Abstract
Circular RNAs (circRNAs) constitute a class of covalently closed RNA molecules. With the continuous advancement of high‐throughput sequencing technology and bioinformatics tools, many circRNAs have been identified in various human tissues and cell lines. Notably, recent studies have indicated that some circRNAs have translational functions. Internal ribosome entry sites and the N6‐methyladenosine modification mediate cap‐independent translation. This review describes these two translation mechanisms and verification methods at the molecular level. Databases (including ORF Finder, Pfam, BLASTp, CircRNADb, CircBase, CircPro, CircCode, IRESite, IRESbase) were used to analyze whether circRNAs have the structural characteristic of translation. CircRNA minigene reporter system containing green fluorescent protein (GFP) confirmed the translation potential of circRNAs. Also, we briefly summarize the roles of proteins/peptides encoded by circRNAs (circFBXW7, circFNDC3B, circLgr4, circPPP1R12A, circMAPK1, circβ‐catenin, circGprc5a, circ‐SHPRH, circPINTexon2, circAKT3) that have been verified thus far in human cancers (triple‐negative breast cancer, colon cancer, gastric cancer, hepatocellular carcinoma, bladder cancer, glioblastoma). Those findings suggest circRNAs have a great implication in translation of the human genome.
Keywords: cancers, circular RNAs, internal ribosome entry sites, N6‐methyladenosine, translation
Mechanism of cap‐independent translation initiated by IRES (eg, circFBXW7 in glioblastoma) and by m6A modification in the 5′‐UTR. USP28, ubiquitin‐specific peptidase 28; YTHDC1, YTH domain‐containing 1; METTL3, methyltransferase‐like 3; METTL14, methyltransferase‐like 14; YTHDF1, YTH domain family protein 1; YTHDF3, YTH domain family protein 3; m6A, N6‐methyladenosine; eIF4G2, eukaryotic translation initiation factor 4 gamma 2; IRES, internal ribosome entry site; and UTR, untranslated region.

1. INTRODUCTION
Circular RNAs (circRNAs) are endogenous RNAs that mostly exist in eukaryotic cells. They are mainly produced by heterogeneous nuclear RNAs (hnRNAs) through the “reverse splicing” mechanism, base pairs in inverted repeat elements (such as ALU), or dimerized RNA‐binding proteins (RBPs) at flanking introns. 1 , 2 The downstream splice donor site and the upstream splice acceptor site are covalently linked to form a loop closed structure without a 5′ end cap and 3′ end polyadenylation tail and no terminal structure. Owing to their highly stable and conservative structure, circRNAs can resist the degradation of ribonuclease. Most circRNAs are derived from protein‐coding genes and classified into three categories: intronic circRNAs (ciRNAs), exonic circRNAs (ecircRNAs), or exonic‐intronic circRNAs (EIciRNAs) 3 , 4 (Figure 1).
FIGURE 1.
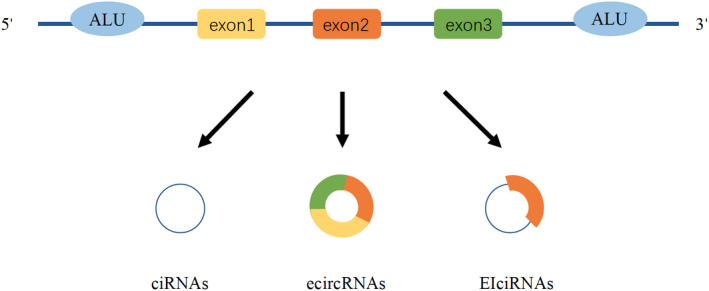
Classification of circRNAs. CircRNAs are divided into circular intronic RNAs (ciRNAs), exonic circRNAs (ecircRNAs), and exon‐intronic circRNAs (EIciRNAs) based on their composition
Moreover, the distribution of circRNAs is specific, mainly in the cytoplasm, 5 , 6 and their expression can be stably detected in serum, saliva, and tissue samples. Additionally, previous experiments have confirmed that circRNAs are differentially expressed in diseases. 7 , 8 Most of these circRNAs have been proposed to have sponging functions and indirectly play a role in promoting/inhibiting tumorigenesis. 9 Previously, circRNAs were classified as noncoding RNAs due to their highly conserved structure. However, recent studies have shown two important mechanisms critical for regulating the translation function of circRNAs. 10 , 11 , 12 One involves internal ribosome entry sites (IRESs), 13 and the other involves the N6‐methyladenosine (m6A) modification 14 ; both are potential mechanisms for cap‐independent translation of circRNAs. In other words, cap‐independent translation of circRNAs greatly expands our understanding of the biological functions of circRNAs and provides new perspectives for cancer treatment.
