Abstract
Background
Circular RNA (circRNA) has been shown to affect the pathological process of osteoarthritis (OA) and is expected to become a potential marker for disease diagnosis. This study aimed to investigate the association between circRNA derived from the gene of runt‐related transcription factor 2 (RUNX2) and OA risk.
Methods
The expression profile of RUNX2‐derived circRNAs in serum of OA patients was detected. Then, the cytological localization of screened differential circRNAs was studied. Luciferase (LUC) reporter assay was used to identify the microRNA (miRNA) sponge capacity of the circRNAs. Bioinformatics analysis was used to construct the functional pathway of this circRNA‐miRNAs network. And then, the diagnostic value of RUNX2‐derived circRNAs in OA was evaluated.
Results
RUNX2‐derived hsa_circ_0005526 (circ_RUNX2) is significantly highly expressed in OA serum and mainly located in the cytoplasm within the cartilage cell by sponging multiple miRNAs (miR‐498, miR‐924, miR‐361‐3p, and miR‐665). Bioinformatics analysis showed ECM‐receptor interaction pathway ranked the most significant pathway of circ_RUNX2‐miRNAs regulatory network in KEGG database. The ROC curve showed that there may be good diagnostic value of serum circ_RUNX2 in OA.
Conclusion
RUNX2‐derived circ_RUNX2 may be involved in OA development via ECM‐receptor interaction pathways and may be used as potential clinical indicator of OA.
Keywords: circular RNA, ECM‐receptor interaction, microRNA, osteoarthritis, RUNX2
RUNX2‐derived hsa_circ_0005526 (circ_RUNX2) was significantly highly expressed in OA serum and sponged by multiple miRNAs (miR‐498, miR‐924, miR‐361‐3p, and miR‐665), which may be served as potential diagnostic marker in OA.
1. INTRODUCTION
Osteoarthritis (OA) is a common chronic degenerative joint disease, which is the main reason for the impairment of activity ability in the elderly. 1 , 2 Osteoarthritis mainly manifested as joint pain, limited movement, joint deformity, and even dysfunction, which seriously damaged the self‐care ability and labor ability of patients. 1 , 2 Currently, the common treatment methods of OA mainly include pain relief and joint replacement surgery for end‐stage patients, but these methods cannot reverse the early disease process or solve many limitations including surgical risk and artificial joint service life. Degeneration of articular cartilage is the main feature of OA. The pathogenesis of OA is complex, and the specific pathological mechanism is still unclear. OA is not a simple aging process. Many pathogenic factors, including matrix metalloproteinase dysfunction, cytokine imbalance, and genetic factors, are involved in the pathogenesis of OA. 3 , 4 These factors interact and influence each other, which makes it more difficult to find out the exact cause of OA.
microRNA (miRNA) is an endogenous non‐coding RNA with a length of about 22 nucleotides. 5 , 6 It can inhibit protein translation by binding with the mRNA of coding gene. At present, it has been found that there are many miRNAs with abnormal expression in OA patients, which can up‐regulate or down‐regulate the gene expression related to the growth of OA chondrocytes, thus interfering with the occurrence and development of OA. 4 , 7 , 8 , 9 For example, down‐regulated miR‐448 can inhibit cartilage degradation induced by IL‐1β‐induced matrix metalloprotein‐13 (MMP‐13) expression. 4 These results suggest that miRNAs are involved in the pathogenesis of OA and are associated with abnormal function of MMPs and cytokines, which may be used as a diagnostic marker and a potential therapeutic target for OA.
Among the many transcriptional components of genes, circular RNA (circRNA) lacks the characteristic "poly(A) tail" molecule of linear transcript and forms a cycled structure through the connection of head and tail ends. 10 , 11 Recent studies have shown that circRNA functions were different from linear RNA, which can bind various miRNAs and isolate them to avoid their interaction with the target mRNA and play the role of miRNA "sponge." 5 Recently, a small number of up‐ or down‐regulated circRNAs have been identified in different cartilage regions. 12 Some of them were found to involve in the development of molecular pathological mechanisms in OA. 12 For instance, the circRNA (has_circ_0005567) derived from EPS15 (epidermal growth factor receptor protein tyrosine kinase substrate 15) gene was identified as a mechanical stress‐related circRNA that involved in mechanical stress, which could control tumor necrosis factor‐alpha (TNF‐α) expression and promotes the degradation of extracellular matrix (ECM). 13
Runt‐related transcription factor 2 (RUNX2) is a key transcription factor associated with osteoblast differentiation, which is responsible for the pathogenesis of OA. 14 , 15 Recently, a number of miRNAs have been identified in the regulation of RUNX2 activity, thus affecting the process of osteogenesis and differentiation. 16 However, little is known about the regulatory network between RUNX2‐derived circRNA and miRNAs in OA. In present study, we investigated the expression of RUNX2‐derived circRNAs between health samples and OA subjects in peripheral blood. This may be helpful to explore the clinical value of RUNX2‐derived circRNAs in OA and identify new biomarkers or therapeutic targets for OA.
