Abstract
The Zika pandemic sparked intense interest in whether immune interactions among dengue viruses 1–4 (DENV1–4) extend to the closely related Zika virus (ZIKV). We investigated prospective pediatric cohorts in Nicaragua that experienced sequential DENV1–3 (2004–15), Zika (2016–17), and DENV2 (2018–20) epidemics. Risk of symptomatic DENV2 infection and severe disease was elevated by one prior ZIKV infection, one prior DENV infection, or one prior DENV infection followed by one ZIKV infection, compared with being flavivirus-naïve. In contrast, multiple prior DENV infections reduced dengue risk. Further, although high preexisting anti-DENV antibody titers protected against DENV1, DENV3, and ZIKV disease, intermediate titers induced by previous ZIKV or DENV infection enhanced future risk of DENV2 disease and severity, as well as DENV3 severity. The observation that prior ZIKV infection can modulate dengue disease severity like a DENV serotype poses challenges to development of dengue and Zika vaccines.
Dengue virus serotypes 1–4 (DENV1–4) and Zika virus (ZIKV) are closely-related mosquito-borne flaviviruses with high global burdens (1, 2). Dengue epidemics often overwhelm health care systems as medical staff respond to life-threatening manifestations of severe dengue disease, including vascular leak syndrome and shock (3). ZIKV spread across the Pacific and Americas in 2013–2017 and caused rare but devastating clinical outcomes, including congenital microcephaly and Guillain-Barré Syndrome in adults (2, 4). Vaccines against both dengue and Zika are undergoing clinical evaluation (4, 5). However, the only licensed dengue vaccine, Dengvaxia®, increases risk of severe dengue in previously DENV-naïve individuals (6). Other dengue vaccine candidates are being evaluated for possible differences in safety and efficacy by DENV infection history. There remains concern that ZIKV infection or Zika vaccines could also enhance subsequent dengue disease.
A prior DENV infection is an established risk factor for future symptomatic and severe dengue during infection with a different serotype (7, 8). A first DENV or ZIKV infection induces antibodies that limit disease upon reinfection with the same virus but also generates non-protective antibodies that bind other serotypes (9). DENV cross-reactive antibodies can facilitate heterologous DENV infection of myeloid cells via antibody-dependent enhancement (ADE) and can increase dengue disease severity in humans (10–14). Many cross-reactive antibodies target epitopes conserved across flaviviruses, including the envelope protein fusion loop (15, 16). CD4+ and CD8+ T cell responses and non-structural protein 1 (NS1) also modulate DENV and ZIKV protection and pathogenesis (17, 18).
Emerging evidence suggests that prior DENV infection may not enhance non-congenital Zika disease, but whether prior ZIKV infection increases future dengue disease in humans is unknown. In vitro and mouse challenge studies showed that antibodies raised against DENV could enhance ZIKV infection (19, 20). However, prior DENV infection was not associated with ZIKV viremia or cytokine expression in experimentally challenged macaques (21–23) or in humans (24–26), nor with fetal demise or congenital Zika syndrome in pregnant women (27). Further, prior DENV infection was protective against uncomplicated Zika in prospective cohort studies (28, 29). Prior DENV infection was also associated with stronger cytotoxic CD8 T cell responses in ZIKV-infected humans and protection against ZIKV in mice (30). In contrast, anti-ZIKV antibodies increased DENV2 infection, viral output, and migration of myeloid cells in skin explants to the same degree as anti-DENV3 antibodies (31). In murine models, transfer of anti-ZIKV antibodies caused greater clinical severity, mortality, pro-inflammatory cytokine levels, and viral load following DENV2 challenge compared to untreated mice (32, 33). In macaques, prior ZIKV infection produced binding but non-neutralizing antibodies to DENV2, and challenge with DENV2 resulted in elevated viral load and hematological changes associated with severe dengue, although not in all studies (23, 34, 35).
The Zika epidemic was followed by several years of low DENV transmission, but in 2019, countries across Latin America reported a major resurgence of dengue cases. Since 2004, we have followed an active cohort of ~3800 children 2–16 years old living in Managua, Nicaragua, for DENV infection and disease (36, 37). As chikungunya virus (CHIKV) and ZIKV were introduced into Nicaragua in 2014 and 2016, respectively, the cohort was extended to capture cases and infections with these emerging arboviruses (28). In 2019–20, Nicaragua experienced the largest dengue epidemic in recorded history (Fig. 1A). An unprecedented number of cohort participants (n=375) were dengue cases. All virologically confirmed dengue cases (n=293) were caused by DENV2.
