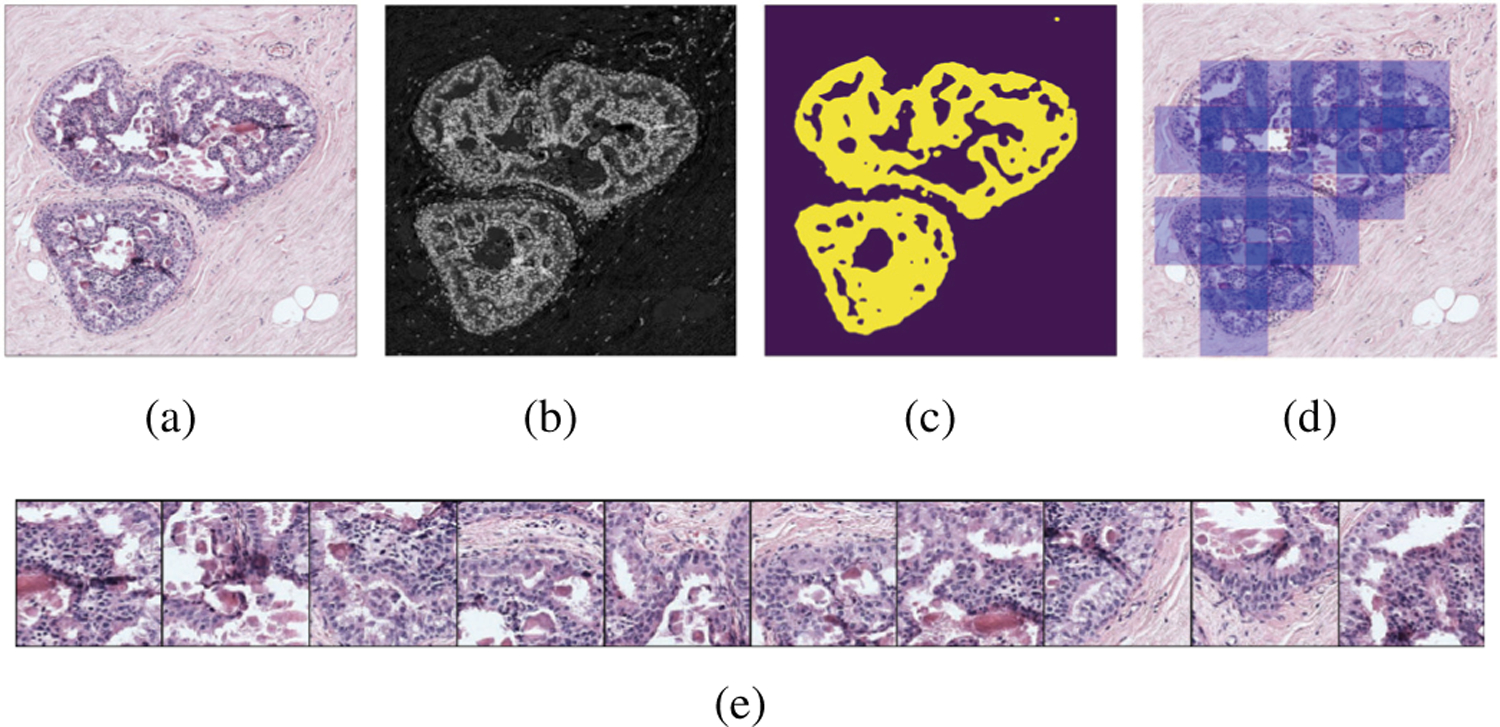
Patch selection for an example ROI. (a) RGB image. (b) Haematoxylin estimate. (c) Nuclei mask. (d) Selected RGB patches. (e) Example patches.
2). CNN Training on Patches:
Due to the limited availability of labeled histopathology images, we opt to fine-tune a pre-trained network. Our first choice for the base CNN architecture is the ImageNet pre-trained VGG16 network [29] due to its relatively large depth and representational capabilities as well as our good experience with an adaptation of this network on the same breast pathology data set [22]. We also use the ResNet-50 network [30] to illustrate the generic applicability of the proposed methodology. The RGB patches sampled in the previous step are used with the same labels as those of the corresponding ROIs to fine-tune both networks. There was no performance improvement when we tried to train all network parameters from scratch due to the limited amount of data available for training. The experiments in this paper use these two specific networks but, as noted earlier, the proposed deep feature representation can be extracted by using any type of deep convolutional architecture.
B. Patch-level Deep Feature Representation
Given a deep network that is trained as in Section III-A2, we use two different methods to extract the initial patch-level feature representations denoted as in the rest of the paper.
1). Penultimate Layer Features:
The first method for obtaining is to directly use the output of the penultimate layer in the patch-level deep network. As the most commonly used approach of employing deep networks for feature extraction, the penultimate layer activations, illustrated for the VGG16 network in Figure 3, provide an overview of the patch content summarized by the fully-connected operations.
Fig. 3.
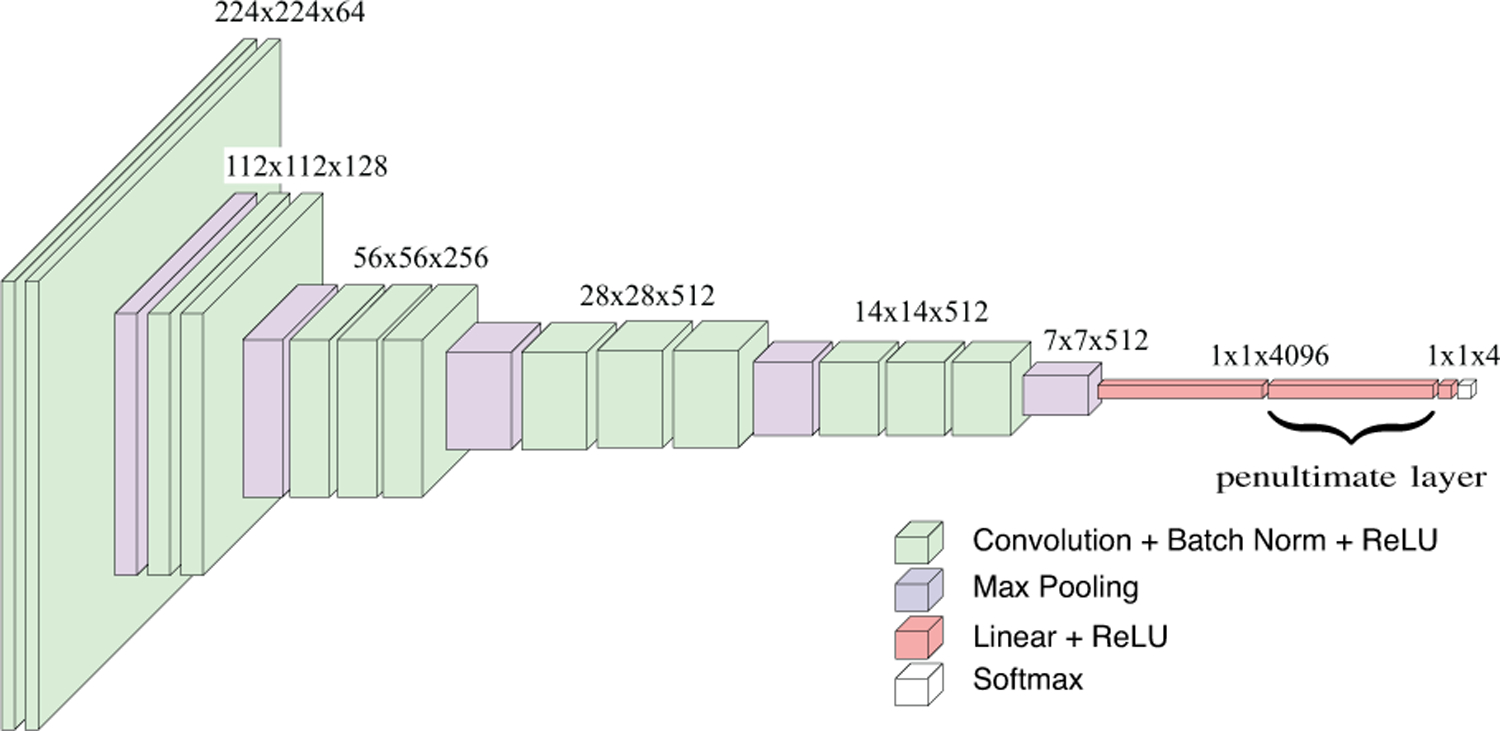
VGG16 network used for patch-level feature representation. This particular network has 13 convolutional and three fully-connected layers where the last layer outputs predictions for each of the K = 4 classes through a softmax activation function. The convolutional layers are denoted as conv{11,12,21,22,31,32,33,41,42,43,51,52,53} and the fully-connected layers are denoted as fc{1,2,3}. The penultimate layer activations correspond to fc2.