2. METHODS
2.1. Clinical specimens
In this study, samples diagnosed as OA in The First Affiliated Hospital of Xiamen University from January 2018 to October 2020 were included with informed consent. The inclusion criteria were as follows: volunteer to participate in the study and agree to take peripheral venous blood samples; meet the diagnostic criteria of OA; age ≥18 years old; subjects were conscious and could communicate correctly, and with no tumor and other serious physical diseases. Exclusion criteria: age <18 years old; acute and chronic pancreatitis; nervous system damage disease; history of psychoactive substance abuse; liver and kidney dysfunction. A total of 60 OA patients (30 males and 30 females) were also included. The mean age of male OA subjects was 70.21 ± 5.20 years (range 45–83 years old) with an average course of disease of 7.1 ± 0.53 years; The mean age of females OA subjects was 71.32 ± 3.15 years (range 51–87 years old) with an average course of disease of 6.5 ± 0.52 years. The healthy control subjects were the healthy population confirmed by routine physical examination in our hospital, 30 males and 30 females. The average age of male and female was 68.56 ± 3.25 years old and 69 ± 3.51 years old respectively. All the contents and implementation process of this study were approved by the Medical Ethics Committee of The First Affiliated Hospital of Xiamen University, and written informed consents were obtained from all subjects.
2.2. Extraction of total RNA from serum and real‐time qPCR analysis
In the morning, 3 ml peripheral blood was collected from the enrolled OA patients and healthy controls and then centrifuged in a frozen centrifuge for 10 min at a speed of 3000 g. Supernatant was collected and stored in a refrigerator at −80°C. The serum RNA was extracted according to the Trizol LS reagent protocol. The concentration and purity of the RNA were evaluated by Multiskan GO1510 spectrophotometer (Thermo Fisher Scientific, Vantaa, FI). 1 μg RNA was used for cDNA synthesis. The circRNA sequence database (http://www.circbase.org/) was used to obtain the transcription initiation sites and sequence information of all known circRNA molecules from RUNX2 gene, and corresponding reverse primers were designed according to the online tool (https://circinteractome.nia.nih.gov/). Real‐time PCR automatic analyzer Mx3005P qPCR System (Stratagene, La Jolla, CA) was used to analyze the expression of circRNA in serum samples. The relative expression level of circRNA was calculated using △Ct method.
2.3. Identification of circRNA and cell sublocalization analysis
In this study, the human normal cartilage cell line C‐28/I2 was cultured in RPMI‐1640 medium containing 1% double resistance (penicillin and streptomycin) and 10% fetal bovine serum. The cell culture chamber was set at 5% CO2 and 37℃. Ribonuclease R (RNase R) is an exonuclease enzyme that can cut and degrade RNA from the 3′‐5′ direction. It can digest almost all linear RNA molecules, but it is not easy to digest circular RNA. Total RNA was digested by RNase R (3 U RNase R/μg RNA) (37℃, 10 min), and the treated RNA was used as template to synthesize cDNA. The expression of linear RUNX2 gene and RUNX2‐derived circRNA were compared with conventional cDNA. Cell sublocalization often determines the type of biological function of circRNA, and we conducted cell sublocalization analysis of the selected circRNA according to the standard procedure of the nucleo‐plasmic separation kit instructions.
2.4. Screening of sponging miRNAs of circRNA derived from RUNX2
The cells were inoculated into 96‐well plates at a cell density of 5×103/ well for 24 h before transfection. The miRNAs that might bind to the selected circRNA were predicted by using circRNA bioinformatics analysis tools (http://www.circbase.org/ and https://circinteractome.nia.nih.gov/index.html), and then their analogues were synthesized. Reporter plasmids containing fragments of candidate circRNA binding sites were co‐transfected into the cells with these miRNA analogues. The activity of luciferase (LUC) was determined after 48 h according to the operating procedure in the manual. Finally, the LUC activity multiples of each miRNA analogue were calculated for comparison with the negative control. For miRNA molecules screened out by the above LUC experiments, cells were transfected with the circRNA overexpression vector and siRNA compounds, respectively, and then the influence of circRNA regulation on these miRNA expression levels was detected.
2.5. Molecular mechanism of RUNX2‐derived circRNAs and potential diagnostic value analysis in OA
Receiver operating characteristic curve (ROC) is a commonly used logistic regression model to evaluate the sensitivity and specificity of clinical indicators. 17 , 18 , 19 In this study, the expression of RUNX2‐derived circRNA was divided into healthy control group and OA patient group, in which the clinical diagnosis of patients was the true criterion in this model. The biological action pathways of the RUNX2‐derived circRNA and its sponging miRNAs were explored and analyzed using multiple bioinformatics database tools, 20 including KEGG (Kyoto Encyclopedia of Genes and Genomes), DIANA‐miRpath, and TargetScan, to reveal the molecular mechanism of a series of complex regulatory networks from circRNA expression to targeted functional gene changes.