Fig. 1. Dengue and Zika cases, DENV- and ZIKV-Ab titers, and infection histories in the Pediatric Dengue Cohort Study (2004–2020).

(A) Confirmed dengue and Zika cases by epidemic season and infecting virus. DENV iELISA titers (B), ZIKV iELISA titers (C), and DENV and ZIKV infection histories (D) for cohort participants, measured at the beginning of each epidemic season. Infection histories: flavivirus-naïve (Naive), entered cohort DENV-immune without subsequent infections (DENV immune), entered flavivirus-naïve with one DENV (DENV) or ZIKV (ZIKV) infection, one prior DENV infection followed by a DENV (DENV-DENV) or ZIKV (DENV-ZIKV) infection, ≥2 prior DENV infections without (2+DENV) or with (2+DENV-ZIKV) a subsequent ZIKV infection.
The epidemiology and longevity of the Nicaraguan cohort allowed us to test whether ZIKV infection modified subsequent risk of dengue disease. All children presenting with suspected dengue, undifferentiated febrile illness, and after 2016, afebrile rash were tested for DENV, ZIKV, and CHIKV infection by real-time RT-PCR in acute-phase samples and by serological assays run on paired acute/convalescent samples. Since 2004, cohort participants have provided healthy annual serum and/or plasma samples that have been tested for DENV-specific antibodies (DENV-Abs) using the DENV inhibition ELISA (iELISA) (n= 8399 children, n=57,963 samples tested, Fig. 1B, table S1) (12, 37). Paired annual samples are tested side-by-side for seroconversion or a ≥4-fold rise in the DENV iELISA (12). Since 2016, samples are screened for ZIKV infection using the ZIKV iELISA (Fig. 1C) and ZIKV NS1 blockade-of-binding assays (n=14,159 and 14,247 measurements, respectively) (37, 38). In each assay, a titer or percent inhibition is derived by measuring the serum antibody concentration required to compete for binding to the antigen of interest with an antigen-specific, labeled antibody. Infection and case information was used to identify complete DENV and ZIKV infection histories for cohort participants (37) (Fig. 1D).
We estimated the probability of symptomatic DENV2 infection in the cohort during the 2019–20 epidemic by ZIKV and DENV infection history using log-binomial generalized linear models (GLMs) adjusted for age and sex. A total of 8.8% of all cohort participants experienced symptomatic DENV infections in 2019–20 (n=302 cases meeting dengue case definition of n=3434 cohort participants with full infection histories). Unexpectedly, children with one prior ZIKV infection had a 12.1% probability of having a symptomatic DENV2 infection in 2019–20 (confidence interval: 9.9–14.5), compared with only 3.5% (2.4–4.6) of flavivirus-naïve children (Fig. 2A, table S2), and similar to 9.2% (4.6–14.5) of children with one prior DENV infection. The increased risk of dengue disease with one prior ZIKV infection remained when confidence intervals were estimated using alternative modeling approaches; after adjustment for years since previous infection, neighborhood-level risk of flavivirus infection, and prior symptomatic ZIKV infection; and when afebrile dengue cases were included in analyses (figs. S1–S5, table S3).
Fig. 2. Probability of symptomatic and severe DENV2 infection by prior DENV and ZIKV infection histories and preexisting antibody titers, 2019–20.