2). Hypercolumn Features:
The second method exploits pixel-level convolutional activations for feature extraction. The activations in the earlier layers provide low-level information such as color, texture, and shape, while the activations in later layers encode contextual information about the input image [31]. The hypercolumn feature representation of a pixel combines these low-level and high-level features and is obtained by concatenating all activations at that pixel location through the layers in the network when pixel-level representations are needed for tasks such as semantic segmentation [32].
Our aim is to extract a patch-level representation from the pixel-level hypercolumn features. A naive concatenation of all pixels’ features will produce a huge vector, e.g., with size over two hundred million for a mildly deep network such as VGG16, and will be prone to overfitting. We designed a procedure that involves statistical operations on a selected set of layers of the convolutional network to obtain the feature representation for an input patch. Figure 4 illustrates these steps for an example layer. The details are provided below.
Fig. 4.
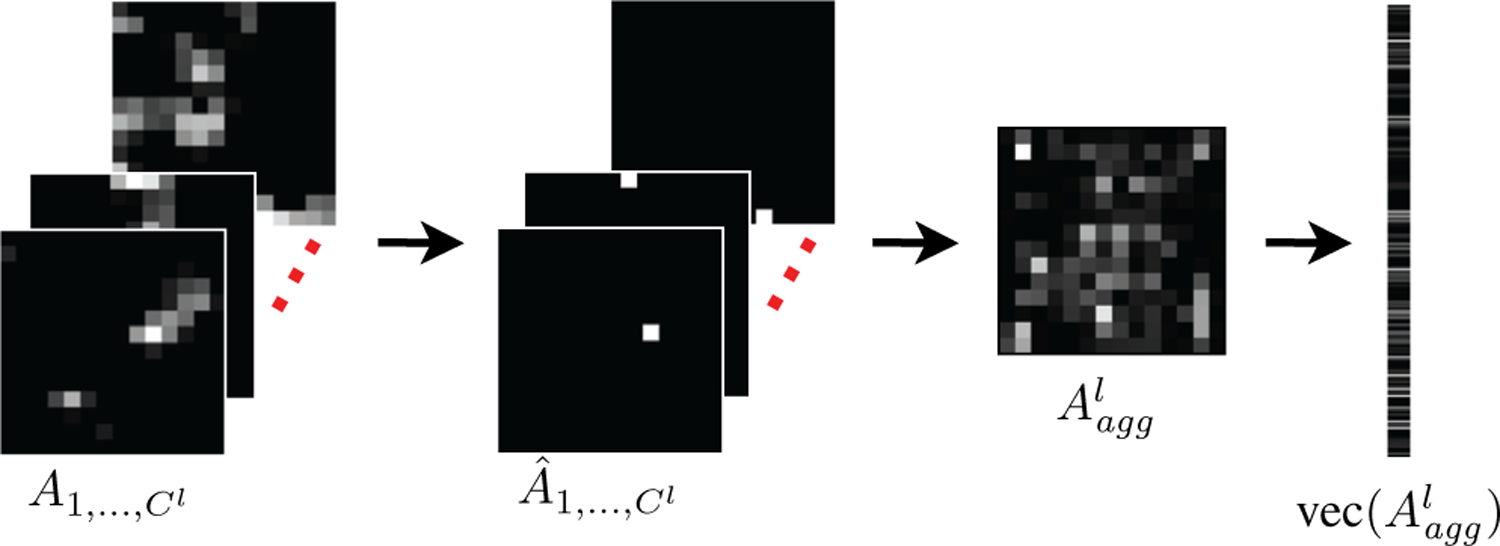
Hypercolumn representation from pixel-level convolutional activations at a particular layer. Given the activation maps for all channels of the layer, the procedure selects the pixels with the maximum activation in each channel , combines these activations in an aggregate activation map by keeping only the selected pixels while suppressing the others , and vectorizes the resulting map as the final representation .
The input for each patch is a set of L layers selected from the deep network. Each layer l ∈{1, …, L} consists of a set of channels that correspond to the convolutional activations , where Cl is the number of channels in layer l and is the matrix that stores the responses of all pixels to the c’th kernel in that layer. For example, for the layer denoted as in Figure 3, the number of channels Cl is 256, and is a matrix of size 56×56. We select the last layers of the last three groups of convolutional layers for both networks. These layers exhibit increasing representational capacity after a sequence of consecutive convolution operations within each group right before the feature map size is decreased with a pooling operation for the next group. For the VGG16 network, the selected layers are . For the ResNet-50 network, we use the last layers of the conv{3_x,4_x,5_x} blocks as described in the experimental setup in Section V-A.
Given the channels in a selected layer l, we first identify the maximum activation in each convolutional channel and turn off the remaining activations as
| (1) |
where (x,y) is the pixel location. The resulting matrix contains a single non-zero pixel that corresponds to the maximum in (1). Then, we combine the top activations by summing over the convolutional channels to obtain an aggregate activation map of the associated layer as
| (2) |
The resulting map, with the same size as the matrices and , preserves the most prominent responses that contain information from different local structures activated by various convolutional kernels in that particular layer. Finally, the resulting maps for all layers are vectorized and concatenated as
| (3) |
where vec(·) denotes the vectorization operation of a given matrix, and is the pixel-level convolutional hypercolumn feature representation of the input patch.