2.6. Statistical analysis
Statistical Program for Social Sciences (SPSS, USA) 17.0 software is used to analyze all the data. The different value between the experimental group and the control group was analyzed by Student's t test; the difference was considered to be statistically significant when p < 0.05 in this study.
3. RESULTS
3.1. RUNX2‐derived circRNA was significantly differentially expressed in serum of OA patients
Through bioinformatics data (http://www.circbase.org/) retrieval, a total of 16 circRNA molecules derived from RUNX2 gene were found in human. The brief information of human circRNAs derived from RUNX2 gene was shown in Table 1. In this study, serum samples of five OA patients and five healthy controls were firstly included to detect the expression of these RUNX2‐derived circRNAs. Three circRNAs (hsa_circ_0005526, hsa_circ_0076685, and hsa_circ_0076684) showed higher levels in OA serum than controls (Figure 1A). Then, the expression statues of these three circRNA molecules were further explored in large sample size (Figure 1B–D). The serum level of hsa_circ_0005526 (circ_RUNX2) of OA patients was significantly higher than that in the control group (Figure 1C, **p < 0.01), and no significant difference was found in the expression levels of the other two circRNAs (Figure 1B,C). Furthermore, we compared the level of Hsa_circ_0005526 between different genders (Figure 1E). The results showed that HSA_ circ_ 0005526 level in male OA patients was significantly higher than that of male healthy controls (Figure 1E, **p < 0.01), and HSA_ circ_ 0005526 level in female OA patients was higher than that of female healthy controls (Figure 1E, **p < 0.01). There was no significant difference in Hsa_circ_0005526 levels between male and female OA patients (Figure 1E, p > 0.05). Moreover, a breakdown analysis by age showed that the significant association between HSA_ circ_ 0005526 level and OA was not significantly driven by age (Figure 1F). The results showed that HSA_circ_0005526 level in OA patients was significantly higher than that of healthy controls in both range of age (45–50 age and 70–80 age) (Figure 1F, **p < 0.01). There was no significant difference in Hsa_circ_0005526 levels between the two OA groups with 45–50 age and 70–80 age (Figure 1F, p > 0.05).
TABLE 1.
The brief information of circRNAs derived from RUNX2 gene in human
| circRNA ID | Position | Spliced length | Annotation |
|---|---|---|---|
| hsa_circ_0131859 | chr6:45296236‐45297465 | 1,229 | ALT_ACCEPTOR, ALT_DONOR, CDS, coding, INTERNAL, intronic, OVCODE, OVERLAPTX, OVEXON, UTR5 |
| hsa_circ_0131860 | chr6:45309315‐45309440 | 125 | ALT_ACCEPTOR, ALT_DONOR, coding, INTERNAL, intronic, OVERLAPTX |
| hsa_circ_0076684 | chr6:45390329‐45405788 | 627 | ANNOTATED, CDS, coding, INTERNAL, OVCODE, OVERLAPTX, OVEXON |
| hsa_circ_0076685 | chr6:45390329‐45480144 | 963 | ANNOTATED, CDS, coding, INTERNAL, OVCODE, OVERLAPTX, OVEXON |
| hsa_circ_0076686 | chr6:45390329‐45518819 | 5,285 | ANNOTATED, CDS, coding, OVCODE, OVERLAPTX, OVEXON, UTR3 |
| hsa_circ_0003563 | chr6:45399599‐45405788 | 262 | ANNOTATED, CDS, coding, INTERNAL, OVCODE, OVEXON |
| hsa_circ_0076687 | chr6:45399599‐45480144 | 598 | ANNOTATED, CDS, coding, INTERNAL, OVCODE, OVERLAPTX, OVEXON |
| hsa_circ_0076688 | chr6:45405683‐45405788 | 105 | ANNOTATED, CDS, coding, INTERNAL, OVCODE, OVEXON |
| hsa_circ_0076689 | chr6:45405683‐45518819 | 4,763 | ANNOTATED, CDS, coding, OVCODE, OVERLAPTX, OVEXON, UTR3 |
| hsa_circ_0076690 | chr6:45459677‐45459851 | 174 | ANNOTATED, CDS, coding, INTERNAL, OVCODE, OVEXON |
| hsa_circ_0005526 | chr6:45459677‐45460699 | 1,022 | ALT_DONOR, CDS, coding, INTERNAL, intronic, OVCODE, OVEXON |
| hsa_circ_0076691 | chr6:45459677‐45480144 | 336 | ANNOTATED, CDS, coding, INTERNAL, OVCODE, OVEXON |
| hsa_circ_0076692 | chr6:45459677‐45513019 | 402 | ANNOTATED, CDS, coding, INTERNAL, OVCODE, OVERLAPTX, OVEXON |
| hsa_circ_0076693 | chr6:45459677‐45518819 | 4,658 | ANNOTATED, CDS, coding, OVCODE, OVERLAPTX, OVEXON, UTR3 |
| hsa_circ_0076694 | chr6:45479982‐45480144 | 162 | ANNOTATED, CDS, coding, INTERNAL, OVCODE, OVEXON |
| hsa_circ_0076695 | chr6:45479982‐45518819 | 4,484 | ANNOTATED, CDS, coding, OVCODE, OVERLAPTX, OVEXON, UTR3 |
FIGURE 1.