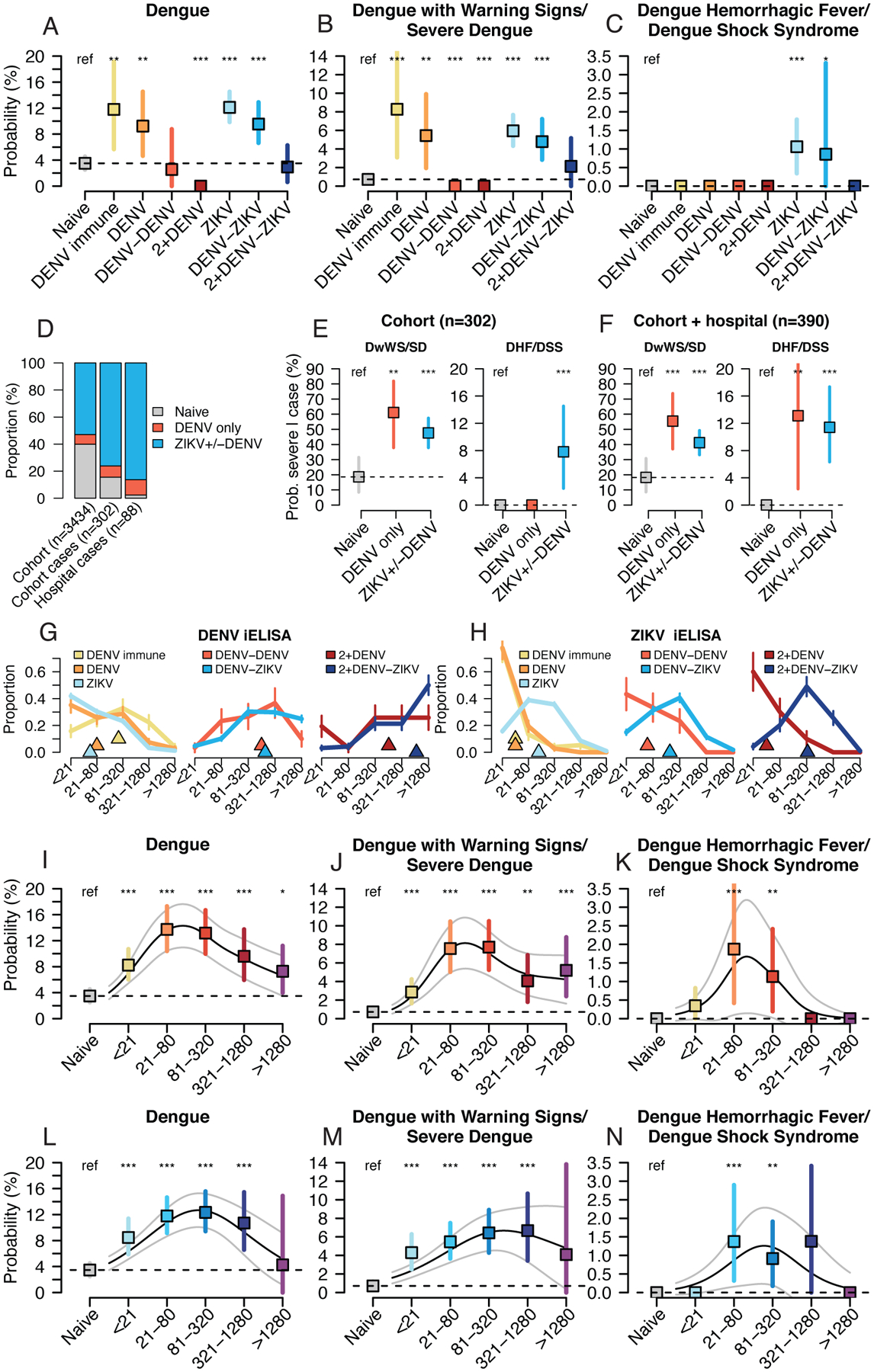
Log-binomial generalized linear models (GLMs) were used to estimate the probability dengue disease in 2019–20 in the cohort study by DENV and ZIKV infection history (A,B,C), preexisting DENV iELISA titers (I,J,K), or preexisting ZIKV iELISA titers (L,M,N), with bootstrap resampling (n=10,000) to construct 95% confidence intervals. Continuous relationships between titers and disease were modeled with log-binomial generalized additive models (GAMs, black lines show probabilities, grey lines show 95% confidence intervals). (D) Infection histories for children in the cohort, cohort dengue cases, and hospital study dengue cases. Differences in the proportion with histories of prior ZIKV+/−DENV or DENV only were tested with a Kruskal-Wallis rank sum test. Probability of severe dengue disease among confirmed dengue cases in cohort (E) or cohort and hospital studies (F) by infection history, estimated using logistic regression. Bootstrap resampling (n=10,000) was used to construct 95% confidence intervals. DENV iELISA (G) and ZIKV iELISA (H) titer distributions for cohort participants in 2019–20 by DENV and ZIKV infection history (triangles show median values, vertical bars show +/−1 standard deviation). All models were adjusted for age and sex, and probabilities are shown for an average study participant (male, age 8). P-values (*, <0.05; **, <0.01; ***, <0.001) indicate significantly different probability estimates from the Naïve group.