C. ROI-level Deep Feature Representation
Our previous research showed that weighted pooling of patches within larger images works well for weakly supervised learning when there is both localization and labeling uncertainty [23]. The ROI-level feature representation proposed here also uses weighted aggregation of patch-level feature vectors. Both the feature vectors and the weights are extracted from the patch-level deep network described earlier.
The input is an ROI R that is modeled as a set of M patches {r1,r2, …, rM}. We assume that each patch is initially mapped to a d-dimensional feature vector as in Section III-B where denotes the vector for patch rm. One of the most widely used representations that are based on aggregation of local features is the bag-of-words (BoW) model [33], where the local instances are quantized into discrete words in a codebook, and average pooling of these words is performed by counting their occurrences into a normalized histogram. In this model, the final representation for a patch becomes a one-hot vector that encodes the codeword assignment for the deep feature representation, , for that patch. The ROI-level feature representation that aggregates all patch encodings is obtained by average pooling that results in a vector whose length is equal to the size of the codebook used.
Another popular approach that can be viewed as a generalization of the BoW model is the Fisher vector framework [34]. By using a Gaussian mixture model that estimates the distribution of the local descriptors, the Fisher vector encoding captures the first and second order differences between the individual descriptors and the mixture components. In this framework, the final representation for a patch is obtained as the concatenation of the gradients computed with respect to the mixture model parameters, and the ROI-level feature representation is also obtained via average pooling that results in a vector whose length is twice the length, d, of the initial patch-level deep feature representation, , times the number of mixture components.
Our proposed representation is based on soft assignments where the patches are associated with the diagnostic classes of interest by using probability estimates that correspond to the confidences in these assignments. Given the K class probabilities estimated for the patch rm by the softmax layer of the deep network with , a new representation for the patch is obtained by concatenating the weighted instances of the original deep feature as
| (4) |
resulting in a patch-level feature representation with length Kd. Here, weighting is influenced by our past work [23] and concatenation can be related to the Fisher vector framework [34]. Then, the final ROI-level feature representation is obtained by average pooling as
| (5) |
In the representation in (4), the feature vector that is computed from the deep network activations contributes differently for each class in according to the class probabilities that act like relevance scores that quantify the significance of that patch for the ROI-level diagnosis. This weighted aggregation illustrated in Figures 5 and 6 results in the class probabilities and the feature activations supporting each other in the learning of class-specific feature vectors.
Fig. 5.
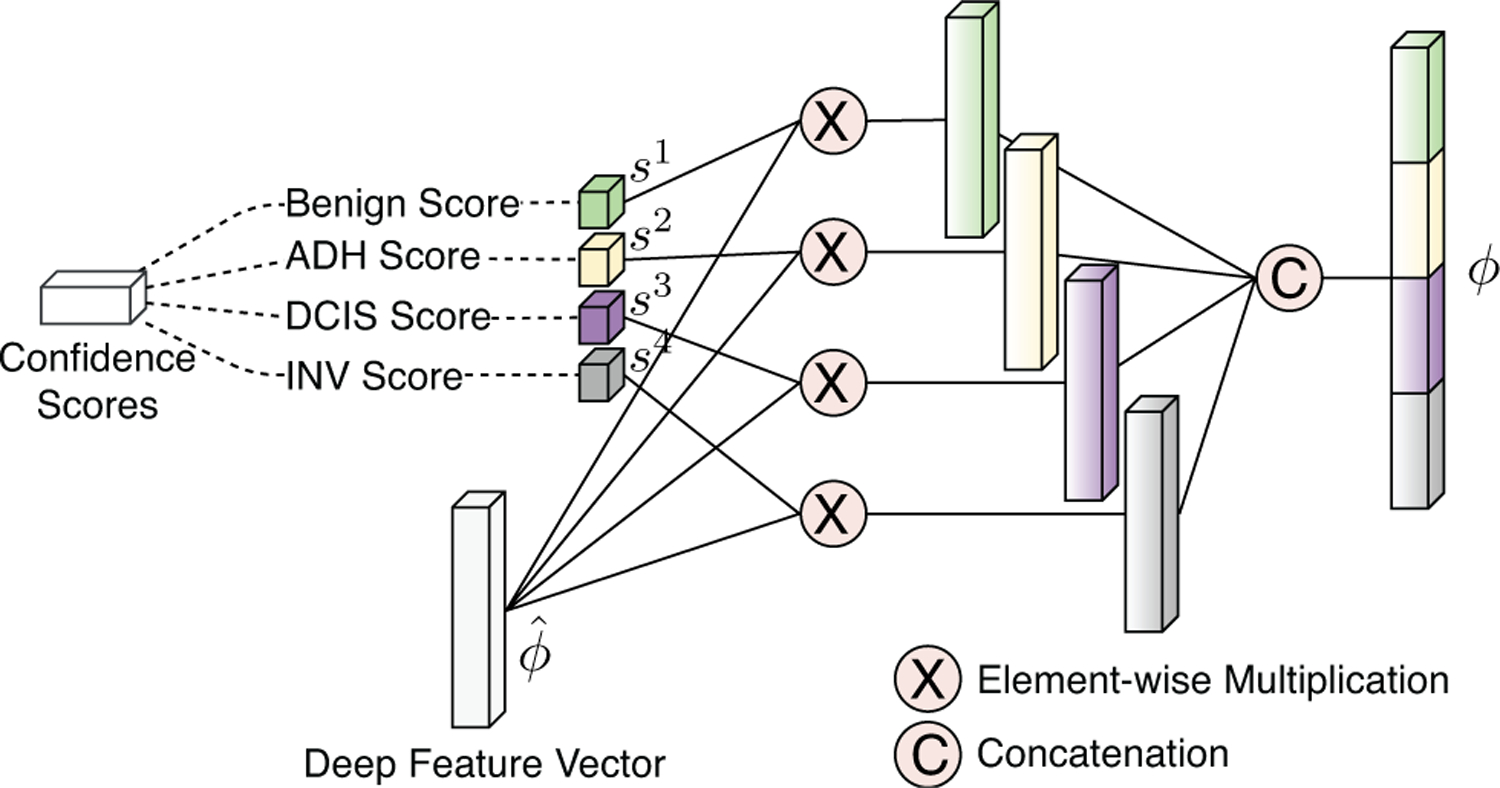
Final patch-level deep feature representation computed from the aggregation of initial deep feature vectors with class-specific network output.
Fig. 6.