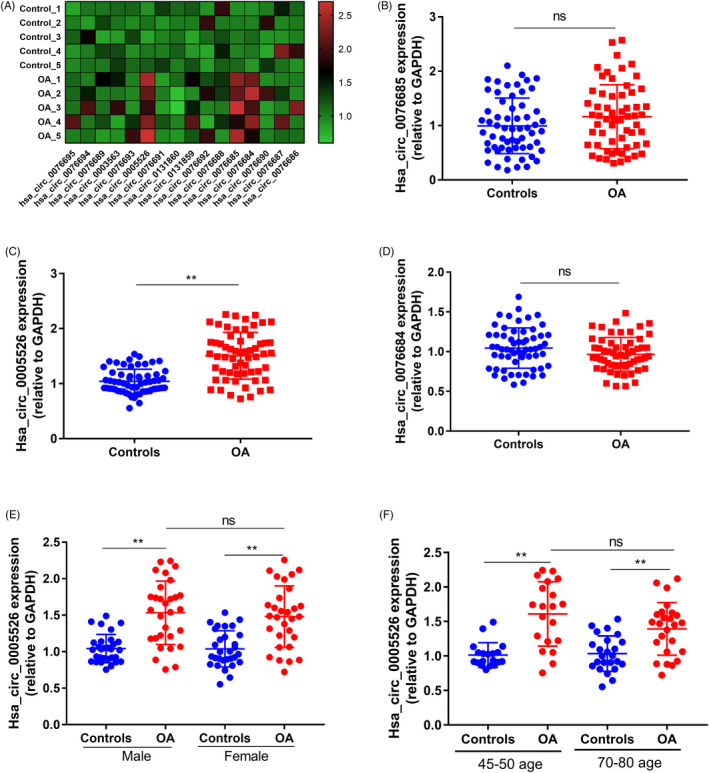
Expression of RUNX2‐derived circRNAs in OA serum. (A) Among 16 circRNA molecules derived from RUNX2, three circRNA molecules showed significantly high expression in OA serum (n = 5, p < 0.001). (B) No significantly differential level of hsa_circ_0076685 was found in serum between healthy controls and OA subjects (n = 60, p > 0.05). (C) The hsa_circ_0005526 (circ_RUNX2) in the serum of OA patients was significantly higher than that in the control group (n = 60, **p < 0.01). (D) There was no statistical difference between the OA samples and healthy subjects regarding hsa_circ_0076684 level in serum (n = 60, p > 0.05). (E) HSA_ circ_ 0005526 level in male OA patients was significantly higher than that of male healthy controls (n = 30, p < 0.01), and HSA_ circ_ 0005526 level in female OA patients was higher than that of female healthy controls (n = 30, p < 0.01). There was no significant difference in Hsa_circ_0005526 levels between male and female OA patients (p > 0.05). (F) HSA_circ_0005526 level in OA patients was significantly higher than that of healthy controls in both range of age (45–50 age and 70–80 age, p < 0.01). There was no significant difference in Hsa_circ_0005526 levels between the two OA groups with 45–50 age and 70–80 age (p > 0.05). *p < 0.05, **p < 0.01, ***p < 0.001
3.2. Subcellular localization of circ_RUNX2
Firstly, bioinformatics tool lncLocator is used to predict the subcellular localization of circ_RUNX2. 21 The analysis showed that most of the circ_RUNX2 located in the cytoplasm (Table 2). Then, the total RNA of C‐28/I2 cell was treated with RNase R. The result showed RUNX2 mRNA was significantly reduced, while the circ_RUNX2 level remained relatively stable (Figure 2A). Nucleocytoplasmic separation experiments showed that the distribution of circ_RUNX2 in cytoplasm was more than that in nucleus (Figure 2B). These results showed circ_RUNX2 had biological characteristics of circRNA resistant to RNase R cleavage and was mainly distributed in the cytoplasm of the cell structure (Figure 2A,B). Bioinformatics sequence analysis showed that multiple miRNAs had potential binding capacity to be sponging by circ_RUNX2 (Figure 2C).
TABLE 2.
Predicted locations of hsa_circ_0005526
| Subcellular locations | Score |
|---|---|
| Cytoplasm | 0.855541101237 |
| Nucleus | 0.0647580322981 |
| Ribosome | 0.015189477325 |
| Cytosol | 0.0487921152041 |
| Exosome | 0.0157192739358 |
FIGURE 2.