Following sequential DENV infections, individuals are thought to be at reduced risk of future dengue disease (9). Consistent with previous findings, children with two prior DENV infections had only a 2.5% (0.0–9.0) probability of symptomatic DENV2 infection. In contrast, children with one DENV infection followed by ZIKV infection remained at significantly elevated risk, with a 9.5% (6.7–13.0) probability of symptomatic DENV2 infection (Fig. 2A). For children with at least two prior DENV infections, the probability of dengue disease was low for those with (2.9%, 0.7–6.2) or without (0.0%, 0.0–0.0) a subsequent ZIKV infection.
A history of ZIKV infection was also a significant risk factor for severe dengue disease. The probability of experiencing Dengue with Warning Signs/Severe Dengue (DwWS/SD, n=144, 2009 World Health Organization [WHO] criteria) was significantly elevated for individuals with one DENV infection (5.4%, 2.0–9.6), one ZIKV infection (5.9%, 4.3–7.7), and one DENV and one ZIKV infection (4.8%, 2.9–7.0) compared to flavivirus-naïve children (0.7%, 0.3–1.2, Fig. 2B, table S4). The risk of Dengue Hemorrhagic Fever/Dengue Shock Syndrome (DHF/DSS) (n=15, 1997 WHO criteria) was also usually high (39) and was significantly greater for children with one prior ZIKV infection (probability: 1.1%, 0.3–1.8) and one DENV and one ZIKV infection (0.9%, 0–3.3) compared to flavivirus-naïve children (0%) (Fig. 2C, table S5). These relationships held for individual manifestations of severe dengue disease (fig. S6). In contrast, multiple DENV infections did not enhance dengue disease severity.
We further tested whether history of ZIKV infection increased risk of dengue disease in a separate study based at the Nicaraguan National Pediatric Reference Hospital. Since 2005, the study has followed 5832 children 6 months to 14 years of age during and after they present to the hospital for suspected dengue, Zika, and chikungunya. In 2018 and 2019, 388 children were enrolled in the hospital study, and all virologically confirmed cases were caused by DENV2 (n=277). For a random subset of hospital study DENV2 cases (n=88), we analyzed acute and convalescent samples (days 1–5 and 12–26 post-symptom onset, respectively) by the ZIKV and DENV E-domain III ELISA and DENV iELISA (40). In combination, these assays had 96% sensitivity and 96% specificity for detecting prior ZIKV infection in a separate set of children from the PDCS (n=53) with known infection histories (37). Compared to the full cohort (53%), significantly more cohort (76%) and hospital study (86%) dengue cases had a prior ZIKV infection, with or without a prior DENV infection (Kruskal-Wallis rank sum test, p<2.2e-16), but not more with prior DENV infection only (p=0.25), suggesting that ZIKV infection history helped explain the higher rate of disease (Fig. 2D). Further, among DENV2 cases in the cohort only or cohort and hospital studies (Fig. 2E and F, table S6, logistic regression adjusted for age and sex), the probability of experiencing DwWS/SD and DHF/DSS was significantly greater for children with histories of ZIKV with or without DENV (DHF/DSS: 11.6%, 6.2–17.2), as compared to naïve children (0.0%) and similar to the rate for those with only prior DENV infection (14.0%, 2.5–31.4).
We have previously shown that the level of preexisting DENV-Abs correlates with subsequent symptomatic and severe dengue disease (12, 14). Here, we found that children with the same number of prior DENV or DENV and ZIKV infections had similar levels of preexisting DENV-Ab titers (Fig. 2G), while preexisting ZIKV-Ab titers were higher for children with prior ZIKV infection, with or without prior DENV infection (Fig. 2H), consistent with previous studies (41–43). We used log-binomial GLMs to test whether preexisting DENV-Abs, including cross-reactive antibodies induced by prior ZIKV infection, and preexisting ZIKV-Abs were associated with dengue disease risk in 2019–20. Children with any level of preexisting cross-reactive DENV-Abs were at significantly greater risk of symptomatic DENV2 than flavivirus-naïve children (Fig. 2I). Those with a range of intermediate preexisting DENV-Abs were at the greatest risk of symptomatic DENV2 disease (1:21–1:80: 13.7%, 10.3–17.4 vs. naïve: 3.5%, 2.4–4.6, Fig. 2I), DwWS/SD (1:21–1:80: 7.5%, 5.0–10.4 vs. naïve: 0.7%, 0.3–1.2, Fig. 2J), and DHF/DSS (1:21–1:80: 1.9%, 0.4–39 vs. naïve: 0%, Fig. 2K). Intermediate preexisting ZIKV-Ab titers were also associated with enhancement of symptomatic and severe dengue (Fig. 2L–N).