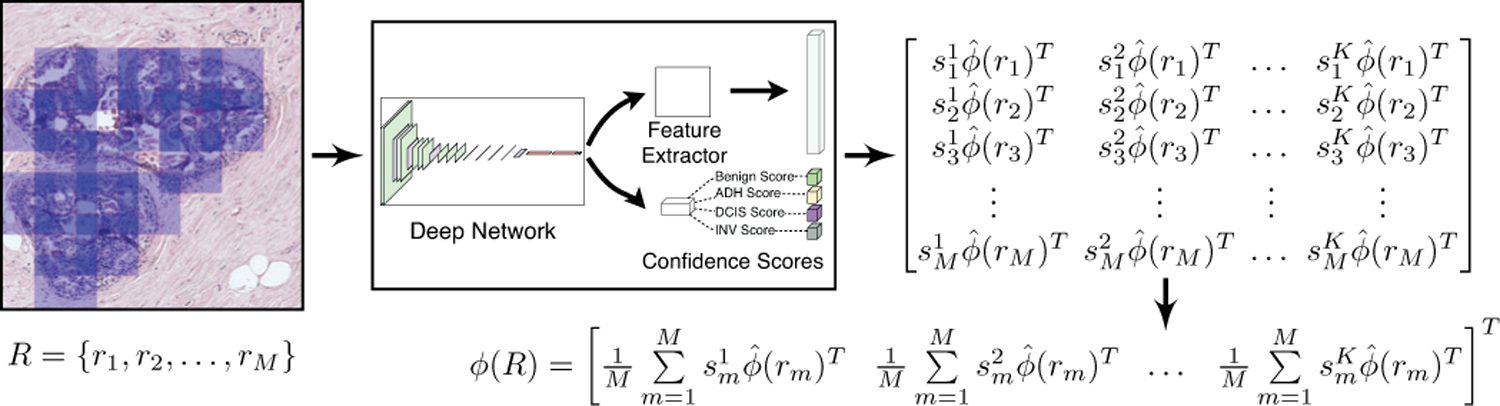
ROI-level deep feature representation computed by pooling the final patch-level deep feature representations.
IV. Classification
The deep feature representations for the ROIs in the training set are used to train a multi-layer perceptron (MLP) to perform multi-class classification on unseen ROIs in the test set whose feature representations are also extracted with the same procedure. We use the consensus ROIs that were manually identified by the experienced pathologists as described in Section II. Alternative approaches that involve classifiers explicitly trained for ROI detection can also be used when no such ROIs are available [21], [22].
V. Experiments
A. Experimental Setup
The deep feature extraction process proposed in this paper is not specific to any network and can be applied to any convolutional architecture. The experiments described here are realized by using the VGG16 and ResNet-50 networks. The patches were sampled as 224×224 pixel windows, particularly due to the input requirement of the VGG16 network. We used 10× magnification, which was empirically decided based on the experiments presented in Section V-C.
During patch-level training described in Section III-A, we applied random rotation, random horizontal/vertical flipping, and random perturbations on the hue channel in the HSV domain as part of the data augmentation routine. We also oversampled Benign and INV patches to reduce the imbalance resulting from intentional oversampling of the ADH and DCIS cases in the data set to study the preinvasive lesions in more detail [1]. We fine-tuned the networks on the same augmented training set using cross-entropy loss. We used batches of 32 patches, and employed Adam optimizer with a learning rate set to 10−4. During patch-level feature extraction in Section III-B, the penultimate layer activations for the VGG16 network are taken from the layer labeled as fc2, resulting in initial feature vectors of length d =4,096. The hypercolumn features are computed from the convolutional activations in the layers labeled as , resulting in initial feature vectors of length d = 4,116 (after vectorization of 56×56, 28×28, and 14×14 pixel maps as shown in Figures 3 and 4). For the ResNet-50 network, the penultimate layer feature representation has length d = 2,048, and the hypercolumn feature vector is obtained from the convolutional activations in the last layers of the conv3_x, conv4_x, and conv5_x blocks, resulting in initial feature vectors of length d = 1,029 (after vectorization of 28×28, 14×14, and 7×7 pixel maps). The final ROI-level features in Section III-C are obtained by average pooling of weighted concatenations of patch-level features, resulting in vectors of length Kd where K = 4. We also evaluated equal representation of all three layers in the hypercolumn vector by upsampling the smaller layers via bilinear interpolation. However, the accuracy decreased by 25% due to the increased dimensionality of the representation.
We use the same training data for all stages of both the proposed methodology and the baseline methods described in Section V-B. For hyperparameter optimization and quantitative evaluation, we further split the test data shown in Table I into two subsets. These subsets correspond to two groups of 60 slides each, that belong to different patients and are randomly selected according to the same class frequency distribution by using stratified sampling. We interchangeably use these two sets, corresponding to 116 and 101 ROIs, respectively, as validation and test data, and report the average accuracy on the test subsets for all experiments in Section V-C. We use normalized accuracy as the performance metric where the per-class accuracy rates are averaged to avoid biases towards classes with larger number of examples.
B. Baselines
The ROI-level feature representations, named Penultimate-Weighted and Hypercolumn-Weighted for the approaches described in Sections III-B1 and III-B2, respectively, are used with a 4-class MLP classifier trained according to the setup in Section V-A. We also evaluated the performances of the following commonly used feature aggregation methods.
Penultimate-Baseline: The initial patch-level feature vectors from the penultimate layer are combined by average pooling (without weighting) for ROI-level features.
Hypercolumn-Baseline: Similarly, the initial hypercolumn features are combined by average pooling.
Majority-Voting: We use the patch-level class probabilities to assign each patch to the most likely class, and apply majority voting to obtain the label of the ROI.
Learned-Fusion [15]: The patch-level class probabilities from the final softmax layer of the network are summed up to create class frequency histograms as ROI features.
Bag-of-Words: We use the initial patch-level feature vectors to compute a codebook for the bag-of-words model. The codebook sizes are selected as 16, 32, and 64 based on our earlier experience on the same data set [21].
Fisher-Vector: We compute the Fisher vector encoding for the initial patch-level vectors using Gaussian mixtures with 16, 32, and 64 components. We apply principal components analysis to improve the accuracy and reduce the memory footprint of the representation [35].
Y-Net [19]: This approach extends the U-Net [36] model for joint training for segmentation and classification using a multi-task loss with 8-class tissue segmentation masks and ROI-level labels. The network produces a tissue-level discriminative segmentation mask after applying a threshold to the local patch probabilities. Histograms of patch class assignments are used as ROI features.
All of these ROI-level feature representations are used with a final MLP classifier for predicting the ROI-level diagnoses (except Majority-Voting that directly outputs the class label).
We also implemented the commonly used operations of cropping, resizing, and pooling. First, we identified the largest square image size that could be fit into the GPU memory for a batch size of 10 as 1120×1120 pixels. Then, for each ROI, we cropped the largest square region that could fit into that ROI’s mask and resized it to 1120×1120 pixels. We used the resulting regions to fine-tune a ResNet-50 network for ROI-level prediction. This procedure is denoted as Crop/resize in the results. We also evaluated replacing the average pooling layer with spatial pyramid pooling [18] using three scales.
Finally, we present the average accuracy from the independent interpretations of all slides by 45 other pathologists that practice breast pathology in their daily routines [1], [19].
C. Results
The first step was the selection of image magnification. We evaluated the proposed representations with patches sampled from 2.5×, 5×, 10×, and 20× magnifications. The results for ResNet-50 are presented in Table III. We determined that 10× magnification provides a good tradeoff for capturing sufficient local context without including irrelevant details.
TABLE III.