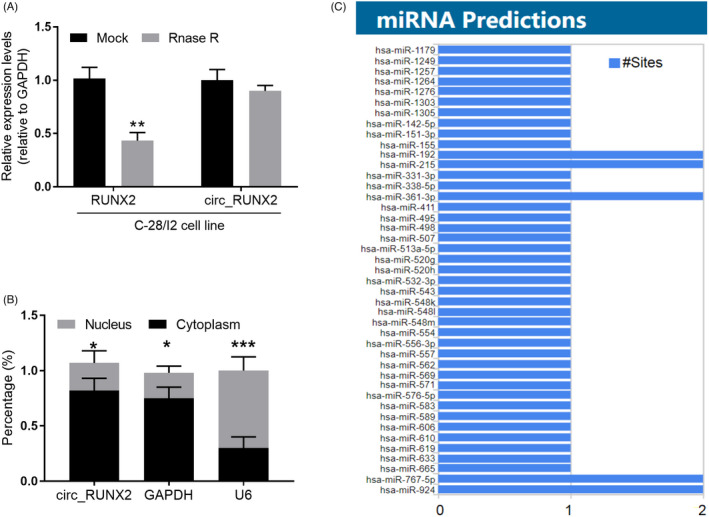
Biological function analysis of circ_RUNX2 in OA. (A) circ_RUNX2 has biological characteristics of resistance to RNA exonuclease. (B) Nucleocytoplasmic separation experiments confirmed that circ_RUNX2 was mainly located in the cytoplasm. (C) The potential sponged miRNAs by circ_RUNX2 in biological information analysis. *p < 0.05, **p < 0.01, ***p < 0.001
3.3. Several miRNAs were sponging by circ_RUNX2
In this study, 43 miRNA molecules that may bind circ_RUNX2 were found in the above analysis (Figure 2C). The LUC report analysis system was used for experimental verification with libraries of 43 miRNA analogues, and it was found that 4 miRNAs (miR‐498, miR‐924, miR‐361‐3p, and miR‐665) had an activity inhibition rate of more than 30% to LUC (Figure 3A). When the circ_RUNX2 overexpressed, it was found that the expressions of these 4 miRNAs were inhibited to varying degrees (Figure 3B). When the circ_RUNX2 was knocked out, the expressions of these 4 miRNAs were significantly up‐regulated (Figure 3C), indicating that circ_RUNX2 could regulate the expression levels of these 4 miRNAs and thus affect their biological functions.
FIGURE 3.
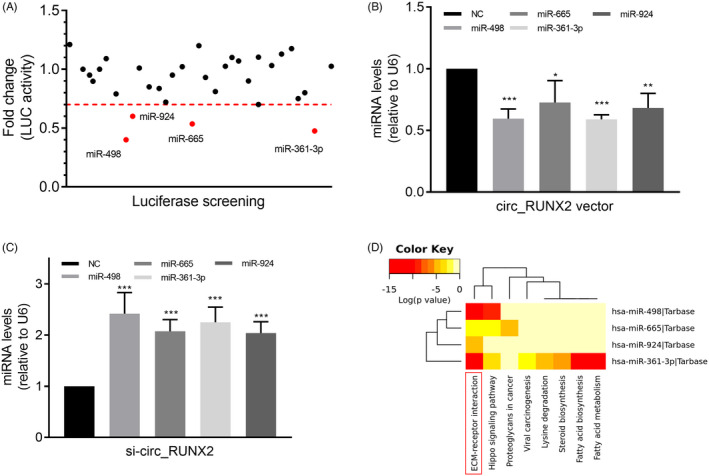
The regulatory role of OA‐related circ_RUNX2 on miRNAs and its biological pathway analysis. (A) The miRNAs with binding ability to circ_RUNX2 were screened by LUC reporter gene assay. (B) Up‐regulated circ_RUNX2 in cells significantly inhibited the expression of 4 miRNAs (miR‐498, miR‐924, miR‐361‐3p, and miR‐665). (C) Down‐regulated circ_RUNX2 expression can significantly increase the levels of the above miRNAs. (D) ECM‐receptor interaction was showed statistically significant correlation between above miRNAs and their mediated pathways by P‐value (log scaled) in heatmap. Red represents high significance. *p < 0.05, **p < 0.01, ***p < 0.01
3.4. ECM‐receptor interaction may be the pathway of the circ_RUNX2‐miRNA‐targets regulatory network in OA
In addition, we analyzed the biological pathway of circ_RUNX2‐miRNA regulatory network in order to expand our understanding of the mechanism of aberrantly expression of circ_RUNX2 in OA. The result showed ECM‐receptor interaction pathway ranked the most significant pathway of circ_RUNX2‐miRNA regulatory network in KEGG database (Figure 3D). In this biological pathway, there are many genes encoding human bone collagen family and regulating cell regeneration, which are potentially regulated by circ_RUNX2‐miRNA regulatory network. Detail information of the result was shown in Table 3.