Epidemiological evidence suggests that prior DENV immunity differentially affects disease caused by each DENV serotype. For instance, DENV2 and DENV4 more commonly manifest as symptomatic/severe disease in secondary infections than DENV1 and in some studies, DENV3 (7, 8, 44, 45). Immune correlate analyses in natural infection and vaccine studies have shown that high preexisting neutralizing antibody titers protect against DENV1 and DENV3 but not necessarily DENV2 (46–48). Consistent with these observations, we found that in the pre-Zika era (2004–2015), children with intermediate preexisting DENV-Ab titers also had increased probability of symptomatic (0.8%, 0.5–1.1 vs. naïve, 0.3%, 0.2–0.4) and severe dengue disease caused by DENV2 (DwWS/SD: 0.3%, 0.1–0.6, vs. naïve: 0.02%, 0.006–0.06, Fig. 3A–C, table S7). Notably, the magnitude of the enhancing effect was greater in the post-Zika era, due in part to the higher incidence of dengue cases in 2019–20 (Fig. 2I–K vs. Fig. 3A–C). In the pre-Zika era, low and intermediate DENV-Ab titers also had an enhancing effect on symptomatic and severe DENV3 infection (0.2%, 0.09–0.4 vs. naïve: 0.03%, 0.01–0.08, Fig. 3G–I) but not on symptomatic or severe DENV1 infection (Fig. 3D–F). In contrast, high preexisting DENV-Abs had a protective effect against symptomatic DENV1 and DENV3 infections (Fig. 3D and G, table S7). When all DENV cases were analyzed simultaneously, a protective effect was observed against symptomatic disease while the enhancing effect increased as the definition of severity narrowed to DHF/DSS (Fig. 3J–L), as we have observed previously (12).
Fig. 3. Probability of disease caused by DENV2, DENV1, DENV3, and ZIKV infection by preexisting DENV and ZIKV iELISA titers, 2004–17.

Each disease outcome was modeled as a function of preexisting antibody titer on both a discrete (colored bars) and continuous (black lines) scale, shown with 95% confidence intervals. Continuous relationships between DENV and ZIKV iELISA titers and each disease outcome were modeled using log-binomial GAMs. Probabilities of Dengue (A,D,G,J), Dengue with Warning Signs/Severe Dengue (B,E,H,K), and Dengue Hemorrhagic Fever/Dengue Shock Syndrome (C,F,I,L) by discrete DENV iELISA titer bins was modeled separately for each infecting serotype (DENV2, A,B,C; DENV1, D,E,F; DENV3, G,H,I) and all serotypes (DENV, J,K,L) in the pre-Zika era (2004–15) using generalized estimating equation (GEE) log-binomial models. Probability of Zika (2016) by pre-infection DENV iELISA (M) and ZIKV iELISA (N) titer bins was modeled using log-binomial GLMs, with 95% confidence intervals constructed using bootstrap resampling (n=10,000). All models were adjusted for age and sex, and model estimates are shown for an average study participant (male, age 8). P-values (*, <0.05, **, <0.01, ***, <0.001) indicate iELISA titer bins significantly different from the Naive group.
Interestingly, and similar to observations for DENV1, high preexisting DENV-Ab and ZIKV-Ab titers were associated with reduced probability of uncomplicated Zika during the 2016 Zika epidemic (Fig. 3M–N, table S8; we did not observe severe Zika in the cohort). Consistent with our previous epidemiological finding (28), one or multiple prior DENV infections were protective against Zika (table S8). However, intermediate DENV-Ab and ZIKV-Ab titers were not significantly protective, possibly indicating more complex relationships (see Fig. S8 for alternative titer bins).
Overall, we find that prior ZIKV infection modulates future dengue disease risk to a similar degree as prior infection with a DENV serotype. A single prior ZIKV infection, like one prior DENV infection, increases the probability of symptomatic and severe dengue disease caused by DENV2. Further, one DENV followed by one ZIKV infection also increased future risk of dengue disease, unlike sequential DENV infections, which reduced future risk, suggesting an important difference between secondary flavivirus infection with ZIKV versus a DENV serotype. Our findings also show that the relationship between preexisting anti-flavivirus antibodies and disease depends on the secondary infecting virus. Intermediate preexisting cross-reactive DENV-Ab or ZIKV-Ab titers enhance risk of DENV2 and DENV3 disease severity, but not DENV1 or ZIKV. High titers protect against symptomatic DENV1, DENV3, and ZIKV, but not DENV2 infection. Thus, we find that asymmetry exists among DENV serotypes and between DENV and ZIKV infections.