Impact of magnification on ROI-level classification (%).
| Method | 2.5× | 5× | 10× | 20× |
|---|---|---|---|---|
| Penultimate-Baseline | 55.50 | 59.74 | 59.79 | 44.57 |
| Hypercolumn-Baseline | 40.04 | 49.85 | 47.17 | 41.98 |
| Penultimate-Weighted | 60.71 | 67.63 | 67.13 | 64.27 |
| Hypercolumn-Weighted | 65.72 | 68.69 | 71.92 | 62.81 |
The next step was the evaluation of the patch sampling strategy for training the networks used as the patch-level feature extractors. All patches sampled from the same ROI are assigned the label of that ROI. This is considered weak supervision because the relevance of each individual patch to the ROI is not truly known. The accuracies of the VGG16 and ResNet-50 networks trained using the patches sampled with the proposed strategy were 51.22% and 51.11%, respectively. These accuracies could not be improved further with additional data augmentation and hyperparameter optimization because of the uncertainty in the patch-level weak labels used for both training and evaluation. We also investigated uniform sampling of patches over a grid on the foreground tissue sections within the ROIs after eliminating the slide background via thresholding of luminosity [9]. The resulting accuracy for the VGG16-based patch classifier was 43.51%. This result shows the effectiveness of sampling of informative patches from the nuclei-dense regions for modeling ductal proliferations.
The final step was the evaluation of the ROI-level classification methods whose performances are summarized in Table IV. The proposed representations, Penultimate-Weighted and Hypercolumn-Weighted, achieved the best performances for both deep networks, with the latter obtaining the top spot while also being above the pathologists’ average performance. When we consider the performances of the baseline representations, Penultimate-Baseline and Hypercolumn-Baseline, we observe that the hypercolumn representation benefits more from the proposed weighted aggregation that learns class-specific features. This result is consistent with the observation in [31] that the further the target classification task (i.e., cancer diagnosis) moves from the original source task of the pre-trained network (i.e., ImageNet), more effective the earlier layers become. The reason could be due to the penultimate layer and the final softmax layer being located close to each other in the network so that their combination results in limited improvement because they encode similar information. Thus, if one has to choose a single layer from the network for feature extraction, the penultimate layer, as commonly used in the literature, is a good choice that provides an effective and compact summary of the image content. On the other hand, the hypercolumn features are obtained from the convolutional units that encode different local characteristics of the input data at different scales, and get a more dramatic boost in performance when fused with the complementary information encoded by the softmax layer. When we consider the remaining baseline methods, we observe that there is no consistent pattern with respect to the commonly used bag-of-words and Fisher vector encoding methods and their parameters (codebook size and number of mixture components, respectively) even after hyperparameter tuning using the validation data. We also observe that simpler aggregation methods, Majority-Voting and Learned-Fusion, behave better than these feature encodings. Another important observation is that all representation-based baselines perform as good as and often better than the transformation-based baselines of cropping, resizing, and pooling.
TABLE IV.
Comparison of ROI-level classification performance (%).
| Method | VGG16 | ResNet-50 |
|---|---|---|
| Majority-Voting | 67.02 | 69.03 |
| Learned-Fusion [15] | 66.45 | 67.77 |
| BoW-16 | 56.00 | 60.40 |
| BoW-32 | 66.49 | 57.90 |
| BoW-64 | 62.53 | 64.53 |
| Fisher-16 | 65.03 | 62.34 |
| Fisher-32 | 65.52 | 60.97 |
| Fisher-64 | 57.75 | 66.75 |
| Crop/resize | – | 58.48 |
| Pyramid-Pooling | – | 61.93 |
| Penultimate-Baseline | 63.10 | 59.79 |
| Hypercolumn-Baseline | 63.86 | 47.17 |
| Penultimate-Weighted | 69.89 | 67.13 |
| Hypercolumn-Weighted | 72.65 | 71.92 |
| Y-Net [19] | 68.20 | |
| Pathologists [19] | 70.00 | |
For more detailed evaluation, confusion matrices and class-specific performances of the proposed representations obtained by using the VGG16 network are given in Tables V and VI, respectively. The numbers in the confusion matrices are accumulated from the two test subsets. The classifier that used the Penultimate-Weighted representation predicted DCIS and ADH better than Benign and INV. For example, the highest recall was achieved for DCIS, where only 11 out of 80 ROIs were misclassified. The highest precision was obtained for ADH, where only 11 of the 61 ROIs predicted as ADH were false positives. The precision for DCIS was relatively lower than that for ADH where the classifier had the tendency to choose DCIS more frequently than any other class. The majority of the Benign ROIs that were misclassified were incorrectly labeled as ADH. ROIs with a consensus diagnosis as INV were correctly classified in 12 cases compared to 7 cases that were wrongly predicted as DCIS, which makes sense given that a large number of cases with INV as the consensus label also had DCIS in their pathology reports. The classifier that used the Hypercolumn-Weighted representation was able to classify more cases of INV and Benign correctly. We observed that inclusion of pixel-level information in the patch-level representation extracted by the hypercolumn features led to an improvement where the classifier learned the characteristics of Benign and INV better, with a small cost of misclassifying more ADH cases as Benign.
TABLE V.
Confusion matrices for ROI-level classification.
| (a)
Penultimate-Weighted |
(b)
Hypercolumn-Weighted |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Predicted | Predicted | ||||||||||
| Benign | ADH | DCIS | INV | Benign | ADH | DCIS | INV | ||||
| Ref. | Benign | 25 | 8 | 4 | 0 | Ref. | Benign | 29 | 5 | 2 | 1 |
| ADH | 13 | 50 | 17 | 1 | ADH | 18 | 44 | 15 | 4 | ||
| DCIS | 2 | 3 | 69 | 6 | DCIS | 3 | 3 | 62 | 12 | ||
| INV | 0 | 0 | 7 | 12 | INV | 0 | 0 | 4 | 15 | ||
TABLE VI.
Class-specific statistics on the performance of ROI-level classification. The number of true positives (TP), false positives (FP), false negatives (FN), and true negatives (TN) are given. Precision, recall (also known as true positive rate and sensitivity), false positive rate (FPR), specificity (also known as true negative rate), and F-measure are also shown.