TABLE 3.
miRNAs sponged by circ_RUNX2 and their targets in ECM‐receptor interaction of KEGG pathway
| miRNAs | p‐value | Genes |
|---|---|---|
| hsa‐miR‐361‐3p | 1.13648578304e‐11 | LAMB2 |
| COL4A2 | ||
| COL6A2 | ||
| COL1A1 a | ||
| DAG1 | ||
| TNC a | ||
| SDC4 | ||
| hsa‐miR‐498 | 1.4823035977e‐20 | COL6A1 |
| COL1A1 a | ||
| TNC a | ||
| hsa‐miR‐665 | 0.00787916536875 | THBS1 |
| COL1A2 | ||
| hsa‐miR‐924 | 3.23516172157e‐05 | COL3A1 |
The common targets of multiple miRNAs in this pathway
3.5. The clinical value of serum circ_RUNX2 in OA
In this study, we found that the serum circ_RUNX2 level was significantly different between OA patients and healthy controls (Figure 1). ROC curve model is an indicator to analyze the specificity and sensitivity of continuous variables. The greater the area under the curve, the greater the diagnostic value of variables. In this study, this model was used to analyze the diagnostic value of circ_RUNX2 in OA, and the results showed that the off‐line area of ROC curve reached 0.82, the sensitivity was 0.78, and the specificity was 0.77 (Figure 4). This indicates that the serum circ_RUNX2 level is expected to be a potential peripheral biomarker with diagnostic value for this disease.
FIGURE 4.
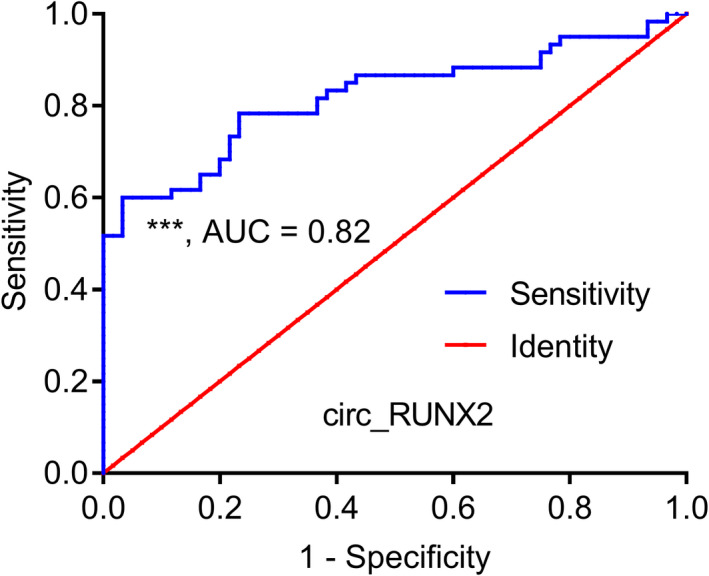
ROC curve analysis showed that the serum level of circ_RUNX2 had good potential diagnostic value in OA.*p < 0.05, **p < 0.01, ***p < 0.01
4. DISCUSSION
RUNX2 plays an important role in cartilage development and guides the formation and invasion of vessels in cartilage. 14 , 22 It is currently recognized that chondrocyte maturation is involved in the pathogenesis of OA, in which RUNX2 is one of the pathogenic genes and is involved in OA cartilage matrix degradation. 14 In this study, we detected the expression profile of 16 circRNAs derived from RUNX2 gene in peripheral blood of OA patients and the results showed that the circ_RUNX2 was significantly highly expressed in the peripheral blood of patients with OA (Figure 1). By using cell biological function analysis, it was found that circ_RUNX2 mainly located in cartilage cytoplasm and could adsorb multiple miRNAs to regulate their biological function (Figure 2). In order to explore the molecular pathology basis of this, we conducted a bioinformatics set analysis of the functions of multiple miRNAs regulated by circ_RUNX2 and found that they could jointly participate in the ECM‐receptor interaction pathway (Figure 3D) and regulate several important functional proteins in this pathway (Table 2). ROC model revealed that the level of circ_RUNX2 in serum has a good value in differentiating OA from healthy people and is expected to serve as a new objective biological indicator of the disease (Figure 4).