Based on our findings and previous literature on DENV1–4 (7, 8, 44–48), we posit that prior ZIKV infection, like prior DENV infection, is particularly capable of enhancing DENV2 disease but that enhancement of other serotypes is possible. Mechanistic studies in animal models and human skin explants have shown that prior primary ZIKV infection induces anti-DENV2 antibodies that facilitate classical ADE of infection and increase disease severity during DENV2 challenge (23, 31–34). In humans, primary ZIKV infection induces lower heterologous neutralizing antibody titers to DENV1–4 than primary DENV1–3 infection (41, 42), suggesting potential for enhancement of multiple serotypes (fig. S9). Further, although secondary ZIKV infection induces highly ZIKV-specific antibodies and boosts heterologous DENV binding antibodies, ZIKV infection does not induce the broadly cross-neutralizing antibodies observed after secondary DENV infection (fig. S9) (41, 42, 49). However, memory B cells/monoclonal antibodies with high DENV1-, DENV2-, and DENV3-ZIKV cross-neutralization have been isolated, suggesting cross-protection is possible (42, 43, 50). The tighter structure of the ZIKV virion may modify the types of antibodies ZIKV induces and limit their neutralizing potency against DENV, as well as reduce the ability of cross-reactive DENV antibodies to enhance ZIKV infection (51, 52). In addition, anti-ZIKV cytotoxic T cells mostly target structural protein epitopes, which may not protect against dengue, as anti-DENV CD8+ T cells mostly target conserved regions of non-structural proteins (53, 54). Finally, whether anti-ZIKV immunity enhances other DENV serotypes may depend on characteristics specific to the secondary infection, including virion structure and maturation state, cell infection mechanism, and fucosylation of IgG (51, 52, 55, 56).
Our findings suggest that protective and pathogenic interactions between DENV1–4 and ZIKV could affect vaccine efficacy and safety. If monovalent Zika vaccines induce cross-reactive DENV antibodies such as those observed following natural ZIKV infection, Zika vaccines could increase risk of subsequent symptomatic and severe dengue disease. In contrast, we find that DENV-Abs induced by natural DENV infection modestly protect against uncomplicated Zika, but the effect on vertically transmitted ZIKV in humans and risk of congenital Zika syndrome requires additional study. Further, differences in efficacy in the Phase 3 trials of multiple dengue vaccines may be explained not only by lack of protection but also by enhancement of certain serotypes (6). Our findings demonstrate why it is important to analyze correlates of protection and risk for dengue caused by specific serotypes separately. Overall, we illustrate critical immunological interactions among ZIKV and the DENV serotypes and reveal fundamental differences in the relationship between preexisting cross-reactive antibodies and serotype-specific risk of symptomatic and severe disease. Elucidating how immunity to DENV1–4, ZIKV, and possibly other flaviviruses modulates future disease risk is of utmost concern for developing and deploying safe, effective flavivirus vaccines and preventing future epidemics.
Supplementary Material
Acknowledgements:
We thank the Pediatric Dengue Cohort Study and Pediatric Dengue Hospital-based Study participants and their families. We are grateful to past and present members of the study team based at the Centro de Salud Sócrates Flores Vivas, Hospital Infantil Manuel de Jesús Rivera, the National Virology Laboratory in the Centro Nacional de Diagnóstico y Referencia, and the Sustainable Sciences Institute in Nicaragua for their dedication and high-quality work. We thank M. Montoya, P. Andrade, and P. Narvekar for providing important data from previous publications for understanding post-ZIKV infection neutralizing antibody titers.
Funding:
This research was funded by National Institutes of Health (NIH) grant P01AI106695 (to E.H.). The Pediatric Dengue Cohort Study was supported by NIH grants P01AI106695 (to E.H.), U19AI118610 (to E.H.), and R01AI099631 (to A.B.), as well as the Pediatric Dengue Vaccine Initiative grant VE-1 (to E.H.) and FIRST grant (to E.H. and J.C.), both from the Bill and Melinda Gates Foundation. L.C.K. was supported in part by the Global Health Equity Scholar FIC/NIH training grant D43TW010540. A.M.dS. was supported by CDC contract 00HVCLJB-2017-04191 (to A.M.dS.) and NIH/NIAID R01AI107731 (to A.M.dS.).