2. CAP‐INDEPENDENT TRANSLATION OF CIRCRNAS
2.1. Verification of circRNAs translation
In general, traditional translation can be initiated not only by the initiation codon (AUG), a suitable translation sequence and open reading frames (ORFs), but also by a dissociative 5′ end. 10 Therefore, novel elements are essential for activating cap‐independent translation. To easily understand the requirements for cap‐independent translation, several databases are listed for reference (Table 1). First, the ORFs with coding potential in circRNAs must be identified. ORF Finder 15 is a graphical sequence analysis tool that can search for all possible ORFs and deduce the translated amino acid sequence. This amino acid sequence is entered into BLASTp or Pfam tools 16 to confirm that the original search result was successful. Second, a comprehensive circRNA database that can better evaluate the coding potential of circRNAs is accessed. CircRNADb 17 includes circRNA genome sequence, IRES, and ORF information. CircBase 18 is constructed through the collection and integration of published circRNA data, which can quickly return circRNA information and supporting evidence. CircPro 19 can also be used to predict and identify circRNAs with coding potential. CircCode 20 recognizes translatable circRNAs in ribose sequence reads (ribo‐seq). Finally, IRES is identified. IRESite 21 contains many published and verified IRESs and provides experimental evidence of these IRESs. IRESbase 22 includes IRESs related to circRNAs and long noncoding RNAs (lncRNAs), and the function of an IRES has been experimentally verified.
TABLE 1.
Databases for verifying proteins/peptides encoded by circRNAs
| Database | Website | Introduction |
|---|---|---|
| ORF Finder | www.ncbi.nlm.nih.gov/gorf/gorf.html | This tool can find all possible ORFs and deduce the translated amino acid sequence. |
| Pfam | http://pfam.xfam.org/ | This tool is used for a homologous search of a protein sequence. |
| BLASTp | https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome" | This tool is used for comparing protein sequences with protein sequences in databases. |
| CircRNADb | http://202.195.183.4:8000/circrnadb/circRNADb.php | This tool includes circRNAs genome sequence, IRES and ORF information. |
| circBase | http://cirbase.org/ | This tool includes circRNAs information and supporting evidence. |
| CircPro | http://bis.zju.edu.cn/CircPro | This tool predicts and identifies circRNAs with coding potential. |
| CircCode | https://github.com/PSSUN/CircCode | This tool recognizes the translatable circRNAs in ribose sequence reads (ribo‐seq). |
| IRESite | http://www.iresite.org | This tool contains a large number of published and verified IRES. |
| IRESbase | http://reprod.njmu.edu.cn/cgi‐bin/iresbase/index.php | This tool includes functional IRES that have been experimentally verified. |
In addition to using bioinformatics tools, Yang Y et al used the circRNA minigene reporter system to identify functional translation mediated by IRES and m6A. 23 , 24 By inserting IRES fragments and fragments containing m6A motifs before the green fluorescent protein (GFP) sequence, the proteins/peptides can be evaluated by Western blotting (WB) or mass spectrometry (MS). In addition, ribosome profiling and polysome fractionation can infer the movement of ribosomal codons, 25 , 26 further confirming that circRNAs have a translational function. To study the biological function of the proteins/peptides encoded by circRNAs, the circRNA transcript is ectopically or endogenously expressed and knocked down or out, and its influence on gene expression and cell phenotype is evaluated. 1
2.2. CircRNA‐encoded proteins/peptides
2.2.1. Translation mediated by IRESs
Internal ribosome entry site‐mediated cap‐independent translation is a relatively mature mechanism currently studied. CircRNAs with ORFs and IRESs upstream can be effectively translated by this mechanism in various human cancers (Figure 2).