RUNX2 is a key transcription factor in the differentiation of osteoblasts and chondrocytes. 22 Its expression is significantly up‐regulated in chondrocytes of osteoarthritis cartilage. 14 The loss of RUNX2 in articular chondrocytes could slow down the progression of OA. 16 Circular RNA and its maternal gene may participate in the pathogenesis of the same disease. This study revealed that circ_RUNX2 can regulate the target genes in the ECM‐receptor interaction pathway by adsorbing multiple miRNAs (miR‐498, miR‐924, miR‐361‐3p, and miR‐665) (Figure 3A–C). These target genes have been proved to play an important role in bone formation, cartilage formation, and joint diseases. 8 , 13 , 23 There are two possible target genes (COL1A1 and TNC) regulated by more than one miRNA in this study. Collagens are the major structural components of the cartilage, which plays an important pathogenic role in OA. 24 The collagen type I alpha 1 chain genes (COL1A1) is one of several collagens showing rich differences in RNA and protein levels of OA. 24 Protein‐protein interaction network analysis showed that COL1A1 is one of the top two OA‐related Hub genes. 24 Tenascin‐C (TNC) is a large multimodular glycoprotein of the ECM structure, which involved in the pathogenesis of OA. 25 The expression of TNC is associated with the development of articular cartilage and promotes cartilage repair. 25 In OA subjects, elevated TNC expression is found in diseased cartilage, synovium, and synovial fluid, which could prevent cartilage damage. 25 , 26 A large number of highly expressed circRNAs have been found in human tissues. For instance, the mechanical stress in chondrocytes could increase the circRNA‐MSR expression which could regulate TNF‐α expression and participated in the chondrocyte ECM. 13 In cartilage tissues, circRNA.33186 directly binds to and inhibits miR‐127‐5p, thereby increasing MMP‐13 expression, and contributes to OA pathogenesis partly by regulating anabolic factor (type II collagen) expression. 23 Thus, it is critical to understanding the molecular mechanisms of OA to reveal the roles of circRNAs and their potential miRNA regulators. 27 , 28 These circRNAs not only have high abundance in cartilage, but also can be dissociated in the peripheral circulation system through various ways.
Because of the closed‐loop structure of circRNAs, they are not easy to be affected by RNA exonuclease and have high expression abundance and certain time sequence and disease specificity, which makes circRNAs have great potential to become new disease diagnosis markers. 11 , 29 The detection of circRNA in peripheral blood has the advantages of non‐invasive, rapid, and low cost. In this study, the high expression of serum circ_RUNX2 in OA patients suggested that circ_RUNX2 may be not only involved in the occurrence and development of OA, but also is expected to have clinical value as a potential biomarker for the diagnosis of the disease. This study is helpful to provide the preliminary theoretical basis for the subsequent development of more specific targeted drugs on the upstream and downstream targets of ECM‐receptor interaction pathway in the process of OA, which has important translational medical value.
In conclusion, this study described the association between circRNA derived from human OA‐related gene RUNX2 and the risk of OA. The circ_RUNX2 is expected to be a potential clinical biomarker of OA, which provides a new idea for the further understanding of the molecular pathological mechanism of OA, and has an important significance for the development of specific targeted therapy strategies for OA.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
Wang C, Li N, Liu Q, et al. The role of circRNA derived from RUNX2 in the serum of osteoarthritis and its clinical value. J Clin Lab Anal. 2021;35:e23858. 10.1002/jcla.23858
Chengyun Wang and Nanzhu Li contributed equally as first authors.
Funding information
This work was supported by Fujian Provincial Natural Science Foundation of China (2018T3017)
DATA AVAILABILITY STATEMENT
The datasets during and/or analyzed during the current study available from the corresponding author on reasonable request.
REFERENCES
- 1. Altman R, Asch E, Bloch D, et al. Development of criteria for the classification and reporting of osteoarthritis. Classification of osteoarthritis of the knee. Diagnostic and Therapeutic Criteria Committee of the American Rheumatism Association. Arthritis Rheum. 1986;29(8):1039‐1049. [DOI] [PubMed] [Google Scholar]
- 2. Altman R, Alarcón G, Appelrouth D, et al. The American College of Rheumatology criteria for the classification and reporting of osteoarthritis of the hip. Arthritis Rheum. 1991;34(5):505‐514. [DOI] [PubMed] [Google Scholar]