Footnotes
Competing interests: EH’s laboratory received research funds from Takeda Vaccines, Inc. to analyze samples from vaccine recipients. EH served on one-time Advisory Boards for Merck and Takeda.
Ethical statement: The Pediatric Dengue Cohort Study and Pediatric Dengue Hospital-based Study were approved by the Institutional Review Boards of the Nicaraguan Ministry of Health, the University of California, Berkeley, and the University of Michigan. Parents or legal guardians of all subjects provided written informed consent, and subjects ≥6 years old provided assent.
Data and materials availability: Data to understand and assess the conclusions of this research are available in the main text and Supplementary Materials. Sequence data are available on GenBank (accession numbers KM204119, KM204118, KU050695, KR011349, and KX421193) and code is available on Zenodo (doi: to be assigned). Individual-level data for reproducing figures may be shared with outside investigators following UC Berkeley IRB approval. Please contact the UC Berkeley Center for the Protection of Human Subjects (ophs@berkeley.edu) and Eva Harris (eharris@berkeley.edu) to arrange for data access. The materials and data used in this study are covered by standard material transfer agreements.
Supplementary Materials
Materials and Methods
Figs. S1–9
Tables S1–8
Other supporting files (IRB protocols)
References 57–82
References and Notes
- 1.Cattarino et al. , Sci. Transl. Med 12, eaax4144 (2020).31996463 [Google Scholar]
- 2.Faria et al. , Nature. 546, 406–410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO/TDR, “Dengue: Guidelines for Diagnosis, Treatment, Prevention and Control” (Geneva, Switzerland, 2009). [PubMed] [Google Scholar]
- 4.Diamond, Ledgerwood, Pierson, Annu. Rev. Med 70, 121–135 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Redoni et al. , Rev. Med. Virol, e2101 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Sridhar et al. , N. Engl. J. Med, NEJMoa1800820 (2018). [Google Scholar]
- 7.Nisalak et al. , 94, 1342–1347 (2016). [Google Scholar]
- 8.Sangkawibha et al. , Am. J. Epidemiol 120, 653–669 (1984). [DOI] [PubMed] [Google Scholar]
- 9.Gallichotte, Baric, de Silva, Adv. Exp. Med. Biol 1062, 63–76 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Halstead, J. Infect. Dis 140, 527–533 (1979). [DOI] [PubMed] [Google Scholar]
- 11.Kliks et al. , Am. J. Trop. Med. Hyg 40, 444–451 (1989). [DOI] [PubMed] [Google Scholar]
- 12.Katzelnick et al. , Science. 358, 929–932 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salje et al. , Nature. 557, 719–723 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waggoner et al. , J. Infect. Dis 221, 1846–1854 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dejnirattisai et al. , Science. 328, 745–8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Alwis et al. , PLoS Pathog. 10, e1004386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elong Ngono Shresta, Annu. Rev. Immunol 36, 279–308 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beatty et al. , Sci. Transl. Med 7, 304ra141 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Bardina et al. , Science. 356, 175–180 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zimmerman et al. , Cell Host Microbe. 24, 731–742.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pantoja et al. , Nat. Commun 8, 15674 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCracken et al. , PLOS Pathog. 13, e1006487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Breitbach et al. , PLOS Pathog. 15, e1007766 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Terzian et al. , Clin. Infect. Dis 65, 1260–1265 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santiago et al. , Open Forum Infect. Dis 6, ofz320 (2019).31363765 [Google Scholar]
- 26.Michlmayr et al. , Cell Rep. 31, 107569 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halai et al. , Clin. Infect. Dis 65, 877–883 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gordon et al. , PLOS Med. 16, e1002726 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez-Barraquer et al. , 610, 607–610 (2019). [Google Scholar]
- 30.Wen et al. , Nat. Commun 8, 1459 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castanha et al. , JCI insight. 5, e133653 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fowler et al. , Cell Host Microbe. 24, 743–750.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watanabe Tan, Chan Vasudevan, J. Infect. Dis 219, 223–233 (2019). [DOI] [PubMed] [Google Scholar]