FIGURE 2.

CircRNA‐encoded proteins/peptides in human cancers. CC, colon cancer; GC, gastric cancer; HCC, hepatocellular carcinoma; TNBC, triple‐negative breast cancer
A circRNA‐translated protein in triple‐negative breast cancer (TNBC)
Breast cancer is a common type of malignant tumor and the second leading cause of cancer mortality among women worldwide; TNBC is known as the subtype with the worst prognosis. 27 Therefore, it is necessary to explore possible mechanisms underlying the progression of TNBC. A study found that circFBXW7 blocks miR‐197‐3p and that the spanning junction ORF of circFBXW7 encodes FBXW7‐185aa, 28 mediated by IRES, regulates the expression of FBXW7 in TNBC, and exhibits a tumor suppression effect. Therefore, circFBXW7 and FBXW7‐185aa may be potential therapeutic targets.
CircRNA‐translated proteins/peptides in colon cancer (CC)
Colon cancer is the second leading cause of cancer‐related death worldwide, and it is a matter of great urgency for researchers to develop more effective molecular targets for CC therapy. 29 A study reported that circFNDC3B can encode a novel protein, circFNDC3B‐218aa. 30 More importantly, this protein can perform biological functions independently and inhibit the proliferation, invasion, and migration of CC cells. CircFNDC3B‐218aa attenuates the inhibitory effect of Snail on fructose‐1,6‐bisphosphatase 1 (FBP1) in CC through the Snail‐FBP1‐EMT axis and enhances cell metabolism switching from glycolysis to oxidative phosphorylation, thereby inhibiting the progression of the epithelial mesenchymal transition (EMT) in CC cells. This finding suggests that circFNDC3B‐218aa may be a potential therapeutic target for CC.
The peptide translated by circLgr4 interacts with and activates Lgr4, which further activates the Wnt/β‐catenin signaling pathway, promoting the self‐renewal and tumorigenesis of CC stem cells. Therefore, medicine targeting the circLgr4‐peptide‐Lgr4 axis may be used to treat CC. 31
Hsa_circ_0000423 (CircPPP1R12A) encodes a peptide, circPPP1R12A‐73aa, which promotes the proliferation, migration, and invasion of CC cells in vivo and in vitro by activating the Hippo‐YAP signaling pathway. 32 YAP1 is a transcriptional activator in the Hippo signaling pathway, and the transcriptional response induced by YAP1 is crucial to the proliferation and metastasis of cancer cells. YAP‐TEAD inhibitor 1 was used to overexpress circPPP1R12A‐73aa, significantly reducing the proliferation, migration, and invasion ability of circPPP1R12A‐73aa‐expressing CC cells.
A circRNA‐translated protein in gastric cancer (GC)
Gastric cancer is the third most common cause of cancer death globally. Due to the lack of specific symptoms of early GC, most patients are diagnosed in the final stage and have poor prognosis. It is urgent to identify biomarkers to improve patient survival. CircMAPK1 (hsa_circ_0004872) encoded a tumor suppressor protein with 109 amino acid length. 33 MAPK1–109aa inhibits the phosphorylation of MAPK1 by competitively binding MEK1, thereby inhibiting the activation of the MAPK1 pathway and its downstream factors and showing the ability to inhibit the proliferation and invasion of GC cells.