| (a)
Penultimate-Weighted | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Class | TP | FP | FN | TN | Precision | Recall/Sensitivity | FPR | Specificity | F-measure |
| Benign | 25 | 15 | 12 | 165 | 0.6250 | 0.6757 | 0.0833 | 0.9167 | 0.6494 |
| ADH | 50 | 11 | 31 | 125 | 0.8197 | 0.6173 | 0.0809 | 0.9191 | 0.7042 |
| DCIS | 69 | 28 | 11 | 109 | 0.7113 | 0.8625 | 0.2044 | 0.7956 | 0.7797 |
| INV | 12 | 7 | 7 | 191 | 0.6316 | 0.6316 | 0.0354 | 0.9646 | 0.6316 |
| (b)
Hypercolumn-Weighted | |||||||||
| Class | TP | FP | FN | TN | Precision | Recall/Sensitivity | FPR | Specificity | F-measure |
| Benign | 29 | 21 | 8 | 159 | 0.58 | 0.7838 | 0.1167 | 0.8833 | 0.6667 |
| ADH | 44 | 8 | 37 | 128 | 0.8462 | 0.5432 | 0.0588 | 0.9412 | 0.6617 |
| DCIS | 62 | 21 | 18 | 116 | 0.747 | 0.775 | 0.1533 | 0.8467 | 0.7607 |
| INV | 15 | 17 | 4 | 181 | 0.4688 | 0.7895 | 0.0859 | 0.9141 | 0.5882 |
Overall, the classifiers trained using the proposed feature representations outperformed the other approaches in comparison. The challenges regarding the categorization of the preinvasive lesions such as ADH and DCIS are mostly consistent with the difficulties faced by the pathologists in comparative studies [1], [25]. However, the automated methods’ accuracies for Benign and INV classes were lower than the typical pathologist’s performance where human observers usually agree in their diagnoses for the cases that are at the extremes of the histologic spectrum. Many recent work in the literature also report higher accuracies for the Benign versus INV classification, but with a major difference in their experimental setup in which samples from atypia classes are not used [9]. Given the original motivation for the preparation of the data set by oversampling the ADH and DCIS cases to study the preinvasive lesions in more detail [1], and in spite of our efforts to decrease the class imbalance by oversampling during the fine-tuning of the patch-level network, the diversity of the extracted patches varied greatly from one class to another due to limited number of ROIs from the minority classes INV and Benign, and resulted in relatively poor performance for these classes. A possible solution in future work is to use classifiers specifically trained for identifying extreme categories such as INV [5] in a hierarchical classification framework [20].
Qualitative results on the local predictions by the fine-tuned VGG16 network are presented in Figure 7. Both the predicted labels of the patches and the class-specific scores are shown for example ROIs. The CNN predictions for the ROI in the first row mostly involved DCIS as almost all patches within the ROI showed the strongest response to that class. The methods involving the proposed feature representations, Penultimate-Weighted and Hypercolumn-Weighted, and the methods we used for comparison, Majority-Voting and Learned-Fusion, were able to correctly classify the ROI as DCIS. Similarly, the patch-level predictions mostly matched the ROI-level consensus diagnoses in the second and third rows. Consequently, the proposed and compared methods all correctly assigned those ROIs to the consensus diagnoses ADH and Benign, respectively. However, when the patch-level predictions of the CNN did not fully represent the consensus diagnosis of the ROI, the comparison methods performed poorly. For example, among all methods, only the proposed representations Penultimate-Weighted and Hypercolumn-Weighted were able to classify the ROI in the fourth row as ADH, whereas only the Hypercolumn-Weighted representation could correctly predict the class label of the fifth ROI as DCIS. Slide-level visualizations are provided in Figure 8. Thresholding on the haematoxylin estimates and connected components analysis were used to obtain ROI proposals on the input slides. Patches were sampled from each region to construct the Hypercolumn-Weighted representation of that region. The ROI-level classifier was used to make predictions for the individual regions. These predictions matched the diagnoses of the consensus ROIs in these slides. Both the quantitative and the qualitative results showed that CNN predictions for individual fixed-sized patches may not be representative enough to perform ROI-level classifications, but the proposed approaches that used weighted aggregations of patch-level image features and score predictions within variable-sized ROIs were able to successfully identify the correct diagnoses.
Fig. 7.
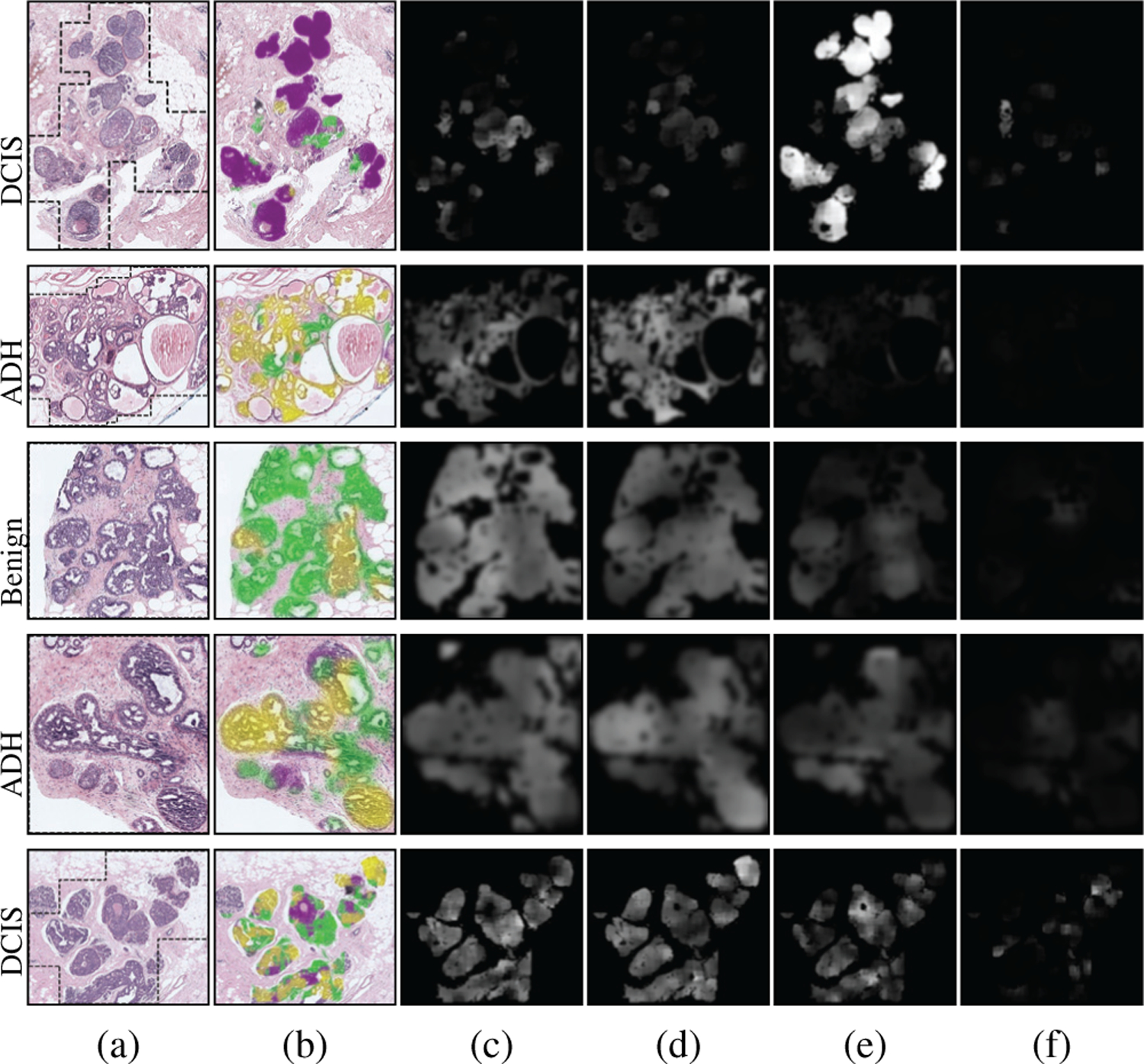
Patch-level outputs by the VGG16 network used for feature extraction. (a) Consensus diagnoses and RGB images for example ROIs with boundaries shown in black. (b) Classes predicted for patches as benign (green), ADH (yellow), DCIS (purple), INV (gray). Scores for individual classes (brighter values indicate higher probability): (c) Benign, (d) ADH, (e) DCIS, (f) INV. These scores are used in (4) as weights for the proposed feature representation.