- 34.George et al. , Sci. Rep 7, 1–10 (2017).28127051 [Google Scholar]
- 35.Pérez-Guzmán et al. , Nat. Commun 10, 4316 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuan et al. , Am. J. Epidemiol 170, 120–129 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.“Supplementary Materials”
- 38.Balmaseda et al. , J. Clin. Microbiol 56, e01785–17 (2018).29305550 [Google Scholar]
- 39.L’Azou et al. , N. Engl. J. Med 374, 1155–1166 (2016).27007959 [Google Scholar]
- 40.Premkumar et al. , J Clin Microbiol 56, 1–13 (2018). [Google Scholar]
- 41.Montoya et al. , J. Infect. Dis 218, 536–545 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andrade et al. , Nat. Commun 10, 938 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robbiani et al. , Cell. 169, 597–609.e11 (2017).28475892 [Google Scholar]
- 44.Guzman, Am. J. Epidemiol 152, 793–799 (2000). [DOI] [PubMed] [Google Scholar]
- 45.Aguas et al. , Sci. Rep 9, 1–12 (2019).30626917 [Google Scholar]
- 46.Endy et al. , J. Infect. Dis 189, 990–1000 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Buddhari et al. , PLoS Negl. Trop. Dis 8, e3230 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moodie et al. , J. Infect. Dis, 1–12 (2018).29506075 [Google Scholar]
- 49.Patel et al. , PLoS Negl. Trop. Dis 11, e0005554 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dussupt et al. , Nat. Med 26, 228–235 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goo et al. , Virology. 515, 191–202 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Gallichotte et al. , J. Infect. Dis 216, 1196–1204 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grifoni et al. , J. Virol 91, 1–19 (2017). [Google Scholar]
- 54.Weiskopf et al. , J. Infect. Dis 212, 1743–1751 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raut et al. , Proc. Natl. Acad. Sci, 201812055 (2018). [Google Scholar]
- 56.Wang et al. , Science. 355, 395–398 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuan et al. , Am. J. Epidemiol 170, 120–129 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Katzelnick et al. , Proc. Natl. Acad. Sci 115, 10762–10767 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Waggoner et al. , J Clin Virol 78, 57–61 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harris et al. , J Clin Microbiol. 36, 2634–2639 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Balmaseda et al. , Am J Trop Med Hyg. 61, 893–897 (1999). [DOI] [PubMed] [Google Scholar]
- 62.Fernández Vázquez, Mem. Inst. Oswaldo Cruz 85, 347–351 (1990). [DOI] [PubMed] [Google Scholar]
- 63.Balmaseda et al. , Trop. Med. Int. Health 11, 935–42 (2006). [DOI] [PubMed] [Google Scholar]
- 64.de Alwis et al. , PLoS Pathog. 10, e1004386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Balsitis et al. , PLoS Pathog. 6, e1000790 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clarke Casals, Am. J. Trop. Med. Hyg 7, 561–573 (1958). [DOI] [PubMed] [Google Scholar]
- 67.Reed Muench, Am. J. Epidemiol 27, 493–497 (1938). [Google Scholar]
- 68.Stettler et al. , Science. 353, 823–6 (2016). [DOI] [PubMed] [Google Scholar]
- 69.Balmaseda et al. , Proc. Natl. Acad. Sci, 201704984 (2017). [Google Scholar]
- 70.Galo et al. , Rev Panam Salud Publica. 41, e56 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gordon et al. , PLoS Negl. Trop. Dis 7, e2462 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.R Core Team, (Vienna, Austria, 2016). [Google Scholar]
- 73.Wood JR Stat. Soc. Ser. B (Statistical Methodol. 73, 3–36 (2011). [Google Scholar]
- 74.Anderson et al. , J. Infect. Dis 209, 360–368 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Montoya et al. , PLoS Negl. Trop. Dis 7, e2357 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reich et al. , J. R. Soc. Interface 10, 20130414 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zambrana et al. , Proc. Natl. Acad. Sci 115, 9294–9299 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Halekoh, Højsgaard, Yan, J. Stat. Softw 15, 1–11 (2006). [Google Scholar]
- 79.Narvaez et al. , PLoS Negl. Trop. Dis 5, 1–8 (2011). [Google Scholar]
- 80.Hammond et al. , Am. J. Trop. Med. Hyg 73, 1063–1070 (2005). [PubMed] [Google Scholar]
- 81.Rocha et al. , Am J Trop Med Hyg. 81, 287–292 (2009). [PubMed] [Google Scholar]
- 82.Alexander et al. , Trop. Med. Int. Heal 16, 936–948 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