A circRNA‐translated protein in hepatocellular carcinoma (HCC)
Hepatocellular carcinoma (HCC) has a high mortality rate, which is attributed to a lack of efficient diagnostic and therapeutic tools. 34 The Wnt/β‐catenin pathway extensively participates in tumor growth, but the reason it is overactivated in HCC is still unknown. 35 A recent study showed that circ‐0004194 (circβ‐catenin) encodes a new protein, β‐catenin‐370aa, which is a subtype of β‐catenin. 36 Additionally, circβ‐catenin regulates the expression of β‐catenin at the protein level, not at the transcription level. β‐catenin‐370aa acts as bait for GSK3β and binds to it, thereby reducing the ubiquitination of β‐catenin by the ubiquitin ligase β‐TrCP, evading the degradation of the proteasome induced by GSK3β. Upregulated β‐catenin enhances the Wnt/β‐catenin pathway and aggravates the malignancy of HCC cells.
A circRNA‐translated peptide in bladder cancer
Bladder cancer stem cells exist in the tumor bulk and can induce tumorigenesis. However, the biology of bladder cancer stem cells is unclear. 37 CircGprc5a is upregulated in bladder cancer stem cells and strengthens the self‐renewal ability of bladder stem cells. In particular, circGprc5a has coding potential and plays a biological role in bladder cancer through the peptide (FDTKPMNLCGR). 38 It participates in bladder tumor cells by relying on the circGprc5a‐peptide‐Gprc5a axis. Blockers of the circGprc5a‐peptide‐Gprc5a axis may be used for targeted therapy of bladder cancer, which is important because patients whose tumors show high expression of circGprc5a have a poor prognosis.
CircRNA‐translated proteins/peptides in glioblastoma
Glioblastoma is one of the most lethal human cancers that may occur at any age and exhibits an extremely poor response to approved therapies. 39 , 40 Obviously, new targets/treatments urgently need to be explored. The FBXW7‐185aa protein encoded by circFBXW7 plays a role not only in the aforementioned TNBC, 28 as mentioned above, but also in glioblastoma. 41 C‐Myc is a key regulator of tumorigenesis. FBXW7‐185aa competitively interacts with USP28 to prevent the binding of USP28 and FBXW7α and then promotes c‐Myc ubiquitination and degradation. Therefore, FBXW7‐185aa inhibits the proliferation and delays cell cycle progression of glioma cells.
Circ‐SHPRH and its encoded protein SHPRH‐146aa are highly expressed in normal human brains. 42 Additionally, SHPRH‐146aa protects SHPRH from the degradation of the ubiquitin proteasome and promotes the ubiquitination of proliferating cell nuclear antigen (PCNA), reducing the malignant behavior of cancer cells in vivo and in vitro.
The peptide PINT87aa encoded by circPINTexon2 43 inhibits the proliferation of glioblastoma cells, and its downregulation induces cell cycle acceleration. Accordingly, the expression of circPINT exon 2 and PINT87a in brain tumor tissues was decreased relative to that in normal brain tissues. It has the same effect in other malignant tumors (including BC, HCC, and gastric cancer), suggesting a poor clinical prognosis. However, a study found that PINT87aa is likely necessary for normal cell survival. 43
AKT3‐174aa, a tumor suppressor protein encoded by circAKT3, is expressed at low levels in glioblastoma tissues. 44 , 45 AKT3‐174aa competitively interacts with phosphorylated PDK1, reduces AKT‐thr308 phosphorylation, and plays a negative regulatory role in modulating the PI3K/AKT signal intensity. Finally, AKT‐thr308 promotes the proliferation and enhances the radiation resistance and tumorigenicity of cancer cells. Furthermore, studies have shown that, in addition to PTEN, AKT‐174aa is also a negative regulator of the RTK/PI3K pathway and may become a potential biomarker for patients with glioblastoma.