Fig. 8.
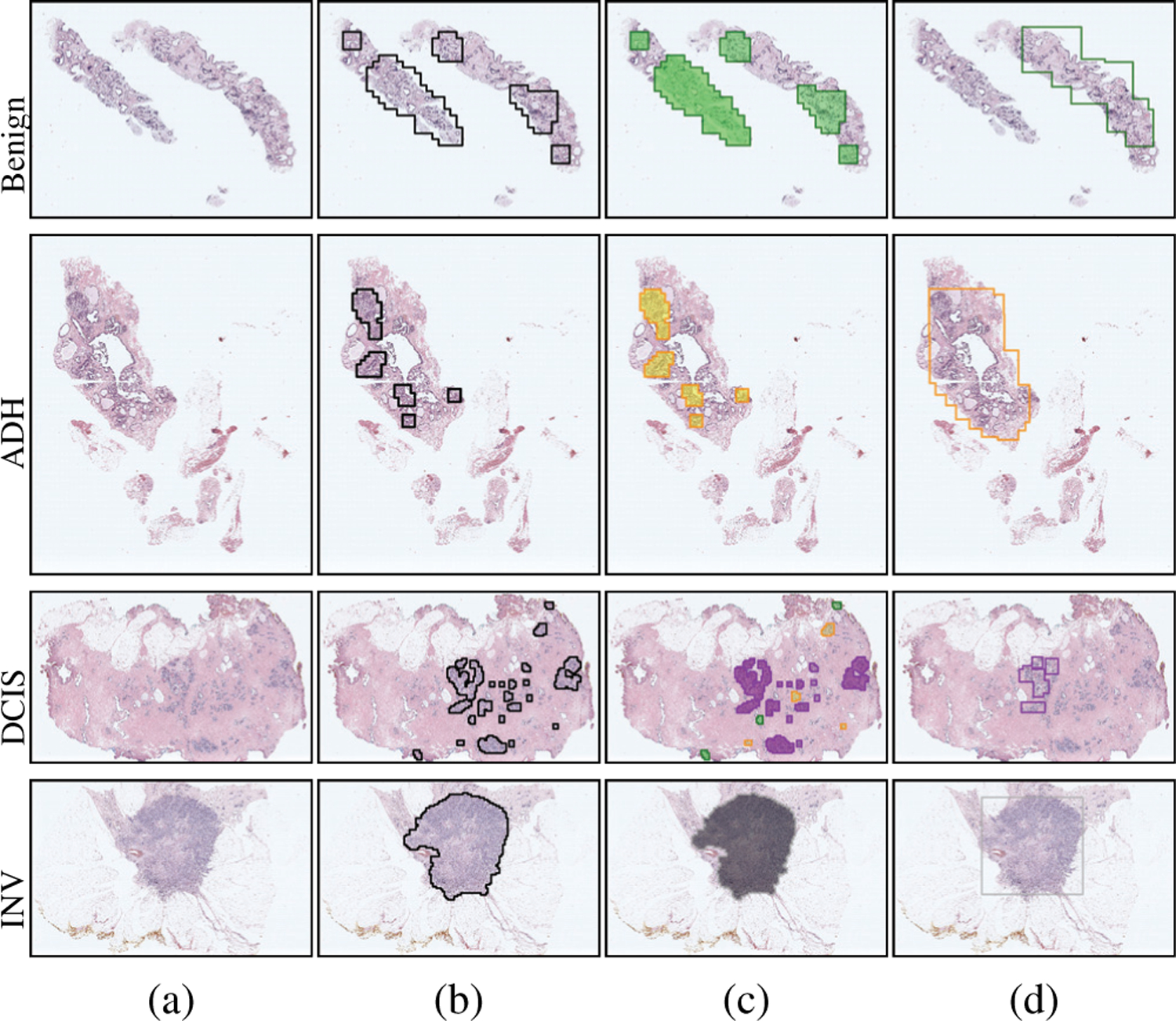
ROI-level outputs for example slides. (a) Consensus diagnoses and RGB images. (b) Regions used as ROI proposals. (c) Classes predicted for these regions as benign (green), ADH (yellow), DCIS (purple), INV (gray). (d) Consensus ROIs and their diagnoses with the same color coding.
VI. Conclusions
Convolutional networks typically operate on fixed-sized inputs and make class predictions on unseen images with the same size. However, ROIs in whole slide images can be drastically different from each other in size, shape, and structure, and it is not straightforward to analyze these ROIs using convolutional networks. We presented an effective generic framework to obtain feature representations for variable-sized ROIs. The proposed method operated on the automatically extracted potentially informative and diverse ROI patches. The local structural information within the patches as well as the class probability distributions of the patches as obtained from the predictions of the deep convolutional network were preserved in the feature representation of the ROI.
We investigated two methods to extract deep feature vectors for a patch. The first approach involved patch-level penultimate layer activations of the network, and the second one used pixel-level features obtained from the convolutional hypercolumn activations. In both approaches, the initial feature vector of the patch was weighted separately by each class probability score from the same network, and concatenation of the weighted vectors formed the final feature representation of the patch. Then, the feature representation of an ROI was obtained by the aggregation of the feature representations of its patches by average pooling. We demonstrated the representational power of the proposed approaches, illustrated using two separate deep network architectures, as they outperformed competing methods in extensive quantitative experiments for ROI-level breast histopathology image classification. Developing an end-to-end framework involving deep architectures for both feature extraction and classification will be studied in future work.
Acknowledgments
C. Mercan, B. Aygunes, and S. Aksoy were supported in part by the Scientific and Technological Research Council of Turkey under Grant No. 117E172. E. Mercan, L. G. Shapiro, D. L. Weaver, and J. G. Elmore were supported in part by the National Cancer Institute of the National Institutes of Health under Awards No. R01-CA172343, R01-140560, and R01-CA225585.
Footnotes
We define ROIs as regions that are identified to be diagnostically relevant by human experts during their interpretation of the slides.
Contributor Information
Caner Mercan, Department of Computer Engineering, Bilkent University, Ankara, 06800, Turkey. He is now with the Radboud University Medical Center, Nijmegen, The Netherlands..
Bulut Aygunes, Department of Computer Engineering, Bilkent University, Ankara, 06800, Turkey..
Selim Aksoy, Department of Computer Engineering, Bilkent University, Ankara, 06800, Turkey..
Ezgi Mercan, Paul G. Allen School of Computer Science & Eng., Univ. of Washington, Seattle, WA 98195, USA. She is now with the Seattle Children’s Hospital, Seattle, WA 98105..
Linda G. Shapiro, Paul G. Allen School of Computer Science & Engineering, University of Washington, Seattle, WA 98195, USA..
Donald L. Weaver, Department of Pathology, University of Vermont, Burlington, VT 05405, USA.
Joann G. Elmore, Department of Medicine, David Geffen School of Medicine, University of California, Los Angeles, CA 90024, USA.