- 3. Castano‐Betancourt MC, Evans DS, Ramos YF, et al. Novel genetic variants for cartilage thickness and hip osteoarthritis. PLoS Genet. 2016;12(10):e1006260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang H, Wu D, Li H, Chen N, Shang Y. Downregulation of microRNA‐448 inhibits IL‐1beta‐induced cartilage degradation in human chondrocytes via upregulation of matrilin‐3. Cell Mol Biol Lett. 2018;23:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cheng J, Zhuo H, Xu M, et al. Regulatory network of circRNA‐miRNA‐mRNA contributes to the histological classification and disease progression in gastric cancer. J Transl Med. 2018;16(1):216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cheng J, Zhuo H, Wang L, et al. Identification of the combinatorial effect of miRNA family regulatory network in different growth patterns of GC. Mol Ther Oncolytics. 2020;17:531‐546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hu G, Zhao X, Wang C, et al. MicroRNA‐145 attenuates TNF‐alpha‐driven cartilage matrix degradation in osteoarthritis via direct suppression of MKK4. Cell Death Dis. 2017;8(10):e3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zheng X, Zhao FC, Pang Y, et al. Downregulation of miR‐221‐3p contributes to IL‐1beta‐induced cartilage degradation by directly targeting the SDF1/CXCR4 signaling pathway. J Mol Med. 2017;95(6):615‐627. [DOI] [PubMed] [Google Scholar]
- 9. Yi J, Liu D, Xiao J. LncRNA MALAT1 sponges miR‐30 to promote osteoblast differentiation of adipose‐derived mesenchymal stem cells by promotion of Runx2 expression. Cell Tissue Res. 2019;376(1):113‐121. [DOI] [PubMed] [Google Scholar]
- 10. Yu X, Ding H, Yang L, et al. Reduced expression of circRNA hsa_circ_0067582 in human gastric cancer and its potential diagnostic values. J Clin Lab Anal. 2020;34(3):e23080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang Y, Li Z, Xu S, Guo J. Novel potential tumor biomarkers: Circular RNAs and exosomal circular RNAs in gastrointestinal malignancies. J Clin Lab Anal. 2020;34(7):e23359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li HZ, Lin Z, Xu XH, Lin N, Lu HD. The potential roles of circRNAs in osteoarthritis: a coming journey to find a treasure. Biosci Rep. 2018;38(5):BSR20180542. 10.1042/BSR20180542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu Q, Zhang X, Hu X, et al. Emerging roles of circRNA related to the mechanical stress in human cartilage degradation of osteoarthritis. Mol Ther Nucleic Acids. 2017;7:223‐230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Komori T. Regulation of proliferation, differentiation and functions of osteoblasts by Runx2. Int J Mol Sci. 2019;20(7):1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Terauchi K, Kobayashi H, Yatabe K, et al. The NAD‐Dependent Deacetylase Sirtuin‐1 regulates the expression of osteogenic transcriptional activator runt‐related transcription factor 2 (Runx2) and production of matrix metalloproteinase (MMP)‐13 in chondrocytes in osteoarthritis. Int J Mol Sci. 2016;17(7):1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Narayanan A, Srinaath N, Rohini M, Selvamurugan N. Regulation of Runx2 by MicroRNAs in osteoblast differentiation. Life Sci. 2019;232:116676. [DOI] [PubMed] [Google Scholar]
- 17. Gao S, Cheng J, Li G, et al. Catechol‐O‐methyltransferase gene promoter methylation as a peripheral biomarker in male schizophrenia. Eur Psychiatry. 2017;44:39‐46. [DOI] [PubMed] [Google Scholar]
- 18. Cheng J, Wang Y, Zhou K, et al. Male‐specific association between dopamine receptor D4 gene methylation and schizophrenia. PLoS ONE. 2014;9(2):e89128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tian H, Li G, Xu G, et al. Inflammatory cytokines derived from peripheral blood contribute to the modified electroconvulsive therapy‐induced cognitive deficits in major depressive disorder. Eur Arch Psychiatry Clin Neurosci. 2020;271(3):475‐485. [DOI] [PubMed] [Google Scholar]
- 20. Vlachos IS, Zagganas K, Paraskevopoulou MD, et al. DIANA‐miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43(W1):W460‐466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cao Z, Pan X, Yang Y, Huang Y, Shen HB. The lncLocator: a subcellular localization predictor for long non‐coding RNAs based on a stacked ensemble classifier. Bioinformatics. 2018;34(13):2185‐2194. [DOI] [PubMed] [Google Scholar]
- 22. Arumugam B, Vishal M, Shreya S, et al. Parathyroid hormone‐stimulation of Runx2 during osteoblast differentiation via the regulation of lnc‐SUPT3H‐1:16 (RUNX2‐AS1:32) and miR‐6797‐5p. Biochimie. 2019;158:43‐52. [DOI] [PubMed] [Google Scholar]
- 23. Zhou ZB, Huang GX, Fu Q, et al. circRNA.33186 contributes to the pathogenesis of osteoarthritis by sponging miR‐127‐5p. Mol Ther. 2019;27(3):531‐541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhu Z, Zhong L, Li R, et al. Study of osteoarthritis‐related hub genes based on bioinformatics analysis. Biomed Res Int. 2020;2020:2379280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hasegawa M, Yoshida T, Sudo A. Tenascin‐C in osteoarthritis and rheumatoid arthritis. Front Immunol. 2020;11:577015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matsui Y, Hasegawa M, Iino T, Imanaka‐Yoshida K, Yoshida T, Sudo A. Tenascin‐C prevents articular cartilage degeneration in murine osteoarthritis models. Cartilage. 2018;9(1):80‐88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen G, Liu T, Yu B, Wang B, Peng Q. CircRNA‐UBE2G1 regulates LPS‐induced osteoarthritis through miR‐373/HIF‐1a axis. Cell Cycle. 2020;19(13):1696‐1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou Z, Du D, Chen A, Zhu L. Circular RNA expression profile of articular chondrocytes in an IL‐1beta‐induced mouse model of osteoarthritis. Gene. 2018;644:20‐26. [DOI] [PubMed] [Google Scholar]
- 29. Lu R, Shao Y, Tao X, Ye G, Xiao B, Guo J. Clinical significances of hsa_circ_0067582 and hsa_circ_0005758 in gastric cancer tissues. J Clin Lab Anal. 2019;33(9):e22984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets during and/or analyzed during the current study available from the corresponding author on reasonable request.