2.2.2. Translation mediated by m6A modification
CircRNAs modified by m6A can also undergo cap‐independent translation. 23 m6A is the most common internal modification of RNAs and has the greatest influence on the regulation of their activity. 46 , 47 m6A modification is realized by a heterodimeric complex composed of methyltransferase‐like 3 (METTL3), acting as where METTL3 is the catalytic subunit, and methyltransferase‐like 14 (METTL14). 14 , 48 , 49 m6a can control pre‐mRNA post‐transcriptional regulation, including splicing, exportation, and translation, by generating conformational changes in local RNA structures or recruiting specific m6A reader proteins. 46 , 50 The protein containing the YTH domain was the first reader to be recognized. YTHDC1 has been proven to regulate the reverse splicing and export of circRNAs. 51 , 52 YTHDF1 was found to increase translation efficiency through the binding of m6A and YTHDF3. 53 , 54 Then, YTHDF3 and eIF4G2 physically associate with endogenous circRNAs to promote the coding of proteins and control cell proliferation. Furthermore, translation from circRNAs is weakened when m6A demethylates fat mass and obesity‐associated protein (FTO). 23 , 55
A study reported that oncogenic human papillomavirus 16 (HPV16) generates circE7, which translated to produce E7 oncoprotein in CaSki cervical carcinoma cells. 56 The expression level of E7 oncoprotein affects cancer cell growth both in vitro and tumor xenografts, but the exact carcinogenic mechanism of E7 peptide remains uncertain.
In summary, in addition to the IRES‐dependent pathway (Table 2), the m6A residues in circRNAs can serve as m6A‐induced internal ribosome‐binding sites (MIRES), thereby promoting cap‐independent translation (Figure 3). 14
TABLE 2.
The role of circRNA‐encoded proteins/peptides in human cancers
| CircRNAs | Protein/Peptide | Cancer | Function | Possible mechanism |
|---|---|---|---|---|
| circFBXW7 | FBXW7‐185aa | Triple‐negative breast cancer | Suppressor | Unknown. |
| circFNDC3B | circFNDC3B‐218aa | Colon cancer | Suppressor | Inhibit the Snail‐FBP1‐EMT axis. |
| circLgr4 | Peptide | Colon cancer | Promote | Promote the activation of the Wnt/β‐catenin signaling pathway. |
| circPPP1R12A | circPPP1R12A‐73aa | Colon cancer | Promote | Promote the activation of the Hippo‐YAP signaling pathway. |
| circMAPK1 | MAPK1‐109aa | Gastric cancer | Suppressor | Play a negative regulatory role in the MAPK signal pathway. |
| circβ‐catenin | β‐catenin‐370aa | Hepatocellular carcinoma | Promote | Enhance the activation of the Wnt/β‐catenin pathway. |
| circGprc5a | FDTKPMNLCGR | Bladder cancer | Promote | Enhance the activation of the circGprc5a‐peptide‐Gprc5a axis. |
| circFBXW7 | FBXW7‐185aa | Glioblastoma | Suppressor | Increase the expression of FBXW7 and induce c‐Myc ubiquitination‐induced degradation. |
| circ‐SHPRH | SHPRH‐146aa | Glioblastoma | Suppressor | Promote the ubiquitination of proliferating cell nuclear antigen (PCNA). |
| circPINTexon2 | PINT87aa | Glioblastoma | Suppressor | Interact with the polymerase‐associated factor complex (PAF1c). |
| circAKT3 | AKT3‐174aa | Glioblastoma | Suppressor | Play a negative regulatory role in the PI3K/AKT signal pathway. |
FIGURE 3.

Mechanism of cap‐independent translation initiated by IRES (eg, circFBXW7 in glioblastoma) and by m6A modification in the 5′‐UTR. eIF4G2, eukaryotic translation initiation factor 4 gamma 2; IRES, internal ribosome entry site; m6A, N6‐methyladenosine; METTL14, methyltransferase‐like 14; METTL3, methyltransferase‐like 3; USP28, ubiquitin‐specific peptidase 28; UTR, untranslated region; YTHDC1, YTH domain‐containing 1; YTHDF1, YTH domain family protein 1; YTHDF3, YTH domain family protein 3
3. PERSPECTIVES
The biological functions of the proteins/peptides encoded by circRNAs have begun to emerge. We summarized the coding functions of circRNAs in human tumors that have been confirmed thus far and the tumor‐promoting or tumor‐inhibiting effects of their products.