References
- [1].Elmore JG et al. , “Diagnostic concordance among pathologists interpreting breast biopsy specimens,” Journal of American Medical Association, vol. 313, no. 11, pp. 1122–1132, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Allison KH et al. , “Histological features associated with diagnostic agreement in atypical ductal hyperplasia of the breast: Illustrative cases from the B-Path study,” Histopathology, vol. 69, pp. 1028–1046, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mercan C et al. , “Multi-instance multi-label learning for multi-class classification of whole slide breast histopathology images,” IEEE Trans. Med. Imag, vol. 37, no. 1, pp. 316–325, January 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Litjens G et al. , “A survey on deep learning in medical image analysis,” Medical Image Analysis, vol. 42, pp. 60–88, December 2017. [DOI] [PubMed] [Google Scholar]
- [5].Cruz-Roa A et al. , “Accurate and reproducible invasive breast cancer detection in whole slide images: A deep learning approach for quantifying tumor extent,” Scientific Reports, no. 46450, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Spanhol FA et al. , “A dataset for breast cancer histopathological image classification,” IEEE Trans. Biomed. Eng, vol. 63, no. 7, pp. 1455–1462, 2016. [DOI] [PubMed] [Google Scholar]
- [7].Sudharshan PJ et al. , “Multiple instance learning for histopathological breast cancer image classification,” Expert Systems With Applications, vol. 117, pp. 103–111, 2019. [Google Scholar]
- [8].Feng Y, Zhang L, and Mo J, “Deep manifold preserving autoencoder for classifying breast cancer histopathological images,” IEEE/ACM Trans. Comput. Biol. Bioinformatics, 2019. [DOI] [PubMed] [Google Scholar]
- [9].Aresta G et al. , “BACH: Grand challenge on breast cancer histology images,” Medical Image Analysis, vol. 56, pp. 122–139, 2019. [DOI] [PubMed] [Google Scholar]
- [10].Araujo T et al. , “Classification of breast cancer histology images usinǵ convolutional neural networks,” PloS ONE, vol. 12, no. 6, p. e0177544, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Roy K et al. , “Patch-based system for classification of breast histology images using deep learning,” Computerized Medical Imaging and Graphics, vol. 71, pp. 90–103, 2019. [DOI] [PubMed] [Google Scholar]
- [12].Vang YS, Chen Z, and Xie X, “Deep learning framework for multi-class breast cancer histology image classification,” in International Conference Image Analysis and Recognition, 2018, pp. 914–922. [Google Scholar]
- [13].Yan R et al. , “A hybrid convolutional and recurrent deep neural network for breast cancer pathological image classification,” in IEEE International Conference on Bioinformatics and Biomedicine, 2018, pp. 957–962. [Google Scholar]
- [14].Kandemir M and Hamprecht FA, “Computer-aided diagnosis from weak supervision: A benchmarking study,” Computerized Medical Imaging and Graphics, vol. 42, pp. 44–50, 2015. [DOI] [PubMed] [Google Scholar]
- [15].Hou L et al. , “Patch-based convolutional neural network for whole slide tissue image classification,” in CVPR, 2016, pp. 2424–2433. [DOI] [PMC free article] [PubMed]
- [16].Xu J et al. , “Stacked sparse autoencoder (SSAE) for nuclei detection on breast cancer histopathology images,” IEEE Trans. Med. Imag, vol. 35, no. 1, pp. 119–130, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zheng Y et al. , “Feature extraction from histopathological images based on nucleus-guided convolutional neural network for breast lesion classification,” Pattern Recognition, vol. 71, pp. 14–25, 2017. [Google Scholar]
- [18].He K et al. , “Spatial pyramid pooling in deep convolutional networks for visual recognition,” IEEE Trans. Pattern Anal. Mach. Intell, vol. 37, no. 9, pp. 1904–1916, September 2015. [DOI] [PubMed] [Google Scholar]
- [19].Mehta S et al. , “Y-Net: Joint segmentation and classification for diagnosis of breast biopsy images,” in MICCAI, 2018.
- [20].Mercan E et al. , “Assessment of machine learning of breast pathology structures for automated differentiation of breast cancer and high-risk proliferative lesions,” JAMA Network Open, vol. 2, no. 8, pp. 1–11, August 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].——, “Localization of diagnostically relevant regions of interest in whole slide images: A comparative study,” Journal of Digital Imaging, vol. 29, no. 4, pp. 496–506, August 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gecer B et al. , “Detection and classification of cancer in whole slide breast histopathology images using deep convolutional networks,” Pattern Recognition, vol. 84, no. 12, pp. 345–356, December 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sumbul G, Cinbis RG, and Aksoy S, “Multisource region attention network for fine-grained object recognition in remote sensing imagery,” IEEE Trans. Geosci. Remote Sens, vol. 57, no. 7, pp. 4929–4937, July 2019. [Google Scholar]
- [24].Mercan C et al. , “From patch-level to ROI-level deep feature representations for breast histopathology classification,” in SPIE Medical Imaging Symposium, San Diego, California, February 17–21 2019. [Google Scholar]
- [25].Elmore JG et al. , “A randomized study comparing digital imaging to traditional glass slide microscopy for breast biopsy and cancer diagnosis,” Journal of Pathology Informatics, vol. 8, no. 1, pp. 1–12, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Brunye TT et al. , “Eye movements as an index of pathologist visual expertise: A pilot study,” PLoS ONE, vol. 9, no. 8, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ruifrok A and Johnston D, “Quantification of histochemical staining by color deconvolution,” Analytical and Quantitative Cytology and Histology, vol. 23, no. 4, pp. 291–299, 2001. [PubMed] [Google Scholar]
- [28].Duda RO, Hart PE, and Stork DG, Pattern Classification John Wiley & Sons, Inc., 2000. [Google Scholar]
- [29].Simonyan K and Zisserman A, “Very deep convolutional networks for large-scale image recognition,” in International Conference on Learning Representations, 2015. [Google Scholar]
- [30].He K et al. , “Deep residual learning for image recognition,” in CVPR, 2016, pp. 770–778.
- [31].Azizpour H et al. , “From generic to specific deep representations for visual recognition,” in CVPR Workshops, 2015, pp. 36–45.
- [32].Hariharan B et al. , “Hypercolumns for object segmentation and fine-grained localization,” in CVPR, 2015, pp. 447–456. [DOI] [PubMed]
- [33].Liu L et al. , “From BoW to CNN: Two decades of texture representation for texture classification,” International Journal of Computer Vision, vol. 127, pp. 74–109, 2019. [Google Scholar]
- [34].Sanchez J et al. , “Image classification with the Fisher vector: Theory and practice,” International Journal of Computer Vision, vol. 105, pp. 222–245, 2013. [Google Scholar]
- [35].Chatfield K et al. , “The devil is in the details: An evaluation of recent feature encoding methods,” in British Machine Vision Conference, 2011, pp. 1–12. [Google Scholar]
- [36].Ronneberger O, Fischer P, and Brox T, “U-net: Convolutional networks for biomedical image segmentation,” in MICCAI, 2015, pp. 234–241.