The idea that circZNF609 57 in myogenesis and circMBL 58 in fly head extracts can encode peptides initially caused great controversy, but after continuous experimental verification, circRNAs were redefined. In addition, studies have found that some circRNAs have both microRNA (miRNA)‐sponging and protein‐encoding functions and can perform the same, opposite, or irrelevant biological functions in a variety of tissues. 59 For example, circFBXW7 inhibits malignant progression by sponging miR‐197‐3p and encoding FBXW7‐185aa in TNBC. 60 Therefore, circRNAs have great potential for use in disease treatment. In the near future, more circRNA‐encoding functions will be verified, and other possible encoding mechanisms will even be discovered.
Although the future of circRNA translation function is very impressive, there are still some problems that need to be considered in depth. For example, the current understanding of the cap‐independent translation mechanism of circRNAs is still unsatisfactory. Are other coding mechanisms possible? The proteins/peptides encoded by circRNAs may serve as new drugs for the treatment of cancers due to their high specificity, low toxicity, and clear biological function. However, their poor stability and short half‐life also greatly limit their clinical application. Therefore, these problems require further research to be solved.
CONFLICT OF INTERESTS
The authors declare that they have no competing interests.
ACKNOWLEDGMENTS
This work was supported by grants from Zhejiang Natural Science Foundation (LY20H130001, LQ21H130001); Ningbo Health Branding Subject Fund (PPXK2018‐02); the Natural Science Foundation of Ningbo (202003N4239); the Ningbo "Technology Innovation 2025" Major Special Project (2020Z097, 2018B10015); the Medical and Health Research Project of Zhejiang Province (2019ZD018, 2021KY307); Ningbo Science and Technology Project (202003N4238); and Zhejiang Province Medicine and Health Technology Plan Project (2017KY133).
Lu Y, Li Z, Lin C, Zhang J, Shen Z. Translation role of circRNAs in cancers. J Clin Lab Anal. 2021;35:e23866. 10.1002/jcla.23866
DATA AVAILABILITY STATEMENT
The data presented in this study can be found in online repositories. The name of repositories and reference number can be found in the review.
REFERENCES
- 1. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Gen. 2019;20(11):675‐691. [DOI] [PubMed] [Google Scholar]
- 2. Ashwal‐Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre‐mRNA splicing. Mol Cell. 2014;56(1):55‐66. [DOI] [PubMed] [Google Scholar]
- 3. Li Z, Ruan Y, Zhang H, Shen Y, Li T, Xiao B. Tumor‐suppressive circular RNAs: Mechanisms underlying their suppression of tumor occurrence and use as therapeutic targets. Cancer Sci. 2019;110(12):3630‐3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang Y, Li Z, Xu S, Guo J. Novel potential tumor biomarkers: Circular RNAs and exosomal circular RNAs in gastrointestinal malignancies. J Clin Lab Anal. 2020;34(7):e23359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang H, Shen Y, Li Z, et al. The biogenesis and biological functions of circular RNAs and their molecular diagnostic values in cancers. Journal of Clinical Laboratory Analysis. 2020;34(1):e23049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huang C, Liang D, Tatomer DC, Wilusz JE. A length‐dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 2018;32(9–10):639‐644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Qu S, Liu Z, Yang X, et al. The emerging functions and roles of circular RNAs in cancer. Cancer Lett. 2018;414:301‐309. [DOI] [PubMed] [Google Scholar]
- 8. Ruan Y, Li Z, Shen Y, Li T, Zhang H, Guo J. Functions of circular RNAs and their potential applications in gastric cancer. Expert Rev Gastroenterol Hepatol. 2020;14(2):85‐92. [DOI] [PubMed] [Google Scholar]
- 9. Lei B, Tian Z, Fan W, Ni B. Circular RNA: a novel biomarker and therapeutic target for human cancers. Int J Med Sci. 2019;16(2):292‐301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Granados‐Riveron JT, Aquino‐Jarquin G. The complexity of the translation ability of circRNAs. Biochim Biophys Acta. 2016;1859(10):1245‐1251. [DOI] [PubMed] [Google Scholar]
- 11. Lei M, Zheng G, Ning Q, Zheng J, Dong D. Translation and functional roles of circular RNAs in human cancer. Mol Cancer. 2020;19(1):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dhamija S, Menon MB. Non‐coding transcript variants of protein‐coding genes ‐ what are they good for? RNA Biol. 2018;15(8):1025‐1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Macejak DG, Sarnow P. Internal initiation of translation mediated by the 5’ leader of a cellular mRNA. Nature. 1991;353(6339):90‐94. [DOI] [PubMed] [Google Scholar]
- 14. Di Timoteo G, Dattilo D, Centrón‐Broco A, et al. Modulation of circRNA metabolism by m(6)A modification. Cell Rep. 2020;31(6):107641. [DOI] [PubMed] [Google Scholar]
- 15. Rombel IT, Sykes KF, Rayner S, Johnston SA. ORF‐FINDER: a vector for high‐throughput gene identification. Gene. 2002;282(1–2):33‐41. [DOI] [PubMed] [Google Scholar]
- 16. Finn RD, Bateman A, Clements J, et al. Pfam: the protein families database. Nucl Acids Res. 2014;42(D1):D222‐D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen X, Han P, Zhou T, Guo X, Song X, Li Y. circRNADb: A comprehensive database for human circular RNAs with protein‐coding annotations. Sci Rep. 2016;6:34985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20(11):1666‐1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Meng X, Chen Q, Zhang P, Chen M. CircPro: an integrated tool for the identification of circRNAs with protein‐coding potential. Bioinformatics (Oxford, England). 2017;33(20):3314‐3316. [DOI] [PubMed] [Google Scholar]
- 20. Sun P, Li G. CircCode: a powerful tool for identifying circRNA coding ability. Front Genet. 2019;10:981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mokrejs M, Masek T, Vopálensky V, Hlubucek P, Delbos P, Pospísek M. IRESite—a tool for the examination of viral and cellular internal ribosome entry sites. Nucl Acids Res. 2010;38:D131‐D136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhao J, Li Y, Wang C, et al. IRESbase: a comprehensive database of experimentally validated internal ribosome entry sites. Genom Proteomics Bioinform. 2020;18(2):129‐139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N(6)‐methyladenosine. Cell Res. 2017;27(5):626‐641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21(2):172‐179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome‐wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science (New York, NY). 2009;324(5924):218‐223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chassé H, Boulben S, Costache V, Cormier P, Morales J. Analysis of translation using polysome profiling. Nucl Acids Res. 2017;45(3):e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394‐424. [DOI] [PubMed] [Google Scholar]
- 28. Ye F, Gao G, Zou Y, et al. circFBXW7 inhibits malignant progression by sponging miR‐197‐3p and encoding a 185‐aa protein in triple‐negative breast cancer. Mol Ther Nucl Acids. 2019;18:88‐98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cassidy S, Syed BA. Colorectal cancer drugs market. Nat Rev Drug Discov. 2017;16(8):525‐526. [DOI] [PubMed] [Google Scholar]
- 30. Pan Z, Cai J, Lin J, et al. A novel protein encoded by circFNDC3B inhibits tumor progression and EMT through regulating Snail in colon cancer. Molecular Cancer. 2020;19(1):71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhi X, Zhang J, Cheng Z, Bian L, Qin J. circLgr4 drives colorectal tumorigenesis and invasion through Lgr4‐targeting peptide. Int J Cancer. 2019. 10.1002/ijc.32549 [DOI] [PubMed] [Google Scholar]
- 32. Zheng X, Chen L, Zhou Y, et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo‐YAP signaling. Mol Cancer. 2021;20(1):42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jiang T, Xia Y, Lv J, et al. A novel protein encoded by circMAPK1 inhibits progression of gastric cancer by suppressing activation of MAPK signaling. Mol Cancer. 2021;20(1):66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lien WH, Fuchs E. Wnt some lose some: transcriptional governance of stem cells by Wnt/β‐catenin signaling. Genes Dev. 2014;28(14):1517‐1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rossi L, Zoratto F, Papa A, et al. Current approach in the treatment of hepatocellular carcinoma. World J Gastrointest Oncol. 2010;2(9):348‐359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liang W‐C, Wong C‐W, Liang P‐P, et al. Translation of the circular RNA circ‐catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019;20(1):84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. 2017;23(10):1124‐1134. [DOI] [PubMed] [Google Scholar]
- 38. Gu C, Zhou N, Wang Z, et al. circGprc5a promoted bladder oncogenesis and metastasis through Gprc5a‐targeting peptide. Mol Ther Nucl Acids. 2018;13:633‐641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987‐996. [DOI] [PubMed] [Google Scholar]
- 40. Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492‐507. [DOI] [PubMed] [Google Scholar]
- 41. Yang Y, Gao X, Zhang M, et al. Novel ROLE of FBXW7 circular RNA in repressing glioma tumorigenesis. JNCI. 2018;110(3):304‐315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Begum S, Yiu A, Stebbing J, Castellano L. Novel tumour suppressive protein encoded by circular RNA, circ‐SHPRH, in glioblastomas. Oncogene. 2018;37(30):4055‐4057. [DOI] [PubMed] [Google Scholar]
- 43. Zhang M, Zhao K, Xu X, et al. A peptide encoded by circular form of LINC‐PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun. 2018;9(1):4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huang X, Li Z, Zhang Q, et al. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR‐198 suppression. Mol Cancer. 2019;18(1):71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xia X, Li X, Li F, et al. Correction to: A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide‐dependent Kinase‐1. Molecular Cancer. 2019;18(1):149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2019;20(10):608‐624. [DOI] [PubMed] [Google Scholar]
- 47. Li S, Mason CE. The pivotal regulatory landscape of RNA modifications. Annu Rev Genomics Hum Genet. 2014;15:127‐150. [DOI] [PubMed] [Google Scholar]
- 48. Liu J, Yue Y, Han D, et al. A METTL3‐METTL14 complex mediates mammalian nuclear RNA N6‐adenosine methylation. Nat Chem Biol. 2014;10(2):93‐95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6‐methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16(2):191‐198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Patil DP, Pickering BF, Jaffrey SR. Reading m(6)A in the transcriptome: m(6)A‐binding proteins. Trends Cell Biol. 2018;28(2):113‐127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hartmann AM, Nayler O, Schwaiger FW, Obermeier A, Stamm S. The interaction and colocalization of Sam68 with the splicing‐associated factor YT521‐B in nuclear dots is regulated by the Src family kinase p59(fyn). Mol Biol Cell. 1999;10(11):3909‐3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xiao W, Adhikari S, Dahal U, et al. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol Cell. 2016;61(4):507‐519. [DOI] [PubMed] [Google Scholar]
- 53. Chen RX, Chen X, Xia LP, et al. N(6)‐methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10(1):4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang X, Zhao BS, Roundtree IA, et al. N(6)‐methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161(6):1388‐1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jia G, Fu Y, Zhao X, et al. N6‐methyladenosine in nuclear RNA is a major substrate of the obesity‐associated FTO. Nat Chem Biol. 2011;7(12):885‐887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhao J, Lee E, Kim J, et al. Transforming activity of an oncoprotein‐encoding circular RNA from human papillomavirus. Nature Communications. 2019;10(1):2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rossi F, Legnini I, Megiorni F, et al. Circ‐ZNF609 regulates G1‐S progression in rhabdomyosarcoma. Oncogene. 2019;38(20):3843‐3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRNAs. Mol Cell. 2017;66(1):9‐21.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen G, Yang J, Chen J, et al. Identifying and annotating human bifunctional RNAs reveals their versatile functions. Sci China Life Sci. 2016;59(10):981‐992. [DOI] [PubMed] [Google Scholar]
- 60. Ye F, Gao G, Zou Y, et al. circFBXW7 inhibits malignant progression by sponging miR‐197‐3p and encoding a 185‐aa protein in triple‐negative breast cancer. Mol Ther Nucleic Acids. 2019;18:88‐98. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study can be found in online repositories. The name of repositories and reference number can be found in the review.


